Abstract
Diagnostics of SARS-CoV-2 infection using real-time reverse-transcription polymerase chain reaction (RT-PCR) on nasopharyngeal swabs is now well-established, with saliva-based testing being lately more widely implemented for being more adapted for self-testing approaches. In this study, we introduce a different concept based on exhaled breath condensate (EBC), readily collected by a mask-based sampling device, and detection with an electrochemical biosensor with a modular architecture that enables fast and specific detection and quantification of COVID-19. The face mask forms an exhaled breath vapor containment volume to hold the exhaled breath vapor in proximity to the EBC collector to enable a condensate-forming surface, cooled by a thermal mass, to coalesce the exhaled breath into a 200–500 μL fluid sample in 2 min. EBC RT-PCR for SARS-CoV-2 genes (E, ORF1ab) on samples collected from 7 SARS-CoV-2 positive and 7 SARS-CoV-2 negative patients were performed. The presence of SARS-CoV-2 could be detected in 5 out of 7 SARS-CoV-2 positive patients. Furthermore, the EBC samples were screened on an electrochemical aptamer biosensor, which detects SARS-CoV-2 viral particles down to 10 pfu mL−1 in cultured SARS-CoV-2 suspensions. Using a “turn off” assay via ferrocenemethanol redox mediator, results about the infectivity state of the patient are obtained in 10 min.
Keywords: Exhaled breath condensate (EBC), Aptamers, SARS-CoV-2, Detection, Electrochemistry, Real-time reverse-transcription polymerase chain reaction (RT-PCR)
1. Introduction
While other human coronaviruses, e.g. HCoV-229 E and HCoV-OC43, have only induced mild common cold effects, the SARS-CoV-2 pandemic has caused more than 2.77 M death worldwide (as for March 27, 2021) with 126 M cases detected. Infection with SARS-CoV-2 is diagnosed worldwide using nasopharyngeal swab samples and more recently saliva samples by detection of SARS-CoV-2 RNA using real-time reverse-transcription polymerase chain reaction (RT-PCR). The procedure to obtain nasal swab samples is not only uncomfortable, but requires specialized personal with risk of contaminating the person performing the test. Saliva tests have the advantage of being simpler to perform, less invasive with limited risks and RT-PCR on saliva specimens has becoming more widely implemented (Ryan et al., 2021; Ter-Ovanesyan et al., 2021; Wyllie et al., 2020). The viscous nature of saliva together with the presence of saliva proteases, responsible for the proteolytic activity of saliva, make the direct application of saliva samples challenging. It is well-known that the major mechanisms of COVID-19 spread are airborne and contact infections primarily due to the high resistance of the virus once in aerosol droplets expelled from infected persons. Given the growing need for sample collection by patients themselves, exhaled breath condensate (EBC) (Khoubnasabjafari et al., 2020; Ryan et al., 2021) might represent an important alternative specimen type for SARS-CoV-2 diagnostic.
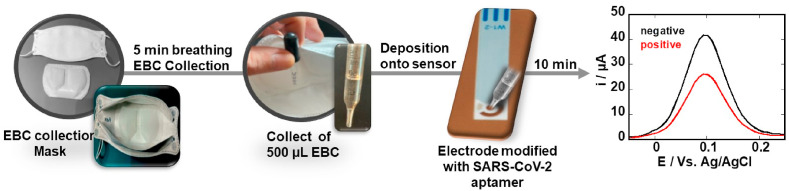
Different from exhaled breath (EB), which is based on exhalation of volatile organic compounds (VOCs NO, CO2, NH3, H2O2, etc.) (Shan et al., 2020), EBC contains lower respiratory droplets which can be analyzed by RT-PCR (Ryan et al., 2021). Indeed, EBC RT-PCR has been already investigated to identify other respiratory viruses, including human coronaviruses with the aim to gain knowledge about the efficiency of a face mask (Leung et al., 2020). Ryan and coworkers reported preliminary data from patients with a positive and negative RT-PCR tests for SARS-CoV-2 where EBC was collected using commercial RTube condensers. RT-PCR of EBC collected samples was positive for SARS-CoV-2 for 21 out of 31 cases (68%) using the E and S proteins assay specific kits and increased to 93.5% using four targets (S, E, NP, ORF1ab) (Ryan et al., 2021). This study strongly supports the hypothesis that EBC collected samples are suitable for SARS-CoV-2 detection. These findings and the possibility to collect EBC from patients during tidal breathing and coughing into a mask prompted us to investigate this non-invasive sample collection method in combination with an electrochemical point-of-care testing (POCT) system for the discrimination between infected SARS-CoV-2 and healthy patients (Fig. 1 ). In this work, we take the opposite approach of that investigated by Cowling et al. (Leung et al., 2020) and propose a face mask to collect EBC. To date only specific devices have been proposed to collect and condensate exhaled breath.
Fig. 1.
Exhaled breath condensate (EBC)-based diagnostic strategy for SARS-CoV-2 infectivity: Laboratory engineered mask allows collection of EBC by first cooling the mask for 30 min in the freezer, putting on the cooled mask and breathing into it for 5 min. EBS formed in the Teflon-lining of the inside of the mask is collected and directly deposited onto an electrochemical sensing modified with SARS-CoV-2 specific aptamer targeting the receptor-binding domain (RBD) region of the S1 spike protein as surface receptor. Using ferrocenemethanol as a redox meditator before and after viral interaction allows discrimination between positive and negative EBC samples.
We show, in this work, the utility of an aptamer-based electrochemical biosensor. Aptamers exhibit many advantages as recognition elements when compared to traditional antibodies due their small size, enhanced chemical stability and low cost of production (Yoo et al., 2020). We report notably on an electrochemical sensing format targeting the spike protein (S) which is embedded in a lipidic membrane forming the SARS-CoV-2 viral outer wall. The spike protein protrudes from the viral membrane, and the viral entry into host cells is mediated by the receptor-binding domain (RBD) region of the spike protein that recognizes the host receptor ACE2. With the spike protein being repeated about 50–200 times on the viral surface (Wrapp et al., 2020), the RBD region of the S protein represents therefore an excellent diagnostic target. The aptamer chosen in this work is a 32-nucleotide aptamer from Base Pair Biotechnologies (Pearland, Texas, USA).
2. Results and discussion
2.1. Collection of exhaled breath condensate
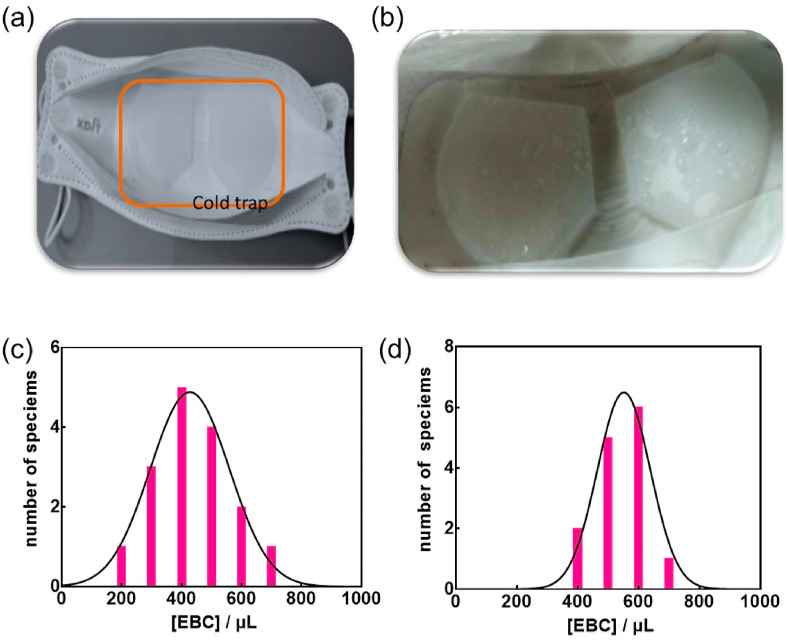
The mask-based EBC collection system uses a commercial face mask fitted with an engineered EBC collector system based on a Teflon coated cooling trap (Fig. 2 a). To increase the EBC collection efficiency, the mask is placed into a freezer at −20 °C for 30 min, before being placed over the mouth of the person to be tested. This polytetrafluoroethylene (PTFE) trap when cooled allows sample liquification on its surface, where the formed droplets can be collected with a pipette and used for analysis directly. The presence of a collection pool at the end of the cold trap allows further collection of EBC (Fig. 2b) without the need of technical expertise (Fig. S1). During the EBC collection, the inside of the mask is not exposed to air and the risk of contamination of the EBC samples is negligible. Using this collection system, 400 ± 150 μL of EBC can be collected within 5 min (Fig. 2c).
Fig. 2.
Mask-based EBC collector: (a) Inside of the laboratory engineered mask showing an exhaled breath condensate (EBC) collector (cold trap, indicated with orange line) for converting breath vapor into a fluid sampe. The EBC collector is made of a Teflon-based condensate-forming surface. (b) Image of EBC formed on the Teflon collector after 5 min breathing into the mask. (c) EBC volume collected in 5 min using the EBC mask (n = 14). (d) EBC volume collected in 5 min using commercial RTube condensers (n = 14). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The collection efficiency was comparable to EBC collected by commercial RTube condensers (Respiratory Research Inc., USA) (Fig. 2d). The collection efficiency is person-dependent as seen in Fig. 2c. However, in most cases, the required 300 μL needed for further analysis was obtained in this manner. While collection of an equal volume of saliva is more efficient at a 5 min time span, saliva is a complex sample matrix containing proteases and other variable components that can impact most assays. This includes the potential degradation of the SARS-CoV-2 S1 protein targeted by the aptamer employed in this test. The much cleaner EBC sample is therefore believed to be more suitable and reliable for rapid testing.
2.2. SARS-CoV-2 aptamer and electrochemical sensor
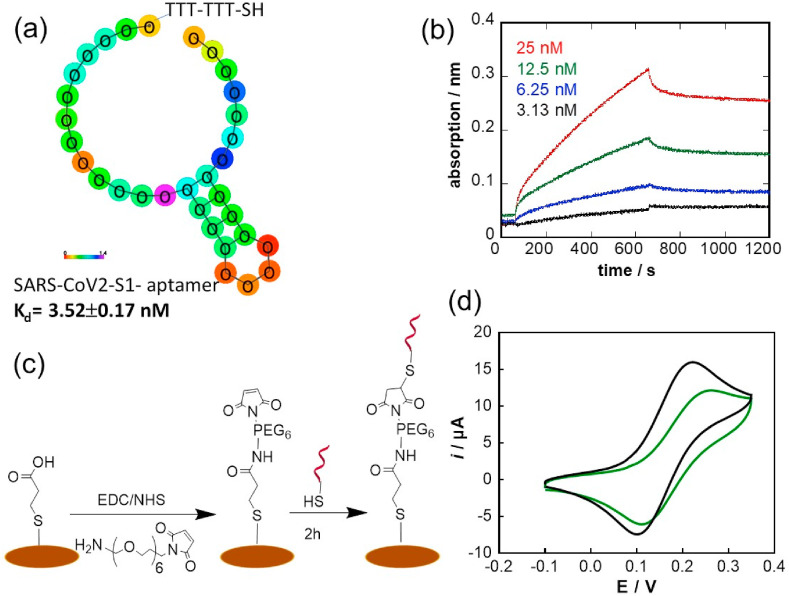
The SARS-CoV-2 aptamer targeting the S1 protein was selected via combinatorial libraries of nucleic acid sequences by the SELEX (systemic evolution of ligands by exponential enrichment) process. As seen in Fig. 3 a, the aptamer investigated in this work is a 20-base aptamer “CFA0688T” (Base Pair Bio) with 1 loop modified on the 5’ end with a thiol-TTT-TTT to give the aptamer some flexibility for its anchoring onto gold interfaces. The binding affinity to the recombinant SARS-CoV-2 S1 spike protein was determined by Biolayer interferometry (BLI) measurements and was determined as KD = 3.52 ± 0.17 nM (R2 = 0.9985) (Fig. 3b). This affinity value is comparable to other reported SARS-CoV-2 aptamers such as the 51-base pair aptamer with 3 hair-pined structures selective to RBD reported by Song et al. (2020) or the 58-base pair aptamer proposed by Torabi at al. (Torabi et al., 2020) with KD values ranging from 5.8 ± 0.8 nM (Song et al., 2020) to 0.49 ± 0.05 nM (Torabi et al., 2020).
Fig. 3.
Electrochemical Aptamer Sensor: (a) 2D structure of DNA aptamer with sequence redacted, (b) Biolayer interferometry (BLI) measurements of biotinylated aptamer linked onto streptavidin-activated BLI sensors with different concentrations of SARS-CoV-2 S protein (3.13 nM, 6.25 nM, 12.5 nM and 25 nM): running buffer: 1x PBS, 1 mM MgCl2, (c) Surface attachment strategy of SARS-Cov-2 aptamer on the gold working electrode of the screen-printed electrode using maleimide-thiol-aptamer linkage. (d) Cyclic voltammograms (CV) of a gold electrode before (black) and after (green) functionalization with SARS-CoV-2 aptamer (10 μg mL−1 for 2 h) using ferrocenemethanol as a redox mediator (1 mM in 0.1 M PBS, pH 7.4, Scan rate = 100 mV s−1). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The attachment of the SARS-CoV-2 aptamer onto screen printed electrodes (SPE) was achieved via a maleimide functionalized poly (ethylene glycol) (PEG) spacer, a commonly employed hydrophilic polymer to avoid biofouling and applied for cysteine-modified aptamer integration by others (Da Pieve et al., 2010). The spacer aids in overcoming any potential steric hindrance in viral detection. The success of the linking strategy was validated using XPS (See SI, Fig. S2).
A key concept in electrochemical systems is the fact that the kinetics of the heterogeneous electron transfer at modified electrodes is strongly dependent on the surface coverage and on the thickness of the modifying layer (Cannes et al., 2003). Fig. 3d depicts the cyclic voltammograms of the gold working electrode before and after modification with the aptamer using ferrocenemethanol as a redox mediator. This small mediator can permeate to a small extent into a monolayer-modified gold electrode or via diffusion through pinholes with electron transfer occurring at the free sites on the electrode. As expected, a decrease in electron transfer is observed in line with the presence of the aptamer on the electrode surface.
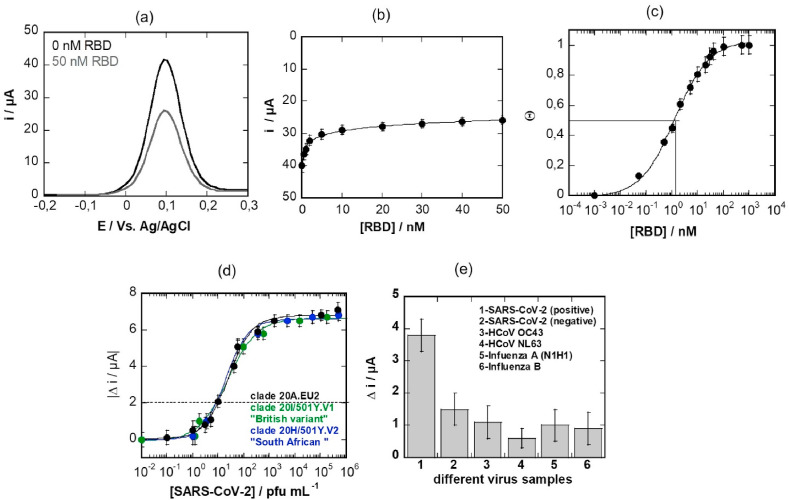
Analysis of 50 nM receptor domain binding from solution using the aptamer-modified electrodes shows a clear decrease in current (Fig. 4 a). This decrease in current is linear up to [RBD] = 10 nM, reaching complete saturation at 50 nM (Fig. 4b). The corresponding curve can be fitted with the Langmuir isotherm (Fig. 4c) using the following equation:
| Θ = j(c0) / j(c∞) = KA × c0 / (1+ KA × c0) |
Fig. 4.
Electrochemical SARS-CoV-2 aptasensor: (a) Differential pulse voltammogram (DPV) of aptamer modified electrode using ferrocenemethanol (1 mM in 0.1 M PBS, pH 7.4) as a redox meditor. Initial signal (black) and after addition 50 nM RBD for 10 min (grey line), washing and recording a new DPV in ferrocenemethanol. The decrease in current is due to RBD binding to the aptamer. DPV conditions: taquis = 3s, Estep = 0.01V, Epulse = 0.06 V, tpulse = 0.02 V, scan rate = 0.06 V s−1. (b) Current response to increasing RBD concentrations using ferrocenemethanol (1 mM PBS, pH 7.4) as a redox probe. (c) Langmuir adsorption isotherm as extracted from Fig. 4b. (d) Dose-dependent response curve toward SARS-CoV-2 virus clade 20A.EU2 (black) as well as clade 20I/501Y.V1, “British variant” (green) and the clade 20H/501Y.V2, “South African variant” (blue) on aptamer-modified electrodes. (e) Selectivity of the aptamer sensors towards other real patient virus samples by comparison to SARS-CoV-2 positive samples. All the virus samples have Ct values between 22–25 and are nasal swab samples. All the values are displayed as means ± SEM (n = 5). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
With Θ being the surface coverage, j (c0) the current density at a given RBD concentration, j (c∞) the current density at infinite bulk analyte concentration; assuming a 1:1 complex between the antigen (RBD) from solution and the aptamer receptor allows estimating the affinity constant KA. From the expected S-shaped curve, a dissociation constant (i.e. a half saturation-constant) KD = 1.6 ± 0.9 nM could be determined, indicating high affinity of the aptamer for RBD, and in line with reported nanomolar dissociation constants for aptamer-protein interactions (Manochehry et al., 2019; Vinkenborg et al., 2012) as well as independent affinity measurements made by Base Pair using biolayer interferometry (Fig. 3b).
These interfaces were investigated for their potential to sense cultured SARS-CoV-2 viral particles (Fig. 4d). Immersion of the sensor into PBS (0.1 M, pH 7 = 4) containing different concentrations of a SARS-CoV-2 isolate shows that the limit of detection (LOD), defined as the lowest level that an analyte can be reliably distinguished from the background, correlates to about 10 pfu mL−1 (correlating to a current difference of 2 μA) with a saturation at 1.5 × 105 pfu mL−1. The detection limit was determined to be about 3 pfu mL−1 from five blank noise signals (95% confidential level). The analytical performance was compared to that of SPE where the thiol-terminated aptamer was directly linked onto the gold surface (Fig. S3). From the analysis of RBD binding to the aptamer, a higher dissociation constant of KD = 6.2 ± 1.2 nM was determined. More importantly, sensing of cultured SARS-CoV-2 viral particles indicates a LOD of about 200 pfu mL−1.
This sensing sensitivity of the maleimide-thiol aptamer sensor is comparable to other electrochemical (Saroglia et al., 2021; Szunerits et al., 2021; Zhang et al., 2020) and electrical (Seo et al., 2020) sensors reported thus far in the literature. In addition, the possibility of detecting the variants 20I/501Y.V1 (called “British variant”) and 20H/501Y.V2 (called “South African variant”) was investigated by using SARS-CoV-2 variant isolates. Fig. 4c indicates that the aptamer-based sensor detects the 20I/501Y.V1e and the 20H/501Y.V2 variants equally well. This is in line with the results recorded using commercial recombinant SARS-CoV-2 S protein considering the different mutations (SI, Fig. S4). Indeed, using BLI measurements, the affinity of the SARS-CoV-2 S protein UK variant to the aptamer is KD = 6.0 ± 3 nM with a kon =(1.52 ± 0.003) × 105 M−1s−1 while in the case of the South African variant the KD is 1.6.0 ± 0.1 nM with a kon =(1.62 ± 0.05) × 105 M−1s−1. This is in the same order as the affinity constant of 3.52 ± 0.17 nM for the wild type (Wuhan) variant ( Fig. 3 b).
The reproducibility of the SARS-CoV-2 aptamer-modified electrodes was expressed in terms of the relative standard deviation, which was determined to be 2.3% at a viral concentration of 103 pfu mL−1 (n = 5). The long-term stability of the sensor when stored in PBS was also evaluated showing a loss of 2.5% in the anodic peak current when testing virus solutions of 103 pfu mL−1 after the electrode has been stored at 4 °C for 1 month. To illustrate the selectivity of the sensor, the aptamer sensor was incubated with other viral samples, obtained by nasal swabs.
Testing other coronaviruses producing symptoms close to those associated to SARS-CoV-2, HCoVOC43 and HCoV NL63, showed a decreased interaction with the aptamer-interface as identified with a current difference smaller compared to the current difference recorded on a positive nasopharyngeal swab sample (Fig. 4e). The same was observed for Influenza A (H1N1) and influenza B samples.
2.3. Exhaled breath condensate analysis
Given that the disease is transmitted via exhaled droplets, and that EBC is the established modality for sampling exhaled aerosol, detection of SARS-CoV-2 in EBC is a promising approach for safe and efficient diagnosis of the disease (Khoubnasabjafari et al., 2020). We validated the possibility of SARS-CoV-2 sensing using EBC collected by commercial Rtube condensers and EBC mask-based system. EBC samples were collected using a cold trap, as shown in Fig. 1. In parallel, nasopharyngeal swab samples were also collected.
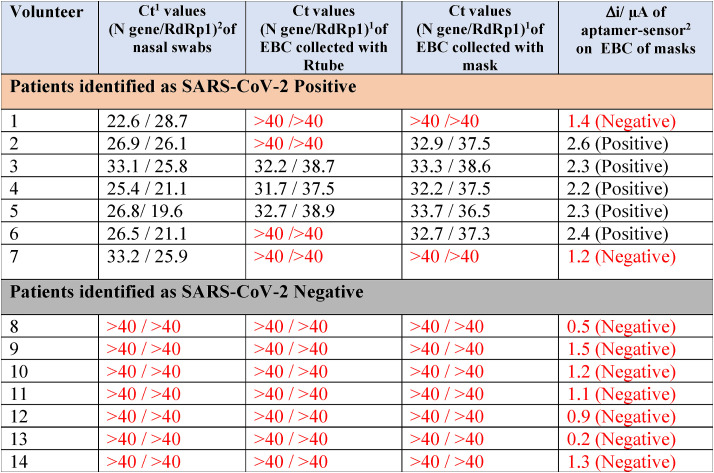
In a proof of principle study, EBC samples of 14 volunteers were collected and analyzed by RT-PCR (Table 1 ). Out of the 14 nasopharyngeal swab samples, seven were identified as SARS-CoV-2 positive and seven as SARS-CoV-2 negative (Cycle threshold (Ct) > 40) by targeting the N structural protein as well as the RNA dependent RNA polymerase (RdRp) nonstructural protein via RT-PCR. Based on our experiments with different dilutions of a SARS-CoV-2 isolate, we estimated that a Ct of 34 approximately corresponds to about 104 copies of viral RNA per milliliter and that this dilution showed no infectivity to Vero cells. The lower Ct values of 22 (Table 1) correlated to about 107 copies of viral RNA per milliliter.
Table 1.
Exhaled breath condensate studies on patients identified by nasopharyngeal swabs RT-PCR as SARS-CoV-2 positive (black numbers) or negative (red numbers).
The results of EBC RT-PCR performed on samples collected by Rtube condensers as well as EBC masks of the SARS-CoV-2 negative patients were in full agreement with those of nasopharyngeal swab samples (7/7, 100%). Testing these samples on the electrochemical sensor, where a current difference higher than 2 μA (Fig. 4d) was considered to be linked to the presence of viral particles, resulted in further identification of these sample as SARS-CoV-2 negative.
In the case of EBC samples, collected from patients identified by nasopharyngeal RT-PCR as SARS-CoV-2 positive, 3 samples out of 7 were identified as SARS-CoV-2 positive using the commercial RTube condenser and 5/7 using the face mask using a Ct of 40 as cut-off. In contrast to the nasopharyngeal samples, the Ct values of the RdRp gene detected in the EBC samples were always considerably lower than the N-gene (Table 1). The difference in the Ct values between nasal swab samples and EBC is linked to the different viral load present in both fluids (Eiche et al., 2020; Giovannini et al., 2021). Indeed, it has been postulated that the viral load of SARS-CoV-2 in aerosol samples is several orders of magnitude below those in nasopharyngeal swabs, which are in the order of 6.41×102–1.34 × 1011 copies/mL (Giovannini et al., 2021). This indicates that the N-gene is likely the more robust gene to target for EBC samples independently of the collection strategy applied. Testing the mask-collected EBC samples on the aptamer-modified electrochemical sensor showed agreement with EBC RT-PCR results using masks. This indicates that such sensors are well-adapted for sensing EBC viral samples and further confirms the presence of active viral particles in exhaled breath of SARS-CoV-2 positive patients.
3. Conclusion
While RT-PCR remains the gold standard method for the detection of SARS-CoV-2 infection, diagnostic methods that allow faster testing in a cost-effective manner and more easily implemented on a larger scale represent a step closer to mass screening with higher frequency. With POC or bedside analysis becoming a global trend in modern diagnostics, simplicity, sensitivity and selectively are the main criteria for choosing an adequate system. Furthermore, reliable, and reproducible sample collection becomes as important as diagnosis in POC testing devices. The intent of this work was to demonstrate the entire system from sampling to sensing and identification. The sensor selectively detects SARS-CoV-2 viral particles down to 10 pfu mL−1 in cultured SARS-CoV-2 suspensions. Additionally, it is shown that converting exhaled breath vapor into EBC provides a convenient and accessible sample source for SARS-CoV-2 viral particles. Our work underlines that EBC can identify SARS-CoV-2 by RT-PCR in patients identified as SARS-CoV-2 positive using nasopharyngeal swab samples. Positive cases of the virus are detectable based on quantitative analysis of EBC samples obtained after 5 min of exhalation into the EBC collecting masks. It remains surprising that Sample 1 with a very low Ct value comes out as negative in EBC RT-PCR and on the aptasensor. The sampling part is directly correlated to viral load in breath and breathing of the patient. Some optimization therefore needs to be implemented to make the sampling step more robust to overcome some false negative issues. Eventual integration of the sensor into the mask itself would likely make the method even more robust and user-friendly (Fig. S1). Nevertheless, the results validate the concept that the detection of SARS-CoV-2 in the breath of COVID-19 patients using a rapid aptasensor is feasible. This opens the path for a rapid personalized screening of the infectious state in a short time and potentially even at home.
4. Experimental part
Materials: 3-mercaptopropionic acid (98%), 1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide hydrochloride) (EDC), N-hydroxysuccinimide (NHS), and ferrocenemethanol were purchased from Sigma-Aldrich and used as-received. Phosphate saline solution (PBS 1 × ) was obtained from Thermo Fisher scientific. Maleimide-PEG6-amine (MW 1 kDa) was purchased from Interchim Uptima. Milli-Q water was used throughout the whole study. Recombinant SARS-CoV-2 Spike Glycoprotein S1 was obtained from Amsbio, USA (Amsbio Cat.# AMS. SPN-C52H4). RBD UK was purchased from Sinobiological (Ref# 40,592-V08H8), and RBD South African from Acrobiosystems (Ref# SPD-C52H).
The screen-printed electrodes used in this work were purchased from PalmSens (The Netherlands, distributed by Hdts in France) and are available under the name AUH3600-IPM-Intelligent Pollutant Monitoring Denmark. They consist of a 3 mm diameter gold working electrode, a silver/silver chloride reference, and a carbon counter electrode.
RTube condensers were purchased from Respiratory Research In, USA.
Mask-based EBC collectors: The mask-based EBC collector is currently fabricated in-house at Diagmetrics and consists of 30 cm wide rolls of 127 μm thick natural virgin Polytetrafluoroethylene (PTFE) sheet (eplastics.com., USA), 10 cm wide 3M 465 double sided adhesive transfer tape (uline.com, USA) as well as a super absorbent polymer (SAP) powder, MediSAP 715 (M2 Polymer Technologies, Inc., Illinois, USA). A stamping jig was constructed from 0.315 cm PTFE plate (eplastics.com, USA) and the jig was cut on a 100 W CO2 laser cutter (Orion Motor Tech, China). A Digital Combo Heat Press (Geo Knight, Massachusetts, USA) was used to stamp the 127 μm thick PTFE sheet using the jig to form a pocket in the PTFE sheet for receiving a thermal mass mixture of water and the SAP. A second layer of the 127 μm PTFE sheet was bonded to the heat stamped 127 μm PTFE sheet using the 3 M 465 adhesive, sandwiching the thermal mass of water/SAP between layers of PTFE sheet. The completed PTFE/thermal mass/3 M 465/PTFE laminated sandwich was hand cut using scissors into the final shape of the EBC collector that is configured and dimensioned to be inserted into a pre-existing face mask or built into a newly constructed face mask. The EBC collector constructed as described was designed for and retro fit into various disposable face masks of different styles and construction, including N95 and KN95 made by 3 M and overseas manufacturers. These masks are currently not commercially available, but were designed with eventual mass production in mind.
Immobilization of thiolated aptamer: All DNA aptamers (Base Pair Biotechnologies) were synthesized by standard phosphoramidite chemistry by IDT (Coralville, IA, USA). Gold electrodes were exposed to 10 μL of 3-mercaptopropionic acid (25 mM) in MQ-water for 30 min at room temperature. The surface was washed with MQ-water and dried in air. Then the acid-terminated surface was activated with EDC/NHS (1:1 M ratio, 15 mM in PBS 1 × , pH 7.4) for 20 min, followed by immersion into NH2-PEG6-maleimide (10 μL, 0.1 mg/mL in PBS 1 × , pH 7.4) for 2 h at 4 °C and washing with MQ-water.
The aptamers were dissolved in 10 mM Tris, 0.1 mM EDTA (pH 7.5) at 100 μg mL−1 and then mixed with 10 mM TCEP in a 1:1 ratio. The PEG6-maleimide modified electrodes were incubated for 2 h with 10 μL of this solution and then washed copiously with MQ-water to remove excess aptamer and unreacted reagents. Finally, the electrodes were incubated 2 h with folding buffer then washed and dried. The surfaces were kept at 4 °C until use.
Electrochemical measurements were performed with a Sensit-Smart smartphone potentiostat (Palmsens, The Netherlands, distributed by Hdts in France). Cyclic voltammograms were recorded at 50 mV s−1 using ferrocenemethanol (1 mM, PBS 1 × , pH 7 = 4) as a redox mediator. The scan direction was from −0.1 to 0.3 V and then back to −0.1 V.
Differential pulse voltammograms (DPV) were acquired in the appropriate potential range using the following DPV parameters: taquis = 3s, Estep = 0.01 V, Epulse = 0.06 V, tpulse = 0.02 V, scan rate = 0.06 V s−1. The diameter of the gold electrode was 3 mm (A = 0.071 cm2).
EBC collection and sensing: Before EBC collection, the engineered masks were placed for 20 min at −20 °C (freezer), and then immediately worn for 5 min. During this 5 min, breathing with open mouth was performed, which resulted in the condensation of the breath on the Teflon-lining of the mask. After 5 min, the mask was carefully removed, and the liquid droplets collected with a plastic pipette. Sensing was performed in two steps. First, a DPV signal was recorded on the aptamer-sensor using ferrocenemethanol (1 mM, PBS 1 × , pH 7 = 4) as a redox mediator. Thereafter, the electrode was immersed into 5 mL of PBS 1 × (pH 7 = 4) 2 times and 200 μL of the collected EBC was deposited onto the working electrode. After 10 min incubation with the EBC samples, the electrode was immersed again into 5 mL of PBS 1 × (pH 7 = 4) for 2 times and then into ferrocenemethanol (1 mM, PBS 1 × , pH 7 = 4). A DPV was recorded using the same conditions as before. The difference in the maximal current before and after EBC contact was used for identification of the sample as positive or negative.
4.1. Virus isolates
Three SARS-CoV-2 patient isolates were used, one clade 20 A.EU2, one clade 20I/501Y.V1 (GISAID: EPI_ISL_1653931) and one clade 20H/501Y.V2 (GISAID: EPI_ISL_1653932).
Virus Titration: Vero E6 cells (ATCC CRL-1586) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% L-glutamine, 1% antibiotics (100 U mL−1 penicillin), in a humidified atmosphere of 5% CO2 at 37 °C. Vero E6 cells were plated in 96-well plates (2.5 × 105 cells/well) 24 h before performing the virus titration. Clinical isolates, obtained from SARS-CoV-2 positive specimens, were cultured on Vero E6 cells. Infected cell culture supernatant was centrifuged for 10 min at 1500 rpm at 4 °C to obtain a virus suspension. The virus suspension was used undiluted and in serial ten-fold dilutions. Virus suspensions were distributed in 6 wells in DMEM supplemented with 2% FBS (Fetal Bovine Serum) to Vero E6 cells, 1% antibiotics (100 U mL−1 penicillin), and 1% L-glutamine. The plates were incubated for 6 days in 5% CO2 atmosphere at 37 °C. The plates were examined daily using an inverted microscope (ZEISS Primovert) to evaluate the extent of the virus-induced cytopathic effect in cell culture. Calculation of estimated virus concentration was carried out by the Spearman and Karber method (Kärber, 1931; Spearman, 1908) and expressed as TCID50/mL (50% tissue culture infectious dose). TCID50/mL values were transformed to PFU/mL by using the formula PFU/mL = TCID50/mL × 0.7.
SARS-CoV-2 RT-PCR: RNA was extracted from 140 μL of EBC using the QIAamp viral RNA mini kit (Qiagen) and eluted in 50 μL of buffer. RT-PCR for the E and RdRp genes was performed using the Eurobio Plex kit (Afzal, 2020). Undetectable SARS-CoV-2 levels were set to Ct > 40. Amplification was performed on 7500 Real-Time PCR System (Applied Biosystems, USA).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Financial support from the Centre National de la Recherche Scientifique (CNRS), the University of Lille, I-SITE via the COVID task force and the Hauts-de-France region via ANR Resilience (CorDial-FLU) is acknowledged. The project is funded by the Horizon 2020 framework programme of the European Union under grant agreement no 101016038. The authors thank all personal at Cerballiance for their valuable help. The authors are thankful all the volunteers who participated in this study, notably the inhabitants of Lille, Aniche and Villeneuve d’Ascq.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113486.
Declarations
Base Pair Biotechnologies authors GWJ and AC have a financial interest in the aptamers used in this study.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J. Adv. Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannes C., Kanoufi F., Bard A.J. Cyclic voltammetry and scanning electrochemical microscopy of ferrocenemethanol at monolayer and bilayer-modified gold electrodes. J. Electroanal. Chem. 2003;547:83–91. [Google Scholar]
- Da Pieve C., Williams P., Haddleton D.M., Palmer R.M.J., Missailidis S. Modification of thiol functionalized aptamers by conjugation of synthetic polymers. Bioconjugate Chem. 2010;21:169–174. doi: 10.1021/bc900397s. [DOI] [PubMed] [Google Scholar]
- Eiche T., Kuster M. Aerosol release by healthy people during speaking: possible contribution to the transmission of SARS-CoV-2. Ijerph. 2020;17:9088. doi: 10.3390/ijerph17239088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannini G., Haick H., Garoli D. Detecting COVID-19 from breath: a game changer for a big challange. ACS Sens. 2021;6:1408–1417. doi: 10.1021/acssensors.1c00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärber G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch F Exp Pathol U Pharmakol. 1931;162:480–483. [Google Scholar]
- Khoubnasabjafari M., Jouyban-Gharamaleki V., Ghanbari R., Jouyban A. Exhaled breath condensate as a potential specimen for diagnosing COVID-19. Bioanalysis. 2020;12:1195–1197. doi: 10.4155/bio-2020-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung N.H.L., Chu D.K.W., Shiu E.Y.C., Chan K.-H., McDevitt J.J., Hau B.J.P, Yen H.-L., Li Y., Ip D.K.M., Peiris J.S.M., Seto W.-H., Leung G.M., Milton D.K., Cowling B.J. Respiratory virus shedding in exhaled breath and efficacy of face masks. Ned. Med. 2020;26:676–680. doi: 10.1038/s41591-020-0843-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manochehry S., McConnell E.M., Li Y. Unraveling determinants of affinity enhancement in dimeric aptamers for a dimeric protein. Sci. Rep. 2019;9:17824. doi: 10.1038/s41598-019-54005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan D.J., Toomey S., Madden S.F., Casey M., Breathnach O.S., Morris P.G., Grogan L., Branagan P., Costello R.W., De Barra E., et al. Use of exhaled breath condensate (EBC) in the diagnosis of SARS-COV-2 (COVID-19) Thorax. 2021;76:86–88. doi: 10.1136/thoraxjnl-2020-215705. [DOI] [PubMed] [Google Scholar]
- Saroglia L.F., Galatà G D.S.R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini A. A reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171:11286. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shan B., Broza Y.Y., Li W., Wang Y., Wu S., Liu Z., Wang J., Gui S., Wang L., Zhang Z., et al. Multiplexed nanomaterial-based sensor arrayfor detection of COVID-19 in exhaled breath. ACS Nano. 2020;14:12125–12132. doi: 10.1021/acsnano.0c05657. [DOI] [PubMed] [Google Scholar]
- Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Discovery of aptamers targeting receptor-binding domain of the SARS-CoV-2 spike Glycoprotein. Anal. Chem. 2020;92:9895–9990. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- Spearman C.T. The method of “right and wrong cases” (constant stimuli) without gauss's formula. Br. J. Psychol. 1908;2:227–242. [Google Scholar]
- Szunerits S., Pagneux Q., Swaidan A., Mishyn V., Roussel A., Cambillau C., Devos D., Engelmann I., Alidjinou E.K., Happy H., et al. The role of the surface ligand on the performance of electrochemical SARS-CoV-2 antigen biosensors. Anal. Bioanal. Chem. 2021:1–11. doi: 10.1007/s00216-020-03137-y. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Ovanesyan D., Gilboa T., Lazarovits R., Rosenthal A., Yu X., Li Z., Church G.M., Walt D.R. Ultrasensitive measurement of both SARS-CoV-2 RNA and antibodies from saliva. Anal. Chem. 2021;93:5365–5370. doi: 10.1021/acs.analchem.1c00515. [DOI] [PubMed] [Google Scholar]
- Torabi R., Ranjbar R., Halaji M., Heiat M. Aptamers. The bivalent agents as probes and therapies for coronavirus infections: a systematic review. Mol. Cell. Probes. 2020;53:101636. doi: 10.1016/j.mcp.2020.101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkenborg J.L., Mayer G., Famulok M. Atamer-based affinity labeling of proteins*. Angew. Chem. Int. Ed. 2012;51:9176–9180. doi: 10.1002/anie.201204174. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Goldsmith J.A., Hsieh C.-L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Warren J.L., Bertie Geng B., Muenker M.C., Moore A.J., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H., Jo H., Oh S.S. Detection and beyond: challenges and advances in aptamer-based biosensors. Mater. Adv. 2020;1:2663–2687. [Google Scholar]
- Zhang L., Fang X., Liu X., Ou H., Zhang H., Wang J., Li Q., Cheng H., Zhang W., Luo Z. Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers. Chem. Commun. 2020;56:10235. doi: 10.1039/d0cc03993d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.