Abstract
Natural products have served human life as medications for centuries. During the outbreak of COVID-19, a number of naturally derived compounds and extracts have been tested or used as potential remedies against COVID-19. Tetradenia riparia extract is one of the plant extracts that have been deployed and claimed to manage and control COVID-19 by some communities in Tanzania and other African countries. The active compounds isolated from T. riparia are known to possess various biological properties including antimalarial and antiviral. However, the underlying mechanism of the active compounds against SARS-CoV-2 remains unknown. Results in the present work have been interpreted from the view point of computational methods including molecular dynamics, free energy methods, and metadynamics to establish the related mechanism of action. Among the constituents of T. riparia studied, luteolin inhibited viral cell entry and was thermodynamically stable. The title compound exhibit residence time and unbinding kinetics of 68.86 ms and 0.014 /ms, respectively. The findings suggest that luteolin could be potent blocker of SARS-CoV-2 cell entry. The study shades lights towards identification of bioactive constituents from T. riparia against COVID-19, and thus bioassay can be carried out to further validate such observations.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00894-021-04833-x.
Keywords: COVID-19, Docking, Luteolin, Metadynamics, SARS-CoV-2
Introduction
Traditionally, drug design and development is quite lengthy, costly, and a challenging process. On average, it takes about 12–15 years, costing $2.6 billion for a new drug to the approval stage [1]. The approval rate of new drugs to enter clinical market is only 12% [1]; unfortunately, some drugs are banned from clinical uses due to undesirable pharmacokinetic profiles. Lack of new chemical entities entering the pharmaceutical companies in recent years and high costs associated with traditional drug design suggest drug repurposing alogside searching for and predicting new drugs entries aided by computational approaches [2, 3].
The recent outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) continues to pose a public health threat. The disease was declared by the World Health Organization (WHO) as a global health emergency, causing more deaths than SARS and MERS [4, 5]. Efforts to find drugs and vaccines to treat the disease are ongoing around the globe, yet there is no single approved drug to treat the disease. Hospitalized patients are generally managed based on the developing symptoms. Many countries have employed different approaches to combat and control the outbreak. The use of plant derived extracts is among many strategies employed by Tanzanian communities to fight COVID-19 outbreak. As an example, Tetradenia riparia (Hochst.) Codd. (Lamiaceae) is among the sources of plant extracts in use for the control and management of the COVID-19 in Tanzania. Based on the local prevalence of the disease, the extract seems to be effective. The previously isolated compounds from the plant were reported to exhibit several potential biological activities including antimalaria, antioxidant, antimicrobial [6], antiinflamatory [7], and antiviral activities [8]. Following the impact of the plant extract in Tanzania during COVID-19 pandemic, it can be hypothesized that the compounds therein could be potential inhibitors for SARS-CoV-2. However, the underlying molecular mechanism of action of the active compounds against SARS-CoV-2 (whether acting on the virus itself or on the human immune system and blocking viral cell entry) remains unknown and is a subject for investigations reported in this paper.
Recent advances in computational power and methods have accelerated the process of drug design, discovery, and development, by studying pharmacological, biological, and pharmacokinetic properties of lead molecules before entering pre-clinical or clinical trials [9–12]. Computational approaches help in identifying molecules demonstrating both drug-like and undesirable properties at an early stage, hence reducing both time and cost related to drug development. Computational methods such as molecular docking, molecular dynamics (MD), and enhanced sampling approaches provide a deeper understanding on the molecular mechanisms of small molecules and have been widely used in drug design, discovery, and development [10, 12–18]. Drugs such as antiviral protease inhibitors have been identified through computational methods and are now available for clinical administration [19].
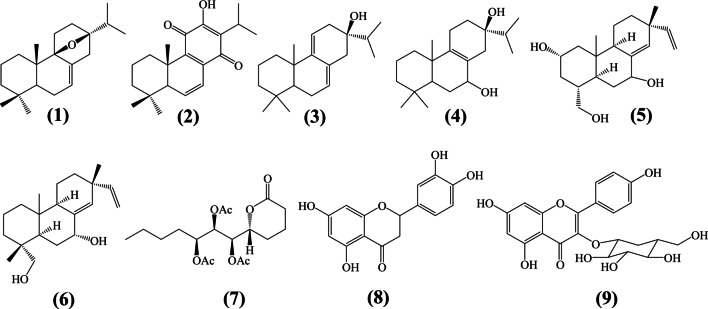
Owing to the advantage of computational methods in drug design, the present study employed different computational approaches, viz. relaxed complex scheme (RCS), molecular dynamics (MD), and metadynamics simulation to investigate the probable mechanism of action and the binding/unbinding processes of the most active compound. Nine previously isolated compounds (displayed in Fig. 1) from T. riparia were screened through computational methods. The work identified luteolin as a blocker of SARS-CoV-2 viral fusion. Coincidentally, the molecule was also previously reported to inhibit viral fusion of the old SARS-CoV, and recently suggested as dietary supplement againt COVID-19 [7].
Fig. 1.
Chemical structures of natural product isolated from Tetradenia riparia
Computational methods
Ensemble-based docking screening
Accommodating protein flexibility in docking calculation is a challenge in drug design; many docking tools can treat ligand as flexible and protein as rigid. Some of the tools like Autodock [20] allow side chain residue fluctuations, however, not the entire protein. In this study, an ensemble structure of the protein from MD simulation was used in order to perform flexible protein-ligand docking screening. The apo protein was subjected to MD simulation as described in next subsection. From the equilibrated structure, a total of 40 snapshots were extracted and subjected to docking calculations. Docking calculations were performed on two structures, i.e, crystal structure and ensemble structures. For crystal structure, five docking runs were performed to investigate the effects of ligand flexibility; the lowest binding energies were selected from each run and averaged. For ensemble structures, the ligands were docked to all generated snapshots to reflect protein flexibility. Ligand and protein preparations, as well as docking screening calculations, were performed using AutoDock Vina [20]. It is important to highlight that our docking calculations were validated using a set of experimental data reported in our recent developed protocol [3, 21] as well as comparison to root mean square deviation (RMSD) values of the redocked molecule which gave an acceptable value of 2.5 Å (Fig. S1).
Here, we present a brief description of the protocol. The ligands displayed in Fig. 1 were obtained from the literature [6, 22, 23]. Avogadro package [24] was used to generate the 3D geometries then brought to minimum conformational energy using universal force field (UFF) available within the code, hydrogen were added at a pH = 7.4. Minimized structures were saved in pdb file format and then converted to pdbqt file format using OpenBabel tool [25]. The protein structures were obtained from the RSCB protein data bank (PDB) with the following PDB IDs: spike RBD-ACE2 (6LZG), 3C-like protease (3CLpro) (6LU7), native human ACE2 (1R42), RdRp (6M71), PLpro (6W9C), Nsp9 RNA-binding protein (6W4B), and nucleocapsid N-terminal RNA-binding protein (6M3M). Missing hydrogen at pH 7.4 and gasteiger charges were added to all protein structures and were converted to pdbqt format. Screening against spike RBD-hACE2 and other protein structures was performed using our customized shell script.
Molecular dynamics simulation
The configuration with the lowest binding free energy from an ensemble-based docking experiment was used as the initial structure for MD simulation. Molecular dynamics for apo and holo proteins was performed using Gromacs 2016 employing OPLS-AA force field [26, 27]. Details on MD simulation are reported in our recent work [21]; here, a brief description is provided. The system was energy minimized using the steepest descent algorithm, followed by equilibration at NVT (constant number of particles, volume, and temperature) ensemble for 500 ps and NPT (constant pressure) ensemble for 1 ns, with position restraints. The equilibrated system was then subjected to MD production run for 100 ns. During the equilibration stage, both temperature and pressure were maintained using Berendsen method [28]. For the production stage, Parinello-Rahman and v-rescale were used for pressure and temperature coupling at 1 bar and 300 K, respectively [29, 30]. Periodic boundary conditions (PBC) were applied in all directions. Particle Mesh Ewald (PME) was used to treat long-range electrostatic interactions with a cutoff distance on 11 Å for both electrostatic and van der Waals interactions, while covalent bonds were constrained using the LINCS algorithm [31, 32]; a time step of 2 fs was used for all calculations. The MD configuration was used for analysis and as the initial structure for metadynamics simulation to investigate drug kinetics and residence time.
MM-PBSA binding free energy calculations
Molecular mechanics Poisson-Boltzmann (MM-PBSA) was calculated using g_mmpbsa tool [33]. The binding free energy (ΔGbind) for protein-ligand interaction is expressed as
| 1 |
where Gcomplex represents the total free energy of the protein-ligand complex while Gprotein and Gligand are the total free energies of the isolated protein and ligand in solvent, respectively. Furthermore, the free energy for each individual entity were derived from
| 2 |
where x stands for the protein or ligand or protein-ligand complex, T and S denote the temperature and entropy, respectively, and TΔS refers to the entropic contribution to the free energy in a vacuum. The average molecular mechanics (MM) energy in vacuum is denoted as ; this term includes bonded and non-bonded interactions. is calculated based on MM force-field parameters as follows
| 3 |
where Ebonded are bonded interactions consisting of bond, angle, dihedral, and improper interactions. The nonbonded interactions Enonbonded include both electrostatic (Eelec) and van der Waals (EvdW) interactions and are modeled using a Coulomb and Lennard-Jones (LJ) potential function, respectively. The free energy of solvation includes Gpolar and Gnon−polar and can be calculated as
| 4 |
where γ is a coefficient related to surface tension, SASA stands for solvent accessible surface area and b is the fitting parameter, and TΔS is the entropic contributions to free energy. In g_mmpbsa, an entropic contribution is not considered [33]. The binding free energy was calculated using a single trajectory, where a total of 200 snapshots were evenly extracted at a predetermined time from the first 40 ns and last 40 ns of the trajectory. The solvent and solute dielectric constants were 80 and 2, respectively, and γ was 0.0227; the PB equation was solved by using the linear PBsolver.
Previous studies have suggested that binding free energy and per-residue energy decomposition are important to establish the underlying mechanism and biological activities of the molecules in protein-ligand or protein-protein interaction [34]. In this study, the per-residue decomposition was performed to provide further insights on the mechanism of ligand under investigation.
Linear interaction energy (LIE)
Linear interaction energy (LIE) is an efficient end-point method suitable in calculating the binding affinity of drugs to receptors with large pockets and conformational flexibility [35]. In order to evaluate the binding affinity of the selected drugs to their active site, LIE was performed. In this approach, the binding free energy (ΔGbind) is calculated by averaging the ligand surrounding potentials (van der Waals and electrostatic contributions) when the ligand is in bound state and when it is free (unbound state) as follows:
| 5 |
A generalized LIE can be written as shown in Eq. 6
| 6 |
where the notation <> represents an ensemble average obtained from MD simulation. α, β are empirical parameters for Coulomb and Lennard-Jones component of energies and were selected as 0.18 and 0.5, respectively. Such values have successfully reproduced experimental results [35, 36]. The offset parameter γ is related to hydrophobicity of the active site and is empirically determined [35, 36]. In this study, LIE was calculated using the lie module embedded in GROMACS.
Well-tempered metadynamics (WT-MetaD)
Drug unbinding kinetics and residence time were investigated using well-tempered metadynamics simulation [16]. In metadynamics (Eq. 7) [17, 37], a history-dependent bias potential, V, constructed along the selected reaction coordinate , called collective variables (CVs), is built as a sum of Gaussian kernels (height (W ) and width (δ)) deposited along the trajectory in the CVs space:
| 7 |
where t is the deposition time and τ is the time interval where Gaussian potential with height is added on the position si (q(kτ)) of the biased molecules.
In WT-MetaD, the Gaussian height decreases with simulation time as shown in Eq. 8.
| 8 |
where W0 is the initial Gaussian height, kB is the Boltzmann constant, and ΔT is the input parameter with the dimension of temperature that controls how the W reduces as the well is filled. To obtain best efficiency, the parameters W0 and ΔT are chosen, and the free energy is computed using Eq. 9.
| 9 |
where T is the temperature of the system. During well-tempered metadynamics, the bias factor γ is defined as the ratio between the temperature of the CVs (T + ΔT) as indicated in Eq. 10.
| 10 |
The choice of collective variables
In the metadynamics simulation, the choice of collective variables (CVs) is not a trivial process. In this work, to better describe luteolin unbinding processes and residence time, the selected set of CVs were employed. Such CVs have also been used in other studies to investigate the ligand unbinding process [38, 39] (Table 1).
| 11 |
Table 1.
Set of CVs used to describe the residence time, kinetics, and unbinding process of luteolin
| CV | Description |
|---|---|
| CV1 | Heavy atoms distances between ligand and residue |
| CV2 | Protein-protein separation distance |
| CV3 | Center of mass (COM) distance between ligand and interface residues |
| CV4 | Coordination number defined in Eq. 11 |
During WT-MetaD, the biasfactor and Gaussian height were set to 15 and 0.6 kJ/mol, respectively, the sigma values were obtained from short unbiased MD simulation, and the value of 0.05 nm for distance and 1 for coordination number yielded good results. All WT-MetaD and free energy analysis were performed using the plumed plugin for MD simulation version 2.4 [40].
Results and discussion
Relaxed complex scheme improves binding affinity and suggests luteolin as a blocker of SARS-CoV-2 viral fusion
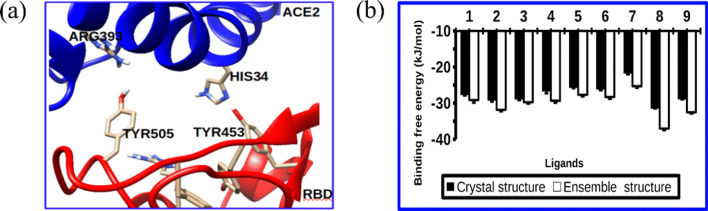
Inhibiting viral cell entry is among the strategies in treating viral diseases. Cell entry inhibitors block the virus’ fusion to host cells and provides an important option for treating viral-related diseases. Compounds designed to block viral cell entry, for example, enfuvirtide [41] and maraviroc [42], are now available for the treatment of HIV which antagonize the interaction between cellular receptors and the viral surface glycoprotein thereby preventing cell entry. SARS-CoV-2 enters human cells by strongly interacting and attaching to the human angiotension-converting enzyme 2 (hACE2) [43]. The interactions of SARS-CoV-2 and hACE2 are now well studied; as an example, in Fig. 2a, we show that the virus uses anchoring amino acids such as TYR453, TYR500, and TYR505 to strongly bind with HIS34, ARG393, and LYS353 from hACE2. Targetting inhibition and blocking viral recognition is a promising approach towards the identification of drugs to treat COVID-19. To provide molecular insights on whether compounds isolated from Tetradenia riparia [6] could work as viral cell entry inhibitors, an ensemble-based virtual screening of nine active compounds was carried out. Figure 2b shows the binding free energies for both ensemble and crystal structure docking of the compounds isolated from the plant. For all compounds 1–9 (Fig. 1), the ensemble structure demonstrates more negative energies when compared to crystal structure. Thus, this observation suggests the improvement of the docking score to an ensemble structure compared to the crystal structure.
Fig. 2.
Relaxed complex scheme improves the binding affinity of luteolin. (a) Residues at the interface involved in SARS-CoV-2 host cell fusion; (b) ensemble and crystal structure docking of the compounds isolated from the plant
Close examination on the binding affinity and mode revealed that ligand 8 (luteolin) bound strongly exhibiting binding energy of about − 36.82 kJ/mol at the interface interacting with residues TYR453, TYR505, and GLY496, and HIS34, LYS353, ASP38, GLN35 from spike RBD, and hACE2 (Fig. 4a). Interaction with these residues is important for weakening the viral spike RBD affinity toward hACE2, hence blocking viral cell entry. The ligands 2, 3, and 9 exhibit comparable crystal structure binding free energies while ligand 7 shows the least binding free energy value.
Fig. 4.
Ligand and protein dynamics. (a) Residues-ligand heavy atom distances. (b) hACE2 spike RBD distance measured for different residues
Accommodating protein flexibility plays an important role in drug design. In this work, a RSC was performed to access whether it could improve the docking scores and eliminate false positive binders. The results suggest that luteolin selectively binds at the interface with high affinity by blocking viral cell entry. The ability of both luteolin and other ligands to bind different SARS-CoV-2 target proteins was investigated as well (see Fig. S1). Similarly, luteolin shows higher affinity with hACE2 (Fig. S2). It was observed that when luteolin was docked to an ensemble structure its binding free energy decreased to − 33.05 kJ/mol compared to − 38.49 kJ/mol of the crystal structure (see Fig. 2). Small binding free energy for ensemble structure is attributed to large hACE2 pocket volume which allows the global flexibility of the residues. The dynamical stability, unbinding kinetics, and residence time of luteolin was investigated by classical MD and metadynamics simulation as discussed in the following section.
Classical MD simulation
Ligand dynamics and stability at spike RBD-hACE2 interface
Docking algorithms are powerful in discriminating good and bad binders; however, they suffer from dynamical and stability issues. To address such challenges, classical MD simulation was performed to understand the dynamics and stability of the ligand at the interface. Root mean square deviation (RMSD) and ligand-protein distances were analyzed to provide insight on the stability and dynamics of the ligand, respectively. Ligand RMSD (distance RMSD and least square fit RMSD) displayed in Figs. 3 and S6 suggest ligand dynamics at the interface. Ligand distance RMSD measures how the ligand changed from its initial docking pose. During the first 5 ns, the ligand remained to its initial docking pose, then changed to second pose where it remained until 38 ns, then it exhibited transition poses until 65 ns. From 65–80 ns, it returned to the second pose and then moved to the third pose where the ligand remained for 5 ns and returned to the second pose where remained stable until the end of the simulation time. The measured RMSD after least square fit indicated two conformation states. First, during the first 30 ns, the ligand remained stable, and then exhibited transition conformation states during 35–50 ns where the RMSD raised from 0.09 to 0.15 nm and remained until 90 ns where it decreased to 0.09 nm. These two RMSD indicate that luteolin exhibited two conformation state; the RMSD of ≤ 0.3 Å obtained after least square fit suggests stability.
Fig. 3.
Ligand dynamics at the ACE2-RBD interface. (a) Ligand distance RMSD and (b) Ligand RMSD obtained after least square fit
Detailed analysis on the dynamics of the ligand at the interface was performed, Fig. 4a shows residues-ligand heavy atom distances and hACE2 spike RBD distance measured for different residues. Furthermore, the snapshots during simulation period are extracted and shown, as indicated from the RMSD profile. It was observed that, luteolin remained in its docking pose with only few fluctuations until 38 ns. Then, two major transitions were observed between 38 and 60 ns before attaining the second pose. The dynamic and binding pose changes of luteolin suggest strong affinity of the anchoring residues between the virus and host cells, which pushes luteolin to another pose. It is interesting to note that luteolin showed stability at the interface which disrupts the recognition between the two proteins as indicated in Fig. 4b. Within the limitation of classical MD simulation, the presented probability distribution suggests that luteolin can disrupt the recognition of the two proteins as discussed in “Metadynamics” section.
Free energy calculations by MM-PBSA and LIE suggest higher binding preference of luteolin to hACE2
Since docking calculation cannot precisely estimate the ligand’s binding affinity to its pocket, end-point free energy methods based on MM-PBSA and LIE which are effective in estimating the binding free energies were performed to evaluate the binding affinity of luteolin. The binding free energy based on MM-PBSA is presented on Table 2. The findings show that van der Waals and electrostatic energy terms contributed significantly to the binding free energy, while polar energy terms unfavorably contributed to the binding free energy. This is attributed to hydrophobic interactions between the residues at the interface and luteolin. Energy decomposition and contribution of each residue (Table 3) suggest residues from hACE2 to have higher affinity with luteolin than those from spike RBD except TYR505. Energy decomposition showed six residues from hACE2 at the interface highly contributed to ligand binding free energy as compared to the two residues from the spike RBD. The total energy contributions to binding free energy (in kJ/mol) of each residue follows the trend; HIS34(− 5.21) > ASP38(− 5.03) > ARG393(− 4.94) > ASN33(− 3.13) from hACE2 and TYR505(− 6.50) > ARG404(− 2.88) from spike RBD (Table 3). From these observations, one may conclude that luteolin exhibits higher affinity with hACE2 residues which weakens the critical node recognition at the interface.
Table 2.
MM-PBSA calculation results (kJ/mol)
| System | EvdW | Eele | Epolar | Enon−polar | ΔGbind |
|---|---|---|---|---|---|
| Complex 1 | − 105.54 | − 50.65 | 99.19 | − 32.93 | − 89.94 |
Table 3.
The calculated per-residue energy contributions (kJ/mol); in parenthesis is standard deviation
| Residue | MM | Polar | Non-polar | ΔGbind |
|---|---|---|---|---|
| hACE2 | ||||
| ASN33 | − 6.16(0.19) | 3.94(0.18) | − 0.92(0.03) | − 3.13(0.12) |
| HIS34 | − 10.35(0.20) | 6.92(0.17) | − 1.79(0.04) | − 5.21(0.17) |
| GLU37 | − 8.79(0.70) | 6.6(0.44) | − 0.39(0.03) | − 2.55(0.36) |
| ASP38 | − 6.43(0.44) | 1.56(0.32) | − 0.17(0.01) | − 5.03(0.22) |
| PRO389 | − 2.81(0.07) | 0.5(0.02) | − 0.45(0.04) | − 2.77(0.11) |
| ARG393 | − 7.65(0.35) | 3.16(0.22) | − 0.46(0.02) | − 4.94(0.23) |
| spike RBD | ||||
| ARG403 | − 11.37(0.51) | 10.56(0.43) | − 2.08(0.05) | − 2.88(0.24) |
| TYR505 | − 7.29(0.23) | 2.18(0.11) | − 1.41(0.04) | − 6.50(0.22) |
Further analysis of the affinity of luteolin at the interface was performed through LIE calculation. The obtained results are as presented in Table 4. The interaction energies were calculated for ligand in bound and free form, the binding free energy obtained from LIE was − 64.16 kJ/mol, suggesting moderate inhibition. Both binding free energies from MM-PBSA and LIE suggest that luteolin possesses inhibitory effect by disrupting critical node recognition at the interface.
Table 4.
Linear interaction energies results (kJ/mol)
| System | ΔGbind | ||||
|---|---|---|---|---|---|
| Complex 1 | − 88.46 | − 93.73 | − 236.47 | − 60.14 | − 64.16 |
Water dynamics at the interface and its role on protein-protein and protein-ligand recognition
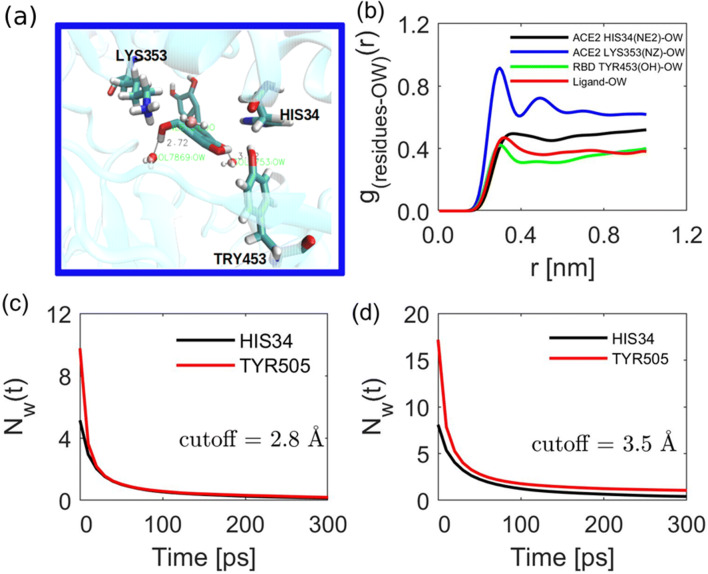
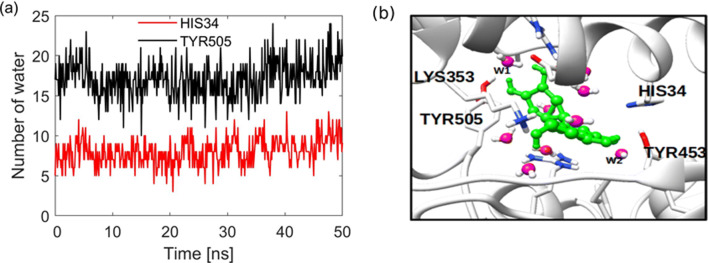
Interfacial water at protein-protein or protein-ligand plays an important role in mediating and stabilization of the interaction. In protein-ligand interactions, water is known to improve the ligand’s recognition and binding affinity [44, 45]. To account for the role of water in the discovery of SARS-CoV-2 cell entry inhibitors, we report water dynamics at the interface and its influence on the binding nature and stability of the ligand. Here, we demonstrate that the ligand binding has a potential role in disrupting this nature of interactions and establish a new interaction pattern. First, the concentration of water in the hydration shells of the anchoring residues with lower binding free energy was analyzed by radial distribution function (RDF) for the bound complex as shown in Fig. 5a-b. One can see that water is peculiarly interacting with amino acid, some residues, for example, LYS353 shows distinct hydration shell as compared to HIS34 and TRY453 (Fig. 5b). This interactions, prompted us to investigate the dynamics of water at the hACE2-spike RBD interface. A recent computational work showed that bridging water at the hACE2-spike RBD interface stabilizes the interaction through formation of hydrogen bonds [46]. It was shown that on average for the apo protein, there are about 20 bridging water at the interface forming hydrogen bonds between the two proteins. We show that the binding of the ligand disrupt the established water networks. To that end, the number of bridging water molecules for the holo protein within 3.5 Å was calculated. It is observed that, as shown in Fig. 6a, the number of water ranges from 8–17 for different residues; in Fig. 6b, we show that water molecule (W2) stabilizes the interaction by mediating the interaction between ligand and TYR453 by forming hydrogen bonds. To gain insights on the dynamics of the bridging water, the survival probability and water residence time near the residues hydration shells were calculated.
Fig. 5.
The role of water in mediating luteolin-residue interaction. (a) Interaction of water with both luteolin and residues at the interface; (b) the radial distribution function (rdf) for water near different residues. Water survival probability at residues for different hydration shells, (c) first hydration shell (0.28 nm), and (d) second hydration shell (0.35 nm)
Fig. 6.
Bridging water molecules. (a) The number of water remaining near HIS34 and TYR505 during the first 50 ns. (b) Water mediating the interaction of luteolin and protein residues
The survival probability of water at interface and near the residues indicates different dynamical behavior upon ligand binding. The number of water molecules near HIS34 and TYR505 within the same hydration shell is different. For example, in the first hydration shell, the number
of water for HIS34 from hACE2 is nearly half to TYR505 from spike RBD. To extract information on water residence time near the residues, the survival probability is fitted to an exponential function as [47].
| 12 |
where np is the number of water present at the residue throughout the simulation time; τs is the residence time for the decay of the stretched exponential. τ2 and τ3 is the residence time for slow biexponential decay for the first and second component, respectively. γ represents a quantitative measure of the deviation of the relaxation curve from classical exponential function. The value of γ = 1 for classical exponential function; large value indicates presence of system temporal disorders [47]. In the present work, residence time of hydration water is defined as the time during which a specific water molecules continuously remain within the volume of 3.5 Å around the residue. The best fit parameters in Fig. 5c and d are presented in Table 5. Here, the average residence time of the stretched exponential is obtained using the expression [47].
| 13 |
where Γ is the gamma function
Table 5.
Residence time of water near residues calculated by fitting the survival probability
| ResID/cutoff | a | γ | b | τ2 | c | τ3 | np | ||
|---|---|---|---|---|---|---|---|---|---|
| HIS34 (2.8 Å) | 3.10283 | 131.581 | 0.31992 | 1.72165 | 7.06034 | 2.38106 | 23.6876 | 0.04441 | |
| TYR505 (2.8 Å) | 2.98469 | 170.691 | 0.29819 | 6.03916 | 5.33047 | 2.79085 | 23.354 | 0.04169 | |
| HIS34 (3.5 Å) | 4.19358 | 162.704 | 0.41879 | 1.87162 | 7.1286 | 4.33655 | 30.3072 | 0.12886 | |
| TYR505 (3.5 Å) | 4.19099 | 134.351 | 0.41927 | 8.21212 | 5.3176 | 6.3524 | 23.1262 | 0.86089 |
As shown in Fig. 5 and Table 5, the residence time and number of water remaining near the surface of each residue is different within the same hydration shell. The residence time of water for HIS34 is
higher than TYR505 for the second hydration shell. HIS34 shows a slow dynamic (higher value of residence time/longer residence time) of water around the residue surface compared to water dynamics near TYR505. The difference in residence time is attributed to the nature of the surface residues, while HIS is polar residue; TYR is an amphipathic due to possession of both polar and non-polar characteristics. The three components residence time for the second hydration shell for HIS34 was observed to be 162.70, 7.12, and 30.30 ps, higher than that of TYR505 which was 134.35, 5.31, and 23.12 ps. The observed slow dynamic could be attributed to the hydrogen bond between the residue surface and water molecules for HIS34 unlikely for the counterpart. The slow dynamics of water near polar/hydrophilic residues has also been reported in other studies [47]. On the other hand, we observe that the number of water remaining near the surface of TYR is 0.86 higher than 0.12 for HIS34, linking the free energy decomposition (Table 3) and the amount of water remaining near the surface, one can infer that, water near TYR505 mediated the interaction by improving the binding free energy. Furthermore, as observed for WT-MetaD (“Metadynamics” section), simulation the unbinding processes proceeded via TYR505 suggesting strong interaction of luteolin and residue. Although there is lack of experimental finding, our observations are in accordance with other computational work [47], where water molecules near polar/hydrophilic were characterized to have slow dynamics.
Metadynamics
Classical MD simulation and binding free energy calculations are able to describe structural dynamics and the major driving forces in complexes at equilibrium. Enhanced sampling methods such as metadynamics are effective and powerful in sampling configuration along the reaction coordinates. In this work, WT-MetaD, a variant of metadynamics which provides better control on convergence and sampling errors, was employed to study the ligand kinetics as well as protein-protein dissociation along the selected CVs.
Ligand unbinding kinetics and residence time
The unbinding processes were obtained from three independent WT-MetaD simulations runs of 80 ns, each starting from the initial configuration obtained from MD simulation. The convergence of each WT-MetaD run is presented in the supporting information which shows all system converged well (Fig. S3-4). It is good to mention that in all simulations, luteolin was observed to exit at the spike RBD vicinity near TYR505 and showed similar unbinding process. This section, therefore, focuses on describing the first unbinding process of luteolin from its native complex state. The unbinding process indicates three binding stages as shown in Fig. 7. The three stages reflects, native complex (binding), pre-complex and unbound states along the selected CV with values of 0 − 0.8, 0.8 − 1.7, and ≥ 1.7 nm. As indicated in Fig. 7a, the native complex is explored for about 18 ns, followed by 6 ns in the pre-complex state. The bound and pre-complex states are separated by the energy barrier of 49.4 kJ/mol as shown in Fig. 7b. To provide further insights into the unbinding process of luteolin, the coordination number was used as another CV. Figure 7c shows a deep minimum at coordinates (0.4,30) for CV3 and CV4 suggesting higher interaction of luteolin with hotspot residues. However, at a distance ≥ 1 nm (pre-complex), the coordination number decreases.
Fig. 7.
Three binding states characterize first unbinding processes of luteolin. (a) The time taken for luteolin to unbind before it comes again to the native state, (b) the 1D free energy of the first unbinding process, (c) 2D free energy surface showing a distinct state of luteolin, and (d) representative snapshots of the unbinding process of luteolin
The representative snapshots taken at (0.4,30), (1.3,5) and (1.8,1) are presented in Fig. 7d, to aid the discussion. At the beginning of the metadynamics simulation, luteolin is in its bound state (Fig. 7d(i)), interacting with the anchoring residues shown in Fig. 2a at the interface. At t = 20 ns, the unbinding proceeds near TYR505, at this stage the ligand is interacting with both LYS353 and TYR505 before it unbound and become full solvated.
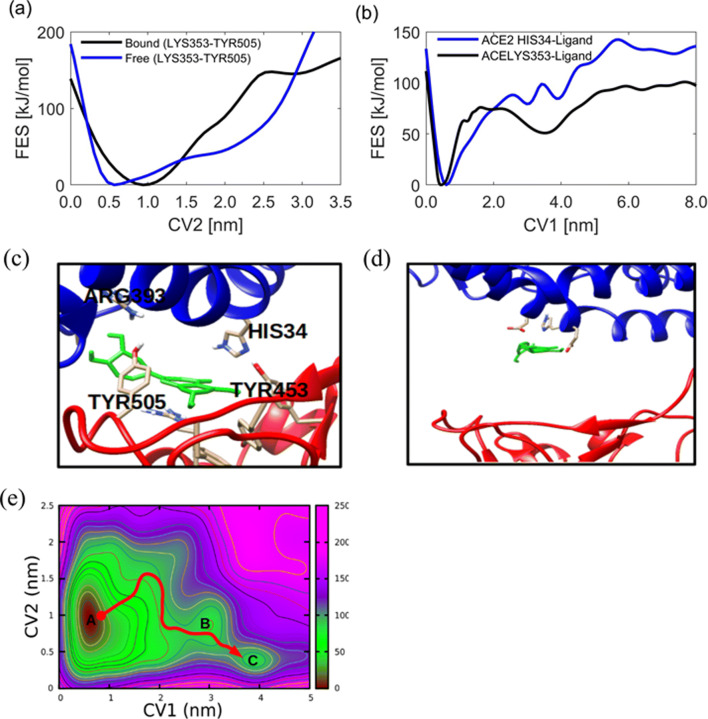
To provide details on the disruptive nature of luteolin to spike RBD-hACE2 recognition, as an example, the free energy for the distance between two hotspot residues TYR505 and LYS353 is monitored. Figure 8a and b shows the 1D free energy surface for the spike RBD-hACE2 as well as residue-ligand distances. One can note that the distance between the two residues without luteolin at the interface shows a minimum free energy at 0.5 nm; however, upon binding of luteolin, the distance increases and shows a minimum free energy at 1 nm. This suggests that the binding of luteolin has a role in weakening the interaction and recognition of the two proteins resulting in low affinity (Fig. 8d) and hence failure to penetrate the human cells. On the other hand, the distance between luteolin with residues from hACE2 and spike RBD shows the minimum free energy at 0.45 nm for two simulations (Fig. 8b).
Fig. 8.
Disruption of hotspot recognition by luteolin. (a) The spike RBD and hACE2 distance before and after binding of luteolin, (b) unbinding free energy of luteolin measured by monitoring heavy atoms from ligand and each residue as the function of CV1, (c–d) the binding and interaction of luteolin at the interface, and (e) 2D free energy for luteolin unbinding and protein-protein dissociation as a function of CV1 and CV2
To gain further insights into the unbinding progress, the 2D free energy surface is presented in Fig. 8e which suggests the existence of three states. The deep minimum (state A) indicates strong affinity of the luteolin at the interface with anchoring residues from virus and hACE2 along with the selected CVs. It is further observed that as the ligand unbind the protein-protein distance decreases as indicated in state B. At state C, luteolin is in unbound state and complete solvation, at this state, protein-protein distance is observed to return to its original distance of 0.5 nm. Generally, the presented free energy shows that, the unbinding pathway of luteolin proceeds through three conformational states from state A via B to state C.
Drug residence time is known to predict in vivo bioactivity [48]; in this study, residence time was calculated using Eq. 14 to quantify how long does luteolin resides at the interface.
| 14 |
where β = (kBT)− 1, B is the Boltzmann constant, and T is temperature. ΔF is the free energy difference between the transition state (TS) and the bound state (FA), as shown in Fig. 7b, the free energy difference between TS and FA is 49.4 kJ/mol, the τex is as an average time taken from bound state to unbound state, in our case τex was found to be 450 ps . The calculated τAB and koff values are 68.86 ms and 0.014/ms, respectively. Although, there is no experimental data at the moment to confirm our observations on the kinetics and inhibition of SARS-CoV-2 cell entry with the studied ligand, the presented residence time and unbinding rates suggest moderate inhibition and hotspot recognition disruption.
Luteolin pharmacological properties
Luteolin, a dietary supplement, is reported to posses several pharmaological properties including antioxidant and anti-inflammatory [49]. Interestingly, previous experimental works have suggested luteolin to block the old SARS-CoV viral cell entry [50, 51]. Since SARS-CoV and SARS-CoV-2 have a higher degree of similarity and have the same mechanisms, they enter host cell by strongly binding to human ACE2. Our finding, like previous experimental work, suggests luteolin could work as SARS-CoV-2 cell entry inhibitor. In addition, the present finding further provides insight into the mechanisms of action and supports the use of the plant for management and controlling COVID-19. The possession of anti-inflammatory activities provides benefits on the treatment of the diseases.
Conclusions
The present work provides insights into the possible mechanisms of the active compounds isolated from Tetradenia riparia as potent SARS-CoV-2 inhibitors. Results obtained from RCS, classical MD simulation, and metadynamics simulations suggest luteolin as blocker of SARS-CoV-2 cell entry. Although there is lack of experimental work on luteolin as SARS-CoV-2, previous experiments showed that luteolin inhibited viral cell fusion of old SARS-CoV with human receptors. Our present finding to some extend supports the use of the plant in managing and controlling COVID-19 and suggests luteolin as a cell entry inhibitor. Possession of antioxidant and anti-inflammatory activities also provides great opportunity of luteolin as inhibitor of SARS-CoV-2. However, other experimental works are still needed to expand our understanding on the other possible mechanisms of action.
Electronic supplementary material
Acknowledgements
We would like to thank the South Africa Centre for High Performance Computing (CHPC) for providing computational resources on their Lengau cluster for this research.
Author contribution
All authors whose names appear on the manuscript
1. Made substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data;
2. Drafted the work or revised it critically for important intellectual content;
3. Approved the version to be published; and
4. Agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
Partial financial support was received from the Data Driven Innovation (DDI) project through the Africa Centre for Research, Agricultural Advancement, Teaching Excellence and Sustainability (CREATES) scheme hosted at the Nelson Mandela African Institution of Science and Technology (NM-AIST) supported DMS, IO, JMV and HS; the University of Dar es Salaam through its Directorate of Research Grant supported GD and SSN, and Kenya Education Network Trust (KENET) for mini-grant supported LWK.
Availability of data and material
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Daniel M. Shadrack, Email: dmssjut@gmail.com
Geradius Deogratias, Email: dgeradius@udsm.ac.tz, Email: dgeradius@gmail.com.
References
- 1.Sullivan T (2019) A tough road: cost to develop one new drug is $2.6 billion; approval rate for drugs entering clinical development is less than 12%. Policy & Medicine
- 2.Deftereos SN, Andronis C, Friedla EJ, Persidis A, Persidis A. Drug repurposing and adverse event prediction using high-throughput literature analysis. Wiley Interdiscip Rev Syst Biol Med. 2011;3(3):323–334. doi: 10.1002/wsbm.147. [DOI] [PubMed] [Google Scholar]
- 3.Shadrack DM, Deogratias G, Kiruri LW, Swai HS, Vianney JM, Nyandoro SS. Ensemble-based screening of natural products and FDA-approved drugs identified potent inhibitors of SARS-CoV-2 that work with two distinct mechanisms. J Mol Graph Model. 2021;105:107871. doi: 10.1016/j.jmgm.2021.107871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, Al-Jabir A, Iosifidis C, Agha R (2020) World Health Organization declares Global Emergency: a review of the 2019 Novel Coronavirus (COVID-19), vol 76 [DOI] [PMC free article] [PubMed]
- 5.Mahase E (2020) Coronavirus: covid-19 has killed more people than sars and mers combined despite lower case fatality rate [DOI] [PubMed]
- 6.Fernandez ACA, Rosa MF, Fernandez CM, Bortolucci WC, Melo UZ, Siqueira VL, Cortez DA, Gonċalves JE, Linde GA, Gazim ZC. Antimicrobial and antioxidant activities of the extract and fractions of Tetradenia riparia (Hochst.) Codd (Lamiaceae) leaves from Brazil. Curr Microbiol. 2017;74(12):1453–1460. doi: 10.1007/s00284-017-1340-9. [DOI] [PubMed] [Google Scholar]
- 7.Ansari WA, Ahamad T, Khan MA, Khan ZA, Khan M.F (2020) Luteolin: A Dietary Molecule as Potential Anti-COVID-19 Agent
- 8.Fan W, Qian S, Qian P, Li X. Antiviral activity of luteolin against Japanese encephalitis virus. Virus Res. 2016;220:112–116. doi: 10.1016/j.virusres.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 9.De Vivo M, Masetti M, Bottegoni G, Cavalli A. Role of molecular dynamics and related methods in drug discovery. J Med Chem. 2016;59(9):4035–4061. doi: 10.1021/acs.jmedchem.5b01684. [DOI] [PubMed] [Google Scholar]
- 10.Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA. Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proc Natl Acad Sci. 2008;105(45):17278–17283. doi: 10.1073/pnas.0805820105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barker JJ, Barker O, Courtney SM, Gardiner M, Hesterkamp T, Ichihara O, Mather O, Montalbetti CA, Müller A, Varasi M, et al. Discovery of a novel Hsp90 inhibitor by fragment linking. ChemMedChem. 2010;5(10):1697–1700. doi: 10.1002/cmdc.201000219. [DOI] [PubMed] [Google Scholar]
- 12.Brooijmans N. Docking methods, ligand design, and validating data sets in the structural genomic era. Structural Bioinformatics. New York, NY, USA: Wiley; 2009. pp. 635–663. [Google Scholar]
- 13.Barakat K, Mane J, Friesen D, Tuszynski J. Ensemble-based virtual screening reveals dual-inhibitors for the p53–MDM2/MDMX interactions. J Mol Graph Model. 2010;28(6):555–568. doi: 10.1016/j.jmgm.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Barakat K, Tuszynski J. Relaxed complex scheme suggests novel inhibitors for the lyase activity of DNA polymerase beta. J Mol Graph Model. 2011;29(5):702–716. doi: 10.1016/j.jmgm.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 15.David L, Nielsen PA, Hedstrom M, Norden B. Scope and limitation of ligand docking: methods, scoring functions and protein targets. Curr Comput Aided Drug Des. 2005;1(3):275–306. doi: 10.2174/1573409054367682. [DOI] [Google Scholar]
- 16.Barducci A, Bussi G, Parrinello M. Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys Rev Lett. 2008;100(2):020603. doi: 10.1103/PhysRevLett.100.020603. [DOI] [PubMed] [Google Scholar]
- 17.Barducci A, Bonomi M, Parrinello M. Metadynamics. Wiley Interdiscip Rev Computat Mol Sci. 2011;1(5):826–843. doi: 10.1002/wcms.31. [DOI] [Google Scholar]
- 18.Cheng LS, Amaro RE, Xu D, Li WW, Arzberger PW, McCammon JA. Ensemble-based virtual screening reveals potential novel antiviral compounds for avian influenza neuraminidase. J Med Chem. 2008;51(13):3878–3894. doi: 10.1021/jm8001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin JH, Perryman AL, Schames JR, McCammon JA. Computational drug design accommodating receptor flexibility: the relaxed complex scheme. J Am Chem Soc. 2002;124(20):5632–5633. doi: 10.1021/ja0260162. [DOI] [PubMed] [Google Scholar]
- 20.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shadrack DM, Swai HS, Hassanali A. A computational study on the role of water and conformational fluctuations in Hsp90 in response to inhibitors. J Mol Graph Model. 2020;96:107510. doi: 10.1016/j.jmgm.2019.107510. [DOI] [PubMed] [Google Scholar]
- 22.Gazim ZC, Rodrigues F, Amorin ACL, Rezende CMd, Soković M, Tešević V, Vučković I, Krstić G, Cortez LER, Colauto NB, et al. New natural diterpene-type abietane from Tetradenia riparia essential oil with cytotoxic and antioxidant activities. Molecules. 2014;19(1):514–524. doi: 10.3390/molecules19010514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Puyvelde L, Liu M, Veryser C, De Borggraeve WM, Mungarulire J, Mukazayire MJ, Luyten W. Active principles of Tetradenia riparia. IV. Anthelmintic activity of 8 (14), 15-sandaracopimaradiene-7α, 18-diol. J Ethnopharmacol. 2018;216:229–232. doi: 10.1016/j.jep.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 24.Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminformatics. 2012;4(1):17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminformatics. 2011;3(1):33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 27.Schmid N, Eichenberger AP, Choutko A, Riniker S, Winger M, Mark AE, van Gunsteren WF. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J. 2011;40(7):843. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 28.Nosé S. An extension of the canonical ensemble molecular dynamics method. Mol Phys. 1986;57(1):187–191. doi: 10.1080/00268978600100141. [DOI] [Google Scholar]
- 29.Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52(12):7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 30.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 31.Cheatham TI, Miller J, Fox T, Darden T, Kollman P. Molecular dynamics simulations on solvated biomolecular systems: the particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J Am Chem Soc. 1995;117(14):4193–4194. doi: 10.1021/ja00119a045. [DOI] [Google Scholar]
- 32.Hess B, Bekker H, Berendsen HJ, Fraaije JG. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 33.Kumari R, Kumar R, Consortium OSDD, Lynn A. gmmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 34.Gupta A, Chaudhary N, Aparoy P. MM-PBSA and per-residue decomposition energy studies on 7-Phenyl-imidazoquinolin-4 (5H)-one derivatives: Identification of crucial site points at microsomal prostaglandin E synthase-1 (mPGES-1) active site. Int J Biol Macromolecules. 2018;119:352–359. doi: 10.1016/j.ijbiomac.2018.07.050. [DOI] [PubMed] [Google Scholar]
- 35.Reddy MR, Erion MD. Free energy calculations in rational drug design. New York: Springer Science & Business Media; 2001. [Google Scholar]
- 36.Aqvist J, Marelius J. The linear interaction energy method for predicting ligand binding free energies. Comb Chem High Throughput Screen. 2001;4(8):613–626. doi: 10.2174/1386207013330661. [DOI] [PubMed] [Google Scholar]
- 37.Laio A, Parrinello M. Escaping free-energy minima. Proc Natl Acad Sci. 2002;99(20):12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tiwary P, Parrinello M. A time-independent free energy estimator for metadynamics. J Phys Chem B. 2014;119(3):736–742. doi: 10.1021/jp504920s. [DOI] [PubMed] [Google Scholar]
- 39.Tiwary P, Mondal J, Morrone JA, Berne B. Role of water and steric constraints in the kinetics of cavity–ligand unbinding. Proc Natl Acad Sci. 2015;112(39):12015–12019. doi: 10.1073/pnas.1516652112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonomi M, Branduardi D, Bussi G, Camilloni C, Provasi D, Raiteri P, Donadio D, Marinelli F, Pietrucci F, Broglia RA, et al. PLUMED: a portable plugin for free-energy calculations with molecular dynamics. Comput Phys Commun. 2009;180(10):1961–1972. doi: 10.1016/j.cpc.2009.05.011. [DOI] [Google Scholar]
- 41.Matthews T, Salgo M, Greenberg M, Chung J, DeMasi R, Bolognesi D. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat Rev Drug Discov. 2004;3(3):215–225. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- 42.Kuritzkes D, Kar S, Kirkpatrick P (2008) Maraviroc
- 43.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 44.Schiebel J, Gaspari R, Wulsdorf T, Ngo K, Sohn C, Schrader TE, Cavalli A, Ostermann A, Heine A, Klebe G. Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes. Nat Commun. 2018;9(1):1–15. doi: 10.1038/s41467-018-05769-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cozzini P, Fornabaio M, Mozzarelli A, Spyrakis F, Kellogg GE, Abraham DJ. Water: How to evaluate its contribution in protein–ligand interactions. Int J Quantum Chem. 2006;106(3):647–651. doi: 10.1002/qua.20812. [DOI] [Google Scholar]
- 46.Malik A, Prahlad D, Kulkarni N, Kayal A (2020) Interfacial water molecules Make RBD of SPIKE protein and human ACE2 to stick together. bioRxiv
- 47.Rani P, Biswas P. Local structure and dynamics of hydration water in intrinsically disordered proteins. J Phys Chem B. 2015;119(34):10858–10867. doi: 10.1021/jp511961c. [DOI] [PubMed] [Google Scholar]
- 48.Copeland RA. The drug–target residence time model: a 10-year retrospective. Nat. Rev. Drug Discov. 2016;15(2):87. doi: 10.1038/nrd.2015.18. [DOI] [PubMed] [Google Scholar]
- 49.Seelinger G, Merfort I, Schempp CM. Anti-oxidant, anti-inflammatory and anti-allergic activities of luteolin. Planta Med. 2008;74(14):1667–1677. doi: 10.1055/s-0028-1088314. [DOI] [PubMed] [Google Scholar]
- 50.Yi L, Li Z, Yuan K, Qu X, Chen J, Wang G, Zhang H, Luo H, Zhu L, Jiang P, et al. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J Virol. 2004;78(20):11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mehla R, Bivalkar-Mehla S, Chauhan A. A flavonoid, luteolin, cripples HIV-1 by abrogation of tat function. Plos ONE. 2011;6(11):e27915. doi: 10.1371/journal.pone.0027915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.