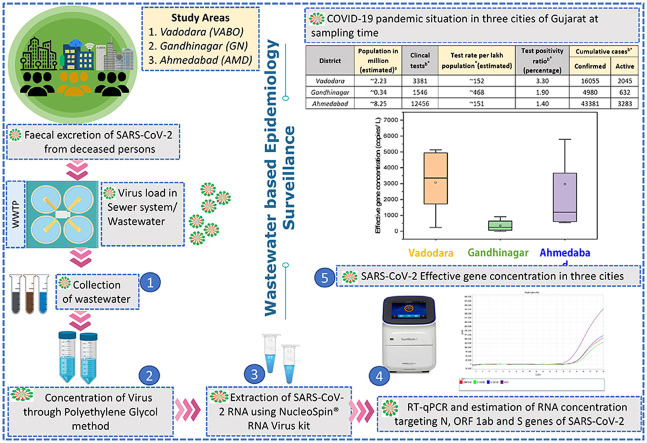
Abstract
Wastewater-based epidemiology (WBE) is a promising approach to understand the actual prevalence of COVID-19 disease at the community level. Different studies have cited the presence of SARS-CoV-2 in wastewater samples. In the present study, eighteen influent wastewater samples from different sewage treatment plants and pumping stations (5 samples from Vadodara city, 4 from Gandhinagar, and 9 from Ahmedabad city) were collected and analyzed for the presence of SARS-CoV-2 RNA in Gujarat state, India. The results showed the highest SARS-CoV-2 effective gene concentration in Vadodara (3078 copies/L), followed by Ahmedabad (2968 copies/L) and Gandhinagar (354 copies/L). On comparing the virus gene concentration in wastewater samples, the SARS-CoV-2 genetic material exhibited a positive relationship with the number of confirmed and active cases in in all three cities. However, a minor variation in SARS-CoV-2 effective gene concentration was seen between Vadodara and Ahmedabad despite a >2.5 and >1.5 folds differences in the cumulative number of confirmed and active cases, respectively. This may occur primarily due to the greater test positivity ratio in Vadodara (3.30%) than Ahmedabad (1.40%) and might be the higher number of asymptomatic patients in Vadodara. The study confirms the potential of the WBE that can be used at a large scale around the globe for better dealing with the pandemic situation.
Keywords: COVID-19, SARS-CoV-2, Wastewater based epidemiology (WBE), Pandemic, Management
Graphical abstract
1. Introduction
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), a beta coronavirus, is more virulent and has caused a larger pandemic than other family members like the SARS-CoV-1 during 2002–2004 and MERS-CoV (Middle East respiratory syndrome coronavirus) in 2012. Coronaviruses (RNA viruses) can rapidly increase their virulence and get acclimatized to novel hosts due to their high mutation rates compared to both DNA viruses and their hosts [1,2]. SARS-CoV-2, like SARS-CoV-1, uses human angiotensin-converting enzyme 2 (ACE2) as a receptor to enter cells [3]. ACE2 is a type I membrane glycoprotein found in the lungs, heart, intestines, and kidneys [4]. ACE2 catalyzes the conversion of angiotensin II to angiotensin-(1–7), and the ACE2/angiotensin-(1–7)/MAS axis resists the adverse effects of the renin-angiotensin system (RAS) that further plays a key role in the body's physiological and pathophysiological balance [4]. Therefore, in the diseased persons, apart from the immunological and inflammatory factors linked to COVID-19 pathogenesis, ACE2 downregulation and imbalance between RAS and ACE2/angiotensin-(1–7)/MAS may also result in multiple organ injuries [5].
The replication of SARS-CoV-2 viral particles in enterocytes of the intestinal lining in humans due to the high expression of the ACE2 receptor leads to gastrointestinal symptoms and virus shedding in the feces [[6], [7], [8]]. The diseased persons have been found to shed the virus for a prolonged period (14–21 days), with viral loads ranging between 1 × 102 to 1 × 106 RNA copies per gram of fecal matter [[9], [10], [11], [12], [13], [14]]. Moreover, many reports suggest virus presence in the feces of diseased persons with acute, mild, and no symptoms, and the patients who have been cured with no further signs [15,16], thus raising doubts over the statistics of COVID-19 pandemic based on clinical analysis. The release of the SARS-CoV-2 virus to the gastrointestinal tract supports the possible fecal-oral transmission route [17]. Likewise, the presence of SARS-CoV-2 RNA in the sewerage, untreated/treated wastewater, and natural water bodies raise the alarm for possible transmission routes via these aqueous matrices [6,[18], [19], [20], [21], [22], [23], [24], [25]]. Amidst various debatable questions related to the origin, transmission, stability, and decay of SARS-CoV-2, it is pertinent to develop a pre-alarming tool for effective COVID-19 preparedness and knowing the actual prevalence of disease at the community level. In this view, wastewater-based epidemiology (WBE) study with a long history in public health concerns, especially for human enteric viruses [26,27], is getting recognition around the globe owing to early and prolonged excretion of coronavirus in feces and subsequent detection in wastewater/sewerage system [6,8,[18], [19], [20], [21], [22], [23], [24], [25],28]. The WBE may give more accurate and unbiased information about the actual extent of COVID-19 compared to the clinical surveillance and covers a larger population, including both asymptomatic and symptomatic patients. Despite the well-proven concept and capabilities of WBE for COVID-19 monitoring, there is a dearth of knowledge related to the impact of other influencing factors, such as population, sewerage system, sampling approach, health infrastructure, clinical testing rate etc. on the SARS-CoV-2 RNA load in wastewater samples. Therefore, to accomplish this knowledge gap, we performed wastewater analysis for the presence of SARS-CoV-2 RNA in three differently planned and demographically dissimilar cities of western India viz. Vadodara (VABO), Gandhinagar (GN), and Ahmedabad (AMD) in Gujarat state with two main objectives; i) detection and estimation of SARS-CoV-2 RNA concentration in untreated wastewater samples from three cities (VABO, GN, AMD) to assess the influence of different contributing variables; ii) unravel the potential of WBE for the monitoring, prediction, and cross-evaluation of reported COVID-19 statistics by comparing the SARS-CoV-2 RNA load in wastewater with the number of cases.
The present study will substantiate the WBE potential and persuade the authorities and policymakers to incorporate WBE surveillance into the regular monitoring program and policy framework to manage current or future COVID-19 like pandemic situations efficiently.
2. Materials and methods
2.1. Sampling location
Vadodara is the third-largest city of Gujarat, located at the banks of the Vishwamitri River, comprising an estimated population of ~2.23 million at present [29]. Vadodara City is one of India's privileged cities with an underground drainage system built in the year 1894. The sewage is collected through a system comprising an underground drainage network, sewage treatment plants (STPs), auxiliary pumping stations (APS), pressure mains, and disposal into the water bodies after treatment. Sewage Disposal Works Department of Vadodara includes 6 STPs & 49 Auxiliary/Main Pumping stations (APS/MPS). Likewise, Ahmedabad is the largest city in Gujarat in terms of population as well as in area. Ahmedabad is also the seventh-largest metropolitan area and third fastest growing city of India, with an estimated population of ~8.25 million in 2020 [29]. The Sabarmati River passes through the center of the city. The Ahmedabad Municipal Corporation (AMC) area is divided into 43 wards and 5 zones. The Ahmedabad Municipal Corporation comprises 9 STPs, 45 Sewage Pumping Stations (SPSs), and an extended sewage network of ~2500 km present in the city. The third study area is Gandhinagar, the capital of Gujarat State, located on the western bank of the Sabarmati River and about 30 km away from Ahmedabad, which is the commercial and cultural heart of Gujarat. The estimated population of Gandhinagar is ~0.33 million in 2020. The entire city's wastewater is first collected in the Sargasan Drainage Pumping Station via the underground pipe network. Thereafter, it is pumped and transferred mainly to the Jaspur and Sargasan STPs, where treatment processes occur.
In all three cities, the sewage collection takes place through a system comprising an underground drainage network, APS/ MPS and STPs, followed by disposal into the natural water bodies and rivers after treatment. Wastewater generated is collected by a network of underground sewers and pumping stations and is conveyed to the STPs for the physical and biological treatment to meet the Gujarat Pollution Control Board (GPCB) guidelines before discharge into the nearest water body.
Details of the sampling locations, such as geospatial positions, the capacity of the treatment plant, the wastewater source, treatment process, and connected residents (approximate) are given in Table S1. Also, the drainage system of Vadodara city has been explained in detail to provide an idea about the sewerage system of the study areas (Fig. S1).
2.2. Sample collection and preparation
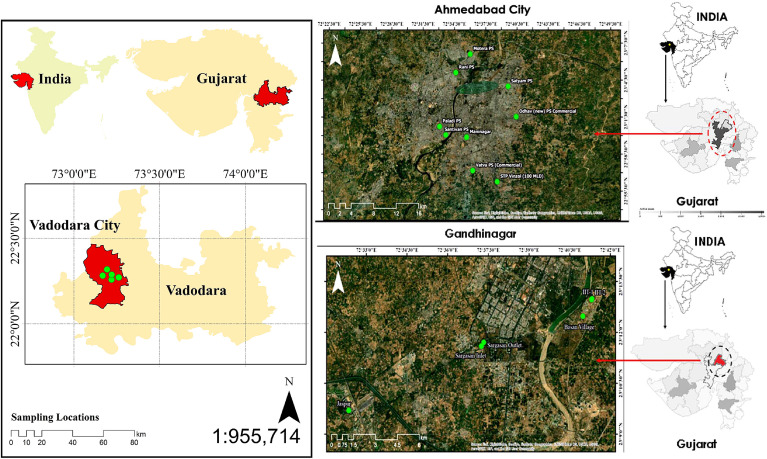
The untreated wastewater was collected from eighteen different locations in three cities, i.e., Vadodara (VABO), Gandhinagar (GN), and Ahmedabad (AMD) of Gujarat state, India. A total of 5 influent samples were collected from five STPs of VABO, 4 samples from different STPs in GN, and 9 samples (8 from SPSs and one from STP) from AMD in the first week of November 2020 (Fig. 1 ). The grab sampling method was used for the sample collection in 500ml polyethylene sterile bottles (Tarsons, PP Autoclavable, Wide Mouth Bottle, Cat No. 582240, India). Collected samples were transferred in an icebox to the laboratory and refrigerated at 4 °C until further process. A sampling blank was also prepared to examine the cross-contamination during transportation. The experiments were performed at Gujarat Biotechnology Research Center (GBRC), an approved laboratory by the Indian Council of Medical Research (ICMR), New Delhi.
Fig. 1.
Geospatial position of sampling locations in three different cities of Gujarat, India.
2.3. SARS-CoV-2 RNA concentration method
The concentration method consisted of a PEG 9000 and NaCl precipitation protocol previously described by Kumar et al., 2020 [6] for wastewater samples. 30ml sample was centrifuged (Model: Sorvall ST 40R, Thermo Scientific) at 4000g for 30 minutes in a 50ml falcon tube followed by the filtration of the supernatant with a syringe filter of 0.2μ (Mixed cellulose esters syringe filter, Himedia). The 25ml sample filtrated was then treated with NaCl (17.5 g/L) and PEG 9000 (80 g/L) and incubated at 17 °C, 100 rpm overnight (Model: Incu-Shaker™ 10LR, Benchmark). The sample was then transferred in an oak ridge tube for further centrifugation (Model: Incu-Shaker™ 10LR, Benchmark) at 14000g for 90 minutes, ultimately forming the pellets. RNase-free water (300 μL) was used for the resuspension of the viral particles after discarding the supernatant. The sample was then stored in a 1.5ml Eppendorf tube at a temperature of −40 °C for RNA isolation.
2.4. Isolation of the SARS-CoV-2 RNA, RT-PCR, gene copy estimation
Using a commercially ready-for-use kit (NucleoSpin® RNA Virus, Macherey-Nagel GmbH & Co. KG, Germany), SARS-CoV-2 RNA isolation was performed. MS2 phage (10 μL), Proteinase K (20 μL) and RAV1 buffer (600 μL) consisting of carrier RNA were mixed with 300 μL of the concentrated viral particles. MS2 phage serves as a molecular process inhibition control [6]. It was used to monitor the efficacy of RNA extraction and PCR inhibition. It should be remembered that MS2 may spontaneously exist in wastewater, so there is a risk that the retrieved MS2 may consist of both the spiked and the background viral material. As per the user manual instructions (Macherey-Nagel GmbH & Co. KG), further procedures were carried out. The last elution was done with 30 μL of kit-supplied elution buffer. Using a Qubit 4 Fluorometer (Invitrogen), RNA concentrations were checked.
The nucleic acid was analyzed to identify the S gene, N gene, and ORF1ab of SARS-CoV-2 and the internal control (MS2) with the help of RT-PCR using the TaqPath™ Covid-19 RT-PCR package (Applied Biosystems). Amplification was conducted in a reaction mixture volume (25 μL) containing 7 μL of RNAs derived from each sample, 10.50 μL Nuclease-free Water, 6.25 μL Master Mix, and 1.25 μL COVID-19 RT-PCR Assay Multiplex. Three controls were used, viz. positive control (TaqPath™ COVID 19 Control), one negative control (from extraction run spiked with MS2), and no template control (NTC). Nuclease-free water was applied as a template-free control in this analysis. Additional process steps were executed, as defined in the product guidebook. The RT-qPCR step consisting of 40 cycles, included UNG incubation (25 °C for 2 min), reverse transcription (53 °C for 10 min), and activation (95 °C for 2 min). The reactions were conducted as instructed in the handbook of Applied BiosystemsTM 7500 Fast Real-Time PCR.
Results were considered inconclusive if less than two genes were detected in a sample. The Ct-value of a given sample was converted to gene copy numbers considering the equivalence of 500 copies of SARS-CoV-2 genes as 26 Ct-value (provided with the kit), and the same was extrapolated to derive approximate copies of each gene. The effective gene concentration was calculated by averaging the gene copies of all three genes in a particular sample. The author's previous publication explained the detailed information related to RT-PCR and gene copy estimation [6,[19], [20], [21], [22]].
2.5. Data visualization
OriginPro 2019b software has been used for data analysis and to draw boxplots.
3. Results and discussion
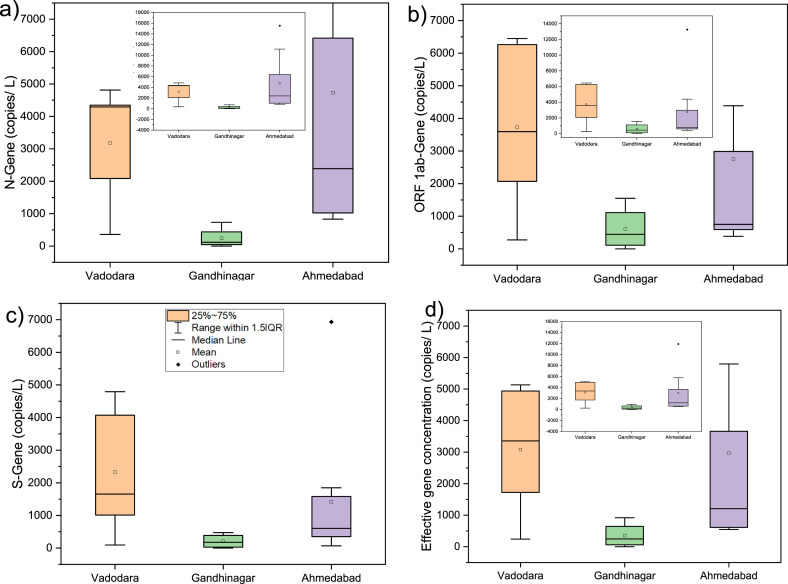
Wastewater samples collected from three cities (VABO, GN, and AMD) of Gujarat, India, showed a clear variation in SARS-CoV-2 RNA load. Comparison of RT-PCR assay findings for the detection of SARS-CoV-2 genes (i.e., N, ORF 1ab, and S genes) in wastewater samples of three cities showed 100% positive samples in Vadodara (5/5), 75% in Gandhinagar (3/4), and 100% in Ahmedabad (9/9). The average N-gene copies were found to be maximum in AMD (4731 copies/L), followed by VABO (3179 copies/L) and GN (243 copies/L). The ORF 1ab-gene copies were found maximum in wastewater samples collected from VABO (3730 copies/L), followed by AMD (2756 copies/L) and GN (611 copies/L). Similarly, the descending order of S-gene copies was: VABO (2325 copies/L)> AMD (1417 copies/L)> GN (207 copies/L). Consequently, maximum SARS-CoV-2 effective gene concentration was noticed in VABO (3078 copies/L), trailed by AMD (2968 copies/L) and GN (354 copies/L). The distribution of SARS-CoV-2 gene copies in wastewater samples collected from three cities is represented in Fig. 2 . Also, the variation in gene copies of the SARS-CoV-2 targeted genes and effective gene concentration in wastewater samples is shown in Table 1 .
Fig. 2.
Distribution of SARS-CoV-2 gene copies, collected from three different cities of Gujarat, India; a.) N gene, b.) ORF 1ab gene, c.) S gene, and d) Effective gene concentration.
Table 1.
Variation in copies of the SARS-CoV-2 targeted genes and effective gene concentration in wastewater samples, collected from three different cities of Gujarat, India.
| Sampling date | Station | Ct value |
Gene copies (copies/L) |
|||||
|---|---|---|---|---|---|---|---|---|
| N | ORF | S | N | ORF | S | Effective gene concentration | ||
| November 02, 2020 | Vadodara | |||||||
| STP-1 Tarsali | 29.9 | 30.31 | 31.43 | 4814 | 3594 | 1656 | 3355 | |
| STP-2 Gajrawad | 31.09 | 31.1 | 32.16 | 2086 | 2069 | 1012 | 1722 | |
| STP-3 Atladara | 30.04 | 29.53 | 29.91 | 4346 | 6261 | 4792 | 5133 | |
| STP-4 Sayaji | 33.76 | 34.18 | 35.96 | 360 | 275 | 93 | 243 | |
| STP-5 Kapurai | 30.06 | 29.49 | 30.13 | 4292 | 6449 | 4072 | 4938 | |
| November 02, 2020 | Gandhinagar | |||||||
| Basan Inlet | 32.65 | 31.53 | 33.32 | 733 | 1551 | 474 | 919 | |
| Jaspur Inlet | 36.00 | 34.53 | 36.91 | 91 | 222 | 53 | 122 | |
| IIT inlet | ND | ND | ND | 0 | 0 | 0 | 0 | |
| Sargasan inlet | 35.20 | 32.78 | 34.04 | 147 | 672 | 301 | 373 | |
| November 05, 2020 | Ahmedabad | |||||||
| Motera PS | 30.63 | 33.65 | 33.73 | 2873 | 386 | 365 | 1208 | |
| Ranip PS | 29.50 | 30.58 | 31.50 | 6415 | 2986 | 1580 | 3661 | |
| Paldi PS | 32.39 | 33.02 | 33.79 | 869 | 577 | 352 | 599 | |
| Santivan PS | 31.57 | 31.88 | 32.95 | 1506 | 1220 | 603 | 1110 | |
| Maninagar PS | 32.46 | 32.98 | 34.57 | 830 | 593 | 217 | 547 | |
| Satyam PS | 32.14 | 32.61 | 36.50 | 1024 | 752 | 68 | 615 | |
| STP Vinzole | 28.75 | 30.03 | 31.27 | 11148 | 4386 | 1848 | 5794 | |
| Odhav PS | 28.31 | 28.52 | 29.39 | 15523 | 13247 | 6930 | 11900 | |
| Vatva PS | 30.90 | 32.83 | 32.53 | 2390 | 654 | 793 | 1279 | |
The SARS-CoV-2 RNA load in wastewater showed a positive connection with the number of confirmed and active cases in all three cities. However, despite having a >2.5 and >1.5 times greater number of confirmed and active cases, respectively in AMD than VABO, a very minimal difference in SARS-CoV-2 effective gene concentration was observed. (Table 2 ). These results contradicted the earlier reports, where SARS-CoV-2 effective gene concentration in wastewater was found following the COVID-19 cases in the two and three-month studies in both GN and AMD, respectively [19,22]. Possible reasons for the disparity between the SARS-CoV-2 viral gene load and cumulative number of confirmed and active cases in VABO and AMD can be explained to, i) sample collection from STPs in VABO, while in the case of AMD, samples mainly collected from SPSs; and the concentration of SARS-CoV-2 RNA might be higher in STPs due to accumulation as compared to the SPSs that reflected in the RT-PCR analyses; ii) around the same number of clinical tests per lakh of the population (~151 individuals) in both cities, but the test positivity ratio was >2 folds in VABO on sampling dates (Table 2); iii) greater existence of asymptomatic patients in VABO led to the low number of reported confirmed cases, also supported by the high test positivity ratio; iv) shedding of virus in the feces by the recovered patients in VABO; v) biases in the reported pandemic statistics owing to the medical infrastructure and capacity to carry out RT-PCR testing in both cities.
Table 2.
Statistics and comparison of COVID-19 pandemic in three different cities of Gujarat, India.
| District | Population in million (estimated)a | Clincal testsb,c | Test rate per lakh populationc(estimated) | Test positivity ratiob,c (percentage) | Cumulative casesb,c |
|
|---|---|---|---|---|---|---|
| Confirmed | Active | |||||
| Vadodara | ~2.23 | 3381 | ~152 | 3.30 | 16055 | 2045 |
| Gandhinagar | ~0.33 | 1546 | ~468 | 1.90 | 4980 | 632 |
| Ahmedabad | ~8.25 | 12456 | ~151 | 1.40 | 43381 | 3283 |
UN world urbanization prospects 2018 [29].
COVID19 INDIA (https://www.covid19india.org/) [32].
Values represent data on the sampling dates.
The results agreed with Kumar et al. [19], who studied weekly temporal variation in SARS-CoV-2 genetic material concentration in wastewater samples targeting N, ORF 1ab, and S genes in a two-month study in Gandhinagar. The results suggested a positive correlation between SARS-CoV-2 effective gene concentration in wastewater and the number of confirmed cases found higher in the month of September compared to August 2020, which corresponded to ~2.2 folds increase in confirmed cases during the study period. Likewise, in another three-month (September to November 2020) weekly analysis of wastewater samples from 9 different locations in Ahmedabad city showed similar trends, and the maximum SARS-CoV-2 effective gene concentration was noticed in November (~10729 copies/L), trailed by September (~3047 copies/L), and October (454 copies/L) in line with a ~ 1.5-fold rise in the confirmed cases during the study period [22]. The decrease in SARS-CoV-2 gene concentration in October subjected to a decline of 20.5% in active cases (~844 cases), while a significant rise in virus effective gene concentration in November 2020 was due to a rise of 1.82% in active cases (~59 cases). Though the rise in active cases was nominal in November, at the same time, a sharp rise of >6000 new cases (14.1%) was reported in November 2020 [22].
Despite the inconsistency between the SARS-CoV-2 effective gene concentration and reported COVID-19 cases when comparing VABO and AMD data, two observations are noteworthy and critical: i) clinical analysis-based data sometimes give false reports of diseased individuals; ii) though the findings of the present study are sufficient enough to substantiate the WBE potential, but a long scale time-series data would provide more accurate estimation and prediction, and also the robustness of WBE approach.
Chakraborty et al. (2021) [30] performed wastewater surveillance for the detection of SARS-CoV-2 RNA in STPs and SPSs of Chennai city, South India, and estimated the actual number of COVID-19 cases. Similarly, Hemalatha et al. (2021) [31] estimated the spread of COVID-19 based on the WBE study in Hyderabad city, India, and endorsed the potential of this technique to study the infection dynamics and efficient management of the pandemic. There are many other studies in the public domain from different parts of the world, such as the Netherlands [33], Spain [24], the USA [34], Paris [35], China [36], India [6,[19], [20], [21], [22],30,31], Australia [37], etc. which support WBE surveillance of COVID-19.
One of the main advantages of WBE is that it includes both asymptomatic and symptomatic individuals. Consequently, it gives a better picture of the pandemic situation compared to clinical analysis-based secondary data, which rely on the number and efficiency of clinical tests. Therefore, under certain circumstances, it is possible that despite an increase in SARS-CoV-2 RNA load in wastewater, no significant change in COVID-19 cases may observe as noticed in VABO in the present study. Therefore, WBE technology can provide the actual extent of the infection at sub-city or zone levels and help in identifying the hot spots within a city [22]. Thus, WBE might offer critical monitoring of SARS-CoV-2 spread within a community, including disease outbreak, tapering, or reemergence.
4. Conclusion
A comparison of SARS-CoV-2 RNA presence in wastewater samples from three cities of Gujarat unveiled the highest load in VABO, followed by AMD and GN. The virus RNA load in wastewater showed a positive correlation with the number of confirmed and active cases in all three cities. However, a small difference in SARS-CoV-2 effective gene concentration between VABO and AMD was noticed, probably due to the greater number of asymptomatic patients and the high test positivity ratio in VABO. Although the rate of clinical testing was very low in the case of both VABO and AMD during the study period, but the WBE results suggested comparatively more pathetic condition in VABO as compared to two other cities. Nevertheless, large time scale temporal dataset is required for more convincing conclusion. The study concludes that regular monitoring of wastewater samples could be used to know the pandemic situation in a particular area and help in tuning the management interventions efficiently. Though WBE has immense potential that must be exploited and included in the policy framework around the globe, long-scale time-series data and epidemiological information are required to substantiate the robustness of this technology. Also, future emphasis should be paid to developing a predictive model using WBE and clinical survey data to convey the situation to the policymakers better and enhance the preparedness and management of epidemic/pandemic situations.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work is funded by UNICEF, Gujarat, and UKIERI. We also acknowledge the help received from Mr. Alok Thakur and other GBRC and GPCB staff who contributed to the sample and data analyses.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cscee.2021.100115.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elena S.F., Sanjuán R. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 2005;79(18):11555–11558. doi: 10.1128/JVI.79.18.11555-11558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.sci378totenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta M.K., Vemula S., Donde R., Gouda G., Behera L., Vadde R. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J. Biomol. Struct. Dyn. 2020;1–11 doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang N., Gong Y., Meng F., Bi Y., Yang P., Wang F. Virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. MedRxiv. 2020 doi: 10.1101/2020.03.28.20043059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amirian E.S. Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int. J. Infect. Dis. 2020;95:363–370. doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARSCoV- 2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26 doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., Enouf V., Houhou-Fidouh N., Valette M., Mailles A., Lucet J.C., Mentre F., Duval X., Descamps D., Malvy D., Timsit J.F., Lina B., van der Werf S., Yazdanpanah Y. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20 doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 15.Tang A., Tong Z., Wang H., Dai Y., Li K., Liu J., Wu W., Yuan C., Yu M., Li P., Yan J. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. J. 2020;26:1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhama K., Patel S.K., Yatoo M.I., Tiwari R., Khan S., Dhama J., Natesan S., Malik Y.S., Singh K.P., Harapan H. SARS-CoV-2 existence in sewage and wastewater: a global public health concern. J. Environ. Manag. 2021;280 doi: 10.1016/j.jenvman.2020.111825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao J., Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venugopal A., Ganesan H., Sudalaimuthu Raja S.S., Govindasamy V., Arunachalam M., Narayanasamy A., Sivaprakash P., Rahman P.K.S.M., Gopalakrishnan A.V., Siama Z., Vellingiri B. Novel wastewater surveillance strategy for early detection of coronavirus disease 2019 hotspots. Curr. Opin. Environ. Sci. Heal. 2020;17:8–13. doi: 10.1016/j.coesh.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar M., Joshi M., Patel A.K., Joshi C.G. Unravelling the early warning capability of wastewater surveillance for COVID-19: a temporal study on SARS-CoV-2 RNA detection and need for the escalation. Environ. Res. 2021 doi: 10.1016/j.envres.2021.110946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar M., Dhangar K., Thakur A.K., Ram B., Chaminda T., Sharma P., Kumar A., Raval N., Srivastava V., Rinklebe J., Kuroda K. Antidrug resistance in the Indian ambient waters of Ahmedabad during the COVID-19 pandemic. J. Hazard Mater. 2021;416 doi: 10.1016/j.jhazmat.2021.126125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar M., Kuroda K., Joshi M., Bhattacharya P., Barcelo D. First comparison of conventional activated sludge versus root-zone treatment for SARS-CoV-2 RNA removal from wastewaters: statistical and temporal significance. Chem. Eng. J. 2021;425 doi: 10.1016/j.cej.2021.130635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar M., Joshi M., Shah A.V., Srivastava V., Dave S. Wastewater surveillance-based city zonation for effective COVID-19 pandemic preparedness powered by early warning: a perspectives of temporal variations in SARS-CoV-2-RNA in Ahmedabad, India. Sci. Total Environ. 2021;792 doi: 10.1016/j.scitotenv.2021.148367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Randazzo W., Truchado P., Cuevas-Ferrando E., Sim´on P., Allende A., S´anchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a Low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asghar H., Diop O.M., Weldegebriel G., Malik F., Shetty S., Bassioni L. El, Akande A.O., Maamoun E. Al, Zaidi S., Adeniji A.J., Burns C.C., Deshpande J., Oberste M.S., Lowther S.A. Environmental surveillance for polioviruses in the global polio eradication initiative. J. Infect. Dis. 2014;210:S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prevost B., Lucas F.S., Goncalves A., Richard F., Moulin L., Wurtzer S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015;79:42–50. doi: 10.1016/j.envint.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Chen Yifei, Chen Liangjun, Deng Qiaoling, Zhang Guqin, Wu Kaisong, Ni Lan, Yang Yibin, et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020 doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 29.UN world urbanization prospects 2018. 2018. https://population.un.org/wup/
- 30.Chakraborty P., Pasupuleti M., Shankar M.J., Bharat G.K., Krishnasamy S., Dasgupta S.C., Sarkar S.K., Jones K.C. First surveillance of SARS-CoV-2 and organic tracers in community wastewater during post lockdown in Chennai, South India: methods, occurrence and concurrence. Sci. Total Environ. 2021;778 doi: 10.1016/j.scitotenv.2021.146252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemalatha M., Kiran U., Kuncha S.K., Kopperi H., Gokulan C.G., Mohan S.V., Mishra R.K. Surveillance of SARS-CoV-2 spread using wastewater-based epidemiology: comprehensive study. Sci. Total Environ. 2021;768 doi: 10.1016/j.scitotenv.2020.144704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.COVID19 India. https://www.covid19india.org/
- 33.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 34.Nemudryi A., Nemudraia A., Surya K., Wiegand T., Buyukyoruk M., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARSCoV- 2 in municipal wastewater. Cell Rep Med. 2020 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wurtzer S., Marechal V., Mouchel J.M., Moulin L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. medRxiv. 2020 [Google Scholar]
- 36.Zhang W., Du R.H., Li B., Zheng X.S., Yang X. Lou, Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.