ABSTRACT
Bacterial outer membrane vesicles (OMVs) enriched with bioactive proteins, toxins, and virulence factors play a critical role in host-pathogen and microbial interactions. The two-component system PhoP-PhoQ (PhoPQ) of Salmonella enterica orchestrates the remodeling of outer membrane lipopolysaccharide (LPS) molecules and concomitantly upregulates OMV production. In this study, we document a novel use of nanoparticle tracking analysis to determine bacterial OMV size and number. Among the PhoPQ-activated genes tested, pagC expression had the most significant effect on the upregulation of OMV production. We provide the first evidence that PhoPQ-mediated upregulation of OMV production contributes to bacterial survival by interfering with complement activation. OMVs protected bacteria in a dose-dependent manner, and bacteria were highly susceptible to complement-mediated killing in their absence. OMVs from bacteria expressing PagC bound to complement component C3b in a dose-dependent manner and inactivated it by recruiting complement inhibitor Factor H. As we also found that Factor H binds to PagC, we propose that PagC interferes with complement-mediated killing of Salmonella in the following two steps: first by engaging Factor H, and second, through the production of PagC-enriched OMVs that divert and inactivate the complement away from the bacteria. Since PhoPQ activation occurs intracellularly, the resultant increase in PagC expression and OMV production is suggested to contribute to the local and systemic spread of Salmonella released from dying host cells that supports the infection of new cells.
KEYWORDS: Salmonella, S. Typhimurium, PagC, Rck, outer membrane vesicles, PhoPQ, C3b, Factor H, complement resistance
INTRODUCTION
Salmonella enterica is a Gram-negative bacterial pathogen that can survive and replicate in both phagocytic and nonphagocytic cells (1, 2) thanks in part to its two-component system PhoP and PhoQ, designated PhoPQ (3). PhoPQ is activated by low Mg2+, acidic pH, and cationic antimicrobial peptides in Salmonella-containing vacuoles and regulates the expression of genes required for intracellular survival (4–6), in part by activating the SPI-2 type III secretion system (T3SS) (3). Activation of PhoPQ also induces the expression of genes encoding outer membrane proteins (OMPs), as well as regulators and enzymes that modify outer membrane (OM) components. Recapitulation of the shift from extracellular to intracellular environmental conditions in vitro leads to activation of the phoPQ regulon and covalent modification of OM lipopolysaccharides (LPSs), thereby destabilizing the highly cross-linked OM (7). These changes increase outer membrane vesicle (OMV) formation and help in the removal of negatively charged LPS detrimental for intracellular survival and in its replacement with modified LPS that is more neutral (7). Accordingly, constitutive expression or induction of PhoPQ-activated genes (pags), such as pagP or pagL which encode OM enzymes that add or remove acyl groups from LPS, result in increased OMV production and concomitant removal of charged LPS, while deletion of these genes reduced the production of OMVs under different experimental conditions (8, 9).
OMVs are spherical (20- to 200-nm diameter) membranous structures primarily composed of LPSs, phospholipids, OMPs, and a lumen filled with cargo that consist mainly of periplasmic proteins (10). OMVs play critical roles in bacterium-bacterium and bacterium-host interactions (11). The production of OMVs allows the bacterium to interact with its environment and mediate pathogenesis through biofilm formation, horizontal gene transfer, intra- and interspecies communication, delivery of toxins, killing of competing microbial cells, resistance to antibiotics, adherence to host cells, and immunomodulation (10, 12–16). Since the proteins of various PhoPQ-activated genes can play a role in OMV production (8, 9, 17), we undertook a systematic analysis of OMVs made by deletion mutants of PhoPQ-regulated genes. We used nanoparticle tracking analysis (NTA) to compare the size and number of OMVs produced. Several mutants showed reduced OMV production, but deletion of pagC had the most significant effect. Thus, we further investigated the role of PagC in the formation of OMVs and assessed the potential implications of these PagC-induced OMVs in Salmonella pathogenesis.
PagC belongs to a family of integral OMPs that form a barrel-shaped transmembrane structure with 8 β-strands and 4 extracellular loops (18, 19). It plays a role in biofilm formation and shares homology with OMPs such as Rck encoded on a Salmonella enterica serovar Typhimurium plasmid and OmpX/Ail of various Enterobacteriaceae (20–27). While both Rck and Ail mediate serum resistance (23, 28–32), the ability of PagC to provide S. Typhimurium with the same protection remains controversial in the literature (33, 34). Therefore, we investigated a possible link between either PagC or OMV production and serum resistance. Notably, we identified a novel PagC-dependent mechanism by which Salmonella evades complement-mediated bacterial killing. Specifically, we described a role for PagC in upregulating OMV production and further demonstrated that OMVs produced by PagC-expressing bacteria attract complement component C3b and inactivate it by recruiting Factor H. As PagC is enriched in OMVs and binds to Factor H, we propose that OMVs triggered by PagC expression serve as complement decoys that trap and inactivate C3b, protecting Salmonella from the bactericidal effect of serum, thereby aiding in local and systemic spread.
RESULTS
PagC is an activator of OMV production.
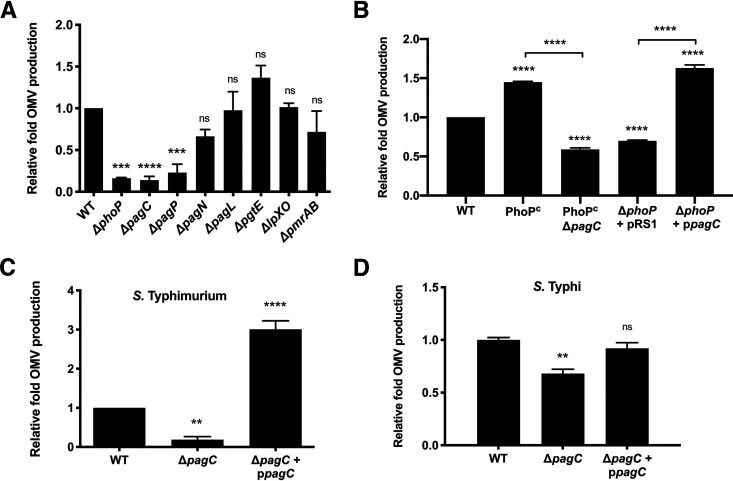
The Salmonella PhoPQ regulator senses the host environment to promote OM remodeling, during which enzymes are activated to modify a “new” LPS, while the “old” unmodified LPS is recycled via OMV production (7). To identify which PhoPQ-activated genes are involved in enhancing OMV production, we isolated OMVs from various pag deletion mutants grown under PhoPQ-activating conditions (5.8L N-minimal media). The number of OMVs as enumerated by NTA decreased by 6.2-fold for ΔphoP, 4.3-fold for ΔpagP, and 7.3-fold for ΔpagC mutants compared with the parental strain S. Typhimurium fliC fljB, designated wild type (WT) in this study (Fig. 1A; see Table S1 in the supplemental material). In contrast, no such significant differences were found in the deletion mutants of other PhoPQ-activated genes, including pagN, pagL, pgtE, lpxO, and pmrAB. Deletion of pagC in a strain that constitutively expressed phoP (PhoPC ΔpagC) reduced the number of OMVs by 2.4-fold compared with the PhoPC mutant, whereas plasmid-based induction of PagC (ppagC) in a ΔphoP mutant increased the number of OMVs produced by 3.2-fold compared with the ΔphoP mutant containing just the empty vector (Fig. 1B and Table S1). We also measured the size (diameter) of the OMVs by both transmission electron microscopy (TEM) (see Fig. S1A in the supplemental material) and NTA (Fig. S1B). The NTA analysis revealed that the diameter of OMVs was not significantly changed by deletion of any of the pags studied (P > 0.05 for each mutant compared with WT) (Fig. S1C). To determine if the effects of pagC on OMV production were recapitulated in a Salmonella serovar that is more virulent for humans, we generated a deletion mutant of pagC in Salmonella enterica serovar Typhi. This mutant also showed a hypovesiculating phenotype compared with the wild-type strain (Fig. 1D; Table S1). Complementation of PagC expression by the ppagC vector in a ΔpagC mutant restored the vesiculation phenotype in both S. Typhi and S. Typhimurium (Fig. 1C and D; Table S1). Taken together, the data indicated that PagC is a significant activator of OMV production in different Salmonella serovars.
FIG 1.
Quantitative analysis of OMV production by Salmonella under PhoPQ-activating conditions (5.8L). All S. Typhimurium strains used were flagella mutants (fliC fljB). (A) Relative fold OMV production by 2 × 109 CFU/ml bacteria was calculated relative to the OMVs produced by parental strain S. Typhimurium fliC fljB (labeled as WT) normalized to 1. NTA analysis showed that the OMV production was significantly downregulated in ΔphoP, ΔpagC, and ΔpagP mutants. (B) A constitutively active phoP mutant (PhoPC) and the ΔphoP mutant induced to express PagC from plasmid ppagC showed a hypervesiculating phenotype. Deletion of pagC in PhoPC and the ΔphoP mutant containing the empty vector (pRS1) decreased OMV production by ∼3-fold. (C) The ΔpagC mutants of S. Typhimurium and S. Typhi showed significant reductions in OMV production compared with their respective WT parental strains. The phenotype of the S. Typhi strain could be restored by complementing the ΔpagC mutants with plasmid ppagC induced to express PagC. The data represent one of three separate, reproducible experiments, expressed as mean ± SEM. Statistical significance was calculated using one-way ANOVA multiple-comparison test with significance set at a P value of <0.05 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; and ns, P > 0.05).
WT and ΔpagC OMVs as analyzed by TEM (A) and nanoparticle tracking analysis (B). (A) Average diameter of OMVs for WT was 138.94 ± 86 nm (mean ± SD) and 114.38 ± 57 nm for ΔpagC, as determined from TEM micrographs and measured by NIH ImageJ software. (B) Peak represents the modal value of the experimental data (154 nm or 149 nm for WT and ΔpagC OMVs, respectively), as calculated by NTA analysis. (C) Mean size (diameter in nm) of OMVs produced by Salmonella grown under PhoPQ-activating conditions, as measured by NTA analysis. The data represent one of three separate, reproducible experiments, expressed as mean ± SEM. Statistical significance was calculated using the one-way ANOVA multiple-comparison test (ns, P > 0.05). Download FIG S1, PDF file, 1.9 MB (1.9MB, pdf) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Number of OMVs/ml from various Salmonella deletion mutants. Download Table S1, DOCX file, 0.02 MB (16.4KB, docx) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To determine whether the level and/or repertoire of OMPs in OMVs were affected in the ΔpagC mutant, we undertook a comparative mass spectrometry analysis of OMPs in OMVs from WT and ΔpagC bacteria. OMPs detected in the OMVs of the SL1344, ATCC14028s, and LT2 (35) wild-type strains were essentially comparable (see Table S2 in the supplemental material). Using OmpA as the baseline standard for OMPs, we found that the relative abundance of most detected OMPs in the OMVs from the ΔpagC mutant only varied slightly compared with those in OMVs from the WT strain (Table S2). However, the relative abundance of some ion transport proteins, such as FoxA, CirA, and FepA, increased significantly in the ΔpagC mutant, as did the porins OmpF and OmpC, the surface receptors FepE and IroN, and the LPS-assembly lipoprotein (LptE). In addition, the relative amounts of FadL, Blc, and the OMP-assembly factor BamA were decreased in the ΔpagC mutant. Thus, the absence of PagC dysregulated OMV production not only by reducing their generation but also by modulating the ratios of selected OMPs.
Outer membrane proteins (OMPs) detected in OMVs from the WT strain and the ΔpagC mutant. Download Table S2, XLSX file, 0.02 MB (23.4KB, xlsx) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
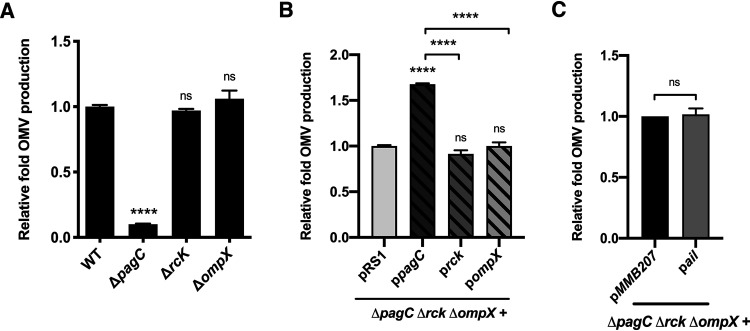
PagC shares sequence similarity with two other S. Typhimurium OMPs, namely, RcK (54%) and OmpX (36%), raising the possibility that they could play a role in OMV production. However, deletion of either rck or ompX, of which both encode OMPs present in OMVs of the wild-type strain (Table S2), did not alter the number of OMVs produced under PhoPQ-activating conditions (Fig. 2A and Table S1). Furthermore, in contrast to PagC, plasmid-driven expression of S. Typhimurium Rck or OmpX did not alter OMV production in a ΔpagC Δrck ΔompX triple deletion mutant (Fig. 2B and Table S1). Additionally, the OMP Ail of Yersinia pestis, which is also similar to PagC (36%), did not affect OMV production when expressed from a plasmid in the ΔpagC Δrck ΔompX triple mutant (Fig. 2C and Table S1). Thus, only PagC among the set of PagC-similar bacterial OMPs (Rck, OmpX, and Ail) activates OMV production.
FIG 2.
Relative fold OMV production by Salmonella grown under PhoPQ-activating conditions (5.8L). (A) Relative fold OMV production by the ΔpagC mutant was significantly reduced compared with WT bacteria or Δrck or ΔompX mutants. Deletion of rck or ompX had no significant effect on OMV production. (B) OMVs produced by the ΔpagC Δrck ΔompX triple mutant expressing PagC, Rck, or OmpX from corresponding plasmids showed that OMV production is upregulated only by the PagC-expressing plasmid in the ΔpagC Δrck ΔompX triple mutant. Plasmid pRS1 is the control empty vector. The data represent one of three separate, reproducible experiments, expressed as mean ± SEM. (C) Relative fold OMV production by ΔpagC Δrck ΔompX triple mutant containing empty vector pMMB207 or pail induced to express Ail from Y. pestis (100 μM isopropyl-β-d-thiogalactopyranoside [IPTG]) showed no significant change in OMV production. Statistical significance was calculated using one-way ANOVA multiple-comparison test and set at a P value of <0.05 (****, P < 0.0001; and ns, P > 0.05).
Salmonella OM protein PagC mediates resistance to serum-dependent bacterial killing.
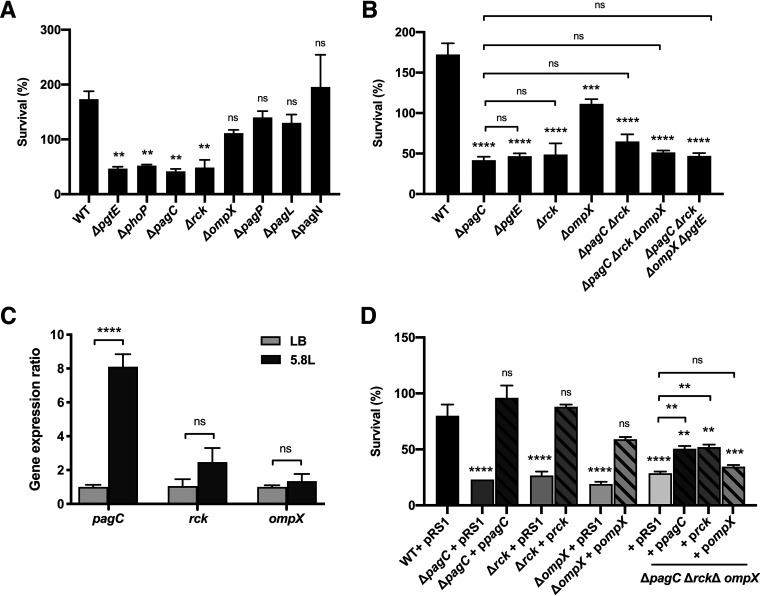
The sequence similarity of PagC with other OMPs, such as the S. Typhimurium Rck and Y. pestis Ail, that provide serum resistance prompted us to reevaluate the role of PagC in resistance to serum-mediated bacterial killing, an issue plagued by conflicting results in the literature (33, 34). For this evaluation, we incubated S. Typhimurium WT and various isogenic deletion mutants of PhoPQ-activated genes with 25% normal human serum (NHS) and compared their resistance to serum-mediated killing. Deletions of pagC and phoP made bacteria more susceptible to serum-mediated killing, as did deletions of the pgtE or rck gene, which are both known to inactivate serum/complement-mediated bacterial killing and serve as PhoPQ-dependent and PhoPQ-independent positive controls, respectively (36, 37). In contrast, deletion of other PhoPQ-activated genes, such as pagP, pagL, and pagN, had no significant effect on bacterial survival (Fig. 3A). The Ail-similar OmpX of Salmonella was only weakly protective (Fig. 3A). Notably, the relative abundance of Rck, OmpX, and PgtE in OMVs from the ΔpagC mutant remained comparable to OMVs from the WT strain (0.8 to 1.7 times) (Table S2), indicating that these proteins were unable to compensate for the absence of PagC in inhibiting serum-mediated killing.
FIG 3.
Serum resistance assay for Salmonella mutants grown under PhoPQ-activating conditions. (A) Survival of Salmonella deletion mutants in 25% NHS incubated at 37°C for 1 h. Deletion of ΔpgtE, Δrck (used as positive controls), ΔphoP, and ΔpagC made the bacteria susceptible to complement-mediated killing. (B) A comparison of serum resistance between Salmonella ΔpagC, Δrck, and ΔompX single, double, and triple mutants. (C) Quantitative RT-PCR-based analysis of gene expression ratio of pagC, rck, and ompX (relative to housekeeping gene rpoB) in wild-type Salmonella grown in LB or under PhoPQ-activating conditions (5.8L) showed the expression of pagC increased significantly under PhoPQ-activating conditions. (D) Resistance to serum-mediated attack was restored in single Salmonella mutants complemented with plasmids induced to express PagC, Rck, and OmpX in the corresponding mutants. Plasmids expressing Rck or PagC complemented best the triple mutant compared with mutants containing empty vector (pRS1). The data represent one of three separate, reproducible experiments, expressed as mean ± SEM. Statistical significance was calculated using one-way ANOVA multiple-comparison test and set at a P value of <0.05 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; and ns, P > 0.05).
To determine if PagC, Rck, and OmpX mediate resistance to complement attack in an additive manner, we compared the serum resistance activity of double and triple mutants with that of the corresponding single mutants (Fig. 3B). Surprisingly, the triple and double mutants were essentially as susceptible as the single mutants, indicating the absence of additive effects under the experimental conditions used. Some level of resistance remained with all the interrogated mutants, as well as a quadruple mutant that included pgtE, suggesting the presence of other potentially protective bacterial components.
Since the absence of additive effects could be related to the suboptimal expression of rck and ompX under PhoPQ-activating conditions, we compared the mRNA levels of pagC, rck, and ompX in Salmonella grown in LB and under 5.8L conditions. Notably, pagC expression alone was significantly upregulated under 5.8L conditions, with the expression of the other two genes being only slightly higher than that in LB (Fig. 3C). However, as the absence of one of the genes could potentially influence the expression of the others, we next used plasmids to individually induce PagC, Rck, or OmpX expression in the single and triple mutants (Fig. 3D). Complementation of the single mutants confirmed that each protein can individually interfere with serum killing effects. In the triple mutant, PagC or Rck expressed from plasmids could compensate for the absence of the other proteins to interfere with serum killing, albeit at a lower level than that in the single complemented mutants or wild-type strain. Thus, in contrast to the data obtained with noncomplemented mutants (Fig. 3B), complementation of the triple mutant with plasmids expressing just one of the proteins suggested potential additive effects of their activities (Fig. 3D). The OmpX-expressing plasmid did not complement the triple mutant, which is in agreement with the weaker effect of serum on the single ompX mutant. Taken together, the data indicated that the protective effect of PagC, Rck, and OmpX depends on their individual level of expression. As expression levels are most likely determined by environmental signals, our findings suggest that each of these proteins uniquely provide bacterial protection in nonoverlapping host compartments.
PhoPQ-induced OMVs interfere with complement-mediated killing of Salmonella in a dose-dependent manner.
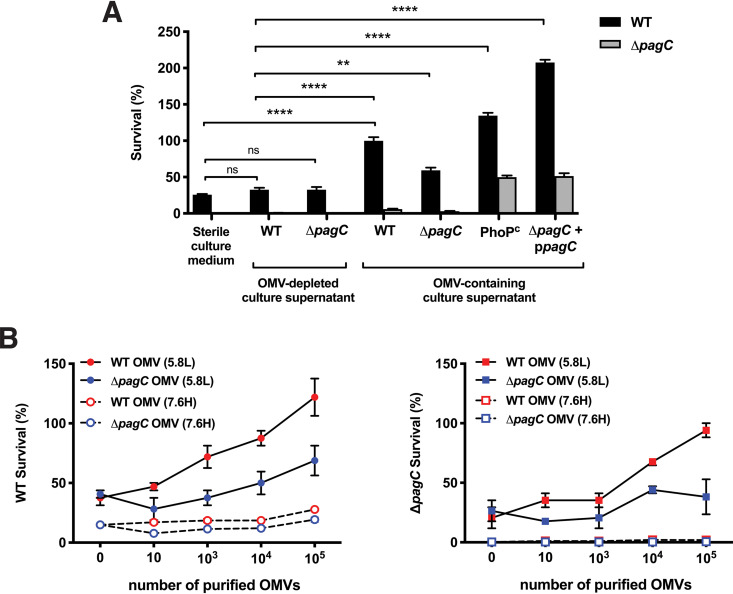
To determine if OMVs induced by PagC activation play a role in resistance to serum-mediated killing, OMV-free WT Salmonella and an isogenic ΔpagC mutant bacteria were resuspended in either sterile culture medium or OMV-containing spent culture supernatants from WT, ΔpagC, ΔphoP, PhoPC, or ΔpagC ppagC mutants before incubating with 25% NHS (Fig. 4A). Bacteria incubated without OMVs were susceptible to serum-mediated killing, whereas the bacteria incubated with OMV-containing culture supernatants from any Salmonella were significantly more resistant to the attack (P < 0.01). The OMV-free WT bacteria supplemented with OMV-containing culture supernatants from the PhoPC or ΔpagC ppagC strains, which overexpress pagC, were significantly more serum resistant than when supplemented with their own OMV-containing culture supernatant, while OMV-containing culture supernatants from the ΔpagC mutant protected significantly less efficiently. Comparable results were obtained with the isogenic ΔpagC mutant (Fig. 4A, gray bars), albeit each separate OMV addition protected significantly less efficiently than in the WT strain (Fig. 4A, black bars). To rule out the potential influence of a hypothetical component from the spent culture supernatants on serum-mediated attack, OMV-free WT bacteria were resuspended in OMV-containing or OMV-depleted spent culture supernatants, before incubating with 25% NHS. The bacteria were significantly better protected when resuspended in OMV-containing spent culture supernatants than in OMV-depleted spent culture supernatants (Fig. 4A). Moreover, bacterial survival was essentially the same when bacteria were resuspended in either sterile culture media or OMV-depleted spent culture supernatants, suggesting that bacterial protection against complement attack was essentially only due to the presence of OMVs.
FIG 4.
Serum protection assay by OMV produced under PhoPQ-activating conditions. (A) Survival of WT and ΔpagC Salmonella incubated with 25% NHS in the presence of sterile culture medium, OMV-depleted culture supernatants, or OMV-containing culture supernatants showed that bacteria incubated with OMV-containing culture supernatants from PagC-expressing bacteria (WT, ΔpagC + ppagC, and PhoPC) were significantly more resistant to serum-mediated attack than bacteria incubated with sterile culture medium or culture supernatant from the ΔpagC mutant. Bacterial survival in sterile culture medium was comparable to the survival in OMV-depleted spent culture supernatants. (B) Survival of WT and ΔpagC Salmonella incubated with 25% NHS in the presence of purified OMVs at indicated concentrations (0 to 105 OMVs), isolated from bacteria grown under PhoPQ-activating (5.8L; solid line) or nonactivating (7.6H; dashed line) conditions. OMVs from bacteria expressing PagC (WT) grown under PhoPQ-activating conditions were more protective than OMVs from bacteria grown under nonactivating conditions. The data represent one of three separate, reproducible experiments, expressed as mean ± SEM. Statistical significance was calculated using Student’s t test and set at a P value of <0.05 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; and ns, P > 0.05).
To bypass confounding effects due to variable OMV numbers in the different spent culture supernatants, we undertook further experiments using equal numbers of purified OMVs prepared from bacteria grown under conditions that do or do not activate PhoPQ (Fig. 4B). Only purified OMVs from bacteria produced under PhoPQ-activating conditions interfered with serum killing of WT bacteria in a dose-dependent manner, with the OMVs from the WT strain being significantly more protective than OMVs from the ΔpagC mutant. The ΔpagC mutant bacteria were protected in a dose-dependent manner only by OMVs from the WT strain produced under PhoPQ activation. Taken together, the data confirmed that under the PhoPQ-activating conditions, OMVs were most protective against serum when they were produced from Salmonella that express PagC.
PhoPQ-induced OMVs block the AP by mediating C3b degradation.
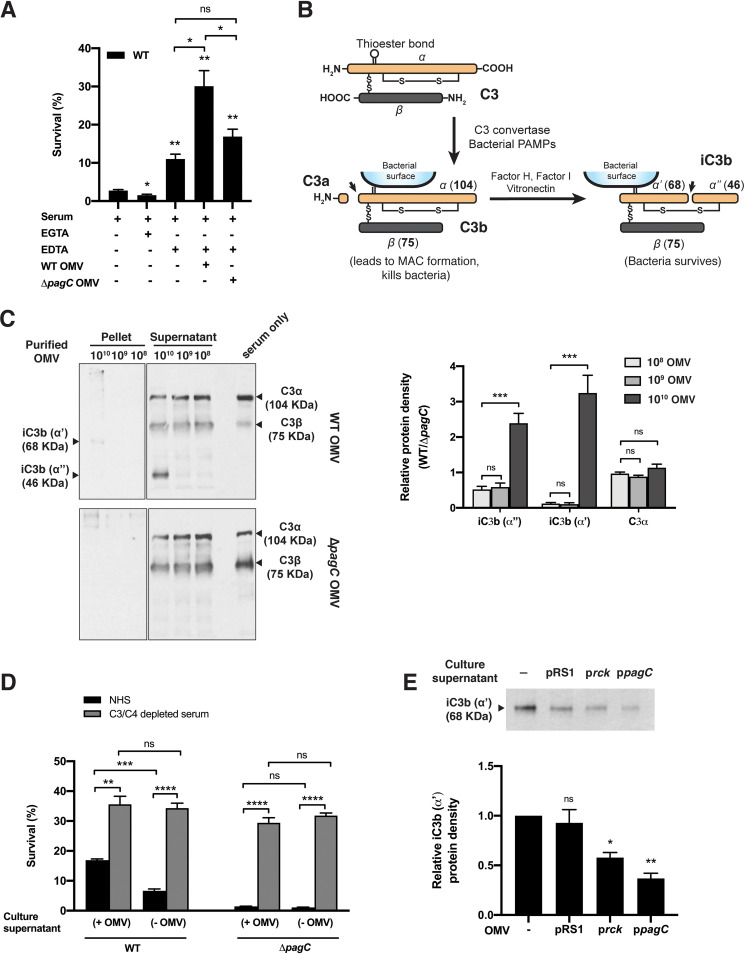
Rck blocks both the alternative pathway (AP) and classical pathway (CP) of the complement by binding to complement inhibitors (29, 31). To identify the complement pathways that are blocked by OMVs, OMV-free Salmonella were reconstituted with purified OMVs from the WT or ΔpagC mutant strains (1 × 109 OMVs) and incubated with human serum treated with EGTA (to block the CP and lectin pathways) or EDTA (to block all 3 complement pathways). As shown in Fig. 5A, WT bacteria without any OMVs were eight times more susceptible to killing when incubated with serum treated with EGTA (AP active) than to the bacteria incubated with EDTA-treated serum (AP blocked). The effects of the AP were also blocked by incubating the bacteria in the presence of OMVs, and the importance of pagC was highlighted by the relatively higher level of protection provided by OMVs from WT bacteria than OMVs from the ΔpagC mutant.
FIG 5.
Blocking of complement activation by OMVs from PagC-expressing bacteria. (A) Survival of WT Salmonella incubated with 25% NHS treated with 5 mM EGTA or EDTA, in the presence or absence of 1 × 109 purified OMVs from WT or ΔpagC mutant. OMVs from PagC-expressing WT bacteria block the alternative pathway of the complement more efficiently than the OMVs from the ΔpagC mutant. Statistical significance was calculated using Student’s t test. (B) Schematic representation of C3 and its cleavage products C3b and iC3b (molecular weight [MW] of the α and β chain and the cleaved or uncleaved products is indicated in the brackets). (C) WT OMVs (108 to 1010 OMVs) incubated with 5% NHS, when immunoblotted with anti-C3 antibody, showed bands for C3b inactivation at 68 kDa (iC3bα′) and 46 kDa (iC3bα″) in OMV pellet and supernatant fractions, respectively. No C3b inactivation bands were found when ΔpagC OMVs were incubated with NHS, suggesting that WT OMVs bind to C3b and inactivate C3b into iC3b in a dose-dependent manner. The Western blot image was spliced to remove an empty lane in between pellet and supernatant fractions. The panel on the right represents relative protein band densities (WT versus ΔpagC OMVs) as analyzed by NIH ImageJ software from 3 separate, reproducible experiments, expressed as mean ± SEM. Statistical significance was calculated using Student’s t test. (D) WT or ΔpagC mutant incubated with C3/C4-depleted human serum (gray bars) survived significantly better than the bacteria incubated with NHS (black bars). Presence of OMV-containing culture supernatant (+ OMV) contributed to bacterial survival when incubated with NHS, compared with bacteria incubated in the presence of OMV-depleted culture supernatants (− OMV). (E) Incubation of the ΔpagC Δrck ΔompX ΔpgtE mutant with NHS in the presence of OMV-containing culture supernatant prevented C3b deposition on bacterial cell surfaces. OMVs from the quadruple mutant expressing PagC prevented C3 deposition most significantly. The bar graph below the blot represents relative protein band densities as analyzed by NIH ImageJ software from 3 separate, reproducible experiments, expressed as mean ± SEM. For iC3bα′, relative protein densities were calculated for bacteria incubated in sterile culture medium (−) with no OMVs versus bacteria incubated with OMV-containing spent culture supernatant. Statistical significance was calculated using the one-way ANOVA multiple-comparison test. The data represent one of three separate, reproducible experiments, expressed as mean ± SEM (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; and ns, P > 0.05).
Since the C3 component of the AP is activated upon interaction with bacterial surface molecules such as LPS (38), we investigated whether OMVs also activate C3. For this investigation, purified WT or ΔpagC mutant OMVs were incubated with 5% NHS and analyzed by immunoblotting with anti-human C3 antibody as described in Materials and Methods. The presence of a 68-kDa (iC3bα′) band in the OMV pellet fraction and a 46-kDa (iC3bα″) band in the supernatant fraction of WT OMVs but not ΔpagC mutant OMVs (Fig. 5C) suggested that OMVs from PagC-expressing bacteria bind to and degrade complement component C3b into iC3bα′ and iC3bα″, the typical fragments of C3b inactivation (Fig. 5B) (39). A decrease in the strength of a band at 104 kDa (C3α) with an increasing number of purified WT OMVs showed that OMVs from PagC-expressing bacteria degrade the complement in a dose-dependent manner. Incubation of the Salmonella WT strain and the ΔpagC mutant with 25% NHS or C3/C4-depleted human serum in the presence of OMV-containing culture supernatant revealed that both strains survived significantly better when incubated with C3/C4-depleted serum and that this enhanced survival was independent of the presence of OMVs (Fig. 5D). As the level of bacterial survival in NHS was OMV dependent, these results further confirm that OMVs interfere with complement-mediated killing of bacteria in a C3-dependent manner. Incubation of the ΔpagC Δrck ΔompX ΔpgtE quadruple mutant with NHS in the presence of sterile culture medium showed a significantly larger amount of C3b (68 kDa) associated with bacterial pellets relative to the bacteria incubated in the presence of OMV-containing culture supernatant from the quadruple mutant induced to express PagC or Rck (Fig. 5E). Taken together, our data suggest that the interaction of C3b with OMVs from PagC-expressing bacteria syphons off and inactivates C3b from the serum, thereby interfering with the bactericidal properties of serum.
OMVs from PagC-expressing bacteria recruit complement inhibitor Factor H.
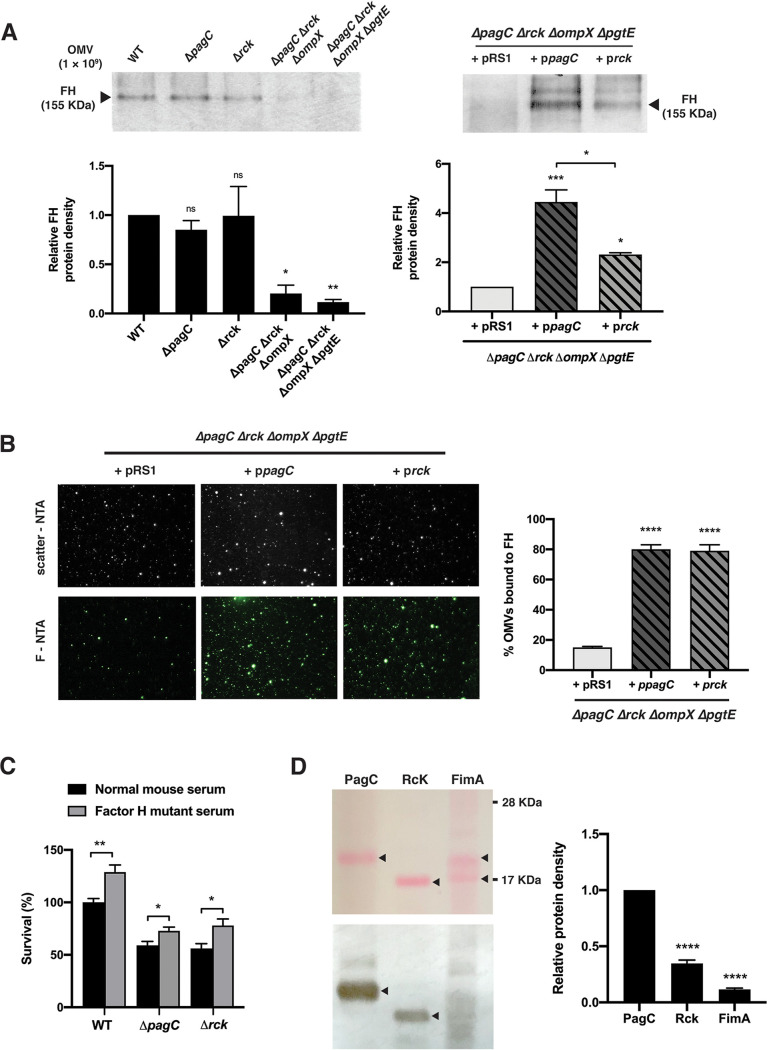
The ability of OMVs from PagC-expressing bacteria to degrade complement component C3b suggests that OMVs can recruit complement inhibitors such as Factor H (FH) or vitronectin. As shown in Fig. 6A, when purified OMVs of Salmonella strains were incubated with purified FH, a significant association was observed, with OMVs from the strains expressing PagC and/or Rck. OMVs from the quadruple ΔpagC Δrck ΔompX ΔpgtE mutant complemented to express PagC bound significantly more FH than OMVs from the same mutant induced to express Rck (Fig. 6A and B). Moreover, when purified OMVs from the quadruple mutant bacteria were incubated in solution with purified FH and analyzed by fluorescence-NTA (F-NTA), significantly fewer OMVs from empty vector-containing bacteria bound FH (15%) than OMVs from ppagC- or prck-carrying bacteria (80% and 79%, respectively) (Fig. 6B; see Fig. S4 to S9 in the supplemental material). These results indicated not only that PagC and Rck present surface-accessible domains on OMVs but also that these domains mediate FH binding to OMVs. In contrast, no significant binding of vitronectin to OMVs could be detected (data not shown). Incubation of WT or ΔpagC or Δrck mutant bacteria with normal mouse serum or FH mutant (W1206R) mouse serum (40) showed that PagC and Rck each contribute independently to bacterial survival in serum in an FH-dependent manner that is additive with the WT strain (Fig. 6C). Furthermore, a Western blot experiment with PagC (18 kDa), Rck (17 kDa), and FimA (18 kDa, type 1 fimbrial subunit used as control) recombinant proteins showed that FH bound to both PagC and Rck, but not FimA, confirming the specific recruitment of FH by the two former proteins (Fig. 6D).
FIG 6.
Factor H interaction with OMVs. (A) 1 × 109 Purified OMVs from WT, ΔpagC, Δrck, ΔpagC Δrck ΔompX, or ΔpagC Δrck ΔompX ΔpgtE mutant bacteria incubated with purified Factor H (FH; 2 μg) showed FH binding by OMVs from PagC- or Rck-expressing bacteria, as detected by Western blotting using anti-FH antibody. The bar graphs below each blot represent relative protein band density as analyzed by NIH ImageJ software from 3 separate, reproducible experiments, expressed as mean ± SEM. The relative protein density was calculated relative to WT bacteria (left) or the ΔpagC Δrck ΔompX ΔpgtE + pRS1 (empty vector)-containing mutant (right). (B) F-NTA analysis showed a significantly high percentage of OMVs from the PagC- or Rck-expressing quadruple mutant bound to FH compared with OMVs from the pRS1-containing quadruple mutant. The % OMVs bound to FH were calculated as the number of OMVs estimated by F-NTA versus the number of OMVs estimated by scatter-NTA × 100. (C) Incubation of WT, ΔpagC mutant, or Δrck mutant bacteria with normal mouse serum or FH mutant (W1206R) serum. Both PagC and Rck contribute to bacterial survival in serum in an FH-dependent manner. Statistical significance was calculated using Student’s t test (A and C). (D) Far-Western analysis of FH binding to recombinant proteins PagC, Rck, and FimA (1 μg each, indicated by arrowheads) showed purified FH (1 μg/ml) bound only to PagC and Rck, with the former showing greater affinity for FH. FimA being only partially reduced shows two bands in the blot stained by Ponceau S. Molecular weight standards are indicated on the right. The graph on the right represents relative protein densities as analyzed by the NIH ImageJ software, averaging 3 separate, reproducible experiments (mean ± SEM). The protein band densities for Rck and FimA were calculated relative to the PagC protein. Statistical significance was calculated using the one-way ANOVA multiple-comparison test (B and D). The data represent one of three separate, reproducible experiments, expressed as mean ± SEM (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; and ns, P > 0.05).
DISCUSSION
The complement system provides a formidable first line of immune attack against any invading pathogenic microbe. Therefore, not surprisingly, many pathogens have evolved various arsenals of defense mechanisms to protect themselves against soluble molecules of the complement system and its attack complex (41). In this study, we demonstrated that the OMP PagC allows S. Typhimurium to evade complement-mediated killing by two distinct mechanisms; it promotes the production of PagC-enriched OMVs that serve as complement traps and also inactivates C3b by recruiting the complement inhibitor Factor H.
In this study, we took advantage of corroborative NTA instruments to quantitatively measure bacterial OMV number and demonstrated that S. Typhimurium SL1344 upregulates OMV production upon PhoPQ activation. We also demonstrated with a single pagC deletion mutant and trans-complementation that PagC alone is a key inducer of OMV production (Fig. 1A). Unlike the PhoPQ-induced PagP and PagL enzymes that are thought to activate OMV production by modifying the acylation profile of LPS and membrane curvature (8, 9, 42–44), PagC, which is transcribed and produced in much larger amounts after PhoPQ activation (35, 45), has no known LPS-modifying properties and can induce Salmonella vesiculation by itself.
A previous report showed that PagC expression under PhoPQ activation induces OMV production in a ΔclpXP S. Typhimurium, a mutant that lacks the ATP protease ClpXP (17). Counterintuitively, and in contrast to our findings, the authors also demonstrated that a ΔclpXP ΔpagC deletion mutant did not affect vesiculation. However, ClpXP targets degradation of the alternative sigma factor RpoS, which coordinates the expression of nearly 23% of Salmonella small RNAs, with each regulating the expression of various proteins, including the major porin OmpD (17, 46–52). Thus, the authors proposed that the clpXP deletion altered OMV production in a manner that might have obscured the pagC deletion phenotype (17). However, this conclusion warrants further investigation, particularly since the authors also mentioned that OMV numbers were not reduced in a single pagC deletion mutant (17), without further discussion of this finding.
In 1994, Heffernan et al. cloned and expressed a pagC gene of S. Typhimurium in Escherichia coli HB101 and showed that it was not required for resistance to serum-mediated bacterial killing (33). In contrast, a later study with S. enterica serovar Choleraesuis by Nishio et al. demonstrated that PagC confers a high level of serum resistance when cloned into E. coli or Salmonella strains (34). The reason for this discrepancy remains unclear since both studies followed essentially the same protocol using S. Typhimurium or S. Choleraesuis pagC genes that encode the same allelic protein. In the present study, we confirmed the observation by Nishio et al., and additionally, we established that PagC interferes with the lethal effects of the human complement toward Salmonella in two synergistic ways.
The ability of PhoPQ-activated PagC to promote Salmonella resistance to complement attack is directly linked, in part, to its ability to induce production of OMVs, which protect bacteria in a dose-dependent manner. We hypothesized that OMVs from PagC-expressing bacteria act as decoys and prevent complement deposition and activation on the bacterial cell surface. In support, we showed that the incubation of OMVs from PagC-expressing bacteria with OMV-free Salmonella significantly reduced C3 deposition on the bacterial cell surface. We also showed that bacterial survival was significantly higher when they were incubated with C3/C4-depleted serum, confirming that bacterial killing by NHS was complement dependent. In addition, PagC itself directly interfered with the alternative complement pathway. We demonstrated that OMVs from PagC-expressing bacteria bind to soluble complement component C3b and recruit complement inhibitor Factor H to inactivate C3b into iC3b (Fig. 5C and 6A, respectively), thus preventing C3 convertase formation. This mechanism interferes with the opsonization of Salmonella and blocks the amplification of the complement pathway that would lead to the formation of the membrane attack complex responsible for bacterial lysis (41).
Interestingly, even though our experiments confirmed that Rck, an OMP homologous to PagC, inhibits complement activity (28, 29, 31), unlike PagC, Rck had no effect on Salmonella vesiculation. Similarly, Salmonella OmpX and Y. pestis Ail proteins, additional PagC-like proteins investigated here, did not affect OMV production, and the OmpX was significantly less protective than PagC or Rck against the bactericidal property of serum. Other bacteria, such as Vibrio cholerae, make OMVs with OMPs that can trap and inactivate complement factors, but these OMPs do not activate OMV production (53, 54). Since PagC seems uniquely able to promote OMV production, it will be important to determine the mechanistic basis for this effect. Given the role of LPS in OMV formation (8) and the reported structural interactions of LPS and the PagC-similar Ail OMP (18), it will be particularly informative to identify the residues of PagC that interact with LPS and determine their potential impact on OMV formation.
Despite the strong inhibitory effects of PagC on serum-mediated killing, OMVs from the ΔpagC mutant still offered some protection, suggesting the potential involvement of alternative protective mechanisms. Additional OMPs, such as Rck, OmpX, PgtE, and TraT, can contribute to Salmonella resistance to the host complement (36, 37, 55). However, the amounts of Rck, OmpX, PgtE, and TraT were comparable or slightly increased in purified OMVs from the ΔpagC mutant versus wild type OMVs, and OMVs from the ΔpagC mutant were only weakly protective. While it remains possible that the reduced abundance of other OMPs, such as PagN, in the ΔpagC mutant (Table S2) contributed to the reduced protective property of the OMVs of this mutant, the pagN mutation was not associated with reduced OMV production (Fig. 1A) or bacterial susceptibility (Fig. 3A). These results suggested that PagC is a key contributor of interference with serum-mediating killing for PhoPQ-activated Salmonella. We confirmed a PgtE effect on serum killing and OMV production but did not investigate TraT, as like Rck, TraT is plasmid encoded and thus absent in strains and serovars that lack this plasmid. Whether TraT alone or yet another surface molecule was responsible for the detected residual survival of bacteria in serum remains to be determined. In addition, S. Typhimurium PhoPQ indirectly activates enzymes that modulate the chain length and complexity of the LPS O-antigen (56, 57), sterically hindering formation of the C9-membrane attack complex on the bacterial surface (58), which may also contribute to the residual protection.
The number of Salmonella surface molecules involved in complement resistance suggests a strong evolutionary selection for protection against this major extracellular pathogen-killing mechanism in host tissues, particularly following inflammation with vascular leakage of serum. Following Salmonella-induced cell death, the bacteria leave the intracellular compartment, either directly from the vacuole or by transiting first through the cytoplasmic compartment. Even though the presence of Salmonella in the extracellular environment might be brief compared with its intracellular residence in hosts, Salmonella remains highly susceptible to complement attack in such an environment. Thus, we propose that the production of protective molecules differentially induced by various regulons ensures that Salmonella is preconditioned for serum resistance in all relevant host compartments, whether Salmonella cycles between host phagocytic cells or through intestinal epithelial cells (59–61). Indeed, our results confirmed that PagC is induced by signals specific to intravacuolar conditions, whereas Rck is upregulated in response to homoserine lactone (62), a quorum sensing signal typically ascribed to the induction of biofilm formation. Thus, Rck likely protects Salmonella where it can multiply to high local density, such as an extravasate-covered intestinal lumen surface. In contrast, PagC induction of OMVs following PhoPQ activation is suggested to provide a cover of protection for the bacteria when they cycle through host cells after being released with their OMVs from dying cells. Specifically, as we demonstrated that Factor H binds PagC, we propose that OMVs released from dying cells bind to and deactivate C3b by recruiting Factor H, thereby promoting further infection by permitting extracellular bacteria to thwart complement-mediated death. The intravacuolar induction of OMV production to prepare Salmonella for the next environment is reminiscent of an evolutionary mechanism of bacterial survival that has been described in the literature as an anticipatory strategy (60, 61).
Few studies have yet investigated the potential role of PagC in bacteria serum resistance in vivo. In one study, a Salmonella pagC-TnphoA mutant that was administered orally or intraperitoneally (i.p.) to BALB/c mice revealed a reduced 50% lethal concentration (LD50) compared with that of the parental wild-type strain (63). However, this mutant was later shown to be attenuated due to the production of a fusion protein, leaving the original question unresolved (64). A second study using competition experiments with wild-type and ΔpagC mutant Salmonella injected simultaneously into mice did not demonstrate any significant effect (65). However, it is likely that the wild-type strain with its large number of OMVs may have masked any potential ΔpagC mutant phenotype linked to reduced vesiculation.
The important effects of PagC on OMV production and complement evasion clearly warrant further noncompetitive in vivo studies for determining its impact on pathogenesis. Notably, Salmonella-infected patients generate antibodies to PagC (66), which would also be expected to be captured by OMVs. Thus, potential therapeutic PagC inhibitors designed to block OMV production could likely compromise Salmonella traps of both host innate and adaptive immune molecules, further highlighting their utility in reducing the pathogenicity of these enteric and systemic bacteria.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All bacterial strains, including mutants and plasmids used in this study are listed in Table S3, along with references 77–82. Unless stated otherwise, all the reagents were procured from MilliporeSigma (St. Louis, MO, USA). The Salmonella strains were grown at 37°C in Luria-Bertani (LB) broth or N-minimal medium [5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, and 0.1% Casamino Acids] with 0.4% glucose and supplemented with 10 μM MgSO4 (buffered with 0.1 M morpholineethanesulfonic acid [MES; pH 5.8]; for PhoPQ-activating, 5.8L conditions) or 10 mM MgSO4 (buffered with 0.1 M morpholinepropanesulfonic acid [MOPS; pH 7.6]; for 7.6H conditions), (7). To grow Salmonella under PhoPQ-activating conditions, bacteria were grown in LB and then sequentially transitioned into 7.6H and 5.8L conditions. A S. Typhimurium SL1344 defective in the production of flagellins (fliC and fljB) was prepared by generalized transduction with phage P22, and this strain (designated wild type [WT]) was used as the parental background strain to engineer all Salmonella deletion mutants. Deletion of fliC and fljB was done to avoid the interference of flagella with OMV purification and analysis. The Salmonella deletion mutants were prepared using Gibson assembly and allelic exchange methods, essentially as described previously (67, 68). The mutants were analyzed for growth defects by growing the bacteria in LB and N-minimal media. No significant difference in growth was found between the wild type or any deletion mutant when adapted to a corresponding medium condition. The primers used in this study are listed in Table S3. To constitutively express PhoPQ-activated genes (PhoPC) in S. Typhimurium SL1344, a phoP- and phoQ-containing amplicon flanking 1,000 bp upstream and downstream of the phoP gene was amplified from a ATCC14028s pho-24 S. Typhimurium mutant strain containing the PhoQ T48I mutation (69) and cloned into a SL1344 ΔphoP mutant by Gibson assembly and allelic exchange methods. For trans-complementation of protein expression, the genes were amplified from S. enterica serovar Typhimurium SL1344 genomic DNA and cloned in a pRS1 plasmid using the Gibson assembly master mix (New England BioLabs Inc., Ipswich, MA, USA). The amplicons were cloned into plasmid pGM81ΔenvZ using Gibson assembly and induced for expression with anhydrotetracycline hydrochloride (AHT; 0.4 μg/ml) for 2 h at 37°C, starting with log-phase cultures (A600 = 0.3). When appropriate, antibiotics were used at the following concentrations: ampicillin at 200 μg/ml, kanamycin at 25 μg/ml, chloramphenicol at 30 μg/ml, and streptomycin at 90 μg/ml.
List of bacterial strains, plasmids, and primers. Download Table S3, DOCX file, 0.03 MB (33.9KB, docx) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
OMV purification.
Salmonella cultures grown overnight in 5.8L or 7.6H N-minimal media were centrifuged with a Beckman Avanti J-E centrifuge at 15,000 × g (JLA-16.250 rotor) for 10 min at 4°C to obtain bacterial cell pellets and OMV-containing culture supernatants. The culture supernatant was centrifuged again at 15,000 × g (JLA-16.250 rotor) for 10 min at 4°C and filtered through 0.45-μm cellulose membrane filters to remove any bacterial cell and cellular debris contamination (70–72). Filtered supernatants were then subjected to ultracentrifugation in a Beckman optima L-90K ultracentrifuge at 300,000 × g (Type 50.2 Ti rotor) for 90 min at 4°C to pellet OMVs. The obtained OMV pellets were resuspended in sterile phosphate-buffered saline (PBS), filtered through a 0.2-μm filter, and checked for bacterial contamination by plating onto LB agar before storing at −20°C for future use.
OMV characterization.
The concentration and size (diameters) of OMVs, normalized by CFU/ml, were quantified by transmission electron microscopy (TEM) and by nanoparticle tracking analysis (NTA) using NanoSight NS300 (Malvern Panalytical, Malvern, UK) or ZetaView (Particle Matrix, Meerbusch, Germany) instruments at the Extracellular Vesicle Core, University of Pennsylvania. For TEM, purified OMVs were placed on 300-mesh copper grids (Electron Microscopy Sciences Hatfield, PA, USA), stained with 1% uranyl acetate for 30 sec, and viewed using FEI Tecnai T12 electron microscope at the EMRL facility of the University of Pennsylvania. The diameter of OMVs observed by TEM analysis was calculated by using the measurement function in the NIH ImageJ v. 1.53 software (http://rsbweb.nih.gov/ij/index.html) for 45 fields per strain at a magnification of 1:100.000. Differences in OMV sizes measured by TEM and NTA can be attributed to overestimations of vesicle size by NTA devices (73).
Proteomic profiling of OMPs.
Identification of OMPs present in the OMVs of the WT strain and ΔpagC mutant was determined by mass spectrometry-based analysis using a Thermo Scientific Orbitrap Fusion instrument, paired to a Thermo Scientific Ultimate 3000 ultra-high-performance liquid chromatography (UHPLC), at the Quantitative Proteomics Research Core facility, University of Pennsylvania. The collected tandem mass spectrometry (MS/MS) spectra were analyzed against the UniProt database of S. enterica serovar Typhimurium SL1344 and compared to OMV-associated OMPs (acidic conditions) from ATCC14028s and LT2 OMVs (35). The abundance of the identified OMPs in the OMVs of the ΔpagC mutant and WT Salmonella strain were each normalized to the abundance of OmpA in the respective strains to calculate a relative abundance for each OMP (ΔpagC versus WT, Table S2).
Serum resistance assays.
For analyzing the sensitivity of Salmonella to serum-mediated killing, bacteria (∼1 × 105 CFU grown in 5.8L N-minimal media (optical density at 600 nm [OD600], 0.5) were incubated with 25% normal human serum (NHS) (Astarte Biologics, Bothell, WA, USA), C3/C4-depleted human serum (Complement Technology Inc., TX, USA), or with normal mouse serum and Factor H mutant serum (40) for 1 hour at 37°C. The number of viable bacteria after incubation was calculated by plating serial dilutions on LB agar. To analyze the role of OMVs in resistance to complement-mediated killing, OMV-free bacteria were obtained by washing the bacteria (grown to an OD600 of 0.5) three times with sterile PBS to remove OMVs. Spent culture supernatant from bacteria grown under 5.8L conditions (OD600, 0.5) was filtered through a 0.2-μ filter to obtain OMV-containing spent culture supernatants or ultracentrifuged to remove OMVs (OMV-depleted spent culture supernatants). The OMV-free bacteria (∼1 × 105 CFU) were resuspended in either sterile culture medium, OMV-depleted, or OMV-containing spent culture supernatants in an equivalent volume ratio. Some experiments were done with OMV-free bacteria incubated with purified OMVs resuspended at specific concentrations in PBS. The resuspended bacteria were then incubated with 25% NHS at 37°C, and the number of viable bacteria after 1 h of incubation was calculated by plating onto LB agar plates. To block CP and LP of the complement, NHS was supplemented with 5 mM EGTA and 10 mM MgCl2 or with 5 mM EDTA to block all 3 pathways (CP, LP, and AP).
Quantitative RT-PCR Assay.
Quantitative reverse transcription-PCR (RT-PCR) was performed as described previously (74). Briefly, total mRNA from the bacterial strains grown in LB or 5.8L N-minimal media (approximately OD600 of 0.5) was extracted using the TRIzol reagent (Gibco BRL, Waltham, MA, USA) according to the manufacturer’s protocol. Residual genomic DNA contamination was removed by treatment with DNase, Turbo DNA-free kit (Ambion, Austin, TX, USA), and the RNA was checked for purity by agarose gel electrophoresis. An absence of DNA contamination was confirmed by PCR without reverse transcription, and RNA concentration was determined with the ND1000 spectrophotometer (NanoDrop products, Wilmington, DE, USA). Four hundred nanograms of RNA was used in reverse transcription reactions using random hexamer primers and a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. Quantitative real-time PCR was carried out using cDNA corresponding to 20 ng RNA, gene-specific primers (Table S3), and the PowerUP SYBR green PCR master mix (Applied Biosystems) with the ABI Fast 7500 real-time PCR system (Applied Biosystems). Expression levels were normalized to the transcription levels for the housekeeping gene rpoB, and relative copy numbers were calculated according to the threshold cycle (2−ΔΔCT) method.
Binding of OMVs to complement components.
OMVs (108 to 1010 OMVs) isolated from bacteria grown in 5.8L N-minimal media were incubated with 5% NHS or 2 μg of purified human Factor H protein (Complement Technology Inc., TX, USA) for 1 hour at 37°C. The mixture was then centrifuged at 200,000 × g (Type 42.2 Ti rotor) for 15 min at 4°C to collect the OMV pellet and the supernatant. The supernatant (containing unbound complement components) and the OMV pellet (containing OMV-bound complement components) were washed twice with sterile PBS and analyzed by Western blotting using horseradish peroxidase (HRP)-conjugated anti-human C3 antibody (1:5,000) (MP Biomedicals, Irvine, CA, USA) or anti-human Factor H antibody (1:500) (Complement Technology Inc., TX, USA). Donkey anti-goat HRP-conjugated antibody at a 1:10,000 dilution was used as the secondary antibody (Jackson ImmunoResearch, PA, USA) followed by probing the blots with ECL substrate (GE Healthcare Life Sciences, MA, USA). Alternatively, interference of C3 deposition on the bacterial cell surface by OMVs was detected by incubating OMV-free ΔpagC Δrck ΔompX ΔpgtE mutant bacteria resuspended in either sterile culture medium or OMV-containing culture supernatants of the ΔpagC ΔrckΔ ompX ΔpgtE mutant carrying plasmids induced to express Rck or PagC with 25% NHS for 1 hour at 37°C. The bacteria were then pelleted and washed twice with PBS to remove OMVs and unbound complement. Complement deposited on bacterial cell surfaces was detected by Western blot analysis of cell lysates using the HRP-conjugated anti-human C3 antibody. Binding of vitronectin to OMV-free WT Salmonella (1 × 107 CFU in the presence of PBS or purified WT or ΔpagC OMVs [1 × 109]) was analyzed by incubating the mixture with 25% NHS for 1 hour at 37°C. The mixture was then centrifuged to collect supernatant, bacterial pellet (15,000 × g, 10 min, 4°C), and OMV pellet (200,000 × g, 15 min, 4°C). Vitronectin binding to the bacteria and OMVs was investigated by Western blotting using anti-human vitronectin (1:500; Complement Technology Inc.) and HRP-conjugated secondary antibody as described above. The images generated from Western blotting were subjected to densitometric analysis by NIH ImageJ software.
Far-Western and Immunolabelling.
A total of 1 μg of recombinant Salmonella PagC, Rck, and FimA proteins (75) were subjected to 15% SDS-PAGE. Proteins were electrotransferred onto a nitrocellulose membrane and blocked with 3% bovine serum albumin (BSA). Subsequently, the membrane was incubated with purified human FH (1 μg/ml, in 0.1% BSA), which is below the physiological concentration of FH in serum (500 μg/ml) (76). Binding of the protein was detected by immunoblotting with goat anti-human FH and donkey anti-goat HRP-conjugated secondary antibody as described above. For immunolabelling the OMVs, purified OMVs from the quadruple mutant induced to express Rck or PagC from corresponding plasmids were incubated with 2 μg of purified human FH, in a final volume of 200 μl. The OMVs were then washed with sterile PBS at 200,000 × g (Type 42.2 Ti rotor) for 15 min at 4°C followed by incubation with anti-human Factor H antibody (1:1,000 dilution) and a 1:2,000 diluted donkey anti-goat Alexa Fluor-488 antibody (Abcam, Cambridge, UK) as the labeled secondary antibody. The fluorescently labeled OMVs were then analyzed by fluorescence NTA (F-NTA) using Zetaview.
Statistics.
One-way analysis of variance (ANOVA) multiple comparisons or Student’s t test were used for statistical calculations using Prism v. 8 (GraphPad Software, San Diego, CA, USA). Statistical significance was set at a P value of <0.05 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001).
Scatter-NTA of OMVs from quadruple mutant (ΔpagC Δrck ΔompX ΔpgtE) containing empty vector (pRS1). Download Movie S1, MOV file, 6.1 MB (6.3MB, mov) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Scatter-NTA of OMVs from quadruple mutant expressing PagC (ppagC). Download Movie S2, MOV file, 6.1 MB (6.2MB, mov) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Scatter-NTA of OMVs from quadruple mutant expressing Rck (prck). Download Movie S3, MOV file, 6 MB (6.1MB, mov) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fluorescence-NTA (F-NTA) showing binding of FH to OMVs from quadruple mutant containing empty vector (pRS1). Download Movie S4, MOV file, 2.1 MB (2.2MB, mov) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
F-NTA showing binding of FH to OMVs from quadruple mutant expressing PagC (ppagC). Download Movie S5, MOV file, 6.2 MB (6.3MB, mov) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
F-NTA showing binding of FH to OMVs from quadruple mutant expressing Rck (prck). Download Movie S6, MOV file, 6.1 MB (6.2MB, mov) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
This work was supported by NIH/NIAID grant AI139982 to D.M.S. We thank Igor Brodsky for the Salmonella LT2 fliC::Cmr strain; Beatrice Claudi, Mark Goulian, and Eric Krukonis for plasmids; Tomoko Yamamoto for the PagC antibodies; Wenchao Song and Takashi Miwa for the Factor H mutant serum; John D. Lambris for anti-human C3 antibodies; Fevzi Daldal for technical support; Meta Kuehn and Mark Goulian for technical discussions; Leslie King for proofreading the manuscript; and University of Pennsylvania QPRC core facility for proteomics profiling.
The opinions expressed in the manuscript are solely the responsibility of the authors and do not necessarily represent the official views and policy of the National Institutes of Health.
We declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Citation Dehinwal R, Cooley D, Rakov AV, Alugupalli AS, Harmon J, Cunrath O, Vallabhajosyula P, Bumann D, Schifferli DM. 2021. Increased production of outer membrane vesicles by Salmonella interferes with complement-mediated innate immune attack. mBio 12:e00869-21. https://doi.org/10.1128/mBio.00869-21.
Contributor Information
Dieter M. Schifferli, Email: dmschiff@vet.upenn.edu.
Samuel I. Miller, University of Washington
REFERENCES
- 1.Geddes K, Cruz F, Heffron F. 2007. Analysis of cells targeted by Salmonella type III secretion in vivo. PLoS Pathog 3:e196. doi: 10.1371/journal.ppat.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richter-Dahlfors A, Buchan AM, Finlay BB. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella Typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J Exp Med 186:569–580. doi: 10.1084/jem.186.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groisman EA. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol 183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia VE, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 5.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 6.Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit RE, Miller SI. 2007. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell 26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Bonnington KE, Kuehn MJ. 2016. Outer membrane vesicle production facilitates LPS remodeling and outer membrane maintenance in Salmonella during environmental transitions. mBio 7:e01532-16. doi: 10.1128/mBio.01532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnington KE, Kuehn MJ. 2017. Breaking the bilayer: OMV formation during environmental transitions. Microb Cell 4:64–66. doi: 10.15698/mic2017.02.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elhenawy W, Bording-Jorgensen M, Valguarnera E, Haurat MF, Wine E, Feldman MF. 2016. LPS remodeling triggers formation of outer membrane vesicles in Salmonella. mBio 7:e00940-16. doi: 10.1128/mBio.00940-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jan AT. 2017. Outer membrane vesicles (OMVs) of Gram-negative bacteria: a perspective update. Front Microbiol 8:1053. doi: 10.3389/fmicb.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulkarni HM, Jagannadham MV. 2014. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology 160:2109–2121. doi: 10.1099/mic.0.079400-0. [DOI] [PubMed] [Google Scholar]
- 13.Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haurat MF, Elhenawy W, Feldman MF. 2015. Prokaryotic membrane vesicles: new insights on biogenesis and biological roles. Biol Chem 396:95–109. doi: 10.1515/hsz-2014-0183. [DOI] [PubMed] [Google Scholar]
- 15.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pathirana RD, Kaparakis-Liaskos M. 2016. Bacterial membrane vesicles: biogenesis, immune regulation and pathogenesis. Cell Microbiol 18:1518–1524. doi: 10.1111/cmi.12658. [DOI] [PubMed] [Google Scholar]
- 17.Kitagawa R, Takaya A, Ohya M, Mizunoe Y, Takade A, Yoshida S, Isogai E, Yamamoto T. 2010. Biogenesis of Salmonella enterica serovar Typhimurium membrane vesicles provoked by induction of PagC. J Bacteriol 192:5645–5656. doi: 10.1128/JB.00590-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh C, Lee H, Tian Y, Schesser Bartra S, Hower S, Fujimoto LM, Yao Y, Ivanov SA, Shaikhutdinova RZ, Anisimov AP, Plano GV, Im W, Marassi FM. 2020. Mutually constructive roles of Ail and LPS in Yersinia pestis serum survival. Mol Microbiol 114:510–520. doi: 10.1111/mmi.14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutta SK, Yao Y, Marassi FM. 2017. Structural insights into the Yersinia pestis outer membrane protein Ail in lipid bilayers. J Phys Chem B 121:7561–7570. doi: 10.1021/acs.jpcb.7b03941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heffernan EJ, Harwood J, Fierer J, Guiney D. 1992. The Salmonella typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J Bacteriol 174:84–91. doi: 10.1128/JB.174.1.84-91.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosselin M, Virlogeux-Payant I, Roy C, Bottreau E, Sizaret PY, Mijouin L, Germon P, Caron E, Velge P, Wiedemann A. 2010. Rck of Salmonella enterica, subspecies enterica serovar Enteritidis, mediates zipper-like internalization. Cell Res 20:647–664. doi: 10.1038/cr.2010.45. [DOI] [PubMed] [Google Scholar]
- 22.Pulkkinen WS, Miller SI. 1991. A Salmonella Typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J Bacteriol 173:86–93. doi: 10.1128/JB.173.1.86-93.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartra SS, Styer KL, O'Bryant DM, Nilles ML, Hinnebusch BJ, Aballay A, Plano GV. 2008. Resistance of Yersinia pestis to complement-dependent killing is mediated by the Ail outer membrane protein. Infect Immun 76:612–622. doi: 10.1128/IAI.01125-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mecsas J, Welch R, Erickson JW, Gross CA. 1995. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J Bacteriol 177:799–804. doi: 10.1128/JB.177.3.799-804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, Huang Q, Cui A, Liu X, Hou B, Zhang L, Liu M, Meng X, Li S. 2018. Overexpression of outer membrane protein X (OmpX) compensates for the effect of TolC inactivation on biofilm formation and curli production in extraintestinal pathogenic Escherichia coli (ExPEC). Front Cell Infect Microbiol 8:208. doi: 10.3389/fcimb.2018.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vica PS, Garcia GO, Paniagua CG. 1997. The lom gene of bacteriophage lambda is involved in Escherichia coli K12 adhesion to human buccal epithelial cells. FEMS Microbiol Lett 156:129–132. [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Li L, Pan F, Zuo G, Yu D, Liu R, Fan H, Ma Z. 2020. PagC is involved in salmonella pullorum OMVs production and affects biofilm production. Vet Microbiol 247:108778. doi: 10.1016/j.vetmic.2020.108778. [DOI] [PubMed] [Google Scholar]
- 28.Heffernan EJ, Reed S, Hackett J, Fierer J, Roudier C, Guiney D. 1992. Mechanism of resistance to complement-mediated killing of bacteria encoded by the Salmonella Typhimurium virulence plasmid gene rck. J Clin Invest 90:953–964. doi: 10.1172/JCI115972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho DK, Jarva H, Meri S. 2010. Human complement factor H binds to outer membrane protein Rck of Salmonella. J Immunol 185:1763–1769. doi: 10.4049/jimmunol.1001244. [DOI] [PubMed] [Google Scholar]
- 30.Ho DK, Skurnik M, Blom AM, Meri S. 2014. Yersinia pestis Ail recruitment of C4b-binding protein leads to factor I-mediated inactivation of covalently and noncovalently bound C4b. Eur J Immunol 44:742–751. doi: 10.1002/eji.201343552. [DOI] [PubMed] [Google Scholar]
- 31.Ho DK, Tissari J, Jarvinen HM, Blom AM, Meri S, Jarva H. 2011. Functional recruitment of human complement inhibitor C4B-binding protein to outer membrane protein Rck of Salmonella. PLoS One 6:e27546. doi: 10.1371/journal.pone.0027546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartra SS, Ding Y, Miya Fujimoto L, Ring JG, Jain V, Ram S, Marassi FM, Plano GV. 2015. Yersinia pestis uses the Ail outer membrane protein to recruit vitronectin. Microbiology 161:2174–2183. doi: 10.1099/mic.0.000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heffernan EJ, Wu L, Louie J, Okamoto S, Fierer J, Guiney DG. 1994. Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella typhimurium and ail from Yersinia enterocolitica. Infect Immun 62:5183–5186. doi: 10.1128/IAI.62.11.5183-5186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishio M, Okada N, Miki T, Haneda T, Danbara H. 2005. Identification of the outer-membrane protein PagC required for the serum resistance phenotype in Salmonella enterica serovar Choleraesuis. Microbiology 151:863–873. doi: 10.1099/mic.0.27654-0. [DOI] [PubMed] [Google Scholar]
- 35.Ramu P, Tanskanen R, Holmberg M, Lahteenmaki K, Korhonen TK, Meri S. 2007. The surface protease PgtE of Salmonella enterica affects complement activity by proteolytically cleaving C3b, C4b and C5. FEBS Lett 581:1716–1720. doi: 10.1016/j.febslet.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 36.Riva R, Korhonen TK, Meri S. 2015. The outer membrane protease PgtE of Salmonella enterica interferes with the alternative complement pathway by cleaving factors B and H. Front Microbiol 6:63. doi: 10.3389/fmicb.2015.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ricklin D, Hajishengallis G, Yang K, Lambris JD. 2010. Complement: a key system for immune surveillance and homeostasis. Nat Immunol 11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parente R, Clark SJ, Inforzato A, Day AJ. 2017. Complement factor H in host defense and immune evasion. Cell Mol Life Sci 74:1605–1624. doi: 10.1007/s00018-016-2418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueda Y, Mohammed I, Song D, Gullipalli D, Zhou L, Sato S, Wang Y, Gupta S, Cheng Z, Wang H, Bao J, Mao Y, Brass L, Zheng XL, Miwa T, Palmer M, Dunaief J, Song WC. 2017. Murine systemic thrombophilia and hemolytic uremic syndrome from a factor H point mutation. Blood 129:1184–1196. doi: 10.1182/blood-2016-07-728253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambris JD, Ricklin D, Geisbrecht BV. 2008. Complement evasion by human pathogens. Nat Rev Microbiol 6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. 2000. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J 19:5071–5080. doi: 10.1093/emboj/19.19.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trent MS, Pabich W, Raetz CR, Miller SI. 2001. A PhoP/PhoQ-induced Lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella Typhimurium. J Biol Chem 276:9083–9092. doi: 10.1074/jbc.M010730200. [DOI] [PubMed] [Google Scholar]
- 43.Kawasaki K, China K, Nishijima M. 2007. Release of the lipopolysaccharide deacylase PagL from latency compensates for a lack of lipopolysaccharide aminoarabinose modification-dependent resistance to the antimicrobial peptide polymyxin B in Salmonella enterica. J Bacteriol 189:4911–4919. doi: 10.1128/JB.00451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SY, Groisman EA. 2014. Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system. Mol Microbiol 91:135–144. doi: 10.1111/mmi.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai J, Kim SI, Ryu S, Yoon H. 2014. Identification and characterization of outer membrane vesicle-associated proteins in Salmonella enterica serovar Typhimurium. Infect Immun 82:4001–4010. doi: 10.1128/IAI.01416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweder T, Lee KH, Lomovskaya O, Matin A. 1996. Regulation of Escherichia coli starvation sigma factor (sigma s) by ClpXP protease. J Bacteriol 178:470–476. doi: 10.1128/JB.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gottesman S. 2003. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol 19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- 48.Maurizi MR. 1992. Proteases and protein degradation in Escherichia coli. Experientia 48:178–201. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- 49.Tu X, Latifi T, Bougdour A, Gottesman S, Groisman EA. 2006. The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc Natl Acad Sci U S A 103:13503–13508. doi: 10.1073/pnas.0606026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein G, Raina S. 2017. Small regulatory bacterial RNAs regulating the envelope stress response. Biochem Soc Trans 45:417–425. doi: 10.1042/BST20160367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colgan AM, Kroger C, Diard M, Hardt WD, Puente JL, Sivasankaran SK, Hokamp K, Hinton JC. 2016. The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet 12:e1006258. doi: 10.1371/journal.pgen.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto T, Sashinami H, Takaya A, Tomoyasu T, Matsui H, Kikuchi Y, Hanawa T, Kamiya S, Nakane A. 2001. Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect Immun 69:3164–3174. doi: 10.1128/IAI.69.5.3164-3174.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aung KM, Sjostrom AE, von Pawel-Rammingen U, Riesbeck K, Uhlin BE, Wai SN. 2016. Naturally occurring IgG antibodies provide innate protection against Vibrio cholerae bacteremia by recognition of the outer membrane protein U. J Innate Immun 8:269–283. doi: 10.1159/000443646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan TT, Morgelin M, Forsgren A, Riesbeck K. 2007. Haemophilus influenzae survival during complement-mediated attacks is promoted by Moraxella catarrhalis outer membrane vesicles. J Infect Dis 195:1661–1670. doi: 10.1086/517611. [DOI] [PubMed] [Google Scholar]
- 55.Pramoonjago P, Kaneko M, Kinoshita T, Ohtsubo E, Takeda J, Hong KS, Inagi R, Inoue K. 1992. Role of TraT protein, an anticomplementary protein produced in Escherichia coli by R100 factor, in serum resistance. J Immunol 148:827–836. [PubMed] [Google Scholar]
- 56.Grossman N, Leive L. 1984. Complement activation via the alternative pathway by purified Salmonella lipopolysaccharide is affected by its structure but not its O-antigen length. J Immunol 132:376–385. [PubMed] [Google Scholar]
- 57.Murray GL, Attridge SR, Morona R. 2006. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J Bacteriol 188:2735–2739. doi: 10.1128/JB.188.7.2735-2739.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grossman N, Schmetz MA, Foulds J, Klima EN, Jimenez-Lucho VE, Leive LL, Joiner KA, Jiminez V. 1987. Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J Bacteriol 169:856–863. doi: 10.1128/JB.169.2.856-863.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geiser P, Di Martino ML, Samperio Ventayol P, Eriksson J, Sima E, Al-Saffar AK, Ahl D, Phillipson M, Webb DL, Sundbom M, Hellstrom PM, Sellin ME. 2021. Salmonella enterica serovar Typhimurium exploits cycling through epithelial cells to colonize human and murine Enteroids. mBio 12:e02684-20. doi: 10.1128/mBio.02684-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tagkopoulos I, Liu YC, Tavazoie S. 2008. Predictive behavior within microbial genetic networks. Science 320:1313–1317. doi: 10.1126/science.1154456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bordi C, Theraulaz L, Mejean V, Jourlin-Castelli C. 2003. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol Microbiol 48:211–223. doi: 10.1046/j.1365-2958.2003.03428.x. [DOI] [PubMed] [Google Scholar]
- 62.Ahmer BM, van Reeuwijk J, Timmers CD, Valentine PJ, Heffron F. 1998. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol 180:1185–1193. doi: 10.1128/JB.180.5.1185-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller SI, Kukral AM, Mekalanos JJ. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A 86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller VL, Beer KB, Loomis WP, Olson JA, Miller SI. 1992. An unusual pagC::TnphoA mutation leads to an invasion- and virulence-defective phenotype in Salmonellae. Infect Immun 60:3763–3770. doi: 10.1128/IAI.60.9.3763-3770.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alix E, Miki T, Felix C, Rang C, Figueroa-Bossi N, Demettre E, Blanc-Potard AB. 2008. Interplay between MgtC and PagC in Salmonella enterica serovar Typhimurium. Microb Pathog 45:236–240. doi: 10.1016/j.micpath.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 66.Harris JB, Baresch-Bernal A, Rollins SM, Alam A, LaRocque RC, Bikowski M, Peppercorn AF, Handfield M, Hillman JD, Qadri F, Calderwood SB, Hohmann E, Breiman RF, Brooks WA, Ryan ET. 2006. Identification of in vivo-induced bacterial protein antigens during human infection with Salmonella enterica serovar Typhi. Infect Immun 74:5161–5168. doi: 10.1128/IAI.00488-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cunrath O, Meinel DM, Maturana P, Fanous J, Buyck JM, Saint Auguste P, Seth-Smith HMB, Korner J, Dehio C, Trebosc V, Kemmer C, Neher R, Egli A, Bumann D. 2019. Quantitative contribution of efflux to multi-drug resistance of clinical Escherichia coli and Pseudomonas aeruginosa strains. EBioMedicine 41:479–487. doi: 10.1016/j.ebiom.2019.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edwards RA, Keller LH, Schifferli DM. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149–157. doi: 10.1016/S0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 69.Gunn JS, Hohmann EL, Miller SI. 1996. Transcriptional regulation of Salmonella virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J Bacteriol 178:6369–6373. doi: 10.1128/JB.178.21.6369-6373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chutkan H, Macdonald I, Manning A, Kuehn MJ. 2013. Quantitative and qualitative preparations of bacterial outer membrane vesicles. Methods Mol Biol 966:259–272. doi: 10.1007/978-1-62703-245-2_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J, Kim OY, Gho YS. 2016. Proteomic profiling of Gram-negative bacterial outer membrane vesicles: current perspectives. Prot Clin Appl 10:897–909. doi: 10.1002/prca.201600032. [DOI] [PubMed] [Google Scholar]
- 72.Qing G, Gong N, Chen X, Chen J, Zhang H, Wang Y, Wang R, Zhang S, Zhang Z, Zhao X, Luo Y, Liang X-J. 2019. Natural and engineered bacterial outer membrane vesicles. Biophys Rep 5:184–198. doi: 10.1007/s41048-019-00095-6. [DOI] [Google Scholar]
- 73.Bachurski D, Schuldner M, Nguyen PH, Malz A, Reiners KS, Grenzi PC, Babatz F, Schauss AC, Hansen HP, Hallek M, Pogge von Strandmann E. 2019. Extracellular vesicle measurements with nanoparticle tracking analysis—an accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J Extracell Vesicles 8:1596016. doi: 10.1080/20013078.2019.1596016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo J, Nair MK, Galvan EM, Liu SL, Schifferli DM. 2011. Tn5AraOut mutagenesis for the identification of Yersinia pestis genes involved in resistance towards cationic antimicrobial peptides. Microb Pathog 51:121–132. doi: 10.1016/j.micpath.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo A, Cao S, Tu L, Chen P, Zhang C, Jia A, Yang W, Liu Z, Chen H, Schifferli DM. 2009. FimH alleles direct preferential binding of Salmonella to distinct mammalian cells or to avian cells. Microbiology 155:1623–1633. doi: 10.1099/mic.0.026286-0. [DOI] [PubMed] [Google Scholar]
- 76.Ferreira VP, Pangburn MK, Cortés C. 2010. Complement control protein factor H: the good, the bad, and the inadequate. Mol Immunol 47:2187–2197. doi: 10.1016/j.molimm.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella Typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 78.Gewirtz AT, Simon PO, Schmitt CK, Taylor LJ, Hagedorn CH, O’Brien AD, Neish AS, Madara JL. 2001. Salmonella Typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J Clin Invest 107:99–109. doi: 10.1172/JCI10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu Y, Hu Q, Dehinwal R, Rakov AV, Grams N, Clemens EC, Hofmann J, Okeke IN, Schifferli DM. 2020. The not so good, the bad and the ugly: differential bacterial adhesion and invasion mediated by Salmonella PagN allelic variants. Microorganisms 8:489. doi: 10.3390/microorganisms8040489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harms A, Liesch M, Korner J, Quebatte M, Engel P, Dehio C. 2017. A bacterial toxin-antitoxin module is the origin of inter-bacterial and inter-kingdom effectors of Bartonella. PLoS Genet 13:e1007077. doi: 10.1371/journal.pgen.1007077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cianfanelli FR, Cunrath O, Bumann D. 2020. Efficient dual-negative selection for bacterial genome editing. BMC Microbiol 20:129. doi: 10.1186/s12866-020-01819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Felek S, Krukonis ES. 2009. The Yersinia pestis Ail protein mediates binding and Yop delivery to host cells required for plague virulence. Infect Immun 77:825–836. doi: 10.1128/IAI.00913-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
WT and ΔpagC OMVs as analyzed by TEM (A) and nanoparticle tracking analysis (B). (A) Average diameter of OMVs for WT was 138.94 ± 86 nm (mean ± SD) and 114.38 ± 57 nm for ΔpagC, as determined from TEM micrographs and measured by NIH ImageJ software. (B) Peak represents the modal value of the experimental data (154 nm or 149 nm for WT and ΔpagC OMVs, respectively), as calculated by NTA analysis. (C) Mean size (diameter in nm) of OMVs produced by Salmonella grown under PhoPQ-activating conditions, as measured by NTA analysis. The data represent one of three separate, reproducible experiments, expressed as mean ± SEM. Statistical significance was calculated using the one-way ANOVA multiple-comparison test (ns, P > 0.05). Download FIG S1, PDF file, 1.9 MB (1.9MB, pdf) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Number of OMVs/ml from various Salmonella deletion mutants. Download Table S1, DOCX file, 0.02 MB (16.4KB, docx) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Outer membrane proteins (OMPs) detected in OMVs from the WT strain and the ΔpagC mutant. Download Table S2, XLSX file, 0.02 MB (23.4KB, xlsx) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of bacterial strains, plasmids, and primers. Download Table S3, DOCX file, 0.03 MB (33.9KB, docx) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Scatter-NTA of OMVs from quadruple mutant (ΔpagC Δrck ΔompX ΔpgtE) containing empty vector (pRS1). Download Movie S1, MOV file, 6.1 MB (6.3MB, mov) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Scatter-NTA of OMVs from quadruple mutant expressing PagC (ppagC). Download Movie S2, MOV file, 6.1 MB (6.2MB, mov) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Scatter-NTA of OMVs from quadruple mutant expressing Rck (prck). Download Movie S3, MOV file, 6 MB (6.1MB, mov) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fluorescence-NTA (F-NTA) showing binding of FH to OMVs from quadruple mutant containing empty vector (pRS1). Download Movie S4, MOV file, 2.1 MB (2.2MB, mov) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
F-NTA showing binding of FH to OMVs from quadruple mutant expressing PagC (ppagC). Download Movie S5, MOV file, 6.2 MB (6.3MB, mov) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
F-NTA showing binding of FH to OMVs from quadruple mutant expressing Rck (prck). Download Movie S6, MOV file, 6.1 MB (6.2MB, mov) .
Copyright © 2021 Dehinwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.