Abstract
Given the severe side effects of the treatments and poor survival, prognostic and predictive biomarkers to guide management of pancreatic cancer are in critical need. We hypothesized that cell proliferation-related pathways are associated with drug response and survival in pancreatic cancer. Six Hallmark cell proliferation-related gene sets (G2M Checkpoint, E2F Targets, MYC Targets V1 and V2, Mitotic Spindle, p53 pathway) defined by MSigDB in gene set variant analysis were evaluated in 3 independent cohorts- TCGA-PAAD (n = 176), GSE57495 (n = 63), and GSE62452 (n = 69). G2M and E2F, as well as Mitotic and p53 pathway correlated highly with other gene sets. All pathways were significantly correlated with MKI67 expression and its proliferation score, but none with cytolytic activity and the rate of pathologically complete resection (R0). All pathways were significantly associated with high alteration and expression of KRAS gene except for MYC v1. G2M, E2F, and p53 pathway were significantly associated with high alteration of TP53 gene. Interestingly, different pathways correlated with the AUC of different cancer therapeutics, such as Gemcitabine (Mitotic: r = 0.706 [P = 0.01]), Paclitaxel (MYC v2: r = -0.636 [P < 0.05]), Apatinib (Mitotic: r = -0.556 [P = 0.03]), Palbociclib (E2F: r = 0.675 [P < 0.01]), and Sorafenib (G2M: r = -0.593 [P = 0.03]). Among all six pathways, only G2M was consistently associated with worse patient survival in all three cohorts. In conclusion, each cell proliferation-related pathway was predictive of a unique agent, and the G2M score alone predicts survival in pancreatic cancer.
Keywords: Biomarker, cell cycle, gene sets, gene expression, GSVA, GSEA, tumor microenvironment, pathway analysis, pancreatic cancer, proliferation, Hallmark gene sets, survival, transcriptome
Introduction
Pancreatic cancer is one of the deadliest malignancies known, with a poor five-year survival rate of less than 5% in the United States [1]. Despite advances in imaging technologies and surgical techniques, the majority of patients present with metastatic disease burden, with only 20% of patients presenting with resectable disease. Surgical resection remains the mainstay of curative treatment for pancreatic cancer. Advancements in patient selection for surgery has allowed for improved median survival in patients [3]. For example, the identification of borderline resectability status has allowed surgeons to choose the appropriate candidates for an attempt at complete curative resection with microscopic tumor clearance (R0). In addition, pancreatic cancer resections that result in positive margins (R1 and R2 [R1/2]) are thought to be influenced by aggressive tumor biology and invasiveness of cancer. These factors appear as influential, if not more, than surgical technique alone in the correlation to survival [2-4].
Understanding the aberrant cell proliferation of pancreas cancer has been heavily studied to identify targeted therapies to cease cancer progression and improve clinical outcomes. Gemcitabine, a cell cycle-specific inhibitor, has been the most common first-line chemotherapy regimen for patients with metastatic pancreatic cancer [5]. CDK inhibitors, such as Palbociclib and Dinaciclib, that target cell proliferation pathways have also been studied as targeted therapies [6,7]. Treatment with combined CDK4/6 and MEK inhibitors has been used for their inhibitory effect on cell cycle progression while also modulating immune features of pancreatic cancer cells [8].
Unfortunately, the proliferation pathway in pancreatic cancer is hard to define. Adding to its complexity, the high amount of genomic heterogeneity of pancreas cancer and its tumor microenvironment have been associated with resistance to therapy [9,10]. Thus far, no reliable molecular targets have been identified that can predict or influence the success of treatment. To this end, there is an urgent need to identify diagnostic, prognostic, and predictive biomarkers, as well as to develop more effective and less toxic therapeutic interventions.
The gene set variation analysis (GSVA) is a computational model that can be used to examine gene expression within a pathway rather than a single gene. Utilization of a pathway or gene-set approach more accurately considers gene coordination, increases model simplicity, and can increase the applicability of prediction models. We had previously reported the utility of a pathway score which was calculated by GSVA with Hallmark gene sets in several cancers [11-13], including pancreatic cancer. A high G2M pathway score in pancreatic cancer showed a link to KRAS or TP53 alteration, which are known to be frequently altered, and are associated with poor prognosis [14]. Given this background, we hypothesized in this study that the other proliferation-related gene sets are also associated with worse survival and have unique characteristics in pancreatic cancer patients. We analyzed the pathways associated with characteristics in pancreatic cancer patients and determined which pathway score had the most prognostic value in patients with pancreatic cancer.
Materials and methods
Pancreatic cancer cohorts and their data
In the study, clinical and mRNA expression data of 176 patients who had a pathological diagnosis of pancreatic cancer were obtained from TCGA pancreatic adenocarcinoma (TCGA_PAAD) cohort through Genomic Data Commons Data Portal (GDC) (https://portal.gdc.cancer.gov) [15]. Mutation data were downloaded through cBioportal [16]. To survival analysis, other three cohorts, Chen et al. (GSE57495; n = 63) [17], and Hussain et al. (GSE62452; n = 69) [18] were obtained from the Gene Expression Omnibus (GEO) repository of the US National Institutes of Health (http://www.ncbi.nlm.ih.gov/geo/). In all analyses, the average gene value with multiple probes and gene expression data, which were transformed for log2 mRNA data for the effect of several drugs on the pancreatic cancer cell line was obtained from the publicly available dataset at CCLE.
Gene set expression analyses
Gene set variation analysis (GSVA) score [19] of the proliferation-related MSigDB Hallmark gene sets [5], including G2M target, E2F checkpoint, MYC v1 and v2, Mitotic, and p53 pathway, were defined as each pathway score using GSVA Bioconductor package (version 3.10), as we previously reported [20-23]. False discovery rate (FDR), as the adjusted p values, less than 0.25 for statistical significance, which was recommended by gene set enrichment analysis (GSEA) software (Lava version 4.0) [24], as we previously reported [25-32].
Statistical analysis
R software (version 4.0.1, R Project for Statistical Computing) and Microsoft Excel (version 16 for Windows) were used in all data analysis and data plotting. To divide the low and high pathway score groups, the median value of each score within cohorts were used. Two-tail Fisher’s exact tests were used to analysis of comparisons between groups, as we described in each figure legends. Cox proportional hazard analyses were used to estimate hazard ratio (HR), 95% CI, and p-value. A p-value of less than 0.05 was taken statistically significant. Tukey type boxplots show median and interquartile level values.
Results
G2M checkpoint and E2F targets scores demonstrated strong correlation and consistency among the 6 cell proliferation-related Hallmark gene sets in pancreatic cancer
Molecular Signatures Database (MSigDB) Hallmark defines 6 gene sets as cell proliferation-related in Gene Set Validation Analysis (GSVA): G2M checkpoint, E2F target, MYC targets v1, MYC targets v2, Mitotic spindle, and p53 pathway [33]. We utilized these cell proliferation-related gene sets as pathway scores and analyzed them in The Cancer Genome Atlas (TCGA) pancreatic adenocarcinoma cohort (TCGA-PAAD, n = 176). The genes included in each score are listed in Table S1. First, we studied how close these 6 scores relate to each other in pancreatic cancer. We found that the correlation of G2M checkpoint pathway score with E2F targets score was the strongest (Figure 1A; r = 0.976). On the other hand, the correlation of Mitotic spindle score with MYC targets v2 score was the weakest (r = 0.564). Next, we studied the pathways that constitute these scores in pancreatic cancer. We investigated the correlation by hallmark gene sets, and we used the categories defined by MSigDB Hallmark gene sets: Cell proliferation, Signaling, Pathway, Metabolic, DNA damage, Development, and Cellular component [33]. The gene sets that showed Spearman’s rank correlation > 0.600 in each analysis were recognized as highly enriched. As expected, G2M checkpoint, E2F targets, and MYC targets v2 pathway scores were strongly correlated with cell proliferation-related gene sets (Figure 1B). On the other hand, Mitotic spindle and p53 pathway scores were highly correlated with signaling gene sets. Immune-related gene sets were not involved in any of the proliferation-related pathway scores. These results suggest that each of the cell proliferation-related pathway scores has unique characteristics.
Figure 1.
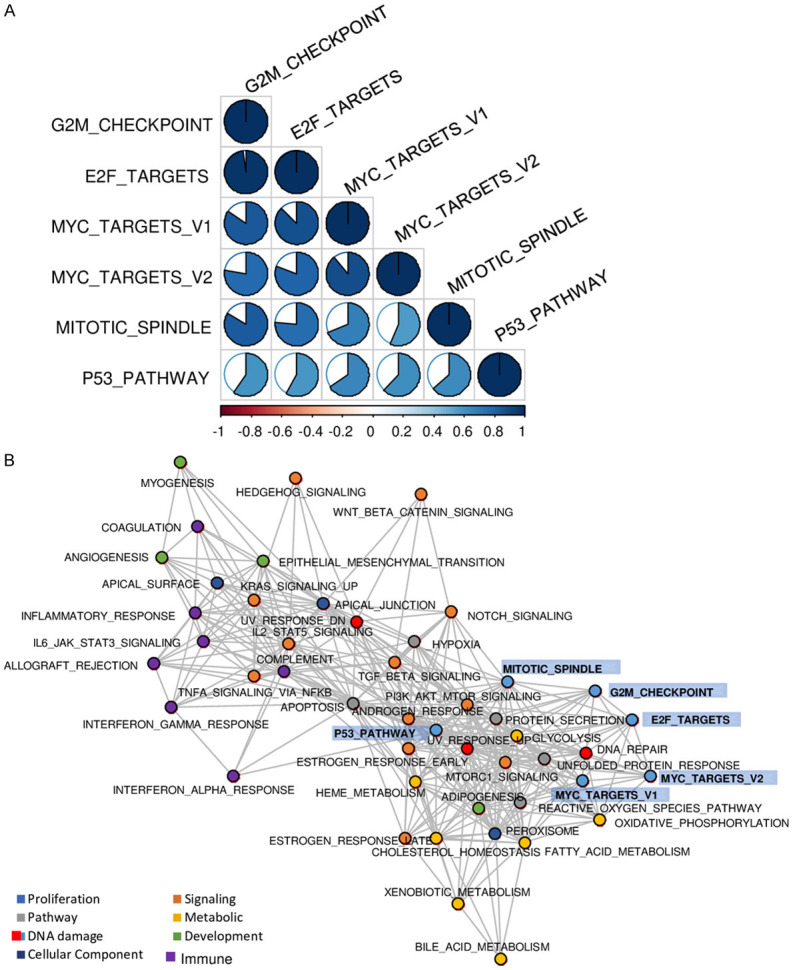
Characteristics of six cell proliferation-related pathway scores in the TCGA pancreatic adenocarcinoma cohort. A. Correlation matrix of six cell proliferation-related pathway scores. The correlation value was indicated by color (blue for positive correlation and red for negative correlation), while the magnitude of correlation was shown with circles. Spearman’s rank correlation was used to the analysis. B. Network plot for highly correlations of hallmark gene sets. Spearman’s rank correlation > 0.600 was defined as highly significantly correlated gene sets.
All six scores were significantly correlated with cell proliferation, but not with immune activity or rate of pathological complete resection (R0)
In clinical practice, cell proliferation is commonly assessed with Ki67 expression, which is encoded by the MKI67 gene. As expected, all six cell proliferation-related pathways were strongly correlated with MKI67 expression (Figure 2A; all spearman rank correlation (r) > 0.500 and all P < 0.001). The proliferation score, calculated by Thorsson et al. [34], was also correlated with all six cell proliferation-related pathways score (Figure 2A). Based upon our previous observation that aggressive breast cancer is counterbalanced with the human immune response [12,13,35], it was of interest to investigate whether highly proliferative pancreatic cancer related with immune activity as estimated by cytolytic activity score (CYT) as established by Rooney et al. [36]. None of the cell proliferation scores were correlated with CYT (Figure 2A). Since pancreatic cancer with aggressive biology is unlikely to result in pathological complete resection (R0) [2,4], we investigated whether any of the six scores correlated with less likelihood of achievement of an R0 resection. Surprisingly, none of the six scores were associated with a decreased R0 resection rate (Figure 2B).
Figure 2.
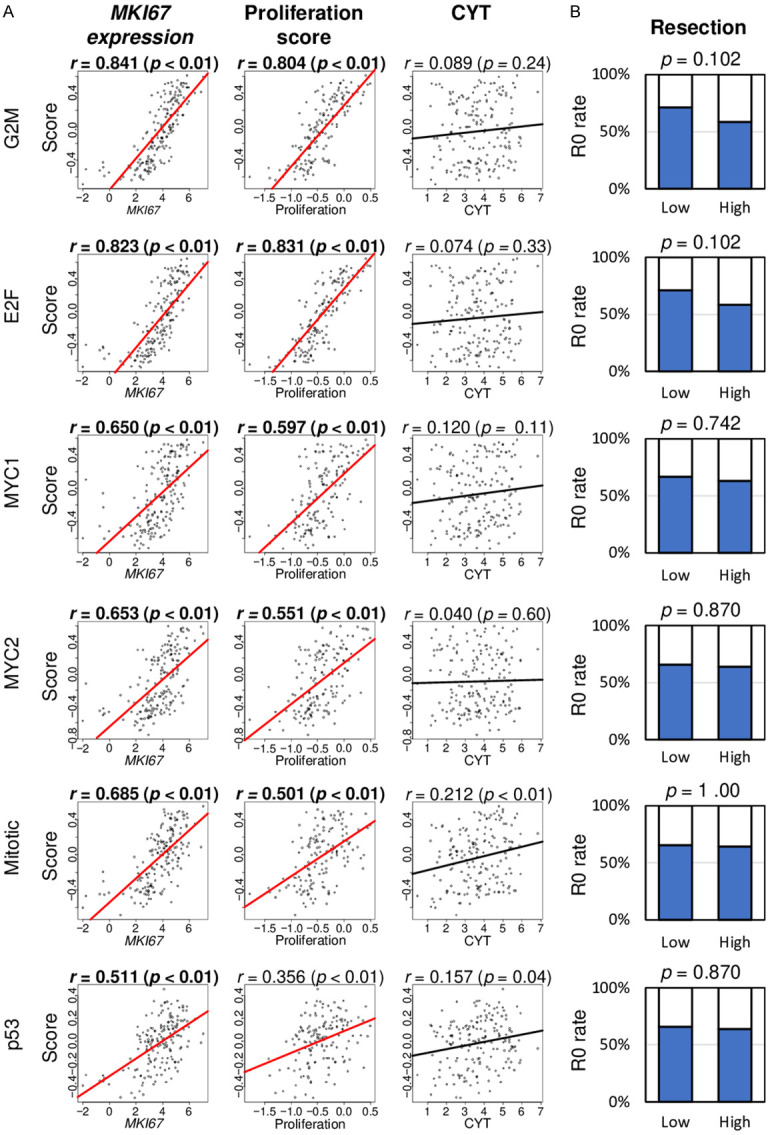
Association of six cell proliferation-related pathway scores with cell proliferation parameters, immune cytolytic activity, and pathological complete resection in the TCGA cohort. A. Correlation plots of cell proliferation-related pathway scores with MKI67 expression, Proliferation score, and Cytolytic Activity score (CYT). Spearman’s rank correlation was used for the analysis. B. Bar plots of the comparison of the low and high score group with R0 resection ratio. Fisher’s test was use for the analysis. The median value was used as a cut-off to divide low and high groups for each pathway. G2M, G2M Checkpoint; E2F, E2F Targets; MYC1, MYC Targets V1; MYC2, MYC Targets V2; Mitotic, Mitotic Spindle; p53, p53 pathway.
High scores in G2M checkpoint, E2F targets and p53 pathways are associated with KRAS and TP53 alteration, and the high score of any cell proliferation-related pathways is correlated with gene expression of KRAS and TP53 in pancreatic cancer
As KRAS and TP53 gene mutations are known to contribute to poor prognosis in pancreatic cancer [37], we hypothesized that the cell proliferation-related pathway scores are associated with alteration and expression of these genes. Indeed, all of the high cell proliferation pathway scores in pancreatic cancer, except for the MYC targets v1 score, were significantly associated with a higher percentage of alteration in the KRAS gene (Figure 3). A high score of the G2M checkpoint, E2F targets, and p53 pathway was significantly associated with a higher percentage of alteration in the TP53 gene (Figure 3; P = 0.026, 0.011, and 0.007, respectively). Furthermore, all six scores were moderately correlated with KRAS gene expression but not with TP53 gene expression (Figure 3).
Figure 3.
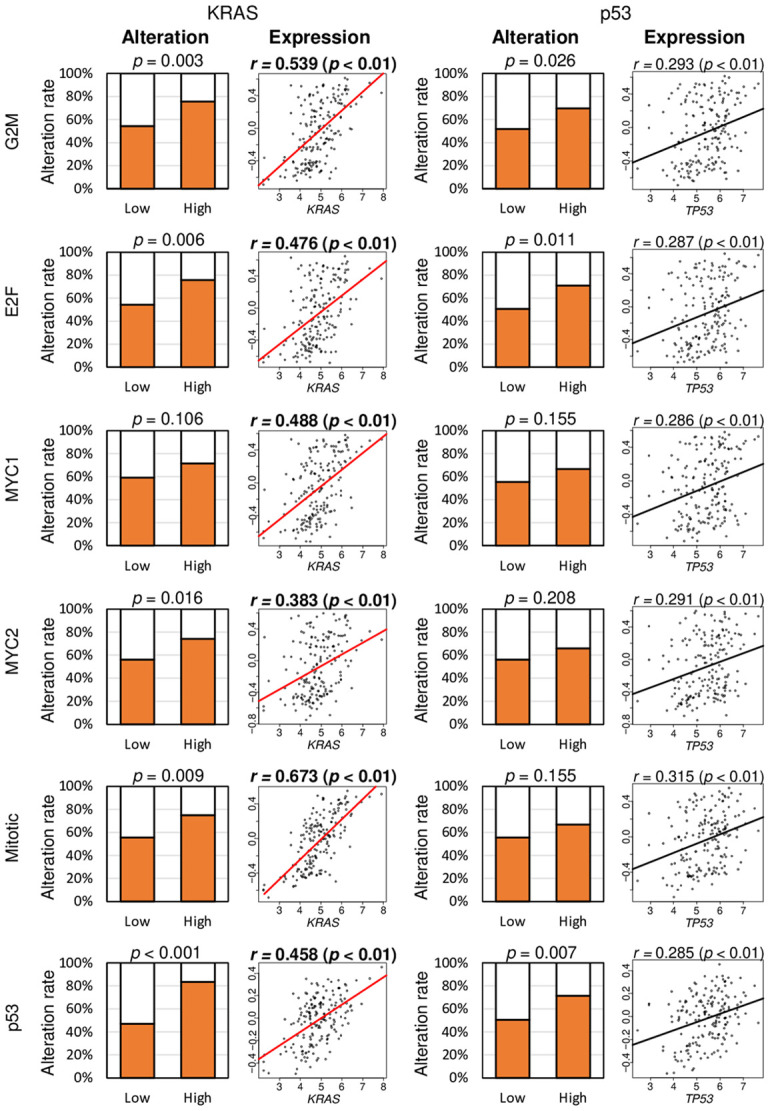
The association of six cell proliferation-related pathway scores with alteration and expression of KRAS and TP53 genes in the TCGA cohort. Bar plots of the percentage of patients with alteration of KRAS and TP53 genes between low and high gene sets. The median value was used to divide low and high score groups as a cut-off. The Fisher’s test was used for the analysis. Correlation plots of cell proliferation-related pathway scores with gene expression of KRAS and TP53. Spearman’s rank correlation was used for the analysis. G2M, G2M Checkpoint; E2F, E2F Targets; MYC1, MYC Targets V1; MYC2, MYC Targets V2; Mitotic, Mitotic Spindle; p53, p53 pathway.
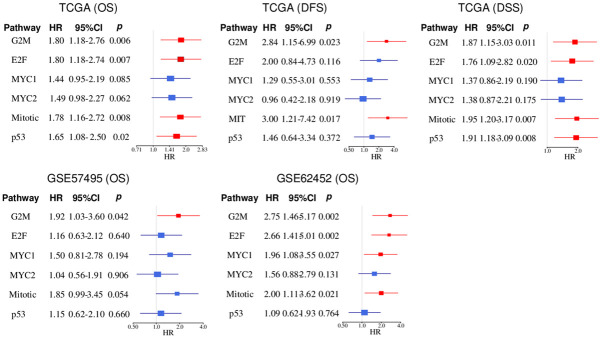
G2M checkpoint pathway score was consistently and strongly associated with worse survival across all three independent cohorts
As we previously reported that enhanced cell proliferation was associated with worse survival in breast [12,13] and pancreatic cancer [14], it was of interest to identify which cell proliferation-related score strongly related with survival in pancreatic cancer patients. We analyzed the hazard ratios (HR) of the six scores related to Overall survival (OS), Disease-free survival (DFS), and Disease-specific survival (DSS) in the TCGA (n = 176), as well as OS in GSE62452 (n = 69) and GSE57495 (n = 63) cohorts. The high score of the G2M checkpoint and Mitotic spindle pathways were associated with poor OS, DFS, and DSS in all cohorts of TCGA and GSE62452; whereas E2F targets and p53 pathway scores were associated with only OS and DSS of TCGA cohort (Figure 4). The MYC targets v1 score was associated with OS in GSE62452 alone. The G2M checkpoint score consistently demonstrated the highest HR in all the survival analyses across all the cohorts (HR; 1.80, 2.84, 1.87, 1.92, 2.75, respectively). None of the other gene sets within the Hallmark gene sets showed significant differences across all cohorts (Tables S2 and S3). These results suggest that the G2M checkpoint pathway score has the highest potential to be used as a prognostic biomarker in pancreatic cancer among the six cell proliferation-related pathway scores.
Figure 4.
Association of six cell proliferation-related scores and patient survival in pancreatic cancer. Forest plots of hazard ratio (HR) for high cell proliferation-related pathway score in the TCGA (n = 176), GSE57495 (n = 63), and GSE62452 (n = 69). The median value was used as a cut-off to divide low and high groups within the cohort. The blue line indicates non-significance and the red line indicated significance. G2M, G2M Checkpoint; E2F, E2F Targets; MYC1, MYC Targets V1; MYC2, MYC Targets V2; Mitotic, Mitotic Spindle; p53, p53 pathway.
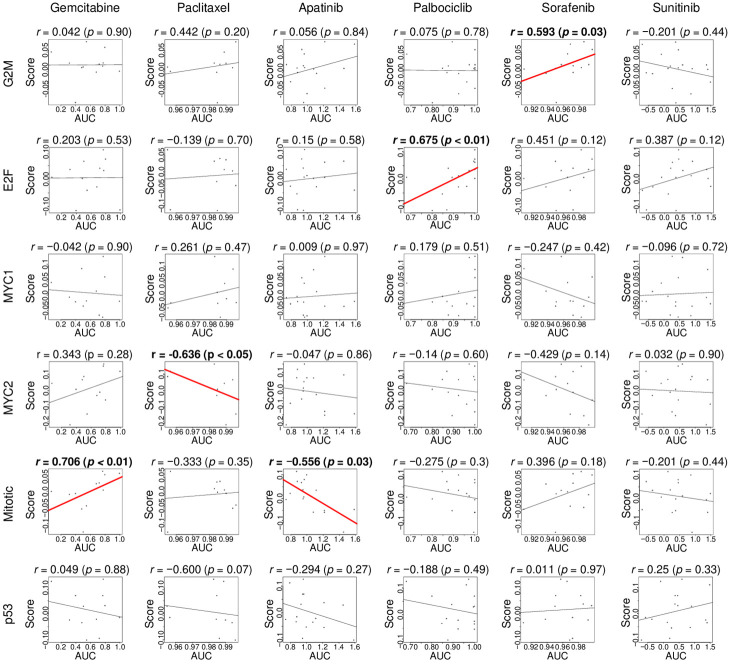
Cell proliferation-related pathway scores predict response for different therapeutic agents used in pancreatic cancer treatment
As the cell proliferation-related pathway scores are associated with survival, we hypothesized that the score also correlates with response to treatment. Since we do not have access to pancreatic cancer patient cohorts with transcriptome and drug response data, we examined the drug sensitivity of pancreatic cancer cell lines, including Gemcitabine, Paclitaxel, Apatinib, Palbociclib, Sorafenib, and Sunitinib, utilizing the Cancer Cell Line Encyclopedia [38]. We found that the G2M checkpoint score correlated moderately with the area under the curve (AUC) of Sorafenib sensitivity (Figure 5; r = 0.593, P = 0.03). The E2F targets score correlated strongly with AUC for Palbociclib (Figure 5; r = 0.675, P < 0.01). The MYC targets v2 score negatively correlated with AUC for Paclitaxel (Figure 5; r = -0.636, P < 0.05). The Mitotic spindle score correlated strongly with AUC for Gemcitabine (Figure 5; r = 0.706, P = 0.01), and negatively with AUC for Apatinib (Figure 5; r = -0.556, P = 0.03). These results suggest that the cell proliferation-related pathway scores may have the potential to predict drug treatment response for pancreatic cancer.
Figure 5.
Correlation between the cell proliferation-related scores and drug response. Correlation plots of the association in human pancreatic cancer cell lines between six proliferation-related gene set score and sensitivity to pancreatic cancer drugs, including Gemcitabine, Paclitaxel, Apatinib, Palbociclib, Sorafenib, and Sunitinib. P-value and rho (r) were analyzed with spearman’s rank correlation coefficient. G2M, G2M Checkpoint; E2F, E2F Targets; MYC1, MYC Targets V1; MYC2, MYC Targets V2; Mitotic, Mitotic Spindle; p53, p53 pathway.
Discussion
In this study, we evaluated six cell proliferation-related pathways (G2M checkpoints, E2F targets, MYC target v1, MYC target v2, Mitotic spindle, and p53 pathway) in pancreatic cancer using the GSVA scoring method with the MSigDB Hallmark gene sets collection. The pathways strongly correlated with each other, especially the G2M checkpoint with E2F target scores. The pathways also highly correlated with gene sets classified into other categories, such as metabolic, DNA damage, cellular component, but the distributions of these components were different in six cell proliferation-related gene sets. All six cell proliferation-related gene sets was strongly correlated with MKI67 gene expression, and the high score of all six cell proliferation-related gene sets was significantly associated with a high proliferation score. Among them, high mitotic spindle score alone was associated with high cytolytic activity. None were associated with the R0 resection rate. A high score of all cell proliferation-related gene sets was significantly associated with a high rate of KRAS gene alteration except for MYC targets v1 score. A high score of G2M checkpoint, E2F targets, and p53 pathway was significantly associated with high rate of TP53 gene alteration. All six gene sets were correlated with KRAS gene expression but not with TP53 gene expression. Interestingly, cell proliferation-related pathway scores predicted response to different therapeutic agents used in pancreatic cancer treatment, such as Gemcitabine, Paclitaxel, Apatinib, Palbociclib, and Sorafenib. A high score of all six gene sets was associated with worse prognosis except for MYC target v2. Finally, only the G2M checkpoint score showed significant association with worse patient survival across all cohorts.
To date, many studies have reported the expression of various genes and signal transduction pathways in cancer. These act in an intricate manner to promote cancer growth and/or treatment resistance. Therefore, it can be difficult to define the complex signaling in cancer by identifying a single gene. GSVA is a useful tool to illustrate a wide perspective of the signaling pathways in cancer. We have previously reported several gene set pathway scores were associated with clinical outcomes in cancer using the GSVA method. KRAS signaling was significantly associated with anti-cancer immune microenvironment as well as improved survival in breast cancer [11]. In pancreatic cancer, a high G2M checkpoint score was significantly associated with worse patient survival, particularly after margin-positive resection [14]. In the majority of cases, pathological margin-negative R0 resection has better survival compared with margin-positive resection, thought to be reflective of the aggressive biology of the cancer in the subset of margin positive patients [2-4]. However, none of the 6 cell proliferation-related scores predicted the R0 resection rate in this study. Proliferation-related gene sets score showed association with tumor aggressiveness, such as pathological grade, AJCC stage, and MKI67 expression. The score of MYC targets v1 and v2 in breast cancer [39] and the G2M checkpoint score in pancreatic cancer strongly correlated with MKI67 gene expression and with worse patient survival [14].
The KRAS and TP53 genes are associated with high rates of alteration in pancreatic cancer [37] and are linked to progression of disease. Among the six cells proliferation-related pathway scores, the E2F checkpoint, G2M targets, and p53 pathway scores were correlated with expression of KRAS and TP53 gene mutations in pancreatic cancer.
Surgical resection remains the mainstay of curative treatment for pancreatic cancer [40]. However, the majority of patients present with metastatic disease at the time of diagnosis, and therefore systemic chemotherapy remains the primary treatment option for most patients. As rates of drug resistance have made chemotherapy less effective, a bigger problem arises. Regimens of FOLFIRINOX (FFX) and Gemcitabine/Nab-paclitaxel (GNP) are used in metastatic patients, and large centers have transitioned these regimens as neoadjuvant treatment for patients with locally advanced pancreatic cancer [41,42]. With this, an increasing number of patients are treated with multiple chemotherapeutic regimens with the intent of downstaging the cancer and enabling complete microscopically negative resection [43], or sparing major surgery altogether. Despite the widespread use of neoadjuvant therapy, it is unclear which regimen is associated with the best possible survival. The recent notion that adjuvant FFX may improve survival in patients already treated with neoadjuvant therapy and tumor resection has been under investigation [44].
Several other pathway-targeted chemotherapies have been studied. Palbociclib, a CDK4 inhibitor, leads to inhibitory effects at different stages of the tumor cell and within the tumor microenvironment, which collectively drives down cancer cell proliferation and invasion [45]. Sorafenib, on the other hand, inhibits signaling pathways such as RAF/MEK/ERK cascades resulting in decreased cell proliferation [46], and target receptors such as VEGFR PDGFR-β for anti-angiogenic effects. Sorafenib also inhibits DNA synthesis and induces tumor cell death in various cancers [47]. Combination therapies have shown potent anti-proliferative and pro-apoptotic efficacy in pancreatic cancer cells [48,49]. Preclinical studies and on-going phase I clinical trials have demonstrated the safety and efficacy of combinatorial radio-chemotherapy plus surgery in pancreatic cancer, including the combination of sorafenib and Vorinostat [50]. Additionally, Sunitinib inhibits endothelial cell proliferation angiogenic proliferation as a broad-spectrum receptor tyrosine kinase (RTK) inhibitor [51]. Though Sunitinib treatment has impressive results in treating neuroendocrine pancreatic tumors, phase II clinical trials in advanced or metastatic pancreatic cancer have shown its consistent failure [52,53].
In our study, proliferation-related gene sets did not show correlation to a sensitivity for Sunitinib or Apatinib, which is a multiple kinase inhibitor with bioactivity against VEGFR-2, PDGFR-β, c-Kit, and c-src [54]. These agents were proven to have a survival benefit in several cancers such as gastric, colon, and breast cancer [55]. Apatinib has demonstrated substantial potential as a new therapeutic option due to its ease of administration, compliance rate, low toxicity, and improved outcomes [56]. Cheng-Ming et al. reported the potential use of Apatinib in the treatment of pancreatic cancer [57]. The ability to target even a minority of pancreatic cancer patients is significant due to the current limitations in treatment options. A biomarker that can predict survival and response to chemotherapy, such as the proliferation-related gene set score, may be able to improve treatment efficacy, reduce toxicity, and improve patients’ quality of life. Based on our study, we speculate that proliferation-related scores can have utility as a predictive biomarker of response to chemotherapies in pancreatic cancer patients.
Our study has its limitations. This study is a retrospective study. In order to establish the utility of cell proliferation-related gene sets as a biomarker in predicting the effectiveness of chemotherapies, a prospective study will be required.
In conclusion, we have demonstrated that the G2M pathway score can serve as a tool for identifying patients who are likely to have poor survival in pancreatic cancer. Our findings support the need for clinical trials to evaluate these gene set scores as a predictive biomarker for response to chemotherapy.
Acknowledgements
This work was supported by US National Institutes of Health/National Cancer Institute grant R01CA160688, R01CA250412, R37CA248018, US Department of Defense BCRP grant W81XWH-19-1-0674, as well as the Edward K. Duch Foundation and Paul & Helen Ellis Charitable Trust to K.T., and US National Cancer Institute cancer center support grant P30-CA016056 to Roswell Park Comprehensive Cancer Center.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Rau BM, Moritz K, Schuschan S, Alsfasser G, Prall F, Klar E. R1 resection in pancreatic cancer has significant impact on long-term outcome in standardized pathology modified for routine use. Surgery. 2012;152:S103–111. doi: 10.1016/j.surg.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Katsuta E, Rashid OM, Takabe K. Fibroblasts as a biological marker for curative resection in pancreatic ductal adenocarcinoma. Int J Mol Sci. 2020;21:3890. doi: 10.3390/ijms21113890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demir IE, Jäger C, Schlitter AM, Konukiewitz B, Stecher L, Schorn S, Tieftrunk E, Scheufele F, Calavrezos L, Schirren R, Esposito I, Weichert W, Friess H, Ceyhan GO. R0 versus R1 resection matters after pancreaticoduodenectomy, and less after distal or total pancreatectomy for pancreatic cancer. Ann Surg. 2018;268:1058–1068. doi: 10.1097/SLA.0000000000002345. [DOI] [PubMed] [Google Scholar]
- 5.Neoptolemos JP, Stocken DD, Friess H, Bassi C, Dunn JA, Hickey H, Beger H, Fernandez-Cruz L, Dervenis C, Lacaine F, Falconi M, Pederzoli P, Pap A, Spooner D, Kerr DJ, Büchler MW. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N Engl J Med. 2004;350:1200–1210. doi: 10.1056/NEJMoa032295. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Chen Z, Li X, He J, Liu Z, Yang L. Baicalein flavone targets cisplatin resistant human pancreatic cancer cells via inducing S-phase cell cycle arrest, inhibition of cell migration and invasion, caspase activation and mitochondrial-dependent apoptosis. J BUON. 2020;25:1947–1953. [PubMed] [Google Scholar]
- 7.Khan T, Seddon AM, Dalgleish AG, Khelwatty S, Ioannou N, Mudan S, Modjtahedi H. Synergistic activity of agents targeting growth factor receptors, CDKs and downstream signaling molecules in a panel of pancreatic cancer cell lines and the identification of antagonistic combinations: Implications for future clinical trials in pancreatic cancer. Oncol Rep. 2020;44:2581–2594. doi: 10.3892/or.2020.7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesinski GB. Braking the cell’s cycle and invigorating T-cell immunity against pancreatic cancer. Gut. 2021;70:4–5. doi: 10.1136/gutjnl-2020-321497. [DOI] [PubMed] [Google Scholar]
- 9.Haeberle L, Steiger K, Schlitter AM, Safi SA, Knoefel WT, Erkan M, Esposito I. Stromal heterogeneity in pancreatic cancer and chronic pancreatitis. Pancreatology. 2018;18:536–549. doi: 10.1016/j.pan.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Cros J, Raffenne J, Couvelard A, Poté N. Tumor heterogeneity in pancreatic adenocarcinoma. Pathobiology. 2018;85:64–71. doi: 10.1159/000477773. [DOI] [PubMed] [Google Scholar]
- 11.Tokumaru Y, Oshi M, Katsuta E, Yan L, Satyananda V, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival. Am J Cancer Res. 2020;10:897–907. [PMC free article] [PubMed] [Google Scholar]
- 12.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Nagahashi M, Matsuyama R, Endo I, Takabe K. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2- breast cancer. Cells. 2020;9:1643. doi: 10.3390/cells9071643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Matsuyama R, Endo I, Takabe K. G2M cell cycle pathway score as a prognostic biomarker of metastasis in estrogen receptor (ER)-positive breast cancer. Int J Mol Sci. 2020;21:2921. [Google Scholar]
- 14.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Katz MHG, Takabe K. High G2M pathway score pancreatic cancer is associated with worse survival, particularly after margin-positive (R1 or R2) resection. Cancers (Basel) 2020;12:2871. doi: 10.3390/cancers12102871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen DT, Davis-Yadley AH, Huang PY, Husain K, Centeno BA, Permuth-Wey J, Pimiento JM, Malafa M. Prognostic fifteen-gene signature for early stage pancreatic ductal adenocarcinoma. PLoS One. 2015;10:e0133562. doi: 10.1371/journal.pone.0133562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang S, He P, Wang J, Schetter A, Tang W, Funamizu N, Yanaga K, Uwagawa T, Satoskar AR, Gaedcke J, Bernhardt M, Ghadimi BM, Gaida MM, Bergmann F, Werner J, Ried T, Hanna N, Alexander HR, Hussain SP. A novel MIF signaling pathway drives the malignant character of pancreatic cancer by targeting NR3C2. Cancer Res. 2016;76:3838–3850. doi: 10.1158/0008-5472.CAN-15-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oshi M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Endo I, Takabe K. Degree of early estrogen response predict survival after endocrine therapy in primary and metastatic er-positive breast cancer. Cancers (Basel) 2020;12:3557. doi: 10.3390/cancers12123557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. Inflammation is associated with worse outcome in the whole cohort but with better outcome in triple-negative subtype of breast cancer patients. J Immunol Res. 2020;2020:5618786. doi: 10.1155/2020/5618786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, Takabe K. Intra-tumoral angiogenesis is associated with inflammation, immune reaction and metastatic recurrence in breast cancer. Int J Mol Sci. 2020;21:6708. doi: 10.3390/ijms21186708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oshi M, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Cherkassky L, Takabe K. Enhanced DNA repair pathway is associated with cell proliferation and worse survival in hepatocellular carcinoma (HCC) Cancers (Basel) 2021;13:323. doi: 10.3390/cancers13020323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oshi M, Tokumaru Y, Patel A, Yan L, Matsuyama R, Endo I, Katz MHG, Takabe K. A novel four-gene score to predict pathologically complete (R0) resection and survival in pancreatic cancer. Cancers (Basel) 2020;12:3635. doi: 10.3390/cancers12123635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oshi M, Tokumaru Y, Asaoka M, Yan L, Satyananda V, Matsuyama R, Matsuhashi N, Futamura M, Ishikawa T, Yoshida K, Endo I, Takabe K. M1 Macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci Rep. 2020;10:16554. doi: 10.1038/s41598-020-73624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Kalinski P, Endo I, Takabe K. Plasmacytoid Dendritic Cell (pDC) infiltration correlate with tumor infiltrating lymphocytes, cancer immunity, and better survival in Triple Negative Breast Cancer (TNBC) more strongly than Conventional Dendritic Cell (cDC) Cancers (Basel) 2020;12:3342. doi: 10.3390/cancers12113342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshi M, Newman S, Murthy V, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. ITPKC as a prognostic and predictive biomarker of neoadjuvant chemotherapy for triple negative breast cancer. Cancers (Basel) 2020;12:2758. doi: 10.3390/cancers12102758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshi M, Katsuta E, Yan L, Ebos JML, Rashid OM, Matsuyama R, Endo I, Takabe K. A novel 4-gene score to predict survival, distant metastasis and response to neoadjuvant therapy in breast cancer. Cancers (Basel) 2020;12:1148. doi: 10.3390/cancers12051148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oshi M, Asaoka M, Tokumaru Y, Yan L, Matsuyama R, Ishikawa T, Endo I, Takabe K. CD8 T cell score as a prognostic biomarker for triple negative breast cancer. Int J Mol Sci. 2020;21:6968. doi: 10.3390/ijms21186968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshi M, Asaoka M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Zsiros E, Ishikawa T, Endo I, Takabe K. Abundance of regulatory T cell (Treg) as a predictive biomarker for neoadjuvant chemotherapy in triple-negative breast cancer. Cancers (Basel) 2020;12:3038. doi: 10.3390/cancers12103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oshi M, Angarita FA, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. High expression of NRF2 is associated with increased tumor-infiltrating lymphocytes and cancer immunity in ER-positive/HER2-negative breast cancer. Cancers (Basel) 2020;12:3856. doi: 10.3390/cancers12123856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The immune landscape of cancer. Immunity. 2019;51:411–412. doi: 10.1016/j.immuni.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, Takabe K. Biologically aggressive phenotype and anti-cancer immunity counterbalance in breast cancer with high mutation rate. Sci Rep. 2020;10:1852. doi: 10.1038/s41598-020-58995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, Quinn MC, Robertson AJ, Fadlullah MZ, Bruxner TJ, Christ AN, Harliwong I, Idrisoglu S, Manning S, Nourse C, Nourbakhsh E, Wani S, Wilson PJ, Markham E, Cloonan N, Anderson MJ, Fink JL, Holmes O, Kazakoff SH, Leonard C, Newell F, Poudel B, Song S, Taylor D, Waddell N, Wood S, Xu Q, Wu J, Pinese M, Cowley MJ, Lee HC, Jones MD, Nagrial AM, Humphris J, Chantrill LA, Chin V, Steinmann AM, Mawson A, Humphrey ES, Colvin EK, Chou A, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Pettitt JA, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Jamieson NB, Graham JS, Niclou SP, Bjerkvig R, Grützmann R, Aust D, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Falconi M, Zamboni G, Tortora G, Tempero MA, Gill AJ, Eshleman JR, Pilarsky C, Scarpa A, Musgrove EA, Pearson JV, Biankin AV, Grimmond SM. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jané-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schulze A, Oshi M, Endo I, Takabe K. MYC targets scores are associated with cancer aggressiveness and poor survival in ER-positive primary and metastatic breast cancer. Int J Mol Sci. 2020;21:8127. doi: 10.3390/ijms21218127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 41.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, Seay T, Tjulandin SA, Ma WW, Saleh MN, Harris M, Reni M, Dowden S, Laheru D, Bahary N, Ramanathan RK, Tabernero J, Hidalgo M, Goldstein D, Van Cutsem E, Wei X, Iglesias J, Renschler MF. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Javed AA, Wright MJ, Siddique A, Blair AB, Ding D, Burkhart RA, Makary M, Cameron JL, Narang A, Herman J, Zheng L, Laheru D, Weiss MJ, Wolfgang C, He J. Outcome of patients with borderline resectable pancreatic cancer in the contemporary era of neoadjuvant chemotherapy. J Gastrointest Surg. 2019;23:112–121. doi: 10.1007/s11605-018-3966-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weniger M, Moir J, Damm M, Maggino L, Kordes M, Rosendahl J, Ceyhan GO, Schorn S. Respect - a multicenter retrospective study on preoperative chemotherapy in locally advanced and borderline resectable pancreatic cancer. Pancreatology. 2020;20:1131–1138. doi: 10.1016/j.pan.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 44.Klaiber U, Hackert T, Neoptolemos JP. Adjuvant treatment for pancreatic cancer. Transl Gastroenterol Hepatol. 2019;4:27. doi: 10.21037/tgh.2019.04.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chou A, Froio D, Nagrial AM, Parkin A, Murphy KJ, Chin VT, Wohl D, Steinmann A, Stark R, Drury A, Walters SN, Vennin C, Burgess A, Pinese M, Chantrill LA, Cowley MJ, Molloy TJ, Waddell N, Johns A, Grimmond SM, Chang DK, Biankin AV, Sansom OJ, Morton JP, Grey ST, Cox TR, Turchini J, Samra J, Clarke SJ, Timpson P, Gill AJ, Pajic M. Tailored first-line and second-line CDK4-targeting treatment combinations in mouse models of pancreatic cancer. Gut. 2018;67:2142–2155. doi: 10.1136/gutjnl-2017-315144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun T, Liu H, Ming L. Multiple roles of autophagy in the sorafenib resistance of hepatocellular carcinoma. Cell Physiol Biochem. 2017;44:716–727. doi: 10.1159/000485285. [DOI] [PubMed] [Google Scholar]
- 47.Keating GM, Santoro A. Sorafenib: a review of its use in advanced hepatocellular carcinoma. Drugs. 2009;69:223–240. doi: 10.2165/00003495-200969020-00006. [DOI] [PubMed] [Google Scholar]
- 48.Wei G, Wang M, Carr BI. Sorafenib combined vitamin K induces apoptosis in human pancreatic cancer cell lines through RAF/MEK/ERK and c-Jun NH2-terminal kinase pathways. J Cell Physiol. 2010;224:112–119. doi: 10.1002/jcp.22099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulivi P, Arienti C, Amadori D, Fabbri F, Carloni S, Tesei A, Vannini I, Silvestrini R, Zoli W. Role of RAF/MEK/ERK pathway, p-STAT-3 and Mcl-1 in sorafenib activity in human pancreatic cancer cell lines. J Cell Physiol. 2009;220:214–221. doi: 10.1002/jcp.21753. [DOI] [PubMed] [Google Scholar]
- 50.Booth L, Roberts JL, Poklepovic A, Dent P. Prior exposure of pancreatic tumors to [sorafenib + vorinostat] enhances the efficacy of an anti-PD-1 antibody. Cancer Biol Ther. 2019;20:109–121. doi: 10.1080/15384047.2018.1507258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendel DB, Laird AD, Xin X, Louie SG, Christensen JG, Li G, Schreck RE, Abrams TJ, Ngai TJ, Lee LB, Murray LJ, Carver J, Chan E, Moss KG, Haznedar JO, Sukbuntherng J, Blake RA, Sun L, Tang C, Miller T, Shirazian S, McMahon G, Cherrington JM. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin Cancer Res. 2003;9:327–337. [PubMed] [Google Scholar]
- 52.O’Reilly EM, Niedzwiecki D, Hall M, Hollis D, Bekaii-Saab T, Pluard T, Douglas K, Abou-Alfa GK, Kindler HL, Schilsky RL, Goldberg RM. A cancer and leukemia group B phase II study of sunitinib malate in patients with previously treated metastatic pancreatic adenocarcinoma (CALGB 80603) Oncologist. 2010;15:1310–1319. doi: 10.1634/theoncologist.2010-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bergmann L, Maute L, Heil G, Rüssel J, Weidmann E, Köberle D, Fuxius S, Weigang-Köhler K, Aulitzky WE, Wörmann B, Hartung G, Moritz B, Edler L, Burkholder I, Scheulen ME, Richly H. A prospective randomised phase-II trial with gemcitabine versus gemcitabine plus sunitinib in advanced pancreatic cancer: a study of the CESAR Central European Society for Anticancer Drug Research-EWIV. Eur J Cancer. 2015;51:27–36. doi: 10.1016/j.ejca.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 54.Xie L, Xu J, Sun X, Tang X, Yan T, Yang R, Guo W. Anorexia, hypertension, pneumothorax, and hypothyroidism: potential signs of improved clinical outcome following apatinib in advanced osteosarcoma. Cancer Manag Res. 2020;12:91–102. doi: 10.2147/CMAR.S232823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott AJ, Messersmith WA, Jimeno A. Apatinib: a promising oral antiangiogenic agent in the treatment of multiple solid tumors. Drugs Today (Barc) 2015;51:223–229. doi: 10.1358/dot.2015.51.4.2320599. [DOI] [PubMed] [Google Scholar]
- 56.Li J, Qin S, Xu J, Xiong J, Wu C, Bai Y, Liu W, Tong J, Liu Y, Xu R, Wang Z, Wang Q, Ouyang X, Yang Y, Ba Y, Liang J, Lin X, Luo D, Zheng R, Wang X, Sun G, Wang L, Zheng L, Guo H, Wu J, Xu N, Yang J, Zhang H, Cheng Y, Wang N, Chen L, Fan Z, Sun P, Yu H. Randomized, double-blind, placebo-controlled phase III trial of apatinib in patients with chemotherapy-refractory advanced or metastatic adenocarcinoma of the stomach or gastroesophageal junction. J. Clin. Oncol. 2016;34:1448–1454. doi: 10.1200/JCO.2015.63.5995. [DOI] [PubMed] [Google Scholar]
- 57.Li CM, Liu ZC, Bao YT, Sun XD, Wang LL. Extraordinary response of metastatic pancreatic cancer to apatinib after failed chemotherapy: a case report and literature review. World J Gastroenterol. 2017;23:7478–7488. doi: 10.3748/wjg.v23.i41.7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.