Abstract
HP1BP3, an ubiquitously expressed nuclear protein belonging to the H1 histone family of proteins, plays an important role in cell growth and viability. Recently, it was reported that HP1BP3 exclusively regulates miRNA biogenesis by enhancing transcriptional miRNA processing. Although HP1BP3 has previously been implicated in common cancer types, the mechanistic functions and effects of HP1BP3 and its role in the prognosis of esophageal squamous cell carcinoma (ESCC) remain unclear. Here, we report that ESCC tissues and cell lines show increased endogenous expression of HP1BP3. Knockdown of HP1BP3 in TE-1 cells significantly inhibited tumor growth and metastasis in vivo emphasizing its role in cell proliferation and invasion. In contrast, overexpression of HP1BP3 significantly enhanced tumor growth and metastasis in Eca-109 cells. Further, we found that HP1BP3 regulates these functions by upregulating miR-23a, which directly binds to the 3’UTR region of TRAF5 downstream to alter cell survival and proliferation. Our findings describe a role for HP1BP3 in promoting tumor growth and metastasis by upregulating miR-23a to target TRAF5 in esophageal cancer. This study provides novel insights into the potential of targeting miRNAs for therapy and as clinical markers for cancer progression.
Keywords: HP1BP3, miR-23a, TRAF5, metastasis, esophageal cancer
Introduction
Esophageal squamous cell carcinoma (ESCC) and adenocarcinoma of the esophagus (AED) are two major types of cancers that affect the gut. ESCC is a multi-step process involving overexpression of cell cycle proteins such as Cyclin D, p53 inactivation and accumulation of genetic mutations [1]. The clinical prognosis of ESCC is not well understood due to its complexity, late presentation and accelerated tumor metastasis. To date it is classified as one of the deadliest cancers with an overall 5-year survival rate of 18% [2]. Transcriptome analysis has been performed on patient tissues to identify novel biomarkers to predict clinical incidence at earlier stages [3-6].
Histone protein complex is a group of proteins that play a role in structural packaging of DNA [7]. Of these, H1 histones are further classified into H1.1 to H1.5 and, H1.0 and H1.x, which are replication independent [8]. Modifications in Histone 1 have been associated with terminal differentiation of cells and promotion of neoplasticity in several types of cancers such as ovarian and colon cancer [9]. Histone modifiers such as, SMYD2 (histone methyltransferase) and KDM4C (histone demethylase), have been previously implicated in the progression of ESCC [10,11].
The nuclear protein, Heterochromatin Protein 1 Binding Protein 3 (HP1BP3), initially described as a novel protein binding to heterochromatin protein 1 (HP1), was later reported to belong to the family of H1 histones [12,13]. HP1BP3 maintains heterochromatin integrity and favors cell proliferation by prolongation of cell cycle phase G1 [14]. It also plays an important role in controlling cell viability and growth, as well as alteration of gene expression [13]. Incidences of HP1BP3 in cancer have been reported in thyroid and prostate cancers, and in certain types of gliomas [15]. HP1BP3 expression in tumors has also been related to acquisition of chemo-resistant traits that favor cell proliferation and tumor progression [16]. HP1BP3 retains most of the functions shared by its family of H1 histones such as chromatin binding, DNA packaging and intranuclear mobility [13]. It has also been shown to play a role in promoting early stages of development including body growth and bone development [17]. Interestingly, it has been reported that HP1BP3 negatively regulates the transcription of heat shock protein Hsp70 and that its expression is regulated by cell stress in Crassostrea hongkongensis [18]. While it is known that HP1BP3 alters gene expression, a recent study by Liu et al. reported that HP1BP3 plays a role in micro-RNA (miRNA) biogenesis by promoting co-transcriptional miRNA processing. HP1BP3 recruits the microprocessor Drosha-DGCR8 complex to miRNA transcription sites. This interferes with the processing but not transcription of several miRNAs including miR-23a [19].
The implications of miRNA in the prognosis of esophageal cancer have been largely described. miR-21, miR-133 and miRNA-138 among others in tissues and miR-21 and miR-233 in blood have been shown to have significant prognostic value [20]. Serum micro-RNA profiles from ESCC patients revealed an upregulation in the expression profile of miR-23a along with other miRNAs such as miR-1246 and miR-3202. miR-1246 was previously validated as a diagnostic and prognostic biomarker for ESCC [21]. miR-23a has been reported to be significantly upregulated in gastric cancer promoting tumor growth by inhibiting apoptosis through interference with translation of related proteins [22]. Further, it has been previously reported that miR-23a can be used as biomarker for chemoresistance in ESCC [23].
Transcriptome analysis of Hp1bp3 knockdown in microglia cells revealed a significant upregulation of immune-related genes involved in, for example, interferon or chemokine signaling [24]. We also previously performed a targeted search for downstream targets of miR-23a using the starBase database (School of Life Science, Sun Yat-sen University, China) that describes miRNA interactors and identified TRAF5 as a putative target [25]. TRAF5 belongs to the tumor necrosis factor receptor-associated factor (TRAF) protein family and is reported to play a role in immune cell activation [26]. miRNA targeting of TRAF5 promotes cell survival during hypoxia in cardiomyocytes [27]. In cancer, several miRNAs target TRAF5 and its implications have been described previously in colorectal cancer, gastric cancer and gliomas [28-30]. Ectopic expression of miR-873 inhibits NF-κB-mediated signaling by targeting TRAF5 in colorectal cancer cells [31]. In ESCC cell lines, miR-26b has been shown to target TRAF5 to promote cell proliferation by interfering with the cell cycle [32].
In this study, we show that HP1BP3 regulates miR-23a expression that selectively targets TRAF5, promoting cell survival, proliferation and metastasis in ESCC cell lines and tumors in vivo.
Material and methods
Patients and tissue samples
Patients involved in the study were recruited from Tong Ren Hospital, Shanghai Jiao Tong University School of Medicine (Shanghai, China) and Ruijin Hospital, Shanghai Jiao Tong University School of Medicine (Shanghai, China). Human ESCC and adjacent non-tumor esophageal tissue samples were collected from 10 patients at the time of surgery after proper informed consent was obtained from each patient enrolled in the study. Tissues were immediately stored at -80°C until use. 21 pairs of ESCC tumor and healthy adjacent samples were collected from patients undergoing esophagectomy and used for immunohistochemical analysis. This study was approved by the ethical committee of Tong Ren Hospital, Shanghai Jiao Tong University School of Medicine and Ruijin Hospital, Shanghai Jiao Tong University School of Medicine.
Cell lines and cell culture
Cell lines used in this study (HEEC, KYSE30, Eca-109, KYSE510, and TE-1) were purchased from the Cell Bank of Shanghai Institute of Cell Biology (Chinese Academy of Medical Sciences, Shanghai, China). KYSE30 and KYSE510 were maintained in RPMI 1640 (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Gaithersburg, MD, USA) and 100 U/ml penicillin/streptomycin. Eca-109, TE-1 and HEEC cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, CA, USA) with 10% FBS and 100 U/ml penicillin/streptomycin. All cell lines were grown under 5% CO2 at 37°C.
Immunohistochemistry
For immunohistochemistry, tissue samples were processed using a routine procedure of deparaffinization and rehydration using alcohol. The sections were treated with 3% hydrogen peroxide and blocked using 5% FBS. After blocking, the samples were incubated with either phosphate buffer saline (PBS) for the negative control or primary antibodies anti-HP1BP3 (1:200, Abcam, Cambridge, MA) or anti-TRAF5 (1:500, Abcam) overnight at 4°C. HRP-conjugated secondary antibodies were used followed by counterstaining with hematoxylin.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent and reverse transcription was performed using Quantscript RT kit (TIANGEN, Beijing, China) according to the manufacturer’s protocol. Samples were run on QuantStudio 6 (Applied Biosystems, Foster City, CA, USA). The primers used are as follows: HP1BP3, forward, 5’-CCACCTGCTACTTCGAGTGA-3’, reverse, 5’-ATCGGTGTTTGTTTCTGGGC-3’; GAPDH, forward, 5’-TCAAGAAGGTGGTGAAGCAGG-3’, reverse, 5’-TCAAAGGTGGAGGAGTGGGT-3’; hsa-miR-23a, stem-loop primer, 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCTAAAGG-3’, forward, 5’-TGCGCATCACATTGCCAGGGA-3’, reverse, 5’-CCAGTGCAGGGTCCGAGGTATT-3’; and U6, stem-loop primer, 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAATAT-3’, forward, 5’-CGCTTCGGCAGCACATATAC-3’, reverse, 5’-AAATATGGAACGCTTCACGA-3’.
Western blot
Cells or tissue samples were lysed using RIPA lysis buffer (Pierce Rockford, IL, USA) with freshly added protease inhibitors (1:1000). The total protein concentration was measured using a BCA protein assay kit (Thermo Fisher, Rockford, IL, USA). Lysates were mixed with loading buffer, run on a 10% SDS-polyacrylamide gel and blotted onto PVDF membranes. The membranes were blocked using 5% milk solution and incubated with primary anti-HP1BP3 (1:500, Abcam), anti-TRAF5 (1:1000, Abcam) or GAPDH (1:1000, Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C. After several washing steps, the membranes were incubated with their corresponding HRP-conjugated secondary antibodies for 30 min and developed using chemiluminescence.
Lentiviral and plasmid transfections
Recombinant lentiviral constructs for shRNA-mediated HP1BP3 knockdown and ectopic expression were constructed by GeneChem (Shanghai, China). The expression system contains the following vectors: pGC-LV, pHelper 2.0 containing VSV-G envelope and pHelper 1.0 containing gag/pol. The following shRNAs were cloned into the vector backbone GV118. Their target sequences are listed as follows (5’→3’): HP1BP3-sh1: GGACATTCCAGCTGAAGAAAT; and HP1BP3-sh2: GGTCCAAACCTGCACCTAAAG. A fragment encoding the human HP1BP3 sequence was amplified and cloned into vector GV342 using the restriction sites AgeI/NheI. The primers used for amplification are as follows: Forward, 5’-ATGGCGACTGATACGTCTCAA-3’, Reverse, 5’-GTCTTTCAGAGTGAAAAAGTAA-3’. Human TRAF5 CDS was cloned and inserted into the pcDNA3.1 plasmid (Invitrogen). miR-23a mimic, inhibitor, or the corresponding negative control were synthesized by GenePharma (Shanghai, China). Polybrene infections were carried out in TE-1 and Eca-109 cells using 2 ml of concentrated lentivirus containing shRNA or cDNA respectively, followed by puromycin selection after 2 weeks. Transfections were performed using LipofectamineTM2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol.
Luciferase reporter assay
The segment of TRAF5 3’UTR (TRAF5 3’UTR-Wt) containing putative miR-23a binding site was PCR amplified and cloned into the pmirGLO vector (Promega, Madison, WI, USA). The mutant TRAF5 3’UTR (TRAF5 3’UTR-Mut) were constructed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA). Wild-type or mutant TRAF5 constructs were cotransfected with miR-23a mimic, miR-23a inhibitor, or the corresponding negative control into Eca-109 cells using LipofectamineTM2000 reagent (Invitrogen). 48 hours post transfection cells were lysed and luciferase activity was measured using a Dual-luciferase Reporter Assay System (Promega) according to manufacturer’s protocol. Luciferase activity was normalized to Renilla luciferase activity.
MTT assay and colony formation assay
Cell viability was assessed by MTT assay. 5 × 103 cells were seeded in 96-well plates and incubated for 1 to 5 days respectively followed by staining with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) dye. Absorbance was measured at 570 nm using a spectrophotometer. Colony formation assays were performed in 6-well plates. 500 cells were seeded and incubated for 14 days. Cells were then fixed with 4% paraformaldehyde and stained with 1% crystal violet. Wells containing an average of 50 cells were counted.
Analysis of apoptosis
Annexin V/propidium iodide (PI) staining was used to determine the frequency of apoptotic cells. Briefly, cells were plated in 6-well plates at a density of 1 × 105 cells per well. Apoptosis was induced by starving the cells of serum for 24 h. The cells were collected by centrifugation, washed with cold PBS, then stained with Annexin V-FITC and PI (Keygentec, Nanjing, China) according to the manufacturer’s instructions. Apoptotic cells were detected using a flow cytometer (CytoFLEX, Beckman Coulter, Brea, CA, USA).
Cell migration and invasion
Cell migration and invasion assays were performed using a transwell system (Becton Dickinson, USA) of 8 mm-pore size membrane filter. The membranes were coated with Matrigel for invasion assays. 1 × 104 cells were resuspended in 200 ml medium (serum-free) and added to the upper chamber of the transwell. The lower chamber was filled with 500 ml of medium containing 10% FBS. The chambers were incubated for 24 hours at 37°C in an atmosphere of 5% CO2. The upper chamber was removed and the migrated fraction of cells in the lower chamber were fixed with methanol and stained with 0.2% crystal violet.
Animal experiments
Four-week old female BALB/c nu/nu mice (n = 6 per group) were used for the subcutaneous xenograft and tail vein metastasis models. For the subcutaneous xenograft model, 2 × 106 ESCC cells were subcutaneously injected into the flanks. The incidence of xenografts and their volume were monitored routinely. The tumor volume was calculated using the formula (cm3) - L × W2 × 0.5 where L and W represents the largest and smallest diameters. The animals were euthanized to harvest the xenografts. For the tail vein metastasis models, 5-6 week old Balb/c nude mice were injected with infected cells through the tail vein. After seven weeks the lungs were harvested. All tissues were immediately fixed and stained with hematoxylin-eosin for analyzing metastasis. All animal experiments were approved by Tong Ren Hospital, Shanghai Jiao Tong University School of Medicine.
Statistical analysis
All statistical analysis was performed using GraphPad Prism 6.0 (GraphPad Software, Inc.). Statistical analysis of the differences between 2 groups was calculated using the Student t-test, and one-way ANOVA was used to analyze the differences among multiple groups. Statistical significance was defined as P < 0.05. Error bars represent SEM of the data.
Results
High HP1BP3 expression in ESCC tissues
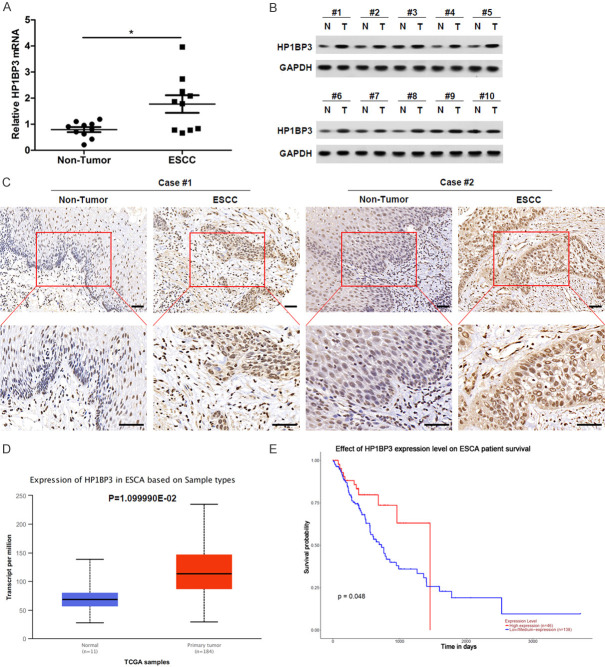
To determine the role of HP1BP3 in ESCC, we first quantified the levels of HP1BP3 mRNA in 10 tumor tissues and compared them with HP1BP3 levels in adjacent non-tumorous tissues using qRT-PCR. HP1BP3 mRNA expression levels were significantly increased in ESCC tissues compared to adjacent non-tumorous tissues (Figure 1A). Furthermore, a consistent increase in HP1BP3 protein levels was observed in tumor samples when compared to their respective adjacent non-tumorous tissues (Figure 1B). Immunohistochemistry on ESCC and adjacent non-tumorous tissues revealed a strong expression of HP1BP3 in ESCC tumor tissues compared to adjacent non-tumorous tissues (Figure 1C). The expression of HP1BP3 in esophageal carcinoma (ESCA) was further validated using the TCGA BCA dataset through the UALCAN web portal (http://ualcan.path.uab.edu/). The data show that the expression of HP1BP3 in ESCA tissues was significantly higher than that of normal tissues (Figure 1D). In addition, Kaplan-Meier curves obtained from the UALCAN web portal reveal a significant increase in the survival rate of patients expressing low levels of HP1BP3 levels (Figure 1E). These results indicate that HP1BP3 may play a role in ESCC.
Figure 1.
HP1BP3 is upregulated in ESCC tissues. A. HP1BP3 mRNA levels were examined in 10 pairs of ESCC tissues and the corresponding adjacent non-tumor tissues. *P < 0.05. B. Western blot showing endogenous protein levels of HP1BP3 in 10 pairs of non-tumor (N) and ESCC (T) tissue samples. C. Representative IHC staining of HP1BP3 in 21 ESCC paraffin-embedding tissues on tissue microarrays. Scale bar = 50 μm. D. The expression of HP1BP3 in esophageal carcinoma (ESCA) from UALCAN based on the TCGA database. E. Kaplan-Meier analysis of overall survival in relation to HP1BP3 expression in ESCA from UALCAN.
HP1BP3 promotes survival and invasion in ESCC cells
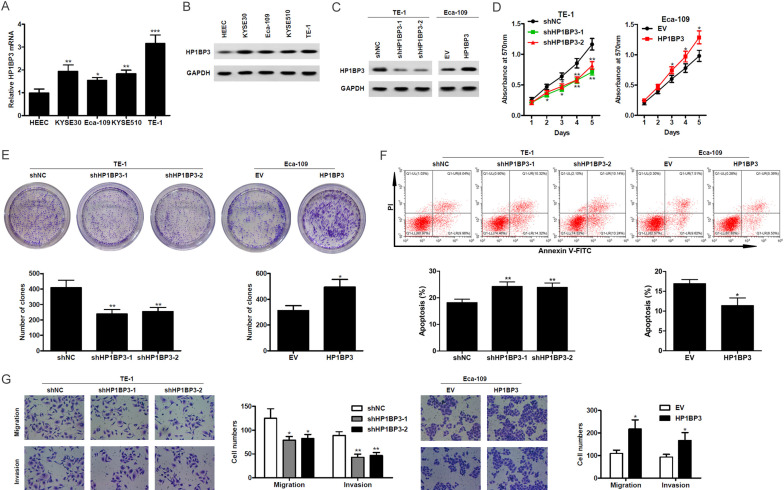
To further study the role of HP1BP3 in ESCC, we used the human normal esophageal epithelial cell line HEEC, as well as several ESCC cell lines (KYSE30, Eca-109, KYSE510, and TE-1). Endogenous mRNA and protein levels of HP1BP3 were quantified by qRT-PCR and western blot. Higher HP1BP3 mRNA and protein expression were observed in ESCC cell lines compared to HEEC cells (Figure 2A and 2B). Next, we performed shRNA knockdown using two different targets against HP1BP3 in TE-1 cells. Reduced endogenous HP1BP3 protein levels were observed upon shRNA treatment (Figure 2C). We also overexpressed HP1BP3 in Eca-109 and verified increased expression at the protein level (Figure 2C). The effect of HP1BP3 on cell viability was measured using an MTT assay. Cell viability decreased over time in HP1BP3 knockdown TE-1 cells compared to non-targeting control cells. In contrast, increased cell viability was observed in HP1BP3-overexpressing Eca-109 cells (Figure 2D). The colony formation assay also showed a decreased number of clones in HP1BP3 knockdown TE-1 cells and an increase in HP1BP3-overexpressing Eca-109 cells (Figure 2E). The effects of HP1BP3 knockdown and overexpression on apoptosis were assessed using Annexin V-PI staining. The frequency of apoptotic TE-1 cells was significantly increased after HP1BP3 knockdown, while HP1BP3 overexpression in Eca-109 cells led to a significant decrease in apoptosis (Figure 2F). To evaluate the role of HP1BP3 in migration and invasion of cells, transwell assays were performed. Upon knockdown of HP1BP3 in TE-1 cells, migration and invasion were significantly reduced as evaluated by the number of cells in the lower chamber of the transwell after 24 hours of incubation (Figure 2G). Upon HP1BP3 overexpression, Eca-109 cells showed significantly higher migration and invasion capabilities compared to cells transfected with an empty-vector control (Figure 2G). Taken together, these findings indicate that HP1BP3 plays a critical role in promoting cell survival, migration and invasion.
Figure 2.
HP1BP3 promotes cell proliferation, migration and invasion, and inhibits apoptosis in vitro in ESCC cell lines. A. Relative mRNA levels of HP1BP3 in HEEC and ESCC cell lines. B. Protein levels of HP1BP3 determined by western blot in HEEC and ESCC cell lines. C. Expression of HP1BP3 in TE-1 cells and stably expressing either non-targeting shRNA control (shNC) or shRNA knockdown of HP1BP3, and in Eca-109 cells stably expressing either HP1BP3 overexpression construct or control vector construct (EV). D. Cell viability measured by MTT assay in HP1BP3-silenced TE-1 cells or HP1BP3-overexpressing Eca-109 cells. E. Representative images and quantification of the number of colonies determined by colony formation assay upon overexpression or knockdown of HP1BP3. F. Apoptosis was induced in HP1BP3-silenced TE-1 cells or HP1BP3-overexpressing Eca-109 cells by serum starvation for 24 hrs. The percentage of apoptotic cells was determined by flow cytometry using annexin V-FITC and propidium iodide (PI) staining. G. Representative images of crystal violet stained cells after transwell migration or Matrigel invasion. Histogram depicting the quantification of cell numbers for each condition. All cell assays were performed in triplicates. *P < 0.05, **P < 0.01, ***P < 0.001.
HP1BP3 upregulates miR-23a expression in ESCC cells
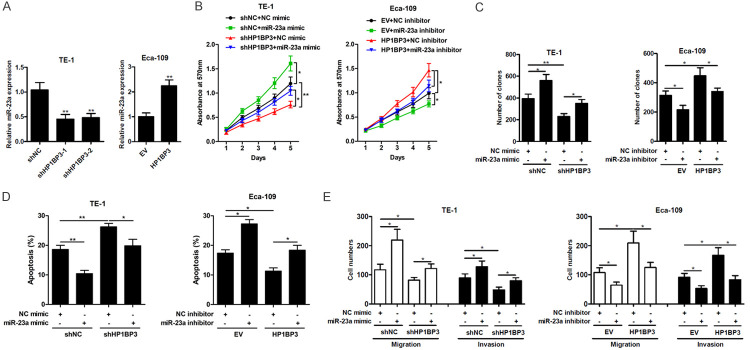
Since it has been reported that HP1BP3 regulates miRNA biogenesis [19], we investigated the expression of miR-23a upon knockdown and overexpression of HP1BP3. In TE-1 cells, knockdown of HP1BP3 achieved a 50% reduction in miR-23a expression while in Eca-109 cells, overexpression of HP1BP3 increased miR-23a expression by 2-fold (Figure 3A), indicating that HP1BP3 positively regulates miR-23a in ESCC cells. To determine whether HP1BP3 promotes survival and invasion in ESCC cells through the induction of miR-23a, a miR-23a mimic or miR-23a inhibitor were transfected into HP1BP3-knockdown TE-1 cells or HP1BP3-overexpressing Eca-109 cells, respectively. As shown in Figure 3B, decreased cell viability in the shHP1BP3-treated TE-1 cells could be reversed by introduction of the miR-23a mimic, while increased cell viability in the HP1BP3-overexpressing Eca-109 cells was reversed in the presence of the miR-23a inhibitor. Similar results were observed in the colony formation assay (Figure 3C). In contrast, increased levels of apoptosis observed in the shHP1BP3-treated TE-1 cells were reduced in the presence of the miR-23a mimic, while the decreased levels of apoptosis in HP1BP3-overexpressing Eca-109 cells were reversed in the presence of the miR-23a mimic (Figure 3D). Furthermore, introduction of the miR-23a mimic and miR-23a inhibitor largely abrogated the suppression and promotion of shHP1BP3 on TE-1 cells and HP1BP3 overexpression on Eca-109 cells migration and invasion, respectively (Figure 3E). Taken together, these results suggest that miR-23a mediates the effects of HP1BP3 on cell viability, proliferation, migration and invasion.
Figure 3.
HP1BP3 upregulates miR-23a expression in ESCC cells. A. qRT-PCR analysis of miR-23a expression in cells with HP1BP3 overexpression or knockdown. B. TE-1 cells with HP1BP3 knockdown were transfected with miR-23a mimic, Eca-109 cells with HP1BP3 overexpression were transfected with miR-23a inhibitor. MTT assay for viability of cells indicated by absorbance at 570 nm. C. Quantification of number of colonies indicating cell proliferation determined by colony formation assay. D. Apoptosis was induced in HP1BP3-silenced TE-1 cells (treated with miR-23a mimic) or HP1BP3-overexpressing Eca-109 cells (treated with miR-23a inhibitor) by serum starvation for 24 hrs. The percentage of apoptotic cells was determined by flow cytometry using annexin V-FITC and propidium iodide (PI) staining. E. Quantification of migration and invasion ability analyzed by transwell migration and invasion assays, respectively. *P < 0.05, **P < 0.01, ***P < 0.001.
miR-23a directly targets and inhibits expression of TRAF5
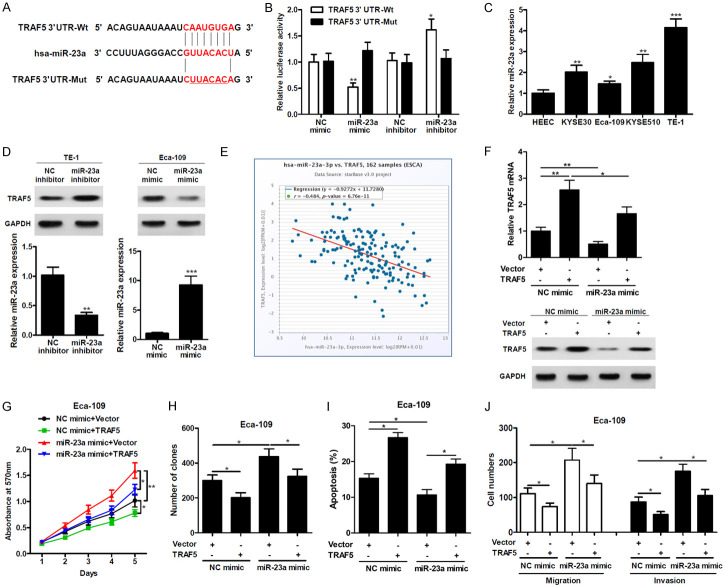
To identify the mechanism by which miR-23a regulates cell viability and proliferation, we investigated a downstream target TRAF5, that was identified using starBase. Sequence comparison revealed a potential miR-23a binding site on the 3’UTR region of TRAF5 (Figure 4A). The mutated version of the construct, which is insensitive to miR-23a targeting is also shown in Figure 4A. Luciferase reporter assays showed that miR-23a expression in Eca-109 cells significantly decreased the luciferase reporter activity of TRAF5 3’UTR-Wt expressing cells, but had no effect on TRAF5 3’UTR-Mut expressing cells (Figure 4B). Relative miR-23a expression levels in the HEEC and other ESCC cell lines were quantified by qRT-PCR. TE-1 cells had the highest expression with a 4-fold increase in miR-23a expression compared to HEEC cells (Figure 4C). TRAF5 protein expression levels were increased in TE-1 cells transfected with the miR-23a inhibitor and decreased in Eca-109 cells transfected with the miR-23a mimic (Figure 4D). Next, we examined the correlation between TRAF5 and miR-23a expression levels from data obtained from starBase of 162 esophageal carcinoma samples. An inverse correlation was observed showing decreased TRAF5 expression with increased miR-23a expression, validating our results that miR-23a directly targets TRAF5 (Figure 4E). We also co-transfected Eca-109 cells with TRAF5 lacking its 3’UTR expression vector and miR-23a mimic or its relevant control. miR-23a decreased endogenous expression of TRAF5, which was overcome when TRAF5 was exogenously introduced into the cells (Figure 4F). The MTT and colony formation assays showed increased cell viability and proliferation in Eca-109 cells when treated with miR-23a. This effect was largely abrogated when TRAF5 was overexpressed (Figure 4G and 4H). While miR-23a reduced the level of apoptosis in Eca-109 cells, overexpression of TRAF5 reversed this effect (Figure 4I). These findings suggest that miR-23a mediated regulation of TRAF5 has a role in cell viability, proliferation and apoptosis. Finally, a TRAF5-mediated reduction in migration and invasion was observed in Eca-109 cells in the presence of the miR-23a mimic compared to control cells (Figure 4J). Together, these results indicate that miR-23a regulates TRAF5 expression to modulate cell viability, proliferation, apoptosis, migration and invasion.
Figure 4.
miR-23a directly targets TRAF5. A. Sequence analysis of 3’UTR of TRAF5 mRNA, miR-23a and the mutant. B. Luciferase activity of wild-type TRAF5-3’UTR (TRAF5 3’UTR-Wt) and mutant-type TRAF5-3’UTR (TRAF5 3’UTR-Mut) reporter genes were measured using the Dual-Luciferase Reporter Assay System in Eca-109 cells transfected with miR-23a mimic or inhibitor. C. Relative miR-23a expression was examined in ESCC cells by qRT-PCR. D. Western blotting analysis of protein levels of TRAF5 (upper) and qRT-PCR analysis of miR-23a (lower) in TE-1 cells or Eca-109 cells treated with miR-23a inhibitor or mimic, respectively. E. Relationship between expression levels of miR-23a and TRAF5 mRNA in 162 Esophageal carcinoma (ESCA) samples obtained from the starBase database. F. qRT-PCR and Western blot analysis of TRAF5 expression in Eca-109 cells co-transfected with miR-23a and TRAF5. G. MTT analysis of cell viability in Eca-109 cells co-transfected with miR-23a and TRAF5. H. Colony formation of Eca-109 cells co-transfected with miR-23a and TRAF5. I. Apoptosis was induced in Eca-109 cells (in the presence of miR-23a mimic and/or TRAF5) by serum starvation for 24 hrs. The percentage of apoptotic cells was determined by flow cytometry using annexin V-FITC and propidium iodide (PI) staining. J. Migration or invasion ability was analyzed in Eca-109 cells co-transfected with miR-23a and TRAF5. *P < 0.05, **P < 0.01, ***P < 0.001.
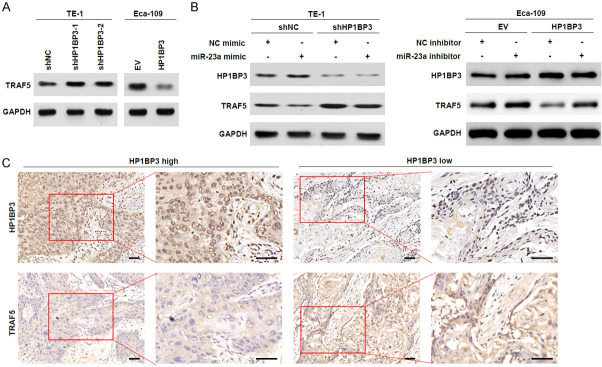
HP1BP3 inhibits TRAF5 expression by upregulating miR-23a
To identify how HP1BP3 regulates miR-23a and TRAF5, we quantified endogenous TRAF5 levels upon knockdown and overexpression of HP1BP3. Knockdown of HP1BP3 in TE-1 cells increased the expression of TRAF5 compared to non-targeting control cells (Figure 5A). The increase in TRAF5 expression upon knockdown of HP1BP3 is reduced when the cells are treated with miR-23a mimic (Figure 5B). In Eca-109 cells, overexpression of HP1BP3 decreased TRAF5 levels, which were rescued when treated with an inhibitor against miR-23a (Figure 5B). Immunohistochemistry analysis of HP1BP3 and TRAF5 expression in high and low HP1BP3-expressing ESCC tissues revealed an inverse correlation between HP1BP3 and TRAF5 expression. Tissues expressing high HP1BP3 levels showed very low TRAF expression, while tissues expressing low HP1BP3 levels had high TRAF expression (Figure 5C). Thus, our data suggest that HP1BP3 reduces TRAF5 expression by increasing the expression of miR-23a.
Figure 5.
HP1BP3 inhibits TRAF5 expression by upregulating miR-23a. A. Western blotting analysis of TRAF5 protein levels in TE-1 cells with HP1BP3 knockdown and Eca-109 cells with HP1BP3 overexpression. B. Western blotting analysis of TRAF5 protein levels in HP1BP3 knockdown TE-1 cells transfected with miR-23a mimic and Eca-109 cells with HP1BP3 overexpression transfected with miR-23a inhibitor. C. Immunohistochemistry to determine protein expression levels of HP1BP3 and TRAF5 in primary human ESCC patient samples. Scale bar = 50 μm.
HP1BP3 promotes esophageal cancer cell growth and metastasis in vivo
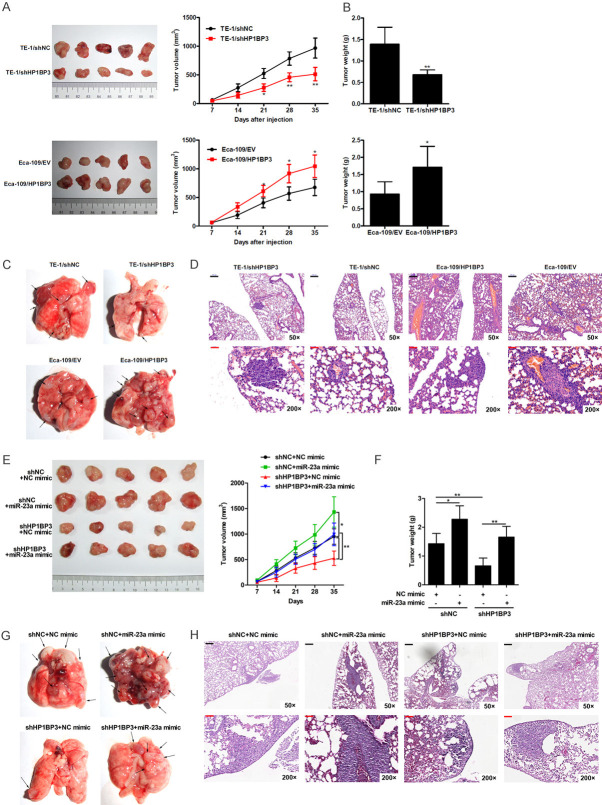
To study the relevance of HP1BP3 expression in vivo, HP1BP3 knockdown TE-1 cells and HP1BP3-overexpressing Eca-109 cells were subcutaneously injected into the right flanks of BALB/c nu/nu mice. The tumor volume was measured every week for five weeks. A significant decrease in tumor volume was observed upon knockdown of HP1BP3, whereas in mice injected with HP1BP3-overexpressing Eca-109 cells, a significant increase in tumor volume was observed (Figure 6A). Similarly, a decrease in tumor weight was observed in mice injected with HP1BP3-knockdown TE-1 cells, while an increase in tumor weight was found in mice injected with HP1BP3-overexpressing Eca-109 cells (Figure 6B). We also found that the number of lung metastatic nodules and size were significantly decreased in the TE-1/shHP1BP3 group, but increased in the Eca-109/HP1BP3 group (Figure 6C, 6D). These findings indicate that HP1BP3 plays an important role in tumor progression in vivo.
Figure 6.
HP1BP3 promotes tumor growth and metastasis in mouse model. A. HP1BP3 knockdown TE-1 cells and HP1BP3 overexpression Eca-109 cells were injected into the right flanks of BALB/c nu/nu mice and monitored for five weeks. The tumor volume was measured every week. B. Averages of tumor weight were calculated at five weeks. C. Representative images of lung tissues with metastatic nodules indicated by arrows are shown. D. Representative images of H&E staining of paraffin-embedded sections of lung metastatic nodules. Black Scale bar = 200 μm, Red Scale bar = 50 μm. E. HP1BP3 knockdown TE-1 cells transfected with miR-23a mimic were injected into the right flanks of BALB/c nu/nu mice and monitored for five weeks. The tumor volume was measured every week. F. Averages of tumor weight were calculated at five weeks post-injection. G. Representative images of lung tissues with metastatic nodules indicated by arrows are shown. H. Representative images of H&E staining of paraffin-embedded sections of lung metastatic nodules. Black Scale bar = 200 μm, Red Scale bar = 50 μm. *P < 0.05, **P < 0.01.
The effects of miR-23a mimic on tumor growth were also studied in control and HP1BP3 knockdown tumors. miR-23a led to an increase in tumor volume and weight in control tumors and partially rescued the tumor growth inhibition induced by shHP1BP3 knockdown (Figure 6E, 6F). In addition, the miR-23a mimic promoted lung metastasis in control tumors, as revealed by elevated numbers and sizes of metastatic nodules (Figure 6G, 6H). Furthermore, miR-23a mimic partially reversed the HP1BP3 knockdown-mediated inhibition of tumor metastasis (Figure 6G, 6H). These findings indicate that the HP1BP3/miR-23a axis plays an important role in tumor progression in vivo.
Discussion
The chromatin binding protein HP1BP3 has been described previously for its role in several types of cancers [19]. In this study, we identify the role of HP1BP3 in ESCC by promoting cell survival and metastasis. We evaluated HP1BP3 in primary tumor tissues which showed a significant upregulation in mRNA and protein levels compared to adjacent non-tumorous tissues. We also showed that HP1BP3 expression was significantly increased in ESCC cell lines. Knockdown of HP1BP3 by shRNA targeting reduced viability in TE-1 cells and overexpression of HP1BP3 in Eca-109 cells promoted cell viability. Further, we confirmed the pro-survival role of HP1BP3 in ESCC cell lines by observing an increase in cell colonies by colony formation assays upon HP1BP3 overexpression and increased invasion and migration capabilities of these cells.
MicroRNAs have been extensively characterized for their role in regulating cell survival in the context of different types of cancers. A global miRNA screen in tumor tissues from patients revealed miR-1246 and miR-23a to be key factors differentially expressed in colorectal and pancreatic tumors, respectively [33]. miR-23a has also been reported to suppress tumor growth in oral squamous cell carcinoma and bladder cancer [34,35]. miR-23a targets the mitotic protein PLK-1 to interfere with the cell cycle and DNA replication in pancreatic cancer [36]. miR-23a was also shown to promote angiogenesis under hypoxic environments commonly observed in tumors [37]. In serum samples from ESCC patients, miR-23a was reported to be significantly upregulated [21]. In another study in serum from ESCC patients being treated with neoadjuvant chemotherapy, miR-23a-5p was upregulated in a group of non-responders [38]. Here, we found that upon knockdown of HP1BP3 there was a significant downregulation of miR-23a expression levels while HP1BP3 overexpression increased miR-23a levels in TE-1 and Eca-109 cells, respectively. Further, we used a miR-23a mimic to overexpress miR-23a and observed an increase in cell viability, which was also associated with cell proliferation, invasion and migration. This effect was reduced upon knockdown of HP1BP3. These results suggest that HP1BP3 promotes miR-23a expression and cell survival. However, the precise mechanisms of regulation are yet to be determined.
Further, we identified TRAF5 as a potential target of miR-23a and functionally tested its regulation. The TRAF family of proteins is prone to genetic alterations in different types of cancers. Of these, TRAF5 modifications are the highest in breast cancer followed by liver, uterine and esophageal cancer [39]. TRAF5 has been previously reported to have a role in promoting colorectal cancer by favoring cell survival and is regulated by miR-141-3p [28]. miR-141-3p acts as a negative regulator of prostate and colorectal cancer, and inhibits the NF-κB pathway via TRAF5, thereby interfering with cell migration and proliferation [28,40]. Using wild-type TRAF5 and a mutant version that is resistant to miR-23a in our reporter assays with Eca-109 cells revealed that TRAF5 is a direct downstream target of miR-23a. TRAF5 expression reduced cell viability and this effect was counteracted upon treatment with the miR-23a mimic in Eca-109 cells. Hence, miR-23a promotes cell viability, proliferation and migration by directly targeting TRAF5. Protein expression assays revealed that upon knockdown of HP1BP3, TRAF5 levels were increased indicating that HP1BP3 regulates TRAF5 expression and in-turn cell survival by increasing the expression of miR-23a.
Our in vitro analysis was further confirmed by performing tumor monitoring studies in vivo in mice models. Although an ideal mouse model is not available for esophageal cancer, subcutaneous xenograft models and more recently the tail vein model are commonly used [41]. Subcutaneous injection of ESCC cell lines with overexpression or knockdown of HP1BP3 favored tumor progression or inhibited tumor metastasis, respectively. Critically, addition of a miR-23a mimic promoted tumor growth and metastasis in the absence or presence of HP1BP3, demonstrating that miR23a acts downstream of HP1BP3 in tumors. Taken together, our study establishes the role of HP1BP3 in regulating miRNA levels of miR-23a, and in-turn TRAF5 expression to promote ESCC progression. Further studies on the mechanism of HP1BP3 regulation of miR-23a levels and possibly other miRNAs could lead to the identification of novel therapies and biomarkers for ESCC.
Acknowledgements
This study was supported by the grant from National Natural Science Foundation of China (81571775, 82001931), Shanghai Key Medical Specialty Program (ZK2019A16), and Multi-center Clinical Research Project of Shanghai Jiao Tong University School of Medicine (DLY201828).
Disclosure of conflict of interest
None.
References
- 1.Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY, Zhao YD, Sun J, Zhou CC, Yao R, Wang SY, Wang P, Sun N, Zhang BH, Dong JS, Yu Y, Luo M, Feng XL, Shi SS, Zhou F, Tan FW, Qiu B, Li N, Shao K, Zhang LJ, Zhang LJ, Xue Q, Gao SG, He J. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46:1097–1102. doi: 10.1038/ng.3076. [DOI] [PubMed] [Google Scholar]
- 2.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, Wolfe C, Hamadeh RR, Moore A, Werdecker A, Gessner BD, Te Ao B, McMahon B, Karimkhani C, Yu C, Cooke GS, Schwebel DC, Carpenter DO, Pereira DM, Nash D, Kazi DS, De Leo D, Plass D, Ukwaja KN, Thurston GD, Yun Jin K, Simard EP, Mills E, Park EK, Catala-Lopez F, deVeber G, Gotay C, Khan G, Hosgood HD 3rd, Santos IS, Leasher JL, Singh J, Leigh J, Jonas JB, Sanabria J, Beardsley J, Jacobsen KH, Takahashi K, Franklin RC, Ronfani L, Montico M, Naldi L, Tonelli M, Geleijnse J, Petzold M, Shrime MG, Younis M, Yonemoto N, Breitborde N, Yip P, Pourmalek F, Lotufo PA, Esteghamati A, Hankey GJ, Ali R, Lunevicius R, Malekzadeh R, Dellavalle R, Weintraub R, Lucas R, Hay R, Rojas-Rueda D, Westerman R, Sepanlou SG, Nolte S, Patten S, Weichenthal S, Abera SF, Fereshtehnejad SM, Shiue I, Driscoll T, Vasankari T, Alsharif U, Rahimi-Movaghar V, Vlassov VV, Marcenes WS, Mekonnen W, Melaku YA, Yano Y, Artaman A, Campos I, MacLachlan J, Mueller U, Kim D, Trillini M, Eshrati B, Williams HC, Shibuya K, Dandona R, Murthy K, Cowie B, Amare AT, Antonio CA, Castaneda-Orjuela C, van Gool CH, Violante F, Oh IH, Deribe K, Soreide K, Knibbs L, Kereselidze M, Green M, Cardenas R, Roy N, Tillmann T, Li Y, Krueger H, Monasta L, Dey S, Sheikhbahaei S, Hafezi-Nejad N, Kumar GA, Sreeramareddy CT, Dandona L, Wang H, Vollset SE, Mokdad A, Salomon JA, Lozano R, Vos T, Forouzanfar M, Lopez A, Murray C, Naghavi M. The global burden of cancer 2013. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau MC, Ng KY, Wong TL, Tong M, Lee TK, Ming XY, Law S, Lee NP, Cheung AL, Qin YR, Chan KW, Ning W, Guan XY, Ma S. FSTL1 promotes metastasis and chemoresistance in esophageal squamous cell carcinoma through NFkappaB-BMP signaling cross-talk. Cancer Res. 2017;77:5886–5899. doi: 10.1158/0008-5472.CAN-17-1411. [DOI] [PubMed] [Google Scholar]
- 4.Ma S, Bao JYJ, Kwan PS, Chan YP, Tong CM, Fu L, Zhang N, Tong AHY, Qin YR, Tsao SW, Chan KW, Lok S, Guan XY. Identification of PTK6, via RNA sequencing analysis, as a suppressor of esophageal squamous cell carcinoma. Gastroenterology. 2012;143:675–686. e612. doi: 10.1053/j.gastro.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Tong M, Chan KW, Bao JY, Wong KY, Chen JN, Kwan PS, Tang KH, Fu L, Qin YR, Lok S, Guan XY, Ma S. Rab25 is a tumor suppressor gene with antiangiogenic and anti-invasive activities in esophageal squamous cell carcinoma. Cancer Res. 2012;72:6024–6035. doi: 10.1158/0008-5472.CAN-12-1269. [DOI] [PubMed] [Google Scholar]
- 6.Tang KH, Dai YD, Tong M, Chan YP, Kwan PS, Fu L, Qin YR, Tsao SW, Lung HL, Lung ML, Tong DK, Law S, Chan KW, Ma S, Guan XY. A CD90(+) tumor-initiating cell population with an aggressive signature and metastatic capacity in esophageal cancer. Cancer Res. 2013;73:2322–2332. doi: 10.1158/0008-5472.CAN-12-2991. [DOI] [PubMed] [Google Scholar]
- 7.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 8.Millan-Arino L, Izquierdo-Bouldstridge A, Jordan A. Specificities and genomic distribution of somatic mammalian histone H1 subtypes. Biochim Biophys Acta. 2016;1859:510–519. doi: 10.1016/j.bbagrm.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Scaffidi P. Histone H1 alterations in cancer. Biochim Biophys Acta. 2016;1859:533–539. doi: 10.1016/j.bbagrm.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Komatsu S, Imoto I, Tsuda H, Kozaki KI, Muramatsu T, Shimada Y, Aiko S, Yoshizumi Y, Ichikawa D, Otsuji E, Inazawa J. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis. 2009;30:1139–1146. doi: 10.1093/carcin/bgp116. [DOI] [PubMed] [Google Scholar]
- 11.Yang ZQ, Imoto I, Fukuda Y, Pimkhaokham A, Shimada Y, Imamura M, Sugano S, Nakamura Y, Inazawa J. Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines. Cancer Res. 2000;60:4735–4739. [PubMed] [Google Scholar]
- 12.Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 13.Garfinkel BP, Melamed-Book N, Anuka E, Bustin M, Orly J. HP1BP3 is a novel histone H1 related protein with essential roles in viability and growth. Nucleic Acids Res. 2015;43:2074–2090. doi: 10.1093/nar/gkv089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta B, Ren Y, Hao P, Sim KH, Cheow E, Adav S, Tam JP, Sze SK. Profiling of the chromatin-associated proteome identifies HP1BP3 as a novel regulator of cell cycle progression. Mol Cell Proteomics. 2014;13:2183–2197. doi: 10.1074/mcp.M113.034975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, Fu XD, Zhang Y. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutta B, Yan R, Lim SK, Tam JP, Sze SK. Quantitative profiling of chromatome dynamics reveals a novel role for HP1BP3 in hypoxia-induced oncogenesis. Mol Cell Proteomics. 2014;13:3236–3249. doi: 10.1074/mcp.M114.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garfinkel BP, Arad S, Le PT, Bustin M, Rosen CJ, Gabet Y, Orly J. Proportionate dwarfism in mice lacking heterochromatin protein 1 binding protein 3 (HP1BP3) is associated with alterations in the endocrine IGF-1 pathway. Endocrinology. 2015;156:4558–4570. doi: 10.1210/en.2015-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu D, Yang Q, Cui M, Zhang Q. The novel transcriptional factor HP1BP3 negatively regulates Hsp70 transcription in Crassostrea hongkongensis. Sci Rep. 2017;7:1401. doi: 10.1038/s41598-017-01573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, Liang C, Kollipara RK, Matsui M, Ke X, Jeong BC, Wang Z, Yoo KS, Yadav GP, Kinch LN, Grishin NV, Nam Y, Corey DR, Kittler R, Liu Q. HP1BP3, a chromatin retention factor for co-transcriptional microRNA processing. Mol Cell. 2016;63:420–432. doi: 10.1016/j.molcel.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao S, Zhao ZY, Zhang ZY, Zhang Y, Wu R. Prognostic value of microRNAs in esophageal carcinoma: a meta-analysis. Clin Transl Gastroenterol. 2018;9:203. doi: 10.1038/s41424-018-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, Komatsu A, Jitsukawa M, Matsubara H. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:644–652. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu X, Wang Y, Liang H, Fan Q, Zhu R, Cui J, Zhang W, Zen K, Zhang CY, Hou D, Zhou Z, Chen X. miR-23a/b promote tumor growth and suppress apoptosis by targeting PDCD4 in gastric cancer. Cell Death Dis. 2017;8:e3059. doi: 10.1038/cddis.2017.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komatsu S, Ichikawa D, Kawaguchi T, Takeshita H, Miyamae M, Ohashi T, Okajima W, Imamura T, Kiuchi J, Arita T, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Otsuji E. Plasma microRNA profiles: identification of miR-23a as a novel biomarker for chemoresistance in esophageal squamous cell carcinoma. Oncotarget. 2016;7:62034–62048. doi: 10.18632/oncotarget.11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuner SM, Ding S, Kaczorowski CC. Knockdown of heterochromatin protein 1 binding protein 3 recapitulates phenotypic, cellular, and molecular features of aging. Aging Cell. 2019;18:e12886. doi: 10.1111/acel.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraus ZJ, Nakano H, Bishop GA. TRAF5 is a critical mediator of in vitro signals and in vivo functions of LMP1, the viral oncogenic mimic of CD40. Proc Natl Acad Sci U S A. 2009;106:17140–17145. doi: 10.1073/pnas.0903786106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Y, Li Y. Upregulation of miR-29b-3p protects cardiomyocytes from hypoxia-induced apoptosis by targeting TRAF5. Cell Mol Biol Lett. 2019;24:27. doi: 10.1186/s11658-019-0151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang Z, Li X, Liu S, Li C, Wang X, Xing J. MiR-141-3p inhibits cell proliferation, migration and invasion by targeting TRAF5 in colorectal cancer. Biochem Biophys Res Commun. 2019;514:699–705. doi: 10.1016/j.bbrc.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Xie Y, Li F, Li Z, Shi Z. miR-135a suppresses migration of gastric cancer cells by targeting TRAF5-mediated NF-kappaB activation. Onco Targets Ther. 2019;12:975–984. doi: 10.2147/OTT.S189976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo W, Sun C, Zhou J, Wang Q, Yu L, Bian XW, Zhou X, Hua D, Wang R, Rao C, Jiang Z, Shi C, Yu S. miR-135a-5p functions as a glioma proliferation suppressor by targeting tumor necrosis factor receptor-associated factor 5 and predicts patients’ prognosis. Am J Pathol. 2019;189:162–176. doi: 10.1016/j.ajpath.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 31.Gong H, Fang L, Li Y, Du J, Zhou B, Wang X, Zhou H, Gao L, Wang K, Zhang J. miR873 inhibits colorectal cancer cell proliferation by targeting TRAF5 and TAB1. Oncol Rep. 2018;39:1090–1098. doi: 10.3892/or.2018.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, Zhao L, Zhao F, Yang G, Wang J. MicroRNA-26b regulates cancer proliferation migration and cell cycle transition by suppressing TRAF5 in esophageal squamous cell carcinoma. Am J Transl Res. 2016;8:1957–1970. [PMC free article] [PubMed] [Google Scholar]
- 33.Piepoli A, Tavano F, Copetti M, Mazza T, Palumbo O, Panza A, di Mola FF, Pazienza V, Mazzoccoli G, Biscaglia G, Gentile A, Mastrodonato N, Carella M, Pellegrini F, di Sebastiano P, Andriulli A. Mirna expression profiles identify drivers in colorectal and pancreatic cancers. PLoS One. 2012;7:e33663. doi: 10.1371/journal.pone.0033663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen F, Qi S, Zhang X, Wu J, Yang X, Wang R. miR-23a-3p suppresses cell proliferation in oral squamous cell carcinomas by targeting FGF2 and correlates with a better prognosis: miR-23a-3p inhibits OSCC growth by targeting FGF2. Pathol Res Pract. 2019;215:660–667. doi: 10.1016/j.prp.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Quan J, Pan X, Zhou J, He A, Lai Y, Zhou G. Suppressing cell growth and inducing apoptosis by inhibiting miR23a5p in human bladder cancer. Mol Med Rep. 2018;18:5256–5260. doi: 10.3892/mmr.2018.9527. [DOI] [PubMed] [Google Scholar]
- 36.Chen B, Zhu A, Tian L, Xin Y, Liu X, Peng Y, Zhang J, Miao Y, Wei J. miR23a suppresses pancreatic cancer cell progression by inhibiting PLK1 expression. Mol Med Rep. 2018;18:105–112. doi: 10.3892/mmr.2018.8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sruthi TV, Edatt L, Raji GR, Kunhiraman H, Shankar SS, Shankar V, Ramachandran V, Poyyakkara A, Kumar SVB. Horizontal transfer of miR-23a from hypoxic tumor cell colonies can induce angiogenesis. J Cell Physiol. 2018;233:3498–3514. doi: 10.1002/jcp.26202. [DOI] [PubMed] [Google Scholar]
- 38.Niwa Y, Yamada S, Sonohara F, Kurimoto K, Hayashi M, Tashiro M, Iwata N, Kanda M, Tanaka C, Kobayashi D, Nakayama G, Koike M, Fujiwara M, Kodera Y. Identification of a serum-based miRNA signature for response of esophageal squamous cell carcinoma to neoadjuvant chemotherapy. J Transl Med. 2019;17:1. doi: 10.1186/s12967-018-1762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu S, Jin J, Gokhale S, Lu AM, Shan H, Feng J, Xie P. Genetic alterations of TRAF proteins in human cancers. Front Immunol. 2018;9:2111. doi: 10.3389/fimmu.2018.02111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang S, Wa Q, Pan J, Peng X, Ren D, Huang Y, Chen X, Tang Y. Downregulation of miR-141-3p promotes bone metastasis via activating NF-kappaB signaling in prostate cancer. J Exp Clin Cancer Res. 2017;36:173. doi: 10.1186/s13046-017-0645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tetreault MP. Esophageal cancer: insights from mouse models. Cancer Growth Metastasis. 2015;8:37–46. doi: 10.4137/CGM.S21218. [DOI] [PMC free article] [PubMed] [Google Scholar]