Abstract
In this study, the medium requirements to increase the production of xylitol by using Candida tropicalis (CT) have been investigated. The technique of single addition or omission of medium components was applied to determine the nutritional requirements. The addition of amino acids such as Asp, Glu, Gln, Asn, Thr, and Gly had no significant effect on CT growth. However, in the absence of other metal ions, there was a higher concentration of cell growth and xylitol production when only Zn2+ was present in the medium. The analysis of various vitamins unveiled that biotin and thiamine were the only vitamins required for the growth of CT. Surprisingly, when only biotin was present in the medium as a vitamin, there was less growth of CT than when the medium was complete, but the amount of xylitol released was significantly higher. Overall, this study will increase the xylitol production using the single omission or addtion technique.
Keywords: Biotin, Thiamine, Urea, Xylitol, Zn2+
Introduction
Xylitol, a five-carbon alcoholic sugar, is popular in the food industry due to its non-carcinogenicity and lack of dependence on insulin [1–4]. People with diabetes and people with low glucose-6-phosphate dehydrogenase do not require insulin and glucose-6-phosphate dehydrogenase when consuming xylitol, which makes it a suitable alternative for these people [5, 6]. Xylitol is currently manufactured from xylose, the five-carbon sugar derived from hemicellulose hydrolysates, through a chemical process using Ni/Al2O3 as a catalyst [7–12].
The type of media utilized during fermentation greatly influences the chemical or nutritional environment, which in turn significantly influences the productivity and the economics of a fermentation process [13–18]. The media used to promote high productivity in commercial/industrial fermentation are mainly developed from complex sources of carbon and nitrogen [19]. However, the rate of growth and the activity of metabolic processes may be affected strongly by the type and ratio of nutrients provided to the culture. Due to this inherent inconsistency of natural origin, the fermentation performance may vary from lot to lot [20].
Yeast extract (YE) is used in the fermentation of xylitol, and thus xylitol is produced using microbial fermentation using YE as one of the medium ingredients. However, the variation yields have been observed due to unknown variations of YE. On top of that, the high price of YE hinders its uses in industrial applications. Moreover, the medium cost is one of the major factors in economic xylitol production [21–26]. Therefore, replacing YE with a synthetic defined medium (SDM) is required to lower the cost of medium and performance variability while maintaining the production yield [27]. Additionally, when producing xylitol, the performance consistency of synthetic-defined media should be comparable to that of YE. When the concentration of nitrogen is high, the recovery and purification of xylitol production become very difficult. Xylitol purification is simplified because no additional contaminants are added to the media, leading to a lower production cost [28]. Hence, it's indeed crucial for the metabolic investigation to have a precise growth medium for the microorganisms, which really supports high yield and productivity. Therefore, SDM which enables exponential growth with high xylitol production while eliminating or adding the need for a single medium component, has been developed in the current study.
Materials and Methods
Microorganism and Media
Candida tropicalis KFCC-10690 was used in this study because it is an established member of the Candida genus [29]. Freezing of the cell stock was done at − 70 °C. This medium consists of 5 g/L YE and 20 g/L glucose. For fermentation, there were 5–33% xylose, 10 g/L YE, and 0–90 g/L glucose in a complex medium that had a concentration of 5 g/L KH2PO4. Finished materials were made up into separate batches of medium and dense components, which consisting of carbohydrates, basal salts, amino acids, vitamins, and metals [30–32]. The components of SDM were sterilized by a membrane filtration method [33] (Millex-GV filter; Millipore Corp., Bedford, Mass), and the working cultures of CT were propagated in SDM. Further, cultures were centrifuged and washed twice in 50 mM potassium phosphate and at the pH of 6.5 to elimination of carryover nutrients. For the inocula, 5% (vol/vol) exponentially growing cells were used.
Fermentation Conditions
Inoculation was carried out on a 500 mL flask with 100 mL of culture medium for 10 h at 30 °C and 250 rpm. The resulting culture broth, diluted to a total volume of 10% (v/v), was transferred to a 500 mL flask and used to inoculate a 5-L jar fermenter, which was filled with 100 mL of production medium until it was 2.8–3.5 L of production medium (Kobiotech. Co., Republic of Korea). Complex media contains 200 g/L of xylose, 17 g/L of glucose, 1.3 g/L of KH2PO4, 2.5 g/L of (NH4)2SO4, 0.13 g/L of MgSO4, and 5 g/L of YE. SDM contains urea 3.1 g/L, xylose 200 g/L, glucose 17 g/L, KH2PO4 1.3 g/L, MgSO4 0.13 g/L, biotin 16.5 g/L, folic acid 7.5 µg/L, thiamine 2.65 mg/L, boric acid 0.5 mg/L, copper sulfate 0.04 mg/L, potassium iodide 0.1 mg/L, ferric chloride 0.2 mg/L, manganese sulfate 0.4 mg/L, sodium molybdate 0.2 mg/L, and zinc sulfate 5.0 mg/L. Experiments in jar fermenters were conducted at 30 °C in a fed-batch mode controlling the pH at 4.8. The peristaltic pump (10–50 mL/h) continuously fed the solution of xylose or the mixture of xylose and glucose, which was aerated at 0.5 vvm. The agitation was increased from 250 to 750 rpm to maintain the percentage of dissolved oxygen above 20 until the cell mass reached 14 g/L; it was then decreased to 340 rpm to limit the concentration of dissolved oxygen.
Enzyme Assay
Cultured cells were collected by centrifuging at 10,000 rpm for 15 min. Washing was carried out with 0.1 M Tris–HCl (pH 7.8), 0.5 mM EDTA, and 5 mM mercaptoethanol. Further, the cells were resuspended in a buffer [34] containing 20 mM Tris–HCl (pH 7.8), 10 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, and 1 mM phenylmethylsulphonylfluoride. Glass beads of 0.5 mm in diameter were used for the suspension. (Sigma, USA). To determine the xylose reductase (XR) activity, a decrease in the absorbance at 340 nm was measured after the addition of D-xylose, a marker for NADPH oxidation (Sigma, USA) [35].
Analytical Methods
A Bradford assay has been used to estimate protein concentration, and bovine serum albumin is being used as a standard [36]. To estimate the concentrations of xylitol, glucose, and xylose, HPLC coupled to an RI detector (Waters 410, USA) and a High-Performance Carbohydrate Column (4.6 mm × 250 mm, Waters, USA) were used. Acetonitrile/water (85:15 v/v) was used as a mobile phase at a 1.5 mL/min flow rate.
Results and Discussion
Influence of Nitrogen Source
A defined medium with a sole carbon source and a sole nitrogen source was designed in order to investigate the effect of the substrates on xylitol production. A shake flask system was developed to explore a range of inorganic and organic nitrogen sources in culturing a defined medium containing 200 g/L xylose and 17 g/L glucose as the carbon source. Ammonia, which is an important component of nitrogen metabolism in yeast, was also tested alongside two other common nitrogen sources, urea and nitrate, and CT grew on all of the nitrogen sources, indicating that they had been consuming it. With the exception of the ammonium acetate experiment, all experiments found that the glucose supply had been depleted after 40 h. Although inorganic nitrogen sources like ammonium tartrate, ammonium nitrate, ammonium acetate, and sodium nitrate were consumed for biomass formation, the production of xylitol was poor after 60 h of cultivation. However, urea has been found to produce xylitol similar to the level achieved by complex media by YE.
Effect of Amino Acids, Nucleic Acids, and Buffers
The cell growth and the amount of xylitol did not change regardless of whether single or multiple amino acids were added, and the same results have been observed when nucleic acids such as guanine, xanthine, adenine, and uracil were omitted from the growth medium. Moreover, no major changes were found in the growth and xylitol production of the strain when tenfold lower levels of buffers, such as phosphate, citrate, and acetate, were added. (Table 1).
Table 1.
Nutrient requirements of C. tropicalis in synthetic defined medium investigated by addition or omission of a single medium component
| Added medium component | OD600a | Omitted medium component | OD600a |
|---|---|---|---|
| None | 14.4 | Phosphate | 11.2 |
| l-Alanine | 14.3 | MgSO4 | 13.0 |
| l-Arginine | 14.7 | H3BO3 | 13.3 |
| l-Asparagine | 14.7 | MnCl2 | 12.4 |
| l-Leucine | 14.4 | ZnCl2 | 5.6 |
| l-Glutamic acid | 14.5 | CuSO4 | 13.1 |
| l-Glutamine | 14.3 | FeCl2 | 13.5 |
| Glycine | 14.4 | NiCl2 | 14.6 |
| l-Lysine | 14.6 | CoCl2 | 14.3 |
| l-Phenylalanine | 14.3 | Na2MoO4 | 13.5 |
| l-Proline | 14.3 | Biotin | 13.4 |
| l-Serine | 14.5 | Inositol | 15.1 |
| l-Tryptophan | 14.7 | Folic acid | 14.5 |
| l-Tyrosine | 14.7 | ρ-Aminobenzoic acid | 14.9 |
| l-Valine | 14.5 | Nicotinic acid | 14.4 |
| l-Histidine | 14.4 | Pantothenate | 14.8 |
| l-Cysteine | 14.6 | Pyridoxamine | 15.0 |
| Adenine | 14.8 | Pyridoxine | 15.2 |
| Guanine | 14.2 | Riboflavin | 10.5 |
| Uracil | 14.5 | Thiamine | 8.7 |
| Xanthine | 14.3 |
Values are the means ± standard deviations of triplicate measurements
aOD measurements were performed after 48 h of incubation
Effect of Metal Ions on the Growth and Xylitol Production
By excluding one metal ion at a time, the metal ion requirement of CT in SDM was determined. The strain grew well when NiCl2 and CoCl2 were omitted individually, and the cell growth was slightly inhibited when FeCl2, CoCl2, H3BO3, Na2MoO4, MnCl2, and CuCl2 were excluded. However, in the absence of ZnCl2, the growth of CT was significantly decreased, and it appeared to be crucial for cell growth (Table 1). Thus, to determine the effect of Zn2+ on cell growth and xylitol production, a 5-L jar fermenter was used. To that end, various concentrations of ZnCl2 were tested in the range of 0 to 10 mg/L. The fermentation conditions were provided in the “Materials and Methods” section. Xylitol gave the greatest yield and productivity at a concentration of 5 mg/L of ZnCl2 (Table 2). Further, an SDM mixture produced by the addition of optimal concentration of ZnSO4 (5 mg/L) was as effective as the complete metal mixture of SDM has been observed to promote cell growth and xylitol production. XR activity of supernatants obtained from cultures grown without zinc were assayed with and without ZnCl2 added to the samples, and no significant activities were found, even when the samples were incubated with zinc for 1 h at 37 °C before the assay. This suggests that zinc does not increase the level of XR activity by an enzyme mechanism. However, it seems to play a metabolic role and is needed during growth to induce significant protease production [37].
Table 2.
Effect of ZnSO4 concentration on the cell growth and xylitol production
| Conc. of ZnSO4 (mg/L) | ||||
|---|---|---|---|---|
| 0 | 1.0 | 5.0 | 10.0 | |
| Cell conc. (OD600) | 47.9 | 47.7 | 43.4 | 38.5 |
| Produced xylitol (g/L) | 186 | 260 | 252 | 246 |
| Yield (Volumetric, %) | 54.3 | 77.1 | 77.2 | 74.7 |
| Productivity (g/L h) | 2.10 | 3.31 | 3.63 | 3.32 |
Effect of Vitamins on the Growth and Xylitol Production
In the absence of any individual vitamin, only riboflavin and thiamine were detected as essential nutrients for growth. In contrast, a similar OD was found when other vitamins were overlooked. Further, we found that folic acid, ρ-Aminobenzoic acid, pantothenate, inositol, niacin, and pyridoxine were unnecessary for cell growth. A single omission of the nonessential vitamins did not change the specific production of xylitol [37, 38], but when biotin was overlooked, the specific xylitol production was significantly decreased, and it emerged to be essential for xylitol production (Table 3). When CT was grown in a medium lacking vitamins except for riboflavin, biotin, and thiamine, the OD of the cultures was getting lower after 48 h. In contrast, the specific xylitol production was increased significantly (Table 3). Biotin limitation decreased the xylose consumption of CT, and the decrease became more significant as the initial concentration of biotin decreased. Biotin acts as a prosthetic group for carboxylases, and it is unclear why its limitation results in more xylitol accumulation.
Table 3.
Effect of individual and multiple omission of essential vitamins on the xylitol production by C. tropicalis in defined medium
| Omission | Xylitol (g/L) | Specific xylitol production (g/g of dry cell weight)a |
|---|---|---|
| None | 114 | 10.6 |
| ρ-Aminobenzoic acid | 117 | 10.8 |
| Biotin | 2.8 | 0.27 |
| Calcium pantothenate | 118 | 11.0 |
| Folic acid | 114 | 10.5 |
| Inositol | 112 | 10.2 |
| Niacin | 118 | 10.8 |
| Pyridoxine | 113 | 10.2 |
| Riboflavin | 108 | 10.1 |
| Thiamine | 103 | 9.90 |
| All essential vitaminsb except riboflavin, biotin, and thiamine | 119 | 11.2 |
aThe specific xylitol production was calculated from a standard curve of OD600 against cell dry weight
bAll essential vitamins: ρ-Aminobenzoic acid, biotin, calcium pantothenate, folic acid, inositol, niacin, pyridoxine hydrochloride, riboflavin, thiamine
Xylitol Production Using a 5-L Jar Fermenter
Finally, a pH-controlled fed-batch culture experiment was carried out to compare the cell growth and xylitol production by CT in the complex medium and the SDM. The composition of the SDM met the nutritional requirements of the strains and took advantage of the beneficial effects in the downstream process. Fermentation of CT in complex medium containing is represented in Fig. 1. During the glucose consumption, pH decreased from 6.4 to 4.8; thereafter, pH increased to 6.8 until the end of the fermentation process. In the SDM, CT grew up with a minimal growth rate of 0.18 h−1 and a final OD of 47.7 and produced 260 g of xylitol per liter with a conversion yield of 81.5% when grown in a pH-controlled fed-batch culture. On the other hand, in the complex medium, the growth rate was 0.23 h−1, and the final OD was 47.1, while the xylitol production was 251 g L−1 with the conversion yield of 78.1% (Table 4). Further, It has been observed that in both the complex medium and the SDM, xylitol production continued after growth had come to an end. Still, beyond the stationary growth phase, more xylitol production was observed in the SDM than in the complex medium. Moreover, the addition to SDM of the ten amino acids (Gln, Leu, Ile, Val, Met, His, Arg, Trp, Pro, and Phe) did not increase the xylitol production of CT.
Fig. 1.
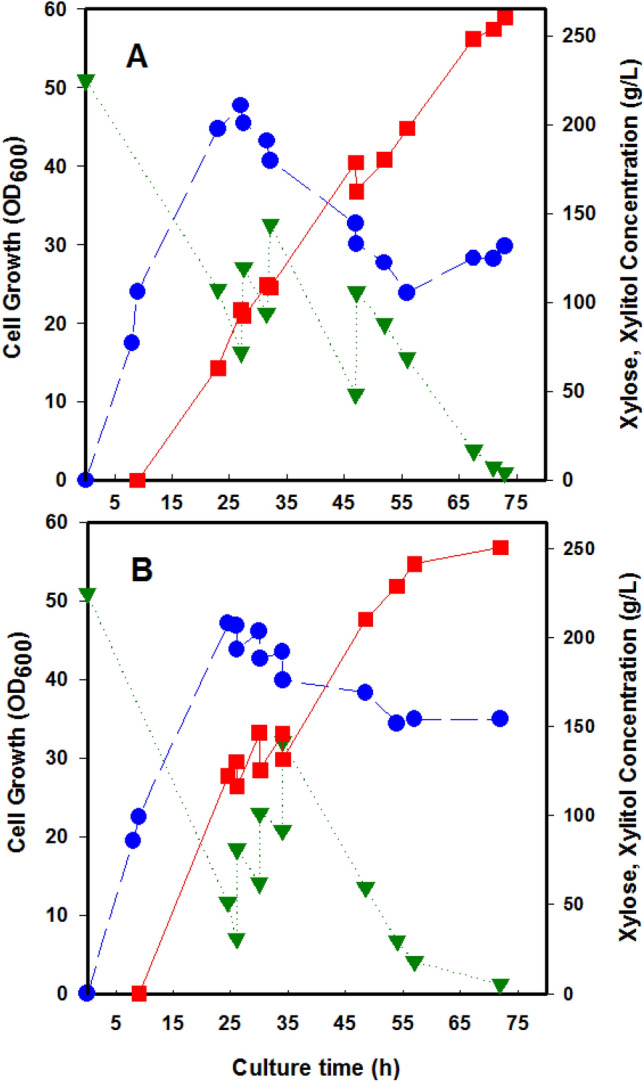
Xylitol production using SDM (A) and complex medium (B) containing urea and yeast extract as a nitrogen source, respectively. Blue circle: cell growth, green inverted triangle: xylose, red square: xylitol
Table 4.
Comparison of xylitol production between the SDM and complex medium containing urea and yeast extract as a nitrogen source, respectively
| Medium | ||
|---|---|---|
| SDM | Complex | |
| Initial pH of medium | 5.6 | 5.0 |
| Culture time (h) | 73 | 72 |
| Maximum cell conc. (OD600) | 47.7 | 47.1 |
| Added conc. of xylose (g/L) | 300 | 300 |
| Final conc. of | ||
| Xylitol (g/L) | 260 | 251 |
| Xylose (g/L) | 4.3 | 5.2 |
| Glycerol (g/L) | 9.5 | 11.0 |
| Yield (Volumetric, %) | 81.5 | 78.1 |
| Productivity (Volumetric, g/L h) | 3.35 | 3.26 |
Conclusions
In this work, we developed a synthetic and cheap medium to allow reproducible xylitol production without variation in yields and productivity. The technique of single addition or omission of medium components revealed that the amino acids such as Asp, Glu, Gln, Asn, Thr, and Gly were slightly affecting the growth of CT. Further, we observed that the amount of cell growth and xylitol production was more significant when Zn2+ ion was present in the medium and other metal ions were not. In addition, it has been observed that CT required only biotin and thiamine as individual vitamins. Surprisingly, when only biotin was present in the medium as a vitamin, the amount of xylitol production was significantly greater than in the complete medium.
Acknowledgements
This research was supported by Basic Science Research Program through the NRF funded by the Ministry of Science, ICT & Future Planning (NRF-2019R1F1A1063131, NRF-2020R1I1A1A01073483, NRF-2019R1C1C1009766). This work was also supported by KU Research Professor program of Konkuk University. This paper was supported by Konkuk University Researcher Fund in 2019.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Gurusamy Muneeswaran and Sanjay K. S. Patel have contributed equally to this work.
Contributor Information
Jung-Kul Lee, Email: jkrhee@konkuk.ac.kr.
Vipin Chandra Kalia, Email: vckaliaku@gmail.com.
In-Won Kim, Email: inwon@konkuk.ac.kr.
References
- 1.Makinen KK. Xylitol and oral health. Adv Food Res. 1979;25:137–157. doi: 10.1016/S0065-2628(08)60236-0. [DOI] [PubMed] [Google Scholar]
- 2.Ylikahri R. Metabolic and nutritional aspects of xylitol. Adv Food Res. 1979;25:159–180. doi: 10.1016/S0065-2628(08)60237-2. [DOI] [PubMed] [Google Scholar]
- 3.Radhakrishnan R, Lee I-J. Foliar treatment of Bacillus methylotrophicus KE2 reprograms endogenous functional chemicals in sesame to improve plant health. Indian J Microbiol. 2017;57:409–415. doi: 10.1007/s12088-017-0666-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soares LCS, Chandel AK, et al. Screening of yeasts for selection of potential strains and their utilization for in situ microbial detoxification (ISMD) of sugarcane bagasse hemicellulosic hydrolysate. Indian J Microbiol. 2016;56:172–181. doi: 10.1007/s12088-016-0573-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pepper T, Olinger PM. Xylitol in sugar-free confections. Food Technol. 1988;42:98–106. [Google Scholar]
- 6.Gao H, Kim TS, Mardina P, Zhou P, Wen F, Lee J-K. Rare sugar production by coupling of NADH oxidase and l-arabinitol dehydrogenase. RSC Adv. 2016;6:66609–66616. doi: 10.1039/C6RA11614K. [DOI] [Google Scholar]
- 7.Dhiman SS, Haw JR, Kalyani D, Kalia VC, Kang YC, Lee J-K. Simultaneous pretreatment and saccharification: green technology for enhanced sugar yields from biomass using a fungal consortium. Bioresour Technol. 2015;179:50–57. doi: 10.1016/j.biortech.2014.11.059. [DOI] [PubMed] [Google Scholar]
- 8.Prabhu P, Doan TN, Tiwari M, Singh R, Kim SC, Hong MK, Kang YC, Kang LW, Lee J-K. Structure-based studies on the metal binding of two-metal-dependent sugar isomerases. FEBS J. 2014;15:3446–3459. doi: 10.1111/febs.12872. [DOI] [PubMed] [Google Scholar]
- 9.Jagtap SS, Dhiman SS, Kim TS, Li J, Lee J-K. Enzymatic hydrolysis of aspen biomass into fermentable sugars by using lignocellulases from Armillaria gemina. Bioresour Technol. 2013;133:307–314. doi: 10.1016/j.biortech.2013.01.118. [DOI] [PubMed] [Google Scholar]
- 10.Jagtap SS, Dhiman SS, Jeya M, Kim I-W, Lee J-K. Characterization of a β-1,4-glucosidase from a newly isolated strain of Pholiota adiposa and its application to the hydrolysis of biomass. Biomass Bioenergy. 2013;54:181–190. doi: 10.1016/j.biombioe.2013.03.032. [DOI] [Google Scholar]
- 11.Hui G, Tiwari M, Jeya M, Lee J-K. Characterization of H2O-forming NADH oxidase from Streptococcus pyogenes and its application in L-rare sugar production. Bioorg Med Chem Lett. 2012;22:1931–1935. doi: 10.1016/j.bmcl.2012.01.049. [DOI] [PubMed] [Google Scholar]
- 12.Jeya M, Nguyen N-P-T, Moon H-J, Kim S-H, Lee J-K. Conversion of woody biomass into fermentable sugars by cellulase from Agaricus arvensis. Bioresour Technol. 2010;101:8742–8749. doi: 10.1016/j.biortech.2010.06.055. [DOI] [PubMed] [Google Scholar]
- 13.Kim JH, Lim B-C, Yeom S-J, Kim Y-S, Kim HJ, Lee J-K, Lee SH, Kim SW, Oh DK. Differential selectivity of the Escherichia coli cell membrane shifts the equilibrium for the enzyme-catalyzed isomerization of galactose to tagatose. Appl Environ Microbiol. 2008;74:2307–2313. doi: 10.1128/AEM.02691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagolu R, Singh R, Shanmugam R, Kondaveeti S, Patel SKS, Kalia VC, Lee J-K. Site-directed lysine modification of xylanase for oriented immobilization onto silicon dioxide nanoparticles. Bioresour Technol. 2021;331:125063. doi: 10.1016/j.biortech.2021.125063. [DOI] [PubMed] [Google Scholar]
- 15.Fernandes AMO, Garcia NFL, Fonseca GG, Leite RSR, Da Paz MF. Evaluation of the fermentative capacity of Saccharomyces cerevisiae CAT-1 and BB9 strains and Pichia kudriavzevii BB2 at simulated industrial conditions. Indian J Microbiol. 2020;60:494–504. doi: 10.1007/s12088-020-00891-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veerasamy M, Venkataraman K, et al. Point of care tuberculosis sero-diagnosis kit for wild animals: combination of proteins for improving the diagnostic sensitivity and specificity. Indian J Microbiol. 2018;58:81–92. doi: 10.1007/s12088-017-0688-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue D, Yao D, You X, Gong C. Green synthesis of the flavor esters with a marine Candida parapsilosis esterase expressed in Saccharomyces cerevisiae. Indian J Microbiol. 2020;60:175–181. doi: 10.1007/s12088-020-00856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalia VC, Patel SKS, Cho B-K, Wood TK, Lee J-K. Emerging applications of bacteria as anti-tumor agents. Sem Cancer Biol. 2021 doi: 10.1016/j.semcancer.2021.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Miller TL, Churchill BW. Substrates for large-scale fermentations. In: Demain AL, Solomon NA, editors. Manual for industrial microbiology and biotechnology. Washington, DC: American Society for Microbiology; 1986. pp. 122–136. [Google Scholar]
- 20.Kumar V, Patel SKS, Gupta RK, Otari SV, Hui G, Lee J-K, Zhang L. Enhanced saccharification and fermentation of agricultural waste using an immobilized enzyme cocktail. Biotechnol J. 2019;14:1800468. doi: 10.1002/biot.201800468. [DOI] [PubMed] [Google Scholar]
- 21.Tiwari M, Moon H-J, Jeya M, Lee J-K. Cloning and characterization of a thermostable xylitol dehydrogenase from Rhizobium etli CFN42. Appl Microbiol Biotechnol. 2010;87:571–581. doi: 10.1007/s00253-010-2478-6. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y-W, Tiwari M, Jeya M, Lee J-K. Covalent immobilization of recombinant Rhizobium etli CFN42 xylitol dehydrogenase onto modified silica nanoparticles. Appl Microbiol Biotechnol. 2011;90:499–507. doi: 10.1007/s00253-011-3094-9. [DOI] [PubMed] [Google Scholar]
- 23.Kalia VC, Gong C, Patel SKS, Lee J-K. Regulation of plant mineral nutrition by signal molecules. Microorganisms. 2021;9:774. doi: 10.3390/microorganisms9040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel SKS, Gupta RK, Kalia VC, Lee J-K. Integrating anaerobic digestion of potato peels to methanol production by methanotrophs immobilized on banana leaves. Bioresour Technol. 2021;323:124550. doi: 10.1016/j.biortech.2020.124550. [DOI] [PubMed] [Google Scholar]
- 25.Hong JH, Kim JH, Park GD, Lee JY, Lee J-K, Kang YC. A strategy for fabricating three-dimensional porous architecture comprising metal oxides/CNT as highly active and durable bifunctional oxygen electrocatalysts and their application. Chem Eng J. 2021;414:128815. doi: 10.1016/j.cej.2021.128815. [DOI] [Google Scholar]
- 26.Kalia VC, Patel SKS, Shanmugam R, Lee J-K. Polyhydroxy alkanoates: trends and advances towards biotechnological applications. Bioresour Technol. 2021;326:124737. doi: 10.1016/j.biortech.2021.124737. [DOI] [PubMed] [Google Scholar]
- 27.Kalyani D, Tiwari M, Li J, Kim SC, Kalia VC, Kang YC, Lee J-K. A highly efficient recombinant laccase from the yeast Yarrowia lipolytica and its application in the hydrolysis of biomass. PLoS ONE. 2015;10:e0120156. doi: 10.1371/journal.pone.0120156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar P, Ray S, Patel SKS, Lee J-K, Kalia VC. Bioconversion of crude glycerol to polyhydroxyalkanoate by Bacillus thuringiensis EGU45 under high nitrogen concentration. Int J Biol Macromol. 2015;78:9–16. doi: 10.1016/j.ijbiomac.2015.03.046. [DOI] [PubMed] [Google Scholar]
- 29.Demirbas A. Comparison study of synthesized red (or blood) orange peels and juice extract-nanoflowers and their antimicrobial properties on fish pathogen (Yersinia ruckeri) Indian J Microbiol. 2021 doi: 10.1007/s12088-021-00945-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews CB, Kuo A, Love KR, Love JC. Development of a general defined medium for Pichia pastoris. Biotechnol Bioeng. 2018;115:103–113. doi: 10.1002/bit.26440. [DOI] [PubMed] [Google Scholar]
- 31.Patel SKS, Ray S, Prakash J, Wee JH, Kim S-Y, Lee J-K, Kalia VC. Co-digestion of biowastes to enhance biological hydrogen process by defined mixed bacterial cultures. Indian J Microbiol. 2019;59:154–160. doi: 10.1007/s12088-018-00777-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel SKS, Kim JH, Kalia VC, Lee J-K. Antimicrobial activity of amino-derivatized cationic polysaccharides. Indian J Microbiol. 2019;59:96–99. doi: 10.1007/s12088-018-0764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short protocols in molecular biology. 3. New York: Wiley; 1986. p. 836. [Google Scholar]
- 34.Chiang C, Knight SG. d-Xylose reductase and xylitol dehydrogenase from Penicillium chrysogenum. Methods Enzymol. 1966;9:188–193. doi: 10.1016/0076-6879(66)09044-X. [DOI] [Google Scholar]
- 35.Singh RK, Singh R, Sivakumar D, Kondaveeti S, Kim T, Li J, Sung BH, Cho B-K, Kim DR, Kim SC, Kalia VC, Zhang Y-HPJ, Zhao H, Kang YC, Lee J-K. Insights into cell-free conversion of CO2 to chemicals by a multienzyme cascade reaction. ACS Catal. 2018;8:11085–11093. doi: 10.1021/acscatal.8b02646. [DOI] [Google Scholar]
- 36.Zhu Y-H, Liu C-Y, Cai S, Guo L-B, Kim I-W, Kalia VC, Lee J-K, Zhang Y-W. Cloning, expression, and characterization of a highly active alcohol dehydrogenase for production of ethyl (S)-4-chloro-3-hydroxybutyrate. Indian J Microbiol. 2019;59:225–233. doi: 10.1007/s12088-019-00795-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondaveeti S, Patel SKS, Woo J, Wee JH, Kim S-Y, Al-Raoush RI, Kim I-W, Kalia VC, Lee J-K. Characterization of cellobiohydrolases from Schizophyllum commune KMJ820. Ind J Microbiol. 2020;60:160–166. doi: 10.1007/s12088-019-00843-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J-K, Patel SKS, Sung BH, Kalia VC. Biomolecules from municipal and food industry wastes: an overview. Bioresour Technol. 2020;298:122346. doi: 10.1016/j.biortech.2019.122346s. [DOI] [PubMed] [Google Scholar]


