Abstract
Helicobacter pylori (H. pylori) plays a crucial role in the pathogenesis of gastritis, peptic ulcer, and gastric cancer. The presence of pathogenicity islands (PAI) genes contributes to the pathogenesis of many gastrointestinal disorders. Cytotoxin-associated gene A (cagA) and vacuolating cytotoxin gene (vacA) are the most known virulence genes in H. pylori. So, our aim was to study H. pylori virulence genes' role in gastric disorders pathogenesis. Our study included 150 adult patients who suffered dyspeptic symptoms and were referred to the GIT endoscopy unit. Gastric biopsies were attained for rapid urease test (RUT) and histopathological examination, and multiplex PCR technique for detection of virulence genes was performed. It was found that 100 specimens were (RUT) positive, of which sixty samples (60%) were PCR positive for H. pylori ureC gene. The vacA and cagA genes were identified in 61.6% and 53% of H. pylori strains, respectively. Only 5 cases were vacA-positive and cagA-negative. The most virulent vacA s1 allele existed in 56.6% of cases. Out of the 60 H. pylori strains, 66% had at least one virulence gene and 34% did not show any virulence gene. H. pylori infection showed significant increase with age. H. pylori are prevalent amid dyspeptic patients in our region. The main genotype combinations were vacA+/cagA+ of s1m1 genotype and they were frequently associated with peptic ulcer diseases, gastritis, and gastroesophageal reflux disease.
1. Introduction
H. pylori infection causes Peptic Ulcer Disease (PUD) and Gastric Carcinoma (GC) and affects almost half of the world's population [1]. H. pylori virulent strains and the host genetic condition were blamed for a wide variety of gastric disorders. H. pylori virulence genes encode the proteins which are responsible for damaging the gastric epithelium.
The H. pylori pathogenicity island (PAI) was initially known as cytotoxin-associated gene (cag), since it was assumed to be related to expression of the vacuolating toxin (vacA). However, it was afterwards demonstrated that both factors, vacA and the PAI, are separate of each other, even though cag-negative strains often do not express vacA [2].
The pathogenicity island (cagA-PAI) is found in the most virulent H. pylori strains. CagA-PAI is about 40 kb region, containing 31 genes that encode a type IV secretion system, related to cagA translocation and the host's inflammatory response [3]. CagA encodes a 120–145 kDa immunodominant protein and is placed at the end of the cagA-PAI [4]. This protein is located on the plasma membrane and is phosphorylated at Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs [5].
The numbers and types of EPIYA motifs at the C-terminal region determine the biological activity of cagA. After translocation, cagA disrupts the signal transduction of gastric epithelial cells producing cytokines which cause chronic gastritis and induce carcinogenesis designating cagA as the first bacterial oncoprotein [6].
Vacuolating cytotoxin A (vacA) can interrupt the endocytic trafficking, release organic anions and HCO3, enhance the immune tolerance, and promote chronic infection by inhibiting immune cells, stimulation of protein kinases, and autophagy modulation [7]. H. pylori strains carrying the vacA gene vary in their vacuolating ability, due to five vacA regions variations: s-region (s1 and s2), i-region (i1, i2, i3), m-region (m1 and m2), d-region (d1 and d2), and c-region (c1 and c2) [8].
VacA gene m region variants affect toxin binding to host cells [9]. vacA s1m1 variants are the most virulent arrangement, s1m2 strains toxin production is modest, and s2m2 strains are rare or not found [10].
Although it is attained in childhood, the highest age groups at risk of H. pylori infection and its mode of transmission are unclear [11]. Identifying the age at which infection may cause gastric symptoms in Egyptian patients would help to appropriately apply preventive plans.
The application of multiplex PCR methods improved our information about H. pylori regulatory genes. In this work, we studied H. pylori virulence genes in infected patients suffering from dyspepsia after gastric biopsy and the association between those genes and endoscopic findings detected.
2. Materials and Methods
This cross-sectional descriptive study involved 150 adult dyspeptic patients who were referred to GIT endoscopy unit, aged between 18 and 75 years. Patients with previous gastric surgery were excluded from the study. Patients receiving antimicrobial agents, bismuth, proton pump inhibitors, or H2 receptor antagonists within the 4 weeks preceding endoscopic examination were also excluded. Informed consents were taken from all participants before starting our study. The ethics committee had reviewed and approved the study.
From each patient enrolled in our study, three gastric biopsies were obtained (two antrum biopsies and one fundus biopsy). One of them was used to screen H. pylori positive specimens by rapid urease test (RUT). Other biopsies were transported to the pathology lab for histopathological evaluation according to the revised Sydney classification for gastritis [12], using both H&E staining and Giemsa stain, and according to the microbiology lab in brucella broth with 0.5% agar as a transport media for polymerase chain reaction (PCR) technique.
In patients with RUT positive, DNA from gastric biopsies was extracted, after being crushed and homogenized well, using QIAamp DNA Mini Kit (50) 51304 from QIAGEN, USA (catalogue number #51304) following the manufacturer's instructions. First, the samples were incubated at 37°C for 12 h in 2 ml brucella broth media and then centrifuged at 10.000 ×g for 5 min. PCR was performed to amplify the ureC (glmM) gene (nt 784–1077, 294 bp) using the following primers: forward primer: 5′-AAGCTTTTA GGGGTGTTAGGGGTTT-3′ and reverse primer: 5′-AAGC TTACTTTCTAACACTAACGC-3′. PCR conditions consisted of 30 amplification cycles (1 min at 93°C, 1 min at 55°C, and 1 min at 72°C) [13].
Multiplex PCR was used to identify vacA and cagA genes in samples positive for ureC (glmM) gene [14] using primers for vacA s1/vacA s2, vacA m1/vacA m2, and cagA (Table 1). Agarose gel (2%) with ethidium bromide was used for separation of the PCR product, using a 100 bp ladder as DNA molecular weight standard.
Table 1.
Primers used for the amplification of vacA alleles and cagA.
| Region amplified | Primers | Primers sequence | Amplicon size | Ref |
|---|---|---|---|---|
| vacA s1/vacA s2 | VAI-F | 5′-ATGGAAATACAACAAACACAC-3′ | 259/286 | Ito et al. [15] |
| VAI-R | 5′-CTGCTTGAATGCGCCAAAC-3′ | |||
| vacA m1/vacA m2 | VAG-F | 5′-CAATCTGTCCAATCAAGCGAG-3′ | 567/642 | Jones et al. [7] |
| VAG-R | 5′-GCGTCAAAATAATTCCAAGG-3′ | |||
| cagA | cagA5c-F | 5′-GTTGATAACGCTGTCGCTTC-3′ | 350 | Graham and Yamaoka [16] |
| cagA3c-R | 5′-GGGTTGTATGATATTTTCCATAA-3′ |
2.1. Statistical Analysis
Statistical analysis was done using IBM Statistical Package for the Social Sciences (SPSS) software, version 20.0, for Windows. Data was presented as mean ± standard deviation or percentages. Chi-squared test was used for categorical variables. P value was significant at < 0.05.
3. Results
150 patients were enrolled from the endoscopy unit. Age ranged from 18 to 75 years (57.2 ± 16.34); 60% of them were females, and 65% of them were from rural area. No one received nonsteroidal anti-inflammatory drugs. Recurrent abdominal pain was the most common presentation followed by nausea, vomiting, dyspepsia, and heart burn (60%, 39%, 24%, and 20%, respectively).
Antral gastritis was the most predominant endoscopic finding followed by Gastroesophageal Reflux Disease (GERD) and pangastritis (36.6%, 34%, and 26.6%, respectively). Normal gastric mucosa was found in only 4 patients (2.6%). Chronic superficial gastritis was the most common finding in histopathological examination, some of which showing mild chronic inflammation with no activity (Figure 1) and others showing moderate chronic inflammation (Figures 2 and 3), followed by atrophic gastritis (Figures 4 and 5), chronic active gastritis (Figure 6), and duodenitis (70%, 20%, 10%, and 6%, respectively).
Figure 1.
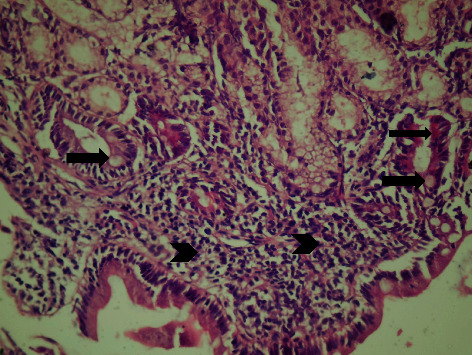
Photomicrograph showing antral chronic superficial gastritis with mixed inflammatory cell infiltrate (arrow heads), mainly lymphocytes and plasma cells; some glandular epithelial cells show intestinal metaplasia, with goblet cells (thick arrow) and Paneth cells (thin arrows), H&E, ×200.
Figure 2.
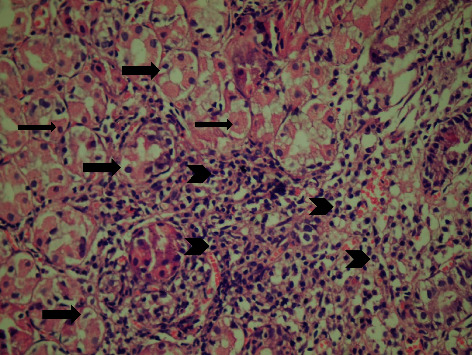
Photomicrograph showing chronic superficial gastritis with mixed inflammatory cell infiltrate (arrow heads), mainly lymphocytes and plasma cells; some glandular epithelial cell shows reactive changes, for example, subnuclear vacuolization (arrows) and parietal cell hyperplasia (thin arrow), H&E, ×200.
Figure 3.
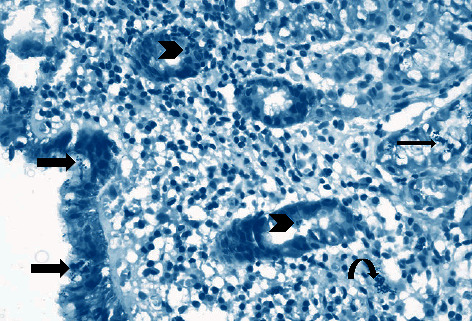
Photomicrograph showing chronic superficial active gastritis (arrow heads pointing to neutrophils attacking glandular epithelium), with mixed inflammatory cell infiltrate, mainly lymphocytes and plasma cells. Helicobacter pylori are present in the superficial mucous layer (thick arrow), within the glands, and in the lamina propria (curved arrow), Giemsa stain, ×200.
Figure 4.
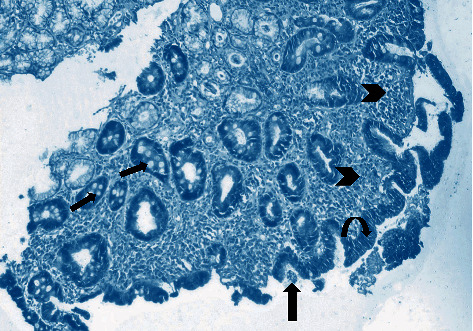
Photomicrograph showing atrophic gastritis with mixed inflammatory cell infiltrate (arrow heads). Helicobacter pylori are present in the superficial mucous layer (thick arrow) and intestinal metaplasia with goblet cells (thin arrow), with marked loss of mucosal glands and mucin depletion in the surface epithelium due to regenerative changes (curved arrow). Giemsa stain, ×100.
Figure 5.
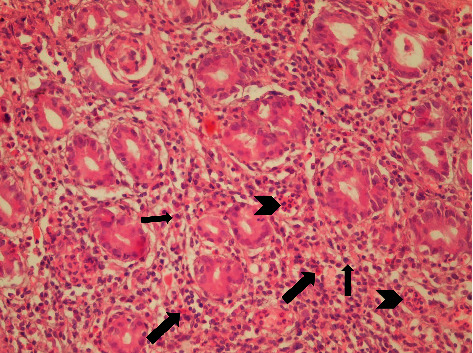
Photomicrograph showing chronic active gastritis with large number of neutrophils (arrow heads) and mixed inflammatory cell infiltrate, namely, plasma cells (thin arrow), lymphocytes (arrow heads), and eosinophils (thick arrow), with moderate atrophy, H&E stain, ×200.
Figure 6.
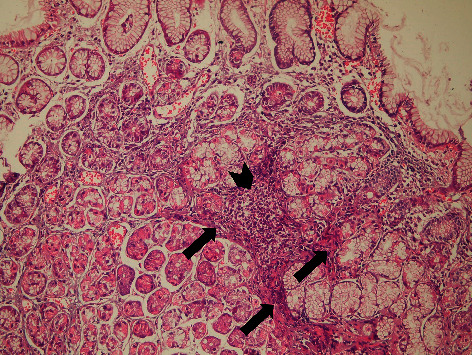
Photomicrograph showing chronic gastritis with mixed inflammatory cell infiltrate and lymphoid follicle formation (arrow head). There is also intramucosal fibrosis (arrows) with moderate glandular atrophy, H&E stain, ×100.
Out of the 150 studied patients, 100 patients (66.6%) were RUT positive. All patients with antral gastritis and combined duodenal and gastric ulcers were RUT positive. Antral gastritis, diffuse erosive gastroduodenitis, duodenal ulcer, gastric ulcer, and combined duodenal and gastric ulcer had statistically significant association with RUT (P < 0.001, <0.001, 0.03, 0.017, and 0.04, respectively).
PCR was performed for all RUT positive samples (100 samples), where 60 samples (60%) were positive for H. pylori ureC (glmM) gene (Figure 7). Antral gastritis represented the most common endoscopic result in patients positive for H. pylori by PCR followed by duodenal ulcer and GERD (33.3%, 28.3%, and 25%, respectively). Pan gastritis, diffuse erosive gastroduodenitis, duodenal ulcer, gastric ulcer, and combined duodenal and gastric ulcer showed statistically significant relation with PCR results (P < 0.001) (Table 2).
Figure 7.
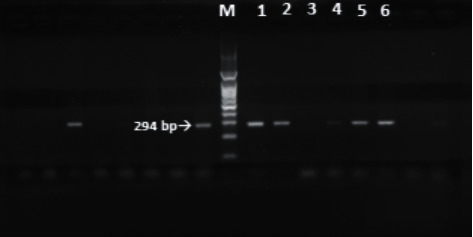
PCR amplification of UreC (glmM) for H. pylori detection: M: 100 bp ladder; lanes 1, 2, and 4–6: positive (at 294 bp); lane 3: negative.
Table 2.
Endoscopic findings in relation to RUT and PCR.
| Findings | RUT∗∗ (100) | P value | OR (95% CI) | PCR∗∗∗ (60) | P value | OR (95% CI) |
|---|---|---|---|---|---|---|
| Antral gastritis (55) | 55 (55%) | <0.001∗ | 20 (33.3%) | 0.489 | 0.8 (0.4–1.6) | |
| Gerd (34) | 36 (36%) | 0.465 | 1.3 (0.6–2.7) | 15 (25%) | 0.057 | 0.5 (0.2–1.0) |
| Pangastritis (40) | 30 (30%) | 0.192 | 1.7 (0.8–3.9) | 5 (8.3%) | <0.001∗ | 0.1 (0.1–0.4) |
| Diffuse gastric mucosal nodularity (40) | 20 (20%) | 0.172 | 0.6 (0.3–1.3) | 10 (16.6%) | 0.115 | 0.5 (0.2–1.2) |
| Diffuse erosive gastroduodenitis (34) | 15 (15%) | <0.001∗ | 0.3 (0.1–0.6) | 8 (13.3%) | <0.001∗ | 0.1 (0.1–0.3) |
| Duodenal ulcer (30) | 25 (25%) | 0.030∗ | 3 (1.1–8.4) | 17 (28.3%) | 0.084 | 0.5 (0.3–1.1) |
| Gastric ulcer (20) | 18 (18%) | 0.017∗ | 5.3 (1.2–23.7) | 8 (13.3%) | <0.001∗ | 0.1 (0.1–0.3) |
| Combined duodenal and gastric ulcers (8) | 8 (8%) | 0.040∗ | 5 (8.3%) | <0.001∗ | 0.1 (0.0–0.2) | |
| Hiatus hernia (3) | 0 (0%) | 0.014∗ | 0 (0%) | <0.001∗ | ||
| Gastric cancer (3) | 2 (2%) | 1.000 | 1 (0.1–11.3) | 0 (0%) | <0.001∗ | |
| Duodenal diverticulum (2) | 0 (0%) | 0 (0%) | <0.001∗ | |||
| Gastric polyp (1) | 0 (0%) | 0.156 | 0 (0%) | <0.001∗ | ||
| Barrette esophagus (1) | 0 (0%) | 0.156 | 0 (0%) | <0.001∗ | ||
| Normal mucosa (4) | 0 (0%) | 0.156 | 0 (0%) | <0.001∗ |
∗ P < 0.001: highly statistically significant; ∗∗RUT: rapid urease test; ∗∗∗PCR: polymerase chain reaction.
vacA and cagA genotypes were identified by multiplex PCR (Figure 8). The vacA and cagA genes were identified in 37 (61.6%) and 32 (53%) of the 60 H. pylori strains, respectively. All strains with cagA gene were vacA gene positive. Only 5 cases were vacA positive and cagA negative.
Figure 8.
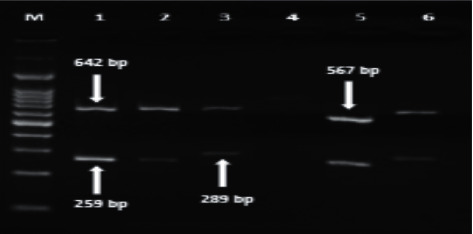
PCR amplification of vacA alleles and cagA: M: 100 bp ladder; lanes 1–3, 5, and 6: vacA+/cagA−; lane 4: vacA−/cagA−; lane 1: s1m2 genotype; lane 2: s1m2 genotype; lane 3: s2m2 genotype; lane 5: s1m1 genotype; lane 6: s2m2 genotype.
H. pylori patients with PUD, gastritis, and GERD (50%, 41.6%, and 25%, respectively) had statistically significant association with cagA genotype (P < 0.001). The most virulent vacA s1 allele was represented in 34 cases (56.6%). The middle region vacA m1 was predominant in 27 cases (45%), while m2 and s2 genotypes were detected in 10 (16.6%) and 3 (5%) cases, respectively (Table 3).
Table 3.
Association between virulence genes and endoscopic findings in H. pylori strains (no. = 60).
| Virulence genes | vacA genotypes (No.) | Endoscopic findings (n = 60) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| PUD∗ (30) No. (%) | P value | Gastritis (25) No. (%) | P value | GERD∗∗ (15) No. (%) | P value | Others (18) No. (%) | P value | ||
| vacA+/cagA+ | s1m1 (26) | 10 (33.3%) | <0.001∗ | 12 (20%) | <0.001∗ | 8 (13.3%) | <0.001∗ | 6 (10%) | <0.001∗ |
| s1m2 (5) | 5 (16.6%) | 4 (6.6%) | 2 (3.3%) | 0 (0.0%) | |||||
| s2m2 (1) | 1 (3.3%) | 0 (0.0%) | 1 (1.6%) | 0 (0.0%) | |||||
| vacA+/cagA− | s1m1 (1) | 0 (0.0%) | 0.392 | 1 (3.3%) | 0.392 | 0 (0.0%) | 0 (0.0%) | ||
| s1m2 (2) | 1 (3.3%) | 1 (3.3%) | 0 (0.0%) | 0 (0.0%) | |||||
| s2m2 (2) | 0 (0.0%) | 2 (6.6%) | 0 (0.0%) | 0 (0.0%) | |||||
| vacA−/cagA− | 13 (23.3%) | 5 (11.6%) | 4 (1.6%) | 12 (16.6%) | |||||
∗PUD: Peptic Ulcer Disease. ∗∗GERD: Gastroesophageal Reflux Disease.
The vacA s1m1/cagA+ was found in 26 cases (43.3%), while vacA s1m2/cagA+ were found in 5 strains only (8.3%). We did not find vac s2m1 genotype, either cagA positive or negative. The vac s2m2 genotype was detected in 3 patients; one of them was cagA positive. Out of the 60 H. pylori strains tested, 39 (66%) had at least one virulence gene and 21 (34%) did not show any virulence genes (Table 3).
The age of the patients positive for H. pylori (60 patients) ranged between 25 and 65 years (±years). It was noticed that H. pylori infection increases significantly with age in most of cases, 45/60 (75%), aged> 45 years with P value <0.001. Genotypes s1m1 and s2m2 showed statistically significant association with patients aged >45 years (P value <0.001), while the association with s1m2 was insignificant (P > 0.05).
4. Discussion
H. pylori is recognized as the most prevalent infectious disease in human being. It causes persistent gastritis that is interrelated to PUD and gastric adenocarcinoma [1]. In all guidelines, H. pylori eradication is accepted as the primary priority strategy for preventing gastric cancer [17].
Dyspeptic patients are more likely to have H. pylori infection than asymptomatic individuals. The burden of H. pylori infection in our patients with dyspepsia was high; about two-thirds of patients tested positive for RUT. Our findings were close to those of an Egyptian study in Mansoura [18]. The prevalence of H. pylori infection in different countries is variable. For instance, it is more than 80% in many areas such as Japan, South America, Turkey, and Pakistan, while in England it is lower (20%) [19].
In this study, antral gastritis was the predominant endoscopic finding, representing 36.6%. Moreover, antral gastritis, diffuse erosive gastroduodenitis, duodenal ulcer, gastric ulcer, and combined duodenal and gastric ulcer showed statistically significant association with RUT (P < 0.001, <0.001, 0.03, 0.017, and 0.04, respectively). Our findings agreed with many studies which reported that most of H. pylori patients developed acute gastritis that resolved spontaneously [20]. We noticed that 38.6% (58/150) of our patients were suffering from PUD with 51.7% (30/58) of them having H. pylori. This agrees with a Kenyan study, which reported that 30% had PUD with H. pylori infection [21].
H. pylori cagA is a highly immunogenic protein encoded by cagA gene. It is associated with cell injury and more severe clinical outcomes, including duodenal ulcer and gastric adenocarcinoma [22]. vacA disrupts epithelial cell tight junctions, alters the host inflammatory response, and suppresses T cell activation [16].
The relation of the vacA and cagA genotypes with clinical outcomes represented by endoscopic findings was examined. About two-thirds (66%) of H. pylori positive specimens had two or at least one of the virulence genes examined in our study. vacA and cagA genotypes were detected in 61.6% and 53%, respectively, of the 60 H. pylori strains. Similarly, a previous study on Cuban patients detected vacA gene in 61.6% of the studied H. pylori strains, considering it the main virulent gene in most of the strains [23]. Another study in Algeria identified the cagA gene in 58% patients [24], and the percentages were as follows in other countries: in Pakistan, 24.2% [25], and, in Japan, 90%, which is correlated with the increased prevalence of gastric cancer in that country [15]. Because of the high degree of geographic variability of H. pylori, certain H. pylori genotypes are possibly associated with severe clinical outcomes in some countries, while presenting as less harmful or even harmless variants in other regions. The observed discrepancies in different studies regarding H. pylori virulence genes may be attributed to difference in facilities or the limitations of PCR methods.
Regarding cagA and VacA genotypes, we found that vacA + cagA+/s1m1 was the most predominant (26/60, 43.3%). Similarly, s1m1 genotype is the most prevalent genotype in Asian population [26]. In our series, we noticed that H. pylori infected patients with PUD, gastritis, and GERD (50%, 41.6%, and 25%, respectively) had a statistically significant association with cagA genotype (P < 0.001). This agrees with a Saudi study which confirmed the association between vacA genotype and severe gastric endoscopic findings [27].
However, cagA+ was not found in our study as a single genotype. It was rather linked to vacA s1m1 in (26/60), vacA s1m2 (5/60), or s2m2 (1/60).
In this study, a lower percentage of vacA s2m2 genotype was noticed, which is considered as a less virulent form as compared with the acutely damaging vacA s1m1 as stated by Falsafi and his colleagues [28]. In our studied population, vacA+/cagA− genotype s1m1 was the least prevalent, and this is consistent with an Iranian study that was carried out in 2015 [28].
5. Conclusion
Helicobacter pylori infection is prevalent among dyspeptic patients. The main genotype combinations in our studied population were vacA+/cagA+ of s1m1 genotype and they were frequently associated with gastritis and GERD, while vacA−/cagA− patients presented mainly with gastritis. The less virulent (s2m2) genotypes were found in vacA+/cagA− and vacA+/cagA+ patients. Eventually, the advanced molecular methods are recently used as dependable tools for characterization of H. pylori virulent strains because of their increasing sensitivity and specificity.
5.1. Main Points
Helicobacter pylori infection is prevalent among dyspeptic patients reflecting the increased risk of gastric disorders including gastric carcinoma.
Knowledge of H. pylori virulence genes can be of clinical significance through improving the clinical prediction of disease risk and identifying those in need to more surveillance and eradication of the infection to prevent serious consequences.
Advanced molecular methods can be used as dependable tools for characterization of H. pylori virulent strains because of their increasing sensitivity and specificity.
Data Availability
The data supporting the findings of this study are available within the article.
Ethical Approval
The study was approved by the Research Ethics Committee, Faculty of Medicine, Suez Canal University.
Consent
A verbal consent was obtained from the patients involved in the study.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
All authors contributed to the study conception and design, in addition to material preparation, data collection, and analysis. All authors commented on previous versions of the manuscript; also, they read and approved the final manuscript.
References
- 1.Chiurillo M. A., Moran Y., Cañas M., et al. Genotyping of Helicobacter pylori virulence-associated genes shows high diversity of strains infecting patients in western Venezuela. International Journal of Infectious Diseases. 2013;17(9):e750–e756. doi: 10.1016/j.ijid.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Tummuru M. K., Cover T. L., Blaser M. J. Mutation of the cytotoxin-associated cagA gene does not affect the vacuolating cytotoxin activity of Helicobacter pylori. Infection and Immunity. 1994;62(6):2609–2613. doi: 10.1128/iai.62.6.2609-2613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitmire J. M., Merrell D. S. Helicobacter pylori genetic polymorphisms in gastric disease development. Advances in Experimental Medicine and Biology. 2019;1149:173–194. doi: 10.1007/5584_2019_365. [DOI] [PubMed] [Google Scholar]
- 4.Kalali B., Mejías-Luque R., Javaheri A., Gerhard M. H. pylori virulence factors: influence on immune system and pathology. Mediators Inflamm. 2014;2014:9. doi: 10.1155/2014/426309.426309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahdavi J., Sondén B., Hurtig M., et al. Helicobacter pylori sabA adhesin in persistent infection and chronic inflammation. Science. 2002;297(5581):573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Backert S., Blaser M. J. The role of cagA in the gastric biology of Helicobacter pylori. Cancer Research. 2016;76(14):4028–4031. doi: 10.1158/0008-5472.can-16-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones K. R., Whitmire J. M., Merrell D. S. A tale of two toxins: Helicobacter pylori cagA and vacA modulate host pathways that impact disease. Frontiers in Microbiology. 2010;1:p. 115. doi: 10.3389/fmicb.2010.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foegeding N. J., Caston R. R., McClain M. S., Ohi M. D., Cover T. L. An overview of Helicobacter pylori vacA toxin biology. Toxins. 2016;8(6) doi: 10.3390/toxins8060173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClain M. S., Beckett A. C., Cover T. L. Helicobacter pylori vacuolating toxin and gastric cancer. Toxins. 2017;9(10) doi: 10.3390/toxins9100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bridge D. R., Merrell D. S. Polymorphism in the Helicobacter pylori cagA and vacA toxins and disease. Gut Microbes. 2013;4(2):101–117. doi: 10.4161/gmic.23797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atherton J. C., Cao P., Peek R. M., Tummuru M. K. R., Blaser M. J., Cover T. L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Journal of Biological Chemistry. 1995;270(30):17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 12.Dixon M. F., Genta R. M., Yardley J. H., Correa P. Classification and grading of gastritis. The American Journal of Surgical Pathology. 1996;20(10):1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Lu J.-J., Perng C.-L., Shyu R.-Y., et al. Comparison of five PCR methods for detection of Helicobacter pylori DNA in gastric tissues. Journal of Clinical Microbiology. 1999;37(3):772–774. doi: 10.1128/jcm.37.3.772-774.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malaty H. M., Graham D. Y., Klein D. G., Adam E. E., Evans D. J., Evans D. J. Transmission of Helicobacter pylori infection studies in families of healthy individuals. Scandinavian Journal of Gastroenterology. 1991;26(9):927–932. doi: 10.3109/00365529108996244. [DOI] [PubMed] [Google Scholar]
- 15.Ito Y., Azuma T., Ito S., et al. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. Journal of Clinical Microbiology. 1997;35(7):1710–1714. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham D. Y., Yamaoka Y. Disease-specific Helicobacter pylori virulence factors: the unfulfilled promise. Helicobacter. 2000;5(S1):S3–S9. doi: 10.1046/j.1523-5378.2000.0050s1003.x. [DOI] [PubMed] [Google Scholar]
- 17.Chattopadhyay S., Patra R., Ramamurthy T., et al. Multiplex PCR assay for rapid detection and genotyping of Helicobacter pylori directly from biopsy specimens. Journal of Clinical Microbiology. 2004;42(6):2821–2824. doi: 10.1128/jcm.42.6.2821-2824.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Serag H. B., Kao J. Y., Kanwal F., et al. Houston consensus conference on testing for Helicobacter pylori infection in the United States. Clinical Gastroenterology and Hepatology. 2018;16(7):992–1002.e6. doi: 10.1016/j.cgh.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suvak B., Dulger A., Suvak O., Aytemiz E., Kemik O. The prevalence of Helicobacter pylori among dyspeptic patients in an earthquake-stricken area. Clinics. 2015;70(1):69–72. doi: 10.6061/clinics/2015(01)12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latif A. H., Shami S. K., Batchoun R., Murad N., Sartawi O. Helicobacter pylori: a jordanian study. Postgraduate Medical Journal. 1991;67(793):994–998. doi: 10.1136/pgmj.67.793.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimang’a A. N., Revathi G., Kariuki S., Sayed S., Devani S. Helicobacter pylori: prevalence and antibiotic susceptibility among Kenyans. South African Medical Journal = Suid-Afrikaanse Tydskrif Vir Geneeskunde. 2010;100(1):53–57. [PubMed] [Google Scholar]
- 22.Graham D. Y., Alpert L. C., Smith J. L., Yoshimura H. H. Iatrogenic Campylobacter pylori infection is a cause of epidemic achlorhydria. The American Journal of Gastroenterology. 1988;83(9):974–980. [PubMed] [Google Scholar]
- 23.Feliciano O., Gutierrez O., Valdés L., et al. Prevalence of Helicobacter pylori vacA, cagA, and iceA genotypes in cuban patients with upper gastrointestinal diseases. BioMed Research International. 2015;2015:6. doi: 10.1155/2015/753710.753710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachir M., Allem R., Tifrit A., et al. Primary antibiotic resistance and its relationship with cagA and vacA genes in Helicobacter pylori isolates from algerian patients. Brazilian Journal of Microbiology. 2018;49(3):544–551. doi: 10.1016/j.bjm.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmad T., Sohail K., Rizwan M., Mukhtar M., Bilal R., Khanum A. Prevalence of helicobacter pyloripathogenicity-associated cagA and vacA genotypes among Pakistani dyspeptic patients. FEMS Immunology & Medical Microbiology. 2009;55(1):34–38. doi: 10.1111/j.1574-695x.2008.00492.x. [DOI] [PubMed] [Google Scholar]
- 26.Memon A. A., Hussein N. R., Miendje Deyi V. Y., Burette A., Atherton J. C. Vacuolating cytotoxin genotypes are strong markers of gastric cancer and duodenal ulcer-associated Helicobacter pylori strains: a matched case-control study. Journal of Clinical Microbiology. 2014;52(8):2984–2989. doi: 10.1128/jcm.00551-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marie M. A. M. Relationship between Helicobacter pylori virulence genes and clinical outcomes in Saudi patients. Journal of Korean Medical Science. 2012;27(2):190–193. doi: 10.3346/jkms.2012.27.2.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falsafi T., Khani A., Mahjoub F., Asgarani E., Sotoudeh N. Analysis of vacA/cagA genotypes/status in Helicobacter pylori isolates from Iranian children and their association with clinical outcome. Turkish Journal of Medical Sciences. 2015;45(1):170–177. doi: 10.3906/sag-1311-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of this study are available within the article.


