Zhao et al. leverage the analysis of local conformational changes in a mutant Hv1 channel to inform the design of new inhibitors. These arginine mimics have improved interaction with the voltage-sensing domain and inhibit the wild-type channel with higher strength than guanidine derivatives.
Abstract
The human voltage-gated proton channel Hv1 is a drug target for cancer, ischemic stroke, and neuroinflammation. It resides on the plasma membrane and endocytic compartments of a variety of cell types, where it mediates outward proton movement and regulates the activity of NOX enzymes. Its voltage-sensing domain (VSD) contains a gated and proton-selective conduction pathway, which can be blocked by aromatic guanidine derivatives such as 2-guanidinobenzimidazole (2GBI). Mutation of Hv1 residue F150 to alanine (F150A) was previously found to increase 2GBI apparent binding affinity more than two orders of magnitude. Here, we explore the contribution of aromatic interactions between the inhibitor and the channel in the presence and absence of the F150A mutation, using a combination of electrophysiological recordings, classic mutagenesis, and site-specific incorporation of fluorinated phenylalanines via nonsense suppression methodology. Our data suggest that the increase in apparent binding affinity is due to a rearrangement of the binding site allowed by the smaller residue at position 150. We used this information to design new arginine mimics with improved affinity for the nonrearranged binding site of the wild-type channel. The new compounds, named “Hv1 Inhibitor Flexibles” (HIFs), consist of two “prongs,” an aminoimidazole ring, and an aromatic group connected by extended flexible linkers. Some HIF compounds display inhibitory properties that are superior to those of 2GBI, thus providing a promising scaffold for further development of high-affinity Hv1 inhibitors.
Introduction
The Hv1 protein (also known as HVCN1 or VSOP) consists of four transmembrane helices, S1 through S4, forming a proton-conducting voltage-sensing domain (VSD; Ramsey et al., 2006; Sasaki et al., 2006) and lacks the pore domain typical of other voltage-gated ion channels (Bayrhuber et al., 2019; Takeshita et al., 2014). Hv1 regulates cellular pH homeostasis and the production of reactive oxygen species by NOX enzymes (DeCoursey, 2013; El Chemaly et al., 2010; Ramsey et al., 2009). Its activity has been reported to increase tumor metastatic potential in different types of cancer (Hondares et al., 2014; Wang et al., 2012; Wang et al., 2013) and to worsen brain damage after ischemic stroke (Li et al., 2019; Wu et al., 2012) in an age-dependent manner (Kawai et al., 2017), motivating the development of Hv1 inhibitors as potential anticancer drugs and neuroprotective agents. The role of microglial Hv1 in exacerbating inflammation in traumatic brain injury (Ritzel et al., 2021) and its contribution to motor deficits after spinal cord injury (Murugan et al., 2020) point to further useful applications of Hv1 inhibitors. Furthermore, Hv1 activity has been found to promote human sperm cell capacitation and motility (Lishko et al., 2010; Musset et al., 2012), suggesting that Hv1 antagonists could be used to aid male contraceptive treatments (Lishko, 2016).
Two major strategies have proved successful in developing ligands targeting the Hv1 VSD: one focused on the use of peptide toxins binding the channel extracellular side (Alabi et al., 2007; Tang et al., 2020; Zhao et al., 2018) and the other focused on small organic molecules targeting the channel intracellular side (Hong et al., 2014; Hong et al., 2013; Kalia and Swartz, 2013; Pupo and Gonzalez León, 2014). Some of these ligands have found applications as pharmacological tools to study the gating and permeation mechanisms of proton-conducting VSDs (Chamberlin et al., 2014; Gianti et al., 2016; Hong et al., 2015; Qiu et al., 2013; Wobig et al., 2020) and the role of Hv1-mediated proton currents in cellular physiology (Asuaje et al., 2017; Mészáros et al., 2020; Rennhack et al., 2017; Zhao et al., 2018). Drug-screening approaches on native proton currents or on other ion channels have led to the identification of additional potential ligands (Kornilov et al., 2014; Shin et al., 2015; Shin and Song, 2014; Song et al., 2012). However, the mechanism of action of these compounds has yet to be determined.
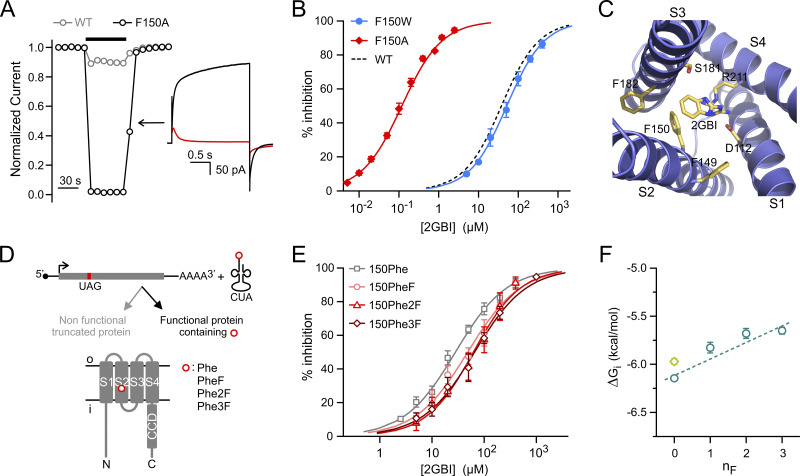
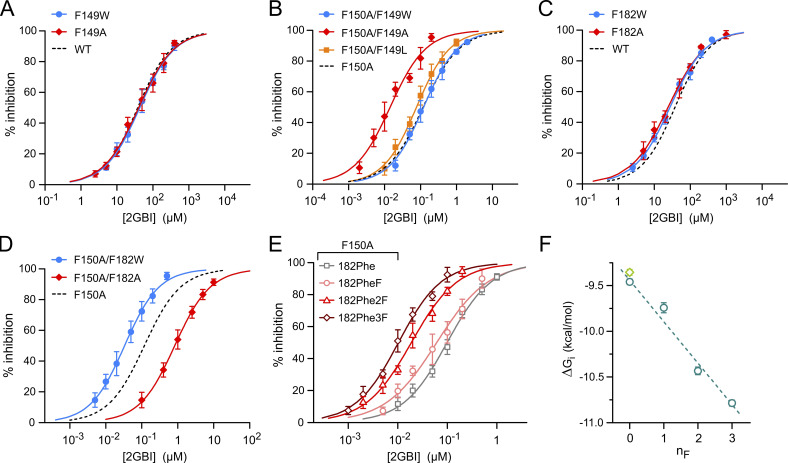
Like the VSDs of other voltage-gated channels, the Hv1 VSD changes conformation in response to membrane depolarization as its S4 helix transitions from a down state to an up state (Chamberlin et al., 2015; Geragotelis et al., 2020; Gonzalez et al., 2013; Okamura et al., 2015). The change in conformation also opens the conduction pathway, allowing protons to reach the selectivity filter located deep in the core of the VSD (Berger and Isacoff, 2011; Musset et al., 2011). The arginine mimic 2-guanidinobenzimidazole (2GBI) inhibits the human Hv1 channel by binding to the intracellular vestibule of the open VSD in the proximity of the selectivity filter (Chamberlin et al., 2014; Geragotelis et al., 2020; Gianti et al., 2016; Hong et al., 2013). Among the residues involved in 2GBI binding, phenylalanine (Phe) 150, located in the S2 helix, was found to play a particularly important role (Hong et al., 2014). The WT channel is inhibited by 2GBI in the micromolar concentration range (Gerdes et al., 2018; Hong et al., 2013). However, when F150 is mutated to alanine, inhibition occurs in the nanomolar range (Hong et al., 2013; Fig. 1, A and B; IC50,WT = 38 ± 2 µM, IC50,150A = 118 ± 7 nM). Understanding the molecular mechanism underlying this difference can lead to the design of new arginine mimics with higher binding affinity for the nonmutated channel.
Figure 1.
Exploring how residue F150 influences Hv1 sensitivity to 2GBI. (A) Drop in normalized current produced by 2 µM 2GBI in Hv1 WT and F150A. Black bar indicates the presence of the inhibitor in the bath solution. Representative traces from Hv1 F150A before (black) and after (red) addition of 2 µM 2GBI (inset). Measurements were performed in inside-out patch configuration. Currents were assessed at the end of a depolarization step to +120 mV from a holding potential of −40 mV, pHin = pHout = 6.0. (B) Concentration dependence of 2GBI-mediated inhibition of Hv1 mutants F150A and F150W compared with WT. Data points are averages of at least six independent measurements. Error bars are SD. (C) Structural model of the VSD of the human Hv1 in the open state from Geragotelis et al. (2020), showing the positions of F149 and F182 relative to F150 and other residues in the binding site (residue-ligand centroid distances in Å: 5.2 for F150, 8.5 for F149, and 9.1 for F182). (D) Schematics of unnatural amino acid substitution approach. (E) Concentration dependences of 2GBI-mediated inhibition of Hv1 constructs in which F150 is substituted with the indicated amino acids using the approach shown in D. Data points are averages from three to seven independent measurements ±SD. Curved lines in B and E represent fits of the data using Eq. 1. See Table S1 for fit parameters. (F) ΔGis measured from IC50s from E as a function of the nF in the Phe ring. Light-green diamond is the value for Hv1 WT. Error bars are SE. Dashed line is the linear fit of the data points in teal.
Here, we explore the effect of the F150A mutation on 2GBI-mediated inhibition using both standard mutagenesis and unnatural amino acid substitutions and find evidence of a local rearrangement in the binding site that leads to the increase in affinity. We then use this information to design more potent inhibitors for the WT channel. We investigate the effect of fluorination on aromatic rings involved in ligand-channel interactions and find that modifications of the six-membered ring of the inhibitor can negatively affect binding of the condensed five-membered ring. In the new Hv1 Inhibitor Flexible (HIF) inhibitors, the two rings are separated by an extended linker. We show that the separation improves the apparent binding to the WT channel over the F150A mutant and allows the fluorination of the six-membered ring to be fully effective at strengthening ligand–channel interactions.
Materials and methods
DNA constructs and channel expression
Constructs containing the sequence of the human HVCN1 channel were generated from cDNA kindly provided by David Clapham (Harvard University, Boston, MA; Ramsey et al., 2006) and from IMAGE clone 5577070 (Open Biosystems). Mutagenesis was performed as in Hong et al. (2013) and Tombola et al. (2008). The monomeric version of Hv1 (Hv1NCVSP) was previously described (Hong et al., 2015). All constructs were subcloned in the pGEMHE vector (Liman et al., 1992) and linearized with NheI or SphI restriction enzymes (New England Biolabs) before in vitro transcription. mRNAs were synthesized using T7 mMessage mMachine transcription kit (Ambion). Xenopus laevis oocytes from Ecocyte Bioscience or Xenopus 1 were injected with mRNAs (50 nl per cell, 0.5–1.5 ng/nl) 1–3 d before the electrophysiological measurements. Injections were performed with a Nanoject II (Drummond Scientific). Cells were kept at 18°C in ND96 medium containing 96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 5 mM pyruvate, and 100 μg/ml gentamycin (pH 7.2).
Unnatural amino acid substitutions
Amber (UAG) stop codons were introduced at specific positions of HVCN1 constructs using site-directed mutagenesis. To ensure proper termination of translation, the native UAG stop codon was replaced by a UAA stop codon. Preparation of synthetic aminoacyl-tRNAs was performed as previously described (Infield et al., 2018a). In brief, N-Boc–protected amino acids were activated as cyanomethyl esters for subsequent coupling to the hybrid dinucleotide phospho-deoxycytidine-phospho-adenosine (pdCpA). Modified tRNA from Tetrahymena thermophila (THG73) containing the CUA anticodon was generated in vitro and ligated to pdCpA carrying the amino acid at the 3′ end. Deprotected aminoacyl-tRNAs were kept at −80°C until use. Right before injection, frozen tRNA aliquots were resuspended in ice-cold ribonuclease-free water. After centrifugation at 4°C for 25 min at 9,600 g, the solutions were transferred in prechilled tubes containing mRNAs with amber stop codons at position F150 or F182. The resulting mixtures had mRNA:tRNA ratios of 1:1 (wt/wt). 50 nl of mixture was injected per cell. Injection of constructs with amber stop codons in the absence of tRNA or in the presence of nonacylated tRNA did not produce measurable currents. Acylated tRNA carried either nonsubstituted Phe or one of the following fluorinated amino acids: 4-fluoroPhe (PheF); 3,4-difluoroPhe (Phe2F); or 3,4,5-trifluoroPhe (Phe3F).
Patch-clamp measurements
Voltage-clamp measurements were performed in inside-out patch configuration using an Axopatch 200B amplifier controlled by pClamp10 software through an Axon Digidata 1440A (Molecular Devices). The signal was lowpass filtered at 1 kHz (Bessel, −80 dB/decade) before digitalization (2-kHz sampling). For display purposes, it was further filtered offline at 200 or 150 Hz (Bessel, −80 dB/decade). Bath and pipette solutions contained 100 mM Mes, 30 mM TEA methanesulfonate, 5 mM TEA chloride, and 5 mM EGTA, adjusted to pH 6.0 with TEA hydroxide. All measurements were performed at 22 ± 1°C. Pipettes had 1–3-MΩ access resistance. Unless otherwise specified, the holding potential was −40 mV, and the depolarization potential was +120 mV. Channel inhibition was determined by isochronal current measurements at the end of the depolarization pulses. Hv1 inhibitors were introduced in the bath using a computer-controlled gravity-fed multivalve perfusion system (Warner Instruments).
Tested Hv1 ligands
All tested compounds were at the highest purity commercially available. 2-aminobenzimidazole (ABI), 2GBI, 2-aminobenzothiazole (ABTA), and 2-guanidino-benzothiazole (GBTA) were from Sigma-Aldrich. 6-fluoro-1H-1,3-benzodiazol-2-amine (ABIFβ); 5,6-difluoro-1H-1,3-benzodiazol-2-amine (ABIF2), and 4,5,6-trifluoro-1H-1, 3-benzodiazol-2-amine (ABIF3) were from Enamine. 4-fluoro-1H-1, 3-benzodiazol-2-amine (ABIFα) was from Combi-Blocks. 3-(2-amino-5-methyl-1H-imidazol-4-yl)-1-(3,5-difluorophenyl)propan-1-one hydrochloride (HIF); 3-(2-amino-5-methyl-1H-imidazol-4-yl)-1-phenylpropan-1-one hydrochloride (HIFNF); N-[(2-amino-5-methyl-1H-imidazol-4-yl)methyl]-3,5-difluorobenzamide (HIFNH); 3-(2-amino-5-methyl-1H-imidazol-4-yl)-1-(3,5-difluorophenyl)propan-1-ol hydrochloride (HIFOH); and (2E)-3-(2-amino-5-methyl-1H-imidazol-4-yl)-1-(3,5-difluorophenyl)prop-2-en-1-one hydrochloride (HIFEN) were custom synthesized by Enamine at a minimum purity of 95% (liquid chromatography–mass spectrometry). See text at the bottom of the PDF for characterization.
Data analysis
Current traces were analyzed using Clampfit10.2 (Molecular Devices) and Origin8.1 (OriginLab). Leak subtraction, rundown correction, and the derivation of concentration dependence curves were performed as in Hong et al. (2013). Concentration dependences were fitted with the Hill equation (Eq. 1):
| (1) |
where %i is the percentage of inhibition at the ligand concentration, [ligand], %i,max is the percentage of maximal inhibition (assumed to be 100%; Hong et al., 2014; Hong et al., 2013), IC50 is the half-maximal inhibitory concentration, and h is the Hill coefficient. The Hill coefficients for all the measured concentration dependences were in the range 0.83–1.50 (see Table S1 for details) in agreement with earlier findings (Hong et al., 2014; Hong et al., 2015). ΔGi free energies were estimated using the relationship: ΔGi = RTlnIC50, where R is the gas constant and T the absolute temperature.
G-V curves were derived from tail currents as previously described (Hong et al., 2013; Tombola et al., 2010). Current rundown was corrected using a reference depolarization step preceding the test depolarization. Conductance was determined from G(Vtest) = (Itest − Itail)/(Vtest − Vtail), where Itail and Vtail are the tail current and voltage (−40 mV) following the depolarization step at Vtest (ranging from −30 mV to +120 mV), and Itest is the current measured at the end of the depolarization step. Gmax was determined from maximal Itail (and corresponding Itest) in the Vtest region in which the tail current saturated. G-V plots were fitted with the Boltzmann equation (Eq. 2):
| (2) |
where V1/2 is the potential of half-maximal activation and s is the slope, all in millivolts. Unless otherwise specified, data are reported as averages from at least four independent measurements, and error bars are SEM. Fitting parameters are shown with standard error (SE). Each average comes from measurements performed on at least two distinct batches of cells. A Welch’s t test was used for statistical analysis of datasets for two compared conditions. A one-way ANOVA test with Tukey’s post hoc correction was used for multiple comparison analysis. No randomization or blinding was applied to this study.
Docking calculations and MD simulations
HIF was parameterized using CGenFF version 2b4 (Vanommeslaeghe and MacKerell, 2012; Vanommeslaeghe et al., 2012). MD simulations of HIF-bound Hv1 VSD embedded in a solvated POPC lipid bilayer were performed using NAMD 2.13 (Phillips et al., 2005). The CHARMM36 force field (Klauda et al., 2010; MacKerell et al., 1998) was used for both protein and lipid, and the TIP3P model was used for water (Jorgensen et al., 1983). The model of the human Hv1 VSD in the up state was from Geragotelis et al. (2020). The final structure of a 25-ns-long trajectory of the Hv1 VSD + 2GBI was used as the initial structure of the Hv1 VSD + HIF system, where 2GBI was substituted by a HIF molecule (aligned and superimposed onto the 2GBI structure). Subsequently, 50 steps of geometry optimization and 1 ns of equilibration under NVT conditions were performed followed by 25 ns of isothermal-isobaric ensemble simulation. Langevin MD (Tuckerman and Berne, 1991) with 1 ps−1 frictional coefficient and Nose-Hoover barostat (Martyna et al., 1994) were used to maintain the temperature and pressure at 300 K and 1 atm, respectively. MD trajectories were obtained with the time step of 2 fs, while all bonds involving H atoms were constrained using the SHAKE algorithm (van Gunsteren and Berendsen, 1977). The system was composed of 10,784, 175, 33, and 38 water, lipid, Na, and Cl molecules, respectively, inside an orthorhombic box unit cell (82.0 × 82.0 × 83.6 Å3) under periodic boundary conditions. The short-range nonbonded interactions were included at the 12 Å cutoff distance, and the long-range electrostatic interactions were incorporated using the Particle Mesh Ewald algorithm (Darden et al., 1993). The system was simulated under 0 mV membrane potential.
In silico absorption, distribution, metabolism, and excretion–toxicity (ADMET) predictions
ADMET properties of 2GBI and HIF compounds were predicted using online tools available at SwissADME (http://www.swissadme.ch), XenoSite (https://swami.wustl.edu/xenosite), and MetaTox (www.way2drug.com/mg2).
SwissADME (Daina et al., 2017) provides predictions on pharmacokinetics, drug-likeness, and medicinal chemistry friendliness. Through its BOILED-Egg model, it predicts gastrointestinal absorption and brain penetration of small molecules (Daina and Zoete, 2016). XenoSite uses a robust neural-network model to predict the atomic sites at which xenobiotics will undergo metabolic modification by Cytochrome P450 enzymes (Zaretzki et al., 2013), whereas MetaTox utilizes the GUSAR algorithm (Zakharov et al., 2016) to estimate the integrated toxicity of xenobiotics and their predicted metabolites expressed as lethal dose, 50% (LD50) for rats with intravenous type of administration (Rudik et al., 2017).
Online supplemental material
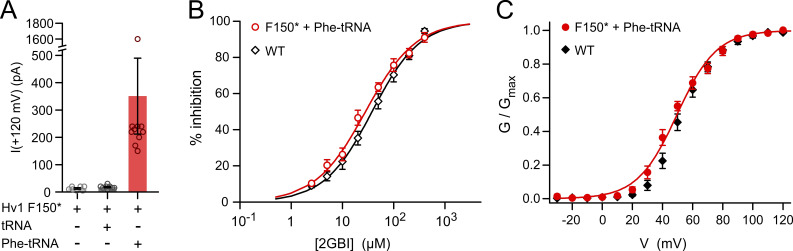
Fig. S1 shows the functional rescue of Hv1 F150* by suppressor Phe-tRNA. Fig. S2 compares the poses of HIF and 2GBI within a structural model of the up state of the Hv1 VSD. Fig. S3 shows the concentration dependences of HIFEN-mediated inhibition of dimeric (WT) and monomeric Hv1. Fig. S4 reports ADMET predictions for HIF compounds and 2GBI. Table S1 shows the fit parameters for the concentration dependences of the Hv1 inhibitors examined in this study. The Supplemental text section reports the chemical characterization of custom-synthesized HIF compounds; see bottom of the PDF.
Figure S1.
Functional rescue of Hv1 F150* by suppressor tRNA. Suppressor Phe-tRNA rescues function of Hv1 mRNA containing UAG codon at position F150 (F150*). (A) Average currents measured from oocytes injected with the indicated combinations of F150* cRNA and tRNAs. Measurements were performed at +120 mV in inside-out patch configuration on 7–10 cells per condition at pHin = pHout = 6.0. Error bars are SEM. (B) Concentration dependences of 2GBI-mediated inhibition of proton currents from the indicated channels. Points are averages from at least three independent measurements ±SD. Curved lines represent fits of the data using Eq. 1. (IC50 = 28 ± 2 µM for F150* mRNA + Phe-tRNA, IC50 = 38 ± 2 µM for Hv1 WT). (C) G-V relationships of proton currents from F150* mRNA + Phe-tRNA and Hv1 WT. Points are averages from at least four independent measurements ±SEM. Data were fitted with Eq. 2. Fit is shown as curved line for F150* mRNA + Phe-tRNA. (V1/2 = 50 ± 3 mV, slope = 13 ± 1 mV for F150* mRNA + Phe-tRNA; V1/2 = 53 ± 3 mV, slope = 11 ± 1 for Hv1 WT).
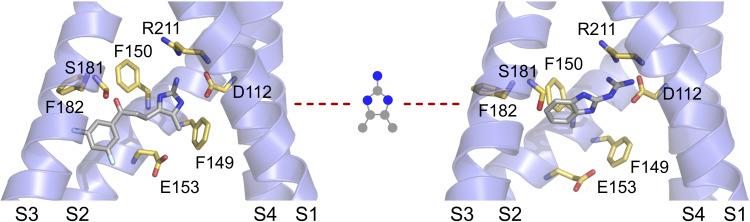
Figure S2.
HIF and 2GBI bound the Hv1 VSD. Comparison of HIF and 2GBI poses within the Hv1 intracellular vestibule in the up state. The 2GBI pose (right panel) is from Geragotelis et al. (2020). The drawing in the center represents the moiety used to align the two inhibitors at the beginning of the HIF simulation. Left panel shows the HIF pose at the end of the simulation.
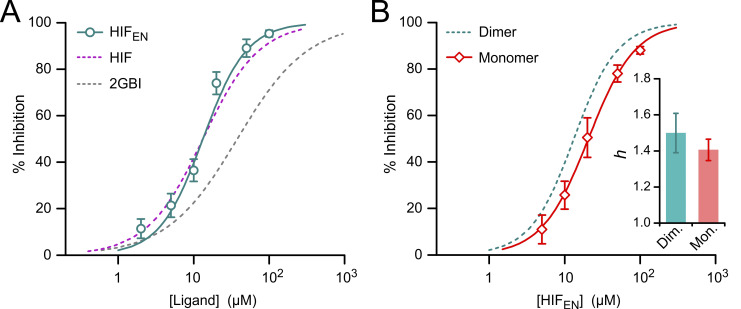
Figure S3.
Hv1 inhibition by HIFEN. (A) Concentration dependence of inhibition of Hv1 WT by HIFEN (teal) compared with HIF (purple) and 2GBI (gray). Each data point represents the mean of five or six independent measurements; error bars are SD. (B) Concentration dependence of HIFEN-mediated inhibition of monomeric (Mon.) Hv1 (red diamonds) compared with dimeric (Dim.) Hv1 (teal dashed line). Monomeric Hv1 was Hv1NCVSP as in Hong et al. (2015). Each data point represents the mean of four or five independent measurements; error bars are SD. Data points in A and B were fitted with Eq. 1. The resulting Hill coefficients (h) are compared in the bar graph. Error bars are SE. IC50 values are reported in Table S1.
Figure S4.
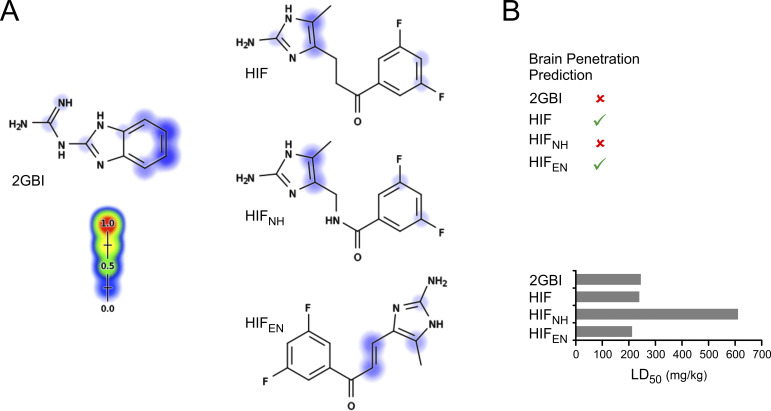
Predicted metabolism and toxicity of HIF compounds versus 2GBI. (A) Sites of cytochrome P450–mediated metabolism for 2GBI, HIF, HIFNH, and HIFEN predicted by XenoSite (https://swami.wustl.edu/xenosite). Scale goes from 0% (blue) to 100% (red) probability of metabolism. (B) Top: Ability to permeate the blood–brain barrier (green checks) for the indicated compounds predicted using BOILED-Egg (http://www.swissadme.ch). Bottom: Prediction of integrated toxicity of the indicated compounds from MetaTox (www.way2drug.com/mg2). Values take into account effects of all first- and second-level metabolites and effect of parent compound on integrated toxicity.
Results
Mutation F150A induces a local rearrangement in the 2GBI binding site
An earlier mutant cycle analysis of 2GBI binding uncovered an interaction between the condensed phenyl ring of the inhibitor and the aromatic ring of F150 (Hong et al., 2014), but the mechanism of this interaction remained unexplored. If either direct steric hindrance or aromatic interactions between rings had a dominant effect on binding, a tryptophan substitution at position 150 would be expected to substantially affect the overall ligand-channel interaction. To test this possibility, we replaced F150 with tryptophan by standard site-directed mutagenesis, expressed the mutant channel in Xenopus oocytes and measured the inhibition curve (IC; concentration dependence of inhibition) of the resulting proton currents in excised membrane patches (Fig. 1 B). The amino acid substitution produced only a minor change in the IC (IC50,150W = 52 ± 2 µM), suggesting that the effect of mutation F150A involves a more complex mechanism.
Compared with Phe, tryptophan is larger in size and can engage in stronger cation–Π or Π–stacking interactions with positively charged or aromatic ligands, respectively. Thus, the F150W substitution may have a mixed effect on 2GBI binding, with a destabilizing component caused by an increase in steric hindrance and a stabilizing component caused by strengthened aromatic interactions. To dissect interactions with opposing effects, we manipulated the Π electron density of F150 by unnatural amino acid substitutions via codon-suppression technology (Infield et al., 2018b; Rodriguez et al., 2007a, 2007b; Fig. 1 D). Due to their high electronegativity and compact size, fluoro-substituents are particularly suitable for altering the Π electron density of aromatic rings without causing large steric perturbations. Fluoro-substitutions affect both cation–Π and Π–stacking interactions and have been previously used to uncover the role of aromatic residues in the stabilization of protein conformations and ligand binding (Pless and Ahern, 2013; Van Arnam and Dougherty, 2014).
We first verified that introducing Phe with no substituents at position 150 using the codon-suppression method produced channels with the same properties as WT Hv1 (Fig. S1). We then replaced the ring of F150 with rings containing one, two, or three fluoro-substituents and measured 2GBI IC50s for each modified channel (Fig. 1 E). We calculated apparent binding free energy (ΔGi) values from IC50s and plotted them as a function of the number of fluoro substituents (nF) in F150 (Fig. 1 F). We observed a positive correlation between ΔGi and nF, suggesting a destabilization of 2GBI apparent binding (ΔGi becomes less negative) with the reduction of the Π-electron density at F150. This result is consistent with the existence of a cation–Π interaction between 2GBI and F150. However, the stabilizing contribution of the interaction appears to be modest, as indicated by the shallow slope of the linear fit in Fig. 1 F (ΔGi/ΔnF = 0.17 ± 0.03 kcal/mol).
The weakly stabilizing interaction between 2GBI and F150 in Hv1 WT must be replaced by stronger interactions between the ligand and the binding site in Hv1 F150A to account for the lower IC50 of the mutant channel. We considered the possibility that the replacement of the aromatic ring of F150 with the small methyl group of alanine could cause rearrangements in the side chains of neighboring residues, resulting in new stabilizing interactions with the inhibitor molecule. We searched for candidate side chains in a structural model of the human Hv1 VSD in the open conformation, built using the 3WKV crystal structure (Takeshita et al., 2014) as initial template (Geragotelis et al., 2020; Fig. 1 C). We noticed that F150 is in proximity to other two aromatic residues, F149 and F182, and hypothesized that one of these residues could be closer to the inhibitor in the F150A mutant, producing a stabilization of 2GBI binding.
Stabilizing aromatic interactions between 2GBI and a Phe in the binding site are expected to be abolished by an alanine substitution at the Phe position, leading to a shift of the IC to higher inhibitor concentrations (right shift). Conversely, the same interactions are expected to be strengthened by a tryptophan substitution, leading to a shift of the IC to lower inhibitor concentrations (left shift). Hence, to determine whether F149 or F182 participates in 2GBI binding in Hv1 F150A, we measured ICs for channels in which F149 or F182 were replaced by either an alanine or a tryptophan, in the presence and absence of the additional mutation F150A.
When F150 was not mutated, alanine or tryptophan substitutions at F149 or F182 produced negligible changes in the IC (Fig. 2, A and C), confirming our previous finding that these Phes are not involved in 2GBI binding in WT Hv1 (Hong et al., 2014). In the presence of F150A, however, the same substitutions perturbed 2GBI-mediated inhibition in different ways (Fig. 2, B and D). The IC was shifted to lower concentrations by the F149A mutation and was unaffected by the F149W mutation (Fig. 2 B; IC50,150A,149A = 14 ± 2 nM, IC50,150A,149W = 123 ± 6 nM). On the other hand, the IC was shifted to higher concentrations by the F182A mutation and to lower concentrations by the F182W mutation (Fig. 2 D; IC50,150A,182A = 826 ± 22 nM, IC50,150A,182W = 33 ± 1 nM).
Figure 2.
Aromatic interactions stabilizing ligand binding in Hv1 F150A. (A and B) Concentration dependences of 2GBI-mediated inhibition of Hv1 WT (A) and F150A (B) with and without the indicated substitutions at position F149. (C and D) Concentration dependences of 2GBI-mediated inhibition of Hv1 WT (C) and F150A (D) with and without the indicated substitutions at position F182. Each data point in A–D is an average from three to nine independent measurements. Error bars are SD. (E) Concentration dependences of 2GBI-mediated inhibition of Hv1 F150A in which F182 is substituted with the indicated amino acids introduced via UAG suppressor tRNA. Each data point is an average from three to eight independent measurements. Curved lines represent fits of the data using Eq. 1. See Table S1 for fit parameters. (F) ΔGis measured from IC50s from E as a function of the nF in the Phe ring. Light-green diamond is the value for Hv1 WT. Error bars are SE. Dashed line is the linear fit of the data points in teal.
The changes in IC50 caused by mutations F182A and F182W in the presence of F150A are consistent with a stabilizing interaction between 2GBI and F182 via aromatic interactions. In contrast, the shift of the IC to lower concentrations observed with the F149A substitution (Fig. 2 B) suggests that additional rearrangements of the binding site are responsible for binding stabilization in the F149A-F150A channel. Furthermore, replacing F149 with leucine, rather than alanine, produced only a negligible shift in the IC (F149L; Fig. 2 B), indicating that the effect of the F149A mutation is most likely due to the reduction in the size of the side chain rather than the loss of the aromatic ring.
Evidence for Π–stacking interactions between 2GBI and F182 in Hv1 F150A
To better understand how F182 stabilizes 2GBI apparent binding in the context of the F150A mutation, we introduced fluoro-substituted Phes at position 182 and measured how the resulting modification in Π-electron density affected the IC (Fig. 2 E). We first verified that the introduction of nonsubstituted Phe at position F182 using the codon-suppression method did not significantly alter 2GBI-mediated inhibition in the F150A background (Fig. 2 F, points at nF = 0). We then replaced F182 with Phes containing rings with one, two, or three fluoro-substituents and measured 2GBI ICs for each modified F150A channel (Fig. 2 E). We calculated ΔGi values from IC50s and plotted them as a function of nF (Fig. 2 F). We observed a negative correlation between ΔGi and nF, suggesting a stabilization of 2GBI apparent binding (ΔGi becomes more negative) with the reduction of the Π-electron density in the F182 ring.
Fluorination of an aromatic side chain can alter protein–ligand interactions in different ways, depending on the orientation of the ligand relative to the side chain, the charge of the ligand, and the polarity of the binding environment. For example, a face-to-face Π–stacking interaction with an electron-rich aromatic ligand in a hydrophobic environment is expected to be strengthened by fluoro-substituents in the aromatic side chain (Cockroft et al., 2005). While our findings are consistent with a Π–stacking interaction between 2GBI and F182 in Hv1 F150A, we cannot rule out the existence of other stabilizing interactions of electrostatic or steric nature. In any case, the interaction seems to provide a stronger contribution to 2GBI-mediated inhibition than the interaction with F150 assessed in Hv1 WT, as indicated by the steeper slope of the linear fit in Fig. 2 F (ΔGi/ΔnF = −0.45 ± 0.04 kcal/mol).
ABI: A simplified 2GBI analogue suitable for investigating binding to the F150A mutant
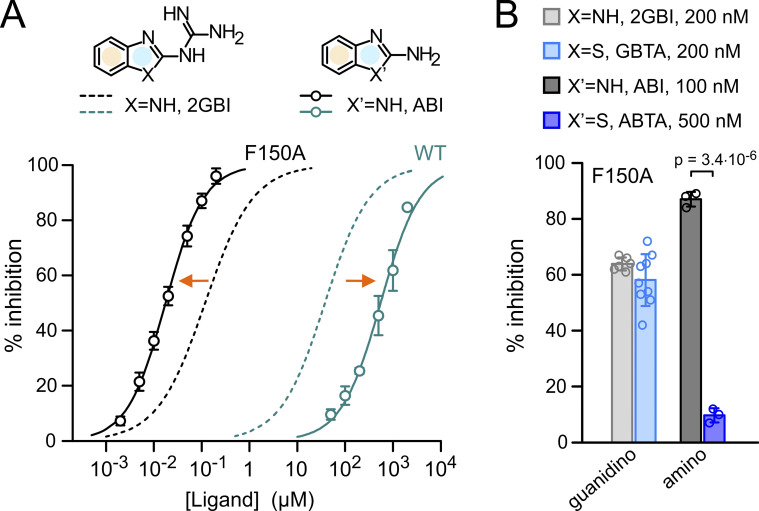
Because addition of fluoro-substituents to the phenyl ring of F182 decreased the IC50 of 2GBI-mediated inhibition of Hv1 F150A, we wondered whether we could create compounds that bind the channel with higher affinity by adding fluoro-substituents to the condensed phenyl ring of the ligand (Fig. 3 A, yellow shade). To investigate this point, we focused on a simplified version of the inhibitor in which the guanidino group linked to the benzimidazole unit is replaced by an amino group. This compound, ABI (Fig. 3 A), was identified as a 2GBI analogue with reduced potency in earlier inhibition assays of Hv1 WT (Hong et al., 2013).
Figure 3.
Simplified ligand for binding optimization. (A) Concentration dependences of inhibition of Hv1 WT and F150A by 2GBI and ABI. Data points are averages from three to five independent measurements ±SD. Curved lines represent fits of the data using Eq. 1. See Table S1 for fit parameters. Orange arrows indicate that Hv1 F150A is more sensitive to ABI than 2GBI, while the situation is reversed for WT. Condensed phenyl and 2-substituted imidazole rings are highlighted in yellow and pale blue, respectively. (B) Percentages of inhibition of Hv1 F150A by guanidino and amino derivatives of benzimidazole and benzothiazole at the indicated concentrations. GBTA and 2GBI showed similar potency, so they could be compared at the same concentration. This was not the case for ABTA and ABI. The minimal concentration at which ABTA-mediated inhibition could be accurately measured was 500 nM, but at that concentration ABI-mediated inhibition was saturated. Despite lowering ABI concentration to 100 nM, this inhibitor still produced a much larger inhibition than ABTA. Error bars are SD (n = 3–9). Welch's t test was used for statistical analysis.
We confirmed that the apparent binding affinity of Hv1 WT for ABI is lower than the apparent binding affinity for 2GBI (IC50 = 553 ± 43 µM for ABI, IC50 = 38 ± 2 µM for 2GBI; Fig. 3 A); however, we found that the situation is reversed for Hv1 F150A. In this case, ABI has the higher apparent binding affinity (IC50 = 18 ± 1 nM for ABI, IC50 = 118 ± 7 nM for 2GBI; Fig. 3 A).
We also found that the inhibition of Hv1 F150A by ABI is more sensitive to modifications of the benzimidazole unit than the inhibition by 2GBI. Replacing the NH group of 2GBI with a sulfur substituent (Fig. 3 B, GBTA) resulted in only a negligible change in inhibitor potency (Fig. 3 B, guanidino). However, when the analogous NH group of ABI was replaced by the same substituent (Fig. 3 B, ABTA), the effect on potency was much larger (Fig. 3 B, amino). We reasoned that the ability of ABI to inhibit Hv1 F150A with high affinity and in a way that is highly sensitive to modifications of the benzimidazole unit would make this ligand a good model to study how fluoro-substituents affect binding.
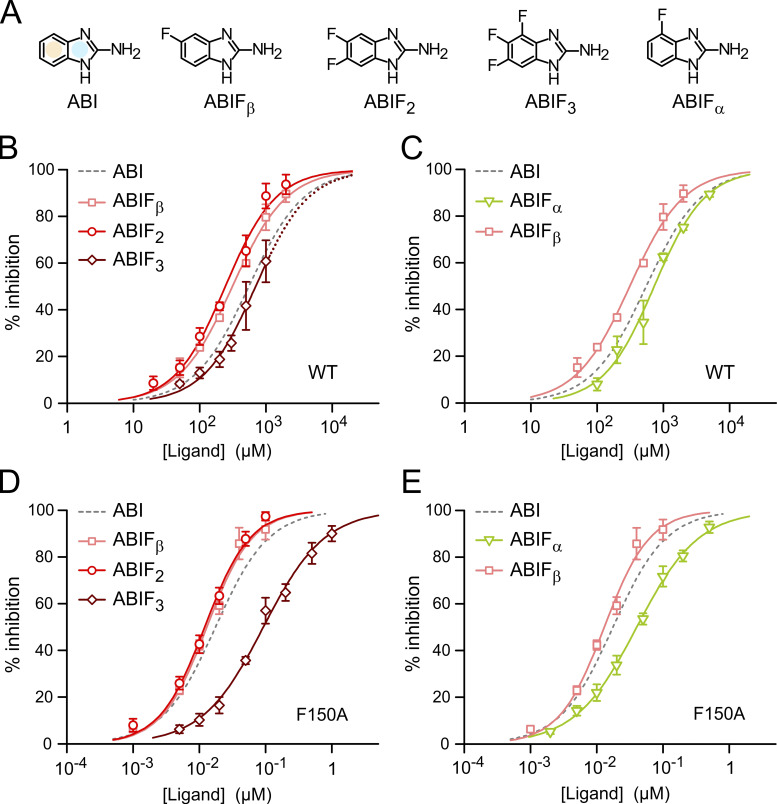
Fluoro-substituted ABIs reveal limitation of ligands with condensed rings
We measured the inhibition of Hv1 WT and F150A by ABI derivatives containing one, two, or three fluoro-substitutions in the condensed phenyl ring of the benzimidazole unit (Fig. 4 A, yellow shade). We found that the addition of one or two substituents produced only small changes in the IC (IC50,ABIFβ = 312 ± 17 µM, IC50,ABIF2 = 244 ± 19 µM for WT; IC50,ABIFβ = 13 ± 1 nM, IC50,ABIF2 = 12 ± 1 nM for F150A), while the addition of the third substituent significantly shifted the IC50 to higher concentrations (IC50,ABIF3 = 699 ± 45 µM for WT; IC50,ABIF3 = 93 ± 6 nM for F150A; Fig. 4, B and D). The trend was the same in Hv1 WT and F150A, but the shift in IC50 with the trifluorinated ligand was more pronounced in Hv1 F150A.
Figure 4.
Impact of phenyl ring fluorination on ABI-mediated inhibition of Hv1 WT and F150A. (A) Structures of tested ABIs showing positions of fluoro substituents in condensed phenyl ring (yellow shade). (B–E) Concentration-dependences of inhibition of Hv1 WT (B and C) and F150A (D and E) by the indicated compounds. Each data point is the average value from three to eight independent measurements. Error bars are SD. Curved lines represent fits of the data using Eq. 1. See Table S1 for fit parameters. Dotted line in B indicates extrapolation of the IC for ABIF3 to concentrations >1 mM (the compound was not soluble at higher concentrations under tested conditions).
Ligand fluorination was expected to strengthen Π–stacking interaction with aromatic residues such as F182 in the F150A channel. So, why did the IC50 shift to higher concentrations with the addition of the third fluorine? We suspected that as the fluoro-substituents withdraw Π electrons from the ligand-condensed phenyl ring, they may also perturb the charge distribution in the adjacent five-membered ring (Fig. 4 A, pale-blue shade), destabilizing its interactions with other parts of the binding site. In this case, a substituent positioned closer to the five-membered ring (Fig. 4 A, ABIFα) should have a stronger destabilizing effect than a substituent positioned farther from that ring (ABIFβ). We tested this hypothesis by comparing the concentration dependences of ABIFα and ABIFβ inhibition (Fig. 4, C and E). Indeed, we found that while the substitution at the β position caused a small increase in apparent affinity, the substitution at the α position caused a decrease. Again, the destabilizing effect was more pronounced in Hv1 F150A (IC50,ABIFα = 729 ± 56 µM, IC50,ABIFβ = 312 ± 17 µM for WT; IC50,ABIFα = 40 ± 1 nM, IC50,ABIFβ = 13 ± 1 nM for F150A).
Design of HIFs: Two-pronged arginine mimics with noncondensed rings
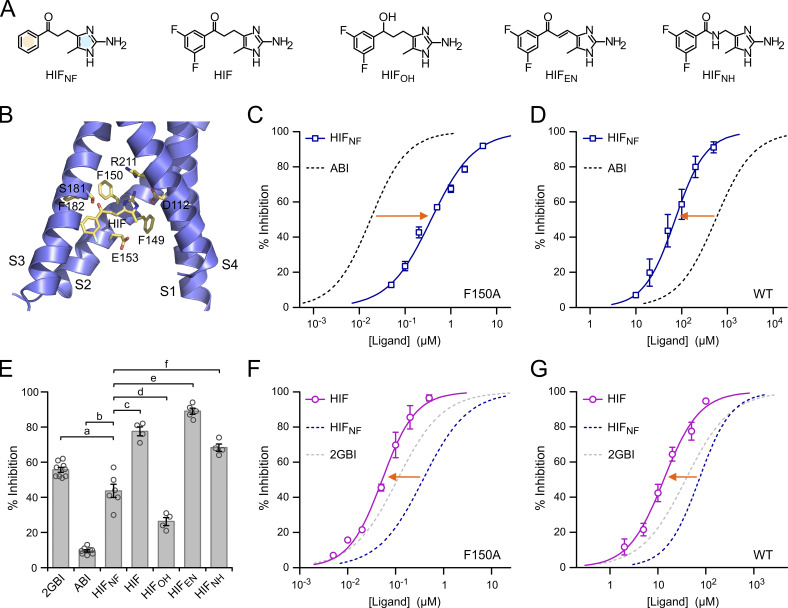
We reasoned that the presence of two condensed aromatic rings in ABI and 2GBI presents a potential vulnerability for the development of analogues with increased binding affinity because substituents intended to stabilize the interaction between one ring and the binding site can interfere with interactions mediated by the other ring. To prevent this interference, we designed a new class of Hv1 inhibitors related to ABI in which the phenyl ring and the 2-aminoimidazole ring are separated by a flexible linker and named them HIFs (Fig. 5 A).
Figure 5.
Hv1 inhibition by HIF compounds. (A) Structures of HIF and related compounds. In HIFNF, the separate phenyl and 2-aminoimidazole rings are highlighted in yellow and blue, respectively. (B) Structural model of the VSD of human Hv1 in the activated conformation (from Geragotelis et al., 2020) interacting with HIF at the end of the MD simulation described in the Materials and methods section and showing residues in the vicinity of the ligand. (C and D) Concentration dependences of HIFNF-mediated inhibition of Hv1 F150A (C) and Hv1 WT (D) compared with ABI. Each data point represents the mean of three to seven independent measurements ±SD. Curved lines represent fits of the data using Eq. 1. See Table S1 for fit parameters. Orange arrows indicate that HIFNF is more effective than ABI at inhibiting Hv1 WT, whereas the situation is reversed for Hv1 F150A. (E) Inhibition of Hv1 WT by the indicated compounds tested at a concentration of 50 µM. Each bar is the mean of four to nine independent measurements. Error bars are SEM. A one-way ANOVA with Tukey’s post hoc test was used for statistical analysis. Comparisons between all pairs of inhibitors were statistically significant (P < 0.05), except for HIF/HIFNH (P > 0.05). For clarity, only comparisons with HIFNF are shown: p(a) = 1.4⋅10−3, p(b) < 1⋅10−9, p(c) = 6.4⋅10−8, p(d) = 1.2⋅10−4, p(e) = 1.0⋅10−9, p(f) = 3.3⋅10−7. (F and G) Concentration dependences of HIF-mediated inhibition of Hv1 F150A (F) and Hv1 WT (G) compared with HIFNF. Each data point represents the mean of three to five independent measurements ±SD. Curved lines represent fits of the data using Eq. 1. See Table S1 for fit parameters. Orange arrows indicate that fluorination of the phenyl ring increases the ligand apparent binding affinity to both Hv1 WT and F150A.
The 2-aminoimidazole moiety was previously identified as an arginine-mimic pharmacophore for Hv1 (Hong et al., 2014; Hong et al., 2013). As a result, this part of the ligand was preserved in HIF compounds. We tested three different ways to connect the phenyl ring to the 2-aminoimidazole moiety. The different connecting linkers were designed to allow the phenyl ring to explore the binding site for potential stabilizing interactions while minimizing the increase in overall hydrophobicity. The carbonyl group connected to the phenyl ring in HIFNF, and HIF is replaced by a hydroxyl group in HIFOH or by an amide group in HIFNH. A double bond in the E-configuration is present in the linker of HIFEN. The unsaturated linker is expected to reduce flexibility and extend the delocalization of Π-electrons.
HIF shares the same core structure of HIFNF, but its phenyl ring is fluorinated. These two compounds were chosen to investigate how ring separation affects the modulation of binding interactions by substituents in the phenyl ring. At pH 6.0, all HIF compounds have predicted LogD values (logarithm of distribution coefficients) lower than 1—LogD (HIF) = 0.37, LogD (HIFEN) = 0.83, LogD (HIFOH) = 0.28, LogD (HIFNF) = 0.09, LogD (HIFNH) = −0.45—and, for the most part, in the range of LogDs of fluorinated ABIs calculated at the same pH (−0.1 − 0.5, ChemAxon LogD Predictor).
We verified that the new inhibitor core structure is compatible with the 2GBI binding site (Hong et al., 2014) by docking HIF within the structural model of the Hv1 VSD in the up state that was previously used for the characterization of 2GBI binding (Geragotelis et al., 2020; Lim et al., 2020). We set the initial position of HIF so that its 2-aminoimidazole ring and the two carbon atoms at positions 4 and 5 would overlap with the corresponding moiety of bound 2GBI. We then ran an unrestrained all-atom MD simulation and followed the movement of the inhibitor for 25 ns. The position of the inhibitor at the end of the simulation is shown in Fig. 5 B. A comparison between HIF and 2GBI bound to the same location within the Hv1 VSD is reported in Fig. S2.
Hv1 inhibition by HIF compounds compared with inhibition by ABI and 2GBI
We tested HIFNF at different concentrations on proton currents measured in inside-out patches from oocytes expressing Hv1 F150A and Hv1 WT. From the resulting concentration curves (Fig. 5, C and D), we found that HIFNF is less potent on Hv1 F150A compared with ABI, (IC50,ABI/IC50,HIFNF ∼0.05), but it is more potent on the WT channel (IC50,ABI/IC50,HIFNF ∼7.7). All HIF compounds were more potent than ABI at inhibiting Hv1 WT, and some of them were even more potent than 2GBI (Fig. 5 E). The different potencies observed with HIF compounds containing different linkers indicate that the connection between the 2-aminoimidazole ring and the phenyl ring affects how the ligands interact with the channel. In particular, HIFOH was less effective at reducing the proton current than HIF, HIFNH, and HIFEN (Fig. 5 E), pointing to an important role for the carbonyl group in the linker.
We compared ICs of HIFNF and its fluorinated analogue, HIF, and found that fluorination of the phenyl ring produced significant increases in potency. The IC50,HIFNF/IC50,HIF was ∼7.0 for Hv1 F150A and ∼5.4 for Hv1 WT (Fig. 5, F and G). These increases were much larger than those observed with similar fluorination of the condensed phenyl ring of ABI (Fig. 4, B–E). Despite lacking the guanidino group of 2GBI, HIF was able to inhibit Hv1 WT with a lower IC50 (Fig. 5 G; IC50,HIF = 13 ± 1 µM, IC50,2GBI = 38 ± 2 µM). HIFNH and HIFEN had similar or larger potency than HIF (Fig. 5 E). Taken together, these findings indicate that the scaffold of HIF compounds interacts more favorably with the WT channel than the ABI/2GBI scaffold, making HIFs promising lead candidates for the development of high-affinity Hv1 inhibitors.
While HIFEN seems slightly more potent than HIF at 50 µM (Fig. 5 E; P = 0.027), there is no significant difference in the IC50 values for the two compounds (Fig. S3 B and Table S1). Because the steepness of the concentration dependence of HIFEN-mediated inhibition appeared to be particularly high, with a Hill coefficient of ∼1.5 (Fig. S3 B, inset; and Table S1), we wondered whether this was due to the dimeric nature of the Hv1 channel. In an earlier study (Hong et al., 2015), we found that the Hill coefficient for the 2GBI analogue GBTA was >1 for dimeric Hv1, but not for the monomeric version of the channel. So, we determined the concentration dependence of HIFEN also for monomeric Hv1 (Fig. S3 B) but found that, in this case, monomerization does not cause significant changes in the Hill coefficient. The potential implications of this result are addressed in Zhao et al. (2021).
Discussion
We investigated the molecular mechanism underlying the increased binding affinity of Hv1 for 2GBI in the presence of the F150A mutation and found evidence of a local rearrangement of the binding site in the mutant channel. As a result of this rearrangement, a stabilizing interaction between F150 and 2GBI is replaced by stronger interactions. We propose that the mutation causes the aromatic side chain of Phe 182 to move closer to the phenyl ring of 2GBI, allowing for a stabilizing Π–stacking interaction, which replaces a weaker cation–Π interaction between F150 and the ligand. We found that introducing fluoro substituents in the phenyl ring of F182 via unnatural amino acid substitutions further stabilized 2GBI apparent binding, suggesting that a similar effect could be observed by fluorinating the phenyl ring of the ligand. Testing this hypothesis on fluorinated ABIs revealed a potential liability of Hv1 inhibitors with two conjugated rings, as substituents introduced in one ring to optimize channel–ligand interactions can destabilize binding by perturbing interactions mediated by the other ring. The separation of the two rings in HIF compounds by an extended linker prevented this liability and allowed the inhibitor to adopt binding poses with higher affinity within the intracellular vestibule of the WT channel.
We interpret the stronger potency of HIF compared with HIFNF (Fig. 5, F and G) as an indication that fluorination of the phenyl ring stabilizes binding without destabilizing electrostatic interactions mediated by the positively charged protonated 2-aminoimidazole group. The finding opens the possibility of further exploiting the derivatization at the phenyl ring to strengthen binding of the HIF scaffold. Changes in the linker could offer another avenue for binding optimization. Our binding model (Fig. 5 B) shows that the methyl group connected to the five-membered ring points toward a relatively wide region within the channel intracellular vestibule, suggesting that the moiety could be expanded to increase ligand–protein contacts.
HIF inhibitors are predicted to have ADMET properties superior to 2GBI and its analogues. When compared with in silico tools available in SwissADME (Daina et al., 2017; Daina and Zoete, 2016) and XenoSite (Zaretzki et al., 2013), both 2GBI and HIF compounds showed good solubility, high gastrointestinal absorption, and favorable P450-mediated metabolism (Fig. S4 A). However, 2GBI and derivatives are predicted to be unable to cross the blood–brain barrier, limiting their utilization as neuroprotective agents in models of stroke and inflammation following central nervous system injury. A similar problem affects inhibitor peptides, such as C6 (Zhao et al., 2018) and AGAP/W38F (Tang et al., 2020). HIF and HIFEN, on the other hand, are predicted to cross the blood–brain barrier, offering opportunities for the development of drugs capable of targeting microglial Hv1 in vivo. When potential toxicity was examined with MetaTox (Rudik et al., 2017), HIF and HIFEN were predicted to have LD50 values similar to 2GBI, whereas HIFNH was predicted to have lower toxicity (Fig. S4 B). While these predictions will need to be tested in animal models, they all point to more favorable features of the HIF scaffold compared with 2GBI.
2GBI has been shown to inhibit Hv1 through an open-channel block mechanism (Hong et al., 2013). Besides F150, other residues were found to interact with the ligand, including D112, S181, and R211 from the S1, S3, and S4 helices, respectively (Chamberlin et al., 2014; Gianti et al., 2016; Hong et al., 2014; Lim et al., 2020). To develop high-affinity inhibitors based on the HIF scaffold, it is important to establish whether the same residues involved in 2GBI binding interact with HIF or whether there are different/additional molecular determinants. This point is investigated in Zhao et al. (2021), which explores the mechanism of HIF-mediated inhibition and the ability of the Hv1 VSD to trap the ligand in the down state.
Supplementary Material
shows fit parameters for concentration dependences of Hv1 inhibitors.
Acknowledgments
Joseph A. Mindell served as editor.
The authors wish to thank members of the Tombola laboratory for useful feedback on the manuscript.
This work was supported by the National Institute of General Medical Sciences and the National Institute of Neurological Disorders and Stroke through grant R01GM098973 to F. Tombola and grant R24NS104617 (The Facility for Atomic Mutagenesis) to C.A. Ahern. The authors thank the Chao Family Comprehensive Cancer Center at the University of California, Irvine for access to shared resources supported by the National Cancer Institute under award P30CA062203. The authors appreciate computational resources from the University of California, Irvine Greenplanet cluster, supported in part by National Science Foundation grant CHE-0840513.
The authors declare no competing financial interests.
Author contributions: F. Tombola oversaw the project; F. Tombola, C. Zhao, L. Hong, J.D. Galpin, and C.A. Ahern designed experiments; C. Zhao, L. Hong, and P.D. Webster performed experiments; S. Riahi, V.T. Lim, and D.J. Tobias designed MD simulations; S. Riahi and V.T. Lim performed MD simulations; F. Tombola, C. Zhao, and L. Hong analyzed data; J.D. Galpin and C.A. Ahern provided key reagents; F. Tombola, C. Zhao, and L. Hong wrote the manuscript; and all authors edited the manuscript.
References
- Alabi, A.A., Bahamonde M.I., Jung H.J., Kim J.I., and Swartz K.J.. 2007. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 450:370–375. 10.1038/nature06266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asuaje, A., Smaldini P., Martín P., Enrique N., Orlowski A., Aiello E.A., Gonzalez León C., Docena G., and Milesi V.. 2017. The inhibition of voltage-gated H+ channel (HVCN1) induces acidification of leukemic Jurkat T cells promoting cell death by apoptosis. Pflugers Arch. 469:251–261. 10.1007/s00424-016-1928-0 [DOI] [PubMed] [Google Scholar]
- Bayrhuber, M., Maslennikov I., Kwiatkowski W., Sobol A., Wierschem C., Eichmann C., Frey L., and Riek R.. 2019. Nuclear Magnetic Resonance Solution Structure and Functional Behavior of the Human Proton Channel. Biochemistry. 58:4017–4027. 10.1021/acs.biochem.9b00471 [DOI] [PubMed] [Google Scholar]
- Berger, T.K., and Isacoff E.Y.. 2011. The pore of the voltage-gated proton channel. Neuron. 72:991–1000. 10.1016/j.neuron.2011.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin, A., Qiu F., Rebolledo S., Wang Y., Noskov S.Y., and Larsson H.P.. 2014. Hydrophobic plug functions as a gate in voltage-gated proton channels. Proc. Natl. Acad. Sci. USA. 111:E273–E282. 10.1073/pnas.1318018111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin, A., Qiu F., Wang Y., Noskov S.Y., and Larsson H.P.. 2015. Mapping the gating and permeation pathways in the voltage-gated proton channel Hv1. J. Mol. Biol. 427:131–145. 10.1016/j.jmb.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockroft, S.L., Hunter C.A., Lawson K.R., Perkins J., and Urch C.J.. 2005. Electrostatic control of aromatic stacking interactions. J. Am. Chem. Soc. 127:8594–8595. 10.1021/ja050880n [DOI] [PubMed] [Google Scholar]
- Daina, A., and Zoete V.. 2016. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem. 11:1117–1121. 10.1002/cmdc.201600182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina, A., Michielin O., and Zoete V.. 2017. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7:42717. 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden, T., York D., and Pedersen L.. 1993. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 98:10089. 10.1063/1.464397 [DOI] [Google Scholar]
- DeCoursey, T.E.2013. Voltage-gated proton channels: molecular biology, physiology, and pathophysiology of the H(V) family. Physiol. Rev. 93:599–652. 10.1152/physrev.00011.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Chemaly, A., Okochi Y., Sasaki M., Arnaudeau S., Okamura Y., and Demaurex N.. 2010. VSOP/Hv1 proton channels sustain calcium entry, neutrophil migration, and superoxide production by limiting cell depolarization and acidification. J. Exp. Med. 207:129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geragotelis, A.D., Wood M.L., Göddeke H., Hong L., Webster P.D., Wong E.K., Freites J.A., Tombola F., and Tobias D.J.. 2020. Voltage-dependent structural models of the human Hv1 proton channel from long-timescale molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 117:13490–13498. 10.1073/pnas.1920943117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes, B., Rixen R.M., Kramer K., Forbrig E., Hildebrandt P., and Steinem C.. 2018. Quantification of Hv1-induced proton translocation by a lipid-coupled Oregon Green 488-based assay. Anal. Bioanal. Chem. 410:6497–6505. 10.1007/s00216-018-1248-7 [DOI] [PubMed] [Google Scholar]
- Gianti, E., Delemotte L., Klein M.L., and Carnevale V.. 2016. On the role of water density fluctuations in the inhibition of a proton channel. Proc. Natl. Acad. Sci. USA. 113:E8359–E8368. 10.1073/pnas.1609964114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, C., Rebolledo S., Perez M.E., and Larsson H.P.. 2013. Molecular mechanism of voltage sensing in voltage-gated proton channels. J. Gen. Physiol. 141:275–285. 10.1085/jgp.201210857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondares, E., Brown M.A., Musset B., Morgan D., Cherny V.V., Taubert C., Bhamrah M.K., Coe D., Marelli-Berg F., Gribben J.G., et al. 2014. Enhanced activation of an amino-terminally truncated isoform of the voltage-gated proton channel HVCN1 enriched in malignant B cells. Proc. Natl. Acad. Sci. USA. 111:18078–18083. 10.1073/pnas.1411390111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, L., Pathak M.M., Kim I.H., Ta D., and Tombola F.. 2013. Voltage-sensing domain of voltage-gated proton channel Hv1 shares mechanism of block with pore domains. Neuron. 77:274–287. 10.1016/j.neuron.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, L., Kim I.H., and Tombola F.. 2014. Molecular determinants of Hv1 proton channel inhibition by guanidine derivatives. Proc. Natl. Acad. Sci. USA. 111:9971–9976. 10.1073/pnas.1324012111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, L., Singh V., Wulff H., and Tombola F.. 2015. Interrogation of the intersubunit interface of the open Hv1 proton channel with a probe of allosteric coupling. Sci. Rep. 5:14077. 10.1038/srep14077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infield, D.T., Lee E.E.L., Galpin J.D., Galles G.D., Bezanilla F., and Ahern C.A.. 2018a. Replacing voltage sensor arginines with citrulline provides mechanistic insight into charge versus shape. J. Gen. Physiol. 150:1017–1024. 10.1085/jgp.201812075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infield, D.T., Matulef K., Galpin J.D., Lam K., Tajkhorshid E., Ahern C.A., and Valiyaveetil F.I.. 2018b. Main-chain mutagenesis reveals intrahelical coupling in an ion channel voltage-sensor. Nat. Commun. 9:5055. 10.1038/s41467-018-07477-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, W.L., Chandrasekhar J., Madura J.D., Impey R.W., and Klein M.L.. 1983. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 79:926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Kalia, J., and Swartz K.J.. 2013. Common principles of voltage-dependent gating for Hv and Kv channels. Neuron. 77:214–216. 10.1016/j.neuron.2013.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, T., Okochi Y., Ozaki T., Imura Y., Koizumi S., Yamazaki M., Abe M., Sakimura K., Yamashita T., and Okamura Y.. 2017. Unconventional role of voltage-gated proton channels (VSOP/Hv1) in regulation of microglial ROS production. J. Neurochem. 142:686–699. 10.1111/jnc.14106 [DOI] [PubMed] [Google Scholar]
- Klauda, J.B., Venable R.M., Freites J.A., O’Connor J.W., Tobias D.J., Mondragon-Ramirez C., Vorobyov I., MacKerell A.D. Jr., and Pastor R.W.. 2010. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 114:7830–7843. 10.1021/jp101759q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornilov, P., Peretz A., Lee Y., Son K., Lee J.H., Refaeli B., Roz N., Rehavi M., Choi S., and Attali B.. 2014. Promiscuous gating modifiers target the voltage sensor of K(v)7.2, TRPV1, and H(v)1 cation channels. FASEB J. 28:2591–2602. 10.1096/fj.14-250647 [DOI] [PubMed] [Google Scholar]
- Li, W., Ward R., Dong G., Ergul A., and O’Connor P.. 2019. Neurovascular protection in voltage-gated proton channel Hv1 knock-out rats after ischemic stroke: interaction with Na+/H+ exchanger-1 antagonism. Physiol. Rep. 7:e14142. 10.14814/phy2.14142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, V.T., Geragotelis A.D., Lim N.M., Freites J.A., Tombola F., Mobley D.L., and Tobias D.J.. 2020. Insights on small molecule binding to the Hv1 proton channel from free energy calculations with molecular dynamics simulations. Sci. Rep. 10:13587. 10.1038/s41598-020-70369-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman, E.R., Tytgat J., and Hess P.. 1992. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 9:861–871. 10.1016/0896-6273(92)90239-A [DOI] [PubMed] [Google Scholar]
- Lishko, P.V.2016. Contraception: Search for an Ideal Unisex Mechanism by Targeting Ion Channels. Trends Biochem. Sci. 41:816–818. 10.1016/j.tibs.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lishko, P.V., Botchkina I.L., Fedorenko A., and Kirichok Y.. 2010. Acid extrusion from human spermatozoa is mediated by flagellar voltage-gated proton channel. Cell. 140:327–337. 10.1016/j.cell.2009.12.053 [DOI] [PubMed] [Google Scholar]
- MacKerell, A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616. 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- Martyna, G.J., Tobias D.J., and Klein M.L.. 1994. Constant-Pressure Molecular-Dynamics Algorithms. J. Chem. Phys. 101:4177. 10.1063/1.467468 [DOI] [Google Scholar]
- Mészáros, B., Papp F., Mocsár G., Kókai E., Kovács K., Tajti G., and Panyi G.. 2020. The voltage-gated proton channel hHv1 is functionally expressed in human chorion-derived mesenchymal stem cells. Sci. Rep. 10:7100. 10.1038/s41598-020-63517-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan, M., Zheng J., Wu G., Mogilevsky R., Zheng X., Hu P., Wu J., and Wu L.J.. 2020. The voltage-gated proton channel Hv1 contributes to neuronal injury and motor deficits in a mouse model of spinal cord injury. Mol. Brain. 13:143. 10.1186/s13041-020-00682-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset, B., Smith S.M., Rajan S., Morgan D., Cherny V.V., and Decoursey T.E.. 2011. Aspartate 112 is the selectivity filter of the human voltage-gated proton channel. Nature. 480:273–277. 10.1038/nature10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset, B., Clark R.A., DeCoursey T.E., Petheo G.L., Geiszt M., Chen Y., Cornell J.E., Eddy C.A., Brzyski R.G., and El Jamali A.. 2012. NOX5 in human spermatozoa: expression, function, and regulation. J. Biol. Chem. 287:9376–9388. 10.1074/jbc.M111.314955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura, Y., Fujiwara Y., and Sakata S.. 2015. Gating mechanisms of voltage-gated proton channels. Annu. Rev. Biochem. 84:685–709. 10.1146/annurev-biochem-060614-034307 [DOI] [PubMed] [Google Scholar]
- Phillips, J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., and Schulten K.. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless, S.A., and Ahern C.A.. 2013. Unnatural amino acids as probes of ligand-receptor interactions and their conformational consequences. Annu. Rev. Pharmacol. Toxicol. 53:211–229. 10.1146/annurev-pharmtox-011112-140343 [DOI] [PubMed] [Google Scholar]
- Pupo, A., and Gonzalez León C.. 2014. In pursuit of an inhibitory drug for the proton channel. Proc. Natl. Acad. Sci. USA. 111:9673–9674. 10.1073/pnas.1408808111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, F., Rebolledo S., Gonzalez C., and Larsson H.P.. 2013. Subunit interactions during cooperative opening of voltage-gated proton channels. Neuron. 77:288–298. 10.1016/j.neuron.2012.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, I.S., Moran M.M., Chong J.A., and Clapham D.E.. 2006. A voltage-gated proton-selective channel lacking the pore domain. Nature. 440:1213–1216. 10.1038/nature04700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, I.S., Ruchti E., Kaczmarek J.S., and Clapham D.E.. 2009. Hv1 proton channels are required for high-level NADPH oxidase-dependent superoxide production during the phagocyte respiratory burst. Proc. Natl. Acad. Sci. USA. 106:7642–7647. 10.1073/pnas.0902761106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennhack, A., Grahn E., Kaupp U.B., and Berger T.K.. 2017. Photocontrol of the Hv1 Proton Channel. ACS Chem. Biol. 12:2952–2957. 10.1021/acschembio.7b00523 [DOI] [PubMed] [Google Scholar]
- Ritzel, R.M., He J., Li Y., Cao T., Khan N., Shim B., Sabirzhanov B., Aubrecht T., Stoica B.A., Faden A.I., et al. 2021. Proton extrusion during oxidative burst in microglia exacerbates pathological acidosis following traumatic brain injury. Glia. 69:746–764. 10.1002/glia.23926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, E.A., Lester H.A., and Dougherty D.A.. 2007a. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 1: minimizing misacylation. RNA. 13:1703–1714. 10.1261/rna.666807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, E.A., Lester H.A., and Dougherty D.A.. 2007b. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 2: evaluating suppression efficiency. RNA. 13:1715–1722. 10.1261/rna.667607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudik, A.V., Bezhentsev V.M., Dmitriev A.V., Druzhilovskiy D.S., Lagunin A.A., Filimonov D.A., and Poroikov V.V.. 2017. MetaTox: Web Application for Predicting Structure and Toxicity of Xenobiotics’ Metabolites. J. Chem. Inf. Model. 57:638–642. 10.1021/acs.jcim.6b00662 [DOI] [PubMed] [Google Scholar]
- Sasaki, M., Takagi M., and Okamura Y.. 2006. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 312:589–592. 10.1126/science.1122352 [DOI] [PubMed] [Google Scholar]
- Shin, H., and Song J.H.. 2014. Antipsychotics, chlorpromazine and haloperidol inhibit voltage-gated proton currents in BV2 microglial cells. Eur. J. Pharmacol. 738:256–262. 10.1016/j.ejphar.2014.05.049 [DOI] [PubMed] [Google Scholar]
- Shin, H., Kim J., and Song J.H.. 2015. Clozapine and olanzapine inhibit proton currents in BV2 microglial cells. Eur. J. Pharmacol. 755:74–79. 10.1016/j.ejphar.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Song, J.H., Marszalec W., Kai L., Yeh J.Z., and Narahashi T.. 2012. Antidepressants inhibit proton currents and tumor necrosis factor-α production in BV2 microglial cells. Brain Res. 1435:15–23. 10.1016/j.brainres.2011.11.041 [DOI] [PubMed] [Google Scholar]
- Takeshita, K., Sakata S., Yamashita E., Fujiwara Y., Kawanabe A., Kurokawa T., Okochi Y., Matsuda M., Narita H., Okamura Y., and Nakagawa A.. 2014. X-ray crystal structure of voltage-gated proton channel. Nat. Struct. Mol. Biol. 21:352–357. 10.1038/nsmb.2783 [DOI] [PubMed] [Google Scholar]
- Tang, D., Yang Y., Xiao Z., Xu J., Yang Q., Dai H., Liang S., Tang C., Dong H., and Liu Z.. 2020. Scorpion toxin inhibits the voltage-gated proton channel using a Zn2+ -like long-range conformational coupling mechanism. Br. J. Pharmacol. 177:2351–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola, F., Ulbrich M.H., and Isacoff E.Y.. 2008. The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron. 58:546–556. 10.1016/j.neuron.2008.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola, F., Ulbrich M.H., Kohout S.C., and Isacoff E.Y.. 2010. The opening of the two pores of the Hv1 voltage-gated proton channel is tuned by cooperativity. Nat. Struct. Mol. Biol. 17:44–50. 10.1038/nsmb.1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckerman, M.E., and Berne B.J.. 1991. Molecular-Dynamics in Systems with Multiple Time Scales - Systems with Stiff and Soft Degrees of Freedom and with Short and Long-Range Forces. J. Chem. Phys. 95:8362. 10.1063/1.461263 [DOI] [Google Scholar]
- Van Arnam, E.B., and Dougherty D.A.. 2014. Functional probes of drug-receptor interactions implicated by structural studies: Cys-loop receptors provide a fertile testing ground. J. Med. Chem. 57:6289–6300. 10.1021/jm500023m [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren, W.F., and Berendsen H.J.C.. 1977. Algorithms for macromolecular dynamics and constraint dynamics. Mol. Phys. 34:1311–1327. 10.1080/00268977700102571 [DOI] [Google Scholar]
- Vanommeslaeghe, K., and MacKerell A.D. Jr. 2012. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 52:3144–3154. 10.1021/ci300363c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe, K., Raman E.P., and MacKerell A.D. Jr. 2012. Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 52:3155–3168. 10.1021/ci3003649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Li S.J., Wu X., Che Y., and Li Q.. 2012. Clinicopathological and biological significance of human voltage-gated proton channel Hv1 protein overexpression in breast cancer. J. Biol. Chem. 287:13877–13888. 10.1074/jbc.M112.345280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Wu X., Li Q., Zhang S., and Li S.J.. 2013. Human voltage-gated proton channel hv1: a new potential biomarker for diagnosis and prognosis of colorectal cancer. PLoS One. 8:e70550. 10.1371/journal.pone.0070550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobig, L., Wolfenstetter T., Fechner S., Bönigk W., Körschen H.G., Jikeli J.F., Trötschel C., Feederle R., Kaupp U.B., Seifert R., and Berger T.K.. 2020. A family of hyperpolarization-activated channels selective for protons. Proc. Natl. Acad. Sci. USA. 117:13783–13791. 10.1073/pnas.2001214117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L.J., Wu G., Akhavan Sharif M.R., Baker A., Jia Y., Fahey F.H., Luo H.R., Feener E.P., and Clapham D.E.. 2012. The voltage-gated proton channel Hv1 enhances brain damage from ischemic stroke. Nat. Neurosci. 15:565–573. 10.1038/nn.3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharov, A.V., Varlamova E.V., Lagunin A.A., Dmitriev A.V., Muratov E.N., Fourches D., Kuz’min V.E., Poroikov V.V., Tropsha A., and Nicklaus M.C.. 2016. QSAR Modeling and Prediction of Drug-Drug Interactions. Mol. Pharm. 13:545–556. 10.1021/acs.molpharmaceut.5b00762 [DOI] [PubMed] [Google Scholar]
- Zaretzki, J., Matlock M., and Swamidass S.J.. 2013. XenoSite: accurately predicting CYP-mediated sites of metabolism with neural networks. J. Chem. Inf. Model. 53:3373–3383. 10.1021/ci400518g [DOI] [PubMed] [Google Scholar]
- Zhao, R., Kennedy K., De Blas G.A., Orta G., Pavarotti M.A., Arias R.J., de la Vega-Beltrán J.L., Li Q., Dai H., Perozo E., et al. 2018. Role of human Hv1 channels in sperm capacitation and white blood cell respiratory burst established by a designed peptide inhibitor. Proc. Natl. Acad. Sci. USA. 115:E11847–E11856. 10.1073/pnas.1816189115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C., Hong L., Riahi S., Lim V.T., Tobias D.J., and Tombola F.. 2021. A novel Hv1 inhibitor reveals a new mechanism of inhibition of a voltage-sensing domain. J. Gen. Physiol. 153:e202012833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows fit parameters for concentration dependences of Hv1 inhibitors.