Abstract
In humans, parity without breastfeeding increases risk of estrogen receptor-negative (ER-) breast cancer and is associated with hypermethylation of FOXA1, a pioneer factor regulating lineage commitment of mammary gland luminal progenitor cells. We postulate that pregnancy-associated repression of FOXA1 results in the accumulation of aberrant, differentiation-arrested luminal progenitor cells which, following additional genetic and epigenetic insults, may give rise to ER- tumors. Consistent with this hypothesis, we show that deletion of Foxa1 in the mouse mammary gland results in a two-fold increase in the proportion of luminal progenitor cells and a reduction in mammary gland epithelial cells that stain positive for ER. These results provide compelling support for the notion that reduced Foxa1 expression is sufficient to alter mammary gland luminal cell fate determination in vivo, which could be a mechanism linking parity with ER- breast cancer.
Keywords: Foxa1, mammary gland, pregnancy, breastfeeding, progenitors
Introduction
Although having children reduces risk of ER+ breast cancer, parity has been shown to increase the risk of ER- disease, particularly among women who do not breastfeed. Mechanisms underlying these relationships are unknown, although inflammation during involution of the breast [1] could be exacerbated with abrupt involution when breastfeeding does not occur or is terminated early [2]. This may be especially relevant for U.S. Black women who have a higher incidence of ER- breast cancer, and who are more likely to be parous and not to breastfeed (reviewed in [2]).
Evidence indicates that both ER+ and ER- breast tumors arise from breast luminal progenitors [3]. This includes BRCA1-deficient cancers, most of which are ER-, where an expanded population of aberrant luminal progenitor cells has been postulated as the cell-of-origin [4]. Our previous studies of breast cancer showed that the majority of differences in DNA methylation between Black and White women were in ER- breast tumors [5,6] and that the FOXA1 gene was one of the most differentially methylated loci between ER- and ER+ cancers. Relationships were most pronounced in tumors from Black women, with FOXA1 more often hypermethylated and silenced in ER- tumors compared to ER+ [6]. When merging the epigenetic results with epidemiological data, we found that the level of methylation at FOXA1 in ER- tumors was positively associated with the number of children born to the patient and, importantly, the association was observed only among women who had not breastfed [6]. Consistent with our DNA methylation and RNA-seq studies, we found that FOXA1 protein expression also was higher in ER+ breast tumors compared to ER- tumors, that increased parity was associated with decreased FOXA1 staining, and that breast feeding partially abrogated this association [7].
Work using breast cancer cell lines has suggested that FOXA1 drives differentiation of luminal progenitors by inducing “luminal” genes and suppressing “basal” genes [8]. Intriguingly, wildtype BRCA1 protein positively regulates FOXA1 by preventing its DNA methylation and silencing [9], suggesting that BRCA1-mutated breast tumors are usually ER- because mutation/inactivation of BRCA1 leads to methylation-induced silencing of FOXA1 in luminal progenitors. We postulate that, analogous to BRCA1 mutations, pregnancy in the absence of breastfeeding can result in DNA methylation and silencing of FOXA1 in luminal progenitors leading to the accumulation of aberrant differentiation-arrested cells which, following additional mutations and/or epimutations, could give rise ER- breast tumors. Consistent with this notion, FOXA1 was shown to become hypermethylated in cancer-free breast tissue obtained from parous women undergoing cosmetic reduction mammoplasty, further suggesting a relationship with reproductive factors [10,11]. Conditional Foxa1 knockouts have been previously reported to affect mammary gland morphogenesis and mammary gland tumor incidence [12,13]. However, there have been no studies describing the effect of Foxa1 deletion on the relative proportions of the different subtypes of mouse mammary gland epithelial cells. To test the hypothesis that deletion of Foxa1 in the mouse mammary gland alters its normal proportions of epithelial subtypes, we knocked out Foxa1 in mouse mammary glands and analyzed the epithelial subtypes by flow cytometry. We observed a 2-fold increase in the proportion of luminal progenitors in Foxa1-depleted mammary glands. We suggest that at least a fraction of these cells is differentiation-arrested, and that in the human breast, analogous cells would be predisposed to develop into ER- mammary gland tumors if transformed.
Material and methods
Mice
Mice were handled in accordance with IACUC/AAALAC and ethics guidelines from the Department of Laboratory Animal Resources (DLAR), Roswell Park Comprehensive Cancer Center. Mice carrying a Foxa1 allele with loxP sites flanking exon 2 (designated as Foxa1 f) [14] were obtained from Dr. Klaus Kaestner at the University of Pennsylvania and crossed with strains carrying a transgene expressing Cre-recombinase under the control of either the mouse mammary tumor virus (MMTV) promoter (MMTV-Cre, line D)(Tg[MMTV-cre]4Mam/J, JAX stock no. 003553) or the keratin 14 promoter (Krt14-Cre)(Tg[KRT14-cre]1Amc/J, JAX stock no. 004782), to obtain experimental genotypes, Foxa1 f/f; MMTV-Cre, Foxa1f/+; MMTV-Cre, and Foxa1f/+; Krt14-Cre as well as controls Foxa1f/f and Foxa1f/+ (Supplementary Figure 1). Exposure to Cre-recombinase results in a 3.5 kb deletion removing approximately 80% of the protein coding sequence [14] (Supplementary Figure 2). Groups of 5-6 virgin females of each genotype produced were sacrificed as 9 weeks old virgins and mammary glands collected.
Mammary gland dissociation and isolation of epithelial cells
Primary mammary epithelial cells (MEC) were isolated from thoracic (the 2nd and 3rd), abdominal (the 4th) and inguinal (the 5th) mammary glands using a protocol provided by Dr. Sung Jin Huh [15]. Chopped tissue was transferred to digestion medium (DMEM/F-12, 10% FBS, 5 mg/ml insulin, pen/strep) containing 2 mg/ml collagenase type 4 (Worthington), incubated at 37°C with shaking for 30 minutes. Following centrifugation, digests were resuspensed in medium and large sized “organoids” collected by filtering through a 40 micron mesh. To obtain single cell suspensions for analysis, organoids were suspended in 0.05% trypsin, incubated at 37°C for 10 minutes, the trypsin inactivated by addition of FBS-containing medium, the cells pelleted and then suspended in medium containing 10 U/ml DNase1 and 5 mM MgCl2.
Flow cytometry analysis
Flow cytometry was carried out using the protocol of Dr. Sung Jin Huh [15]. Freshly prepared single-cell suspensions of MECs were resuspended in 0.5% BSA, 2 mM EDTA in PBS. Approximately 1 × 106 cells were incubated for 30 minutes with 5 antibodies (biotin-conjugated rat anti-mouse CD31 [BD Pharmingen, cat no. 558737], biotin-conjugated rat anti-mouse CD45 [BD Pharmingen, cat no. 553077], APC-labeled anti-mouse CD24 [BioLegend, cat no. 101814], PE-labeled hamster anti-mouse CD61 [BD Pharmingen, cat no. 553347], FITC-labeled rat anti-mouse CD29, [BioLegend, cat no. 102206]) and DAPI. Following washing in PBS, cells were resuspended in PBS containing Alexa750-conjugated streptavidin (Molecular Probes, cat no. S21384). DAPI facilitates removal of dead cells; anti-CD31-biotin and anti-CD45-biotin, together with Alexa750-streptavidin, excludes endothelial cells and leukocytes from the analysis. CD31- CD45- cells were further selected with anti-CD24, and these cells analyzed for CD29 and CD61 signals to determine the proportions of the 3 epithelial cell populations (Supplementary Figure 3). Flow cytometry was performed using a BD LSRII Flow Cytometer available in the Roswell Park Flow Cytometry and Imaging Core.
PCR genotyping
Genomic DNA was extracted using the Mouse Tail Direct PCR Kit (Creative Biogene, Shirley, NY) and single-cell suspensions of mammary epithelial cells (described above) using the Wizard Genomic DNA Purification kit (Promega). PCR was carried out using Direct PCR kit and run on a BioRad C1000 Touch Thermal Cycler. For Foxa1, the following primers were used: 5’-CTGTGGATTATGTTCCTGAT-3’ and 5’-GTGTCAGGATGCCTATCTGGT-3’, amplify the wild-type and the floxed Foxa1 alleles, giving rise to PCR products of 244 and 260 bp, respectively [14]. For the Cre gene, 5’-CGACCAGGTTCGTTCACTCA-3’ (forward primer), and 5’-CAGCGTTTTCGTTCTGCCAA-3’ (reverse primer) resulting in a PCR product of 184 bp. To confirm deletion of Foxa1, primers 5’-GATCACTCAAGCAGGCCTGT-3’ and 5’-TCCCCCTTTGCCACGTTTTA-3’ were used which flank the deleted region, and amplify the deleted Foxa1 allele, yielding a 530 bp PCR product.
Immunohistochemistry and image analysis
Mammary glands were fixed with 10% buffered formalin overnight, embedded in paraffin, sectioned, and stained with an antibody against estrogen receptor-α (ERα) (Abcam, cat. no. ab3575). Stained slides were digitally imaged at 20x magnification using the Aperio ScanScope XT (Aperio Technologies, Vista, CA). Whole section slides were annotated manually to identify mammary gland epithelial regions for the image analysis. The percent of cells stained was recorded in each intensity category: 0, 1+ (only partial or weak staining), 2+ (moderate and complete staining), and 3+ (intense and complete staining). A histological score (H-score) was calculated by the formula: [1 × (% cells 1+) + 2 × (% cells 2+) + 3 × (% cells 3+)] × 100 [16].
Statistical analysis
The flow cytometry profiles were compared between mouse groups in a pairwise fashion using two-sided Wilcoxon rank sum (WRS) exact tests. Analyses were conducted in SAS v9.4 (Cary, NC) at a nominal significance level of 0.05.
Results
Floxed Foxa1 alleles were deleted early in mammary gland development using two Cre-driver mouse strains, MMTV-Cre (D line) and Krt14-Cre, both of which function in the mammary gland [12]. Only virgin females were used in this study to avoid confounding factors associated with changes in mammary gland epithelial subpopulations during pregnancy, lactation, and involution (reviewed in [17]). Mammary glands were collected from 9 week-old virgin mice of 5 different genotypes (Supplementary Figure 1), the cells dissociated, tested for deletion of Foxa1 (Supplementary Figure 2) and analyzed by flow cytometry (Methods). Following heterozygous deletion of Foxa1 using Krt14-Cre, there was an increase in the proportion of luminal progenitor cells (CD24+CD61+CD29lo) from approximately 17% in control mice (Foxa1 +/f) to approximately 42% in Foxa1 +/f; Krt14-Cre +/- mice (P = 0.004, Holm-Bonferroni adjusted t-test) (Figure 1A, 1B). A similar trend was observed when Foxa1 was heterozygously deleted using the MMTV-Cre transgenic line, however the increase in luminal progenitors was not statistically significant, possibly due to the mosaic nature of Cre-recombinase excision in the mammary gland by MMTV-Cre [12]. On the other hand, when Foxa1 was homozygously deleted in mammary epithelial cells using MMTV-Cre (Foxa1 f/f; MMTV-Cre+/- mice), the fraction of luminal progenitors was also significantly increased compared to its control (Foxa1 f/f) (P = 0.016, Holm-Bonferroni adjusted t-test) (Figure 1C, 1D). Homozygous deletion of Foxa1 using Krt14-Cre results in the complete absence of mammary ducts [12,13] precluding the analysis of mammary epithelial cells. As would be predicted by the postulated blockage of differentiation of luminal progenitors, there was a corresponding decrease in the proportion of mature luminal cells (CD24+CD61-CD29lo) (P = 0.004, Holm-Bonferroni adjusted t-test) in Foxa1 +/f; Krt14-Cre mammary glands, although this was not observed in the Foxa1 +/f; MMTV-Cre line. However, we also observed a reduction in the proportion of basal/stem cells in both Foxa1 +/f; Krt14-Cre and Foxa1 +/f; MMTV-Cre lines, although the difference was not statistically significant in the latter mouse strain. Thus, at least for basal/stem cells, the apparent decrease in relative proportion may be indirect due to the expansion of the luminal progenitors in these mice (see discussion).
Figure 1.
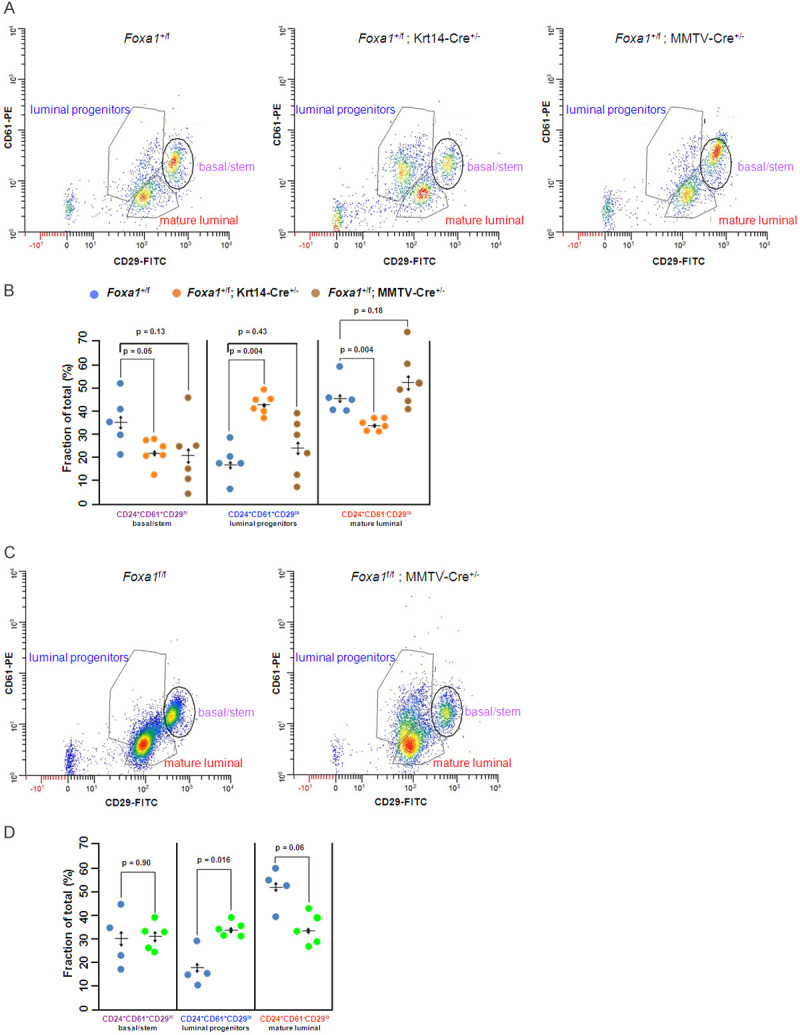
Analysis of epithelial cell populations from Foxa1 depleted mammary glands. A. Representative flow cytometry profiles of the 3 major epithelial cell types characterized by CD24, CD61, and CD29 in mouse mammary gland from wild type mice (Foxa1 +/f) and mice heterozygously deleted for Foxa1 by Krt14-Cre (Foxa1 +/f; Krt14-Cre +/-) or MMTV-Cre (Foxa1 +/f; MMTV-Cre +/-). B. Summary of results for 5-6 independent mouse mammary glands (colored points) showing means and SD, (p values determined by WRS exact test). C. Flow cytometry profile of 3 major epithelial cell types in mouse mammary gland from wild type mice (Foxa1 f/f) and mice homozygously deleted for Foxa1 by MMTV-Cre (Foxa1f/f; MMTV-Cre +/-). D. Summary of results for 5-6 mice showing means and SD, (p values determined WRS exact test).
Since Foxa1 is indispensable for expression of ERα [18], we assessed the effect of Foxa1 deletion on the prevalence of ER-positive cells in mammary gland sections by immunohistochemistry. We observed a marked decrease in number of ERα-positive cells in the mammary ducts of Foxa1 +/f; Krt14-Cre +/-, and to lesser extent in Foxa1 +/f; MMTV-Cre +/- mice, compared to the wildtype control Foxa1 +/f mice (Figure 2, Supplementary Figure 4).
Figure 2.
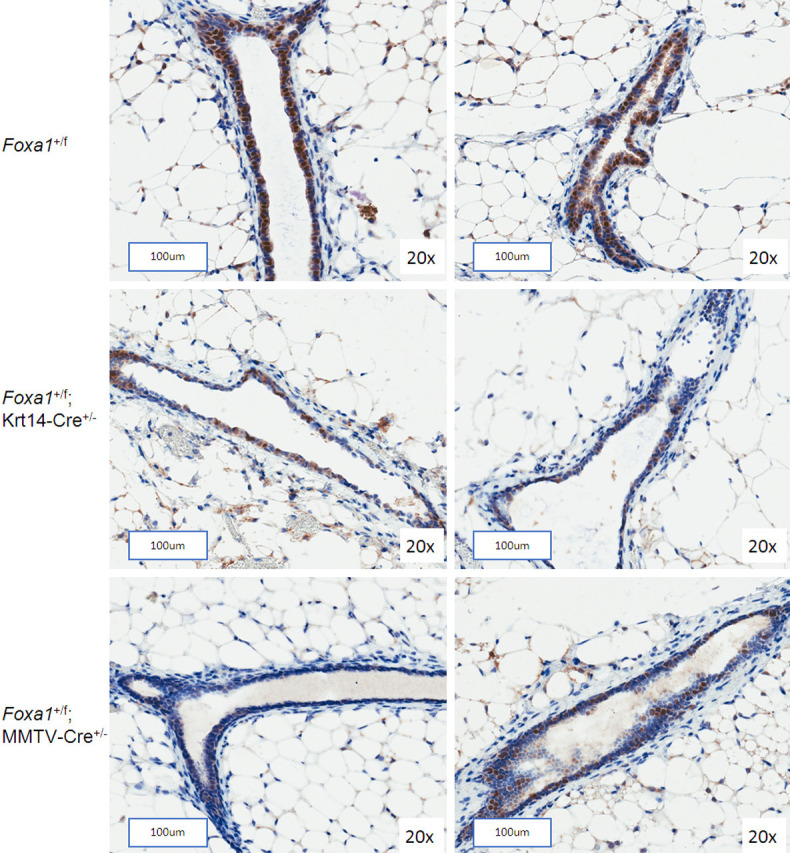
ERα immunostaining of mammary gland sections from conditional Foxa1 knockout mice. Expression of ERα in the ductal epithelial cells of mammary glands from 9-week-old virgin Foxa1 +/f; Krt14-Cre +/-, Foxa1 +/f; MMTV-Cre +/- or control Foxa1 +/f mice determined by IHC. The scale bars represent 100 μm.
Discussion
As a surrogate for DNA methylation-induced silencing, we conditionally deleted Foxa1 in the mouse mammary gland using a floxed-Foxa1 allele in combination with two Cre-recombinase drivers. Results demonstrate that depletion of Foxa1 leads to dramatic changes in the proportions of mammary gland epithelial cell populations, resulting in an abnormal accumulation of luminal progenitors. Similar increases in luminal progenitors occur following deletion of Gata3, a gene upstream of Foxa1 [19]. However, we did not find GATA3 to be differentially methylated in ER+ versus ER- tumors in our study of human breast cancer [6], supporting instead the importance of FOXA1 in the etiology of ER- breast tumors. There are at least two different types of luminal progenitor cells in the mouse mammary gland, a large ER- subset that are precursors to the secretory alveolar cells, and a smaller ER+ subset of cells that express high levels of Foxa1 [17,20]. These ER+ progenitors probably give rise to ductal luminal cells because they express Foxa1 and other transcription factors required for ductal development [18,20], and Foxa1 ablation leads to complete absence of ductal tree formation [12,13]. It is therefore likely that ER+ progenitor cells were the cells affected by Foxa1 depletion in the present study, resulting in blocked differentiation and accumulation in the luminal progenitors in mutant mice. Through its function as a “pioneer factor”, Foxa1 regulates a plethora of genes by binding to transcriptionally repressed chromatin and facilitating subsequent binding by multiple transcription factors including ERα [21]. Thus, its depletion in these cells (or their precursors) may lead to an accumulation of differentiation-arrested luminal progenitors with increased proliferative capacity due to disruption of the FOXA1 and ERα transcriptomes. The overall effect of Foxa1 deletion on differentiation of luminal epithelial cells remains to be fully elucidated but will become clearer by analysis of mammary glands from conditional Foxa1 knockouts using single-cell RNA-seq approaches [22-24].
Unexpectantly, we also found a reduction in the proportion of basal/stem cells in some of the Foxa1 knockout mammary glands. Foxa1 is not known to be expressed in the basal cell compartment of the mouse mammary gland and therefore its deletion in these cells is not expected to produce a phenotype. Hence, the finding of a reduced proportion of cells in the basal/stem cell compartment may simply reflect a greater absolute number of epithelial cells in the mammary glands in these two mouse lines as a consequence of the increased number of highly proliferative, differentiation-blocked, luminal progenitor cells.
Our results provide support for the notion that reduced Foxa1 expression alters luminal cell fate determination in vivo and proof-of-principle that pregnancy-linked methylation and silencing of FOXA1 may lead to the accumulation of aberrant differentiation-arrested luminal progenitors in the human breast, which following additional mutations and/or epimutations, could give rise ER- breast tumors. For the most part, a mechanistic link between parity and breastfeeding and increased methylation of FOXA1 remaine to be elucidated [25]. However, with the convincing evidence in the literature that parity increases risk of ER- breast cancer, particularly among women who do not breastfeed, coupled with our molecular results showing that parity without breastfeeding results in hypermethylation of FOXA1, particularly among Black women, these animal experiments provide a reasonable mechanism for these associations; e.g., through abnormal accumulation of luminal progenitor cells. This work is potentially relevant to the important question of why ER- breast cancer is more common in Black women who are more likely to have children and not to breastfeed. Identification of key genes such as FOXA1 may provide potential targets for cancer prevention, e.g. through the development of targeted epigenetic-modifying agents, to reverse silencing of pro-luminal genes.
Acknowledgements
We thank Satrajit Sinha for the Krt14-Cre mouse, Klaus Kaestner for the floxed-Foxa1 mouse, and Sung Jin Huh and Kornelia Polyak (Dana-Farber Cancer Institute, Boston, MA) for protocols to carry out isolation and flow cytometry analysis of mouse mammary epithelial cells. This work was supported by grants from the Roswell Park Alliance Foundation (M. Higgins), the Breast Cancer Research Foundation (BCRF, C. Ambrosone) and the NCI/NIH R01 CA225947 (Ambrosone, Higgins, Palmer). Use of the Shared Resources was supported by Roswell Park Comprehensive Center and National Cancer Institute (NCI) grant P30CA016056.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Schedin P. Pregnancy-associated breast cancer and metastasis. Nat Rev Cancer. 2006;6:281–291. doi: 10.1038/nrc1839. [DOI] [PubMed] [Google Scholar]
- 2.Palmer JR, Viscidi E, Troester MA, Hong CC, Schedin P, Bethea TN, Bandera EV, Borges V, McKinnon C, Haiman CA, Lunetta K, Kolonel LN, Rosenberg L, Olshan AF, Ambrosone CB. Parity, lactation, and breast cancer subtypes in African American women: results from the AMBER Consortium. J Natl Cancer Inst. 2014;106:dju237. doi: 10.1093/jnci/dju237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gross K, Wronski A, Skibinski A, Phillips S, Kuperwasser C. Cell fate decisions during breast cancer development. J Dev Biol. 2016;4:4. doi: 10.3390/jdb4010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, Feleppa F, Huschtscha LI, Thorne HJ kConFab. Fox SB, Yan M, French JD, Brown MA, Smyth GK, Visvader JE, Lindeman GJ. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 5.Ambrosone CB, Young AC, Sucheston LE, Wang D, Yan L, Liu S, Tang L, Hu Q, Freudenheim JL, Shields PG, Morrison CD, Demissie K, Higgins MJ. Genome-wide methylation patterns provide insight into differences in breast tumor biology between American women of African and European ancestry. Oncotarget. 2014;5:237–248. doi: 10.18632/oncotarget.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espinal AC, Buas MF, Wang D, Cheng DT, Sucheston-Campbell L, Hu Q, Yan L, Payne-Ondracek R, Cortes E, Tang L, Gong Z, Zirpoli G, Khoury T, Yao S, Omilian A, Demissie K, Bandera EV, Liu S, Ambrosone CB, Higgins MJ. FOXA1 hypermethylation: link between parity and ER-negative breast cancer in African American women? Breast Cancer Res Treat. 2017;166:559–568. doi: 10.1007/s10549-017-4418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng TD, Yao S, Omilian AR, Khoury T, Buas MF, Payne-Ondracek R, Sribenja S, Bshara W, Hong CC, Bandera EV, Davis W, Higgins MJ, Ambrosone CB. FOXA1 protein expression in ER(+) and ER(-) breast cancer in relation to parity and breastfeeding in black and white women. Cancer Epidemiol Biomarkers Prev. 2020;29:379–385. doi: 10.1158/1055-9965.EPI-19-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernardo GM, Bebek G, Ginther CL, Sizemore ST, Lozada KL, Miedler JD, Anderson LA, Godwin AK, Abdul-Karim FW, Slamon DJ, Keri RA. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene. 2013;32:554–563. doi: 10.1038/onc.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong C, Fujino K, Monteiro LJ, Gomes AR, Drost R, Davidson-Smith H, Takeda S, Khoo US, Jonkers J, Sproul D, Lam EW. FOXA1 repression is associated with loss of BRCA1 and increased promoter methylation and chromatin silencing in breast cancer. Oncogene. 2015;34:5012–5024. doi: 10.1038/onc.2014.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh S, Gu F, Wang CM, Lin CL, Liu J, Wang H, Ravdin P, Hu Y, Huang TH, Li R. Genome-wide DNA methylation profiling reveals parity-associated hypermethylation of FOXA1. Breast Cancer Res Treat. 2014;147:653–659. doi: 10.1007/s10549-014-3132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zendehbad Z, Izadi P, Daraei A, Yekaninejad MS, Nafissi N, Younosi N, Khorasani G, Tavakkoly Bazzaz J. Early parity epigenetic footprint of FOXA1 gene body in normal breast tissue of iranian women. Iran Biomed J. 2019;23:99–106. doi: 10.29252/.23.2.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Zhao Y, Skerry B, Wang X, Colin-Cassin C, Radisky DC, Kaestner KH, Li Z. Foxa1 is essential for mammary duct formation. Genesis. 2016;54:277–285. doi: 10.1002/dvg.22929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng D, McLaughlin SA, Kaestner KH, Li Z. Genetic ablation of mammary ducts through foxa1 prevents breast cancer occurrence. Am J Cancer Res. 2019;9:424–428. [PMC free article] [PubMed] [Google Scholar]
- 14.Gao N, LeLay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22:3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huh SJ, Clement K, Jee D, Merlini A, Choudhury S, Maruyama R, Yoo R, Chytil A, Boyle P, Ran FA, Moses HL, Barcellos-Hoff MH, Jackson-Grusby L, Meissner A, Polyak K. Age- and pregnancy-associated DNA methylation changes in mammary epithelial cells. Stem Cell Reports. 2015;4:297–311. doi: 10.1016/j.stemcr.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirsch FR, Varella-Garcia M, Bunn PA Jr, Di Maria MV, Veve R, Bremmes RM, Baron AE, Zeng C, Franklin WA. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J. Clin. Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 17.Fu NY, Nolan E, Lindeman GJ, Visvader JE. Stem cells and the differentiation hierarchy in mammary gland development. Physiol Rev. 2020;100:489–523. doi: 10.1152/physrev.00040.2018. [DOI] [PubMed] [Google Scholar]
- 18.Bernardo GM, Lozada KL, Miedler JD, Harburg G, Hewitt SC, Mosley JD, Godwin AK, Korach KS, Visvader JE, Kaestner KH, Abdul-Karim FW, Montano MM, Keri RA. FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development. 2010;137:2045–2054. doi: 10.1242/dev.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, Lindeman GJ, Visvader JE. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 20.Shehata M, Teschendorff A, Sharp G, Novcic N, Russell IA, Avril S, Prater M, Eirew P, Caldas C, Watson CJ, Stingl J. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernardo GM, Keri RA. FOXA1: a transcription factor with parallel functions in development and cancer. Biosci Rep. 2012;32:113–130. doi: 10.1042/BSR20110046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bach K, Pensa S, Grzelak M, Hadfield J, Adams DJ, Marioni JC, Khaled WT. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat Commun. 2017;8:2128. doi: 10.1038/s41467-017-02001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pal B, Chen Y, Vaillant F, Jamieson P, Gordon L, Rios AC, Wilcox S, Fu N, Liu KH, Jackling FC, Davis MJ, Lindeman GJ, Smyth GK, Visvader JE. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat Commun. 2017;8:1627. doi: 10.1038/s41467-017-01560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giraddi RR, Chung CY, Heinz RE, Balcioglu O, Novotny M, Trejo CL, Dravis C, Hagos BM, Mehrabad EM, Rodewald LW, Hwang JY, Fan C, Lasken R, Varley KE, Perou CM, Wahl GM, Spike BT. Single-cell transcriptomes distinguish stem cell state changes and lineage specification programs in early mammary gland development. Cell Rep. 2018;24:1653–1666. e1657. doi: 10.1016/j.celrep.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambrosone CB, Higgins MJ. Relationships between breast feeding and breast cancer subtypes: lessons learned from studies in humans and in mice. Cancer Res. 2020;80:4871–4877. doi: 10.1158/0008-5472.CAN-20-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


