Abstract
Stroke constitutes the second leading cause of death and a major cause of disability worldwide. Stroke is normally classified as either ischemic or hemorrhagic stroke (HS) although 87% of cases belong to ischemic nature. Approximately 700,000 individuals suffer an ischemic stroke (IS) in the US each year. Recent evidence has denoted a rather pivotal role for defective macroautophagy/autophagy in the pathogenesis of IS. Cellular response to stroke includes autophagy as an adaptive mechanism that alleviates cellular stresses by removing long-lived or damaged organelles, protein aggregates, and surplus cellular components via the autophagosome-lysosomal degradation process. In this context, autophagy functions as an essential cellular process to maintain cellular homeostasis and organismal survival. However, unchecked or excessive induction of autophagy has been perceived to be detrimental and its contribution to neuronal cell death remains largely unknown. In this review, we will summarize the role of autophagy in IS, and discuss potential strategies, particularly, employment of natural compounds for IS treatment through manipulation of autophagy.
Keywords: Adaptive autophagy, Cell death, Cerebral I/R injury, Ischemic stroke, Maladaptive autophagy
1. Introduction - an overview of autophagy machinery in neuronal cells
Macroautophagy typically referred to as “autophagy”, is a cellular mechanism to sequester damaged/aged organelles, superfluous proteins, and cellular components (Klionsky, et al., 2021; Mizushima & Levine, 2020; Y. Yang & Klionsky, 2020; Yingmei Zhang, Sowers, & Ren, 2018). In this process, cargos are enwrapped by a transient double-membrane structure termed a phagophore, which then expands and matures into a double-membrane autophagosome. The formed autophagosome subsequently fuses with a lysosome, leading to the breakdown of cargo contents and recycling of building blocks of autophagic substrates (Ajoolabady, Aghanejad, et al., 2020; A. Ajoolabady, et al., 2021; Klionsky, et al., 2021; Levine & Kroemer, 2019; S. Wang & Ren, 2018). Neurons are particularly vulnerable to various intrinsic and extrinsic insults including exposure to metabolic derangement, ischemia/reperfusion (I/R), energy crisis, neurotoxins, neurodegeneration, physical trauma, and inflammation (A. Ajoolabady, et al., 2021; L. Galluzzi, Bravo-San Pedro, Blomgren, & Kroemer, 2016). Under these stressful conditions, autophagy is thought to be activated to various degrees by stress to secure survivability of neuronal cells and the overall wellbeing of central nervous system through maintenance of neuronal homeostasis, clearance of protein aggregates and damaged mitochondria, preservation of energy balance via recycling of amino acids, fatty acids, and glucose, as well as alleviation of endoplasmic reticulum (ER) stress (Peker & Gozuacik, 2020; Ren, Bi, Sowers, Hetz, & Zhang, 2021). To this end, mild to moderate induction of autophagy may act as a pro-survival mechanism in neuronal cells, a process often being referred to as “adaptive/mild/cytoprotective autophagy”. However, increases in autophagy are not always favorable for neuronal health (Lorenzo Galluzzi, et al., 2012; R. Shi, et al., 2012). Excessive rises in autophagic activity might lead to cytosolic accumulation of autophagosomes and enhanced degradation of essential cellular components (Lorenzo Galluzzi, et al., 2012; Ren & Taegtmeyer, 2015; R. Shi, et al., 2012). While the concept of autophagic cell death (ACD) as a maladaptive feature of autophagy has been supported multiple studies, although this concept is still debatable (Button, Luo, & Rubinsztein, 2015). To test whether excessive induction of autophagy is indeed a causative factor for cell death, interpretations must be rooted from manipulating autophagy to the normal physiological state (maintaining baseline autophagy) rather than the null state (complete depletion of autophagy) (Berry & Baehrecke, 2007). Unfortunately, many conclusions and reports for autophagy are essentially based on inhibiting autophagy completely (null state) rather than maintaining autophagy at the baseline level (physiological state). In another word, excessive induction of autophagy might be a salvage effort to restore cellular homeostasis under lethal stresses, which might take part in cell death execution rather than direct induction of cell death, in a fashion similar to the role of ATP in apoptosis (albeit not directly inducing apoptosis) (S. Shen, Kepp, & Kroemer, 2012). Thus, in mammalian cells, multiple mechanisms may contribute to cell death upon excessive autophagy, and dependency on the cellular context, although duration and extent of autophagy necessary for cell death induction remain largely elusive (A. Ajoolabady, et al., 2021; Das, Shravage, & Baehrecke, 2012). Therefore, unrestrained induction of autophagy (or unchecked autophagy) may function as a pro-death mechanism and is referred to as “maladaptive/excessive/unchecked autophagy” (Bar-Yosef, Damri, & Agam, 2019).
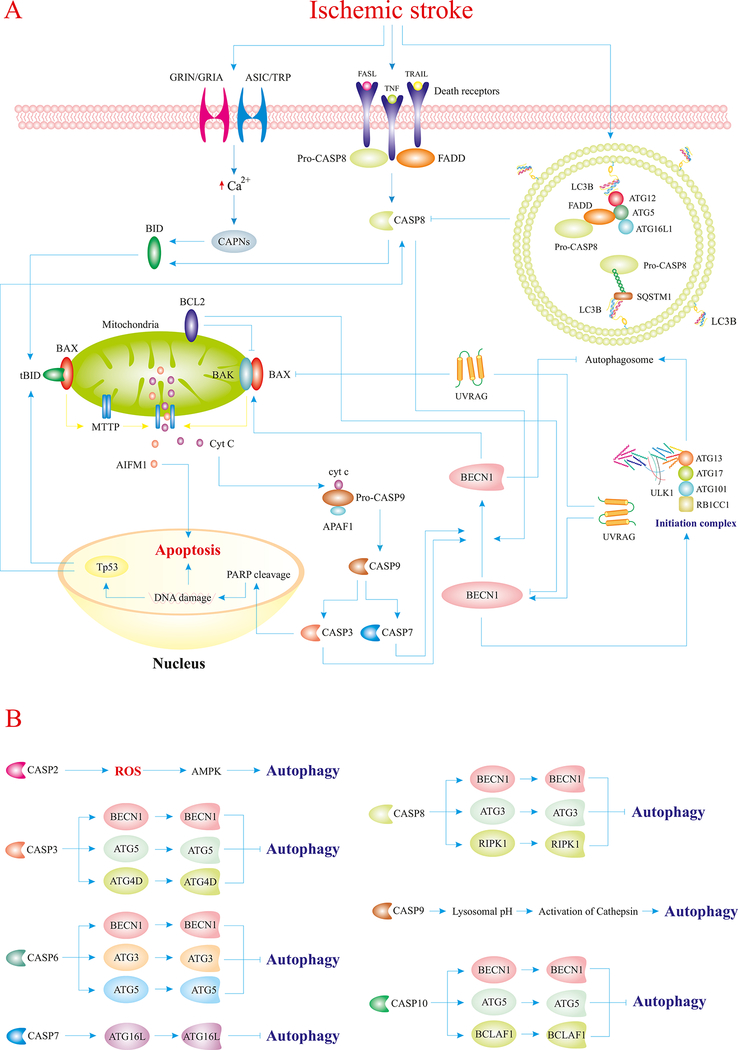
Both adaptive and maladaptive autophagy are regulated by a hierarchy of proteins; namely, the autophagy-related (ATG) protein family. To date, over 30 mammalian ATG proteins have been identified, which regulate different steps of the autophagy pathway encompassing the formation and nucleation of autophagosomes, as well as the organization of autolysosomes (Fig. 1) (Yingmei Zhang, et al., 2018). mTORC1 is a master regulator of autophagy that inhibits autophagy initiation via phosphorylation of ULK1 (Ajoolabady, Aghanejad, et al., 2020). Depending on the nature of stress, various molecules and regulators form a complex network of signaling pathways that regulate autophagy. For instance, in the setting of IS, cerebral hypoperfusion causes nutrient deprivation, oxidative stress, and ER stress, all of which trigger multiple signaling cascades leading to autophagy induction in neurons (Fig 1, 2, 3) (Jiaojiao Pang, et al., 2016; Ren, Sowers, & Zhang, 2018; Tan, Gong, Dong, Pei, & Ren, 2019; Yingmei Zhang, et al., 2018).
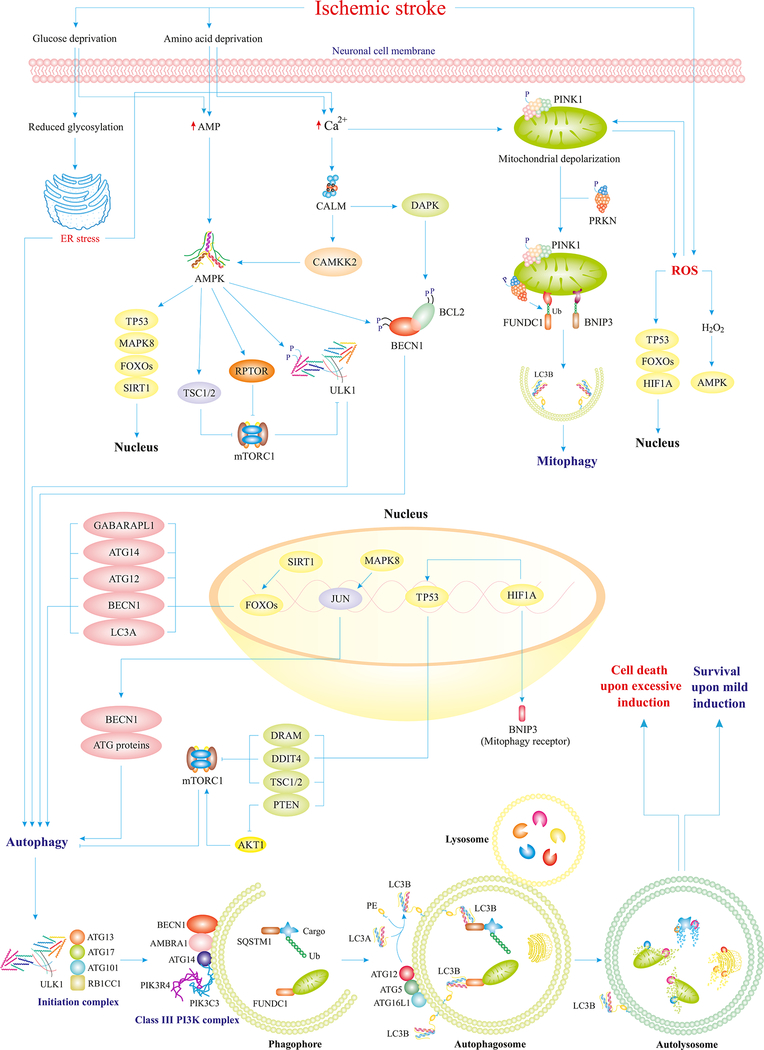
Fig. 1.
IS perturbs autophagy homeostasis through distinct mechanisms. IS provokes deprivation of oxygen and nutrient as a result of interrupted blood flow. Loss of glucose and amino acids overtly promotes accumulation of AMP, Ca2+, and ROS in the cytosol of neurons. Overproduction of AMP turns on the master regulator of autophagy, AMPK, which facilitates upregulation of ATG proteins, inhibition of mTORC1, and activation of ULK1. Elevated levels of Ca2+ also activate AMPK and depolarize mitochondrial membrane, which would in turn evoke mitophagy and ROS production. In response to ROS generation, AMPK and transcriptional factors are activated to further promote ATG proteins and mitophagy receptors. Ultimately, activated ATG proteins pave the way for the initiation of autophagy to foster the clearance of long-lived or damaged organelles, and superfluous proteins through the autophagosomal-lysosomal fusion. ULK1-ATG13-ATG101-RB1CC1/FIP200 form the autophagy initiation complex, BECN1-NRBF2-ATG14-PIK3R4/VPS15-PIK3C3/VPS34 form the class III phosphatidylinositol 3-kinase complex, which aids the initiation complex in the formation of phagophores, and the ATG12–ATG5-ATG16L1 complex mediates lipidation of LC3A and formation of autophagosomes around damaged organelles or certain cargos. Excessive induction of autophagy results in cell death (ACD/autosis), whereas normal/mild induction improves neuronal survival in IS (Ajoolabady, Aghanejad, et al., 2020; Ghislat & Knecht, 2015; Mrakovcic & Fröhlich, 2018; J. Ren, et al., 2020; Simon, Friis, Tait, & Ryan, 2017).
Abbreviations: CAMKK2 (calcium/calmodulin dependent protein kinase kinase 2), DAPK (death associated protein kinase), Ub (ubiquitin), FUNDC1 (FUN14 domain containing 1), RPTOR/raptor (regulatory associated protein of mTOR complex 1), JUN/C-Jun (Jun proto-oncogene, AP-1 transcription factor subunit), GABARAPL1 (GABA type A receptor associated protein like 1), DRAM (DNA damage regulated autophagy modulator), DDIT4 (DNA damage inducible transcript 4), RB1CC1/FIP200 (RB1 inducible coiled-coil 1), NRBF2 (nuclear receptor binding factor 2), PIK3R4/VPS15 (phosphoinositide-3-kinase regulatory subunit 4), UVRAG (UV radiation resistance associated gene), PE (phosphatidylethanolamine).
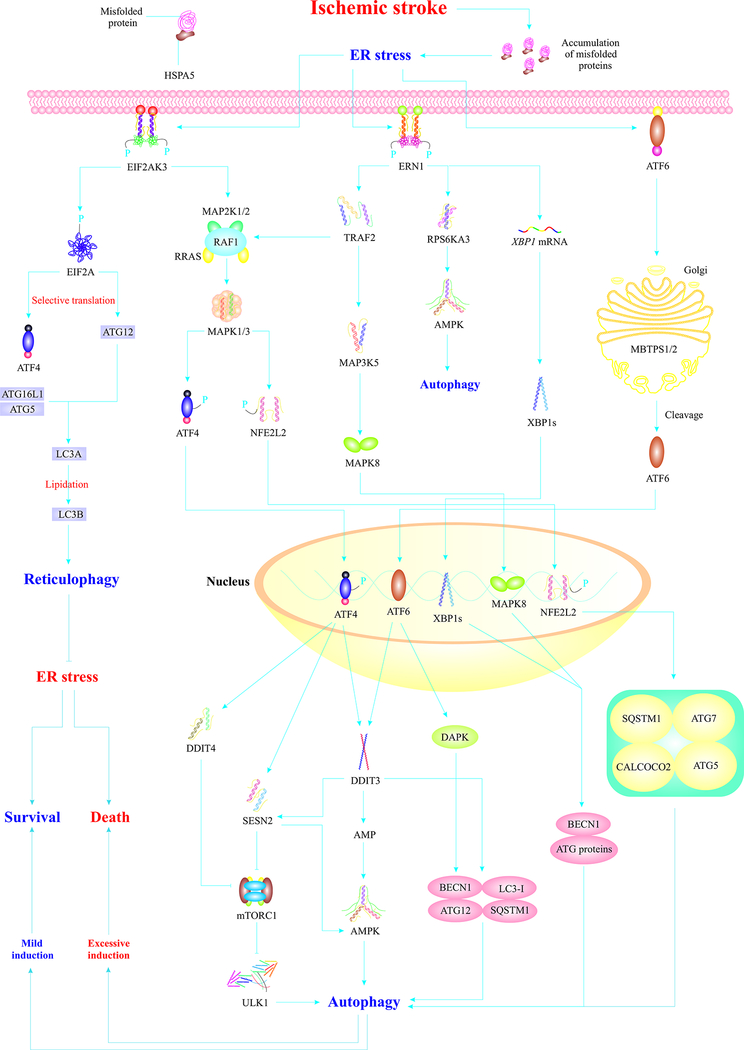
Fig. 2.
IS induces ER stress, which triggers autophagy through three major ER stress sensors. IS-induced ER stress mediates EIF2AK3 dimerization and activation. EIF2AK3 phosphorylates/activates EIF2A/eIF2α, which induces reticulophagy (selective autophagy of the ER) via selective translation of ATG12 leading to alleviation of ER stress. EIF2AK3 primarily induces adaptive autophagy via activation of ATF4 and NFE2L2/Nrf-2 proteins, which translocate to the nucleus and induce autophagy via distinct machineries. AMPK activation, upregulation of BECN1 and ATG genes, and mTORC1 inhibition are the main mechanisms of ATF4-mediated autophagy. NFE2L2/Nrf-2 also induces autophagy through upregulation of autophagy-associated proteins. Upon ER stress insult, ERN1 also undergoes autophosphorylation and homodimerization. Activated ERN1 induces activation of MAPK8 which translocates to the nucleus and induces autophagy via the upregulation of BECN1 and ATG genes. The endoribonuclease domain of ERN1 cleaves XBP1 mRNA, which produces XBP1s that translocates to the nucleus and induces autophagy through upregulation of BECN1 and ATG genes. The third branch of UPR, ATF6, also translocates to the Golgi and gets cleaved by MBTPS1/S1P and MBTPS2/S2P, which produces activated ATF6 which translocates to the nucleus, triggers adaptive responses, and upregulates DAPK, to evoke autophagy. Autophagy initially acts as an adaptive response element that relieves ER stress. However, severe ER stress induces maladaptive autophagy, leading to neuronal cell death (Amir Ajoolabady, et al., 2021; Fusakio, et al., 2016; Jurkin, et al., 2014; Papaioannou, et al., 2018; Radanovic, et al., 2020; Urra, et al., 2018; D. Wang, et al., 2018; Yucel, et al., 2019)
Abbreviations: HSPA5/Bip (heat shock protein family A (Hsp70) member 5), MAP2K½(mitogen-activated protein kinase kinase ½), RRAS (RAS related), RAF1 (Raf-1 proto-oncogen, serine/threonine kinase), EIF2A/eIF2α (eukaryotic translation initiation factor 2A), TRAF2 (TNF receptor associated factor 2), MAP3K5 (mitogen-activated protein kinase kinase kinase 5), RPS6KA3 (ribosomal protein S6 kinase A3), XBP1 (X-box binding protein 1), MBTPS½ (membrane bound transcription factor peptidase, site ½), MAPK3/ERK1 (mitogen-activated protein kinase 3), XBP1s (spliced X-box binding protein 1), ATF4 (activating transcription factor 4), DDIT3/CHOP (DNA damage inducible transcript 3), SESN2 (sestrin 2), AMP (adenosine monophosphate).
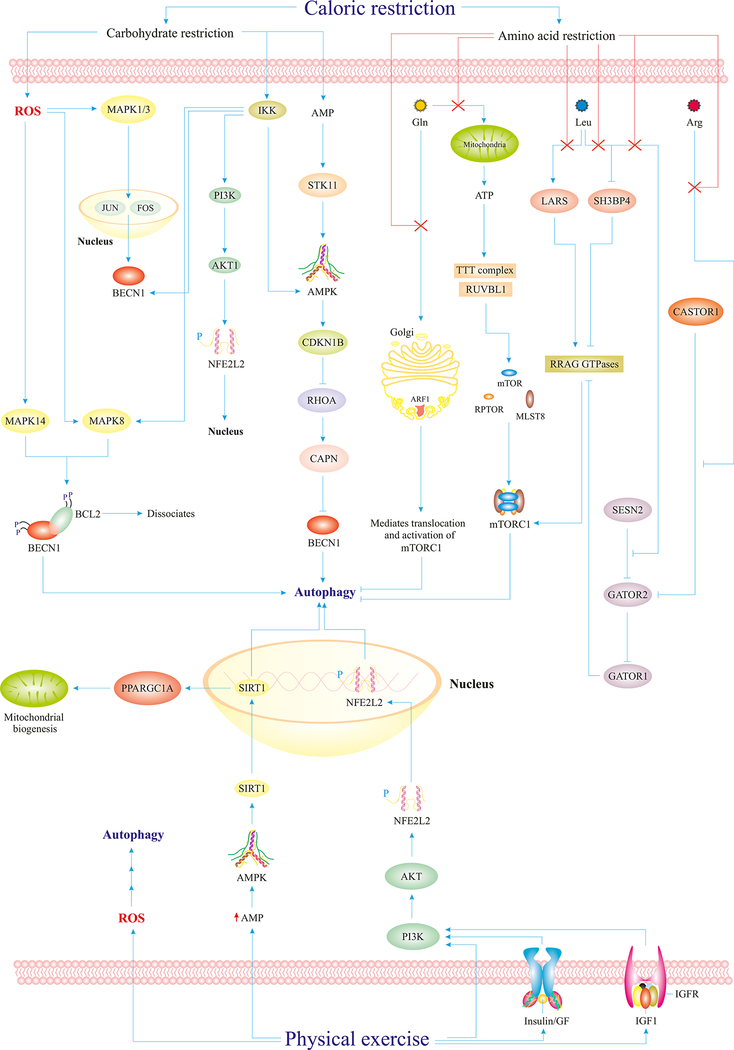
Fig. 3.
Caloric restriction and physical exercise maintain adaptive autophagy. Carbohydrate restriction induces mild generation of ROS in the cytosol, which activates the MAPK1/ERK2-MAPK3/ERK1 pathways leading to the upregulation of BECN1. Besides, IKK activation mediates activation of potential autophagy regulators and elements such as AMPK, BECN1, and MAPK8. The PI3K-AKT1-NFE2L2/Nrf-2 pathway also leads to nuclear translocation of NFE2L2/Nrf-2 and autophagy activation. Both carbohydrate and amino acid restriction culminate in accelerated AMP levels in the cytosol and subsequent AMPK activation. Reduced Gln uptake blocks Golgi-mediated mTORC1 activation and ATP-mediated formation of mTORC1 components. Leu restriction mediates SH3BP4 activation and LARS/LRS and GATOR2 inhibition all of which culminates in autophagy induction via suppression of the RRAG GTPases-mTORC1 cascade. Similarly, Arg restriction also places a stop codon on GATOR2, which results in GATOR1 activation and subsequent blockade of RRAG GTPases and mTORC1, and, thus, autophagy activation. Physical exercise also mediates autophagy activation via reduced cytosolic AMP level, activation of the PI3K-AKT1-NFE2L2/Nrf-2 pathway, and mild ROS generation.
Abbreviations: IKK (inhibitor of nuclear factor-kappa B kinase subunit beta), Gln (Glutamine), Leu (leucine), Arg (arginine), SH3BP4 (SH3 domain-binding protein 4), LARS/LRS (leucyl-tRNA synthetase), STK11/LKB1 (serine/threonine kinase 11), FOS/C-Fos (Fos proto-oncogene, transcription factor subunit), CDKN1B/p27/Kip1 (cyclin dependent kinase inhibitor 1B), TTT (Tel2-Tti1-Tti2) complex, RUVBL1 (RuvB like AAA ATPase 1), CASTOR1 (cytosolic arginine sensor for mTORC1 subunit 1), RRAG (Ras related GTPl binding), MLST8 (mTOR associated protein, LST8 homolog), ARF1 (ADP ribosylation factor 1), MAPK14/p38 (mitogen-activated protein kinase 14), CAPN (calpains protein family), GATOR2 (GTPase activating proteins toward Rags subcomplex 2), GATOR1 (Gap Activity Toward Rags 1), NFE2L2/Nrf-2 (nuclear factor, erythroid 2 like 2), PPARGC1A/PGC1-α (PPARG coactivator 1 alpha), GF (growth factor), IGFR (insulin like growth factor 1 receptor).
Of note, numerous studies have indicated the role of autophagy in the pathogenesis and treatment of cardiovascular, metabolic, neurodegenerative, and infectious diseases (A. Ajoolabady, et al., 2021; Deretic, Saitoh, & Akira, 2013; J.-G. Li, Chu, & Praticò, 2018; J. Pang, et al., 2019; Ren & Zhang, 2018; S. Wang & Ren, 2018; S. Wang, et al., 2020). In the setting of cardiac ischemia, autophagy protects myocardium and cardiomyocytes from ischemia-induced damage and apoptosis (Gustafsson & Gottlieb, 2009). In addition, induction of basal autophagy is a cytoprotective mechanism to attenuate the onset and progression of neurodegeneration in mouse models of Parkinson’s, Huntington’s, and Alzheimer’s diseases (Nixon, 2013; S. Wang, et al., 2020). Despite technological and biological advances in cerebrovascular research, tPA is approved by FDA and remains a major treatment option for IS (Fukuta, et al., 2017). Yet, many IS patients are at high risk of serious cerebral damage due to narrowed therapeutic window, reperfusion damage, and rehemorrhagic complications (Rostami, et al., 2019; Wei, Wang, & Miao, 2012). Thus, exploring autophagy in neural tissues in the face of ischemia challenge should provide novel neuroprotective information for both prevention and treatment of stroke.
IS often triggers maladaptive autophagy. Therefore, post-ischemic period is accompanied by maladaptive autophagy. In this regard, pre-ischemic administration of melatonin inhibited ER stress-induced excessive (maladaptive) autophagy (Fig. 2), leading to alleviation of acute neural injury following IS challenge (Feng, et al., 2017). This finding suggests the utility of pre-ischemic interventions in combating post-ischemic maladaptive autophagy and neural damage, and also consolidates the maladaptive nature of autophagy in the post-ischemic period. Besides, DNM1L-dependent mitophagy (selective autophagy of mitochondria) was activated in an MCAO rat model and OGD PC12 cells during early ischemia phase and assisted removal of damaged mitochondria (Fig. 1). In this context, adaptive autophagy may also occur at the early stage of ischemia (Zuo, et al., 2014).
However, limb remote ischemic preconditioning 30 minutes after ischemia induced AKT1-mediated BCL2/BECN1 disruption (Fig. 1) and consequently adaptive autophagy, contributing to alleviation of mitochondrial damage in IS. Therefore, limb remote ischemic preconditioning is an effective remedy to activate adaptive autophagy in the face of IS (Qi, et al., 2015). Also, in MCAO rats, DEX postconditioning alleviated memory dysfunction and boosted learning via amelioration of inflammation and maladaptive autophagy through inhibition of MAPK8/JNK1 (Fig. 1) (Y. Zhu, et al., 2019). Thus, DEX postconditioning plays an essential role in IS treatment and maladaptive autophagy cessation. Besides, cerebral ischemia postconditioning also suppresses maladaptive autophagy and HMGB1 secretion (a mediator of injury) in cellular OGD (8h) and reperfusion (24h) models, indicating the protective role of ischemic postconditioning in IS treatment (J. Wang, Han, Sun, & Feng, 2016).
Furthermore, subjecting Sprague-Dawley rats to ischemic preconditioning was shown to turn on AMPK (Fig. 1), adaptive autophagy and consequently reduced infarct volume, apoptosis, neurological deficits, and cerebral ischemia. This study indicates that ischemic preconditioning renders neuroprotection through induction of adaptive autophagy (T. Jiang, et al., 2015). Cortical spreading depression preconditioning, which mimics peri-infarct depolarization, significantly suppressed apoptosis, attenuated neurological deficits, and infarct size via activation of AMPK-mediated adaptive autophagy in cortical penumbra in rat models (P. Shen, et al., 2017). Therefore, cortical spreading depression preconditioning and adaptive autophagy induction may be potential strategies to jack up ischemic tolerance of the brain. Indeed, preconditioning and postconditioning interventions have shown promises as new neuroprotective measures. In this regard, examination of ischemic preconditioning also revealed the upregulation of TSC1 (Fig. 1) in hippocampal CA1 neurons, which protected these neurons against ischemia both in vitro and in vivo. TSC1-rendered neuroprotection was attributed to induction of “productive autophagy” (i.e., effective completion of autophagic flux) through a mTORC1-dependent inhibitory mechanism (Fig. 1) (Papadakis, et al., 2013; Rajanikant & Sugunan, 2013). Therefore, maintaining productive autophagy, which can be achieved by ischemic preconditioning, is of paramount importance for neural survival. Here in this review, we will try to decipher the Janus faces of autophagy (adaptive or maladaptive) in neurons in IS and summarize potential therapeutic strategies for IS via autophagy manipulation.
2. An overview of pathophysiology in stroke
Ischemia in the central nervous system leads to pronounced glucose and oxygen deficiency and disturbance of ionic gradients in neural cells. In consequence, depolarized neurons experience intense glutamate efflux and Ca2+ influx, which trigger cell death mechanisms including necrosis and apoptosis (Fang, Wu, & Ren, 2006; Lipton, 1999; A. Sun & Ren, 2013; H. Xu, Zhang, & Ren, 2019). Thus, it is proposed that excitotoxicity is one of the underlying mechanisms of ischemia-mediated neuronal damage. Although the role of Ca2+ in excitotoxicity is still enigmatic, intracellular accumulation of Ca2+ induces mitochondrial damage, production of ROS and activation of proteases and phospholipases, leading to the fate of cell death (Bano & Nicotera, 2007). In addition, Wang and coworkers reported that NMDARs play a vital role in cell death or survival fate upon ischemia. Furthermore, they found that neuronal gap junctions are vital for excitotoxicity, mediated by NMDARs, denoting the importance of gap junctions in IS-induced neuronal cell death (Y. Wang, et al., 2010). Therefore, therapeutic intervention of excitotoxicity should potentiate the recovery of stroke insult.
Mitochondrial dysfunction upon ischemia challenge induces generation of free radicals and exacerbates oxidative stress (Steinberg, et al., 2014). Among various culprit factors triggering mitochondrial ROS production, NOX5 is known to evoke accumulation of ROS during ischemia (Moskowitz, Lo, & Iadecola, 2010). Meanwhile, suppression of TRPM7 in hippocampal CA1 neurons was reported to potentiate neuronal resistance to ischemia. Such neuroprotection is believed to related to TRPM7-induced Ca2+ accumulation in the cytosol and subsequently activation of maladaptive machineries (H.-S. Sun, et al., 2009). Therefore, implementing novel strategies to scavenge ROS and free radicals may alleviate ischemia-induced damages in the brain.
ER organelle stores a vast quantity of intracellular Ca2+ and mediates protein folding in ER lumen and protein synthesis on ER membrane (Ren, et al., 2021; Roussel, et al., 2013). However, ischemia induces onset of ER stress (Fig. 2) to disturb these physiological processes, resulting in the accumulation of misfolded proteins. Extensive or maladaptive ER stress activates ER stress-responding proteins including EIF2AK3/PERK, ERN1/IRE1α, and ATF6, leading to cell death (probably due to excessive autophagy or apoptosis) and neural injury (Sano & Reed, 2013). Here in this review, we aim to delineate the role of excessive/maladaptive autophagy in the execution of cell death and neural injury ensuing IS.
3. Autophagy in IS
As mentioned above, autophagy process serves as a double-edged sword that can heal or deteriorate injured neurons upon ischemia insult. Excessive or persistent activation of autophagy is detrimental to neurons, by way of activation of cell death mechanisms. Although a number of researchers believe that maladaptive autophagy does not necessarily execute cell death via massive degradation of cellular components. Rather, excessive autophagy can contribute to the execution of other cell death mechanisms such as apoptosis (Button, et al., 2015). Thus, in this section, we will attempt to compare maladaptive versus adaptive autophagy machineries under ischemia and the association between maladaptive autophagy and apoptosis. Furthermore, we briefly explain the fine-tune of autophagy in different regions of the brain and the impact of age and gender in various forms of cell deaths upon maladaptive autophagy.
3.1. Maladaptive autophagy
Maladaptive autophagy (excessive autophagy induction) has been widely reported in the current literatures with regards to the role of autophagy in IS. Disruption of BBB contributes to neurological impairment upon IS, which demonstrates the importance of BBB integrity for IS alleviation (Changjun Yang, Hawkins, Doré, & Candelario-Jalil, 2019). OCLN is a 65-kDa integral protein that plays an important role in intracellular tight junctions stabilization and thereby integrity of BBB (Cummins, 2012). In this regard, findings from in vivo and in vitro IS models suggested that disruption of BBB integrity is partly driven by maladaptive autophagy, which is expected to promote OCLN sequestration via autophagosomes, and subsequently degradation of OCLN, resulting in elevated permeability for toxic substances in BBB. Thus, maladaptive autophagy is an important venue of IS-associated BBB damage, and a promising target to increase BBB integrity and neuronal survival in IS (K.-A. Kim, et al., 2020).
In murine and in vitro models of IS, SNHG3, a long non-coding RNA, is upregulated, resulting in downregulation of MIR485, leading to upregulation of ATG7 and BECN1, enhanced formation of MAP1LC3B/LC3B, and ultimately boosted autophagy, which would trigger neuronal cell apoptosis (Cao, Pan, Zhang, Guo, & Huang, 2020). Despite the pitfall of employing 3-MA for the evaluation of the contribution of autophagy to cell death, data from other studies have indicated that ATG7 might be relevant to apoptotic cell death in neurons and other mammalian cells. For instance, chloroquine (CQ) and bafilomycin A1 (Baf A1) treatment of neural precursor cells may induce lysosome dysfunction and subsequently accumulation of autophagosomes, which prompt TP53/p53 phosphorylation, CASP3 activation, and apoptotic cell death (Walls, et al., 2010). Meanwhile, ATG7 knockdown was shown to significantly inhibit apoptotic cell death, suggesting the ATG7-dependency of neuronal apoptosis upon autophagosome accumulation. However, ATG7 knockdown did not affect starvation-induced autophagosome accumulation and cell death (Walls, et al., 2010). Another piece of evidence for possible involvement of ATG7 in apoptotic cell death was from acute kidney injury models where ATG7 was found to mediate renal cell apoptosis through interacting/activating PRKCD upon a vancomycin-induced accumulation of autophagosomes and thus acute kidney injury in vivo and in vitro (X. Xu, et al., 2019).
More evidence has suggested that upregulation of FKBP5 serves as a potential contributor for maladaptive autophagy upon IS through disruption of AKT1-FOXO3 complex and hyper-activation of FOXO3 (S. Yu, Yu, Bu, He, & Feng, 2020). FKBP5 was considered to interact with AKT1 and thus block its phosphorylation/activation. In consequence, deactivated AKT1 would not be able to phosphorylate/inactivate FOXO3, leaving FOXO3 active to upregulate autophagy-associated genes (Fig. 1) (Boonying, et al., 2019; S. Yu, et al., 2020). Furthermore, MTMR14 is a phosphoinositide 3-phosphatase and its deficiency mediated Ca2+ leakage from the ER due to the accumulation of MTMR14 substrates including PtdIns(3,5)P2 and PtdIns(3,4)P2 in muscle cells (J. Shen, et al., 2009). Pan and coworkers reported downregulation of MTMR14 in an in vitro model of cerebral I/R injury, resulting in abundant formation of LC3B, and expression of BECN1, and PTEN, ultimately provoking excessive induction of autophagy and neuronal cell death. These authors also showed that MTMR14 protected against cerebral I/R injury by interacting with PTEN to dampen its expression (Pan, Liu, Wang, Wen, & Wang, 2020). However, exactly how MTMR14 modulates PTEN expression is not entirely clear. Given that PTEN is also a phosphoinositide 3-phosphatase (Matsuoka & Ueda, 2018), it might be speculated that MTMR14 and its products might allosterically modulate PTEN activation (Campbell, Liu, & Ross, 2003). Data of this study also revealed that excessive autophagy may contribute to neuronal cell death, based on beneficial response of autophagy inhibition using 3-MA. Nonetheless, neuronal cell death might be explained by other means such as apoptosis considering the upregulation of PTEN and BECN1. In this regard, PTEN was reported to downregulate/deactivate the PI3K/PIK3C3-AKT1 signaling cascade (Weng, Brown, & Eng, 2001) that not only culminates in autophagy but also provokes apoptosis (H. Yu, et al., 2020). Besides, PTEN-induced apoptotic signals would activate caspases which can cleave the upregulated BECN1 molecules, leading to cleaved BECN1-mediated apoptosis (R. Kang, Zeh, Lotze, & Tang, 2011). Taken together, when excessive autophagy and apoptosis coexist under cerebral I/R conditions, neuronal cell death might be the ultimate fate from a complex interplay between autophagy and apoptosis, although further verification is still warranted. This complex interplay of cell death was also observed in neonatal hypoxia-ischemia (H/I), accompanied by excessive autophagy and autophagosome/autolysosome accumulation, resulting in apoptotic cell death in cortical neurons. ATG7 silencing (a gold-standard for autophagy ablation) and 3-MA treatment substantially attenuated CASP3 activation, indicating excessive autophagy-triggered apoptosis in cortical neurons under H/I condition. By contrast, cell death was noted in hippocampal neurons independent of apoptosis and likely through ACD (Ginet, Puyal, Clarke, & Truttmann, 2009). Moreover, murine neonatal cerebral H/I dramatically boosted autophagy and autophagosome formation, resulting in both CASP3-dependent and - independent hippocampal neuronal cell death. ATG7 deficiency in mice inhibited H/I-induced hippocampal neuronal cell death, revealing a role for excessive autophagy in both apoptotic and non-apoptotic cell deaths. Also, H/I induced excessive autophagy and exclusively CASP3-independent cell death in neonatal mice (Koike, et al., 2008). Therefore, one can hypothesize that excessive autophagy predisposes to apoptosis through yet undefined mechanisms. Furthermore, the influence of age and gender on different forms of cell death in the face of ischemic injury must be taken into consideration, as apoptotic mechanisms are more noticeable in neonatal murine brains, necrosis and oxidative stress are alike at all ages, and excessive autophagy is apparently more pronounced in adult mouse brains. Besides, AIFM1 translocation is more prominent in fetal male brains, whereas CASP3 activation is more accentuated in female brains (C Zhu, et al., 2005; Changlian Zhu, et al., 2006).
In PC12 cells, cortical neurons and murine IS models, ischemia was shown to facilitate excessive autophagy through upregulation of MYH9/NMMHC-IIA, which, by interacting with F-actin, promotes trafficking of ATG9A, a core autophagy protein involved in lipid transfer to the expanding phagophore (G. Wang, et al., 2020). Using 3-MA, data from this study suggested that excessive autophagy culminated in ACD, although inhibiting the interaction between MYH9 and F-actin or blocking F-actin polymerization suppressed excessive autophagy and thus mitigated IS-induced neurological defects in mice (G. Wang, et al., 2020). Adverse effects of inhibiting F-actin polymerization to block excessive autophagy should be investigated. Moreover, enhanced ATG9A trafficking might trigger other yet unknown signals, contributing to the ultimate cell death independent of excessive autophagy. Future work should try to reveal how increased ATG9A trafficking affects transport of other molecules, particularly ATG proteins, on cytoskeleton scaffolds, which might lead to perturbation of cellular trafficking and initiation of death pathways.
IS also induced overexpression of RTN4 and RTN4 receptors, which mediate unchecked autophagy via enhanced expression of LC3A, BECN1, and SQSTM1/p62, and reduced expression of RHOA and ROCK1, leading to secondary neuronal damage in murine ipsilateral thalamus (W. Xu, et al., 2020). However, further studies are required to determine the exact nature of cell death and possible involvement of autophagy (W. Xu, et al., 2020). Examination of NOD1 in cerebral I/R injury in rat cortical neurons revealed upregulation of NOD1 and inhibition of its ubiquitination, which triggered irreversible ER stress and consequently excessive autophagy and apoptosis (X. Ma, et al., 2020). Therefore, it is conceivable that IS induces excessive autophagy and apoptosis through mediating irreversible (likely “maladaptive”) ER stress. Moreover, IS may contribute to neuronal death via heme-induced irreversible ER stress. Irreversible ER stress was also found to induce excessive autophagy which contributed to apoptotic cell death through a mechanism likely dependent on BECN1 and ATG5. These findings have offered compelling evidence that excessive autophagy induces cell death through apoptosis rather than ACD (Z. Yang, et al., 2020). Of note, irreversible ER stress also directly triggers apoptosis without engagement of autophagy signaling (Deniaud, et al., 2008).
Luo and coworkers demonstrated maladaptive autophagy induction in a murine MCAO model as well as in an in vitro OGD IS model. Their data showed that MEG3 blocks the inhibitory effect of MIR378 (microRNA 378) on GRB2, resulting in GRB2 upregulation, attenuated phosphorylation of the AKT1-mTORC1 signaling axis, BECN1 activation, accumulation of autophagosomes, and ultimately neuronal cell death (Luo, et al., 2020). Moreover, such excessive autophagy culminated in ACD as evidenced by the fact that 3-MA offered amelioration in neuronal cell death through autophagy ablation.
Although 3-MA is widely utilized for autophagy inhibition, results based on this compound should be evaluated and interpreted with caution (Y.-T. Wu, et al., 2010). 3-MA, LY294002, and wortmannin are PI3K inhibitors, which block Class I and III PI3K, leading to both activation and suppression of autophagy respectively (Kong & Yamori, 2008). This is because Class I PI3K mediates AKT1-mTORC1 axis activation and autophagy suppression, whereas Class III PI3K induces autophagy through forming complexes with ATG proteins (Itakura, Kishi, Inoue, & Mizushima, 2008).
Therefore, another interpretation for GRB2-mediated neuronal cell death could be that excessive autophagy was accompanied by apoptosis, which derived neuronal cell death. GRB2-mediated inactivation of AKT1, which triggers apoptosis and accelerates autophagy through mTORC1 inactivation, resulting in a maladaptive accumulation of autophagosomes and pro-apoptotic signals (Chung, et al., 2019; Song, et al., 2016). It is also important to examine the possible bi-directional regulation between excessive autophagy and apoptosis, from their initiation until cell death induction using real-time monitoring, in order to find novel crosstalk molecules or common pathways that link these two processes.
Of note, there is a controversy between finding from Luo and colleagues and data of others in terms of the role of GRB2 on AKT activation. A number of studies have indicated that GRB2 upregulation activates PI3K, which, in turn, phosphorylates/activates AKT1 in mammalian cells (tumor cells) (Ijaz, et al., 2017; Yuan, et al., 2020). Thus, GRB2-mediated activation of AKT1 can lead to phosphorylation mTORC1 and consequently suppression of autophagy (Fig. 1). However, report from Luo and associates showed that GRB2 upregulation induced excessive autophagy via attenuation of AKT and mTORC1 phosphorylation. In this context, more evidence is needed to discern this controversy noted between tumor cells and neurons under ischemia challenge.
In N2a cells and rat IS models, upregulation of MIR202–5p dramatically downregulated EIF4E, leading to decreased levels of LC3B and autophagosomes, and activation of the AKT1-GSK-3β axis which blocks apoptosis (B. Li, Huang, Meng, Yu, & Yang, 2020). However, IS evoked downregulation of MIR202–5p, upregulation of EIF4E, and suppression of the AKT1-GSK-3β axis, which provoked excessive autophagy and apoptosis (B. Li, et al., 2020). Suppression of AKT1-GSK-3β axis mediates GSK-3β activation that leads to initiation of autophagy via TSC1-mediated inhibition of mTORC1 (Guo, Liu, Cai, & Le, 2018). Additionally, GSK-3β activation triggers apoptosis via BCL2 inhibition (C. Zhang, et al., 2016). Thus, in this context, the AKT1- GSK-3β axis is a critical junction between autophagy and apoptosis. Although these studies failed to examine autophagy-dependency on IS-induced neurological deficits, the neuroprotective role of MIR202–5p might likely be attributed to suppression of excessive autophagy or apoptosis or a combination of both. Moreover, data of this study have shown that downregulation of MIR202–5p triggered EIF4E upregulation, which, in turn, attenuated phosphorylation of AKT1 and GSK-3β. Nonetheless, a controversy exists for the role of EIF4E in AKT1 activation. For instance, EIF4E mediated activation/phosphorylation of AKT1 via upregulation of Nibrin in fibroblasts (Culjkovic, et al., 2008). Also, EIF4E upregulated downstream effectors of AKT1, prompting amplification of AKT1 signaling (Culjkovic, et al., 2008). Therefore, more studies are warranted to shed more light on EIF4E-mediated regulation of AKT1 and GSK-3β in the context of IS. Taken together, IS can result in maladaptive autophagy through multiple pathways including GRB2-AKT1-mTORC1, MIR202–5p-EIF4E-AKT1- GSK-3β, SNHG3-MIR485-ATG7, RTN4-RTN4R, and MTMR14-PTEN signaling axis, all of which might predispose neurons to the fate of cell death.
3.2. Adaptive autophagy
Adaptive or cytoprotective autophagy also plays a cardinal role in the pathophysiology and management of IS. Timely removal of long-lived, superfluous proteins and damaged organelles by adaptive autophagy plays an essential role to attenuate IS-associated cerebral injury (L. Galluzzi, et al., 2016). Growing evidence suggests that dysfunction of adaptive autophagy contributes to neuronal apoptosis and necrosis in cerebral ischemia (Rao, et al., 2020). Blocking autophagy using ATG7 knockdown reinforced cerebral I/R injury-induced brain damage, cytochrome c release, and apoptosis both in vitro and in vivo (X. Zhang, et al., 2013). Besides, knockdown of PRKN, a protein involved in mitophagy, exacerbated I/R-induced neuronal cell death, indicating the cytoprotective nature of mild to moderate autophagy in neuronal cells under I/R (X. Zhang, et al., 2013). Wang and colleagues reported that autophagic degradation of GJA1/connexin-43 is mediated by SRC, PRKC/PKC, PINK1, OPTN, and CALCOCO2/NDP52, which averts apoptosis, supporting an adaptive role of autophagy in cerebral I/R. These researchers utilized rapamycin to confirm that specific autophagic degradation of GJA1 hindered apoptosis, although it is noteworthy that rapamycin is a poor choice to simulate specific autophagic degradation (X. Wang, et al., 2020). Also, increased MIR187–3p levels have been found in IS, resulting in the attenuation of BSCL2 protein and adaptive autophagy impairment in PC12 cells (Z. Ren, et al., 2020). In rat IS models, BSCL2 protein suppression is one of the possible mechanisms through which IS impairs adaptive autophagy (Z. Ren, et al., 2020). Report from an in vivo model of cerebral I/R injury revealed a mechanism for induction of adaptive autophagy in response to neuronal cell injury. Zhang and coworkers demonstrated that activation of CLCN3/CIC-3 leads to enhanced interaction and complex formation between BECN1 and PIK3C3/VPS34 which culminates in adaptive autophagy (B. Zhang, Deng, Zhou, & Fang, 2020). Finally, STX17 is upregulated in response to cerebral I/R injury, which facilitates the fusion between lysosomes and autophagosomes, leading to induction of adaptive autophagy and alleviation of ER stress (L. Chen, et al., 2020). Overall, adaptive autophagy tends to reverse undesired and deleterious effects evoked by IS although excessive or persistent induction of autophagy might lead to maladaptive effects of autophagy in neuronal cells.
3.3. Autophagy in focal cerebral ischemia (FCI)
The characteristics of autophagy appears to vary in distinct regions of brain lesions. For example, BECN1 was dramatically upregulated in the penumbra region 6 hours after ischemia in a rat model of FCI. Also, CASP3 upregulation was reported in BECN1-overexpressing cells, suggesting possible activation of apoptosis during excessive autophagy in the penumbra region (Rami, Langhagen, & Steiger, 2008). Ischemic postconditioning also initiated neuroprotection in the MCAO rat model of FCI through suppression of maladaptive autophagy (L. Gao, et al., 2012). Enhanced formation of autophagosomes, upregulation of LC3A, BECN1, and LAMP2, were noted in rat models of FCI and cultured primary astrocytes, which ultimately led to astrocyte injury and cell death. This study further denoted the maladaptive nature of autophagy during FCI, and thereby, inhibiting autophagy can confer neuroprotection (Qin, et al., 2010). Moreover, BECN1 and LC3A were upregulated after reperfusion under hyperbaric oxygen (HBO) preconditioning or ischemia in a MCAO-induced Sprague-Dawley rat model of transient FCI, to confer neuroprotection. Nonetheless, autophagy upregulation was more accentuated under HBO preconditioning (Yan, et al., 2011). Therefore, HBO preconditioning enhances adaptive autophagy in transient FCI, suggesting that adaptive or maladaptive nature of autophagy might be determined by transient versus prolonged FCI (Yan, et al., 2011). Hence, different parts of ischemic brain lesions or different phases of ischemia should be dissected with regards to autophagy targeting maneuvers.
3.4. Autophagy in global cerebral ischemia (GCI)
Autophagy might follow a different pattern upon GCI. In the context of GCI, function and level of mTORC1 were attenuated in CA1 neurons of the hippocampus, resulting in the upregulation of autophagy. This study reported for the first time that GCI induces autophagic degradation of mTORC1 to rescue hippocampal neurons against cell death (Hwang, et al., 2017). Besides, transient GCI induced maladaptive autophagy, which subsequently triggered apoptosis in hippocampal CA1 in an animal model of transient GCI. However, FGF2 upregulation attenuated GCI-induced apoptosis and autophagy, through inhibition of mitochondrial translocation of TP53 and mTORC1, respectively (D. Sun, et al., 2018). However, maintenance of adaptive autophagy is also deemed neuroprotective for GCI, as OPRD1-elicited neuroprotection was attributed to induction of autophagy via the AMPK/mTOR1/ULK1 pathway (Fig. 1) 3 days after ischemia (Lai, et al., 2020). Also, intermittent hypoxia preconditioning exacerbated neuronal damage upon GCI challenge via activation of excessive autophagy through mTORC1 inhibition in the hippocampus of Wistar rats (Y.-N. Zhao, et al., 2017). Thus, inhibition of maladaptive autophagy in the face of GCI should be considered a novel promising therapeutic approach. Furthermore, loss of mitochondrial membrane potential (MMP), mitochondrial swelling, maladaptive autophagy, necroptosis, and apoptosis were all reported upon GCI insult (Fakharnia, Khodagholi, Dargahi, & Ahmadiani, 2017). Therefore, it can be inferred that GCI induces excessive autophagy, which might contribute to cell death mechanisms including necroptosis and apoptosis. Overall, upon GCI, inhibition of maladaptive autophagy, while maintenance of basic autophagy seems to be the most preferred intervention, though, such approach remains rather challenging.
3.5. Autophagy in the ischemic penumbra and ischemic core
Examination of autophagy in the penumbra and core regions has offered new perspectives with regards to therapeutics of IS. For example, autophagy was upregulated 12–24 hours following the onset of ischemia, as manifested by elevated levels of BECN1 and LC3A in penumbra regions and ischemic core, in a murine model of FCI with NFKB1 knockout (W.-L. Li, et al., 2013). Mechanistically, the decline of the AKT1/mTORC1 axis contributed to autophagy upregulation, which was deemed maladaptive for its contribution to vascular and neuronal cell injury. Thus, NFKB1 knockout might play a role in cell death induced by excessive autophagy in the barrel cortex (W.-L. Li, et al., 2013). In a MCAO rat model of non-transient IS, LC3A and CASP3 were upregulated in the penumbra 5 hours following onset of ischemia prior to declination. It is believed that upregulation of LC3A and CASP3 contributed to the increment of cerebral infarction and neurological deficit (Deng, He, Yang, & Zhang, 2016). Thus, this study implies an obligatory role for maladaptive autophagy in apoptotic cell death in penumbra as an aftermath of ischemia. Moreover, in FCI induced by MCAO, pinocembrin (flavanone) treatment protected the penumbra area against I/R injury, the effect of which was attributed to adaptive autophagy and inhibition of apoptosis (Zhao, Zhang, Li, Wu, & Du, 2014). Therefore, inhibition of excessive/maladaptive autophagy and activation of adaptive autophagy jointly confer optimal neuroprotection against I/S in penumbra and core areas of the brain.
3.6. Autophagy in ischemic microglial and endothelial cells
Reminiscent of neurons, microglial and endothelial cells are heavily impacted by changes in autophagic flux in ischemia. Exosomes containing MIR-30d-5p obtained from adipose-derived stem cells (ADSC) exhibited neuroprotection against acute IS through suppression of autophagy and enhanced polarization of M2 microglia/macrophage in primary microglia subjected to OGD and a murine model of acute IS. Suppression of autophagy attenuated inflammatory response, cerebral infarction, and polarization of microglia to M1 (M. Jiang, et al., 2018), indicating the maladaptive autophagy nature of IS in microglial cells. Similarly, DEX postconditioning blocked maladaptive autophagy in microglia and hippocampal neurons following hypoxic-ischemic brain damage in neonatal rats (H. Xue, et al., 2021). Besides, scrutiny of tissue samples from ischemic penumbra revealed appearance of maladaptive autophagy following IS onset to evoke microglial inflammation through downregulation of CX3CL1 (a cytokine) in the MCAO rat model (H.-Y. He, Ren, Guo, & Deng, 2019). Moreover, autophagy facilitated microglial inflammatory damage and cerebral edema via downregulation of CX3CL1 in tissues obtained from ischemic penumbra in a rat model of MCAO (H.-Y. He, et al., 2019). Therefore, autophagy activation contributes to microglial injury in response to IS. Similarly, autophagy activation in microglial cells exacerbated edema formation, infarction size, inflammation, and ultimately, neurological deficits upon FCI triggered by permanent MCAO (Z. Yang, Zhong, Zhong, Xian, & Yuan, 2015). These findings further suggested maladaptive nature of autophagy in FCI in microglial cells. The neuroprotective effect of minocycline (an antibiotic) against IS has been reported recently. Subjecting HUVECs to OGD/R showed that a small dosage of minocycline upregulated autophagy and elicited neuroprotection (Dong, Xiao, Cheng, Ye, & Zheng, 2016). This finding implies that minocycline can be used as a therapeutic agent to induce adaptive autophagy in brain endothelial cells in the face of IS. Besides, in BMECs subjected to OGD/R, MALAT1 (a long noncoding RNA) conferred neuroprotection, courtesy of adaptive autophagy activation. Mechanistically, MALAT1 downregulated MIR-26b, and thereby, upregulated ULK2 and autophagy (Z. Li, Li, & Tang, 2017). Also, as mentioned above, maladaptive autophagy contributed to the disruption of BBB integrity and increased BBB permeability for toxic substances in brain endothelial cells (K.-A. Kim, et al., 2020). Therefore, IS triggers maladaptive autophagy in brain endothelial cells. To this end, it is rational to engage strategies to inhibit maladaptive autophagy in these cells.
3. 7. The vulnerability of autophagy associated genes to a high mutation rate
Many of genes displayed in Fig. 1 such as PINK1, AMPK, CALM, HIF1A, and FOXOs are turned on in response to IS challenge and are prone to mutations. For instance, homozygous nonsense mutation (Trp 437→X) in PINK1 gene contributed to mitochondrial respiratory defect in fibroblasts and early onset of parkinsonism (Piccoli, et al., 2008). In myocardial ischemia, mutation (Thr 400→Asn) in the PRKAG2 gene, which encodes γ2 subunit of AMPK, resulted in increased infarct size and apoptosis in a murine I/R model (Banerjee, et al., 2007). Besides, a mutation (phe 90→leu) in the CALM1 gene contributed to sudden arrhythmic death via modulation of Ca2+ release from sarcoplasmic reticulum (Kryshtal, et al., 2014), which, in turn, modulates autophagy (Law, et al., 2010). Moreover, Cerychova and coworkers showed that haploinsufficiency in HIF1A gene (HIF1A+/−) increased the probability of developing cardiac dysfunction in offspring whose mothers afflicted gestational diabetes. Thus, HIF1A is a pivotal factor in the programming of fetal cardiovascular system while a global decline of HIF1A level (haploinsufficiency in HIF1A gene) can drastically disturb fetal cardiovascular system (Cerychova, et al., 2018). Furthermore, FOXO mutations were demonstrated to modulate lifespan in Drosophila (Yamamoto & Tatar, 2011). Although these studies have not been performed per se in IS models or neuron cells, it can be postulated that these genes are prone to various mutations. Therefore, genetic mutations might underlie autophagy defect or overactivation upon IS. Table. 3 summarized vascular impairment/stroke/hypoxia, phenotype, and animal models associated with the relevant gene mutations in IS.
Table 3.
Vascular impairment/stroke associated with the relevant target gene mutation
| Human stroke equivalent | Relevant genes | Vascular impairment | Refs |
|---|---|---|---|
| A neonatal rat model of brain hypoxic/ischemia | HIF1A and its target gene VEGFA upregulate at 8 hours of reperfusion and decline at 24 hours | After mild I/R, VEGFA may be a potential target to confer neuroprotection and vascular survival | (D. Mu, et al., 2003) |
| MCAO rat models | Early ischemic cascade induces upregulation of VEGF and KDR in the cortex | Downregulation of VEGF and KDR reduces vascular impairment under hypoxia | (Ma, Lovekamp-Swan, Bekele, Dohi, & Schreihofer, 2013) |
| A rat model of photothrombotic ring stroke | Upregulation of VEGFA and VEGFC and their receptors | Promote early angiogenesis after the stroke | (Gu, Brännström, Jiang, Bergh, & Wester, 2001) |
| Neonatal rodent MCAO models | Upregulation of HIF1A and its downstream signaling after a hypoxic insult | Enhance angiogenesis, neurogenesis, neuronal survival, and endothelial cell proliferation after the stroke | (Shimotake, Derugin, Wendland, Vexler, & Ferriero, 2010) |
4. Therapeutic manipulation of autophagy in the management of IS
According to the aforementioned studies, therapeutic manipulation of autophagy appears to be a possible approach in the management of IS. To-date, many potentially relevant therapies have been explored for IS management. Fibrinolytic therapy, also known as thrombolysis, refers to lysis of blood clots formed in blood vessels using tPA or other therapeutic measures, to achieve early reperfusion of ischemic brain tissues (Demchuk, Felburg, & Alexandrov, 1999). Intravenous thrombolysis is an approved treatment for acute IS within a 3-hour time window following stroke attack (D. Zhang, et al., 2014). However, blood pressure variability 24 hours after thrombolysis may negatively impact long-term outcomes and reperfusion in patients (Liu, Chen, Yan, Zhang, & Lou, 2015). Thrombectomy is also a therapeutic intervention aimed to remove a blood clot in blood vessels, commonly employed in brain arteries too (Saver, et al., 2015). It is well perceived that thrombus composition has an impact on the efficacy of thrombolysis and thrombectomy procedures in acute IS, with clots rich in fibrin exhibiting enhanced recanalization maneuvers (the number of times that recanalization can take place) and unfavorable outcomes compared to clots rich in red blood cells (Jolugbo & Ariëns, 2021). In sum, thrombectomy and intravenous thrombolysis are potential recanalization therapies. Nonetheless, time-window dependency and heterogeneity of thrombus are considered main limitations for these therapies. The aforementioned examples have highlighted the importance of maintaining adaptive autophagy, and retardation of maladaptive autophagy in therapeutic approaches against IS-associated complications. Given that IS induces maladaptive autophagy, targeted inhibition on autophagy after IS and in the prehospital setting should render neuroprotection prior to definitive therapy for in-patient settings. Therefore, combinatory interventions including (A) maintaining adaptive autophagy prior to IS, (B) inhibiting maladaptive autophagy following IS in prehospital setting (Fig. 4), and (C) using proper treatments such as thrombectomy and intravenous thrombolysis within hospital may render overall benefit and neuroprotection against IS. The following section briefly summarizes potential strategies to maintain adaptive autophagy while suppressing excessive or maladaptive autophagy.
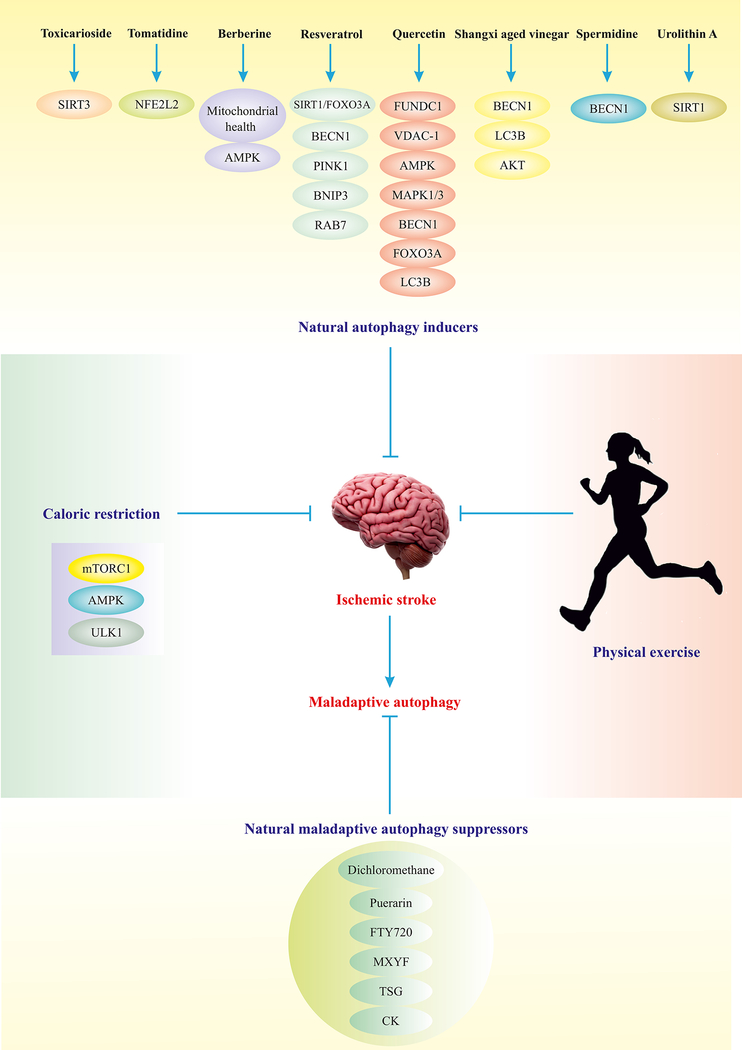
Fig. 4.
Strategies for manipulation of autophagy in IS management. Depicted natural compounds have the potential to induce adaptive autophagy or block maladaptive autophagy via manipulation of autophagy elements. Certain natural autophagy inducers have been reported to mildly turn on several autophagy regulators, leading to adaptive autophagy and alleviation of IS. Physical exercise and caloric restriction are the most suitable, conventional and applicable measures to maintain adaptive autophagy and alleviate IS.(Ajoolabady, Aslkhodapasandhokmabad, Aghanejad, Zhang, & Ren, 2020)
Abbreviations: SIRT3 (sirtuin 3), RAB7 (RAB7, member RAS oncogene family), VDAC1 (voltage dependent anion channel 1).
4. 1. Potential regulators of maladaptive autophagy
Potential regulators of maladaptive autophagy refer to certain molecules that can suppress excessive induction of autophagy. For instance, the neuroprotective potential of neuroprotectin D1 (a derivative of the polyunsaturated fatty acid docosahexaenoic acid) was demonstrated in an in vitro model of cerebral I/R injury, the effect of which was attributed to suppression of oxidative stress and maladaptive autophagy via enhanced expression of the E3 ubiquitin-protein ligase RNF146, and activation of WNT-CTNNB1. RNF146 downregulation potentiates maladaptive autophagy, denoting the therapeutic potential of targeting RNF146 in promoting neuronal resistance against maladaptive autophagy in IS (Q. Mu, et al., 2020). In a rat model of IS, long non-coding RNAs (lncRNAs) were found to drive cerebral protection for ischemic postconditioning through retarding excessive autophagy (Jie, et al., 2020). Moreover, TIGAR resists maladaptive autophagy thus resulting in reduced infarct volume and improved neurological deficit (D.-M. Zhang, et al., 2019). Overall, novel regulators of maladaptive autophagy could be considered for drug development of pharmaceutical or natural compounds in the therapeutic interventions against IS.
Table. 1 summarizes recently identified candidate therapeutic agents with the potential to suppress maladaptive autophagy evoked by IS. Among these enlisted compounds, DEX is capable of rendering neuroprotection after IS through inhibition of neuronal maladaptive autophagy. In the cultured OGD model and MCAO mouse model, DEX upregulated CCND1, SQSTM1, and HIF1A, and downregulated LC3A and BECN1, leading to suppression of unchecked excessive autophagy and apoptosis (C. Luo, et al., 2017). Moreover, DEX protected the brain in developing rats via downregulation of DNM1L and BAX, and promotion of autophagic flux. DNM1L and BAX downregulation intensified mitochondrial integrity and suppressed apoptosis, respectively (Shan, Sun, Yang, Shang, & Liu, 2018). In a murine model of traumatic brain injury, DEX also inhibited NLRP3 and CASP1 while suppressing maladaptive autophagy through modulation of circLrp1b (a circular RNA)/MIR27a-3p/DRAM2 pathway (H. Li, et al., 2020). Mechanistically, in traumatic brain injury, circLrp1b interrupted MIR27a-3p, and thereby, increased levels of DRAM2 (Fig. 1), which triggered excessive/maladaptive autophagy (H. Li, et al., 2020). Overall, DEX is a potential compound perceived to maintain adaptive autophagy and hinder excessive autophagy induction following IS.
Table 1.
Therapeutic compounds suppressing maladaptive autophagy in IS
| Compounds | Name/Source | Therapeutic Effects/Mechanisms | Ref |
|---|---|---|---|
| CK | Ginsenoside monomer compound K (an intestinal derivative of ginseng) | Suppresses maladaptive autophagy and apoptosis via regulating the formation of autophagosomes and inhibiting the AMPK-mTORC1 axis, elicits neuroprotection against cerebral I/R injury, attenuates ROS and Ca2+ overload in the mitochondria | (Q. Huang, et al., 2020) |
| DEX | Anxiety-reducing pharmaceutical agent | Substantially attenuates brain injury via inhibition of maladaptive autophagy upon IS | (C. Luo, et al., 2017) |
| Dichloromethane | Extracted from Piper longum L. and Piper nigrum L. | Suppresses maladaptive autophagy via activation of AKT1/mTORC1 and elicits neuroprotection | (Yiwei Zhang, et al., 2020) |
| HBO | Hyperbaric oxygen therapy | Promotes learning and memory, attenuates inflammation and cytosolic Ca2+ overload, and blocks maladaptive autophagy via activation of mTORC1 and reduction of ATG5 and LC3B upon cerebral I/R injury | (Chunxia Chen, et al., 2020) |
| Icariside II | Icariin II/Baohuoside I | Attenuates maladaptive autophagy in rat IS model via direct interaction/inhibition of PDE5, which enhances cGMP level and PRKG1/PKG activation, leading to GSK-3β inactivation and autophagy inhibition | (J. Gao, et al., 2020) |
| MXYF | Mu-Xiang-You-Fang (Chinese herbal compound) | Alleviates IS, boosts cell viability and mitochondrial integrity, and inhibits maladaptive autophagy via downregulation of ULK1, AMPK, BECN1, and LC3A and upregulation of mTORC1 in PC12 cells | (H.-x. Ma, et al., 2020) |
| PPF | Diprivan | Significantly diminishes SNCA/α-synuclein aggregation, potentiates mTORC1-RPS6K/S6K signaling cascade, and suppresses maladaptive autophagy in murine IS models | (Y. Wang, et al., 2020) |
| Puerarin | A traditional Chinese herb | Alleviates cerebral dysfunction via inhibiting the expression of AMPK and ULK1 thus, blocking maladaptive autophagy | (J.-F. Wang, et al., 2018) |
| Sevoflurane | A methyl isopropyl ether | Retards memory loss and reduces infarct volume, apoptosis, and maladaptive autophagy via BECN1, LC3B/A, BAD, and CASP3 downregulation and BCL2 upregulation in murine cerebral I/R injury. Also inhibits NFKB1 phosphorylation and NFKBIA/kBα activation | (C. X. Shi, et al., 2020) |
| SIGMAR1 agonist | SIGMAR1 agonist | Inhibits maladaptive autophagy and apoptosis in pericytes thereby, improves their survivability and IS recovery | (Yuan Zhang, et al., 2020) |
| STS | Sodium tanshinone IIA sulfonate | Elicits remarkable neuroprotection against IS via downregulation of autophagy elements including SIRT6, BECN1, and LC3B. Inhibits inflammation | (L. Wang, et al., 2020) |
| TSG | Tetrahydroxystilbene glucoside derived from Fallopia multiflora | Alleviates neurobehavioral deficits and reduces infarct volume via maladaptive autophagy inhibition in the murine IS model | (F. Yu, et al., 2019) |
Propofol (PPF) is another compound capable of preventing neuronal injury in murine cerebral cortical neurons following exposure to OGD/re-oxygenation (B. Sun, et al., 2018). PPF-induced neuroprotection was attributed to suppression of maladaptive autophagy via inhibition of the Ca2+/CAMKK2/AMPK axis (Fig. 1) (B. Sun, et al., 2018). In addition, PPF treatment inhibited cerebral I/R-induced upregulation of LC3A, TP53, DRAM, BECN1, and BBC3 in hippocampus, suggesting the benefit of PPF in protecting neurons against maladaptive autophagy and apoptosis following cerebral I/R injury (Cui, et al., 2013). In murine hippocampal neurons, PPF also inhibited TNF-triggered excessive autophagy via blockade of intracellular Ca2+ accumulation (Fig. 1) and CALM2 activation (Y. Li, He, Lv, Chen, & Chen, 2020). Collectively, PPF has exhibited some promising translational applicability following the incidence of IS.
Puerarin, extracted from Radix puerariae (a traditional Chinese herb), suppressed cerebral I/R-induced excessive autophagy via modulating the AMPK/mTORC1/ULK1 axis (Fig. 1) in a rat model of IS (J.-F. Wang, et al., 2018). Meanwhile, puerarin alleviated cerebral infarct size, edema, and neurological deficiency through attenuation of hyperactivated autophagy in neurons but not astrocytes in infarct penumbra in a rat MCAO model (Hongyun, Tao, Pengyue, Liqiang, & Yihao, 2017). These data favor that puerarin exhibits neuroprotection against IS via regulation of maladaptive/excessive autophagy, indicating its translational potential for IS management.
Sevoflurane is an FDA-approved anesthesia drug, and protects neurons against hypoxic-ischemia by blocking hyperactivated autophagy through the PTEN/AKT1/mTORC1 modulating EZH2 activation (Fig. 1) (H. Xue, et al., 2019). Besides, sevoflurane postconditioning alleviated brain injury in a rat model of neonatal hypoxic-ischemia through MAPK1/ERK signaling (Shuo Wang, Xue, Xu, Niu, & Zhao, 2019). Although the precise underlying mechanism remains unclear, it is believed that MAPK1 inhibits the TSC complex, which activates RHEB and mTORC1, leading to autophagy inhibition (Meng, Frank, & Jewell, 2018). Overall, sevoflurane postconditioning may alleviate maladaptive autophagy in the realm of IS.
Icariside II (ICS II) was shown to protect neurons against maladaptive autophagy in MCAO rat and OGD models through suppression of excessive autophagy and downregulation of LC3A, BECN1, ATG5, and ATG7 (J. Gao, et al., 2019). Indeed, ICS II suppressed maladaptive autophagy through deactivation of the PRKG1/GSK-3β axis, which turned on mTORC1 (Fig. 1) to suppress autophagy (J. Gao, et al., 2019).
Table. 2 also summarizes various compounds with promises for the maintenance of adaptive autophagy in the context of IS. Among these compounds, anthocyanin (a potential antioxidant) improved the survival of human glial cells upon OGD through maintaining adaptive autophagy (Y. K. Kim, et al., 2012). Moreover, ezetimibe (a lipidemia therapeutic compound) reduced neuronal apoptosis and brain infarct size, as well as improved neurological function via activation of AMPK-dependent adaptive autophagy (Fig. 1) in a rat model of MCAO (J. Yu, et al., 2018). This finding revealed merits behind ezetimibe for further exploration in the management of IS. Similarly, metformin (a drug used for the treatment of diabetes) improved movement disability and psychological disorders evoked by global cerebral ischemia through activation of AMPK-dependent autophagy, indicating the possible translational capacity of metformin for IS alleviation (Sarkaki, et al., 2015).
Table 2.
Therapeutic compounds with the potential to induce adaptive autophagy in IS
| Compounds | Name/Source | Therapeutic Effects/Mechanisms | Ref |
|---|---|---|---|
| Anthocyanin | Found in black rice, blueberry, and raspberry | Considerably boosts autophagy, prevents oxidative stress damage, attenuates inflammation, and blocks apoptosis in SH-SY5Y cells IS model | (Cai, et al., 2020) |
| Astragaloside IV | A pentacyclic triterpenoid | Activates autophagy and attenuates neuronal apoptosis in HT22 cells IS model | (Y. Zhang, et al., 2019) |
| BCP | β-caryophyllene | Upregulates BECN1, LC3B, and BCL2 and increases autophagosomes, downregulates SQSTM1, resulting in the alleviation of neurological deficit and infarct volume via adaptive autophagy activation | (Rao, et al., 2020) |
| DEX | Anxiety-reducing pharmaceutical agent | Induces adaptive autophagy via TSC2-mTOR signaling cascade in OGD-induced IS in astrocytes | (Chen Zhu, Zhou, Luo, & Chen, 2020) |
| Eugenol | Derived from Acorus gramineus | Reduces neurological deficits and infarct volume by restraining apoptosis and restoring adaptive autophagy via manipulating the AMPK-mTORC1-RPS6K pathway both in HT22 cells and animal IS models | (X. Sun, D. Wang, et al., 2020) |
| Ezetimibe | A compound used for the treatment of high blood cholesterol | Lessens infarct volume and neurobehavioral deficiency prevents apoptosis and activates autophagy in the rat IS model | (J. Yu, et al., 2018) |
| Geniposide | Found in medicinal herbs | Reinvigorates adaptive autophagy, suppresses NLRP3 inflammasome in BV-2 microglial model of cerebral I/R injury | (Fu, et al., 2020) |
| GK | Ginkogolide K | Induces autophagy and enhances the proliferation and migration of astrocytes upon IS | (Ying Zhang & Miao, 2018) |
| GRb1 | Ginsenoside Rb1 | Enhances adaptive autophagy upon cerebral I/R injury thus, alleviates cerebral damage | (Lu, et al., 2011) |
| Metformin | Glucophage | Lessens IS risk via activating autophagy in an AMPK-dependent manner | (T. Jiang, et al., 2014) |
| NAMPT | nicotinamide phosphoribosyl transferase | Promotes cell survival via maintaining adaptive autophagy | (P. Wang, et al., 2012) |
| Progesterone | A female hormone | Reduces the production and activation of HMGB1 and NLRP3 inflammasome, respectively, and activates adaptive autophagy in microglia and animal IS models | (Espinosa-Garcia, et al., 2020) |
| Rapamycin | A natural compound | Induces autophagy via mTORC1 inhibition, resulting in attenuated infarct volume, alleviated neurologic deficiency, and boosted mitochondrial functions | (M. Wu, et al., 2018) |
| Resveratrol | A natural polyphenol | Attenuates infarct volume via activating AMPK-mediated autophagy/mitophagy leading to the removal of superoxide anion and prevention of cytosolic Ca+2 overload and mitochondrial failure in the rat IS model | (Pineda-Ramírez, et al., 2020) |
| Triptolide | Found in thunder god vine | Induces adaptive autophagy and suppresses apoptosis following ischemia | (Y. Yang, et al., 2015) |
| VX-765 | A selective inhibitor of CASP1 | Activated adaptive autophagy via the AMPK pathway, thus ameliorated inflammation and apoptosis in ameliorated IS-induced in murine transplanted HUMSCs | (Z. Sun, et al., 2020) |
Rapamycin is widely examined in the context of IS, with capacity to overtly augment the neurological score and attenuate lesion size in embolic MCAO rat models (K. M. Buckley, et al., 2014). Besides, both pre- and post-administration of rapamycin in the setting of MCAO model activated autophagy through mTORC1 inhibition, and downregulated CASP3 and CASP9 in the ischemic penumbra, leading to attenuated neurological dysfunction and infarct size in MCAO rat models (M. Wu, et al., 2018). Moreover, in HT22 murine hippocampal neurons, rapamycin ameliorated OGD-induced injury via activation of autophagy (K. Buckley, 2015). Likewise, resveratrol alleviated cerebral I/R injury and reduced brain fluid content and infarct size via inhibition of NLRP3 through SIRT1-dependent regulation of autophagy (Fig. 1) (Q. He, et al., 2017). To this end, rapamycin and resveratrol may possess promising translational applicability to maintain adaptive autophagy in IS challenge. Of note, using nanomedicine and drug delivery strategies can also move therapeutics forward (Alizadeh, Zarebkohan, Salehi, Ajjoolabady, & Rahmati-Yamchi, 2019; Hashemi, et al., 2020; Khiavi, et al., 2020).
4. 2. MicroRNAs governing both adaptive and maladaptive autophagy
MicroRNAs (miRNAs) are well known to modulate differentiation, metabolism, apoptosis, and cell proliferation through regulating levels of target genes (Henninger & Mayasi, 2019). As mentioned above, IS triggers changes in the level of miRNAs, leading to alteration in selective genes vital for autophagy (X. Chen, et al., 2019). In an attempt to unravel the neuroprotective role of remote ischemic preconditioning (RIPC), RIPC was found to promote upregulation of MIR144, which further upregulates and downregulates AKT1 and PTEN, respectively. AKT1 upregulation and PTEN downregulation may account for suppression of autophagy and apoptosis, suggesting a role for inhibition of apoptosis, excessive autophagy, or both, in the neuroprotection of MIR144 (Zhong, et al., 2020). In contrast, a non-coding RNA, MIR30D-5p, negatively regulated BECN1 levels, resulting in dampened adaptive autophagy and neuronal death in hypoxic-ischemic rats (F. Zhao, et al., 2017). Likewise, MIR497 was shown to inhibit adaptive autophagy induction, whereas suppression of MIR497 enhanced neuronal autophagy and ameliorated IS-induced injury in murine models (X. Chen, et al., 2019). These findings denote the therapeutic potential of several miRNAs as both positive and negative regulators for adaptive and maladaptive autophagy in IS.
Ample evidence has uncovered regulation of miRNAs by various organismal and cellular stress. On the other side of the coin, both up- and down-regulation of miRNAs can activate ample stress responses in mammalian cells (Akkoc & Gozuacik, 2020). Recent focus on autophagy regulation by way of miRNAs has denoted an emerging field in the molecular biology aspect of autophagy. Human body contains nearly 2000 miRNAs and 30000 genes that produce proteins. These miRNAs regulate almost 60% of these 30000 genes (Friedman, Farh, Burge, & Bartel, 2009). Moreover, a growing body of evidence has indicated that more than one miRNA (several miRNAs) concurrently regulate the expression of genes encoding proteins involved in autophagy or encoding autophagy regulators such as AKT1, AMPK, and mTOR (H. Zhu, et al., 2009). In other words, several miRNAs can target one given autophagy gene, and therefore, creating a rather diverse responses in autophagy. Hence, miRNAs may serve as potential tools for the regulation of adaptive and maladaptive autophagy. To overcome these apparent disadvantages, novel approaches should focus on manipulating the entire network of miRNAs rather than a single miRNA to ensure optimized or specific regulation of autophagy under ischemia. Successful development of these novel approaches should enable us to apply miRNAs as new clinical tools for the management of IS.
4. 3. Role of stem cells in the regulation of adaptive and maladaptive autophagy
Transplantation of BMSCs was explored in recent years as a potential therapeutic avenue in IS. Findings from murine models of IS have revealed that BMSCs transplantation attenuated maladaptive autophagy while promoting UPS in IS (Tadokoro, et al., 2020b). Following IS insult, sEV secreted by human-induced pluripotent stem cells-derived mesenchymal stem cells (iPSC-iMSC) were reported to shrink cerebral infarct volume, increase angiogenesis, and mitigate neurological impairments through undermining maladaptive autophagy in a STAT3-dependent manner in vitro and in vivo. This finding revealed the therapeutic potential of iPSC-iMSC-secreted sEV against cerebral ischemia (Xia, et al., 2020). Given that sEV contains both RNA and protein molecules, further experiments are desired to unravel how these molecules interact to modulate STAT3 signaling and autophagy. In particular, emphasis should be focus on possible autophagy-dependency of sEV-induced neuroprotection using loss-of-function experimental approaches for autophagy (such as ATG genes knockdown rather than using 3-MA) (Xia, et al., 2020).
Co-culturing ADMSCs in an in vitro model of IS also yielded neuroprotective effects by way of sEV. It was revealed that sEV-residing MIR25–3p attenuated maladaptive autophagy via interacting with TP53 resulting in blockade of the TP53-BNIP3 signaling cascade in both in vitro and in vivo settings of IS, suggesting that MIR25–3p is a key factor in the anti-autophagic property of ADMSCs-produced sEV (Kuang, et al., 2020). Moreover, astrocyte-secreted exosomes inhibit maladaptive autophagy by transferring MIR190B to suppress autophagy in HT22 cells under OGD conditions (Pei, Li, Zhu, & Zhou, 2020). Additionally, cerebral I/R injury-induced activation of the NLRP3 inflammasome underlies pyroptosis-mediated neuronal cell death, which is negatively controlled by mild induction of autophagy. BMSCs-secreted exosomes, which trigger adaptive autophagy through AMPK and suppress inflammation, lead to inhibition of pyroptosis in cerebral I/R injury (Zeng, et al., 2020). In this context, stem cells hold the promise as a therapeutic avenue to induce adaptive autophagy and suppress maladaptive autophagy in neuronal cells upon IS challenge.
One of the limitations associated with BMSC transplantation is that it regulates autophagy and UPS within 30 minutes following IS challenge. To this end, the effect of the transplantation might decline after 30 minutes (Tadokoro, et al., 2020a). More importantly, iMSC-secreted sEV not only suppresses maladaptive autophagy, but also remarkably enhances blood vessel formation (angiogenesis) (Xia, et al., 2020). Ample clinical and experimental evidence has consolidated a close correlation among angiogenesis, neurovascular remodeling, remyelination, and lessened neural injury, resulting in improvement of functional recovery ensuing IS (Brumm & Carmichael, 2012; Krupinski, Kaluza, Kumar, Kumar, & Wang, 1994). However, extensive preparation of MSCs is not readily available with the current techniques, while donors-obtained MSCs are usually insufficient. Moreover, multiple factors (e.g., age and health status for donors) might also affect the differentiation and growth potential of these cells, which greatly hinder their therapeutic and clinical efficacies (Hu, et al., 2015). Besides, MSC-sEV-mediated suppression of autophagy relies heavily on substances secreted by these MSC-sEV vesicles, mainly, miRNAs. Thus, as mentioned above, application of miRNAs for autophagy regulation should represent an emerging field, which would require further explorations to pave the way for optimal clinical application of MSC-derived sEV.
4. 4. Electroacupuncture (EA) as a potential solution for maladaptive autophagy
Over the past years, exploration for the impact of EA on permanent pathological insults such as inflammation, neuropathy, and cancer has grown dramatically. EA, particularly, in the frequency range of 2–10 Hz, activates various chemicals such as the pain relief opioids attenuate proinflammatory cytokines, norepinephrine, and serotonin (R. Zhang, Lao, Ren, & Berman, 2014). In addition to pharmacotherapy, EA may also benefit neurological deficits in IS patients following brain I/R injury through suppression of CASP3, ATG7, and LC3B/A, as well as elevated PI3K expression and phosphorylation of mTORC1, AKT1, LAMP1, and SQSTM1, resulting in concerted attenuation of apoptosis and maladaptive autophagy (M.-M. Wang, et al., 2020). Besides, EA averts maladaptive autophagy via potentiating the WNT-GSK-3β signaling cascade, resulting in GSK-3β inhibition in conjunction with mTORC1 activation (Chengyu Chen, et al., 2020). In a rat model of cerebral I/R injury, EA was associated with improved neuronal survival, reduced infarct size, and improved dendritic spine density via activation of a SIRT1-FOXO1 axis, resulting in autophagy inhibition, implying that potentiating SIRT1-mediated inhibition of autophagy might lessen unrestrained induction of autophagy (Mei, et al., 2020). Moreover, EA was shown to inhibit maladaptive autophagy under central post-stroke pain (CPSP) in hippocampus through a PTGS2-CTNNB1-dependent manner (L. Zheng, et al., 2020). EA treatment offers neuroprotection, the effect of which is attributed to the inhibition of ER stress-induced maladaptive autophagy and apoptosis in cerebral I/R injury (X. Sun, H. Liu, et al., 2020). How EA protects from neuronal damage in stroke, beyond simply a correlation with autophagy pathways, remains largely elusive.
Taken together, EA alleviates neurological injuries and symptoms, as well as attenuates cerebral infarction size in rat models (Xing, Wang, Feng, Dong, & Zhang, 2018). EA pretreatment downregulates apoptotic genes and blocks apoptosis in ischemic penumbra (X. Xue, et al., 2014). Also, EA preconditioning is capable of alleviating IS-mediated cerebral impairment through autophagy suppression (Z. Wu, Zou, Zou, Zhou, & Cui, 2015). However, some evidence indicated that the neuroprotective function of EA preconditioning was attributed to autophagy upregulation (Z.-Q. Wu, Cui, Zhu, & Zou, 2016). These controversies can be explained by different time points of EA implementation or different stages of pathology prior to application of EA. Therefore, the difference in the timing of intervention and pathological stage of stroke may impact the clinical efficacy of EA as a tool for autophagy regulation, suggesting the necessity of optimized treatment modality (timing of intervention and disease stage) for proper application of EA in clinics.
4. 5. Physical exercise and caloric restriction on the maintenance of adaptive autophagy
Besides genetic, environmental factors including diet, physical exercise, and age can potentially influence predisposition, prevention, and recovery of IS (X. Gao, Yang, & ZhiPing, 2006). Exploration of the implication of total antioxidant capacity in the predisposition to HS and IS revealed that antioxidants contributed to the alleviation of IS but had no effect on HS in a cohort study, while high-dose of vitamin E uptake potentiated the risk of HS (Del Rio, et al., 2011). These findings suggest that diets rich in antioxidants should help to rescue or prevent cerebral ischemia in vulnerable individuals. Besides, a Mediterranean-style diet, which includes olive oil, fish, whole grains, vegetables, and fruits, was demonstrated to lower IS incidence and vascular events, and improve cardiovascular health in the northern Manhattan cohort study (Gardener, et al., 2011). Based on this observation, the Mediterranean-style diet may hinder the outbreak of IS among populations. Moreover, enhanced uptake of flavanone subclass (like citrus fruit) attenuated the risk of incidence in a study that included 69622 women and followed up for 14 years (Cassidy, et al., 2012). Also, scrutiny of the impact of magnesium-rich diet on IS revealed that low serum level of magnesium enhanced IS risk through its effect on diabetes mellitus and hypertension among white and black participants, suggesting a fundamental role of magnesium-rich diet in the prevention of IS (Ohira, et al., 2009). Moreover, seven cohort studies substantiated that overconsumption of red meat, particularly, processed meat enhanced the risk of developing IS (Cuili Yang, et al., 2016). Furthermore, another cohort study revealed a positive correlation between whole milk intake and intracerebral hemorrhage risk, as well as between subarachnoid hemorrhage and yogurt usage (Larsson, et al., 2009). Interestingly, Morgenstern and team observed a remarkable correlation between sum of fast-food restaurants and stroke incidence in the neighborhoods in a community-based investigation (Morgenstern, et al., 2009). Therefore, abstaining from overconsumption of fast foods, red meat, and certain dairy products can substantially decrease the risk of stroke. Taken together, dietary attitudes or desire can determine how much an individual is at risk of IS and also prognosis for recovery after the stroke attack.
Age is another risk factor that can influence IS incidence or recovery among populations. Elderly individuals are at more risk of IS and usually suffer severe post-stroke effects (R.-L. Chen, Balami, Esiri, Chen, & Buchan, 2010). In a group of 2219 stroke patients in a cohort study, the association between recovery and age was analyzed and results indicated that young patients (<55 years of age) exhibited maximum improvement and faster functional recovery compared to older populations (>55 years of age). Nonetheless, age should not be considered as a limiting factor in rehabilitation among stroke patients (Kugler, Altenhöner, Lochner, & Ferbert, 2003). Also, in another independent cohort study, neurological defects and disabilities observed in stroke patients six months after stroke attack were more prominent in elderly individuals, particularly, in elder women (with adjusted stroke subtypes) (Kelly-Hayes, et al., 2003). Examining the correlation between age and functional recovery in stroke patients during the post-ischemic period (3 months after stroke attack) revealed that age would inversely predict the outcome independent of gender, thrombolysis, and severity/etiology of stroke attack (Knoflach, et al., 2012).
Physical exercise also exhibits some rehabilitative potentials. Nonetheless, it is imperative to know when to engage regular exercise following stroke attack. In an MCAO rat model, 30 minutes of Rota-Rod exercise 6 hours after the reperfusion exacerbated apoptotic cell death, as manifested by upregulation of BAX and CASP3. However, 30 minutes of exercise 24 hours or 3 days after reperfusion did not elicit such effect, indicating that early physical exercise should not be practiced as it may exacerbate stroke outcomes (F. Li, et al., 2017). Resistance exercise for 12 weeks (each week for three times) improved state and trait anxiety in stroke patients one year after the stroke in a pilot study (Aidar, et al., 2012). Besides, physical exercise remarkably attenuated infarct volume augmented neural function, reduced neural apoptosis, and increased the number of neural progenitor cells through activation of IGF1/AKT1 signaling cascade following IS insult (H.-Q. Zheng, et al., 2014). Moreover, Sprague-Dawley rats undergoing exercise before MCAO, retrieved more functional cells in the infarct arena, had superior angiogenesis and exhibited better motor performance after MCAO compared with the non-exercising rats (H. Zhang, Lee, Borlongan, & Tajiri, 2019). These data convincingly support the notion that lack of physical exercise enhances the incidence of IS in all populations. Overall, these observations imply that maintenance of an active lifestyle and moderate physical exercise should help to prevent IS or alleviate post-stroke injuries.
Of note, in an ARIC (atherosclerosis risk in communities) study, diabetes mellitus was deemed a strong risk factor for IS (Folsom, et al., 1999). Also, insulin resistance, increased levels of fasting insulin, and larger waist-to-hip ratio greatly escalated IS risk (Folsom, et al., 1999). In another ARIC study, elevated levels of factor XI were correlated with enhanced risk of IS. Therefore, individuals with elevated levels of factor XI are at a much higher risk for IS (Suri, Yamagishi, Aleksic, Hannan, & Folsom, 2010). Exploration of the correlation among intake of vegetables, fruits, refined-grain, and whole-grain, and coronary artery disease (CAD), total mortality, and IS in an ARIC cohort study exhibited a tie between consumption of these foods and CAD but not IS, raising the controversy for food consumption and prevalence of IS (Steffen, et al., 2003). In sum, various environmental factors can contribute to IS, and atherosclerosis, which is associated with dietary attitudes and lifestyle, is a potential contributor to IS. In the following sections, we aim to discuss the correlation between environmental factors and autophagy in IS.
Physical exercise and caloric restriction are two essential components for a healthy lifestyle. Physical exercise is referred to as regular, scheduled, and well-balanced physical activity that maintains normal brain physiology and health (Andreotti, et al., 2020; Haili Tian, Chen, & Ren, 2019; N. N. Wu, et al., 2019). Ample evidence suggests that mild physical exercise promotes autophagy and prevents maladaptive autophagy (Fig. 3) (H. Tian, et al., 2020). Mild exercise also activated autophagy and mitophagy, leading to the prevention of neurodegeneration in substantia nigra in aged murine models (Almeida, et al., 2018). It was revealed that physical exercise triggers AMPK activation, to inhibit mTORC1 and activate autophagy. Activated AMPK also triggered FOXOs to upregulate ATG proteins and LC3B (Fig. 1) (Batatinha, Diniz, de Souza Teixeira, Krüger, & Rosa-Neto, 2019; Schwalm, et al., 2015).
Moreover, treadmill exercise suppressed maladaptive autophagy, impeded translocation and binding of HMGB1 to BECN1, reduced apoptosis and infarct size, and improved IS recovery, indicating an essential role of treadmill exercise in ameliorating neurological deficits in IS (G. Pan, et al., 2020). In addition, treadmill exercise following IS overtly inhibited maladaptive autophagy and autophagy-mediated SQSTM1 degradation, thus, ameliorating cerebral infarction (L. Zhang, et al., 2014). The late Beth Levine’s group also showed that treadmill exercise triggered adaptive autophagy in cerebral cortex, which boosted neurological cognitive function in adult rats (C. He, Sumpter, & Levine, 2012). These data have suggested that treadmill exercise induces adaptive autophagy while suppressing maladaptive autophagy. Moreover, 12 weeks of treadmill exercise, induced adaptive autophagy via increased phosphorylation/inactivation of GSK-3β, leading to reduced hyperphosphorylation of MAPT in the cerebral cortex, and thereby, attenuation of cognitive decline in rat models (E.-B. Kang & Cho, 2015). Furthermore, endurance exercise triggered autophagy, boosted anabolic state, and enhanced mitochondrial turnover via mitophagy induction in a manner dependent upon AMPK and MAPK8 activation (Fig. 1) in murine brain cortex (Kwon, Jang, & Lee, 2021). Moreover, endurance exercise upregulated autophagy-associated genes such as BECN1, LC3A, and SQSTM1, and enhanced neurogenesis via AKT1/mTORC1/RPS6KB1 axis in the hippocampus, suggesting that both autophagy and neurogenesis are activated by endurance exercise (Jang, 2020). Besides, ultraendurance exercise (long-distance running) upregulated BNIP3 (Fig. 1) and enhanced autophagic flux in human muscles, favoring the benefit of endurance exercises on cytoprotective autophagic flux (Jamart, et al., 2012). Acute exercise also instigated autophagy in cardiac and skeletal tissues through a mechanism dependent upon BCL2/BECN1 dissociation (Fig. 1) (C. He, Bassik, et al., 2012). Luo and co-workers found that freestyle swimming exercise (10 weeks [5 days per week, each day for 30 minutes]) promotes autophagy and lysosomal degradation in the hippocampus, leading to improved cognitive function in rats (L. Luo, et al., 2017). Also, running exercise, particularly, prolonged exercise (5 days per week for 8 weeks, wheel running 18 rpm for 1 hour) promoted autophagy through TFEB (transcription factor EB ) activation, leading to upregulation of autophagy and lysosomal genes in the brain cortex, which improved its functions (J. Huang, et al., 2019).
Caloric restriction, or reduced caloric uptake without malnutrition, protects against cerebral ischemia by inducing adaptive autophagy. Jeong and associates explored the effects of two-week intermittent fasting after cerebral ischemia in a rat IS model and noted that intermittent fasting lessens cerebral edema, infarct size, neuronal apoptosis, and autophagosome accumulation to ameliorate defective adaptive autophagy (Jeong, et al., 2016). Caloric restriction enhances ghrelin in the plasma and NPY in the hypothalamus, to impose neuroprotection via autophagy activation in murine cortical neurons. Besides, NPY and ghrelin exhibit synergistic effects (Ferreira-Marques, et al., 2016). Similarly, short-term fasting overtly triggers adaptive autophagy in Purkinje cells and cortical neurons (Alirezaei, et al., 2010). Fisetin is also a caloric restriction mimetic, which is reported to upregulate autophagic genes such as BECN1, ATG3, and SIRT1, and attenuates inflammation in aged brain models (Singh, Singh, Garg, & Rizvi, 2018). Overall, physical exercise and caloric restriction are strategies to maintain mild autophagy. Nonetheless, further studies are warranted to determine optimal intensity and duration of physical exercise, the extent of caloric restriction, and limitations of generalizing from animal studies to human for the most appropriate autophagy management (Fig. 3, 4).
5. Challenges of targeting autophagy in the management of IS
Although studies to delineate and classify types of dysfunctions in adaptive autophagy of neuronal cells upon IS are missing, it can be assumed that autophagy dysfunctions might occur at different stages of autophagy such as initiation, elongation, and maturation, or in pre-autophagic signaling pathways (Mizushima & Levine, 2020; Yingmei Zhang, et al., 2018; Y. Zhang, Whaley-Connell, Sowers, & Ren, 2018). Given the supposed complexity of autophagy deficits, activating autophagy via manipulating a certain molecule or cell signaling pathway such as inhibition of mTORC1 might not fully and properly reinvigorate adaptive autophagy in ischemia. To this end, in an effort to maintain efficient management of IS by manipulating autophagy (either during or after stroke attack), future studies might reveal various defects of autophagy in its sequential stages or upstream signaling pathways. In line with this, we offer “holistic targeting” intervention which in our terminology refers to developing therapeutics or pharmaceutical agents which specifically target/modulate a wide spectrum of autophagy regulators/defects to reinvigorate adaptive autophagy in neuronal cells. Developing agents that combat defective autophagy in various diseases including IS requires novel technologies and collaboration of scientists in almost all fields.
We also propose that mammalian cells might possess the “intrinsic autophagy repairing systems” to reinitiate autophagy once it becomes defective. In other words, mammalian cells might maintain an intrinsic capacity to counteract autophagy defects or induce autophagy via alternative pathways, via hitherto unidentified mechanisms or processes. Scientific proof for this scenario is that master regulators of autophagy such as AMPK are subjected to many other regulations (Lee & Kim, 2010). For instance, intracellular Ca2+, CAMKK2, CALM (Fig. 1), PRKACA, STK11, and SESN1 are among essential AMPK regulators (K. Wu, et al., 2016). We believe that neuron cells can reinitiate autophagy by controlling these regulators. However, further investigations are warranted to demonstrate such mechanisms in neuron cells. Thus, deciphering these mechanisms; then, potentiating neuronal cells to engage these mechanisms in the aftermath of IS, might be a new era in therapeutic manipulation of autophagy. Regarding the complexity of autophagy during starvation (Walls, et al., 2010), the starvation-induced autophagy might unveil or disclose possible “intrinsic autophagy repairing systems” in neuronal cells under ischemia.
Considering crosstalk between autophagy and apoptosis (Fig. 5), a number of studies have shown adaptive autophagy induction while circumventing apoptosis. For instance, under nutrient deprivation, the STK11-AMPK signaling cascade was activated leading to phosphorylation and stabilization of CDKN1B at Thr 198 which then potentiated mammalian cells to suppress apoptosis to combat metabolic crisis through autophagy induction (Liang, et al., 2007). Therefore, the STK11-AMPK-CDKN1B pathway is a potential link between autophagy induction and apoptosis inhibition. As mentioned above, inhibition of AKT1 triggers both autophagy and apoptosis, and AKT1 activation blocks both autophagy and apoptosis. In this regard, under nutrient deprivation, SPHK1 was overexpressed which accounted for S1P production and subsequent induction of autophagy through mTORC1 inhibition, independent of AKT1, and any apparent participation of BECN1. Silencing of ATG7 and SK1 resulted in autophagy ablation during starvation and induced apoptotic cell death (Lavieu, et al., 2006). Taken together, future studies might teach us how to induce adaptive autophagy from pathways that circumvent apoptosis.
Fig. 5.
Crosstalk between autophagy and apoptosis in IS
Abbreviations: FADD (Fas associated via death domain), CASP8 (caspase 8), BID (BH3 interacting domain death agonist), MTTP (microsomal triglyceride transfer protein), CASP9 (caspase 9), APAF1 (apoptotic peptidase activating factor), GRIN (glutamate ionotropic receptor NMDA type subunits), GRIA (glutamate ionotropic receptor AMPA type subunits), CASP7 (caspase 7), CASP2 (caspase 2), CASP10 (caspase 10).
As aforementioned, excessive autophagy is characterized by accumulated autophagosomes. Although accumulated autophagosomes may result in massive degradation of cellular components and ACD onset, the underlying causes of autophagosome accumulation and their role in cell death induction require further scrutiny. From the cellular perspective, the number of autophagosomes exceeds that of lysosomes in the realm of unchecked autophagy (Malicdan, Noguchi, Nonaka, Saftig, & Nishino, 2008), leading to the accumulation of autophagosomes. However, this might not be the only explanation for autophagosome accumulation for maladaptive autophagy. The below questions are also needed to be answered in future studies:
Is excessive or persistent induction the only mode of maladaptivity in autophagy? Which processes or mechanisms might constitute maladaptive autophagy?
What non-canonical pathways might trigger autophagy from pathways that are detrimental to the survival of mammalian/neuronal cells?
Does affinity of interactions between BECN1 and ATG proteins or pre-apoptotic proteins (such as caspases) determine to what degree BECN1 molecules are cleaved and thereby contribute to apoptosis? Or are there other factors that affect the fate of upregulated BECN1 molecules?
How to define an appropriate threshold beyond which autophagy would be irreversible? Do mathematical models for quantifying active ATG levels (the number of active ATG molecules that participate in autophagy) work?
6. Concluding remarks
Accumulating evidence has convincingly noted presence of both adaptive and maladaptive autophagy following IS challenge, resulting in either alleviation or exacerbation, respectively, of brain injury. Given that much of the supporting evidence is originated from animal studies, human confirmation is needed to corroborate the experimental findings. As aforementioned, the main strategy for the management of maladaptive or unchecked autophagy is to employ compounds that suppress autophagy genes or signaling components responsible for autophagy induction. However, this strategy has failed to fully suppress maladaptive autophagy in the face of IS, indicating the complexity of managing maladaptive autophagy. That is because maladaptive autophagy does not necessarily occur via excessive activation of a certain autophagy regulator such as AMPK, but rather might be the result of excessive activation of multiple molecules and pathways involved in autophagy. Thus, developing multipotential agents to impose desired effects on autophagy might allow for entirely new therapeutic interventions in IS management.
8. Acknowledgements
JR was supported by the National Key R&D Program of China (2017YFA0506000), University of Wyoming Faculty Grant-in-Aid, Natural Science Foundation of China (81521001), and the American Diabetes Association (7-13-BS-142-BR). SW was supported by Natural Science Foundation of China (82000351). NH is supported by K08NS091499 from the National Institute of Neurological Disorders and Stroke of NIH. JMP is supported by the Austrian Federal Ministry of Education, Science and Research, the Austrian Academy of Sciences and the City of Vienna and grants from the Austrian Science Fund (FWF) Wittgenstein award (Z 271-B19), the T. von Zastrow foundation, and a Canada 150 Research Chairs Program (F18-01336). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. All authors have full access to the study and accept responsibility to submit for publication. We wish to express our sincere apology for those authors whose important work was unable to be cited due to space limitation.
List of abbreviations in alphabetical order:
- ADMSCs
(adipose-derived MSCs)
- AKT1
(AKT serine/threonine kinase 1)
- AIFM1
(apoptosis inducing factor mitochondria associated 1)
- AMPK
(5’ adenosine monophosphate-activated protein kinase)
- ATF6
(activating transcription factor 6)
- ATG3
(autophagy related 3)
- ATG5
(autophagy related 5)
- ATG7
(autophagy related 7)
- ATG9A
(autophagy related 9A)
- BAD
(BCL2 associated agonist of cell death)
- BBB
(blood-brain barrier)
- BBC3
(BCL2 binding component 3)
- BAX
(BCL2 associated X, apoptosis regulator)
- BCL2
(BCL2 apoptosis regulator)
- BECN1
(beclin 1)
- BMECs
(brain microvascular endothelial cells)
- BMSCs
(bone marrow stromal cells)
- BNIP3
(BCL2 interacting protein 3)
- BSCL2
(BSCL2 lipid droplet biogenesis associated, seipin)
- CALCOCO2/NDP52
(calcium binding and coiled-coiled domain 2)
- CALM1
(Calmodulin 1)
- CALM2
(calmodulin 2)
- CASP1
(caspase 1)
- CASP3
(caspase 3)
- CASP9
(caspase 9)
- CCND1
(cyclin D1)
- cGMP
(cyclic guanosine monophosphate)
- CLCN3/CIC-3
(chloride voltage-gated channel 3)
- CDKN1B
(cyclin dependent kinase inhibitor 1B)
- CTNNB1
(catenin beta 1)
- CX3CL1
(C-X3-C motif chemokine ligand 1)
- DEX
(dexmedetomidine)
- DNM1L
(dynamin 1 like)
- DRAM2
(DNA damage regulated autophagy modulator 2)
- EIF4E
(eukaryotic translation initiation factor 4E)
- EIF2AK3/PERK
(eukaryotic translation initiation factor 2 alpha kinase 3)
- ERN1/IRE1α
(endoplasmic reticulum to nucleus signaling 1)
- EZH2
(enhancer of zeste 2 polycomb repressive complex 2 subunit)
- FGF2
(fibroblast growth factor 2)
- FKBP5
(FKBP prolyl isomerase 5)
- FOXO3
(forkhead box O3)
- GJA1/connexin-43
(gap junction protein alpha 1)
- GSK-3β/GSK3B
(glycogen synthase kinase 3 beta)
- GRB2
(growth factor receptor bound protein 2)
- HIF1A/HIF1α,
hypoxia inducible factor 1 subunit alpha
- HMGB1
(high mobility group box 1)
- HUMSCs
(human umbilical mesenchymal stem cells)
- HUVECs
(human umbilical vein endothelial cells)
- IGF1
(insulin like growth factor 1)
- KDR
(kinase insert domain receptor)
- LAMP1
(lysosomal associated membrane protein 1)
- LAMP2
(lysosomal associated membrane protein 2)
- LPS
(lipopolysaccharide)
- 3-MA
(3-methyladenine)
- MALAT1
(metastasis associated lung adenocarcinoma transcript 1)
- MAPK1
(mitogen-activated protein kinase 1)
- MAPK8/JNK1
(mitogen-activated protein kinase 8)
- MAP1LC3B/LC3B
(microtubule associated protein 1 light chain 3 beta)
- MAPT
(microtubule associated protein tau)
- MCAO
(middle cerebral artery occlusion)
- MEG3
(maternally expressed 3)
- MMP
(mitochondrial membrane potential)
- MTMR14
(myotubularin related protein 14)
- mTORC1
(mechanistic target of rapamycin kinase complex 1)
- MYH9/NMMHC-IIA
(myosin heavy chain 9)
- NFKB1/NF-κB
(nuclear factor kappa B subunit 1)
- NFKBIA/kBα
(NFKB inhibitor alpha)
- NLRP3
(NLR Family Pyrin Domain Containing 3)
- NMDARs
(N-methyl-d-aspartate receptors)
- NOD1
(nucleotide binding oligomerization domain containing 1)
- NOX5
(NADPH oxidase 5)
- OCLN
(occludin)
- NPY
(neuropeptide Y)
- OGD
(oxygen-glucose deprivation)
- OPRD1
(opioid receptor delta 1)
- OPTN
(optineurin)
- PDE5A
(phosphodiesterase 5A)
- PI3K
(phosphatidylinositol 3-kinase catalytic subunit type 3)
- PIK3C3/VPS34
(phosphatidylinositol 3-kinase catalytic subunit type 3)
- PINK1
(PTEN induced kinase 1)
- PRKAG2
(protein kinase AMP-activated non-catalytic subunit gamma 2)
- PRKCD
(protein kinase C delta)
- PRKC/PKC
(protein kinase C)
- PRKG1
(protein kinase cGMP-dependent 1)
- PKG
(protein kinase G)
- PPP1R12A
(protein phosphatase 1 regulatory subunit 12A)
- PRKACA
(protein kinase cAMP-activated catalytic subunit alpha)
- PRKN
(parkin RBR E3 ubiquitin protein ligase)
- PTGS2
(prostaglandin-endoperoxide synthase 2)
- PtdIns(3,5)P2
(phosphatidylinositol 3,5-bisphosphate)
- PtdIns(3,4)P2
(phosphatidylinositol 3,4-bisphosphate)
- PTEN
(phosphatase and tensin homolog)
- RHEB
(Ras homolog, mTORC1 binding)
- RHOA
(ras homolog family member A)
- RNF146
(ring finger protein 146)
- RPS6KB1
(ribosomal protein S6 kinase B1)
- ROCK1
(Rho associated coiled-coil containing protein kinase 1)
- ROS
(reactive oxygen species)
- RTN4
(reticulon 4)
- S1P
(sphingosine 1-phosphate)
- sEV
(small extracellular vesicles)
- SESN1
(Sestrin 1)
- SIRT1
(sirtuin 1)
- SNHG3
(small nucleolar RNA host gene 3)
- SPHK1
(sphingosine kinase 1)
- SQSTM1/p62
(sequestosome 1)
- SRC
(SRC proto-oncogene, non-receptor tyrosine kinase)
- STAT1
(signal transducer and activator of transcription 1)
- STAT3
(signal transducer and activator of transcription 3)
- STK11
(serine/threonine kinase 11)
- STX17
(syntaxin 17)
- TIGAR
(TP53 induced glycolysis regulatory phosphatase)
- TNF
(tumor necrosis factor)
- TP53/p53
(tumor protein p53)
- tPA
(tissue plasminogen activator)
- TRPM7
(transient receptor potential cation channel subfamily M member 7)
- TSC1
(TSC complex subunit 1)
- TSC2
(TSC complex subunit 2)
- ULK1
(unc-51 like autophagy activating kinase 1)
- ULK2
(unc-51 like autophagy activating kinase 2)
- UPS
(ubiquitin-proteasome system)
- VEGFA/C
(vascular endothelial growth factor A/C)
- WNT
(Wnt family member)
Footnotes
DISCLOSURE STATEMENT: NH serves as a consultant to Astrocyte Pharmaceuticals, Inc. All other authors have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aidar FJ, de Oliveira RJ, Silva AJ, de Matos DG, Mazini Filho ML, Hickner RC, & Machado Reis V (2012). The influence of resistance exercise training on the levels of anxiety in ischemic stroke. Stroke research and treatment, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajoolabady A, Aghanejad A, Bi Y, Zhang Y, Aslkhodapasand HH, Abhari A, & Ren J (2020). Enzyme-based autophagy in anti-neoplastic management: From molecular mechanisms to clinical therapeutics. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer, 188366. [DOI] [PubMed] [Google Scholar]
- Ajoolabady A, Aslkhodapasandhokmabad H, Aghanejad A, Zhang Y, & Ren J (2020). Mitophagy Receptors and Mediators: Therapeutic Targets in the Management of Cardiovascular Ageing. Ageing Research Reviews, 101129. [DOI] [PubMed] [Google Scholar]
- Ajoolabady A, Aslkhodapasandhokmabad H, Henninger N, Demillard LJ, Nikanfar M, Nourazarian A, & Ren J (2021). Targeting Autophagy in Neurodegenerative Diseases: from Molecular Mechanisms to Clinical Therapeutics. Clin Exp Pharmacol Physiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajoolabady A, Wang S, Kroemer G, Klionsky DJ, Uversky VN, Sowers JR, Aslkhodapasandhokmabad H, Bi Y, Ge J, & Ren J (2021). ER stress in Cardiometabolic Diseases: from Molecular Mechanisms to Therapeutics. Endocrine Reviews. [DOI] [PubMed] [Google Scholar]
- Akkoc Y, & Gozuacik D (2020). MicroRNAs as major regulators of the autophagy pathway. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1867, 118662. [DOI] [PubMed] [Google Scholar]
- Alirezaei M, Kemball CC, Flynn CT, Wood MR, Whitton JL, & Kiosses WB (2010). Short-term fasting induces profound neuronal autophagy. Autophagy, 6, 702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh L, Zarebkohan A, Salehi R, Ajjoolabady A, & Rahmati-Yamchi M (2019). Chitosan-based nanotherapeutics for ovarian cancer treatment. Journal of drug targeting, 27, 839–852. [DOI] [PubMed] [Google Scholar]
- Almeida MF, Silva CM, Chaves RS, Lima NC, Almeida RS, Melo KP, Demasi M, Fernandes T, Oliveira EM, & Netto LE (2018). Effects of mild running on substantia nigra during early neurodegeneration. Journal of sports sciences, 36, 1363–1370. [DOI] [PubMed] [Google Scholar]
- Andreotti DZ, do Nascimento Silva J, Matumoto AM, Orellana AM, de Mello PS, & Kawamoto EM (2020). Effects of physical exercise on autophagy and apoptosis in aged brain: human and animal studies. pathways, 29, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee SK, Ramani R, Saba S, Rager J, Tian R, Mathier MA, & Ahmad F (2007). A PRKAG2 mutation causes biphasic changes in myocardial AMPK activity and does not protect against ischemia. Biochemical and Biophysical Research Communications, 360, 381–387. [DOI] [PubMed] [Google Scholar]
- Bano D, & Nicotera P (2007). Ca2+ signals and neuronal death in brain ischemia. Stroke, 38, 674–676. [DOI] [PubMed] [Google Scholar]
- Bar-Yosef T, Damri O, & Agam G (2019). Dual role of autophagy in diseases of the central nervous system. Frontiers in cellular neuroscience, 13, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batatinha HAP, Diniz TA, de Souza Teixeira AA, Krüger K, & Rosa-Neto JC (2019). Regulation of autophagy as a therapy for immunosenescence-driven cancer and neurodegenerative diseases: The role of exercise. Journal of cellular physiology, 234, 14883–14895. [DOI] [PubMed] [Google Scholar]
- Berry DL, & Baehrecke EH (2007). Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell, 131, 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonying W, Joselin A, Huang E, Qu D, Safarpour F, Iyirhiaro GO, Gonzalez YR, Callaghan SM, Slack RS, & Figeys D (2019). Pink1 regulates FKBP 5 interaction with AKT/PHLPP and protects neurons from neurotoxin stress induced by MPP+. Journal of Neurochemistry, 150, 312–329. [DOI] [PubMed] [Google Scholar]
- Brumm AJ, & Carmichael ST (2012). Not just a rush of blood to the head. Nature medicine, 18, 1609–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley K (2015). Rapamycin, an evolving role in up-regulation of autophagy to improve stroke outcome and increase neuronal survival to stroke type injuries.
- Buckley KM, Hess DL, Sazonova IY, Periyasamy-Thandavan S, Barrett JR, Kirks R, Grace H, Kondrikova G, Johnson MH, & Hess DC (2014). Rapamycin up-regulation of autophagy reduces infarct size and improves outcomes in both permanent MCAL, and embolic MCAO, murine models of stroke. Experimental & translational stroke medicine, 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button RW, Luo S, & Rubinsztein DC (2015). Autophagic activity in neuronal cell death. Neuroscience bulletin, 31, 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Li X, Pan Z, Zhu Y, Tuo J, Meng Q, Dai G, Yang G, & Pan Y (2020). Anthocyanin ameliorates hypoxia and ischemia induced inflammation and apoptosis by increasing autophagic flux in SH-SY5Y cells. European Journal of Pharmacology, 883, 173360. [DOI] [PubMed] [Google Scholar]
- Campbell RB, Liu F, & Ross AH (2003). Allosteric activation of PTEN phosphatase by phosphatidylinositol 4, 5-bisphosphate. Journal of Biological Chemistry, 278, 33617–33620. [DOI] [PubMed] [Google Scholar]
- Cao Y, Pan L, Zhang X, Guo W, & Huang D (2020). LncRNA SNHG3 promotes autophagy-induced neuronal cell apoptosis by acting as a ceRNA for miR-485 to upregulate ATG7 expression. Metabolic Brain Disease, 1–9. [DOI] [PubMed] [Google Scholar]
- Cassidy A, Rimm EB, O’Reilly ÉJ, Logroscino G, Kay C, Chiuve SE, & Rexrode KM (2012). Dietary flavonoids and risk of stroke in women. Stroke, 43, 946–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerychova R, Bohuslavova R, Papousek F, Sedmera D, Abaffy P, Benes V, Kolar F, & Pavlinkova G (2018). Adverse effects of Hif1a mutation and maternal diabetes on the offspring heart. Cardiovascular diabetology, 17, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen W, Nong Z, Nie Y, Chen X, Pan X, Guo Y, Yao M, & Deng W (2020). Hyperbaric oxygen alleviated cognitive impairments in mice induced by repeated cerebral ischemia-reperfusion injury via inhibition of autophagy. Life Sciences, 241, 117170. [DOI] [PubMed] [Google Scholar]
- Chen C, Yu Q, Xu K, Cai L, Felicia BM, Wang L, Zhang A, Dai Q, Geng W, & Wang J (2020). Electroacupuncture Pretreatment Prevents Ischemic Stroke and Inhibits Wnt Signaling-Mediated Autophagy Through the Regulation of GSK-3β Phosphorylation. Brain Research Bulletin. [DOI] [PubMed] [Google Scholar]
- Chen L, Xia Y-F, Shen S-F, Tang J, Chen J-L, Qian K, Chen Z, Qin Z-H, & Sheng R (2020). Syntaxin 17 inhibits ischemic neuronal injury by resuming autophagy flux and ameliorating endoplasmic reticulum stress. Free Radical Biology and Medicine. [DOI] [PubMed] [Google Scholar]
- Chen R-L, Balami JS, Esiri MM, Chen L-K, & Buchan AM (2010). Ischemic stroke in the elderly: an overview of evidence. Nature Reviews Neurology, 6, 256–265. [DOI] [PubMed] [Google Scholar]
- Chen X, Lin S, Gu L, Zhu X, Zhang Y, Zhang H, Shao B, Zhuge Q, & Jin K (2019). Inhibition of miR-497 improves functional outcome after ischemic stroke by enhancing neuronal autophagy in young and aged rats. Neurochemistry international, 127, 64–72. [DOI] [PubMed] [Google Scholar]
- Chung Y-P, Yen C-C, Tang F-C, Lee K-I, Liu S-H, Wu C-C, Hsieh S-S, Su C-C, Kuo C-Y, & Chen Y-W (2019). Methylmercury exposure induces ROS/Akt inactivation-triggered endoplasmic reticulum stress-regulated neuronal cell apoptosis. Toxicology, 425, 152245. [DOI] [PubMed] [Google Scholar]
- Cui D, Wang L, Jiang W, Qi A, Zhou Q, & Zhang X (2013). Propofol prevents cerebral ischemia-triggered autophagy activation and cell death in the rat hippocampus through the NF-κB/p53 signaling pathway. Neuroscience, 246, 117–132. [DOI] [PubMed] [Google Scholar]
- Culjkovic B, Tan K, Orolicki S, Amri A, Meloche S, & Borden KL (2008). The eIF4E RNA regulon promotes the Akt signaling pathway. The Journal of cell biology, 181, 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins PM (2012). Occludin: one protein, many forms. Molecular and cellular biology, 32, 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G, Shravage BV, & Baehrecke EH (2012). Regulation and function of autophagy during cell survival and cell death. Cold Spring Harbor perspectives in biology, 4, a008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio D, Agnoli C, Pellegrini N, Krogh V, Brighenti F, Mazzeo T, Masala G, Bendinelli B, Berrino F, & Sieri S (2011). Total antioxidant capacity of the diet is associated with lower risk of ischemic stroke in a large Italian cohort. The Journal of nutrition, 141, 118–123. [DOI] [PubMed] [Google Scholar]
- Demchuk AM, Felburg RA, & Alexandrov AV (1999). Clinical recovery from acute ischemic stroke after early reperfusion of the brain with intravenous thrombolysis. New England Journal of Medicine, 340, 894–895. [DOI] [PubMed] [Google Scholar]
- Deng Y. h., He H. y., Yang L. q., & Zhang P. y. (2016). Dynamic changes in neuronal autophagy and apoptosis in the ischemic penumbra following permanent ischemic stroke. Neural regeneration research, 11, 1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniaud A, Maillier E, Poncet D, Kroemer G, Lemaire C, & Brenner C (2008). Endoplasmic reticulum stress induces calcium-dependent permeability transition, mitochondrial outer membrane permeabilization and apoptosis. Oncogene, 27, 285–299. [DOI] [PubMed] [Google Scholar]
- Deretic V, Saitoh T, & Akira S (2013). Autophagy in infection, inflammation and immunity. Nature Reviews Immunology, 13, 722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Xiao S, Cheng M, Ye X, & Zheng G (2016). Minocycline induces protective autophagy in vascular endothelial cells exposed to an in vitro model of ischemia/reperfusion‑induced injury. Biomedical reports, 4, 173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Garcia C, Atif F, Yousuf S, Sayeed I, Neigh GN, & Stein DG (2020). Progesterone Attenuates Stress-Induced NLRP3 Inflammasome Activation and Enhances Autophagy Following Ischemic Brain Injury. International journal of molecular sciences, 21, 3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakharnia F, Khodagholi F, Dargahi L, & Ahmadiani A (2017). Prevention of cyclophilin D-mediated mPTP opening using cyclosporine-A alleviates the elevation of necroptosis, autophagy and apoptosis-related markers following global cerebral ischemia-reperfusion. Journal of Molecular Neuroscience, 61, 52–60. [DOI] [PubMed] [Google Scholar]
- Fang CX, Wu S, & Ren J (2006). Intracerebral hemorrhage elicits aberration in cardiomyocyte contractile function and intracellular Ca2+ transients. Stroke, 37, 1875–1882. [DOI] [PubMed] [Google Scholar]
- Feng D, Wang B, Wang L, Abraham N, Tao K, Huang L, Shi W, Dong Y, & Qu Y (2017). Pre-ischemia melatonin treatment alleviated acute neuronal injury after ischemic stroke by inhibiting endoplasmic reticulum stress-dependent autophagy via PERK and IRE 1 signalings. Journal of pineal research, 62, e12395. [DOI] [PubMed] [Google Scholar]
- Ferreira-Marques M, Aveleira CA, Carmo-Silva S, Botelho M, de Almeida LP, & Cavadas C (2016). Caloric restriction stimulates autophagy in rat cortical neurons through neuropeptide Y and ghrelin receptors activation. Aging (Albany NY), 8, 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsom AR, Rasmussen ML, Chambless LE, Howard G, Cooper LS, Schmidt MI, & Heiss G (1999). Prospective associations of fasting insulin, body fat distribution, and diabetes with risk of ischemic stroke. The Atherosclerosis Risk in Communities (ARIC) Study Investigators. Diabetes care, 22, 1077–1083. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK-H, Burge CB, & Bartel DP (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome research, 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Zhang X, Lu Y, Wang F, Xu Z, Liu S, Zheng H, & Liu X (2020). Geniposide inhibits NLRP3 inflammasome activation via autophagy in BV-2 microglial cells exposed to oxygen–glucose deprivation/reoxygenation. International immunopharmacology, 84, 106547. [DOI] [PubMed] [Google Scholar]
- Fukuta T, Asai T, Yanagida Y, Namba M, Koide H, Shimizu K, & Oku N (2017). Combination therapy with liposomal neuroprotectants and tissue plasminogen activator for treatment of ischemic stroke. The FASEB Journal, 31, 1879–1890. [DOI] [PubMed] [Google Scholar]
- Fusakio ME, Willy JA, Wang Y, Mirek ET, Al Baghdadi RJ, Adams CM, Anthony TG, & Wek RC (2016). Transcription factor ATF4 directs basal and stress-induced gene expression in the unfolded protein response and cholesterol metabolism in the liver. Mol Biol Cell, 27, 1536–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Bravo-San Pedro JM, Blomgren K, & Kroemer G (2016). Autophagy in acute brain injury. Nat Rev Neurosci, 17, 467–484. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams J, Alnemri E, Baehrecke E, Blagosklonny M, Dawson TM, Dawson V, El-Deiry W, & Fulda S (2012). Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death & Differentiation, 19, 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Li F, Long L, Xu F, Feng L, Liu Y, Shi J, & Gong Q (2019). Icariside II, a phosphodiesterase 5 inhibitor, attenuates brain injury in focal cerebral ischemia/reperfusion rats: Involvement of inhibition of GSK-3β-mediated activation of autophagy. The FASEB Journal, 33, 500.513–500.513. [Google Scholar]
- Gao J, Long L, Xu F, Feng L, Liu Y, Shi J, & Gong Q (2020). Icariside II, a phosphodiesterase 5 inhibitor, attenuates cerebral ischaemia/reperfusion injury by inhibiting glycogen synthase kinase-3β-mediated activation of autophagy. British journal of pharmacology, 177, 1434–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Jiang T, Guo J, Liu Y, Cui G, Gu L, Su L, & Zhang Y (2012). Inhibition of autophagy contributes to ischemic postconditioning-induced neuroprotection against focal cerebral ischemia in rats. PLoS One, 7, e46092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Yang H, & ZhiPing T (2006). Association studies of genetic polymorphism, environmental factors and their interaction in ischemic stroke. Neuroscience letters, 398, 172–177. [DOI] [PubMed] [Google Scholar]
- Gardener H, Wright CB, Gu Y, Demmer RT, Boden-Albala B, Elkind MS, Sacco RL, & Scarmeas N (2011). Mediterranean-style diet and risk of ischemic stroke, myocardial infarction, and vascular death: the Northern Manhattan Study. The American journal of clinical nutrition, 94, 1458–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislat G, & Knecht E (2015). Regulation of Autophagy by Amino Acid Starvation Involving Ca2+. In Autophagy: Cancer, Other Pathologies, Inflammation, Immunity, Infection, and Aging (pp. 69–79): Elsevier. [Google Scholar]
- Ginet V, Puyal J, Clarke PG, & Truttmann AC (2009). Enhancement of autophagic flux after neonatal cerebral hypoxia-ischemia and its region-specific relationship to apoptotic mechanisms. The American journal of pathology, 175, 1962–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Brännström T, Jiang W, Bergh A, & Wester P (2001). Vascular endothelial growth factor-A and-C protein up-regulation and early angiogenesis in a rat photothrombotic ring stroke model with spontaneous reperfusion. Acta neuropathologica, 102, 216–226. [DOI] [PubMed] [Google Scholar]
- Guo F, Liu X, Cai H, & Le W (2018). Autophagy in neurodegenerative diseases: pathogenesis and therapy. Brain pathology, 28, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson A. s. B., & Gottlieb RA (2009). Autophagy in ischemic heart disease. Circulation research, 104, 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi V, Farhadi S, Chaleshtari MG, Seashore-Ludlow B, Masjedi A, Hojjat-Farsangi M, Namdar A, Ajjoolabady A, Mohammadi H, & Ghalamfarsa G (2020). Nanomedicine for improvement of dendritic cell-based cancer immunotherapy. International immunopharmacology, 83, 106446. [DOI] [PubMed] [Google Scholar]
- He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, & Sun Q (2012). Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature, 481, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Sumpter J, Rhea, & Levine B (2012). Exercise induces autophagy in peripheral tissues and in the brain. Autophagy, 8, 1548–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H-Y, Ren L, Guo T, & Deng Y-H (2019). Neuronal autophagy aggravates microglial inflammatory injury by downregulating CX3CL1/fractalkine after ischemic stroke. Neural regeneration research, 14, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Li Z, Wang Y, Hou Y, Li L, & Zhao J (2017). Resveratrol alleviates cerebral ischemia/reperfusion injury in rats by inhibiting NLRP3 inflammasome activation through Sirt1-dependent autophagy induction. International immunopharmacology, 50, 208–215. [DOI] [PubMed] [Google Scholar]
- Henninger N, & Mayasi Y (2019). Nucleic acid therapies for ischemic stroke. Neurotherapeutics, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongyun H, Tao G, Pengyue Z, Liqiang Y, & Yihao D (2017). Puerarin provides a neuroprotection against transient cerebral ischemia by attenuating autophagy at the ischemic penumbra in neurons but not in astrocytes. Neuroscience letters, 643, 45–51. [DOI] [PubMed] [Google Scholar]
- Hu G. w., Li Q, Niu X, Hu B, Liu J, Zhou S. m., Guo S. c., Lang H. l., Zhang C. q., & Wang Y (2015). Exosomes secreted by human-induced pluripotent stem cell-derived mesenchymal stem cells attenuate limb ischemia by promoting angiogenesis in mice. Stem cell research & therapy, 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang X, Zhu Y, Li Z, Zhu YT, Wu JC, Qin ZH, Xiang M, & Lin F (2019). Exercise activates lysosomal function in the brain through AMPK-SIRT1-TFEB pathway. CNS neuroscience & therapeutics, 25, 796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Lou T, Wang M, Xue L, Lu J, Zhang H, Zhang Z, Wang H, Jing C, & Zhao D (2020). Compound K inhibits autophagy-mediated apoptosis induced by oxygen and glucose deprivation/reperfusion via regulating AMPK-mTOR pathway in neurons. Life Sciences, 117793. [DOI] [PubMed] [Google Scholar]
- Hwang J-Y, Gertner M, Pontarelli F, Court-Vazquez B, Bennett MVL, Ofengeim D, & Zukin RS (2017). Global ischemia induces lysosomal-mediated degradation of mTOR and activation of autophagy in hippocampal neurons destined to die. Cell Death & Differentiation, 24, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijaz M, Shahabz M, Jiang W, Fathy A, Nesa E, Wang D, & Wang F (2017). Oncogenic role of Grb2 in breast cancer and Grb2 antagonists as therapeutic drugs. Canc Therapy & Oncol Int J, 3, 1–7. [Google Scholar]
- Itakura E, Kishi C, Inoue K, & Mizushima N (2008). Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Molecular biology of the cell, 19, 5360–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamart C, Benoit N, Raymackers J-M, Kim HJ, Kim CK, & Francaux M (2012). Autophagy-related and autophagy-regulatory genes are induced in human muscle after ultraendurance exercise. European journal of applied physiology, 112, 3173–3177. [DOI] [PubMed] [Google Scholar]
- Jang Y (2020). Endurance exercise-induced expression of autophagy-related protein coincides with anabolic expression and neurogenesis in the hippocampus of the mouse brain. NeuroReport, 31, 442–449. [DOI] [PubMed] [Google Scholar]
- Jeong JH, Yu KS, Bak DH, Lee JH, Lee NS, Jeong YG, Kim DK, Kim JJ, & Han SY (2016). Intermittent fasting is neuroprotective in focal cerebral ischemia by minimizing autophagic flux disturbance and inhibiting apoptosis. Experimental and therapeutic medicine, 12, 3021–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Wang H, Jin M, Yang X, Ji H, Jiang Y, Zhang H, Wu F, Wu G, & Lai X (2018). Exosomes from MiR-30d-5p-ADSCs reverse acute ischemic stroke-induced, autophagy-mediated brain injury by promoting M2 microglial/macrophage polarization. Cellular Physiology and Biochemistry, 47, 864–878. [DOI] [PubMed] [Google Scholar]
- Jiang T, Yu J-T, Zhu X-C, Zhang Q-Q, Tan M-S, Cao L, Wang H-F, Shi J-Q, Gao L, & Qin H (2015). Ischemic preconditioning provides neuroprotection by induction of AMP-activated protein kinase-dependent autophagy in a rat model of ischemic stroke. Molecular neurobiology, 51, 220–229. [DOI] [PubMed] [Google Scholar]
- Jiang T, Yu JT, Zhu XC, Wang HF, Tan MS, Cao L, Zhang QQ, Gao L, Shi JQ, & Zhang YD (2014). Acute metformin preconditioning confers neuroprotection against focal cerebral ischaemia by pre-activation of AMPK-dependent autophagy. British journal of pharmacology, 171, 3146–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie Z, Luo W, Xiong J, Wang Y, Jia W, & Han J (2020). Ischemic postconditioning plays a protective role in brain through regulating autophagy by LncRNAs. Neuropsychiatric Sciences and Molecular Biology, 1, 11–22. [Google Scholar]
- Jolugbo P, & Ariëns RA (2021). Thrombus Composition and Efficacy of Thrombolysis and Thrombectomy in Acute Ischemic Stroke. Stroke, STROKEAHA. 120.032810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkin J, Henkel T, Nielsen AF, Minnich M, Popow J, Kaufmann T, Heindl K, Hoffmann T, Busslinger M, & Martinez J (2014). The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J, 33, 2922–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang E-B, & Cho J-Y (2015). Effect of treadmill exercise on PI3K/AKT/mTOR, autophagy, and Tau hyperphosphorylation in the cerebral cortex of NSE/htau23 transgenic mice. Journal of exercise nutrition & biochemistry, 19, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Zeh H, Lotze M, & Tang D (2011). The Beclin 1 network regulates autophagy and apoptosis. Cell Death & Differentiation, 18, 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly-Hayes M, Beiser A, Kase CS, Scaramucci A, D’Agostino RB, & Wolf PA (2003). The influence of gender and age on disability following ischemic stroke: the Framingham study. Journal of Stroke and Cerebrovascular Diseases, 12, 119–126. [DOI] [PubMed] [Google Scholar]
- Khiavi MA, Safary A, Barar J, Ajoolabady A, Somi MH, & Omidi Y (2020). Multifunctional nanomedicines for targeting epidermal growth factor receptor in colorectal cancer. Cellular and Molecular Life Sciences, 77, 997–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-A, Kim D, Kim J-H, Shin Y-J, Kim E-S, Akram M, Kim E-H, Majid A, Baek S-H, & Bae O-N (2020). Autophagy-mediated occludin degradation contributes to blood–brain barrier disruption during ischemia in bEnd. 3 brain endothelial cells and rat ischemic stroke models. Fluids and Barriers of the CNS, 17, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Yoon HH, Lee YD, Youn D-Y, Ha TJ, Kim H-S, & Lee J-H (2012). Anthocyanin extracts from black soybean (Glycine max L.) protect human glial cells against oxygen-glucose deprivation by promoting autophagy. Biomolecules & therapeutics, 20, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, Abeliovich H, Abildgaard MH, Abudu YP, Acevedo-Arozena A, Adamopoulos IE, Adeli K, Adolph TE, Adornetto A, Aflaki E, Agam G, Agarwal A, Aggarwal BB, Agnello M, Agostinis P, Agrewala JN, Agrotis A, Aguilar PV, Ahmad ST, Ahmed ZM, Ahumada-Castro U, Aits S, Aizawa S, Akkoc Y, Akoumianaki T, Akpinar HA, Al-Abd AM, Al-Akra L, Al-Gharaibeh A, Alaoui-Jamali MA, Alberti S, Alcocer-Gomez E, Alessandri C, Ali M, Alim Al-Bari MA, Aliwaini S, Alizadeh J, Almacellas E, Almasan A, Alonso A, Alonso GD, Altan-Bonnet N, Altieri DC, Alvarez EMC, Alves S, Alves da Costa C, Alzaharna MM, Amadio M, Amantini C, Amaral C, Ambrosio S, Amer AO, Ammanathan V, An Z, Andersen SU, Andrabi SA, Andrade-Silva M, Andres AM, Angelini S, Ann D, Anozie UC, Ansari MY, Antas P, Antebi A, Anton Z, Anwar T, Apetoh L, Apostolova N, Araki T, Araki Y, Arasaki K, Araujo WL, Araya J, Arden C, Arevalo MA, Arguelles S, Arias E, Arikkath J, Arimoto H, Ariosa AR, Armstrong-James D, Arnaune-Pelloquin L, Aroca A, Arroyo DS, Arsov I, Artero R, Asaro DML, Aschner M, Ashrafizadeh M, Ashur-Fabian O, Atanasov AG, Au AK, Auberger P, Auner HW, Aurelian L, Autelli R, Avagliano L, Avalos Y, Aveic S, Aveleira CA, Avin-Wittenberg T, Aydin Y, Ayton S, Ayyadevara S, Azzopardi M, Baba M, Backer JM, Backues SK, Bae DH, Bae ON, Bae SH, Baehrecke EH, Baek A, Baek SH, Baek SH, Bagetta G, Bagniewska-Zadworna A, Bai H, Bai J, Bai X, Bai Y, Bairagi N, Baksi S, Balbi T, Baldari CT, Balduini W, Ballabio A, Ballester M, Balazadeh S, Balzan R, Bandopadhyay R, Banerjee S, Banerjee S, Banreti A, Bao Y, Baptista MS, Baracca A, Barbati C, Bargiela A, Barila D, Barlow PG, Barmada SJ, Barreiro E, Barreto GE, Bartek J, Bartel B, Bartolome A, Barve GR, Basagoudanavar SH, Bassham DC, Bast RC Jr., Basu A, Batoko H, Batten I, Baulieu EE, Baumgarner BL, Bayry J, Beale R, Beau I, Beaumatin F, Bechara LRG, Beck GR Jr., Beers MF, Begun J, Behrends C, Behrens GMN, Bei R, Bejarano E, Bel S, Behl C, Belaid A, Belgareh-Touze N, Bellarosa C, Belleudi F, Bello Perez M, Bello-Morales R, Beltran JSO, Beltran S, Benbrook DM, Bendorius M, Benitez BA, Benito-Cuesta I, Bensalem J, Berchtold MW, Berezowska S, Bergamaschi D, Bergami M, Bergmann A, Berliocchi L, Berlioz-Torrent C, Bernard A, Berthoux L, Besirli CG, Besteiro S, Betin VM, Beyaert R, Bezbradica JS, Bhaskar K, Bhatia-Kissova I, Bhattacharya R, Bhattacharya S, Bhattacharyya S, Bhuiyan MS, Bhutia SK, Bi L, Bi X, Biden TJ, Bijian K, Billes VA, Binart N, Bincoletto C, Birgisdottir AB, Bjorkoy G, Blanco G, Blas-Garcia A, Blasiak J, Blomgran R, Blomgren K, Blum JS, Boada-Romero E, Boban M, Boesze-Battaglia K, Boeuf P, Boland B, Bomont P, Bonaldo P, Bonam SR, Bonfili L, Bonifacino JS, Boone BA, Bootman MD, Bordi M, Borner C, Bornhauser BC, Borthakur G, Bosch J, Bose S, Botana LM, Botas J, Boulanger CM, Boulton ME, Bourdenx M, Bourgeois B, Bourke NM, Bousquet G, Boya P, Bozhkov PV, Bozi LHM, Bozkurt TO, Brackney DE, Brandts CH, Braun RJ, Braus GH, Bravo-Sagua R, Bravo-San Pedro JM, Brest P, Bringer MA, Briones-Herrera A, Broaddus VC, Brodersen P, Brodsky JL, Brody SL, Bronson PG, Bronstein JM, Brown CN, Brown RE, Brum PC, Brumell JH, Brunetti-Pierri N, Bruno D, Bryson-Richardson RJ, Bucci C, Buchrieser C, Bueno M, Buitrago-Molina LE, Buraschi S, Buch S, Buchan JR, Buckingham EM, Budak H, Budini M, Bultynck G, Burada F, Burgoyne JR, Buron MI, Bustos V, Buttner S, Butturini E, Byrd A, Cabas I, Cabrera-Benitez S, Cadwell K, Cai J, Cai L, Cai Q, Cairo M, Calbet JA, Caldwell GA, Caldwell KA, Call JA, Calvani R, Calvo AC, Calvo-Rubio Barrera M, Camara NO, Camonis JH, Camougrand N, Campanella M, Campbell EM, Campbell-Valois FX, Campello S, Campesi I, Campos JC, Camuzard O, Cancino J, Candido de Almeida D, Canesi L, Caniggia I, Canonico B, Canti C, Cao B, Caraglia M, Carames B, Carchman EH, Cardenal-Munoz E, Cardenas C, Cardenas L, Cardoso SM, Carew JS, Carle GF, Carleton G, Carloni S, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carosi JM, Carra S, Carrier A, Carrier L, Carroll B, Carter AB, Carvalho AN, Casanova M, Casas C, Casas J, Cassioli C, Castillo EF, Castillo K, Castillo-Lluva S, Castoldi F, Castori M, Castro AF, Castro-Caldas M, Castro-Hernandez J, Castro-Obregon S, Catz SD, Cavadas C, Cavaliere F, Cavallini G, Cavinato M, Cayuela ML, Cebollada Rica P, Cecarini V, Cecconi F, Cechowska-Pasko M, Cenci S, Ceperuelo-Mallafre V, Cerqueira JJ, Cerutti JM, Cervia D, Cetintas VB, Cetrullo S, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chakrabarti O, Chakraborty T, Chakraborty T, Chami M, Chamilos G, Chan DW, Chan EYW, Chan ED, Chan HYE, Chan HH, Chan H, Chan MTV, Chan YS, Chandra PK, Chang CP, Chang C, Chang HC, Chang K, Chao J, Chapman T, Charlet-Berguerand N, Chatterjee S, Chaube SK, Chaudhary A, Chauhan S, Chaum E, Checler F, Cheetham ME, Chen CS, Chen GC, Chen JF, Chen LL, Chen L, Chen L, Chen M, Chen MK, Chen N, Chen Q, Chen RH, Chen S, Chen W, Chen W, Chen XM, Chen XW, Chen X, Chen Y, Chen YG, Chen Y, Chen Y, Chen YJ, Chen YQ, Chen ZS, Chen Z, Chen ZH, Chen ZJ, Chen Z, Cheng H, Cheng J, Cheng SY, Cheng W, Cheng X, Cheng XT, Cheng Y, Cheng Z, Chen Z, Cheong H, Cheong JK, Chernyak BV, Cherry S, Cheung CFR, Cheung CHA, Cheung KH, Chevet E, Chi RJ, Chiang AKS, Chiaradonna F, Chiarelli R, Chiariello M, Chica N, Chiocca S, Chiong M, Chiou SH, Chiramel AI, Chiurchiu V, Cho DH, Choe SK, Choi AMK, Choi ME, Choudhury KR, Chow NS, Chu CT, Chua JP, Chua JJE, Chung H, Chung KP, Chung S, Chung SH, Chung YL, Cianfanelli V, Ciechomska IA, Cifuentes M, Cinque L, Cirak S, Cirone M, Clague MJ, Clarke R, Clementi E, Coccia EM, Codogno P, Cohen E, Cohen MM, Colasanti T, Colasuonno F, Colbert RA, Colell A, Colic M, Coll NS, Collins MO, Colombo MI, Colon-Ramos DA, Combaret L, Comincini S, Cominetti MR, Consiglio A, Conte A, Conti F, Contu VR, Cookson MR, Coombs KM, Coppens I, Corasaniti MT, Corkery DP, Cordes N, Cortese K, Costa MDC, Costantino S, Costelli P, Coto-Montes A, Crack PJ, Crespo JL, Criollo A, Crippa V, Cristofani R, Csizmadia T, Cuadrado A, Cui B, Cui J, Cui Y, Cui Y, Culetto E, Cumino AC, Cybulsky AV, Czaja MJ, Czuczwar SJ, D'Adamo S, D'Amelio M, D'Arcangelo D, D'Lugos AC, D'Orazi G, da Silva JA, Dafsari HS, Dagda RK, Dagdas Y, Daglia M, Dai X, Dai Y, Dai Y, Dal Col J, Dalhaimer P, Dalla Valle L, Dallenga T, Dalmasso G, Damme M, Dando I, Dantuma NP, Darling AL, Das H, Dasarathy S, Dasari SK, Dash S, Daumke O, Dauphinee AN, Davies JS, Davila VA, Davis RJ, Davis T, Dayalan Naidu S, De Amicis F, De Bosscher K, De Felice F, De Franceschi L, De Leonibus C, de Mattos Barbosa MG, De Meyer GRY, De Milito A, De Nunzio C, De Palma C, De Santi M, De Virgilio C, De Zio D, Debnath J, DeBosch BJ, Decuypere JP, Deehan MA, Deflorian G, DeGregori J, Dehay B, Del Rio G, Delaney JR, Delbridge LMD, Delorme-Axford E, Delpino MV, Demarchi F, Dembitz V, Demers ND, Deng H, Deng Z, Dengjel J, Dent P, Denton D, DePamphilis ML, Der CJ, Deretic V, Descoteaux A, Devis L, Devkota S, Devuyst O, Dewson G, Dharmasivam M, Dhiman R, di Bernardo D, Di Cristina M, Di Domenico F, Di Fazio P, Di Fonzo A, Di Guardo G, Di Guglielmo GM, Di Leo L, Di Malta C, Di Nardo A, Di Rienzo M, Di Sano F, Diallinas G, Diao J, Diaz-Araya G, Diaz-Laviada I, Dickinson JM, Diederich M, Dieude M, Dikic I, Ding S, Ding WX, Dini L, Dinic J, Dinic M, Dinkova-Kostova AT, Dionne MS, Distler JHW, Diwan A, Dixon IMC, Djavaheri-Mergny M, Dobrinski I, Dobrovinskaya O, Dobrowolski R, Dobson RCJ, Dokic J, Dokmeci Emre S, Donadelli M, Dong B, Dong X, Dong Z, Dorn Ii GW, Dotsch V, Dou H, Dou J, Dowaidar M, Dridi S, Drucker L, Du A, Du C, Du G, Du HN, Du LL, du Toit A, Duan SB, Duan X, Duarte SP, Dubrovska A, Dunlop EA, Dupont N, Duran RV, Dwarakanath BS, Dyshlovoy SA, Ebrahimi-Fakhari D, Eckhart L, Edelstein CL, Efferth T, Eftekharpour E, Eichinger L, Eid N, Eisenberg T, Eissa NT, Eissa S, Ejarque M, El Andaloussi A, El-Hage N, El-Naggar S, Eleuteri AM, El-Shafey ES, Elgendy M, Eliopoulos AG, Elizalde MM, Elks PM, Elsasser HP, Elsherbiny ES, Emerling BM, Emre NCT, Eng CH, Engedal N, Engelbrecht AM, Engelsen AST, Enserink JM, Escalante R, Esclatine A, Escobar-Henriques M, Eskelinen EL, Espert L, Eusebio MO, Fabrias G, Fabrizi C, Facchiano A, Facchiano F, Fadeel B, Fader C, Faesen AC, Fairlie WD, Falco A, Falkenburger BH, Fan D, Fan J, Fan Y, Fang EF, Fang Y, Fang Y, Fanto M, Farfel-Becker T, Faure M, Fazeli G, Fedele AO, Feldman AM, Feng D, Feng J, Feng L, Feng Y, Feng Y, Feng W, Fenz Araujo T, Ferguson TA, Fernandez AF, Fernandez-Checa JC, Fernandez-Veledo S, Fernie AR, Ferrante AW Jr., Ferraresi A, Ferrari MF, Ferreira JCB, Ferro-Novick S, Figueras A, Filadi R, Filigheddu N, Filippi-Chiela E, Filomeni G, Fimia GM, Fineschi V, Finetti F, Finkbeiner S, Fisher EA, Fisher PB, Flamigni F, Fliesler SJ, Flo TH, Florance I, Florey O, Florio T, Fodor E, Follo C, Fon EA, Forlino A, Fornai F, Fortini P, Fracassi A, Fraldi A, Franco B, Franco R, Franconi F, Frankel LB, Friedman SL, Frohlich LF, Fruhbeck G, Fuentes JM, Fujiki Y, Fujita N, Fujiwara Y, Fukuda M, Fulda S, Furic L, Furuya N, Fusco C, Gack MU, Gaffke L, Galadari S, Galasso A, Galindo MF, Gallolu Kankanamalage S, Galluzzi L, Galy V, Gammoh N, Gan B, Ganley IG, Gao F, Gao H, Gao M, Gao P, Gao SJ, Gao W, Gao X, Garcera A, Garcia MN, Garcia VE, Garcia-Del Portillo F, Garcia-Escudero V, Garcia-Garcia A, Garcia-Macia M, Garcia-Moreno D, Garcia-Ruiz C, Garcia-Sanz P, Garg AD, Gargini R, Garofalo T, Garry RF, Gassen NC, Gatica D, Ge L, Ge W, Geiss-Friedlander R, Gelfi C, Genschik P, Gentle IE, Gerbino V, Gerhardt C, Germain K, Germain M, Gewirtz DA, Ghasemipour Afshar E, Ghavami S, Ghigo A, Ghosh M, Giamas G, Giampietri C, Giatromanolaki A, Gibson GE, Gibson SB, Ginet V, Giniger E, Giorgi C, Girao H, Girardin SE, Giridharan M, Giuliano S, Giulivi C, Giuriato S, Giustiniani J, Gluschko A, Goder V, Goginashvili A, Golab J, Goldstone DC, Golebiewska A, Gomes LR, Gomez R, Gomez-Sanchez R, Gomez-Puerto MC, Gomez-Sintes R, Gong Q, Goni FM, Gonzalez-Gallego J, Gonzalez-Hernandez T, Gonzalez-Polo RA, Gonzalez-Reyes JA, Gonzalez-Rodriguez P, Goping IS, Gorbatyuk MS, Gorbunov NV, Gorgulu K, Gorojod RM, Gorski SM, Goruppi S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graef M, Graler MH, Granatiero V, Grasso D, Gray JP, Green DR, Greenhough A, Gregory SL, Griffin EF, Grinstaff MW, Gros F, Grose C, Gross AS, Gruber F, Grumati P, Grune T, Gu X, Guan JL, Guardia CM, Guda K, Guerra F, Guerri C, Guha P, Guillen C, Gujar S, Gukovskaya A, Gukovsky I, Gunst J, Gunther A, Guntur AR, Guo C, Guo C, Guo H, Guo LW, Guo M, Gupta P, Gupta SK, Gupta S, Gupta VB, Gupta V, Gustafsson AB, Gutterman DD, H BR, Haapasalo A, Haber JE, Hac A, Hadano S, Hafren AJ, Haidar M, Hall BS, Hallden G, Hamacher-Brady A, Hamann A, Hamasaki M, Han W, Hansen M, Hanson PI, Hao Z, Harada M, Harhaji-Trajkovic L, Hariharan N, Haroon N, Harris J, Hasegawa T, Hasima Nagoor N, Haspel JA, Haucke V, Hawkins WD, Hay BA, Haynes CM, Hayrabedyan SB, Hays TS, He C, He Q, He RR, He YW, He YY, Heakal Y, Heberle AM, Hejtmancik JF, Helgason GV, Henkel V, Herb M, Hergovich A, Herman-Antosiewicz A, Hernandez A, Hernandez C, Hernandez-Diaz S, Hernandez-Gea V, Herpin A, Herreros J, Hervas JH, Hesselson D, Hetz C, Heussler VT, Higuchi Y, Hilfiker S, Hill JA, Hlavacek WS, Ho EA, Ho IHT, Ho PW, Ho SL, Ho WY, Hobbs GA, Hochstrasser M, Hoet PHM, Hofius D, Hofman P, Hohn A, Holmberg CI, Hombrebueno JR, Yi-Ren Hong CH, Hooper LV, Hoppe T, Horos R, Hoshida Y, Hsin IL, Hsu HY, Hu B, Hu D, Hu LF, Hu MC, Hu R, Hu W, Hu YC, Hu ZW, Hua F, Hua J, Hua Y, Huan C, Huang C, Huang C, Huang C, Huang C, Huang H, Huang K, Huang MLH, Huang R, Huang S, Huang T, Huang X, Huang YJ, Huber TB, Hubert V, Hubner CA, Hughes SM, Hughes WE, Humbert M, Hummer G, Hurley JH, Hussain S, Hussain S, Hussey PJ, Hutabarat M, Hwang HY, Hwang S, Ieni A, Ikeda F, Imagawa Y, Imai Y, Imbriano C, Imoto M, Inman DM, Inoki K, Iovanna J, Iozzo RV, Ippolito G, Irazoqui JE, Iribarren P, Ishaq M, Ishikawa M, Ishimwe N, Isidoro C, Ismail N, Issazadeh-Navikas S, Itakura E, Ito D, Ivankovic D, Ivanova S, Iyer AKV, Izquierdo JM, Izumi M, Jaattela M, Jabir MS, Jackson WT, Jacobo-Herrera N, Jacomin AC, Jacquin E, Jadiya P, Jaeschke H, Jagannath C, Jakobi AJ, Jakobsson J, Janji B, Jansen-Durr P, Jansson PJ, Jantsch J, Januszewski S, Jassey A, Jean S, Jeltsch-David H, Jendelova P, Jenny A, Jensen TE, Jessen N, Jewell JL, Ji J, Jia L, Jia R, Jiang L, Jiang Q, Jiang R, Jiang T, Jiang X, Jiang Y, Jimenez-Sanchez M, Jin EJ, Jin F, Jin H, Jin L, Jin L, Jin M, Jin S, Jo EK, Joffre C, Johansen T, Johnson GVW, Johnston SA, Jokitalo E, Jolly MK, Joosten LAB, Jordan J, Joseph B, Ju D, Ju JS, Ju J, Juarez E, Judith D, Juhasz G, Jun Y, Jung CH, Jung SC, Jung YK, Jungbluth H, Jungverdorben J, Just S, Kaarniranta K, Kaasik A, Kabuta T, Kaganovich D, Kahana A, Kain R, Kajimura S, Kalamvoki M, Kalia M, Kalinowski DS, Kaludercic N, Kalvari I, Kaminska J, Kaminskyy VO, Kanamori H, Kanasaki K, Kang C, Kang R, Kang SS, Kaniyappan S, Kanki T, Kanneganti TD, Kanthasamy AG, Kanthasamy A, Kantorow M, Kapuy O, Karamouzis MV, Karim MR, Karmakar P, Katare RG, Kato M, Kaufmann SHE, Kauppinen A, Kaushal GP, Kaushik S, Kawasaki K, Kazan K, Ke PY, Keating DJ, Keber U, Kehrl JH, Keller KE, Keller CW, Kemper JK, Kenific CM, Kepp O, Kermorgant S, Kern A, Ketteler R, Keulers TG, Khalfin B, Khalil H, Khambu B, Khan SY, Khandelwal VKM, Khandia R, Kho W, Khobrekar NV, Khuansuwan S, Khundadze M, Killackey SA, Kim D, Kim DR, Kim DH, Kim DE, Kim EY, Kim EK, Kim HR, Kim HS, Hyung-Ryong K, Kim JH, Kim JK, Kim JH, Kim J, Kim JH, Kim KI, Kim PK, Kim SJ, Kimball SR, Kimchi A, Kimmelman AC, Kimura T, King MA, Kinghorn KJ, Kinsey CG, Kirkin V, Kirshenbaum LA, Kiselev SL, Kishi S, Kitamoto K, Kitaoka Y, Kitazato K, Kitsis RN, Kittler JT, Kjaerulff O, Klein PS, Klopstock T, Klucken J, Knaevelsrud H, Knorr RL, Ko BCB, Ko F, Ko JL, Kobayashi H, Kobayashi S, Koch I, Koch JC, Koenig U, Kogel D, Koh YH, Koike M, Kohlwein SD, Kocaturk NM, Komatsu M, Konig J, Kono T, Kopp BT, Korcsmaros T, Korkmaz G, Korolchuk VI, Korsnes MS, Koskela A, Kota J, Kotake Y, Kotler ML, Kou Y, Koukourakis MI, Koustas E, Kovacs AL, Kovacs T, Koya D, Kozako T, Kraft C, Krainc D, Kramer H, Krasnodembskaya AD, Kretz-Remy C, Kroemer G, Ktistakis NT, Kuchitsu K, Kuenen S, Kuerschner L, Kukar T, Kumar A, Kumar A, Kumar D, Kumar D, Kumar S, Kume S, Kumsta C, Kundu CN, Kundu M, Kunnumakkara AB, Kurgan L, Kutateladze TG, Kutlu O, Kwak S, Kwon HJ, Kwon TK, Kwon YT, Kyrmizi I, La Spada A, Labonte P, Ladoire S, Laface I, Lafont F, Lagace DC, Lahiri V, Lai Z, Laird AS, Lakkaraju A, Lamark T, Lan SH, Landajuela A, Lane DJR, Lane JD, Lang CH, Lange C, Langel U, Langer R, Lapaquette P, Laporte J, LaRusso NF, Lastres-Becker I, Lau WCY, Laurie GW, Lavandero S, Law BYK, Law HK, Layfield R, Le W, Le Stunff H, Leary AY, Lebrun JJ, Leck LYW, Leduc-Gaudet JP, Lee C, Lee CP, Lee DH, Lee EB, Lee EF, Lee GM, Lee HJ, Lee HK, Lee JM, Lee JS, Lee JA, Lee JY, Lee JH, Lee M, Lee MG, Lee MJ, Lee MS, Lee SY, Lee SJ, Lee SY, Lee SB, Lee WH, Lee YR, Lee YH, Lee Y, Lefebvre C, Legouis R, Lei YL, Lei Y, Leikin S, Leitinger G, Lemus L, Leng S, Lenoir O, Lenz G, Lenz HJ, Lenzi P, Leon Y, Leopoldino AM, Leschczyk C, Leskela S, Letellier E, Leung CT, Leung PS, Leventhal JS, Levine B, Lewis PA, Ley K, Li B, Li DQ, Li J, Li J, Li J, Li K, Li L, Li M, Li M, Li M, Li M, Li M, Li PL, Li MQ, Li Q, Li S, Li T, Li W, Li W, Li X, Li YP, Li Y, Li Z, Li Z, Li Z, Lian J, Liang C, Liang Q, Liang W, Liang Y, Liang Y, Liao G, Liao L, Liao M, Liao YF, Librizzi M, Lie PPY, Lilly MA, Lim HJ, Lima TRR, Limana F, Lin C, Lin CW, Lin DS, Lin FC, Lin JD, Lin KM, Lin KH, Lin LT, Lin PH, Lin Q, Lin S, Lin SJ, Lin W, Lin X, Lin YX, Lin YS, Linden R, Lindner P, Ling SC, Lingor P, Linnemann AK, Liou YC, Lipinski MM, Lipovsek S, Lira VA, Lisiak N, Liton PB, Liu C, Liu CH, Liu CF, Liu CH, Liu F, Liu H, Liu HS, Liu HF, Liu H, Liu J, Liu J, Liu J, Liu L, Liu L, Liu M, Liu Q, Liu W, Liu W, Liu XH, Liu X, Liu X, Liu X, Liu X, Liu Y, Liu Y, Liu Y, Liu Y, Liu Y, Livingston JA, Lizard G, Lizcano JM, Ljubojevic-Holzer S, ME LL, Llobet-Navas D, Llorente A, Lo CH, Lobato-Marquez D, Long Q, Long YC, Loos B, Loos JA, Lopez MG, Lopez-Domenech G, Lopez-Guerrero JA, Lopez-Jimenez AT, Lopez-Perez O, Lopez-Valero I, Lorenowicz MJ, Lorente M, Lorincz P, Lossi L, Lotersztajn S, Lovat PE, Lovell JF, Lovy A, Low P, Lu G, Lu H, Lu JH, Lu JJ, Lu M, Lu S, Luciani A, Lucocq JM, Ludovico P, Luftig MA, Luhr M, Luis-Ravelo D, Lum JJ, Luna-Dulcey L, Lund AH, Lund VK, Lunemann JD, Luningschror P, Luo H, Luo R, Luo S, Luo Z, Luparello C, Luscher B, Luu L, Lyakhovich A, Lyamzaev KG, Lystad AH, Lytvynchuk L, Ma AC, Ma C, Ma M, Ma NF, Ma QH, Ma X, Ma Y, Ma Z, MacDougald OA, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, Maday S, Madeo F, Madesh M, Madl T, Madrigal-Matute J, Maeda A, Maejima Y, Magarinos M, Mahavadi P, Maiani E, Maiese K, Maiti P, Maiuri MC, Majello B, Major MB, Makareeva E, Malik F, Mallilankaraman K, Malorni W, Maloyan A, Mammadova N, Man GCW, Manai F, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manjili MH, Manjithaya R, Manque P, Manshian BB, Manzano R, Manzoni C, Mao K, Marchese C, Marchetti S, Marconi AM, Marcucci F, Mardente S, Mareninova OA, Margeta M, Mari M, Marinelli S, Marinelli O, Marino G, Mariotto S, Marshall RS, Marten MR, Martens S, Martin APJ, Martin KR, Martin S, Martin S, Martin-Segura A, Martin-Acebes MA, Martin-Burriel I, Martin-Rincon M, Martin-Sanz P, Martina JA, Martinet W, Martinez A, Martinez A, Martinez J, Martinez Velazquez M, Martinez-Lopez N, Martinez-Vicente M, Martins DO, Martins JO, Martins WK, Martins-Marques T, Marzetti E, Masaldan S, Masclaux-Daubresse C, Mashek DG, Massa V, Massieu L, Masson GR, Masuelli L, Masyuk AI, Masyuk TV, Matarrese P, Matheu A, Matoba S, Matsuzaki S, Mattar P, Matte A, Mattoscio D, Mauriz JL, Mauthe M, Mauvezin C, Maverakis E, Maycotte P, Mayer J, Mazzoccoli G, Mazzoni C, Mazzulli JR, McCarty N, McDonald C, McGill MR, McKenna SL, McLaughlin B, McLoughlin F, McNiven MA, McWilliams TG, Mechta-Grigoriou F, Medeiros TC, Medina DL, Megeney LA, Megyeri K, Mehrpour M, Mehta JL, Meijer AJ, Meijer AH, Mejlvang J, Melendez A, Melk A, Memisoglu G, Mendes AF, Meng D, Meng F, Meng T, Menna-Barreto R, Menon MB, Mercer C, Mercier AE, Mergny JL, Merighi A, Merkley SD, Merla G, Meske V, Mestre AC, Metur SP, Meyer C, Meyer H, Mi W, Mialet-Perez J, Miao J, Micale L, Miki Y, Milan E, Milczarek M, Miller DL, Miller SI, Miller S, Millward SW, Milosevic I, Minina EA, Mirzaei H, Mirzaei HR, Mirzaei M, Mishra A, Mishra N, Mishra PK, Misirkic Marjanovic M, Misasi R, Misra A, Misso G, Mitchell C, Mitou G, Miura T, Miyamoto S, Miyazaki M, Miyazaki M, Miyazaki T, Miyazawa K, Mizushima N, Mogensen TH, Mograbi B, Mohammadinejad R, Mohamud Y, Mohanty A, Mohapatra S, Mohlmann T, Mohmmed A, Moles A, Moley KH, Molinari M, Mollace V, Moller AB, Mollereau B, Mollinedo F, Montagna C, Monteiro MJ, Montella A, Montes LR, Montico B, Mony VK, Monzio Compagnoni G, Moore MN, Moosavi MA, Mora AL, Mora M, Morales-Alamo D, Moratalla R, Moreira PI, Morelli E, Moreno S, Moreno-Blas D, Moresi V, Morga B, Morgan AH, Morin F, Morishita H, Moritz OL, Moriyama M, Moriyasu Y, Morleo M, Morselli E, Moruno-Manchon JF, Moscat J, Mostowy S, Motori E, Moura AF, Moustaid-Moussa N, Mrakovcic M, Mucino-Hernandez G, Mukherjee A, Mukhopadhyay S, Mulcahy Levy JM, Mulero V, Muller S, Munch C, Munjal A, Munoz-Canoves P, Munoz-Galdeano T, Munz C, Murakawa T, Muratori C, Murphy BM, Murphy JP, Murthy A, Myohanen TT, Mysorekar IU, Mytych J, Nabavi SM, Nabissi M, Nagy P, Nah J, Nahimana A, Nakagawa I, Nakamura K, Nakatogawa H, Nandi SS, Nanjundan M, Nanni M, Napolitano G, Nardacci R, Narita M, Nassif M, Nathan I, Natsumeda M, Naude RJ, Naumann C, Naveiras O, Navid F, Nawrocki ST, Nazarko TY, Nazio F, Negoita F, Neill T, Neisch AL, Neri LM, Netea MG, Neubert P, Neufeld TP, Neumann D, Neutzner A, Newton PT, Ney PA, Nezis IP, Ng CCW, Ng TB, Nguyen HTT, Nguyen LT, Ni HM, Ni Cheallaigh C, Ni Z, Nicolao MC, Nicoli F, Nieto-Diaz M, Nilsson P, Ning S, Niranjan R, Nishimune H, Niso-Santano M, Nixon RA, Nobili A, Nobrega C, Noda T, Nogueira-Recalde U, Nolan TM, Nombela I, Novak I, Novoa B, Nozawa T, Nukina N, Nussbaum-Krammer C, Nylandsted J, O'Donovan TR, O'Leary SM, O'Rourke EJ, O'Sullivan MP, O'Sullivan TE, Oddo S, Oehme I, Ogawa M, Ogier-Denis E, Ogmundsdottir MH, Ogretmen B, Oh GT, Oh SH, Oh YJ, Ohama T, Ohashi Y, Ohmuraya M, Oikonomou V, Ojha R, Okamoto K, Okazawa H, Oku M, Olivan S, Oliveira JMA, Ollmann M, Olzmann JA, Omari S, Omary MB, Onal G, Ondrej M, Ong SB, Ong SG, Onnis A, Orellana JA, Orellana-Munoz S, Ortega-Villaizan MDM, Ortiz-Gonzalez XR, Ortona E, Osiewacz HD, Osman AK, Osta R, Otegui MS, Otsu K, Ott C, Ottobrini L, Ou JJ, Outeiro TF, Oynebraten I, Ozturk M, Pages G, Pahari S, Pajares M, Pajvani UB, Pal R, Paladino S, Pallet N, Palmieri M, Palmisano G, Palumbo C, Pampaloni F, Pan L, Pan Q, Pan W, Pan X, Panasyuk G, Pandey R, Pandey UB, Pandya V, Paneni F, Pang SY, Panzarini E, Papademetrio DL, Papaleo E, Papinski D, Papp D, Park EC, Park HT, Park JM, Park JI, Park JT, Park J, Park SC, Park SY, Parola AH, Parys JB, Pasquier A, Pasquier B, Passos JF, Pastore N, Patel HH, Patschan D, Pattingre S, Pedraza-Alva G, Pedraza-Chaverri J, Pedrozo Z, Pei G, Pei J, Peled-Zehavi H, Pellegrini JM, Pelletier J, Penalva MA, Peng D, Peng Y, Penna F, Pennuto M, Pentimalli F, Pereira CM, Pereira GJS, Pereira LC, Pereira de Almeida L, Perera ND, Perez-Lara A, Perez-Oliva AB, Perez-Perez ME, Periyasamy P, Perl A, Perrotta C, Perrotta I, Pestell RG, Petersen M, Petrache I, Petrovski G, Pfirrmann T, Pfister AS, Philips JA, Pi H, Picca A, Pickrell AM, Picot S, Pierantoni GM, Pierdominici M, Pierre P, Pierrefite-Carle V, Pierzynowska K, Pietrocola F, Pietruczuk M, Pignata C, Pimentel-Muinos FX, Pinar M, Pinheiro RO, Pinkas-Kramarski R, Pinton P, Pircs K, Piya S, Pizzo P, Plantinga TS, Platta HW, Plaza-Zabala A, Plomann M, Plotnikov EY, Plun-Favreau H, Pluta R, Pocock R, Poggeler S, Pohl C, Poirot M, Poletti A, Ponpuak M, Popelka H, Popova B, Porta H, Porte Alcon S, Portilla-Fernandez E, Post M, Potts MB, Poulton J, Powers T, Prahlad V, Prajsnar TK, Pratico D, Prencipe R, Priault M, Proikas-Cezanne T, Promponas VJ, Proud CG, Puertollano R, Puglielli L, Pulinilkunnil T, Puri D, Puri R, Puyal J, Qi X, Qi Y, Qian W, Qiang L, Qiu Y, Quadrilatero J, Quarleri J, Raben N, Rabinowich H, Ragona D, Ragusa MJ, Rahimi N, Rahmati M, Raia V, Raimundo N, Rajasekaran NS, Ramachandra Rao S, Rami A, Ramirez-Pardo I, Ramsden DB, Randow F, Rangarajan PN, Ranieri D, Rao H, Rao L, Rao R, Rathore S, Ratnayaka JA, Ratovitski EA, Ravanan P, Ravegnini G, Ray SK, Razani B, Rebecca V, Reggiori F, Regnier-Vigouroux A, Reichert AS, Reigada D, Reiling JH, Rein T, Reipert S, Rekha RS, Ren H, Ren J, Ren W, Renault T, Renga G, Reue K, Rewitz K, Ribeiro de Andrade Ramos B, Riazuddin SA, Ribeiro-Rodrigues TM, Ricci JE, Ricci R, Riccio V, Richardson DR, Rikihisa Y, Risbud MV, Risueno RM, Ritis K, Rizza S, Rizzuto R, Roberts HC, Roberts LD, Robinson KJ, Roccheri MC, Rocchi S, Rodney GG, Rodrigues T, Rodrigues Silva VR, Rodriguez A, Rodriguez-Barrueco R, Rodriguez-Henche N, Rodriguez-Rocha H, Roelofs J, Rogers RS, Rogov VV, Rojo AI, Rolka K, Romanello V, Romani L, Romano A, Romano PS, Romeo-Guitart D, Romero LC, Romero M, Roney JC, Rongo C, Roperto S, Rosenfeldt MT, Rosenstiel P, Rosenwald AG, Roth KA, Roth L, Roth S, Rouschop KMA, Roussel BD, Roux S, Rovere-Querini P, Roy A, Rozieres A, Ruano D, Rubinsztein DC, Rubtsova MP, Ruckdeschel K, Ruckenstuhl C, Rudolf E, Rudolf R, Ruggieri A, Ruparelia AA, Rusmini P, Russell RR, Russo GL, Russo M, Russo R, Ryabaya OO, Ryan KM, Ryu KY, Sabater-Arcis M, Sachdev U, Sacher M, Sachse C, Sadhu A, Sadoshima J, Safren N, Saftig P, Sagona AP, Sahay G, Sahebkar A, Sahin M, Sahin O, Sahni S, Saito N, Saito S, Saito T, Sakai R, Sakai Y, Sakamaki JI, Saksela K, Salazar G, Salazar-Degracia A, Salekdeh GH, Saluja AK, Sampaio-Marques B, Sanchez MC, Sanchez-Alcazar JA, Sanchez-Vera V, Sancho-Shimizu V, Sanderson JT, Sandri M, Santaguida S, Santambrogio L, Santana MM, Santoni G, Sanz A, Sanz P, Saran S, Sardiello M, Sargeant TJ, Sarin A, Sarkar C, Sarkar S, Sarrias MR, Sarkar S, Sarmah DT, Sarparanta J, Sathyanarayan A, Sathyanarayanan R, Scaglione KM, Scatozza F, Schaefer L, Schafer ZT, Schaible UE, Schapira AHV, Scharl M, Schatzl HM, Schein CH, Scheper W, Scheuring D, Schiaffino MV, Schiappacassi M, Schindl R, Schlattner U, Schmidt O, Schmitt R, Schmidt SD, Schmitz I, Schmukler E, Schneider A, Schneider BE, Schober R, Schoijet AC, Schott MB, Schramm M, Schroder B, Schuh K, Schuller C, Schulze RJ, Schurmanns L, Schwamborn JC, Schwarten M, Scialo F, Sciarretta S, Scott MJ, Scotto KW, Scovassi AI, Scrima A, Scrivo A, Sebastian D, Sebti S, Sedej S, Segatori L, Segev N, Seglen PO, Seiliez I, Seki E, Selleck SB, Sellke FW, Selsby JT, Sendtner M, Senturk S, Seranova E, Sergi C, Serra-Moreno R, Sesaki H, Settembre C, Setty SRG, Sgarbi G, Sha O, Shacka JJ, Shah JA, Shang D, Shao C, Shao F, Sharbati S, Sharkey LM, Sharma D, Sharma G, Sharma K, Sharma P, Sharma S, Shen HM, Shen H, Shen J, Shen M, Shen W, Shen Z, Sheng R, Sheng Z, Sheng ZH, Shi J, Shi X, Shi YH, Shiba-Fukushima K, Shieh JJ, Shimada Y, Shimizu S, Shimozawa M, Shintani T, Shoemaker CJ, Shojaei S, Shoji I, Shravage BV, Shridhar V, Shu CW, Shu HB, Shui K, Shukla AK, Shutt TE, Sica V, Siddiqui A, Sierra A, Sierra-Torre V, Signorelli S, Sil P, Silva BJA, Silva JD, Silva-Pavez E, Silvente-Poirot S, Simmonds RE, Simon AK, Simon HU, Simons M, Singh A, Singh LP, Singh R, Singh SV, Singh SK, Singh SB, Singh S, Singh SP, Sinha D, Sinha RA, Sinha S, Sirko A, Sirohi K, Sivridis EL, Skendros P, Skirycz A, Slaninova I, Smaili SS, Smertenko A, Smith MD, Soenen SJ, Sohn EJ, Sok SPM, Solaini G, Soldati T, Soleimanpour SA, Soler RM, Solovchenko A, Somarelli JA, Sonawane A, Song F, Song HK, Song JX, Song K, Song Z, Soria LR, Sorice M, Soukas AA, Soukup SF, Sousa D, Sousa N, Spagnuolo PA, Spector SA, Srinivas Bharath MM, St Clair D, Stagni V, Staiano L, Stalnecker CA, Stankov MV, Stathopulos PB, Stefan K, Stefan SM, Stefanis L, Steffan JS, Steinkasserer A, Stenmark H, Sterneckert J, Stevens C, Stoka V, Storch S, Stork B, Strappazzon F, Strohecker AM, Stupack DG, Su H, Su LY, Su L, Suarez-Fontes AM, Subauste CS, Subbian S, Subirada PV, Sudhandiran G, Sue CM, Sui X, Summers C, Sun G, Sun J, Sun K, Sun MX, Sun Q, Sun Y, Sun Z, Sunahara KKS, Sundberg E, Susztak K, Sutovsky P, Suzuki H, Sweeney G, Symons JD, Sze SCW, Szewczyk NJ, Tabecka-Lonczynska A, Tabolacci C, Tacke F, Taegtmeyer H, Tafani M, Tagaya M, Tai H, Tait SWG, Takahashi Y, Takats S, Talwar P, Tam C, Tam SY, Tampellini D, Tamura A, Tan CT, Tan EK, Tan YQ, Tanaka M, Tanaka M, Tang D, Tang J, Tang TS, Tanida I, Tao Z, Taouis M, Tatenhorst L, Tavernarakis N, Taylor A, Taylor GA, Taylor JM, Tchetina E, Tee AR, Tegeder I, Teis D, Teixeira N, Teixeira-Clerc F, Tekirdag KA, Tencomnao T, Tenreiro S, Tepikin AV, Testillano PS, Tettamanti G, Tharaux PL, Thedieck K, Thekkinghat AA, Thellung S, Thinwa JW, Thirumalaikumar VP, Thomas SM, Thomes PG, Thorburn A, Thukral L, Thum T, Thumm M, Tian L, Tichy A, Till A, Timmerman V, Titorenko VI, Todi SV, Todorova K, Toivonen JM, Tomaipitinca L, Tomar D, Tomas-Zapico C, Tomic S, Tong BC, Tong C, Tong X, Tooze SA, Torgersen ML, Torii S, Torres-Lopez L, Torriglia A, Towers CG, Towns R, Toyokuni S, Trajkovic V, Tramontano D, Tran QG, Travassos LH, Trelford CB, Tremel S, Trougakos IP, Tsao BP, Tschan MP, Tse HF, Tse TF, Tsugawa H, Tsvetkov AS, Tumbarello DA, Tumtas Y, Tunon MJ, Turcotte S, Turk B, Turk V, Turner BJ, Tuxworth RI, Tyler JK, Tyutereva EV, Uchiyama Y, Ugun-Klusek A, Uhlig HH, Ulamek-Koziol M, Ulasov IV, Umekawa M, Ungermann C, Unno R, Urbe S, Uribe-Carretero E, Ustun S, Uversky VN, Vaccari T, Vaccaro MI, Vahsen BF, Vakifahmetoglu-Norberg H, Valdor R, Valente MJ, Valko A, Vallee RB, Valverde AM, Van den Berghe G, van der Veen S, Van Kaer L, van Loosdregt J, van Wijk SJL, Vandenberghe W, Vanhorebeek I, Vannier-Santos MA, Vannini N, Vanrell MC, Vantaggiato C, Varano G, Varela-Nieto I, Varga M, Vasconcelos MH, Vats S, Vavvas DG, Vega-Naredo I, Vega-Rubin-de-Celis S, Velasco G, Velazquez AP, Vellai T, Vellenga E, Velotti F, Verdier M, Verginis P, Vergne I, Verkade P, Verma M, Verstreken P, Vervliet T, Vervoorts J, Vessoni AT, Victor VM, Vidal M, Vidoni C, Vieira OV, Vierstra RD, Vigano S, Vihinen H, Vijayan V, Vila M, Vilar M, Villalba JM, Villalobo A, Villarejo-Zori B, Villarroya F, Villarroya J, Vincent O, Vindis C, Viret C, Viscomi MT, Visnjic D, Vitale I, Vocadlo DJ, Voitsekhovskaja OV, Volonte C, Volta M, Vomero M, Von Haefen C, Vooijs MA, Voos W, Vucicevic L, Wade-Martins R, Waguri S, Waite KA, Wakatsuki S, Walker DW, Walker MJ, Walker SA, Walter J, Wandosell FG, Wang B, Wang CY, Wang C, Wang C, Wang C, Wang CY, Wang D, Wang F, Wang F, Wang F, Wang G, Wang H, Wang H, Wang H, Wang HG, Wang J, Wang J, Wang J, Wang J, Wang K, Wang L, Wang L, Wang MH, Wang M, Wang N, Wang P, Wang P, Wang P, Wang P, Wang QJ, Wang Q, Wang QK, Wang QA, Wang WT, Wang W, Wang X, Wang X, Wang Y, Wang Y, Wang Y, Wang YY, Wang Y, Wang Y, Wang Y, Wang Y, Wang Z, Wang Z, Wang Z, Warnes G, Warnsmann V, Watada H, Watanabe E, Watchon M, Wawrzynska A, Weaver TE, Wegrzyn G, Wehman AM, Wei H, Wei L, Wei T, Wei Y, Weiergraber OH, Weihl CC, Weindl G, Weiskirchen R, Wells A, Wen RH, Wen X, Werner A, Weykopf B, Wheatley SP, Whitton JL, Whitworth AJ, Wiktorska K, Wildenberg ME, Wileman T, Wilkinson S, Willbold D, Williams B, Williams RSB, Williams RL, Williamson PR, Wilson RA, Winner B, Winsor NJ, Witkin SS, Wodrich H, Woehlbier U, Wollert T, Wong E, Wong JH, Wong RW, Wong VKW, Wong WW, Wu AG, Wu C, Wu J, Wu J, Wu KK, Wu M, Wu SY, Wu S, Wu SY, Wu S, Wu WKK, Wu X, Wu X, Wu YW, Wu Y, Xavier RJ, Xia H, Xia L, Xia Z, Xiang G, Xiang J, Xiang M, Xiang W, Xiao B, Xiao G, Xiao H, Xiao HT, Xiao J, Xiao L, Xiao S, Xiao Y, Xie B, Xie CM, Xie M, Xie Y, Xie Z, Xie Z, Xilouri M, Xu C, Xu E, Xu H, Xu J, Xu J, Xu L, Xu WW, Xu X, Xue Y, Yakhine-Diop SMS, Yamaguchi M, Yamaguchi O, Yamamoto A, Yamashina S, Yan S, Yan SJ, Yan Z, Yanagi Y, Yang C, Yang DS, Yang H, Yang HT, Yang H, Yang JM, Yang J, Yang J, Yang L, Yang L, Yang M, Yang PM, Yang Q, Yang S, Yang S, Yang SF, Yang W, Yang WY, Yang X, Yang X, Yang Y, Yang Y, Yao H, Yao S, Yao X, Yao YG, Yao YM, Yasui T, Yazdankhah M, Yen PM, Yi C, Yin XM, Yin Y, Yin Z, Yin Z, Ying M, Ying Z, Yip CK, Yiu SPT, Yoo YH, Yoshida K, Yoshii SR, Yoshimori T, Yousefi B, Yu B, Yu H, Yu J, Yu J, Yu L, Yu ML, Yu SW, Yu VC, Yu WH, Yu Z, Yu Z, Yuan J, Yuan LQ, Yuan S, Yuan SF, Yuan Y, Yuan Z, Yue J, Yue Z, Yun J, Yung RL, Zacks DN, Zaffagnini G, Zambelli VO, Zanella I, Zang QS, Zanivan S, Zappavigna S, Zaragoza P, Zarbalis KS, Zarebkohan A, Zarrouk A, Zeitlin SO, Zeng J, Zeng JD, Zerovnik E, Zhan L, Zhang B, Zhang DD, Zhang H, Zhang H, Zhang H, Zhang H, Zhang H, Zhang H, Zhang H, Zhang HL, Zhang J, Zhang J, Zhang JP, Zhang KYB, Zhang LW, Zhang L, Zhang L, Zhang L, Zhang L, Zhang M, Zhang P, Zhang S, Zhang W, Zhang X, Zhang XW, Zhang X, Zhang X, Zhang X, Zhang X, Zhang XD, Zhang Y, Zhang Y, Zhang Y, Zhang YD, Zhang Y, Zhang YY, Zhang Y, Zhang Z, Zhang Z, Zhang Z, Zhang Z, Zhang Z, Zhang Z, Zhao H, Zhao L, Zhao S, Zhao T, Zhao XF, Zhao Y, Zhao Y, Zhao Y, Zhao Y, Zheng G, Zheng K, Zheng L, Zheng S, Zheng XL, Zheng Y, Zheng ZG, Zhivotovsky B, Zhong Q, Zhou A, Zhou B, Zhou C, Zhou G, Zhou H, Zhou H, Zhou H, Zhou J, Zhou J, Zhou J, Zhou J, Zhou K, Zhou R, Zhou XJ, Zhou Y, Zhou Y, Zhou Y, Zhou ZY, Zhou Z, Zhu B, Zhu C, Zhu GQ, Zhu H, Zhu H, Zhu H, Zhu WG, Zhu Y, Zhu Y, Zhuang H, Zhuang X, Zientara-Rytter K, Zimmermann CM, Ziviani E, Zoladek T, Zong WX, Zorov DB, Zorzano A, Zou W, Zou Z, Zou Z, Zuryn S, Zwerschke W, Brand-Saberi B, Dong XC, Kenchappa CS, Li Z, Lin Y, Oshima S, Rong Y, Sluimer JC, Stallings CL, & Tong CK (2021). Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy, 1–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoflach M, Matosevic B, Rücker M, Furtner M, Mair A, Wille G, Zangerle A, Werner P, Ferrari J, & Schmidauer C (2012). Functional recovery after ischemic stroke—a matter of age: data from the Austrian Stroke Unit Registry. Neurology, 78, 279–285. [DOI] [PubMed] [Google Scholar]
- Koike M, Shibata M, Tadakoshi M, Gotoh K, Komatsu M, Waguri S, Kawahara N, Kuida K, Nagata S, & Kominami E (2008). Inhibition of autophagy prevents hippocampal pyramidal neuron death after hypoxic-ischemic injury. The American journal of pathology, 172, 454–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, & Yamori T (2008). Phosphatidylinositol 3-kinase inhibitors: promising drug candidates for cancer therapy. Cancer science, 99, 1734–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupinski J, Kaluza J, Kumar P, Kumar S, & Wang JM (1994). Role of angiogenesis in patients with cerebral ischemic stroke. Stroke, 25, 1794–1798. [DOI] [PubMed] [Google Scholar]
- Kryshtal D, Hwang HS, Johnson C, Chazin W, George AL, & Knollmann B (2014). Calmodulin Mutation (CALM-F90L) Associated With Familial Idiopathic Ventricular Fibrillation Disrupts L-type Ca Channel Inactivation and Activates Sarcoplasmic Reticulum Ca Release in Ventricular Myocytes. Circulation, 130, A17878–A17878. [Google Scholar]
- Kuang Y, Zheng X, Zhang L, Graf I, Bähr M, & Doeppner TR (2020). Abstract TP270: Adipose Derived Mesenchymal Stem Cells Reduce Autophagic Flux After Ischemic Stroke in Mice via Extracellular Vesicle Transfer of MiR-25–3p. Stroke, 51, ATP270–ATP270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler C, Altenhöner T, Lochner P, & Ferbert A (2003). Does age influence early recovery from ischemic stroke? Journal of neurology, 250, 676–681. [DOI] [PubMed] [Google Scholar]
- Kwon I, Jang Y, & Lee Y (2021). Endurance Exercise-Induced Autophagy/Mitophagy Coincides with a Reinforced Anabolic State and Increased Mitochondrial Turnover in the Cortex of Young Male Mouse Brain. Journal of Molecular Neuroscience, 71, 42–54. [DOI] [PubMed] [Google Scholar]
- Lai Z, Gu L, Yu L, Chen H, Yu Z, Zhang C, Xu X, Zhang M, Zhang M, & Ma M (2020). Delta opioid peptide [d-Ala2, d-Leu5] enkephalin confers neuroprotection by activating delta opioid receptor-AMPK-autophagy axis against global ischemia. Cell & bioscience, 10, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson SC, Männistö S, Virtanen MJ, Kontto J, Albanes D, & Virtamo J (2009). Dairy foods and risk of stroke. Epidemiology (Cambridge, Mass.), 20, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, & Codogno P (2006). Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. Journal of Biological Chemistry, 281, 8518–8527. [DOI] [PubMed] [Google Scholar]
- Law BY, Wang M, Ma D-L, Al-Mousa F, Michelangeli F, Cheng S-H, Ng MH, To K-F, Mok AY, & Ko RY (2010). Alisol B, a novel inhibitor of the sarcoplasmic/endoplasmic reticulum Ca2+ ATPase pump, induces autophagy, endoplasmic reticulum stress, and apoptosis. Molecular cancer therapeutics, 9, 718–730. [DOI] [PubMed] [Google Scholar]
- Lee WH, & Kim SG (2010). AMPK-dependent metabolic regulation by PPAR agonists. PPAR research, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, & Kroemer G (2019). Biological Functions of Autophagy Genes: A Disease Perspective. Cell, 176, 11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Huang Z, Meng J, Yu W, & Yang H (2020). MiR-202–5p attenuates neurological deficits and neuronal injury in MCAO model rats and OGD-induced injury in Neuro-2a cells by targeting eIF4E-mediated induction of autophagy and inhibition of Akt/GSK-3β pathway. Molecular and cellular probes, 51, 101497. [DOI] [PubMed] [Google Scholar]
- Li F, Shi W, Zhao EY, Geng X, Li X, Peng C, Shen J, Wang S, & Ding Y (2017). Enhanced apoptosis from early physical exercise rehabilitation following ischemic stroke. Journal of neuroscience research, 95, 1017–1024. [DOI] [PubMed] [Google Scholar]
- Li H, Lu C, Yao W, Xu L, Zhou J, & Zheng B (2020). Dexmedetomidine inhibits inflammatory response and autophagy through the circLrp1b/miR-27a-3p/Dram2 pathway in a rat model of traumatic brain injury. Aging (Albany NY), 12, 21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-G, Chu J, & Praticò D (2018). Downregulation of autophagy by 12/15Lipoxygenase worsens the phenotype of an Alzheimer’s disease mouse model with plaques, tangles, and memory impairments. Molecular Psychiatry, 1–10. [DOI] [PubMed] [Google Scholar]
- Li W-L, Yu S, Chen D, Yu S, Jiang Y-J, Genetta T, & Wei L (2013). The regulatory role of NF-κB in autophagy-like cell death after focal cerebral ischemia in mice. Neuroscience, 244, 16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, He Z, Lv H, Chen W, & Chen J (2020). Calpain-2 plays a pivotal role in the inhibitory effects of propofol against TNF-α-induced autophagy in mouse hippocampal neurons. Journal of Cellular and Molecular Medicine, 24, 9287–9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Li J, & Tang N (2017). Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience, 354, 1–10. [DOI] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu Z-X, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, & Walker CL (2007). The energy sensing LKB1–AMPK pathway regulates p27 kip1 phosphorylation mediating the decision to enter autophagy or apoptosis. Nature cell biology, 9, 218–224. [DOI] [PubMed] [Google Scholar]
- Lipton P (1999). Ischemic cell death in brain neurons. Physiological reviews, 79, 1431–1568. [DOI] [PubMed] [Google Scholar]
- Liu K, Chen Q, Yan S, Zhang S, & Lou M (2015). Relationship between early blood pressure variability and reperfusion in acute ischemic stroke patients with intravenous thrombolysis. Zhejiang da xue xue bao. Yi xue ban= Journal of Zhejiang University. Medical Sciences, 44, 603–610, 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Jiang Y, Zhou Z, Yue X, Wei N, Chen Z, Ma M, Xu G, & Liu X (2011). Intranasal ginsenoside Rb1 targets the brain and ameliorates cerebral ischemia/reperfusion injury in rats. Biological and Pharmaceutical Bulletin, 34, 1319–1324. [DOI] [PubMed] [Google Scholar]
- Luo C, Ouyang M-W, Fang Y-Y, Li S-J, Zhou Q, Fan J, Qin Z-S, & Tao T (2017). Dexmedetomidine protects mouse brain from ischemia-reperfusion injury via inhibiting neuronal autophagy through up-regulating HIF-1α. Frontiers in cellular neuroscience, 11, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H-C, Yi T-Z, Huang F-G, Wei Y, Luo X-P, & Luo Q-S (2020). Role of Long Noncoding RNA MEG3/MiR-378/GRB2 Axis in Neuronal Autophagy and Neurological Functional Impairment in Ischemic Stroke. Journal of Biological Chemistry, jbc. RA119. 010946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Dai J-R, Guo S-S, Lu A-M, Gao X-F, Gu Y-R, Zhang X-F, Xu H-D, Wang Y, & Zhu Z (2017). Lysosomal proteolysis is associated with exercise-induced improvement of mitochondrial quality control in aged hippocampus. Journals of Gerontology Series A: Biomedical Sciences and Medical Sciences, 72, 1342–1351. [DOI] [PubMed] [Google Scholar]
- Ma H. x., Hou F, Chen A. l., Li T. t., Zhu Y. f., & Zhao Q. p. (2020). Mu-Xiang-You-Fang protects PC12 cells against OGD/R-induced autophagy via the AMPK/mTOR signaling pathway. Journal of Ethnopharmacology, 252, 112583. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang W, Xu C, Zhang S, Zhao J, Pan Q, & Wang Z (2020). Nucleotide-binding oligomerization domain protein 1 enhances oxygen-glucose deprivation and reperfusion injury in cortical neurons via activation of endoplasmic reticulum stress-mediated autophagy. Experimental and Molecular Pathology, 104525. [DOI] [PubMed] [Google Scholar]
- Ma Y, Lovekamp-Swan T, Bekele W, Dohi A, & Schreihofer DA (2013). Hypoxia-inducible factor and vascular endothelial growth factor are targets of dietary soy during acute stroke in female rats. Endocrinology, 154, 1589–1597. [DOI] [PubMed] [Google Scholar]
- Malicdan MC, Noguchi S, Nonaka I, Saftig P, & Nishino I (2008). Lysosomal myopathies: an excessive build-up in autophagosomes is too much to handle. Neuromuscular Disorders, 18, 521–529. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, & Ueda M (2018). Mutual inhibition between PTEN and PIP3 generates bistability for polarity in motile cells. Nature communications, 9, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Z-G, Huang Y-G, Feng Z-T, Luo Y-N, Yang S-B, Du L-P, Jiang K, Liu X-L, Fu X-Y, & Deng Y-H (2020). Electroacupuncture ameliorates cerebral ischemia/reperfusion injury by suppressing autophagy via the SIRT1-FOXO1 signaling pathway. Aging (Albany NY), 12, 13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D, Frank AR, & Jewell JL (2018). mTOR signaling in stem and progenitor cells. Development, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, & Levine B (2020). Autophagy in Human Diseases. N Engl J Med, 383, 1564–1576. [DOI] [PubMed] [Google Scholar]
- Morgenstern LB, Escobar JD, Sánchez BN, Hughes R, Zuniga BG, Garcia N, & Lisabeth LD (2009). Fast food and neighborhood stroke risk. Annals of Neurology: Official Journal of the American Neurological Association and the Child Neurology Society, 66, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz MA, Lo EH, & Iadecola C (2010). The science of stroke: mechanisms in search of treatments. Neuron, 67, 181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrakovcic M, & Fröhlich LF (2018). p53-mediated molecular control of autophagy in tumor cells. Biomolecules, 8, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu D, Jiang X, Sheldon RA, Fox CK, Hamrick SE, Vexler ZS, & Ferriero DM (2003). Regulation of hypoxia-inducible factor 1α and induction of vascular endothelial growth factor in a rat neonatal stroke model. Neurobiology of disease, 14, 524–534. [DOI] [PubMed] [Google Scholar]
- Mu Q, Zhou H, Xu Y, He Q, Luo X, Zhang W, & Li H (2020). NPD1 inhibits excessive autophagy by targeting RNF146 and wnt/β-catenin pathway in cerebral ischemia-reperfusion injury. Journal of Receptors and Signal Transduction, 1–8. [DOI] [PubMed] [Google Scholar]
- Nixon RA (2013). The role of autophagy in neurodegenerative disease. Nature medicine, 19, 983–997. [DOI] [PubMed] [Google Scholar]
- Ohira T, Peacock JM, Iso H, Chambless LE, Rosamond WD, & Folsom AR (2009). Serum and dietary magnesium and risk of ischemic stroke: the Atherosclerosis Risk in Communities Study. American journal of epidemiology, 169, 1437–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Jin L, Shen W, Zhang J, Pan J, Cheng J, Xie Q, Hu Q, Wu S, & Zhang H (2020). Treadmill exercise improves neurological function by inhibiting autophagy and the binding of HMGB1 to Beclin1 in MCAO juvenile rats. Life Sciences, 243, 117279. [DOI] [PubMed] [Google Scholar]
- Pan Q, Liu Y, Wang G, Wen Z, & Wang Y (2020). MTMR14 protects against cerebral stroke through suppressing PTEN-regulated autophagy. Biochemical and Biophysical Research Communications, 529, 1045–1052. [DOI] [PubMed] [Google Scholar]
- Pang J, Fuller ND, Hu N, Barton LA, Henion JM, Guo R, Chen Y, & Ren J (2016). Alcohol dehydrogenase protects against endoplasmic reticulum stress-induced myocardial contractile dysfunction via attenuation of oxidative stress and autophagy: role of PTEN-Akt-mTOR signaling. PLoS One, 11, e0147322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J, Peng H, Wang S, Xu X, Xu F, Wang Q, Chen Y, Barton LA, Chen Y, Zhang Y, & Ren J (2019). Mitochondrial ALDH2 protects against lipopolysaccharide-induced myocardial contractile dysfunction by suppression of ER stress and autophagy. Biochim Biophys Acta Mol Basis Dis, 1865, 1627–1641. [DOI] [PubMed] [Google Scholar]
- Papadakis M, Hadley G, Xilouri M, Hoyte LC, Nagel S, McMenamin MM, Tsaknakis G, Watt SM, Drakesmith CW, & Chen R (2013). Tsc1 (hamartin) confers neuroprotection against ischemia by inducing autophagy. Nature medicine, 19, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou A, Higa A, Jegou G, Jouan F, Pineau R, Saas L, Avril T, Pluquet O, & Chevet E (2018). Alterations of EDEM1 functions enhance ATF6 pro-survival signaling. FEBS J, 285, 4146–4164. [DOI] [PubMed] [Google Scholar]
- Pei X, Li Y, Zhu L, & Zhou Z (2020). Astrocyte-derived exosomes transfer miR-190b to inhibit oxygen and glucose deprivation-induced autophagy and neuronal apoptosis. Cell Cycle, 19, 906–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peker N, & Gozuacik D (2020). Autophagy as a cellular stress response mechanism in the nervous system. Journal of molecular biology, 432, 2560–2588. [DOI] [PubMed] [Google Scholar]
- Piccoli C, Sardanelli A, Scrima R, Ripoli M, Quarato G, D’Aprile A, Bellomo F, Scacco S, De Michele G, & Filla A (2008). Mitochondrial respiratory dysfunction in familiar parkinsonism associated with PINK1 mutation. Neurochemical research, 33, 2565–2574. [DOI] [PubMed] [Google Scholar]
- Pineda-Ramírez N, Alquisiras-Burgos I, Ortiz-Plata A, Ruiz-Tachiquín M-E, Espinoza-Rojo M, & Aguilera P (2020). Resveratrol activates neuronal autophagy through AMPK in the ischemic brain. Molecular neurobiology, 57, 1055–1069. [DOI] [PubMed] [Google Scholar]
- Qi Z, Dong W, Shi W, Wang R, Zhang C, Zhao Y, Ji X, Liu KJ, & Luo Y (2015). Bcl-2 phosphorylation triggers autophagy switch and reduces mitochondrial damage in limb remote ischemic conditioned rats after ischemic stroke. Translational stroke research, 6, 198–206. [DOI] [PubMed] [Google Scholar]
- Qin A-P, Liu C-F, Qin Y-Y, Hong L-Z, Xu M, Yang L, Liu J, Qin Z-H, & Zhang H-L (2010). Autophagy was activated in injured astrocytes and mildly decreased cell survival following glucose and oxygen deprivation and focal cerebral ischemia. Autophagy, 6, 738–753. [DOI] [PubMed] [Google Scholar]
- Radanovic T, Gecht M, Covino R, Hummer G, Niwa M, & Ernst R (2020). Mechanistic Dissection of Sphingolipid Binding to the ER Stress Transducer ATF6-Insights into the Coordination of Sphingolipid and Protein Production. Biophysical Journal, 118, 243a.31883614 [Google Scholar]
- Rajanikant G, & Sugunan S (2013). Commentary: linking productive autophagy to neuroprotection: potential implications for anti-ischemic therapy. CNS & neurological disorders drug targets, 12, 298–299. [DOI] [PubMed] [Google Scholar]
- Rami A, Langhagen A, & Steiger S (2008). Focal cerebral ischemia induces upregulation of Beclin 1 and autophagy-like cell death. Neurobiology of disease, 29, 132–141. [DOI] [PubMed] [Google Scholar]
- Rao J-Y, Wang Q, Wang Y-C, Xiang F, Tian X-C, Liu D-H, & Dong Z (2020). β-caryophyllene alleviates cerebral ischemia/reperfusion injury in mice by activating autophagy. Zhongguo Zhong yao za zhi= Zhongguo zhongyao zazhi= China journal of Chinese materia medica, 45, 932–936. [DOI] [PubMed] [Google Scholar]
- Ren J, Bi Y, Sowers JR, Hetz C, & Zhang Y (2021). Endoplasmic reticulum stress and unfolded protein response in cardiovascular diseases. Nat Rev Cardiol. [DOI] [PubMed] [Google Scholar]
- Ren J, Sowers JR, & Zhang Y (2018). Metabolic stress, autophagy, and cardiovascular aging: from pathophysiology to therapeutics. Trends in Endocrinology & Metabolism, 29, 699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Sun M, Zhou H, Ajoolabady A, Zhou Y, Tao J, Sowers JR, & Zhang Y (2020). FUNDC1 interacts with FBXL2 to govern mitochondrial integrity and cardiac function through an IP3R3-dependent manner in obesity. Science advances, 6, eabc8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, & Taegtmeyer H (2015). Too much or not enough of a good thing--The Janus faces of autophagy in cardiac fuel and protein homeostasis. J Mol Cell Cardiol, 84, 223–226. [DOI] [PubMed] [Google Scholar]
- Ren J, & Zhang Y (2018). Targeting autophagy in aging and aging-related cardiovascular diseases. Trends in pharmacological sciences, 39, 1064–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Xie P, Lv J, Hu Y, Guan Z, Chen L, & Yu W (2020). miR‑187‑3p inhibitor attenuates cerebral ischemia/reperfusion injury by regulating Seipin‑mediated autophagic flux. International journal of molecular medicine, 46, 1051–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostami N, Nikkhoo A, Ajjoolabady A, Azizi G, Hojjat-Farsangi M, Ghalamfarsa G, Yousefi B, Yousefi M, & Jadidi-Niaragh F (2019). S1PR1 as a novel promising therapeutic target in cancer therapy. Molecular diagnosis & therapy, 23, 467–487. [DOI] [PubMed] [Google Scholar]
- Roussel BD, Kruppa AJ, Miranda E, Crowther DC, Lomas DA, & Marciniak SJ (2013). Endoplasmic reticulum dysfunction in neurological disease. The Lancet Neurology, 12, 105–118. [DOI] [PubMed] [Google Scholar]
- Sano R, & Reed JC (2013). ER stress-induced cell death mechanisms. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1833, 3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaki A, Farbood Y, Badavi M, Khalaj L, Khodagholi F, & Ashabi G (2015). Metformin improves anxiety-like behaviors through AMPK-dependent regulation of autophagy following transient forebrain ischemia. Metabolic Brain Disease, 30, 1139–1150. [DOI] [PubMed] [Google Scholar]
- Saver JL, Goyal M, Bonafe A, Diener H-C, Levy EI, Pereira VM, Albers GW, Cognard C, Cohen DJ, & Hacke W (2015). Stent-retriever thrombectomy after intravenous t-PA vs. t-PA alone in stroke. New England Journal of Medicine, 372, 2285–2295. [DOI] [PubMed] [Google Scholar]
- Schwalm C, Jamart C, Benoit N, Naslain D, Prémont C, Prévet J, Van Thienen R, Deldicque L, & Francaux M (2015). Activation of autophagy in human skeletal muscle is dependent on exercise intensity and AMPK activation. The FASEB Journal, 29, 3515–3526. [DOI] [PubMed] [Google Scholar]
- Shan Y, Sun S, Yang F, Shang N, & Liu H (2018). Dexmedetomidine protects the developing rat brain against the neurotoxicity wrought by sevoflurane: role of autophagy and Drp1–Bax signaling. Drug design, development and therapy, 12, 3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Yu W-M, Brotto M, Scherman JA, Guo C, Stoddard C, Nosek TM, Valdivia HH, & Qu C-K (2009). Deficiency of MIP/MTMR14 phosphatase induces a muscle disorder by disrupting Ca 2+ homeostasis. Nature cell biology, 11, 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P, Hou S, Zhu M, Zhao M, Ouyang Y, & Feng J (2017). Cortical spreading depression preconditioning mediates neuroprotection against ischemic stroke by inducing AMP-activated protein kinase-dependent autophagy in a rat cerebral ischemic/reperfusion injury model. Journal of Neurochemistry, 140, 799–813. [DOI] [PubMed] [Google Scholar]
- Shen S, Kepp O, & Kroemer G (2012). The end of autophagic cell death? In: Taylor & Francis. [DOI] [PubMed] [Google Scholar]
- Shi CX, Jin J, Wang XQ, Song T, Li GH, Li KZ, & Ma JH (2020). Sevoflurane attenuates brain damage through inhibiting autophagy and apoptosis in cerebral ischemia‑reperfusion rats. Molecular medicine reports, 21, 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Weng J, Zhao L, Li XM, Gao TM, & Kong J (2012). Excessive autophagy contributes to neuron death in cerebral ischemia. CNS neuroscience & therapeutics, 18, 250–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotake J, Derugin N, Wendland M, Vexler ZS, & Ferriero DM (2010). Vascular endothelial growth factor receptor-2 inhibition promotes cell death and limits endothelial cell proliferation in a neonatal rodent model of stroke. Stroke, 41, 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon H-U, Friis R, Tait SW, & Ryan KM (2017). Retrograde signaling from autophagy modulates stress responses. Science signaling, 10. [DOI] [PubMed] [Google Scholar]
- Singh S, Singh AK, Garg G, & Rizvi SI (2018). Fisetin as a caloric restriction mimetic protects rat brain against aging induced oxidative stress, apoptosis and neurodegeneration. Life Sciences, 193, 171–179. [DOI] [PubMed] [Google Scholar]
- Song F, Wang Y, Jiang D, Wang T, Zhang Y, Ma H, & Kang Y (2016). Cyclic compressive stress regulates apoptosis in rat osteoblasts: involvement of PI3K/Akt and JNK MAPK signaling pathways. PLoS One, 11, e0165845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen LM, Jacobs DR Jr, Stevens J, Shahar E, Carithers T, & Folsom AR (2003). Associations of whole-grain, refined-grain, and fruit and vegetable consumption with risks of all-cause mortality and incident coronary artery disease and ischemic stroke: the Atherosclerosis Risk in Communities (ARIC) Study. The American journal of clinical nutrition, 78, 383–390. [DOI] [PubMed] [Google Scholar]
- Steinberg GK, Kondziolka D, Schwartz NE, Wechsler L, Lunsford L, Coburn ML, Billigen JB, Keren-Gill H, McGrogan M, & Case C (2014). A novel phase ½A study of intraparenchymal transplantation of human modified bone marrow derived cells in patients with stable ischemic stroke. Stroke, 45, A149–A149. [Google Scholar]
- Sun A, & Ren J (2013). ALDH2, a novel protector against stroke? Cell Res, 23, 874–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Ou H, Ren F, Huan Y, Zhong T, Gao M, & Cai H (2018). Propofol inhibited autophagy through Ca 2+/CaMKKβ/AMPK/mTOR pathway in OGD/R-induced neuron injury. Molecular Medicine, 24, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Wang W, Wang X, Wang Y, Xu X, Ping F, Du Y, Jiang W, & Cui D (2018). bFGF plays a neuroprotective role by suppressing excessive autophagy and apoptosis after transient global cerebral ischemia in rats. Cell Death & Disease, 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H-S, Jackson MF, Martin LJ, Jansen K, Teves L, Cui H, Kiyonaka S, Mori Y, Jones M, & Forder JP (2009). Suppression of hippocampal TRPM7 protein prevents delayed neuronal death in brain ischemia. Nature neuroscience, 12, 1300–1307. [DOI] [PubMed] [Google Scholar]
- Sun X, Liu H, Sun Z, Zhang B, Wang X, Liu T, Pan T, Gao Y, Jiang X, & Li H (2020). Acupuncture Protects Against Cerebral Ischemia-Reperfusion Injury Via Suppressing Endoplasmic Reticulum Stress-Mediated Autophagy and Apoptosis. [DOI] [PMC free article] [PubMed]
- Sun X, Wang D, Zhang T, Lu X, Duan F, Ju L, Zhuang X, & Jiang X (2020). Eugenol Attenuates Cerebral Ischemia-Reperfusion Injury by Enhancing Autophagy via AMPK-mTOR-P70S6K Pathway. Frontiers in Pharmacology, 11, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Gu L, Wu K, Wang K, Ru J, Yang S, Wang Z, Zhuge Q, Huang L, & Huang S (2020). VX-765 enhances autophagy of human umbilical cord mesenchymal stem cells against stroke - induced apoptosis and inflammatory responses via AMPK/mTOR signaling pathway. CNS neuroscience & therapeutics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri MFK, Yamagishi K, Aleksic N, Hannan PJ, & Folsom AR (2010). Novel hemostatic factor levels and risk of ischemic stroke: the Atherosclerosis Risk in Communities (ARIC) Study. Cerebrovascular Diseases, 29, 497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro K, Fukui Y, Yamashita T, Liu X, Tsunoda K, Shang J, Morihara R, Nakano Y, Tian F, & Sasaki R (2020a). Bone Marrow Stromal Cell Transplantation Drives Molecular Switch from Autophagy to the Ubiquitin-Proteasome System in Ischemic Stroke Mice. Journal of Stroke and Cerebrovascular Diseases, 29, 104743. [DOI] [PubMed] [Google Scholar]
- Tadokoro K, Fukui Y, Yamashita T, Liu X, Tsunoda K, Shang J, Morihara R, Nakano Y, Tian F, & Sasaki R (2020b). Bone Marrow Stromal Cell Transplantation Drives Molecular Switch from Autophagy to the Ubiquitin-Proteasome System in Ischemic Stroke Mice. Journal of Stroke and Cerebrovascular Diseases, 104743. [DOI] [PubMed] [Google Scholar]
- Tan Y, Gong Y, Dong M, Pei Z, & Ren J (2019). Role of autophagy in inherited metabolic and endocrine myopathies. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease, 1865, 48–55. [DOI] [PubMed] [Google Scholar]
- Tian H, Chen P, & Ren J (2019). Physical exercise, autophagy and cardiometabolic stress in aging. Aging (Albany NY), 11, 5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Liu S, Ren J, Lee JKW, Wang R, & Chen P (2020). Role of Histone Deacetylases in Skeletal Muscle Physiology and Systemic Energy Homeostasis: Implications for Metabolic Diseases and Therapy. Front Physiol, 11, 949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urra H, Henriquez DR, Canovas J, Villarroel-Campos D, Carreras-Sureda A, Pulgar E, Molina E, Hazari YM, Limia CM, Alvarez-Rojas S, Figueroa R, Vidal RL, Rodriguez DA, Rivera CA, Court FA, Couve A, Qi L, Chevet E, Akai R, Iwawaki T, Concha ML, Glavic A, Gonzalez-Billault C, & Hetz C (2018). IRE1alpha governs cytoskeleton remodelling and cell migration through a direct interaction with filamin A. Nat Cell Biol, 20, 942–953. [DOI] [PubMed] [Google Scholar]
- Walls KC, Ghosh AP, Franklin AV, Klocke BJ, Ballestas M, Shacka JJ, Zhang J, & Roth KA (2010). Lysosome dysfunction triggers Atg7-dependent neural apoptosis. Journal of Biological Chemistry, 285, 10497–10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hou C, Cao Y, Cheng Q, Zhang L, Li H, Feng L, & Shen Y (2018). XBP1 activation enhances MANF expression via binding to endoplasmic reticulum stress response elements within MANF promoter region in hepatitis B. The international journal of biochemistry & cell biology, 99, 140–146. [DOI] [PubMed] [Google Scholar]
- Wang G, Wang T, Hu Y, Wang J, Wang Y, Zhang Y, Li F, Liu W, Sun Y, & Yu B (2020). NMMHC IIA triggers neuronal autophagic cell death by promoting F-actin-dependent ATG9A trafficking in cerebral ischemia/reperfusion. Cell Death & Disease, 11, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-F, Mei Z-G, Fu Y, Yang S-B, Zhang S-Z, Huang W-F, Xiong L, Zhou H-J, Tao W, & Feng Z-T (2018). Puerarin protects rat brain against ischemia/reperfusion injury by suppressing autophagy via the AMPK-mTOR-ULK1 signaling pathway. Neural regeneration research, 13, 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Han D, Sun M, & Feng J (2016). Cerebral ischemic post‑conditioning induces autophagy inhibition and a HMGB1 secretion attenuation feedback loop to protect against ischemia reperfusion injury in an oxygen glucose deprivation cellular model. Molecular medicine reports, 14, 4162–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xiong X, Zhang X, Ye Y, Jian Z, Gao W, & Gu L (2020). Sodium tanshinone IIA sulfonate protects against cerebral ischemia-reperfusion injury by inhibiting autophagy and inflammation. Neuroscience. [DOI] [PubMed] [Google Scholar]
- Wang M-M, Zhang M, Feng Y-S, Xing Y, Tan Z-X, Li W-B, Dong F, & Zhang F (2020). Electroacupuncture Inhibits Neuronal Autophagy and Apoptosis via the PI3K/AKT Pathway Following Ischemic Stroke. Frontiers in cellular neuroscience, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Guan Y-F, Du H, Zhai Q-W, Su D-F, & Miao C-Y (2012). Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy, 8, 77–87. [DOI] [PubMed] [Google Scholar]
- Wang S, & Ren J (2018). Role of autophagy and regulatory mechanisms in alcoholic cardiomyopathy. Biochim Biophys Acta Mol Basis Dis, 1864, 2003–2009. [DOI] [PubMed] [Google Scholar]
- Wang S, Wang L, Qin X, Turdi S, Sun D, Culver B, Reiter RJ, Wang X, Zhou H, & Ren J (2020). ALDH2 contributes to melatonin-induced protection against APP/PS1 mutation-prompted cardiac anomalies through cGAS-STING-TBK1-mediated regulation of mitophagy. Signal Transduct Target Ther, 5, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Xue H, Xu Y, Niu J, & Zhao P (2019). Sevoflurane postconditioning inhibits autophagy through activation of the extracellular signal-regulated kinase cascade, alleviating hypoxic-ischemic brain injury in neonatal rats. Neurochemical research, 44, 347–356. [DOI] [PubMed] [Google Scholar]
- Wang X, Feng L, Xin M, Hao Y, Wang X, Shang P, Zhao M, Hou S, Zhang Y, & Xiao Y (2020). Mechanisms underlying astrocytic connexin-43 autophagy degradation during cerebral ischemia injury and the effect on neuroinflammation and cell apoptosis. Biomedicine & Pharmacotherapy, 127, 110125. [DOI] [PubMed] [Google Scholar]
- Wang Y, Denisova JV, Kang KS, Fontes JD, Zhu BT, & Belousov AB (2010). Neuronal Gap Junctions Are Required for NMDA Receptor–Mediated Excitotoxicity: Implications in Ischemic Stroke. Journal of neurophysiology, 104, 3551–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tian D, Wei C, Cui V, Wang H, Zhu Y, Wu A, & Yue Y (2020). Propofol Attenuates α-Synuclein Aggregation and Neuronal Damage in a Mouse Model of Ischemic Stroke. Neuroscience bulletin, 36, 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K, Wang P, & Miao CY (2012). A double-edged sword with therapeutic potential: an updated role of autophagy in ischemic cerebral injury. CNS neuroscience & therapeutics, 18, 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L-P, Brown JL, & Eng C (2001). PTEN induces apoptosis and cell cycle arrest through phosphoinositol-3-kinase/Akt-dependent and - independent pathways. Human molecular genetics, 10, 237–242. [DOI] [PubMed] [Google Scholar]
- Wu K, Huang C, Shi X, Chen F, Xu Y-H, Pan Y-X, Luo Z, & Liu X (2016). Role and mechanism of the AMPK pathway in waterborne Zn exposure influencing the hepatic energy metabolism of Synechogobius hasta. Scientific reports, 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Zhang H, Kai J, Zhu F, Dong J, Xu Z, Wong M, & Zeng LH (2018). Rapamycin prevents cerebral stroke by modulating apoptosis and autophagy in penumbra in rats. Annals of clinical and translational neurology, 5, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NN, Tian H, Chen P, Wang D, Ren J, & Zhang Y (2019). Physical exercise and selective autophagy: benefit and risk on cardiovascular health. Cells, 8, 1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y-T, Tan H-L, Shui G, Bauvy C, Huang Q, Wenk MR, Ong C-N, Codogno P, & Shen H-M (2010). Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. Journal of Biological Chemistry, 285, 10850–10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z-Q, Cui S. y., Zhu L, & Zou Z. q. (2016). Study on the mechanism of mTOR-mediated autophagy during electroacupuncture pretreatment against cerebral ischemic injury. Evidence-Based Complementary and Alternative Medicine, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Zou Z, Zou R, Zhou X, & Cui S (2015). Electroacupuncture pretreatment induces tolerance against cerebral ischemia/reperfusion injury through inhibition of the autophagy pathway. Molecular medicine reports, 11, 4438–4446. [DOI] [PubMed] [Google Scholar]
- Xia Y, Ling X, Hu G, Zhu Q, Zhang J, Li Q, Zhao B, Wang Y, & Deng Z (2020). Small extracellular vesicles secreted by human iPSC-derived MSC enhance angiogenesis through inhibiting STAT3-dependent autophagy in ischemic stroke. Stem cell research & therapy, 11, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Wang M-M, Feng Y-S, Dong F, & Zhang F (2018). Possible involvement of PTEN signaling pathway in the anti-apoptotic effect of electroacupuncture following ischemic stroke in rats. Cellular and molecular neurobiology, 38, 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhang Y, & Ren J (2019). ALDH2 and Stroke: A Systematic Review of the Evidence. Adv Exp Med Biol, 1193, 195–210. [DOI] [PubMed] [Google Scholar]
- Xu W, Xiao P, Fan S, Chen Y, Huang W, Chen X, Liu G, Dang C, Zeng J, & Xing S (2020). Blockade of Nogo-A/Nogo-66 receptor 1 (NgR1) Inhibits Autophagic Activation and Prevents Secondary Neuronal Damage in the Thalamus after Focal Cerebral Infarction in Hypertensive Rats. Neuroscience, 431, 103–114. [DOI] [PubMed] [Google Scholar]
- Xu X, Pan J, Li H, Li X, Fang F, Wu D, Zhou Y, Zheng P, Xiong L, & Zhang D (2019). Atg7 mediates renal tubular cell apoptosis in vancomycin nephrotoxicity through activation of PKC-δ. The FASEB Journal, 33, 4513–4524. [DOI] [PubMed] [Google Scholar]
- Xue H, Wu Z, Xu Y, Gao Q, Zhang Y, Li C, & Zhao P (2021). Dexmedetomidine post-conditioning ameliorates long-term neurological outcomes after neonatal hypoxic ischemia: The role of autophagy. Life Sciences, 270, 118980. [DOI] [PubMed] [Google Scholar]
- Xue H, Xu Y, Wang S, Wu Z-Y, Li X-Y, Zhang Y-H, Niu J-Y, Gao Q-S, & Zhao P (2019). Sevoflurane post-conditioning alleviates neonatal rat hypoxic-ischemic cerebral injury via Ezh2-regulated autophagy. Drug design, development and therapy, 13, 1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, You Y, Tao J, Ye X, Huang J, Yang S, Lin Z, Hong Z, Peng J, & Chen L (2014). Electro-acupuncture at points of Zusanli and Quchi exerts anti-apoptotic effect through the modulation of PI3K/Akt signaling pathway. Neuroscience letters, 558, 14–19. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, & Tatar M (2011). Insulin receptor substrate chico acts with the transcription factor FOXO to extend Drosophila lifespan. Aging cell, 10, 729–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Zhang H, Bai X, Lu Y, Dong H, & Xiong L (2011). Autophagy activation is involved in neuroprotection induced by hyperbaric oxygen preconditioning against focal cerebral ischemia in rats. Brain research, 1402, 109–121. [DOI] [PubMed] [Google Scholar]
- Yang C, Hawkins KE, Doré S, & Candelario-Jalil E (2019). Neuroinflammatory mechanisms of blood-brain barrier damage in ischemic stroke. American Journal of Physiology-Cell Physiology, 316, C135–C153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Pan L, Sun C, Xi Y, Wang L, & Li D (2016). Red meat consumption and the risk of stroke: a dose–response meta-analysis of prospective cohort studies. Journal of Stroke and Cerebrovascular Diseases, 25, 1177–1186. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gao K, Hu Z, Li W, Davies H, Ling S, Rudd JA, & Fang M (2015). Autophagy upregulation and apoptosis downregulation in DAHP and triptolide treated cerebral ischemia. Mediators of Inflammation, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, & Klionsky DJ (2020). Autophagy and disease: unanswered questions. Cell Death Differ, 27, 858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhong L, Zhong S, Xian R, & Yuan B (2015). Hypoxia induces microglia autophagy and neural inflammation injury in focal cerebral ischemia model. Experimental and Molecular Pathology, 98, 219–224. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhou C, Shi H, Zhang N, Tang B, & Ji N (2020). Heme Induces BECN1/ATG5-Mediated Autophagic Cell Death via ER Stress in Neurons. Neurotoxicity Research, 1–12. [DOI] [PubMed] [Google Scholar]
- Yu F, Xue W, Dong L, Hu X, Huang D, & Wang K (2019). Tetrahydroxystilbene glucoside suppresses NAPDH oxidative stress to mitigate apoptosis and autophagy induced by cerebral ischemia/reperfusion injury in mice. Evidence-Based Complementary and Alternative Medicine, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Shao J, Huang R, Guan Y, Li G, Chen S, Zhou F, Yao Q, & Shen J (2020). Targeting PTEN to regulate autophagy and promote the repair of injured neurons. Brain Research Bulletin, 165, 161–168. [DOI] [PubMed] [Google Scholar]
- Yu J, Li X, Matei N, McBride D, Tang J, Yan M, & Zhang JH (2018). Ezetimibe, a NPC1L1 inhibitor, attenuates neuronal apoptosis through AMPK dependent autophagy activation after MCAO in rats. Experimental neurology, 307, 12–23. [DOI] [PubMed] [Google Scholar]
- Yu S, Yu M, Bu Z, He P, & Feng J (2020). FKBP5 Exacerbates Impairments in Cerebral Ischemic Stroke by Inducing Autophagy via the AKT/FOXO3 Pathway. Frontiers in cellular neuroscience, 14, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Fan Y, Gao Z, Sun X, Zhang H, Wang Z, Cui Y, Song W, Wang Z, & Zhang F (2020). SHP2 promotes proliferation of breast cancer cells through regulating Cyclin D1 stability via the PI3K/AKT/GSK3β signaling pathway. Cancer biology & medicine, 17, 707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel SS, Stelzer W, Lorenzoni A, Wozny M, Langosch D, & Lemberg MK (2019). The Metastable XBP1u Transmembrane Domain Defines Determinants for Intramembrane Proteolysis by Signal Peptide Peptidase. Cell Rep, 26, 3087–3099 e3011. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Zhou Y, Liang D, He H, Liu X, Zhu R, Zhang M, Luo X, Wang Y, & Huang G (2020). Exosomes Secreted From Bone Marrow Mesenchymal Stem Cells Attenuate Oxygen-Glucose Deprivation/Reoxygenation-Induced Pyroptosis in PC12 Cells by Promoting AMPK-Dependent Autophagic Flux. Frontiers in cellular neuroscience, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Deng F, Zhou C, & Fang S (2020). ClC-3 induction protects against cerebral ischemia/reperfusion injury through promoting Beclin1/Vps34-mediated autophagy. Human Cell, 1–10. [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu S, Yuan X, Hu Z, Li H, Wu M, Yuan J, Zhao Z, Su J, & Wang X (2016). Valproic acid promotes human glioma U87 cells apoptosis and inhibits glycogen synthase kinase-3β through ERK/Akt signaling. Cellular Physiology and Biochemistry, 39, 2173–2185. [DOI] [PubMed] [Google Scholar]
- Zhang D-M, Zhang T, Wang M-M, Wang X-X, Qin Y-Y, Wu J, Han R, Sheng R, Wang Y, & Chen Z (2019). TIGAR alleviates ischemia/reperfusion-induced autophagy and ischemic brain injury. Free Radical Biology and Medicine, 137, 13–23. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zou X, Sy C, Qin H, Wang Y, Liao X, & Liu L (2014). Thrombolysis and reperfusion: advanced understanding of early management strategies in acute ischemic stroke. Neurological research, 36, 391–396. [DOI] [PubMed] [Google Scholar]
- Zhang H, Lee J-Y, Borlongan CV, & Tajiri N (2019). A brief physical activity protects against ischemic stroke. Brain circulation, 5, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Niu W, He Z, Zhang Q, Wu Y, Jiang C, Tang C, Hu Y, & Jia J (2014). Autophagy suppression by exercise pretreatment and p38 inhibition is neuroprotective in cerebral ischemia. Brain research, 1587, 127–132. [DOI] [PubMed] [Google Scholar]
- Zhang R, Lao L, Ren K, & Berman BM (2014). Mechanisms of acupuncture–electroacupuncture on persistent pain. Anesthesiology, 120, 482–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yan H, Yuan Y, Gao J, Shen Z, Cheng Y, Shen Y, Wang R-R, Wang X, & Hu W-W (2013). Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy, 9, 1321–1333. [DOI] [PubMed] [Google Scholar]
- Zhang Y, He Q, Yang M, Hua S, Ma Q, Guo L, Wu X, Zhang C, Fu X, & Liu J (2020). Dichloromethane extraction from Piper nigrum L. and P. longum L. to mitigate ischemic stroke by activating the AKT/mTOR signaling pathway to suppress autophagy. Brain research, 147047. [DOI] [PubMed] [Google Scholar]
- Zhang Y, & Miao J-M (2018). Ginkgolide K promotes astrocyte proliferation and migration after oxygen-glucose deprivation via inducing protective autophagy through the AMPK/mTOR/ULK1 signaling pathway. European Journal of Pharmacology, 832, 96–103. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sowers JR, & Ren J (2018). Targeting autophagy in obesity: from pathophysiology to management. Nature Reviews Endocrinology, 14, 356–376. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Whaley-Connell AT, Sowers JR, & Ren J (2018). Autophagy as an emerging target in cardiorenal metabolic disease: From pathophysiology to management. Pharmacol Ther, 191, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Wei Q, Leng S, Li C, Han B, Bai Y, Zhang H, & Yao H (2020). Activation of Sigma-1 Receptor Enhanced Pericyte Survival via the Interplay Between Apoptosis and Autophagy: Implications for Blood–Brain Barrier Integrity in Stroke. Translational stroke research, 11, 267–287. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang Y, Jin X. f., Zhou X. h., Dong X. h., Yu W. t., & Gao W. j. (2019). The role of astragaloside IV against cerebral ischemia/reperfusion injury: suppression of apoptosis via promotion of P62-LC3-autophagy. Molecules, 24, 1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Qu Y, Zhu J, Zhang L, Huang L, Liu H, Li S, & Mu D (2017). miR-30d-5p Plays an Important Role in Autophagy and Apoptosis in Developing Rat Brains After Hypoxic–Ischemic Injury. Journal of Neuropatholgy & Experimental Neurology, 76, 709–719. [DOI] [PubMed] [Google Scholar]
- Zhao G, Zhang W, Li L, Wu S, & Du G (2014). Pinocembrin protects the brain against ischemia-reperfusion injury and reverses the autophagy dysfunction in the penumbra area. Molecules, 19, 15786–15798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y-N, Guo X-F, Li J-M, Chen C-X, Li S-X, & Xu C-J (2017). mTOR/autophagy pathway in the hippocampus of rats suffering intermittent hypoxia preconditioning and global cerebral ischemia-reperfusion. Oncotarget, 8, 23353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H-Q, Zhang L-Y, Luo J, Li L-L, Li M, Zhang Q, & Hu X-Q (2014). Physical exercise promotes recovery of neurological function after ischemic stroke in rats. International journal of molecular sciences, 15, 10974–10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Li X-Y, Huang F-Z, Zhang X-T, Tang H-B, Li Y-S, Zhang WK, Li X-J, & Tian G-H (2020). Effect of electroacupuncture on relieving central post-stroke pain by inhibiting autophagy in the hippocampus. Brain research, 1733, 146680. [DOI] [PubMed] [Google Scholar]
- Zhong S-J, Cui M-M, Gao Y-T, Cao X-Y, Chen B, & Wen X-R (2020). MicroRNA-144 promotes remote limb ischemic preconditioning-mediated neuroprotection against ischemic stroke via PTEN/Akt pathway. Acta Neurologica Belgica, 1–12. [DOI] [PubMed] [Google Scholar]
- Zhu C, Wang X, Xu F, Bahr B, Shibata M, Uchiyama Y, Hagberg H, & Blomgren K (2005). The influence of age on apoptotic and other mechanisms of cell death after cerebral hypoxia–ischemia. Cell Death & Differentiation, 12, 162–176. [DOI] [PubMed] [Google Scholar]
- Zhu C, Xu F, Wang X, Shibata M, Uchiyama Y, Blomgren K, & Hagberg H (2006). Different apoptotic mechanisms are activated in male and female brains after neonatal hypoxia–ischaemia. Journal of Neurochemistry, 96, 1016–1027. [DOI] [PubMed] [Google Scholar]
- Zhu C, Zhou Q, Luo C, & Chen Y (2020). Dexmedetomidine Protects Against Oxygen–Glucose Deprivation-Induced Injury Through Inducing Astrocytes Autophagy via TSC2/mTOR Pathway. Neuromolecular medicine, 22, 210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, Liu C-G, & Yang J-M (2009). Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy, 5, 816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li S, Liu J, Wen Q, Yu J, Yu L, & Xie K (2019). Role of JNK signaling pathway in dexmedetomidine post-conditioning-induced reduction of the inflammatory response and autophagy effect of focal cerebral ischemia reperfusion injury in rats. Inflammation, 42, 2181–2191. [DOI] [PubMed] [Google Scholar]
- Zuo W, Zhang S, Xia C-Y, Guo X-F, He W-B, & Chen N-H (2014). Mitochondria autophagy is induced after hypoxic/ischemic stress in a Drp1 dependent manner: the role of inhibition of Drp1 in ischemic brain damage. Neuropharmacology, 86, 103–115. [DOI] [PubMed] [Google Scholar]