See the article by Benitez et al., pp. 1072–1086.
Cells not only need to know how to synthesize proteins but also how to degrade them. This task is taken over by the ubiquitin-proteasome system, which eliminates misfolded, damaged, or simply unneeded proteins. Adequate protein degradation is of utmost importance for cell homeostasis, in particular, in fast proliferating cancer cells with a high protein turnover. The idea of targeting the proteasome to interfere with protein homeostasis in cancer has therefore emerged over the last two decades.
The proteasome is made up of a 20S core and two 19S regulatory subunits, together forming the 26S proteasome in mammals.1 The core subunit constitutes the catalytic part, containing several enzymatic sites with different proteolytic specificities. Proteins are tagged for degradation by a small protein called ubiquitin, which is added in multiple steps to form a polyubiquitinated protein. The polyubiquitin chain attaches the tagged protein to the regulatory subunit of the proteasome and starts the degradation process, leading to small peptides or amino acid sequences that can be reutilized by the cell for new protein synthesis. This is similar to autophagy, the cell’s route for orderly degradation and recycling of large cellular components. Both pathways are tightly regulated by multiple oncogenes and tumor suppressors, centering around PI3K/mTOR signaling. PI3K/mTOR is one of the most important intracellular pathways that is often deregulated in cancer.2 In glioblastoma, increased pathway activity is generally due to loss of the tumor suppressor and negative regulator PTEN.
Inhibitors of the proteasome target one or all of the catalytic sites in a reversible or irreversible manner. The best-known and first-in-class inhibitor, Bortezomib, was introduced in clinical practice in 2003 and is highly active in hematological malignancies, including multiple myeloma and mantle cell lymphoma. Unfortunately, its success was not reproduced in solid tumors and new generations of inhibitors are actively being pursued, including for brain tumors.3 Carfilzomib, used in a study in this issue,4 is a selective, potent, irreversible inhibitor of the β5 subunit that does not cross the blood-brain barrier (BBB). The new kid on the block, Marizomib, inhibits all three catalytic sites, crosses the BBB and has been positively evaluated in phase I/II clinical trials in glioblastoma patients.5 Currently, a phase III trial on newly diagnosed glioblastoma is ongoing (EORTC-1709, MIRAGE, NCT 03345095).5
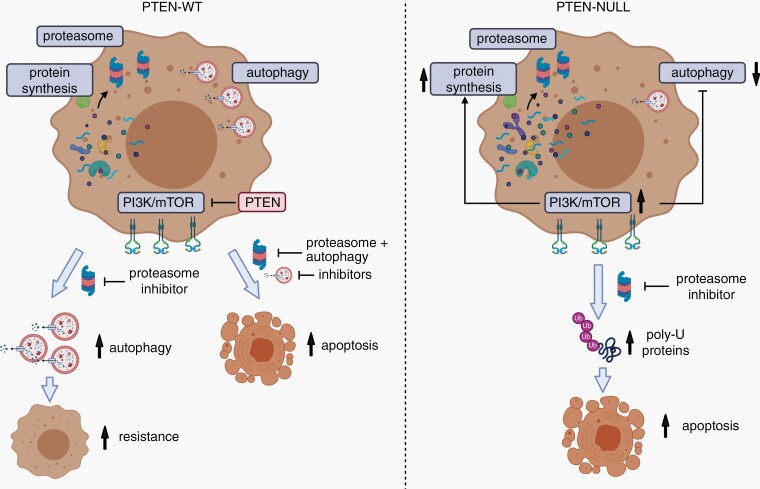
The work by Benitez et al.4 strengthens a role for proteasome inhibition in the treatment of glioblastoma and suggests that patients with PTEN deficiency and high PI3K/mTOR pathway activity may be specifically sensitive to such treatment (Figure 1). Starting from a 320-compound library screen in PTEN-expressing (PTEN-wt) vs PTEN-null glioblastoma stem-like cells (GSCs), Carfilzomib was identified as the only drug showing specificity in the PTEN context. PTEN-null cells were more sensitive to Carfilzomib, which was also true in cells expressing an enzymatically dead PTEN mutant. The data were consolidated in multiple GSCs and with several proteasome inhibitors (Ixazomib, Delanzomib, Oprozomib, and Marizomib). The higher sensitivity can be explained by increased protein synthesis in PTEN-deficient tumor cells via increased PI3K/mTOR activity. In line, the authors show that concomitant application of PI3K or mTOR inhibitors reduced the sensitivity to proteasome inhibition. Interestingly, the effect was not due to a difference in baseline proteasome activity, rather it correlated with a low autophagic flux in the PTEN-null context, resulting in increased protein ubiquitination followed by cell death through apoptosis. Surprisingly, Carfilzomib also showed activity in an orthotopic GSC xenograft model in mice. Since only one model was used and no histological data were provided, it is unclear whether this result can be translated into multiple patient-derived models with defined PTEN status. In this context, the use of brain-penetrant Marizomib would be of higher value.
Fig. 1.
Schematic representation of proteasome inhibition in PTEN-wt and PTEN-null glioblastoma cells, as described in the study of Benitez et al.4 (Illustration created with Biorender.com).
In an attempt to provide a mean to stratify responders from nonresponders to proteasome inhibition, the authors analyze differentially expressed proteins in PTEN-null and PTEN-wt cells upon treatment, and propose a 102-protein signature representing proteins uniquely activated upon proteasome inhibition in PTEN-null cells. Refinement to a 48-gene signature and a ROC bioinformatics analysis of TCGA gene expression data asked if the signature could discriminate glioblastoma with high or low PI3K/mTOR pathway activity. This was indeed the case, based on prominent expression of genes like mTOR, AKT1, and PIK3CA, however, it did not discriminate according to PTEN expression. Interrogating PTEN at the genetic level may be more reliable than gene expression. On the other hand, PI3K/mTOR activity can be deregulated through a variety of other mechanisms including mutation or amplification of PI3K and activation of tyrosine kinase receptors upstream of PI3K.
Overall, these data are critical to guide novel treatment options against glioblastoma, while they also raise additional puzzles in the grand scenery of PI3K/mTOR signaling. At first sight, the data suggest that clinical trials with proteasome inhibitors might benefit from stratification based on PTEN status and/or PI3K/mTOR activity, with increased activity in PTEN-deficient and PI3K/mTOR high patients (Figure 1). However, whether the distinction should be based on PTEN deficiency or pathway activity remains an open question. If available, molecular information from upcoming clinical trials may provide insight into selective efficacy in patient subgroups. In this context, another study recently linked the impact of proteasome inhibition in glioblastoma to the p53 pathway.6
Importantly, concomitant inhibition of proteasome activity and autophagy led to increased cell death in all GSCs, which was at least partially independent of PTEN status. This may not be too surprising since even PTEN-proficient glioblastoma is highly proliferative and thus likely to display high protein synthesis rates. Autophagy is considered a resistance mechanism against proteasome inhibition, thus supporting a dual-targeting strategy for glioblastoma patients. However, glioblastoma cells differ in their baseline activity of autophagy,7 and the sensitivity to proteasome inhibition may depend on the balance between protein synthesis and autophagy in each tumor. Despite multiple remaining questions, the present work warrants a cautiously optimistic view on the outcome of the ongoing EORTC-1709 trial.
Acknowledgments
This text is the sole product of the authors and no third party had input or gave support to its writing.
References
- 1. Wang X, Meul T, Meiners S. Exploring the proteasome system: a novel concept of proteasome inhibition and regulation. Pharmacol Ther. 2020;211:107526. [DOI] [PubMed] [Google Scholar]
- 2. Yang J, Nie J, Ma X, et al. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer. 2019;18:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thibaudeau TA, Smith DM. A practical review of proteasome pharmacology. Pharmacol Rev. 2019;71:170–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benitez JA, Finlay D, Castanza A, et al. PTEN deficiency leads to proteasome addiction, a novel vulnerability in glioblastoma. Neuro Oncol. 2021;23:1072–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roth P, Mason WP, Richardson PG, Weller M. Proteasome inhibition for the treatment of glioblastoma. Expert Opin Investig Drugs. 2020;29:1133–1141. [DOI] [PubMed] [Google Scholar]
- 6. Johansson P, Krona C, Kundu S, et al. A patient-derived cell atlas informs precision targeting of glioblastoma. Cell Rep. 2020;32:107897. [DOI] [PubMed] [Google Scholar]
- 7. Abdul Rahim SA, Dirkse A, Oudin A, et al. Regulation of hypoxia-induced autophagy in glioblastoma involves ATG9A. Br J Cancer. 2017;117:813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]