ABSTRACT
Spotted fever group rickettsioses (SFRs) are devastating human infections. Vascular endothelial cells (ECs) are the primary targets of rickettsial infection. Edema resulting from EC barrier dysfunction occurs in the brain and lungs in most cases of lethal SFR, but the underlying mechanisms remain unclear. The aim of the study was to explore the potential role of Rickettsia-infected, EC-derived exosomes (Exos) during infection. Using size exclusion chromatography (SEC), we purified Exos from conditioned, filtered, bacterium-free media collected from Rickettsia parkeri-infected human umbilical vein ECs (HUVECs) (R-ECExos) and plasma of Rickettsia australis- or R. parkeri-infected mice (R-plsExos). We observed that rickettsial infection increased the release of heterogeneous plsExos, but endothelial exosomal size, morphology, and production were not significantly altered following infection. Compared to normal plsExos and ECExos, both R-plsExos and R-ECExos induced dysfunction of recipient normal brain microvascular ECs (BMECs). The effect of R-plsExos on mouse recipient BMEC barrier function is dose dependent. The effect of R-ECExos on human recipient BMEC barrier function is dependent on the exosomal RNA cargo. Next-generation sequencing analysis and stem-loop quantitative reverse transcription-PCR (RT-qPCR) validation revealed that rickettsial infection triggered the selective enrichment of endothelial exosomal mir-23a and mir-30b, which potentially target the endothelial barrier. To our knowledge, this is the first report on the functional role of extracellular vesicles following infection by obligately intracellular bacteria.
KEYWORDS: exosome, extracellular vesicle, endothelial cell, endothelial barrier function, spotted fever group rickettsial infection, barrier function, rickettsial infection
INTRODUCTION
Spotted fever group (SFG) rickettsioses (SFRs) are devastating human infections (1). A licensed vaccine is not available. It is forecasted that increased ambient temperatures under conditions of global climate change will lead to more widespread distribution of rickettsioses (2). These arthropod-borne diseases are caused by obligately intracellular bacteria of the genus Rickettsia, including Rickettsia rickettsii (3, 4) and R. parkeri (5–7), which cause Rocky Mountain spotted fever and R. parkeri rickettsiosis (8), respectively, in the United States and Latin America; R. conorii, the causative agent of Mediterranean spotted fever endemic to southern Europe, North Africa, and India (9); and R. australis, which causes Queensland tick typhus in Australia (10). Vascular endothelial cells (ECs) are the primary targets of infection, and EC tropism plays a central role during pathogenesis (1, 3, 11). Edema resulting from EC barrier dysfunction occurs in the brain and lungs in most cases of lethal SFR (12). Typically, rickettsial infection is controlled by appropriate broad-spectrum antibiotic therapy if diagnosed early (3, 4). However, rickettsial infections can cause nonspecific signs and symptoms, rendering early clinical diagnosis difficult (13, 14). Untreated or misdiagnosed rickettsial infections are frequently associated with severe morbidity and mortality (1, 4, 15–17). A fatality rate as high as 32% has been reported for hospitalized patients with Mediterranean spotted fever (17). Although doxycycline is the antibiotic of choice for rickettsial infections, it only stops bacteria from reproducing and does not kill the rickettsiae. Comprehensive understanding of rickettsial pathogenesis is urgently needed for the development of novel therapeutics (7, 16, 18–22).
Eukaryotic cell-to-cell communication is critical for maintaining homeostasis and responding quickly to environmental stimuli (23–51). Besides direct intercellular contact, this communication is often mediated by soluble factors that can convey signals to a large repertoire of responding cells, either locally or remotely. Extracellular vesicles (EVs) transfer functional mediators to neighboring and distant recipient cells (33). EVs are broadly classified into two categories, exosomes (Exos) (50 to 150 nm) and microvesicles (100 to 1,000 nm), owing to their endocytic or plasma membrane origin (38, 52–66). Exos and microvesicles are also termed small and large EVs, respectively (55). Exo biogenesis begins with the formation of intraluminal vesicles, the intracellular precursors of Exos, after the inward budding of the membranes of late endosomes (37, 54). Intraluminal vesicles are internalized into a multivesicular body, which transits toward and fuses with the plasma membrane, before releasing intraluminal vesicles into the extracellular environment as Exos (53). An Exo contains many types of biomolecules, including proteins, nucleic acids, and lipids (67). Once bound to the plasma membrane of the recipient cell, Exos can induce functional responses by multiple mechanisms, e.g., activating receptors on recipient cells or releasing their bioactive cargos after internalization (67, 68). In infectious biology, EVs from infected donor cells contain cargos that are associated with the virulence of the pathogen or the activation of host self-defense mechanisms (33–38, 69–71). EVs released from macrophages infected by intracellular bacteria, such as Mycobacterium tuberculosis and Salmonella enterica serovar Typhimurium, have been shown to stimulate a proinflammatory response in noninfected macrophages in a Toll-like receptor-dependent manner (70). Unfortunately, the role(s) of EVs in the pathogenesis of obligately intracellular bacterial infections remains unknown.
Although small noncoding RNA (sncRNA) species (<150 nucleotides) are relatively stable compared with other RNA molecules, they remain vulnerable to RNase-mediated digestion (72). The discovery of extracellular sncRNAs in the blood, despite the abundant presence of RNases, led to the proposal of a scenario in which sncRNAs are encapsulated in EVs (55, 72–74) or form circulating ribonucleoproteins (75, 76). Extracellular RNAs are enriched in sncRNAs (77). A growing number of reports have established that many, if not all, of the effects of EVs are mediated by microRNA (52, 55–60, 63) or tRNA fragment (61, 78) cargos, which remain functional to regulate cellular behaviors of the recipient cells (79). Recent studies provide emerging evidence that microRNAs are selectively sorted into EVs independently of their cellular levels (52, 55–62). We reported that R. conorii infection induces significant upregulation of specific tRNA-derived RNA fragments in host cells, but no global changes of microRNAs in perfusion-rinsed mouse lung tissues were observed (80). Information regarding the potential role of extracellular RNAs during rickettsial infections is still lacking.
The aim of this study was to explore the potential role of rickettsia-infected, EC-derived Exos following infection. Using size exclusion chromatography (SEC), we purified Exos from conditioned, filtered, bacterium-free media collected from R. parkeri-infected human umbilical vein ECs (HUVECs) (R-ECExos) and plasma of R. australis- or R. parkeri-infected mice (R-plsExos). We observed that compared to noninfectious normal mouse plsExos and normal HUVEC-derived Exos, both R-plsExos and R-ECExos induced dysfunction of normal brain microvascular ECs (BMECs). The effect of R-plsExos on mouse recipient BMEC barrier function is dose dependent. The effect of R-ECExos on human recipient BMEC barrier function is dependent upon exosomal RNA cargos. Saponin-assisted active exosomal permeabilization pretreatment (81–83) of R-ECExos with RNase mitigated the effect of R-ECExos on human recipient BMEC barrier function. Next-generation sequencing analysis and stem-loop quantitative reverse transcription-PCR (RT-qPCR) validation revealed that R. parkeri infection triggered the selective enrichment of endothelial exosomal mir-23a and mir-30b, which potentially target the endothelial barrier.
RESULTS
Quality assessment of bacterium-free R-plsExos and media of R-ECExos.
Using SEC, we isolated small EVs (50 to 150 nm) from rickettsia-infected mouse plasma and HUVECs from culture media; both were passed through two 0.2-μm filters. Quantitative real-time PCR validated that no rickettsial DNA copies were detected in either R-plsExos isolated from R. australis- or R. parkeri-infected mice infected with 2 50% lethal doses (LD50) of bacteria (80, 84–87) on day 4 postinfection (p.i.) or the R-ECExos that were purified 72 h p.i. from R. parkeri-infected HUVECs (6) using a multiplicity of infection (MOI) of 10 (Fig. S1).
Quantities of rickettsiae in plasma-derived Exos (plsExos) (n = 5/group) and HUVEC culture media-derived Exos (ECExos) (n = 3/group), determined by quantitative real-time PCR. Data are presented as means ± standard errors. Statistical significance was determined using one-way analysis of variance. **, P < 0.01. Download FIG S1, JPG file, 0.1 MB (115.5KB, jpg) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
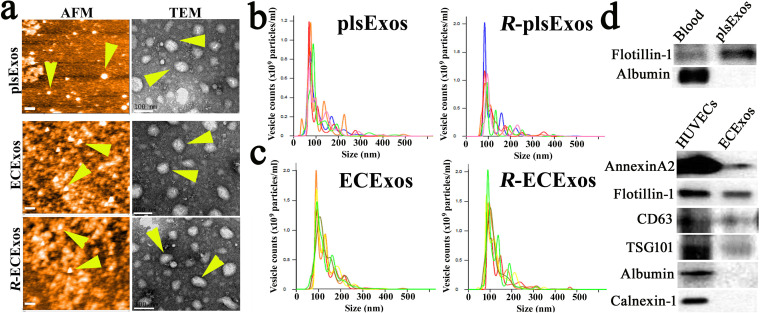
Sizes and morphologies of isolated EVs from mouse plasma and EC culture media, respectively, were initially evaluated using transmission electron microscopy (TEM) (52, 88) or atomic force microscopy (AFM) (89). The images captured using TEM and AFM show particles with typical exosomal morphology (arrowheads in Fig. 1a), as published previously (52, 88, 90). Using nanoparticle tracking analysis (NTA), the size distribution of isolated EVs was also confirmed to be in the range of 50 to 150 nm, which is the expected size of Exos (Fig. 1b and c). We also verified the purity of isolated Exos using Western immunoblotting to detect traditional exosomal markers as shown in Fig. 1d (64, 73, 90).
FIG 1.
Characterization of plsExos and ECExos after SEC isolation. (a) plsExos and ECExos morphologies were verified using atomic force microscopy (AFM) (left; scale bars, 200 nm) and transmission electronic microscopy (TEM) (right; scale bars, 100 nm). (b and c) The vesicle size distribution of isolated EVs was analyzed using nanoparticle tracking analysis (NTA) (n = 5 per group). (d) Expressions of indicated protein markers in 100 μg of proteins of plsExos (upper portion) and ECExos (lower portion) were examined using Western immunoblotting.
These data demonstrate that purified EVs from rickettsia-infected mouse plasma or culture media used in these studies were free of bacteria or bacterial DNA, were intact and did not aggregate, and fell within the expected size range of Exos.
Exos are differentially induced and detected in mouse plasma and EC culture media in response to rickettsial infection.
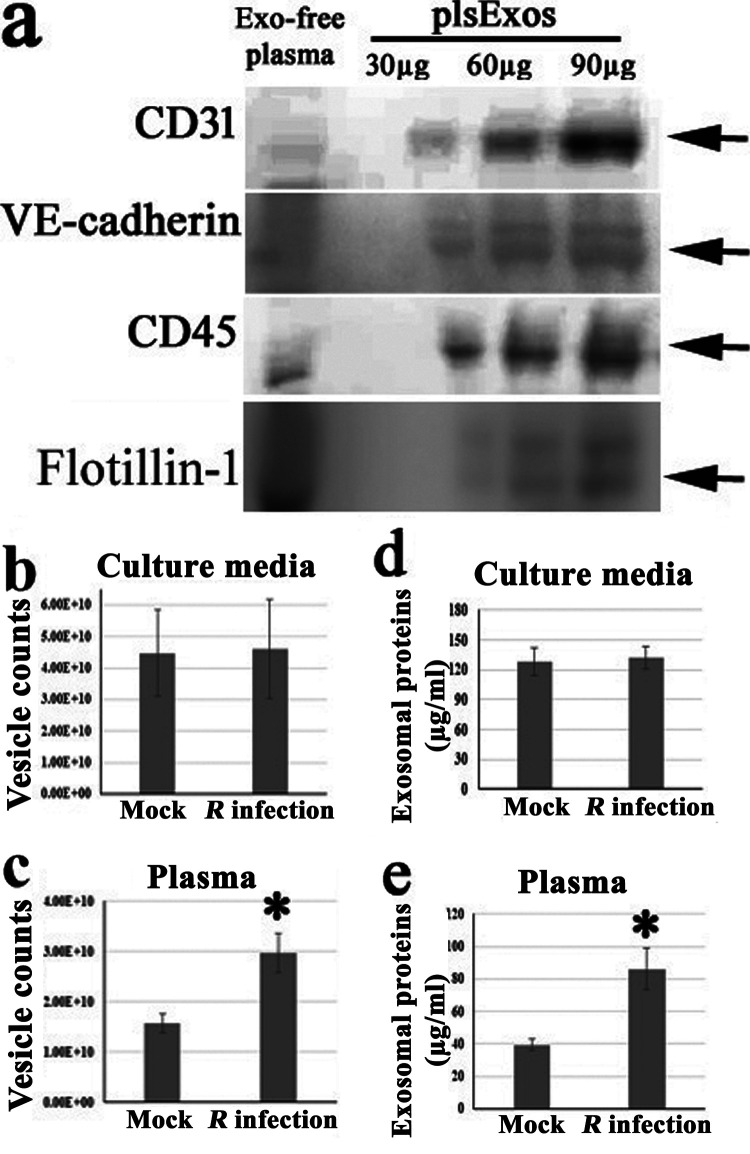
Exos from circulating blood have been identified as being heterogeneous and derived from multiple cell types, including ECs. Using Western immunoblotting, we detected Exo and EC markers (CD31 and VE-cadherin [91, 92]) in mouse plsExos, as well as markers of other cells (CD45) (Fig. 2a), suggesting that the mouse plsExos used in these studies were derived from different types of cells, including ECs.
FIG 2.
Exos are differentially induced and detected in mouse plasma and EC culture media in response to rickettsial infection. (a) Expression of indicated protein markers (i.e., 30, 60, and 90 μg of plsExos proteins) was examined using Western immunoblotting. (b and c) The concentration of plsExos and ECExos was analyzed using NTA (n = 5 per group). (d and e) The concentration of exosomal total protein was determined using the micro-bicinchoninic acid (microBCA) protein assay (n = 5 per group). Statistical significance was determined using Student’s t test. *, P < 0.05.
Exosomal particle counts were measured using NTA and showed that similar numbers of endothelial Exos were produced by mock and R. parkeri infection groups in vitro (Fig. 2b). However, the number of mouse R-plsExos was upregulated on day 4 p.i. (P = 0.02) in vivo (Fig. 2c) in the model of R. australis infection. Exos were also assessed using exosomal total protein content (Fig. 2d and e) (88). The generation of R-plsExos was significantly upregulated on day 4 p.i. in both in vivo models of R. australis (P = 0.016) (Fig. 2e) and R. parkeri (P = 0.014) (Fig. S2a) infections. No difference in exosomal total protein content was observed between ECExos and R-ECExos in the R. parkeri infection model (Fig. 2d). Furthermore, we also compared the morphology of EC-derived Exos using TEM and AFM, which demonstrated no significant differences between normal ECExos and R-ECExos (Fig. 1a).
(a) Quantities of plsExos on day 4 postinfection from mock-treated mice or mice infected with 2 LD50 of R. parkeri given by the intravenous route were assessed using the total protein concentration of plsExos (n = 5 per group). Statistical significance was determined using Student’s t test. (b) The TEER value of normal mouse recipient BMECs was measured after treatment with normal plsExos (mock) or R-plsExos (from an R. parkeri-infected mouse) at 400, 2,000, or 8,000 particles per cell for 72 h. Statistical significance was determined using one-way analysis of variance. *, P < 0.05. Download FIG S2, JPG file, 0.08 MB (86.7KB, jpg) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Collectively, these data suggest that R. australis and R. parkeri infections increase heterogeneous plsExo release. However, endothelial Exo size, morphology, and production were not significantly altered after R. parkeri infection in vitro.
Recipient cells efficiently take up Exos.
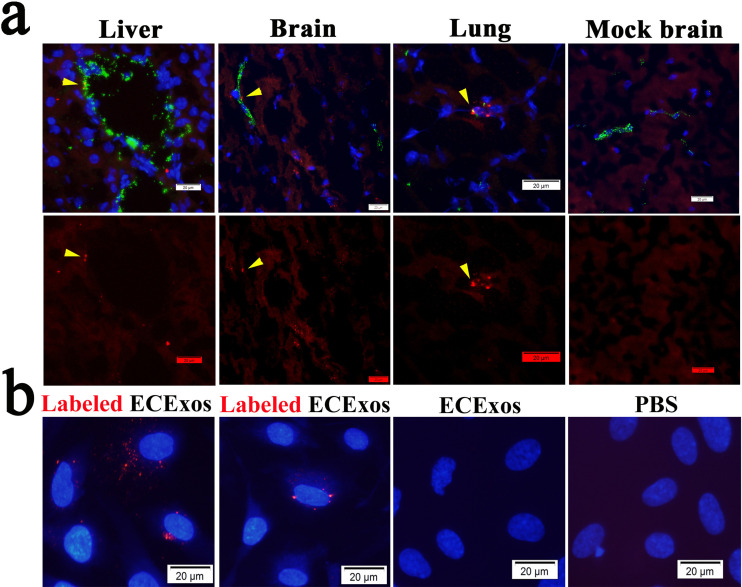
ECs are directly exposed to circulating substances and Exos, which are abundant in blood and are taken up by ECs (93, 94). To confirm that ECs take up Exos in vivo, we intravenously delivered fluorescent PKH26-prelabeled normal plsExos (1 × 1011 particles per mouse in 100 μl of phosphate-buffered saline [PBS]) to normal mice as described previously (90). As shown in Fig. 3a, colocalization between PKH26 (red) and CD31 (green, a marker of EC lineage) (arrowheads) were identified in multiple organs in mice, which were extensively perfused with PBS at 6 h postinjection, prior to fixation. These data suggest that plsExos directly interact with ECs in vivo.
FIG 3.
Recipient cells take up Exos. (a) Purified plsExos (5 × 1010 particles in 50 μl PBS) labeled with PKH26 were administered to normal C57BL/6J mice intravenously (n = 3). After 4 h, organs were dissected for frozen sectioning after euthanasia and perfusion via the right ventricle. Representative immunofluorescent staining of ECs from liver, brain, and lung using an antibody against CD31 (an EC marker) is shown. Normal rabbit serum was used as an antibody during immunofluorescent staining (Fig. S3). The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells with red fluorescence indicate the uptake of PKH26 labeled Exos. Scale bars, 20 μm. (b) Purified ECExos were labeled with PKH26 (red) and added to the culture medium of human BMECs (2,000 particles per cell) as indicated. Pictures were taken using fluorescence microscopy after 2 h of ECExo incubation. Scale bars, 20 μm.
Normal rabbit serum was used as a control antibody during immunofluorescence (IF) staining of a mouse lung tissue sample treated in same way. Scale bar indicates 20 μm. Download FIG S3, JPG file, 0.1 MB (137.2KB, jpg) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, we examined HUVEC-derived ECExo uptake using normal recipient cells (i.e., human BMECs) in vitro. PKH26-prelabeled ECExos and nonlabeled controls were added to the cultured human BMECs. As early as 2 h after incubation, the uptake of PKH26-prelabeled ECExos by BMECs was visualized using fluorescence microscopy (Fig. 3b).
These data suggest that vascular ECs efficiently take up Exos in our models.
Effect of mouse R-plsExos on normal mouse recipient ECs.
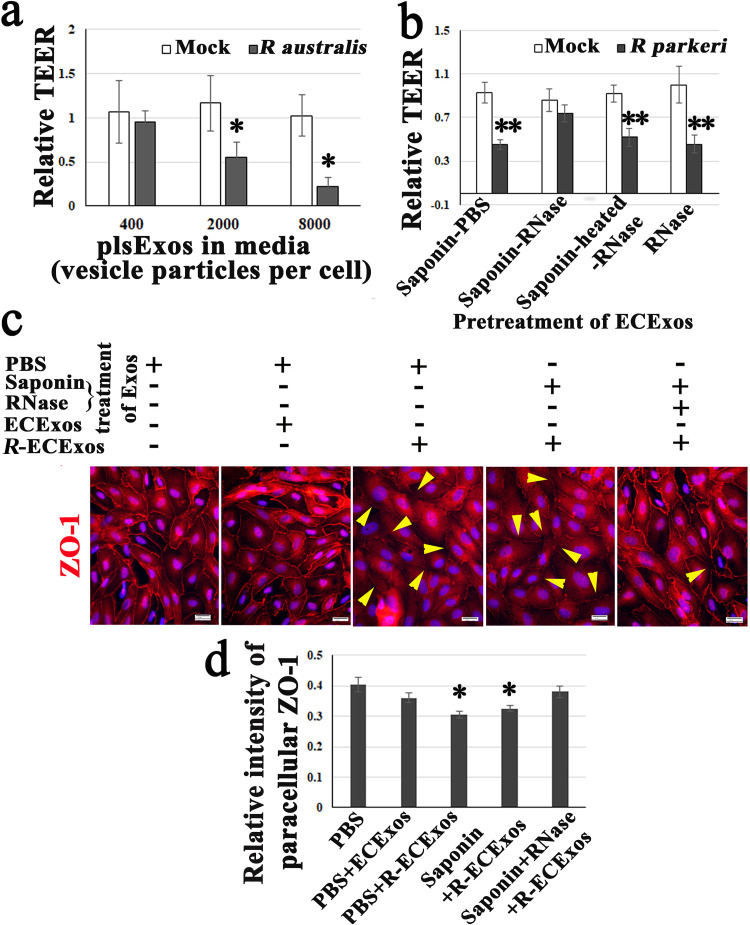
We next sought to evaluate the potential effect of R-plsExo on normal recipient ECs during rickettsial infection. Using SEC, R-plsExos from a mouse that was intravenously infected with a 2-LD50 dose of R. australis (86, 87) or R. parkeri (6, 95) were isolated. Normal mouse recipient BMECs were treated with normal plsExos or R-plsExos at different doses (i.e., 400, 2,000, or 8,000 Exo particles/per cell) for 72 h before measurement of the transendothelial electrical resistance (TEER), an indicator for endothelial paracellular barrier function (96). We found that compared to normal mouse plsExos, mouse R-plsExos derived on day 4 p.i. with R. australis or R. parkeri reduced the TEER in normal mouse recipient ECs in different dose-dependent manners (Fig. 4a and Fig. S2b).
FIG 4.
Effect of R-plsExos or R-ECExos on normal recipient ECs. (a) The transendothelial electrical resistance (TEER) values of normal mouse recipient BMECs were measured after treatment with normal plsExos (mock) or R-plsExos at 400, 2,000, or 8,000 Exo particles per cell for 72 h. *, P < 0.05. (b) The TEER values of normal human recipient BMECs were measured after a 72-h treatment with normal ECExos (mock) or R-ECExos (2,000 Exo particles per cell), which were pretreated with 20 μg/ml of RNase in the presence or absence of 0.1% saponin. **, P < 0.01. (c) Immunofluorescence staining of tight junctional protein ZO-1 (red) in normal human recipient BMECs that were treated with different Exos for 72 h. The yellow arrowheads indicate the decreased signals of paracellular ZO-1. Nuclei of human recipient BMECs were counterstained with DAPI (blue). (d) Relative fluorescent intensities of paracellular ZO-1. Normal rabbit serum was used as negative reagent control during immunofluorescent staining (Fig. S4). Scale bars, 20 μm. Statistical significance was determined using one-way analysis of variance. *, P < 0.05.
Normal rabbit serum was used as a control antibody during IF staining of human BMECs treated in same way. Scale bar indicates 20 μm. Download FIG S4, JPG file, 0.06 MB (64KB, jpg) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
This evidence suggests that mouse R. australis- or R. parkeri-plsExos induce dysfunction in normal recipient ECs in a dose-dependent manner.
Human R-ECExos induced dysfunction of normal human recipient ECs in an exosomal RNA-dependent manner.
Endothelial markers were detected in plsExos (Fig. 2a). Given that ECs are the major target cells during rickettsial infection, HUVEC-derived ECExos were used to explore their effect on normal human recipient BMEC function.
It was first found that compared with normal (mock) ECExos (2,000 Exo particles), R-ECExos (2,000 Exo particles) reduced TEER in normal human recipient BMECs (Fig. 4b). Furthermore, R-ECExos (2,000 Exo particles/cell) weakened the tight junctional protein ZO-1 (arrowheads in Fig. 4c) of normal human recipient BMECs.
Exos contain many types of biomolecules, including proteins and nucleic acids, which contribute to disease pathogenesis (68). Active encapsulation techniques have been widely employed in the field of EV research, showing no significant impairment of exosomal constitution, integrity, or functionality (81–83). To identify the functional exosomal cargos during R. parkeri infection, we employed saponin-assisted active permeabilization (81–83) to pretreat exosomal cargos with 20 μg/ml of RNase in the presence of 0.1 mg/ml of saponin. Such pretreatment of R-ECExos mitigated the effect on TEER in normal human recipient BMECs compared to RNase in the absence of permeabilization or heat-treated RNase in the presence of saponin (Fig. 4b). Similar pretreatment of R-ECExos with RNase in the presence of saponin also mitigated the effect on the tight junctional protein ZO-1 in normal human recipient BMECs (Fig. 4c and d).
We also isolated Exos from HUVEC culture media at 72 h after treatment with heat-inactivated R. parkeri (MOI, 10) and observed no differences in exosomal total protein contents compared to the mock group (Fig. S5a). Such ECExos (2,000 particles/cell) induced no effect on the paracellular ZO-1 of normal human recipient BMECs (Fig. S5b and c).
(a) Quantities of ECExos from mock-, heat-inactivated R. parkeri-, or R. parkeri-infected HUVECs 72 h p.i. at an MOI of 10 were assessed using the total protein concentration of ECExos (n = 4 per group). (b and c) Immunofluorescence staining of tight junctional protein ZO-1 (red) and relative fluorescent intensities of paracellular ZO-1. Statistical significance was determined using one-way analysis of variance. Scale bar indicates 20 μm. *, P < 0.05. Download FIG S5, JPG file, 0.4 MB (419.7KB, jpg) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
These data suggest that R-ECExos can induce normal human recipient EC barrier dysfunction in an exosomal RNA cargo-dependent manner.
R. parkeri infection upregulates exosomal mir-23a and mir-30b.
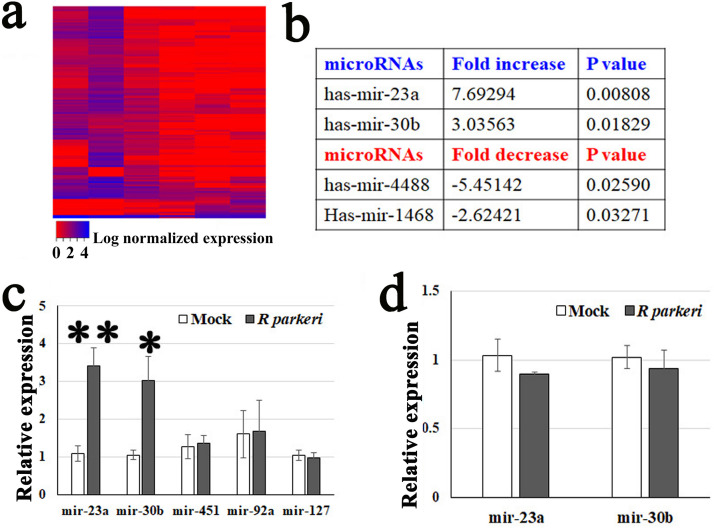
EV RNA cargo mostly consists of sncRNAs, mainly microRNAs and tRNA-derived fragments (61, 77, 78). A growing number of reports have established that many effects of EVs are mediated by microRNAs (52, 55–60, 63). We characterized the exosomal microRNA cargo using next-generation sequencing (Fig. 5a). RNAs were isolated from Exos released from HUVECs infected with R. parkeri (at an MOI of 10) for 72 h or mock infected. R. parkeri is a biosafety level 2 (BSL2) pathogen, which lends itself to mechanistic studies. There were no differences in total sncRNAs (<150 nucleotides) per Exo obtained from normal ECExos compared with R-ECExos. Seventy-two hours after R. parkeri infection, mir-23a and mir-30b exhibited the greatest induction of expression in R-ECExos, reaching 7.69-fold and 3.04-fold increases compared to controls, respectively (Fig. 5b).
FIG 5.
Rickettsial infection alters microRNA expression in ECExos. (a) Heat map clustering of microRNAs in normal ECExos versus R-ECExos (n = 3). (b) microRNA expression in R-ECExos versus normal ECExos (n = 3). (c) Stem-loop RT-qPCR analysis of microRNAs obtained from normal ECExos (mock) and R-ECExos (rickettsial). **, P < 0.01; *, P < 0.05. (d) Stem-loop RT-qPCR analysis of microRNAs obtained from normal (mock) and rickettsia-infected donor HUVECs. Statistical significance was determined using one-way analysis of variance.
We next validated the enhanced expression of mir-23a and mir-30b in Exos using stem-loop RT-qPCR, which is a common method for detecting sncRNAs in EVs (52, 61, 97, 98). In Fig. 5c, exosomal mir-23a was upregulated after rickettsial infection with a 3-fold increase in expression compared to the mock group (P < 0.01). Similarly, mir-30b had a nearly 3-fold increase (P < 0.05). However, the levels of mir-127 (99), mir-451, and mir-92a were stable between mock ECExos and R-ECExos (Fig. 5c). Furthermore, we did not detect different levels of these miRNAs in cell samples between groups (Fig. 5d). mir-23a (100–103) and mir-30b (103, 104) have been documented to target endothelial barrier functions. Analysis of the interactions among mir-23a and mir-30b and potential mRNA targets (Tables S1 and S2) suggests putative mRNA candidates potentially associated with vascular endothelial barrier functions.
GO enrichment analysis of mir-23a-3p targeted genes. aCount is the number of genes in the user-provided lists with membership in the given ontology term. Log10(P) is the P value in log base 10. Download Table S1, DOCX file, 0.01 MB (15.2KB, docx) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
GO enrichment analysis of mir-30b-5p targeted genes. aCount is the number of genes in the user-provided lists with membership in the given ontology term. Log10(P) is the P value in log base 10. Download Table S2, DOCX file, 0.01 MB (15.3KB, docx) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Collectively, our data suggest that mir-30b and mir-23a are selectively sorted into R-ECExos following R. parkeri infection.
We have performed an additional experiment to examine mouse mir-23a and mouse mir-30b in mouse plsExos using stem-loop RT-qPCR. As shown in Fig. S6, levels of mouse plsExo mir-23a (P = 0.413) and mir-30b (P = 0.237) increased after R. parkeri infection, but the results were not statistically significant.
Stem-loop RT-qPCR analysis of microRNA expression in plsExos. microRNA expression in normal plsExos versus R-plsExos (n = 6). R-plsExos were isolated from mice on day 4 after infection with 2 LD50 of R. parkeri given by the intravenous route. Statistical significance was determined using one-way analysis of variance. No statistical differences were found using a P value of <0.05. Download FIG S6, JPG file, 0.05 MB (47.7KB, jpg) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
ECs are the primary mammalian host target cells of SFR infection (2, 5, 6). The most prominent pathophysiological effect during SFR infections is increased microvascular permeability, followed by vasogenic cerebral edema and noncardiogenic pulmonary edema with potentially fatal outcomes (2, 5). Cellular and molecular mechanisms underlying endothelial barrier dysfunction in rickettsiosis remain largely unknown (7–9). The novel findings in the present study are that R. australis or R. parkeri infection increases the release of heterogeneous plsExos, but endothelial Exo size, morphology, and production are not significantly altered following infection. Mouse R-plsExos induced dysfunction of normal mouse recipient BMECs in a dose-dependent manner, and human R-ECExos induced dysfunction of normal human recipient BMECs in an exosomal RNA cargo-dependent manner. Next-generation sequencing and stem-loop RT-qPCR analyses suggested that mir-23a and mir-30b are selectively sorted into R-ECExos after R. parkeri infection. To our knowledge, this is the first report involving EVs in obligately intracellular bacterial infections.
Exos are in a size range similar to that of viruses (33, 94) and contain many types of biomolecules, including proteins and nucleic acids, which contribute to diseases pathogenesis. Exos are being actively investigated in cancers, as biomarkers, and as potential therapeutics (33–38, 68, 70). Exos have been studied in the context of different infections (33–38, 70, 71). During infection, EVs released from the host can be derived from the pathogen or the host. It has been reported that pathogens can utilize different mechanisms to hijack host Exos to maintain their survival and increase their pathogenicity (33). Mycobacterium tuberculosis releases lipoarabinomannan into Exos to decrease the interferon response of the recipient macrophage (33). Exosomal gp63 from Leishmania has been shown to downregulate proinflammatory genes in dendritic cells and macrophages (105). Exos released from Leishmania donovani-infected macrophages block the formation of microRNA-122 in recipient hepatocytes, resulting in a higher parasite burden (106). However, most enveloped virions are the same size as Exos, and major exosomal surface markers CD63 and CD81 are enriched in enveloped viruses (54). Such similarities make the separation of virions and Exos in infected samples particularly challenging (54). Rickettsiae are strictly intracellular bacteria that are about 2.0 μm in length (22, 107, 108). We succeeded in isolating and purifying bacterium-free plsExos and ECExos from rickettsia-infected mouse plasma and cell culture media, respectively, taking advantage of SEC technology, which now serves as the technical foundation for studying the potential role of Exos in the pathogenesis of rickettsiosis.
Differential centrifugation has been employed for Exo isolation for many years, but the technique suffers from aggregation and decreased integrity of Exos (38, 53, 64–66). Recently, single-step SEC was employed successfully for Exo purification, with improved integrity, yield, and no aggregation (38, 53, 64–66). In the present study, using SEC technology, we have successfully isolated Exos from plasma and culture media in BSL2/3 facilities and validated Exo quality using multiple EV-specific assays (Fig. 1) demonstrating size, purity, and morphological integrity without aggregation. Exo size and morphology were not significantly changed after rickettsial infection (Fig. 1). Generation of plsExos was significantly upregulated after R. australis or R. parkeri infection, while no difference was detected in plasma protein concentrations. However, our in vitro endothelial R. parkeri infection model demonstrated no significant difference in exosomal generation between normal and rickettsia-infected ECs. Circulating Exos have been identified as heterogeneous and derived from multiple different types of cells, including ECs, epithelial cells, leukocytes, erythrocytes, and platelets (90). Our data suggest that ECExo size, morphology, and production were not significantly altered after infection. However, quantitative information regarding the production of SFG rickettsial infection-induced cell-type Exos in vivo remains elusive and requires further research.
Research on most species of lethal human SFG rickettsial infections, including R. rickettsii (3, 4), R. conorii (9), and R. australis (10), is restricted to BSL3 facilities. One key experimental method in our study was to employ novel SEC technology to isolate high-quality Exos from plasma and culture media. Vascular ECs are exceedingly thin (109). Preliminary experiments revealed that we obtained smaller amounts of Exo particles from ECs cultured in the same-size culture vessel at the same confluence for the same duration compared with other cell types. It is practical and feasible to isolate endothelial Exos from media using SEC on a relatively larger scale following infection with R. parkeri (5–7), which requires a lower biocontainment level than R. australis. R. parkeri, the cause of R. parkeri rickettsiosis (8), is proposed as an experimental pathogen for studying the pathogenesis of SFG rickettsiosis and can be experimentally handled at BSL2 (6, 95). In a murine model of intravenous inoculation of R. parkeri (Atlantic Rainforest strain) using C3H/HeN mice, signs of illness begin on day 3 and fatalities occur on day 6 p.i. (95), during which time microvascular damage is detected in multiple organs (95); this is similar to the pathology observed in our murine model of R. australis infection (85, 87). However, differences among various pathogen species must be considered in future mechanistic studies. Furthermore, current technological hurdles hamper us from isolating ECExos from plasma on a large scale for downstream experiments. To collect sufficient ECExos using SEC technology for mechanistic studies, culturing cells in media is the sole practical source for ECExo isolation. To investigate the functional role of ECExos in R. parkeri infection, we used HUVECs as the source of Exo-producing cells and BMECs as the recipient cells. Research to develop feasible cell-type Exo isolation using the same pathogens in the same host models is warranted for future mechanistic studies.
Exos can induce functional responses using multiple mechanisms, including releasing bioactive components after internalization (67, 68). In infectious disease biology, EVs from infected donor cells are associated with virulence of the pathogens in recipient cells (33–38, 69–71). In the present study, both R-plsExos and R-ECExos weakened the barrier function of the normal ECs. Concomitantly, human R-ECExos induced disruption of the tight junctional protein ZO-1 in recipient human BMECs in an exosomal RNA-dependent manner. However, the underlying mechanism remains unclear.
The discovery of extracellular sncRNAs in the blood, despite the abundant presence of RNases, led to the proposal of a scenario in which sncRNAs are encapsulated in EVs (55, 72–74) or in the form of circulating ribonucleoproteins (75, 76). EV-enclosed mRNAs are mostly fragmented, and extracellular RNAs are enriched in sncRNAs (77). Despite a previous report that the average copy number of miRNAs in each EV is low (110), accumulating evidence suggests a critical function of EV-containing miRNAs. EV RNA cargo mostly consists of sncRNAs (77). A growing number of reports have established that many, if not all, of the effects of EVs are mediated by microRNAs (52, 55–60, 63), which remain functional to regulate cellular behaviors of the recipient cell (79). Exosomal microRNAs are of particular interest due to their participation in posttranslational regulation of gene expression. A single microRNA can regulate many target genes to affect biological function (111). Recent studies provide evidence that microRNAs are selectively sorted into EVs, independent of their cellular levels (52, 55–62). We observed no significant differences in total sncRNAs (<150 nucleotides) per Exo between normal ECExo and R-ECExo. Seventy-two hours after R. parkeri infection, expression levels of mir-23a and mir-30b were remarkably upregulated in R-ECExos, but no change was observed in the mock-infected cells. These data suggest that mir-23a and mir-30b are selectively sorted into R-ECExos during R. parkeri infection. The underlying mechanism is yet to be elucidated.
Analysis of the interactions among enriched exosomal microRNAs and potential mRNA targets will provide putative mRNA candidates for future studies. Given that the molecular and functional effects of mir-23a (100–103) and mir-30b (103, 104) have been documented to target endothelial barrier functions, further research into the selective sorting mechanism(s) and functional roles of exosomal mir-23a and mir-30b may provide new insights into the pathogeneses of SFR. Furthermore, additional research may validate specific exosomal microRNAs as impactful druggable targets for the prevention and treatment of fatal human diseases caused by Rickettsia and other pathogens.
MATERIALS AND METHODS
Mouse models of R. australis and R. parkeri infections.
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of the University of Texas Medical Branch (UTMB). C57BL/6J mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Mice used for R. australis infections were 8- to 12-week-old males. C57BL/6J mice are highly susceptible to R. australis, becoming ill on day 3 p.i. and succumbing to infection on day 6. Therefore, this organism was chosen as the SFG rickettsial agent for the in vivo studies (10). Male C57BL/6 mice infected with R. australis are an established animal model of human SFG rickettsiosis because the pathology involves disseminated endothelial infection and pathological lesions, including vasculitis in multiple organs, similar to what is observed in human SFG rickettsiosis (10, 87). However, use of R. australis requires BSL3 containment. An ordinarily lethal dose of 2 LD50 of R. australis (the LD50 is 1 × 106 PFU) was injected through the tail vein (87), and whole-blood samples were collected on day 4 p.i. for plasma isolation.
R. parkeri is proposed as an experimental pathogen for studying the pathogenesis of SFG rickettsiosis and is similar to R. australis (10, 84, 86, 87), but it can be experimentally handled using BSL2 containment (6, 95). For isolation of endothelial Exos from infected media on a large scale, it is more practical and feasible to use R. parkeri. In a murine model of intravenous inoculation of R. parkeri (Atlantic Rainforest strain) using C3H/HeN mice, signs of illness began on day 3 and fatalities occurred on day 6 p.i. (95), during which time microvascular damage was detected in multiple organs (95), which is similar to the pathology observed in the murine model of R. australis (85, 87). C3H/HeJ mice (8- to 12-week-old males) were obtained from the Jackson Laboratory. An ordinarily lethal dose of 2 LD50 of R. parkeri (Atlantic Rainforest strain) was injected through the tail vein (87), and blood samples were collected on day 4 p.i. for plasma isolation.
NTA.
To determine the size and concentration of EVs, nanoparticle tracking analysis (NTA) was performed at the Nanomedicines Characterization Core Facility (University of North Carolina, Chapel Hill, NC). Briefly, isolated Exo samples were diluted to a concentration of 5 × 109 to 1 × 1011 particles/ml in filtered PBS. The samples were then processed on a NanoSight NS500 (NanoSight, Malvern Instruments, Westborough, MA) to capture particles moving by way of Brownian motion (camera type, scientific complementary metal oxide semiconductor [sCMOS]; camera level, 16; detection threshold, 5). The hydrodynamic diameters were calculated using the Stokes-Einstein equation. The 100-nm standard particles and the diluent PBS alone were used for reference.
MicroRNA quantification in HUVEC Exos.
As previously reported (52), RNAs were extracted from purified HUVEC Exos. Small RNAs (6 to 150 nucleotides) and microRNA fractions (10 to 40 nucleotides) were quantified using high-resolution small RNA analysis (2100 Bioanalyzer system; Agilent, Santa Clara, CA) at the Biopolymer Facility (Harvard Medical School, Cambridge, MA). To determine the concentration of small RNAs and microRNAs per Exo, the quantified sncRNA/microRNA value was normalized to the Exo count, which was evaluated using NTA.
Bioinformatic analysis of sequencing data.
Sequencing was done using an Illumina NextSeq as single-end 75-bp reads generating between 4.8 and 40.9 million reads per sample. Quality control of the samples was performed using Qiagen CLC Genomics Workbench 20.0. Raw sequencing reads were trimmed to remove Qiagen 3′-AACTGTAGGCACCATCAAT and 5′-GTTCAGAGTTCTACAGTCCGACGATC adapters, as well as filtered based on initial quality assessment. Reads dominated by low-quality base calls, and longer than 55 nucleotides, were excluded from the downstream analyses. Filtered data underwent further transcriptome sequencing (RNA-Seq) analysis using the CLC Genomics Workbench 20.0 RNA-Seq Analysis 2.2 module with RNAcentral noncoding human RNA (112, 113) (downloaded 16 April 2020), miRbase 22.1, and the ENSMBL GRCh38 noncoding RNA gene collection (114) (downloaded 20 November 2019). Differential expression analysis was performed using the Differential Expression in Two Groups 1.1 module. The differential expression module uses multifactorial statistics based on a negative binomial generalized linear model (GLM) to correct for differences in library size between the samples and the effects of confounding factors. The Wald test was used to compare the expression of noncoding RNA between the groups.
Atomic force microscopy (AFM).
The purified and concentrated EV samples were diluted at 1:10, 1:100, and 1:1,000 with molecular-grade water. Glass coverslips were cleaned three times with ethanol and acetone and then three times with molecular-grade water. Each coverslip was correctly labeled, placed in the hood to dry under laminar flow for 1 h, and subjected to coating with the diluted EV samples on the designated area for 30 min. EV samples were washed away gently with molecular-grade water, and the coverslip was dried for 1 h.
The coverslip coated with EV samples was examined using an AFM (CoreAFM; Nanosurf AG, Liestal, Switzerland) using contact mode in the air. A PPP-FMR-50 probe (0.5 to 9.5 N/m, 225 μm in length, and 28 μm in width; Nanosensors, Neuchatel, Switzerland) was used. The parameters of the cantilever were calibrated using the default script from the CoreAFM program using the method of Sader et al. (115). The cantilever was approached to the sample under the set point of 20 nN (115), and topography scanning was done using the following parameters: 256 points per line and 1.5 s per line in a 5-μm by 5-μm image.
Rickettsiae, cell culture, and rickettsial infections.
R. australis (Cutlack strain) (87) and R. parkeri (Atlantic Rainforest strain) (95) were prepared as described previously. Uninfected Vero cells were processed as mock-infected control material using the same procedure. All biosafety level (BSL)2/3 and animal BSL2/3 experiments were performed in CDC-certified facilities in the Galveston National Laboratory at UTMB, Galveston, TX, using established procedures.
A standard protocol to isolate brain microvascular endothelial cells (BMECs) from C57BL/6 and C3H/HeN mice (116) was used. Human umbilical vein endothelial cells (HUVECs; Cell Applications, Inc.) or BMECs were cultivated in 5% CO2 at 37°C on type I rat tail collagen-coated round glass coverslips (12-mm diameter; Ted Pella, Redding, CA) until 90% confluence was observed. HUVECs were infected with R. parkeri at an MOI of 10. Uninfected ECs were used as mock controls and were subjected to the same procedure. All experiments were performed in triplicate. Normal mouse or rabbit IgGs were used as negative controls.
ECExo and plsExo isolation, concentration, and permeabilization.
(i) ECExo isolation and concentration. Donor HUVECs in T75 flasks were infected using R. parkeri at an MOI of 10 or were mock infected for 72 h, and 11 ml of medium was collected. The media were passed through 0.2-μm syringe filters twice. Following the manufacturer’s instructions, 10 ml of filtered medium was subjected to the qEV10 column (Izon, New Zealand) for SEC isolation. Four fractions (the number 7 to 10 fractions) were collected as the Exo-enriched fractions, which were concentrated using 100,000-molecular-weight-cutoff (MWCO) polyethersulfone (PES) Vivaspin centrifugal filters (Thermo Fisher Scientific). Exo samples (in 200 μl of PBS) were stored at −80°C prior to use in downstream assays.
(ii) plsExo isolation and concentration. For plasma isolation, blood samples were collected in anticoagulation tubes 4 days following infection with R. australis, R. parkeri, or saline (mock infected). The plasma sample (200 μl) was passed through 0.2-μm syringe filters twice. Following the manufacturer’s instructions, filtered plasma was placed onto the qEVoriginal column (Izon, New Zealand) for SEC isolation. The number 7 to 9 fractions were collected as the Exo-enriched fractions, which were concentrated using 100,000-MWCO PES Vivaspin centrifugal filters (Thermo Fisher Scientific). Exo samples (in 200 μl of PBS) were stored at −80°C prior to use in downstream assays. Following the manufacturer’s instructions, three fractions (the number 13 to 15 fractions) from the qEVoriginal column isolation were collected as the Exo-free, high-protein plasma fractions, which were concentrated using centrifugal filters and used in Western immunoblotting as a control.
For saponin-assisted active exosomal permeabilization pretreatment (81–83) of Exos using RNase, Exo samples (1 × 109 particles/ml) and RNase (20 μg/ml) (Thermo Fisher Scientific) were incubated with 0.1 mg/ml of saponin (Thermo Fisher Scientific) at room temperature for 15 min. After being rinsed using PBS, Exo samples were concentrated using 100,000-MWCO PES Vivaspin centrifugal filters.
Distribution of Exos in vivo and in vitro.
Using a published approach, recipient cell uptake of Exos was assessed in vivo and in vitro (90). Briefly, following incubation using materials from the PKH26 red fluorescent cell linker kit (Millipore Sigma, St. Louis, MO), the PKH26 red-prelabeled Exos were washed three times with PBS prior to ultracentrifugation at 100,000 × g for 20 min at 4°C using a Beckman L7-80 and SW41 rotor (Beckman Coulter, Indianapolis, IN) to remove unbound stain. PBS without Exos was processed using the same steps as the mock PKH26-labeled tracer. A single injection of PKH26-labeled exosomes (about 1 × 1011 particles in 100 μl of PBS) via the tail vein of a normal mouse was done to observe the distribution of Exos in the lungs, liver, and brain 6 h after injection. Immunofluorescence staining was done using frozen sections with rabbit antibodies to CD31. For the in vitro assessment, living BMECs were exposed to PKH26-prelabeled ECExos (2,000 particles/cell) in the culture media of normal human BMECs. After 2 h, cells were fixed and immediately subjected to fluorescence microscopy. All solutions of PKH26-labeled ECExos were filtered with a 0.2-μm filter. Fluorescent images were analyzed using Olympus BX51 epifluorescence and a Nikon A1R MP ECLIPSE Ti confocal microscope with NIS-Elements imaging software version 4.50.00 (Nikon, Tokyo, Japan).
Stem-loop real-time PCR.
Total RNA was extracted from EVs by using TRIzol (Invitrogen). An exogenous synthetic microRNA, namely, cel-mir-39, was diluted in TRIzol before extraction to act as a normalizer. The concentration of total RNA was measured by NanoDrop (ND-2000). Primers for cel-mir-39, has-mir-23a, has-mir-30b, has-mir-92a, has-mir-451, has-mir-127, mmu-mir-23a, mmu-mir-30b, mmu-mir-92a, and mmu-mir-127 for TaqMan microRNA assay INV SM 10 were purchased from Thermo Fisher Scientific. A TaqMan microRNA reverse transcription kit (Applied Biosystems) was used for reverse transcription reactions. The 15-μl RT reaction mixtures contained 5 ng of total RNA template, 3 μl of RT primer (5×), 0.15 μl of deoxynucleoside triphosphate (dNTPs; 100 mM), 1 μl of MultiScribe reverse transcriptase (50 U/μl), 1.5 μl of reverse transcription buffer (10×), 0.19 μl of RNase inhibitor (20 U/μl), and 4.16 μl of nuclease-free water. Reverse transcription conditions were 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. For PCR amplification, the 10-μl PCRs included 0.7 μl of cDNA template acquired as described above, 0.5 μl of TaqMan small RNA assay mix (20×), 5 μl of PCR master mix, and 3.8 μl of nuclease-free water. qPCR conditions were 50°C for 2 min and 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 65°C for 30 s. The relative expression of each miRNA was expressed as 2−ΔΔCT by the CFX Connect real-time system (Bio-Rad, Hercules, CA).
Statistics.
Statistical significance was determined using Student’s t test or one-way analysis of variance. Results were regarded as significant if two-tailed P values were <0.05. All data are expressed as means ± standard errors of the means.
Supplemental Materials and Methods. Download Text S1, DOCX file, 0.03 MB (28.2KB, docx) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We gratefully acknowledge Pragnesh Patel for his contributions establishing the capacity of the exosomal size distribution analysis. We gratefully acknowledge Kimberly Schuenke for her critical review and editing of the manuscript. We thank Hugo Samano for input during the planning phases of the experiments.
This work was supported by NIH grant R01AI121012 (B.G.), R21AI137785 (B.G.), R21AI154211 (B.G.), R03AI142406 (T.B.S. and B.G.), and R21AI144328 (T.B.S. and B.G.).
The sponsors had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
B.G. and T.B.S. designed the study, performed experiments, analyzed data, and wrote the manuscript. Y.L., C.Z., Z.S., Q.C., and K.K. performed experiments, analyzed data, and the prepared the manuscript. Y.Q., J.B., A.G., J.X., and A.D. analyzed data. Y.J. and G.G. designed the study and analyzed data.
We declare that we have no conflicts of interest.
Footnotes
Citation Liu Y, Zhou C, Su Z, Chang Q, Qiu Y, Bei J, Gaitas A, Xiao J, Drelich A, Khanipov K, Jin Y, Golovko G, Saito TB, Gong B. 2021. Endothelial exosome plays a functional role during rickettsial infection. mBio 12:e00769-21. https://doi.org/10.1128/mBio.00769-21.
Contributor Information
Tais B. Saito, Email: tais.berellisaito@nih.gov.
Bin Gong, Email: bigong@utmb.edu.
Daniel E. Voth, University of Arkansas for Medical Sciences
Jimmy D. Ballard, University of Oklahoma Health Sciences Center
REFERENCES
- 1.Dumler JS, Walker DH. 2005. Rocky Mountain spotted fever—changing ecology and persisting virulence. N Engl J Med 353:551–553. doi: 10.1056/NEJMp058138. [DOI] [PubMed] [Google Scholar]
- 2.Parola P, Socolovschi C, Jeanjean L, Bitam I, Fournier PE, Sotto A, Labauge P, Raoult D. 2008. Warmer weather linked to tick attack and emergence of severe rickettsioses. PLoS Negl Trop Dis 2:e338. doi: 10.1371/journal.pntd.0000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman AS, Murphy SM, Demma LJ, Holman RC, Curns AT, McQuiston JH, Krebs JW, Swerdlow DL. 2006. Rocky mountain spotted fever in the United States, 1997–2002. Ann N Y Acad Sci 1078:154–155. doi: 10.1196/annals.1374.026. [DOI] [PubMed] [Google Scholar]
- 4.Walker DH, Paddock CD, Dumler JS. 2008. Emerging and re-emerging tick-transmitted rickettsial and ehrlichial infections. Med Clin North Am 92:1345–1361, x. doi: 10.1016/j.mcna.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Drexler NA, Dahlgren FS, Heitman KN, Massung RF, Paddock CD, Behravesh CB. 2016. National surveillance of spotted fever group rickettsioses in the United States, 2008–2012. Am J Trop Med Hyg 94:26–34. doi: 10.4269/ajtmh.15-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saito TB, Bechelli J, Smalley C, Karim S, Walker DH. 2019. Vector tick transmission model of spotted fever rickettsiosis. Am J Pathol 189:115–123. doi: 10.1016/j.ajpath.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamason RL, Bastounis E, Kafai NM, Serrano R, Del Álamo JC, Theriot JA, Welch MD. 2016. Rickettsia Sca4 reduces vinculin-mediated intercellular tension to promote spread. Cell 167:670–683.e10. doi: 10.1016/j.cell.2016.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitman TJ, Richards AL, Paddock CD, Tamminga CL, Sniezek PJ, Jiang J, Byers DK, Sanders JW. 2007. Rickettsia parkeri infection after tick bite. Emerg Infect Dis 13:334–336. doi: 10.3201/eid1302.061295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan YG, Riley SP, Martinez JJ. 2010. Adherence to and invasion of host cells by spotted fever group rickettsia species. Front Microbiol 1:139. doi: 10.3389/fmicb.2010.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng HM, Wen J, Walker DH. 1993. Rickettsia australis infection: a murine model of a highly invasive vasculopathic rickettsiosis. Am J Pathol 142:1471–1482. [PMC free article] [PubMed] [Google Scholar]
- 11.Walker DH, Ismail N. 2008. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol 6:375–386. doi: 10.1038/nrmicro1866. [DOI] [PubMed] [Google Scholar]
- 12.Sahni A, Fang R, Sahni SK, Walker DH. 2019. Pathogenesis of rickettsial diseases: pathogenic and immune mechanisms of an endotheliotropic infection. Annu Rev Pathol 14:127–152. doi: 10.1146/annurev-pathmechdis-012418-012800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valbuena G, Walker DH. 2009. Infection of the endothelium by members of the order Rickettsiales. Thromb Haemost 102:1071–1079. doi: 10.1160/TH09-03-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paris DH, Dumler JS. 2016. State of the art of diagnosis of rickettsial diseases: the use of blood specimens for diagnosis of scrub typhus, spotted fever group rickettsiosis, and murine typhus. Curr Opin Infect Dis 29:433–439. doi: 10.1097/QCO.0000000000000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Openshaw JJ, Swerdlow DL, Krebs JW, Holman RC, Mandel E, Harvey A, Haberling D, Massung RF, McQuiston JH. 2010. Rocky mountain spotted fever in the United States, 2000–2007: interpreting contemporary increases in incidence. Am J Trop Med Hyg 83:174–182. doi: 10.4269/ajtmh.2010.09-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botelho-Nevers E, Socolovschi C, Raoult D, Parola P. 2012. Treatment of Rickettsia spp. infections: a review. Expert Rev Anti Infect Ther 10:1425–1437. doi: 10.1586/eri.12.139. [DOI] [PubMed] [Google Scholar]
- 17.de Sousa R, Nóbrega SD, Bacellar F, Torgal J. 2003. Mediterranean spotted fever in Portugal: risk factors for fatal outcome in 105 hospitalized patients. Ann N Y Acad Sci 990:285–294. doi: 10.1111/j.1749-6632.2003.tb07378.x. [DOI] [PubMed] [Google Scholar]
- 18.Kim HK, Premaratna R, Missiakas DM, Schneewind O. 2019. Rickettsia conorii O antigen is the target of bactericidal Weil-Felix antibodies. Proc Natl Acad Sci U S A 116:19659–19664. doi: 10.1073/pnas.1911922116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rennoll SA, Rennoll-Bankert KE, Guillotte ML, Lehman SS, Driscoll TP, Beier-Sexton M, Rahman MS, Gillespie JJ, Azad AF. 2018. The cat flea (Ctenocephalides felis) immune deficiency signaling pathway regulates Rickettsia typhi infection. Infect Immun 86:e00562-17. doi: 10.1128/IAI.00562-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hillman RD, Baktash YM, Martinez JJ. 2013. OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with α2β1 integrin. Cell Microbiol 15:727–741. doi: 10.1111/cmi.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsheim RF, Kurtti TJ, Munderloh UG. 2009. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS One 4:e8361. doi: 10.1371/journal.pone.0008361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suwanbongkot C, Langohr IM, Harris EK, Dittmar W, Christofferson RC, Macaluso KR. 2019. Spotted fever group Rickettsia infection and transmission dynamics in Amblyomma maculatum. Infect Immun 87:e00804-18. doi: 10.1128/IAI.00804-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung LK, Park YH, Zheng Y, Brodsky IE, Hearing P, Kastner DL, Chae JJ, Bliska JB. 2016. The Yersinia virulence factor YopM hijacks host kinases to inhibit type III effector-triggered activation of the pyrin inflammasome. Cell Host Microbe 20:296–306. doi: 10.1016/j.chom.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao X, Kajino-Sakamoto R, Doss A, Aballay A. 2017. Distinct roles of sensory neurons in mediating pathogen avoidance and neuropeptide-dependent immune regulation. Cell Rep 21:1442–1451. doi: 10.1016/j.celrep.2017.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behera AK, Hildebrand E, Uematsu S, Akira S, Coburn J, Hu LT. 2006. Identification of a TLR-independent pathway for Borrelia burgdorferi-induced expression of matrix metalloproteinases and inflammatory mediators through binding to integrin alpha 3 beta 1. J Immunol 177:657–664. doi: 10.4049/jimmunol.177.1.657. [DOI] [PubMed] [Google Scholar]
- 26.Allen LH, Criss AK. 2019. Cell intrinsic functions of neutrophils and their manipulation by pathogens. Curr Opin Immunol 60:124–129. doi: 10.1016/j.coi.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darville T, Hiltke TJ. 2010. Pathogenesis of genital tract disease due to Chlamydia trachomatis. J Infect Dis 201(Suppl 2):S114–S125. doi: 10.1086/652397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visvabharathy L, Freitag NE. 2017. Propofol sedation exacerbates kidney pathology and dissemination of bacteria during Staphylococcus aureus bloodstream infections. Infect Immun 85:e00097-17. doi: 10.1128/IAI.00097-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paudel S, Baral P, Ghimire L, Bergeron S, Jin L, DeCorte JA, Le JT, Cai S, Jeyaseelan S. 2019. CXCL1 regulates neutrophil homeostasis in pneumonia-derived sepsis caused by Streptococcus pneumoniae serotype 3. Blood 133:1335–1345. doi: 10.1182/blood-2018-10-878082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maldonado-Arocho FJ, Green C, Fisher ML, Paczosa MK, Mecsas J. 2013. Adhesins and host serum factors drive Yop translocation by yersinia into professional phagocytes during animal infection. PLoS Pathog 9:e1003415. doi: 10.1371/journal.ppat.1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Portal-Celhay C, Tufariello JM, Srivastava S, Zahra A, Klevorn T, Grace PS, Mehra A, Park HS, Ernst JD, Jacobs WR, Philips JA. 2016. Mycobacterium tuberculosis EsxH inhibits ESCRT-dependent CD4. Nat Microbiol 2:16232. doi: 10.1038/nmicrobiol.2016.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sia JK, Rengarajan J. 2019. Immunology of Mycobacterium tuberculosis infections. Microbiol Spectr 7:GPP3-0022-2018. doi: 10.1128/microbiolspec.GPP3-0022-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schorey JS, Harding CV. 2016. Extracellular vesicles and infectious diseases: new complexity to an old story. J Clin Invest 126:1181–1189. doi: 10.1172/JCI81132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coelho C, Brown L, Maryam M, Vij R, Smith DFQ, Burnet MC, Kyle JE, Heyman HM, Ramirez J, Prados-Rosales R, Lauvau G, Nakayasu ES, Brady NR, Hamacher-Brady A, Coppens I, Casadevall A. 2019. Virulence factors, including listeriolysin O, are secreted in biologically active extracellular vesicles. J Biol Chem 294:1202–1217. doi: 10.1074/jbc.RA118.006472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hui WW, Hercik K, Belsare S, Alugubelly N, Clapp B, Rinaldi C, Edelmann MJ. 2018. Salmonella enterica serovar Typhimurium alters the extracellular proteome of macrophages and leads to the production of proinflammatory exosomes. Infect Immun 86:e00386-17. doi: 10.1128/IAI.00386-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nandakumar R, Tschismarov R, Meissner F, Prabakaran T, Krissanaprasit A, Farahani E, Zhang BC, Assil S, Martin A, Bertrams W, Holm CK, Ablasser A, Klause T, Thomsen MK, Schmeck B, Howard KA, Henry T, Gothelf KV, Decker T, Paludan SR. 2019. Intracellular bacteria engage a STING-TBK1-MVB12b pathway to enable paracrine cGAS-STING signalling. Nat Microbiol 4:701–713. doi: 10.1038/s41564-019-0367-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones LB, Bell CR, Bibb KE, Gu L, Coats MT, Matthews QL. 2018. Pathogens and their effect on exosome biogenesis and composition. Biomedicines 6:79. doi: 10.3390/biomedicines6030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis CN, Phillips H, Tomes JJ, Swain MT, Wilkinson TJ, Brophy PM, Morphew RM. 2019. The importance of extracellular vesicle purification for downstream analysis: a comparison of differential centrifugation and size exclusion chromatography for helminth pathogens. PLoS Negl Trop Dis 13:e0007191. doi: 10.1371/journal.pntd.0007191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orchard RC, Kittisopikul M, Altschuler SJ, Wu LF, Süel GM, Alto NM. 2012. Identification of F-actin as the dynamic hub in a microbial-induced GTPase polarity circuit. Cell 148:803–815. doi: 10.1016/j.cell.2011.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor BD, Zheng X, Darville T, Zhong W, Konganti K, Abiodun-Ojo O, Ness RB, O'Connell CM, Haggerty CL. 2017. Whole-exome sequencing to identify novel biological pathways associated with infertility after pelvic inflammatory disease. Sex Transm Dis 44:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velarde JJ, Ashbaugh M, Wessels MR. 2014. The human antimicrobial peptide LL-37 binds directly to CsrS, a sensor histidine kinase of group A Streptococcus, to activate expression of virulence factors. J Biol Chem 289:36315–36324. doi: 10.1074/jbc.M114.605394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elgharably H, Mann E, Awad H, Ganesh K, Ghatak PD, Gordillo G, Sai-Sudhakar CB, Roy S, Wozniak DJ, Sen CK. 2013. First evidence of sternal wound biofilm following cardiac surgery. PLoS One 8:e70360. doi: 10.1371/journal.pone.0070360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruckert WM, Price CT, Abu Kwaik Y. 2014. Rapid nutritional remodeling of the host cell upon attachment of Legionella pneumophila. Infect Immun 82:72–82. doi: 10.1128/IAI.01079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orihuela CJ, Mahdavi J, Thornton J, Mann B, Wooldridge KG, Abouseada N, Oldfield NJ, Self T, Ala’Aldeen DA, Tuomanen EI. 2009. Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. J Clin Invest 119:1638–1646. doi: 10.1172/JCI36759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green ER, Juttukonda LJ, Skaar EP. 2020. The manganese-responsive transcriptional regulator MumR protects Acinetobacter baumannii from oxidative stress. Infect Immun 88:e00762-19. doi: 10.1128/IAI.00762-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mooney B, Torres-Velez FJ, Doering J, Ehrbar DJ, Mantis NJ. 2019. Sensitivity of Kupffer cells and liver sinusoidal endothelial cells to ricin toxin and ricin toxin-Ab complexes. J Leukoc Biol 106:1161–1176. doi: 10.1002/JLB.4A0419-123R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paumet F, Wesolowski J, Garcia-Diaz A, Delevoye C, Aulner N, Shuman HA, Subtil A, Rothman JE. 2009. Intracellular bacteria encode inhibitory SNARE-like proteins. PLoS One 4:e7375. doi: 10.1371/journal.pone.0007375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Boyer MA, Holmgren AM, Shin S. 2020. Legionella-infected macrophages engage the alveolar epithelium to metabolically reprogram myeloid cells and promote antibacterial inflammation. Cell Host Microbe 28:683–698.e6. doi: 10.1016/j.chom.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sixt BS, Bastidas RJ, Finethy R, Baxter RM, Carpenter VK, Kroemer G, Coers J, Valdivia RH. 2017. The Chlamydia trachomatis inclusion membrane protein CpoS counteracts STING-mediated cellular surveillance and suicide programs. Cell Host Microbe 21:113–121. doi: 10.1016/j.chom.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roxas JL, Vedantam G, Viswanathan VK. 2019. Epithelial maturity influences EPEC-induced desmosomal alterations. Gut Microbes 10:241–245. doi: 10.1080/19490976.2018.1506669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mishra M, Ressler A, Schlesinger LS, Wozniak DJ. 2015. Identification of OprF as a complement component C3 binding acceptor molecule on the surface of Pseudomonas aeruginosa. Infect Immun 83:3006–3014. doi: 10.1128/IAI.00081-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee H, Li C, Zhang Y, Zhang D, Otterbein LE, Jin Y. 2019. Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J Exp Med 216:2202–2220. doi: 10.1084/jem.20182313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meldolesi J. 2018. Exosomes and ectosomes in intercellular communication. Curr Biol 28:R435–R444. doi: 10.1016/j.cub.2018.01.059. [DOI] [PubMed] [Google Scholar]
- 54.Mathieu M, Martin-Jaular L, Lavieu G, Théry C. 2019. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 55.Temoche-Diaz MM, Shurtleff MJ, Nottingham RM, Yao J, Fadadu RP, Lambowitz AM, Schekman R. 2019. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. Elife 8:e47544. doi: 10.7554/eLife.47544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. 2016. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 5:e19276. doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukherjee K, Ghoshal B, Ghosh S, Chakrabarty Y, Shwetha S, Das S, Bhattacharyya SN. 2016. Reversible HuR-microRNA binding controls extracellular export of miR-122 and augments stress response. EMBO Rep 17:1184–1203. doi: 10.15252/embr.201541930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. 2013. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A, Tripodi M. 2016. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep 17:799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 60.Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N, Levy S, Zhang B, Coffey RJ, Patton JG. 2015. KRAS-dependent sorting of miRNA to exosomes. Elife 4:e07197. doi: 10.7554/eLife.07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiou NT, Kageyama R, Ansel KM. 2018. Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep 25:3356–3370.e4. doi: 10.1016/j.celrep.2018.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MAJ, Sadek P, Sie D, Zini N, Middeldorp JM, Ylstra B, de Menezes RX, Würdinger T, Meijer GA, Pegtel DM. 2014. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep 8:1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 63.Alexander M, Hu R, Runtsch MC, Kagele DA, Mosbruger TL, Tolmachova T, Seabra MC, Round JL, Ward DM, O’Connell RM. 2015. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat Commun 6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, Möller A. 2015. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles 4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stranska R, Gysbrechts L, Wouters J, Vermeersch P, Bloch K, Dierickx D, Andrei G, Snoeck R. 2018. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J Transl Med 16:1. doi: 10.1186/s12967-017-1374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Böing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, Nieuwland R. 2014. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles 3:23430. doi: 10.3402/jev.v3.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Niel G, D’Angelo G, Raposo G. 2018. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 68.Soekmadji C, Hill AF, Wauben MH, Buzás EI, Di Vizio D, Gardiner C, Lötvall J, Sahoo S, Witwer KW. 2018. Towards mechanisms and standardization in extracellular vesicle and extracellular RNA studies: results of a worldwide survey. J Extracell Vesicles 7:1535745. doi: 10.1080/20013078.2018.1535745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Silverman JM, Reiner NE. 2011. Exosomes and other microvesicles in infection biology: organelles with unanticipated phenotypes. Cell Microbiol 13:1–9. doi: 10.1111/j.1462-5822.2010.01537.x. [DOI] [PubMed] [Google Scholar]
- 70.Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. 2007. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood 110:3234–3244. doi: 10.1182/blood-2007-03-079152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee H, Groot M, Pinilla-Vera M, Fredenburgh LE, Jin Y. 2019. Identification of miRNA-rich vesicles in bronchoalveolar lavage fluid: insights into the function and heterogeneity of extracellular vesicles. J Control Release 294:43–52. doi: 10.1016/j.jconrel.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang D, Lee H, Zhu Z, Minhas JK, Jin Y. 2017. Enrichment of selective miRNAs in exosomes and delivery of exosomal miRNAs in vitro and in vivo. Am J Physiol Lung Cell Mol Physiol 312:L110–L121. doi: 10.1152/ajplung.00423.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hagiwara K, Katsuda T, Gailhouste L, Kosaka N, Ochiya T. 2015. Commitment of annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Lett 589:4071–4078. doi: 10.1016/j.febslet.2015.11.036. [DOI] [PubMed] [Google Scholar]
- 74.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 75.Duss O, Stepanyuk GA, Grot A, O’Leary SE, Puglisi JD, Williamson JR. 2018. Real-time assembly of ribonucleoprotein complexes on nascent RNA transcripts. Nat Commun 9:5087. doi: 10.1038/s41467-018-07423-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Herster F, Bittner Z, Archer NK, Dickhöfer S, Eisel D, Eigenbrod T, Knorpp T, Schneiderhan-Marra N, Löffler MW, Kalbacher H, Vierbuchen T, Heine H, Miller LS, Hartl D, Freund L, Schäkel K, Heister M, Ghoreschi K, Weber ANR. 2020. Neutrophil extracellular trap-associated RNA and LL37 enable self-amplifying inflammation in psoriasis. Nat Commun 11:105. doi: 10.1038/s41467-019-13756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei Z, Batagov AO, Schinelli S, Wang J, Wang Y, El Fatimy R, Rabinovsky R, Balaj L, Chen CC, Hochberg F, Carter B, Breakefield XO, Krichevsky AM. 2017. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun 8:1145. doi: 10.1038/s41467-017-01196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baglio SR, Rooijers K, Koppers-Lalic D, Verweij FJ, Pérez Lanzón M, Zini N, Naaijkens B, Perut F, Niessen HW, Baldini N, Pegtel DM. 2015. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther 6:127. doi: 10.1186/s13287-015-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patton JG, Franklin JL, Weaver AM, Vickers K, Zhang B, Coffey RJ, Ansel KM, Blelloch R, Goga A, Huang B, L’Etoille N, Raffai RL, Lai CP, Krichevsky AM, Mateescu B, Greiner VJ, Hunter C, Voinnet O, McManus MT. 2015. Biogenesis, delivery, and function of extracellular RNA. J Extracell Vesicles 4:27494. doi: 10.3402/jev.v4.27494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gong B, Lee YS, Lee I, Shelite TR, Kunkeaw N, Xu G, Lee K, Jeon SH, Johnson BH, Chang Q, Ha T, Mendell NL, Cheng X, Bouyer DH, Boor PJ, Ksiazek TG, Walker DH. 2013. Compartmentalized, functional role of angiogenin during spotted fever group rickettsia-induced endothelial barrier dysfunction: evidence of possible mediation by host tRNA-derived small noncoding RNAs. BMC Infect Dis 13:285. doi: 10.1186/1471-2334-13-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fuhrmann G, Serio A, Mazo M, Nair R, Stevens MM. 2015. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J Control Release 205:35–44. doi: 10.1016/j.jconrel.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 82.Ibsen SD, Wright J, Lewis JM, Kim S, Ko SY, Ong J, Manouchehri S, Vyas A, Akers J, Chen CC, Carter BS, Esener SC, Heller MJ. 2017. Rapid isolation and detection of exosomes and associated biomarkers from plasma. ACS Nano 11:6641–6651. doi: 10.1021/acsnano.7b00549. [DOI] [PubMed] [Google Scholar]
- 83.Kojima M, Gimenes-Junior JA, Chan TW, Eliceiri BP, Baird A, Costantini TW, Coimbra R. 2018. Exosomes in postshock mesenteric lymph are key mediators of acute lung injury triggering the macrophage activation. FASEB J 32:97–110. doi: 10.1096/fj.201700488R. [DOI] [PubMed] [Google Scholar]
- 84.Liu Y, Xiao J, Zhang B, Shelite TR, Su Z, Chang Q, Judy B, Li X, Drelich A, Bei J, Zhou Y, Zheng J, Jin Y, Rossi SL, Tang SJ, Wakamiya M, Saito T, Ksiazek T, Kaphalia B, Gong B. 2020. Increased talin-vinculin spatial proximities in livers in response to spotted fever group rickettsial and Ebola virus infections. Lab Invest 100:1030–1041. doi: 10.1038/s41374-020-0420-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Su Z, Chang Q, Drelich A, Shelite T, Judy B, Liu Y, Xiao J, Zhou C, He X, Jin Y, Saito T, Tang S, Soong L, Wakamiya M, Fang X, Bukreyev A, Ksiazek T, Russell W, Gong B. 2020. Annexin A2 depletion exacerbates the intracerebral microhemorrhage induced by acute rickettsia and Ebola virus infections. PLoS Negl Trop Dis 14:e0007960. doi: 10.1371/journal.pntd.0007960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.He X, Zhang W, Chang Q, Su Z, Gong D, Zhou Y, Xiao J, Drelich A, Liu Y, Popov V, Zhao X, Wakamiya M, Gaitas A, Lu F, Gong B. 2019. A new role for host annexin A2 in establishing bacterial adhesion to vascular endothelial cells: lines of evidence from atomic force microscopy and an in vivo study. Lab Invest 99:1650–1660. doi: 10.1038/s41374-019-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gong B, Shelite T, Mei FC, Ha T, Hu Y, Xu G, Chang Q, Wakamiya M, Ksiazek TG, Boor PJ, Bouyer DH, Popov VL, Chen J, Walker DH, Cheng X. 2013. Exchange protein directly activated by cAMP plays a critical role in bacterial invasion during fatal rickettsioses. Proc Natl Acad Sci U S A 110:19615–19620. doi: 10.1073/pnas.1314400110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee H, Zhang D, Laskin DL, Jin Y. 2018. Functional evidence of pulmonary extracellular vesicles in infectious and noninfectious lung inflammation. J Immunol 201:1500–1509. doi: 10.4049/jimmunol.1800264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sharma S, LeClaire M, Gimzewski JK. 2018. Ascent of atomic force microscopy as a nanoanalytical tool for exosomes and other extracellular vesicles. Nanotechnology 29:132001. doi: 10.1088/1361-6528/aaab06. [DOI] [PubMed] [Google Scholar]
- 90.Zhang D, Lee H, Wang X, Rai A, Groot M, Jin Y. 2018. Exosome-mediated small RNA delivery: a novel therapeutic approach for inflammatory lung responses. Mol Ther 26:2119–2130. doi: 10.1016/j.ymthe.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gong B, Ma L, Liu Y, Gong Q, Shelite T, Bouyer D, Boor PJ, Lee YS, Oberhauser A. 2012. Rickettsiae induce microvascular hyperpermeability via phosphorylation of VE-cadherins: evidence from atomic force microscopy and biochemical studies. PLoS Negl Trop Dis 6:e1699. doi: 10.1371/journal.pntd.0001699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Noda K, Zhang J, Fukuhara S, Kunimoto S, Yoshimura M, Mochizuki N. 2010. Vascular endothelial-cadherin stabilizes at cell-cell junctions by anchoring to circumferential actin bundles through alpha- and beta-catenins in cyclic AMP-Epac-Rap1 signal-activated endothelial cells. Mol Biol Cell 21:584–596. doi: 10.1091/mbc.e09-07-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang H, Liu J, Qu D, Wang L, Wong CM, Lau C-W, Huang Y, Wang YF, Huang H, Xia Y, Xiang L, Cai Z, Liu P, Wei Y, Yao X, Ma RCW, Huang Y. 2018. Serum exosomes mediate delivery of arginase 1 as a novel mechanism for endothelial dysfunction in diabetes. Proc Natl Acad Sci U S A 115:E6927–E6936. doi: 10.1073/pnas.1721521115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yáñez-Mó M, Siljander PR-M, Andreu Z, Zavec AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colás E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NHH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Krämer-Albers E-M, Laitinen S, Lässer C, Lener T, Ligeti E, Linē A, Lipps G, Llorente A, Lötvall J, Manček-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-‘t Hoen ENM, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pállinger É, Del Portillo HA, Reventós J, Rigau M, Rohde E, Sammar M, et al. 2015. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Londoño AF, Mendell NL, Walker DH, Bouyer DH. 2019. A biosafety level-2 dose-dependent lethal mouse model of spotted fever rickettsiosis: Rickettsia parkeri Atlantic Rainforest strain. PLoS Negl Trop Dis 13:e0007054. doi: 10.1371/journal.pntd.0007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilhelm I, Fazakas C, Krizbai IA. 2011. In vitro models of the blood-brain barrier. Acta Neurobiol Exp (Wars) 71:113–128. [DOI] [PubMed] [Google Scholar]
- 97.Tang F, Hajkova P, Barton SC, Lao K, Surani MA. 2006. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res 34:e9. doi: 10.1093/nar/gnj009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moltzahn F, Olshen AB, Baehner L, Peek A, Fong L, Stöppler H, Simko J, Hilton JF, Carroll P, Blelloch R. 2011. Microfluidic-based multiplex qRT-PCR identifies diagnostic and prognostic microRNA signatures in the sera of prostate cancer patients. Cancer Res 71:550–560. doi: 10.1158/0008-5472.CAN-10-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Link F, Krohn K, Schumann J. 2019. Identification of stably expressed housekeeping miRNAs in endothelial cells and macrophages in an inflammatory setting. Sci Rep 9:12786. doi: 10.1038/s41598-019-49241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li J, Zhao Y, Lu Y, Ritchie W, Grau G, Vadas MA, Gamble JR. 2016. The poly-cistronic miR-23-27-24 complexes target endothelial cell junctions: differential functional and molecular effects of miR-23a and miR-23b. Mol Ther Nucleic Acids 5:e354. doi: 10.1038/mtna.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Oikawa S, Wada S, Lee M, Maeda S, Akimoto T. 2018. Role of endothelial microRNA-23 clusters in angiogenesis in vivo. Am J Physiol Heart Circ Physiol 315:H838–H846. doi: 10.1152/ajpheart.00742.2017. [DOI] [PubMed] [Google Scholar]
- 102.Lee Y, Kim SJ, Choo J, Heo G, Yoo JW, Jung Y, Rhee SH, Im E. 2020. miR-23a-3p is a key regulator of IL-17C-induced tumor angiogenesis in colorectal cancer. Cells 9:1363. doi: 10.3390/cells9061363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lopez-Ramirez MA, Reijerkerk A, de Vries HE, Romero IA. 2016. Regulation of brain endothelial barrier function by microRNAs in health and neuroinflammation. FASEB J 30:2662–2672. doi: 10.1096/fj.201600435RR. [DOI] [PubMed] [Google Scholar]
- 104.Bridge G, Monteiro R, Henderson S, Emuss V, Lagos D, Georgopoulou D, Patient R, Boshoff C. 2012. The microRNA-30 family targets DLL4 to modulate endothelial cell behavior during angiogenesis. Blood 120:5063–5072. doi: 10.1182/blood-2012-04-423004. [DOI] [PubMed] [Google Scholar]
- 105.Hassani K, Olivier M. 2013. Immunomodulatory impact of leishmania-induced macrophage exosomes: a comparative proteomic and functional analysis. PLoS Negl Trop Dis 7:e2185. doi: 10.1371/journal.pntd.0002185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ghosh J, Bose M, Roy S, Bhattacharyya SN. 2013. Leishmania donovani targets Dicer1 to downregulate miR-122, lower serum cholesterol, and facilitate murine liver infection. Cell Host Microbe 13:277–288. doi: 10.1016/j.chom.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Teysseire N, Boudier JA, Raoult D. 1995. Rickettsia conorii entry into Vero cells. Infect Immun 63:366–374. doi: 10.1128/IAI.63.1.366-374.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Walker DH. 1988. Diagnosis of rickettsial diseases. Pathol Annu 23(Part 2):69–96. [PubMed] [Google Scholar]
- 109.Roberts K, Alberts B, Johnson A, Walter P, Hunt T. 2002. Molecular biology of the cell. Garland Science, New York, NY. [Google Scholar]
- 110.Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M. 2014. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A 111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Manna I, Iaccino E, Dattilo V, Barone S, Vecchio E, Mimmi S, Filippelli E, Demonte G, Polidoro S, Granata A, Scannapieco S, Quinto I, Valentino P, Quattrone A. 2018. Exosome-associated miRNA profile as a prognostic tool for therapy response monitoring in multiple sclerosis patients. FASEB J 32:4241–4246. doi: 10.1096/fj.201701533R. [DOI] [PubMed] [Google Scholar]
- 112.The RNAcentral Consortium, Petrov AI, Kay SJE, Kalvari I, Howe KL, Gray KA, Bruford EA, Kersey PJ, Cochrane G, Finn RD, Bateman A, Kozomara A, Griffiths-Jones S, Frankish A, Zwieb CW, Lau BY, Williams KP, Chan PP, Lowe TM, Cannone JJ, Gutell R, Machnicka MA, Bujnicki JM, Yoshihama M, Kenmochi N, Chai B, Cole JR, Szymanski M, Karlowski WM, Wood V, Huala E, Berardini TZ, Zhao Y, Chen R, Zhu W, Paraskevopoulou MD, Vlachos IS, Hatzigeorgiou AG, Ma L, Zhang Z, Puetz J, Stadler PF, McDonald D, Basu S, Fey P, Engel SR, Cherry JM, Volders PJ, Mestdagh P, Wower J, Clark MB, Quek XC, Dinger ME. 2017. RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res 45:D128–D134. doi: 10.1093/nar/gkw1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kozomara A, Birgaoanu M, Griffiths-Jones S. 2019. miRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, Bennett R, Bhai J, Billis K, Boddu S, Cummins C, Davidson C, Dodiya KJ, Gall A, Girón CG, Gil L, Grego T, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, Kay M, Laird MR, Lavidas I, Liu Z, Loveland JE, Marugán JC, Maurel T, McMahon AC, Moore B, Morales J, Mudge JM, Nuhn M, Ogeh D, Parker A, Parton A, Patricio M, Abdul Salam AI, Schmitt BM, Schuilenburg H, Sheppard D, Sparrow H, Stapleton E, Szuba M, Taylor K, Threadgold G, Thormann A, Vullo A, et al. 2019. Ensembl 2019. Nucleic Acids Res 47:D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sader JE, Sanelli JA, Adamson BD, Monty JP, Wei X, Crawford SA, Friend JR, Marusic I, Mulvaney P, Bieske EJ. 2012. Spring constant calibration of atomic force microscope cantilevers of arbitrary shape. Rev Sci Instrum 83:103705. doi: 10.1063/1.4757398. [DOI] [PubMed] [Google Scholar]
- 116.Drelich A, Judy B, He X, Chang Q, Yu S, Li X, Lu F, Wakamiya M, Popov V, Zhou J, Ksiazek T, Gong B. 2018. Exchange protein directly activated by cAMP modulates Ebola virus uptake into vascular endothelial cells. Viruses 10:563. doi: 10.3390/v10100563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantities of rickettsiae in plasma-derived Exos (plsExos) (n = 5/group) and HUVEC culture media-derived Exos (ECExos) (n = 3/group), determined by quantitative real-time PCR. Data are presented as means ± standard errors. Statistical significance was determined using one-way analysis of variance. **, P < 0.01. Download FIG S1, JPG file, 0.1 MB (115.5KB, jpg) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(a) Quantities of plsExos on day 4 postinfection from mock-treated mice or mice infected with 2 LD50 of R. parkeri given by the intravenous route were assessed using the total protein concentration of plsExos (n = 5 per group). Statistical significance was determined using Student’s t test. (b) The TEER value of normal mouse recipient BMECs was measured after treatment with normal plsExos (mock) or R-plsExos (from an R. parkeri-infected mouse) at 400, 2,000, or 8,000 particles per cell for 72 h. Statistical significance was determined using one-way analysis of variance. *, P < 0.05. Download FIG S2, JPG file, 0.08 MB (86.7KB, jpg) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Normal rabbit serum was used as a control antibody during immunofluorescence (IF) staining of a mouse lung tissue sample treated in same way. Scale bar indicates 20 μm. Download FIG S3, JPG file, 0.1 MB (137.2KB, jpg) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Normal rabbit serum was used as a control antibody during IF staining of human BMECs treated in same way. Scale bar indicates 20 μm. Download FIG S4, JPG file, 0.06 MB (64KB, jpg) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(a) Quantities of ECExos from mock-, heat-inactivated R. parkeri-, or R. parkeri-infected HUVECs 72 h p.i. at an MOI of 10 were assessed using the total protein concentration of ECExos (n = 4 per group). (b and c) Immunofluorescence staining of tight junctional protein ZO-1 (red) and relative fluorescent intensities of paracellular ZO-1. Statistical significance was determined using one-way analysis of variance. Scale bar indicates 20 μm. *, P < 0.05. Download FIG S5, JPG file, 0.4 MB (419.7KB, jpg) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
GO enrichment analysis of mir-23a-3p targeted genes. aCount is the number of genes in the user-provided lists with membership in the given ontology term. Log10(P) is the P value in log base 10. Download Table S1, DOCX file, 0.01 MB (15.2KB, docx) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
GO enrichment analysis of mir-30b-5p targeted genes. aCount is the number of genes in the user-provided lists with membership in the given ontology term. Log10(P) is the P value in log base 10. Download Table S2, DOCX file, 0.01 MB (15.3KB, docx) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Stem-loop RT-qPCR analysis of microRNA expression in plsExos. microRNA expression in normal plsExos versus R-plsExos (n = 6). R-plsExos were isolated from mice on day 4 after infection with 2 LD50 of R. parkeri given by the intravenous route. Statistical significance was determined using one-way analysis of variance. No statistical differences were found using a P value of <0.05. Download FIG S6, JPG file, 0.05 MB (47.7KB, jpg) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental Materials and Methods. Download Text S1, DOCX file, 0.03 MB (28.2KB, docx) .
Copyright © 2021 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.