Abstract
Colorectal cancer (CRC), one of the major health problems worldwide, mostly develops from colorectal adenomas. Advanced adenomas are generally considered as precancerous lesions and patients are recommended to remove the adenomas. Screening for colorectal cancer is usually performed by fecal tests (FOBT or FIT) and colonoscopy, however, their benefits are limited by uptake and adherence. Most CRC develops from colorectal advanced adenomas, but there is currently a lack of effective noninvasive screening method for advanced adenomas. N-glycans in human serum hold the great potentials as biomarker for diagnosis of human cancers. Our aim was to discover blood-based markers for screening and diagnosis of advanced adenomas and CRC, and to ascertain their efficiency in classifying healthy controls, patients with advanced adenomas and CRC by incorporating machine learning techniques with reliable and simple quantitative method with “Bionic Glycome” as internal standard based on the high-throughput Matrix-assisted Laser Desorption/Ionization Mass Spectrometry (MALDI-MS). The quantitative results showed that there is a positive correlation between multi-antennary, sialylated N-glycans and CRC progress, while bi-antennary core-fucosylated N-glycans are negatively correlated with CRC progress. Machine learning is a powerful classification tool, suitable for mining big data, especially the large amount of data generated by high-throughput technologies. Using the predictive model constructed by machine learning, we obtained the classification accuracy of 75% for classification of 189 samples including CRC, advanced adenomas and healthy controls, and the accuracy of 87% for detection of the disease group that required treatment, including CRC and advanced adenomas. To our delight, the model successfully applied to the prediction of 176 samples collected a few months later, and five samples were wrongly predicted in the disease group. Overall, this diagnostic model we constructed here has valuable potential in the clinical application of detecting advanced adenomas and colorectal cancer and could compensate for the limitations of the current screening methods for detection of CRC and advanced adenomas.
Keywords: Colorectal cancer, advanced adenoma, serum N-glycome quantification, internal standard, machine learning, mass spectrometry, biomarker
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related death worldwide [1]. CRC mostly develops from advanced adenomas. Advanced adenomas are considered as precancerous lesions with a high risk of carcinogenesis [2]. Patients who are detected and treated in the precancerous lesions or in the early stages of the disease have significantly higher survival rates, and the survival rates can exceed 90% [3]. Thus, effective screening of CRC is helpful to reduce the burden of CRC by preventing the development of cancer or detecting it at a curable stage [4].
Colonoscopy is the gold standard method for CRC detection. And this method has the disadvantages of invasiveness, a bowel preparation requirement, a risk for bowel rupture, high cost, and it is not suitable for the patients with anorectal stenosis, peritoneal irritation, severe cardiopulmonary function and other diseases. These considerations limit the widespread use of colonoscopy. Fecal occult blood test is a non-invasive test for CRC screening, but it exhibits low sensitivity and specificity for CRC and advanced adenomas [5]. Now the fecal immunochemical test (FIT) is replacing the fecal occult blood test in CRC screening [6,7]. However, the diagnostic performance of the FIT depends on the cutoff value for a positive result and other factors also influence the result, including male sex, older age, obesity, smoking, aspirin use and newly detected nonneoplastic bowel disease [8,9]. The sensitivity of FIT for detecting CRC is 79% and even poor (31%) for advanced adenomas screening [10]. As the most common serum tumor biomarkers, carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9), are beneficial in monitoring of cancer progression but not suitable for screening advanced adenomas and CRC due to their low sensitivity and specificity [11]. Fecal DNA testing shows higher sensitivity but it is unfit for population screening due to its high cost [12]. Therefore, there is an urgent need to develop more effective, inexpensive and non-invasive screening and diagnosis methods for advanced adenomas and CRC.
Glycosylation is one of the most common post-translation modifications of proteins. Abnormal glycosylation has been observed in many types of diseases, including various cancers [13,14]. Serum glycomic profiling is an emerging non-invasive screening tool that can be used to find potential biomarkers in the diagnosis of early stage cancer and disease surveillance [15-18]. And some studies have demonstrated the potential clinical application value of serum glycans in CRC. Zhao et al. found that serum core-fucosylated di-antennary N-glycans decreased in CRC patients by DNA sequencer-assisted/fluorophore-assisted carbohydrate electrophoresis (DSA-FACE) [19]. Stefan W. de Vroome et al. [20] evaluated the changes of total serum N-glycome (TSNG) in CRC patients and showed the potential value of TSNG as a prognostic biomarker (panel) for CRC. In those previously published studies, researchers usually focused on investigating the glycan alterations between CRC patients and healthy controls by analyzing a limited number of identified N-glycans, and further constructed relative models for CRC diagnosis. While the glycan alterations in precancerous lesions stage of CRC have been rarely reported, and few models were established for the diagnosis of advanced adenomas.
Machine learning “learns” a specific pattern from past data and utilizes this pattern to predict future data. It has been used to interpret speech [21], process images [22]. Recently, machine learning algorithms have attracted people’s attention in the field of health care. Machine learning can interpret massive and complicated data sets such as matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) spectra and have the ability to correctly evaluate complex patterns. Various machine learning algorithms, including Random Forest and Support Vector Machine (SVM) are robust and widely used algorithms for disease diagnosis [23-27].
Our study aims to evaluate the performance of N-glycans in screening and diagnosis of advanced adenomas and CRC. First, we quantified N-glycome from 186 samples including 39 advanced adenomas, 90 CRC and 57 healthy controls by using quantification method with “Bionic Glycome” as internal standard we previously developed [28], and found the differences of N-glycans among these three groups. Second, machine learning was applied to mine the data and to find the optimal model to classify advanced adenomas, CRC and healthy controls. Finally, the model was applied to an independent validation cohort to predict the patients. Our reliable quantification method combined with machine learning can find more effective model for CRC and advanced adenomas screening and diagnosis based on identified total serum N-glycans. To the best of our knowledge, this is the first time to combined serum N-glycome with machine learning to differentiation CRC and advanced adenomas from healthy controls.
Materials and methods
Chemicals and reagents
Milli-Q water was prepared by a Milli-Q system (Milford, MA, USA). Sodium dodecyl sulfate (SDS), 1-hydroxybenzotriazole monohydrate (HOBt), sodium borodeuteride (NaBD4), trifluoroacetic acid (TFA), sodium hydroxide (NaOH), super-2,5-dihydroxybenzoic acid (super-2,5-DHB), and 10 × phosphate buffered solution (PBS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). 1-Ethyl-3-(3-(dimethylamino)propyl)-carbodiimide (EDC) was purchased from Fluorochem (Hadfield, U.K.). Peptide-N-glycosidase F (PNGase F) and Nonidet P-40 (NP-40) were purchased from New England Biolabs (Ipswich, MA, USA). HPLC-grade acetonitrile (ACN), ethanol (EtOH), and formic acid (FA) were purchased from Merck (Darmstadt, Germany). Peptide calibration standard (TOFMix) was purchased from LaserBio Laboratories (LaserBio Laboratories, France).
Serum samples
All serum samples were collected from Shanghai East Hospital, Tongji University, Shanghai, China, from April 2019 to January 2021. The exclusion criteria were: (1) patients who ever received chemotherapy or radiation therapy; (2) patients who had simultaneously developed other tumors. The training cohort collected from April 2019 to April 2020 including a subset of 39 patients with pathologically confirmed advanced adenomas (diameter >1 cm, with severe high-grade neoplasia, tubulovillous, villious, sessile serrated or traditional serrated histology or more than three adenomas of any size), 90 patients with pathologically confirmed colorectal cancer and 57 age- and gender-matched healthy volunteers. And the samples in validation set were collected from May 2020 to January 2021, which included a subset of 59 advanced adenomas patients, 73 colorectal cancer patients and 44 age- and gender-matched healthy controls. Clinical data from the patients are summarized in Table 1. The cut-off values recommended by the manufacturer for CEA, CA19-9, AFP levels were 5.0 μg/L, 37 U/L and 20 U/mL respectively. The venous blood samples were obtained preoperatively during the morning fasting state. The serum samples were collected by centrifuging blood samples at 2000 × g for 10 min after a 30 min clotting at ambient temperature. The serum samples were aliquot and stored at -80°C until analysis. In addition, a pooled serum sample used for production of Bionic Glycome as internal standard was prepared by mixing 5 μL serum from each sample and stored at -80°C before usage. Approvals were obtained from the Institutional Review Board and informed written consents from all participants were acquired.
Table 1.
The clinical characteristics of the participants
| Training cohort (N = 186) | Validation cohort (N = 176) | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| AA | CRC | Healthy Control | AA | CRC | Healthy Control | ||
| Total N = 362 | 39 | 90 | 57 | 59 | 73 | 44 | |
| Age (min-max) | 61 (33-85) | 61 (24-88) | 59 (32-81) | 59 (22-84) | 62 (35-84) | 57 (33-77) | |
| Gender (male/female) | 27/12 | 59/31 | 40/17 | 39/20 | 46/27 | 24/20 | |
| Tumor location | colon | 27 | 51 | 45 | 27 | ||
| rectum | 12 | 39 | 14 | 46 | |||
| Differentiation | High | 6 | 3 | ||||
| Moderate | 56 | 47 | |||||
| Poor | 28 | 23 | |||||
| CEA | Mean (Min-Max) | 3.24 (0.98-7.45) | 41.62 (0.83-594) | 2.21 (1.03-4.78) | 1.74 (0.10-4.94) | 49.68 (0.7-1804) | 2.15 (0.57-4.56) |
| <5 ng/mL | 36 | 48 | 57 | 59 | 40 | 44 | |
| ≥5 ng/mL | 3 | 42 | 0 | 0 | 33 | 0 | |
| CA19-9 | Mean (Min-Max) | 17.60 (5.41-46.8) | 105.70 (2-1863) | 14.96 (0.99-35.12) | 7.75 (1.06-36.6) | 157.46 (1.53-1869) | 13.12 (1.0-35.86) |
| <37 U/ml | 38 | 62 | 57 | 59 | 60 | 44 | |
| ≥37 U/mL | 1 | 38 | 0 | 0 | 13 | 0 | |
| AFP | Mean (Min-Max) | 2.97 (0.98-5.55) | 7.87 (0.908-322) | 3.58 (0.90-7.85) | 3.36 (0.45-10.3) | 3.12 (0.99-12.3) | 3.58 (0.91-10.3) |
| <20 ng/ml | 39 | 88 | 57 | 59 | 73 | 44 | |
| ≥20 ng/ml | 0 | 2 | 0 | 0 | 0 | 0 | |
| Stage | I | 14 | 10 | ||||
| II | 24 | 22 | |||||
| III | 22 | 25 | |||||
| IV | 30 | 16 | |||||
Colorectal cancer (CRC); Advanced adenomas (AA); Carcinoembryonic antigen (CEA); Carbohydrate antigen 19-9 (CA19-9); Alpha-fetoprotein (AFP).
N-glycan release
Five microliter serum was denatured at 60°C for 10 min by adding 10 μL 2% SDS. When cooling to room temperature, the denatured sample was added 10 μL of glycobuffer (4% NP-40, 5 × PBS, PH 7.5) and 1 μL PNGase F and incubated at 37°C overnight.
Bionic Glycome preparation
The detailed procedure was described in a previous report [28]. Briefly, N-glycans released from the pooled serum sample was added two volumes of ethanol to precipitate proteins. After centrifugation at 4°C for 15 min (13000 g), the supernatant was collected and mixed with 1% volume of FA and incubated at 37°C for 2 h. Then the solution was reduced by gently adding a 50% volume of fresh prepared 2 M NaBD4 with a 2 h incubation at 65°C. The reduced N-Glycans were purified using a HILIC SPE.
Sialic acid derivatization
The released N-glycans and purified reduced N-glycans were derivatized by freshly prepared ethylation reagent (250 mM EDC and 250 mM HOBt in ethanol) with a 1 h incubation at 37°C. Before purification, ACN was added to the derived glycans to prepare the sample for cleanup according to the procedure reported previously [29].
MALDI-MS analysis
For MALDI-MS analysis, we used TOFMix containing an eight-peptide calibration standard to calibrate the MS. One microliter of enriched ethyl-esterified glycans was spotted on a MALDI plate and allowed to dry by air. Then, 1 μL super-2,5-DHB (5 mg/mL) 1 mM NaOH in 50% ACN was added onto the plate and allowed to dry by air.
The spots were added 0.2 μL ethanol to form homogeneity of the spot surface by re-crystallization. Each sample was spotted in triplicate. The samples were interrogated automatically in a “batch mode” by AXIMA Resonance MALDI-quadrupole ion trap-time of flight MS (Shimadzu Corp. JP) equipped with a 337 nm nitrogen laser in reflector positive ionization mode. The m/z range was set at 1000 to 4000. The spectrum of each spot was generated by accumulating 200 profiles with 2 laser shots per profile.
Data processing and statistical analysis
The MALDI-MS data were exported as ASCII file, and then pre-processed, normalized and extracted by software of Progenesis MALDI. The glycan compositions were assigned according to the previous reports. The GlycoWorkbench software was used for the annotation of MS spectra. The quantification for relative abundance of each N-glycan was calculated by the signal intensity ratio (light vs heavy), measuring the most abundant isotopic peak area ratio (sample vs internal standard) [28]. The results for glycans were assessed by calculating the mean, SD, and the coefficient of variation (CV). Only the N-glycans with CVs less than 25% were used as predictors in the model. The derived glycosylation traits were calculated according to the structure features of glycans. Intergroup differences were evaluated by performing t-test. For the multiple corrections, the false discovery rate (FDR) was used based on the Benjamini-Hochberg procedure, and adjusted P<0.05 was considered statistically significant. Receiver-operator-characteristics (ROC) test was used to assess the discriminant ability of the glycan traits. The significance of the resulting values of area-under-the-curve (AUC) was assigned. Support vector machine were used in this study. The ten-fold cross validation method was used during the training process, and then the classification accuracy was computed to represent the overall performance for each model with 49 N-glycans. The statistical analyses were performed using R 4.0.2 software and GraphPad Prism 6 software.
Results
Serum N-glycan profile
Figure 1 shows the whole pipeline for the construction of N-glycome based model for advanced adenomas and CRC screening. Serum N-glycan profiles were analyzed in 98 patients with advanced adenomas, 163 patients with CRC and 101 healthy people. Among these samples, 186 samples were used as training set and the 176 samples were used as the validation set. Their characteristics are presented in Table 1. All the samples were individually mixed with the Bionic Glycome and detected by MALDI-MS [28]. The representative mass spectrum was shown in Figure 2A. The glycan structures and compositions were proposed based on their mass and tandem MS information in our previous literatures [28]. According to the quantification results, 54 N-glycans were detected and 49 N-glycans with the CVs less than 25% as listed in Table S1 were further analyzed in the following study. And derived traits calculation based on their number of antennae, bisection, galactosylation, fucosylation, and (linkage-specific) sialylation are listed in Table S2.
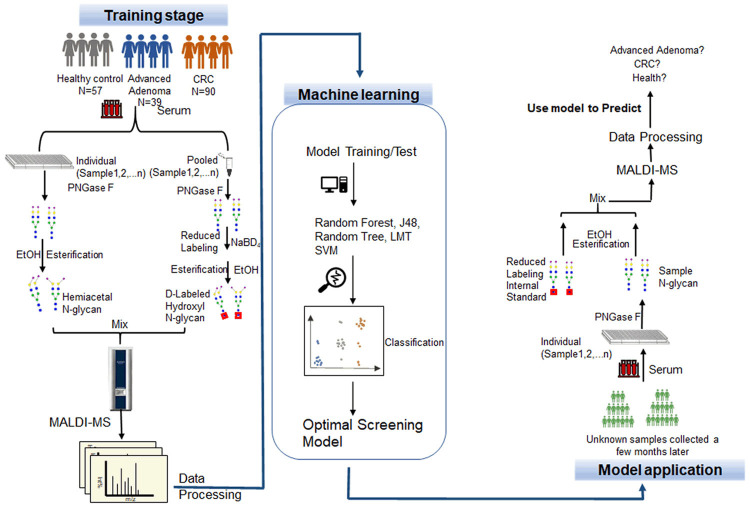
Figure 1.
Overview of the whole workflow of the study. Screening and diagnosis of colorectal cancer and advanced adenoma by Bionic Glycome method and machine learning. CRC, colorectal cancer; LMT, logistic model trees; SVM, support vector machine.
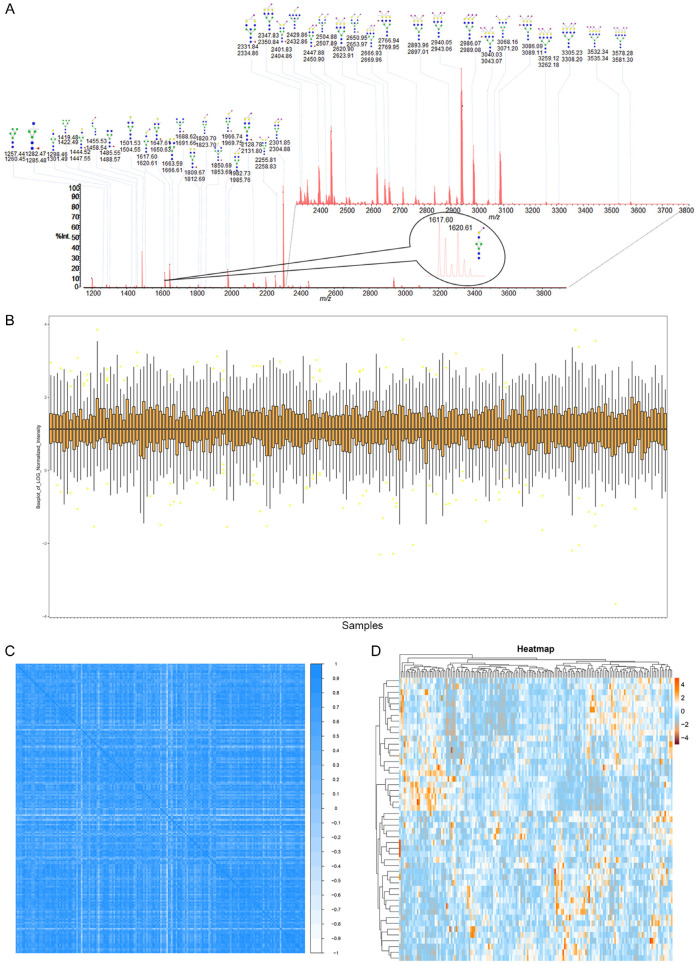
Figure 2.
Serum N-glycan information in the training cohort. A. The presentative MALDI-MS spectrum of the mixture of N-glycome from human serum and its Bionic Glycome as internal standard. All the m/z values of glycan peaks were single sodium adducts ([M+Na]+). A total of 49 doublets with a 3 Da mass difference was detected and the inset is an enlarged spectrum of one doublet at m/z 2301.85/2304.88. Green circle, Man; yellow circle, Gal; blue square, GlcNAc; red triangle, Fuc; clockwise purple diamond, α2,6-linked sialic acid; anticlockwise purple diamond, α2,3-linked sialic acid. B. Relative intensity box plot of N-glycome for all samples in the training cohort. C. Person correlation analysis of N-glycome for all samples in the training cohort. D. Heatmap of serum N-glycans in Healthy control, Advanced Adenoma and Colorectal Cancer.
The high quantitative accuracy of the quantitative method based on the “Bionic Glycome” internal standards allowed us to explore the differences among patients with advanced adenomas, colorectal cancer and healthy people. We first checked the quality of our data to verify the reliability of the quantitative method and the quantitative results. The normalized data were log2 transformed and the relative intensity distribution of N-glycan of all samples are basically in the same range of variation (Figure 2B). Pearson’s correlation coefficient between the glycome in all samples were 0.62 (Figure 2C). The heatmap of hierarchical clustering (Figure 2D) shows that there is no missing data and no algorithm is needed to fill in the data. The results above prove that the data is stable, with high credibility and quality.
Changes of serum N-glycome in patients with advanced adenomas and CRC
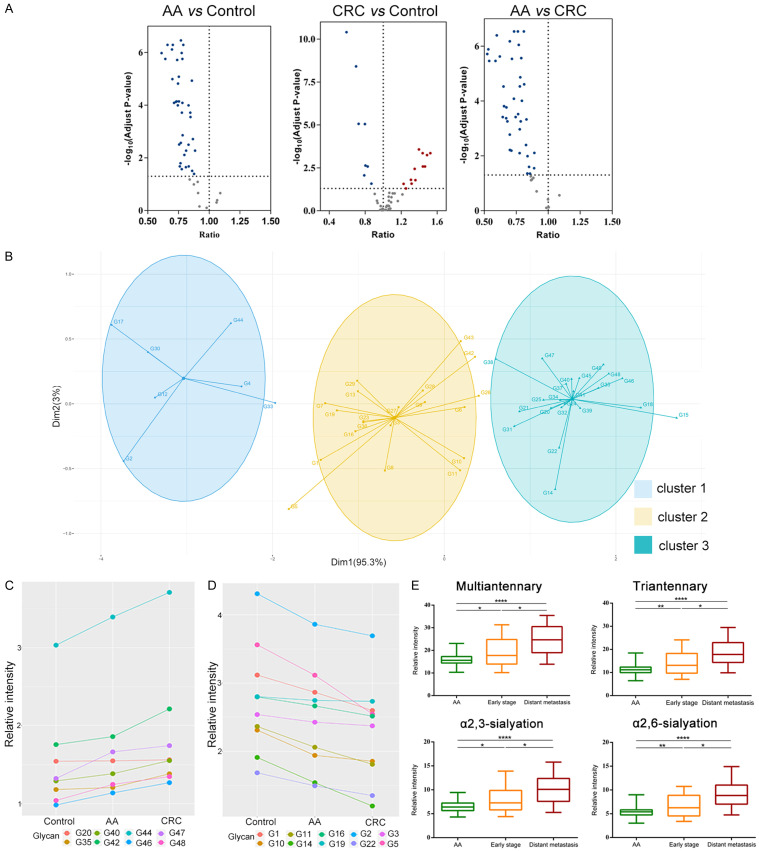
We quantified 49 N-glycans in order to find the alternation of N-glycan in advanced adenomas and CRC. Relative quantitative data of the 49 N-glycans of 186 samples in the training cohort were listed in Table S3. Among them, 39 N-glycan levels decreased in Advanced Adenomas vs Healthy Controls; 12 N-glycan levels increased and 8 N-glycan levels decreased in CRC vs Healthy Controls and 40 N-glycan levels decreased in Advanced Adenomas vs CRC (Figure 3A). Next, we explored the connections between these N-glycans in all the samples. Unsupervised cluster analysis grouped these N-glycans into 3 distinct clusters by the K-means classifier (Figure 3B and Table S4). We found the N-glycans in the cluster 2 were mainly N-glycans in IgG and most high-mannose glycans were clustered into cluster 1, while the most multi-antennary (tri-antennary and tetra-antennary) N-glycans were clustered into cluster 3. It is suggested that different types of N-glycans may play different roles in these three groups, which indicated that there are sophisticated and precise regulatory mechanisms for N-glycans synthesis. Then we analyzed the changes of each N-glycan in the healthy controls and patients with advanced adenoma and CRC. Correlations between the average expression level of these N-glycans in the three groups and the known vector were analyzed using Pearson’s correlation coefficient and applied a threshold to the correlation coefficient at r>0.9. The results showed that 8 N-glycans had a positive correlation with the progression of the disease in the three groups of healthy controls, patients with advanced adenomas and patients with colorectal cancer, while 10 N-glycans had a negative correlation (Figure 3C, 3D). The eight increasing N-glycans in these three sets were mainly sialylated multi-antennary N-glycans, while the decreasing N-glycans were mainly galactosylated and fucosylated. The derived traits were also compared among these three sets of samples (Figure S1), we discovered that the multiantennary, tri-antennary, α2,3-sialylated and α2,6-sialylated N-glycans changed as disease progressed, and their levels increased from advanced adenomas to early stage (TNM = 1) and distant metastasis (TNM = 4) (Figure 3E). These results indicated that N-glycans synthesis may change in human body during the development of CRC.
Figure 3.
Changes of serum N-glycome in patients with CRC and advanced adenomas. (A) Volcano plots showed the differentially expressed N-glycans in three groups (AA vs Control, CRC vs Control, AA vs CRC; AA, Colorectal Advanced Adenoma; CRC, Colorectal Cancer; x axis, ratio of the N-glycans between two groups; y axis, adjust p-value). Adjust p-value <0.05 was considered statistically significant and the ratio between the two groups less than 1 was considered to be a decrease while greater than 1 was considered to be an increase. The dotted lines show the threshold for statistical significance. Red dots represent the increased, and blue dots represent the decreased. (B) Unsupervised cluster analysis of the N-glycome in three groups by K-means clustering algorithm. (C, D) The correlations between the glycans and CRC progress. The glycans that were positively (C) or negatively (D) related to the progression of CRC. The threshold of the Pearson’s correlation coefficient was set to be r>0.9. (E) Changes of the relative intensity of four derived glycosylation traits (multiantennary, triantennary, α2,3-sialyation, α2,6-sialyation) in advanced adenoma, early stage and late stage CRC (Early stage, TNM = 1; Distant metastasis, TNM = 4. * The equivalent of P<0.05, ** the equivalent of P<0.01, *** the equivalent of P<0.001, **** the equivalent of P<0.0001).
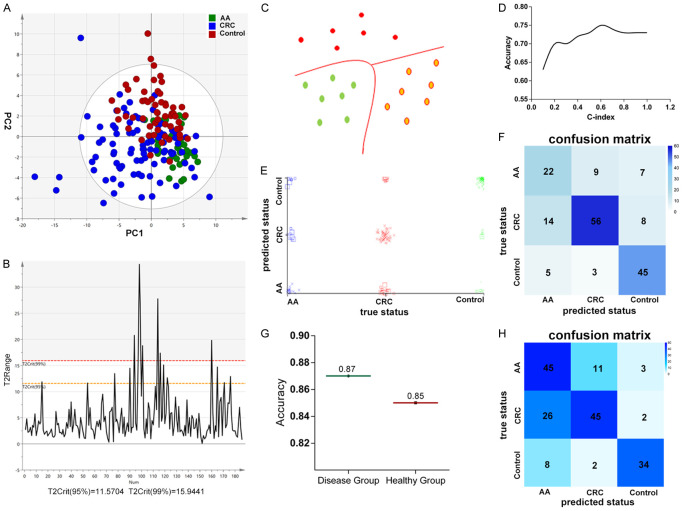
Screening and diagnosis of CRC and advanced adenoma using model constructed by machine learning
Through the N-glycan structure abundance analysis, we found that compared with healthy controls, 14 types of glycans have significant differences in the serum of patients with advanced adenoma and CRC (Figure 4A) and AUC above 0.8 were shown in Figure 4B-D; Table S5. Due to the limited ability of a single feature as a biomarker in discriminating samples, and in order to provide more comprehensive understanding of the data while mining the data more deeply, we combined machine learning to find the optimal model to classify advanced adenomas, CRC and healthy controls. Before building the model, we use O2PLS-DA to optimize visualization of the N-glycosylation changes in advanced adenomas and CRC (Figure 5A), and eliminate samples that exceed 95% T2Crit (Figure 5B). The O2PLS-DA showed that patients (advanced adenomas, CRC) could be differentiated from healthy controls, but there will be some overlap between advanced adenomas and colorectal cancer (Figure 5A). Then we used sequential minimal optimization training Support Vector Machines (SVM) (Figure 5C) to classify advanced adenoma, CRC, and healthy controls with the accurate relative quantitative data of total serum N-glycome and other clinical data including age, gender and common tumor markers (CEA, AFP and CA19-9) (54 features in total). The tenfold cross-validation was performed. In general, by selecting a polynomial kernel function and optimizing the parameters (Figure 5D), the accuracy of the model for classify colorectal cancer, advanced adenomas and healthy controls was 75%. The three classification results were shown in Figure 5E, 5F. Specifically, the discriminative accuracy of advanced adenomas, colorectal cancer and healthy controls were 58%, 72% and 85%, respectively. Most of the incorrectly samples were between advanced adenomas and CRC for nine of sixteen incorrectly classified advanced adenomas were classified as colorectal cancer, and fourteen of twenty-two incorrectly classified colorectal cancers were classified as advanced adenomas (Figure 5F). This was consistent with the results of O2PLS-DA, with some overlap between advanced adenomas and CRC (Figure 5A). Advanced adenoma has a high risk of becoming cancerous and these patients are recommended for surgery. Whether it is recognized as advanced adenoma or CRC by the model, more in-depth examination such as colonoscopy and treatment such as surgery should be performed. Therefore, people who were classified as advanced adenomas and CRC by the model are considered to be true-positive results. And in this premises, the diagnostic accuracy of the disease group (advanced adenoma and colorectal cancer) was 87%, while the correct identification of the healthy group was 85% (Figure 5G).
Figure 4.
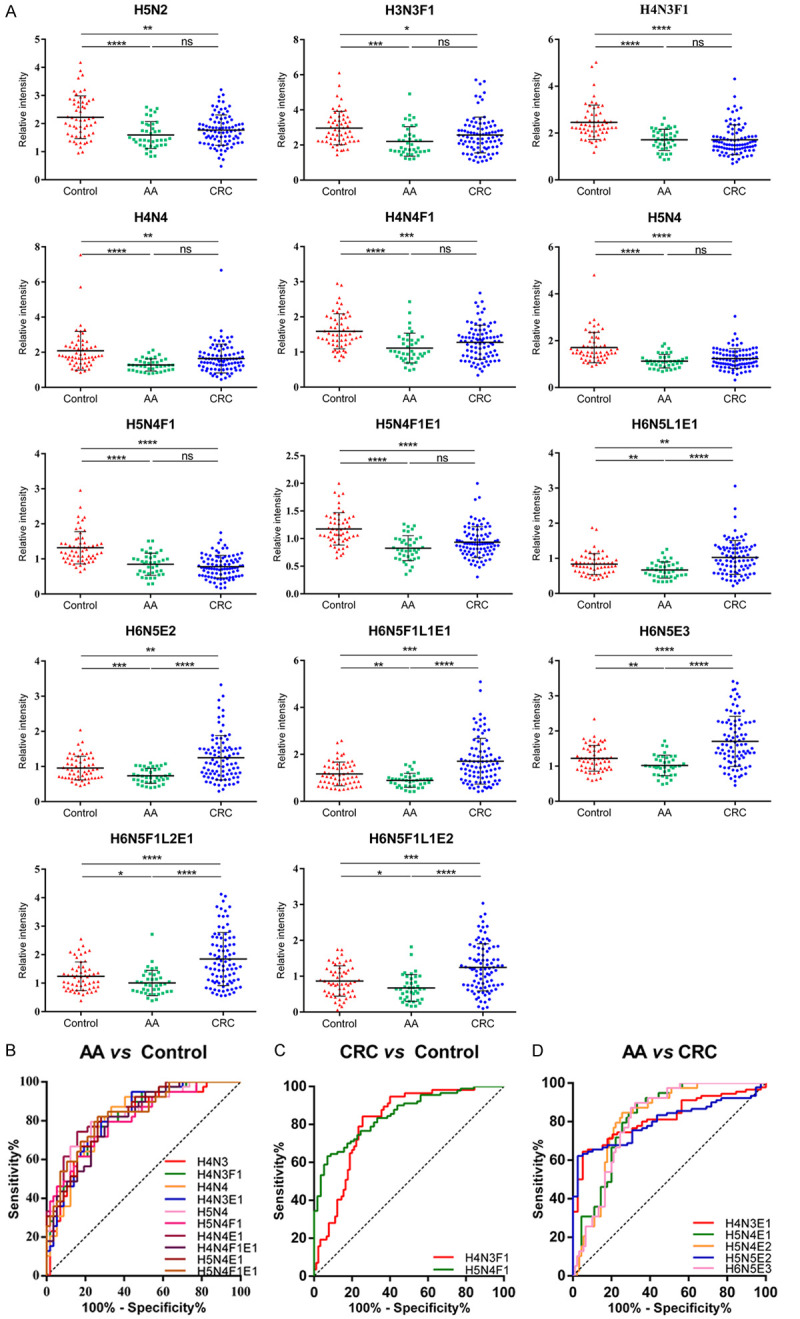
Relative intensity of the N-glycans with significant differences and their ROC analysis. (A) Scatter plot depicting the different relative intensity of fourteen N-glycans structure in AA and CRC compared with healthy controls. (B-D) Receiver operating characteristic (ROC) curve analyses for the the N-glycans with AUC above 0.8 of AA vs Control (B), CRC vs Control (C) and AA vs CRC (D). H = hexose, N = N-acetylhexosamine, F = fucose, L = lactonized N-acetylneuraminic acid (α2,3-linked), E = ethyl esterified N-acetylneuraminic acid (α2,6-linked). * The equivalent of P<0.05, ** the equivalent of P<0.01, *** the equivalent of P<0.001, **** the equivalent of P<0.0001.
Figure 5.
Disease classification with N-glycome combining machine learning. A. O2PLS-DA of the three groups. B. Hotelling’s T2Range Line Plot of the samples. C. Schematic diagram of SVM. D. Schematic diagram of optimizing c-index. E, F. Classification performance of the model constructed by machine learning with Support Vector Machine in training cohort. E. Scatter plot depicting the prediction results of the model on samples, with each point representing one sample. F. Confusion matrix indicating classification result. G. The diagnostic accuracy of the disease group (advanced adenoma and colorectal cancer) and the healthy group in training cohort. H. Confusion matrix indicating the classification performance in validation cohort.
Evaluation of our glycan-based model
In order to further evaluate the classification effect of the model, we used this model to predict the newly obtained samples. Among these 176 samples, only five samples in disease group were incorrectly predicted to healthy people. The discriminative accuracy of disease group and healthy controls were 96.2% and 77.3% respectively. The specific classification results were shown in Figure 5H. The results demonstrated the great potential of the model as a screening biomarker for advanced adenomas and colorectal cancer.
Discussion
We have successfully achieved the relative quantification of the absolute abundances of the serum N-glycans based on the novel N-glycome quantitation method with Bionic Glycome as internal standard. Our Bionic Glycome method can be used to prepare the corresponding internal standard for samples from any source to achieve accurate glycome quantification. Recently, we also applied it to the study of mouse serum glycome [30]. By one step of glycan reducing and isotope labeling (Glycan-RAIL), each glycan to be identified in the sample can have a corresponding bionic glycan with 3Da increment as its specific internal standard. Only one step of glycan reducing will reduce the complexity of internal standard preparation. The samples did not need to be reduced and isotope labeled, therefore it would reduce the loss of N-glycan content and make the quantitative results more credible. The “Bionic Glycome” internal standard was produced using N-glycome from pooled samples thus has the same glycan structure and composition and similar abundance of glycome profile with the N-glycome to be analyzed in the biological samples which make the quantification more accurate. And by esterifying sialic acid [29], we can discriminate α2,3-linked and α2,6-linked sialic acid. All above make our results more reliable. We have explained this method in detail in our previous article [28]. And we used this reliable method to explore the N-glycome changes in advanced adenoma and CRC. We found that the all glycans positively correlated with the development of CRC were sialylated N-glycans (Figure 3C, G20 (H5N4E1), G35 (H6N5L1E1), G40 (H6N5L2E1), G42 (H6N5E3), G44 (H5N5L1E1), G46 (H7N6L2E1), G47 (H7N6L1E2), and G48 (H7N6L3E1)), and most of them were multi-antennary glycans. Specifically, from advanced adenoma, early stage to late metastasis stage, tri-antennary, multi-antennary, α2,3-sialylation and α2,6-sialylation N-glycans were increasing as the CRC progresses. Our findings were in line with previous study [20], higher levels of branching and sialylation N-glycans were associated with the promotion of invasion and metastasis of CRC. Multi-branch N-glycans have been reported to be involved in the regulation of cell proliferation and differentiation in tissue and cell line, and have been suggested as markers to predict the aggressiveness of CRC [31]. The GlcNAc transferase V is responsible for the increase of multi-antennary N-glycans. Cells expressing high levels of GlcNAc transferase V have decreased cell-cell adhesion. All of these findings indicated that GlcNAc transferase V may be involved in the pathogenesis of malignant transformation.
As shown in Figure 3D, the G1 (oligomannosidic N-glycan, Man5, m/z = 1257.44), was negatively correlated with the progression of CRC, and the relative content in CRC was significantly lower than that in advanced adenoma and healthy controls. Interestingly, compared with the control, in the total serum proteins of mouse, Man5 glycan was significantly decreased in cancer [32], indicating that N-mannosylation may play a vital role in cancer progression for both human and mice.
Among other glycans that were negatively related to the development of CRC, most of them are di-antennary core-fucosylated glycans (G2 (H3N3F1), G5 (H4N3F1), G10 (H4N4F1), G14 (H5N4F1), G16 (H4N5F1), G19 (H4N4F1E1), G22 (H5N4F1E1)) and terminal mono-galactose or di-galactose glycans (G3 (H4N3), G5 (H4N3F1), G10 (H4N4F1), G11 (H5N4), G14 (H5N4F1), G16 (H4N5F1)). The alteration of galactosylation in serum N-glycans has been found in a variety of tumors [17,33,34], especially the alteration in IgG has been proven to be a cancer marker with great potential [35-38]. The previous studies also have consistent results that the expression level of core-fucosylated di-antennary glycan was significantly decreased in CRC [19,20]. Abnormal fucosylation can affect the adhesion ability of tumor cells, which in turn affects cells proliferation, differentiation and migration. The alteration of core fucosylation further demonstrated that the decreased core fucosylation contributes to CRC progression. However, it seems that this change is tumor-specific. Studies have shown that core fucosylation level increased in prostate cancer [39], endometrial cancer [40], pancreatic cancer [41]. The core fucosylation is regulated by Fut8 and fucosidase, and the decrease of fucosylation may be caused by the down-regulation of Fut8 expression [19] and/or up-regulation of fucosidase expression [42]. From a biological point of view, specific glycan structures may reflect specific disease-related pathways. Therefore, further studies of the specific mechanisms behind these changes are needed.
It is interesting to note that there are some N-glycans whose relative contents are not linearly correlated with the progression of the CRC (Figure S2), indicating that the alteration of the N-glycosylation during the development of CRC are complex. The glycome profile in serum has changed in advanced adenoma stage before the formation of malignant tumors. In general, among the glycans we detected, some of them were positively or negatively correlated with the progression of the CRC, which could imply the progression of the disease and provide a new basis for the development of CRC; while most of the changes in serum glycans were complicated. This is also one of the difficulties in glycobiology research and we still have a lot of work to do.
The alterations of the N-glycan level in cancer patients serum may reflect the alterations of the glycoproteins in cancer patients serum. Antibodies (IgG, IgA and IgM), transferrin and alpha-2-macroglobulin represent together approximately 75% of all serum glycoproteins [43]. The remaining part mainly consists of alpha-1-antitrypsin, alpha-1-acid glycoprotein, haptoglobin, ceruloplasmin, the complement system, and apolipoproteins-making the liver and plasma B-cells the main source of serum glycoproteins [43,44]. Based on the observed changes in combination with our recent review mapping N-glycan contributions to serum glycoproteins, IgG might be responsible for the decrease of terminal mono-galactose or di-galactose N-glycans [43,45]. We know that chronic inflammation is one of the characteristics of cancer. It is likely that the changes of N-glycan level observed in serum of CRC are related to the inflammatory response. And during inflammation, the production of acute phase proteins is increased which leads to their higher abundance in the serum, but also cytokines produced by the tumor microenvironment can stimulate the hepatocytes in the liver [46]. In our study, we observed an increase in multi-antennary N-glycans as well as α2,3-sialylation and α2,6-sialylation N-glycans. Increased expression of multi-branching and sialic acids N-glycans has been reported on haptoglobin, alpha-1-acid glycoprotein and other acute phase proteins for inflammatory conditions as well as cancer [43,46,47]. These results suggest that changes of N-glycan level are associated with the presence of cancer and further studies are needed to determine the possible role of such changed glycosylation in cancers.
In the traditional screening methods of differential glycans, researchers use parametric test or non-parametric test to determine whether the sample has statistical difference, and then use multiple linear regression, logistic regression and other regression models to evaluate the classification effect of glycan biomarkers. However, these methods may have the following issues: 1) The biological problem that biomarkers can be used to distinguish disease from normal control can be regarded as a classification problem in mathematics. The traditional regression models are more suitable for dealing with linear separable classification problems with single boundary, while taking the glycomic data as an example, the omics data are often non-linearly separable and the application of linear regression models alone may lead to poor classification results. 2) Traditional regression analysis lacks intuitive visualization means, and it is difficult to express the linear hyperplane in high-dimensional space through images. Fortunately, the use of machine learning can solve the above problems well. For glycomic data, the consistent heterogeneity that exists on any glycosylated proteins is one of its unique features which different from genomic and proteomic data. The glycosylation signal of the target protein is distributed in many different glycoforms. And this glycan heterogeneity provides a unique opportunity in the field of biomarker discovery for researchers to use multiple features to classify samples. In the field of glycan biomarker screening, glycomics researchers rarely use all the glycosylation information of samples, but tend to find a single feature [48] or several features [49,50] as biomarkers to discriminate the samples in the healthy or disease group. However, this method will definitely eliminate information that may be useful for classifying samples. Recently, Gordan et al. [27] using machine learning to construct a non-linear model to investigate the ability of the plasma N-glycome to predict incidence of type 2 diabetes and cardiovascular diseases. In our research, we used machine learning to classify the disease group and the healthy group on the basis of the MS quantitative data. As for as we know, this is the first time to build the model to screening and diagnosis CRC and advanced adenomas based on the total serum protein N-glycome using machine learning.
Colorectal cancer screening has gained increasing attention as an important means to reduce the incidence and mortality of colorectal cancer. However, the number of people who prefer colonoscopy for screening and prevention of colorectal cancer is very limited. And most people are reluctant to perform colonoscopy due to the invasiveness and discomfort of colonoscopy. As the high-risk type of adenomas, advanced adenomas are recommended for increased surveillance and/or excision [2,51]. Here, using our model, we can identify not only patients with colorectal cancer but also patients with advanced adenoma. And we found a simple, non-invasive, inexpensive and relatively sensitive screening method to identify people who would benefit from colonoscopy. In the training cohort, the diagnostic accuracy of our model was 58% for advanced adenoma, 72% for CRC, and 87% for the overall disease group, whereas the screening accuracy of CEA for the disease group was 34.8%, of which the diagnostic accuracy for advanced adenoma was only 7.6%. To further evaluate the classification ability of the model, the model was used to classify the newly collected samples in the validation cohort. The diagnostic accuracy of the disease group (patients with advanced adenomas and CRC) was as high as 96.2%, and only five samples in disease group were incorrectly predicted to healthy people. In practical terms, the classification accuracy was 76.3% for advanced adenomas and 61.6% for CRC. The diagnostic accuracy of the model has been significantly higher than other markers: fecal occult blood test (50% sensitivity for cancers and 17%-46% for larger (diameter >1 cm) polyps) [5,52], Fecal Immunochemical Test (FIT) (73.8% sensitivity for cancers and 23.8% for precancerous lesions) [8,10,53], FIT combined with DNA mutation and methylation (51.6-92.3% sensitivity for cancer and 42.4% for larger (diameter >1 cm) polyps) [53,54] and the test for SEPT9 DNA in blood (48.2% sensitivity for cancer and 11.2% for adenoma) [55]. The carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) are the most widely applied serum biomarkers in clinics. Increased serum level of CEA or CA19-9 indicates the presence of CRC [56,57]. However, both lack sensitivity and specificity for CRC which precludes the use for diagnosis, but may be useful in staging evaluation and monitoring after treatment [58,59]. CEA is a highly heterogeneous glycoprotein that contains 60% carbohydrate. Individual differences because of various glycoprotein patterns are to be expected. And 15%-40% of CRC patients have perpetually non-elevated CEA levels [60]. CA19-9 is not useful in the diagnosis of CRC. A systematic review by Acharya et al. involving 156 studies demonstrated its low sensitivity (0.471) despite adequate specificity (0.924) [61]. Comparatively, CA19-9 fares poorer than CEA (0.533 and 0.864 for sensitivity and specificity, respectively [61], in which CEA has already been established to be ineffective for diagnosing CRC [58]. In our study, the screening accuracy of CEA for the disease group was 34.8% (45/129), of which the diagnostic accuracy for CRC was 46.7% (42/90) and for advanced adenoma was only 7.6% (3/39) in the training cohort. And the screening and diagnostic accuracy of CA19-9 for the disease group was 30.2% (39/129), for CRC was 42.2% (38/90), for advanced adenoma was 2.6% (1/39). Therefore, we believed that N-glycan profiling would be more effective than the detection of individual glycosylated molecules. Screening and detection of CRC and its precancerous lesion, advanced adenomas, is important to improve CRC incidence and prognosis. Our model has potential value in clinical applications, because whether it is classified as advanced adenoma or CRC, timely treatment and intervention must be performed to block the progression of the disease.
Conclusion
Using the N-glycome quantification method with “Bionic Glycome” as the internal standard, we analyzed the expression levels of N-glycans in the three subsets of healthy controls, advanced adenomas and CRC. The results revealed that total serum N-glycome changed specifically as the CRC progresses. These change characteristics provide important reference data for the mechanism of serum N-glycosylation in the initiation and progression of CRC. The further research is still required to identify the mechanisms behind these changes in order to deepen our understanding of the relationship between serum N-glycome and CRC. And this research proved the value of serum total protein N-glycome in CRC diagnosis. By combining MALDI-MS data and machine learning method, we constructed a N-glycome-based model for the detection of CRC and advanced adenomas with high efficiency and specificity, and we successfully applied this model in CRC and advanced adenomas screening. The positive results indicate its broad application prospect in disease diagnosis. And we will further prove our model in larger sample groups and prospective studies in the future.
Acknowledgements
We gratefully acknowledge financial support from the National Key Research and Development Program of China (2018YFC0910303), National Natural Science Foundation of China (32071276, 31770858, 81702961), the Outstanding Clinical Discipline Project of Shanghai Pudong (PWYgy2018-02) and Shanghai Municipal Science and Technology Major Project (Grant No. 2017SHZDZX01).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Bjerrum A, Lindebjerg J, Andersen O, Fischer A, Lynge E. Long-term risk of colorectal cancer after screen-detected adenoma: experiences from a Danish gFOBT-positive screening cohort. Int J Cancer. 2020;147:940–947. doi: 10.1002/ijc.32850. [DOI] [PubMed] [Google Scholar]
- 3.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. The case for early detection. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 4.Malila N, Senore C, Armaroli P. European guidelines for quality assurance in colorectal cancer screening and diagnosis. First edition--organisation. Endoscopy. 2012;44(Suppl 3):SE31–48. doi: 10.1055/s-0032-1309783. [DOI] [PubMed] [Google Scholar]
- 5.Bretthauer M. Colorectal cancer screening. J Intern Med. 2011;270:87–98. doi: 10.1111/j.1365-2796.2011.02399.x. [DOI] [PubMed] [Google Scholar]
- 6.Moss S, Mathews C, Day TJ, Smith S, Seaman HE, Snowball J, Halloran SP. Increased uptake and improved outcomes of bowel cancer screening with a faecal immunochemical test: results from a pilot study within the national screening programme in England. Gut. 2017;66:1631–1644. doi: 10.1136/gutjnl-2015-310691. [DOI] [PubMed] [Google Scholar]
- 7.Hol L, van Leerdam ME, van Ballegooijen M, van Vuuren AJ, van Dekken H, Reijerink JC, van der Togt AC, Habbema JD, Kuipers EJ. Screening for colorectal cancer: randomised trial comparing guaiac-based and immunochemical faecal occult blood testing and flexible sigmoidoscopy. Gut. 2010;59:62–68. doi: 10.1136/gut.2009.177089. [DOI] [PubMed] [Google Scholar]
- 8.Lee JK, Liles EG, Bent S, Levin TR, Corley DA. Accuracy of fecal immunochemical tests for colorectal cancer systematic review and meta-analysis. Ann Int Med. 2014;160:171. doi: 10.7326/M13-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amitay EL, Cuk K, Niedermaier T, Weigl K, Brenner H. Factors associated with false-positive fecal immunochemical tests in a large German colorectal cancer screening study. Int J Cancer. 2019;144:2419–2427. doi: 10.1002/ijc.31972. [DOI] [PubMed] [Google Scholar]
- 10.de Wijkerslooth TR, Stoop EM, Bossuyt PM, Meijer GA, van Ballegooijen M, van Roon AH, Stegeman I, Kraaijenhagen RA, Fockens P, van Leerdam ME, Dekker E, Kuipers EJ. Immunochemical fecal occult blood testing is equally sensitive for proximal and distal advanced neoplasia. Am J Gastroenterol. 2012;107:1570–1578. doi: 10.1038/ajg.2012.249. [DOI] [PubMed] [Google Scholar]
- 11.Vatandoost N, Ghanbari J, Mojaver M, Avan A, Ghayour-Mobarhan M, Nedaeinia R, Salehi R. Early detection of colorectal cancer: from conventional methods to novel biomarkers. J Cancer Res Clin Oncol. 2016;142:341–351. doi: 10.1007/s00432-015-1928-z. [DOI] [PubMed] [Google Scholar]
- 12.Ahlquist DA. Multi-target stool DNA test: a new high bar for noninvasive screening. Dig Dis Sci. 2015;60:623–633. doi: 10.1007/s10620-014-3451-5. [DOI] [PubMed] [Google Scholar]
- 13.Hakomori SI. Aberrant glycosylation in tumors and tumor-associated carbohydrate antigens. Adv Cancer Res. 1989;52:257–331. doi: 10.1016/s0065-230x(08)60215-8. [DOI] [PubMed] [Google Scholar]
- 14.An HJ, Kronewitter SR, de Leoz ML, Lebrilla CB. Glycomics and disease markers. Curr Opin Chem Biol. 2009;13:601–607. doi: 10.1016/j.cbpa.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, Lebrilla CB. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst. 2011;136:3663–3671. doi: 10.1039/c1an15093f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saldova R, Fan Y, Fitzpatrick JM, Watson RW, Rudd PM. Core fucosylation and alpha 2-3 sialylation in serum N-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia. Glycobiology. 2011;21:195–205. doi: 10.1093/glycob/cwq147. [DOI] [PubMed] [Google Scholar]
- 17.Qin R, Zhao J, Qin W, Zhang Z, Zhao R, Han J, Yang Y, Li L, Wang X, Ren S, Sun Y, Gu J. Discovery of non-invasive glycan biomarkers for detection and surveillance of gastric cancer. J Cancer. 2017;8:1908–1916. doi: 10.7150/jca.17900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saldova R, Haakensen VD, Rødland E, Walsh I, Stöckmann H, Engebraaten O, Børresen-Dale AL, Rudd PM. Serum N-glycome alterations in breast cancer during multimodal treatment and follow-up. Mol Oncol. 2017;11:1361–1379. doi: 10.1002/1878-0261.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao YP, Ruan CP, Wang H, Hu ZQ, Fang M, Gu X, Ji J, Zhao JY, Gao CF. Identification and assessment of new biomarkers for colorectal cancer with serum N-glycan profiling. Cancer. 2012;118:639–650. doi: 10.1002/cncr.26342. [DOI] [PubMed] [Google Scholar]
- 20.de Vroome SW, Holst S, Girondo MR, van der Burgt YEM, Mesker WE, Tollenaar RAEM, Wuhrer M. Serum N-glycome alterations in colorectal cancer associate with survival. Oncotarget. 2018;9:30610–30623. doi: 10.18632/oncotarget.25753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nassif AB, Shahin I, Attili I, Azzeh M, Shaalan K. Speech recognition using deep neural networks: a systematic review. IEEE Access. 2019;7:19143–19165. [Google Scholar]
- 22.Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM, Chang YC, Huang CS, Shen D, Chen CM. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci Rep. 2016;6:24454. doi: 10.1038/srep24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics. 2018;15:41–51. doi: 10.21873/cgp.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie J, Zhang L, Chen Z, Hu A, Liu S, Lu D, Xia Y, Qian J, Yang P, Shen H. Protein-protein correlations based variable dimension expansion algorithm for high efficient serum biomarker discovery. Anal Chim Acta. 2020;1119:25–34. doi: 10.1016/j.aca.2020.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Tyanova S, Albrechtsen R, Kronqvist P, Cox J, Mann M, Geiger T. Proteomic maps of breast cancer subtypes. Nat Commun. 2016;7:10259. doi: 10.1038/ncomms10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chocholova E, Bertok T, Jane E, Lorencova L, Holazova A, Belicka L, Belicky S, Mislovicova D, Vikartovska A, Imrich R, Kasak P, Tkac J. Glycomics meets artificial intelligence - potential of glycan analysis for identification of seropositive and seronegative rheumatoid arthritis patients revealed. Clin Chim Acta. 2018;481:49–55. doi: 10.1016/j.cca.2018.02.031. [DOI] [PubMed] [Google Scholar]
- 27.Wittenbecher C, Stambuk T, Kuxhaus O, Rudman N, Vuckovic F, Stambuk J, Schiborn C, Rahelic D, Dietrich S, Gornik O, Perola M, Boeing H, Schulze MB, Lauc G. Plasma N-glycans as emerging biomarkers of cardiometabolic risk: a prospective investigation in the EPIC-potsdam cohort study. Diabetes Care. 2020;43:661–668. doi: 10.2337/dc19-1507. [DOI] [PubMed] [Google Scholar]
- 28.Qin WJ, Zhang ZJ, Qin RH, Han J, Zhao R, Gu Y, Pan YQ, Gu JX, Ren SF. Providing Bionic Glycome as internal standards by glycan reducing and isotope labeling for reliable and simple quantitation of N-glycome based on MALDI-MS. Anal Chim Acta. 2019;1081:112–119. doi: 10.1016/j.aca.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Reiding KR, Blank D, Kuijper DM, Deelder AM, Wuhrer M. High-throughput profiling of protein N-glycosylation by MALDI-TOF-MS employing linkage-specific sialic acid esterification. Anal Chem. 2014;86:5784–5793. doi: 10.1021/ac500335t. [DOI] [PubMed] [Google Scholar]
- 30.Han J, Pan YQ, Qin WJ, Gu Y, Xu XY, Zhao R, Sha JC, Zhang RR, Gu JX, Ren SF. Quantitation of sex-specific serum N-glycome changes in expression level during mouse aging based on Bionic Glycome method. Exp Gerontol. 2020;141:111098. doi: 10.1016/j.exger.2020.111098. [DOI] [PubMed] [Google Scholar]
- 31.de Freitas Junior JC, Morgado-Diaz JA. The role of N-glycans in colorectal cancer progression: potential biomarkers and therapeutic applications. Oncotarget. 2016;7:19395–19413. doi: 10.18632/oncotarget.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Leoz ML, Young LJ, An HJ, Kronewitter SR, Kim J, Miyamoto S, Borowsky AD, Chew HK, Lebrilla CB. High-mannose glycans are elevated during breast cancer progression. Mol Cell Proteomics. 2011;10:M110.002717. doi: 10.1074/mcp.M110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wieczorek M, Braicu EI, Oliveira-Ferrer L, Sehouli J, Blanchard V. Immunoglobulin G subclass-specific glycosylation changes in primary epithelial ovarian cancer. Front Immunol. 2020;11:654. doi: 10.3389/fimmu.2020.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, Westhrin M, Bondt A, Wuhrer M, Standal T, Holst S. Serum protein N-glycosylation changes in multiple myeloma. Biochim Biophys Acta Gen Subj. 2019;1863:960–970. doi: 10.1016/j.bbagen.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Zhong AL, Qin RH, Qin WJ, Han J, Gu Y, Zhou L, Zhang HQ, Ren SF, Lu RQ, Guo L, Gu JX. Diagnostic significance of serum IgG galactosylation in CA19-9-negative pancreatic carcinoma patients. Front Oncol. 2019;9:114. doi: 10.3389/fonc.2019.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren SF, Zhang ZJ, Xu CJ, Guo L, Lu RQ, Sun YH, Guo JM, Qin RH, Qin WJ, Gu JX. Distribution of IgG galactosylation as a promising biomarker for cancer screening in multiple cancer types. Cell Res. 2016;26:963–966. doi: 10.1038/cr.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin W, Pei H, Qin R, Zhao R, Han J, Zhang Z, Dong K, Ren S, Gu J. Alteration of serum IgG galactosylation as a potential biomarker for diagnosis of neuroblastoma. J Cancer. 2018;9:906–913. doi: 10.7150/jca.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin R, Yang Y, Chen H, Qin W, Han J, Gu Y, Pan Y, Cheng X, Zhao J, Wang X, Ren S, Sun Y, Gu J. Prediction of neoadjuvant chemotherapeutic efficacy in patients with locally advanced gastric cancer by serum IgG glycomics profiling. Clin Proteomics. 2020;17:4. doi: 10.1186/s12014-020-9267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark DJ, Schnaubelt M, Hoti N, Hu Y, Zhou Y, Gooya M, Zhang H. Impact of increased FUT8 expression on the extracellular vesicle proteome in prostate cancer cells. J Proteome Res. 2020;19:2195–2205. doi: 10.1021/acs.jproteome.9b00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimoyama H, Shibata TK, Ito M, Oda T, Itoh T, Mukai M, Matsuya-Ogawa M, Adachi M, Murakami H, Nakayama T, Sugihara K, Itoh H, Suzuki T, Kanayama N. Partial silencing of fucosyltransferase 8 gene expression inhibits proliferation of Ishikawa cells, a cell line of endometrial cancer. Biochem Biophys Rep. 2020;22:100740. doi: 10.1016/j.bbrep.2020.100740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tada K, Ohta M, Hidano S, Watanabe K, Hirashita T, Oshima Y, Fujnaga A, Nakanuma H, Masuda T, Endo Y, Takeuchi Y, Iwashita Y, Kobayashi T, Inomata M. Fucosyltransferase 8 plays a crucial role in the invasion and metastasis of pancreatic ductal adenocarcinoma. Surg Today. 2020;50:767–777. doi: 10.1007/s00595-019-01953-z. [DOI] [PubMed] [Google Scholar]
- 42.Szajda SD, Snarska J, Puchalski Z, Zwierz K. Lysosomal exoglycosidases in serum and urine of patients with colon adenocarcinoma. Hepatogastroenterology. 2008;55:921–925. [PubMed] [Google Scholar]
- 43.Clerc F, Reiding KR, Jansen BC, Kammeijer GS, Bondt A, Wuhrer M. Human plasma protein N-glycosylation. Glycoconj J. 2016;33:309–343. doi: 10.1007/s10719-015-9626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozcan S, Barkauskas DA, Ruhaak LR, Torres J, Cooke CL, An HJ, Hua S, Williams CC, Dimapasoc LM, Kim JH, Camorlinga-Ponce M, Rocke D, Lebrilla CB, Solnick JV. Serum glycan signatures of gastric cancer. Cancer Prev Res (Phila) 2014;7:226–35. doi: 10.1158/1940-6207.CAPR-13-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arnold JN, Saldova R, Hamid UM, Rudd PM. Evaluation of the serum N-linked glycome for the diagnosis of cancer and chronic inflammation. Proteomics. 2008;8:3284–3293. doi: 10.1002/pmic.200800163. [DOI] [PubMed] [Google Scholar]
- 46.Weiz S, Wieczorek M, Schwedler C, Kaup M, Braicu EI, Sehouli J, Tauber R, Blanchard V. Acute-phase glycoprotein N-glycome of ovarian cancer patients analyzed by CE-LIF. Electrophoresis. 2016;37:1461–1467. doi: 10.1002/elps.201500518. [DOI] [PubMed] [Google Scholar]
- 47.Kim JH, Lee SH, Choi S, Kim U, Yeo IS, Kim SH, Oh MJ, Moon H, Lee J, Jeong S, Choi MG, Lee JH, Sohn TS, Bae JM, Kim S, Min YW, Lee H, Lee JH, Rhee PL, Kim JJ, Lee SJ, Kim ST, Lee J, Park SH, Park JO, Park YS, Lim HY, Kang WK, An HJ, Kim JH. Direct analysis of aberrant glycosylation on haptoglobin in patients with gastric cancer. Oncotarget. 2017;8:11094–11104. doi: 10.18632/oncotarget.14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colhoun HO, Treacy EP, MacMahon M, Rudd PM, Fitzgibbon M, O’Flaherty R, Stepien KM. Validation of an automated ultraperformance liquid chromatography IgG N-glycan analytical method applicable to classical galactosaemia. Ann Clin Biochem. 2018;55:593–603. doi: 10.1177/0004563218762957. [DOI] [PubMed] [Google Scholar]
- 49.Liu S, Cheng L, Fu Y, Liu BF, Liu X. Characterization of IgG N-glycome profile in colorectal cancer progression by MALDI-TOF-MS. J Proteomics. 2018;181:225–237. doi: 10.1016/j.jprot.2018.04.026. [DOI] [PubMed] [Google Scholar]
- 50.Russell AC, Simurina M, Garcia MT, Novokmet M, Wang Y, Rudan I, Campbell H, Lauc G, Thomas MG, Wang W. The N-glycosylation of immunoglobulin G as a novel biomarker of Parkinson’s disease. Glycobiology. 2017;27:501–510. doi: 10.1093/glycob/cwx022. [DOI] [PubMed] [Google Scholar]
- 51.Loberg M, Kalager M, Holme O, Hoff G, Adami HO, Bretthauer M. Long-term colorectal-cancer mortality after adenoma removal. N Engl J Med. 2014;371:799–807. doi: 10.1056/NEJMoa1315870. [DOI] [PubMed] [Google Scholar]
- 52.Winawer SJ, Fletcher RH, Miller L, Godlee F, Stolar MH, Mulrow CD, Woolf SH, Glick SN, Ganiats TG, Bond JH, Rosen L, Zapka JG, Olsen SJ, Giardiello FM, Sisk JE, Van Antwerp R, Brown-Davis C, Marciniak DA, Mayer RJ. Colorectal cancer screening: clinical guidelines and rationale. Gastroenterology. 1997;112:594–642. doi: 10.1053/gast.1997.v112.agast970594. [DOI] [PubMed] [Google Scholar]
- 53.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 54.Ahlquist DA, Taylor WR, Mahoney DW, Zou H, Domanico M, Thibodeau SN, Boardman LA, Berger BM, Lidgard GP. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol. 2012;10:272–277. e271. doi: 10.1016/j.cgh.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, Castaños-Vélez E, Blumenstein BA, Rösch T, Osborn N, Snover D, Day RW, Ransohoff DF PRESEPT Clinical Study Steering Committee, Investigators and Study Team. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317–325. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drake PM, Cho W, Li BS, Prakobphol A, Johansen E, Anderson NL, Regnier FE, Gibson BW, Fisher SJ. Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin Chem. 2010;56:223–236. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thirunavukarasu P, Sukumar S, Sathaiah M, Mahan M, Pragatheeshwar KD, Pingpank JF, Zeh H 3rd, Bartels CJ, Lee KK, Bartlett DL. C-stage in colon cancer: implications of carcinoembryonic antigen biomarker in staging, prognosis, and management. J Natl Cancer Inst. 2011;103:689–697. doi: 10.1093/jnci/djr078. [DOI] [PubMed] [Google Scholar]
- 58.Stiksma J, Grootendorst DC, van der Linden PW. CA19-9 as a marker in addition to CEA to monitor colorectal cancer. Clin Colorectal Cancer. 2014;13:239–244. doi: 10.1016/j.clcc.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 59.Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Cell surface protein glycosylation in cancer. Proteomics. 2014;14:525–546. doi: 10.1002/pmic.201300387. [DOI] [PubMed] [Google Scholar]
- 60.Duffy MJ, van Dalen A, Haglund C, Hansson L, Klapdor R, Lamerz R, Nilsson O, Sturgeon C, Topolcan O. Clinical utility of biochemical markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines. Eur J Cancer. 2003;39:718–727. doi: 10.1016/s0959-8049(02)00811-0. [DOI] [PubMed] [Google Scholar]
- 61.Acharya A, Markar SR, Matar M, Ni M, Hanna GB. Use of tumor markers in gastrointestinal cancers: surgeon perceptions and cost-benefit trade-off analysis. Ann Surg Oncol. 2017;24:1165–1173. doi: 10.1245/s10434-016-5717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.