ABSTRACT
Macrotermitine termites have domesticated fungi in the genus Termitomyces as their primary food source using predigested plant biomass. To access the full nutritional value of lignin-enriched plant biomass, the termite-fungus symbiosis requires the depolymerization of this complex phenolic polymer. While most previous work suggests that lignocellulose degradation is accomplished predominantly by the fungal cultivar, our current understanding of the underlying biomolecular mechanisms remains rudimentary. Here, we provide conclusive omics and activity-based evidence that Termitomyces employs not only a broad array of carbohydrate-active enzymes (CAZymes) but also a restricted set of oxidizing enzymes (manganese peroxidase, dye decolorization peroxidase, an unspecific peroxygenase, laccases, and aryl-alcohol oxidases) and Fenton chemistry for biomass degradation. We propose for the first time that Termitomyces induces hydroquinone-mediated Fenton chemistry (Fe2+ + H2O2 + H+ → Fe3+ + •OH + H2O) using a herein newly described 2-methoxy-1,4-dihydroxybenzene (2-MH2Q, compound 19)-based electron shuttle system to complement the enzymatic degradation pathways. This study provides a comprehensive depiction of how efficient biomass degradation by means of this ancient insect’s agricultural symbiosis is accomplished.
KEYWORDS: symbiosis, lignin degradation, Termitomyces, metabolites, redox chemistry, biodegradation, lignocellulose, redox proteins, secondary metabolism
INTRODUCTION
Among the different types of nutritional symbiosis, crop agriculture represents one of the most sophisticated systems. Beyond examples from humans, only a few insect lineages maintain and manure external symbiotic partners (1). Fungus-growing termites (Macrotermitinae) underwent a major transition ca. 30 million years ago, when they started to domesticate the mutualistic fungus Termitomyces (Agaricales, Lyophyllaceae) as their main food source (2, 3). Since then, fungus-growing termites have become major biomass decomposers of dead plant material, resulting in a substantial ecological footprint in the Old World (sub)tropics (4, 5).
Termitomyces is manured by termite workers in a cork-like structure termed the “fungus comb,” which is found within the underground chambers of the termite mound and is comprised of predigested plant material (Fig. 1A and B) (6). Old termite workers collect and transport a mix of dead plant material (7), while younger workers macerate and ingest the plant material along with asexual Termitomyces spores and enzymes, which are produced in fungal nodules on the mature parts of the fungal comb (1, 2). The resulting lignocellulose and spore-enriched feces is then used to craft fresh fungus comb. After spore germination, the fungus matures within 15 to 20 days, and energy-rich fungal nodules are formed to serve as the major food source for younger workers (8). During an average turnover time of 45 to 50 days, the remains of the comb material serve as the major nutrition of older workers, resulting overall in the nearly wasteless decomposition and recycling of plant material (9).
FIG 1.
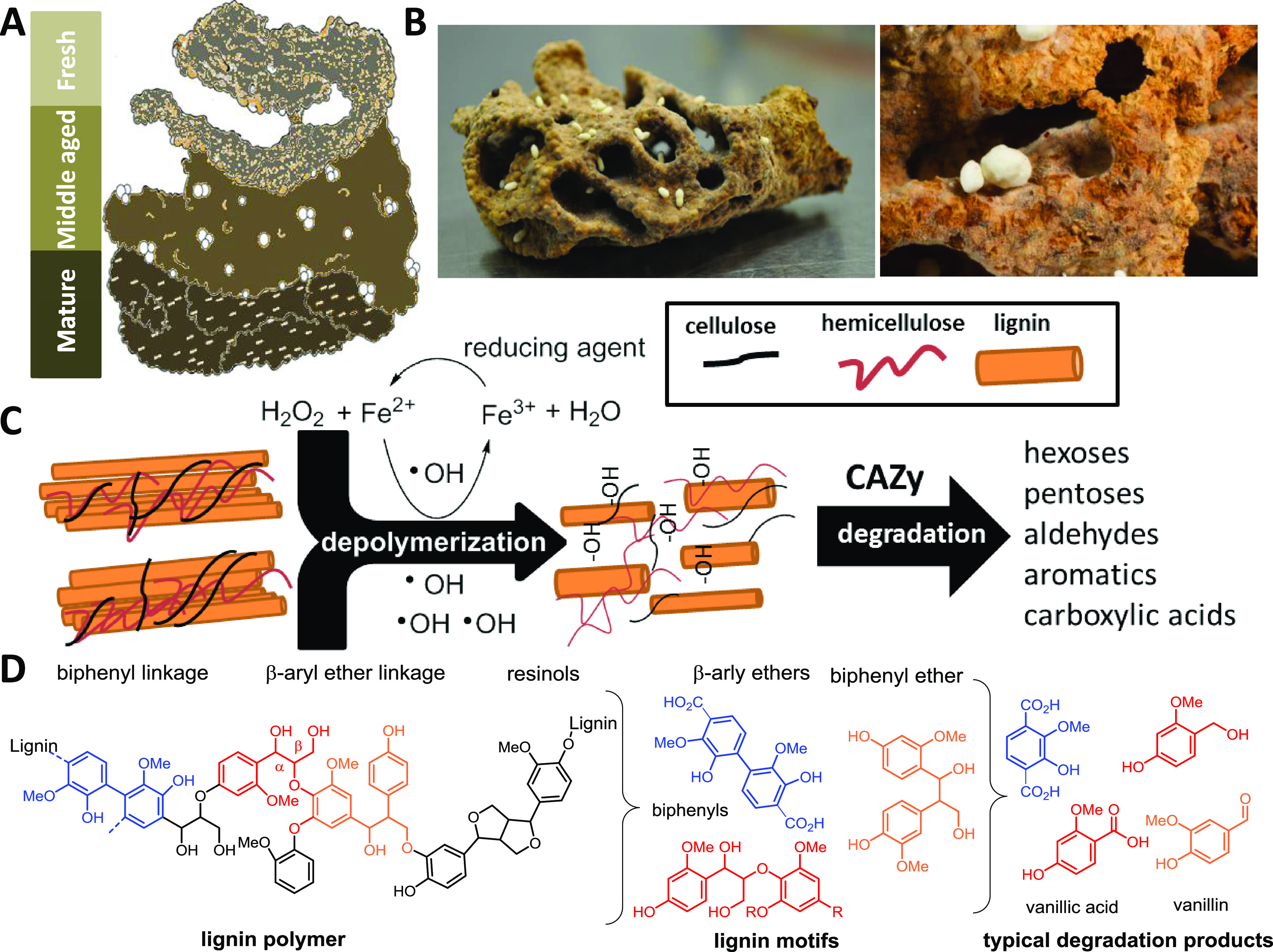
(A) Schematic representation of fungus comb at different maturation stages; (B) freshly collected mature fungus comb carrying fungal nodules; (C) schematic representation of lignin depolymerization via hydroxylation and oxidative cleavage with subsequent degradation by CAZy enzymes to smaller metabolites; (D) schematic structure of lignin biopolymer, lignin motifs, and lignin-derived degradation products.
Although the feeding behavior of termites has been studied in detail for decades (10), the underlying biochemical mechanisms for degrading the foraged plant biomass have remained largely unresolved and are the topic of intense discussion (1, 11). Plant biomass consists mostly of lignocellulose, a complex matrix consisting of cell wall polysaccharides: cellulose (40 to 50%), hemicellulose (25 to 30%), and the structurally complex and inhomogeneous phenolic polymer lignin (15 to 20%) (12). The depolymerization and degradation of lignin provides an enormous energetic burden to any microorganism due to lignin’s recalcitrant nature and the strong covalent carbon-carbon and carbon-oxygen linkages between hydroxycinnamoyl alcohol-derived monomers that are covalently cross-linked to plant polysaccharides (Fig. 1C and D) (13, 14). However, once oxidative mechanisms have broken up the dense lignin structure, degrading enzymes are able to diffuse into the material and access the embedded biphenylic, phenolic, and carbohydrate reservoirs for further biomass conversion (15–17).
Although the degradation process appears to be a necessary endeavor to manure the product of the complex fungus-termite-bacterium symbiosis, the fate of lignin within termite fungus combs still remains unclear. A recent study on fungus comb pretreatment in Odontotermes formosanus by Li et al. indicated that lignin is partly cleaved during the first gut passage (18). Additionally, it was hypothesized that Termitomyces might have lost key delignification potential throughout its evolutionary history with the termites. However, previous and more recent transcriptomic and analytically guided studies of other Macrotermitinae species by da Costa and coworkers showed that fresh comb from Odontotermes spp. and Macrotermes natalensis is lignin rich (7), suggesting that the role of gut passage in lignin cleavage may differ between termite species (9). Based on fungal transcriptome sequencing (RNA-seq) analysis and enzymatic assays, the study reasoned that maturation of the fungus comb causes the decomposition of the lignocellulose-rich biomass through the actions of fungal and/or bacterial enzymes.
These partially contradictory results led us to investigate whether Termitomyces has the capacity to depolymerize or even degrade lignin-rich biomass. Hence, we commenced our analysis by a comparative genome analysis of nine Termitomyces species and an assessment of their capacity to produce ligninolytic enzymes (e.g., laccase [EC 1.10.3.2], lignin peroxidase [LiP; EC 1.11.1.14], manganese peroxidase [MnP], oxygenase [unspecific peroxygenases [UPOs]; EC 1.11.2.1], and versatile peroxidases [VPs] [19] as well as other enzymes supporting degradative pathways and protecting against oxidative stress) (20–22). Here, we show that all investigated Termitomyces have similar repertoires of carbohydrate-active enzymes (CAZymes), including a conserved set of ligninolytic enzymes (a MnP, a dye-decolorizing peroxidase [DyP], UPOs, and laccases) with a broad aromatic and phenolic substrate spectrum (23, 24), but lack other class II peroxidases (e.g., LiPs, VPs) that are known to readily oxidize the more recalcitrant nonphenolic moieties of lignin, as described for other basidiomycete white-rot fungi (13, 14, 16). Our findings were supported by the analysis of gene expression levels in RNA-seq data sets obtained from fungus comb at different maturation stages (7). Additional in silico and biochemical studies led us to the conjecture that Termitomyces might employ hydroquinone-mediated Fenton chemistry (Fe2+ + H2O2 + H+ → Fe3+ + •OH + H2O) using a herein newly described 2-methoxy-1,4-dihydroxybenzene (2-MH2Q; compound 19)-based electron shuttle system to complement enzymatic lignin degradation pathways. We further deduced that the presence of small dicarboxylic acids produced by Termitomyces not only allows the fungus to solubilize necessary metal ions but also mediates Fenton-based redox chemistry, making the system one of the most effective farming insect symbioses.
RESULTS
Genomic and transcriptomic analysis of lignocellulolytic capacity.
First, we subjected two Termitomyces species, excavated in South Africa in 2011 and 2015, to whole-genome sequencing using Illumina sequencing technology (LGC Genomics [Berlin, Germany]) and RNA sequencing using the BGISeq-500 platform (BGI, Hong Kong). Annotated genomes of both species were obtained using AUGUSTUS 3.3.3 after RNA-seq data were mapped to the genomes and used for algorithm training. The resulting draft genome of Termitomyces sp. strain T153 (Macrotermes natalensis) had an estimated size of 84.1 Mb (scaffold N50 = 23.88 kb), with more than 13,000 genes (GenBank accession no. JACKQL000000000). Similarly, the draft genome of Termitomyces sp. strain T112 (Macrotermes natalensis) had an estimated size of 79.8 Mb (scaffold N50 = 33.34 kb) and also >13,000 genes (accession no. JACKQM000000000). For further analysis, we also reannotated seven Termitomyces genomes deposited in GenBank, including our previously reported Termitomyces sp. strain J132 (alias P5) from Macrotermes natalensis (3), using the same settings in AUGUSTUS 3.3.3 (Tables S2 and S3 at https://doi.org/10.5281/zenodo.4431413). To gain insights into the functional capacity for biomass degradation, we first identified CAZyme families within each genome using a local installation of the dbCAN2 server (25–27).
Comparison of all nine Termitomyces genomes revealed that all species had relatively similar predicted proteomes with comparable numbers of polysaccharide-degrading enzymes, such as exo-cellobiohydrolases, endoglucanases assigned to different glycoside hydrolase (GH) families, and lytic polysaccharide monooxygenase (LPMOs), but no particular enrichment or reduction of CAZy families compared to those of other basidiomycete reference genomes was found (Fig. 2; see Fig. S1 and S2 at https://doi.org/10.5281/zenodo.4431413) (28).
FIG 2.
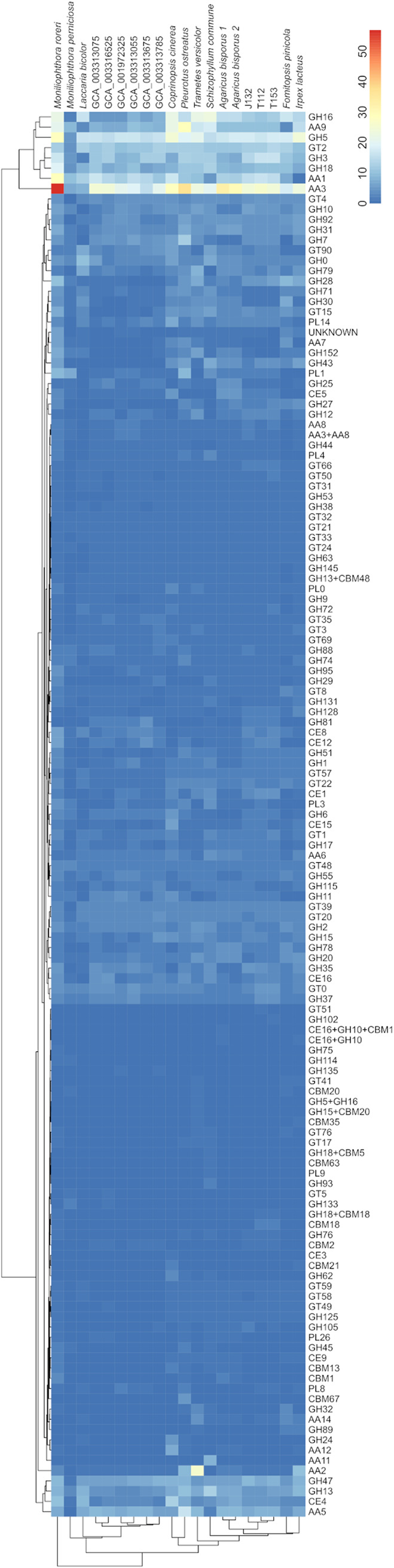
Heatmap of the numbers of hits for representatives of different CAZy families in the predicted proteomes of Termitomyces spp. (T112, T153, J132, GCA_001972325, GCA_003313055, GCA_003313075, GCA_003313675, GCA_003313785, GCA_003316525) and other selected basidiomycete fungi (Laccaria bicolor, Moniliophthora perniciosa, Moniliophthora roreri, Agaricus bisporus var. Burnettii [Agaricus bisporus 1], Agaricus bisporus var. Bisporus [Agaricus bisporus 2], Coprinopsis cinerea, Schizophyllum commune, Fomitopsis pinicola, Trametes versicolor, Pleurotus ostreatus, Irpex lacteus). The vertical axis shows clustering of enzymes based on abundance.
We then specifically searched Termitomyces genomes for the presence/absence of gene sequences encoding highly oxidizing enzymes that could contribute to the depolymerization and catabolic degradation of lignin (Fig. 2; see Table S4 at https://doi.org/10.5281/zenodo.4431413) (18). It is worth noting that Termitomyces genomes contained, on average, 16 gene sequences encoding laccases (AA1) (29–32), oxidases with low redox potential that use diphenols and related substances as electron donors and oxygen as the acceptor, thereby creating reactive C and O-based radical species in the process. In addition, we identified one putative MnP (AA2) per genome, an enzyme that generates highly reactive Mn3+ species that, once chelated, are able to diffuse through the dense network of lignocellulose, causing oxidation degradation due to their higher redox potential (23). Furthermore, a subset of gene sequences encoding alcohol oxidases and dehydrogenases (AA3 and AA5) are known to catalyze the oxidation of (aryl)-alcohols or carbohydrates with the concomitant formation of hydroquinones and/or H2O2 (33, 34). Unlike in previous studies (1), we also identified sequences encoding a DyP and an UPO (Tables S5 and S6 at https://doi.org/10.5281/zenodo.4431413), both of which are known for their versatile substrate spectrum. However, homologous sequences to other class II peroxidases (PODs), such as LiPs, were not detectable. Along these lines, iron reductase domains (AA8, EC 1.16.1) and putative benzoquinone reductases (AA6, EC 1.6.5.6) that are key to maintaining efficient Fenton chemistry-based redox cycles by reductive Fe2+ sequestration and regeneration of organic benzoquinone-based redox shuttles were identified.
Subsequently, the expression levels of candidate genes related to lignin depolymerization were analyzed in RNA-seq data obtained from three regions in the fungus comb (Fig. 3) (7): fresh comb (within which most plant biomass decomposition is likely to occur), old comb (where decomposition might still occur but to a lesser extent), and nodules as (which feed young workers and serve for fungal spore and enzyme transport) (Fig. 3; Table S27 at https://doi.org/10.5281/zenodo.4431413).
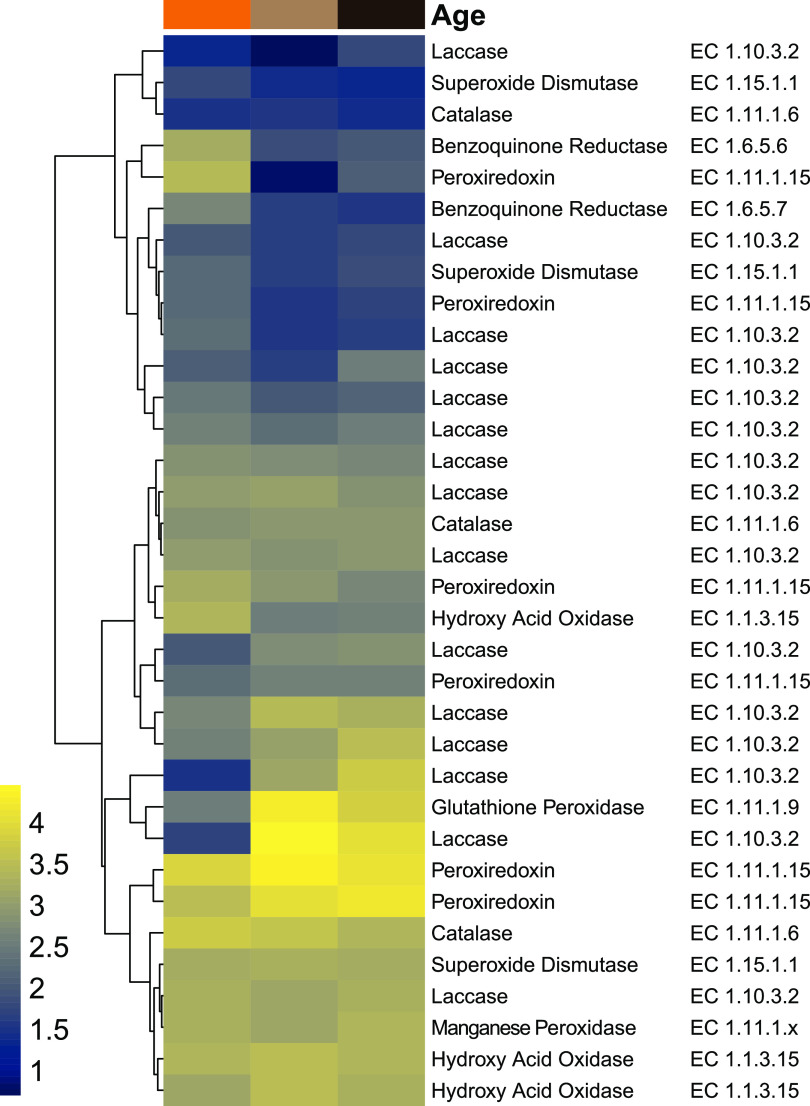
FIG 3.
Heatmap of redox enzyme transcription levels based on RNA-seq data of fresh comb (light brown), old comb (black) and nodules (orange) from Macrotermes colony Mn156 (7). Transcript abundances are depicted as log10 gene expression values, and color schemes were generated by viridis (version 0.5.1) (68, 71).
Here, we found differentiating transcription levels of genes encoding oxidative enzymes (such as laccases, a MnP, and a UPO) and enzymes of the CAZy families AA3 and AA5, as well as enzymes that protect against reactive intermediates (e.g., benzoquinone reductase [EC 1.6.5.7], superoxide dismutase [EC 1.15.1.1], glutathione peroxidase [EC1.11.1.9], and peroxiredoxin [EC 1.11.1.15]) across all three data sets. The combined genetic and transcriptomic survey revealed that Termitomyces has the capacity to produce lignocellulolytic enzymes and may even be able to induce and catalyze Fenton chemistry (35).
Fenton chemistry of Termitomyces.
Fenton chemistry involves the reaction between Fe2+ and H2O2, yielding Fe3+ and a highly reactive hydroxyl radical (•OH), a powerful oxidant (E0 = 2.8 V versus that of a normal hydrogen electrode) that is able to unselectively oxidize hydrocarbons and nonphenolic aromatic units within lignocellulose-rich material. Brown-rot fungi are known to make use of Fenton chemistry to depolymerize lignocellulose biomass (36) and modulate the redox potential of Fe2+/3+ species by secretion of dicarboxylic acids that act as chelators to form diffusible Fe complexes and as proton donors for catalytic degradation processes (37). Additionally, redox-active fungal quinones (Q) and hydroxyquinones (H2Q), such as 2,5-dimethoxy-1,4-benzoquinone (2,5-DMQ), 2,5-dimethoxy-1,4-hydroquinone (2,5-DMH2Q), and its regioisomer 4,5-dimethoxy-1,2-benzendiol (4,5-DMH2Q), have been discussed to serve as redox shuttles (3 H2Q + 2 O2 → 3 Q + 2 H2O + 2 HO•) in the Fenton chemistry of rotting fungi (e.g., Serpula lacrymans, the Gloeophyllales, and the Polyporales) (38–40), as they have the ability to switch between oxidation states via one-electron transfer reactions that allow for the concomitant formation of Fe2+ from Fe3+ and hydroxyl radicals (HO•) from H2O2 and O2 (see Fig. 5 and 6).
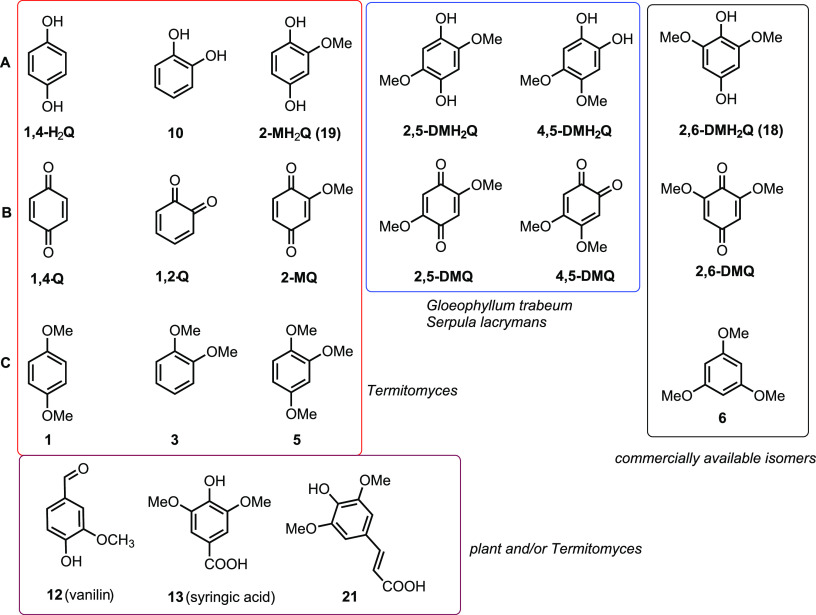
FIG 5.
Structures of redox-active compounds discussed in this work. (A) Hydroxyquinones (H2Q); (B) corresponding quinones (Q); (C) methoxylated derivatives of H2Q. Compounds identified from Termitomyces are highlighted in a red box, compounds identified from other rotting fungi are marked with a blue box, derivatives isolated from Termitomyces and of plant origin are highlighted in a purple box, and commercial derivatives for comparison are highlighted in a black box.
FIG 6.
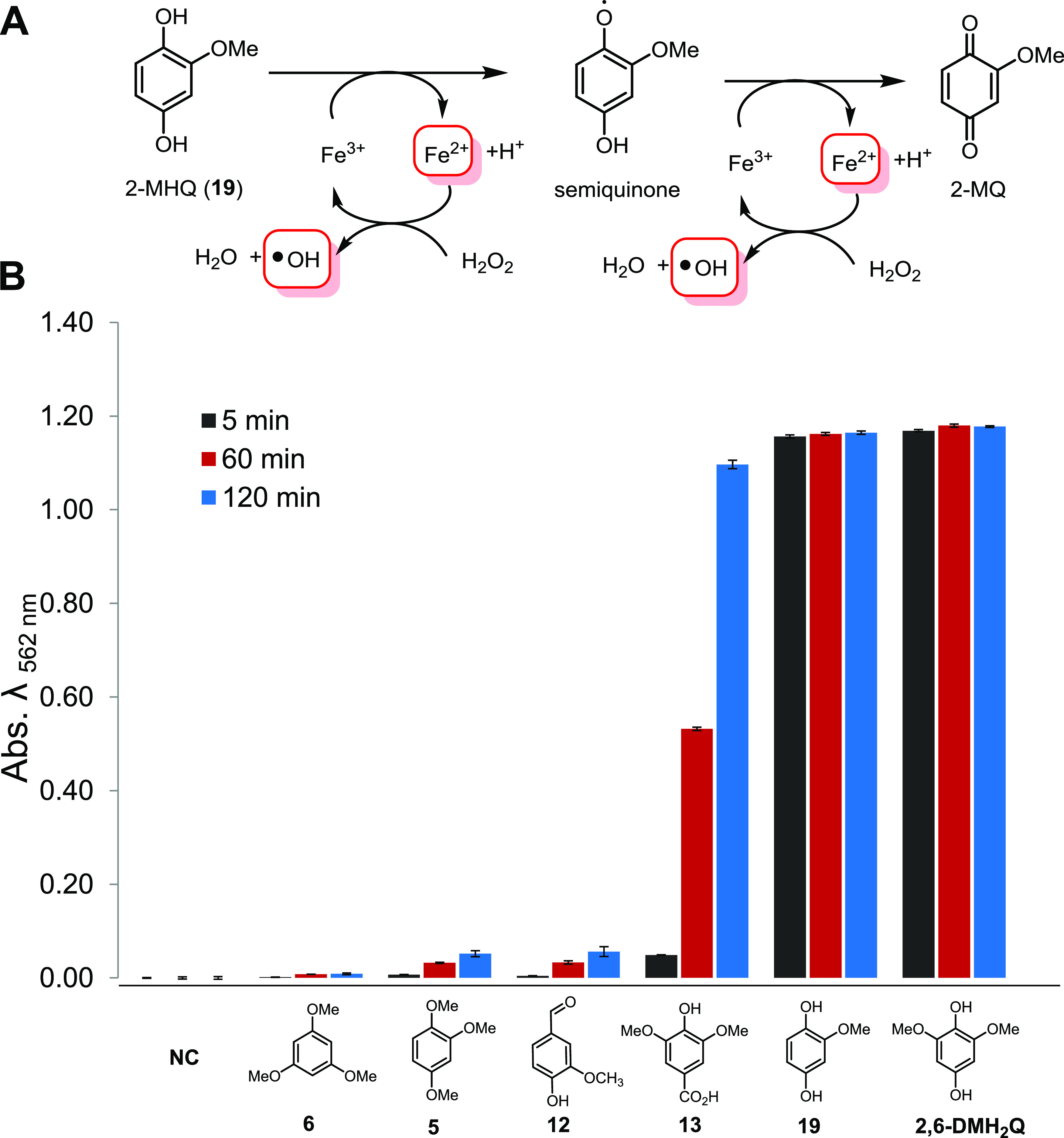
(A) Mechanistic depiction of the 2-MH2Q-initiated Fenton reaction via the formation of a radical semiquinone species and oxidation to 2-MQ; (B) quantification of Fe3+ reduction by H2Q using a colorimetric ferrozine-based assay (sodium acetate [NH4OAc] buffer, pH 4). Error bars indicate ±0.5 standard deviation (n = 3). Abs. λ562 nm, absorbance at a wavelength of 562 nm; NC, negative control.
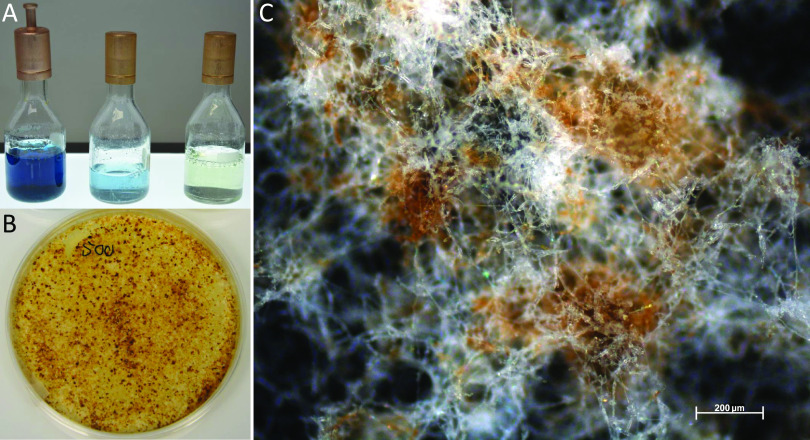
Thus, we evaluated whether Termitomyces employs any of those measures to enable lignin depolymerization by using Termitomyces sp. T153 and P5 as model strains. First, we employed a standardized colorimetric ferrozine assay to determine if extracellular Fe3+ is reduced to Fe2+ within the surrounding mycelium, a prerequisite to initiate Fenton chemistry (41, 42). As depicted in Fig. 4A, topical application of a ferrozine solution caused a clear color change within minutes, which was indicative of the immediate reduction of Fe3+ to Fe2+. Next, we determined the pH range within the fungal mycelium, as enzyme activities, the redox potential of H2O2, and metal complexes are strongly pH dependent (35). We found indications that Termitomyces acidifies the surrounding medium (Fig. 4D), which would benefit enzyme activities of lignin-degrading enzymes with a pH optimum of 4.5 to 5.0 (14, 21). As the Fenton reaction also requires H2O2, we tested if Termitomyces generates sufficient extracellular H2O2 to initiate the reaction. Based on an H2O2-dependent colorimetric assay, we found that Termitomyces generates approximately 4 to 6 μg extracellular H2O2 per gram mycelium during growth on solid support (mycelium age, 7 to 21 days) (Tables S18 to S20 at https://doi.org/10.5281/zenodo.4431413).
FIG 4.
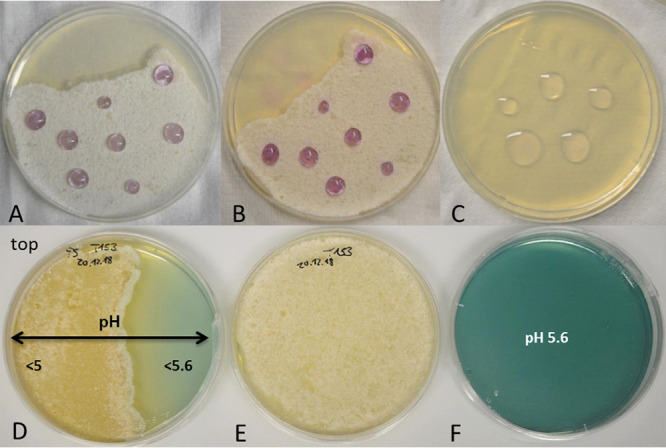
(A and B) Ferrozine solution added to a Termitomyces sp. T153 culture grown on PDA (18 days) and incubated for 5 min (A) and 30 min (B); (C) ferrozine solution on a PDA plate (negative control); (D and E) Termitomyces sp. T153 grown on PDA (28 days) containing bromocresol green as a pH indicator (D) and without the indicator (E); (F) PDA plate containing bromocresol green (negative control).
In a next step, we evaluated if Termitomyces produces redox-active H2Q/Q using gas chromatography coupled with mass spectrometry (GC-MS). Although the formation of previously reported 2,5-DM(H2)Q was not observed, we were intrigued to detect 2-methoxy-1,4-benzoquinone (2-MQ), its reduced H2Q named 2-methoxy-1,4-dihydroxybenzene (2-MH2Q), and the fully methylated derivative 1,2,4-trimethoxybenzene (compound 5), as well as other structurally related (di)methoxylated hydroxybenzenes (e.g., compounds 1, 3, and 12) (Fig. 5). Additionally, we verified the identities of the newly detected quinone derivatives 2-MQ and 2-MH2Q by synthesis and comparison of GC-MS retention times (Fig. S20 and S21 and Tables S16, S17, and S24 at https://doi.org/10.5281/zenodo.4431413).
To evaluate the ability of H2Qs to reduce Fe3+ to Fe2+, we employed the established ferrozine-based Fe3+ reduction assay (43). Overall, 2,6-DMH2Q (compound 18), a regioisomer of 2,5-DMH2Q, was the most reactive derivative that was able to reduce Fe3+ to Fe2+ within seconds and was therefore used as positive control in further experiments (Fig. 6). In comparison, 2-MH2Q (compound 19) showed a reduced reactivity, which is likely a reflection of the decreasing electron density of the aromatic system due to the lack of one additional electron-donating –OCH3 group. Other tested (methoxylated) hydroxybenzenes showed a reduced reactivity compared to those of compounds 18 and 19. Subsequently, we expanded our studies to combinations of redox-active derivatives and were able to observe in most cases the superposition of redox activities but no indications of synergistic activity (Fig. S9 and Table S21 at https://doi.org/10.5281/zenodo.4431413).
As Fenton chemistry produces highly reactive hydroxyl radicals (•OH), we then confirmed the presence of these short-lived radicals in our H2Q-mediated Fenton reactions using a fluorometric assay based on the reaction with terephthalic acid (TPA). As in literature reports for 2,6-DMH2Q (compound 18) (37–40), the newly identified and structurally related H2Q compound 19 catalyzed the formation of •OH in the presence of H2O2 and Fe3+ within seconds. In contrast, derivatives such as 1,2-dihydroxybenzene (compound 10) and syringic acid (compound 13) caused the formation of hydroxyl radicals with lower initial reactivities, but they formed over a period of more than 90 min (Fig. S5 at https://doi.org/10.5281/zenodo.4431413). Having verified that Termitomyces produces reactive H2Qs that are able to induce the formation of Fenton reagents (Fe2+, H2O2, and •OH), we then elaborated on the influence of fungus-derived dicarboxylic acids (oxalic acid, tartaric acid, malic acid, fumaric acid, and succinic acid) (44–46) on the Fenton reaction (Fig. 7). While at low concentrations of oxalic acid (0.1 mM) most H2Qs were still able to reduce the formed Fe3+ complexes, increasing concentrations caused the formation of stable Fe complexes with altered redox potentials, such that only the most reactive, 2,6-DMH2Q (compound 18), was able to reduce Fe3+ to Fe2+ (Fig. S10 to S12 at https://doi.org/10.5281/zenodo.4431413) (46). At 10 mM oxalic acid, a significant amount of autoxidation-related Fe3+ reduction was observed. A similar reactivity trend, albeit with a stronger autoxidation effect, was observed when tartaric acid was investigated as a chelating agent (47). In contrast, the presence of malic, fumaric, or succinic acid only moderately altered the redox potential of the Fe-complexes, and only low rates of autoxidation were observed (Fig. S14 to S16 at https://doi.org/10.5281/zenodo.4431413).
FIG 7.
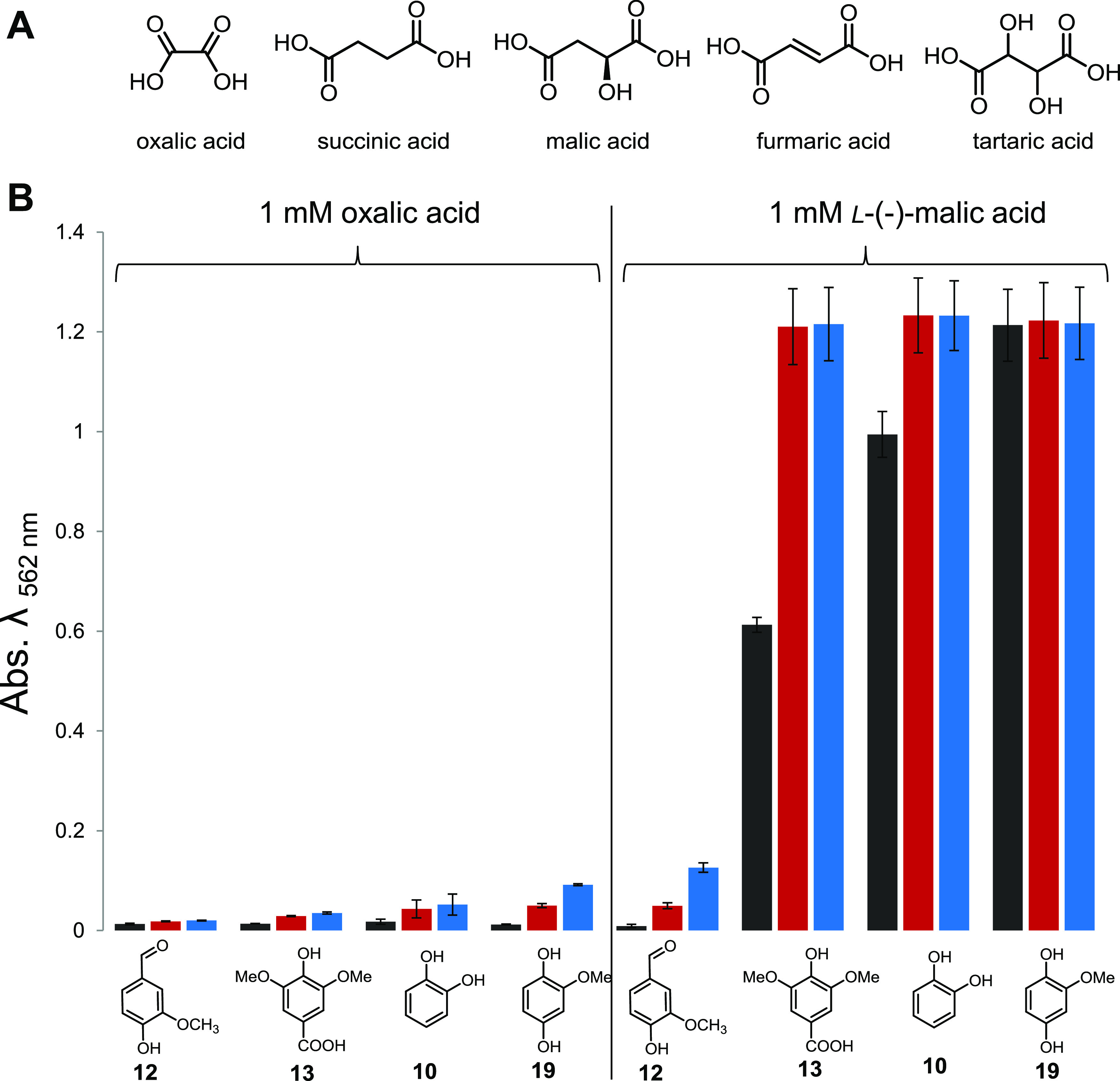
(A) Structures of metal-chelating dicarboxylic acids; (B) quantification of Fe3+ reduction by H2Q using a colorimetric ferrozine-based assay in in the presence of 1 mM oxalic acid and 1 mM L-(-)-malic acid after 5 min (black), 60 min (red) and 120 min (blue). Error bars indicate ±0.5 standard deviation (n = 3).
While laboratory culture conditions generally supply sufficient Fe concentrations for growth, we questioned whether or not the natural fungal comb environment provides the necessary metal ions for Fenton chemistry (48). To answer this question, we analyzed the element composition of fungus comb, gut fluids of termite workers, and soil samples derived from within and outside termite colonies from different locations using inductively coupled plasma atomic emission spectrometry (ICP-AES) (49). All tested samples contained Al, Fe, and Ti as some of the most abundant main elements, in addition to significant amounts of Mn. However, amounts of elements important for growth (C, H, P, K, Ca, Mg) were low in all soil samples, with a particularly strong depletion of phosphorus, but potassium was enriched compared to levels in comb and gut samples (Fig. S22 to S29 and Tables S15, S25, and S26 at https://doi.org/10.5281/zenodo.4431413). Sequential ion extraction of soil samples was performed to analyze the soluble metal ion content, and only low concentrations of most metal ions were detectable (50, 51). Although these findings indicate that fungus comb and the gut environment accommodate large amounts of insoluble Fe/Al oxides, the nano- and microscopic surface areas of these minerals may act as the necessary catalytic centers for Fenton-like redox chemistry (52).
Enzyme activity tests catalyzing the degradation of model lignin compounds.
We then questioned if enzymatic degradation of lignin or lignin-like model substances by Termitomyces is measurable using colorimetric assays or MS-based analytical tools (53). For a first test, we supplemented the culture medium of Termitomyces sp. T153 with the pigment-based model substance Azure B (54), previously used to measure the redox activity of LPs due to its stability toward oxidative activities of MnPs. Over a time course of seven days, we were able to monitor the decolorization of Azure B by Termitomyces, an effect which became more pronounced with the increasing biomass and age of the fungus culture (Fig. S16 at https://doi.org/10.5281/zenodo.4431413). To evaluate if the degrading activity was due to the activity of secreted oxidative enzymes and/or H2Q-mediated Fenton-based chemistry, we tested both effectors separately and in combination. While quantification of enzymatic effects was hampered by technical challenges due to intrinsic light absorption of enzymes concentrates, H2Q-mediated Fenton chemistry clearly induced the degradation of Azure B within 5 to 10 min in a comparison with the control (Fenton reagents without H2Qs) (Fig. S16 at https://doi.org/10.5281/zenodo.4431413) (55). We then evaluated whether or not laccase activity was detectable within the secretome using a syringaldazine-based assay and compared the activity to the reactivity of a commercial laccase from Trametes versicolor (56). However, only residual laccase activity was detectable compared to the activity in the positive control and thus was unlikely accountable for the degradation of Azure B.
Lastly, we evaluated if Termitomyces exhibits MnP enzymatic activity, which is marked by the oxidation of Mn2+ to Mn3+ and the release of the highly reactive oxidant as a carboxylic acid chelate, using a previously reported leukoberbelin blue test (57). As shown in Fig. 8, leukoberbelin-containing Termitomyces cultures and cell-free culture supernatant resulted in the formation of the blue leukoberbelin complex within minutes, which indicated the formation of Mn3+/4+ species. When Termitomyces was grown on potato dextrose agar (PDA) plates containing both elevated Mn2+ concentrations (200 to 500 μM) and indicator dye, the formation of blue leukoberbelin-Mn3+/4+ complexes was detectable within a few days, and longer incubation times resulted in macroscopic MnOx precipitates forming around fungal hyphae within 10 to 17 days (Fig. 4C). We further confirmed the expression of the gene encoding the putative MnP by reverse transcription-PCR (RT-PCR) (Fig. S18 and S19 at https://doi.org/10.5281/zenodo.4431413).
FIG 8.
(A) PDB containing Leukoberbelin blue (left to right, culture of Termitomyces sp. 153, cell-free supernatant, and PDB broth as a control); (B) Termitomyces sp. T153 cultivated on PDA containing 500 μM MnCl2 after 28 days; (C) microscopic image of fungal mycelium after 24 days showing brown MnO2 deposits.
Proteomic analysis.
Building on our enzymatic studies and to link the observed activities with their putative enzymatic origins, we conducted a liquid chromatography tandem mass spectrometry (LC-MS/MS)-based proteomic analysis of secreted enzymes of Termitomyces culture supernatants, which were prepared in two different buffer systems (NaOAc, pH 4.5; KH2PO4, pH 6.5). Overall, a total of 255/303 secreted proteins were detectable, which were mostly assigned to fungal carbohydrate metabolism groups, such as glucosidases, glucanases, or chitinases (Tables S29 to S32 at https://doi.org/10.5281/zenodo.4431413). Interestingly, a potential lignin-degrading aromatic peroxygenase (8th/13th) and one MnP (13th/11th) ranked among the top 15 most abundant protein sequences, while two other yet-unassigned peroxidases were also detectable (31st, 141th/17th, 142nd), albeit at lower abundances. In total, five laccases were detectable, albeit in minor abundances (starting from 76th/99th).
DISCUSSION
In the two major fungus-growing termite genera, Macrotermes and Odontotermes, the decomposition of plant biomass by the fungal cultivar Termitomyces is based on the intricate interactions between the predigestive gut passage and the external fungus comb bioreactor. Although a series of studies have elaborated on the functional roles of Termitomyces in plant biomass degradation (1–3), experimental insights into the biochemical mechanisms necessary for plant biomass degradation have remained sparse.
Which ligninolytic enzymes are produced by Termitomyces?
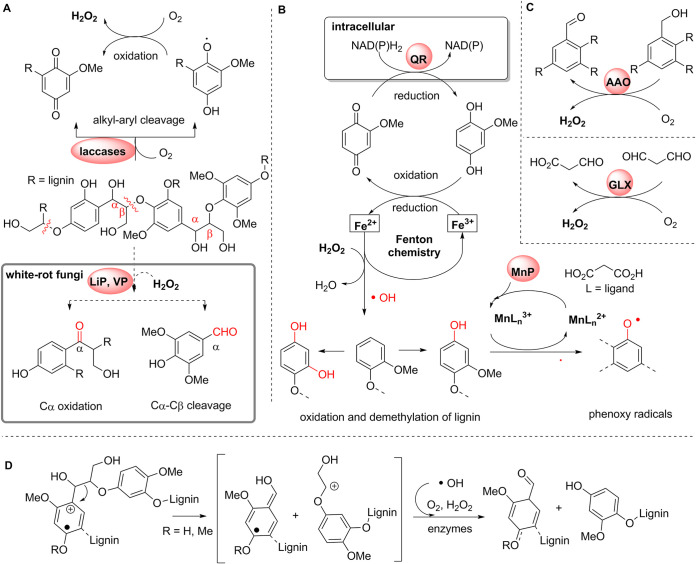
Our omics-based analysis clearly shows that Termitomyces has the capability to produce a set of extracellular lignocellulose‐degrading enzymes, most of which generate diffusible extracellular oxidants (superoxide O2−, hydroxyl radicals •OH, H2O2, redox-active Mn3+/4+ species, or phenoxy radicals) that oxidize the aromatic polymeric three-dimensional (3D) structure of lignin (Fig. 1D and Fig. 9).
FIG 9.
Lignin modifications and oxidation pathways by Termitomyces. (A) Schematic depiction of lignin oxidation by, e.g., laccases (Lac) in contrast to degradation by LiP and VP typically found in white-rot fungi (gray box); (B) oxidation and oxidative demethylation of lignin substructures by 2-MH2Q-catalyzed Fenton chemistry via the formation of short-lived hydroxyl radicals and regeneration of H2Q by (intracellular) benzoquinone reductases (QR); (C) formation of H2O2 by (aryl)-alcohol oxidases (AAO) and glyoxal oxidases (GLX); (D) oxidative C-C cleavage of lignin substructures via phenoxy and methoxy radicals derived from radicals and/or enzymatic processes.
It is particularly intriguing that Termitomyces encodes, on average, 16 different laccases that are differentially transcribed and might differ in their reactivities and substrate spectra. Although laccases are considered not to be essential for lignin degradation (22, 23), their presence likely assists in partial oxidation of phenolic and nonphenolic aromatic moieties that facilitate further fragmentation and depolymerization (Fig. 9). Here, it is also worth highlighting that produced (aryl)-alcohol oxidases are able to efficiently oxidize and cleave β-ether units present within lignin substructures via single-electron transfer reactions (22, 23). Our study also provides conclusive genomic and biochemical evidence that Termitomyces secretes not only a reactive MnP, a class II peroxidase, but also a DyP and a UPO, both of which are known for oxidizing a broad-substrate spectrum. While none of these enzymes are capable of degrading lignin alone, their combined enzymatic actions should allow for lignin’s partial depolymerization, which is necessary for other enzymes of microbial or termite origin to overcome physical barriers of the polymer and access their target substrates in the interior of the dense polymer.
Does Fenton chemistry play a role?
Following up on the idea that Termitomyces utilizes complementary Fenton chemistry for breaking chemical bonds in the dense lignocellulose, we evaluated the presence and absence of metabolic and enzymatic factors necessary to drive the radical process. Here, we provide collective evidence that Termitomyces employs Fenton chemistry by the secretion of high levels of (extracellular) H2O2 and the production of H2Qs that reduce Fe3+ to Fe2+. For the first time, we document that the Termitomyces-derived metabolite 2-MH2Q (compound 19) acts as a redox shuttle for Fenton chemistry and induces the formation of Fe2+, as with 4,5-DMH2Q and 2,5-DMH2Q (21, 35). Genomic and transcriptomic analyses also showed that Termitomyces produces two benzoquinone reductases that may reduce MQ to MH2Q and thereby close the H2Q/Qbased redox shuttle cycle. Considering that Fenton chemistry produces several strong oxidants, we evaluated the influence of fungal dicarboxylic acids on the H2Q-based reduction of Fe3+ complexes and found that complexation with oxalic acid renders the metal ion less available for reduction in a concentration-dependent manner. Thus, we hypothesize that Termitomyces actively applies these protective measures in the proximity of its fungal hyphae, which at the same time allows the sequestering of Fe3+ for intracellular processes.
Considering the observation that fungus comb material is crafted from macerated plant material and is interspersed with metal oxide-rich soil, it appears likely that Fenton-based degradation pathways play a major role in the overall biomass conversion during fungal maturation. The importance of Fenton chemistry in plant degradation was recently demonstrated by Schiøtt and Boomsma (58), who showcased that the combination of enzymatic degradation and Fenton chemistry plays an important role in the coevolved leafcutter ant symbiosis, where ants create spatially isolated substrate pellets, called Fenton pellets, that might function as small contained bioconversion reactors.
Conclusions.
Collectively, our genomic, transcriptomic, metabolomic, and proteomic studies document that Termitomyces utilizes a specific set of oxidative enzymes as well as Fenton chemistry to cope with the challenging task of degrading the lignin-rich plant biomass and presumably applies the same mechanism to detoxify xenobiotic compounds present within the comb, such as plant and microbial natural products, which often have structures similar to those of lignin monomers (48, 50, 52). Our findings increase our general understanding of the role of Fenton chemistry within the symbiosis of termites and their cultivar and shed light on the molecular synergy mechanisms that may have been decisive for integrating the complementary contributions of termites and their cultivar. Whether or not symbiotic and lignocellulolytic bacteria present within the comb might also contribute and complement fungal-lignin degradation capabilities is the topic of current investigations (59, 60) that will further elaborate on the question why the Termitomyces-termite symbiosis has become the most successful path for the termite cultivar.
MATERIALS AND METHODS
Genome sequencing and processing.
DNA was extracted from laboratory-grown heterocaryotic Termitomyces strains T112 and T153, and genome sequences were produced at LGC Genomics (Berlin, Germany) using the Illumina MiSeq V3 platform with 300-bp paired-end reads and approximately 12 million read pairs per sequencing. All library groups were demultiplexed using the Illumina bcl2fastq 2.17.1.14 software (RAW folder, Group subfolders). Up to two mismatches, or N’s, were allowed in the barcode read when the barcode distances between all libraries on the lane allowed for it. Sequencing adapters were clipped from all raw reads, and reads with final lengths of <20 bases were discarded. Afterwards, reads were quality trimmed by removing reads containing more than one N, deleting reads with sequencing errors, trimming reads at the 3′ end to get a minimum average Phred quality score of 10 over a window of 10 bases, and discarding reads with final lengths of less than 20 bp. From the final set of reads, FastQC reports were created for all FASTQ files. SPAdes version 3.13.0 was used for assembly. Prior to annotation, the genomes were soft masked with RepeatMasker 4.0.9 (61). RNA-seq data were mapped to the genomes with STAR 2.7.3a (62) and used to train the AUGUSTUS gene predictor with BRAKER 2.1.5 (63). Finally, the genomes of T112 and T153 were annotated with AUGUSTUS 3.3.3 (64). Protein and mRNA hints were used for the annotation. For details, see https://doi.org/10.5281/zenodo.4431413.
RNA sequencing.
RNA was obtained from mycelia of Termitomyces strains T153 and T112 cultivated on different growth media for 10 days at room temperature. Mycelium was harvested by scraping it from agar plates with a scalpel, freezing it in liquid nitrogen, and storing it at −80°C until RNA extraction. RNA extracts underwent 100-bp paired-end BGISeq-500 sequencing at BGI (Hong Kong). For details, see https://doi.org/10.5281/zenodo.4431413.
RNA-seq data acquisition and processing.
RNA-seq data for fresh comb (NCBI accession no. SRR5944783), old comb (accession no. SRR5944781), and nodules (accession no. SRR5944782) of Termitomyces strains from Macrotermes colony Mn156 were downloaded from the European Nucleotide Archive (65). The raw RNA-seq data were mapped to the annotated genes of T153 using HiSat2 with spiced alignments disabled (version 4.8.2) (66). Transcript abundance was then estimated using HTSeq-count (version 0.11.2) (67). Count data from HTSeq were imported into R using the DESeq2 package (version 1.22.2) (67). Genes with low transcript abundance (<10) were filtered out, and the remaining genes were log10 transformed (68). A heatmap for the identified redox enzymes was generated using the pheatmap package (version 1.0.12) (69) in R (70) with color schemes generated by viridis (version 0.5.1) (71). For details, see https://doi.org/10.5281/zenodo.4431413.
CAZy analysis.
Identification of CAZymes in the predicted proteomes of Termitomyces and other Basidiomycetes strains was performed using a local installation of the dbCAN2 server and all three included tools (HMMER, DIAMOND, and Hotpep searches against the databases included in dbCAN2) (72). For a reliable analysis, we kept only matches that were independently identified by at least two of three annotation strategies and only genes and transcripts classified by their substrate target and thus putative enzymatic functions. EC numbers were assigned using peptide-based functional annotation (http://www.cazy.org/). For details, see https://doi.org/10.5281/zenodo.4431413.
GC-MS analysis.
The fungal isolates Termitomyces sp. P5 and T153 were cultivated on solid media containing different carbon sources. GC-MS analyses of biosamples were carried out with an Agilent (Santa Clara, CA, USA) HP 7890B gas chromatograph fitted with an HP5-MS silica capillary column (30 m, 0.25-mm internal diameter, 0.50-μm film) connected to an HP 5977A inert-mass detector. For details, see https://doi.org/10.5281/zenodo.4431413.
Activity studies of Termitomyces sp. T153.
Detection and quantification of H2O2 in the culture medium of Termitomyces sp. T153 was performed using a fluorimetric hydrogen peroxide assay kit (Sigma-Aldrich). For details, see https://doi.org/10.5281/zenodo.4431413.
Detection of hydroxyl radicals.
Concentrations of hydroxyl radicals were measured using a fluorometric assay based on the reaction with terephthalic acid (TPA), yielding the fluorescent oxidation product hydroxy-terephthalic acid (hTPA) (for details, see https://doi.org/10.5281/zenodo.4431413).
Ferrozine assay.
Fe2+ concentrations were evaluated using a standardized ferrozine assay. For details, see https://doi.org/10.5281/zenodo.4431413.
Proteomic analysis.
Termitomyces sp. T153 was cultured in potato-dextrose broth (25 ml) for 12 days (20°C, 150 rpm), and secreted enzymes were collected and digested according to a standardized protocol (for details, see https://doi.org/10.5281/zenodo.4431413). LC-MS/MS analysis was performed on an UltiMate 3000 RSLCnano system connected to a Q Exactive Plus mass spectrometer (both from Thermo Fisher Scientific, Waltham, MA, USA). Tandem mass spectra were searched against the UniProt database record for Termitomyces sp. J132 (https://www.uniprot.org/proteomes/UP000053712; 26 November 2019) using Proteome Discoverer (PD) 2.4 (Thermo) and the algorithms of Mascot 2.4, Sequest HT (version PD2.2) and MS Amanda 2.0. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (73) partner repository with the dataset identifier PXD025936. For details, see 10.5281/zenodo.4431413.
Protein analysis and activity tests.
Proteomic analysis and experimental details of laccase and MnP activity tests are deposited at https://doi.org/10.5281/zenodo.4431413.
Supporting information can be accessed free of charge at Zenodo (https://doi.org/10.5281/zenodo.4781753) and figshare (https://doi.org/10.6084/m9.figshare.14073491) and contains information regarding culture conditions, isolation procedures, structure elucidation, activity assays, expression-level data, CAZy counts, and a proteomic hit list.
ACKNOWLEDGMENTS
This study was funded by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation project ID 239748522-SFB 1127 [project A6] grant to C.B. and CRC/TR 124 FungiNet project no. 210879364 [projects A1 and Z2] grant to A.A.B. and O.K.), by the Danish Council for Independent Research (grant DFF-7014-00178), and by a European Research Council consolidator grant (771349) to M.P. C.G. and N.G.-C. acknowledge financial support from the state budget of the Slovenian Research Agency (research project J4-2549, research programs P1-0198 and P1-0170, Infrastructural Centre Mycosmo).
Help with microscopy pictures by David Zopf is greatly appreciated (SFB 1127/2 ChemBioSys project no. 239748522 [project Z]).
This paper was written with contributions from all authors. All authors have approved the final version.
There are no conflicts of interest to declare.
Footnotes
Citation Schalk F, Gostinčar C, Kreuzenbeck NB, Conlon BH, Sommerwerk E, Rabe P, Burkhardt I, Krüger T, Kniemeyer O, Brakhage AA, Gunde-Cimerman N, de Beer ZW, Dickschat JS, Poulsen M, Beemelmanns C. 2021. The termite fungal cultivar Termitomyces combines diverse enzymes and oxidative reactions for plant biomass conversion. mBio 12:e03551-20. https://doi.org/10.1128/mBio.03551-20.
Contributor Information
Christine Beemelmanns, Email: Christine.Beemelmanns@hki-jena.de.
Nicole Dubilier, Max Planck Institute for Marine Microbiology.
REFERENCES
- 1.Li H, Young SE, Poulsen M, Currie CR. 2021. Symbiont-mediated digestion of plant biomass in fungus-farming insects. Annu Rev Entomol 66:297–316. doi: 10.1146/annurev-ento-040920-061140. [DOI] [PubMed] [Google Scholar]
- 2.Wisselink M, Aanen DK, van ’t Padje A. 2020. The longevity of colonies of fungus-growing termites and the stability of the symbiosis. Insects 11:527. doi: 10.3390/insects11080527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poulsen M, Hu H, Li C, Chen Z, Xu L, Otani S, Nygaard S, Nobre T, Klaubauf S, Schindler PM, Hauser F, Pan H, Yang Z, Sonnenberg ASM, de Beer ZW, Zhang Y, Wingfield MJ, Grimmelikhuijzen CJP, de Vries RP, Korb J, Aanen DK, Wang J, Boomsma JJ, Zhang G. 2014. Complementary symbiont contributions to plant decomposition in a fungus-farming termite. Proc Natl Acad Sci U S A 111:14500–14505. doi: 10.1073/pnas.1319718111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones JA. 1990. Termites, soil fertility and carbon cycling in dry tropical Africa: a hypothesis. J Trop Ecol 6:291–305. doi: 10.1017/S0266467400004533. [DOI] [Google Scholar]
- 5.Kirk PM, Cannon PF, David JC, Stalpers JA. 2001. Ainsworth and Bisby’s dictionary of the fungi. CAB International, Wallingford, Oxfordshire, United Kingdom. [Google Scholar]
- 6.Rohrmann GF. 1978. The origin, structure, and nutritional importance of the comb in two species of Macrotermitinae. Pedobiologia 18:89–98. [Google Scholar]
- 7.da Costa RR, Hu H, Pilgaard B, Vreeburg SME, Schückel J, Pedersen KSK, Kračun SK, Busk PK, Harholt J, Sapountzis P, Lange L, Aanen DK, Poulsen M. 2018. Enzyme activities at different stages of plant biomass decomposition in three species of fungus-growing termites. Appl Environ Microbiol 84:e01815-17. doi: 10.1128/AEM.01815-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu H, da Costa RR, Pilgaard B, Schiøtt M, Lange L, Poulsen M. 2019. Fungiculture in termites is associated with a mycolytic gut bacterial community. mSphere 4:e00165-19. doi: 10.1128/mSphere.00165-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyodo F, Tayasu I, Inoue T, Azuma JI, Kudo T, Abe T. 2003. Differential role of symbiotic fungi in lignin degradation and food provision for fungus-growing termites (Macrotermitinae: Isoptera). Funct Ecol 17:186–193. doi: 10.1046/j.1365-2435.2003.00718.x. [DOI] [Google Scholar]
- 10.Geib SM, Filley TR, Hatcher PG, Hoover K, Carlson JE, del Mar Jimenez-Gasco M, Nakagawa-Izumi A, Sleighter RL, Tien M. 2008. Lignin degradation in wood-feeding insects. Proc Natl Acad Sci U S A 105:12932–12937. doi: 10.1073/pnas.0805257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyauchi S, Kiss E, Kuo A, Drula E, Kohler A, Sánchez-García M, Morin E, Andreopoulos B, Barry KW, Bonito G, Buée M, Carver A, Chen C, Cichocki N, Clum A, Culley D, Crous PW, Fauchery L, Girlanda M, Hayes RD, Kéri Z, LaButti K, Lipzen A, Lombard V, Magnuson J, Maillard F, Murat C, Nolan M, Ohm RA, Pangilinan J, de Freitas Pereira M, Perotto S, Peter M, Pfister S, Riley R, Sitrit Y, Stielow JB, Szöllősi G, Žifčáková L, Štursová M, Spatafora JW, Tedersoo L, Vaario L-M, Yamada A, Yan M, Wang P, Xu J, Bruns T, Baldrian P, Vilgalys R, et al. 2020. Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits. Nat Commun 11:5125. doi: 10.1038/s41467-020-18795-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. 2010. Lignin biosynthesis and structure. Plant Physiol 153:895–905. doi: 10.1104/pp.110.155119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dashtban M, Schraft H, Syed TA, Qin W. 2010. Fungal biodegradation and enzymatic modification of lignin. Int J Biochem Mol Biol 1:36–50. [PMC free article] [PubMed] [Google Scholar]
- 14.Bugg TDH, Ahmad M, Hardiman EM, Rahmanpour R. 2011. Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep 28:1883–1896. doi: 10.1039/c1np00042j. [DOI] [PubMed] [Google Scholar]
- 15.Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, Asiegbu FO, Baker SE, Barry K, Bendiksby M, Blumentritt M, Coutinho PM, Cullen D, de Vries RP, Gathman A, Goodell B, Henrissat B, Ihrmark K, Kauserud H, Kohler A, LaButti K, Lapidus A, Lavin JL, Lee Y-H, Lindquist E, Lilly W, Lucas S, Morin E, Murat C, Oguiza JA, Park J, Pisabarro AG, Riley R, Rosling A, Salamov A, Schmidt O, Schmutz J, Skrede I, Stenlid J, Wiebenga A, Xie X, Kües U, Hibbett DS, Hoffmeister D, Högberg N, Martin F, Grigoriev IV, Watkinson SC. 2011. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 333:762–765. doi: 10.1126/science.1205411. [DOI] [PubMed] [Google Scholar]
- 16.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martínez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Górecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kües U, Kumar TKA, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, et al. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 17.Gaskell J, Blanchette RA, Stewart PE, BonDurant SS, Adams M, Sabat G, Kersten P, Cullen D. 2016. Transcriptome and secretome analyses of the wood decay fungus Wolfiporia cocos support alternative mechanisms of lignocellulose conversion. Appl Environ Microbiol 82:3979–3987. doi: 10.1128/AEM.00639-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Yelle DJ, Li C, Yang M, Ke J, Zhang R, Liu Y, Zhu N, Liang S, Mo X, Ralph J, Currie CR, Mo J. 2017. Lignocellulose pretreatment in a fungus-cultivating termite. Proc Natl Acad Sci U S A 114:4709–4714. doi: 10.1073/pnas.1618360114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz-Dueñas FJ, Barrasa JM, Sánchez-García M, Camarero S, Miyauchi S, Serrano A, Linde D, Babiker R, Drula E, Ayuso-Fernández I, Pacheco R, Padilla G, Ferreira P, Barriuso J, Kellner H, Castanera R, Alfaro M, Ramirez L, Pisabarro AG, Riley R, Kuo A, Andreopoulos W, LaButti K, Pangilinan J, Tritt A, Lipzen A, He G, Yan M, Ng V, Grigoriev IV, Cullen D, Martin F, Rosso M-N, Henrissat B, Hibbett D, Martínez AT. 2020. Genomic analysis enlightens agaricales lifestyle evolution and increasing peroxidase diversity. Mol Biol Evol 38:1428–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ayuso-Fernandes I, Ruiz DF, Martinez AT. 2018. Evolutionary convergence in lignin-degrading enzymes. Proc Natl Acad Sci U S A 115:6428–6433. doi: 10.1073/pnas.1802555115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt-Dannert C. 2016. Biocatalytic portfolio of Basidiomycota. Curr Opin Chem Biol 31:40–49. doi: 10.1016/j.cbpa.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Yao B, Su X. 2018. Linking enzymatic oxidative degradation of lignin to organics detoxification. Int J Mol Sci 19:3373. doi: 10.3390/ijms19113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin X, Sun X, Huang H, Bai Y, Wang Y, Luo H, Yao B, Zhang X, Su X. 2017. Oxidation of a non-phenolic lignin model compound by two Irpex lacteus manganese peroxidases: evidence for implication of carboxylate and radicals. Biotechnol Biofuels 10:103. doi: 10.1186/s13068-017-0787-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Wu S, Ma X, Chen W, Zhang J, Duan S, Gao Y, Kui L, Huang W, Wu P, Shi R, Li Y, Wang Y, Li J, Guo X, Luo X, Li Q, Xiong C, Liu H, Gui M, Sheng J, Dong Y. 2018. The genome sequences of 90 mushrooms. Sci Rep 8:9982. doi: 10.1038/s41598-018-28303-2.(Author Correction, 10:8460, doi:). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Busk PK, Lange L. 2013. Function-based classification of carbohydrate-active enzymes by recognition of short, conserved peptide motifs. Appl Environ Microbiol 79:3380–3391. doi: 10.1128/AEM.03803-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kameshwar AKS, Qin W. 2016. Recent developments in using advanced sequencing technologies for the genomic studies of lignin and cellulose degrading microorganisms. Int J Biol Sci 12:156–171. doi: 10.7150/ijbs.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taprab Y, Johjima T, Maeda Y, Moriya S, Trakulnaleamsai S, Noparatnaraporn N, Ohkuma M, Kudo T. 2005. Symbiotic fungi produce laccases potentially involved in phenol degradation in fungus combs of fungus-growing termites in Thailand. Appl Environ Microbiol 71:7696–7704. doi: 10.1128/AEM.71.12.7696-7704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johjima T, Ohkuma M, Kudo T. 2003. Isolation and cDNA cloning of novel hydrogen peroxide-dependent phenol oxidase from the basidiomycete Termitomyces albuminosus. Appl Microbiol Biotechnol 61:220–225. doi: 10.1007/s00253-003-1236-4. [DOI] [PubMed] [Google Scholar]
- 31.Bose S, Mazumder S, Mukherjee M. 2007. Laccase production by the white-rot fungus Termitomyces clypeatus. J Basic Microbiol 47:127–131. doi: 10.1002/jobm.200610206. [DOI] [PubMed] [Google Scholar]
- 32.Thurston CF. 1994. The structure and function of fungal laccases. Microbiology 140:19–26. doi: 10.1099/13500872-140-1-19. [DOI] [Google Scholar]
- 33.Kersten P, Cullen D. 2014. Copper radical oxidases and related extracellular oxidoreductases of wood-decay Agaricomycetes. Fungal Genet Biol 72:124–130. doi: 10.1016/j.fgb.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 34.Sützl L, Laurent CVFP, Abrera AT, Schütz G, Ludwig R, Haltrich D. 2018. Multiplicity of enzymatic functions in the CAZy AA3 family. Appl Microbiol Biotechnol 102:2477–2492. doi: 10.1007/s00253-018-8784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen KA, Houtman CJ, Ryan ZC, Hammel KE. 2001. Pathways for extracellular Fenton chemistry in the brown rot basidiomycete Gloeophyllum trabeum. Appl Environ Microbiol 67:2705–2711. doi: 10.1128/AEM.67.6.2705-2711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halliwell B, Gutteridge JMC. 2015. Free radicals in biology and medicine. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 37.Arantes V, Milagres AMF. 2007. The synergistic action of ligninolytic enzymes (MnP and Laccase) and Fe3+-reducing activity from white-rot fungi for degradation of Azure B. Enzyme Microb Technol 42:17–22. doi: 10.1016/j.enzmictec.2007.07.017. [DOI] [Google Scholar]
- 38.Suzuki MR, Hunt CG, Houtman CJ, Dalebroux ZD, Hammel KE. 2006. Fungal hydroquinones contribute to brown rot of wood. Environ Microbiol 8:2214–2223. doi: 10.1111/j.1462-2920.2006.01160.x. [DOI] [PubMed] [Google Scholar]
- 39.Korripally P, Timokhin VI, Houtman CJ, Mozuch MD, Hammel KE. 2013. Evidence from Serpula lacrymans that 2,5-dimethoxyhydroquinone is a lignocellulolytic agent of divergent brown rot basidiomycetes. Appl Environ Microbiol 79:2377–2383. doi: 10.1128/AEM.03880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah F, Schwenk D, Nicolás C, Persson P, Hoffmeister D, Tunlid A. 2015. Involutin is an Fe3+-reductant secreted by the ectomycorrhizal fungus Paxillus involutus during Fenton-based decomposition of organic matter. Appl Environ Microbiol 81:8427–8433. doi: 10.1128/AEM.02312-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeitner TM. 2014. Optimized ferrozine-based assay for dissolved iron. Anal Biochem 454:36–37. doi: 10.1016/j.ab.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 42.Verschoor MJ, Molot LA. 2013. A comparison of three colorimetric methods of ferrous and total reactive iron measurement in freshwaters. Limnol Oceanogr Methods 11:113–125. doi: 10.4319/lom.2013.11.113. [DOI] [Google Scholar]
- 43.Haddou M, Benoit-Marquié F, Maurette M-T, Oliveros E. 2010. Oxidative degradation of 2,4-dihydroxybenzoic acid by the Fenton and photo-Fenton processes: kinetics, mechanisms, and evidence for the substitution of H2O2 by O2. Helvetica 93:1067–1080. doi: 10.1002/hlca.200900380. [DOI] [Google Scholar]
- 44.Otani S, Challinor VL, Kreuzenbeck NB, Kildgaard S, Krath CS, Larsen LLM, Aanen DK, Rasmussen SA, Beemelmanns C, Poulsen M. 2019. Disease-free monoculture farming by fungus-growing termites. Sci Rep 9:8819. doi: 10.1038/s41598-019-45364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Utset B, Garcia J, Casado J, Domènech X, Peral J. 2000. Replacement of H2O2 by O2 in Fenton and photo-Fenton reactions. Chemosphere 41:1187–1192. doi: 10.1016/s0045-6535(00)00011-4. [DOI] [PubMed] [Google Scholar]
- 46.Varela E, Tien M. 2003. Effect of pH and oxalate on hydroquinone-derived hydroxyl radical formation during brown rot wood degradation. Appl Environ Microbiol 69:6025–6031. doi: 10.1128/AEM.69.10.6025-6031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fenton H, Horstman J. 1894. LXXIII.—Oxidation of tartaric acid in presence of iron. J Chem Soc Trans 65:899–910. doi: 10.1039/CT8946500899. [DOI] [Google Scholar]
- 48.Arshad MA, Schnitzer M. 1987. The chemistry of a termite fungus comb. Plant Soil 98:247–256. doi: 10.1007/BF02374828. [DOI] [Google Scholar]
- 49.Deke AL, Adugna WT, Fite AT. 2016. Soil physic-chemical properties in termite mounds and adjacent control soil in Miyo and Yabello Districts of Borana zone, southern Ethiopia. Am J Agric For 4:69–74. doi: 10.11648/j.ajaf.20160404.11. [DOI] [Google Scholar]
- 50.Jouquet P, Tessier D, Lepage M. 2004. The soil structural stability of termite nests: role of clays in Macrotermes bellicosus (Isoptera, Macrotermitinae) mound soils. Eur J Soil Biol 40:23–29. doi: 10.1016/j.ejsobi.2004.01.006. [DOI] [Google Scholar]
- 51.Lyngsie G, Krumina L, Tunlid A, Persson P. 2018. Generation of hydroxyl radicals from reactions between a dimethoxyhydroquinone and iron oxide nanoparticles. Sci Rep 8:10834–10834. doi: 10.1038/s41598-018-29075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krumina L, Lyngsie G, Tunlid A, Persson P. 2017. Oxidation of a dimethoxyhydroquinone by ferrihydrite and goethite nanoparticles: iron reduction versus surface catalysis. Environ Sci Technol 51:9053–9061. doi: 10.1021/acs.est.7b02292. [DOI] [PubMed] [Google Scholar]
- 53.Owen BC, Haupert LJ, Jarrell TM, Marcum CL, Parsell TH, Abu-Omar MM, Bozell JJ, Black SK, Kenttämaa HI. 2012. High-performance liquid chromatography/high-resolution multiple stage tandem mass spectrometry using negative-ion-mode hydroxide-doped electrospray ionization for the characterization of lignin degradation products. Anal Chem 84:6000–6007. doi: 10.1021/ac300762y. [DOI] [PubMed] [Google Scholar]
- 54.Casciello C, Tonin F, Berini F, Fasoli E, Marinelli F, Pollegioni L, Rosini E. 2017. A valuable peroxidase activity from the novel species Nonomuraea gerenzanensis growing on alkali lignin. Biotechnol Rep 13:49–57. doi: 10.1016/j.btre.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng S, Zhang W, He J, Yang X, Wang D, Zeng G. 2016. Enhancement of Fenton oxidation for removing organic matter from hypersaline solution by accelerating ferric system with hydroxylamine hydrochloride and benzoquinone. J Environ Sci (China) 41:16–23. doi: 10.1016/j.jes.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 56.Krumbein WE, Altmann HJ. 1973. A new method for the detection and enumeration of manganese oxidizing and reducing microorganisms. Helgolander Wiss Meeresunters 25:347–356. doi: 10.1007/BF01611203. [DOI] [Google Scholar]
- 57.Rashid GMM, Zhang X, Wilkinson RC, Fülöp V, Cottyn B, Baumberger S, Bugg TDH. 2018. Sphingobacterium sp. T2 manganese superoxide dismutase catalyzes the oxidative demethylation of polymeric lignin via generation of hydroxyl radical. ACS Chem Biol 13:2920–2929. doi: 10.1021/acschembio.8b00557. [DOI] [PubMed] [Google Scholar]
- 58.Schiøtt M, Boomsma JJ. 2021. Proteomics reveals synergy between biomass degrading enzymes and inorganic Fenton chemistry in leaf-cutting ant colonies. Elife 10:e61816. doi: 10.7554/eLife.61816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown ME, Chang MCY. 2014. Exploring bacterial lignin degradation. Curr Opin Chem Biol 19:1–7. doi: 10.1016/j.cbpa.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 60.Murphy R, Benndorf R, de Beer ZW, Vollmers J, Kaster AK, Beemelmanns C, Poulsen M. 2021. Comparative genomics reveals prophylactic and catabolic capabilities of Actinobacteria within the fungus-farming termite symbiosis. mSphere 6:e01233-20. doi: 10.1128/mSphere.01233-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smit A, Hubley R, Green P. 2013–2015. http://www.repeatmasker.org.
- 62.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoff KJ, Lomsadze A, Borodovsky M, Stanke M. 2019. Whole-genome annotation with BRAKER. Methods Mol Biol 1962:65–95. doi: 10.1007/978-1-4939-9173-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stanke M, Morgenstern B. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33:465–467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.S96. [DOI] [PubMed] [Google Scholar]
- 69.Raivo K. 2019. pheatmap: pretty heatmaps. R package version 1.0.12. R Foundation for Statistical Computing, Vienna, Austria. https://CRAN.R-project.org/package=pheatmap. [Google Scholar]
- 70.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 71.Simon G. 2018. viridis: default color maps from ‘matplotlib.’ R package version 0.5.1. R Foundation for Statistical Computing, Vienna, Austria. https://CRAN.R-project.org/package=viridis. [Google Scholar]
- 72.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PL, Xu Y, Yin Y. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:95–101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]