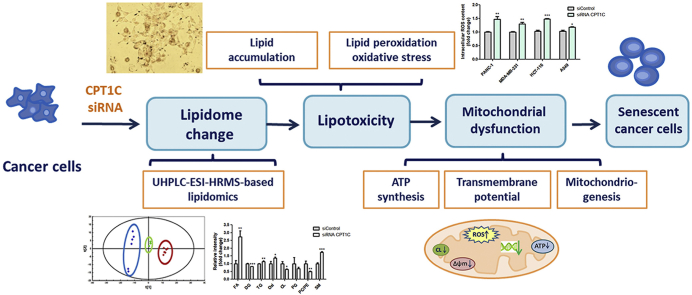
Abstract
Lipotoxicity, caused by intracellular lipid accumulation, accelerates the degenerative process of cellular senescence, which has implications in cancer development and therapy. Previously, carnitine palmitoyltransferase 1C (CPT1C), a mitochondrial enzyme that catalyzes carnitinylation of fatty acids, was found to be a critical regulator of cancer cell senescence. However, whether loss of CPT1C could induce senescence as a result of lipotoxicity remains unknown. An LC/MS-based lipidomic analysis of PANC-1, MDA-MB-231, HCT-116 and A549 cancer cells was conducted after siRNA depletion of CPT1C. Cellular lipotoxicity was further confirmed by lipotoxicity assays. Significant changes were found in the lipidome of CPT1C-depleted cells, including major alterations in fatty acid, diacylglycerol, triacylglycerol, oxidative lipids, cardiolipin, phosphatidylglycerol, phosphatidylcholine/phosphatidylethanolamine ratio and sphingomyelin. This was coincident with changes in expressions of mRNAs involved in lipogenesis. Histological and biochemical analyses revealed higher lipid accumulation and increased malondialdehyde and reactive oxygen species, signatures of lipid peroxidation and oxidative stress. Reduction of ATP synthesis, loss of mitochondrial transmembrane potential and down-regulation of expression of mitochondriogenesis gene mRNAs indicated mitochondrial dysfunction induced by lipotoxicity, which could further result in cellular senescence. Taken together, this study demonstrated CPT1C plays a critical role in the regulation of cancer cell lipotoxicity and cell senescence, suggesting that inhibition of CPT1C may serve as a new therapeutic strategy through induction of tumor lipotoxicity and senescence.
Keywords: Lipidomics, Lipid accumulation, Lipid peroxidation, Oxidative stress, Mitochondrial dysfunction, Anticancer target
Graphical abstract
Highlights
-
•
LC/MS-based lipidomic analysis of cancer cells was conducted after siRNA depletion of CPT1C.
-
•
Significant changes were found in the lipidome of CPT1C-depleted cells, including major alterations in fatty acid, diacylglycerol, triacylglycerol, oxidative lipids, cardiolipin, phosphatidylglycerol, phosphatidylcholine/phosphatidylethanolamine ratio and sphingomyelin.
-
•
Knockdown of CPT1C induces lipid accumulation, lipotoxicity and mitochondrial dysfunction and senescence.
-
•
CPT1C plays a critical role in the regulation of cancer cell lipotoxicity and cell senescence, suggesting that inhibition of CPT1C may serve as a new therapeutic strategy.
1. Introduction
The concept of cellular senescence was originally proposed in 1961 by Hayflick and Moorhead [1] who discovered that cells could not proliferate indefinitely under in vitro culture conditions. Currently, cellular senescence refers to the irreversible arrest of cell proliferation, characterized by inhibited proliferation ability, DNA replication, increased senescence-associated-β-gal (SA-β-gal) activity and senescence-associated secretory phenotype (SASP) [2,3]. Senescence response has profound implications in the suppression of cancer development, and it is a complex process caused by numerous triggers, including telomere shortening, genomic damage, and mitochondrial dysfunction [[4], [5], [6]]. Recently, a close link between cellular senescence and lipotoxicity was reported [7].
Lipotoxicity is caused by the accumulation of lipids, particularly fatty acids, in nonadipose cells or tissues that are not capable of metabolizing or storing lipids [8,9]. Disruption of lipid metabolism and lipotoxicity could lead to cellular dysfunction, which is involved in the development of various diseases, such as diabetes, Alzheimer’s disease and cancer [10,11]. A collective review of recent studies has proven that the main mechanism of senescence is lipid peroxidation and oxidative stress [12,13]. Excess lipids are vulnerable to the attack by reactive oxygen species (ROS) and conversion into toxic reactive lipid peroxides and oxidized lipids (oxi-lipid), which could have lipotoxic effects on mitochondrial DNA (mtDNA), RNA and proteins of mitochondrial machinery, and oxidation equilibrium, further leading to mitochondrial dysfunction and senescence [8,14,15]. In addition, it was reported that senescence could be a physiological degenerative process accelerated by lipotoxic insults, while conversely, the process of senescence can make cells more vulnerable to lipotoxicity [16].
Carnitine palmitoyltransferase 1C (CPT1C), a member of the CPT1 family, is an enzyme located in the outer mitochondrial membrane that catalyzes the acylation of long-chain fatty acids and their entry into mitochondria for fatty acid β-oxidation [17]. The CPT1 family consists of three members, namely, CPT1A, mainly expressed in the liver; CPT1B highly distributed in muscle; and CPT1C specifically expressed in brain and tumors [18]. Despite the great similarity of gene sequences to that of CPT1A and CPT1B, CPT1C exerts unique physiological functions, such as participating in learning and cognition, regulating the emotional state, and it is necessary for energy homeostasis as well as food intake [[19], [20], [21]]. Recently, it was found that compared with paracancerous tissues, CPT1C expression was abnormally increased in malignant tumors such as human lung cancer and breast cancer, and CPT1C could promote the proliferation and growth of tumor cells under metabolic pressure such as glucose deprivation and hypoxia, suggesting that CPT1C may serve as a new therapeutic target for anti-tumor treatment [22,23].
Most recently, extended culturing of pancreatic carcinoma PANC-1 cells was found to cause growth arrest and severe cellular senescence [24,25]. Low CPT1C expression is involved in replicative senescence and CPT1C is a novel regulator of cancer cell senescence. Moreover, CPT1C exerts a significant effect on tumor cellular senescence among the other members of the CPT family [26]. In addition, abnormal lipid metabolism was preliminarily found in replicative senescent PANC-1 cells [27]. However, whether loss of CPT1C could induce senescence via lipotoxicity remains unknown. Therefore, the current study aimed to elucidate whether knockdown of CPT1C could induce lipotoxicity resulting in cancer cell senescence. These results demonstrated that CPT1C plays a crucial role in the regulation of cancer cell lipotoxicity and cell senescence, suggesting that inhibition of CPT1C may serve as a new therapeutic strategy through induction of tumor lipotoxicity and senescence.
2. Materials and methods
2.1. Chemicals and reagents
Dulbecco's modified Eagle's medium (DMEM) and Roswell Park Memorial Institute (RPMI) 1640 medium were purchased from Corning Inc. (New York, USA) and HyClone Laboratories, Inc. (Los Angeles, CA, USA). Fetal bovine serum (FBS), penicillin sodium, and streptomycin sulfate solution were purchased from Gibco Life Technologies (Grand Island, NE, USA). All the other chemicals and solvents were of analytical grade or HPLC grade when appropriate and commercially available.
2.2. Cell culture and transfections with siRNA
Human pancreatic cancer PANC-1 cells were provided by the American Type Culture Collection (Manassas, VA, USA), human breast cancer cell line MDA-MB-231, human colorectal carcinoma cell line HCT-116 and human lung cancer cell line A549 were obtained from Dr. Jun Du (Sun Yat-sen University, China). Cells were cultured in DMEM or RPMI 1640 medium containing 10% FBS, 100 IU/mL of penicillin sodium, and 100 μg/mL of streptomycin sulfate at 37 °C in a 5% CO2 atmosphere with saturated humidity.
PANC-1, MDA-MB-231, HCT-116 and A549 cells were transfected with 5 nM small interfering RNA (siRNA) (RiboBio, Guangzhou, China) using Lipofectamine RNAiMAX transfection reagent (Invitrogen Corporation, Carlsbad, CA, USA) according to a previous report [24]. The siRNAs sequences are listed in Table S1. Each cancer cell line was transfected with siRNAs for 72 h prior to the following experiments.
2.3. Bromodeoxyuridine (BrdU), colony formation, and SA-β-gal assay
As previously reported [24], for BrdU incorporation measurement, a cell proliferation ELISA BrdU kit (Roche, CA, USA) was applied according to the manufacturer's protocol. Briefly, after adding 10 μL of BrdU labeling solution, the cells were incubated for 2 h at 37 °C. A specific antibody anti-BrdU-POD working solution was employed and the absorbance wavelength was set at 370 nm. For colony formation assay, different densities (5000, 2500, and 1250 cells/well) of cells were seeded into 6-well plates and cultured at 37 °C for 10–14 days. 4% formaldehyde was used to fix the cells and 1% crystal violet was used to stain the cell colonies. For senescence-associated β-galactosidase (SA-β-gal) activity assay, following the manufacturer's protocol (Beyoutime, China), cells were fixed with gluteraldehyde for 10–15 min and then washed twice with PBS before staining with X-gal staining solution overnight. Inverted fluorescence microscope was used to obtain photograph images.
2.4. Real-time PCR analysis
Total cellular RNAs from each cancer cell line were isolated using Trizol reagent (Takara Bio Inc., Shiga, Japan). Extracted RNA was reversely transcribed to cDNA by using Prime Script RT Reagent Kit (Takara Bio Inc., Shiga, Japan). Real-time PCR analysis was carried out in Applied Biosystems 7500 real-time PCR System by using SYBR Premix Ex-Taq II Kit (Takara Bio Inc., Shiga, Japan). Data were analyzed using the ΔΔCt method. The specific primers used for real-time PCR analysis were demonstrated in Table S2.
2.5. Lipid extraction, chromatographic and mass spectrometric conditions
For lipidomic analysis, the cells were grown in 6-well plates to approximately 90% confluence as previously described [27]. Briefly, the cells were quenched with liquid nitrogen and detached using cell scrapers. After two cycles of freeze–thaw, 1.2 mL of chilled extraction solvent (methanol/MTBE/H2O, 4:5:5, V/V/V) was added into 200 μL of cell suspension. After centrifugation at 1,000 rpm for 5 min at 4 °C, the mixtures were vortexed vigorously for 2 min and left on ice for 10 min. The supernatants were collected, and 200 μL of each sample was dried under nitrogen flow at room temperature, and stored at −20 °C until analysis. Finally, the residuals were resuspended with 100 μL of isopropanol/methanol (1:1, V/V) and injected for further UHPLC-ESI-MS-based lipodomics analysis.
Lipidomic chromatographic separation was carried out by using a reverse-phase Ascentis Express C18 100 mm × 2.1 mm 2.7 μm column (Sigma-Aldrich, St. Louis, MO, USA) on a Thermo Scientific Dionex Ultimate 3000 UHPLC system. Flow rate was set at 300 μL/min and column temperature was maintained at 45 °C. A gradient elution was employed with 50% acetonitrile in water with 5 mM ammonium formate and 0.1% formic acid (mobile phase A) and 5% acetonitrile in isopropanol with 5 mM ammonium formate and 0.1% formic acid (mobile phase B): 80% mobile phase A for 0.5 min, 20%–50% mobile phase B in 7 min, 50%–80% mobile phase B in the next 10 min, 80%–100% mobile phase B in 10 min and maintained 100% mobile phase B for 1.9 min, following 100%–20% mobile phase B for 0.1 min and 80% mobile phase A for 3 min. The quality controls for each cancer cell line were generated by pooling 20 μL of each sample and were injected intermittently during the whole process of sample injections.
Analysis of metabolites was performed with a Thermo Scientific Q ExactiveTM benchtop Orbitrap mass spectrometer with a heated ESI source in both positive and negative modes. Untargeted lipidomic profiling analyses acquired the data in the full MS-ddMS2 mode (100–1000 m/z), followed by top-10 data-dependent MS/MS at 17,500 FWHM resolution. The other main parameters for MS/MS included AGC target 1e6; maximum IT 65 ms; normalized step collision energy 25 and 35 for positive ion mode and 20, 30, and 40 for negative ion mode; apex trigger 5–10 s; and dynamic exclusion 10 s. The spray voltage was operated at 3.5 kV, and ion transfer capillary was set to 300 °C. Chromatograms and mass spectra from the UHPLC-ESI-MS runs were processed as raw files in Xcalibur (Thermo Scientific, San Jose, CA, USA).
2.6. Lipidomic data processing
Identification and relative quantification of the lipid molecular species were assisted by use of LipidSearch software (Thermo Scientific, San Jose, CA, USA) as reported previously [27]. The precursor tolerance was set to 5 ppm mass window while 10 ppm mass window was chosen as the product tolerance. The m-score threshold was set to 5.0. For multivariate principal component analysis (PCA), the raw data containing aligned peak areas with matched mass-to-charge ratios and retention times were transformed into a multivariate matrix and analyzed by SIMCA 13.0 software (Umetrics, Kinnelon, NJ, USA). Then supervised orthogonal partial least squares-discriminant analysis (OPLS-DA) was performed. The major lipid ions that contributed to the separation of sample groups were shown in the loading scatter S-plots.
2.7. Oil red O staining and Nile red fluorescence measurement
For oil red O staining, the cells were washed with PBS and then fixed with 10% formaldehyde for 30 min at room temperature. Stock solution of oil red O (0.5%) was diluted to 60% isopropyl alcohol with water (3:2, V/V) and used to stain cells for 15 min in the dark, followed by washing carefully with water twice. After dyeing, images were obtained immediately under an inverted fluorescence microscope (Olympus, Tokyo, Japan) at 200 × magnification.
For Nile red fluorescence measurement, the cells were transfected for 72 h, washed with PBS and then fixed with 4% paraformaldehyde for 10 min. After adding 100 μL of 5 μg/mL Nile red solution and incubating for 15 min, the cells were washed gently with PBS twice and the fluorescence intensity was determined immediately (excitation wavelength at 543 nm and emission wavelength at 598 nm).
2.8. Malondialdehyde (MDA) and ROS assay
MDA assay was performed according to the manufacturer's protocol (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Seventy -two h after transfection, a specified extraction solvent was added and cells were harvested after centrifugation and the protein was quantified. Glacial acetic acid and other reagents in the kit were added before heating for 40 min in a 95 °C water bath. The absorbance was measured at 530 nm wavelength. ROS assay was conducted using a DCFH-DA fluorescent probe (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) and the final concentration was 10 μM. After incubation at 37 °C for 60 min, the fluorescence intensity was detected. The excitation wavelength was set at 485 nm while the emission wavelength was 525 nm.
2.9. ATP luminescent cell viability assay and mitochondrial transmembrane potential experiment
For ATP production measurements, 72 h after siRNA transfection, the cell plates were taken out at room temperature for 30 min and then 100 μL of CellTiter-Glo was added according to the manufacturer's protocol (Promega Corporation, Madison, MI, USA). Cells were fully cracked by slowly shaking for 2 min and incubated for 10 min at room temperature to stabilize the luminescence signal. Luminescence intension was recorded by a Flex Station 3 multifunctional enzyme marker device. The mitochondrial transmembrane potential was analyzed by fluorescence dequenching of rhodamine (rh) 123 (Sigma, St. Louis, MO, USA) with ArrayScanVTI High Content Application (Thermo Fisher, Waltham, MA, USA). The excitation wavelength was set at 485 nm while the emission wavelength was 535 nm.
2.10. Statistical analysis
All values of this study were expressed as mean ± SEM. Statistical analysis was conducted by SPSS and Two-tailed Student's t tests. Graphs were performed using GraphPad Prism v6.0c software (San Diego, CA, USA). Significance was presented by ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 vs. the control.
3. Results
3.1. Knockdown of CPT1C induces cellular senescence in cancer cells
Transfection with siRNA for 72 h was applied to knock down CPT1C in PANC-1, MDA-MB-231, HCT-116 and A549 cells and significant reduction of CPT1C mRNA level was found in all cell lines (Fig. S1). Combined with previous interfering efficiency results [24], CPT1C siRNA-3 for PANC-1, HCT-116 and A549 cells and CPT1C siRNA-1 for MDA-MB-231 cells were chosen for the following experiments.
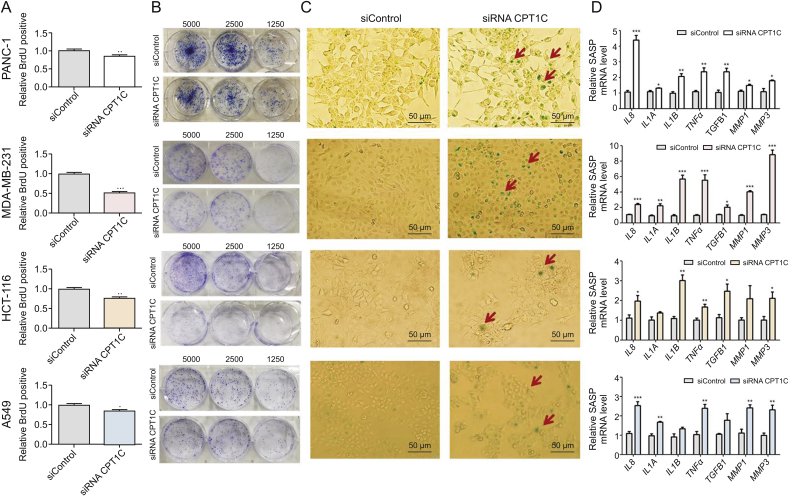
Cellular senescence was monitored by BrdU incorporation, colony forming ability, SA-β-gal activity and mRNA expression of SASP. Lower colorimetric immunoassay, weaker capacity to form colonies, and enhanced SA-β-gal activity were observed in CPT1C siRNA-treated group (Fig. 1). Additionally, the expression levels of typical SASP factors, including interleukin 8 (IL8), IL1A, IL1B, matrix metallopeptidase 1 (MMP1) and MMP3 mRNAs were markedly increased when CPT1C was knocked down in cancer cells. Taken together, these results confirmed that knockdown of CPT1C induced cellular senescence in cancer cells.
Fig. 1.
CPT1C knockdown induces cellular senescence in PANC-1, MDA-MB-231, HCT-116 and A549 cells. (A) BrdU incorporation activity during DNA synthesis. Data are represented as mean ± SEM, n = 5/group (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. the siControl group). (B) Cell colony formation ability by staining with crystal violet after cultured at indicated dilutions. (C) Senescence associated β-gal activity assay. (D) qPCR analysis of senescence-associated secretory phenotype (SASP) mRNAs. Data are represented as mean ± SEM, n = 4/group (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. the siControl group). BrdU: bromodeoxyuridine; CPT1C: carnitine palmitoyltransferase 1C; SASP: senescence-associated secretory phenotype; IL: interleukin; TNFα: tumor necrosis factor alpha; TGFB1: transforming growth factor beta; MMP: matrix metallopeptidase.
3.2. Knockdown of CPT1C significantly changes the lipidome of cancer cells
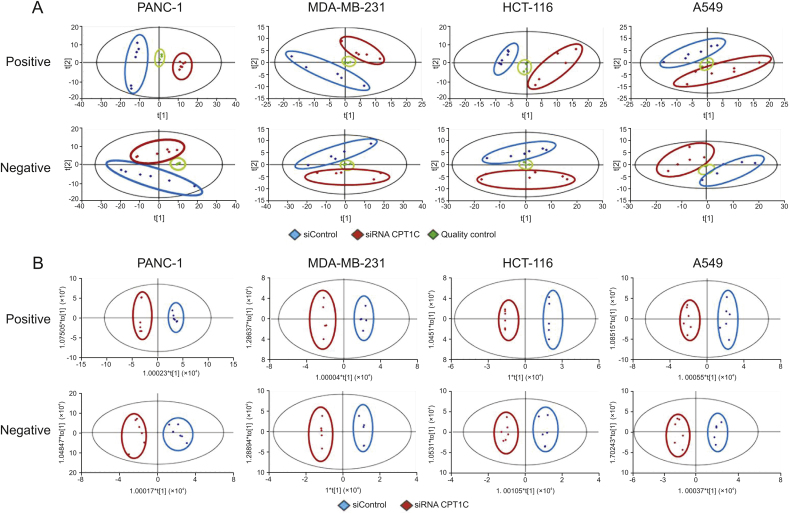
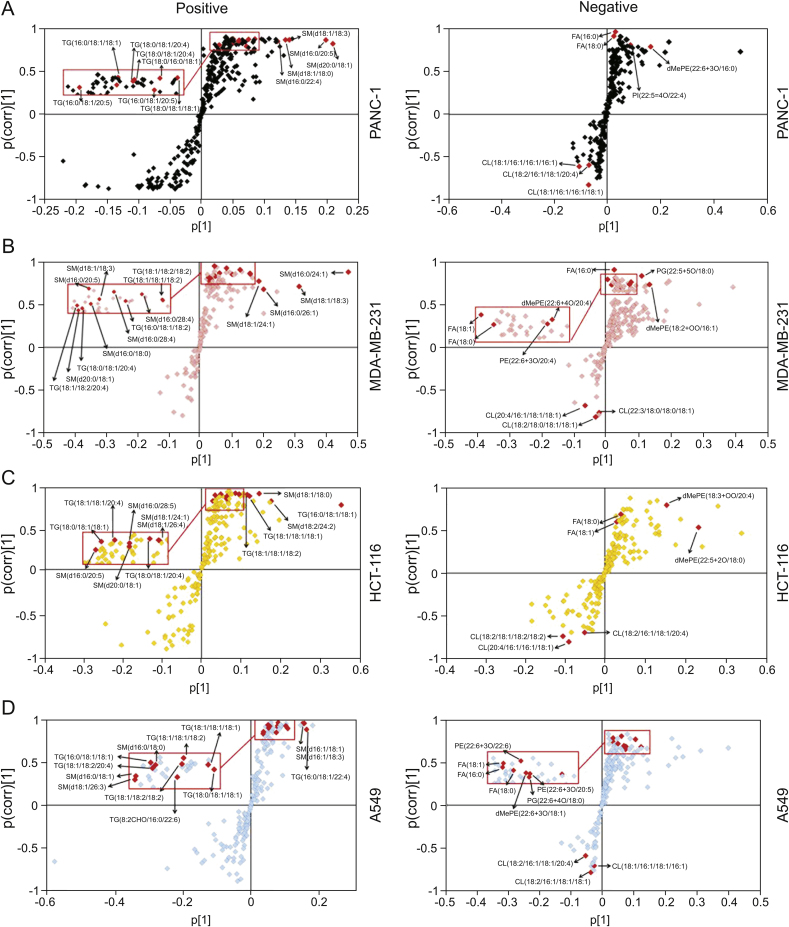
To comprehensively investigate whether knockdown of CPT1C could affect the lipidome of these cell lines, lipidomic analysis was carried out in CPT1C siRNA-treated cancer cells. PCA was performed to uncover the differences in the lipidomic profiles under both positive and negative ion modes. Data points of quality controls coalesced, which indicated stability and reproducibility of the UHPLC-ESI-HRMS operation system (Fig. 2A). PCA showed clear separation between siControl-treated and CPT1C siRNA-treated cancer cells, indicating distinct discrimination in the lipidomes between the two groups. OPLS-DA and loading S-plots were conducted to reveal the contribution of each detected ion to the lipidomic distinctions. OPLS-DA separated well the siRNA CPT1C group from the siControl group among all samples (Fig. 2B). In the S-plot, the ions that contributed most to the discrimination were selected and identified with the aid of Lipidsearch software (Fig. 3). The major lipid ions were identified as fatty acids (FA), triacylglycerol (TG), sphingomyelin (SM) and cardiolipin (CL).
Fig. 2.
Principal component analysis (PCA) and OPLS-DA score plots of PANC-1, MDA-MB-231, HCT-116 and A549 cells after knockdown of CPT1C under positive and negative ion modes. (A) PCA score plots of lipidomic profiles obtained from LC-MS/MS, n = 6/group. (B) Orthogonal partial least squares-discriminant analysis (OPLS-DA) score plots of lipidomic profiles obtained from LC-MS/MS, n = 6/group.
Fig. 3.
OPLS-DA loading S-plots of PANC-1 (A), MDA-MB-231 (B), HCT-116 (C) and A549 (D) cells after knockdown of CPT1C. Data were obtained from LC-MS/MS under positive and negative ion modes, n = 6/group. Each point represents an individual lipid ion. The major significant lipid ions are labeled in the S-plots.
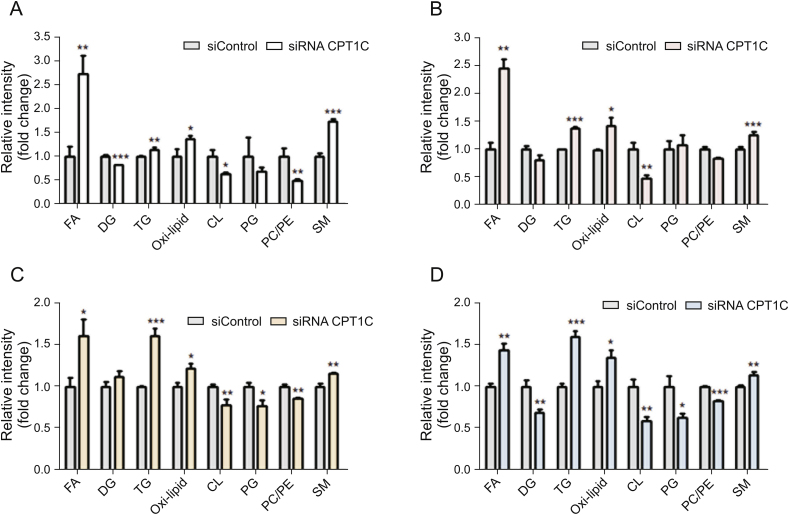
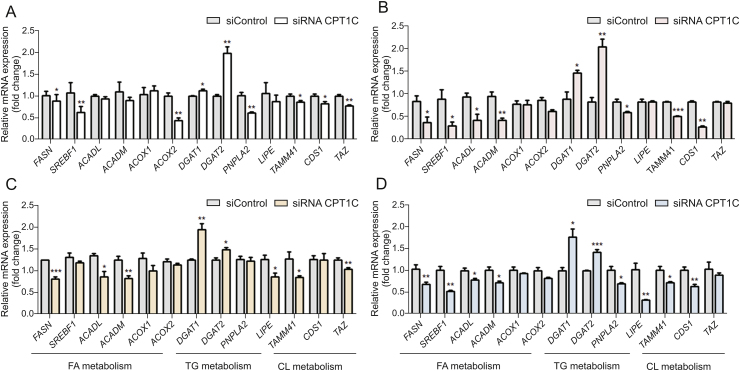
The levels of specific lipid species and mRNA involved in lipid metabolism were measured and compared with the negative control. The total FA contents were remarkably increased in cancer cells when knocking down CPT1C (Fig. 4). The mRNA levels of genes related to FA synthesis and oxidation, including fatty acid synthase (FASN), sterol regulatory element-binding transcription factor 1 (SREBF1), acyl-CoA dehydrogenase long chain (ACADL), acyl-CoA dehydrogenase medium chain (ACADM), acyl-CoA oxidase 1 (ACOX1) and ACOX2, were decreased in different degrees (Fig. 5). The levels of diacylglycerol (DG) were decreased whereas the levels of TG were increased, which was consistent with the elevated expressions of diacylgycerol acyltransferase 1 (DGAT1) and diacylgycerol acyltransferase 2 (DGAT2) and decreased expressions of patatin-like phospholipase domain-containing protein 2 (PNPLA2) and lipase E (LIPE). Higher levels of FA and TG revealed lipid accumulation after knockdown of CPT1C. Furthermore, total contents of oxi-lipid showed trends of increase, suggesting possible lipid peroxidation and oxidative stress. Moreover, reduced levels of mitochondrial characteristic lipids, CL and phosphatidylglycerol (PG), were found in the CPT1C siRNA group, indicating impaired mitochondrial function. In addition, lower ratios of phosphatidylcholine/phosphatidylethanolamine (PC/PE) and higher levels of SM were also observed in CPT1C knockdown cells (Fig. 4). The detailed information of FA, TG, oxi-lipid, CL and SM (different FA chains of carbon number and degree of unsaturation) with significant difference between the siRNA CPT1C group and the siControl group in four cancer cell lines is listed in Tables S3-S6.
Fig. 4.
Comparison of cellular content of lipid species in PANC-1 (A), MDA-MB-231 (B), HCT-116 (C) and A549 (D) cells after CPT1C knockdown. Data are represented as mean ± SEM, n = 6/group (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. siControl group). Fold change means the siRNA CPT1C vs. the siControl group. FA: fatty acids; DG: diglyacylglycerol; TG: triacylglycerol; oxi-lipid: oxidized lipids; CL: cardiolipin; PG: phosphatidylglycerol; PC/PE: phosphatidylcholine/phosphatidylethanolamine; SM: sphingomyelin.
Fig. 5.
Gene expressions related to the homeostasis of the above lipids in PANC-1 (A), MDA-MB-231 (B), HCT-116 (C) and A549 cells (D) upon knockdown of CPT1C. Data are represented as mean ± SEM, n = 4/group (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. siControl group). Fold change means the siRNA CPT1C vs. the siControl group. FASN: fatty acid synthase; SREBF1: sterol regulatory element-binding transcription factor 1; ACADL: acyl-CoA dehydrogenase long chain; ACADM: acyl-CoA dehydrogenase medium chain; ACOX: acyl-CoA oxidase; DGAT: diacylgycerol acyltransferase; PNPLA2: patatin-like phospholipase domain containing protein 2; LIPE: lipase E; TAMM41: TAM41 mitochondrial translocator assembly and maintenance homolog; CDS1: CDP-diacylglycerol synthase 1; TAZ: tafazzin.
Overall, these data suggested that knockdown of CPT1C could significantly change the lipidome and affect cancer cell lipid homeostasis. The dysregulation of lipid homeostasis could impact the normal physiological function of cells.
3.3. Knockdown of CPT1C induces lipid accumulation, lipotoxicity and mitochondrial dysfunction
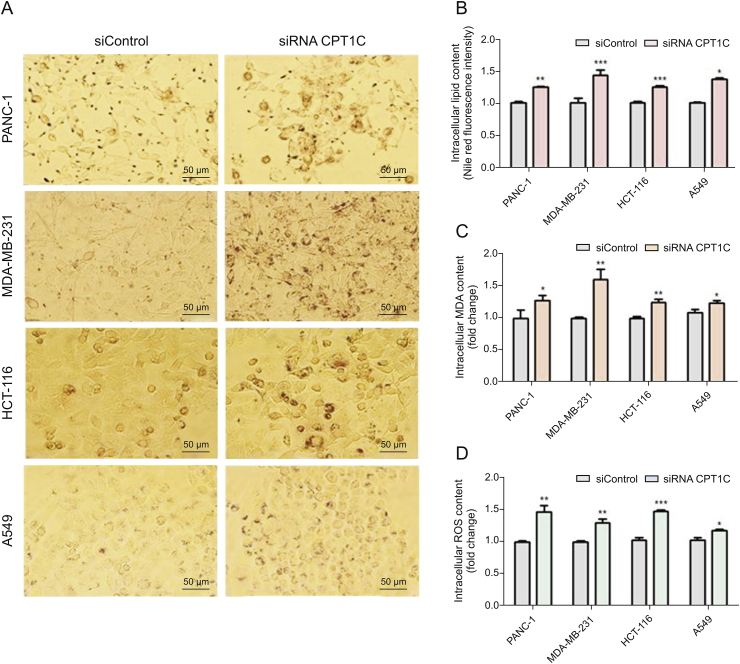
Since the lipidomic data indicated that knockdown of CPT1C changed lipid metabolism, cellular lipotoxic assays were performed to further understand the relationship between CPT1C and lipotoxicity. Oil red O staining showed a significant increase of lipid droplets in CPT1C siRNA-treated cancer cells (Fig. 6A). Nile red fluorescence intensity measurements revealed elevated lipid contents in CPT1C siRNA-treated cells as compared with the siControl-treated cells (Fig. 6B). Contents of the typical toxic lipid marker malondialdehyde (MDA) were increased significantly, suggesting higher levels of lipid peroxidation (Fig. 6C). Also, higher level of ROS was observed, which indicated disturbed balance of oxidation and reduction, and oxidative stress (Fig. 6D). In brief, these results demonstrated that knockdown of CPT1C could induce lipid accumulation and lipotoxicity.
Fig. 6.
Knockdown of CPT1C induces lipid accumulation and lipotoxicity in PANC-1, MDA-MB-231, HCT116 and A549 cells. (A) Oil red O staining of CPT1C knockdown cancer cells, n = 3/group. (B) Nile red fluorescence intensity measurement of cancer cells. Data are represented as mean ± SEM, n = 5/group (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. siControl group). (C) MDA concentration of cancer cells. Data are represented as mean ± SEM, n = 5/group (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. siControl group). (D) Intracellular ROS content measurement of cancer cells upon knockdown of CPT1C. Data are represented as mean ± SEM, n = 5/group (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. siControl group). Fold change means the siRNA CPT1C vs. the siControl group.
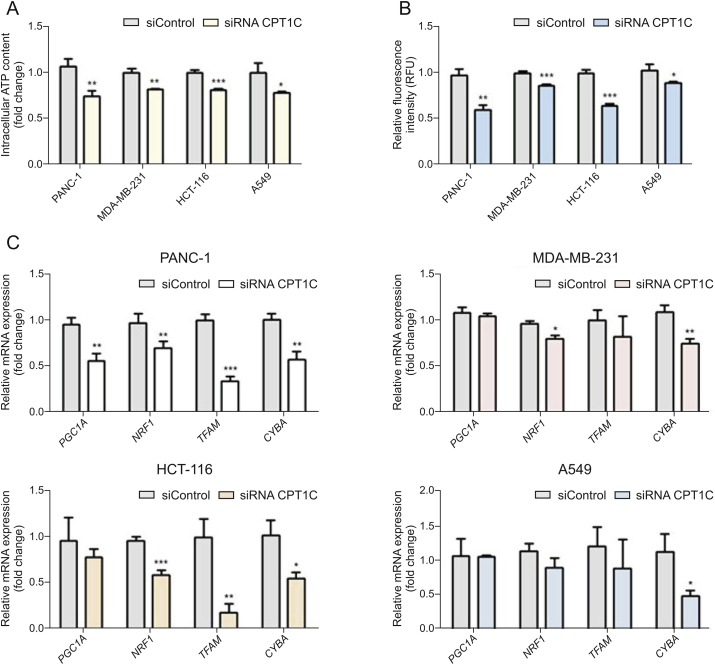
Since mitochondria is the main site of lipid metabolism and ROS production, the accumulation of lipid peroxides in mitochondria and the occurrence of oxidative stress can cause damage to mitochondrial structure and function [14]. ATP is the most direct energy source of cells and most of ATP is generated in mitochondria, while mitochondrial membrane potential is a necessary prerequisite to guarantee mitochondrial respiratory function [28]. Therefore, ATP production and mitochondrial membrane potential are important indicators of mitochondrial function. Lower levels of ATP and decreased mitochondrial membrane potential indicated mitochondrial dysfunction (Figs. 7A and B). Furthermore, lipid peroxides could have lipotoxic effects on mitochondrial DNA (mtDNA). Thus, mRNAs encoded by genes related to mtDNA synthesis were measured. Expressions of peroxisome-proliferator-activated receptor γ coactivator-1α (PGC1A), nuclear respiratory factor 1 (NRF1), transcription factor A mitochondrial (TFAM) and cytochrome b (CYBA) mRNAs were decreased to different extents after knockdown of CPT1C, indicating impaired mtDNA function (Fig. 7C).
Fig. 7.
CPT1C knockdown induces mitochondrial dysfunction in PANC-1, MDA-MB-231, HCT-116 and A549 cells. (A) ATP synthesis in cancer cells upon knockdown of CPT1C. (B) Loss of mitochondrial transmembrane potential measured by the rh123 dequenching method upon CPT1C siRNA. Data are represented as mean ± SEM, n = 5/group (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. siControl group). (C) Mitochondriogenesis mRNA expression in CPT1C knockdown cells. Data are represented as mean ± SEM, n = 4/group (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. siControl group). Fold change means the siRNA CPT1C vs. the siControl group. PGC1A: peroxisome-proliferator-activated receptor γ coactivator-1α; NRF1: nuclear respiratory factor 1; TFAM: transcription factor A mitochondrial; CYBA: cytochrome b.
4. Discussion
Cellular senescence can suppress the proliferation of tumor cells and inhibit the occurrence and development of tumors [29]. Our previous studies revealed that knockdown of CPT1C could induce cellular senescence in PANC-1, MDA-MB-231, HCT-116 and A549 cells [24,26]. In the current study, lipidomic analysis of these four cancer cell lines after CPT1C knockdown was performed. Cellular lipotoxicity was detected through a series of lipotoxic assays. The results demonstrated that loss of CPT1C could significantly change the lipidome, induce lipotoxicity, and further lead to mitochondrial dysfunction and senescence, suggesting universality in relationship between CPT1C, lipotoxicity and senescence in different cancer cells. CPT1C depletion may serve as a new therapeutic strategy through induction of cellular lipotoxicity and senescence.
It has been reported that CPT1C is an outer integral membrane protein enriched in hypothalamus with mitochondrial localization [30]. In our previous study [24], co-localization of CPT1C with mitochondria in PANC-1 cells was detected with immunofluorescence assays, which supports our conclusion that CPT1C is a key regulator of mitochondrial dysfunction and cellular senescence [24,25]. However, it is also found that CPT1C is localized in the endoplasmic reticulum (ER) of neurons [30]. Although the protein sequences of CPT1C containing necessary catalytic reaction sites share high similarity with CPT1A and CPT1B, the subcellular localization results showed CPT1C is also likely to be located in ER and microsomal structure [17]. Furthermore, whether CPT1C has CPT capacity remains controversial. When large amounts of CPT classic catalytic substrates such as carnitine and acyl-CoA esters were added into CPT1C-high expressed yeast, no acyltransferase activity was observed [31]. However, it is proved that CPT activity could be detected in overexpression of CPT1C PC12 cells and acyl-CoA is the catalytic substrate of CPT1C, but the catalytic activity of CPT1C was much lower than that of CPT1A and CPT1B [32]. Therefore, compared with CPT1A and CPT1B, CPT1C may possess other unique physiological functions, which needs to be further clarified.
Senescent cells exert several recognized features such as enhanced SA-β-gal activity, lower proliferation capacity, and weaker capacity to form colonies. However, not all aging cells exhibit all known senescent characteristics. Therefore, in general, at least three indicators should be combined to investigate whether cell senescence occurs [3]. In the present study, lower BrdU proliferation capacity, weaker capacity to form colonies, and enhanced SA-β-gal activity and SASP expression levels were observed in PANC-1, MDA-MB-231, HCT-116 and A549 cells after CPT1C knockdown.
Compared with that of normal cells, lipid homeostasis of senescent cells also changes significantly [7]. Earlier studies have shown that abnormal lipid metabolism can serve as a new mechanism of metabolic transformation of tumor cells, and importantly, small changes in abundance, composition, or location of lipids can have profound effects on cell viability and function [33]. Lipidomics can systematically and comprehensively analyze the species and content of most lipid molecules, and thus has been widely used in the discovery of disease biomarkers, drug targets and related mechanisms [34]. Given the close relationship between senescence, CPT1C, fatty acid oxidation and transportation, a more extensive lipidomic analysis was carried out to elucidate the lipids affected by CPT1C levels.
PCA and OPLS-DA showed that knockdown of CPT1C could change the cancer cell lipidomes. S-plot analysis further revealed that the major lipid ions contributing to the difference between control and CPT1C siRNA-treated cells were FA, TG, SM and CL. Free FA refers to non-esterified FA which can be released by hydrolysis of TG, phospholipid and other lipids. FA can also be synthesized by FASN, which is regulated by SREBF1, and SREBF1 could be a potential gatekeeper for lipotoxicity [35]. FA oxidation is mainly carried out in mitochondria by ACADL, ACADM, ACOX1, ACOX2 and other FA oxidases. The major physiological functions of FA include energy supply via β-oxidation, structural composition of biofilm and formation of lipid signaling molecules [33,36]. Although at present, while it is not known whether CPT1C participates in FA oxidation, there are data supporting the view that CPT1C can affect FA metabolism. For example, after 14 weeks of high-fat diet, Cpt1c knockout mice gained weight and their non-esterified FA contents in serum increased significantly as compared to wild-type mice. Under starvation, the FA oxidation rate of wild-type and Cpt1c null mice was both increased, but the increase rate in cpt1c-null group was much lower than that in wild-type group [19,37]. Moreover, it was found that the FA oxidation rate of MCF7 cells was significantly increased after overexpression of CPT1C [22]. The current study showed that total FA content of tumor cells increased dramatically after CPT1C was knocked down, particularly of FA (16:0), FA (18:0) and FA (18:1). The accumulation of these saturated FA and monounsaturated FA in cells may cause mitochondrial damage. Excess FA and oxidative damage could induce biofilm damage and associated aging and metabolic diseases [38].
On the other hand, FASN, SREBF1, ACADL, ACADM, ACOX1 and ACOX2 mRNAs related to FA synthesis and oxidative metabolism were significantly reduced in the CPT1C siRNA cells, consistent with the report that excess saturated FA could inhibit the synthesis of endogenous FA through negative feedback regulation and disrupt FA generation and metabolic balance [36]. TG is a conjugate of glycerol with FA of various lengths and compositions that store and provide energy for cells. When cellular FA exceeds the requirement of synthesis and catabolism, FA can be converted into TG by esterification enzymes and stored in lipid droplets. Therefore, the increase of TG content can also indicate the accumulation of FA in cells to some extent [39]. TG accumulation during replicative senescence and the observation of excessive TGs in a nonadipose tissue were identified as markers of lipotoxicity [7]. CPT1C promotes the storage of acyl-CoA of TG in neurons, rather than the oxidation process [40]. A previous study reported that FA and TG accumulated in senescent PANC-1 cells [27]. In the present study, the total TG contents in CPT1C knockdown cells showed an increasing trend while lower levels of DG were observed. After CPT1C knockdown, the DGAT1 and DGAT2 mRNAs involved in synthesis were increased, while PNPLA2 and LIPE mRNAs encoding hydrolytic enzymes were decreased, suggesting that knockdown of CPT1C promotes the synthesis of TG and inhibits its hydrolysis, resulting in the accumulation of TG in cells. Furthermore, Oil red O staining and Nile red fluorescence intensity measurement results indicated increased lipid droplets and elevated lipid contents in cancer cells upon knockdown of CPT1C. These data demonstrated that knockdown of CPT1C could significantly affect the metabolism of FA and TG, and induce lipid accumulation in cancer cells.
Lipidomic data showed that the oxidative lipids in tumor cells treated with CPT1C siRNA were higher than those in the siControl-treated group. The significant increase of lipid peroxide content suggested that tumor cells have increased oxidative damage after CPT1C knockdown. Lipid peroxidation is the result of hydroxyl radical attack of fatty acyl chains of lipids, and has widespread toxic effects on cellular function. Although almost all intracellular organelles and compartments produce ROS, hydrogen peroxide generated by mitochondria is generally considered to be the main source [41]. Under normal circumstances, there is a dynamic balance between ROS production and defense system. When intracellular lipids accumulate, ROS may react with these lipids, induce lipid peroxidation and then produce a series of lipid peroxidation metabolites, including MDA. With the continuous production and accumulation of lipid peroxides, ROS also rapidly increases, while the clearance capacity is significantly reduced. The balance of oxidation and antioxidant capacity is disturbed, followed by oxidative stress, which causes oxidative damage to cell macromolecules and eventually leads to lipotoxicity, cell dysfunction and senescence [8]. CPT1C is also related to lipid peroxidation and oxidative stress. Takotsubo syndrome patients show accumulation of lipids and lipid peroxidation reactions, and the levels of CPT1C mRNA are significantly reduced [42]. In an untargeted metabolomic study of Cpt1c-null mice, the content of oxidized glutathione in Cpt1c-null group was significantly increased, suggesting the generation of oxidative stress in the brain [43]. To further investigate the effects of lipid accumulation on lipid toxicity, total MDA and ROS contents of CPT1C knockdown cells were examined. Consistently, compared with those in the control group, significantly higher levels of MDA and ROS were observed, indicating that knockdown of CPT1C could induce lipid peroxidation and oxidative stress.
Since mitochondria are the main site of FA oxidation and ROS production, the accumulation of lipid peroxides in mitochondria can cause serious lipid toxic damage to mitochondrial structure and function, and lead to cell dysfunction [14]. In the current study, a series of assays were conducted to evaluate mitochondrial function. A reduction of mitochondrial characteristic lipids, CL and PG, were found in the siRNA CPT1C-treated cells. Diphosphatidyl glycerol (CL), also known as cardiac phospholipids, is a characteristic lipid associated with mitochondrial function (oxidative phosphorylation, respiratory chain) and apoptosis [44]. Reduction of CL in senescent cells was also reported, indicating that CL content is related to cell senescence [45]. PG is a precursor molecule for the synthesis of CL and is responsible for the mitochondrial respiratory chain activity. Since CL is located in the mitochondrial inner membrane, it is particularly susceptible to lipid peroxidation. After the peroxidation of the most common mitochondrial CL-tetramethylacyl CL in mammalian tissues, not only the lipid hydrogen peroxide in the membrane accumulated, but also the mitochondrial toxic metabolite 4-hydroxynonenal was produced. Peroxidation of CL is often accompanied by a decrease in the activity of complexes I and IV, resulting in impaired mitochondrial respiration [46]. CL synthesis is mainly catalyzed by TAM41 mitochondrial translocator assembly and maintenance homolog (TAMM41), CDP-diacylglycerol synthase 1 (CDS1), and the spatial configuration arrangement of CL needs to be catalyzed by tafazzin (TAZ) [44]. The current study showed that after CPT1C knockdown, TAMM41, CDS1 and TAZ mRNAs were decreased, which was consistent with significantly decreased CL contents. Lower levels of ATP and decreased mitochondrial membrane potential were also found upon CPT1C knockdown. Furthermore, genes related to mtDNA synthesis were detected. Biosynthesis of mtDNA was mainly regulated by the PGC1A-NRF1-TFAM pathway. When the expressions of PGC1A was inhibited, the expressions of its downstream signaling molecules, such as NRF1 and TFAM, were inhibited, which affected mtDNA replication. Cytochrome b, a representative subunit of mtDNA encoding gene, can reflect the copy capacity of mtDNA. The position of mtDNA in mitochondria is relatively bare and vulnerable to oxidative damage, and therefore the decreased biosynthesis capacity of mtDNA indicates mtDNA damage [47,48]. In the present study, the expressions of PGC1A, NRF1, TFAM and CYBA (cytochrome b) mRNAs were decreased to various extents, reflecting impaired mtDNA function. Taken together, these results showed that knockdown of CPT1C could induce lipotoxicity and mitochondrial dysfunction, which is consistent with the results of a previous finding that CPT1C regulates cancer cell senescence through mitochondria-associated metabolic reprograming [24].
In addition, a lower ratio of PC/PE and higher levels of SM were also observed in CPT1C knockdown cells. PC and PE, the most common phospholipids within cells, are the main structural lipids in biofilms. Specifically, membrane fluidity and permeability can be damaged by oxidation of lipid membranes. Alteration of membrane components (PC/PE) can reduce membrane mobility, increase stiffness and permeability, resulting in senescence [49,50]. SM is also the major structural component of cell membrane, and its metabolites, ceramide and sphingosine, are bioactive signaling molecules [51]. It was reported that the SM content was increased during senescence, but the specific relationship and mechanism of SM and senescence remains unknown [52]. Ceramide can regulate cell proliferation and apoptosis, and neuronal ceramide metabolism can be regulated by CPT1C [53]. However, in this study, there was no statistical difference of the ceramide after CPT1C knockdown, suggesting that silencing CPT1C had little effect on tumor cell ceramide metabolism.
Knockdown of other CPT members also has been investigated. Lipid accumulation was related to CPT1A mRNA expression in skeletal cells [54]. Lipids accumulated and abnormal mitochondrial function was noted in cardiac muscle of Cpt1b+/- mice, demonstrating that absence of Cpt1b can aggravate the cardiac hypertrophy possibly due to lipotoxicity [55]. While in cancer cells, combined with previous results [26], CPT1C exerted a significant role in mitochondria-associated senescence via lipotoxicity compared with other CPT isoforms.
In summary, the present study demonstrates that CPT1C plays a crucial role in the regulation of cancer cell lipotoxicity and cell senescence. For the reason that the role of Cpt1c in FA metabolism remains largely unresolved, further studies are warranted to provide more information about the relationship and molecular mechanisms of CPT1C, lipotoxicity and senescence. This study suggests that inhibition of CPT1C may serve as a new therapeutic strategy through induction of tumor lipotoxicity and senescence.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
The work was supported by the National Key Research and Development Program of China (Grant No. 2017YFE0109900), the National Natural Science Foundation of China (Grant Nos. 82025034 and 81973392), the Shenzhen Science and Technology Program (Grant No. KQTD20190929174023858), the Natural Science Foundation of Guangdong (Grant No. 2017A030311018), the 111 project (Grant No. B16047), the Key Laboratory Foundation of Guangdong Province (Grant No. 2017B030314030), the Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (Grant No. 2017BT01Y093), and the National Engineering and Technology Research Center for New drug Druggability Evaluation (Seed Program of Guangdong Province, Grant No. 2017B090903004).
Footnotes
Peer review under responsibility of Xi’an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.04.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Hayflick L., Moorhead P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Salama R., Sadaie M., Hoare M. Cellular senescence and its effector programs. Genes Dev. 2014;28:99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Otin C., Blasco M.A., Partridge L. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muñoz-Espín D., Serrano M. Cellular senescence: from physiology to pathology. Nat. Rev. Mol. Cell Biol. 2014;15:482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 5.Campisi J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Lizardo D.Y., Lin Y.L., Gokcumen O. Regulation of lipids is central to replicative senescence. Mol. Biosyst. 2017;13:498–509. doi: 10.1039/c6mb00842a. [DOI] [PubMed] [Google Scholar]
- 8.Hauck A.K., Bernlohr D.A. Oxidative stress and lipotoxicity. J. Lipid Res. 2016;57:1976–1986. doi: 10.1194/jlr.R066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y., Hirose H., Ohneda M. Beta-cell lipotoxicity in the pathogenesis of non-insulin-dependent diabetes mellitus of obese rats: impairment in adipocyte-beta-cell relationships. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10878–10882. doi: 10.1073/pnas.91.23.10878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaffer J.E. Lipotoxicity: many roads to cell dysfunction and cell death: introduction to a thematic review series. J. Lipid Res. 2016;57:1327–1328. doi: 10.1194/jlr.E069880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biden T.J., Boslem E., Chu K.Y. Lipotoxic endoplasmic reticulum stress, beta cell failure, and type 2 diabetes mellitus. Trends Endocrinol. Metabol. 2014;25:389–398. doi: 10.1016/j.tem.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Bilinski T., Paszkiewicz T., Zadrag-Tecza R. Energy excess is the main cause of accelerated aging of mammals. Oncotarget. 2015;6:12909–12919. doi: 10.18632/oncotarget.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sies H. Oxidative stress: a concept in redox biology and medicine. Redox Biol. 2015;4:180–183. doi: 10.1016/j.redox.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schrauwen P., Schrauwen-Hinderling V., Hoeks J. Mitochondrial dysfunction and lipotoxicity. Biochim. Biophys. Acta. 2010;1801:266–271. doi: 10.1016/j.bbalip.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Gallage S., Gil J. Mitochondrial dysfunction meets senescence. Trends Biochem. Sci. 2016;41:207–209. doi: 10.1016/j.tibs.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Slawik M., Vidal-Puig A.J. Lipotoxicity, overnutrition and energy metabolism in aging. Ageing Res. Rev. 2006;5:144–164. doi: 10.1016/j.arr.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Casals N., Zammit V., Herrero L. Carnitine palmitoyltransferase 1C: from cognition to cancer. Prog. Lipid Res. 2016;61:134–148. doi: 10.1016/j.plipres.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Price N.T., van der Leij F.R., Jackson V.N. A novel brain-expressed protein related to carnitine palmitoyltransferase I. Genomics. 2002;80:433–442. doi: 10.1006/geno.2002.6845. [DOI] [PubMed] [Google Scholar]
- 19.Wolfgang M.J., Kurama T., Dai Y. The brain-specific carnitine palmitoyltransferase-1c regulates energy homeostasis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7282–7287. doi: 10.1073/pnas.0602205103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao X.F., Chen W., Kong X.P. Enhanced susceptibility of Cpt1c knockout mice to glucose intolerance induced by a high-fat diet involves elevated hepatic gluconeogenesis and decreased skeletal muscle glucose uptake. Diabetologia. 2009;52:912–920. doi: 10.1007/s00125-009-1284-0. [DOI] [PubMed] [Google Scholar]
- 21.Carrasco P., Jacas J., Sahun I. Carnitine palmitoyltransferase 1C deficiency causes motor impairment and hypoactivity. Behav. Brain Res. 2013;256:291–297. doi: 10.1016/j.bbr.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Zaugg K., Yao Y., Reilly P.T. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25:1041–1051. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez-Macedo N., Feng J., Faubert B. Depletion of the novel p53-target gene carnitine palmitoyltransferase 1C delays tumor growth in the neurofibromatosis type I tumor model. Cell Death Differ. 2013;20:659–668. doi: 10.1038/cdd.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Chen Y., Guan L. Carnitine palmitoyltransferase 1C regulates cancer cell senescence through mitochondria-associated metabolic reprograming. Cell Death Differ. 2018;25:735–748. doi: 10.1038/s41418-017-0013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y., Wang Y., Huang Y. PPARalpha regulates tumor cell proliferation and senescence via a novel target gene carnitine palmitoyltransferase 1C. Carcinogenesis. 2017;38:474–483. doi: 10.1093/carcin/bgx023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan L., Chen Y., Wang Y. Effects of carnitine palmitoyltransferases on cancer cellular senescence. J. Cell. Physiol. 2019;234:1707–1719. doi: 10.1002/jcp.27042. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H., Gao Y., Sun J. Optimization of lipid extraction and analytical protocols for UHPLC-ESI-HRMS-based lipidomic analysis of adherent mammalian cancer cells. Anal. Bioanal. Chem. 2017;409:5349–5358. doi: 10.1007/s00216-017-0483-7. [DOI] [PubMed] [Google Scholar]
- 28.Chinopoulos C., Adam-Vizi V. Mitochondria as ATP consumers in cellular pathology. Biochim. Biophys. Acta. 2010;1802:221–227. doi: 10.1016/j.bbadis.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Campisi J. Aging and cancer: the double-edged sword of replicative senescence. J. Am. Geriatr. Soc. 1997;45:482–488. doi: 10.1111/j.1532-5415.1997.tb05175.x. [DOI] [PubMed] [Google Scholar]
- 30.Dai Y., Wolfgang M.J., Cha S.H. Localization and effect of ectopic expression of CPT1c in CNS feeding centers. Biochem. Biophys. Res. Commun. 2007;359:469–474. doi: 10.1016/j.bbrc.2007.05.161. [DOI] [PubMed] [Google Scholar]
- 31.Price N., van der Leij F., Jackson V. A novel brain-expressed protein related to carnitine palmitoyltransferase I. Genomics. 2002;80:433–442. doi: 10.1006/geno.2002.6845. [DOI] [PubMed] [Google Scholar]
- 32.Sierra A.Y., Gratacos E., Carrasco P. CPT1c is localized in endoplasmic reticulum of neurons and has carnitine palmitoyltransferase activity. J. Biol. Chem. 2008;283:6878–6885. doi: 10.1074/jbc.M707965200. [DOI] [PubMed] [Google Scholar]
- 33.Currie E., Schulze A., Zechner R. Cellular fatty acid metabolism and cancer. Cell Metabol. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han X., Gross R.W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Williams K.J., Argus J.P., Zhu Y. An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity. Canc. Res. 2013;73:2850–2862. doi: 10.1158/0008-5472.CAN-13-0382-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calder P.C. Functional roles of fatty acids and their effects on human health. JPEN J. Parenter. Enter. Nutr. 2015;39:18S–32S. doi: 10.1177/0148607115595980. [DOI] [PubMed] [Google Scholar]
- 37.Wolfgang M.J., Cha S.H., Millington D.S. Brain-specific carnitine palmitoyl-transferase-1c: role in CNS fatty acid metabolism, food intake, and body weight. J. Neurochem. 2008;105:1550–1559. doi: 10.1111/j.1471-4159.2008.05255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ford J.H. Saturated fatty acid metabolism is key link between cell division, cancer, and senescence in cellular and whole organism aging. Age. 2010;32:231–237. doi: 10.1007/s11357-009-9128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong Y., Dou L.J., Liang J. Link between obesity and cancer: role of triglyceride/free fatty acid cycling. Eur. Rev. Med. Pharmacol. Sci. 2014;18:2808–2820. [PubMed] [Google Scholar]
- 40.Rinaldi C., Schmidt T., Situ A.J. Mutation in CPT1C associated with pure autosomal dominant spastic paraplegia. JAMA Neurol. 2015;72:561–570. doi: 10.1001/jamaneurol.2014.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farmer E.E., Mueller M.J. ROS-mediated lipid peroxidation and RES-activated signaling. Annu. Rev. Plant Biol. 2013;64:429–450. doi: 10.1146/annurev-arplant-050312-120132. [DOI] [PubMed] [Google Scholar]
- 42.Borchert T., Hübscher D., Guessoum C.I. Catecholamine-dependent β-adrenergic signaling in a pluripotent stem cell model of Takotsubo cardiomyopathy. J. Am. Coll. Cardiol. 2017;70:975–991. doi: 10.1016/j.jacc.2017.06.061. [DOI] [PubMed] [Google Scholar]
- 43.Lee J., Wolfgang M.J. Metabolomic profiling reveals a role for CPT1c in neuronal oxidative metabolism. BMC Biochem. 2012;13:23. doi: 10.1186/1471-2091-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlame M., Greenberg M.L. Biosynthesis, remodeling and turnover of mitochondrial cardiolipin. Biochim. Biophys. Acta. 2017;1862:3–7. doi: 10.1016/j.bbalip.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monteiro J.P., Oliveira P.J., Jurado A.S. Mitochondrial membrane lipid remodeling in pathophysiology: a new target for diet and therapeutic interventions. Prog. Lipid Res. 2013;52:513–528. doi: 10.1016/j.plipres.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Kiebish M.A., Han X., Cheng H. Cardiolipin and electron transport chain abnormalities in mouse brain tumor mitochondria: lipidomic evidence supporting the Warburg theory of cancer. J. Lipid Res. 2008;49:2545–2556. doi: 10.1194/jlr.M800319-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S., Fan Q., Li A. Dynamic mobilization of PGC-1alpha mediates mitochondrial biogenesis for the protection of RGC-5 cells by resveratrol during serum deprivation. Apoptosis. 2013;18:786–799. doi: 10.1007/s10495-013-0837-3. [DOI] [PubMed] [Google Scholar]
- 48.Hildenbeutel M., Hegg E.L., Stephan K. Assembly factors monitor sequential hemylation of cytochrome b to regulate mitochondrial translation. J. Cell Biol. 2014;205:511–524. doi: 10.1083/jcb.201401009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibellini F., Smith T.K. The Kennedy pathway--De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life. 2010;62:414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- 50.Fajardo V.A., McMeekin L., LeBlanc P.J. Influence of phospholipid species on membrane fluidity: a meta-analysis for a novel phospholipid fluidity index. J. Membr. Biol. 2011;244:97–103. doi: 10.1007/s00232-011-9401-7. [DOI] [PubMed] [Google Scholar]
- 51.Bienias K., Fiedorowicz A., Sadowska A. Regulation of sphingomyelin metabolism. Pharmacol. Rep. 2016;68:570–581. doi: 10.1016/j.pharep.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 52.Mielke M.M., Bandaru V.V., Han D. Factors affecting longitudinal trajectories of plasma sphingomyelins: the Baltimore Longitudinal Study of Aging. Aging Cell. 2015;14:112–121. doi: 10.1111/acel.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carrasco P., Sahun I., McDonald J. Ceramide levels regulated by carnitine palmitoyltransferase 1C control dendritic spine maturation and cognition. J. Biol. Chem. 2012;287:21224–21232. doi: 10.1074/jbc.M111.337493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meng Z.X., Yin Y., Lv J.H. Aberrant activation of liver X receptors impairs pancreatic beta cell function through upregulation of sterol regulatory element-binding protein 1c in mouse islets and rodent cell lines. Diabetologia. 2012;55:1733–1744. doi: 10.1007/s00125-012-2516-2. [DOI] [PubMed] [Google Scholar]
- 55.He L., Kim T., Long Q. Carnitine palmitoyltransferase-1b deficiency aggravates pressure overload-induced cardiac hypertrophy caused by lipotoxicity. Circulation. 2012;126:1705–1716. doi: 10.1161/CIRCULATIONAHA.111.075978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.