Abstract
Background
The current reference standard to diagnose a SARS-CoV-2 infection is real-time reverse transcriptase polymerase chain reaction (RT-PCR). This test poses substantial challenges for large-scale community testing, especially with respect to the long turnaround times. SARS-CoV-2 antigen tests are an alternative, but typically use a lateral flow assay format rendering them less suitable for analysis of large numbers of samples.
Methods
We conducted an evaluation of the Diasorin SARS-CoV-2 antigen detection assay (DAA) compared to real-time RT-PCR (Abbott). The study was performed on 248 (74 qRT-PCR positive, 174 qRT-PCR negative) clinical combined oro-nasopharyngeal samples of individuals with COVID-19-like symptoms obtained at a Municipal Health Service test centre. In addition, we evaluated the analytical performance of DAA with a 10-fold dilution series of SARS-CoV-2 containing culture supernatant and compared it with the lateral flow assay SARS-CoV-2 Roche/SD Biosensor Rapid Antigen test (RRA).
Results
The DAA had an overall specificity of 100% (95%CI 97.9%–100%) and sensitivity of 73% (95%CI 61.3%–82.7%) for the clinical samples. Sensitivity was 86% (CI95% 74.6%–93.3%) for samples with Ct-value below 30. Both the DAA and RRA detected SARS-CoV-2 up to a dilution containing 5.2 × 102 fifty-percent-tissue-culture-infective-dose (TCID50)/ml.
Discussion
The DAA performed adequately for clinical samples with a Ct-value below 30. Test performance may be further optimised by lowering the relative light unit (RLU) threshold for positivity assuming the in this study used pre-analytical protocol . The test has potential for use as a diagnostic assay for symptomatic community-dwelling individuals early after disease onset in the context of disease control.
Keywords: SARS-CoV-2, Evaluation, Antigen detection assay
Keywords: Abbreviations. Real-time RT-PCR, Real-time reverse transcriptase polymerase chain reaction; LFA, Lateral flow assay; VTM, Virus transport medium; DAA, Diasorin SARS-CoV-2 Antigen detection assay; TCID50, Fifty-percent-tissue-culture-infective-dose; RRA, SARS-CoV-2 Roche/SD Biosensor Rapid Antigen test; GLY-medium, Gelatin-lactalbumin-yeast virus transport medium (Mediaproducts; Groningen, The Netherlands); MHS, Municipal Health Services; LOD, Limit of detection
1. Background
Accurate and sustainable test strategies are key in the control of SARS-CoV-2 community spread [[1], [2], [3]]. The current reference standard to diagnose a SARS-CoV-2 infection is real-time reverse transcriptase polymerase chain reaction (real-time RT-PCR). Real-time RT-PCR is a highly sensitive and specific test, but raises substantial challenges when applied for large-scale community testing due to the long turnaround time (6–8 h after arrival of the specimen in the lab) and the need for a highly specialised laboratory environment. Furthermore, the testing capacity is limited by availability of extraction and PCR reagents and disposables. SARS-CoV-2 antigen lateral flow assays (LFA) have been proposed as an alternative for large-scale community testing of symptomatic individuals in the context of disease control [[2], [3], [4]]. Multiple LFA platforms have been evaluated in this context and showed satisfactory clinical performance for application as a diagnostic test in symptomatic community dwelling individuals within a limited number of days after symptom onset [[5], [6], [7]]. The performance of the LFA on large numbers of samples, however, is labour intensive and creates specific logistic challenges due to the strict time intervals to be respected when performing the test and the absence of automatic processing and registration.
SARS-CoV-2 antigen assays that can be performed on existing automated analysers could potentially form an alternative as they are less dependant on manual labour and allow large numbers of samples to be processed in a shorter period of time. Moreover, some of these SARS-CoV-2 antigen assays can be performed on oro-/nasopharyngeal swabs suspended in virus transport medium (VTM) which can also be used to perform a confirmatory real-time RT-PCR for SARS-CoV-2 or in-depth genetic typing when needed [8,9].
The objective of this study was to evaluate the test performance of the ‘Diasorin SARS-CoV-2 Antigen detection assay’ (DAA), a 96-well microtiterplate based two-step sandwich chemiluminescence immunoassay for the quantitative determination of SARS-CoV-2 nucleocapsid antigen performed on high-throughput platform, compared to the real-time RT-PCR performed on the Alinity M (Abbott) as the reference method.
2. Study design and objectives
2.1. Analytical performance evaluation with virus culture supernatant
Analytical sensitivity and repeatability were evaluated by diluting a cell cultured SARS-CoV-2 strain (SARS hCoV-19/Netherlands/NoordBrabant_10003/2020 SARS-CoV-2; heat inactivated for 2 h at 60 °C in a biosafety level 3 laboratory before use for the current experiments at BSL-2 level) with 50% tissue culture infectious dose (TCID50) of 5.62 × 107 per mL in a 10-fold dilution series (10−1 – 10−8) in gelatin-lactalbumin-yeast virus transport (GLY) medium (Mediaproducts, Groningen, The Netherlands) with an end-volume of 9 mL. The dilutions were used to compare the sensitivity of the DAA (Diasorin, Saluggia, Italy) with the lateral flow assay SARS-CoV-2 Roche/SD Biosensor Rapid Antigen test (RRA) (Roche, Basel, Switzerland) and real-time RT-PCR (Alinity M, Abbott, Chicago, United States). The antigen tests were performed in triplicate, real-time RT-PCR in duplicate and a blank sample was included for DAA background signal evaluation.
2.2. Clinical samples
COVID-19 testing of non-hospitalized symptomatic patients in The Netherlands is coordinated by Municipal Health Services (MHS). A person with COVID-19-like symptoms (rhinitis, cough, elevated temperature (not further specified), shortness of breath or sudden loss of sense of taste or smell) makes an appointment at a regional MHS test centre. In the MHS test centre where the clinical samples were collected, a combined oropharyngeal and nasopharyngeal flocked swab was obtained and suspended in 3 mL GLY medium for routine real-time RT-PCR in accordance with the Dutch national COVID-19 testing protocol. The swab was removed just before further execution of real-time RT-PCR. All qRT-PCR positive samples obtained at the MHS test centre in Tilburg, The Netherlands during one day in November 2021 were included; in addition 174 qRT-PCR negative samples obtained the same day were selected.
2.3. Real-time reverse transcriptase PCR (real-time RT-PCR)
Real-time reverse transcriptase PCR for SARS-CoV-2 was performed with the CE-IVD labelled ‘Alinity M SARS-CoV-2 Assay’ (Alinity M, Abbott, Chicago, United States) with N-gene and RdRP-gene targets, according to the manufacturer's instructions. The assay uses 500 µl of sample, which consisted of 500 µl dilution of heat inactivated SARS-CoV-2 culture supernatant or 500 µl of GLY medium from the clinical samples. The real-time RT-PCR was performed within 4 h of collection of clinical samples.
2.4. The Diasorin SARS-CoV-2 Antigen detection assay (DAA)
‘The Diasorin SARS-CoV-2 Antigen detection assay’ (DAA) (Diasorin, Saluggia, Italy) is a direct two-step sandwich chemiluminescence immunoassay for the quantitative determination of SARS-CoV-2 nucleocapsid antigen. Before testing, 1 mL of GLY medium from clinical samples and 1 mL of the dilutions of SARS-CoV-2 culture supernatant SARS-CoV-2 were inactivated within 8 h of collection by adding 1 ml Diasorin lysis buffer to the test tubes. DAA was performed on the Liaison XL automated chemiluminescence analyser (Diasorin) according to the manufacturer's instructions within 30 min - 2 h after inactivation of the samples. The cut-off for a positive result was 200 relative light units (RLU) (after subtraction for the blank background signal) as determined by the manufacturer.
2.5. SARS-CoV-2 Roche/SD Biosensor Rapid Antigen test (RRA)
The SARS-CoV-2 Roche/SD Biosensor Rapid Antigen test (RRA) (Roche, Basel, Switzerland) is a chromatographic lateral flow immunoassay for the qualitative detection of SARS-CoV-2 specific antigens in respiratory specimens. 350 μl of diluted culture supernatant or a swab soaked in supernatant (during 3 h with shaft) were mixed with 320 μl RRA buffer. The LFA was subsequently performed according to the manufacturer's instructions: 3 drops of the suspension were applied to the small testing well of the test device. Following 15 min of incubation the results of the test were read. Tests were interpreted: as positive when a purple-red line appeared at both the positive control and test site; as negative when a purple-red line appeared at the positive control site only; as inconclusive when a purple-red line did not appear at the positive control site or more than two purple-red lines appeared.
2.6. Statistical analysis
Adjusted-Wald confidence intervals were calculated of the specificity and sensitivity on the selected clinical samples of the DAA compared to real-time RT-PCR, overall and stratified by real-time RT-PCR Ct-value (Ct-value <30 and Ct-value <25) using pandas v1.1.5, statsmodels v0.12.1, and numpy v1.19.4. Additionally, a simple linear regression model was created and a Pearson's correlation coefficient was calculated to describe the relation between the Ct-value of the real-time RT-PCR positive samples and 10log transformed RLU value in the DAA using scipy v 1.5.4 and statsmodels v0.12.1. Only samples with a RLU value higher than the limit of measurement (22 RLU) were included in the comparison.
3. Results
3.1. Analytical performance
A positive result in the DAA was obtained from dilution 10−1 until dilution 10−5 of the cell cultured SARS-CoV-2 strain in all three series (Table 1 ). The RLU progressively decreased up to a dilution of 10−5 after which the signal became indistinguishable from the background of the blank sample (Table 1). The average decrease in Ct-value of the real-time RT-PCR was 3.6 for every step of sample dilution, starting at a mean Ct-value of 10.23 in step 10−1 until a Ct-value of 35.32 in step 10−8. The highest Ct-value at which the DAA in every dilution series still had a positive result was dilution step 10−4 (Ct-value 20.20) (Table 1).
Table 1.
Relationship between real-time RT-PCR, DAA and RRA results for each step in the dilution series of the cultured SARS-CoV-2 virus stock. Both SARS-CoV-2 Ag assays were performed in triplicate on nasopharynx swabs soaked in each dilution.
| Dilution step | TCID50/ml | Ct-value real-time RT-PCR* | DAA (RLU signal range)** | RRA** | |
|---|---|---|---|---|---|
| 10−1 | 5,62.10 [6] | 10.23 | 3/3 | (>100,000) | 3/3 |
| 10−2 | 5,62.10 [5] | 13.17 | 3/3 | (14,436–16,112) | 3/3 |
| 10−3 | 5,62.104 | 16.75 | 3/3 | (2247–2437) | 3/3 |
| 10−4 | 5,62.10 [3] | 20.20 | 3/3 | (272–328) | 3/3 |
| 10−5 | 5,62.10 [2] | 24.33 | 0/3 | (86–89) | 0/3 |
| 10−6 | 5,62.10 | 28.02 | 0/3 | (53–62) | 0/3 |
| 10−7 | 5,62 | 31.21 | 0/3 | (52–60) | 0/3 |
| 10−8 | 5,62.10−1 | 35.32 | 0/3 | (56–65) | 0/3 |
| blank | 0 | > 50 | 0/3 | (48–58) | 0/3 |
* Based on 500 μl of the dilution series, real-time RT-PCR was performed in duplicate, the Ct-value is the mean, ** The column shows the number of replicates in which the antigen test was positive, the cut-off for positivity is 200 RLU.
3.2. Test performance on clinical samples
A total of 248 samples were included of which 74 (29,9%) had detectable SARS-CoV-2 RNA. DAA was positive in 54 out of the 74 SARS-CoV-2 real-time RT-PCR positive samples (sensitivity: 73% (95%CI: 61.3%−82.7%)) No positive DAA results were observed in the SARS-CoV-2 real-time RT-PCR negative samples (specificity: 100% (95%CI 97.9%−100%)) (Table 2 ). In all real-time RT-PCR negative samples the RLU value of DAA was 60 or less. (Table 3 ).
Table 2.
A) DAA results and real-time RT-PCR results of corresponding clinical samples (n = 248) overall B) DAA results and real-time RT-PCR results of real-time RT-PCR positive clinical samples (n = 74) stratified by real-time RT-PCR Ct-value category (Ct-value >30, Ct-value 25–30 and Ct-value < 25).
| real-time RT-PCR result | |||||
|---|---|---|---|---|---|
| positive | negative | ||||
| DAA result | positive | 54 (Ct-value 12.2–27.7) | 0 | ||
| negative | 20 (Ct-value 22.6–39.5) | 174 | |||
| Real-time RT-PCR Ct-value | |||||
| Ct > 30 | Ct: 25–30 | Ct <25 | |||
| DAA result | positive | 0 | 1 | 53 | |
| negative | 11 | 5 | 4 |
Table 3.
Distribution of DAA RLU values for real-time RT-PCR positive and negative samples.
| RLU signal in DAA | real-time RT-PCR result | |
|---|---|---|
| positive | negative | |
| <22 | 11 | 156 |
| 22–40 | 1 | 17 |
| 40–60 | 2 | 1 |
| 60–80 | 1 | 0 |
| 80–100 | 1 | 0 |
| 100–120 | 0 | 0 |
| 120–140 | 1 | 0 |
| 140–160 | 0 | 0 |
| 160–180 | 3 | 0 |
| 180–200 | 0 | 0 |
| >200 | 54 | 0 |
Sensitivity and specificity stratified by CT-value category are presented in Table 2 .
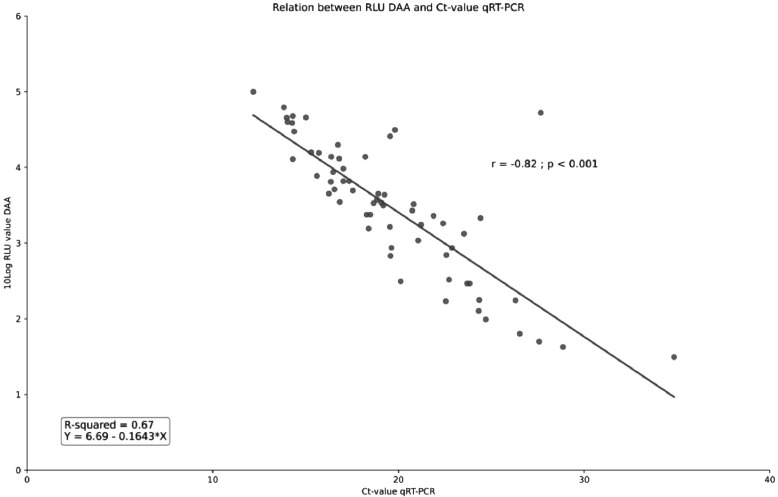
The 10log transformed DAA RLU values were inversely correlated with the Ct-values of the real-time RT-PCR (Pearsons's correlate r: −0.82; p < 0.001). Based on the linear regression model every increase in Ct-value of 1 resulted in an expected decrease of 0.1643 in 10log DAA RLU (95%CI −0.194 - −0.135) (Fig. 1 ).
Fig. 1.
Relation between the log transformed RLU values of the DAA and Ct-values of the real-time RT-PCR (concordant results only).
4. Discussion
We found an overall specificity of 100% (95%CI 97.9%−100%) and sensitivity of 73% (95%CI 61.3%−82.7%) of the DAA compared to the RRA when performed on clinical samples obtained in community-dwelling symptomatic individuals. The sensitivity was 86% (95%CI:75,5%−93.3%) for samples with a Ct-value below 30 and 93% (CI95% 83.0%−98.1%) for samples with a Ct-value below 25. Furthermore, DAA RLU were found to inversely correlate with real-time RT-PCR Ct-values. The analytical performance of the DAA was comparable with the lateral flow Rapid Antigen Test from Roche.
The performance of a number of SARS-CoV-2 LFA for use amongst symptomatic community-dwelling individuals has been evaluated and is in line with the results found in this study [5], [6], [7]. Clinical specificity of the ‘BD Veritor System for Rapid Detection of SARS-CoV-2′, the RRA and ‘Abbott PanbioTM COVID-19 Ag Rapid Test’ varied from 99.5% to 100% and the observed sensitivity on clinical samples ranged between 73% and 85% overall. All LFA had a higher sensitivity for samples with lower real-time RT-PCR Ct-values, varying from 94.3% for samples with a Ct-value below 30 for the Roche/SD Biosensor test to 98.0% for samples with a Ct-value below 32 for the Abbott Panbio test [5], [6], [7].
Studies evaluating the performance of currently available automated SARS-CoV-2 Antigen assays widely vary in study design (composition of the clinical cohort, pre-analytical protocols) and gain very different results. The LUMIPULSE SARS-CoV-2 Ag kit (Fujirebio, Tokyo, Japan) and VITROS Immunodiagnostic Products SARS-CoV-2 Antigen test (Ortho Clinical Diagnostics, Raritan, USA) were found to have respectively a 55.2% and 83.3% sensitivity and 99.6% and 100% specificity compared to the used real-time RT-PCR when performed on clinical samples. Analytical sensitivity was not determined [10,11]. The mariPOC SARS-CoV-2 Antigen Test (ArcDia International, Turku, Finland) gained a sensitivity of 100.0% when directly performed on clinical swab specimens (84.4% in undefined transport mediums) and a specificity 100.0%. The observed limit of detection was 2.7 TCID50/test [12].
The manufacturer of the DAA (Diasorin) reports a clinical specificity of 99.5% (95%CI 97.3% - 99.9%) and an overall sensitivity of 94.6% (95%CI 82.3% - 98.5%) compared to RT-PCR when performed on nasopharyngeal specimen within 10 days after symptom onset. For samples with a real-time RT-PCR Ct-value below 33, the manual of the manufacturer reports a 97.1% (95%CI 85.5%−99.5%) sensitivity. For analytical sensitivity the reported lower limit of detection is 22 TCID50/ml [8]. The sensitivity on the selected clinical samples, overall and for samples with a real-time RT-PCR value below 30, and analytical sensitivity (LOD 5620 TCID50/mL) observed in this evaluation are substantially lower. Although real-time RT-PCR Ct-values are known to vary depending on the assays and platforms used, this is unlikely to completely explain the observed discrepancy – given the magnitude. Our study has several limitations which may have led to an underestimation of the sensitivity on clinical samples. First, information about the number of days between symptom onset and sampling was not available. As a number of performance evaluation studies of SARS-CoV-2 LFA have shown a substantial difference in test performance correlated with the time since symptom onset, further research on the DAA performance in relation to the timing of sampling is needed [5,6]. Furthermore, the swabs were not directly preserved in DAA inactivation buffer: they were kept into 3 ml of GLY-medium, of which 1 ml was added to 1 ml of DAA inactivation buffer before testing. This dilution could theoretically reduce the assay signal by more than 50%. Differences between types of swabs and quantity and quality of absorbed clinical material could also account for differences in sensitivity.
One of the strengths of the study is the correlation of real-time RT-PCR Ct-values and test performance with TCID50. Real-time RT-PCR Ct-values are known to vary as they depend on the real-time RT-PCR platform and assay used, correlation with the TCID50 increases the generalisability of the study results. However, viral culture, PCR and antigen tests each have their specific target and are unlikely to correlate perfectly under all circumstances.
The World Health Organisation (WHO) and European Centre for Disease Control (ECDC) set the minimum performance requirements for the use of SARS-CoV-2 rapid antigen tests at 80% sensitivity and 97% specificity compared to nucleic acid amplification tests [3,13].
The principal aim of testing symptomatic community dwelling individuals is to efficiently implement disease control measures (quarantine, source and contract tracing) in order to control SARS-CoV-2 community spread. In this context, detecting individuals with high viral loads – corresponding to lower Ct-values – is paramount [2,6,14,15]. For samples in lower Ct-value categories, the clinical sensitivity observed in this study does meet the WHO and ECDC minimum requirement of an 80% sensitivity. It depends, in other words, on the distribution of Ct-values in the target population whether the DAA meets the international requirements. Periodic monitoring of characteristics of the target population, such as disease severity (symptomatic / asymptomatic testing), time between sampling and symptom onset and Ct-value distribution may prevent otherwise unnoticed decreases of sensitivity.
If the cut-off for positivity of the DAA would be lowered, for example from 200 to 60 RLU, the overall sensitivity in our clinical study sample would rise from 73% (95%CI 61.3% −82.7%) to 81% (95%CI 70.3%−89.3%) without affecting the 100% specificity. ( Table 3 ) Although lowering the RLU cut-off resulted in an improvement of test performance when conducting the pre-analytical steps as described above, we also found that ‘background’ signal varied between different types of swabs (data not shown).
Thus, the distribution of the RLU values among patients without SARS-CoV-2 infection may be affected by changes in the pre-analytical procedures, or by changes in the population tested. We therefore emphasize that any modification in the test cut-off should be validated in each specific test-setting.
In conclusion, our quantitative data confirmed the close relation between Ct-values and RLU and the DAA showed an adequate sensitivity compared with real-time RT-PCR among clinical samples with Ct-value below 30. The DAA has potential for use as a diagnostic test for symptomatic community-dwelling individuals in the context of transmission control.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.European Centre for Disease Prevention and Control. COVID-19 testing strategies and objectives. ECDC. 2020 Sep 15.
- 2.Mina M.J., Parker R., Larremore D.B. Rethinking Covid-19 test sensitivity - a strategy for containment. N. Engl. J. Med. 2020 doi: 10.1056/NEJMp2025631. Sep 30. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organisation . WHO; 2020. Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection Using Rapid Immunoassays – Interim Guidance. September 11. [Google Scholar]
- 4.Guglielmi Giorgia. Fast coronavirus tests: what they can and can’t do. Nature. 2020 doi: 10.1038/d41586-020-02661-2. Sep 16. [DOI] [PubMed] [Google Scholar]
- 5.Van der Moeren N., Zwart V.F., Lodder E.B., Van den Bijllaardt W., Van Esch H.J.R.M., et al. Performance evaluation of a SARS-CoV-2 rapid antigentest: test performance in the community in the Netherlands. Medarchives. 2020 doi: 10.1101/2020.10.19.20215202. Octoberhttps://doi.org/ [DOI] [Google Scholar]
- 6.Iglὁi Z., Velzing J., van Beek J., van de Vijver D., Aron G., Ensing R., et al. Clinical evaluation of the Roche/SD Biosensor rapid antigen test with symptomatic, non- hospitalized patients in a municipal health service drive-through testing. Medarchives. 2020 doi: 10.1101/2020.11.18.20234104. Novemberhttps://doi.org/ [DOI] [Google Scholar]
- 7.Gremmels H., Winkel B.M.F., Schuurman R., Rosingh A., Rigter N.A.M., Rodriguez O., et al. Real-life validation of the Panbio COVID-19 Antigen Rapid test (Abbott) in community- dwelling subjects with symptoms of potential SARS-CoV-2 infection. Medarchives. 2020 doi: 10.1101/2020.10.16.20214189. Novemberhttps://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diasorin. LIAISON® SARS-CoV-2 Ag ([REF] 311490). Diasorin. 2020.
- 9.Gorzynski J.E., De Jong H.N., Amar D., Hughes C., Ioannidis A., Bierman R., et al. High-throughput SARS-CoV-2 and host genome sequencing from single nasopharyngeal swabs. Medarchives. 2020 September. [Google Scholar]
- 10.Hirotsua Y., Maejimab M., hibusawab M., Nagakubob Y., Hosakab K., Amemiyac K., et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngealswabs, including from seven serially followed patients. Int. J. Infect. Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Favresse J., Gillot C., Oliveira M., Cadrobbi J., Elsen M., Eucher C., et al. Head-to-head comparison of rapid and automated antigen detection tests for the diagnosis of SARS-CoV-2 infection. J. Clin. Med. 2021;10:265. doi: 10.3390/jcm10020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koskinen J.M., Antikainen P., HotakainenK Haveri A, Ikonen N., Savolainen-Kopra C., et al. Clinical validation of automated and rapid mariPOC SARS-CoV-2 Antigen test. Medarchives. 2021 doi: 10.1038/s41598-021-99886-6. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Centre for Disease Control . ECDC; 2020. Options for the Use of rapid Antigen Tests for COVID-19 in the EU/EEA and the UK. Technical report19 Nov. [Google Scholar]
- 14.Widders A., Broom A., Broom J. SARS-CoV-2: the viral shedding vs infectivity dilemma. Infect. Dis. Health. 2020;25(3):210–215. doi: 10.1016/j.idh.2020.05.002. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26(5):672–675. doi: 10.1038/s41591-020-0869-5. May. [DOI] [PubMed] [Google Scholar]