ABSTRACT
Chlamydia are obligate intracellular Gram-negative bacteria distinguished by a unique developmental biology confined within a parasitophorous vacuole termed an inclusion. The chlamydial plasmid is a central virulence factor in the pathogenesis of infection. Plasmid gene protein 4 (Pgp4) regulates the expression of plasmid gene protein 3 (Pgp3) and chromosomal glycogen synthase (GlgA), virulence factors secreted from the inclusion to the host cytosol by an unknown mechanism. Here, we identified a plasmid-dependent secretion system for the cytosolic delivery of Pgp3 and GlgA. The secretion system consisted of a segregated population of globular structures originating from midcycle reticulate bodies. Globular structures contained the Pgp4-regulated proteins CT143, CT144, and CT050 in addition to Pgp3 and GlgA. Genetic replacement of Pgp4 with Pgp3 or GlgA negated the formation of globular structures, resulting in retention of Pgp3 and GlgA in chlamydial organisms. The generation of globular structures and secretion of virulence factors occurred independently of type 2 and type 3 secretion systems. Globular structures were enriched with lipopolysaccharide but lacked detectable major outer membrane protein and heat shock protein 60, implicating them as outer membrane vesicles. Thus, we have discovered a novel chlamydial plasmid-dependent secretion system that transports virulence factor cargo from the chlamydial inclusion to the host cytosol.
KEYWORDS: Chlamydia, plasmid, secretion, virulence factors
INTRODUCTION
Chlamydia trachomatis is an obligate intracellular bacterial pathogen which causes blinding trachoma and sexually transmitted infections (STI) that afflict millions of people worldwide (1, 2). C. trachomatis is unique among intracellular bacteria because it undergoes a specialized biphasic developmental growth cycle involving two distinct morphological forms: the infectious nonreplicative elementary body (EB) and the noninfectious replicative reticulate body (RB) (3). The EBs attach to host cells and are then internalized into a parasitophorous vacuole, termed an inclusion, that fails to fuse with the host lysosome. Within this protected niche, EBs differentiate into RBs, which replicate by binary fission or a polarized budding process (4) and differentiate back to EBs. Following lysis of host cells, or extrusion of inclusions, EBs are released and initiate a new round of infection. During its intracellular development C. trachomatis interacts with the host cell to obtain nutrients and biosynthetic precursors and evade innate host defenses (5). Many of these interactions are aided by chlamydial virulence factors which are secreted using a conventional type 2 secretion system (T2SS) and a type 3 secretion system (T3SS) (6–8).
C. trachomatis has a 7.5-kb virulence-associated plasmid (9, 10) including 8 open reading frames (ORF) encoding plasmid gene proteins 1 (Pgp1) to Pgp8. Pgp4 is a master positive regulator of the expression of Pgp3 and a set of conserved chromosomal genes, including those encoding glycogen synthase (GlgA), CT143, CT144, and CT050. Plasmid-deficient strains (11, 12) and Pgp3- and Pgp4-null mutant strains (13, 14) exhibit attenuated infection characteristics in murine and nonhuman primate infection models, demonstrating the critical role the plasmid plays in chlamydial pathogenesis. Pgp3 and GlgA are secreted into the host cytosol (15, 16). Pgp3 plays a role in the establishment of persistent infection by evading antimicrobial peptide host defenses (14, 17). Although the precise role of GlgA in chlamydial pathogenesis remains poorly defined, nonisogenic Chlamydia muridarum strains with GlgA mutations produce fewer infectious progeny in cell culture and display reduced shedding in the urogenital and gastrointestinal tracts in mouse models (18–20). Importantly, while it is firmly established that Pgp3 and GlgA are located in the host cytosol (15, 16), the mechanism of secretion has not been identified. Here, we describe a novel plasmid-regulated secretion system for the delivery of Ppg3 and GlgA to the host cytosol.
RESULTS
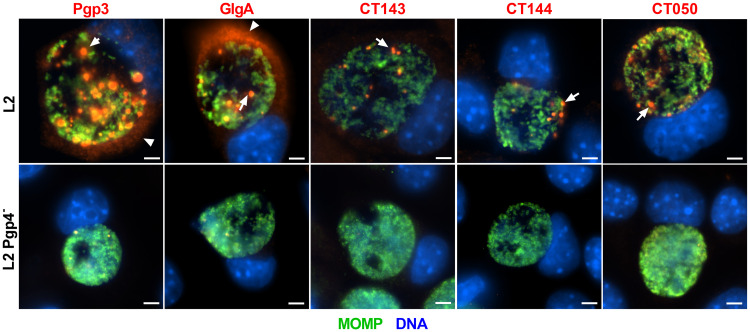
Pgp4-regulated proteins exhibit a globular staining pattern localized to the inclusion lumen.
To investigate the expression pattern of Pgp4-regulated proteins Pgp3, GlgA, CT143, CT144, and CT050, we infected McCoy cells with C. trachomatis L2 or a L2 Pgp4-null mutant strain. Infected cells were fixed and stained with antibodies against Pgp3, GlgA, CT143, CT144, CT050, and the chlamydial major outer membrane protein (MOMP) (Fig. 1). Within C. trachomatis wild-type L2, Pgp3, GlgA, CT143, CT144, and CT050 displayed a globular staining pattern in the lumen of the inclusion. There was a differential staining pattern, with Pgp3 and GlgA but not CT143, CT144, and CT050 being detected in the host cell cytosol. There was minimal labeling of globular structures in the Pgp4-null strain, a result consistent with previous findings that expression of these proteins is Pgp4 dependent (21). The globular staining patten was also observed in L2-infected cells fixed with methanol (see Fig. S1 in the supplemental material), a result indicating that staining pattern is not due to a fixation effect (22). Importantly, a similar globular immunofluorescent-antibody (IFA) staining pattern was observed in cells infected with different C. trachomatis genital and ocular serovars (Fig. S2), indicating that the pattern is not unique to serovar L2.
FIG 1.
Plasmid-regulated proteins exhibit a globular staining pattern localized to the inclusion lumen of infected cells. McCoy cells infected with C. trachomatis wild-type L2 strain or an L2 pgp4-null strain were fixed with PFA at 40 hpi and stained with antibodies against Pgp3, GlgA, CT143, CT144, or CT050, together with the chlamydial major outer membrane protein (MOMP). All 5 proteins displayed a globular staining pattern found in the lumen of the chlamydial inclusion (arrows). Pgp3 and GlgA were detected in the host cell cytosol (arrowheads). Bar, 10 μm.
Globular staining pattern is not affected by fixation strategy. McCoy cells infected with wild-type L2 were fixed at 40 hpi with methanol and stained with antibodies against Pgp3, GlgA, CT143, CT144, or CT050, together with the chlamydial major outer membrane protein (MOMP). All 5 proteins displayed globular staining pattern found in the lumen of the chlamydial inclusion (arrow). Bar, 10 μm. Download FIG S1, TIF file, 2.4 MB (2.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Globular staining pattern of Pgp3 and GlgA in cells infected with different chlamydial strains. (A) McCoy cells infected with C. trachomatis wild-type (WT) or plasmid-deficient strains of different serovars were fixed with PFA at 40 hpi and stained with antibodies against Pgp3 and the chlamydial major outer membrane protein (MOMP). Pgp3 displayed a distinctive globular staining pattern (arrow) found primarily in the lumen of the chlamydial inclusion and was also detected in the cytosol of infected cells (arrowhead). The globular and host cell cytosol staining was not detectable in cells infected with plasmid-deficient strains. (B) Like Pgp3, GlgA also displayed a distinctive globular staining pattern found primarily in the lumen of the chlamydial inclusion and was detected in the cytosol of infected cells but was absent in cells infected with plasmid-deficient strains. Bar, 10 μm. Download FIG S2, TIF file, 2.9 MB (2.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
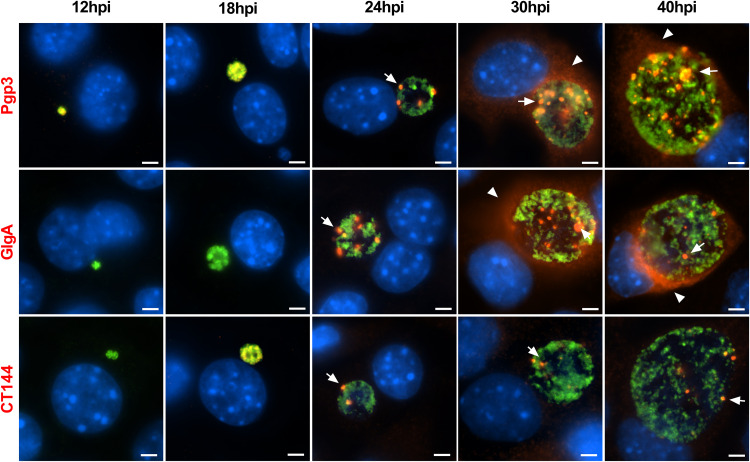
Temporal kinetics of globular staining.
We next investigated the temporal kinetics of globular structure appearance by immunofluorescence staining (Fig. 2). In keeping with transcriptomics data, Pgp3 and CT144 were detected in inclusions as early as 12 and 18 h postinfection (hpi), whereas GlgA staining occurred later (23). All antibodies similarly stained globular structures in inclusions at 24 hpi. In contrast, there was a marked difference in Pgp3 and GlgA staining at 30 and 40 hpi compared to that of CT144. Pgp3 and GlgA antibodies reacted with a greater number of globular structures, and the staining was more intense. This was in contrast to CT144 staining, which did not change noticeably from 24 to 40 hpi. Notably, the distinct changes in Pgp3 and GlgA globular staining coincided with the detection of Pgp3 and GlgA in the host cytosol. Whether the different numbers of globular structures among these proteins account for the functional difference remains unknown.
FIG 2.
Temporal kinetics of Pgp3, GlgA, and CT144 expression in infected McCoy cells. McCoy cells infected with the C. trachomatis wild-type L2 strain were harvested at different times postinfection, as indicated, and processed for IFA assay. Pgp3 was detected as early as 12 hpi. The globular staining pattern (arrowheads) of Pgp3, GlgA, and CT144 was evident at 24 hpi. At 30 hpi, Pgp3 and GlgA staining was markedly stronger and more abundant than CT144 staining, which did not significantly change from the 24-h time point. The increase expression of Pgp3 and GlgA at 30 and 40 hpi coincided with protein secretion into the host cell cytosol (arrows). Bar, 10 μm.
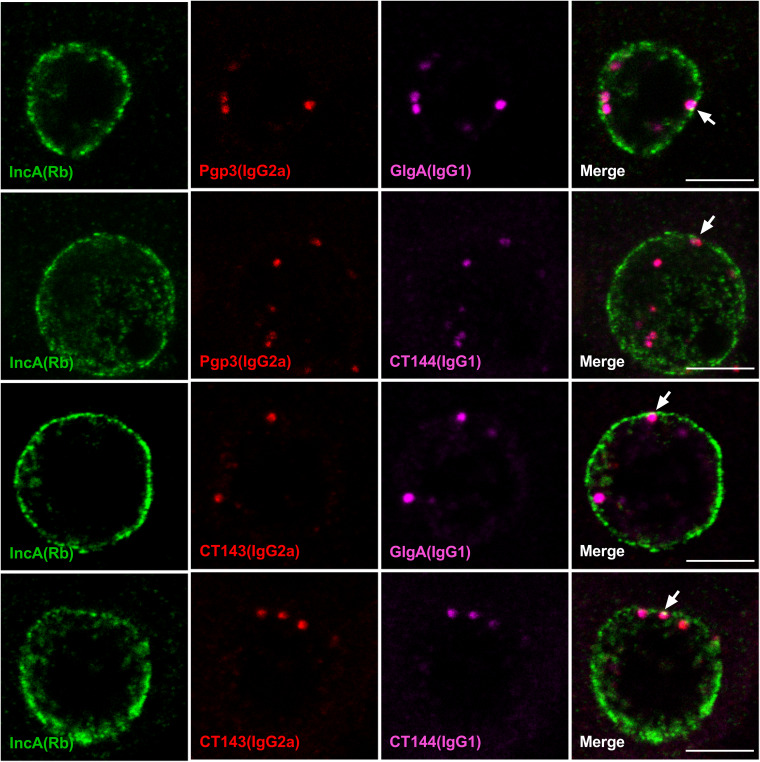
Pgp3, GlgA, CT143, and CT144 colocalize in the same globular structure.
Although the plasmid-regulated proteins Pgp3, GlgA, CT143, and CT144 exhibited similar globular staining characteristics, we wanted to determine whether they colocalized to the same globular structures. Since the globular staining pattern was first observed at 24 hpi, we used confocal microscopy of infected cells at this time point to address this question. Infected-cell cultures were stained with pairs of isotype-compatible monoclonal antibodies, together with a rabbit anti-IncA antibody. Anti-Pgp3, -GlgA, -CT143, and -CT144 antibodies displayed clear overlapping globular structure staining (Fig. 3). We also observed a close association of globular structures with the luminal side of the inclusion membrane, suggesting that they would be in position to interact with the membrane to facilitate delivery of Pgp3 and GlgA to the host cell cytosol. The Pearson's correlation coefficient values of each stained protein pair were greater than 0.5 (Fig. S3), confirming colocalization of the proteins to the same globular structures. Collectively, these results demonstrate that these plasmid-regulated proteins colocalized in the same globular structure.
FIG 3.
Pgp3, GlgA, CT143, and CT144 colocalize to the same globular structure. WT L2-infected McCoy cells were fixed with PFA at 24 hpi, stained with different combination of anti-Pgp3, GlgA, CT143, CT144, and IncA, and analyzed by confocal microscopy. Anti-IncA was used to identify the boundary of the chlamydial inclusion membrane. Bar, 5 μm. Note that each of the plasmid-regulated proteins is contained within the same globular structure. Bar, 5 μm.
Colocalization analysis. The Pearson’s efficiency was calculated using Coloc2 in ImageJ. Download FIG S3, TIF file, 0.8 MB (819.1KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
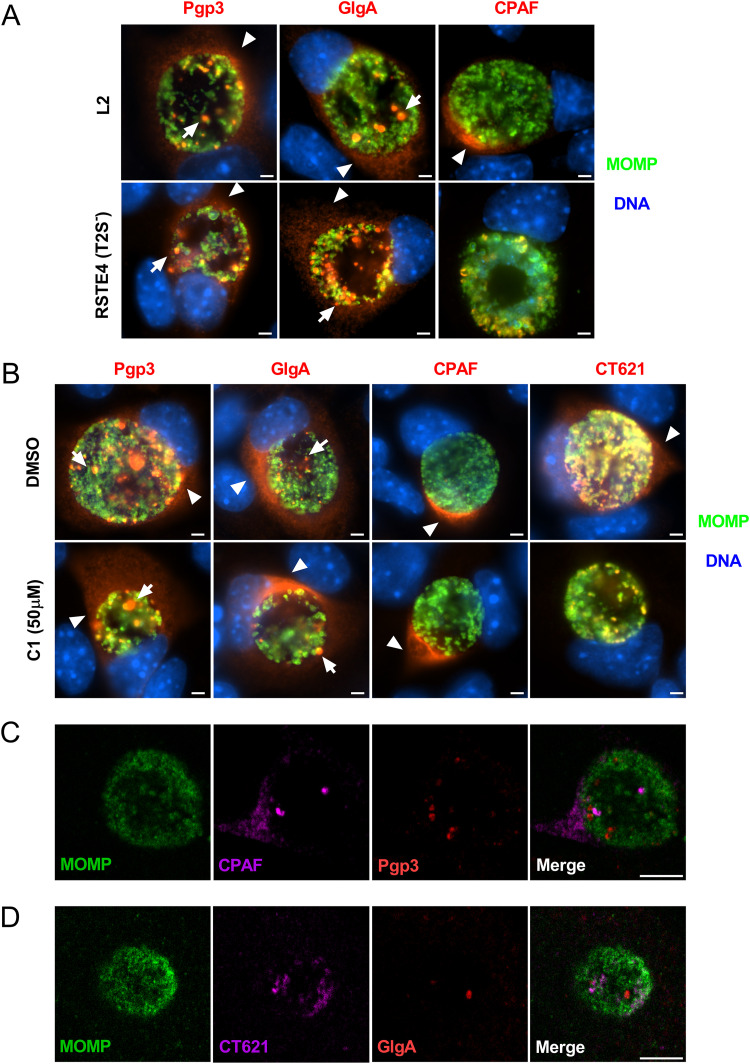
Globular structure staining and secretion of Pgp3 and GlgA into the host cell cytosol are independent of T2SS and T3SS.
Chlamydia possesses both T2SS and T3SS. The T2SS is required for secretion of CPAF, CT795, and CT311 into the host cell cytosol (6, 24, 25). To investigate whether Pgp3 and GlgA secretion was T2SS dependent, we used the T2SS-deficient L2 strain RSTE4 (26) and monitored the localization of Pgp3, GlgA, and CPAF. At 40 hpi, Pgp3 and GlgA were detected in the chlamydial inclusion and host cell cytosol in wild-type- and RSTE4-infected cells (Fig. 4A). In contrast, CPAF, a known T2SS substrate (6), was detected in cytosol of wild-type-L2-infected cells but not in the cytosol of RSTE4-infected cells. Taken together, these results demonstrated that the secretion of Pgp3 and GlgA into the host cell cytosol was not dependent on T2SS. To investigate whether the T3SS was required for the secretion of Pgp3 and GlgA, we treated L2-infected cells with the T3SS inhibitor C1 and analyzed cytosolic Pgp3 and GlgA staining by IFA assay. As controls, we stained for CPAF and CT621, known T2SS and T3SS substrates, respectively (6, 27). Ppg3 and GlgA globular staining and cytosolic staining were observed in C1-treated cells (Fig. 4B). In contrast, C1 treatment inhibited secretion of CT621, a known T3SS effector, but had no effect on CPAF secretion. In addition, we found that CPAF and Pgp3 displayed distinct staining patterns which did not colocalize with each other, suggesting that these two proteins utilized different secretion processes (Fig. 4C). Similarly, CT621 did not colocalize with GlgA (Fig. 4D). Taken together, the results support the conclusion that secretion of Pgp3 and GlgA occurs by a mechanism independent of T2SS or T3SS.
FIG 4.
The formation of globular structures and GlgA and Pgp3 secretion to the host cytosol are independent of T2SS and T3SS. (A) WT-L2- or T2SS-deficient-L2 (RSTE4)-infected McCoy cells were fixed with PFA at 40 hpi and immunolabeled with anti-Pgp3, anti-GlgA, or anti-CPAF, together with anti-MOMP. The globular (arrows) and host cell cytosol (arrowheads) staining of Pgp3 and GlgA was detected in both L2- and RSTE4-infected cells. In contrast, the secretion of CPAF into the host cell cytosol was detected only in cells infected with WT L2, not in cells infected with RSTE4. Bar, 10 μm. (B) McCoy cells were infected with WT L2, and at 18 hpi, the T3SS inhibitor C1 was added to the culture medium. Cells were fixed at 40 hpi and immunolabeled with anti-Pgp3, anti-GlgA, anti-CPAF, or anti-CT621, together with anti-MOMP. Bar, 10 μm. (C) McCoy cells infected with WT L2 were fixed at 24 hpi and immunolabeled with anti-Pgp3 MAb and anti-CPAF MAb together with anti-MOMP. Staining of Pgp3 did not overlap staining of CPAF. Bar, 5 μm. (D) McCoy cells infected with WT L2 were fixed at 24 hpi and immunolabeled with anti-CT621 serum and anti-MOMP, with relevant secondary antibodies. Then cells were immunolabeled with Alexa Fluor 568-conjugated anti-GlgA MAb. Staining of CT621 did not overlap staining of GlgA. Bar, 5 μm.
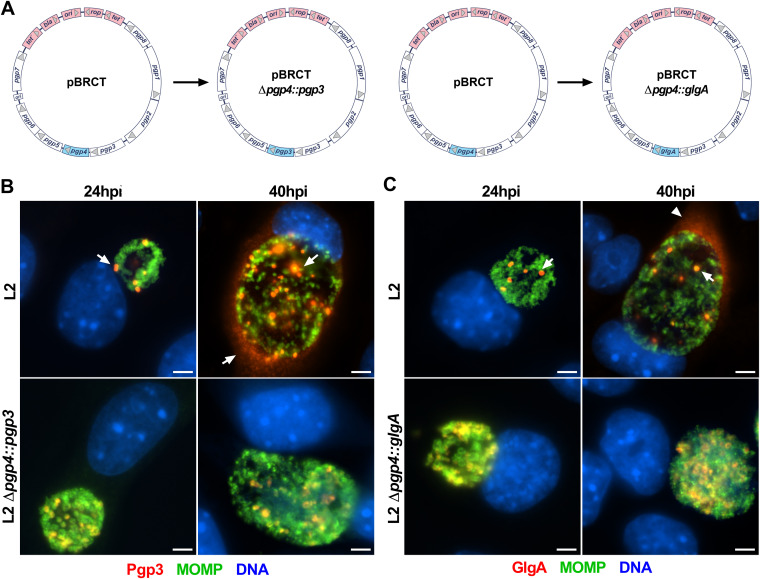
The formation of globular structures and secretion of Pgp3 and GlgA are dependent on Pgp4.
Since Pgp4 regulates the expression of Pgp3 and GlgA (21, 28), it is not possible to determine if Pgp4 is also required for the secretion of these two proteins. To address this question, we constructed two pBRCT plasmids in which the pgp4 ORF was replaced with the pgp3 ORF (pBRCT Δpgp4::pgp3) or the glgA ORF (pBRCT Δpgp4::glgA) (Fig. 5A). With these constructs, the expression of Pgp3 and GlgA was Pgp4 independent. Cells were infected with plasmid-transformed organisms and then analyzed for expression and localization of Pgp3 and GlgA. Immunostaining showed that Pgp3 was expressed in L2 Δpgp4::pgp3-infected cells, indicating that the pgp4 promoter drove the expression of Pgp3 in the absence of Pgp4. Importantly, in L2 Δpgp4::pgp3-infected cells, Pgp3 colocalized with MOMP and there was no detectable Pgp3 in the host cell cytosol (Fig. 5B). Similarly, in L2 Δpgp4::glgA-infected cells, GlgA colocalized with MOMP and was not secreted into the host cell cytosol (Fig. 5C). Thus, the formation of globular structures and the secretion of Pgp3 and GlgA into the host cell cytosol are Pgp4 dependent.
FIG 5.
The formation of globular structures and GlgA and Pgp3 secretion to the host cytosol are dependent on Pgp4. (A) The Pgp4 ORF was replaced by the Pgp3 or GlgA ORF. (B) The Pgp4 promoter drove the expression of Pgp3. In the absence of Pgp4, Pgp3 was highly expressed but there was no cytosolic secretion observed. (C) The expression of GlgA was driven by the Pgp4 promoter. In the absence of Pgp4, GlgA was highly expressed but expression was also restricted to chlamydial organisms. Bar, 10 μm.
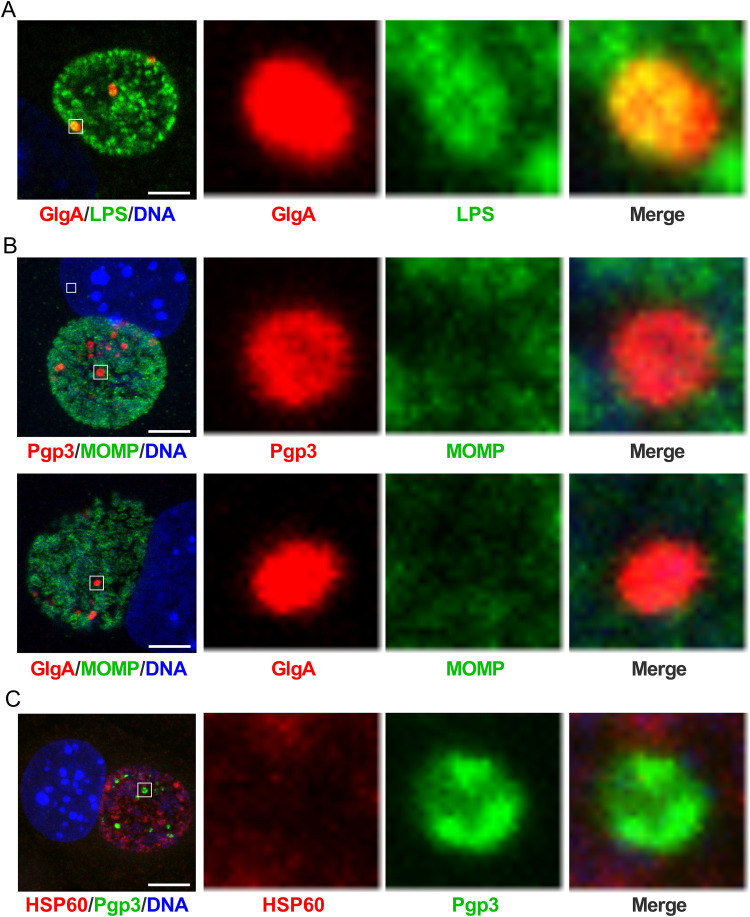
The globular structures contain LPS but not MOMP or HSP60.
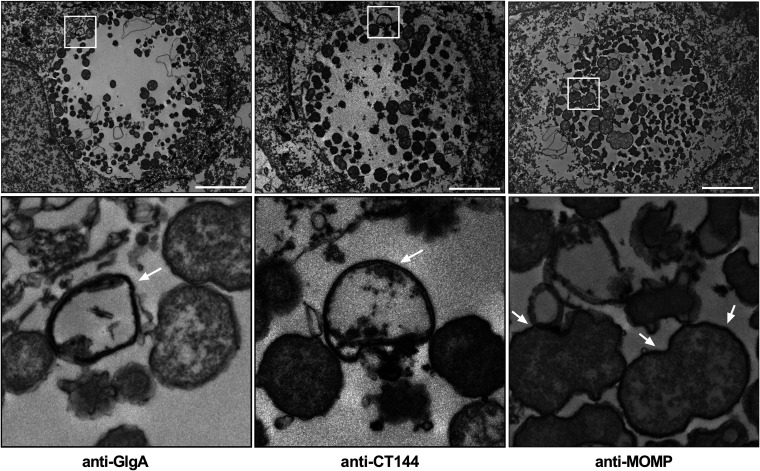
To investigate whether the globular structures were a unique population of chlamydial developmental forms or specialized structures distinct from chlamydial organisms, we costained infected cells with anti-Pgp3 or anti-GlgA, together with antilipopolysaccharide (anti-LPS), MOMP, or HSP60 antibodies based on the compatibility of antibody isotypes and analyzed images by confocal microscopy. GlgA staining overlapped the LPS staining (Fig. 6A). In contrast, Pgp3 and GlgA staining did not overlap MOMP staining (Fig. 6B), and Pgp3 staining did not overlap HSP60 staining (Fig. 6C). Similar results were observed by costaining of CT143 or CT144 with MOMP and of CT143 with Pgp3 (Fig. S3). Immunoelectron microscopy of L2-infected McCoy cells was performed at 24 hpi using anti-GlgA, anti-CT144, and anti-MOMP for ultrastructural identification of globular structures. Globular structures, approximately 0.5 μm in size, stained with anti-GlgA and -CT144 antibodies but not anti-MOMP antibodies were found in L2 inclusions (Fig. 7). These results demonstrated that the globular structures are distinct from chlamydial organisms, as they lack MOMP and HSP60. Based on these staining characteristics we conclude that the globular structures are most likely outer membrane vesicles originating from RBs.
FIG 6.
The globular structures contain chlamydial LPS but do not costain with chlamydial MOMP and HSP60. (A) WT L2-infected McCoy cells were fixed at 24 hpi, stained with anti-GlgA together with anti-LPS, and analyzed by confocal microscopy. GlgA colocalized with the LPS. Bar, 5 μm. (B) WT L2-infected McCoy cells were fixed at 24 hpi and costained with anti-Pgp3 and anti-MOMP or with anti-GlgA and anti-MOMP. Pgp3 and GlgA did not colocalize with MOMP. (C) WT L2-infected McCoy cells were fixed at 24 hpi and costained with anti-Pgp3 and anti-HSP60. Pgp3 did not colocalize with HSP60. Bar, 5 μm. In each row, the area highlighted by the white box in the leftmost panel is magnified in the second, third, and fourth panels.
FIG 7.
Transmission immunoelectron microscopy shows globular structures in the inclusion lumen. WT L2-infected McCoy cells were fixed at 24 hpi, immunolabeled with anti-GlgA, anti-CT144, or anti-MOMP, and prepared for TEM. Anti-GlgA and -CT144 antibodies specifically recognize globular structures in the inclusion lumen (arrows). Anti-MOMP antibodies labeled chlamydial EB and RB developmental forms (arrows). The globular structures did not react with anti-MOMP antibodies. Bars, 5 μm. The bottom row shows magnifications of the areas highlighted by the white boxes in the top row.
DISCUSSION
Pgp3 and GlgA are two chlamydial plasmid-regulated virulence factors which are secreted into the host cell cytosol by an unknown mechanism. Here, we demonstrated that Pgp3 and GlgA, together with other plasmid-regulated proteins, including CT143 and CT144, are colocalized in the globular structures, which are often associated with the chlamydial inclusion membrane. The formation of the globular structures and the secretion of Pgp3 and GlgA to the host cell cytosol are not dependent on either T2SS or T3SS but rather are dependent on Pgp4. These globular structures do not contain chlamydial MOMP or HSP60 but have LPS. Collectively, our findings describe a novel plasmid-regulated system for delivery of plasmid-regulated virulence factors Pgp3 and GlgA to the host cell cytosol.
Previous studies described the globular structure staining pattern of CT143, CT144, and CT050 (29, 30); the present study confirms those observations and, importantly, describes for first time that CT143, CT144, Pgp3, and GlgA colocalize to the same globular structure. Results from experiments using a T2SS mutant strain and the T3SS inhibitor C1 indicate that neither of these secretion systems is required for globular structure formation or secretion of Pgp3 and GlgA to the host cell cytosol. Jorgensen and Valdivia previously showed that the localization of CT050 to globular structures was not affected by C1 (29). CPAF is secreted by the T2SS; in this process, CPAF is translocated into the periplasm space. It has been proposed that CPAF is secreted via outer membrane vesicles (OMV) (31). Consistent with previous studies (15), we showed that Pgp3 does not colocalize with CPAF, indicating that Pgp3 is not in the same secretion vesicle as CPAF. Similarly, GlgA does not colocalize with the T3SS substrate CT621, which shows that the translocation of GlgA is different from that of CT621 and is not dependent on T3SS. Of note, GlgA, CT143, and CT144 were previously identified as T3SS substrates (18, 30), a finding not supported by our results. One possible explanation for this discrepancy could be that surrogate T3SS were used in those studies. Recently, Yanatori et al. showed that bioinformatic predictions of Chlamydia T3S effectors followed by experimental validation of secretion in Yersinia or Shigella surrogate systems can give high frequencies of false positives (32). Their observations reinforce the idea that caution is warranted when evaluating if chlamydial proteins are secreted by T3SS and also suggest that T3SS-independent pathways may play a critical role in the delivery of chlamydial virulence factors (33). Using Pgp4 replacement mutants, we showed that globular structure staining and secretion of Pgp3 and GlgA were dependent on Pgp4. This shows that the formation of globular structures in the inclusion lumen is Pgp4 dependent or might require one or more Pgp4-regulated chromosomal gene products. The globular structure components CT143 and CT144 are small conserved hypothetical proteins with unknown function (34). The precise role of CT143 and CT144 in globular structure formation and secretion of Pgp3 and GlgA needs to be investigated.
To reach the host cell cytosol, Pgp3 and GlgA need to cross the inclusion membrane. By costaining Pgp3 and GlgA with the inclusion membrane protein IncA, we found that the globular structures were closely associated with the inclusion membrane, suggesting that interactions between these structures and inclusion membrane may facilitate the secretion of Pgp3 and GlgA to the host cell cytosol. Phospholipase D (PLD) (CT084) is a Pgp4-regulated chromosomal gene (21). PLD catalyzes the hydrolysis of phospholipids and affects membrane fusion and permeability (35). It is unknown if PLD is a component of globular structures, but if it is, its interaction with the inclusion membrane might facilitate delivery of Pgp3 and GlgA to the host cell cytosol. It will be important to experimentally test this hypothesis in future studies.
The unique composition of the globular structures and their distinct physical separation from typical replicating chlamydial organisms provide some insight into what the origin and biogenesis of the structures might be. We hypothesize that these structures are derived from RBs, or alternatively OMV generated from RBs. If globular structures originate from RBs, one would expect them to contain MOMP and HSP60, which they did not. Therefore, we favor the latter possibility, because the globular structures stained for chlamydial LPS but not MOMP and HSP60. A common characteristic of Gram-negative OMV is that they are often enriched with LPS with minimal amounts of outer membrane pore proteins, which is consistent with our results (36). The globular structures contain Pgp3 and GlgA as cargo, sharing the characteristic of Gram-negative OMV, which are selectively loaded with virulence factor cargo (36). Therefore, we propose that the globular structures originate from RB OMV. Clearly, for globular structures to be identified as OMV, they need to be isolated and characterized biochemically. However, the limited abundance of these structures, together with chlamydiae’s obligate intracellular growth requirement, makes this goal especially challenging.
MATERIALS AND METHODS
Cell culture and chlamydial strains.
McCoy and L929 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Corning, Inc., Corning, NY, USA) supplemented with 10% fetal bovine serum at 37°C with 5% CO2. C. trachomatis L2 (L2/LGV-434/Bu), L2R, D (UW-3/Cx), plasmid-deficient D, A2497, plasmid-deficient A2497, and H (UW-4/Cx) strains were described previously (11, 12, 14). The plasmid-deficient H strain was isolated from plasmid-positive H strain using novobiocin as described previously (37).
Plasmid constructs.
To replace pgp4 ORF with either glgA or pgp3 ORF, two DNA fragments lacking pgp4 ORF were amplified using CloneAmp HiFi PCR Premix (TaKaRa, Mountain View, CA, USA) and pBRCT (21) as a template. A third PCR product containing the glgA or pgp3 ORF was generated using C. trachomatis L2 DNA as the template. PCR products were purified using a QIAquick gel extraction kit (Qiagen, Germantown, MD, USA) and then subjected to fusion by using an In-Fusion HD cloning kit (TaKaRa, USA) according to the manufacturer’s instructions. The fusion products were transformed into Stellar competent cells (TaKaRa, USA), and transformants were selected for ampicillin resistance on LB agar plates. Bacterial colonies were screened for the presence of pgp3 or glgA in the correct orientation by PCR. Plasmids extracted from bacterial colonies with the desired PCR screening results were subjected to DNA sequencing. Plasmid with correct DNA sequence was transformed to Δdam Δdcm competent E. coli cells (New England Biolabs, Ipswich, MA, USA) for amplification. The final plasmids constructs were used to transform chlamydiae. Primers are shown in Table S1.
List of primer sequences used for making genetic replacement constructs in this study. Download Table S1, XLSX file, 0.009 MB (9.9KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Transformation of C. trachomatis L2R.
Transformation of C. trachomatis L2R was performed as described previously (21). Briefly, density gradient-purified EB and plasmid DNAs were added to CaCl2 buffer (10 mM Tris [pH 7.4] and 50 mM CaCl2) to a final volume of 275 μl. After gentle pipetting, the mixture was incubated at room temperature for 30 min. The mixture was suspended in 12 ml ice-cold sucrose phosphate glutamate (SPG) buffer and added to 6-well plates seeded with confluent L929 cells. Plates were centrifuged at 545 × g at room temperature for 1 h, and the inoculum was replaced with 3 ml complete medium (DMEM containing 10% fetal bovine serum) and incubated for 5 h at 37°C. The medium was replaced with medium containing 1 μg/ml cycloheximide and 10 IU/ml penicillin G. The strains were plaque cloned twice and expanded.
Monoclonal antibody production.
The glgA ORF was amplified by PCR, and the amplicon was inserted into pET-26b (Sigma, St. Louis, MO, USA) using NdeI and XhoI sites to express the fusion protein with a C-terminal His tag. Primers used for PCR were as follows: forward, 5′-GGGAATTCCATATGAAAATTATTCACACAGCTATC-3′; reverse, 5′-CCGCTCGAGTTGTTTATAAATTTCTAAATATTTATTG-3′. Expression of GlgA/His fusion protein was induced with isopropyl-β-d-thiogalactoside (IPTG) for 6 h at room temperature. Fusion proteins were extracted by lysing bacteria in BugBuster extraction reagent containing 1 mM phenylmethylsulfonyl fluoride, 75 U/ml aprotinin, 20 μM leupeptin, and 1.6 μM pepstatin (Sigma, USA). After centrifuge, the supernatant was passed through a His GraviTrap column (Global Life Sciences Solutions, Pittsburgh, PA, USA) for protein purification. The purified protein was used to immunize mice for making monoclonal antibodies as described previously (38).
Effect of T3SS C1 inhibitor and T2SS mutation on secretion of Pgp3 and GlgA.
At 18 hpi, 50 μM C1 was added to C. trachomatis-infected cells. C1- or DMSO-treated infected cells were harvested at 40 hpi. The T2SS mutant strain RSTE4 was kindly provided by Raphael Valdivia (Duke University, NC, USA). McCoy cells were infected with RSTE4 strain and were fixed at 40 hpi for IFA staining.
Antibody labeling.
The anti-GlgA monoclonal antibody (MAb) was directly labeled with Alexa Fluor 568 using an Alexa Fluor 568 antibody labeling kit (Thermo Fisher, USA) following the manufacturer’s protocols.
Immunofluorescence.
McCoy cells grown on coverslips (2 × 105 cells) were infected with C. trachomatis strains at a multiplicity of infection (MOI) of 0.2. Infected cells were fixed with 4% paraformaldehyde (PFA) (Santa Cruz, Dallas, TX, USA) for 30 min at room temperature and then permeabilized with 0.1% Triton X-100 for 15 min at room temperature or fixed with 100% cold methanol for 10 min. After blocking with 2% bovine serum albumin (BSA) in PBS for 30 min, coverslips were incubated with primary antibodies at 37°C for 1 h. Primary antibodies were rabbit anti-MOMP serum, mouse anti-MOMP MAb (L2I-45, IgG3), mouse anti-CT143 MAb (IgG2a), mouse anti-CT144 MAb (IgG1), mouse anti-Pgp3 serum, mouse anti-Pgp3 MAb (2H4, IgG2a), mouse anti-GlgA MAb (F443G, IgG1), mouse anti-CPAF MAb (100a, IgG1), mouse anti-HSP60 MAb (IgG1), mouse anti-LPS MAb (EVI-H1, IgG2a), rabbit anti-CT050 serum, mouse anti-CT621 serum, and rabbit anti-IncA serum. Coverslips were washed with PBS and incubated with DAPI (4′,6-diamidino-2-phenylindole) and corresponding secondary antibodies labeled with Alexa Fluor 488, 568, or 647 (Thermo Fisher, Waltham, MA, USA). In certain stainings, anti-GlgA MAb directly labeled with Alex Fluor 568 was used. After mounting with ProLong Gold antifade mountant (Thermo Fisher, USA), coverslips were imaged with a Nikon Eclipse 80i microscope or Leica SP8 confocal microscope. All images were processed using ImageJ. Pearson's correlation coefficient values were calculated using the Coloc2 program in ImageJ.
Immunoelectron microscopy.
C. trachomatis serovar L2-infected McCoy cells were fixed in 2% paraformaldehyde–0.25% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) in 0.1 M phosphate buffer for 2 h at room temperature. Cells were permeabilized with 0.01% saponin for 5 min at room temperature. Fixed cells were incubated with anti-GlgA, anti-CT144, and anti-MOMP antibodies, followed by peroxidase-conjugated secondary antibodies, and prepared for transmission electron microscopy (TEM) as described previously (39).
Statistical analyses.
Data are presented as means and standard deviations (SD). GraphPad Prism 8.0 software was used for data analysis.
CT143 and CT144 do not colocalize with MOMP or HSP60. (A) WT L2-infected McCoy cells were fixed at 24 hpi, stained with anti-CT143 and CT144 together with anti-MOMP, and analyzed by confocal microscopy. CT143 and CT144 did not colocalize with the MOMP. Bar, 5 μm. (B) WT L2-infected McCoy cells were fixed at 24 hpi and costained with anti-CT143 and anti-HSP60. CT143 did not colocalize with HSP60. Bar, 5 μm. The images on the right are magnifications of the areas highlighted by white boxes on the left. Download FIG S4, TIF file, 2.7 MB (2.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ACKNOWLEDGMENTS
We thank Guangming Zhong (University of Texas Health Science Center at San Antonio) for the generous contribution of anti-Pgp3, anti-CPAF and anti-CT621 antibodies; Raphael Valdivia (Duke University) for the generous contribution of anti-CT050 antibody and the RSTE4 strain; Huizhou Fan (Rutgers University) for the generous contribution of C1 compound; and Ted Hackstadt (National Institute of Allergy and Infectious Diseases) for the generous contribution of anti-IncA antibody.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
We declare no competing interests.
Footnotes
This article is a direct contribution from Harlan D. Caldwell, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Dan Rockey, Oregon State University, and Richard Morrison, University of Arkansas for Medical Sciences.
Citation Lei L, Yang C, Patton MJ, Smelkinson M, Dorward D, Ma L, Karanovic U, Firdous S, McClarty G, Caldwell HD. 2021. A chlamydial plasmid-dependent secretion system for the delivery of virulence factors to the host cytosol. mBio 12:e01179-21. https://doi.org/10.1128/mBio.01179-21.
Contributor Information
Harlan D. Caldwell, Email: hcaldwell@niaid.nih.gov.
Scot P. Ouellette, University of Nebraska Medical Center
REFERENCES
- 1.Burton MJ, Mabey DC. 2009. The global burden of trachoma: a review. PLoS Negl Trop Dis 3:e460. doi: 10.1371/journal.pntd.0000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. 2016. WHO Guidelines for the Treatment of Chlamydia trachomatis. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 3.Moulder JW. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol Rev 55:143–190. doi: 10.1128/MR.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ouellette SP, Lee J, Cox JV. 2020. Division without binary fission: cell division in the FtsZ-less Chlamydia. J Bacteriol 202:e00252-20. doi: 10.1128/JB.00252-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elwell C, Mirrashidi K, Engel J. 2016. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol 14:385–400. doi: 10.1038/nrmicro.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D, Lei L, Lu C, Flores R, DeLisa MP, Roberts TC, Romesberg FE, Zhong G. 2010. Secretion of the chlamydial virulence factor CPAF requires the Sec-dependent pathway. Microbiology (Reading) 156:3031–3040. doi: 10.1099/mic.0.040527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hower S, Wolf K, Fields KA. 2009. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol Microbiol 72:1423–1437. doi: 10.1111/j.1365-2958.2009.06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bugalhao JN, Mota LJ. 2019. The multiple functions of the numerous Chlamydia trachomatis secreted proteins: the tip of the iceberg. Microb Cell 6:414–449. doi: 10.15698/mic2019.09.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comanducci M, Ricci S, Cevenini R, Ratti G. 1990. Diversity of the Chlamydia trachomatis common plasmid in biovars with different pathogenicity. Plasmid 23:149–154. doi: 10.1016/0147-619x(90)90034-a. [DOI] [PubMed] [Google Scholar]
- 10.Palmer L, Falkow S. 1986. A common plasmid of Chlamydia trachomatis. Plasmid 16:52–62. doi: 10.1016/0147-619x(86)90079-x. [DOI] [PubMed] [Google Scholar]
- 11.Carlson JH, Whitmire WM, Crane DD, Wicke L, Virtaneva K, Sturdevant DE, Kupko JJ, 3rd, Porcella SF, Martinez-Orengo N, Heinzen RA, Kari L, Caldwell HD. 2008. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect Immun 76:2273–2283. doi: 10.1128/IAI.00102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kari L, Whitmire WM, Olivares-Zavaleta N, Goheen MM, Taylor LD, Carlson JH, Sturdevant GL, Lu C, Bakios LE, Randall LB, Parnell MJ, Zhong G, Caldwell HD. 2011. A live-attenuated chlamydial vaccine protects against trachoma in nonhuman primates. J Exp Med 208:2217–2223. doi: 10.1084/jem.20111266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Huang Y, Yang Z, Sun Y, Gong S, Hou S, Chen C, Li Z, Liu Q, Wu Y, Baseman J, Zhong G. 2014. Plasmid-encoded Pgp3 is a major virulence factor for Chlamydia muridarum to induce hydrosalpinx in mice. Infect Immun 82:5327–5335. doi: 10.1128/IAI.02576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C, Kari L, Lei L, Carlson JH, Ma L, Couch CE, Whitmire WM, Bock K, Moore I, Bonner C, McClarty G, Caldwell HD. 2020. Chlamydia trachomatis plasmid gene protein 3 is essential for the establishment of persistent infection and associated immunopathology. mBio 11:e01902-20. doi: 10.1128/mBio.01902-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li ZY, Chen D, Zhong YM, Wang SP, Zhong GM. 2008. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect Immun 76:3415–3428. doi: 10.1128/IAI.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu CX, Lei L, Peng B, Tang LL, Ding HL, Gong SQ, Li ZY, Wu YM, Zhong GM. 2013. Chlamydia trachomatis GlgA is secreted into host cell cytoplasm. PLoS One 8:e68764. doi: 10.1371/journal.pone.0068764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou SP, Dong XH, Yang ZS, Li ZY, Liu QZ, Zhong GM. 2015. Chlamydial plasmid-encoded virulence factor Pgp3 neutralizes the antichlamydial activity of human cathelicidin LL-37. Infect Immun 83:4701–4709. doi: 10.1128/IAI.00746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gehre L, Gorgette O, Perrinet S, Prevost MC, Ducatez M, Giebel AM, Nelson DE, Ball SG, Subtil A. 2016. Sequestration of host metabolism by an intracellular pathogen. Elife 5:e12552. doi: 10.7554/eLife.12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao L, Zhang T, Melero J, Huang Y, Liu Y, Liu Q, He C, Nelson DE, Zhong G. 2017. The genital tract virulence factor pGP3 is essential for Chlamydia muridarum colonization in the gastrointestinal tract. Infect Immun 86:e00429-17. doi: 10.1128/IAI.00429-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu C, Wu H, Sun Y, Kong J, Shao L, Chen X, Liu Q, Liu Y. 2020. GlgA plays an important role in the induction of hydrosalpinx by Chlamydia muridarum. Pathog Dis 78:ftaa027. doi: 10.1093/femspd/ftaa027. [DOI] [PubMed] [Google Scholar]
- 21.Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, Watkins H, Zhou B, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD. 2013. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect Immun 81:636–644. doi: 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kokes M, Valdivia RH. 2015. Differential translocation of host cellular materials into the Chlamydia trachomatis inclusion lumen during chemical fixation. PLoS One 10:e0139153. doi: 10.1371/journal.pone.0139153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci U S A 100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi M, Lei L, Gong S, Liu Q, DeLisa MP, Zhong G. 2011. Chlamydia trachomatis secretion of an immunodominant hypothetical protein (CT795) into host cell cytoplasm. J Bacteriol 193:2498–2509. doi: 10.1128/JB.01301-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lei L, Qi M, Budrys N, Schenken R, Zhong G. 2011. Localization of Chlamydia trachomatis hypothetical protein CT311 in host cell cytoplasm. Microb Pathog 51:101–109. doi: 10.1016/j.micpath.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snavely EA, Kokes M, Dunn JD, Saka HA, Nguyen BD, Bastidas RJ, McCafferty DG, Valdivia RH. 2014. Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog Dis 71:336–351. doi: 10.1111/2049-632X.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong S, Lei L, Chang X, Belland R, Zhong G. 2011. Chlamydia trachomatis secretion of hypothetical protein CT622 into host cell cytoplasm via a secretion pathway that can be inhibited by the type III secretion system inhibitor compound 1. Microbiology (Reading) 157:1134–1144. doi: 10.1099/mic.0.047746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong S, Yang Z, Lei L, Shen L, Zhong G. 2013. Characterization of Chlamydia trachomatis plasmid-encoded open reading frames. J Bacteriol 195:3819–3826. doi: 10.1128/JB.00511-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen I, Valdivia RH. 2008. Pmp-like proteins Pls1 and Pls2 are secreted into the lumen of the Chlamydia trachomatis inclusion. Infect Immun 76:3940–3950. doi: 10.1128/IAI.00632-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.da Cunha M, Pais SV, Bugalhao JN, Mota LJ. 2017. The Chlamydia trachomatis type III secretion substrates CT142, CT143, and CT144 are secreted into the lumen of the inclusion. PLoS One 12:e0178856. doi: 10.1371/journal.pone.0178856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong G. 2011. Chlamydia trachomatis secretion of proteases for manipulating host signaling pathways. Front Microbiol 2:14. doi: 10.3389/fmicb.2011.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanatori I, Miura K, Chen YS, Valdivia RH, Kishi F. 2021. Application of a C. trachomatis expression system to identify C. pneumoniae proteins translocated into host cells. J Bacteriol 203:e00511-20. doi: 10.1128/JB.00511-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan JD, Nelson DE. 2021. A same-genus screening approach reveals novel effectors and new possibilities for investigating Chlamydia pathogenesis. J Bacteriol 203:e00157-21. doi: 10.1128/JB.00157-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connell CM, AbdelRahman YM, Green E, Darville HK, Saira K, Smith B, Darville T, Scurlock AM, Meyer CR, Belland RJ. 2011. Toll-like receptor 2 activation by Chlamydia trachomatis is plasmid dependent, and plasmid-responsive chromosomal loci are coordinately regulated in response to glucose limitation by C. trachomatis but not by C. muridarum. Infect Immun 79:1044–1056. doi: 10.1128/IAI.01118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selvy PE, Lavieri RR, Lindsley CW, Brown HA. 2011. Phospholipase D: enzymology, functionality, and chemical modulation. Chem Rev 111:6064–6119. doi: 10.1021/cr200296t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Connell CM, Ingalls RR, Andrews CW, Jr, Scurlock AM, Darville T. 2007. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J Immunol 179:4027–4034. doi: 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- 38.Chronopoulou E, Uribe-Benninghoff A, Corbett CR, Berry JD. 2014. Hybridoma technology for the generation of rodent mAbs via classical fusion. Methods Mol Biol 1131:47–70. doi: 10.1007/978-1-62703-992-5_4. [DOI] [PubMed] [Google Scholar]
- 39.Taylor LD, Nelson DE, Dorward DW, Whitmire WM, Caldwell HD. 2010. Biological characterization of Chlamydia trachomatis plasticity zone MACPF domain family protein CT153. Infect Immun 78:2691–2699. doi: 10.1128/IAI.01455-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Globular staining pattern is not affected by fixation strategy. McCoy cells infected with wild-type L2 were fixed at 40 hpi with methanol and stained with antibodies against Pgp3, GlgA, CT143, CT144, or CT050, together with the chlamydial major outer membrane protein (MOMP). All 5 proteins displayed globular staining pattern found in the lumen of the chlamydial inclusion (arrow). Bar, 10 μm. Download FIG S1, TIF file, 2.4 MB (2.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Globular staining pattern of Pgp3 and GlgA in cells infected with different chlamydial strains. (A) McCoy cells infected with C. trachomatis wild-type (WT) or plasmid-deficient strains of different serovars were fixed with PFA at 40 hpi and stained with antibodies against Pgp3 and the chlamydial major outer membrane protein (MOMP). Pgp3 displayed a distinctive globular staining pattern (arrow) found primarily in the lumen of the chlamydial inclusion and was also detected in the cytosol of infected cells (arrowhead). The globular and host cell cytosol staining was not detectable in cells infected with plasmid-deficient strains. (B) Like Pgp3, GlgA also displayed a distinctive globular staining pattern found primarily in the lumen of the chlamydial inclusion and was detected in the cytosol of infected cells but was absent in cells infected with plasmid-deficient strains. Bar, 10 μm. Download FIG S2, TIF file, 2.9 MB (2.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Colocalization analysis. The Pearson’s efficiency was calculated using Coloc2 in ImageJ. Download FIG S3, TIF file, 0.8 MB (819.1KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of primer sequences used for making genetic replacement constructs in this study. Download Table S1, XLSX file, 0.009 MB (9.9KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
CT143 and CT144 do not colocalize with MOMP or HSP60. (A) WT L2-infected McCoy cells were fixed at 24 hpi, stained with anti-CT143 and CT144 together with anti-MOMP, and analyzed by confocal microscopy. CT143 and CT144 did not colocalize with the MOMP. Bar, 5 μm. (B) WT L2-infected McCoy cells were fixed at 24 hpi and costained with anti-CT143 and anti-HSP60. CT143 did not colocalize with HSP60. Bar, 5 μm. The images on the right are magnifications of the areas highlighted by white boxes on the left. Download FIG S4, TIF file, 2.7 MB (2.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.