Abstract
Ferroptosis is primarily triggered by a failure of the glutathione (GSH)-glutathione peroxidase 4 (GPX4) reductive system and associated overwhelming lipid peroxidation, in which enzymes regulating polyunsaturated fatty acid (PUFA) metabolism, and in particular acyl-CoA synthetase long chain family member 4 (ACSL4), are central. Here, we found that exogenous oxygen radicals generated by photodynamic therapy (PDT) can directly peroxidize PUFAs and initiate lipid autoxidation, coinciding with cellular GSH depletion. Different from canonical ferroptosis induced by RSL3 or erastin, PDT-initiated lipid peroxidation and ferroptotis-like cell death is independent of lipoxygenase (ALOXs) and ACSL4. Especially, this form of cell death modality can be triggered in malignant cells insensitive to or acquired resistance to canonical ferroptosis inducers. We also observed a distinct iron metabolism pathway in this PDT-triggered cell death modality, in which cytosolic labile iron is decreased probably due to its relocation to mitochondria. Inhibition of the mitochondrial Ca2+ and Fe2+ uniporter (MCU) effectively prevented PDT-triggered lipid peroxidation and subsequent cell death. Therefore, we tentatively term this distinct ferroptosis-like cell death as liperoptosis. Moreover, using the clinically approved photosensitizer Verteporfin, PDT inhibited tumor growth through inducing prevailing ferroptosis-like cell death in a mouse xenograft model. With its site-specific advantages, these findings highlight the value of using PDT to trigger lipid peroxidation and ferroptosis-like cell death in vivo, and will benefit exploring the exact molecular mechanism of immunological effects of PDT in cancer treatment.
Keywords: Lipid peroxidation, Ferroptosis, Liperoptosis, PDT, Cell death
Ferroptosis, as a non-apoptotic form of cell death that can be pharmaceutically and genetically modulated [1], has been implicated in multiple pathologies and may offer yet-unprecedented therapeutic strategies for effective cancer treatment [2,3]. For instance, it has been demonstrated that cancer cells in a high mesenchymal therapy-resistant state and drug-tolerant ‘persister’ cancer cells acquire a high vulnerability to ferroptosis inducing agents [[4], [5], [6]]. Moreover, CD8+ T cells harness ferroptosis to increase antitumor efficacy during cancer immunotherapy by downregulation of the cystine/glutamate antiporter (also known as system xc−) [7]. The main hallmark of ferroptosis is an accumulation of iron-dependent lipid peroxidation, which is usually kept at a physiological level by the glutathione peroxidase 4 (GPX4) -glutathione (GSH)-cysteine axis and the ferroptosis suppressor protein-1 (FSP1)-coenzyme Q10-NAD(P)H system [[8], [9], [10]]. Perturbation of any part of these protective systems, e.g., through inhibition of system xc− by erastin or covalent inhibition of GPX4 by RSL3, may cause uncontrolled lipid peroxidation and consequently ferroptotic cell death.
Despite substantial progress on understanding the molecular mechanisms of ferroptosis, mysteries and controversies still remain [11]. One of these mysteries includes the precise role of iron in the process of lipid peroxidation, as it is still being debated whether this peroxidation occurs in an enzymatic or non-enzymatic way [[12], [13], [14]]. It is widely accepted that enzymes vital to polyunsaturated fatty acids (PUFA) metabolism, in particular acyl-CoA synthetase long-chain family member 4 (ACSL4) and perhaps arachidonic acid lipoxygenases (ALOXs), are central players of ferroptosis [15,16]. For instance, Acsl4 knockout cells or those with low expression are highly resistant or insensitive to ferroptosis [15]. However, a few recent studies point to more complicated molecular mechanisms of ferroptosis in diverse contexts without using canonical inducers (i.e., erastin and RSL3). For instance, FINO2 can induce lipid peroxidation independent of ALOX activity [17], and ACSL4 was reported to be dispensable for ALOX12-mediated p53-dependent ferroptosis [18].
In the other avenue, PDT is a therapeutic modality that produces a large amount of site-specific reactive oxygen species (ROS, e.g., singlet oxygen) in the light-exposed area (Fig. 1a) to destroy cancer cells, pathogenic microbes et al. [19,20]. Although it can result in membrane lipid peroxidation [21], multiple cell death pathways are usually present in PDT [22], which hinders the understanding of its immunogenic activity. Here, we have examined the factors that determine the cell death pathways in PDT and identified the conditions under which PDT dominantly incites non-enzymatic lipid peroxidation and subsequently triggers a ferroptosis-like cell death pathway. This distinct cell death was tentatively termed as “liperoptosis”, because of its dependence on accumulation of lipid peroxidation, which is efficiently blocked by typical ferroptosis inhibitors, liproxstatin-1 (Lip-1) and ferrostatin-1 (Fer-1). However, we observed preliminary evidences of a different iron metabolism pathway during liperoptosis, in which cytosolic iron is translocated into mitochondria. This type of cell death is interestingly insensitive to both ACSL4 and ALOXs, exhibiting the therapeutic potential to combat a variety of cancer cells insensitive to or acquired resistance to canonical ferroptosis inducers.
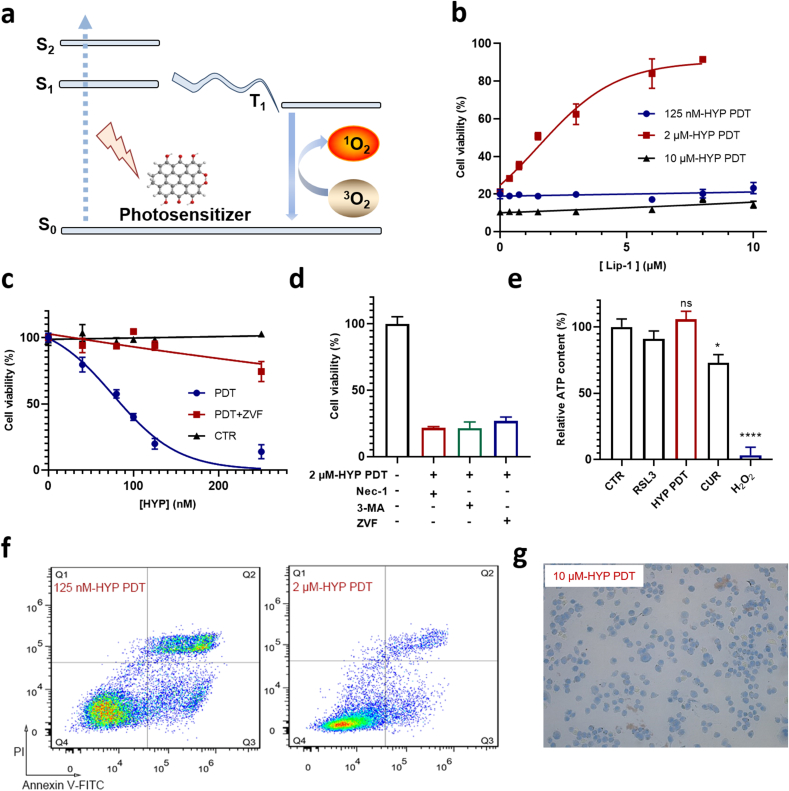
Fig. 1.
PDT triggers apoptosis, necrosis and ferroptosis-like cell death in A549 cells. a, Mechanistic scheme of the generation of ROS in PDT. S0, S1 and S2 stand for the singlet ground state, the first excited singlet state and the second excited singlet state, respectively. T1 stands for the first excited triplet state. b, Dose-dependent effect of Lip-1 on cell death triggered under three different HYP-PDT conditions (125 nM-30 min, 2 μM-2 min, 10 μM-2 min). c, Ability of the pan-caspase inhibitor ZVF to prevent A549 cell death induced by PDT using low HYP concentrations. d, Effect of cell-death-suppressing compounds on PDT (2 μM-2min) induced A549 cell death. RIPK1 inhibitor Nec-1 (20 μM), autophagic inhibitor 3-MA (30 mM), pan-caspase inhibitor ZVF (100 μM). e, Relative ATP level in A549 cells treated with RSL3 (10 μΜ, for 8 h), or PDT (2 μM-2 min, 8 h after), or curcumin (CUR, 50 μM, for 8 h), or H2O2 (20 mM, for 2 h). PDT induced cell death in A549 cells analyzed by flow cytometry (125 nM-30 min, 2 μM-2 min) in f and trypan blue staining (10 μM-2 min, 30 min after PDT) in g. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
1. Results
1.1. PDT triggers ferroptosis-like cell death
Utilizing hypericin (HYP) as photosensitizer, an orphan drug approved by the European Medicine Agency to treat cutaneous T-cell lymphoma [23], we performed PDT in cancer cell lines to identify the underlying cell death pathways. HYP-PDT can trigger multiple forms of cell death including apoptosis, necrosis and autophagy [24,25]. In the human lung adenocarcinoma A549 cell line, which shows medium sensitivity to canonic ferroptosis inducer RSL3, we firstly tested the rescue effects of the canonical ferroptosis inhibitor Lip-1 26, 27 under three different PDT conditions (typically 125 nM-30 min, 2 μM-2 min and 10 μM-2 min). Surprisingly, Lip-1 almost completely blocked the cell death process when using 2 μM-HYP PDT, while it showed no rescue effects under the two other conditions (Fig. 1b). Apoptosis is the major cell death pathway in the low HYP concentration regime (up to 250 nM in our experiments), which was confirmed by flow cytometry stained with Annexin V-FITC/PI (Fig. 1f), western blotting of cleaved caspase-3 (Fig. S1a) and application of the pan-caspase inhibitor Z-VAD-FMK (ZVF) (Fig. 1c). In the high HYP concentration regime (i.e., 10 μM) most cells underwent necrosis as characterized by a rapid loss of membrane integrity (Fig. 1g) and enlarged but intact nuclei (Fig. S1d), while neither ferroptosis nor apoptosis inhibitors could block this form of cell death (Fig. S2).
In the middle HYP concentration regime (i.e., 2 μM), we noticed that the cell death is different from both apoptosis and necrosis. While both Lip-1 (Fig. 1b) and Fer-1 (Fig. S1c) efficiently inhibited this form of cell death, effects of other inhibitors including ZVF, necrostatin-1 (Nec-1, an inhibitor of receptor interacting protein kinase 1, RIPK1), and 3-methyladenine (3-MA, an inhibitor of autophagy) were negligible (Fig. 1d). Using transmission electron microscopy (TEM), we found that cells treated with 2 μM-HYP PDT exhibited similar morphologic features to those observed in RSL3-treated cells, i.e., shrunken mitochondria, and were distinct from cells treated with 10 μM-HYP PDT (Fig. S1e). Moreover, cells treated with 2 μM HYP-PDT failed to show a distinct population of Annexin V-FITC-positive/PI-negative cells, different from cells treated with 125 nM HYP-PDT (Fig. 1f). Consistently, cleaved caspase-3 was not detected using western blotting in cells treated with 2 μM HYP-PDT in contrast to cells treated with staurosporine or 125 nM HYP-PDT (Fig. S1a). Additionally, cells treated with 2 μM-HYP PDT, as well as those with RSL3, showed no significant decrease in intracellular ATP content, different from necrotic cell death resulting from H2O2 treatment (Fig. 1e). From these findings we concluded that the main form of cell death induced by 2 μM-HYP PDT (thereafter the default condition for PDT unless specified) resembles ferroptosis.
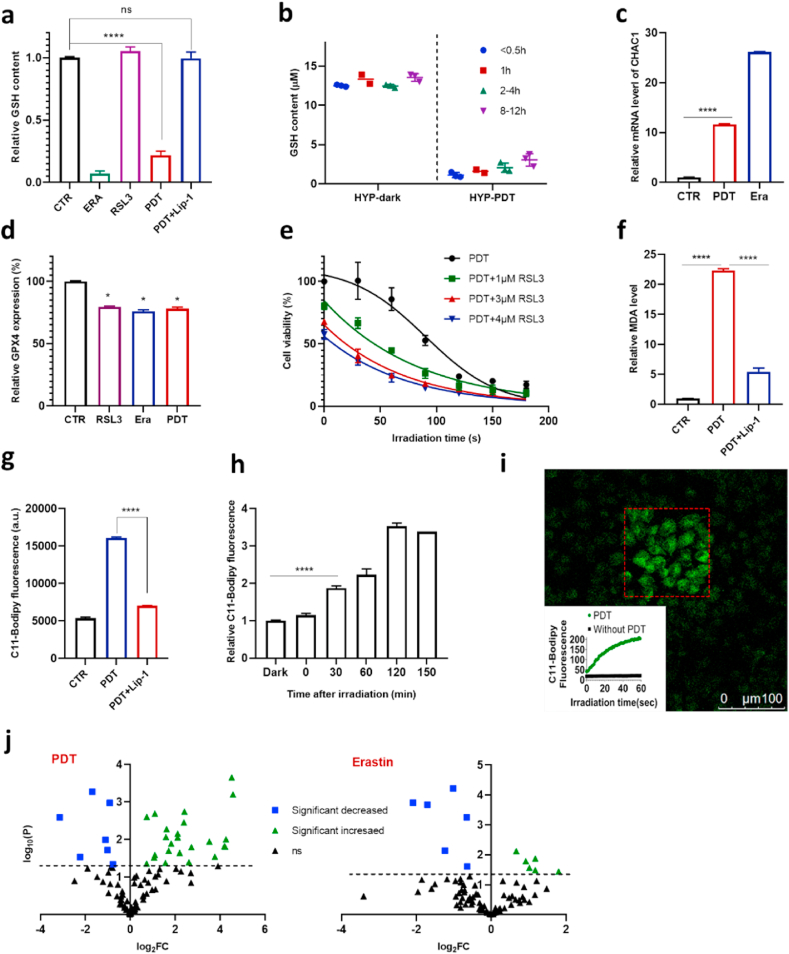
To identify the molecular mechanisms of PDT induced cell death, we firstly examined intracellular GSH levels. HYP-PDT decreased the GSH level by approximately 70%, in a manner almost as strongly as erastin (Fig. 2a). It is known that erastin results in GSH depletion through irreversibly inhibiting system xc−, while PDT did not affect the activity of system xc− as measured by detecting glutamate levels in glutamate-free medium [17,26] supernatant using LC-MS (Fig. S3b). Consistently, we found that β-mercaptoethanol (β-ME), which could promote cystine uptake through a pathway alternative to system xc− [27], could not rescue cells from death in the PDT treatment (Fig. S3c). We furthermore longitudinally monitored the GSH levels at various time points after PDT. A dramatic drop of GSH level was observed almost immediately after PDT (< 0.5 h), and the level was kept low up to 12 h (Fig. 2b). Such an immediate depletion of GSH could result from the quick rise of cytosolic ROS induced by HYP-PDT as measured by DCFH fluorescence (Fig. S4). To support this hypothesis, we applied electron paramagnetic resonance (EPR) to detect the PDT generated ROS (mainly singlet oxygen) and found that GSH could efficiently reduce ROS levels in vitro (Fig. S5a). Since cytosolic ROS decreased to basal levels at about 2 h after PDT (Fig. S4), the lowered GSH level was sustained through PDT probably due to its fast consumption in GPX4-mediated reduction of increased lipid hydroperoxides as determined by C11-BODIPY staining (Fig. 2g and h). We used Lip-1 to inhibit lipid peroxidation and observed that it could effectively preserve cellular GSH levels (Fig. 2a). Moreover, the mRNA level of CHAC1 was also increased significantly in PDT treated cells (Fig. 2c), similar to those treated with erastin [26]. As a marker of the endoplasmic reticulum (ER) stress [26], the upregulation of CHAC1 expression is consistent with the abundant distribution of HYP in ER (Fig. S11).
Fig. 2.
PDT depletes GSH and triggers lipid peroxidation in A549 cells. a, Intracellular GSH content in A549 cells treated with RSL3 (10 μM, for 8 h), or erastin (15 μM, for 8 h), or PDT (8 h after). Lip-1 concentration is 8 μM b, Changes of intracellular GSH content over time post PDT treatment. c, CHAC1 mRNA levels after erastin (15 μM, for 8 h) or PDT (4 h after) treatment. d, GPX4 protein abundance in A549 cells treated with RSL3 (10 μM), erastin (15 μM) or PDT for 8 h measured by Elisa. e, Effect of combination of PDT and RSL3 on cell viability (HYP concentration was 2 μM). f, Ability of Lip-1 (8 μM) to reduce MDA level elevated by PDT (8 h after). g, Ability of Lip-1 to prevent C11-BODIPY oxidation in cells treated with RSL3 (10 μM) or PDT. 250 nM Lip-1 is used in RSL3 treatment and 8 μM in PDT treatment. h, Time dependence of C11-BODIPY oxidation in cells after PDT treatment. i, Increase of C11-BODIPY oxidation in PDT treated cells with irradiation time measured with confocal laser scan microscope. The confocal image shows the distribution of oxidized C11-BODIPY after 60 s irradiation where the light area roughly coincides with the irradiated area (red square). j, Volcano plots showing the change in abundance of oxidized PE species in erastin or PDT treated cells (for 8 h), FC stands for fold of change, and the dotted line represents P=0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We further examined the impact of PDT on expression levels of GPX4, the key enzyme that prevents lipid peroxidation in lipid bilayers along with GSH [28]. A slight decrease in GPX4 protein abundance and mRNA level was detected (Fig. 2d, Fig. S6), similar to that observed in RSL3- or erastin-treated cells. We also measured the activity of GPXs based on the reduction of peroxides. Like erastin, PDT did not affect GPXs activity in contrast to RSL3 treatment (Fig. S7). Additionally, we observed that both RSL3 and erastin sensitize cells to PDT (Fig. 2e, Fig. S8b), indicating a vital role of the GSH-GPX4 system in the cell death triggered by PDT.
1.2. PDT directly initiates lipid peroxidation and cell death independent of ACSL4
Since elevated lipid peroxidation is the hallmark of ferroptosis, we next studied in more detail the levels of lipid peroxides using C11-BODIPY by flow cytometry upon PDT. Remarkably, the lipid hydroperoxide level increased immediately after light exposure and continued to increase over 2 h (Fig. 2h). Consistently, the level of malondialdehyde (MDA), a breakdown product of lipid peroxidation [29], was also strongly increased in PDT treated cells (Fig. 2f). Both the lipid peroxidation level and MDA level were efficiently suppressed by Lip-1 (Fig. 2f and g).
The immediately elevated lipid peroxidation in PDT treated cells was distinct from the slower elevation in cells with dysfunctional GSH-GPX4 axis (several hours), indicating a different oxidation mechanism in PDT. It is known that ROS (especially singlet oxygen) produced during PDT can directly oxidize phospholipids containing PUFAs [21,30]. By taking advantage of the multiple channels of confocal laser scanning microscopy (CLSM), we adapted the fluorescence recovery after photobleaching (FRAP) method to detect lipid peroxidation in situ during PDT irradiation. HYP in selected cells were firstly irradiated with a laser pulse of 633 nm to mimic PDT, and then fluorescence of the oxidized C11-BODIPY (λex 488 nm) in the whole field of view (including those cells without 633 nm irradiation) was detected. Indeed, C11-BODIPY fluorescence from the selected area increased immediately in response to laser irradiation, while it kept unchanged in the non-irradiated area (Fig. 2i, Fig. S12a). Notably, lipid radicals were also detected by EPR after irradiation in extracted cell membranes, as well as in linoleic acid, mixed with iron (tens of μM) and HYP, demonstrating the capability of PDT to initiate lipid peroxidation and subsequent autoxidation in vitro (Fig. S13). Addition of Lip-1 reduced, but not completely suppressed the fluorescence intensity during light irradiation (Fig. S12a). Because Lip-1 did not effectively react with singlet oxygen (Fig. S12b), it reduced the fluorescence intensity of C11-BODIPY mainly through quenching lipid ROS (Fig. S12d), consistent with the notion that Lip-1 is mainly localized in lipid bilayers.
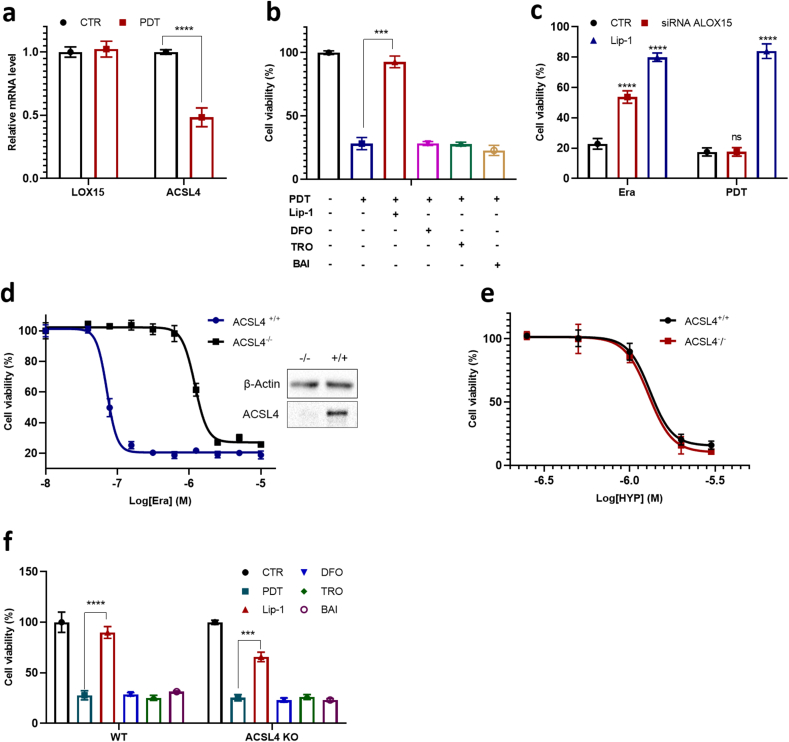
Lipidomic analysis has been used to characterize altered patterns of oxidized phospholipids in cells and tissues undergoing ferroptosis. As expected, HYP-PDT induced extensive remodeling of lipids (Table S1). Specifically, while erastin caused an increase of 6 oxidized PE species after treatment, consistent with previous reports [15], HYP-PDT caused an increase of 21 oxidized PE species (Fig. 2j). This difference was also found in oxidized PC species (Fig. S14). The diversified oxidation pattern we observed here is similar to that found in FINO2-induced ferroptosis, where a large subset of lipidome is oxidized independent of ALOX activity [17]. Moreover, we observed that the ALOX15 mRNA level did not change apparently in PDT treated cells (Fig. 3a), and baicalein (BAI, a nonspecific lipoxygenase inhibitor and weak radical trapping antioxidant) did not block the cell death process in PDT (Fig. 3b). We then used siRNA to knockdown ALOX15 in another ferroptosis-sensitive HT1080 fibrosarcoma cells (Fig. S9a), but it exhibited no discernible effect on PDT-induced cell death (Fig. 3c).
Fig. 3.
PDT triggers cell death independent of ALOXs and ACSL4. a, mRNA level of ALOX15 and ACSL4 in A549 cells treated with PDT (4 h after). b, Effects of different ferroptosis inhibitors on PDT induced cell death in A549 cells. Lip-1 (8 μM), DFO (100 μM), TRO (100 μM), and BAI (100 μM) is incubated 4 h before PDT. c, Cell viability of HT1080 cells transfected with control siRNA (CTR) or the ALOX15-specific siRNA (pre-incubated for 24 h), then treated with PDT or erastin for 24 h. d, Dose-dependent cytotoxicity of erastin in WT (ACSL4+/+) and ACSL4 KO (ACSL4−/−) MEF cells for 24 h and western blotting analysis of ACSL4 expression. e, Dose-dependent cytotoxicity of PDT in WT (ACSL4+/+) and Acsl4 KO (ACSL4−/−) MEF cells (2 min irradiation). f, Effects of ferroptosis inhibitors (8 μM Lip-1, 100 μM DFO, 100 μM TRO, and 100 μM BAI) on PDT-treated A549 cells.
In PDT, the mRNA level of ACSL4 was significantly decreased (Fig. 3a), which was likely a cellular response to elevated lipid peroxidation. But neither troglitazone (TRO) nor rosiglitazone (ROSI) could significantly rescue cells from death in PDT (Fig. 3b, Fig. S15). Furthermore, we applied PDT in a ACSL4−/− mouse embryonic fibroblast (MEF) cell line (Fig. 3d). While ACSL4 KO MEFs were expectedly resistant to RSL3 and erastin in comparison with wilt type MEFs (Fig. 3d, Fig. S10), these two types of cells exhibited nearly the same response to PDT (Fig. 3e), and to inhibitors including Lip-1, TRO and BAI (Fig. 3f). In summary, PDT is distinct from canonical ferroptosis inducers such as RSL3 erastin, by triggering lipid peroxidation and subsequent cell death independent of ALOXs and ACSL4.
1.3. PDT-triggered cell death exhibits distinct iron dependence and metabolism
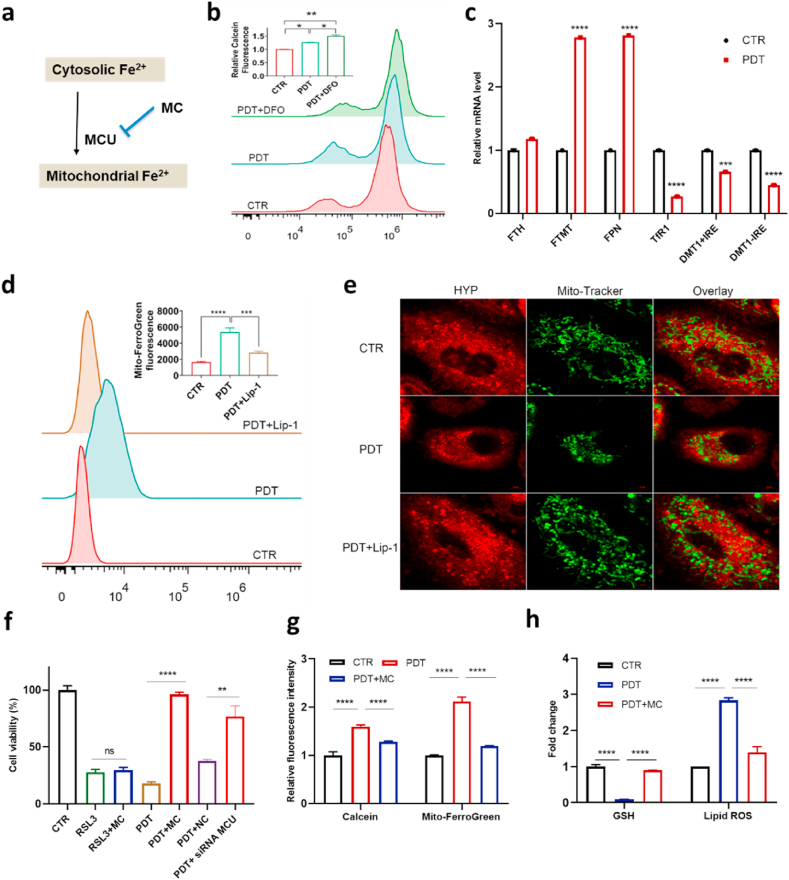
An increase of the labile iron pool (LIP) level, localized primarily in the cytosol [31], is one of the characteristic features of ferroptosis. Accordingly, the iron chelator deferoxamine (DFO) efficiently suppresses lipid ROS and the lethality of ferroptosis inducers [32] including RSL3 and erastin as well as genetic knockout of GPX4 [33]. Surprisingly, PDT-induced cell death was not suppressed by DFO and was rather aggravated to some extent (Fig. 3b, Fig. S16). This counterintuitive observation prompted us to further investigate the role of iron in PDT. We first measured the LIP levels after light irradiation based on Calcein [34], whose fluorescence intensity correlates inversely with labile iron content. LIP of A549 dropped significantly after PDT and decreased further in the presence of DFO (Fig. 4b). To further explore the mechanism of the observed LIP decrease, we then studied the effects of PDT on the expression of genes regulating iron metabolism. While the mRNA level of TfR1 (transferrin receptor 1) and DMT1 (divalent metal transporter 1) decreased significantly, the expression of FPN (Ferroportin) and FTMT (mitochondrial ferritin) increased (Fig. 4c). The decreased expression of TfR1 and DMT1 suggested lowered iron uptake rate [35] and consequently reduced LIP level [36]. The increased expression level of FPN and FTMT suggested elevated iron export from cytosol and likely iron storage in mitochondria, respectively.
Fig. 4.
Relocation of cytosolic iron into mitochondria in response to PDT. a, Scheme of iron translocation in PDT. b, Cytosolic labile iron pool (LIP) level (Calcein was used as indicator) in PDT-treated A549 cells measured by flow cytometry; c, Relative mRNA level of TfR1, DMT1+IRE, DMT1-IRE, FTH, FTMT, and FPN in PDT-treated A549 cells. d, Ferrous iron level in mitochondria in PDT-treated A549 cells measured by Mito-FerroGreen. e, Representative confocal microscopy images of PDT treated A549 (4 h after) stained with MitoTracker Green. Lip-1 is 8 μM. f, Effect of minocycline (MC, 100 μM) or siRNA MCU (25 nM) on cell death triggered by PDT in A549 cells (NC = Negative Control). g, Ability of MC to prevent the decrease of cytosolic LIP level and an increase of mitochondrial ferrous iron level in PDT-treated A549 cells as measured by flow cytometry. h, Ability of MC to prevent depletion of intracellular GSH level and elevation of lipid peroxidation level in PDT-treated A549 cells. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We then examined the fate of labile iron after PDT. FTH, which encodes a cytosol protein to store excessive iron, failed to show altered expression upon PDT, in contrast to its mitochondrial counterpart, FTMT (Fig. 4c). Therefore, we hypothesized that PDT could result in translocation of cytosolic iron into mitochondria. Using a highly specific fluorescent dye, Mito-FerroGreen [37], whose fluorescence intensity correlates with ferrous iron content, a significant increase of mitochondrial iron was observed in response to PDT (Fig. 4d). The mitochondrial morphology in A549 cells was also examined using MitoTracker Green. We observed an increased condensation of mitochondria especially around the nucleus, indicating pronounced damage of mitochondria 4 h after PDT treatment (Fig. 4e). By contrast, no changes in mitochondrial membrane potential (Fig. S17a) and mitochondrial ROS (Fig. S17c) were detectable using Rh123 and MitoSOX, respectively. In accordance, the mitochondria-targeted antioxidant and superoxide scavenger Mito-tempo [2] failed to inhibit the cell death triggered by PDT (Fig. S18). It is thus reasonable to assume that at an early stage after PDT, excessive Fe2+ entering mitochondria was stored in FTMT, which can diminish oxidative damage under conditions associated with mitochondrial iron overload [38]. Additionally, PDT would not be expected to directly oxidize the mitochondrial membrane due to the low abundance of HYP in mitochondria (Fig. S11). Hence, mitochondrial damage might be a secondary event in PDT triggered ferroptosis.
Translocation of cytosolic labile iron into mitochondria can be achieved via mitochondrial Ca2+ and Fe2+ uniporter (MCU) [39], which is the primary transporter of iron into mitochondria under oxidative stress [40]. We applied the MCU inhibitor minocycline (MC) before PDT and found that it could efficiently rescue cells from death (Fig. 4f). Such an effect was unique to PDT-induced cell death, as no effect on RSL3-induced cell death was observed. To exclude any off-target effects of MC, we used siRNA to knockdown MCU expression (Fig. S19). In line with the application of MC, considerable protection against HYP-PDT induced cell death was achieved by MCU knockdown (Fig. 4f). Moreover, application of MC blunted both the reduction of cytosolic LIP and increased Fe2+ in mitochondria in response to PDT (Fig. 4g), indicating a key role of MCU in translocating irons during PDT. Consistent with the cell viability results, a drop in GSH level in PDT-treated cells could be prevented by MC (Fig. 4h). Moreover, while elevated lipid ROS was significantly reduced by inhibiting iron translocation into mitochondria using MC (Fig. 4h), elevated mitochondrial iron level could also be alleviated by reduction of lipid ROS using Lip-1 (Fig. 4d). This interdependence between lipid peroxidation and mitochondrial iron in PDT is the same as that between lipid ROS and cytosol iron in erastin or RSL3 induced ferroptosis. Taken together, our findings demonstrate that PDT induced cell death involves an iron metabolism pathway different from canonical ferroptosis (Fig. 4a).
1.4. The generality of PDT to trigger ferroptosis-like cell death
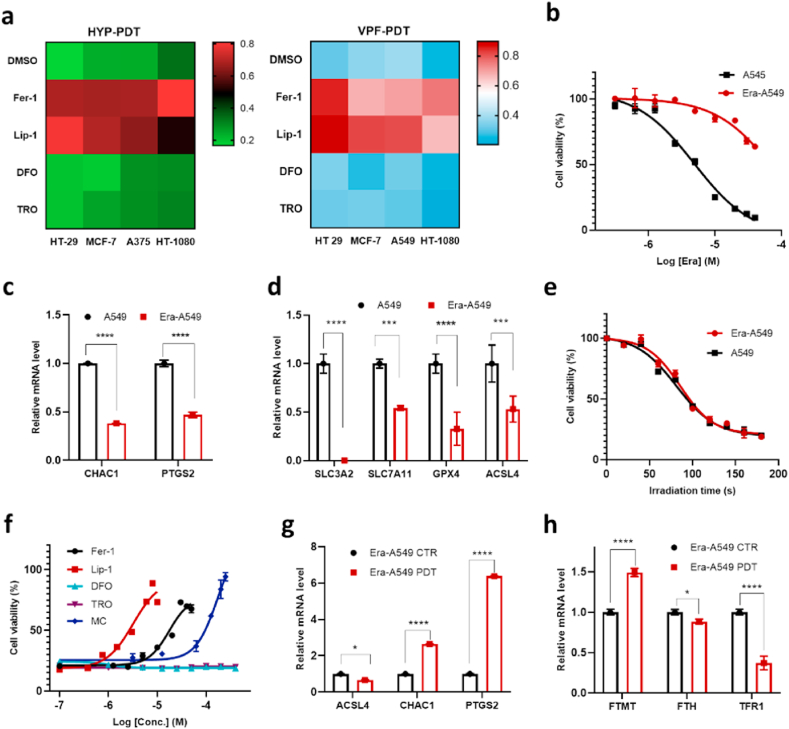
To evaluate whether PDT-induced ferroptosis-like cell death is a general phenomenon, we applied different photosensitizers, including Verteporfin (VPF, an FDA-approved PDT drug [23]) and rose bengal (RB), in multiple cancer cell lines. Cell death induced by these PSs exhibited a similar responsive pattern to four ferroptosis inhibitors (Fig. 5a, Fig. S20), where the death could all be effectively blocked by Lip-1 and Fer-1.
Fig. 5.
Generality of PDT triggered ferroptosis-like cell death. a, Effect of ferroptosis-suppressing compounds (30 μM Fer-1, 8 μM Lip-1, 100 μM DFO, 100 μM TRO) on various cell lines treated with PDT using HYP (2 μM-2 min) or VPF (4 μM-2 min). The relative cell viability is represented by color (deep blue and deep red represent 0 and 1, respectively). b, The dose-dependent cytotoxicity of erastin on parental or erastin-resistant A549 cells (Era-A549). c-d, Relative mRNA level of CHAC1, PTGS2, SLC3A2, SLC7A11, GPX4, and ACSL4 in parental and erastin-resistant cells. e, Dependence of cell viability on PDT irradiation duration in parental and erastin-resistant A549 cells. f, Effect of different ferroptosis inhibitors on the viability of PDT treated Era-A549 cells. g-h, Relative mRNA level of CHAC1, PTGS2, ACSL4, FTMT, FTH, and TFR1 after PDT treatment in Era-A549 cells (4 h after). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Notably, PDT was able to induce ferroptosis-like cell death in MCF-7 (Fig. 5a), which lacks ACSL4 expression and is therefore resistant to both erastin and RSL3. This finding inspired us to analyse whether PDT might kill cancer cells with acquired resistance to canonical ferroptosis inducers. Hence, 'persister' A549 cells, which were more resistant to erastin (Fig. 5b), were derived from treatment of cells with a series of graded concentrations of erastin. In these erastin-resistant A549 cells (Era-A549), the mRNA expression level of both subunits of system xc− (SLC3A2 and SLC7A11) decreased dramatically, so did the positive regulator of ferroptosis ACSL4 (Fig. 5d, Fig. S21a). Interestingly, CHAC1 and PTGS2, whose upregulation is considered as makers of ferroptosis [26], were downregulated in these erastin-resistant cells (Fig. 5c). We then examined the sensitivity of these resistant cells to PDT. Firstly, Era-A549 cells showed the same response trend as parental A549 cells towards PDT irradiation duration (Fig. 5e), as well as towards inhibitors including Fer-1, Lip-1, MC, TRO and DFO (Fig. 5f). Secondly, CHAC1 and PTGS2 in Era-A549 cells were both dramatically upregulated after PDT, while ACSL4 was slightly downregulated (Fig. 5g). Thirdly, the increased FTMT and decreased TFR1 (Fig. 5h) expression levels, along with the effect of MC to block cell death (Fig. 5f), indicated a similar translocation of cytosolic iron into mitochondria in Era-A549 cells. In summary, PDT triggers a similar ferroptotis-like cell death pathway in both Era-A549 and A549, as well as in ACSL4 KO MEF cells.
1.5. PDT triggers ferroptosis-like cell death in vivo
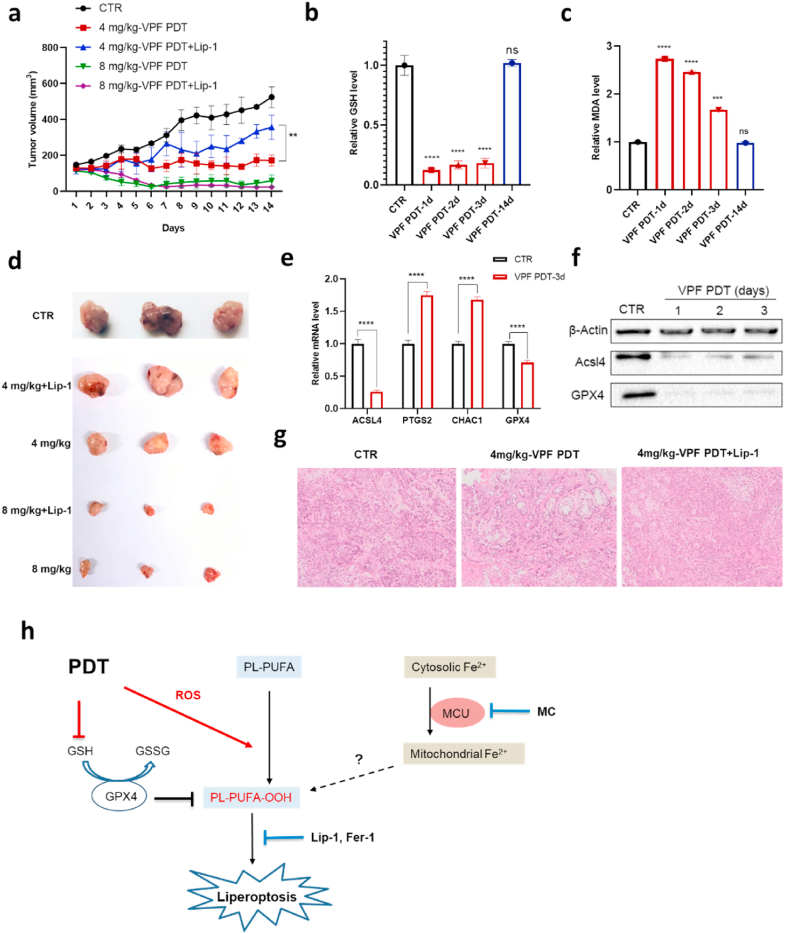
Spurred by these findings, we asked whether PDT could result in ferroptosis-like cell death in vivo. BALB/c nude mice with A549 subcutaneous xenografts were treated by single intravenous injection of Verteporfin, whose longer absorption wavelength would increase the penetration depth of light into tumors. Tumors were then exposed to 660 nm laser light and the tumor volumes were monitored over 2 weeks (Fig. S24). PDT treatment significantly inhibited the tumor growth (Fig. 6a), and significant differences were also observed between the two groups receiving different Verteporfin doses. In the 8 mg/kg group, the tumor surface turned to dark brown on the same day after light exposure, and the tumor sizes began to decrease sharply to approximately 30 mm3 later. In the 4 mg/kg group, the tumor surface had no color change in the first 2 days, and the final tumor sizes were just slightly larger than the initial size of 100 mm3.
Fig. 6.
PDT treatment causes ferroptosis-like cell death and tumor regression in A549 bearing mice. a, Tumor growth inhibition by PDT and effect of Lip-1. b-c, Quantification of the GSH level (b) and MDA level (c) in frozen tumor sections obtained at different times after PDT with 4 mg/kg Verteporfin. d, Images of resected tumors obtained 14 days after PDT treatment. e, Changes of mRNA expressions in frozen tumor sections obtained 3 days after PDT treatment. f, Expression of ACSL4 and GPX4 by Western blotting analysis in tumors at different times upon PDT with 4 mg/kg Verteporfin. g, Histological examination of tumors by H&E staining. h, Mechanistic scheme of PDT triggered lipid peroxidation and liperoptosis.
Moreover, the two groups exhibited different responses to the ferroptosis inhibitor Lip-1 (Fig. 6a). While it did not significantly affect the PDT efficacy in the 8 mg/kg group, Lip-1 counteracted the PDT efficacy in the 4 mg/kg group. Examination of the histological sections of tumors also showed that tissue damages caused by PDT with 4 mg/kg Verteporfin, instead of PDT with 8 mg/kg Verteporfin (Fig. S24d), could be largely prevented by Lip-1 (Fig. 6g). These differences are consistent with the in vitro results, where the cell death induced by medium PS dose, instead of high dose PS, could be effectively blocked by Lip-1 (Fig. 1b).
We next sought to investigate the biochemical changes in the tumor tissues treated with 4 mg/kg Verteporfin. The GSH level in tumors was significantly depleted at the first days after PDT treatment and recovered to normal 14 days after treatment (Fig. 6b). Consistently, the MDA level was elevated in the first days after treatment and returned to normal later (Fig. 6c). Additionally, the mRNA levels of both PTGS2 and CHAC1 were increased, although the fold change was modest 3 days after PDT treatment (Fig. 6e). Interestingly, the abundance of GPX4 was more drastically lowered by PDT in vivo than in vitro (Fig. 6e), which, together with the depletion of GSH, could promote lipid peroxidation during the first few days after light irradiation. As a response to the elevated lipid peroxidation, both the protein and mRNA expression of ACSL4, were significantly lowered in the PDT-treated tumors (Fig. 6e and f). The changes in GPX4 and ACSL4 abundance were both diminished 14 days after PDT treatment (Fig. S24c), so did the levels of GSH and MDA. These results demonstrate that PDT with an appropriate dose could induce the ferroptotis-like cell death in mouse xenograft models in the first few days after light irradiation, which is sufficient to inhibit the tumor growth thereafter.
2. Discussion
The accumulation of a lethal amount of lipid hydroperoxides in ferroptosis reflects a disruption of redox homeostasis in membrane lipids [13]. It can be triggered by the declined lipid ROS reductive capacity when one or more of the lipid antioxidant systems are impaired, especially the cysteine-GSH-GPX4 axis and the FSP1–Q10–NAD(P)H system [[8], [9], [10]]. Thereby, the elevated lipid peroxidation results from endogenous iron-dependent oxidation processes, although the exact mechanisms remain to be clarified [13]. On the other hand, exogenous oxidizing compounds, particularly FINO2 and artemisinin derivatives, can also oxidize lipids and induce ferroptosis [17]. Similarly, the exogenous ROS generated by PDT directly oxidize lipids and results in lipid peroxidation while the activity of system xc− and GPX4 is not affected despite immediate depletion of GSH (summarized in Fig. 6h). In contrast to the usually slow accumulation of lipid peroxidation in ferroptosis resulting from a failure of the antioxidant systems (within hours) [41], PDT generates a significantly high concentration of lipid hydroperoxides immediately after light irradiation within minutes (Fig. 2h and i). According to the proposed lipid autoxidation model [42], positive enzyme regulators of ferroptosis, e.g. ALOXs, POR and ACSL4 [16,43,44], sensitize cells to ferroptosis through facilitating the autoxidation process, and a concentration ‘threshold’ of lipid hydroperoxides should be reached for lipid autoxidation and hence ferroptotic cell death to proceed inevitably. Despite the difficulty to quantify this threshold, roughly 20 μM is proposed for the HEK293 cells [42] with a lower concentration estimated for the more sensitive MEF cells. We have observed that micromolar concentration of the antioxidants Lip-1 or Fer-1 (Fig. 1b, Fig. S1c) is required to prevent PDT-triggered cell death, which is nearly an order of magnitude higher than in RSL3 or erastin induced ferroptosis [45]. Therefore, we believe that PDT can produce sufficient amounts of lipid ROS to incite the lipid autoxidation cascade without any significant contribution from ALOXs and ACSL4. Consistently, FINO2 has been also reported to drive lipid peroxidation independent of ALOXs [17]. Such an autoxidation triggered by PDT proceeds inevitably while the activity of system xc− and GPXs is intact (Fig. S3b, Fig. S7). This explains the continuously lowered cellular GSH level (Fig. 2b) and the ineffectiveness of supplement with β-ME (Fig. S3c).
Intracellular labile iron increases during canonical ferroptosis, mainly through ferritin degradation (ferritinophagy) [46,47] or heme oxygenase-1 (HO-1)-mediated heme catabolism [2,48]. In PDT- triggered cell death, we have observed a different iron metabolism pathway. Cytosolic iron is translocated into mitochondria via MCU and results in increased mitochondrial iron level. This pathway is also different from a previous observation in PDT where iron was firstly released into the cytosol from lysosomes before its transportation to mitochondria [49]. In that case, the LIP level is enhanced and DFO can effectively suppress the PDT-induced cell death. Furthermore, we have demonstrated that this intracellular iron relocation instead of absorption of extracellular iron [50], is interdependent with lipid peroxidation. Although mechanism of this iron translocation and its interaction with lipid peroxidation require further investigation, we provide a new perspective to understand the iron metabolism pathway in cells undergoing lipid peroxidation. Interestingly, recent reports on the involvement of iron-sulfur cluster synthesis in ferroptosis [51] and the DHODH-mediated mitochondria ferroptosis defense system [52], also point to a potential role of mitochondrial iron metabolism in ferroptosis.
Based on the above differences, we tentatively term this PDT-triggered cell death as liperoptosis. This type of cell death is also caused by accumulation of lethal amount of lipid hydroperoxides, which is characteristically blocked by lipid specific antioxidants, e.g. Lip-1 and Fer-1. However, its source of lipid peroxidation is distinct from that of ferroptosis resulting from the ACSL4-related enzymatically oxidizing process. Interestingly, a distinct ferroptosis pathway mediated by p53 has also been reported to be independent of ACSL4, where an exogenous oxidant, tert-butyl hydroperoxide, is indispensable to initiate this type of cell death [18]. Moreover, we noted a recent report that the PDT-triggered cell death in MDA-MB-231, a cell line sensitive to RSL3, could be also partially blocked by Fer-1 [53].
Triggering lipid peroxidation by PDT may be particularly advantageous to combat ferroptosis-resistant or ferroptosis-insensitive cancer cells. Some malignant cells are insensitive to canonical ferroptosis and some sensitive cells may also acquire resistance to ferroptosis inducers, through lowering gene expression of positive regulators of ferroptosis, in particular ACSL4 [15]. Moreover, because regulation of tumor ferroptosis is a strategy exploited by CD8+ T cells to enhance their anti-tumor efficacy [7], it is reasonable to assume that tumors may acquire ferroptosis-resistance to achieve immune evasion. Hence, ferroptosis-resistance may impose challenges to clinical exploration of ferroptosis as a potential cancer therapeutic strategy. In this sense, PDT provides a new avenue to face these challenges with its proven clinical applicability. Additionally, PDT exhibits another advantage of its high spatial and temporal accuracy to target the tumors in vivo, which may mitigate the potential side effects of ferroptosis inducers (e.g. by inhibiting GPX4) to normal tissues. Conclusively, our findings should guide in the design of in vivo applicable strategies based on PDT to exploit the full therapeutic potential of triggering lipid peroxidation-dependent cell death in difficult-to-treat tumor.
3. Methods
3.1. Chemicals
All chemicals were purchased from Selleck unless stated otherwise. Minocycline hydrochloride (T1101) was obtained from TargetMol. Hypericin (HYP) was obtained from Chengdu Purify Technology.
Cell lines and media. HT29, A549, A375, MCF-7, HT1080, MEF cells were obtained from National Infrastructure of Cell Line Resource of China, and Acsl4 KO MEF were obtained as published [15]. HT29, A549, MEF, and Acsl4 KO MEF cells were grown in DMEM with 10% fetal bovine serum (FBS), 1% penicillin and streptomycin (Macgene). A375, MCF-7, and MCF-7 resistant cells were cultured in 1640 medium with 10% FBS. HT1080 cells were grown in MEM with 10% FBS, 1% non-essential amino acids (Gibco) and 1% penicillin-streptomycin mix. All cells were cultured in a 37 °C incubator (ESCO) with humidified atmosphere of 5% CO2. Persister A549 cells (erastin resistant A549 cells) were established by exposing A549 cells to graded concentrations of (5–20 μM) erastin over about 4 months. Cells that survived eventually in the presence of 20 μM erastin proliferated as the erastin resistant cells.
3.2. PDT treatment and cell viability assay
Cells were seeded in 96-well plates (5000 cells per well) and allowed to adhere overnight. The medium was replaced on the following day with 100 μL of growth medium containing a required concentration of HYP and the cells were incubated in the dark for 4 h. Cells were exposed to red light irradiation using a 300 W Xenon arc lamp (600 nm band filter, 1.5 mW/cm2, CEAULIGHT, China) for designated time and were further incubated thereafter. The designated modulating compound (e.g. Z-VAD-FMK, Lip-1, Fer-1, Nec-1, 2-MA, baicalein, troglitazone etc.) was usually added simultaneously with HYP. Cell viability was assessed typically 24 h after light irradiation using CCK-8 kit. All cell viability data were normalized to the DMSO vehicle condition. Measurement were performed in three independent replicates.
3.3. Western blotting
Total protein was extracted from cells using the RIPA lysis buffer (H1107020, Macgene) according to manufacturer's protocols. An equal amount of proteins was loaded in each lane. Proteins were separated on 12% gradient Tris-glycine gels (10 μl/well, 10 wells) (Sigma Aldrich), and electrically transferred to a PVDF membrane (Bio-Rad). After blocking the membrane with 5% skim milk, the target proteins were immune-blotted with their corresponding antibody at 1:1000 dilution after overnight incubation at 4 °C. Antibodies used were GPX4 (ab125066, Abcam), cleaved caspase-3 antibody (ab2302, Abcam), ACSL4 (22401-1-AP, Proteintech) and beta-actin (ab8226, Abcam). Thereafter, the horseradish peroxidase (HRP)-conjugated anti-rabbit IgG H&L (E030120, Earthox, 1:2000 dilution) and anti-mouse IgG H&L (E030110, Earthox, 1:2000 dilution) were applied as the secondary antibodies, and bands were detected with western blotting detection reagents (Bio-rad) using ECL (S4904300, YEASEN).
3.4. Transmission electron microscopy
1.0 × 106 cells were plated in 100 mm dishes and allowed to adhere overnight. After 24 h, cells were treated with vehicle (DMSO, 8 h), or RSL3 (10 μM, 10 h), or HYP (2 μM, 8 h after irradiation), or HYP (10 μM, 0.5 h after irradiation), and then washed with pre-cooled PBS and digested. The cells were collected and fixed in a mixture of 2.0% PFA and 2.5% glutaraldehyde overnight at 4 °C. Thereafter, they were fixed in 1% osmium tetroxide for 1.5 h, and then dehydrated using a graded series of ethanol (50%, 70%, 80%, 90%, 95%, and 100%), as well as pure acetone for 10 min, and then embedded in Epon 812 + 815 medium. TEM samples were prepared using an ultrathin slicer and observed in a transmission electron microscopy (JEM-100).
3.5. Annexin V-FITC/PI staining
Treated cells were trypsinized, harvested and washed with PBS, and stained with Annexin V/PI staining kit (Solarbio) according to manufacturer's protocols. Briefly, 500 μl binding buffer was added to each well and 5 × 104 cells were transferred to a 1.5 ml centrifuge tube. Then, 5 μl of Annexin V-FITC and 5 μl of propidium iodide (PI) were added, and the cells were gently vortexed. Cells were incubated for 5 min at room temperature in the dark, and analyzed by flow cytometry (FACSCalibur, BD). Data were collected from at least 10,000 cells. The results were analyzed by FlowJo 7.6.2 software (Tree Star Inc., USA).
3.6. Measurement of ATP levels
ATP levels were measured using a commercially available ATP assay (Beyotime, China). Briefly, cells treated with RSL3 (10 μΜ, for 8 h), or PDT (2 μM-2min, 8 h after), or curcumin (CUR, 50 μM, for 8 h), or H2O2 (20 mM, for 2 h), or vehicle (DMSO, 8 h) were washed with PBS and lysed. The collected lysate was incubated with ATP Monitoring Enzyme, and luminescence was measured using a Victor3 plate-reader. Protein content was determined by BCA assay, and this value was used to normalize ATP levels to protein.
3.7. Measurement of reduced glutathione
Cells treated with 15 μM erastin, or 10 μM RSL3, or PDT (2 μM-2 min HYP), or vehicle, were harvested at certain time after treatment, and were then lysed using ice-cold RIPA buffer (Macgene). Samples were centrifuged for 15 min at 4 °C at 14,000 g. The resulting supernatant was collected. Part of the supernatant was used to detect protein content by BCA assay, and part was deproteinized to measure the reduced glutathione levels using GSH/GSSG Ratio Detection Assay Kit (S0053, Beyotime) by following the manufacturer's protocol.
3.8. GPXs activity measurement
The PDT (2 μM HYP, 2 min irradiation) treated cells were harvested at a certain time after irradiation, washed twice with PBS, and then re-suspended in 200 μl assay buffer. The assay buffer contained 0.1 M Tris-HCl (pH 8.0), 0.5 mM EDTA and 1.25% Triton X-100. Cells were lysed by sonication and cleared of insoluble components by centrifugation at 14,000 g for 10 min at 4 °C. Collected supernatant was transferred to a clean tube and kept on ice. The protein concentration of each sample was determined using the BCA assay. A total of 200 μg protein was added to the reaction well and the volume was adjusted to 50 μl with assay buffer. 40 μl reaction mix containing 33 μl assay buffer, 3 μl 60 mM NADPH, 2 μl glutathione reductase (GR, 1 U/ml), and 2 μl 3 mM GSH solution) was added and well mixed. 10 μl cumene hydroperoxide solution was then added to measure the GPXs activity by monitoring the absorption at 340 nm.
3.9. Quantitative RT-PCR
Total RNA was extracted using the TRleasy reagent (10606ES60, YEASEN) by following the manufacturer's protocols. Briefly, 1 μg total RNA for each sample was reverse-transcribed into cDNA using the Hieff First Strand cDNA Synthesis Super Mix (11103ES70, YEASEN). The cDNA was then analyzed by qRT-PCR using Hieff qPCR SYBR Green Master Mix (11203ES03, YEASEN) on ABI Quant Studio 6 system (Applied Biosystems). For the quantitative RT–PCR analysis of human transcripts, the following primers were used: ChaC1 forward 5′-CCTGAAGTACCTGAATGTGCGAGA-3′, ChaC1 reverse 5′-GCAGCAAGTATTCAAGGTTGTGGC-3'; PTGS2 forward 5′-CGGTGAAACTCTGGCTAGACAG-3′, PTGS2 reverse 5′-GCAAACCGTAGATGCTCAGGGA-3'; TfR1 forward 5′-ACCCATTCGTGGTGATCAAT-3′, TfR1 reverse 5′-CGTTTCCAACTGCCCTATGA-3'; DMT1+IRE forward 5′-ACCTTAGAACTGGATTAGGGT-3′, DMT1+IRE reverse 5′-AAGGATAAACTGAGCTCGCCC-3'; DMT1-IRE forward 5′-AGTGGTTTATGTCCGGGACC-3′, DMT1-IRE reverse 5′-TTTAACGTAGCCACGGGTGG-3'; Acsl4 forward 5′-GAATGGATGATTGCAGCACAGA-3′, Acsl4 reverse 5′-CCTCAGATTCATTTAGCCCATGAAC-3′, Gpx4 forward 5′-CGCTGTGGAAGTGGATGAAG-3′, Gpx4 reverse 5′-GAGCTAGAAATAGTGGGGCAGGT-3'; Alox 15 forward 5′-TTGGTTATTTCAGCCCCCATC-3′, Alox 15 reverse 5′-TGTGTTCACTGGGTGCAGAGA-3′ SLC7A11 forward 5′-TGCTGGGCTGATTTTATCTTCG-3′, SLC7A11 reverse 5′-GAAAGGGCAACCATGAAGAGG-3'; SLC3A2 forward 5′-CTGGTGCCGTGGTCATAATC-3′, SLC3A2 reverse 5′-GCTCAGGTAATCGAGACGCC-3'; mitochondrial ferritin (FTMT) forward 5′-AAGGTGACCCCCATTTGTGC-3′, mitochondrial ferritin (FTMT) reverse 5′-GGGGCCCCCATCTTCACTAA-3'; ferritin heavy chain (FTH) forward 5′-GAGGTGGCCGAATCTTCCTTC-3′, ferritin heavy chain (FTH) reverse 5′-TCAGTGGCCAGTTTGTGCAG-3'; FPN forward 5′- CCTGTTAACAAGCACCTCAGC -3′, FPN reverse 5′-TTGCAGAGGTCAGGTAGTCG-3'; GAPDH forward 5′-ACCCAGAAGACTGTGGATGG-3′, GAPDH reverse 5′-TCTAGACGGCAGGTCAGGTC-3′.
3.10. C11-BODIPY lipid peroxidation assay
Lipid peroxidation was measured by the C11-BODIPY 581/591 (D3861, Invitrogen, USA). The treated cells were incubated with the kit reagent (10 μM in DMEM without FBS) for 30 min. Thereafter, the harvested cells were washed twice with PBS and re-suspended in 500 μl PBS. Lipid peroxidation levels were measured using flow cytometry (BD FACSCalibur) (λex: 488 nm), and the data were analyzed using FlowJo. A total of 10,000 cells in each sample was analyzed.
In addition, the lipid peroxidation was also visualized and measured simultaneously during PDT treatment using confocal laser scan microscope (CLSM) by adapting the fluorescence recovery after photo-bleaching (FRAP) method. Briefly, 150,000 cells were plated at a 35 mm glass-bottom dish and allowed to adhere overnight. Cells were then pretreated with vehicle (DMSO, 4 h) or HYP (2 μM, 4 h) in the dark, before the dish was put on the stage of an inverted confocal microscope equipped with a chamber containing 5% CO2 atmosphere and heated to 37 °C (Leica TCS SP8). Imaging was carried out using a water-immersion objective HC PL APO 63 × /1.20 W heated to 37 °C. Basel lipid peroxidation level was firstly measured by detecting 500–530 nm emission after 488 nm excitation of oxidized C11-BODIPY. The HYP incubated cells were then excited with the 633 nm laser line using the single iteration of bleaching pulse focused on the selected area of a single cell or a group of cells, to mimic the PDT irradiation. Right after the 633 nm pulse, fluoresce of the oxidized C11-BODIPY was detected with 488 nm excitation and 500–530 nm collection. The area with elevated oxidized C11-BODIPY fluorescence was roughly overlapped with the selected area excited with 633 nm laser. Usually 60 images of oxidized C11-BODIPY were acquired during 60 s or 180 s. Fluorescence intensity from the selected area under 633 nm excitation was then used to quantify the change of oxidized C11-BODIPY level.
3.11. MDA measurement
Cells were seeded in 6-well plates and pretreated with HYP with or without 8 μM Lip-1. 2 × 106 cells were harvested and washed twice by cold PBS 8 h later after irradiation, and then lysed with RIPA reagent and centrifuged 12000g for 10 min at 4 °C. The collected supernatant was kept on ice. The protein content was determined by BCA assay and equilibrated. 200 μL of 0.37% TBA reagent (diluted with 5% trichloroacetic acid) were added into each well containing 100 μL sample, and the mixture was incubated at 95 °C for 60 min. After cooling down to room temperature in an ice bath for 10 min, 200 μL supernatant (containing MDA-TBA adduct) was taken and added into a 96-well microplate. The plate was immediately measured on a microplate reader at OD 532 nm for colorimetric assay or at Ex/Em = 532/553 nm for fluorometric assay.
3.12. EPR spectroscopy
Detection of reactive radicals generated by HYP, HYP and linoleic acid mixture, HYP and cell membrane mixture during photoexcitation was performed on an X-band Bruker ER A200 spectrometer. About 500 μL sample in a 1.5 ml centrifuge tube was irradiated by a 300 W Xenon arc lamp with a 450 nm cutoff filter (~5 mW/cm2) for designated time. To detect singlet oxygen, 200 mM final concentration of 4-OH-TEMP was added to the HYP solution (20 μM) before irradiation. After irradiation, 30 μL aliquot was immediately aspirated into a glass capillary and transferred to the EPR resonator. To detect lipid radicals, mixture of HYP (20 μM) with linoleic acid (3 mM) or lysed cell membrane (diluted 1:1 with H2O) was further mixed with DTPA (0.1 mM), POBN (400 mM) and with or without ferrous ion (1 mM) before light irradiation. The typical settings applied for EPR detection were: scan range, 100 G; sweep time 60 s; modulation amplitude, 1 G; modulation frequency, 100 kHz; and microwave power, 19.23 mW.
3.13. Mass spectrometry-based untargeted lipidomics
Lipids were extracted by the Folch procedure [54]. Briefly, 10 million cells treated with DMSO, or 15 μM erastin, or 2 μM dark HYP (without irradiation), or 2 μM irradiated HYP (for 2 min) were collected 8 h after treatment and washed twice with PBS, then the precipitate was sonicated with 2: 1 chloroform-methanol mixture (v/v) to a final dilution 20-fold the volume. The crude extract was mixed thoroughly with water (0.2 of its volume) and shaken for 1 h. The sample was centrifuged for 10 min at 3000 g at 4 °C. After removing the upper phase as much as possible, the interface was carefully rinsed for three times with small amounts of pure solvents. Finally, the lower phase and remaining rinsing fluid were fused into one solution by addition of methanol. The solution was dried under vacuum and stored at −80 °C. The protein pellet was used to measure the protein concentration through BCA assay. For LC-MS analysis, the sample was reconstituted in a solution containing chloroform/methanol/water mixture (60:30:4.5, v/v/v). A quality control sample was also prepared by combining 40 μL of each sample and used to assess the reproducibility of the features through the runs. LC-MS was performed in a Q Exactive HF Orbitrap LC-MS/MS System (Thermo Scientific). The LC part is Ultimate 3000 UHPLC with an InertSustain C18 HP column (150 × 2.1 mm; 3 μm). Mobile phases were acetonitrile:water (60:40, v/v)+ 10 mM ammonium formate and 0.1% formic acid (A) and acetonitrile:water (90:10, v/v) + 10 mM ammonium formate and 0.1% formic acid (B). The gradient elution time was 25 min with the following gradient elution programs of B: 0 min, 40% B; 2 min, 43% B; 2.1 min, 50% B; 12 min, 70% B; 18 min, 99% B; 20 min, 99% B; 20.5 min, 40% B; 25 min, 40% B with the flow rate of 0.25 mL/min. The MS parameters were set as following: the auxiliary gas is 10; ion source sheath gas is 40; electrospray voltage is 3.3 kV (ESI+) and 3 kV (ESI-); heating temperature of ion source gas is 300 °C, and the ion transfer tube temperature is 320 °C, scan range was from 200 to 1200 m/z. MS-DIAL was used for baseline correction, retention correction, peak identification and alignment, and statistical analysis of the raw data. Orthogonal partial least-squares discriminant analysis (OPLS-DA) was performed to examine significant differences between control and PDT treated cells using SIMCA 13.0 software (Umetrics). Variable Importance in the Project (VIP) value in PLS-DA model was used for selecting and identifying biomarkers.
3.14. Mitochondrial morphology and ROS measurement
2 × 105 cells were seeded in 35 mm glass-bottom dishes and allowed to adhere overnight. On the next day, cells were pretreated with dark HYP (2 μM, without irradiation), or irradiated HYP (2 μM, for 2 min), irradiated HYP (2 μM, for 2 min) +Lip-1 (8 μM). 2 h after irradiation, cells were stained with Mito Tracker Green FM (200 nM, Invitrogen) for 30 min at 37 °C., Images were acquired using a LSM800 inverted microscope (Zeiss) with a 63 × 1.4 NA oil immersion objective. 488 nm and 561 nm laser lines were used to excite the Mito Tracker Green FM and HYP, respectively.
To detect mitochondrial ROS, the PDT treated cells were first stained with Mito Tracker Green FM for 30 min at 37 °C, and then washed twice with PBS before staining with 1.25 μM MitoSOX red (Invitrogen) for another 30 min at 37 °C. Images were acquired using CLSM with 488 nm and 561 nm laser lines to excite Mito Tracker Green FM and MitoSOX, respectively.
3.15. MMP measurement
Rh123 kit (BestBIO) was used to detect changes in mitochondrial membrane potential. Briefly, cells were seeded in 24-well plates with 40,000–60,000 cells/well. The treated cells were harvested and stained with Rh123 according to the manufacturer's protocol for 30 min at 37 °C. After washing with PBS cells were re-suspended in 200 μL of assay buffer and Rh123 fluorescence was assessed by flow cytometry (Ex 488 nm, Em 520 nm). Data were collected from at least 10,000 cells.
3.16. Iron assay
Cells were seeded in 24-well plates with 40,000–60,000 cells/well overnight. On the next day, the treated cells were stained with 0.1 μM Calcein AM (C131116, Aladdin) for cytosolic LIP or 5 μM Mito-FerroGreen (M489, Dojindo) for mitochondrial LIP according to the manufacturer's protocols, respectively. The harvested cells were washed twice with PBS and then analyzed by flow cytometer cytometry. Ex 488 nm/Em 530 nm for Calcein and Ex 505 nm/Em 535 nm for Mito-FerroGreen.
3.17. SiRNA transfection
Cells (100,000 per well) were seeded in a 6-well plate or a 96-well plate (2500 per well) and allowed to adhere overnight. On the next day, cells were transfected with 10 nM of siRNAs (final concentration/well) using Hieff Trans liposomal transfection reagent (YEASEN) according to the manufacture's protocols. Parallel cultures were assayed for gene expression in 6-well plate after 36 h using RT-qPCR and in 96-well plate for cell viability in response to drug treatment 24 h post transfection. The Mitochondria electrogenic Ca2+, Fe2+ uniporter (MCU) siRNA was obtained from Sangon Biotech (China) and the siRNA sequences were: sense 5′-CGGCUUACCUGGUGGGAAUTT-3′, antisense 5′-AUUCCCACCAGGUAAGCCGTT-3'.
The ALOX15 siRNA were obtained from Dharmacon (China) and the siRNA sequences were: sense 5′-CGAUACACCCUGGAAAUUA-3′, antisense 5′-AAACUGGAAGGACGGGUUA-3′, sense 5′-GAAGGAAGUUGUACCGGUG-3′, antisense 5′-CACCGGUACAACUUCCUUC-3′, sense 5′-CAUCCUAUCUUCAAGCUUA-3′, antisense 5′-UAAGCUUGAAGAUAGGAUG-3'.
3.18. In vivo xenograft mouse study
BALB/c nude mice (male, 5-week old and ~25 g, purchased from Department of Animal Science of Peking University Health Science Center, China) were subcutaneously injected with A549 cells (5 × 105 cells) in the axilla. During the treatment, body weight of the mice was recorded every day, and the tumor volume was calculated using the formula: 0.5 × length × width [2]. Once the tumor size reached ~100 mm3, the mice were randomly divided into group of 3 mice per cage. 100 μl of VPF (4 mg/kg or 8 mg/kg) or saline was intravenously injected and the tumor surface was then irradiated for twice by 660 nm laser (50 mW/cm2, irradiation duration 15 min) at 1.5 h and 4 h after injection. To examine the effect of Lip-1, a dose of 10 mg/kg was intraperitoneally administrated two days before the irradiation and then once a day for 2 weeks after the irradiation. Mice were euthanatized at the designated time or the end of treatment, and tumor tissues were frozen and stored at −80 °C for further analysis. In histological examination, the dissected tumor tissues were fixed with 10% buffered formalin, dehydrated in ethanol, embedded with paraffin, and stained with hematoxylin and eosin (H&E). In western blotting, GSH and MDA analysis, ~50 mg sample was collected and lysed with RIPA lysis buffer supplemented with the proteinase inhibitor Cocktail (YEASEN, China) at a ratio of 100:1 (v/v). The tissues were homogenized in a bullet blender at speed 4 for 4 min, and the sample was then centrifuged (12,000 rpm) for 10 min at 4 °C. The supernatant was then obtained for corresponding measurement. To extract RNA, RNase-free tubes were used to homogenize the tumor tissues (~50 mg) for supernatants.
3.19. Statistical analysis
All statistical analyses except those in lipidomics were performed using GraphPad Prism 8.0. Data were typically plotted as the mean ± s.d., n = 3 biologically independent samples. All P values were calculated using the two-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
Acknowledgements
G.L. is supported by the National Natural Science Foundation of China (31971173, 91643112). M.C. is supported by the Deutsche Forschungsgemeinschaft (CO 291/7–1, CO 291/9-1, CO 291/10-1), the German Federal Ministry of Education and Research (BMBF) VIP + program NEUROPROTEKT (03VP04260), the Ministry of Science and Higher Education of the Russian Federation (075-15-2019-1933), and the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement No. GA 884754).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102056.
Conflicts of interest
M.C. is an inventor on patents for some of the compounds described herein, and co-founder and shareholder of ROSCUE Therapeutics GmbH.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Dixon S.J. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang X.X. Ferroptosis as a target for protection against cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 2019;116:2672–2680. doi: 10.1073/pnas.1821022116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad M. Regulation of lipid peroxidation and ferroptosis in diverse species. Genes Dev. 2018;32:602–619. doi: 10.1101/gad.314674.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viswanathan V.S. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hangauer M.J. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247–250. doi: 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedmann Angeli J.P., Krysko D.V., Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Canc. 2019;19:405–414. doi: 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- 7.Wang W. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doll S. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 9.Bersuker K. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seiler A. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent-and AIF-mediated cell death. Cell Metabol. 2008:237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Feng H.Z., Stockwell B.R. Unsolved mysteries: how does lipid peroxidation cause ferroptosis? PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyurina Y.Y. Redox lipidomics technology: looking for a needle in a haystack. Chem. Phys. Lipids. 2019;221:93–107. doi: 10.1016/j.chemphyslip.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conrad M., Pratt D.A. The chemical basis of ferroptosis. Nat. Chem. Biol. 2019;15:1137–1147. doi: 10.1038/s41589-019-0408-1. [DOI] [PubMed] [Google Scholar]
- 14.Doll S., Conrad M. Iron and ferroptosis: a still ill-defined liaison. IUBMB Life. 2017;69:423–434. doi: 10.1002/iub.1616. [DOI] [PubMed] [Google Scholar]
- 15.Kagan V.E. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017;13:81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doll S. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaschler M.M. FINO2 initiates ferroptosis through GPX4 inactivation and iron oxidation. Nat. Chem. Biol. 2018;14:507–515. doi: 10.1038/s41589-018-0031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu B. ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat. Cell Biol. 2019;21:579–591. doi: 10.1038/s41556-019-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsher D.W. Cancer revoked: oncogenes as therapeutic targets. Nat. Rev. Canc. 2003;3:375–380. doi: 10.1038/nrc1070. [DOI] [PubMed] [Google Scholar]
- 20.Yu W. Autophagy inhibitor enhance ZnPc/BSA nanoparticle induced photodynamic therapy by suppressing PD-L1 expression in osteosarcoma immunotherapy. Biomaterials. 2019;192:128–139. doi: 10.1016/j.biomaterials.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Girotti A.W. Photosensitized oxidation of membrane lipids: reaction pathways, cytotoxic effects, and cytoprotective mechanisms. J. Photochem. Photobiol., B. 2001;63:103–113. doi: 10.1016/s1011-1344(01)00207-x. [DOI] [PubMed] [Google Scholar]
- 22.Donohoe C., Senge M.O., Arnaut L.G., Gomes-da-Silva L.C. Cell death in photodynamic therapy: from oxidative stress to anti-tumor immunity. Biochim. Biophys. Acta Rev. Canc. 2019;1872 doi: 10.1016/j.bbcan.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Van Straten D., Mashayekhi V., De Bruijn H.S., Oliveira S., Robinson D.J. Oncologic photodynamic therapy: basic principles, current clinical status and future directions. Cancers. 2017;9:19. doi: 10.3390/cancers9020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Andrade G.P. Comparative in vitro study of photodynamic activity of hypericin and hypericinates in MCF-7 cells. J. Photochem. Photobiol., B. 2017;175:89–98. doi: 10.1016/j.jphotobiol.2017.08.025. [DOI] [PubMed] [Google Scholar]
- 25.Vantieghem A. Hypericin-induced photosensitization of HeLa cells leads to apoptosis or necrosis - involvement of cytochrome c and procaspase-3 activation in the mechanism of apoptosis. FEBS Lett. 1998;440:19–24. doi: 10.1016/s0014-5793(98)01416-1. [DOI] [PubMed] [Google Scholar]
- 26.Dixon S.J. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife. 2014;3 doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conrad M.S. Hideyo the oxidative stress-inducible cystine/glutamate antiporter, system x c−: cystine supplier and beyond. Amino Acids. 2012;42:231–246. doi: 10.1007/s00726-011-0867-5. [DOI] [PubMed] [Google Scholar]
- 28.Ursini F.M.M., Valente M., Ferri L., Gregolin C. Purification from pig liver of a protein which protects liposomes and biomembranes from peroxidative degradation and exhibits glutathione peroxidase activity on phosphatidylcholine hydroperoxides. Biochim. Biophys. Acta. 1982;710:197–211 I. doi: 10.1016/0005-2760(82)90150-3. [DOI] [PubMed] [Google Scholar]
- 29.Wang H. Characterization of ferroptosis in murine models of hemochromatosis. Hepatology. 2017;66:449–465. doi: 10.1002/hep.29117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alves E. Photodynamic oxidation of Escherichia coli membrane phospholipids: new insights based on lipidomics. Rapid Commun. Mass Spectrom. 2013;27:2717–2728. doi: 10.1002/rcm.6739. [DOI] [PubMed] [Google Scholar]
- 31.Su L.J. Reactive oxygen species-induced lipid peroxidation in apoptosis, autophagy, and ferroptosis. Oxid Med Cell Longev. 2019 doi: 10.1155/2019/5080843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimada K. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat. Chem. Biol. 2016;12:497–503. doi: 10.1038/nchembio.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedmann Angeli J.P. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miotto G. Insight into the mechanism of ferroptosis inhibition by ferrostatin-1. Redox Biol. 2020;28:101328. doi: 10.1016/j.redox.2019.101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian Z.M., Tang P.L., Wang Q. Iron crosses the endosomal membrane by a carrier-mediated process. Prog. Biophys. Mol. Biol. 1997;67:1–15. doi: 10.1016/s0079-6107(97)00009-6. [DOI] [PubMed] [Google Scholar]
- 36.Lam-Yuk-Tseung S., Gros P. Distinct targeting and recycling properties of two isoforms of the iron transporter DMT1 (NRAMP2, slc11A2) Biochemistry. 2006;45:2294–2301. doi: 10.1021/bi052307m. [DOI] [PubMed] [Google Scholar]
- 37.Issitt T. Neuropilin-1 controls endothelial homeostasis by regulating mitochondrial function and iron-dependent oxidative stress. iScience. 2019;11:205–223. doi: 10.1016/j.isci.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nie G.J., Sheftel A.D., Kim S.F., Ponka P. Overexpression of mitochondrial ferritin causes cytosolic iron depletion and changes cellular iron homeostasis. Blood. 2005;105:2161–2167. doi: 10.1182/blood-2004-07-2722. [DOI] [PubMed] [Google Scholar]
- 39.Hu J. Translocation of iron from lysosomes to mitochondria during acetaminophen-induced hepatocellular injury: protection by starch-desferal and minocycline. Free Radic. Biol. Med. 2016;97:418–426. doi: 10.1016/j.freeradbiomed.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uchiyama A. Translocation of iron from lysosomes into mitochondria is a key event during oxidative stress-induced hepatocellular injury. Hepatology. 2008;48:1644–1654. doi: 10.1002/hep.22498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torii S. An essential role for functional lysosomes in ferroptosis of cancer cells. Biochem. J. 2016;473:769–777. doi: 10.1042/BJ20150658. [DOI] [PubMed] [Google Scholar]
- 42.Shah R., Shchepinov M.S., Pratt D.A. Resolving the role of lipoxygenases in the initiation and execution of ferroptosis. ACS Cent. Sci. 2018;4:387–396. doi: 10.1021/acscentsci.7b00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan B. Membrane damage during ferroptosis is caused by oxidation of phospholipids catalyzed by the oxidoreductases POR and CYB5R1. Mol. Cell. 2020;81:355–369. doi: 10.1016/j.molcel.2020.11.024. [DOI] [PubMed] [Google Scholar]
- 44.Zou Y. Cytochrome P450 oxidoreductase contributes to phospholipid peroxidation in ferroptosis. Nat. Chem. Biol. 2020;16:302–309. doi: 10.1038/s41589-020-0472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zilka O. On the mechanism of cytoprotection by ferrostatin-1 and liproxstatin-1 and the role of lipid peroxidation in ferroptotic cell death. ACS Cent. Sci. 2017;3:232–243. doi: 10.1021/acscentsci.7b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hou W. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12:1425–1428. doi: 10.1080/15548627.2016.1187366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao M.H. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26:1021–1032. doi: 10.1038/cr.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwon M.Y., Park E., Lee S.J., Chung S.W. Heme oxygenase-1 accelerates erastin-induced ferroptotic cell death. Oncotarget. 2015;6:24393–24403. doi: 10.18632/oncotarget.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hung H.I., Schwartz J.M., Maldonado E.N., Lemasters J.J., Nieminen A.L. Mitoferrin-2-dependent mitochondrial iron uptake sensitizes human head and neck squamous carcinoma cells to photodynamic therapy. J. Biol. Chem. 2013;288:677–686. doi: 10.1074/jbc.M112.422667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao M.H., Monian P., Quadri N., Ramasamy R., Jiang X.J. Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terzi E.M., Sviderskiy V.O., Alvarez S.W., Whiten G.C., Possemato R. Iron-sulfur cluster deficiency can be sensed by IRP2 and regulates iron homeostasis and sensitivity to ferroptosis independent of IRP1 and FBXL5. Science Advances. 2021;7 doi: 10.1126/sciadv.abg4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao C. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593:586–590. doi: 10.1038/s41586-021-03539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dos Santos A.F. Distinct photo-oxidation-induced cell death pathways lead to selective killing of human breast cancer cells. Cell Death Dis. 2020;11 doi: 10.1038/s41419-020-03275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Folch J., Lees M., Stanley G.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.