Abstract
COVID-19 is a global crisis of unimagined dimensions. Currently, Remedesivir is only fully licensed FDA therapeutic. A major target of the vaccine effort is the SARS-CoV-2 spike-hACE2 interaction, and assessment of efficacy relies on time consuming neutralization assay. Here, we developed a cell fusion assay based upon spike-hACE2 interaction. The system was tested by transient co-transfection of 293T cells, which demonstrated good correlation with standard spike pseudotyping for inhibition by sera and biologics. Then established stable cell lines were very well behaved and gave even better correlation with pseudotyping results, after a short, overnight co-incubation. Results with the stable cell fusion assay also correlated well with those of a live virus assay. In summary we have established a rapid, reliable, and reproducible cell fusion assay that will serve to complement the other neutralization assays currently in use, is easy to implement in most laboratories, and may serve as the basis for high throughput screens to identify inhibitors of SARS-CoV-2 virus-cell binding and entry.
Author summary
COVID-19 continues to be a global public health concern. Currently, the in vitro assessment of efficacy of any vaccine or therapeutic is based on time-consuming neutralization assays using live or pseudotyped virus at BSL2/BSL3 biocontainment. There are many factors which will affect the neutralization assay, including quality of plasmid, transfection efficiency, virus titer, etc. Here, we developed a novel cell fusion assay based upon spike-hACE2 interaction, which demonstrated excellent correlation with standard spike pseudotyping for inhibition by convalescent sera, cloned antibodies, and biologics, after 16–24 hours co-incubation. It also correlated well with a live virus assay. Other than two stable cell lines, our cell fusion assay requires no specialized research reagents or laboratory equipment and should be easy to adapt for use in most investigative and clinical settings. It will allow for the testing of sera after vaccination or infection, to assess for level of immune protection, and it could be used for high throughput screening for monoclonal antibodies, compounds, and biologics that interfere with virus-cell binding and entry.
Introduction
A novel severe acute respiratory syndrome coronavirus (SARS-CoV-2) has caused pandemic disease after its emergence at the end of 2019. It was declared as a major global public health issue by WHO in March 2020 [1]. As of May 18th 2021, SARS-CoV-2 has infected nearly 164 million people and has caused close to 3.4 million deaths world-wide (https://coronavirus.jhu.edu/). Currently, many prophylactic and therapeutic strategies are under development to stem this global crisis [2–5], including use of small molecule drugs [6], biologics including interferon [7,8], convalescent sera [9], monoclonal antibodies [10,11], oligonucleotides [12], peptides [13], and vaccines [14,15]. So far, Remdesivir is the only FDA-approved drug for the treatment for COVID-19 patients (https://www.fda.gov/drugs/drug-safety-and-availability/fdas-approval-veklury-remdesivir-treatment-covid-19-science-safety-and-effectiveness), and three vaccines have Emergency Used Authorization (EUA) from the FDA (https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines), and four vaccines have been formally approved by the EMA (https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines/covid-19-vaccines). However, no other specific drug against the novel coronavirus has been formally approved by the FDA or EMA.
SARS-CoV-2 belongs to the genus Coronavirus, in the family Coronaviridae. It is an enveloped, non-segmented, positive-sense single-stranded RNA virus [16,17]. Genomic sequences of SARS-CoV-2 and SARS-CoV show high similarity, with amino acid sequence identity being >76% [18]. The genome of SARS-CoV-2 is nearly 30 kb in length, including many open reading frames (ORFs) which express at least 27 proteins [17,19]. Among them, the surface spike glycoprotein (S) plays a key role in viral entry into target cells [20–22]. The receptor-binding subunit S1 attaches to the host cell via the cellular receptor human Angiotensin-Converting Enzyme 2 (hACE2), triggering proteolytic activation of S and subsequent conformational change of the S2 subunit, which facilitates the fusion of viral and cellular membranes [23–25].
An essential element of developing any prophylactic or therapeutic antiviral or vaccine is quantitative measurement of viral replication. The current gold standards for SARS-CoV-2 neutralization include pseudotyping using S and a suitable virus core encoding a reporter [26–29], or inhibition of live virus replication in vitro [30,31] or in animal models [32,33]. Pseudotyping requires production of vector supernatants at BSL2 or BSL2+ biocontainment that are then cryostored until use, with assay readout on susceptible cells after a few days; live virus requires BSL3 laboratory and readout by plaque reduction or similar assay 3–5 days after cell or animal infection.
To circumvent some of these issues we developed a cell fusion assay that utilizes S-expressing and hACE2-expressing cells that when mixed together provides a rapid and quantitative readout, based upon human immunodeficiency virus type 1 (HIV) Tat transactivation of an integrated HIV long terminal repeat (LTR)-firefly luciferase gene. Sera from COVID-19+ patients and anti-spike monoclonal antibodies inhibited cell fusion, which correlated highly with pseudotyping and use of live virus results. Soluble receptor binding domain (RBD) of S also inhibited cell fusion to a lesser extent, as did soluble hACE2. This assay is rapid and can be easily modified for high-throughput format, which will facilitate vaccine development, potent monoclonal antibodies screening, and in vitro drug testing against the virus.
Results
Development of a quantitative assay for the measurement of S-hACE2-mediated cell fusion in transiently transfected cells
Spike or S protein mediates cell entry into susceptible target cells expressing hACE2 [24,28,34–36]. We first tested whether co-expression of S and hACE2 could result in cell fusion. 293T cells were transiently co-transfected by calcium phosphate co-precipitation with CMV expression plasmids encoding S and hACE2. At 48 h cells were fixed and stained with crystal violet, and there was obvious multinucleate cell formation that was dependent upon both S and hACE2 (S1 Fig). To test whether this occurred when cells were individually transfected, plasmids encoding S and hACE2 were introduced separately into 293Ts along with CMV-Tat and HIV LTR-FFLUC, respectively, and cells mixed at 48 h. The next day cells were fixed, and cell syncytia were observed with higher amounts of plasmid transfected, up to 1 μg (S2A–S2E Fig). In parallel, cells were lysed 24 h after mixing, and we observed a marked increase in RLU (S2F Fig).
Because enumeration of multinucleate cells is at best semi-quantitative, we decided to focus on development of a quantitative cell fusion assay, based upon HIV LTR activation by HIV Tat. As an initial experiment 293T cells were transiently co-transfected with a plasmid encoding either empty vector (EV), S driven by CMV promoter with or without protease TMPRSS2, or VSV-G along with HIV long terminal repeat driving firefly luciferase (HIV LTR-FFLUC). After 48 hours, these cells were incubated with target 293T cells that had been co-transfected with a plasmid encoding EV or hACE2 with or without TMPRSS2, along with HIV Tat, each driven by the CMV promoter. After another 48 h cells were lysed and FFLUC activity quantified by luminometry. We observed a ~100-fold increase in RLU when both S and hACE2 were each separately transfected, consistent with cell fusion (S3 Fig). As expected, when VSV-G was introduced the increase in RLU occurred independent of expression of hACE2. Interestingly, co-transfection of TRMPRSS2, the protease thought to activate S for cell fusion [34,37], did not further increase RLU activity, when introduced along with either S or hACE2 in the producers or targets, respectively (S3 Fig). This may be because 293Ts already express this protease or presence of this specific protease is not required for cell fusion in these cells.
Studies with soluble hACE2 and Spike ectodomain
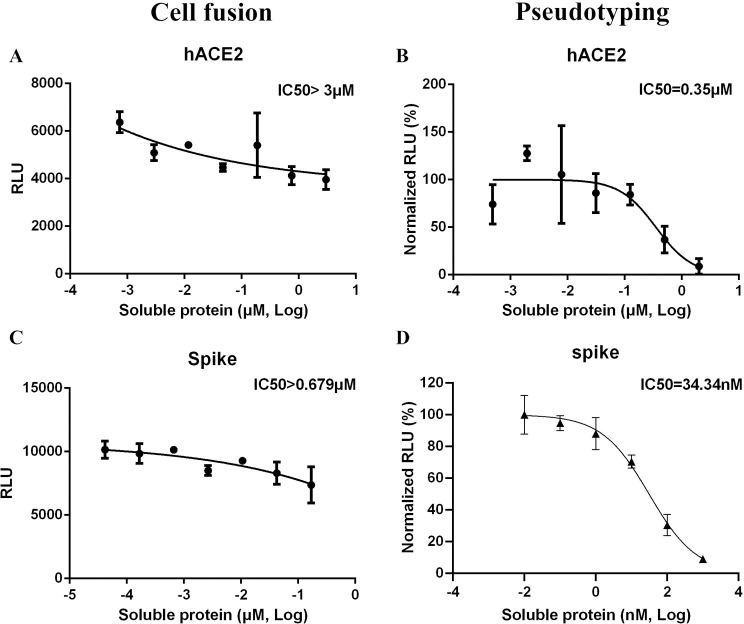
We wished to test the validity of the cell fusion assay by using putative biological inhibitors. To characterize the cell fusion assay, increasing concentrations of soluble hACE2 protein were pre-incubated with S-expressing cells for 1 h prior to mixing with hACE2-expressing cells. Soluble hACE2 was able to inhibit cell fusion at a relatively high concentration, with calculated IC50 of >3μM (Fig 1A). Entry of pseudotyped particles was also inhibited by soluble hACE2, with an IC50 of 350 nM (Fig 1B).
Fig 1. Soluble proteins inhibit cell fusion in the transient system.
Increasing concentrations of soluble hACE2 (A) and soluble Spike RBD (C) were pre-incubated with spike-expressing cells or hACE2-expressing target cells as in S3 Fig, and RLU quantified the next day. Spike pseudotyping in the presence of soluble hACE2 (B) and soluble Spike RBD (D) was performed using 293T-hACE2 targets, with RLU measured at ~72 h.
We performed similar experiments with purified ectodomain of S protein (Fig 1C and 1D). Soluble ectodomain of S was pre-incubated with S-expressing cells prior to mixing with hACE2 expressing cells, with cell fusion measured the next day. Calculated IC50 for soluble ectodomain of S was >679 nM (Fig 1C), whereas for pseudotyped particles IC50 was ~34 nM (Fig 1D). This suggests that, compared to pseudotyping, cell fusion is more difficult to inhibit by at least 10- to 20-fold, using either of these biomolecules.
Serum from COVID-19+ patients inhibit cell fusion and syncytia formation
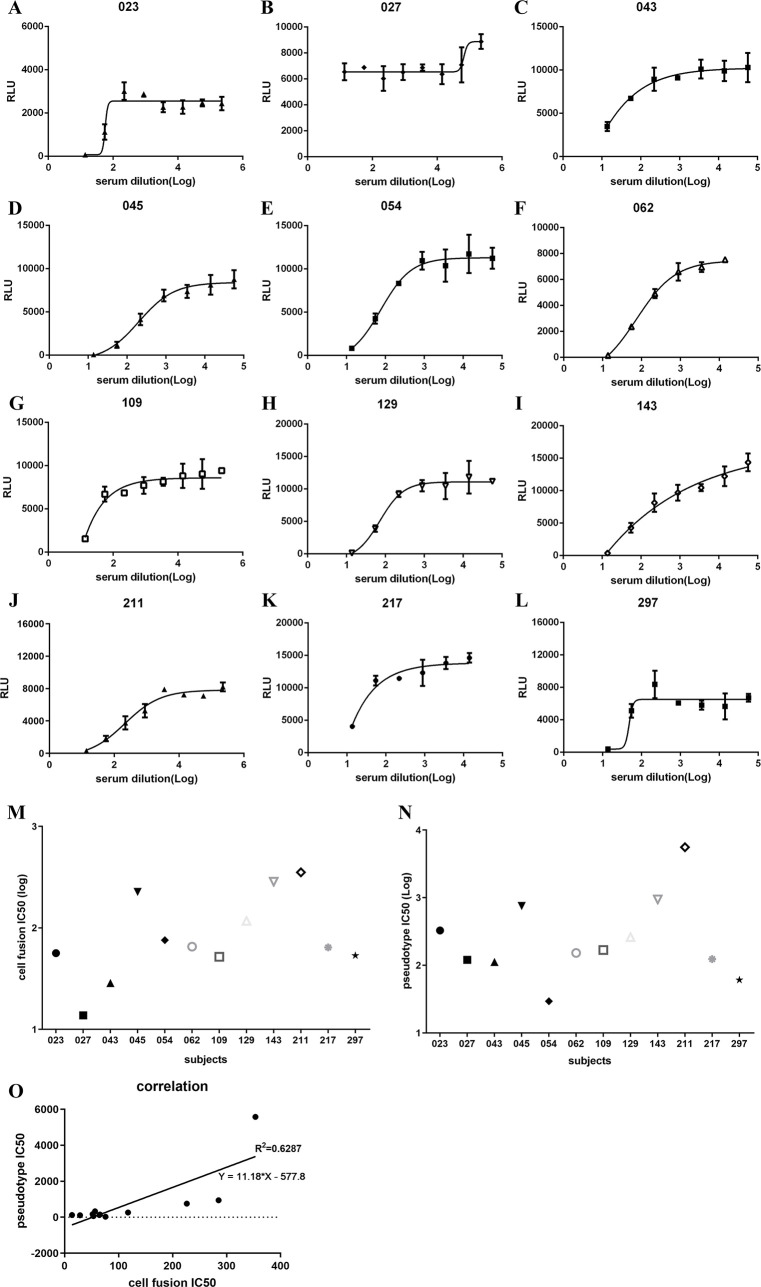
A convenience sampling of convalescent and acute illness sera from 12 COVID-19+ subjects, some with acute infection and some during recovery phase, were also tested in the cell fusion assay. Demographic and clinical characteristics of the subjects are shown in S1 Table. With a single exception, all tested sera inhibited cell fusion at varying titers (Fig 2A–2L). Setting aside outlier 027, IC50 titers for the cell fusion assay varied between 13.75 and 353.5 (Fig 2M). Serum was also tested in the pseudotyping assay (Fig 2N), and there was a high degree of correlation between cell fusion and pseudotyping IC50 titers (Fig 2O). Similar to what we had observed with soluble hACE2, inhibition of cell fusion required more sera than inhibition of pseudotyping. This suggests that, depending on the biologic or drug being tested, compared to pseudotyping the cell fusion assay may be a more rigorous test of viral inhibition.
Fig 2. COVID-19+ convalescent and acute illness sera inhibit cell fusion in the transient system.
Four-fold serially-diluted serum samples were pre-incubated with spike-expressing producer cells for 1 hour, then hACE2-expressing target cells were added, as per S3 Fig. RLU was measured the next day (A-L), and IC50 values calculated for each (M). Similarly, IC50 values were calculated for pseudotyping, performed as in Fig 1 (N). Correlation between IC50 values for the transient cell fusion and S-pseudotyping (O).
Development of stable cell lines to quantify S-hACE2 cell fusion
The data presented above demonstrate quantitation of cell fusion in transiently transfected cells. Because transient transfection is relatively complicated, occasionally unreliable, and not amenable to high throughput use, we decided to develop stable cell lines that could quantify cell fusion rapidly, reliably, and reproducibly. First, cell lines stably expressing S (TZMbl-Spike) or hACE2 (HOS-3734 or 3742) were generated, as described in Materials and Methods. TZMbls, based upon HeLa cells, have integrated HIV LTR-FFLUC and HIV LTR-LacZ cassettes that are both Tat-responsive; they have been widely used in the HIV field for titering virus stocks and performing pseudotyping assays, especially to measure neutralization by sera and cloned antibodies. The HOS cells used here had been transduced with a third-generation HIV vector in which tat remains intact. Importantly, unlike 293Ts, HOS cells do not naturally fuse with TZMbl cells. Expression of the expected 160 kD hACE2 protein in both HOS-3734 and HOS-3742 cells was verified by immunoblotting (S4A Fig). Expression of S in TZMbl-S cells was observed by using anti-FLAG antibody on immunoblots (S4B Fig). Syncytia formation was observed 24 h post incubation when these two cell types were mixed together (S5A–S5F Fig). The syncytia were LacZ+, as expected (S6 Fig). Similarly, the extent of cell fusion could be quantified by measuring RLU (S5G Fig). Only background levels of RLU were observed when TZMbls without S were used. Markedly increased RLU were measured when the hACE2 expressing cells (either HOS-3734 or HOS-3742) were co-cultured with TZMbl-S cells. Interestingly, there was detectable RLU above background when TZMbl-S cells were mixed with HOS cells carrying empty vector, indicating that either i) HOS cells express another protein that can function as a receptor, ii) HOS cells express a very low, undetectable level of hACE2, or iii) limited cell fusion can occur in the presence of S, independent of the expression of a specific cellular receptor. A time course experiment demonstrated that RLU clearly increased by 20 h and peaked at around 30 h after co-culture (S5H Fig).
Further characterization of the stable cell line cell fusion system
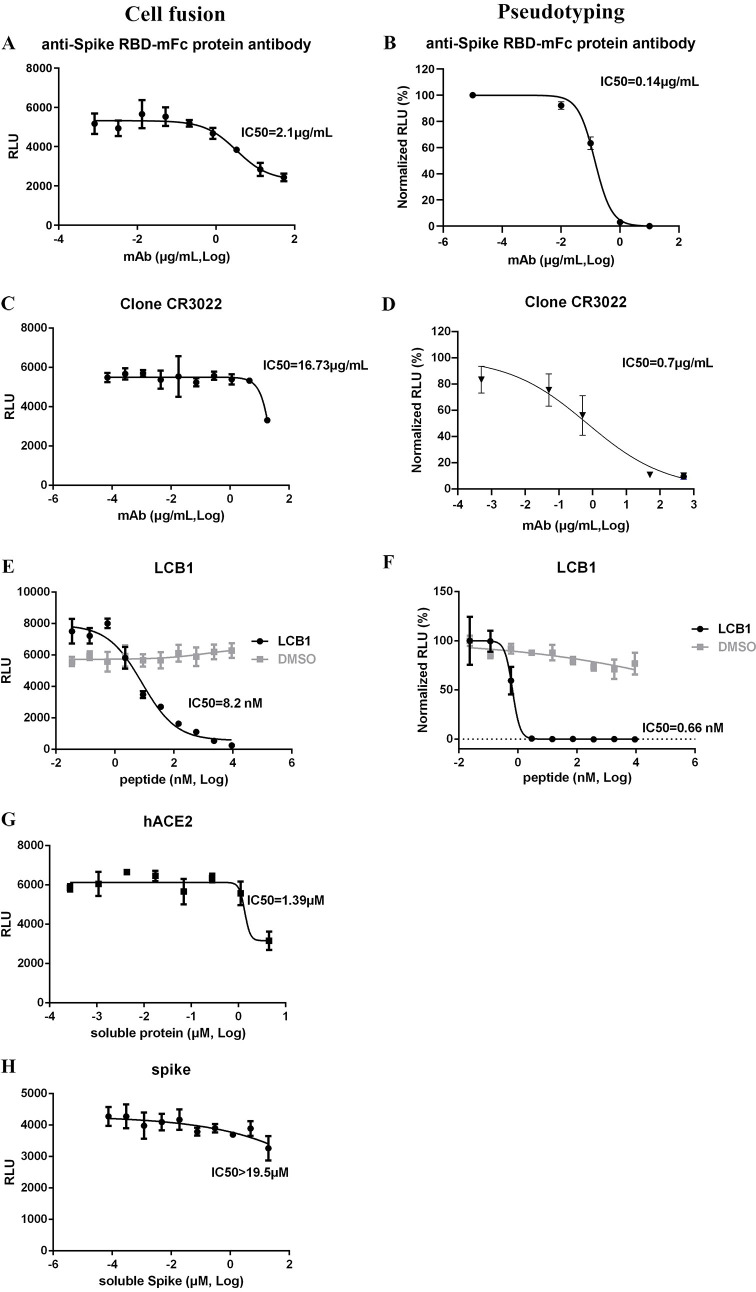
Anti-spike monoclonal antibodies, soluble hACE2 and spike RBD, and convalescent/acute illness human sera were tested in the stable cell line cell fusion system. Both commercial monoclonal antibody against Spike RBD mFc protein (Fig 3A) and clone CR3022 (Fig 3C) were able to inhibit cell fusion in the stable system, and the IC50 values were 2.1 and 16.73 μg/mL, respectively, whereas in the pseudotyping assay IC50 values were 15–20 fold less at 0.14 and 0.70 μg/mL, respectively (Fig 3B and 3D). Peptide LCB1 was reported to efficiently neutralize pseudotyped Sars-Cov-2 virus entry [38]. When we tested its ability to inhibit cell fusion the IC50 was 8.2 nM (Fig 3E), which is 12-fold greater than the IC50 value for inhibiting pseudotyping (0.66 nM; Fig 3F). Furthermore, a time of addition experiment demonstrated that LCB1 could efficiently inhibit cell fusion at -1 and 0 h before target-producer cell co-incubation (S7 Fig). Soluble hACE2 inhibited cell fusion at higher concentrations, with an IC50 of 1.39 μM (Fig 3G), whereas soluble spike RBD was virtually inactive (IC50 >19.5 μM, Fig 3H).
Fig 3. Monoclonal antibodies, peptide LCB1, and soluble proteins inhibit cell fusion in the stable system.
Serially diluted anti-Spike RBD-mFc protein antibody(A), Clone CR3022 (C), peptide LCB1 (E), soluble hACE2 (G) and soluble Spike protein (H) were incubated with producer cells (TZMbl-Spike) for 1 h. Target cells (HOS-3734) were then added, with RLU measured the next day. Serially diluted Spike RBD-mFc antibody (B), Clone CR3022 (D) or peptide LCB1 (F) were incubated with S-pseudotyped viral particles, and then added to target 293T-hACE2 cells, with RLU measured at ~72 h. IC50 values are shown.
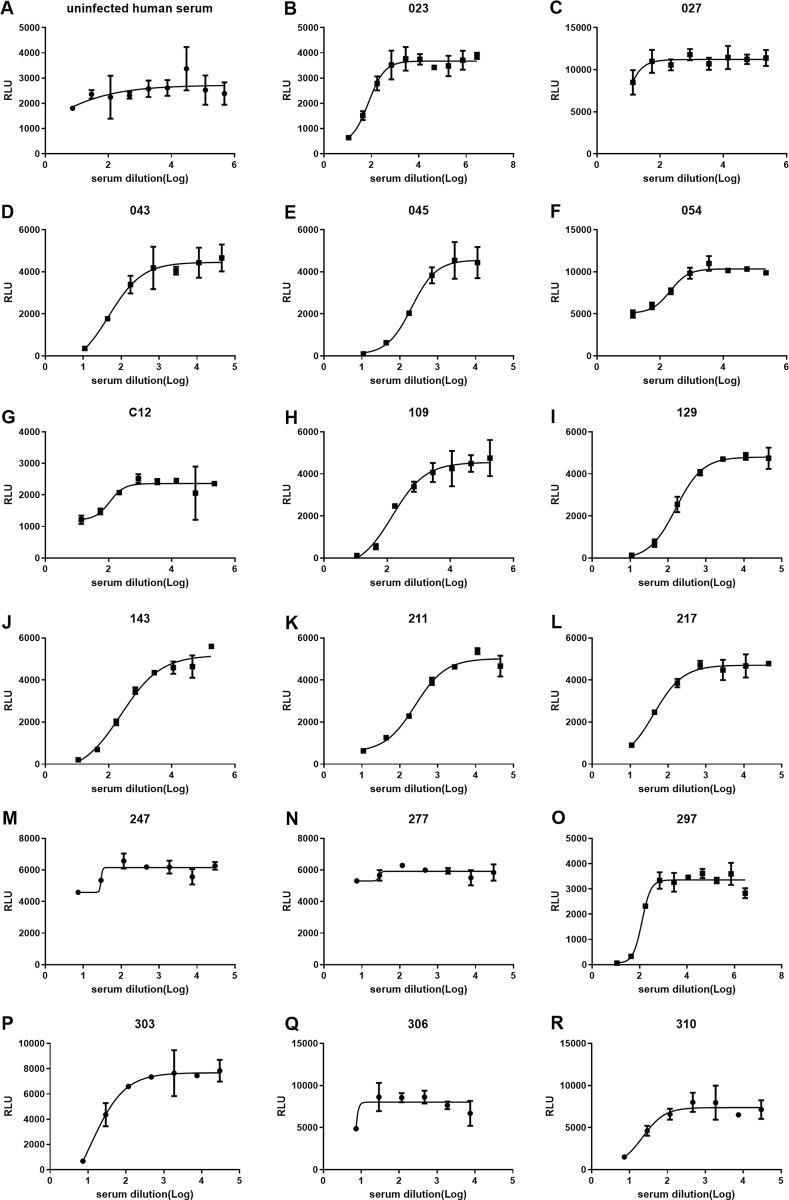
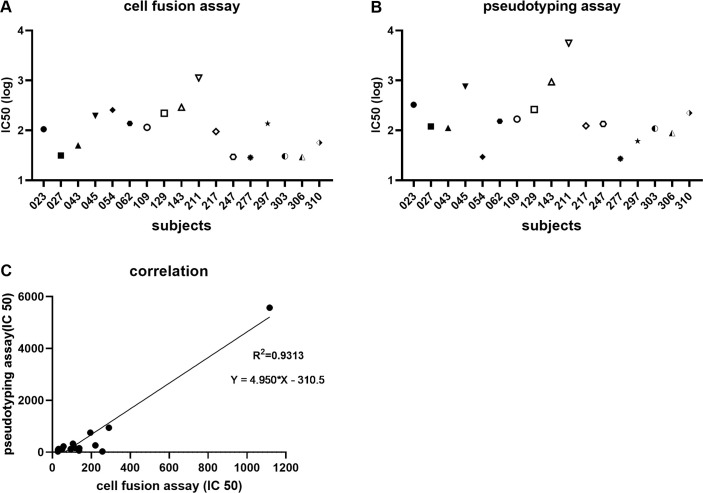
We randomly picked another 5 acute and convalescent sera such that 17 were tested using stable cell line system (HOS-3734). Clinical and demographic characteristics of these subjects are included in S1 Table. Again, 13 out of the 17 convalescent sera were able to inhibit cell fusion, with the exception of subjects 027, 247, 277, and 306 (Fig 4); IC50 values are shown in Fig 5A. And IC50 values for pseudotyping assay are shown in Fig 5B. For these samples there was an extremely robust correlation between IC50 values of the stable cell fusion and pseudotyping assays (R2 = 0.93; Fig 5C). Again, for all the tested sera the cell fusion IC50 titers were lower than those of pseudotyping. Additionally 12 sera were tested on the HOS-3742 stable cell line system (S8 Fig), and results were consistent with those using HOS-3734 cells.
Fig 4. COVID-19+ convalescent and acute illness sera inhibit cell fusion in the stable system.
Four-fold serially-diluted sera were pre-incubated with TZMbl-Spike producer cells for 1 h, then HOS-3734 target cells were added. RLU was measured the next day. (A) uninfected human serum. (B-R) convalescent and acute phase sera from COVID 19 (+) patients.
Fig 5. Correlation between the cell fusion and pseudotyping assays.
IC50 values of the 17 serum samples to inhibit cell-cell fusion between TZMbl-Spike producer and HOS-3734 target cells are shown; note log scale of Y-axis (A). IC50 values of the 17 serum samples to inhibit pseudotying virus entry are shown (B). Correlation between cell fusion and pseudotyping IC50 values (C).
A total of 7 sera were selected at random to test neutralization activity using replication-competent SARS-CoV-2-nLuc-GFP reporter virus (S9 Fig) on VeroE6 cells, and IC50 values were calculated. Based on those results IC50 values were correlated between the stable cell fusion and pseudotying assays (R2 = 0.997; S10A Fig), between pseudotyping and replication-competent virus assays (R2 = 0.82; S10B Fig), and between cell fusion and replication-competent virus assays (R2 = 0.79; S10C Fig).
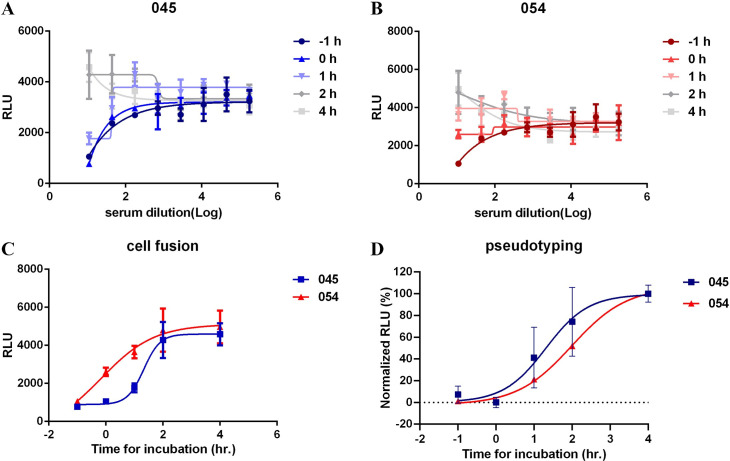
Furthermore, a time of addition experiment demonstrated that convalescent sera 045 could efficiently inhibit cell fusion at -1 and 0 h before target-producer cell co-incubation (Fig 6A and 6C), and convalescent sera 054 could efficiently inhibit cell fusion at -1 h (Fig 6B and 6C). Beyond 2 h of co-incubation inhibition of cell fusion by sera 045 or 054 was minimal to non-existent (Fig 6C). As shown in Fig 6D, data from the cell fusion assay is largely consistent with that of pseudotyping, although it appears that even at time of addition = 2 h of both sera there is some degree of inhibition of pseudotyping.
Fig 6. Addition of sera after co-culture minimally inhibits cell fusion.
Four-fold serially-diluted sera 045 (A) or 054 (B) was added 1 h before or at 0, 1, 2, or 4 h after co-culture of HOS-3734 and TZMbl-S cells. RLU was measured the next day. (C) Time course assay of inhibiting cell fusion at highest concentration of sera. (D) Times course assay of inhibiting S-pseudotyped viral particles entry, also using a high concentration of sera.
In order to test the reproducibility of the cell fusion assay, LCB1 peptide and murine mAb against Spike protein (details of this mAb to be published separately) were each tested 5 times in 5 independent experiments performed at separate, discrete times over a 6 month period, with respective, aggregate IC50s of 0.096±0.04 μg/mL and 5.52±1.64 nM (see S11 Fig for data for LCB1 peptide). Thus, the data suggest that the stable cell lines are reliably well-behaved and may be a suitable platform for high-throughput screening of sera, monoclonal antibodies,biologics, and small molecules that inhibit SARS-CoV-2 virus entry into cells.
Discussion
Current prophylactic and therapeutic efforts to stem the global COVID-19 pandemic include vaccines, biologics, and small molecules [2,15,39–42]. Virtually all of the vaccines target SARS-CoV-2 spike-hACE2 interaction, as do many of the other agents, in order to prevent virus binding and entry into target, receptor-bearing cells [15,43,44]. Current ‘gold standards’ to quantify the efficacy of such measures include inhibition of target cell infection by pseudotyped particles and fully replication-competent virus [6,28]. Both of these methods require consistent and reliable production and cryostorage of viral particles, use under BSL2+ or BSL3 biocontainment, and readout is typically performed several days later (although there are exceptions; see [45]). Herein we borrowed a page from the HIV playbook and report the development of stable cell lines in which quantification of cell fusion is available overnight after co-incubating the cells.
We were impressed with the degree of syncytia formation after co-expression of S and hACE2 (S1 Fig), which also occurred when the protein products were separately expressed and the cells subsequently mixed (S2 Fig). Because counting multinucleate giant cells is at best semi-quantitative, we next used a reporter assay after transient transfection that relies upon HIV Tat trans-activating the HIV LTR [46]. Although transient transfection did give somewhat reliable and rapid results which correlated with S pseudotyping using HIV cores (Figs 1 and 2), it required repeated, fresh cell transfections of multiple plasmids, which is inconvenient, variable, and may not be reproducible if widely employed. The stable cell lines that we have developed, on the other hand, are facile to use and very reliable. They behave very consistently over many months, even in the absence of antibiotic selection. The actual RLU values obtained are quite high for cells plated in 96-well format and the dynamic range coupled with low standard deviations, based upon inhibition by sera and monoclonal antibodies tested, should be sufficient for a high throughput screen to identify inhibitors of S and hACE2 interaction.
At present it is unknown precisely how SARS-CoV-2 spreads and replicates in human tissues, including the respiratory tract [47]. Whether cell-free versus cell-to-cell transmission of this virus occurs in vivo is an open question; generally speaking cell-to-cell transmission of virus via a virological synapse is much more efficient than infection by cell free virus [48–53]. It is conceivable that SARS-CoV-2 causes cell fusion between S and hACE2-expressing cells in man [28,54]. There is increasing histopathological evidence of giant, multinucleate respiratory epithelial and intra-alveolar cells in COVID-19+ patients [55–57]. Although some of those cells had features suggestive of being virally-infected, definitive evidence is lacking. It should be made clear that our results here are not meant to address or answer that interesting question, but rather to demonstrate that a quantitative assay based upon cell fusion has been established for spike-hACE2 interaction.
That the cell fusion and pseudotyping IC50 values for the convalescent and acute illness COVID-19+ sera (Figs 2 and 4 and 5 and S10) correlated highly suggests that the cell fusion assay is indeed measuring the ability of S and hACE2 to interact. Unsurprisingly, there was also significant correlation between the cell fusion, pseudotyping, and live virus assays in terms of IC50titers, although certainly with additional serum testing discrepancies may arise which might be worth scientific pursuit. It is also not surprising that more sera and antibody is needed to inhibit the cells from fusing. Although we have not attempted to quantify the numbers of spike and hACE2 proteins on the stable cell lines, typical expression levels would be >10,000 molecules of each, whereas an intact SARS-CoV-2 virion may have just a few dozen spike trimers [58–60], depending upon its source and how it is made. Nor is it known how many S-hACE2 interactions are required to trigger irreversible pore formation and subsequent cell-cell or cell-virus fusion. Although we do not know the relative cell surface density of either S or receptor on either stable cell line, based upon stoichiometry alone it is quite likely that inhibition of cell-cell fusion is much more difficult than inhibition of virus-cell fusion, whether the latter is due to pseudotyped or replication-competent virus. Here, this is borne out in the much higher amounts of sera and antibody required to achieve 50% inhibition of fusion (Figs 2–5). More importantly, however, the IC50 values for both assays are highly correlated (Figs 2 and 5 and S10), suggesting that the cell fusion assay has utility and in fact may be a more rigorous test for the inhibitory power of any antibody, serum, biologic, or even small molecule, when compared to other cell-based assays that rely on production of virus.
In addition to sera and monoclonal antibodies, we tested two other biologics—purified, soluble hACE2 and RBD of spike (Figs 1 and 3). Soluble hACE2, stabilized by an Fc domain for a longer half-life in plasma, has been proposed as a potential therapeutic agent, especially since the S-hACE2 interaction has a KD in the low nanomolar range [61,62]. hACE2 has now been subjected to saturation mutagenesis of the RBD-interacting residues, and several hACE2 variants have been identified that have even greater affinity for RBD (KD in the picomolar range) [63]. Here, we tested wt, soluble hACE2 (Figs 1A and 3G), and although it inhibited cell fusion the amounts required to do so were relatively high. Whether soluble or stabilized hACE2 variants are more potent against cell fusion will require further testing. At the moment soluble RBD and variants thereof have only been proposed and tested as potential vaccine candidates [64], not as therapeutics, so RBD testing here was purely academic. As anticipated, RBD did interfere with both pseudotyping and cell fusion, although for the latter the degree of inhibition was marginal at best. The fact that both soluble hACE2 and RBD inhibited cell fusion further corroborates the validity of the assay. A third biologic, peptide LCB1, also had low nM IC50 inhibitory activity in the cell fusion assay, as would be predicted based upon pseudotyping results.
The cell fusion assay was highly reproducible in that repeated testing of both an inhibitory peptide and a murine mAb gave extremely consistent results over a period of many months, with at most a 2-fold variance in IC50 values. This suggests that the cell fusion assay, when performed by other investigators throughout the world, should also behave similarly, since all that needs to be done is co-culture of two stable cell lines, without the need for making, storing, or testing any virus, live or pseudotyped. This will allow straightforward comparisons of the potency of various virus entry inhibitors between disparate laboratories.
In summary, we have developed a novel SARS-CoV-2 spike-hACE2 cell fusion assay that is rapid, reliable, and reproducible. Other than two stable, well-behaved cell lines it requires no specialized research reagents or laboratory equipment and should be easy to adapt for use in most investigative and clinical settings. It will allow for the testing of sera after vaccination or infection, to assess for level of immune protection, and it could be used for high throughput screening for compounds and biologics that interfere with virus-cell binding and entry.
Material and methods
Ethics statement
COVID-19+ convalescent serum was obtained from YNHH hospital (IMPACT research team). IMPACT study used the Yale university institutional review board (IRB) or Biomedical (HIC). And the approval was granted by the Yale HIC. All informed, written consent was obtained from all subjects.
Vectors and plasmids
CMV-driven expression plasmid for S that was also FLAG-tagged at the COOH terminus (pcDNA-SARS-CoV-2-S) was a kind gift of Craig Wilen (Yale). Plasmid encoding human ACE2 (hACE2) was obtained from Addgene (hACE2; catalog #1786). The hACE2 2.6 kbp ORF was blunt-subcloned into pShuttle (Clontech) to make pShuttle-hACE2. It was also blunt-cloned into a third generation HIV vector 3’ of CMV promoter and 5’ of an IRES-puror cassette to generate pHIV-CMV-hACE2-IRES-Puro. A separate third generation HIV vector pLV-EF1a-hACE2-cMYC-FLAG-IRES-Puro was obtained from Craig Wilen. pSV-Tat, pCMV-Tat, and pLTR-LUC were kind gifts of Andrew Rice (Baylor College of Medicine). Spike from pcDNA-SARS-CoV-2-S was inserted into a piggybac transposon (originally obtained from Matt Wilson of Baylor, along with the transposase plasmid pCMV-piggybac) that had been modified to encode a CMV-IRES-bsdr cassette; resultant plasmid was named pT-PB-SARS-CoV-2-Spike-IRES-Blasti.
Cell lines
Human embryonic kidney cell line 293T (#CRL-3216), human bone cell line HOS (#CRL-1543) were originally purchased from ATCC. Africa green monkey kidney cell line VeroE6 (#CRL-1586) were purchased from ATCC. TZMbl cells (#JC53BL-13) were obtained from the NIH AIDS Reagent Program. The HOS cells were stably transduced with a third generation HIV vector encoding tat, along with eGFP, mRFP, and bleomycin resistance gene; they were maintained in 200–400 μg/mL phleomycin (Invivogen) and were eGFP and mRFP-positive by flow cytometry. hACE2 was subsequently introduced by VSV G-mediated HIV-based transduction using pLV-EF1a-hACE2-cMYC-FLAG-IRES-Puro and pHIV-CMV-hACE2-IRES-Puro, respectively, to produce HOS-3734 and HOS-3742 cells, both cell lines maintained in selection using 10 μg/mL puromycin (Sigma-Aldrich). Those two vectors were also introduced into 293T cells to produce 293T-hACE2 cells for use with pseudotyped particles. Control HOS-2072 cells were created by transducing them with the empty vector HIV-CMV-IRES-puro and maintaining them in 10 μg/mL puromycin. TZMbl cells stably expressing S were created by co-transfecting TZMbl cells with pT-PB-SARS-CoV-2-Spike-IRES-Blasti along with pCMV-piggybac and resistant cells selected with 10 μg/mL blasticidin (Invivogen). The control stable cell line not expressing S was generated by co-transfecting pCMV-piggybac with pT-pB-IRES-Blasti and selecting for blasticidin-resistant cells.
Serum, antibodies, soluble RBD, and soluble hACE2
COVID-19+ convalescent and acute phase sera were obtained from YNHH (IMPACT research team). Human uninfected serum was purchased from Sigma-Aldrich. Goat anti-hACE2 polyclonal antisera (#AF933) was purchased from R&D Systems and was used at 1:2000. Rabbit anti-goat antisera was purchased from Sigma-Aldrich and used at 1:20000. Mouse-anti-FLAG monoclonal antibody was purchased from Sigma-Aldrich, as was goat anti-mouse IgG. Monoclonal human IgG1 antibody against SARS-CoV and SARS-CoV-2 Spike (clone CR3022) were purchased from InvivoGen. Murine anti-SARS-CoV-2 Spike antibody (SARS-CoV-2 Spike RBD-mFc protein) was purchased from Sino Biological (# 40592-MM57). 56-mer peptide LCB1 (DKEWILQKIYEIMRLLDELGHAEASMRVSDLIYEFMKKGDERLLEEAERLLEEVER) was synthesized by ABI Scientific and dissolved in DMSO and stored at -80°C prior to use.
Spike RBD domain plasmid was a gift of David Veesler (University of Washington) and the human ACE2 ectodomain plasmid was a gift of Jason McLellan (University of Texas at Austin). Both constructs have histidine tags to allow Ni column purification. Plasmids were transfected into Expi293F cells (ThermoFisher) using the manufacturer’s protocol. Culture supernatant containing the secreted protein was harvested after 3–4 days, and dialyzed against its Ni-NTA binding buffer. Protein was then purified through Ni-NTA affinity chromatography (Qiagen), and size exclusion chromatography (SEC) using a Superdex 200 column (GE Life Sciences) equilibrated with its SEC buffer (listed below). SDS-PAGE was used to monitor purification steps and ensure protein homogeneity. Peak fractions were concentrated, flash frozen and stored at -80°C for future use.
Cell fusion assay
For the transient transfection cell fusion assay producer 293T cells were co-transfected with pSV-TAT or pCMV-Tat and pcDNA-SARS-CoV-2-S, while target 293T cells were co-transfected with pLTR-LUC and pShuttle-hACE2 or hACE2 plasmids. At 48 h transfected cells were lifted, mixed 1:1, and after another 16–24 h cells were lysed and RLU measured by plate reader in 96-well format as described. Images of cell syncytia were captured with a Nikon TE2000 epifluorescence microscope running MetaMorph software. For the stable cell fusion assay HOS cell lines stably expressing HIV Tat and hACE2 (termed HOS-3734 and HOS-3742) were mixed 1:1 with TZMbl cells stably expressing S. After 16–24 h FFLUC activity was measured and syncytia images captured as described above. To observe LacZ activity, after cell fixation X-gal substrate was used as described. All experiments were performed with biological duplicates and repeated at least twice.
Cell fusion inhibition by serum or antibodies
Producer and target cells were generated as described above. With regards to the transient system, producer cells were lifted 48 h post transfection and 104 cells were resuspended in 100 μL medium per well in 96-well plates. Serial dilutions of antibody or serum were then added to producer cells and incubated at 37°C for 1 hour. At that point 104 target cells (50 μL per well) that had been transfected 48 h prior were added to producer cells, and after another 24 h cells were lysed in 0.1 mL and RLU measured. With regards to the stable cell lines, 104 producer cells (TZMbl-Spike) in 100 μL of medium in the absence of blasticidin were seeded in 96 well plates. After 24 h, 70 μL of four-fold serially diluted antibody or serum was added into producer cells and incubated at 37°C for 1 hour. Serum and antibody concentrations were the same as above. At that time 104 target cells (HOS-3734 or HOS-3742) in 50 μL medium were then added to the producer cells, and after another 24 h cells were lysed in 0.1 mL and RLU measured. Data were analyzed with non-linear regression using GraphPad Prism to determine the neutralization curve and the IC50 values calculated.
Inhibition of cell fusion with soluble hACE2, Spike RBD and peptide LCB1
Seventy μL of four-fold serially diluted, purified soluble hACE2, spike RBD or peptide LCB1 were added to 96-well plates, which were seeded with 104 producer or target cells in 100 μL per well, respectively. hACE2 dilutions began at 10 μM, spike RBD at 43 μM, and peptide LCB1 at 9.176 μM. After 1 h 0.5×104 target or producer cells in 50 μL were added per well. After 16–24 h 100 μL of lysis buffer was added to each well and RLU measured. The assay was performed for both transient and stable cell lines.
Time course experiment
96 well plates were seeded with 104 HOS-3734 cells in 100 μL per well. Four-fold serially diluted serum (045 or 054) or LCB1 were added at -1, 0, +1, +2, and +4 h relative to TZMbl-S cells addition. After 16–24 h cells were lysed and RLU measured.
Pseudotyped virus neutralization assay
Pseudotyped HIV-FFLUC was produced as previously described [65] but using pcDNA-SARS-CoV-2-S instead of VSV G or HIV Env plasmid. If necessary, pseudotyped particles were concentrated by ultracentrifugation. Serum from clinical samples, antibodies, or soluble proteins (hACE2 or Spike RBD) were serially diluted as indicated and pre-incubated with SARS-CoV-2 pseudotyped particles for 1 h at 37°C before inoculation onto 293T-hACE2 target cells. After an overnight incubation fresh medium was added. After another 48 h cells were lysed and RLU measured. IC50 values of sera, antibodies or soluble proteins were calculated using GraphPad Prism software. Non-linear regression with normalized response model was applied.
Replication-competent SARS-CoV-2 Nano luciferase neutralization assay
Experiments using infectious SARS-CoV-2 were performed in a Biosafety Level 3 facility, licensed by the State of Connecticut and Yale University. Nano luciferase expressing SARS-CoV-2 infectious clone (“icSARS-CoV-2-nLuc-GFP”) was previously described and generously provided by Ralph Baric (UNC) [30]. P3 viral stock was generated in VeroE6 cells (cultured in DMEM containing 5%FBS, 1% sodium pyruvate, and 1% Penicillin-Streptomycin) by infecting at a MOI 0.01 for two-three days to generate a working stock. After incubation the supernatant was clarified by centrifugation (500g × 5min) and filtered through a 0.45-micron filter, and virus titer was determined by plaque assay on VeroE6 cells as previously described [66,67]. VeroE6 cells were plated at 3000 cells/well in 384 well clear bottom black cell culture plate (Greiner Bio-One). Twenty-four h post seeding, human sera, which were heat-inactivated at 56°C for 30 min, were diluted 1:20 (starting dilution), then serially diluted 2-fold for 8 dilutions in DMEM (2% FBS, 1% sodium pyruvate, and 1% Penicillin-Streptomycin). 50 μL dilutions were added to 50 μL of icSARS-CoV-2-nLuc-GFP (MOI 0.01, ~2PFU/ μL), and incubated for 1 hour at 37°C. Media was then removed from cells and replaced with 20 μL of virus-sera mixture, and incubated for 48 h at 37°C. 5 μL of Nano-Glo Luciferase substrate (Promega) was added to each well and luciferase signal was measured using Cytation 5 plate reader (BioTek). Values of serum-treated samples were normalized to non-serum controls. Assay was performed in triplicate and averages of these normalized values were plotted in Prism 9 (GraphPad). IC50 values were calculated.
Western blotting
Expression of SARS-CoV-2 spike and hACE2 proteins in cells were verified by immunoblotting. In the transient system, cells transfected with plasmids encoding Spike or hACE2 were lysed with RIPA buffer 48 h post transfection. Stable cell lines expressing spike or hACE2 were similarly lysed. Samples were boiled for 10 min in the presence of SDS and DTT and size-separated on a pre-made SDS-PAGE gradient gel (Bio-Rad), which was then transferred onto PVDF filter membranes as described [65]. hACE2 and Spike proteins were detected by goat anti-hACE2 and anti-FLAG primary and rabbit anti-goat-HRP and rabbit anti-mouse-HRP secondary antibodies, respectively.
Supporting information
293T cells were transiently co-transfected with increasing amounts of CMV expression plasmids separately encoding S and hACE2, along with CMV-Tat and HIV LTR-FFLUC. At 48 h cells were fixed, stained with crystal violet (A-E), and photomicrographed, with semi-quantification of cell syncytia indicated (- rare or no syncytia; +++ most cells are in syncytia). In parallel RLU was measured, +/- SD (F).
(TIF)
293T cells were transiently transfected either with CMV expression plasmids encoding S and CMV-Tat or hACE2 and LTR-FFLUC in increasing amounts (0.5–4.0 μg per well). Cells were mixed 48 h post transfection. Plates were microphotographed 24 h post co-incubation after fixation and crystal violet staining (A-E); in parallel RLU +/- SD was measured at 24 h post co-incubation (F). Quantification of syncytia as per S1 Fig.
(TIF)
293Ts were transfected with CMV-Tat and either mock (-), CMV-hACE2 (A), or CMV-TMPRSS2 (P) [indicated at bottom of each bar to left of forward slash (/)], or HIV LTR-FFLUC and either mock (-), CMV-Spike (S), CMV-TMPRSS2 (P) VSV-G (V) [indicated at bottom of each bar to right of forward slash (/)]. At 48 h cells were mixed 1:1 and 48 h later lysed and RLU measured. Background was ~1000 RLU but increased several orders of magnitude in presence of both Spike and hACE2. Addition of protease had no effect or was inhibitory. Note log scale of ordinate axis.
(TIF)
(A) hACE2 immunoblots. Lane 1: 293Ts, 2: 293T-hACE2 cells, 3: HOS-3742, 4: HOS-3734, 5: HOS cells. Below is shown immunoblot for GAPDH as a loading control. (B) Spike immunoblots. Lane 1:293Ts, 2: 293Ts transfected with pcDNA-SARS-CoV-2-S, 3: TZMbl-Spike cells, 4: TZMbl-EV cells. Below is shown immunoblot for GAPDH as a loading control.
(TIF)
(A-F) Producer cells stably expressing spike protein (TZMbl-Spike) or control cells with empty vector (TZMbl-EV) were mixed with target cells stably expressing hACE2 (HOS-3734/HOS-3742) or control cells (HOS-EV). After 24 h, cells were photomicrographed and syncytia semi-quantified. In parallel, cells were lysed and RLU +/- SD measured (G). Producer TZMbl-Spike and target HOS-3734 cells were mixed in triplicate in 96-well plates, and RLU measured at different time points after co-culture (H). Semi-quantification of cell syncytia as per S1 Fig.
(TIF)
Producer cells stably expressing spike protein (TZMbl-Spike) or control cells with empty vector (TZMbl-EV) were mixed with target cells stably expressing hACE2 (HOS-3734/HOS-3742) or control cells (HOS-EV). After 24 h, cells were fixed in formaldehyde-glutaraldehyde, stained using X-gal at 37°C overnight, and photomicrographed. All syncytia were blue. Semi-quantification of cell syncytia as per S1 Fig.
(TIF)
Four-fold serially diluted LCB1 was added 1 h before (A) or at 0 (B), 1 (C), 2 (D), or 4 h (E) after co-culture of HOS-3734 and TZMbl-S cells. RLU was measured the next day.
(TIF)
(A-F) Four-fold serially-diluted sera were pre-incubated with TZMbl-Spike producer cells for 1 h, then HOS-3734 or HOS-3742 target cells were added. RLU was measured the next day. The red and blue curves represent fitting to data obtained from HOS-3734 and HOS-3742, respectively.
(TIF)
Fifty μL of indicated two-fold serially diluted sera was mixed with 50 μL of icSars-Cov-2-nLuc-GFP virus (MOI 0.01, ~2 PFU/μL) 1 h before incubation with VeroE6 cells. Nano-Glo Luciferase activity was quantified at 48 h and normalized to no serum control.
(TIF)
(A) Correlation between pseudotyping and cell fusion assays (R2 = 0.9968). (B) Correlation between pseudotyping and replication-competent virus assays (R2 = 0.8206). (C) Correlation between cell fusion and replication-competent virus assays (R2 = 0.788). In all cases plotted values reflect serum titer required to achieve 50% inhibition of RLU activity.
(TIF)
(A-E) Inhibition of cell fusion by LCB1 was performed in 5 independent experiments and in each case IC50 values were calculated. (F) Overall reproducibility of IC-50 values of the cell fusion assay using LCB1 peptide.
(TIF)
(DOCX)
Acknowledgments
We thank Drs. Rice, Wilen, Veesler, McLellan, and Wilson for generous reagent gifts. We thank the Yale IMPACT Research Team Membership (Abeer Obaid, Akiko Iwasaki, Allison Nelson, Arnau Casanovas-Massana, Angela Nunez, Anjelica Martin, Bertie Geng, Codruta Todeasa, Denise Shepard, Elizabeth B. White, Erin Silva, Giuseppe DeIuliis, Harold Rahming, Hong-Jai Park, Irene Matos, Jessica Nouws, Kadi-Ann Rose, Kelly Anastasio, Kristina Brower, Laura Glick, Lokesh Sharma, Maksym Minasyan, Maria Batsu, Maxine Kuang, Melissa Linehan, Michael H. Askenase, Mikhail Smolgovsky, Nicole Sonnert, Pavithra Vijayakumar, Santos Bermejo, Sofia Velazquez, Tyler Rice, William Khoury-Hanold, Xiaohua Peng, Yiyun Cao, John Fournier, M. Catherine Muenker, Adam J. Moore, Molly L. Bucklin, David McDonald, Camila Odio, & Yvette Strong). The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl cells (Cat# 8129) from Dr. John C. Kappes, and Dr. Xiaoyun Wu. The nLuc SARS-CoV-2 reporter virus was obtained from Ralph Baric.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Funding for this study was provided by the National Institutes of Health (awards R01 AI150334 to RES, and T32AI007517 to BI). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of manuscript.
References
- 1.Vogel L. What’s next now that the WHO has declared a COVID-19 pandemic? Can Med Assoc J. 2020;192(13):E349–E50. doi: 10.1503/cmaj.1095855 WOS:000524529500009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li GD, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov. 2020;19(3):149–50. doi: 10.1038/d41573-020-00016-0 WOS:000518151800001. [DOI] [PubMed] [Google Scholar]
- 3.Klasse PJ, Moore JP. Antibodies to SARS-CoV-2 and their potential for therapeutic passive immunization. Elife. 2020;9. ARTN e57877 doi: 10.7554/eLife.57877 WOS:000543717900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarma P, Prajapat M, Avti P, Kaur H, Kumar S, Medhi B. Therapeutic options for the treatment of 2019-novel coronavirus: An evidence-based approach. Indian J Pharmacol. 2020;52(1):1–5. doi: 10.4103/ijp.IJP_119_20 WOS:000521111100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lythgoe MP, Middleton P. Ongoing Clinical Trials for the Management of the COVID-19 Pandemic. Trends Pharmacol Sci. 2020;41(6):363–82. doi: 10.1016/j.tips.2020.03.006 WOS:000533609600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, Pache L, et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020. doi: 10.1038/s41586-020-2577-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shalhoub S. Interferon beta-1b for COVID-19. Lancet. 2020;395(10238):1670–1. doi: 10.1016/S0140-6736(20)31101-6 WOS:000561610000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park A, Iwasaki A. Type I and Type III Interferons—Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe. 2020;27(6):870–8. doi: 10.1016/j.chom.2020.05.008 WOS:000540760600007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloch EM, Shoham S, Casadevall A, Sachals BS, Shaz B, Winters JL, et al. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J Clin Invest. 2020;130(6):2757–65. doi: 10.1172/JCI138745 WOS:000544332600011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sajna KV, Kamat S. Antibodies at work in the time of SARS-CoV-2. Cytotherapy. 2020. doi: 10.1016/j.jcyt.2020.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang CY, Li WT, Drabek D, Okba NMA, van R, Osterhaus ADME, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020;11(1). ARTN 2251 doi: 10.1038/s41467-020-16256-y WOS:000533953700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rehman SU, Tabish M. Alternative splicing of ACE2 possibly generates variants that may limit the entry of SARS-CoV-2: a potential therapeutic approach using SSOs. Clin Sci. 2020;134(10):1143–50. doi: 10.1042/CS20200419 WOS:000535971700003. [DOI] [PubMed] [Google Scholar]
- 13.Sonya VanPatten MH, Ahmad Altiti, Kai F Cheng, Mustafa H Ghanem & Yousef Al-Abed1. <Evidence supporting the use of peptides and peptidomimetics as potential SARS-CoV-2 (COVID-19) therapeutics.pdf>. 2020. [DOI] [PMC free article] [PubMed]
- 14.Corey L, Mascola JR, Fauci AS, Collins FS. A strategic approach to COVID-19 vaccine R&D. Science. 2020;368(6494):948–50. doi: 10.1126/science.abc5312 WOS:000538332600030. [DOI] [PubMed] [Google Scholar]
- 15.Graham BS. Rapid COVID-19 vaccine development. Science. 2020;368(6494):945–6. doi: 10.1126/science.abb8923 WOS:000538332600028. [DOI] [PubMed] [Google Scholar]
- 16.Li G, Fan YH, Lai YN, Han TT, Li ZH, Zhou PW, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424–32. doi: 10.1002/jmv.25685 WOS:000511535900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan JFW, Kok KH, Zhu Z, Chu H, To KKW, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan (vol 23, pg 2511, 2019). Emerg Microbes Infec. 2020;9(1):540–. doi: 10.1080/22221751.2020.1737364 WOS:000518959700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang HH, Li XM, Li T, Zhang SB, Wang LZ, Wu X, et al. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur J Clin Microbiol. 2020;39(9):1629–35. doi: 10.1007/s10096-020-03899-4 WOS:000528517000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–+. doi: 10.1038/s41586-020-2008-3 WOS:000518098100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol. 2016;3:237–61. doi: 10.1146/annurev-virology-110615-042301 WOS:000389586100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosch BJ, van der Zee R, de Haan CAM, Rottier PJM. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801–11. doi: 10.1128/jvi.77.16.8801-8811.2003 WOS:000184462800022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wrapp D, Wang NS, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–+. doi: 10.1126/science.abb2507 WOS:000520023800044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li YW, Zhou W, Yang L, You R. Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor. Pharmacol Res. 2020;157. ARTN 104833 doi: 10.1016/j.phrs.2020.104833 WOS:000538778100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li WH, Moore MJ, Vasilieva N, Sui JH, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–4. doi: 10.1038/nature02145 WOS:000186800800040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–+. doi: 10.1038/s41586-020-2012-7 WOS:000518098100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt F, Weisblum Y, Muecksch F, Hoffmann H-H, Michailidis E, Lorenzi JCC, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. 2020. doi: 10.1101/2020.06.08.140871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses-Basel. 2020;12(5). ARTN 513 doi: 10.3390/v12050513 WOS:000565860200030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou XY, Liu Y, Lei XB, Li P, Mi D, Ren LL, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1). ARTN 1620 doi: 10.1038/s41467-020-15562-9 WOS:000522438200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang SB, Hillyer C, Du LY. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses (vol 41, pg 355, 2020). Trends Immunol. 2020;41(6):545–. doi: 10.1016/j.it.2020.04.008 WOS:000537375400009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH, 3rd, et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182(2):429–46 e14. doi: 10.1016/j.cell.2020.05.042 ; PubMed Central PMCID: PMC7250779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanderheiden A, Edara VV, Floyd K, Kauffman RC, Mantus G, Anderson E, et al. Development of a Rapid Focus Reduction Neutralization Test Assay for Measuring SARS-CoV-2 Neutralizing Antibodies. Curr Protoc Immunol. 2020;131(1):e116. doi: 10.1002/cpim.116 ; PubMed Central PMCID: PMC7864545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johansen MD, Irving A, Montagutelli X, Tate MD, Rudloff I, Nold MF, et al. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. 2020. doi: 10.1038/s41385-020-00340-z WOS:000561265600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bao LN, Deng W, Huang BY, Gao H, Liu JN, Ren LL, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–+. doi: 10.1038/s41586-020-2312-y WOS:000551400800001. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–+. doi: 10.1016/j.cell.2020.02.052 WOS:000526184900011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses-Basel. 2012;4(6):1011–33. doi: 10.3390/v4061011 WOS:000305801900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shang J, Wan YS, Luo CM, Ye G, Geng QB, Auerbach A, et al. Cell entry mechanisms of SARS-CoV-2. P Natl Acad Sci USA. 2020;117(21):11727–34. doi: 10.1073/pnas.2003138117 WOS:000536797100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glowacka I, Bertram S, Muller MA, Allen P, Soilleux E, Pfefferle S, et al. Evidence that TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J Virol. 2011;85(9):4122–34. doi: 10.1128/JVI.02232-10 WOS:000289618600007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao LX, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020;370(6515):426–+. doi: 10.1126/science.abd9909 WOS:000581077200038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeyanathan M, Afkhami S, Smaill F, Miller MS, Lichty BD, Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol. 2020. doi: 10.1038/s41577-020-00434-6 WOS:000566053600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y, Li J, Pang Z. Recent insights for the emerging COVID-19: Drug discovery, therapeutic options and vaccine development. Asian Journal of Pharmaceutical Sciences. 2020. doi: 10.1016/j.ajps.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, Carter LJ, et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Central Science. 2020;6(3):315–31. doi: 10.1021/acscentsci.0c00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seyedpour S, Khodaei B, Loghman AH, Seyedpour N, Kisomi MF, Balibegloo M, et al. Targeted therapy strategies against SARS-CoV-2 cell entry mechanisms: A systematic review of in vitro and in vivo studies. J Cell Physiol. 2020. doi: 10.1002/jcp.30032 WOS:000567435500001. [DOI] [PubMed] [Google Scholar]
- 43.Gordon DE, Jang GM, Bouhaddou M, Xu JW, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–+. doi: 10.1038/s41586-020-2286-9 WOS:000546557400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ravichandran S, Coyle EM, Klenow L, Tang JJ, Grubbs G, Liu SF, et al. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci Transl Med. 2020;12(550). ARTN eabc3539 doi: 10.1126/scitranslmed.abc3539 WOS:000546369500005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nie J, Li Q, Wu J, Zhao C, Hao H, Liu H, et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):680–6. doi: 10.1080/22221751.2020.1743767 PubMed Central PMCID: PMC7144318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazurov D, Ilinskaya A, Heidecker G, Lloyd P, Derse D. Quantitative Comparison of HTLV-1 and HIV-1 Cell-to-Cell Infection with New Replication Dependent Vectors. Plos Pathog. 2010;6(2). ARTN e1000788 doi: 10.1371/journal.ppat.1000788 WOS:000275295900038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milewska A, Kula-Pacurar A, Wadas J, Suder A, Szczepanski A, Dabrowska A, et al. Replication of Severe Acute Respiratory Syndrome Coronavirus 2 in Human Respiratory Epithelium. J Virol. 2020;94(15). ARTN e00957-20 doi: 10.1128/JVI.00957-20 WOS:000550174300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong P, Agosto LM, Munro JB, Mothes W. Cell-to-cell transmission of viruses. Curr Opin Virol. 2013;3(1):44–50. doi: 10.1016/j.coviro.2012.11.004 WOS:000315759800007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aubert M, Yoon M, Sloan DD, Spear PG, Jerome KR. The Virological Synapse Facilitates Herpes Simplex Virus Entry into T Cells. J Virol. 2009;83(12):6171–83. doi: 10.1128/JVI.02163-08 WOS:000266237900021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudnicka D, Feldmann Jrm, Porrot Fo, Wietgrefe S, Guadagnini Sp, Prévost M-C, et al. Simultaneous Cell-to-Cell Transmission of Human Immunodeficiency Virus to Multiple Targets through Polysynapses. J Virol. 2009;83(12):6234–46. doi: 10.1128/JVI.00282-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weng J, Krementsov DN, Khurana S, Roy NH, Thali M. Formation of Syncytia Is Repressed by Tetraspanins in Human Immunodeficiency Virus Type 1-Producing Cells. J Virol. 2009;83(15):7467–74. doi: 10.1128/JVI.00163-09 WOS:000267747400012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belser JA, Creager HM, Sun XJ, Gustin KM, Jones T, Shieh WJ, et al. Mammalian Pathogenesis and Transmission of H7N9 Influenza Viruses from Three Waves, 2013–2015. J Virol. 2016;90(9):4647–57. doi: 10.1128/JVI.00134-16 WOS:000375125400035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mothes W, Sherer NM, Jin J, Zhong P. Virus Cell-to-Cell Transmission. J Virol. 2010;84(17):8360–8. doi: 10.1128/JVI.00443-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia S, Liu MQ, Wang C, Xu W, Lan QS, Feng SL, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–55. doi: 10.1038/s41422-020-0305-x WOS:000522380000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bradley BT, Maioli H, Johnston R. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series (vol 396, pg 320, 2020). Lancet. 2020;396(10247):312–. WOS:000561615700025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tian SF, Xiong Y, Liu H, Niu L, Guo JC, Liao MY, et al. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Modern Pathol. 2020;33(6):1007–14. doi: 10.1038/s41379-020-0536-x WOS:000526338100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polak SB, Van Gool IC, Cohen D, von der Thusen JH, van Paassen J. A systematic review of pathological findings in COVID-19: a pathophysiological timeline and possible mechanisms of disease progression. Modern Pathol. 2020. doi: 10.1038/s41379-020-0603-3 WOS:000542079500006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu C, Yang Y, Gao Y, Shen C, Ju B, Liu C, et al. Viral Architecture of SARS-CoV-2 with Post-Fusion Spike Revealed by Cryo-EM. 2020. doi: 10.1101/2020.03.02.972927 [DOI] [Google Scholar]
- 59.Bar-On YM, Flamholz A, Phillips R, Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9. ARTN e57309 doi: 10.7554/eLife.57309 WOS:000535259000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C, et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020. doi: 10.1016/j.cell.2020.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu P, Wysocki J, Souma T, Ye M, Ramirez V, Zhou B, et al. Novel ACE2-Fc chimeric fusion provides long-lasting hypertension control and organ protection in mouse models of systemic renin angiotensin system activation. Kidney International. 2018;94(1):114–25. doi: 10.1016/j.kint.2018.01.029 [DOI] [PubMed] [Google Scholar]
- 62.Zhou QA, Kato-Weinstein J, Li Y, Deng Y, Granet R, Garner L, et al. Potential Therapeutic Agents and Associated Bioassay Data for COVID-19 and Related Human Coronavirus Infections. ACS Pharmacology & Translational Science. 2020. doi: 10.1021/acsptsci.0c00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kui K. Chan DD, Preeti Sharma, Shawn A. Abbasi, John M. Dye, David M. Kranz, Andrew S. Herbert, Erik Procko. <Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2.pdf>. [DOI] [PMC free article] [PubMed]
- 64.Moore JP, Klasse PJ, Silvestri G. COVID-19 Vaccines: “Warp Speed” Needs Mind Melds, Not Warped Minds. J Virol. 2020;94(17). doi: 10.1128/jvi.01083-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang YJ, Chapman JH, Ulcay A, Sutton RE. Neutralization Synergy between HIV-1 Attachment Inhibitor Fostemsavir and Anti-CD4 Binding Site Broadly Neutralizing Antibodies against HIV. J Virol. 2019;93(4). ARTN e01446-18 doi: 10.1128/JVI.01446-18 WOS:000457744600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL, et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell. 2021;184(1):76–91 e13. doi: 10.1016/j.cell.2020.10.028 ; PubMed Central PMCID: PMC7574718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med. 2020;217(12). doi: 10.1084/jem.20201241 ; PubMed Central PMCID: PMC7401025 with SARS-CoV2 spike N-terminal domain pending; reports, "Unrelated to the subject of the work, I have founded, co-founded, and/or hold equity in biotechnology companies including Simcha Therapeutics, Forty Seven Inc., and ALX Oncology. I have also consulted for Medicenna Therapeutics, a company that licensed patents I invented in immuno-oncology. None of these companies are in the SARS-CoV-2 space or work on infectious disease to my knowledge. Broadly related to the subject of this work, within the past year, I have purchased and disposed shares in Gilead Sciences and Vir Biotechnology, which are working on therapeutics in the coronavirus space. I currently hold no shares in either of those companies." No other disclosures were reported. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
293T cells were transiently co-transfected with increasing amounts of CMV expression plasmids separately encoding S and hACE2, along with CMV-Tat and HIV LTR-FFLUC. At 48 h cells were fixed, stained with crystal violet (A-E), and photomicrographed, with semi-quantification of cell syncytia indicated (- rare or no syncytia; +++ most cells are in syncytia). In parallel RLU was measured, +/- SD (F).
(TIF)
293T cells were transiently transfected either with CMV expression plasmids encoding S and CMV-Tat or hACE2 and LTR-FFLUC in increasing amounts (0.5–4.0 μg per well). Cells were mixed 48 h post transfection. Plates were microphotographed 24 h post co-incubation after fixation and crystal violet staining (A-E); in parallel RLU +/- SD was measured at 24 h post co-incubation (F). Quantification of syncytia as per S1 Fig.
(TIF)
293Ts were transfected with CMV-Tat and either mock (-), CMV-hACE2 (A), or CMV-TMPRSS2 (P) [indicated at bottom of each bar to left of forward slash (/)], or HIV LTR-FFLUC and either mock (-), CMV-Spike (S), CMV-TMPRSS2 (P) VSV-G (V) [indicated at bottom of each bar to right of forward slash (/)]. At 48 h cells were mixed 1:1 and 48 h later lysed and RLU measured. Background was ~1000 RLU but increased several orders of magnitude in presence of both Spike and hACE2. Addition of protease had no effect or was inhibitory. Note log scale of ordinate axis.
(TIF)
(A) hACE2 immunoblots. Lane 1: 293Ts, 2: 293T-hACE2 cells, 3: HOS-3742, 4: HOS-3734, 5: HOS cells. Below is shown immunoblot for GAPDH as a loading control. (B) Spike immunoblots. Lane 1:293Ts, 2: 293Ts transfected with pcDNA-SARS-CoV-2-S, 3: TZMbl-Spike cells, 4: TZMbl-EV cells. Below is shown immunoblot for GAPDH as a loading control.
(TIF)
(A-F) Producer cells stably expressing spike protein (TZMbl-Spike) or control cells with empty vector (TZMbl-EV) were mixed with target cells stably expressing hACE2 (HOS-3734/HOS-3742) or control cells (HOS-EV). After 24 h, cells were photomicrographed and syncytia semi-quantified. In parallel, cells were lysed and RLU +/- SD measured (G). Producer TZMbl-Spike and target HOS-3734 cells were mixed in triplicate in 96-well plates, and RLU measured at different time points after co-culture (H). Semi-quantification of cell syncytia as per S1 Fig.
(TIF)
Producer cells stably expressing spike protein (TZMbl-Spike) or control cells with empty vector (TZMbl-EV) were mixed with target cells stably expressing hACE2 (HOS-3734/HOS-3742) or control cells (HOS-EV). After 24 h, cells were fixed in formaldehyde-glutaraldehyde, stained using X-gal at 37°C overnight, and photomicrographed. All syncytia were blue. Semi-quantification of cell syncytia as per S1 Fig.
(TIF)
Four-fold serially diluted LCB1 was added 1 h before (A) or at 0 (B), 1 (C), 2 (D), or 4 h (E) after co-culture of HOS-3734 and TZMbl-S cells. RLU was measured the next day.
(TIF)
(A-F) Four-fold serially-diluted sera were pre-incubated with TZMbl-Spike producer cells for 1 h, then HOS-3734 or HOS-3742 target cells were added. RLU was measured the next day. The red and blue curves represent fitting to data obtained from HOS-3734 and HOS-3742, respectively.
(TIF)
Fifty μL of indicated two-fold serially diluted sera was mixed with 50 μL of icSars-Cov-2-nLuc-GFP virus (MOI 0.01, ~2 PFU/μL) 1 h before incubation with VeroE6 cells. Nano-Glo Luciferase activity was quantified at 48 h and normalized to no serum control.
(TIF)
(A) Correlation between pseudotyping and cell fusion assays (R2 = 0.9968). (B) Correlation between pseudotyping and replication-competent virus assays (R2 = 0.8206). (C) Correlation between cell fusion and replication-competent virus assays (R2 = 0.788). In all cases plotted values reflect serum titer required to achieve 50% inhibition of RLU activity.
(TIF)
(A-E) Inhibition of cell fusion by LCB1 was performed in 5 independent experiments and in each case IC50 values were calculated. (F) Overall reproducibility of IC-50 values of the cell fusion assay using LCB1 peptide.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.