Abstract
Background
Histone acetylation/deacetylase process is one of the most studied epigenetic modifications. Histone deacetylase inhibitors (HDACis) have shown clinical benefits in haematological malignancies but failed in solid tumours due to the lack of biomarker-driven stratification.
Methods
We perform integrative pharmaco-transcriptomic analysis by correlating drug response profiles of five pan-HDACis with transcriptomes of solid cancer cell lines (n=659) to systematically identify generalizable gene signatures associated with HDACis sensitivity and resistance. The established signatures are then applied to identify cancer subtypes that are potentially sensitive or resistant to HDACis, and drugs that enhance the efficacy of HDACis. Finally, the reproductivity of the established HDACis signatures is evaluated by multiple independent drug response datasets and experimental assays.
Findings
We successfully delineate generalizable gene signatures predicting sensitivity (containing 46 genes) and resistance (containing 53 genes) to all five HDACis, with their reproductivity confirmed by multiple external sources and independent internal assays. Using the gene signatures, we identify low-grade glioma harbouring isocitrate dehydrogenase 1/2 (IDH1/2) mutation and non-YAP1-driven subsets of small-cell lung cancer (SCLC) that particularly benefit from HDACis monotherapy. Further, based on the resistance gene signature, we identify clinically-approved Dasatinib as a synthetic lethal drug with HDACi, synergizing in inducing apoptosis and reactive oxygen species on a panel of SCLC. Finally, Dasatinib significantly enhances the therapeutic efficacy of Vorinostat in SCLC xenografts.
Interpretation
Our work establishes robust gene signatures predicting HDACis sensitivity/resistance in solid cancer and uncovers combined Dasatinib/HDACi as a synthetic lethal combination therapy for SCLC.
Keywords: Histone deacetylase, Gene signature, Dasatinib, Small-cell lung cancer, Isocitrate dehydrogenase, YAP1
Research in context.
Evidence before this study
Histone acetylation/deacetylation represents a major epigenetic regulatory process and plays an essential role in human cancers. Upregulation of HDACs has been reported in several human cancer types, making these regulative enzymes a very interesting therapeutic target. However, HDAC inhibitors show clinical success in the treatment of patients with haematological malignancies, but not those with solid tumours leading to the highly heterogeneous responsiveness. The major reason is the lack of biomarkers to stratify patients for HDAC targeted therapies.
Added value of this study
Our work systematically identified generalizable gene signatures associated with HDACis sensitivity and resistance with reproductivity. More importantly, based on the established signatures, we identified non-YAP1-driven small-cell lung cancer (SCLC) and brain tumours with IDH1/2 mutations as the cancer subsets suitable for HDAC targeted therapies. Further, we identified that the effect of HDAC inhibitors in SCLC could be further synergistically enhanced by combining clinically-approved Dasatinib.
Implications of all the available evidence
Our findings may be instrumental for the precise management of HDAC targeted therapies for a subset of patients with solid tumours.
Alt-text: Unlabelled box
1. Introduction
Histone modifications, particularly histone acetylation/deacetylation, represent a major epigenetic regulatory process and play an essential role in a number of human cancers. Histone deacetylation is a critical regulator of gene transcription involving the removal of acetyl groups and is regulated by a group of enzymes termed histone deacetylases (HDACs). HDACs are divided into four classes: class I (HDAC 1, 2, 3, and 8), II (IIa: HDAC 4, 5, 7, and 9; IIb: HDAC 6 and 10), III (SIRT1, 2, 3, 4, 5, 6, and 7) and IV (HDAC11) [1]. HDACs also regulate the acetylation of a variety of non-histone proteins, substrates through different mechanisms [2,3], resulting in multiple biological consequences involved in cell cycle arrest, apoptosis, autophagy, and generation of reactive oxygen species (ROS) [4,5].
Dysregulation of HDACs in multiple types of human cancer is associated with poor prognosis [6], making these enzymes an important therapeutic target [1]. Currently, most chemically-developed HDACis are pan-HDACis rather than being HDAC class member-specific [7]. To date, four pan-HDACis, Vorinostat, Romidepsin, Belinostat, and Panobinostat, have been approved by the Food and Drug Administration for the treatment of haematological malignancies [7], and show particular benefit in cutaneous T-cell lymphoma, but failed to demonstrate therapeutic benefit in solid tumours. Additionally, as single agents, HDACis have shown poor response duration with the rapid development of therapy resistance [4]. Together, these data suggest an urgent need to develop a biomarker-guided stratification strategy, as well as evaluating the success of combinatorial therapy aimed at improving the effectiveness of HDACis while delaying highlighting the importance of combined therapy in delaying the emergence of treatment resistance [7].
Dysregulated Hippo-YAP signalling pathway has been frequently observed in cancer cells developing resistance to various treatments, including drug compounds modulating epigenetic process, e.g. BET [8] and HDAC inhibitors [9]. Hippo signalling transduction converges on the LATS1/2-dependent phosphorylation of the transcriptional master YAP (Yes-associated protein, encoded by YAP1) and its co-activator TAZ. Phosphorylation YAP/TAZ by LATS kinases promotes their translocation from the nucleus to the cytoplasm, where it is sequestered by 14-3-3 proteins, leading to subsequently ubiquitin-mediated proteolysis. Following the inactivation of the Hippo pathway, unphosphorylated YAP and its co-activator TAZ translocate to the nucleus to control the expression of their downstream target genes, which have been shown to control cell proliferation and inhibit cell death, underpinning the tumorigenic potential of YAP/TAZ. Additionally, we and other groups have demonstrated a close link between the YAP activity and the effect of BCR/ABL-SRC inhibitors, e.g. Dasatinib [10], [11], [12], [13]. Interestingly, multiple lines of evidence show that HDACis can overcome resistance to BCR/ABL-SRC inhibitors [14], [15], [16]. Collectively, the above evidence suggests a potentially reciprocal compensation between HDAC- and YAP-regulated signalling pathways.
In this study, we sought to systematically delineate the gene networks correlated with HDACi sensitivity and resistance in solid tumour cells to identify the responsive biomarkers to HDAC targeted therapies. To generalize the results and minimize potential off-target effects, multiple clinically-approved HDACis, namely Vorinostat, Entinostat, and Panobinostat, and clinically-advanced HDACis Belinostat and Apicidin, were integrated. Based on comprehensive correlation analysis of drug response profiles with genome-wide transcriptomics across solid cancer cell lines (n=659), we established HDACis sensitive and resistant signatures, which leads to identifying cancer subtypes that are suitable for HDACis-based therapy. More importantly, the integration of the HDACis resistant signature informed Dasatinib as an ideally synergistic combination treatment to enhance the effect of HDACis.
2. Methods
2.1. Cell culture, drug treatment
SCLC (DMS-114 [ATCC Cat# CRL-2066, RRID:CVCL_1174], DMS-53 [ATCC Cat# CRL-2062, RRID:CVCL_1177], GLC-15 [KCB Cat# KCB 90028YJ, RRID:CVCL_6904]) and NSCLC cell lines (H1650 [KCLB Cat# 91650, RRID:CVCL_1483], H4006 [ATCC Cat# CRL-2871, RRID:CVCL_1269], H1792 [ATCC Cat# CRL-5895, RRID:CVCL_1495], H1944 [ATCC Cat# CRL-5907, RRID:CVCL_1508]) were cultured in RPMI-1640 (Sigma-Aldrich, R8758), supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin (Sigma-Aldrich) at 37 °C in a humid incubator with 5% CO2. All cancer cell lines were obtained from ATCC (American Type Culture Collection, Manassas, VA, USA) and have been authenticated using STR profiling within the last three years and are confirmed free from mycoplasma contamination. Clinically-approved Dasatinib (Cat. #S1021), Vorinostat (Cat. #S1047), Bosutinib (Cat. #S1014), and Panobinostat (Cat. #S1030) were purchased from Selleck (China).
2.2. Ethics statements for animal experiments
All animal studies were conducted according to the guidelines for the care and use of laboratory animals and were approved (#KS20210429) by the Institutional Animal Care and Use Committee of Shanghai Institute of Nutrition and Health.
2.3. Cell viability assay and quantitative analysis of drug synergy
Cells were seeded in 96-well plates (1,000 – 1,500 cells/well), and treated with DMSO (control), Dasatinib or Bosutinib (dual Src/Abl inhibitor inhibitors), HDAC inhibitors (Vorinostat and Panobinostat), alone or in combination for 96 h, and cell viability was quantified by the Acid Phosphatase Assay Kit (ab83367; Abcam, Cambridge, UK) according to the manufacturer's protocol. The median inhibitory concentration (IC50) was calculated using GraphPad Prism 7. Drug synergism was determined by CompuSyn software, which is based on the median-effect principle and the combination index–isobologram theorem [17], [18], [19], [20]. CompuSyn software generates fraction affected (Fa) and combination index (CI) values. CI < 1.0, synergism; CI = 1.0, additive effects; CI > 1.0, antagonism.
2.4. Flow cytometry-based measurement of apoptosis, lipid ROS, and cell cycle
SCLC cells (GLC-15) treated for 24 h with DMSO (control), Dasatinib (0.5 µM), and Vorinostat (HDAC inhibitor, 1 µM), alone and in combination, were subsequently subjected for flow cytometry-based measurement of apoptosis, lipid ROS, and cell cycle, respectively.
For apoptosis analysis, the above cells were stained with the Annexin V-FITC/PI apoptosis detection kit (Cat. #A211-01; Vazyme Biotech Co., Ltd) according to the manufacturer's instruction and as previously described [12].
Concerning the lipid ROS analysis, the cells were stained (20 min, 37°C) with 2.5 µM of BODIPY™ 581/591 C11 (Invitrogen, D3861). After washing with PBS, the cells were harvested and resuspended in 2% FBS-containing PBS.
Regarding the cell cycle analysis, cells were washed twice with cold PBS and resuspended in PBS to get single-cell suspensions, which were then dropped in ice-cold 70% ethanol for at least 2 hours. After that, cells were stained with 1 µg/ml DAPI. For the details of cell cycle analysis, a protocol available at https://flowcytometry-embl.de/wp-content/uploads/2016/12/DAPIstaining-.pdf was used.
Subsequently, the cells were subjected to flow cytometry analysis performed by BD LSRII, with data analysis (n=3 biological repeats) using FlowJo v10.
2.6. Animal experiments
Age- and gender-matched NSG (NOD-SCID IL2Rγnull (IMSR Cat# NM-NSG-012, RRID:IMSR_NM-NSG-012)) mice (n=5 each group) were used for animal experiments with human small-cell lung cancer cell line (GLC-15). For GLC-15 xenografts, tumour cells mixed with Matrigel (356231; Corning) were subcutaneously inoculated in left and right flanks (2 million/injection). Treatment was initiated when tumor volume was around 100 mm3, and drugs (Vorinostat, 25 mg/kg, i.p.; Dasatinib, 15 mg/kg, i.p.) were prepared in the following solvent: 2% DMSO + 40% PEG300 + 5% Tween-80 + 53% saline administered 7 days/week*2 weeks. Tumour size/volume was measured every 3 days and calculated by the formula: (D × d2)/2, where “D” refers to the long tumour diameter and “d” the short one [18].
2.7. Public databases
Processed and standardized drug screening and genome-wide gene expression data across a set of small-molecule compounds (n=481) and solid cancer cell lines (n=659) from the supplementary files (Supplementary Data Set 3–6) of a published study [21] were downloaded for reanalysis. Correlation data across all 481 small molecules against individual transcriptomes that are significantly correlated with response to at least one small molecule were included for analysis [21]. The area under the curve (AUC), determined by fitted concentration-response curves (2-fold dilution, over a 16-point concentration range), is used as a measure of sensitivity. Fisher's z-transformation was applied to the correlation coefficients (Pearson) to adjust for (normalize) variations in cancer cell line number across small molecules and contexts [21], with significance corresponding to a Bonferroni-corrected, two-tailed distribution with family-wise error-rate α < 0.025 in each tail (|z| > 5.83). The frequency of significant compound–target pairs were compared to all compound–transcript pairs exceeding this |z| threshold using a chi-squared test [21]. For validation analysis, the drug response profiles to different HDACis from independent CTRP (V1) (https://portals.broadinstitute.org/ctrp.v1/) [22], GDSC (Genomics of Drug Sensitivity in Cancer, https://www.cancerrxgene.org/) dataset and NCI-60 cancer cell cohort (n=41, 19 hematopoietic/lymphoid-derived cancer cell lines were excluded) [23] were employed. The normalized RNA-sequencing data of identified genes across The Cancer Genome Atlas (TCGA) Pan-cancer cohort was downloaded from cBioPortal (https://www.cbioportal.org/). Normalized transcriptomic data of cancer cell lines were downloaded from Cancer Cell Line Encyclopedia (CCLE) project (https://portals.broadinstitute.org/ccle). The protein level of YAP and TAZ across cancer cell lines was extracted from the normalized level 3 data of reverse phase protein array (RPPA) were downloaded from The Cancer Proteome Atlas (TCPA) database (https://tcpaportal.org/tcpa/) of the TCGA project [24]. DNA methylation profiles across the TCGA pan-cancer cohort were downloaded from the UCSC Xena portal (https://xena.ucsc.edu/). Protein interactions and pathway enrichment analyses were based on STRING databases (version 11.0; https://string-db.org/). Characterization of SCLC subtypes was based on a previously curated public resource [25].
2.8. Immune subtype models
Immune subtype models (C1-C6) were based on a previous study [26]. The genes contained in each signature were evaluated using model-based clustering by the “mclust” R package. Each sample was finally to be grouped based on its predominance with the C1-C6 signature. R software (version 3.6.3) was used for statistical analyses and data presentation.
2.9. Gene signature scores
2.9.1. HDACis sensitive and resistant signature score
HDACis sensitive and resistant signature scores were calculated by summarizing the Z-normalized log2RSEM (RNA-Seq by Expectation-Maximization, downloaded from cBioPortal (https://www.cbioportal.org/)) of the expression data for the genes in the HDACis sensitive and resistant signatures, based on the RNA-sequencing data of TCGA pan-solid patients` tumour and pan-solid cancer cell lines from CCLE. Gene signature sore calculation: after scaling the genes expression value by Apply function in R, a sum of gene expression of the selected genes within the HDACi sensitivity and resistance gene signatures was then summarised as a single score for each sample.
2.9.2. YAP-TAZ target gene signature score
A curated 22-gene YAP-TAZ target gene signature (MYOF, AMOTL2, LATS2, CTGF, CYR61, ANKRD1, ASAP1, AXL, F3, IGFBP3, CRIM1, FJX1, FOXF2, GADD45A, CCDC80, NT5E, DOCK5, PTPN14, ARHGEF17, NUAK2, TGFB2, RBMS3) were utilized based on a previous study [27], which used published RNA-sequencing and ChIP-sequencing data across various cancer types. YAP-TAZ target score was calculated by summarizing the Z-normalized log2RSEM (RNA-Seq by Expectation-Maximization) of the expression data for the 22 curated YAP/TAZ downstream transcription target genes, based on the TCGA MPM RNA-sequencing data.
2.10. Survival analysis
Survival analysis was performed using “survminer” and “survival” R packages. Tumour samples within the TCGA Pan-cancer, LGG, and MESO cohorts were divided into two groups, based on the best-separation cut-off value of indicated genes to plot the Kaplan–Meier survival curves.
2.11. Statistical analysis
Data were presented as mean ± s.d., with the indicated sample size (n) representing biological replicates. Data analysis was performed by GraphPad Prism 7 (GraphPad Software, Inc., San Diego, CA, USA). The group size was determined based on preliminary experiments but no statistical method was used to predetermine sample size. Group allocation was performed in a blinded manner. Gene expression and survival data derived from the public database, as well as the correlation coefficient (Pearson), were analyzed using R (version 3.6.3) [28]. Statistical significance was determined by one-way/two-way analysis of variance (ANOVA), Bonferroni's multiple comparison test, and Student's t-test using GraphPad Prism 8, unless otherwise indicated. P < 0.05 was considered statistically significant.
2.12. Role of the funding source
This work was supported by the National Natural Science Foundation of China (82072570 to F. Yao; 82002941 to B. Sun). The funder had no any role in study design, data collection, data analysis, interpretation, or writing of report.
3. Results
3.1. Systematic correlation analysis identifies common gene signatures predicting the sensitivity and resistance to different pan-HDAC inhibitors
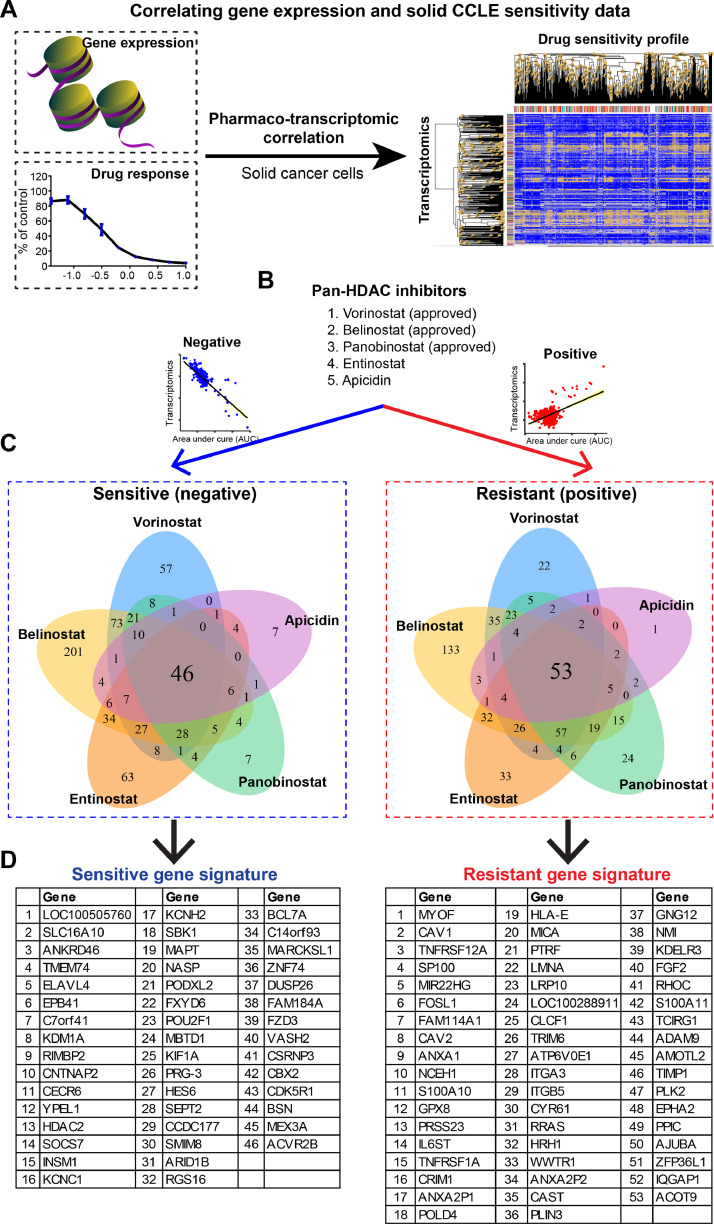
To systematically identify common gene signatures that predict HDACi response, we correlated the sensitivity profiling of five different HDACis (Vorinostat, Entinostat, Panobinostat, Belinostat, and Apicidin) against the transcriptomes of a cohort of pan-solid cancer cell lines (n=659), based on a previously curated small-molecule compound library screen dataset (Fig. 1A) [21]. The results revealed two gene signatures that were significantly correlated (empirical p-value < 0.05), positively or negatively, with the area under the curve (AUC), a measure of drug sensitivity determined by fitted concentration-response curves, of all five HDACis (Fig. 1B–D; Suppl. Table 1). Of note, the positively correlated genes indicate that higher gene expression correlates with a higher AUC value of HDACis and is resistant to HDACis. In contrast, the negatively correlated genes indicate that higher gene expression correlates with lower AUC value of HDACis and are sensitive to HDACis. Thus, we defined the positively correlated genes as HDACis resistant signature, containing 53 genes, and the negatively correlated genes as HDACis sensitive signature, containing 46 genes (Fig. 1D).
Fig. 1.
Systematic correlation identifies generalized gene signatures predictive of therapeutic response to HDACis. A, Schematic model shows the pharmacogenomic analysis that integrates transcriptomic data and drug response profiles across pan-solid cancer cell lines (n=659). B-D, Generalized HDACis sensitive (in blue, left in B-D) and resistant (in red, right in B-D) signatures were established based on five HDAC inhibitors (B). Pearson correlation was performed, and genome-wide transcriptomes were included. Bonferroni-corrected significance cutoff of |z| > 5.83, representing a two-tailed distribution with family-wise error-rate α < 0.025 in each tail, was used.
3.2. Validity of the established HDACis sensitivity and resistance gene signatures
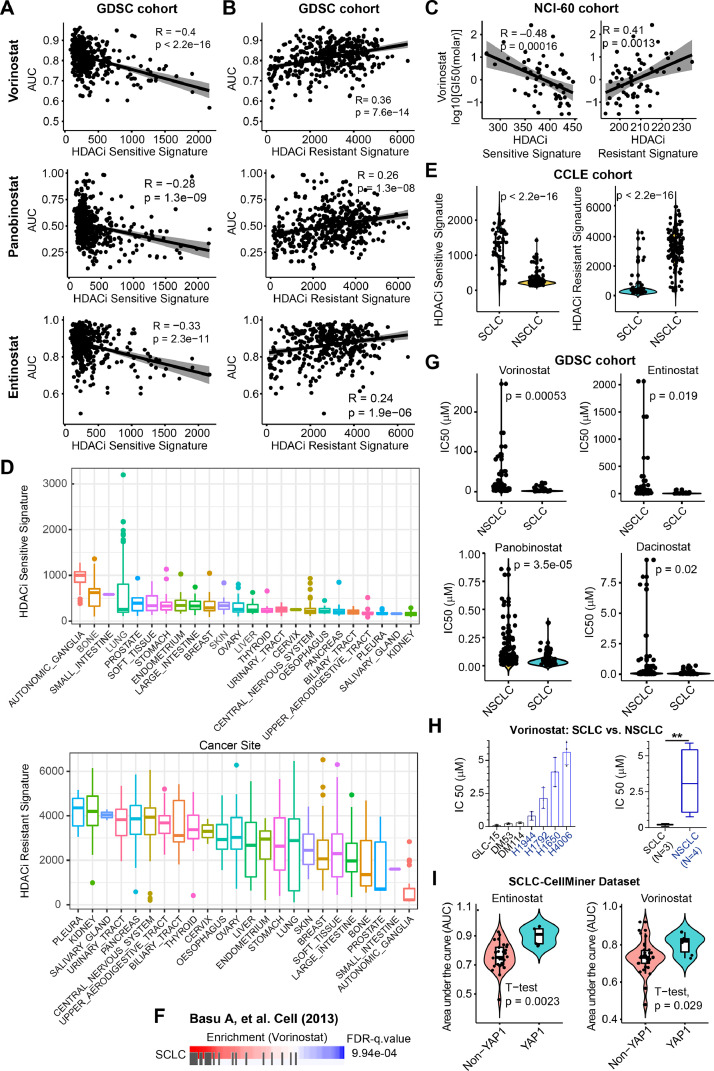
To confirm the reliability and validity of our established gene signatures, we applied them to an independent cohort GDSC (Genomics of Drug Sensitivity in Cancer, https://www.cancerrxgene.org/), which provides sensitivity data of hundreds of drug compounds, including various clinically-approved HDACis, e.g. Vorinostat, Entinostat, and Panobinostat, assayed on hundreds of cancer cell lines from CCLE (Cancer Cell Line Encyclopedia) cohort. We first calculated HDACi sensitive and resistance gene signature scores across pan-solid cancer cell lines (Suppl. Table 2), which were then merged with the drug sensitivity profiles from GDSC. The results showed that the AUC values of the HDACis Vorinostat, Entinostat, and Panobinostat were all significantly negatively correlated with the sensitive gene signature score (Fig. 2A), and positively correlated with the resistant gene signature score (Fig. 2B), confirming the reliability and validity of our established HDACis signatures. Additionally, we applied the HDACis signatures to a second independent NCI-60 cell line cohort consisting of 41 solid cancer cell lines treated with Vorinostat. The results confirmed a significant correlation between the HDACis signatures and the drug effects of Vorinostat (Fig. 2C).
Fig. 2.
Generalized HDACis gene signatures reveal SCLC as potential subsets for therapeutic response to HDACis. A-B, An independent data cohort validating the correlation of HDACis sensitive (A) and resistant (B) signatures with the AUC (area under the curve) values of three HDACis (Vorinostat, Panobinostat, and Entinostat). Drug response profiles were downloaded from the GDSC (Genomics of Drug Sensitivity in Cancer; https://www.cancerrxgene.org/) database. C, An independent NCI-60 data cohort validating the correlation of HDACis sensitive (left) and resistant (right) signatures with –log10 [GI50(molar)] of Vorinostat. Drug response profiles were downloaded from CellMinerCBD (version 1.2) (https://discover.nci.nih.gov/cellminercdb/), and then merged with the gene expression array of the tested cancer cell lines. D, Prospective analysis of HDACis sensitive (upper panel) and resistant (lower panel) signature scores across CCLE (Cancer Cell Line Encyclopedia) pan-solid cancer lines. HDACis sensitive and resistant signature scores were calculated by summarizing the Z-normalized log2RSEM (RNA-Seq by Expectation-Maximization) of the expression data for the genes in the HDACis sensitive and resistant signatures, based on the RNA-sequencing data of pan-solid cancer cell lines from CCLE. E, Significant difference (by Welch's t-test) of HDACis sensitive (upper panel) and resistant (lower panel) signature scores between small cell lung cancer (SCLC) and NSCLC cells. F, Cancer cells with high sensitivity (in red) to HDACi Vorinostat are significantly enriched for SCLC. Data were from Basu A, et al. Cell (2013). G, Violin plots showing that SCLC cells are more sensitive to four HDACis reflected by the IC50 values, compared with NSCLC cells. H, Experimental validation of the differential sensitivity (reflected by IC50 value) of SCLC and NSCLC cells in response to HDACi Vorinostat. A significant difference was calculated by Welch's t-test. **p < 0.01 (by Welch's t-test). I, Independent datasets CellMiner-SCLC validating the association of non-YAP1-driven SCLC with therapeutic responses to two clinically-approved HDACis. Data were from a curated public dataset (https://discover.nci.nih.gov/SclcCellMinerCDB/).
3.3. The HDACis sensitivity and resistance gene signatures facilitate the identification of cancer subtypes vulnerable to HDACis
Next, we applied HDACis sensitivity and resistance gene signatures to the cancer cell line cohort to identify cancer subtypes that are potentially vulnerable to HDACi. The results showed that cancer cells originating from the neuronal system, small intestine, and bone display the highest HDACi sensitive score and lowest resistant score (Fig. 2D), suggesting that cancer cells with these lineages are more likely to be sensitive to HDACi treatment. By contrast, cancer cells originating from the pleural tissue exhibit the highest HDACi resistant and low HDACi sensitive signature score, indicating that pleural mesothelioma (MESO) cells are generally resistant to HDACis (Fig. 2D). Of note, there is a wide distribution of HDACi sensitivity and resistance scores across lung cancer cells (Fig. 2D), implying that response to HDACis in these lineage-derived tumours is highly heterogeneous. Interestingly, the top 50 cell lines with high HDACi sensitive scores were mainly derived from small-cell lung cancer (SCLC) (Suppl. Fig. 1A). In addition, SCLC has a significantly higher HDACi sensitive score but a lower resistant score compared with non-SCLC (NSCLC) (Fig. 2E). In support of this, mining the data source from Basu, A. et al. [22] showed that SCLC is more sensitive to Vorinostat, compared with NSCLC (Fig. 2F). Along these lines, IC50 of the four HDACis is much lower for SCLC compared with NSCLC based on the independent GDSC cohort (Fig. 2G), which was further experimentally validated by cell viability assay using 3 SCLC and 3 NSCLC cell lines in response to the clinically-approved HDACi Vorinostat (Fig. 2H). Along the same line, Crisanti, et al. measured the IC50 value of a panel of thoracic cancer cell lines, including NSCLC, SCLC and MESO, in response to Panobinostat, demonstrating that SCLC cells display the highest sensitivity, whereas MESO cells exhibit the most resistance to HDACi Panobinostat [29]. Taken together, these results suggest that HDACi may represent a promising treatment for SCLC cells. Of important note, SCLC can be broadly divided into 4 molecular subtypes by differential expression of four key transcription regulators: achaete-scute homolog 1 (ASCL1), neurogenic differentiation factor 1 (NEUROD1), yes-associated protein 1 (YAP1), and POU class 2 homeobox 3 (POU2F3) [30]. We further found that only YAP1 expression is significantly correlated with the HDACi sensitive (negatively) and resistant (positively) signatures (Suppl. Fig. 1B), suggesting that the YAP1-driven SCLC subtype may be resistant to HDACi. In support of this, the YAP1-driven SCLC subtype is more resistant to HDACis, compared with other SCLC subtypes, as indicated by significantly higher AUC value of YAP1-driven SCLC cells in response to the HDACis Entinostat and Vorinostat (Fig. 2I). Additionally, using the previously curated YAP-TAZ target score that reflects the activity of the dysregulated Hipp-YAP pathway [27], we observed a strong negative correlation between HDACi sensitivity and YAP-TAZ target signature scores in cancer cell lines from the CCLE (Suppl. Fig. 2A).
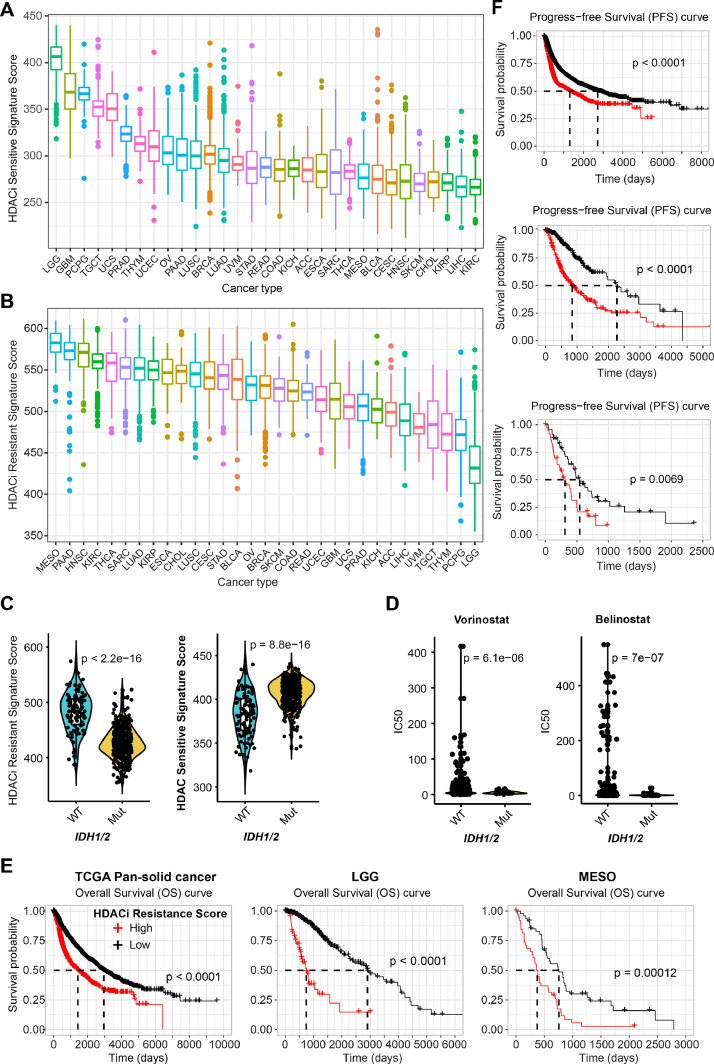
In addition to solid cancer cell lines, we also applied the established HDACi signatures to Pan-cancer patient samples provided by The Cancer Genome Atlas (TCGA) project. Of note, the TCGA does not contain SCLC samples, mainly because most SCLC patients are diagnosed by fine-needle aspiration that leads to a lack of readily accessible and adequate tumour tissue. Clinically, HDACi sensitive and resistant signatures are significantly negatively correlated (Suppl. Figure 1B, C). More importantly, in line with the observations on cell lines (Fig. 2D), patient samples from the neuronal system display the highest HDACi sensitive and lowest HDACi resistant signatures, whereas MESO samples exhibit the highest HDACi resistant signature (Fig. 3A, B). Also, there is a significantly negative correlation between HDACi sensitivity and YAP-TAZ target signature scores across low-grade glioma (LGG) and glioblastoma (GBM) tumours (Suppl. Fig. 2B). Notably, genes negatively regulating the Hippo-YAP signalling pathway (e.g., NF2, LATS1/2) are mostly observed in MESO tumours (Suppl. Fig. 2C) [11], and activation of YAP oncoprotein, the terminal effector of the Hippo pathway, has been reported in more than 70% of MESO samples. Accordingly, MESO tumours display the highest YAP-TAZ target score across TCGA pan-solid tumour cohorts (Suppl. Fig. 2D). Given that the YAP signalling pathway correlates with the resistance to HDACis (Suppl. Figure 1B, 2A; Fig. 2H), these multiple lines of evidence support the exclusion of MESO as a potential tumour type for HDACi monotherapy, highlighting the failure of a previous phase III clinical trial investigating Vorinostat in patients with advanced MESO [31].
Fig. 3.
Generalized HDACis gene signatures reveal IDH1/2-mutant LGG as potential subsets for therapeutic response to HDACis. A-B, Prospective analysis of HDACis sensitive (A) and resistant (B) signature scores across TCGA pan-solid cancer cohort. C, Significant difference (by Welch's t-test) of HDACis sensitive signature score between IDH1/2-mutant (mut) and wild-type (WT) LGG (low-grade glioma). D, Significant difference (by Welch's t-test) in the sensitivity (reflected by IC50 [median inhibitory concentration (IC50)] value) of IDH1/2-mutant (mut) and -wildtype (WT) solid cancer cells in response to two HDACis (Vorinostat, Belinostat). E-F, Kaplan-Meier survival analyses of pan-solid cancer (left in E; upper in F), LGG (middle in E and F), and MESO (right in E; lower in F) stratified by the HDACis resistant gene signatures.
It is well-known that mutations in isocitrate dehydrogenase 1 or 2 (IDH1/2), which results in the production of the oncometabolite 2-hydroxglutarate (2-HG), an inhibitor of α-ketoglutarate-dependent DNA demethylases, are the most common genetic alterations in glioma [32]. Interestingly, we observed that IDH1/2 mutations were significantly associated with high HDACi sensitive and low resistant scores (Fig. 3C), suggesting that IDH1/2-mutant tumours are likely to be more sensitive to HDACis. Supporting our findings, IDH1/2-mutant solid cancer cells have significantly lower IC50 values compared with wild-type (WT) in response to HDACis Vorinostat and Belinostat, based on an independent GDSC data cohort (Fig. 3D). It was recently reported that IDH-mutant GBM displays higher sensitivity to the HDACi panobinostat, compared with IDH-WT GBM [33]. Furthermore, the HDACi resistant signature predicts a poor prognosis in terms of overall survival (OS) and progression-free survival (PFS) in pan-solid cancer types, LGG (highest HDACi sensitive signature), and MESO (high HDACi resistant signature) (Fig. 3E, F). Given the role of IDH1/2 in regulating DNA methylation profiles, we sought to know whether IDH1/2 mutations were associated with altered DNA methylation levels in tumours. Our analysis showed that overall, DNA methylation profiles of gene sets in the HDACi sensitive and resistant signatures across TCGA pan-cancer patient cohort displayed an agreement with the transcriptomic data (Fig. 3A, B; Suppl. Fig. 3A, 3B). For instance, LGG tumours have the lowest transcriptomic level and highest DNA methylation level in terms of the HDACi resistant gene signature (Suppl. Fig. 3A). Besides, compared with MESO that has the highest HDACi resistant signature score (Fig. 3B), LGG tumours exhibit significantly higher DNA methylation level of the HDACi resistant genes (Suppl. Fig. 3B).
3.4. Identifying Dasatinib as a synergistic combination treatment with HDACi
The efficacy of HDACi treatment as a monotherapy is not durable, and resistance is common in response to HDACi treatment. Thus, it is critical to identify additional targets to enhance the efficacy of HDACi treatment.
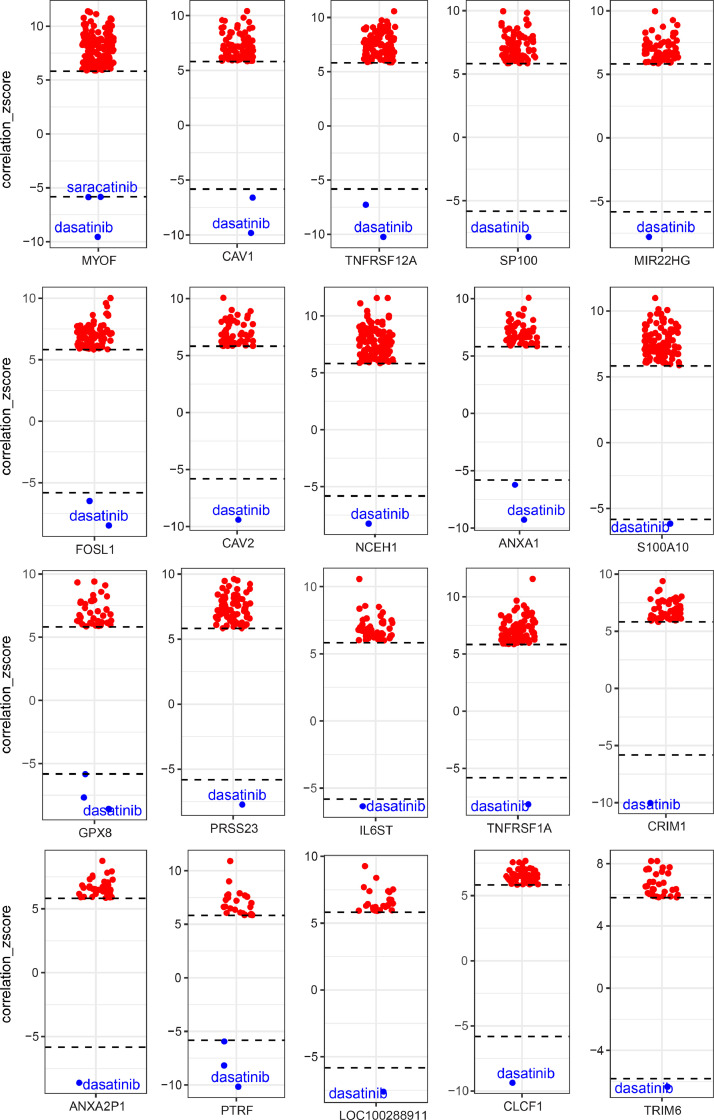
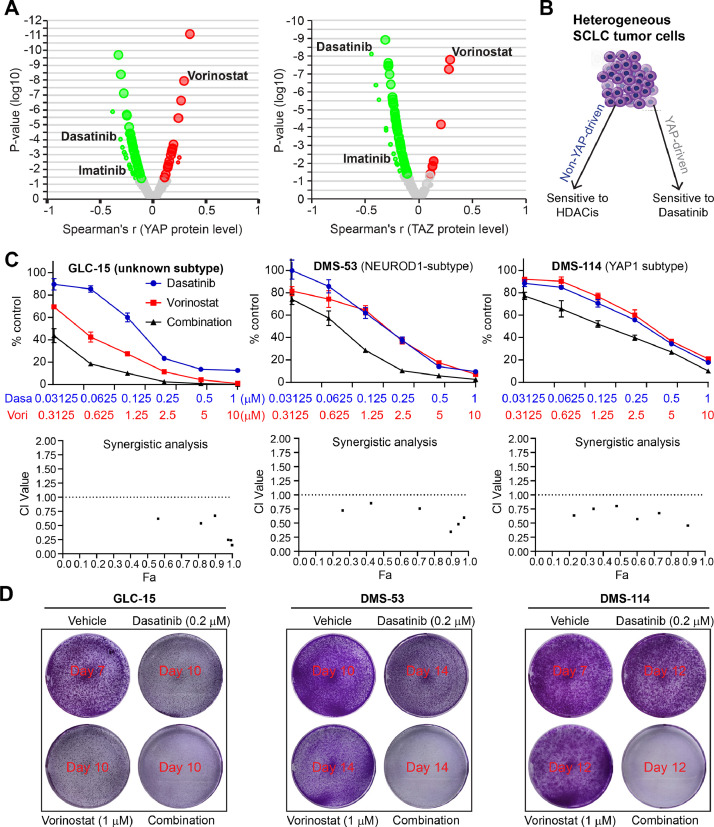
The genes in the established HDACi resistant signature represent potential effectors that mediate the resistance to HDACi targets, thus leading to the assumption that targeting these genes might sensitize cancer cells to HDAC targeted therapy. Next, we sought to identify drug candidates whose effects correlated with the expression of these genes. Through individually correlating the 53 genes of the HDACi resistant signature (Fig. 1D) with the drug compounds in the library, we observed that 38 of 53 (71.7%) genes whose higher expression significantly correlate with lower AUC values of Dasatinib (Fig. 4; Suppl. Figure 4), suggesting cancer cells with high expression of these HDACi resistant genes are more sensitive to Dasatinib treatment. These data suggest that combining BCR/ABL-SRC inhibitor Dasatinib with HDACis may augment the effect of HDACis while helping overcome resistance. Since YAP1 expression is highly correlated with HDACi sensitivity (negatively) and resistance (positively) signatures (Suppl. Figure 1B), we further correlated the AUC values of the above drug compounds library with the protein level of YAP and TAZ (a co-activator of YAP) across cancer cell lines, demonstrating Dasatinib as one of the most significantly negatively (sensitive) correlated drugs, while Vorinostat as the most positively (resistant) correlated drugs (Fig. 5A). These data suggest that cancer cells with activated YAP are more sensitive to Dasatinib treatment but resistant to Vorinostat. In support of this, previous evidence demonstrated that tumours with YAP activation display high sensitivity to Dasatinib [10], [11], [12], [13]. Further, in an independent GDSC drug sensitivity dataset, we observed a significant negative correlation between the effect of Dasatinib and HDACis on solid cancer cell lines (Suppl. Figure 5). Collectively, these data indicate a reciprocal interplay between YAP- and HDAC-involved signalling pathways. As a consequence, combined HDACi and dasatinib therapies are likely to broadly inhibit both YAP- and non-YAP-driven tumours, particularly in SCLC tumours (Fig. 5B).
Fig. 4.
Genes in the HDACis resistant signature correlate with the sensitivity to Dasatinib. Box-and-whisker plots show the extent of correlation between cytotoxic effects (reflected by the area under the curve [AUC] value) of drug compounds in the library (n=481) and expression level of genes in the HDACi resistant signature. The y-axis indicates z scored Pearson's correlation coefficients; line, median; box, 25–75th percentile; whiskers, 2.5th and 97.5th percentile expansion; Here, significantly (p < 0.05) correlated inhibitors were shown (red dots: positively correlated; blue dots: negatively correlated). Note that cells with smaller AUC values are more sensitive to the tested drugs. Dasatinib is listed as the most negatively correlated drug. Pearson correlation was performed, and genome-wide transcriptomes were included. Bonferroni-corrected significance cutoff of |z| > 5.83, representing a two-tailed distribution with family-wise error-rate α < 0.025 in each tail, was used.
Fig. 5.
A reciprocal interaction between Dasatinib and HDACis. A, Volcano plots showing the extent of correlation between cytotoxic effects (reflected by the area under the curve [AUC] value) of drug compounds in the library and protein levels of YAP and TAZ. The protein data of YAP and TAZ were extracted from an independent protein array dataset assayed on hundreds of cancer cell lines from the TCPA portal (https://tcpaportal.org/mclp/#/), which was then integrated with the drug sensitivity data (AUC values). Green dots indicate the significantly negatively correlated genes while the red the positively correlated ones. Note that cells with smaller AUC values are more sensitive to the tested drugs. Dasatinib is listed as the most negatively (sensitive) correlated drug, while Vorinostat is the most positively (resistant) correlated one. B, Schematic models showing the rationale of combined HDACis and Dasatinib in both YAP-driven and non-YAP-driven tumours, particularly SCLC (small-cell lung cancer). C, Dose-response curves (A) and synergy analysis (B) of three SCLC (small cell lung cancer) cells treated for 96h with Dasatinib and Vorinostat (HDACi), alone or in combination. Data are shown as mean ± s.d. (n=3). Fraction affected (Fa) and combination index (CI) were determined by the CompuSyn software. CI < 1 indicates synergism. D, Clonogenic assay of SCLC cells (50000 cells/6-well) treated with Dasatinib and Vorinostat (HDACi), alone or in combination at the indicated drug concentrations for 7-14 days, depending on the doubling time of each cell line. Cells in the vehicle groups were stained immediately once reaching confluence, and cells in the treated groups were stained several days later.
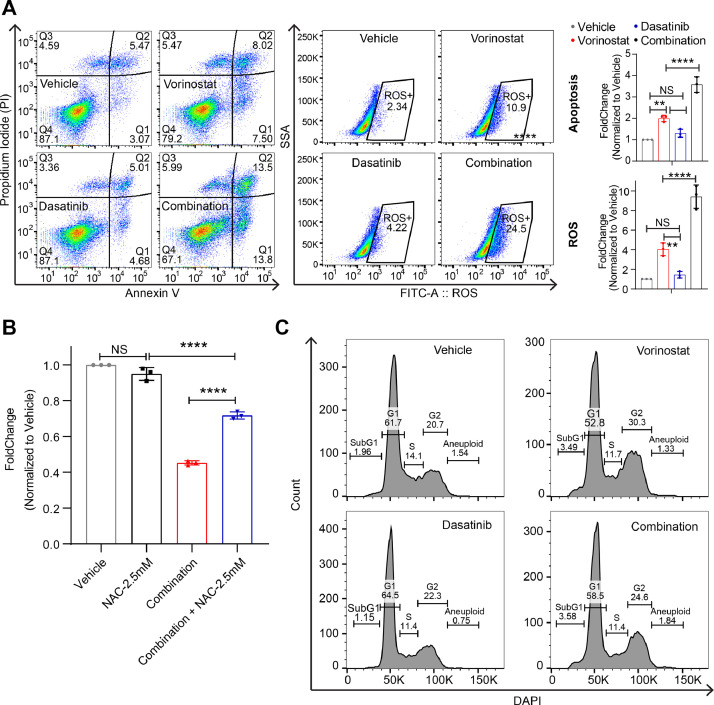
The above evidence reveals the SCLC cells are highly primed for HDACi treatment. To validate this, we treated three SCLC cells with Vorinostat and Dasatinib, alone or in combination, which showed that the combination treatment synergistically inhibited all tested SCLC cell lines (Fig. 5C, 5D). To exclude the potential off-target effect of Vorinostat (HDACi) and Dasatinib (SRC/ABLi), we tested one other clinically-approved HDACi (Panobinostat) and SRC/ABLi (Bosutinib) in the experimental system, which recapitulated our finds with Vorinostat and Dasatinib (Suppl. Figure 6A, 6B), confirming the synergism between HDACi and SRC/ABLi. We and other groups previously established the link of the effect of HDACi with apoptosis, elevated ROS level, and cell cycle arrest [5,12,34,35]. Flow cytometry-based analyses showed that combined Vorinostat and Dasatinib significantly induced apoptosis and elevated ROS level compared with Vorinostat or Dasatinib treatment alone (Fig. 6A). More importantly, the addition of the ROS scanger N-Acetylcysteine (2.5 mM) rescued the inhibitory effect of the combination treatment (Fig. 6B). Additionally, HDACi alone mainly induced cell cycle arrest at the G2/M phase, while Dasatinib alone predominantly arrested the cells at the G1 phase (Fig. 6C). Together, these data suggest that the combination treatment exerts multiple effects on SCLC cells.
Fig. 6.
The effects of combined HDACi and Dasatinib on SCLC cells. A, Flow cytometry-based analyses of apoptosis (Annexin V positive cells; left) and lipid reactive oxygen species (ROS; right) assayed on SCLC cells (GLC-15) treated for 48h with Dasatinib (0.5 μM) and Vorinostat (HDACi; 1 μM), alone or in combination. The corresponding quantifications (n=3) were shown on the right. *p < 0.05, **p < 0.01, ****p < 0.0001 by ANOVA test. NS, non-significance. B, Small–cell lung cancer (SCLC) cells (GLC-15) were treated for 72h with combined Vorinostat (HDACi; 1 μM) and Dasatinib (0.5 μM) with or without 2.5 mM N-acetyl-L-cysteine (NAC). Cells were seeded 1,000 cells per well in 96-well plates. ****p < 0.0001 (n=3) by ANOVA test. NS, non-significance. C, Flow cytometry-based analyses of apoptosis (Annexin V positive cells; left) and lipid reactive oxygen species (ROS; right) assayed on SCLC cells (GLC-15) treated for 24h with Dasatinib (0.5 μM) and Vorinostat (HDACi; 1 μM), alone or in combination. The assay was repeated twice.
3.5. Dasatinib promotes the therapeutic efficacy of Vorinostat in SCLC tumour xenografts
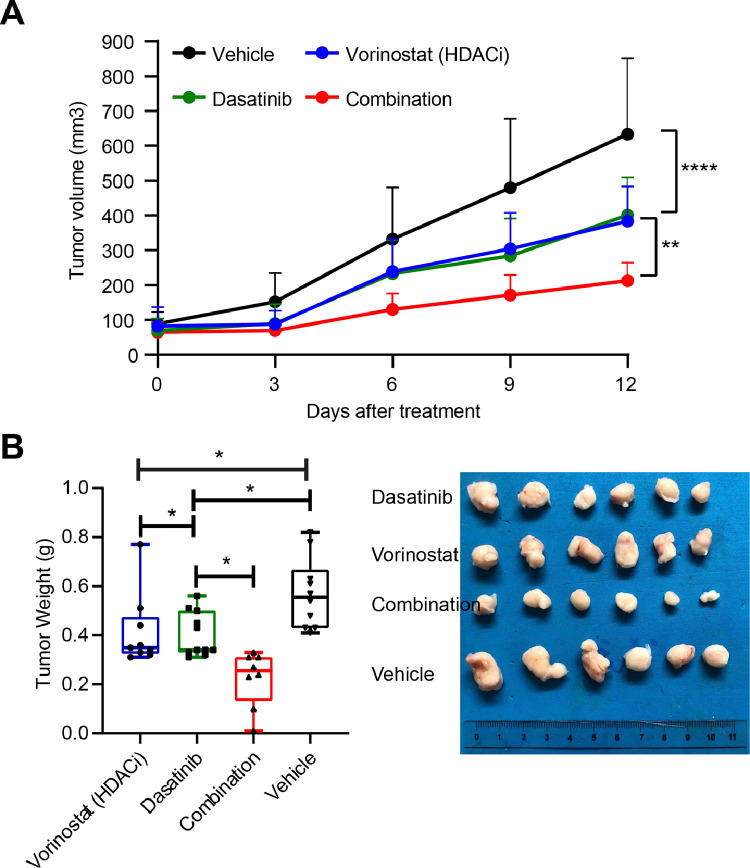
We further investigated the therapeutic potential of combined Dasatinib and Vorinostat for treating SCLC tumours in vivo. In this preclinical model, we observed that Dasatinib significantly enhanced the therapeutic efficacy of Vorinostat in SCLC xenografts (Fig. 7A, 7B).
Fig.7.
Dasatinib enhances antitumor effects of Vorinostat in preclinical SCLC mouse models. A, Volumes of GLC15 xenografts treated with vehicle or the indicated drugs. **p < 0.01, ***p < 0.001 by two-way ANOVA. B, Tumor weights at the end of the treatment. Representative images of tumour size in each group were shown to the right. *p < 0.05 by one-way ANOVA.
4. Discussion
HDACs are promising therapeutic targets and the development of HDAC inhibitors is a hot topic in the treatment of cancer [7]. However, the treatment response to HDACis is highly heterogeneous [36], and identifying the right subset candidates for HDACis-based therapy remains a major challenge in solid tumours, despite the demonstrated effect in haematological tumours. Exacerbating this dilemma is the rapid development of resistance to HDACi treatment [4,37,38]. In this study, common HDACis sensitive and resistance signatures were established by integrated pharmacogenomics of five HDACis, which showed the robustness to identify two patient subsets for HDACis monotherapy: IDH1/2-mutant LGG and non-YAP1-featured SCLC. More importantly, we further identified Dasatinib, which preferentially inhibits cancer cells with YAP activation, as an ideal targeted therapy to synergistically enhance the efficacy of HDACi in SCLC.
4.1. HDACis response signatures
Histone acetylation/deacetylation is one of the most widely investigated epigenetic processes. HDACs regulate the acetylation of a variety of histone and nonhistone proteins, regulating the transcription of diverse genes involved in cell proliferation, survival, differentiation, and tumour immunity [39], [40], [41]; however, the exact molecular mechanism affecting the efficacy currently remains unclear [38], despite the successful applicability of HDACis for treatment of haematological tumours.
Few studies have investigated the molecular biomarkers to predict the therapeutic responses to HDACi in cancer. Thus, clinical trials have been largely based on unselected patients for HDACis treatment, leading to their repeated failures [4,38]. Given the pleiotropic but heterogeneous effects of HDACis on cancer cells, it becomes critical to identify the biomarkers for their true therapeutic benefits in cancer. In this study, we for the first time established generalized gene signatures to identify cancer subsets that are potentially sensitive to HDACis. Intriguingly, we found that IDH1/2, which is frequently mutated in LGG, has a high HDACis sensitive signature, and solid cancer cells with IDH1/2 mutation display a high sensitivity to two clinically-approved HDACis, compared with WT IDH1/2. In support of this Zhang, et al. recently demonstrated that in leukaemia harbouring mutated IDH1/2, which is a frequent event and plays a key role in its pathogenesis, HDACis rapidly reversed the imbalance of H3K9 methylation/acetylation in diseased hematopoietic stem and progenitor cells, which significantly reduced the leukemic burden [42]. The evidence from this study reveals new therapeutic opportunities for leukaemia patients carrying IDH1/2 mutations. Thus, HDACis may be also a promising treatment in solid tumours, particularly LGG and GBM that are predominantly characterized with IDH1/2 mutations [43], which has been recently reported in an abstract publication [33] and thus worths further pursuit in the future design of clinical trials.
Another interesting finding in this study is that the non-YAP1-driven SCLC subtype is vulnerable to HDACis single treatment. Recent studies also demonstrated the effective inhibition of SCLC with HDACis [29,44,45], which could be further enhanced through combined chemotherapies in preclinical models [34,46]. In a non-randomized phase 2 trial, pan-HDACi panobinostat monotherapy demonstrated tumour shrinkage in 2 out of 19 previously treated SCLC patients. However, the study was based on unselected SCLC patients and prematurely closed because of a lack of activity, arguing the need for biomarkers-guided stratification of HDACis for the treatment of SCLC patients. Typically, SCLC is largely divided into 4 molecular subtypes: ASCL1, NEUROD1, YAP1, and POU2F3 [8]. Intriguingly, in this study, we found that YAP1 was the only subtyped SCLC highly correlated with HDACi sensitive (negatively) and resistant (positively) signatures (Fig. 2F; Suppl. Figure 1B), suggesting that HDACi as monotherapy preferentially inhibits non-YAP1-featured SCLC subtype. Rewired YAP activation, which is extensively involved in various therapy resistance [47,48], might mediate the resistance to HDACis in SCLC or other solid tumours.
Additionally, genes in the established HDACis resistant signature are significantly enriched with the immune system (Fig. 5B). Supporting our findings, emerging evidence has shown the role of HDACs in modulating tumour immunity and efficacy to ICB [41,[49], [50], [51]]. In contrast to genetic alterations, epigenetic plasticity enables rapid, dynamic, and flexible changes that favour immune escape, and resistance to therapies [52,53]. Therefore, targeting epigenetic alterations represents a promising therapeutic approach and epigenetic-based therapies could have a major clinical impact, notably in combination with conventional therapies and immune therapies [54].
Thus, defining common HDACis response signatures provides additional insights that allow for more effective and precise management of HDAC inhibitors.
4.2. Combined targeted therapy with HDACis
As single agents, treatment with HDAC inhibitors has demonstrated limited clinical benefit for patients with solid tumours, prompting the investigation of novel treatment combinations with other cancer therapeutics [7]. Treatment strategies combining HDACis with approved anticancer drugs could dramatically improve the therapeutic success rate through inhibition of the tumour resistance mechanism. The rationales of therapeutic combinations with HDAC inhibitors have been discussed, mainly including combined DNA-damaging chemotherapeutic agents, radiotherapy, hormonal therapies, DNA methyltransferase inhibitors, and immunotherapies, as well as various other small-molecule inhibitors [39,40,[55], [56], [57], [58]].
Additionally, we and other groups have recently demonstrated that Dasatinib treatment modulates host anti-tumour immunity, and enhance the efficacy of immune checkpoint blockade (ICBs) [28,59,60]. Intriguingly, functional pathway analysis of genes in the HDACi resistant signature revealed enrichment of the immunity-mediated pathways (Suppl. Figure 7A, 7B). Moreover, LGG tumours (high HDACi sensitive signature) display a dramatic difference in the immune subtypes, mainly immunologically quiet and lymphocyte depleted subtypes, compared with MESO (high HDACi resistant signature) (Suppl. Figure 7C). In support of this, previous evidence also showed the potential of HDACi to boost the effect of immunotherapy [50]. Notably, these genes were also significantly enriched with a reference publication about the predictive biomarkers for Dasatinib treatment in melanoma (Suppl. Figure 7B) [61]. Together, these analyses further support an interplay between HDACi and Dasatinib treatment and provide the rationale for combined HDACi and Dasatinib targeted therapies.
In a recent phase III trial for advanced hormone receptor-positive breast cancer, the HDAC inhibitor Chidamide in combination with exemestane, increased the median progression-free survival to 7.4 months in comparison to 3.8 months with placebo [62], leading to the approval of this combination for the treatment of breast cancer. Other HDAC inhibitors are undergoing trials in solid tumours, including entinostat in phase III trials for locally advanced or metastatic recurrent hormone receptor-positive breast cancer (NCT02115282). In this study, we found that Dasatinib together with HDACis might be an ideal combined targeted therapy for certain solid tumours. First, genes in the HDACis resistant signature whose high expression were mostly correlated with the sensitivity profile of Dasatinib. Second, high expression of YAP1 displays a strongly positive correlation with HDACis resistant signature (Fig. 2H), and predict resistance to HDACis but sensitivity to Dasatinib (Fig. 5A) [28]. Finally, Dasatinib synergizes with HDACis in inhibiting SCLC cells (Fig. 5C-E). In line with our findings, previous studies showed a synergistic treatment effect between HDACis and imatinib, another clinically-approved BCR/ABL-SRC inhibitor as Dasatinib, in leukaemia patients [15,63]. Also, a previous study showed that targeting HDAC could dramatically enhance the efficacy of Dasatinib on pancreatic cancer cells [16]. Together, combined Dasatinib and HDACi represent a promising combination treatment in solid tumours, in particular in SCLC which is characterized by the highest HDACi sensitivity signature score across different cancer types. Of note, although SCLC generally has the highest HDACi sensitivity signature score, heterogeneity also exists, in that among the four molecular subtypes of SCLC, YAP-driven SCLC is associated with resistance to HDACi treatment alone. Given that Dasatinib treatment could preferentially inhibit SCLC with YAP activation [10], [11], [12], [13], thus combinatorial Dasatinib and HDACi may represent a promising treatment for SCLC, independent of the four molecular subtypes.
4.3. Potential molecular mechanisms underpinning the synergism between combined SRC/Abl targeted therapies with HDACis
Despite the therapeutic efficacy of HDAC inhibition for certain tumour subtypes, it remains unclear about the underlying molecular mechanisms underpinnings the activities of HDAC inhibitors. Since the main functions of HDAC are involved in the process of histone deacetylation, altered chromatin remodelling activities or resulting chromatin mark changes might play a role. Recent evidence showed that the most prevalent changes associated with transcriptional regulation caused by HDACi occur at distal enhancer elements [64], [65], [66]. Accordingly, accumulated evidence reveals that HDACi modulates chromatin mark changes of acetylated H3K27 (H3K27ac) [64,66], a typical marker of super-enhancers [67]. Consequently, HDACi increases the acetylation level of the targeted genes, resulting in enhanced gene transcription. Based on the importance of HDAC in regulating super-enhancers, it is likely that the potential mechanisms of the synergism between Dasatinib and HDACi might be due to the upregulated activity of genes participating in the SRC-signalling pathway, which is the main target of Dasatinib. Further studies are warranted.
Overall, the use of surrogate biomarkers to predict the HDACis therapy response is essential. Identification of subsets that are suitable for HDACis, and rational combinations, particularly with clinically-approved inhibitors, that would lead to increased efficacy while overcoming resistance will ensure the optimal clinical application of these agents. Pharmacogenomics enables robust identification of molecular signatures that may help to predict HDACi therapeutic response and cancer subtypes that are suitable for HDACi treatment. Dasatinib is a synergistic combination to enhance HDAC-targeted therapy in SCLC.
Contributors
Conceptualisation: H.Y. and F.Y.; Data curation: Y.H., K.X. and T.Z.; Formal analysis: H.Y. and B.S.; Investigation: H.Y., B.S., Y.H., K.X. and T.Z. This study was conceived, designed, and interpreted by H.Y., and F.Y.. H.Y., B.S.; Methodology: H.Y.; Project administration: F.Y. and G.H.; Software: H.Y.; Supervision: F.Y. and G.H.; Writing – original draft: H.Y.; S.R.R.H. critically reviewed the manuscript; H.Y., F.Y. and G.H. had primary responsibility for the final content; All authors read, revised and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgments
Acknowledgements
We thank Haizhen Jin (Central Laboratory, Shanghai Chest Hospital, Shanghai Jiao Tong University) for the flow cytometry analysis. This study used TCGA Program database. The interpretation and reporting of these data are the sole responsibility of the authors. The authors acknowledge the efforts of the National Cancer Institute. This work was supported by the National Natural Science Foundation of China (82072570 to F. Yao; 82002941 to B. Sun).
Data Sharing Statement
Not applicable.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103457.
Contributor Information
Haitang Yang, Email: xyxm8889@163.com.
Feng Yao, Email: feng.yao@shchest.org.
Appendix. Supplementary materials
References
- 1.Verza FA, Das U, Fachin AL, Dimmock JR, Marins M. Roles of Histone Deacetylases and Inhibitors in Anticancer Therapy. Cancers (Basel) 2020;12(6) doi: 10.3390/cancers12061664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nalawansha DA, Zhang Y, Herath K, Pflum MKH. HDAC1 Substrate Profiling Using Proteomics-Based Substrate Trapping. ACS Chem Biol. 2018;13(12):3315–3324. doi: 10.1021/acschembio.8b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh BN, Zhang G, Hwa YL, Li J, Dowdy SC, Jiang SW. Nonhistone protein acetylation as cancer therapy targets. Expert Rev Anticancer Ther. 2010;10(6):935–954. doi: 10.1586/era.10.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robey RW, Chakraborty AR, Basseville A, Luchenko V, Bahr J, Zhan Z. Histone deacetylase inhibitors: emerging mechanisms of resistance. Mol Pharm. 2011;8(6):2021–2031. doi: 10.1021/mp200329f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckschlager T, Plch J, Stiborova M, Hrabeta J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int J Mol Sci. 2017;18(7) doi: 10.3390/ijms18071414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weichert W. HDAC expression and clinical prognosis in human malignancies. Cancer Lett. 2009;280(2):168–176. doi: 10.1016/j.canlet.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 7.Ho TCS, Chan AHY, Ganesan A. Thirty Years of HDAC Inhibitors: 2020 Insight and Hindsight. J Med Chem. 2020;63(21):12460–12484. doi: 10.1021/acs.jmedchem.0c00830. [DOI] [PubMed] [Google Scholar]
- 8.Gobbi G, Donati B, Do Valle IF, Reggiani F, Torricelli F, Remondini D. The Hippo pathway modulates resistance to BET proteins inhibitors in lung cancer cells. Oncogene. 2019;38(42):6801–6817. doi: 10.1038/s41388-019-0924-1. [DOI] [PubMed] [Google Scholar]
- 9.Basu D, Reyes-Mugica M, Rebbaa A. Histone acetylation-mediated regulation of the Hippo pathway. PLoS One. 2013;8(5):e62478. doi: 10.1371/journal.pone.0062478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J, Wang X, Tang B, Liu H, Zhang M, Wang Y. A tightly controlled Src-YAP signaling axis determines therapeutic response to dasatinib in renal cell carcinoma. Theranostics. 2018;8(12):3256–3267. doi: 10.7150/thno.23964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, Xu D, Schmid RA, Peng RW. Biomarker-guided targeted and immunotherapies in malignant pleural mesothelioma. Ther Adv Med Oncol. 2020;12 doi: 10.1177/1758835920971421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H, Zhao L, Gao Y, Yao F, Marti TM, Schmid RA. Pharmacotranscriptomic Analysis Reveals Novel Drugs and Gene Networks Regulating Ferroptosis in Cancer. Cancers (Basel) 2020;12(11) doi: 10.3390/cancers12113273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamar JM, Xiao Y, Norton E, Jiang ZG, Gerhard GM, Kooner S. SRC tyrosine kinase activates the YAP/TAZ axis and thereby drives tumor growth and metastasis. J Biol Chem. 2019;294(7):2302–2317. doi: 10.1074/jbc.RA118.004364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rauzan M, Chuah CT, Ko TK, Ong ST. The HDAC inhibitor SB939 overcomes resistance to BCR-ABL kinase Inhibitors conferred by the BIM deletion polymorphism in chronic myeloid leukemia. PLoS One. 2017;12(3) doi: 10.1371/journal.pone.0174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen T, Dai Y, Attkisson E, Kramer L, Jordan N, Nguyen N. HDAC inhibitors potentiate the activity of the BCR/ABL kinase inhibitor KW-2449 in imatinib-sensitive or -resistant BCR/ABL+ leukemia cells in vitro and in vivo. Clin Cancer Res. 2011;17(10):3219–3232. doi: 10.1158/1078-0432.CCR-11-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chien W, Sudo M, Ding LW, Sun QY, Wuensche P, Lee KL. Functional Genome-wide Screening Identifies Targets and Pathways Sensitizing Pancreatic Cancer Cells to Dasatinib. J Cancer. 2018;9(24):4762–4773. doi: 10.7150/jca.25138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 18.Yang H, Liang SQ, Xu D, Yang Z, Marti TM, Gao Y. HSP90/AXL/eIF4E-regulated unfolded protein response as an acquired vulnerability in drug-resistant KRAS-mutant lung cancer. Oncogenesis. 2019;8(9):45. doi: 10.1038/s41389-019-0158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu D, Yang H, Yang Z, Berezowska S, Gao Y, Liang SQ. Endoplasmic Reticulum Stress Signaling as a Therapeutic Target in Malignant Pleural Mesothelioma. Cancers (Basel) 2019;11(10) doi: 10.3390/cancers11101502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu D, Liang SQ, Yang H, Bruggmann R, Berezowska S, Yang Z. CRISPR Screening Identifies WEE1 as a Combination Target for Standard Chemotherapy in Malignant Pleural Mesothelioma. Mol Cancer Ther. 2020;19(2):661–672. doi: 10.1158/1535-7163.MCT-19-0724. [DOI] [PubMed] [Google Scholar]
- 21.Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12(2):109–116. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154(5):1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1-2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W. TCPA: a resource for cancer functional proteomics data. Nat Methods. 2013;10(11):1046–1047. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tlemsani C, Pongor L, Elloumi F, Girard L, Huffman KE, Roper N. SCLC-CellMiner: A Resource for Small Cell Lung Cancer Cell Line Genomics and Pharmacology Based on Genomic Signatures. Cell Rep. 2020;33(3) doi: 10.1016/j.celrep.2020.108296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH. The Immune Landscape of. Cancer. Immunity. 2018;48(4):812–830. doi: 10.1016/j.immuni.2018.03.023. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Xu X, Maglic D, Dill MT, Mojumdar K, Ng PK. Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer. Cell Rep. 2018;25(5):1304–1317. doi: 10.1016/j.celrep.2018.10.001. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Xu D, Yang Z, Yao F, Zhao H, Schmid RA. Vol. 12. 2020. Systematic Analysis of Aberrant Biochemical Networks and Potential Drug Vulnerabilities Induced by Tumor Suppressor Loss in Malignant Pleural Mesothelioma. (Cancers (Basel)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crisanti MC, Wallace AF, Kapoor V, Vandermeers F, Dowling ML, Pereira LP. The HDAC inhibitor panobinostat (LBH589) inhibits mesothelioma and lung cancer cells in vitro and in vivo with particular efficacy for small cell lung cancer. Mol Cancer Ther. 2009;8(8):2221–2231. doi: 10.1158/1535-7163.MCT-09-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer. 2019;19(5):289–297. doi: 10.1038/s41568-019-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buikhuisen WA, Burgers JA, Vincent AD, Korse CM, van Klaveren RJ, Schramel FM. Thalidomide versus active supportive care for maintenance in patients with malignant mesothelioma after first-line chemotherapy (NVALT 5): an open-label, multicentre, randomised phase 3 study. Lancet Oncol. 2013;14(6):543–551. doi: 10.1016/S1470-2045(13)70125-6. [DOI] [PubMed] [Google Scholar]
- 32.Zhang C, Moore LM, Li X, Yung WK, Zhang W. IDH1/2 mutations target a key hallmark of cancer by deregulating cellular metabolism in glioma. Neuro Oncol. 2013;15(9):1114–1126. doi: 10.1093/neuonc/not087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sears TK, Woolard KD. Abstract 1917: The HDAC inhibitor panobinostat elicits preferential cytotoxic and antiproliferative effects in IDH mutant glioblastoma. Cancer Research. 2020;80(16 Supplement):1917. - [Google Scholar]
- 34.Bruzzese F, Rocco M, Castelli S, Di Gennaro E, Desideri A, Budillon A. Synergistic antitumor effect between vorinostat and topotecan in small cell lung cancer cells is mediated by generation of reactive oxygen species and DNA damage-induced apoptosis. 2009;8(11):3075-87. [DOI] [PubMed]
- 35.Wang L, Leite de Oliveira R, Huijberts S, Bosdriesz E, Pencheva N, Brunen D. An Acquired Vulnerability of Drug-Resistant Melanoma with Therapeutic Potential. Cell. 2018;173(6):1413–1425. doi: 10.1016/j.cell.2018.04.012. e14. [DOI] [PubMed] [Google Scholar]
- 36.McClure JJ, Li X, Chou CJ. Advances and Challenges of HDAC Inhibitors in Cancer Therapeutics. Adv Cancer Res. 2018;138:183–211. doi: 10.1016/bs.acr.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Mrakovcic M, Frohlich LF. Molecular Determinants of Cancer Therapy Resistance to HDAC Inhibitor-Induced Autophagy. Cancers (Basel) 2019;12(1) doi: 10.3390/cancers12010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rana Z, Diermeier S, Hanif M, Rosengren RJ. Understanding Failure and Improving Treatment Using HDAC Inhibitors for Prostate Cancer. Biomedicines. 2020;8(2) doi: 10.3390/biomedicines8020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fantin VR, Richon VM. Mechanisms of resistance to histone deacetylase inhibitors and their therapeutic implications. Clin Cancer Res. 2007;13(24):7237–7242. doi: 10.1158/1078-0432.CCR-07-2114. [DOI] [PubMed] [Google Scholar]
- 40.Bots M, Johnstone RW. Rational combinations using HDAC inhibitors. Clin Cancer Res. 2009;15(12):3970–3977. doi: 10.1158/1078-0432.CCR-08-2786. [DOI] [PubMed] [Google Scholar]
- 41.Zhao LM, Zhang JH. Histone Deacetylase Inhibitors in Tumor Immunotherapy. Curr Med Chem. 2019;26(17):2990–3008. doi: 10.2174/0929867324666170801102124. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Wang X, Wang XQD, Su J, Putluri N, Zhou T. Dnmt3a loss and Idh2 neomorphic mutations mutually potentiate malignant hematopoiesis. Blood. 2020;135(11):845–856. doi: 10.1182/blood.2019003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen R, Zhang M, Zhou Y, Guo W, Yi M, Zhang Z. The application of histone deacetylases inhibitors in glioblastoma. J Exp Clin Cancer Res. 2020;39(1):138. doi: 10.1186/s13046-020-01643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia D, Augert A, Kim DW, Eastwood E, Wu N, Ibrahim AH. Crebbp Loss Drives Small Cell Lung Cancer and Increases Sensitivity to HDAC Inhibition. Cancer Discov. 2018;8(11):1422–1437. doi: 10.1158/2159-8290.CD-18-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun L, He Q, Tsai C, Lei J, Chen J, Vienna Makcey L. HDAC inhibitors suppressed small cell lung cancer cell growth and enhanced the suppressive effects of receptor-targeting cytotoxins via upregulating somatostatin receptor II. Am J Transl Res. 2018;10(2):545–553. [PMC free article] [PubMed] [Google Scholar]
- 46.Pan CH, Chang YF, Lee MS, Wen BC, Ko JC, Liang SK. Vorinostat enhances the cisplatin-mediated anticancer effects in small cell lung cancer cells. BMC Cancer. 2016;16(1):857. doi: 10.1186/s12885-016-2888-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13(4):246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 48.Nilsson MB, Sun H, Robichaux J, Pfeifer M, McDermott U, Travers J. A YAP/FOXM1 axis mediates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components. Sci Transl Med. 2020;12(559) doi: 10.1126/scitranslmed.aaz4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bretz AC, Parnitzke U, Kronthaler K, Dreker T, Bartz R, Hermann F. Domatinostat favors the immunotherapy response by modulating the tumor immune microenvironment (TIME) J Immunother Cancer. 2019;7(1):294. doi: 10.1186/s40425-019-0745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kroesen M, Gielen P, Brok IC, Armandari I, Hoogerbrugge PM, Adema GJ. HDAC inhibitors and immunotherapy; a double edged sword? Oncotarget. 2014;5(16):6558–6572. doi: 10.18632/oncotarget.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Waschke BC, Woolaver RA, Chen SMY, Chen Z, Wang JH. HDAC inhibitors overcome immunotherapy resistance in B-cell lymphoma. Protein Cell. 2020;11(7):472–482. doi: 10.1007/s13238-020-00694-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aspeslagh S, Morel D, Soria JC, Postel-Vinay S. Epigenetic modifiers as new immunomodulatory therapies in solid tumours. Ann Oncol. 2018;29(4):812–824. doi: 10.1093/annonc/mdy050. [DOI] [PubMed] [Google Scholar]
- 53.Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357(6348) doi: 10.1126/science.aal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17(10):630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 55.Grant S, Dai Y. Histone deacetylase inhibitors and rational combination therapies. Adv Cancer Res. 2012;116:199–237. doi: 10.1016/B978-0-12-394387-3.00006-9. [DOI] [PubMed] [Google Scholar]
- 56.Huan S, Gui T, Xu Q, Zhuang S, Li Z, Shi Y. Combination BET Family Protein and HDAC Inhibition Synergistically Elicits Chondrosarcoma Cell Apoptosis Through RAD51-Related DNA Damage Repair. Cancer Manag Res. 2020;12:4429–4439. doi: 10.2147/CMAR.S254412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyamoto K, Watanabe M, Boku S, Sukeno M, Morita M, Kondo H. xCT Inhibition Increases Sensitivity to Vorinostat in a ROS-Dependent Manner. Cancers (Basel) 2020;12(4) doi: 10.3390/cancers12040827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang T, Sun B, Zhong C, Xu K, Wang Z, Hofman P. Targeting histone deacetylase enhances the therapeutic effect of Erastin-induced ferroptosis in EGFR -activating mutant lung adenocarcinoma. Translational Lung Cancer Research. 2021;10(4):1857–1872. doi: 10.21037/tlcr-21-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tu MM, Lee FYF, Jones RT, Kimball AK, Saravia E, Graziano RF. Targeting DDR2 enhances tumor response to anti-PD-1 immunotherapy. Sci Adv. 2019;5(2):eaav2437. doi: 10.1126/sciadv.aav2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen R, Lee WC, Fujimoto J, Li J, Hu X, Mehran R. Evolution of genomic and T cell repertoire heterogeneity of malignant pleural mesothelioma under dasatinib treatment. Clin Cancer Res. 2020 doi: 10.1158/1078-0432.CCR-20-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eustace AJ, Kennedy S, Larkin AM, Mahgoub T, Tryfonopoulos D, O'Driscoll L. Predictive biomarkers for dasatinib treatment in melanoma. Oncoscience. 2014;1(2):158–166. doi: 10.18632/oncoscience.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang Z, Li W, Hu X, Zhang Q, Sun T, Cui S. Tucidinostat plus exemestane for postmenopausal patients with advanced, hormone receptor-positive breast cancer (ACE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20(6):806–815. doi: 10.1016/S1470-2045(19)30164-0. [DOI] [PubMed] [Google Scholar]
- 63.Lernoux M, Schnekenburger M, Losson H, Vermeulen K, Hahn H, Gerard D. Novel HDAC inhibitor MAKV-8 and imatinib synergistically kill chronic myeloid leukemia cells via inhibition of BCR-ABL/MYC-signaling: effect on imatinib resistance and stem cells. Clin Epigenetics. 2020;12(1):69. doi: 10.1186/s13148-020-00839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen TTT, Zhang Y, Shang E, Shu C, Torrini C, Zhao J. HDAC inhibitors elicit metabolic reprogramming by targeting super-enhancers in glioblastoma models. J Clin Invest. 2020;130(7):3699–3716. doi: 10.1172/JCI129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagaraja S, Vitanza NA, Woo PJ, Taylor KR, Liu F, Zhang L. Transcriptional Dependencies in Diffuse Intrinsic Pontine Glioma. Cancer Cell. 2017;31(5):635–652. doi: 10.1016/j.ccell.2017.03.011. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanchez GJ, Richmond PA, Bunker EN, Karman SS, Azofeifa J, Garnett AT. Genome-wide dose-dependent inhibition of histone deacetylases studies reveal their roles in enhancer remodeling and suppression of oncogenic super-enhancers. Nucleic Acids Res. 2018;46(4):1756–1776. doi: 10.1093/nar/gkx1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.