Abstract
Microorganisms are present in nearly every oil or bitumen sample originating from temperate reservoirs. Nevertheless, it is very difficult to obtain reliable estimates about microbial processes taking place in deep reservoirs, since metabolic rates are rather low and differ strongly during artificially cultivation. Here, we demonstrate the importance and impact of microorganisms entrapped in microscale water droplets for the overall biodegradation process in bitumen. To this end, we measured degradation rates of heavily biodegraded bitumen from the Pitch Lake (Trinidad and Tobago) using the novel technique of reverse stable isotope labeling, allowing precise measurements of comparatively low mineralization rates in the ng range in microcosms under close to natural conditions. Freshly taken bitumen samples were overlain with artificial brackish water and incubated for 945 days. Additionally, three-dimensional distribution of water droplets in bitumen was studied with computed tomography, revealing a water bitumen interface of 1134 cm2 per liter bitumen, resulting in an average mineralization rate of 9.4–38.6 mmol CO2 per liter bitumen and year. Furthermore, a stable and biofilm-forming microbial community established on the bitumen itself, mainly composed of fermenting and sulfate-reducing bacteria. Our results suggest that small water inclusions inside the bitumen substantially increase the bitumen–water interface and might have a major impact on the overall oil degradation process.
Keywords: asphalt, oil reservoirs, micro habitats, water droplets, isotope dilution, anaerobic biodegradation, petroleum mineralization, biofilm formation
Short abstract
Tiny water droplets in natural bitumen reservoirs substantially increase the bitumen−water interface and potentially impact the entire biodegradation process.
Introduction
Crude oil is one of the most important natural resources in our modern life but a major fraction of economically available oil exists as biodegraded heavy oil or bitumen.1−3 During the degradation process, n-alkanes, monocyclic alkanes, and alkyl benzenes disappear first, which leads to a lower oil quality and finally to natural bitumen mostly consisting of saturated and aromatic hydrocarbons, resins, and asphaltene.4−6 However, our knowledge about in situ biological processes and degradation rates in anoxic oil reservoirs is scarce due to the limited access to the deep subsurface.1,2 The most common metabolisms found in oil reservoirs are sulfate reduction, fermentation, acetogenesis, and methanogenesis.3,5,7,8 A broad range of metabolic pathways like iron(III)-, manganese(IV)-, nitrate-, or nitrite reduction have been detected in oil reservoirs as well.9,10 Nevertheless, their influence still remains unclear, since the microbial degradation of hydrocarbons in the environment is mostly limited by the availability of electron acceptors and the bioavailability of the hydrocarbons.3,11 Since electron acceptors in oil reservoirs are quickly depleted by microorganisms and do not get replenished unless water flooding is implemented, fermentation and methanogenesis are believed to be mainly responsible for the oil degradation in most reservoirs.9,10,12 Fermentative, syntrophic microorganisms degrade complex hydrocarbons like fatty acids, alcohols, sugars, amino acids, and aromatic compounds stepwise into smaller molecules such as hydrogen, acetate, and carbon dioxide.12−17 In addition, acetate is alternatively oxidized by syntrophic acetate oxidation.15−17 Many of these reactions are endergonic and are only energetically feasible if the products are kept at low concentrations.12,16 Acetate is commonly converted by acetoclastic methanogens into methane and carbon dioxide, whereas hydrogen and carbon dioxide are used by hydrogenotrophic methanogens to produce methane.12−17 However, sulfate-reducing bacteria (SRB) are present in practically every oil reservoir and are known for complete alkane (C3–C20) oxidation to carbon dioxide in the presence of sulfate.7,18 They are also capable of mineralizing aromatic hydrocarbon degradation such as benzene, toluene, naphthalene, 2-methylnaphthalene, phenanthrene, ethylbenzene, and xylene carbon dioxide.5,18−20
Even if forced in experimental setups, anaerobic biodegradation of oil is an extremely slow process which is difficult to assess. In the natural environment, biodegradation is assumed to be even slower or sometimes completely inhibited due to temperature and other abiotic factors. Therefore, we applied the relatively new method of reverse stable isotope labeling, enabling the measurement of degradation of unknown carbon sources even at extremely low rates.21−24 This robust and sensitive method is based on the dilution of a 13C-labeled bicarbonate buffer with nonlabeled carbon dioxide from biodegradation of hydrocarbons at natural isotopic abundance.21
The second major limitation for biodegradation of hydrocarbons is the poor bioavailability (i.e., water solubility and dissolution kinetics). Hence, biodegradation rates depend to a large extent on the mass transfer of hydrocarbons into the aqueous phase and the surface area of the oil–water interface is a limiting factor for oil degradation. This is supported by the common concept of biodegradation in crude oil reservoirs, indicating that microbial degradation takes place at the bottom of the oil-bearing leg at the oil–water-transition-zone and is, thus, limited by the interface area.25 However, it was discovered that bitumen contains small water droplets and pockets populated with oil-degrading microbial communities.26−28 Assuming that the area of the oil–water interface is a limiting factor for biodegradation in oil reservoirs, such water droplets might potentially increase the overall degradation kinetics by increasing the total oil–water surface area. When the transfer of hydrocarbons into the water phase is rate-limiting, microorganisms often accumulate at the substrate surface, here the oil–water interface, to overcome the transfer limitations. Several studies showed the formation of biofilms in the presence of oil, oil sands, or bitumen when incubated in artificial media containing additives like vitamins or trace elements with pure- or enrichment cultures.29−32
The aim of this study was to determine the impact of microbially populated microscale water droplets enclosed in natural bitumen on the overall biodegradation process. Therefore, surface dependent biodegradation rates of bitumen were analyzed and combined with size and distribution measurements of enclosed water droplets in bitumen. Our findings allowed us to calculate a theoretical degradation rate per intrabitumen interface area and indicate a notable amplification of the bitumen–water transition zone through microscale water droplets in bitumen phase.
Materials and Methods
Bitumen Smpling
Bitumen was sampled from the Pitch Lake (N 10°14′0.6882″, W 61°37′44.5638″) located in Trinidad, Trinidad and Tobago. Liquid bitumen freshly seeping on the Pitch Lake was sampled with decapitated 50 mL syringes and directly transferred into sterile microcosms. To avoid oxygen-contaminated samples, the bitumen samples were taken from 10 to 15 cm beneath the surface, after the spring was refilled by fresh bitumen from the subsurface. The microcosms were then flushed directly on site with N2 (5.0 grade; Massy Gas Products, Trinidad and Tobago) and sealed with butyl stoppers. After shipping, each microcosm was flushed again for 15 min with N2 (5.0 grade; sterile through upstream filters). The surface temperature of the sampled bitumen was ∼33.5 °C as measured on site with a PL-120-digital thermometer T2 (Voltcraft, Switzerland). The four microcosms 1–4 contained 109.03, 116.39, 64.42, and 130.03 g bitumen, respectively. Additional bitumen from the same seep was sampled in sterile glass jars for further analysis.
Microcosm Setup
To analyze the geochemistry of the enclosed water droplets, bitumen from additional glass jars was heated to 40–45 °C for ∼30 min. Aliquots of ∼15 g were transferred into 50 mL centrifugation tubes and centrifuged for 120 min at 3214g. The water on top of the bitumen was collected and the procedure was repeated until no water appeared after centrifugation. The anion and cation contents of the water were determined by ion chromatography (Dionex, CA, U.S.A.; Metrohm, Filderstadt, Germany) (for detailed sample preparation and measurement conditions see S1 of the Supporting Information, SI). The pH of extracted water was 7.9.
On the basis of the geochemical composition of the droplet water (see S2), an anoxic brackish water was prepared with a final concentration of 16.3 mM NH4Cl, 5.4 mM KCl, 1.1 mM CaCl2·2H2O, 1.3 mM MgCl2·6H2O, 173 mM NaCl, 9.7 mM NaHSO4, and 31.7 mM NaH2PO4·2H2O. In addition, 1 mM resazurin was added as redox indicator. Then, the brackish water was boiled to remove O2 and CO2. Subsequently, the bottle was sealed with a butyl stopper, flushed with Biogon (CO2/N2 = 20/80 (v/v); Air Liquide, Germany) and autoclaved. Na2S (sterile, anaerobic) was added as reducing agent to a final concentration of 0.5 mM (according to Widdel et al., 1981) to ensure an oxygen free environment.33 The brackish medium was buffered with a NaHCO3 buffer solution (sterile, anaerobic; final concentration 30 mM) and adjusted to pH 7, after injection into the microcosms the pH rose to ∼8 (close to the natural measured 7.9). The carbonate buffer was 13C-labeled with x(13C) = 10 atom % as a mixture of regular NaHCO3x(13C) = 1.11% (Carl Roth, Germany) and 13C-labeled NaHCO3x(13C) = 98% (Sigma-Aldrich, MO, U.S.A.).33 In total, 140 mL of the buffered and reduced brackish water was added without further supplements to each bitumen-containing microcosms 1–4 to artificially increase the present natural water volume. All microcosms were incubated at ∼32 °C.
Bitumen sterilization: In contrast to aerobic, abiotic bitumen oxidation (<125–135 °C), anoxic thermal alteration of bitumen starts to be quantitatively relevant at temperatures >200 °C. However, there are strong indications that decomposition of minor fraction occurs even at temperatures <200 °C.34−37 Most of the bitumen (∼90%) boils at temperatures >350 °C, nonetheless, even much lower temperatures (40–120 °C) strongly affect the bitumen viscosity which decreases while heating.38−41 A lower viscosity enables entrapped inorganic compounds with low boiling points, such as carbon dioxide, carbon monoxide, hydrogen sulfide, or carbonyl sulfide and smaller organic compounds present in bitumen such as neopentane, methane, acetaldehyde, propane, propylene, pyridine, and benzene, to degas from the bitumen.37,42−44 Therefore, autoclaving the bitumen likely leads to compound decompositions and changes of the matrix and certainly to a degassing of various compounds necessary for microbial growth. Another frequently used method for sterilization is γ radiation. However, several studies showed that bitumen is affected by γ radiation, resulting in radiolytic gas production (hydrogen) and swelling of the bitumen body causing changes of the overall composition.45−48 Chemical sterilization would also affect and change the bitumen matrix. Additionally, spore formers with unknown germination times are present in many oil fields which are difficult to monitor within the bitumen.49−51 Because of these unknown matrix effects, we did not setup sterile controls.
Assessing Bitumen Mineralization
The parameters carbon dioxide development, cell density, sulfate concentration, and pH were monitored during the 945 days of incubation at days 0, 42, 93, 140, 259, 470, 604, 877, and 945. All samples were taken anaerobically with N2 flushed syringes (5.0 grade; sterile Hungate needle).
For analyzing CO2-development, aqueous samples (0.5 mL) were taken with syringes through the stoppers and directly transferred into 12 mL Labco Exertainer vials (Labco Limited, U.K.) which were preamended with 100 μL of 85% phosphoric acid, closed with screw caps containing butyl rubber septa, and flushed with CO2-free synthetic air (6.0 grade; Air Liquide, Germany).21−23,52 Samples were analyzed with a Delta Ray CO2 Isotope Ratio Infrared Spectrometer (Thermo Fisher Scientific, MA, U.S.A.) with Universal Reference Interface Connect for measuring carbon isotope compositions of CO2.21−23,53 CO2-free synthetic air (Air Liquide, Germany) was used as carrier gas. CO2 in synthetic air at 414.2 ppm (Air Liquide, Germany) was used for CO2 concentration calibration. CO2 reference gases used for calibration of carbon isotope ratios had δ13C values of −9.7‰ (Thermo Fisher, Bremen, Germany) and x(13C) = 10% (Sigma-Aldrich, Taufkirchen, Germany). Pure CO2 gas with x(13C) = 10% was used as working reference gas. The CO2 concentration for reference and sample gas entering the analyzer was set to 380 ppm for optimal precision. Each sample was measured for 5 min and the obtained δ13C values were averaged. The stable carbon isotope data were received as delta values and converted into isotope-amount fraction according to Coplen et al., (2011).54
To determine the isotope amount fraction x(13C) of the electron donor bitumen, 0.779, 0.861, and 0.467 mg bitumen were transferred into tin vials and measured with a Pyro Cube element analyzer (Elementar Analysensysteme, Langenselbold, Germany) using the CN mode coupled to an IsoPrime 100 isotope ratio mass spectrometer (IRMS, (Elementar Analysensysteme, Langenselbold, Germany), as described in Köster et al., (2018).55
Microbial growth in the aqueous phase was monitored by cell counting with light microscopy (DMLS, Leica, Germany), equipped with a 40x/0.65 ocular and a Thoma counting chamber (Brand GmbH + Co KG, Germany).
Sulfate was measured with ion chromatography (IC) (see S7).
Microbial Community Composition
For DNA extraction of planktonic cells, 1.5 mL (time points 0 and 4) and 0.5 mL (time points 6 and 8) water samples were extracted with the DNeasy PowerLyser Power Soil Kit (Qiagen GmbH, Germany) according to the manufacturer’s instructions with modified steps as described in the SI (see S2).
At the end of the incubation, the remaining water was removed from each microcosm and the microcosms containing the bitumen were frozen at −70 °C for 4 h. To extract DNA from cells on the bitumen surface, thin pieces were flaked off the frozen bitumen surface with a scalpel, 0.4–0.7 g were transferred into triplicate bead-beating tubes per microcosm. Bitumen samples were extracted as described in Himmelberg (2018).56 A further cleaning step for bitumen samples after the extraction was modified from the described purification step in the Illumina protocol to remove humic acids and other matrix components which inhibit the PCR (see S3). Amplification of the 16S rRNA genes, library preparation, and sequencing were performed as described in Pannekens et al., (2020).27,57 Raw sequencing reads were deposited in the NCBI database under BioProject PRJNA645080.
Biofilm Visualization on Bitumen Surface
Biofilms were visualized with seven fluorescence-labeled lectins including Concanavalin A (Con A), Peanut agglutinin (PNA), Ulex Europaeus Agglutinin I (UEA), Griffonia simplicifolia lectin II (GS II), wheat germ agglutinin (WGA), soybean agglutinin (SBA), Oncorhynchus keta Lectin (CSL 3) to cover a broad range of target sugars which might be part of the extracellular polymeric substances (EPS) matrix.58−61 The lectins (10 μg mL–1 each final concentration) were mixed in a buffer solution consisting of 0.01 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 0.15 M NaCl, 0.1 mM CaCl2·2H2O, 0.1 mM MnCl2·4H2O, 0.1 mM MgCl2·6H2O, pH 7.5, and 10 μM Syto9 as counter stain for microbial cells. The staining mixture (50 μL) was added to each sample and incubated for 30 min in the dark. Afterward, the samples were gently rinsed three times with deionized water to remove the staining mix.61 Drinking water biofilms grown on ethylene propylene diene monomer (EPDM) coupons served as staining controls. An epifluorescence microscope was used for sample analysis. For detailed information see S5.
3D Imaging of Bitumen–Droplet Distribution
The water droplet distribution was determined via quantitative image analysis of CT scans. To this end, Pitch Lake bitumen was either filled directly in the field (3 columns, filled by pulling) or in the laboratory (3 columns, transferred by syringe after incubation at 45 °C) into round plastic columns (15 cm in length with a diameter of 1.5 or 1 cm, respectively). The top and bottom were sealed with rubber stoppers in the field and exchanged with hot glue in the laboratory. The sealed columns were placed on plastic stands and stored at 4 °C until analysis. 3D images were taken using X-ray computed tomography (CT) at a spatial resolution of 12.5 μm. Quantitative image analysis was used to segment the images and quantify the size destruction of the water droplets. For detailed information on scanning condition and image analysis steps please see S9.
Results and Discussion
Mineralization of Bitumen
Surprisingly, sulfate was present in all extracted water samples (9.7 ± 3.9 mM on average, see S2) and fermenting and sulfate-reducing bacteria were the most abundant members of the original community. Therefore, we focused mainly on bitumen mineralization to carbon dioxide by fermenting and sulfate-reducing bacteria (for detailed equations see S10), since sulfate reducers often outcompete or inhibit methanogenesis if sulfate is available in quantitative amounts.6,12,32,62,63 Sulfate reduction and CO2 production were monitored in four microcosms to assess the mineralization and degradation rates of bitumen. Reproducible sulfate reduction occurred in all microcosms (Figure 1A) with 6.7, 9.5, 8.7, and 9.0 mM consumed over 945 days, respectively (average sulfate reduction of 8.5 ± 1.1 mM).
Figure 1.
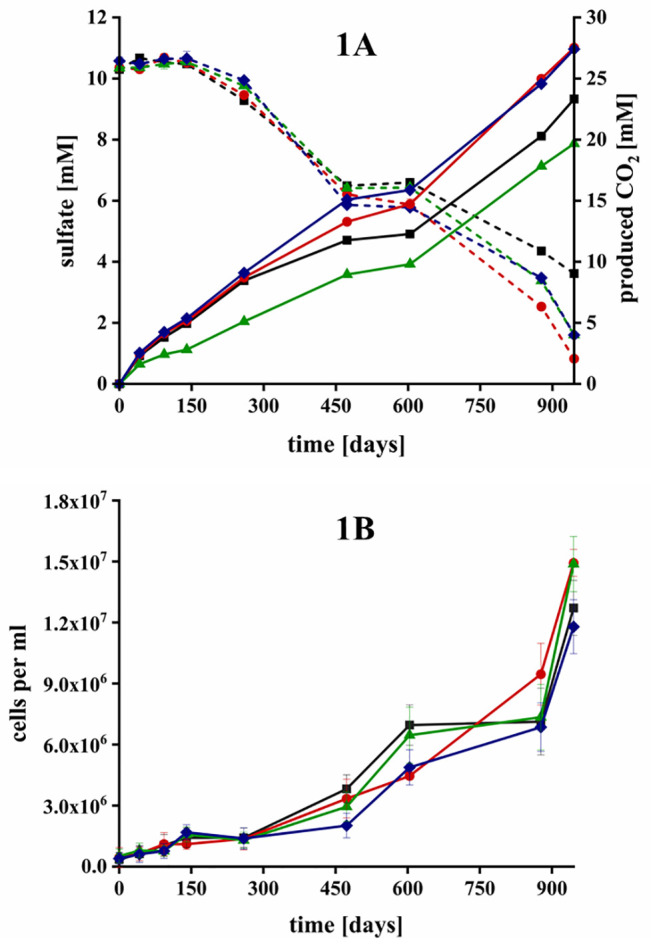
Degradation of bitumen in microcosms with sulfate as electron acceptor in microcosm 1 (black ■), 2 (red ●), 3 (green ▲), and 4 (blue ⧫) over 945 days of incubation. (1A) Degradation was measured by formation of carbon dioxide (solid lines) and depletion of sulfate (dashed lines). Error bars depict the standard deviation of triplicate analytical measurements but are barely visible because they are smaller than the symbol size. (1B) Microbial growth was assessed with light microscopy and a Thoma counting-chamber. Error bars depict the standard deviation of a minimum of six countings per sample.
Correspondingly, the CO2 concentrations increased by 23.3, 27.5, 19.7, and 27.4 mM, respectively, with an average production of 24.5 ± 3.2 mM CO2 during 945 days of incubation. Considering the used water volume leads to an average absolute production of 2.93 mmol CO2 during the incubation or 1.13 mmol CO2 per year. The values were calculated based on the measured isotopic signature of the bitumen of x(13C) = 1.075%. The pH values fluctuated slightly between 7.8–8.3 during the entire incubation, which ensured that the majority of CO2 was dissolved as bicarbonate in the aqueous phase. Previous analyses of the bitumen composition showed that short chain alkanes and alkenes with a carbon oxidation state of -II to -III had been already depleted.19,26 On the basis of the C/H ratio of Pitch Lake bitumen measured by Meckenstock et al., (2014), we estimated a carbon oxidation state of approximately −1.38 in the bitumen.26 Hence, we calculated an average electron balance for the four microcosms assuming that sulfate was reduced to sulfide (8.5 mM × 8e–). Only ∼12.6 mM of the produced CO2 (∼52%) could be linked to sulfate reduction based on the produced CO2 (24.5 mM × 5.38e–).26 A possible explanation are fermenters, which are present in numerous oil reservoirs and play an important role in the overall hydrocarbon degradation. Fermenters often belong to the most abundant microorganisms after incubation of oil, heavy oil, or water from oil reservoirs.5,64−67 Among others, several members of the phylum Chloroflexi, the order Desulfobacterales, the family Deferribacteraceae, or the genus Thermovirga were present in our communities. These microorganisms are either known fermenters or possess at least the ability for hydrocarbons fermentation in oil reservoirs.5,63−65,67−70 In addition, many sulfate reducers present in oil reservoirs can also ferment in the absence of sulfate or when sulfate reduction is inhibited.12 Other fermenting microorganisms often live in syntrophic communities by providing metabolites such as hydrogen and acetate to microorganisms like SRB or methanogens8,24,49,62 Therefore, it is likely that the remaining carbon and electron equivalents were not evolved as CO2 but were present as fermentation products.
Cell Growth and Community Evolution over Time
To assess the microbial growth in the presence of bitumen as sole carbon and electron source, we monitored the cell numbers in the aqueous phase. There was no other inoculum added and all cells originated from the bitumen itself, most likely from the small water droplets that ascended to the surface upon incubation in the microcosms (Figure 1B). During the first 150 days, the cell numbers in the planktonic phase tripled in all four microcosms from 4.3 × 105 ± 3.7 × 105 to 1.4 × 106 ± 3.7 × 105 cells mL–1 on average. The growth rates increased after ∼500 days, resulting in an average cell count of 1.4 × 107 ± 1.2 × 106 cells after 945 days of incubation.
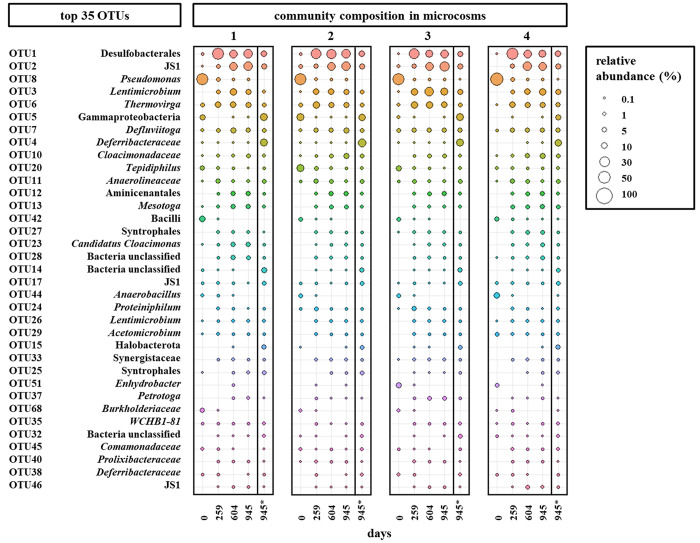
A community analysis based on the V3–4 region of 16S rRNA gene sequences indicated similar oil-degrading communities developed in the four microcosms over the time (Figure 2).
Figure 2.
Microbial community compositions of the water phase and the surface biofilm of the four microcosms showing the relative abundance of the 35 most abundant operational taxonomic units (OTUs) on 97% similarity level in all four microcosms. Planktonic communities were analyzed at days 0, 259, 604, and 945 and the community of the water-bitumen interface (*) at day 945, only.
At the start of the incubation, the microbial communities in all four microcosms were mainly dominated by the genera Pseudomonas (OTU 8), Tepidiphilus (OTU 20), Anaerobacillus (OTU 44), and members of the class Bacilli (OTU 42) at relative abundances of 54.9 ± 8.2%, 7.4 ± 5.0%, 4.1 ± 3.1%, and 4.4 ± 2.2% respectively. The low cell count and low number of rRNA genes after extraction of these samples agree with the assumption that the cells originate from the little water droplets enclosed in the oil sample. However, the four originally dominating OTUs almost disappeared after 259 days of incubation. The most abundant OTU 1 after 259 days belonged to the order Desulfobacterales with a relative abundance of 29.2 ± 11.5% averaged over all four microcosms indicating a succession within the microbial community which might be correlated to the availability of sulfate as electron acceptor instead of methanogenesis in the original samples. Members of the order Desulfobacterales are very versatile comprising strictly anaerobic sulfate-reducing, chemoorganotrophic, chemolithoheterotrophic, or chemolithoautotrophic bacteria, and some are able to ferment.63,71,72 Other abundant members of the community belonged to the candidate class JS1 (OTU 2), and the genera Lentimicrobium (OTU 3), Thermovirga (OTU 6), and Defluviitoga (OTU 7). Many prevalent community members such as representatives of Desulfobacterales, Comamonadaceae, Synergistaceae, Petrotogaceae, Deferribacteraceae, and Tepidiphilus were also earlier found to be present in Pitch Lake water droplets. However, the evolved abundances in our setups are not reflecting the natural abundances due to artificial influences such as an enlarged water volume favoring faster growing microorganisms, the dilution of toxic or growth inhibiting compounds such as selenate (sulfate analog inhibiting SRB growth) or an increased overall amount of available sulfate24,27 Nevertheless, representatives of these microorganisms were also found in other oil reservoirs and are known to be fermentative and sulfate-reducing oil degraders.65,69,70
After 945 days of incubation, the bitumen–water interface samples showed different microbial communities compared to those in the planktonic phase. Besides the abundant OTUs 1 (9.6 ± 1.4%), 2 (7.1 ± 0.9%), 3 (4.6 ± 2.4%), 6 (3.5 ± 0.9%), and 7 (4.1 ± 1.0%) that were also found in the water phase, the biofilm community was dominated by OTUs 4 and 5 belonging to the Deferribacteraceae and Gammaproteobacteria with an abundance of 17.9 ± 3.0% and 13.8 ± 3.7%, respectively, but the abundance of 0.04 ± 0.03% and 0.02 ± 0.02% in the water phase, respectively, was minor. The family Deferribacteraceae is known for anaerobic Fe(III), Mn(IV), or nitrate reduction, but also fermentation. Deferribacteriaceae were also found to be part of biofilms, previously.68,73 Similarly, OTUs 14, 15, and 17 (unclassified bacteria, Halobacterota, and candidate class JS1) were mainly present at the bitumen–water interface. Nevertheless, Fe(III) or Mn(IV) oxides as electron acceptors are generally only abundant in low concentration in oil reservoirs around the globe and are not known to have a prevalent impact on the oil degradation process.3,8
Colonization and Biofilm Formation on Bitumen
Biofilms are a very successful form of life and make microorganisms resistant to stress, for instance protection against toxic compounds, protection against dehydration, or helping to overcome mass transfer limitations for nutrient transport and many more.74−76 We stained pieces of the bitumen surface with the DNA stain Syto9 to visualize attached microbial cells in biofilms. Additionally, we used fluorescence-labeled lectins targeting mono- and polysaccharides which may be part of the EPS matrix (Figure 3).74,77,78
Figure 3.
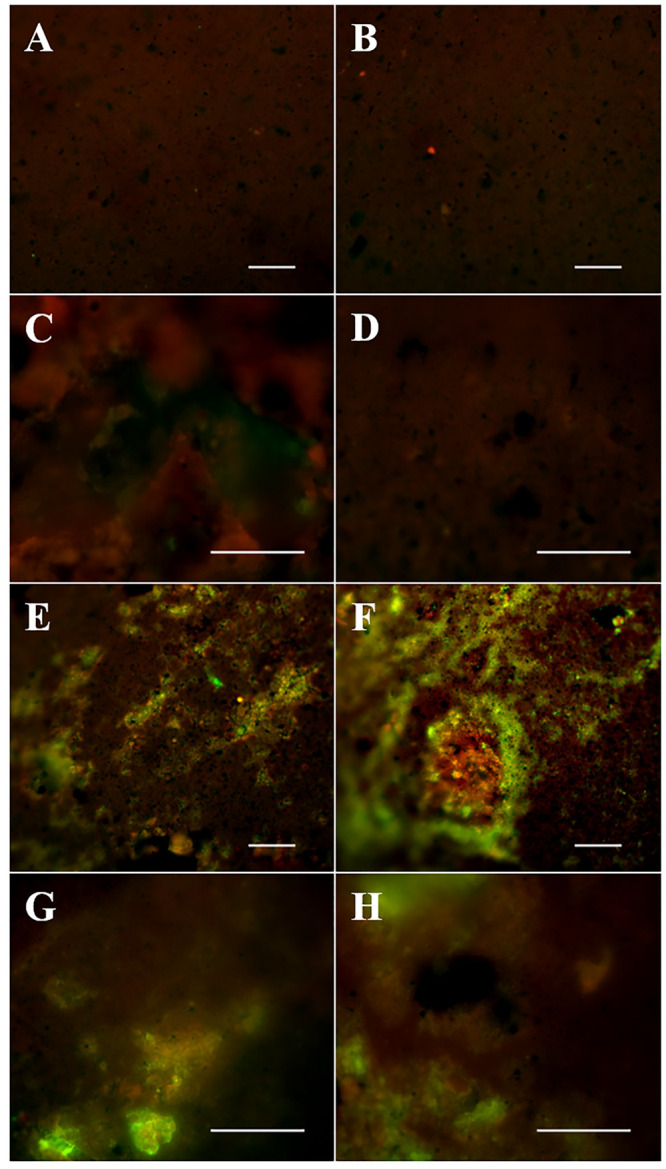
Fluorescence images of bitumen surfaces after 945 days of incubation in the microcosms. Images A and B show bitumen controls stained with the Syto9-lectin mix that were stored at 4 °C without incubation. Images C (microcosm 3) and D (microcosm 4) depict unstained controls of the bitumen surface in our microcosms after 945 days incubation. Images E–H show the bitumen surface of microcosms 1 (E and F), 3 (G), and 4 (H) after 945 days of incubation stained with the Syto9-lectin mix. All scale bars indicate 50 μm.
The chosen lectins covered a broad range of the most common EPS polysaccharides. Positive controls with drinking water biofilms showed the functioning of the lectin staining and no evidence for lectin–lectin binding or unspecific inhibition of the staining (Figure S1).59,77,79 The nonincubated but stained bitumen controls (Figure 3, A and B) showed a weak fluorescence and heterogeneity of the bitumen surface. Only occasionally, strongly fluorescent dots were observed, most likely from stained single microbial cells on the surface, but no significant fluorescence of the added lectins was visible. However, images of nonstained bitumen after 945 days incubation showed the formation of structures on the surface (Figure 3, C and D) and these structures did not appear in any nonincubated control. In contrast to unstained sample controls, stained bitumen surfaces in our microcosms after 945 days of incubation (Figure 3, E–H) showed prominent red and green fluorescent structures and aggregates, which are located in cavities and cracks along the uneven bitumen surface. The green fluorescent parts (presumably Syto9-stained cells) are mostly surrounded by a red fluorescent corona (presumably lectin-stained EPS). The weaker red signal was overlain by the green signal and is therefore less dominant in the merged pictures. Unfortunately, we were not able to take pictures at a larger magnification because of the spiky character of the surface and a strong bitumen shifting while excitation. However, the arrangement and shape of the present structures strongly indicated the formation of biofilms at the bitumen–water interface. Cell numbers were only determined from liquid samples, but the biofilms indicate that very likely a large part of the cells was attached to the bitumen surface which escaped counting in the liquid samples.
Water Content and Distribution of Droplets in the Bitumen
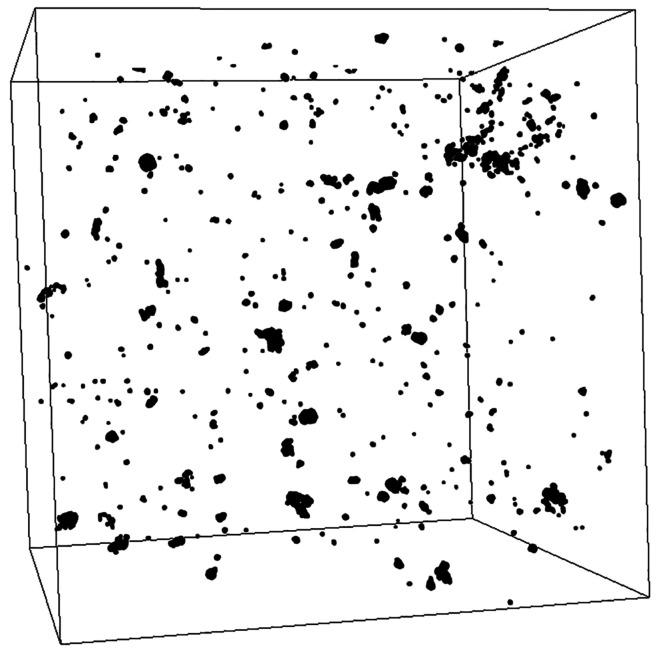
Seven CT scans with a total volume of 8.45 mL bitumen revealed 6330 water droplets (Figure 4). Most of these droplets were rather small with an average size of 2.18 nL. Only about 0.2% of the water droplets were larger than 0.1 μL including the three largest with 0.42, 0.55, and 2.33 μL.
Figure 4.
Partial section of a scan of a bitumen column, showing the spatial distribution of water droplets (black) in the bitumen (white). The cube’s edge length is 10 mm. The largest depicted droplet here has a volume of 54 nL. The image was created with Fiji using a data set from a single CT scan.
The total droplet volume accounted for an average water content of 2.5 μL per mL bitumen with an average bitumen–water surface area of 1.13 cm2 mL–1, which is ∼50-times less than the overall water content of the bitumen described by Meckenstock et al., (2014).26 Only a minor part of the total water content was present in larger water droplets which can serve as a micro habitat. Our measurements show the size and distribution of enclosed water droplets, however it remains still unclear how fast these droplets ascent through the bitumen and if they merge or split during their way to the surface under natural conditions, since the exact temperature, pressure, and pore size in the deep subsurface reservoir remain rather unknown. Nonetheless, considering the bitumen density of 1.15 g cm–3 and the mean bitumen mass (104.97 g corresponds to 91.28 mL) filled into the microcosms, the overall bitumen–water surface of our microcosms was enlarged by additional 310.5% (103.1 cm2) of the visible bitumen–water surface (33.2 cm2).
On the basis of the assumption that the bitumen–water surface area is a limiting parameter for the overall degradation process (bioavailability of organic compounds by diffusion and dissolution), we calculated the degradation rates per surface area by using the average amount of produced CO2 from the microcosms (1.13 mmol year–1). However, former experiments showed that heating lowers the bitumen viscosity and enhanced the buoyance density-driven up-rise of water droplets toward the bitumen surface within days. Therefore, it is likely that most droplets reached the overlaying water phase within a few days after the setup.26,27 Hence, we calculated the degradation rate per bitumen-water surface area either based on the visible surface area only (33.2 cm2) or based on the visible surface area plus the assumed droplet-water surface area (136.3 cm2) in our microcosms, resulting in 8.3–34.1 μmol CO2 cm–2 year–1, respectively.
We then estimated the theoretical degradation rate in a regular bitumen sample based on the biodegradation rates per surface area and the assumption that the degradation would only take place in the water droplets dispersed in the bitumen. Considering the water droplet surface area per volume of bitumen (1.13 cm2 mL–1) led to a degradation rate of 9.4–38.6 mmol CO2 L–1 bitumen year–1. Assuming an average chemical formula of the hydrocarbons in the bitumen of CxH1.4x (Mw ≈ 13.5 g mol–1), the CO2-production accounts for a biodegradation of 0.13–0.52 g carbon L–1 bitumen year–1. However, the relatively big volume of the overlaying water added to the microcosms might have affected the microbial activity to a certain degree. Furthermore, the cohesive oil–water transition zone and the available sulfate amount artificially increased the degradation rates in our experimental microcosms and are probably leading to an overestimation of the degradation rates. Nonetheless, the degradation rates resulted in decay times of around 2200–9000 years for a liter of bitumen, which is indeed on a geological time scale, although it is still up to 15-times higher compared to methanogenesis as final electron-accepting process in crude oil.64,80 We conclude that the overall bioavailable oil–water interface area is notably enlarged by dispensed water droplets in oil. Our results suggest that this facet of microbial life in oil reservoirs has the potential to substantially contribute to the overall biodegradation of oil over millions of years in addition to degradation at the oil–water-transition-zone. Furthermore, we could show that the reverse stable isotope measurements allowed to assess such slow biodegradation rates in the range of geological time scales.
Acknowledgments
We thank Christian Thyssen for his support with sample preparation for ion chromatography. Till Bornemann supported with advice for the cleaning steps during DNA extraction. We thank Riad Hossein for assisting the sampling campaigns in Trinidad.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.1c00596.
Details about geochemical measurements, DNA extraction, biofilm visualization, sulfate determination, 3D-imaging of bitumen, and additional tables and figures (PDF)
This work was supported by the ERC advanced grant EcOILogy [grant number 666952] funded by the European Research Council and the German Research Foundation [grant number BR 5493/1–1], and the EU Horizon 2020 project Reground [grant number 641768].
The authors declare no competing financial interest.
Supplementary Material
References
- Head I. M.; Jones D. M.; Larter S. R. Biological activity in the deep subsurface and the origin of heavy oil. Nature 2003, 426, 344. 10.1038/nature02134. [DOI] [PubMed] [Google Scholar]
- Aitken C. M.; Jones D. M.; Larter S. Anaerobic hydrocarbon biodegradation in deep subsurface oil reservoirs. Nature 2004, 431, 291–294. 10.1038/nature02922. [DOI] [PubMed] [Google Scholar]
- Youssef N.; Elshahed M. S.; McInerney M. J. Microbial processes in oil fields: culprits, problems, and opportunities. Adv. Appl. Microbiol. 2009, 66, 141–251. 10.1016/S0065-2164(08)00806-X. [DOI] [PubMed] [Google Scholar]
- Kayukova G.; Uspensky B.; Abdrafikova I.; Musin R. Characteristic features of the hydrocarbon composition of Spiridonovskoe (Tatarstan) and Pitch Lake (Trinidad and Tobago) asphaltites. Pet. Chem. 2016, 56, 572–579. 10.1134/S0965544116070082. [DOI] [Google Scholar]
- Mbadinga S. M.; Wang L.-Y.; Zhou L.; Liu J.-F.; Gu J.-D.; Mu B.-Z. Microbial communities involved in anaerobic degradation of alkanes. Int. Biodeterior. Biodegrad. 2011, 65, 1–13. 10.1016/j.ibiod.2010.11.009. [DOI] [Google Scholar]
- Larter S.; Wilhelms A.; Head I.; Koopmans M.; Aplin A.; Di Primio R.; Zwach C.; Erdmann M.; Telnaes N. The controls on the composition of biodegraded oils in the deep subsurface - part 1: biodegradation rates in petroleum reservoirs. Org. Geochem. 2003, 34, 601–613. 10.1016/S0146-6380(02)00240-1. [DOI] [Google Scholar]
- Widdel F.; Rabus R. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 2001, 12, 259–276. 10.1016/S0958-1669(00)00209-3. [DOI] [PubMed] [Google Scholar]
- Lovley D. R.; Chapelle F. H. Deep subsurface microbial processes. Rev. Geophys. 1995, 33, 365–381. 10.1029/95RG01305. [DOI] [Google Scholar]
- Pannekens M.; Kroll L.; Müller H.; Mbow F. T.; Meckenstock R. U. Oil reservoirs, an exceptional habitat for microorganisms. New Biotechnol. 2019, 49, 1–9. 10.1016/j.nbt.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-X.; Mbadinga S. M.; Liu J.-F.; Zhou L.; Yang S.-Z.; Gu J.-D.; Mu B.-Z. Microbiota and their affiliation with physiochemical characteristics of different subsurface petroleum reservoirs. Int. Biodeterior. Biodegrad. 2017, 120, 170–185. 10.1016/j.ibiod.2017.02.005. [DOI] [Google Scholar]
- Meckenstock R. U.; Elsner M.; Griebler C.; Lueders T.; Stumpp C.; Aamand J.; Agathos S. N.; Albrechtsen H.-J.; Bastiaens L.; Bjerg P. L.; Boon N.; Dejonghe W.; Huang W. E.; Schmidt S. I.; Smolders E.; Sørensen S. R.; Springael D.; van Breukelen B. M. Biodegradation: updating the concepts of control for microbial cleanup in contaminated aquifers. Environ. Sci. Technol. 2015, 49, 7073–7081. 10.1021/acs.est.5b00715. [DOI] [PubMed] [Google Scholar]
- Jiménez N.; Richnow H. H.; Vogt C.; Treude T.; Krüger M. Methanogenic hydrocarbon degradation: evidence from field and laboratory studies. J. Mol. Microbiol Biotechnol 2016, 26, 227–242. 10.1159/000441679. [DOI] [PubMed] [Google Scholar]
- Gieg L. M.; Davidova I. A.; Duncan K. E.; Suflita J. M. Methanogenesis, sulfate reduction and crude oil biodegradation in hot Alaskan oilfields. Environ. Microbiol. 2010, 12, 3074–3086. 10.1111/j.1462-2920.2010.02282.x. [DOI] [PubMed] [Google Scholar]
- Gieg L. M.; Fowler S. J.; Berdugo-Clavijo C. Syntrophic biodegradation of hydrocarbon contaminants. Curr. Opin. Biotechnol. 2014, 27, 21–29. 10.1016/j.copbio.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Jones D. M.; Head I. M.; Gray N. D.; Adams J. J.; Rowan A. K.; Aitken C. M.; Bennett B.; Huang H.; Brown A.; Bowler B. F. J.; Oldenburg T.; Erdmann M.; Larter S. R. Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 2008, 451, 176–180. 10.1038/nature06484. [DOI] [PubMed] [Google Scholar]
- Xu D.; Zhang K.; Li B.-G.; Mbadinga S. M.; Zhou L.; Liu J.-F.; Yang S.-Z.; Gu J.-D.; Mu B.-Z. Simulation of in situ oil reservoir conditions in a laboratory bioreactor testing for methanogenic conversion of crude oil and analysis of the microbial community. Int. Biodeterior. Biodegrad. 2019, 136, 24–33. 10.1016/j.ibiod.2018.10.007. [DOI] [Google Scholar]
- Head I.; Larter S.; Gray N.; Sherry A.; Adams J.; Aitken C.; Jones D.; Rowan A.; Huang H.; Röling W.. Hydrocarbon degradation in petroleum reservoirs. In Handbook of Hydrocarbon and Lipid Microbiology; Springer: New York, 2010; pp 3097–3109. [Google Scholar]
- Heider J.; Spormann A. M.; Beller H. R.; Widdel F. Anaerobic bacterial metabolism of hydrocarbons. FEMS microbiology reviews 1998, 22, 459–473. 10.1111/j.1574-6976.1998.tb00381.x. [DOI] [Google Scholar]
- Meckenstock R. U.; Boll M.; Mouttaki H.; Koelschbach J. S.; Tarouco P. C.; Weyrauch P.; Dong X.; Himmelberg A. M. Anaerobic degradation of benzene and polycyclic aromatic hydrocarbons. J. Mol. Microbiol Biotechnol 2016, 26, 92–118. 10.1159/000441358. [DOI] [PubMed] [Google Scholar]
- Aeckersberg F.; Bak F.; Widdel F. Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch. Microbiol. 1991, 156, 5–14. 10.1007/BF00418180. [DOI] [Google Scholar]
- Schulte S.; Köster D.; Jochmann M.; Meckenstock R. Applying reverse stable isotope labeling analysis by mid-infrared laser spectroscopy to monitor BDOC in recycled wastewater. Sci. Total Environ. 2019, 665, 1064–1072. 10.1016/j.scitotenv.2019.02.061. [DOI] [PubMed] [Google Scholar]
- Dong X.; Jochmann M. A.; Elsner M.; Meyer A. H.; Bäcker L. E.; Rahmatullah M.; Schunk D.; Lens G.; Meckenstock R. U. Monitoring microbial mineralization using reverse stable isotope labeling analysis by mid-infrared laser spectroscopy. Environ. Sci. Technol. 2017, 51, 11876–11883. 10.1021/acs.est.7b02909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X.; Bäcker L. E.; Rahmatullah M.; Schunk D.; Lens G.; Meckenstock R. U. Quantification of microbial degradation activities in biological activated carbon filters by reverse stable isotope labelling. AMB Express 2019, 9, 109. 10.1186/s13568-019-0827-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeva M. K.; Coates J. D. Specific inhibitors of respiratory sulfate reduction: towards a mechanistic understanding. Microbiology 2019, 165, 254–269. 10.1099/mic.0.000750. [DOI] [PubMed] [Google Scholar]
- Bennett B.; Adams J.; Gray N.; Sherry A.; Oldenburg T.; Huang H.; Larter S.; Head I. The controls on the composition of biodegraded oils in the deep subsurface-Part 3. The impact of microorganism distribution on petroleum geochemical gradients in biodegraded petroleum reservoirs. Org. Geochem. 2013, 56, 94–105. 10.1016/j.orggeochem.2012.12.011. [DOI] [Google Scholar]
- Meckenstock R. U.; von Netzer F.; Stumpp C.; Lueders T.; Himmelberg A. M.; Hertkorn N.; Schmitt-Kopplin P.; Harir M.; Hosein R.; Haque S.; Schulze-Makuch D. Water droplets in oil are microhabitats for microbial life. Science 2014, 345, 673–676. 10.1126/science.1252215. [DOI] [PubMed] [Google Scholar]
- Pannekens M.; Voskuhl L.; Meier A.; Müller H.; Haque S.; Frösler J.; Brauer V.; Meckenstock R. Densely populated water droplets in heavy oil seeps. Appl. Environ. Microbiol. 2020, 86, 1–12. 10.1128/AEM.00164-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S.; Crowley D. E. Microbial diversity in natural asphalts of the rancho la brea tar pits. Appl. Environ. Microbiol. 2007, 73, 4579–4591. 10.1128/AEM.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Langomazino N.; Sellier R.; Jouquet G.; Trescinski M. Microbial degradation of bitumen. Experientia 1991, 47, 533–539. 10.1007/BF01949873. [DOI] [Google Scholar]
- Schneiker S.; dos Santos V. A. P. M.; Bartels D.; Bekel T.; Brecht M.; Buhrmester J.; Chernikova T. N.; Denaro R.; Ferrer M.; Gertler C.; Goesmann A.; Golyshina O. V.; Kaminski F.; Khachane A. N.; Lang S.; Linke B.; McHardy A. C.; Meyer F.; Nechitaylo T.; Puhler A.; Regenhardt D.; Rupp O.; Sabirova J. S.; Selbitschka W.; Yakimov M. M.; Timmis K. N.; Vorholter F. J.; Weidner S.; Kaiser O.; Golyshin P. N. Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat. Biotechnol. 2006, 24, 997–1004. 10.1038/nbt1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golby S.; Ceri H.; Gieg L. M.; Chatterjee I.; Marques L. L.; Turner R. J. Evaluation of microbial biofilm communities from an Alberta oil sands tailings pond. FEMS Microbiol. Ecol. 2012, 79, 240–250. 10.1111/j.1574-6941.2011.01212.x. [DOI] [PubMed] [Google Scholar]
- Wolf M.; Bachofen R. Microbial legradation of bitumen. Experientia 1991, 47, 542–548. 10.1007/BF01949875. [DOI] [Google Scholar]
- Widdel F.; Pfennig N. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. Arch. Microbiol. 1981, 129, 395–400. 10.1007/BF00406470. [DOI] [PubMed] [Google Scholar]
- Babu D. R.; Cormack D. E. Effect of low-temperature oxidation on the composition of Athabasca bitumen. Fuel 1984, 63, 858–861. 10.1016/0016-2361(84)90080-2. [DOI] [Google Scholar]
- Kapadia P. R.; Kallos M. S.; Gates I. D. A review of pyrolysis, aquathermolysis, and oxidation of Athabasca bitumen. Fuel Process. Technol. 2015, 131, 270–289. 10.1016/j.fuproc.2014.11.027. [DOI] [Google Scholar]
- Jia N.; Moore R.; Mehta S.; Van Fraassen K.; Ursenbach M.; Zalewski E.. In Compositional changes for Athabasca bitumen in the presence of oxygen under low temperature conditions, Canadian International Petroleum Conference, 2004; Petroleum Society of Canada: 2004.
- Strausz O. P.; Jha K. N.; Montgomery D. S. Chemical composition of gases in Athabasca bitumen and in low-temperature thermolysis of oil sand, asphaltene and maltene. Fuel 1977, 56, 114–120. 10.1016/0016-2361(77)90128-4. [DOI] [Google Scholar]
- Merdrignac I.; Espinat D. Physicochemical characterization of petroleum fractions: the state of the art. Oil Gas Sci. Technol. 2007, 62, 7–32. 10.2516/ogst:2007002. [DOI] [Google Scholar]
- Selucky M. L.; Chu Y.; Ruo T.; Strausz O. P. Chemical composition of Athabasca bitumen. Fuel 1977, 56, 369–381. 10.1016/0016-2361(77)90061-8. [DOI] [Google Scholar]
- Mehrotra A. K.; Svrcek W. Y. Viscosity of compressed Athabasca bitumen. Can. J. Chem. Eng. 1986, 64, 844–847. 10.1002/cjce.5450640520. [DOI] [Google Scholar]
- Badamchi-Zadeh A.; Yarranton H.; Svrcek W.; Maini B. Phase behaviour and physical property measurements for VAPEX solvents: Part I. Propane and Athabasca bitumen. J. Can. Pet Technol. 2009, 48, 54–61. 10.2118/09-01-54. [DOI] [Google Scholar]
- Del Bianco A.Composition and analysis of heavy petroleum fractions Appl. Catal., A. 1994; Vol. 114.N29–N30. 10.1016/0926-860X(94)80190-8. [DOI] [Google Scholar]
- Mckenna A. M.; Purcell J. M.; Rodgers R. P.; Marshall A. G. Heavy petroleum composition. 1. Exhaustive compositional analysis of Athabasca bitumen HVGO distillates by Fourier transform ion cyclotron resonance mass spectrometry: A definitive test of the Boduszynski model. Energy Fuels 2010, 24, 2929–2938. 10.1021/ef100149n. [DOI] [Google Scholar]
- Hayes M.; Stacey M.; Standley J.; Entwistle A. Studies on bitumen: Part 3. Experiments on the biodegradation of bitumen by soil micro-organisms. Fuel 1972, 51, 146–149. 10.1016/0016-2361(72)90065-8. [DOI] [Google Scholar]
- Phillips D.; Hitchon J.; Johnson D.; Matthews J. The radiation swelling of bitumens and bitumenised wastes. J. Nucl. Mater. 1984, 125, 202–218. 10.1016/0022-3115(84)90546-4. [DOI] [Google Scholar]
- Gehringer P.; Proksch E.; Szinovatz W. The gamma radiolysis of bitumen. Pt. 1. Atomkernenergie Kerntechnik 1981, 39, 12–15. [Google Scholar]
- Mello M.; Braz D.; Motta L.; Leite L.. Investigation on the effects of gamma irradiation on bitumen. 2011. [Google Scholar]
- Mouazen M.; Poulesquen A.; Bart F.; Masson J.; Charlot M.; Vergnes B. Rheological, structural and chemical evolution of bitumen under gamma irradiation. Fuel Process. Technol. 2013, 114, 144–153. 10.1016/j.fuproc.2013.03.039. [DOI] [Google Scholar]
- Christman G. D.; León-Zayas R. I.; Zhao R.; Summers Z. M.; Biddle J. F. Novel clostridial lineages recovered from metagenomes of a hot oil reservoir. Sci. Rep. 2020, 10, 1–11. 10.1038/s41598-020-64904-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cunha C. D.; Rosado A. S.; Sebastián G. V.; Seldin L.; Von der Weid I. Oil biodegradation by Bacillus strains isolated from the rock of an oil reservoir located in a deep-water production basin in Brazil. Appl. Microbiol. Biotechnol. 2006, 73, 949–959. 10.1007/s00253-006-0531-2. [DOI] [PubMed] [Google Scholar]
- Rosnes J. T.; Torsvik T.; Lien T. Spore-forming thermophilic sulfate-reducing bacteria isolated from North Sea oil field waters. Appl. Environ. Microbiol. 1991, 57, 2302–2307. 10.1128/aem.57.8.2302-2307.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assayag N.; Rivé K.; Ader M.; Jézéquel D.; Agrinier P. Improved method for isotopic and quantitative analysis of dissolved inorganic carbon in natural water samples. Rapid Commun. Mass Spectrom. 2006, 20, 2243–2251. 10.1002/rcm.2585. [DOI] [PubMed] [Google Scholar]
- van Geldern R.; Nowak M. E.; Zimmer M.; Szizybalski A.; Myrttinen A.; Barth J. A.; Jost H.-J. R. Field-based stable isotope analysis of carbon dioxide by mid-infrared laser spectroscopy for carbon capture and storage monitoring. Anal. Chem. 2014, 86, 12191–12198. 10.1021/ac5031732. [DOI] [PubMed] [Google Scholar]
- Coplen T. B. Guidelines and recommended terms for expression of stable-isotope-ratio and gas-ratio measurement results. Rapid Commun. Mass Spectrom. 2011, 25, 2538–2560. 10.1002/rcm.5129. [DOI] [PubMed] [Google Scholar]
- Köster D.; Wolbert J.-B.; Schulte M. S.; Jochmann M. A.; Schmidt T. C. Origin of xylitol in chewing gum: A compound-specific isotope technique for the differentiation of corn-and wood-based xylitol by LC-IRMS. J. Agric. Food Chem. 2018, 66, 2015–2020. 10.1021/acs.jafc.7b05212. [DOI] [PubMed] [Google Scholar]
- Himmelberg A.Life in Pitch-Bacteria in a Natural Oil Emitting Lake Help to Understand Anaerobic Biodegradation of Polycyclic Aromatic Hydrocarbons, PhD-Thesis; Technical University of Munich, 2019. [Google Scholar]
- Tebbe C. C.; Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl. Environ. Microbiol. 1993, 59, 2657–2665. 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyssen C.Biofilm Formation by the Manganese-Oxidizing Bacterium Leptothrix discophora Strain SS-1 and Corrosion of Stainless Steel, PhD-thesis; University of Duisburg-Essen, 2018. [Google Scholar]
- Strathmann M.; Wingender J.; Flemming H.-C. Application of fluorescently labelled lectins for the visualization and biochemical characterization of polysaccharides in biofilms of Pseudomonas aeruginosa. J. Microbiol. Methods 2002, 50, 237–248. 10.1016/S0167-7012(02)00032-5. [DOI] [PubMed] [Google Scholar]
- Shiina N.; Tateno H.; Ogawa T.; Muramoto K.; Saneyoshi M.; Kamiya H. Isolation and characterization of L-rhamnose-binding lectins from chum salmon (Oncorhynchus keta) eggs. Fish. Sci. 2002, 68, 1352–1366. 10.1046/j.1444-2906.2002.00575.x. [DOI] [Google Scholar]
- Neu T. R.; Swerhone G. D.; Lawrence J. R. Assessment of lectin-binding analysis for in situ detection of glycoconjugates in biofilm systems. Microbiology 2001, 147, 299–313. 10.1099/00221287-147-2-299. [DOI] [PubMed] [Google Scholar]
- Muyzer G.; Stams A. J. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- Kuever J.; Rainey F. A.; Widdel F. Desulfobacterales ord. nov.. Bergey’s Manual of Systematics of Archaea and Bacteria 2015, 1–7. 10.1002/9781118960608.gbm01019. [DOI] [Google Scholar]
- Gieg L. M.; Duncan K. E.; Suflita J. M. Bioenergy production via microbial conversion of residual oil to natural gas. Appl. Environ. Microbiol. 2008, 74, 3022–3029. 10.1128/AEM.00119-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollivier B.; Borgomano J.; Oger P. Petroleum: from formation to microbiology. Microbial Life of the Deep Biosphere 2014, 8, 161–185. 10.1515/9783110300130.161. [DOI] [Google Scholar]
- Zhou Z.; Liang B.; Wang L.-Y.; Liu J.-F.; Mu B.-Z.; Shim H.; Gu J.-D. Identifying the core bacterial microbiome of hydrocarbon degradation and a shift of dominant methanogenesis pathways in the oil and aqueous phases of petroleum reservoirs of different temperatures from China. Biogeosciences 2019, 16, 4229–4241. 10.5194/bg-16-4229-2019. [DOI] [Google Scholar]
- Toth C. R.; Gieg L. M. Time course-dependent methanogenic crude oil biodegradation: dynamics of fumarate addition metabolites, biodegradative genes, and microbial community composition. Front. Microbiol. 2018, 8, 2610. 10.3389/fmicb.2017.02610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H.; Stetter K. O. Deferribacteraceae fam. nov. Bergey’s Manual of Systematics of Archaea and Bacteria 2015, 1–1. 10.1002/9781118960608.fbm00095. [DOI] [Google Scholar]
- Dahle H.; Birkeland N.-K. Thermovirga lienii gen. nov., sp. nov., a novel moderately thermophilic, anaerobic, amino-acid-degrading bacterium isolated from a North Sea oil well. Int. J. Syst. Evol. Microbiol. 2006, 56, 1539–1545. 10.1099/ijs.0.63894-0. [DOI] [PubMed] [Google Scholar]
- Sun L.; Toyonaga M.; Ohashi A.; Tourlousse D. M.; Matsuura N.; Meng X.-Y.; Tamaki H.; Hanada S.; Cruz R.; Yamaguchi T.; Sekiguchi Y. Lentimicrobium saccharophilum gen. nov., sp. nov., a strictly anaerobic bacterium representing a new family in the phylum Bacteroidetes, and proposal of Lentimicrobiaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 2635–2642. 10.1099/ijsem.0.001103. [DOI] [PubMed] [Google Scholar]
- Kuever J., The Family Desulfobulbaceae. In The Prokaryotes; Springer: New York, 2014; pp 75–86. [Google Scholar]
- Zapata-Peñasco I.; Salazar-Coria L.; Saucedo-García M.; Villa-Tanaka L.; Hernández-Rodríguez C. Bisulfite reductase and nitrogenase genes retrieved from biocorrosive bacteria in saline produced waters of offshore oil recovery facilities. Int. Biodeterior. Biodegrad. 2013, 81, 17–27. 10.1016/j.ibiod.2012.07.005. [DOI] [Google Scholar]
- Daghio M.; Gandolfi I.; Bestetti G.; Franzetti A.; Guerrini E.; Cristiani P. Anodic and cathodic microbial communities in single chamber microbial fuel cells. New Biotechnol. 2015, 32, 79–84. 10.1016/j.nbt.2014.09.005. [DOI] [PubMed] [Google Scholar]
- Flemming H.-C. The perfect slime. Colloids Surf., B 2011, 86, 251–259. 10.1016/j.colsurfb.2011.04.025. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L.; Costerton J. W.; Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Johnsen A.; Karlson U. Evaluation of bacterial strategies to promote the bioavailability of polycyclic aromatic hydrocarbons. Appl. Microbiol. Biotechnol. 2004, 63, 452–459. 10.1007/s00253-003-1265-z. [DOI] [PubMed] [Google Scholar]
- Sutherland I. W. Polysaccharases for microbial exopolysaccharides. Carbohydr. Polym. 1999, 38, 319–328. 10.1016/S0144-8617(98)00114-3. [DOI] [Google Scholar]
- Flemming H. C.; Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–33. 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Czaczyk K.; Myszka K. Biosynthesis of extracellular polymeric substances (EPS) and its role in microbial biofilm formation. Pol J. Environ. Stud 2007, 16, 799–806. [Google Scholar]
- Berdugo-Clavijo C.; Gieg L. M. Conversion of crude oil to methane by a microbial consortium enriched from oil reservoir production waters. Front. Microbiol. 2014, 5, 197. 10.3389/fmicb.2014.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.