Abstract
Prostate cancer (PCa) is the most commonly diagnosed male malignancy worldwide. Early diagnosis and metastases detection are crucial features to diminish patient mortality. High fat diet (HFD) and metabolic syndrome increase PCa risk and aggressiveness. Our goal was to identify miRNAs-based biomarkers for PCa diagnosis and prognosis associated with HFD. Mice chronically fed with a HFD or control diet (CD) were subcutaneously inoculated with androgen insensitive PC3 cells. Xenografts from HFD-fed mice showed increased expression of 7 miRNAs that we named “candidates” compared to CD-fed mice. These miRNAs modulate specific metabolic and cancer related pathways. Using bioinformatic tools and human datasets we found that hsa-miR-19b-3p and miR-101-3p showed more than 1,100 validated targets involved in proteoglycans in cancer and fatty acid biosynthesis. These miRNAs were significantly increased in the bloodstream of PCa patients compared to non-PCa volunteers, and in prostate tumors compared to normal adjacent tissues (NAT). Interestingly, both miRNAs were also increased in tumors of metastatic patients compared to tumors of non-metastatic patients. Further receiver-operating characteristic (ROC) analysis determined that hsa-miR-19b-3p and hsa-miR-101-3p in serum showed poor predictive power to discriminate PCa from non-PCa patients. Hsa-miR-19b-3p showed the best score to discriminate between tumor and NAT, while hsa-miR-101-3p was useful to differentiate between metastatic and non-metastatic PCa patients. Hsa-miR-101-3p was increased in exosomes isolated from blood of PCa patients. Although more detailed functional exploration and validation of the molecular mechanisms are required, we identified hsa-miR-19b-3p and hsa-miR-101-3p with high potential for PCa diagnosis and prognosis.
Keywords: High fat diet, prostate cancer, miR-19b-3p, miR-101-3p, biomarker
Introduction
Prostate cancer (PCa) is the second most diagnosed cancer and the fifth leading cause of cancer death in men worldwide [1]. It is a heterogeneous disease that can appear as an indolent tumor without the need for treatment, as well as an aggressive disease that spreads at an early stage and can lead to death. Currently, PCa is stratified into low or high risk based on Gleason score, the level of prostate specific antigen (PSA) and other clinical-pathological parameters. However, this classification is insufficient to predict the progression of the disease, in particular the presence of metastases that leads to an incurable disease and confers a high level of morbidity [2]. Therefore, it is important to find novel biomarkers with high potential for PCa diagnosis and prognosis.
Metabolic syndrome (MeS) is a pathophysiological disorder whose diagnosis requires the detection of at least three of the following factors: low high density lipoprotein (HDL) cholesterol levels and increased visceral adiposity, triglycerides, blood pressure and fasting glucose levels [3]. Interestingly, several epidemiological studies demonstrated that MeS increases PCa aggressiveness and progression, the incidence of high-grade tumors, and biochemical recurrence [4-7]. Nonetheless, the molecular mechanism responsible for the effect of MeS on the progression and aggressiveness of PCa tumors is yet to be fully determined. Previously, we generated a PCa and MeS-like disease mice model by chronically feeding animals with a high fat diet (HFD). Thus, we identified several molecular links associating PCa and MeS, including the transcription co-regulator C-terminal binding protein 1 (CTBP1) [8-11].
MicroRNAs (miRNAs) are non-coding small RNAs involved in the post-transcriptional regulation of gene expression [12,13]. Functional studies have revealed that miRNAs play a critical role in carcinogenesis, tumor progression, and metastasis, including PCa [13-15]. Also, miRNAs can be released from tumors to the bloodstream through several mechanisms, including exosomes, which act as a communication vehicle between healthy and tumorigenic cells. These vesicles are protected from RNAses, remaining stable for a long time [16]. Therefore, miRNAs emerge as good candidate biomarkers for PCa diagnosis and/or prognosis.
Recently, using an androgen-sensitive PCa and MeS-like mice model, we found a signature of 5 miRNAs with a key role in the interaction between white adipose tissue (WAT) and PCa in HFD-fed mice [17].
The aim of this work was to identify a signature of oncogenic miRNAs regulated by MeS in prostate tumors and to explore their use as possible biomarkers for PCa diagnosis and/or prognosis.
Materials and methods
Cell culture
PC3 cell line (ATCC: CRL-1435) was grown in RPMI 1640 (Invitrogen) supplemented with 10% of fetal bovine serum and antibiotics in a 5% CO2 humidified atmosphere at 37°C. This human cell line has been authenticated using STR profiling within the last three years.
PCa xenografts and MeS murine model
NOD Scid Gamma (NSG) mice (N = 12) were used to develop MeS and PC3 xenografts as previously described [8]. Four-week-old male mice were maintained under pathogen free conditions keeping the IBYME’s animal care guidelines.
RNA isolation and stem-loop RT-qPCR
Total RNA was isolated using TriReagent (Molecular Research Center) from plasma and xenografts. Before RNA isolation, cel-miR-39 synthetic miRNA (20 fmol) was spiked in the plasma. miRNAs were retrotranscribed using the stem-loop method as previously described [8]. Briefly, 100 ng (xenografts) or 4 μl (plasma) of total RNA and 0.07 μM of stem-loop primer were preheated (70°C, 5 min). Retrotranscription (RT) was performed using M-MLV reverse transcriptase (Promega) and incubated in MyGenie96 Thermal Block (Bioneer) (30 min 16°C, 60 min 42°C, 2 min 70°C). qPCRs were run in 10 μl with 0.1 μM of each primer and 5 μl of PowerUp™ SYBR™ Green Master Mix (Thermo Fisher), in StepOne Plus Real Time PCR (Applied Biosystems) (50°C 2 min, 95°C 10 min, 40 cycles: 95°C 15 s, annealing temperature 15 s, 60°C 1 min and 95°C 15 s) as previously described [17]. All reactions were run in duplicate. The expression levels of miRNAs were calculated using ΔΔCT method normalizing to hsa-miR-103a-3p levels and control. The expression levels of plasma miRNAs were normalized to cel-miR-39. Primer sequences for miRNA RT-qPCR are listed in Table 1.
Table 1.
Primer sequences used for stem-loop RT-qPCR
| Primer | Sequence (5’-3’) | T ann (°C) |
|---|---|---|
| RT-Stem-loop-Rv | TGGTGCAGGGTCCGAGGTATT | - |
| RT-hsa-miR-19b-3p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACTCAGTT | - |
| RT-hsa-miR-19b-3p Fw | GGCGGTGTGCAAATCCATGC | 65 |
| RT-hsa-miR-320a-5p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACCGCCGG | - |
| RT-hsa-miR-320a-5p Fw | GGCGGGCCAACACAACCCGGA | 65 |
| RT-hsa-miR-101-3p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACTTCAGT | - |
| RT-hsa-miR-101-3p Fw | CGCGCGTACAGTACTGTGATA | 62 |
| RT-hsa-miR-3613-3p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACCGCGGT | - |
| RT-hsa-miR-3613-3p Fw | CCGGCCACAAAAAAAAAAGCCCA | 65 |
| RT-hsa-miR-1207-5p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACCCCCTC | - |
| RT-hsa-miR-1207-5p Fw | GGCGGTGGCAGGGAGGCTGG | 65 |
| RT-hsa-miR-5095-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACCGCGGT | - |
| RT-hsa-miR-5095 Fw | GGCGCTTACAGGCGTGAACC | 65 |
| RT-hsa-miR-2277-5p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACGACTGG | - |
| RT-hsa-miR-2277-5p Fw | GGCGGAGCGCGGGCTGAGCGCTG | 65 |
| RT-hsa-miR-4497-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACGCCCAG | - |
| RT-hsa-miR-4497 Fw | CGCGCTCCGGGACGG | 65 |
| RT-hsa-miR-320e-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACCCTTCT | - |
| RT-hsa-miR-320e Fw | GCGCGGAAAGCTGGGTTG | 65 |
| RT-hsa-miR-4532-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACCGCCGG | - |
| RT-hsa-miR-4532 Fw | GAGACCCCGGGGAGC | 65 |
| RT-hsa-miR-4668-5p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACGACAAA | - |
| RT-hsa-miR-4668-5p Fw | GGCGGAGGGAAAAAAAAAAGGA | 65 |
| RT-hsa-miR-30b-5p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACGCTGAG | - |
| RT-hsa-miR-30b-5p Fw | CCGCGCTGTAAACATCCTACAC | 70 |
| RT-hsa-miR-221-3p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACGAAACC | - |
| RT-hsa-miR-221-3p Fw | GGCGGAGCTACATTGTCTGCTG | 65 |
| RT-hsa-miR-195-5p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACGCCAAT | - |
| RT-hsa-miR-195-5p Fw | GGGGGGTAGCAGCACAGAAAT | 65 |
| RT-hsa-miR-21-5p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACTCAACA | - |
| RT-hsa-miR-21-5p Fw | CGGGGGGTAGCTTATCAGACTG | 65 |
| RT-hsa-miR-16-5p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACCGCCAA | - |
| RT-hsa-miR-16-5p Fw | GGCCCGTAGCAGCACGTAAATA | 65 |
| RT-hsa-miR-103a-3p-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACTCATAG | - |
| RT-hsa-miR-103a-3p Fw | GGGGAGCAGCATTGTACAGGG | 67 |
| RT-miR-Cel39-STEM | GTCTCCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGGAGACCAAGCT | - |
| RT-miR-Cel39 Fw | CGGGGTCACCGGGTGTAAATC | 65 |
Functional enrichment analyses
To investigate the functional mechanisms of selected miRNAs, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses and Gene Ontology (GO) annotation using DIANA-miRPath v3 tool (http://snf-515788.vm.okeanos.grnet.gr/), from a list of experimentally validated target genes derived from DIANA-TarBase v7. The top 25 of the statistically significant terms (p-value < 0.01) were selected. The relevance of miRNAs in each term was studied through the heat maps returned by DIANA-miRPath v3.
To explore the functional role of selected miRNA target genes and identify the regulated pathways, a KEGG pathway enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics tool (https://david.ncifcrf.gov/tools.jsp). Only significantly enriched terms (p-value < 0.05) were analyzed.
Circulating miRNAs profile analysis
Circulating miRNA expression profile was analyzed in the serum of a cohort of 809 PCa and 282 non-cancer patients (negative PCa patients + non-cancer controls) using microarrays data available from Urabe and Matsuzaki group [18]. Normalized signal intensity data (GSE112264) was downloaded from the Gene Expression Omnibus (GEO) public functional genomics data repository (https://www.ncbi.nlm.nih.gov/geo/) and analyzed using the ggpubr R package. Shapiro-Wilk and Levene tests were used to assess normality and homogeneity of variances. Student’s t test was applied for data that fulfill the requirements. Otherwise, Mann-Whitney test was performed using R.
GSE112264 was generated by 3D-Gene® Human miRNA Oligo Chip (Toray Industries, Inc.).
TCGA dataset analysis
Mature miRNA and gene expression of prostate tumors of patients was obtained from the TCGA Prostate Cancer (PRAD) cohort available in the UCSC Xena bioinformatics tool [19] (https://xenabrowser.net/). 52 PCa samples paired with 52 normal adjacent tissues (NAT) were included in the present study excluding patients without this information. The miRNA-Seq (IlluminaHiSeq_miRNASeq) and RNAseq (IlluminaHiSeq) data was downloaded as log2 (RPM+1) values. Data normalization and homogeneity of variances was assessed using Shapiro-Wilk test and boxplot respectively. Paired Sample t-Test was applied for data that fulfill the requirements. Otherwise, Sign median test was performed using the signmedian.test R package.
miRNA expression profile analysis in tumors of metastatic PCa patients
miRNA expression profile in metastatic PCa patients was analyzed in a cohort of 19 patients with metastases diagnosis after radical prostatectomy and 19 patients without evidence of disease recurrence, using next generation whole miRNome sequencing results from Nam’s work available from GEO dataset (GSE117674) [20]. GSE117674 was generated by Ion Torrent S5 XL (Homo sapiens) high-throughput sequencing. Student’s t test or Mann-Whitney test were applied as appropriate.
Exosomes miRNA database analysis
miRNA expression profiles in exosomes was obtained from The Extracellular Vesicles miRNA (EVmiRNA) database [21] (http://bioinfo.life.hust.edu.cn/EVmiRNA). The expression levels of hsa-miR-101-3p in exosomes from bloodstream of PCa patients (n = 23) and healthy donors (HD) (n = 58) were downloaded and represented in a heat map using heatmap.plus, gplots and RColorBrewer R packages.
Receiver-operating characteristic (ROC) analysis
To evaluate the power of hsa-miR-19b-3p and miR-101-3p to distinguish between PCa and non-PCa, and between metastatic and non-metastatic PCa patients, we performed a receiver-operating characteristic (ROC) analysis in tumor and serum dataset described above. The ROC curve and the area under the curve (AUC) were plotted and calculated for both miRNAs using the GraphPad Prism 8 software.
Correlation matrix
Expression levels of hsa-miR-19b-3p, hsa-miR-101-3p and their respective target genes in prostate tumors from patients were obtained from TCGA PRAD data available in UCSC Xena. For miRNA mature strand and gene expression, the miRNA-Seq (IlluminaHiSeq_miRNASeq) and RNAseq (IlluminaHiSeq) data was downloaded as log2 (RPM+1) values. Only PCa samples (close to 500 patients) were included in the present analysis. Using the Hmisc R package, we generated a correlation matrix for each miRNA, applying the Spearman correlation coefficient. For hsa-miR-19b-3p, target genes that showed a negative correlation with the miRNA and correlation coefficient rho < -0.4 plus p-value < 0.05 were selected for further analysis. In addition, target genes that negatively correlated with hsa-miR-101-3p with a correlation coefficient rho < -0.2 and a p-value < 0.05 were selected. These target genes were entered into DAVID database to explore their functional role and identify the regulated pathways (See Functional enrichment analysis).
Clustering and STRING database
To study the relationship between selected target genes of hsa-miR-19b-3p and miR-101-3p, a hierarchical cluster analysis with “average” as an agglomeration method was performed using the R package. Additionally, a gene interaction analysis was performed using the STRING database (https://string-db.org/).
Statistical analysis
For the in vivo (N = 6 per group) experiments results are given as mean and standard deviation (SD). Student’s t-Test was performed. Shapiro-Wilk and Levene tests were used to assess normality and homogeneity of variances. *P < 0.05; **P < 0.01; ***P < 0.001.
Results
HFD induces miRNA expression in PC3 xenograft
Previously, we reported different mice models of PCa and MeS [8,9,17]. Here, NSG mice chronically fed with a HFD or control diet (CD) were subcutaneously inoculated with androgen insensitive PC3 cells. Seven weeks after cell inoculation, mice were sacrificed, plasma was collected and tumor excised for RNA isolation. As we previously described [8], no differences were observed regarding tumor growth and body weight, between treatments (Data not shown).
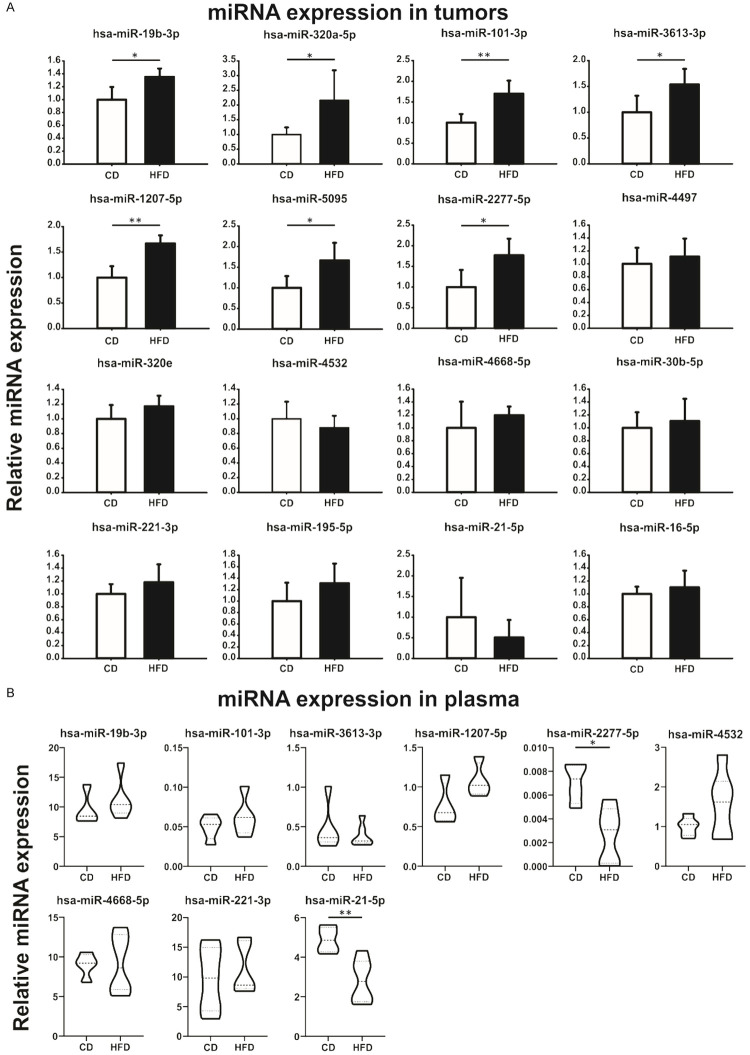
A miRNA set was selected from literature and our previous work [8,17] and assessed by stem-loop RT-qPCR. As shown in Figure 1A, PC3 xenograft from HFD-fed mice showed significantly increased the expression of 7 miRNAs that we named “candidates”: hsa-miR-19b-3p, miR-320a-5p, miR-101-3p, miR-3613-3p, miR-1207-5p, miR-5095 and miR-2277-5p, compared to CD-fed mice. No changes were detected in hsa-miR-4497, miR-320e, miR-4532, miR-4668-5p, miR-30b-5p, miR-221-3p, miR-195-5p, miR-21-5p and miR-16-5p (Figure 1A).
Figure 1.
HFD modulates the expression of miRNAs in androgen-insensitive prostate tumors and serum from mice. A. Stem-loop RT-qPCR from PC3 xenografts obtained from CD- or HFD-fed NSG mice using specific primers for the indicated miRNAs is shown. Data were normalized to hsa-miR-103a-3p and control. Statistical analysis was performed using t-test. B. Mice plasma samples were analyzed by stem-loop RT-qPCR for the indicated miRNAs and normalized to spike-in cel-miR-39 synthetic miRNA. 2ΔCt is graphed (ΔCt value = Ctcel-39-Ctsample). Data were analyzed by t-test.
Additionally, we determined the expression profile of the candidate miRNAs in mice bloodstream. As shown in Figure 1B, we detected hsa-miR-19b-3p, miR-101-3p, miR-3613-3p, miR-1207-5p, miR-2277-5p, miR-4532, miR-4668-5p, miR-221-3p and miR-21-5p. Interestingly, hsa-miR-2277-5p and miR-21-5p were significantly down-regulated in plasma from HFD mice compared to control animals (Figure 1B). Hsa-miR-320a-5p, miR-5095, miR-4497 and miR-320e were undetectable in mice plasma.
Candidate miRNAs modulate specific metabolic and cancer related pathways
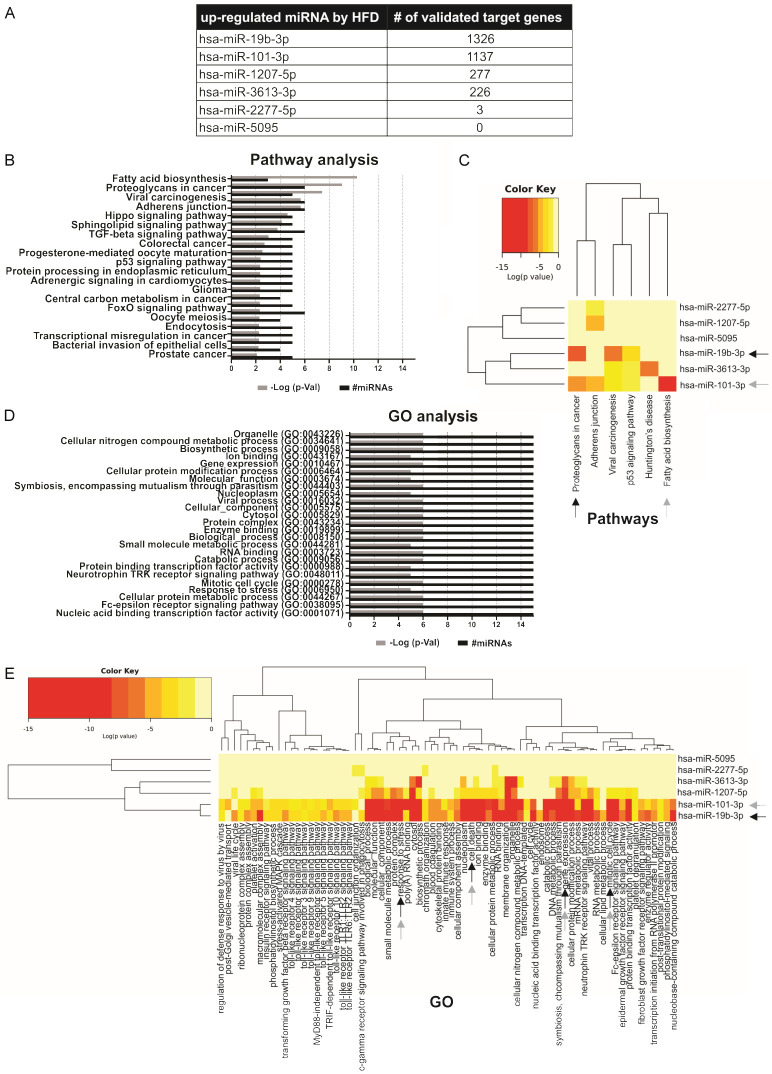
To investigate the functional mechanisms of the 7 candidate miRNAs, we used the DIANA-miRPath v3 tool. Since not all the target genes predicted by the bioinformatics algorithms finally turn out to be true targets, we used the DIANA-TarBase v7 of the miRPath bioinformatics tool, which has the information of the target genes experimentally validated for each miRNA. Five of the 7 candidate miRNAs showed validated target genes, while hsa-miR-5095 had no targets and miR-320a-5p was not found in DIANA-miRPath v3 tool and was not considered in further analyses (Figure 2A). hsa-miR-19b-3p and miR-101-3p miRNAs were the miRNAs that showed the highest number of target genes, with more than 1,100 validated targets (Figure 2A). Then, with all the validated target genes, we performed a KEGG pathway enrichment analysis using a p-value < 0.01. This analysis revealed that target genes of candidate miRNAs were associated with processes such as Fatty acid biosynthesis, Proteoglycans in cancer, Viral carcinogenesis, Adherens junction, p53 signaling pathway, Central carbon metabolism in cancer, Transcriptional misregulation in cancer and PCa, among others (Figure 2B). In particular, hsa-miR-19b-3p target genes were mostly involved in proteoglycans in cancer, while miR-101-3p target genes were involved in fatty acid biosynthesis. To a lesser extent, hsa-miR-3613-3p target genes were mostly associated with Huntington’s disease and miR-1207-5p and miR-2277-5p shared adherens junction KEGG pathway (Figure 2C).
Figure 2.
Functional enrichment analysis of candidate miRNAs. A. Number of validated target genes of candidate miRNAs. B. Barplot representation of the top significant KEGG pathways associated to the target genes of candidate miRNAs. C. Heat map of miRNAs and their related pathways. Arrows indicate the 2 miRNAs with the highest number of validated target genes and the pathways where they are most involved. D. Barplot representation of the top 25 significant GO terms associated with the target genes of candidate miRNAs. E. Heat map of miRNAs and their related GO terms. Arrows indicate the 2 miRNAs with the highest number of validated target genes and the GO terms where they are most involved.
Furthermore, using DIANA-miRPath v3, a GO analysis was performed at three levels: biological processes, cellular components and molecular function. The top 25 of the statistically significant terms (p-value < 0.01) were shown in Figure 2D. Target genes of miRNAs up-regulated by HFD were enriched in processes associated with Gene expression, Mitotic cell cycle and Response to stress (Figure 2D). Interestingly, hsa-miR-19b-3p and miR-101-3p were the most involved miRNAs in these terms in addition to the cell death term (Figure 2E).
In summary, up-regulated miRNAs by HFD modulated specific metabolic and cancer related pathways.
Hsa-miR-19b-3p, miR-101-3p, miR-1207-5p and miR-5095 are increased in the bloodstream of PCa patients
To determine the use of the candidate miRNAs as potential biomarkers for PCa diagnosis or prognostic, we explored expression data available in GEO dataset. Urabe and Matsuzaki group, using microarray technologies, performed a serum miRNA profile of 1591 male samples, including 809 PCa, 241 negative PCa patients, 41 non-cancer controls and 500 other solid cancers (GSE112264) [18]. Hence, we used these microarrays results to investigate the levels of the 7 candidate miRNAs in serum samples of a cohort of 809 PCa and 282 non-cancer patients (negative PCa patients + non-cancer controls). As shown in Figure 3A, hsa-miR-19b-3p, miR-101-3p, miR-1207-5p and miR-5095 were increased in the bloodstream of PCa patients compared to non-PCa patients. No differences were found in circulating levels of hsa-miR-3613-3p and miR-2277-5p, while hsa-miR-320a-5p was not detected in this cohort of patients (Figure 3A).
Figure 3.
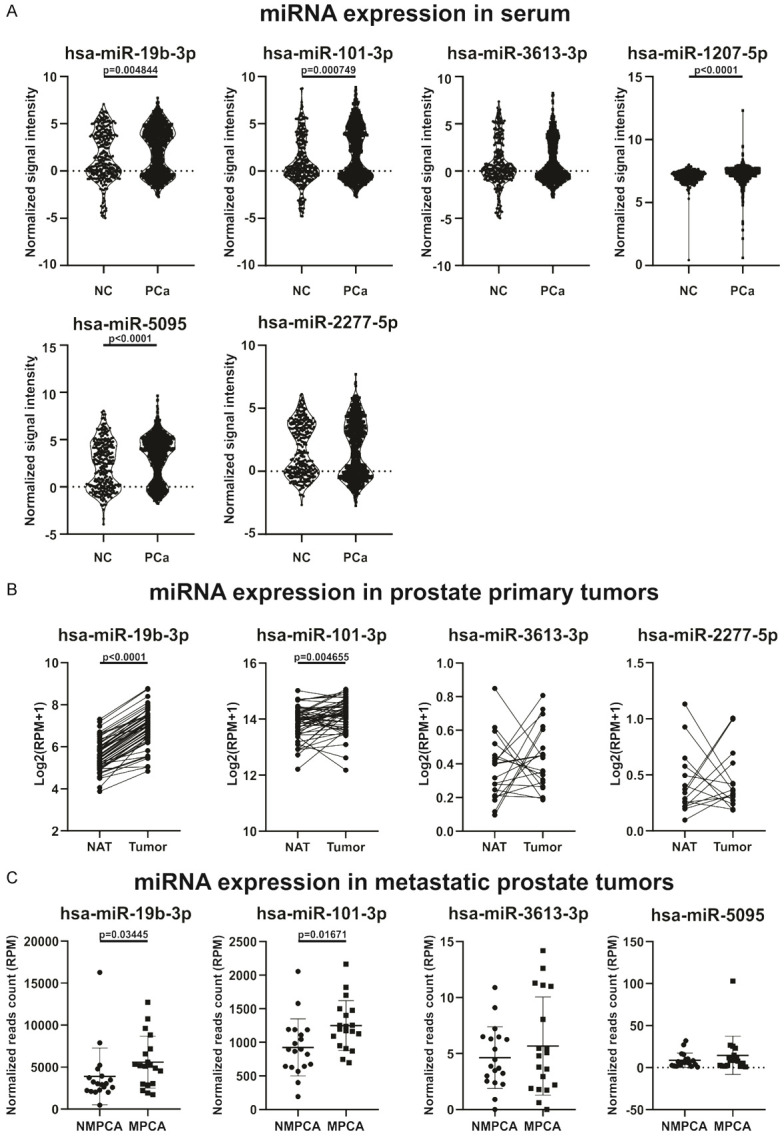
Candidate miRNAs expression in tumor tissue and circulation of PCa patients. A. Circulating miRNA expression levels of hsa-miR-19b-3p, miR-101-3p, miR-3613-3p, miR-1207-5p, miR-5095 and miR-2277-5p in plasma of PCa and non-PCa (NC) patient samples. Normalized signal intensity values are plotted. B. Expression levels of hsa-miR-19b-3p, miR-101-3p, miR-3613-3p and miR-2277-5p in prostate primary solid tumor and NAT. Log2 (RPM+1) are graphed. C. Expression levels of hsa-miR-19b-3p, miR-101-3p, miR-3613-3p and miR-5095 in nonmetastatic PCa (NMPCA) and metastatic PCa (MPCA). Normalized reads count are plotted.
Hsa-miR-19b-3p and miR-101-3p are up-regulated in prostate tumors from patients and increased in metastatic PCa tumors
To analyze the expression profile of the candidate miRNAs in prostate tumors from patients, we employed the TCGA PRAD cohort available in the UCSC Xena bioinformatics tool [19]. This data set has the information of the mature miRNAs expression from tumor tissue and NAT, obtained by RNAseq. We found that hsa-miR-19b-3p and miR-101-3p were significantly increased in prostate tumor compared to NAT (Figure 3B). No significant differences were found regarding the expression of hsa-miR-3613-3p and miR-2277-5p, while no expression data were found for hsa-miR-320a-5p, miR-1207-5p and miR-5095 (Figure 3B).
We explored the next generation whole miRNome sequencing results from Nam’s work available from GEO dataset (GSE117674) [20]. Nam’s group analyzed the expression profile of miRNAs in prostate tumors of patients who had a radical prostatectomy. They identified those patients who had been diagnosed with metastases after surgery (n = 19) and compared their expression levels with those patients who did not show evidence of disease recurrence (n = 19). Within this data set, we analyzed the candidate miRNAs. As shown in Figure 3C, hsa-miR-19b-3p and miR-101-3p were increased in tumors of metastatic patients (after surgery) compared to tumors of non-metastatic patients. There were no differences in the expression of hsa-miR-3613-3p and miR-5095 (Figure 3C). Furthermore, hsa-miR-320a-5p, miR-1207-5p, miR-2277-5p were not detected in this cohort of patients.
Hsa-miR-19b-3p and miR-101-3p are good biomarkers in tissue but not in serum to distinguish between PCa from non-PCa patients
To assess whether hsa-miR-19b-3p and miR-101-3p are good diagnosis biomarkers to distinguish between PCa patients from non-PCa, we performed a ROC analysis. As shown in Figure 4A, poor predictive power to discriminate between PCa and non-PCa patients was obtained when we calculated the area under the curve (AUC) for hsa-miR-19b-3p and hsa-miR-101-3p in serum (0.5563 and 0.5886, respectively). Regarding tumor tissue, AUC for hsa-miR-19b-3p and hsa-miR-101-3p were 0.861 and 0.6307, respectively (Figure 4B), which shows that both miRNAs are useful to distinguish between tumor tissue and NAT. Additionally, we found that both, hsa-miR-19b-3p and hsa-miR-101-3p, were able to differentiate between metastatic and non-metastatic PCa disease due to AUC were 0.7008 and 0.7590, respectively (Figure 4C). In particular, hsa-miR-19b-3p showed the best score to discriminate between tumor tissue and NAT, while hsa-miR-101-3p was more useful to differentiate between metastatic and non-metastatic PCa patients.
Figure 4.
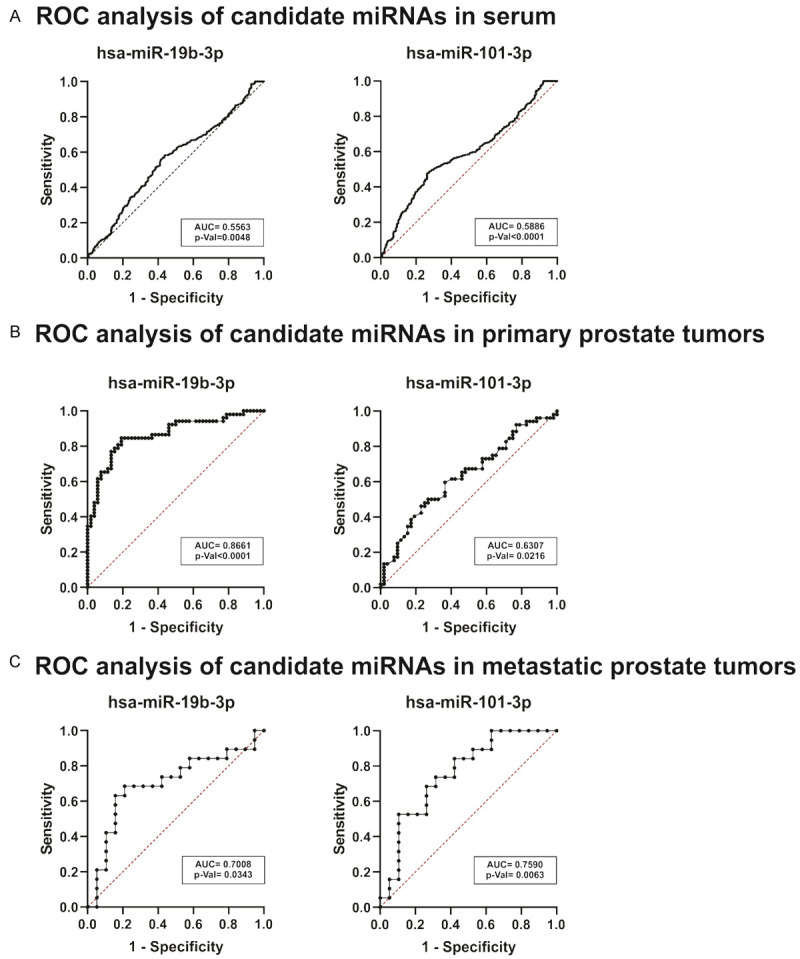
ROC analysis in tumor and serum dataset. (A-C) ROC curve and the AUC were plotted and calculated for hsa-miR-19b-3p and miR-101-3p in serum (A), primary prostate tumors (B) and metastatic prostate tumors (C).
Hsa-miR-101-3p is increased in exosomes isolated from blood of PCa patients
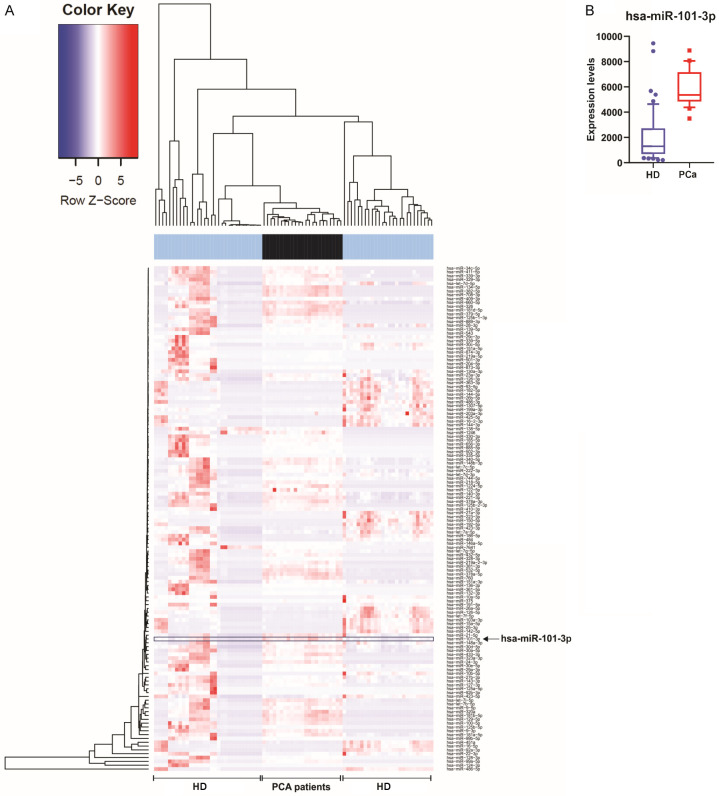
As miRNAs can be released into the bloodstream associated to proteins, HDL or wrapped into exosomes, we analyzed the presence of hsa-miR-19b-3p and miR-101-3p in exosomes from blood samples of PCa patients (n = 23) and HD (n = 58), using EVmiRNA database [21]. As shown in Figure 5, hsa-miR-101-3p was increased in exosomes of PCa patients compared to HD. Hsa-miR-19b-3p was not found in this dataset.
Figure 5.
MiRNAs expression in exosomes from blood of PCa patients. A. Heat map representation of miRNAs in exosomes from bloodstream of PCa patients and healthy donors. Arrows indicate the hsa-miR-101-3p. Hsa-miR-19b-3p was not found in this dataset. B. Box plot of hsa-miR-101-3p expression.
These results suggest that hsa-miR-101-3p is released into the bloodstream of PCa patients through exosomes, disseminating to other organs which reinforce the miRNA role in metastasis.
Hsa-miR-19b-3p is involved in proteoglycans in cancer and focal adhesion pathways
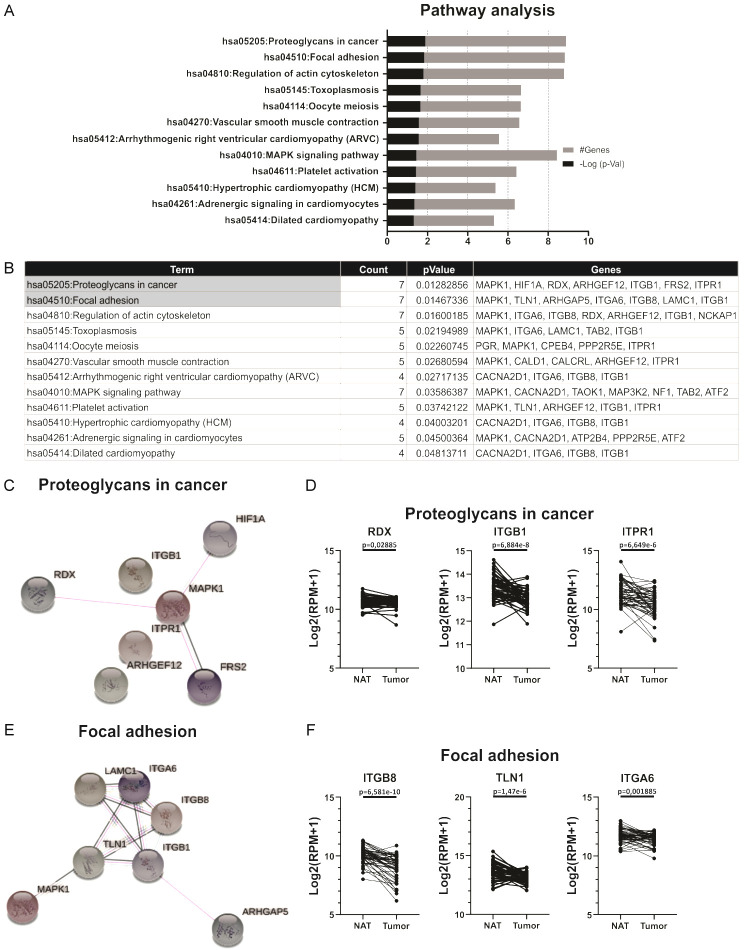
As we previously mentioned, we found that hsa-miR-19b-3p showed 1,326 validated target genes (Figure 2A). MiRNAs regulate gene expression at the post-transcriptional level mostly by decreasing the expression of their target gene. In order to found relevant target genes for hsa-miR-19b-3p, we selected targets that negatively correlated with the expression of hsa-miR-19b-3p. A correlation matrix with R packages was performed, using the expression data of hsa-miR-19b-3p and its target genes in the tumors of PCa patients available in TCGA PRAD data present in UCSC Xena. We selected target genes that showed a negative correlation with hsa-miR-19b-3p, and correlation coefficient rho < -0.4 plus p-value < 0.05 for subsequently analysis (Data not shown). To explore the functional role of these 160 selected target genes and to identify the regulated pathways, a KEGG pathway enrichment analysis was performed using the DAVID bioinformatics tool. The significantly enriched terms (p-value < 0.05) were plotted and listed in Figure 6A and 6B, respectively. The most significant enriched KEGG terms were Proteoglycans in cancer and Focal adhesion (Figure 6A, 6B). Seven of the selected target genes (MAPK1, HIF1A, RDX, ARHGEF12, ITGB1, FRS2, ITPR1) were involved in Proteoglycans in cancer, while another 7 (MAPK1, TLN1, ARHGAP5, ITGA6, ITGB8, LAMC1, ITGB1) were involved in Focal adhesion (Figure 6B). Both pathways share only two genes: MAPK1 and ITGB1.
Figure 6.
Functional enrichment analysis of selected target genes of hsa-miR-19b-3p. A. Barplot representation of the top significant KEGG pathways associated to the selected target genes of hsa-miR-19b-3p. B. Top significant KEGG pathways and the target genes involved in each of them. C. Interactional network of the 7 genes involved in “Proteoglycans in cancer”. D. Expression levels of RDX, ITGB1 and ITPR1 in prostate primary solid tumor and NAT. Log2 (RPM+1) values are graphed. E. Interactional network of the 7 genes involved in “Focal adhesion”. F. Expression levels of ITGB8, TLN1 and ITGA6 in prostate primary solid tumor and NAT. Log2 (RPM+1) values are graphed.
To analyze the relationship and interaction between the genes belonging to these two pathways selected in Figure 6B, a clustering analysis with R packages was performed. Genes that were enriched in the Proteoglycan in cancer pathway are all grouped in the same cluster except FRS2 (Data not shown). Additionally, genes enriched in the Focal adhesion pathway were found in the same cluster except ITGB8 (Data not shown). These results suggest that the genes belonging to each pathway are related to each other. Using the STRING database, an interaction analysis between the genes belonging to each pathway was performed. Only 4 of the genes involved in proteoglycans in cancer interact with each other (Figure 6C), while in focal adhesion, the interaction occurs between all the genes involved (Figure 6E).
Using gene expression data available in TCGA PRAD cohort present in Xena, we found that 3 of the genes involved in proteoglycans in cancer pathway (RDX, ITGB1, and ITPR1) were down-regulated in tumors compared to NAT (Figure 6D). Furthermore, 4 of the genes involved in focal adhesion (ITGB8, TLN1, ITGA6 and ITGB1) showed a decreased expression in tumors relative to NAT (Figure 6F). These results suggest that hsa-miR-19b-3p might function on proteoglycan in cancer and focal adhesion pathways in PCa by regulating the expression of these target genes.
Analysis of hsa-miR-101-3p validated target genes and pathways with TarBase v7 showed 1,137 validated target genes (Figure 2A). We next performed the correlation matrix between hsa-miR-101-3p expression data and its validated target genes available in TCGA PRAD cohort present in UCSC Xena. Target genes that negatively correlated with hsa-miR-101-3p with a correlation coefficient rho < -0.2 and a p-value < 0.05 were selected (Data not shown). Using DAVID bioinformatics tool, KEGG pathway enrichment analysis of six target genes selected was performed. The results showed only one significant term (p-value < 0.05): Proteasome with two target genes: PSMD1 and POMP (Data not shown). A clustering analysis revealed that both genes were related in a small cluster (Data not shown). Furthermore, through an analysis with the STRING database, we have found that both genes interact with each other (Data not shown).
Finally, using TCGA PRAD project present in Xena, no significant differences were found in PSMD1 and POMP expression levels between prostate tumors and NAT (Data not shown).
Discussion
In this work we provide evidence for hsa-miR-19b-3p and hsa-miR-101-3p as biomarkers for the diagnosis and prognosis of PCa associated with HFD. We found that HFD increases the expression of 7 miRNAs (hsa-miR-19b-3p, miR-320a-5p, miR-101-3p, miR-3613-3p miR-1207-5p, miR-5095 and miR-2277-5p) in PCa xenografts, which are involved in metabolic and cancer related pathways. Only 4 of these miRNAs (hsa-miR-19b-3p, miR-101-3p, miR-1207-5p and miR-5095) were increased in the bloodstream of PCa patients compared to non-PCa patients. Moreover, we found that hsa-miR-19b-3p and miR-101-3p were significantly increased in prostate tumors of metastatic patients compared to tumors of non-metastatic patients. In addition, ROC analysis showed that both miRNAs are promising biomarker candidates to distinguish between PCa and NAT, and metastatic and non-metastatic patients; however, the level of these miRNAs at serum level seems to be useless as PCa biomarkers.
Osip’yants et al. performed a classification of PCa patients using plasma samples from non-metastatic and metastatic PCa based on miRNAs [22]. They found that metastatic PCa patients showed higher plasma levels of hsa-miR-19b-3p paired with miR-297 compared to non-metastatic patients [22]. Another study showed that hsa-miR-19b-3p (also called hsa-miR-19b) was up-regulated in BPH-1, PC3 and 22Rv1 PCa cell lines, and down-regulated in DU-145 cells, compared to the normal prostate epithelial cell line PNT1B [23]. The same group demonstrated that hsa-miR-19b along with 3 other miRNAs promoted cell proliferation of PCa in vitro by targeting regulation of PTEN and its downstream signals, PI3K/Akt and cyclin D1 [23]. The hsa-miR-19b-3p belongs to the miR-17-92 cluster, which possesses oncogenic properties due to its participation in the regulation of cell survival, proliferation, differentiation and the cell cycle. Stimulation of the hyperexpression of this cluster in DU-145 cells leads to an increase in proliferative, migratory and invasive activities [24]. MiR-19 has been considered the key oncogenic component of the aforementioned cluster, because miR-19 (including hsa-miR-19a and hsa-miR-19b) partially suppressed PTEN expression, activating the AKT-mTOR signaling pathway and promoting cell survival [25]. Altogether, these results strongly support hsa-miR-19b-3p as a potential biomarker for PCa diagnosis and prognosis.
Hsa-miR-101-3p was proposed as a good biomarker for diagnosis, progression and therapy response in PCa. MiR-101-3p (also called hsa-miR-101) was found to be decreased in the DU-145 and PC3 cells compared to the non-tumorigenic prostate epithelial cell line PWR-1E, while it was increased in the LNCaP cell line [26]. Another report showed that hsa-miR-101 induced apoptosis in PCa cells through decreased RLIP76 expression followed by concomitant suppressions of the PI3K/AKT/Bcl-2 pathway [27].
Recent studies, in opposition to our results, have shown that hsa-miR-101-3p is negatively regulated in tumors and it is less abundant in the bloodstream of metastatic PCa compared to non-metastatic PCa patients [28,29]. These results may be due to the heterogeneity of PCa, and the different cohorts of patients analyzed. Additionally, we found that hsa-miR-101-3p was increased in exosomes from PCa patients compared to exosomes from HD. Hence, hsa-miR-101-3p might play a role in distant metastatic sites, reassessing it as an essential miRNA to investigate in the future in PCa metastasis samples from patients.
Then, we decided to investigate the possible effects of these 2 candidate miRNA biomarkers on prostate tumors and their aggressiveness. We established that 12 target genes that negatively correlate with hsa-miR-19b-3p in prostate tumor samples from patients were involved in Proteoglycan in cancer and Focal adhesion pathways. We found that RDX, ITGB1, ITPR1, ITGB8, TLN1 and ITGA6 were down-regulated in tumors compared to NAT. Immunhistochemical staining of RDX (Radixin) showed a higher intensity in prostatic intraepithelial neoplasia and normal prostates, with the lowest levels of staining in prostatic adenocarcinoma [30]. These results might support our data. However, in vitro studies demonstrated that RDX is required for PC3 cell migration, and that its depletion by RNA interference increased cell spreading area and cell-cell adhesion mediated by adherens junctions [31].
Knockdown of ITGB1 significantly inhibited cancer cell migration and invasion in PCa cells by regulating downstream signaling [32]. Moreover, ITGB1 was highly expressed in prostate tumors [32] and exosomes derived from urine of metastatic PCa patients [33].
On the other hand, little is known about the involvement of ITPR1 (also called IP3R1) in PCa. It was found that DU-145 cells contain about 10% ITPR1 and 90% ITPR3 [34]. ITPR1 depletion prevented apoptosis induction in colorectal cancer DLD1 cells, ovarian cancer A2780 cells, and clear cell renal cell carcinoma RCC4 cells, compared to apoptosis in cells treated with scrambled siRNA [35]. Moreover, ITPR1 is involved in SFN (sulforaphane)-induced apoptosis through the depletion of reticular calcium and modulation of transcription factors through nuclear calcium up-regulation [36]. Altogether, these results indicate that ITPR1 has a pro-apoptotic effect.
ITGB8 was highly expressed in prostatic intraepithelial neoplasia in a in silico analysis of a PCa progression microarray available in Oncomine. Furthermore, ITGB8 knockdown decreased PC3 and 22Rv1 cell migration and invasion [37]. However, similar to our findings, He et al. demonstrated that ITGB8 was down-regulated in PCa compared with matched para-cancerous tissue [38].
It was reported that TLN1 overexpression enhanced PCa cell adhesion, migration and invasion by activating survival signals and conferring resistance to anoikis, and shRNA-mediated TLN1 loss led to significant inhibition of PCa metastasis in vivo [39]. Also, TLN1 protein was up-regulated in PCa tissues compared with benign prostatic hyperplasia tissue, and it was an independent predictor for lymph node metastasis and biochemical recurrence of PCa [40]. Furthermore, the cytoplasmic expression of TLN1 was higher in metastatic tissue compared to primary prostate tumors [39]. Nevertheless, Betts et al. revealed that TLN1 mRNA expression decreased in primary prostatic stromal cells in response to the exposure of conditioned medium from androgen treated epithelial cells [41]. In this sense, the authors suggest that this reduction in the expression of TLN1 could be part of a mechanism that leads to the loss of cell adhesion in epithelial cells mediated by the action of androgens. Their studies showed that TLN1 is expressed in the stroma of prostate tissue sections and at higher levels in epithelial cells. Therefore, they suggest that the expression of TLN1 in epithelial cells may also play an important role in the regulation of cell adhesion in both, benign and malignant prostates [41]. Thus, TLN1 expression in patient tumors may be affected by androgen levels, being higher in androgen-insensitive PCa.
Finally, ITGA6 showed a marked decrease in its expression in malignant WPE-NB26 cells grown in Matrigel compared to immortalized non-malignant RWPE-1 cells [42]. Furthermore, PC3 cells exhibited a down regulation of ITGA6 compared to the RWPE cell line [43]. Moreover, low Gleason score correlated with increased ITGA6 expression, while high Gleason score with low and negative ITGA6 expression [44].
Although we found that 6 (RDX, ITGB1, ITPR1, ITGB8, TLN1 and ITGA6) of the 12 targeted genes by hsa-miR-19b-3p were downregulated in tumors from PCa patients compared to NAT, the expression of some of them (ITGB1, TLN1) was positively correlated with tumor metastasis. This contradicts the results indicating that hsa-miR-19b-3p was found to be up-regulated in metastatic PCa. This conflict may be due to several reasons; including that the analysis of the target genes has been performed in-silico, that target genes of miRNAs can differ according to the tissue, and also not all the target genes have been studied and validated experimentally in most tissues. Thus, to carry out a more exhaustive analysis, we selected those target genes that have been experimentally validated in at least one tissue. None of the hsa-miR-19b-3p target genes has been experimentally validated in prostate tissue or PCa cell lines. Thus, those genes which are negatively correlated with hsa-miR-19b-3p and whose expression in PCa tumors is contradictory to the literature are not direct targets of the aforementioned miRNA in prostate tissue and are being targeted by one or more other miRNAs. Further in vitro experiment, such as a luciferase assay in PCa cells, is needed to confirm whether these genes are indeed targets for hsa-miR-19b-3p in prostate tissue.
No significant changes in the expression of hsa-miR-101-3p target genes were found in tumor samples compared with NAT. Therefore, we believe that hsa-miR-101-3p might play a relevant role in the metastatic niches but not in the primary tumor.
Our results propose hsa-miR-19b-3p and hsa-miR-101-3p as oncomiRs at tumor and metastatic site level, respectively. Furthermore, these miRNAs were increased by HFD in our murine model. Labbé et al., 2019 showed that HFD exacerbates the MYC transcriptional program by MYC overexpression in murine prostate [45]. Thus, we performed a correlation matrix with R packages between the expression data of hsa-miR-19b-3p and miR-101-3p and selected genes from tumors of PCa patients (TCGA PRAD data present at UCSC Xena). We found that there is no direct correlation between candidate miRNAs and the MYC transcriptional program, suggesting that HFD exacerbates the MYC transcriptional program by MYC overexpression in normal prostate and induces the expression of hsa-miR-19b-3p and miR-101-3p in PCa, by independent mechanisms. Therefore, patients under HFD conditions could have a more aggressive PCa due to increased expression of hsa-miR-19b-3p, hsa-miR-101-3p and the MYC transcriptional program.
Taken together, we identified 2 miRNAs (miR-19b-3p and miR-101-3p) with high potential for diagnosis and prognosis to distinguish between PCa and NAT, and between metastatic from non-metastatic patients (see hypothetical model in Figure 7). However, these miRNAs might be useful at tumor level but not in the serum of patients. More detailed functional exploration and validation of the molecular mechanisms associated with these miRNAs are indispensable, but our findings might furnish some meaningful insights into diagnosis and prognosis of PCa patients.
Figure 7.
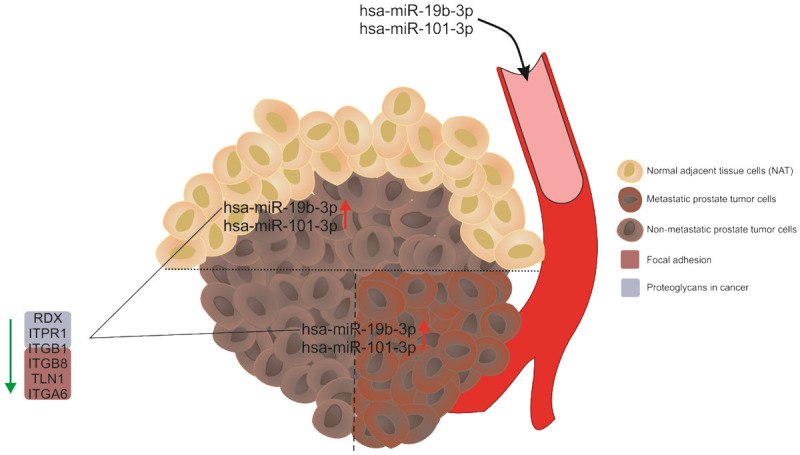
Hypothetical model. Hsa-miR-19b-3p and hsa-miR-101-3p are induced in prostate tumors from patients compared to NAT, and in tumors of metastatic patients versus tumors of non-metastatic patients. Both miRNAs are increased in the bloodstream of PCa patients compared to those without cancer. Hsa-miR-19b-3p negatively regulates the expression of genes involved in the proteoglycan in cancer and focal adhesion pathways, which could contribute to a more aggressive tumor phenotype.
Acknowledgements
This research was supported by the Argentinean Agency of Science and Technology (ANPCyT PICT 2014-324; PICT 2015-1345, PICT 2018-1304, PICT START UP-2019-21). The authors thank Fundación Williams (Argentina) for their support, and NIH/NCI Cancer Center Support Grant P30CA013696 (K.G.), R01 1R01CA253368 (K.G.). This work was part of Ph.D. thesis of Rocío Belén Duca supported by the CONICET fellowship from Argentina. The authors declare no conflict of interest.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Fu Q, Liu X, Liu Y, Yang J, Lv G, Dong S. MicroRNA-335 and -543 suppress bone metastasis in prostate cancer via targeting endothelial nitric oxide synthase. Int J Mol Med. 2015;36:1417–1425. doi: 10.3892/ijmm.2015.2355. [DOI] [PubMed] [Google Scholar]
- 3.National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) Third Report of the National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel III) final report. Communication. 2002;106:3143–421. [PubMed] [Google Scholar]
- 4.Albanes D, Weinstein SJ, Wright ME, Männistö S, Limburg PJ, Snyder K, Virtamo J. Serum insulin, glucose, indices of insulin resistance, and risk of prostate cancer. J Natl Cancer Inst. 2009;101:1272–1279. doi: 10.1093/jnci/djp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gacci M, De Nunzio C, Sebastianelli A, Salvi M, Vignozzi L, Tubaro A, Morgia G, Serni S. Meta-analysis of metabolic syndrome and prostate cancer. Prostate Cancer Prostatic Dis. 2017;20:146–155. doi: 10.1038/pcan.2017.1. [DOI] [PubMed] [Google Scholar]
- 6.Grundmark B, Garmo H, Loda M, Busch C, Holmberg L, Zethelius B. The metabolic syndrome and the risk of prostate cancer under competing risks of death from other causes. Cancer Epidemiol Biomarkers Prev. 2010;19:2088–2096. doi: 10.1158/1055-9965.EPI-10-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammarsten J, Damber JE, Peeker R, Mellström D, Högstedt B. A higher prediagnostic insulin level is a prospective risk factor for incident prostate cancer. Cancer Epidemiol. 2010;34:574–579. doi: 10.1016/j.canep.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Dalton GN, Massillo C, Scalise GD, Duca R, Porretti J, Farré PL, Gardner K, Paez A, Gueron G, De Luca P, De Siervi A. CTBP1 depletion on prostate tumors deregulates miRNA/mRNA expression and impairs cancer progression in metabolic syndrome mice. Cell Death Dis. 2019;10:1–13. doi: 10.1038/s41419-019-1535-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massillo C, Dalton GN, Porretti J, Scalise GD, Farré PL, Piccioni F, Secchiari F, Pascuali N, Clyne C, Gardner K, De Luca P, De Siervi A. CTBP1/CYP19A1/estradiol axis together with adipose tissue impacts over prostate cancer growth associated to metabolic syndrome. Int J Cancer. 2019;144:1115–1127. doi: 10.1002/ijc.31773. [DOI] [PubMed] [Google Scholar]
- 10.Porretti J, Dalton GN, Massillo C, Scalise GD, Farré PL, Elble R, Gerez EN, Accialini P, Cabanillas AM, Gardner K, De Luca P, De Siervi A. CLCA2 epigenetic regulation by CTBP1, HDACs, ZEB1, EP300 and miR-196b-5p impacts prostate cancer cell adhesion and EMT in metabolic syndrome disease. Int J Cancer. 2018;143:897–906. doi: 10.1002/ijc.31379. [DOI] [PubMed] [Google Scholar]
- 11.Moiola CP, De Luca P, Zalazar F, Cotignola J, Rodriguez-Seguí SA, Gardner K, Meiss R, Vallecorsa P, Pignataro O, Mazza O, Vazquez ES, De Siervi A. Prostate tumor growth is impaired by CtBP1 depletion in high-fat diet-fed mice. Clin Cancer Res. 2014;20:4086–4095. doi: 10.1158/1078-0432.CCR-14-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018;9:1–12. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massillo C, Dalton GN, Farré PL, De Luca P, De Siervi A. Implications of microRNA dysregulation in the development of prostate cancer. Reproduction. 2017;154:R81–R97. doi: 10.1530/REP-17-0322. [DOI] [PubMed] [Google Scholar]
- 14.Sharma N, Baruah MM. The microRNA signatures: aberrantly expressed miRNAs in prostate cancer. Clin Transl Oncol. 2019;21:126–144. doi: 10.1007/s12094-018-1910-8. [DOI] [PubMed] [Google Scholar]
- 15.Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285:116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 16.Schwarzenbach H, Gahan PB. MicroRNA shuttle from cell To-cell by exosomes and its impact in cancer. Noncoding RNA. 2019;5:1–22. doi: 10.3390/ncrna5010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massillo C, Duca RB, Lacunza E, Dalton GN, Farré PL, Taha N, Piccioni F, Scalise GD, Gardner K, De Siervi A. Adipose tissue from metabolic syndrome mice induces an aberrant miRNA signature highly relevant in prostate cancer development. Mol Oncol. 2020;14:2868–2883. doi: 10.1002/1878-0261.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urabe F, Matsuzaki J, Yamamoto Y, Kimura T, Hara T, Ichikawa M, Takizawa S, Aoki Y, Niida S, Sakamoto H, Kato K, Egawa S, Fujimoto H, Ochiya T. Large-scale circulating microRNA profiling for the liquid biopsy of prostate cancer. Clin Cancer Res. 2019;25:3016–3025. doi: 10.1158/1078-0432.CCR-18-2849. [DOI] [PubMed] [Google Scholar]
- 19.Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, Zhu J, Haussler D. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nam RK, Wallis CJD, Amemiya Y, Benatar T, Seth A. Identification of a novel MicroRNA panel associated with metastasis following radical prostatectomy for prostate cancer. Anticancer Res. 2018;38:5027–5034. doi: 10.21873/anticanres.12821. [DOI] [PubMed] [Google Scholar]
- 21.Liu T, Zhang Q, Zhang J, Li C, Miao YR, Lei Q, Li Q, Guo AY. EVmiRNA: a database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019;47:D89–D93. doi: 10.1093/nar/gky985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osip’yants AI, Knyazev EN, Galatenko AV, Nyushko KM, Galatenko VV, Shkurnikov MY, Alekseev BY. Changes in the level of circulating hsa-miR-297 and hsa-miR-19b-3p miRNA are associated with generalization of prostate cancer. Bull Exp Biol Med. 2017;162:379–382. doi: 10.1007/s10517-017-3620-6. [DOI] [PubMed] [Google Scholar]
- 23.Tian L, Fang YX, Xue JL, Chen JZ. Four microRNAs promote prostate cell proliferation with regulation of PTEN and its downstream signals in vitro. PLoS One. 2013;8:1–19. doi: 10.1371/journal.pone.0075885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou P, Ma L, Zhou J, Jiang M, Rao E, Zhao Y, Guo F. miR-17-92 plays an oncogenic role and conveys chemo-resistance to cisplatin in human prostate cancer cells. Int J Oncol. 2016;48:1737–1748. doi: 10.3892/ijo.2016.3392. [DOI] [PubMed] [Google Scholar]
- 25.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao P, Deng Z, Wan M, Huang W, Cramer SD, Xu J, Lei M, Sui G. MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1α/HIF-1β. Mol Cancer. 2010;9:1–12. doi: 10.1186/1476-4598-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Song Q, Cai Y, Wang P, Wang M, Zhang D. RLIP76-dependent suppression of PI3K/AKT/Bcl-2 pathway by miR-101 induces apoptosis in prostate cancer. Biochem Biophys Res Commun. 2015;463:900–906. doi: 10.1016/j.bbrc.2015.06.032. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y, Chen F, Shen L, Tang X, Du C, Sun Z, Ding H, Chen J, Shen B. Biomarker microRNAs for prostate cancer metastasis: screened with a network vulnerability analysis model. J Transl Med. 2018;16:1–16. doi: 10.1186/s12967-018-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antognelli C, Cecchetti R, Riuzzi F, Peirce MJ, Talesa VN. Glyoxalase 1 sustains the metastatic phenotype of prostate cancer cells via EMT control. J Cell Mol Med. 2018;22:2865–2883. doi: 10.1111/jcmm.13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartholow TL, Chandran UR, Becich MJ, Parwani AV. Immunohistochemical staining of radixin and moesin in prostatic adenocarcinoma. BMC Clin Pathol. 2011;11:1. doi: 10.1186/1472-6890-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valderrama F, Thevapala S, Ridley AJ. Radixin regulates cell migration and cell-cell adhesion through Rac1. J Cell Sci. 2012;125:3310–3319. doi: 10.1242/jcs.094383. [DOI] [PubMed] [Google Scholar]
- 32.Kurozumi A, Goto Y, Matsushita R, Fukumoto I, Kato M, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa T, Seki N. Tumor-suppressive microRNA-223 inhibits cancer cell migration and invasion by targeting ITGA3/ITGB1 signaling in prostate cancer. Cancer Sci. 2016;107:84–94. doi: 10.1111/cas.12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bijnsdorp IV, Geldof AA, Lavaei M, Piersma SR, van Moorselaar RJ, Jimenez CR. Exosomal ITGA3 interferes with non-cancerous prostate cell functions and is increased in urine exosomes of metastatic prostate cancer patients. J Extracell Vesicles. 2013;2:22097. doi: 10.3402/jev.v2i0.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanoverberghe K, Mariot P, Vanden Abeele F, Delcourt P, Parys JB, Prevarskaya N. Mechanisms of ATP-induced calcium signaling and growth arrest in human prostate cancer cells. Cell Calcium. 2003;34:75–85. doi: 10.1016/s0143-4160(03)00024-1. [DOI] [PubMed] [Google Scholar]
- 35.Rezuchova I, Hudecova S, Soltysova A, Matuskova M, Durinikova E, Chovancova B, Zuzcak M, Cihova M, Burikova M, Penesova A, Lencesova L, Breza J, Krizanova O. Type 3 inositol 1,4,5-trisphosphate receptor has antiapoptotic and proliferative role in cancer cells. Cell Death Dis. 2019;10:186. doi: 10.1038/s41419-019-1433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudecova S, Markova J, Simko V, Csaderova L, Stracina T, Sirova M, Fojtu M, Svastova E, Gronesova P, Pastorek M, Novakova M, Cholujova D, Kopacek J, Pastorekova S, Sedlak J, Krizanova O. Sulforaphane-induced apoptosis involves the type 1 IP3 receptor. Oncotarget. 2016;7:61403–61418. doi: 10.18632/oncotarget.8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mertens-Walker I, Fernandini BC, Maharaj MS, Rockstroh A, Nelson CC, Herington AC, Stephenson SA. The tumour-promoting receptor tyrosine kinase, EphB4, regulates expression of Integrin-β8 in prostate cancer cells. BMC Cancer. 2015;15:1–10. doi: 10.1186/s12885-015-1164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Z, Tang F, Lu Z, Huang Y, Lei H, Li Z, Zeng G. Analysis of differentially expressed genes, clinical value and biological pathways in prostate cancer. Am J Transl Res. 2018;10:1444–1456. [PMC free article] [PubMed] [Google Scholar]
- 39.Sakamoto S, McCann RO, Dhir R, Kyprianou N. Talin1 promotes tumor invasion and metastasis via focal adhesion signaling and anoikis resistance. Cancer Res. 2010;70:1885–1895. doi: 10.1158/0008-5472.CAN-09-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu N, Chen HJ, Chen SH, Xue XY, Chen H, Zheng QS, Wei Y, Li XD, Huang JB, Cai H, Sun XL. Upregulation of Talin-1 expression associates with advanced pathological features and predicts lymph node metastases and biochemical recurrence of prostate cancer. Medicine (Baltimore) 2016;95:e4326. doi: 10.1097/MD.0000000000004326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Betts AM, Collett GP, Neal DE, Robson CN. Paracrine regulation of talin mRNA expression by androgen in human prostate. FEBS Lett. 1998;434:66–70. doi: 10.1016/s0014-5793(98)00953-3. [DOI] [PubMed] [Google Scholar]
- 42.Bello-DeOcampo D, Kleinman HK, Deocampo ND, Webber MM. Laminin-1 and α6β1 integrin regulate acinar morphogenesis of normal and malignant human prostate epithelial cells. Prostate. 2001;46:142–153. doi: 10.1002/1097-0045(20010201)46:2<142::aid-pros1018>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 43.Windus LC, Kiss DL, Glover T, Avery VM. In vivo biomarker expression patterns are preserved in 3D cultures of Prostate Cancer. Exp Cell Res. 2012;318:2507–2519. doi: 10.1016/j.yexcr.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Schmelz M, Cress AE, Scott KM, Bürgery F, Cui H, Sallam K, McDaniel KM, Dalkin BL, Nagle RB. Different phenotypes in human prostate cancer: α6 or α3 integrin in cell-extracellular adhesion sites. Neoplasia. 2002;4:243–254. doi: 10.1038/sj.neo.7900223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Labbé D, Zadra G, Yang M, Reyes J, Lin C, Cacciatore S, Ebot E, Creech A, Giunchi F, Fiorentino M, Elfandy H, Syamala S, Karoly E, Alshalalfa M, Erho N, Ross A, Schaeffer E, Gibb E, Takhar M, Den R, Lehrer J, Karnes R, Freedland S, Davicioni E, Spratt D, Ellis L, Jaffe J, D’Amico A, Kantoff P, Bradner J, Mucci L, Chavarro J, Loda M, Brown M. High-fat diet fuels prostate cancer progression by rewiring the metabolome and amplifying the MYC program. Nature Communications. 2019;10:1–14. doi: 10.1038/s41467-019-12298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]