Abstract
Metabolic responses to food influence cardiometabolic disease risk, but large-scale high-resolution studies are lacking. We recruited n = 1,002 twins and unrelated healthy adults in the UK into the PREDICT1 study and assessed postprandial metabolic responses in a clinic setting and at home. We observed large inter-individual variability (population coefficient of variation [SD/mean]%) in postprandial blood triglyceride (103%), glucose (68%), and insulin (59%) responses to identical meals. Person-specific factors, such as the gut microbiome, had a greater influence (7.1% of variance) than meal macronutrients (3.6%) for postprandial lipemia, but not for postprandial glycemia (6.0% and 15.4% respectively); genetic variants had a modest impact on predictions (9.5% for glucose, 0.8% for triglyceride, 0.2% for c-peptide). Findings were independently validated in a US cohort (n = 100). We developed a machine learning model that predicted both triglyceride (r = 0.47) and glycemic (r = 0.77) responses to food intake. These findings may be informative for developing personalized diet strategies. ClinicalTrials.gov registration: NCT03479866.
Introduction
Effective prevention strategies are required to reduce the immense global burden of nutrition-related non-communicable diseases (NCD)1. Nutritional research and the corresponding guidelines2–4 focus on population averages. However, the high between-person variability in response to foods and weight-loss diets5 demands development of more personalized approaches. Empirically-based precision nutrition requires research using multi-dimensional, high-resolution time-series data from adequately powered studies6. The application of technologies to accurately and precisely quantify many postprandial (non-fasting) traits in large cohorts and in real-world settings is extending capabilities in this field of research.
Although fasting blood assays are used in many clinical diagnoses, most people are predominantly in the postprandial state during waking hours. Postprandial lipid, glucose and insulin dyshomeostasis are independent risk factors for NCDs and obesity7,8,9. Postprandial hyperglycemia raises risk of cardiovascular disease (CVD), coronary heart disease (CHD)10 and cardiovascular mortality, even in individuals with normal fasting glucose11, and postprandial triglyceride is more predictive of CVD than fasting concentrations12,13, highlighting the relevance of diet and its metabolic consequences in cardiovascular risk.
A person’s unique postprandial glycemic and lipiaemic responses are likely attributable their biological (e.g. microbiome and nuclear DNA variation) and lifestyle characteristics2,14, as demonstrated previously for specific meals5. While postprandial glycemic responses are important health determinants, glycemic control is just one part of a more complex metabolic equation involving triglyceride (the primary alternative energy substrate to glucose) and insulin (regulating glucose and triglyceride transport and metabolism)15. Thus, also characterizing postprandial regulation of lipids and identifying the factors responsible for individual variations could help optimize diet recommendations targeting broader improvements in cardiometabolic health.
The PREDICT 1 clinical trial (NCT03479866) was designed to quantify and predict individual variations in postprandial triglyceride, glucose and insulin responses to standardized meals. PREDICT 1 enrolled twins and unrelated adults from the UK in whom genetic, metabolic, microbiome composition, meal composition and meal context data were obtained to distinguish predictors of individual responses to meals. These predictions were validated in an independent cohort of adults from the USA.
Our findings show wide variations in postprandial responses between people, even identical twins, attributable in large part to modifiable factors. We found that people who experience poor metabolic responses to a given meal are likely to respond poorly to other meals of the same macronutrient profile, and the overall correlation between postprandial glucose and triglyceride responses is weak. The postprandial prediction models we have developed could help to optimize personalized diet recommendations.
Results:
1002 healthy adults from the UK completed baseline clinic measurements consisting of postprandial metabolic responses (0–6h; blood triglyceride, glucose and insulin concentrations) to sequential mixed-nutrient dietary challenges. Findings were validated a US cohort of 100 healthy adults. Additional data was collected over the subsequent 13-day period at home, where postprandial responses to eight meals (seven in duplicate) of different macronutrient (fat, carbohydrate, protein and fiber) content were measured using continuous glucose monitors (CGM) and dried blood spot (DBS) analysis. The study design is described in detail in the Methods and Figure 1, the inclusion criteria and descriptive characteristics of study subjects are presented in Supplemental Table 1. Further information on the research design is available in the Life Sciences Reporting Summary linked to this article.
Figure 1. Experimental design.
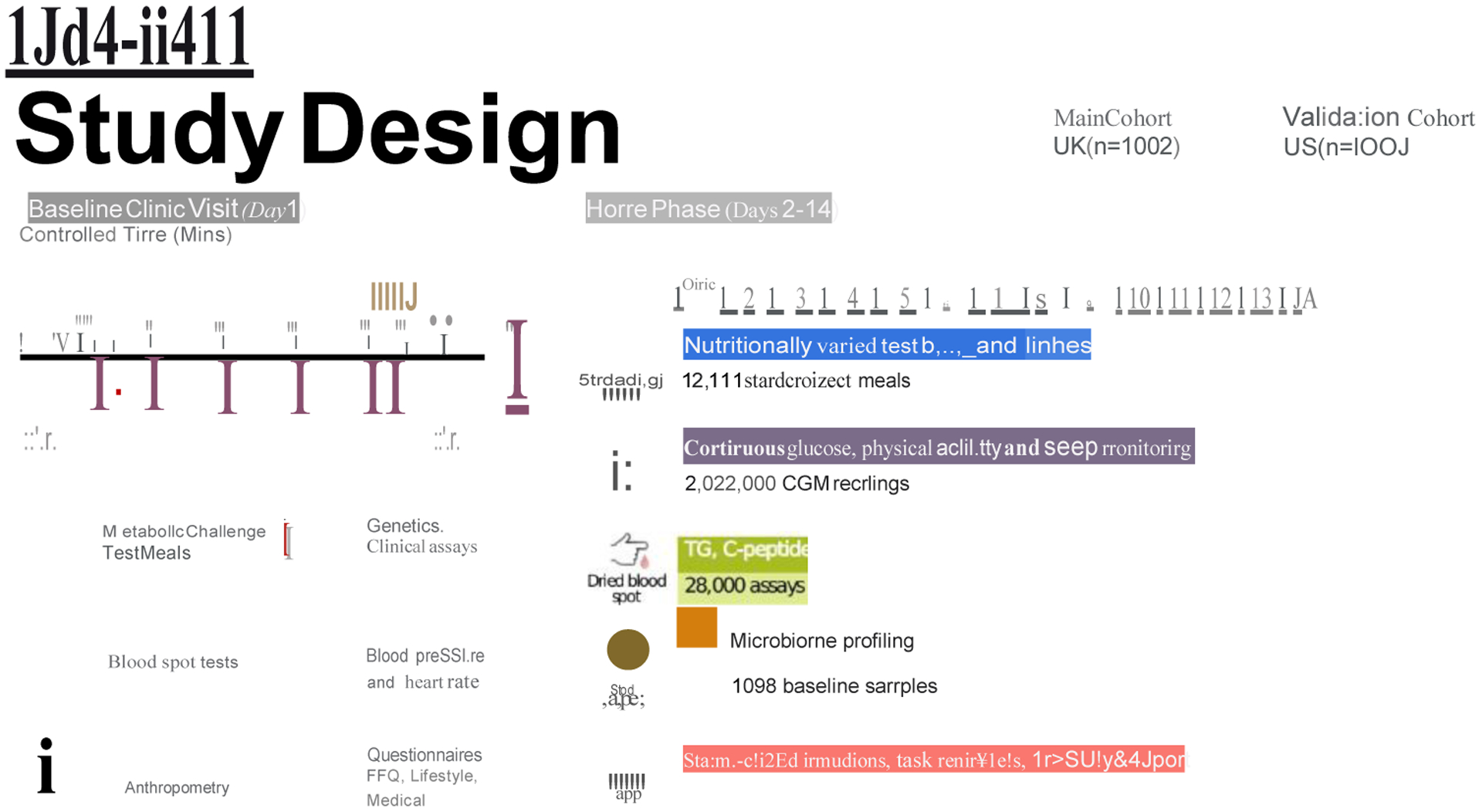
The PREDICT 1 study comprised a primary UK-based cohort (nmax = 1,002) and an independent US-based validation cohort (nmax = 100).
Inter- and intra-individual variation in postprandial responses
Inter-individual variability in postprandial responses was examined in a tightly controlled clinic setting following the sequential standardized test meal challenge after fasting (Figure 2a). The inter-individual patterns of response for each outcome was assessed using Levene’s test of variance. Heterogeneity across all postprandial time-points (fasting to 6-hrs) varied greatly for triglyceride (p = 3.931e-11), glucose (p = 2.91e-194) and insulin (p = 2.45e-17) concentrations. In serum, the population coefficient of variation was higher for postprandial triglyceride6hr-rise (103%) and glucoseiAUC0-2h (68%) compared with fasting values (50% and 10%, respectively). This was not true for insuliniAUC0-2h (59%) compared to fasting (69%; Figure 2a), suggesting that these measures of postprandial triglyceride and glucose concentrations, but not insulin, provide better discrimination of an individual’s metabolic tolerance than fasting values.
Figure 2. Variation in postprandial responses.
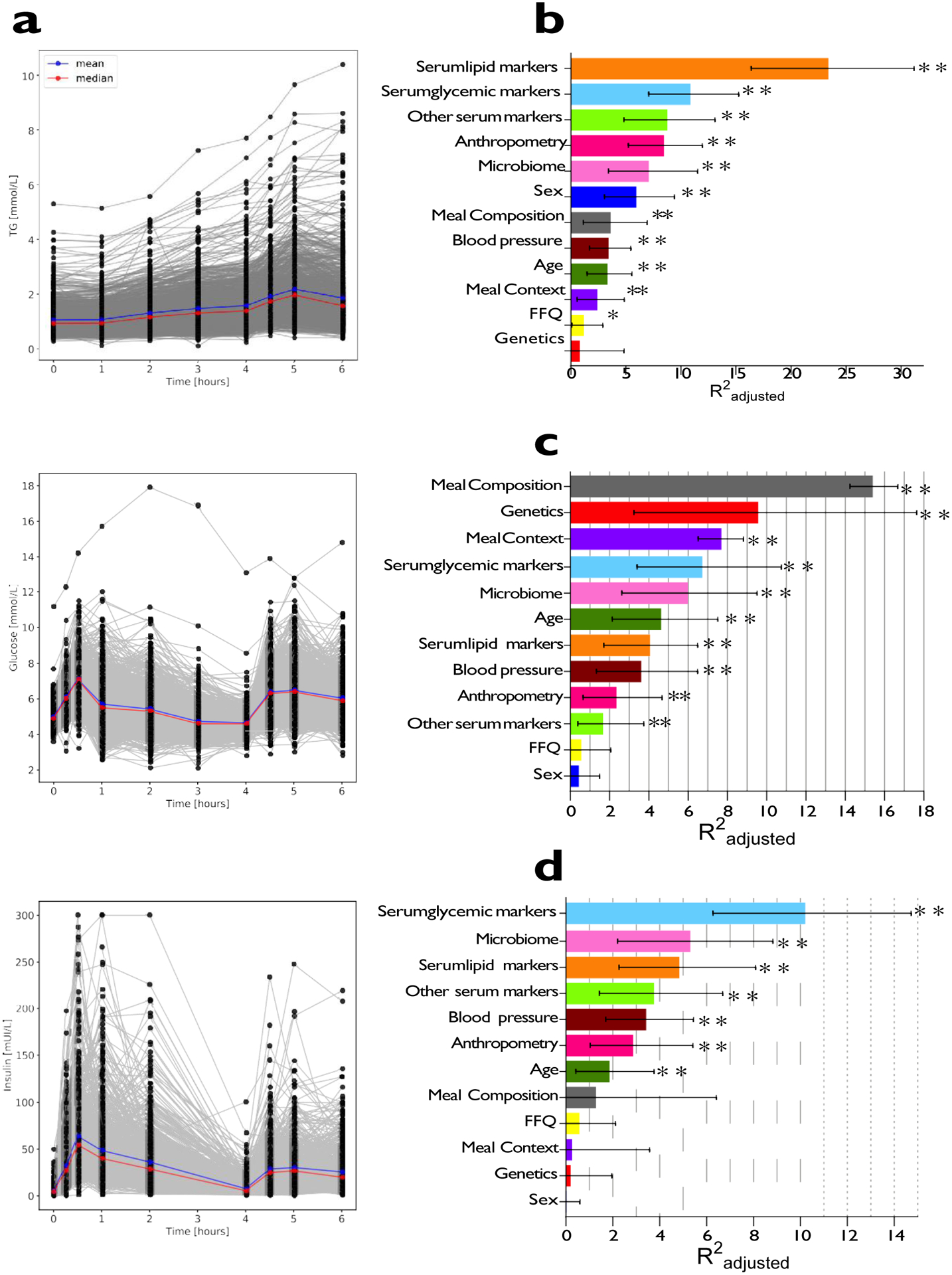
a. Inter-individual variation in triglyceride, glucose and insulin postprandial responses to the breakfast and lunch meal challenges in the clinic (n = 1002). b. Determinants of triglyceride6h-rise measured from DBS (comparison of meals 1 and 7). c. Determinants of glucoseiAUC0-2h measured by CGM (comparison of 7 test meals; 1, 2, 4, 5, 6, 7 and 8). d. Determinants of C-peptide1h-rise measured from DBS as a proxy for insulin (comparison of meals 2 and 3). Trait variations explained for each input variable are derived from separate (non-hierarchical) regression models. Values represent adjusted-R2 and error bars reflect 95% confidence intervals. Meal composition and Meal context adjusted-R2 values were derived from meal sample sizes as follows; triglyceride6h-rise, n = 712; glucoseiAUC0-2h, n = 9102; C-peptide1h-rise, n = 186. All other determinant values were derived from meal sample sizes as follows; triglyceride6h-rise, n = 920; glucoseiAUC0-2h, n = 958; C-peptide1h-rise, n = 960. TG = triglyceride, DBS = dried blood spots, CGM = continuous glucose monitor. * p<0.05, ** p<0.01, *** p<0.001 using multivariable linear regression.
A key assumption when developing personalized prediction algorithms is that an individual’s unique response to the same meal is reproducible. Much of the between-person phenotypic variability observed in studies examining response to diet interventions that include only a single test–response scenario could be a result of regression to the mean and other sources of error. Repeated-measures (multiple measures taken within individual at a single time-point and across multiple time points) can be used to partition error from true biological variability, thereby improving the precision of the estimate. Accordingly, we administered test meals of varying macronutrient composition in duplicate per participant, under similar conditions (see Methods and Supplemental Table 2 for details). We also used continual glucose monitors (CGMs), which provided sequential measures of blood glucose at 5 minute intervals during the study period. Intra-individual variability (repeatability) was assessed using intra-class correlation coefficients (ICC) for triglyceride, C-peptide (from DBS assays) and glucose (from CGM) measurements. The ICCs were: triglyceride6h-rise = 0.46 [95% CI 0.37, 0.54]; glucoseiAUC0-2h = 0.74 [95% CI 0.72, 0.75]; C-peptide2h-rise = 0.62 [95% CI 0.54, 0.69] (Supplemental Table 3). The differences in ICCs between triglyceride, C-peptide and glucose measurements partly reflect the different assays used (DBS and CGM) (see Methods).
Predicting individual postprandial responses within a population
We assessed the overall extent to which input variables (Supplemental Table 3) predict personal postprandial responses (Figure 2b–d), initially using multivariable linear regression. Input variables include: i) baseline characteristics (age, sex, clinical biochemistry (lipid, glycaemic and other measures), anthropometry)); ii) genetics (single nucleotide polymorphisms (SNPs)); iii) gut microbiome features); iv) habitual diet (from Food Frequency Questionnaire (FFQ)); v) meal context (sleep, previous meals, physical activity, meal sequence and /or timing); vi) meal composition (energy from carbohydrate, sugar, fat, protein and fiber). Postprandial glycemic responses were determined from serum and CGM measurements in the clinic and at home (from 7 standardized meals and 6,616 readings; see Methods). Postprandial C-peptide and triglyceride were determined (from two standardized meals) from serum and DBS assays collected during the clinic and home phases. We also tested the correlation between fasting and postprandial characteristics and found that the correlation between postprandial triglyceride, with regards to postprandial glucose and postprandial C-peptide measures was low (Figure 3a).
Figure 3. Relationship of baseline values, genetic and microbiome factors to postprandial responses.
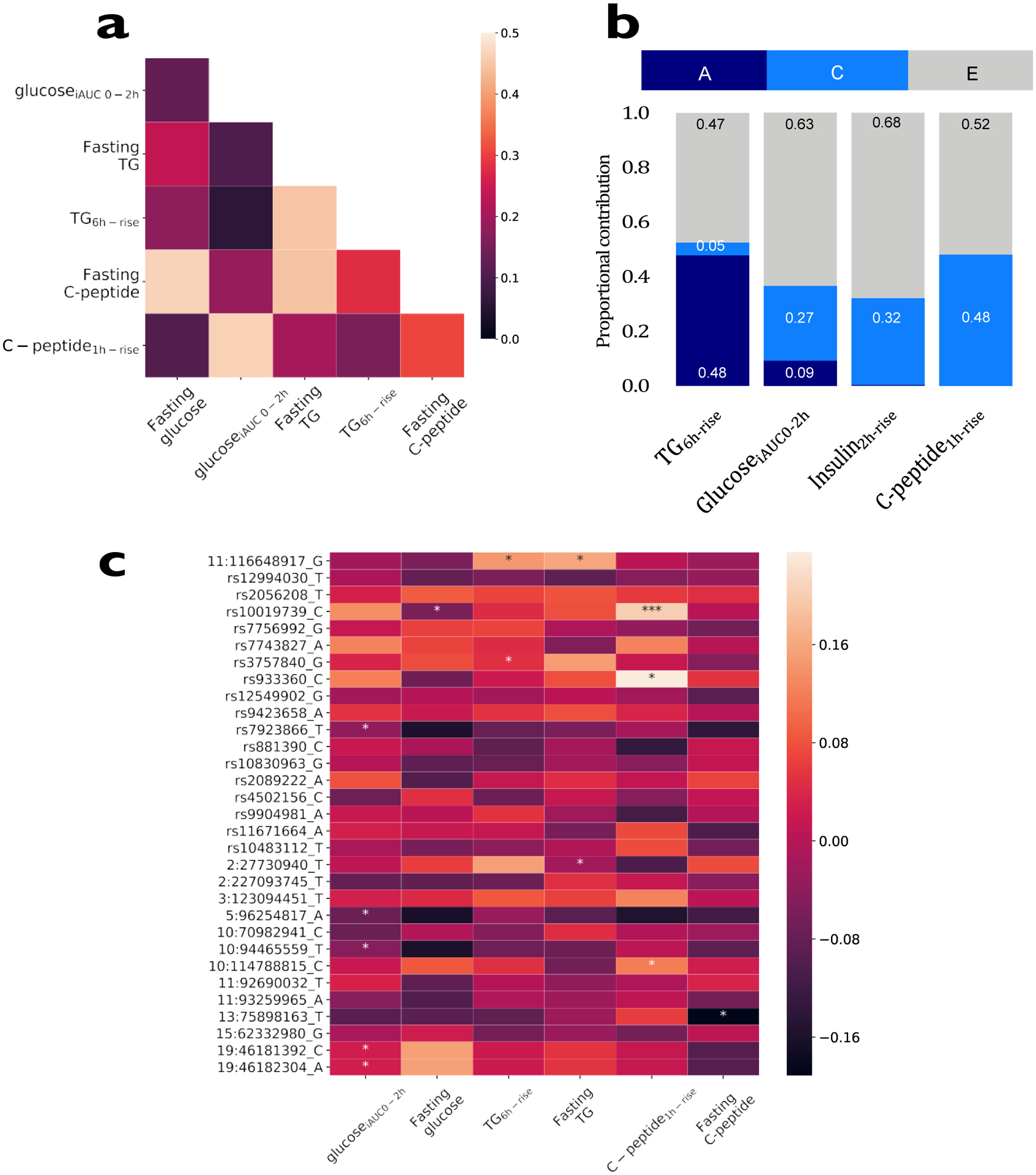
a. Pearson correlations between baseline values and postprandial prediction measures of 980 participants from the UK cohort. b. Heritability of postprandial responses (the ACE model was fitted on log-scaled postprandial responses for triglyceride, glucose, insulin and C-peptide) in 183 MZ and 47 DZ twin pairs. A; additive genetic component, C; shared environmental component, E; individual environmental component. c. SNP associations with postprandial measures focusing on SNPs identified in published postprandial trait GWAS17–21 (n = 241; * p<0.05, *** p<0.001, using two-sided chi-squared test).
Individual baseline characteristics.
The proportions of trait variance explained by individual baseline characteristics are shown in Figures 2b, c and d for triglyceride6h-rise, glucoseiAUC0-2h,and C-peptide1h-rise respectively (Supplemental Table 3).
Genetic factors.
The heritability of postprandial responses in the UK cohort was examined using classical twin methods (variance components analyses) to establish the upper bound of what might be predicted by directly measured genetic variation. Two-thirds of the cohort was recruited from the TwinsUK registry16, of which 230 twin pairs (n = 460; 183 MZ and 47 DZ) were studied for heritability. Additive genetic factors explained 30% of the variance in glucoseiAUC0-2h, whereas only 4% of the variance in triglyceride6h-rise and 9% of the variance in insulin2h-rise were explained in this way (Figure 3b). The estimated genetic variances in insulin1h-rise and C-peptide1h-rise were close to zero (Supplemental Table 4).
SNP-based genetic factors.
In a subgroup of participants who are part of the TwinsUK cohort and had genome wide genotyping previously measured with available GWAS data (n = 241), we tested whether 32 SNPs derived from previous genome-wide scans of postprandial glucose, insulin or triglyceride concentrations17–21 were also associated with the postprandial variables studied here. Several SNPs were significantly (p<0.05) associated with these variables (Figure 3c and Supplemental Table 4), but collectively explained only ~9% of observed variation in glucoseiAUC0-2h (Figure 2c), and less than 1% of variation for postprandial triglyceride and postprandial C-peptide (Figure 2b and 2d).
Gut microbiome (16S rRNA).
We estimated the contribution of gut microbiome composition using relative bacterial taxonomic abundances and measures of community diversity and richness, derived from 16S rRNA high-throughput sequencing of baseline stool specimens (Supplemental Table 4). We found that without adjusting for any other individual characteristics the gut microbiome composition explained 7.5% of postprandial triglyceride6h-rise, 6.4% of postprandial glucoseiAUC0-2h and 5.8% of postprandial C-peptide1h-rise.
Meal composition, habitual diet and meal context.
To determine the impact of the macronutrient composition of meals, we measured triglyceride6h-rise and C-peptide1h-rise for two standardized home phase meals of contrasting macronutrient compositions (for triglyceride, comparison of meals 1 and 7: 85 vs 28g of carbohydrate and 50 vs 40 g of fat at breakfast, both followed by a lunch of 71g carbohydrate and 22g fat; for C-peptide, comparison of meal 2 and 3: 71 vs 41 g of carbohydrate and 22 vs 35 g of fat; Supplement Table 2) in subsets of participants (n = 712 and n = 186, respectively). GlucoseiAUC0-2h was measured for seven standardized meals (comparison of meals 1, 2, 4, 5, 6, 7 and 8: 28 – 95 g carbohydrate; 0 – 53 g fat) totalling 9,102 meals in 920 individuals. The proportions of variance explained by meal composition, habitual diet, and by meal context are shown for triglyceride6h-risein Figure 2b, for glucoseiAUC0-2hin Figure 2c, and for C-peptide1h-risein Figure 2d. A multivariate regression model (meals 1, 2, 4, 5, 6, 7 and 8) revealed that the GlucoseiAUC0-2h (mmol/L*s) was significantly (P<0.001) reduced by 79, 142 and 185 for every 1g fat, fiber and protein respectively, after adjustment for carbohydrate consumption.
Machine learning model.
To estimate the unbiased predictive utility of the factors analysed, we used a machine learning approach robust to overfitting22. Random Forest regression models23 were fitted using all the informative features (meal composition, habitual diet, meal context, anthropometry, genetics, microbiome, clinical and biochemical parameters) to predict triglyceride6h-rise, glucoseiAUC0-2h and C-peptide1h-rise in the UK cohort dataset. The predicted values were compared with the observed values for each trait using Pearson correlation coefficients (r); these correlations were r = 0.47, r = 0.77 and r = 0.30 for triglyceride6h-rise, glucoseiAUC0-2h and C-peptide1h-rise, respectively. Similar correlations were observed in the held-out validation set (US cohort) and the model predictions for triglyceride6h-rise and glucoseiAUC0-2h were r = 0.42 and r = 0.75, respectively, but much weaker for C-peptide1h-rise (r = 0.14) (Figure 4). The features used to fit the models are reported in Supplemental Table 5. The repeatability and robustness of the machine-learning model is presented in the Extended Data Figure 4.
Figure 4 -. Machine learning models fitted in to postprandial measures.
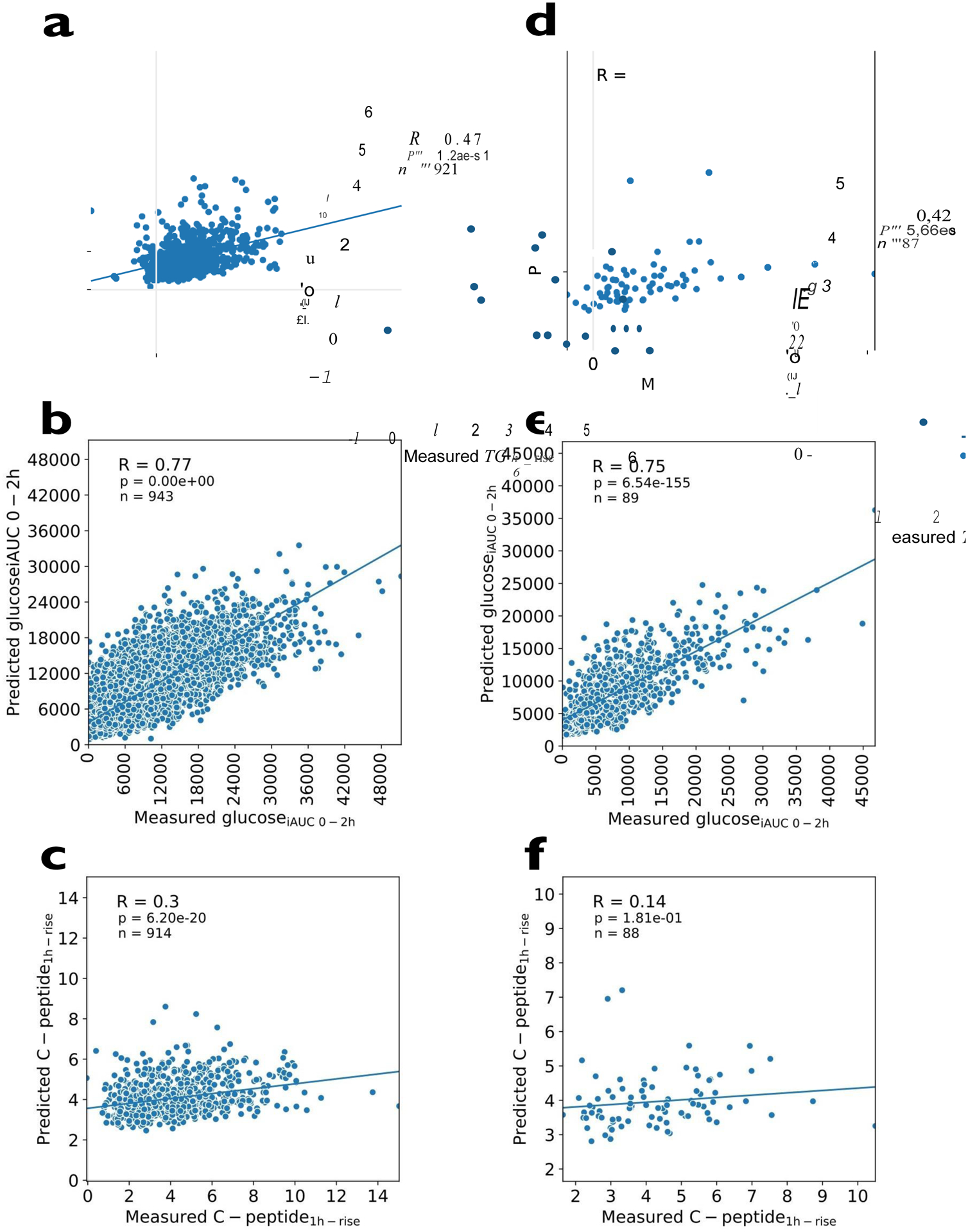
a. Machine learning model for TG6h-rise in the UK cohort. b. Machine learning model for glucoseiAUC0-2h in the UK cohort. c. Machine learning model for C-peptide1h-rise postprandial responses in the UK cohort. The machine learning models in the US validation cohort are shown in Figures 4 d-f. The relationship between variables is expressed as Pearson’s correlation coefficient (r) and denoted with a regression line; n represents participant number; the features used to predict each value are the same as those listed in the linear models in Figure 2b–d.
Postprandial responses in relation to surrogate scores of clinical outcomes.
We compared the extent to which fasting and postprandial concentrations for the different biomarkers could be used to predict impaired glucose tolerance (7.8–11.0 mmol/L 2 hours after an OGTT) and atherosclerotic cardiovascular disease (ASCVD) 10-yr risk score (Methods) by comparing the area under the receiver operator characteristics (ROC-AUC) curves; Figure 5. We found that fasting triglyceride and triglyceride6h-rise contributed similarly to the ROC-AUC for ASCVD risk, and that including both was more informative than including only one of them (Figure 5a). We also found that, although postprandial glucose was not as informative as fasting glucose, adding glucoseiAUC0-2h to fasting glucose resulted in a slightly higher ROC-AUCs (0.72 vs 0.69) for ASCVD 10-yr risk. Fasting C-peptide and fasting glucose were as effective (ROC AUC = 0.69) as fasting triglyceride in ASCVD prediction, whereas postprandial C-peptide (ROC AUC= =0.63) and postprandial glucose were weaker (ROC AUC = 0.62) than postprandial triglyceride (ROC AUC = 0.71). Fasting and postprandial triglyceride concentrations were weakly predictive (ROC AUC = 0.55 and 0.59, respectively) of impaired glucose tolerance (IGT), whereas fasting and postprandial C-peptide were moderately predictive (ROC AUC = 0.64 and 0.65 respectively), although with no added predictive value in combination. We did not include here the prediction of IGT using CGM glucose. This is because IGT is defined solely based on the blood glucose concentration at 2hrs during an OGTT, which is captured by the CGM glucose recording, and so the derivation of the predictor and the clinical score variables would be heavily dependent upon one another. Results were similar in the UK and US cohorts (Figure 5).
Figure 5. Associations between fasting and postprandial values for TG, C-peptide and glucose concentrations with clinical measures in the UK cohort.
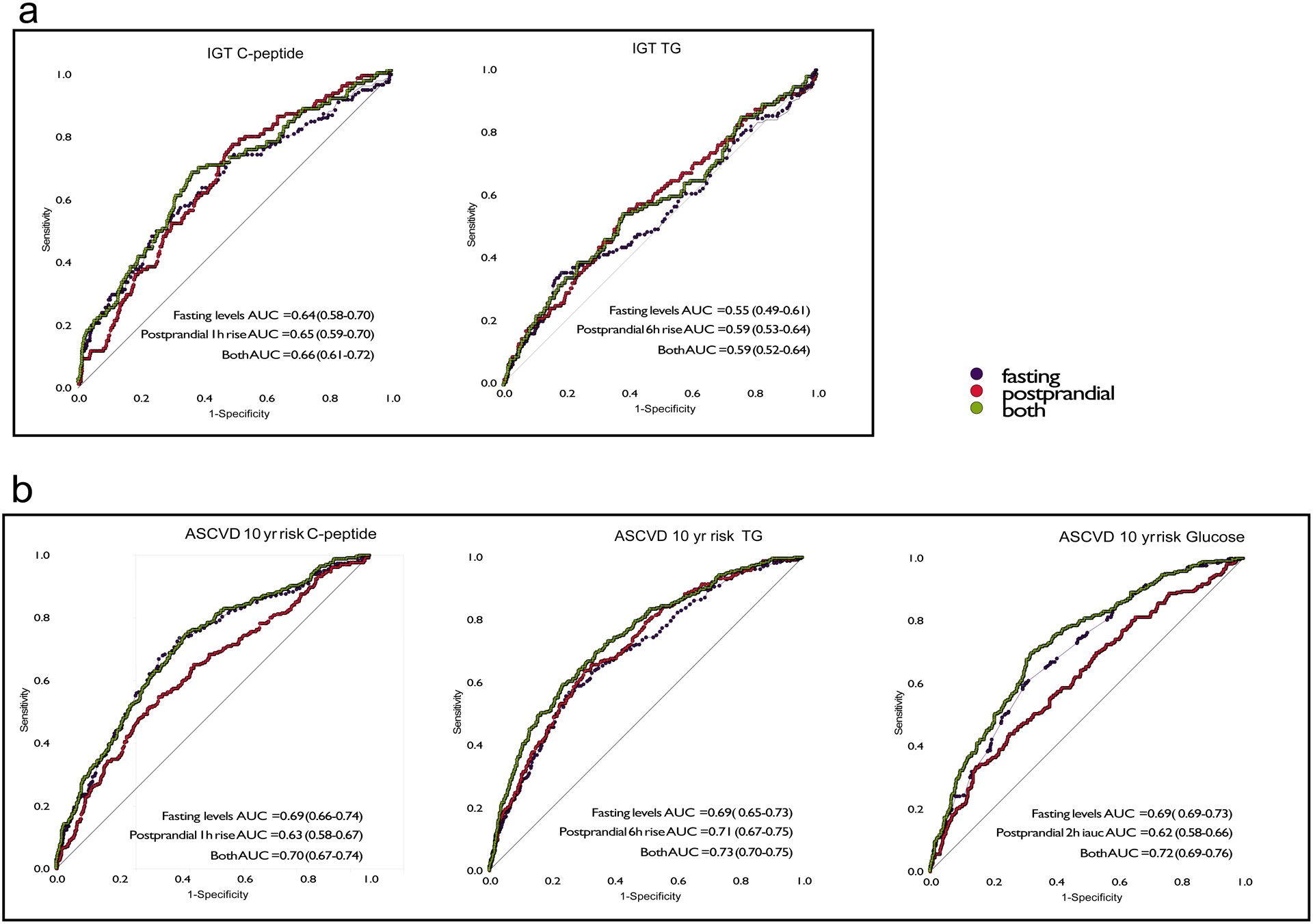
Receiver operator characteristics curves illustrating the predictive utility of fasting and postprandial TG, glucose and C-peptide measures to discriminate the bottom 70% from the top 30% of the cohort (cut-off ASCVD 10 year risk of 0.0183) for a. atherosclerotic cardiovascular disease (ASCVD) 10-year risk n = 951 independent samples from the UK and b. impaired glucose tolerance (IGT) n = 826 independent samples from the UK. The same analyses were performed in the US cohort (n = 92 independent samples) resulting in ROC AUC (95%CI) values for ASCVD 10 year risk of: C-peptide fasting AUC = 0.68 (0.56–0.80), postprandial AUC = 0.66 (0.54–0.77), both AUC = 0.69 (0.58–0.81); TG fasting AUC = 0.73(0.63–0.84), postprandial AUC = 0.75 (0.65–0.85), both AUC = 0.77 (0.67–0.88); and glucose fasting AUC = 0.74-(0.63–0.85), postprandial AUC = 0.64 (0.52–0.76), both AUC = 0.76 (0.64–0.85). For impaired glucose tolerance values were: C-peptide fasting AUC = 0.66 (0.53–0.80), postprandial AUC = 0.59 (0.46–0.72), both AUC = 0.67 (0.54–0.80); and Triglyceride fasting AUC = 0.66 (0.53–0.80), postprandial AUC = 0.59 (0.46–0.72), both AUC = 0.61 (0.54–0.80).
Decoding individual responses
Having investigated postprandial responses within the population, we then explored the responses at the individual level. We examined glycemic responses, as the granular CGM data collected during the at-home phase enabled us to assess real-world effects in detail, which was not possible for triglyceride or C-peptide. We investigated how much of an individual’s postprandial response is a attributable to a meal’s glycemic properties, compared with how the variation resulting from other modifiable factors such as meal timing, exercise and sleep.
We first examined the contribution of the meal. Although it is a widely held notion that, for an individual, variations in meal composition are primarily responsible for the variation in responses to food and that ranking of meal responses should be the same for all people24–25, we explored whether meal-specific responses unique to the individual exists. We ranked the order of each participant’s glucoseiAUC0-2h for every possible pair of standardized meals consumed at home. We then determined how frequently these rankings differed for each participant. For most pairs of meals, the ranking was the same for all individuals (e.g. OGTT has a higher glucoseiAUC0-2h than high-fiber muffins in all participants, Figure 6a). However, for select pairs of meals, the ranking was reversed in up to 48% of participants, such as between the medium fat and carbohydrate at lunch vs high carbohydrate breakfast (350 of 727 participants) (meal 2 vs. meal 4; Supplemental Table 2). In 186 out of 498 (37.3%) participants, discrepancies were also seen between the high fat and the high protein meals (meals 7 and 8). The distribution of how these meals were ranked for the participants of the PREDICT study is presented in Extended Data 2.
Figure 6. Person-specific diversity in postprandial response.
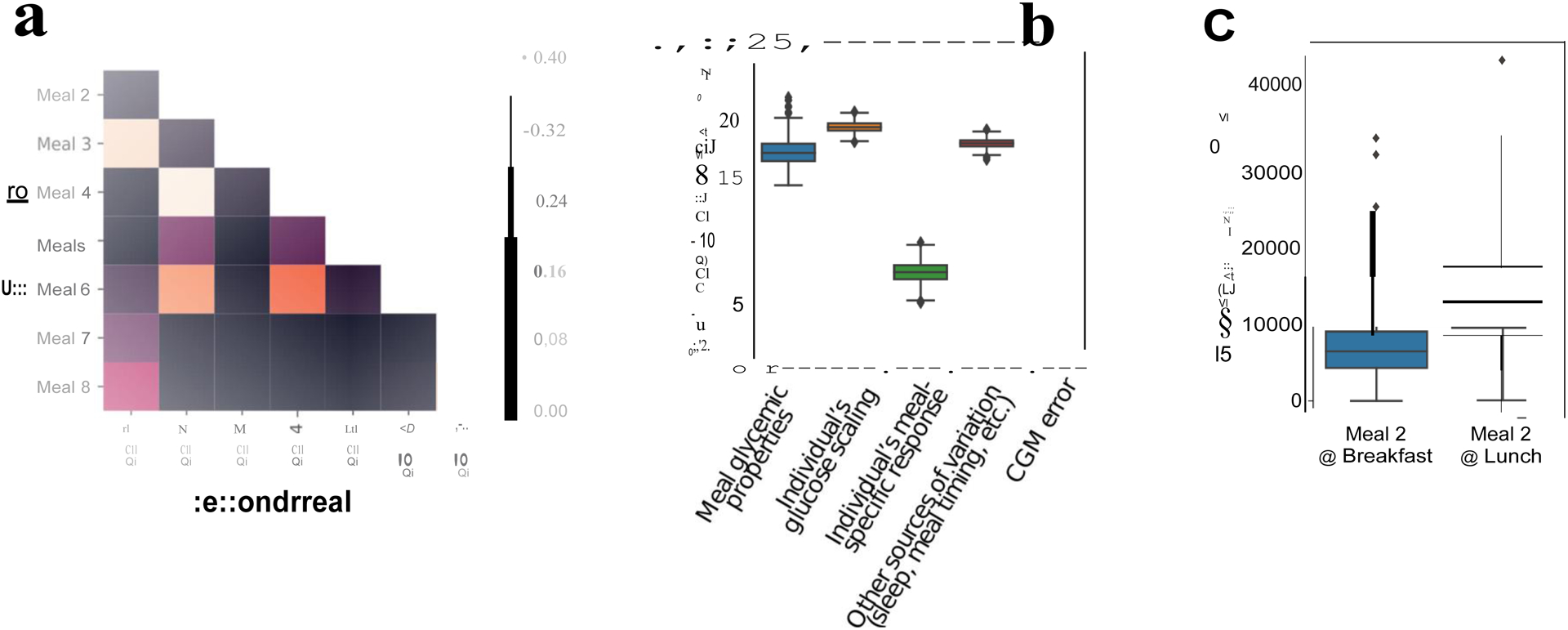
a. Proportion of times in the PREDICT 1 study that the ranking of the glycemic response (glucoseiAUC0-2h) to pairs of set meals was altered (n = 828, UK cohort). b. Effect size for factors explaining glycemic response. The different sources of variation were estimated using ANOVA, as described in Supplemental Table 3. The x-axis can be approximately interpreted as percent increase (or decrease) in iAUC attributable to the model parameters (n = 483 individuals) c. Time of day effects. (n = 920, UK cohort). Boxes show quartiles (25th, 50th, 75th percentiles); whiskers show the 95% interval.
We note that the reordering of meal rankings could have been the result of noise. We therefore used ANOVA to estimate the effect size for the different factors explaining glycemic response (Figure 6b), including person-specific effects (effects that vary between people but not between meals). As described in the Methods, we considered not only the effect of the meal macronutrient and energy content in the response (meal composition), but also considered how each individual responded on average to all their set meals relative to the population (individual glucose scaling), as well as the effect of the individual’s meal-specific response, the error attributable to the glucose measurement and other sources of variation (including modifiable sources of variation such as sleep, circadian rhythm and exercise).
We found that, consistent with the linear models described earlier, the ANOVA models show that there are three meal-related factors explaining individual glycemic responses. Meal macronutrient composition alters iAUC by 16.73% (95%CI 15.37 – 18.92%), but the individual glucose scaling is larger, altering iAUC by roughly 18.74% (17.96% – 19.46%), while the individual’s meal-specific response is much smaller, affecting the final meal iAUC by 7.63% (6.11% – 8.96%). Other modifiable sources of variation not directly related to the meal composition, such as meal timing, exercise and sleep, contributed similar amounts of variance as the meal’s composition (Figures 6b and c). To investigate whether modifying the order in which meals are consumed and time of the day affect glycemic responses, we looked at participants eating an identical meal (meal 2) for breakfast and lunch. The average glycemic response for the same individuals was on average 2-fold higher (t-statistic = −35.7, 2721 d.f.; P< 0.001) when the meal was ingested for lunch (mean glucose 2h iAUC = 14254 SD = 6593) (4h following the metabolic challenge breakfast) than when ingested for breakfast (mean glucose 2h iAUC = 7216, SD = 4157), although with wide individual variation (Figure 6c).
Discussion:
Nutrition and health are intimately linked. Each day people make diet-related decisions that are influenced by perceived enjoyment and satiation, as well as health benefits and harm attributed to specific foods and beverages. Standard nutritional guidelines2–4 are typically based on population averages. However, it is increasingly evident that one-size nutritional recommendations do not fit all, which is exemplified by the variable efficacy of tightly controlled lifestyle intervention trials26–29. To address these challenges, we undertook a two-week interventional trial, including a tightly-controlled in-clinic day and a two-week at-home phase, where postprandial metabolic responses to a series of standardized meals were obtained in more than 1,000 healthy adults from the UK and USA. The primary aim was to derive algorithms that predict an individual’s postprandial metabolic responses to specific foods. The core outcomes were variations in blood concentrations of triglyceride, glucose and insulin (or C-peptide), as these biomarkers work in concert to affect cardiometabolic risk8,30.
In many cases, we observed responses that contrast with those reported in traditional clinic-based studies, thereby reshaping conclusions about the key factors influencing responses to foods. For example, genetic influence was less than expected, especially for triglyceride, while modifiable factors like meal timing conveyed larger effects than anticipated.
Meal composition has large effects on postprandial insulinemic and lipidemic response31Some small studies suggest that meals with high-fat and/or protein content elicit very different postprandial responses than lower-fat and/or protein meals with identical carbohydrate content (reviewed in31). The type of fat in a meal also alters the lipemic response32. However, measuring postprandial triglyceride and C-peptide at-home in large cohorts is both logistically challenging and places a considerable burden on. Thus, for pragmatic reasons, only two pairs of meals (high fat and high carbohydrate, respectively) were used to calculate postprandial triglyceride and C-peptide responses and the difference in macronutrient content of these meals was low. This limited number of different meals and their relatively similar macronutrient content might explain why the effects seen for postprandial triglyceride and C-peptide were lower than expected.
In addition to fasting concentrations of triglyceride and glucose, we found that postprandial triglyceride and glucose concentrations were informative for IGT and CVD risk determination. However, postprandial C-peptide measurements provided no additional information over fasting concentrations. We found that although postprandial triglyceride and glucose responses were highly variable between individuals, a person’s response to the same meals was often similar and therefore predictable. Any given individual generally responds comparably to different meals of the same macronutrient profile, with some people experiencing large postprandial excursions across most meals, whereas others consistently experience modest responses. This is important for individualized prediction and recommendations, as it suggests that once one has learned about an individual’s postprandial response to specific foods, their response to other foods could be inferred.
We show that a person’s glycemic response is the result not only of individual-specific glucose scaling, which determines whether a person is a high or low responder to all meals, but that there are also meal-specific responses unique to an individual. Possible explanations include individual genetic differences in the ability to digest high-starch meals33. Zeevi and co-workers5 reported an example where one participant had an exaggerated glycemic response to a banana but not to a cookie, whereas the second participant had the opposite response. We assessed this phenomenon in our data and found that individual glucose scaling and meal-specific responses both exist, but individual meal-specific responses are generally much more effective than scaling.
People differ greatly in their responses to diet interventions. The DIETFITS study, for example, randomised 609 people to either a healthy low fat or a healthy low carbohydrate diet for 12-months34. By study end, average weight loss was similar between groups (~5–6kg), but wide variations were seen within groups (−30kg to +10kg). Elsewhere, the Diabetes Prevention Program showed that although a standardized intensive lifestyle intervention focusing on changes in diet (tailored only to the energy requirements of the individual) lowered diabetes risk substantially28, its efficacy varied greatly across the study population26,27, and was determined to some extent by genetic factors29. While response to diet interventions will depend partly on adherence, findings from the PREDICT trial and elsewhere35,36 suggest that even in highly-adherent participants, substantial response variations exist, which might be predictable. In PREDICT, non-food-specific factors (e.g., meal timing, sleep, activity) were highly informative of these person-specific responses.
Previous large-scale studies of postprandial responses have focused solely on glycemic outcomes because assessing postprandial triglyceride and insulin concentrations in free-living conditions is challenging2,25. Here, we assessed glycemic responses with CGMs, but also assessed triglyceride and C-peptide concentrations during the at-home period of the study using a validated DBS method and support from a specifically designed mobile app (Methods).The low correlation between triglyceride and glucose suggests that prediction algorithms relying solely on glucose would be insufficient for the detection of dysregulated triglyceride responses.
The prediction algorithms we developed are likely to have been strengthened by the use of randomized, mixed meals, containing combinations of macronutrients reflective of those seen in real-world settings, rather than supra-physiological lipid or carbohydrate challenges, as used in previous studies.
In general, genetics, contrary to our expectations, was not a predominant determinant of these responses; we found that the heritable fraction (the trait variance explained by additive genetic factors) of C-peptide and/or insulin concentrations at 1 hr was very low (0.3%) and at 2 hrs remained low (9.1%). The heritable fractions for postprandial triglyceride (6hr rise) and glucose (2hr iAUC) responses were higher, but still modest (16% and 30%, respectively). Despite the wealth of publicly available SNP data (see: www.type2diabetesgenetics.org), there is no robust data for these specific postprandial traits, as almost all published GWAS of serological traits have focused on fasting values. Nevertheless, in exploratory analyses, we examined the predictive value of loci previously linked to post-challenge triglyceride, glucose or insulin concentrations17–21 but found that the predictive utility of these variants was poor, particularly for triglyceride and C-peptide (Figure 3c). The modest heritability of postprandial traits means that even in an unrealistically optimistic scenario, where most of this trait variance is explained by known DNA variants, it is unlikely that prediction algorithms using DNA variant data alone, which many direct-to-consumer nutrigenomics companies advocate, would succeed.
The lack of a major genetic component to these traits highlights the likely involvement of modifiable environment exposures. Indeed, we found that meal composition and context (e.g. meal timing, exercise, sleep and circadian rhythm) were core determinants of postprandial metabolism. These predictions were strengthened using data on gut microbiome diversity. Using machine learning combining all relevant data, an individual’s postprandial triglyceride and glycemic responses could be meaningfully predicted, with similar results in the US validation cohort. For C-peptide, the prediction was much weaker in the validation cohort (r = 0.30 UK, r = 0.14 US), possibly reflecting the lower number of test meals relative to the number of input variables, which could adversely affect the reliability of the prediction37. The postprandial glycemic predictions were similar to those reported by Zeevi and colleagues5, although the analysis methods and input features are not directly comparable.
Despite having developed these prediction algorithms, there is scope for improvement, such as inclusion of a more diverse array of meal interventions and with more detailed assessments of contextual factors than in the current study. Technological advances could also help to improve predictions. For example, although glucose can be continuously assessed with CGMs, no commercially available devices suitable for free-living assessments of continuous insulin and triglyceride concentrations currently exist. Moreover, owing to the differences in tolerability and the lower limit of detectable responses of dietary carbohydrates compared with fats38, our trial suggests that the prediction of postprandial glucose is methodologically superior to triglyceride responses (see Fig 2b–d). Difficulties in directly comparing changes in triglyceride and glucose were a limitation of our study. Continuous, accurate measures of these traits could substantially improve predictions owing to reductions in model error and the ability to study non-linear patterns of response, which may be important. The inclusion of deep ‘-omics’ data may further enhance the predictive ability of these algorithms; for example, here we used microbiome data derived from 16S RNA sequencing, which, whilst proving valuable for prediction (explaining 6.4% and 7.5% of the variances for glucose and triglyceride responses, respectively), may prove even more informative if derived from higher-resolution metagenomic sequencing. The nutritional signatures detectable within the metabolome, both in blood39 and feces40, suggest that including a larger metabolomics panel and quite probably other -omics data, e.g. meta-transcriptomics, transcriptomics or proteomics, in our algorithms would add costs but also enhance predictions. Using FFQs, we found that habitual diet explains a small proportion (<2%) of an individual’s postprandial responses. However, FFQs have well-known limitations, and other objective approaches may be considerably less biased and error prone27. Pairing this with short-term assessments, like the weighed dietary record included in the PREDICT study app, may help mitigate these limitations. More comprehensive challenge tests might also reveal new aspects of postprandial metabolism; here, we used a 6-hr meal tolerance test, as this was deemed the maximum duration that most participants were likely to accept. Data from longer duration challenge tests (up to 8hrs), for both glucose and triglyceride responses, may provide valuable information.
For postprandial triglyceride and glucose responses, the prediction models derived in the UK cohort performed almost as well in the independent US validation cohort, which is reassuring given differences in environmental factors; nevertheless, both cohorts were comprised of younger healthy adults of European ancestry. Thus, the generalization of our findings would require validation in people of non-European ancestry, older adults, and in people with diseases that affect metabolism such as diabetes. The clinical implications of our predictions will require appropriately powered longitudinal studies.
In conclusion, this is the most comprehensive assessment to date of metabolic responses to nutritional challenges in a rigorous intervention setting. We observed considerable inter-individual differences in postprandial metabolic responses to the same meals, challenging the logic of standardized diet recommendations. These findings, in addition to the scalability of the assessment methods and the accuracy of the prediction algorithms described here, mean that, at least from a cardiometabolic health perspective, population-wide personalized nutrition has potential as a strategy for disease prevention.
Methods
Study population, study design, recruitment criteria, meal challenges and Zoe app.
Study population
The PREDICT 1 study (Personalised Responses to DIetary Composition Trial) was a multinational study conducted between 5th June 2018 and 8th May 2019. The primary cohort was recruited at St. Thomas’ Hospital in London, UK and a validation cohort (that underwent the same profiling as in the UK) assessed at Massachusetts General Hospital (MGH) in Boston, Massachusetts as described in the detail in the protocol41. In the UK, participants (target enrolment = 1,000) were recruited from the TwinsUK cohort, an ongoing research cohort described elsewhere16 and online advertising (Extended Data Figure 1a). In the US, participants (target enrolment = 100) were recruited through online advertising, research participant databases and Rally for Research (https://rally.partners.org/), an online recruiting portal for research trials (Extended Data Figure 1b). Ethical approval for the study was obtained in the UK from the Research Ethics Committee and Integrated Research Application System (IRAS 236407) and in the US from the Institutional Review Board (Partners Healthcare IRB 2018P002078). The trial was registered on ClinicalTrials.gov (registration number: NCT03479866), as part of the registration for the PREDICT Programme of research, which also includes 2 other study protocol cohorts. The trial was run in accordance with the Declaration of Helsinki and Good Clinical Practice.
Study participants were healthy individuals aged between 18–65 years and able to provide written informed consent. Criteria used to assess eligibility are listed in Extended Data Table 1. Exclusion criteria included; ongoing inflammatory disease; cancer in the last three years (excluding skin cancer); long term gastrointestinal disorders including IBD or Coeliac disease (gluten allergy), but not including IBS; taking the following daily medications: immunosuppressants, antibiotics within the last three months; capillary glucose level of >12mmol/L (or 216 mg/dL), or Type I diabetes mellitus, or taking medications for type II diabetes mellitus; currently suffering from acute clinically diagnosed depression; heart attack (myocardial infarction) or stroke in the last 6 months; pregnant; vegan, suffering from an eating disorder or unwilling to take foods that are part of the study.
Study design
1,002 generally healthy adults from the United Kingdom (UK) (non-twins, and identical [monozygotic; MZ] and non-identical [dizygotic; DZ] twins) and 100 healthy adults from the United States (US) (non-twins; validation cohort) were enrolled and completed baseline clinic measurements. Key outcomes include postprandial metabolic responses (0–6h; blood triglyceride, glucose and insulin concentrations) to sequential mixed-nutrient dietary challenges (containing 86g carbohydrate and 53g fat at 0h; 71g carbohydrate and 22g fat at 4h) administered in a tightly controlled clinic setting on day 1 (Figure 1). A second set of outcomes were assessed over the subsequent 13-days at home. Lipemic and C-peptide responses (as a surrogate for insulin) to two standard meals differing in fat and carbohydrate composition were assessed at home using dried blood spot (DBS) assays collected at three postprandial time-points. Glycemic responses to eight meals (seven in duplicate) of different macronutrient (fat, carbohydrate, protein and fiber) content were assessed using continuous glucose monitors (CGM). In addition, participants wore physical activity and sleep monitors for the duration of the study and provided stool samples for microbiome profiling.
We selected specific timepoints and increments for triglyceride, glucose, insulin and C-peptide to reflect the different pathophysiological processes for each measure. To monitor compliance, all test meals consumed by participants were logged in the Zoe app (with an accompanying picture) and reviewed in real time by the study nutritionists. Only test meals that were consumed according to the standardized meal protocol were included in the analysis.
Baseline clinic visit (Day 1):
Participants in the UK were mailed a pre-visit study pack with a stool collection kit and a health and lifestyle (amended Twins Research health and lifestyle questionnaire 42and food frequency questionnaire (European Prospective Investigation into Cancer and Nutrition (EPIC) Food-Frequency Questionnaire (FFQ)43). In the US, minor modifications were made to the health and lifestyle questionnaires to conform to a US population and the Harvard Semi-quantitative FFQ, a validated US instrument, was substituted for the EPIC FFQ. Stool collection and questionnaires were completed at home and returned to study staff at the baseline visit. Participants were asked to refrain from exercise and to limit fat, fiber and alcohol intake for 24 hours beforehand and to abstain from caffeine from 6pm the night before the baseline visit. Participants arrived at 8:30am for their visit, having fasted from 9pm the night before, and were cannulated in the forearm (antecubital vein) to collect a fasted blood sample, before being fitted with wearable devices (continuous glucose monitor (CGM; Freestyle Libre Pro, Abbott, Abbott Park, IL, US) and wrist-based triaxial accelerometer (AX3, Axivity, Newcastle, UK)). Heart rate and blood pressure were measured using an automated blood pressure monitor while fasted (in triplicate, with mean of second and third measurements recorded). Participant weight, height, hip and waist circumference were measured using standard clinical techniques. Fasting blood glucose level was checked using HemoCue Glucose 201 + System (Radiometer, Crawley, UK) or Stat Strip (Nova Biomedical, Waltham, MA, US) in the UK and US, respectively.
Following the baseline blood draw, participants consumed a breakfast (muffins and milkshake at 0 min) and lunch (muffins at 240 min) test meal (Supplemental Table 2), each to be consumed within 10 minutes. Additional venous blood was collected via cannula at 15, 30, 60, 120, 180, 240, 270, 300 and 360 minutes. Participants had access to water to sip throughout the visit. Between blood sampling, participants were trained in how to complete the study at home, including when and how to consume standardised test meals, perform DBS, and use the Zoe study app. Upon completing their baseline visit, participants received all the components necessary to complete the home-phase.
Home-phase (Days 2–14):
During the study home-phase, participants consumed multiple standardised test meals for breakfast and lunch over a 9–11-day period, differing in macronutrient composition (carbohydrate, fat, protein and fiber) while wearing the CGM and accelerometer. Participants recorded all of their dietary intake and exercise on the Zoe study app throughout the study. DBS tests were completed on 4 days before and after test meals, as outlined in the online protocol41. Following completion of the home-phase, participants returned all study samples and devices to study staff via standard mail.
Test meal preparation, nutrient composition, timings and standardised participant test meal instructions
Upon completing their baseline visit, participants received a home-phase meal pack containing test meal components (nutrient composition; Supplemental Table 2) which they consumed according to standardised instructions for breakfast and, on some days, lunch. Test meals consisted of either an oral glucose tolerance test (OGTT; on 2 days) or muffins, which were consumed on their own or paired with chocolate milk, protein shake, or commercial fiber bars and ordered according to one of 3 protocol groups described in Supplemental Table 2. Meal order for the 3 protocol groups was randomised using Microsoft Access for each participant, using a 2-block randomisation and 1 non-randomised block.
Participants were instructed to fast for a minimum of 8 hours prior to consuming a test breakfast meal, and to fast for 3 or 4 hours after meal consumption (depending on test meal; in protocol 1, fasting period was 3 hours for Meal 5 and 4 hours for all other meals; in protocols 2 and 3, the fasting period was 3 hours for all breakfast meals, excluding combinations of breakfast and lunch, where fasting periods were 4 and 2 hours, respectively). They were advised to limit exercise and drink only plain, still water during fasting periods. When fasting was completed, participants could eat, drink and exercise as they liked for the rest of the day. Participants were asked to consume all muffin-based meals within 10 minutes and the OGTT within 5 minutes and to notify study staff if this was not achieved, in which case the data was excluded from analysis. If the participant chose to accompany their home-phase muffin-based test meals with a tea or coffee (with up to 40ml of 0.1% fat cow’s milk, but no sugar or sweeteners), they were instructed to consume this drink consistently, in the same strength and amount, alongside all muffin-based test meals throughout the study. Participants were instructed to not consume any food or drink other than water alongside the OGTT, and to avoid physical activity during the 3-hour fasting period that followed it.
Test meals and any dietary intake consumed within fasting periods, including accompanying drinks, were recorded in the Zoe app by participants with the exact time at consumption and ingredient quantities so that compliance could be monitored by study staff. Only test meals that were completed according to instructions were included in analysis.
Test meals were prepared and packaged in the Dietetics Kitchen (Department of Nutritional Sciences, King’s College London, London, UK) using standard ingredients; plain flour, sugar, baking powder, vanilla essence, milk, egg, salt, high-oleic sunflower oil, whey protein powder, chocolate milkshake powder (Nesquik, Nestle, Gatwick, UK), and commercially available fiber bars (Chocolate Fudge Brownie, Fiber One, General Mills, MN, US; Goodness Bar Apple & Walnut, The Food Doctor, Hessle, UK). Test meals were shipped frozen, under temperature controlled conditions, to the US to limit variability of the intervention. Participants were instructed to freeze their muffins at home and defrost each set of muffins in the fridge the night before consuming them. Test meal drinks were prepared by the participant at home by mixing pre-portioned powder sachets with long-life milk provided (Meal 1, 220ml 0.1% fat milk; Meal 8, 200ml 1.6% fat milk). Powder sachets and fiber bars were stored at room temperature until consumption. The OGTT (Meal 5) consisted of a pre-portioned powdered glucose sachet which participants mixed with 300ml water in the UK. In the US participants were provided with pre-mixed OGTTs ready for consumption (Cat# 82028-512; VRW, US).
Zoe study app and dietary assessment methodology
The Zoe app was developed to support the PREDICT 1 study by serving as an electronic notebook of study tasks, a tool for recording all dietary intake and a portal for communication with study staff. The app sent participants notifications and reminders to complete tasks at certain time-points, such as when their test lunch meals and DBS were due, and asked participants to report their hunger and alertness levels on visual analogue scales truncated from Flint et al44. Participants were asked to log in the app any exercise which would not be well captured by a wrist-affixed accelerometer, such as cycling. Participants logged their full dietary intake using the app over the 14-day study period, including all standardized test meals and free-living foods, beverages (including water) and medications. Data logged into the app was uploaded onto a digital dashboard in real time and reviewed and assessed for logging accuracy and study guideline compliance by study staff.
‘Study staff trained all participants at their baseline clinic visit on how to accurately weigh and record dietary intake through the Zoe study app, using photographs, product barcodes, product-specific portion sizes, and digital scales. Study nutritionists also reviewed food logging data by comparing the photographs uploaded by subjects with the items they logged on the app. Any uncertainties were clarified actively with the participant through the app messaging system or via phone while the participant was on the study.
Protocol versions and amendments
Protocol amendments for the PREDICT study, post-commencement of the study and participant enrolment, are as follows: The first amendment (approved by UK IRAS 1st August 2018) allowed additional test meals to be included in the home-phase and participants’ logging of gut transit time by using a Metabolic Challenge Breakfast (Meal 1) on the clinic day dyed blue with food coloring. The DBS protocol was also changed according to physiological peaks in biomarkers (triglyceride or C-peptide). Starting on 28 Aug 2018, triglyceride was measured on Days 2–3 at fasting, 300 and 360 minutes post-prandially, while C-peptide was quantified on Days 4–5 at fasting, 30 and 120 minutes post-prandially as described for Protocol Group 2. A second saliva sample collection was also added on the clinic day, at 30 minutes after the metabolic challenge breakfast, to measure salivary amylase production post prandially and provide a comparison to fasted amylase levels. The second amendment (approved by UK IRAS 2nd September 2018) was a change in the lower BMI limit for eligibility to 16.5kg/m2 (originally 20 kg/m2). Minor meal changes were made, not requiring ethical approval, which resulted in Protocol Group 3 (implemented in January 2019). In the US, on 3 January 2019, the IRB approved an amendment (PREDICT-US v2.0) to address meal changes introduced in the UK for Group 3 and to allow the use of multiple CGMs on the same participant. No other major amendments to the intervention protocol were made during the study period in the US.
Outcome variables and sample collection, handling and analysis
Dried blood spot collection, method validation and analysis.
Dried blood spot collection:
Triglyceride and C-peptide were quantified from DBS tests completed by participants at the baseline visit (at fasted baseline and 300 minutes post-breakfast; for method validation) and on the first 4 days of the home-phase while consuming test meals (test timings and associated meals are outlined in the online protocol41).
The Zoe app sent participants reminders to complete their DBS tests at due times, which participants then logged in the app by recording the time at testing and a photo of the completed card for quality assessment by study staff. Test cards not meeting the quality protocol (multiple small spots or inadequate coverage) were not included in analysis. Test cards were stored in aluminium sachets with desiccant once completed and placed in the fridge at the end of the study day or until participants mailed them back to the study site. DBS cards were then frozen (−80 °C) and shipped for analysis (Vitas Analytical Services, Oslo, Norway).
Dried blood spot method validation:
DBS C-peptide and triglyceride concentrations were validated during PREDICT, against venous serum concentrations collected during the baseline clinic visit at 0 and 300 minutes post breakfast test meals. Correlations between the two methods were found to be high; for triglyceride (1,772 pairs) Pearson’s r = 0.94; for C-peptide (1,679 pairs) Pearson’s r = 0.91.
Quantification of total triglyceride from DBS:
From the DBS sample, 2 punches were taken and transferred into a HPLC vial and lipids extracted with methanol at 600 rpm and 25 °C for 3 hours. The resulting extract was processed with a triglyceride kit (FUJIFILM Wako Chemicals GmbH, Neuss, Germany) at 600 rpm and 37 °C for 2.5 hours and the reaction products were subsequently analyzed by HPLC-UV. HPLC was performed with a HP 1260/1290 infinity liquid chromatograph (Agilent Technologies, Palo Alto, CA, US) using UV detection. The analyte was separated from matrix components on a 4.6 mm × 100 mm reversed phase column at 40 °C. A one-point calibration curve was made from analysis of triglyceride standard after enzymatic reaction with the kit. The analytical method is linear from 0.5–6 mmol/L with a quantification limit of 0.3 mmol/L.
Quantification of C-peptide from DBS:
C-peptide in DBS were assayed using a Mercodia solid phase two-site enzyme immunoassay (ELISA; Mercodia AB, Uppsala, Sweden). Three spots were punched into the kit plate with anti-C-peptide antibodies bound to the well. Assay buffers were added and C-peptide extracted from the spots at 4 °C. After washing, peroxidase-conjugated anti-C-peptide antibodies were added and after the second incubation and a washing step, the bound conjugate was detected by reaction with 3,3’,5,5’-tetramethylbenzidine (TMB). The reaction was stopped by adding acid to give a colorimetric endpoint that was read spectrophotometrically at 450 nm.
Stool sample collection, method validation and microbial analysis
Stool sample collection:
Participants collected a stool sample at home prior to their clinical visit. Samples were collected using the EasySampler collection kit (ALPCO, NH, US) into fecal collection tubes containing DNA/RNA Shield buffer (Zymo Research, CA, US). Upon receipt in the laboratory, samples were homogenised, aliquoted and stored at −80 °C in Qiagen PowerBeads 1.5 mL tubes (Qiagen, Germany). The sample collection procedure was tested and validated internally comparing different storage conditions (fresh, frozen, buffer), different DNA extraction kits (PowerSoilPro, FastDNA, ProtocolQ, Zymo), and different sequencing technologies (16S rRNA and arrays), data not shown.
Microbiome 16S rRNA gene sequencing and analysis:
The DNA was isolated by QIAGEN Genomic Services using DNeasy® 96 PowerSoil® Pro. Optical density measurement was done using Spectrophotometer Quantification (Tecan Infinite 200). The V4 hyper-variable region of the 16S rRNA gene was then amplified at Genomescan, Leiden, Netherlands. Libraries were sequenced for 300 bp paired-end reads using the Illumina NovaSeq6000 platform. In total, 9.6 Pbp were generated and raw reads were rarefied to 360k reads per sample. Rarefied reads were analyzed using the DADA2 pipeline45. Quality control of the reads was performed using the “filterAndTrim” function from the DADA2 package truncating eight nucleotides from each read to remove barcodes, discarding all reads with quality less than 20, discarding all reads with at least one N, and removing the phiX Illumina spike-in. Only paired-end reads with at least 120 bp and with an expected DADA2 error less than 4 were retained for downstream analyses. Error rates were inferred from the cleaned set of reads (“learnErrors” function) and used in the DADA2 algorithm (“mergePairs” function) for merging the reads, after dereplication (“derepFastq” function). Merged reads were further processed retaining only reads within 280 and 290 bp, representing the majority of the distribution of the lengths. Reads were further processed to remove chimeras using the “removeBimeraDenovo” function with a consensus method. Finally, taxonomy was assigned using the SILVA database (version 132) using the “assignTaxonomy” function and requiring a minimum bootstrap value of 80 obtaining a table of relative abundances of operational taxonomic units (OTUs). To address the issue of compositionality in the microbiome data set46 the relative abundance values were normalized using the (arcsin-sqrt) transformation as described in47. Measures of alpha diversity were computed (see47). The distributions of the Simpson and Shannon indices of alpha diversity on the transformed 16S abundance data are presented in Supplemental Table 4.
Venous blood sample collection
Participants came into the clinical research facilities at 8:30am and were cannulated in the forearm antecubital vein. Venous blood was collected at 0 minutes (prior to a test breakfast) and at 9 timepoints postprandially (15, 30, 60, 120, 180, 240, 270, 300, and 360 minutes). Plasma glucose was analyzed from blood samples collected into fluoride oxalate tubes and centrifuged at 1900 g for 10 min at 4 °C. Serum C-peptide, insulin, triglyceride, fasting lipid profile, thyroid stimulating hormone, alanine aminotransferase, and liver function panel were analyzed from blood samples collected into gel separator serum tubes and allowed to stand at room temperature before centrifuging at 1900 g for 10 min at 4 °C. Samples were aliquoted and stored at −80 °C. Blood, for complete blood count (CBC) analysis, was collected into EDTA tubes, kept at 4 °C and analyzed within 12 hours of collection.
Serum biomarkers
In the UK, insulin, glucose, triglyceride and C-peptide analysis was conducted by Affinity Biomarkers Labs (London, UK). Glucose and triglyceride analyses were conducted on a Siemens ADVIA 1800 using Siemens assay kits (Siemens Healthcare Diagnostics Ltd, Surrey, UK). Triglyceride was analyzed using the ADVIA chemistry triglyceride method based on the Fossati three-step enzymatic reaction with a Trinder endpoint. Glucose was analyzed using the ADVIA chemistry glucose oxidase (GLUO) method (based on the modified method of Keston). C-peptide and insulin were analyzed using the Siemens ADVIA Centaur XP systems using a two-site sandwich immunoassay. Complete blood count (CBC) was measured by Viapath (London, UK) for the UK cohort using standard automated clinical chemistry techniques. The inter-assay coefficient of variation for PREDICT samples analyzed by Affinity were: insulin 3.4%, C-peptide 7.9%, triglyceride 3.7%, and glucose 2.6%.
In the US, CBC was established using fresh blood samples in the MGH Core Laboratory. Hb1AC tests were performed by the MGH Diabetes A1c lab. Glucose, insulin, triglyceride, and C-peptide were conducted by Quest Diagnostics (Boston, MA) using standard automated clinical chemistry techniques.
Upon completion of the US study, frozen serum and plasma samples were sent from the US to the UK and the entire cohort had liver function panel, full lipids (TC, HDL-C LDL-C and triglyceride), thyroid stimulating hormone and alanine aminotransferase measurements performed by Affinity Biomarkers Labs. Details described elsewhere48.
Glucose using continuous glucose monitoring
Interstitial glucose was measured every 15 minutes using Freestyle Libre Pro continuous glucose monitors (Abbott, Abbott Park, IL, US). Monitors were fitted by trained nurses on the upper, non-dominant arm at participants’ baseline visit and covered with Opsite Flexifix adhesive film (Smith & Nephew Medical Ltd, Hull, England) for improved durability, and worn for the entire study duration (14 days). Data collected 12 hours and onwards after activating the device was used for analysis. For a subgroup of participants (n = 377), we fitted two monitors on their arms and calculated the Coefficient of Variation (CV = 11.75%) and correlation (r = 0.97) of their iAUC responses to standardized meals (Extended Data Figure 2b).
Time points for analyses:
Glucose:
The 2-hour glucose iAUC was used for both clinic and at-home analyses.
Insulin and C-peptide:
C-peptide was measured at home as a surrogate for insulin secretion, because the reliability of C-peptide measured from DBS is higher than that of insulin (see49) and C-peptide remains stable on paper filters for up to 6 months49. C-peptide was measured at 60 minutes postprandially to coincide with the peak in C-peptide seen in healthy individuals in clinic, and again at 120 minutes to coincide with the strong decline in insulin level (Extended Data Figure 2c). However, because previous genetic studies have tested the heritability of postprandial insulin at 120 minutes, this time point was included for our own heritability analyses (Figures 2b–c). All other analyses refer to the 1-hour rise for C-peptide.
Triglyceride:
The rise in triglyceride at 6 hours postprandially (triglyceride6h-rise) was selected to represent postprandial lipemic response from serum collected at clinic and home-based DBS tests. This is a measure of lipemia most closely correlated with atherogenic lipoproteins compared to iAUC0-6h, Cmax and 4h triglyceride concentration (see.50–52).
Activity and sleep
Energy expenditure was measured using a triaxial accelerometer (AX3, Axivity, UK) fitted by nurses at the baseline visit on the non-dominant wrist and worn for the duration of the study (except during water-based activities, including showers and swimming). Accelerometers were programmed to measure acceleration at 50 Hz with a dynamic range of ±8 g (where g refers to local gravitational force equal to 9.8 m/s2). Non-wear periods were defined as windows of at least 1 hour with less than 13mg for at least 2 out of 3 axes, or where 2 out of 3 axes measured less than 50mg. Windows of sleep were measured using methods described elsewhere53.
Genotyping
Whole genome genotyping was available for 241 individuals from the UK cohort from previous TwinsUK studies. Genotyping was performed with the Illumina Infinium HumanHap610. Normalised GWAS intensity data were pooled and genotypes called on the basis of the Illuminus algorithm. No calls were assigned if the most likely call was less than a posterior probability of 0.95. Validation of pooling was done by visual inspection of 100 random, shared SNPs for overt batch effects (none were observed). SNPs that had a low call rate (≤90%), Hardy-Weinberg p values <10−6 and minor allele frequencies <1% were excluded, and samples with call rates <95% were removed. Genotype imputations were performed to increase the coverage. Imputation of genotypes for all polymorphic SNPs that passed the quality control stage were performed on the Michigan Imputation Server (https://imputationserver.sph.umich.edu) using the 1000G Phase3 v5 reference panel54. SNPs previously reported to be associated with postprandial glycemia, triglyceride or insulin GWAS17–20 were extracted from the full set of genome wide genotypes using PLINK and tested for association with postprandial measures using linear regression methods.
Processing of habitual diet information
UK nutrient intakes were determined using FETA software to calculate macro- and micro- nutrient data43. Submitted FFQs were excluded if greater than 10 food items were left unanswered, or if the total energy intake estimate derived from FFQ as a ratio of the subject’s estimated basal metabolic rate (determined by the Harris-Benedict equation)43 was more than two standard deviations outside the mean of this ratio (<0.52 or >2.58).
Statistical analysis
Basic analyses
The descriptive characteristics of study participants are summarized in Supplemental Table 1 In order to reduce the dimension of the data, principal component analysis (PCAs) with orthogonal transformation (varimax procedure) was applied to derive principal components (PC) representative of individual characteristics (20 PCAs), microbiome (40 PCAs), meal composition (1 PCA), habitual diet (5 PCAs) and meal context (5 PCAs) (see Supplemental Table 3 for full list of input variables). All the necessary prerequisites of PC analysis including linearity, Kaiser–Meyer–Olkin measure of 0.88, and the significant Bartlett’s test of sphericity (p < 0.001) were met. Each participant received a score for each category mentioned above. To investigate the association between each outcome (iAUC, triglyceride6h-rise, C-peptide1h-rise) and our exposures (individual baseline characteristics, microbiome (16S), meal content, habitual diet and meal context) multivariable regressions were applied and R2 reported. Further, we derived PCAs for the anthropometrics, biochemical/clinical factors, physical activity and sleep features separately to investigate their role. Multi-collinearity for the multiple linear regressions was assessed with variance inflation factors (VIF) at each step55. Multi-collinearity was considered high when the VIF was >1038. Receiver operating characteristic (ROC) curves were constructed and the area under the curve (AUC) was calculated to assess the discriminatory power of (fasting blood glucose vs. 2h glucose iAUC), (fasting triglyceride vs. triglyceride6h-rise) and (fasting C-peptide vs. C-peptide1h-rise) to detect impaired glucose tolerance, and ASCVD 10 year risk (70% applied as a cut-off point). Values of AUC range from 0.5 and 1, with 0.5 indicating no discrimination, and 1 indicating perfect discrimination (2). A p-value ⩽0.05 was considered statistically significant. All analyses were performed using R (version 3.4.2 R Core Team (2017)).
Meal composition
To estimate macronutrient effects on glycemic response, we fitted a multivariate regression model with carbohydrates, fats, fiber and protein as predictors on meals 1, 2, 4, 5, 6, 7 and 8.
Multicollinearity was assessed for these predictors through VIF and we concluded that it was non existent (VIF < 10). The regression coefficients were all significant (p < 0.001) with values −79.23 mmol/L*s, −142.41 mmol/L*s and −185.49 mmol/L*s for fat, fiber and protein respectively, after having adjusted by carbohydrates.
Heritability and ACE model
To estimate the heritability, we analyzed the data according to the classical ACE model. In this model, heritability is an approximation of the relative importance of additive genetic differences for variance of postprandial responses in the population56. Shared or familial environmental influences reflect experiences that contribute to twin similarity. Non-shared or individual-specific environmental influences refer to the contribution of environmental experiences not shared by family members. Information concerning shared genetic and environmental influences is best estimated by structural equation modelling techniques that fit models of twins by zygosity in order to describe the 154 causes of the variance in OA. Therefore, the total variance in the trait can be partitioned into genetic variance (A), shared (familial) environmental variance (C), and individual-specific environmental variance (E). The level of statistical significance was set at p<0.05 in all analyses, and the R software (version 3.0.2) together with the “mets” (Multivariate Event Times) package (https://rdrr.io/cran/mets/src/R/methodstwinlm.R) was used for all statistical analyses.
Meal ranking
Six different type of meals were ranked for each individual as being the one with the highest glucose 2h iAUC for that person (rank 6), the one with the second highest glucose iAUC (rank 5). ….down the the one with the lowest glucose 2h iAUC (rank 1). The distribution of these “in-person rankings” is presented in Extended Data Figure 3.
Multilinear ANOVA to assess role of individualized responses to meals
The different sources of variation in glycemic response for Meal 2,3,4,6 and 8 (described in Supplemental Table 3) were analysed using the Multilevel Linear ANOVA40 model and were analysed using a multilevel (hierarchical) linear Bayesian ANOVA model as described by Gelman and Hill57.
The different sources of variation in glycemic response for Meal 2,3,4,6 and were analysed using a multilevel (hierarchical) Linear Bayesian ANOVA model as described by (Gelman & Hill 2007).
Hierarchical Bayes models can accommodate non-normal dependent variables that are difficult to incorporate in classical ANOVA and multilevel linear models. The approach consists of sub-models at two levels: at level 1 the parameters of individuals, meals and person-meal interactions, and at level 2 the moments of the distributions from which level 1 parameters are drawn. Level 2 imposes some homogeneity on level 1 parameters, for example
i.e. the meal terms are are distributed normally with the same standard deviation aa, ensuring homogeneity.
aa ~ HalfCauchy(5) i.e. the standard deviation of the above distribution has a particular prior (a half cauchy distribution with a scale factor of 5).
The other terms ({3p, ym,p, Em,p,k, Em,p,k,n) have similar hierarchical distributions (though the standard deviations of Em,p,k, Em,p,k,n have uniform prior as opposed to a half cauchy).
The parameters at both levels (i.e. all the am ’s and aa and analogously for the other parameters) are sampled using an Markov Chain Monte-Carlo routine in pymc358 and we plot the sampled values of aa, a/3, ay, aE and in Figure 6b.
where:
-
log(iAUC) = ym,p,k,n: the 2 hour iAUC for person p, eating meal m, for the k th time measured on cgm n (given the availability of data with 2 CGMs for a subset as described in below.
the 2 hour iAUC for person p, eating meal m, for the k th time measured on cgm n (given that we have 2 cgms for many people)
am : meal content (across all people) for meal m, e.g. high and low carbohydrate meals
{3p : individual glucose scaling (across all meals) for person p, e.g. overall high and low responding people
ym,p : the meal-specific response for individual p to meal m, e.g. a specific person responds particularly strongly to a specific meal
Em,p,k,n : error stemming from the cgm (participants selected for this analysis wore 2 CGM devices, so n indexes the device providing the measurement)
Em,p,k : other sources of variation, including meal timing, exercise, sleep and circadian rhythm
This Bayesian ANOVA model is a Bayesian hierarchical model attempts to explain the observed log(iAUC) of a meal as a sum of categorical terms., i.e. individuals are not classified according to any characteristics but are included as unique individuals with log(iAUC 2h glucose) for various different meals. If this was an extended Glycemic Index model it would correspond to expressing the log(iAUC) as the sum of a meal term (analogous to the glycemic load of the meal) and an individualized term. This “individual glucose scaling” is not a linear function of a person’s characteristics (such as age, sex or BMI) but rather it is how each individual ranks overall given the log(iAUC) values for the various meals. This allowed us to test whether there was an interaction term between meals and persons, i.e. an individualized response component to particular meals that was not merely due to a person being a high, average or low responder and to a meal having on average a higher glycemic response (e.g. OGTT) than another meal (e.g. a high fat muffin). Given the availability of data concerning repeated occurrences of a person eating a particular meal and multiple CGMs measurements for the same meal we were able to extend the model to include a person-meal interaction and a CGM error and, analogously, infer the error due to the CGMs and the degree to which a person’s response to a particular meal is consistently higher or lower than one would expect from the glycemic index model .i.e. a personalized glycemic load. The person-meal interaction effects allow different people to have different ordering of glycemic responses to meals, so one person might respond more strongly to meal A than meal B, whilst another person might respond more strongly to meal B than meal A. Figure 6c shows show 50% and 95% intervals on standard deviations of the effects in the model. These can be approximately interpreted as percent increase (or decrease) in iAUC contributed by the various effects in the model.
CGM repeatability.
A subset of participants (n = 483) wore two continuous glucose measurement devices simultaneously, providing duplicate measurements for the meals they consumed and therefore allowing us to distinguish CGM error from unexplained sources of variation. Postprandial glucose measurements for 3280 meals eaten collectively by 483 participants from UK were used in this analysis. (Extended Data Figure 2b).
Computation of clinical indices
Atherosclerotic Cardiovascular Disease 10 year risk:
(AHA/JACC ASCVD 10 year risk) The 10-year atherosclerotic cardiovascular disease (ASCVD)59 risk score is a gender and race specific single multivariable risk assessment tool used to estimate the 10-year CVD risk of an individual, and has clinically replaced the Framingham-10 year cardiovascular risk score. It is based on the age, sex, ethnicity, total and HDL cholesterol, systolic blood pressure, smoking status, use of blood pressure lowering medications, and the presence of type 2 diabetes (T2D).
Impaired glucose tolerance:
We used the standard definition from the American Diabetes Association60 (Fasting plasma glucose < 7.0 mmol/l and OGTT 2-hour value >= 7.8 mmol/l but < 11.1 mmol/l).
Validation of Machine learning model cross validation and difference (Bland-Altman plots)
To further illustrate the reliability of the machine learning predictions, we conducted a leave-one-out cross validation procedure and generated Bland-Altman plots to analyze the agreement between two. To generate the Bland-Altman plots we used the Predict UK and US data showing Predicted vs Measured postprandial responses. We generated Bland-Altman plots for predicted and measured postprandial responses for each biomarker (Triglycerides, C-peptide and Glucose). (Extended Data Figure 4a).
Leave-one-out cross-validated Pearson R scores in Predict UK
To perform k-fold cross validation, the entire dataset is split into k groups. Treating each group as a test set and the remaining groups as the training set, the model is fitted k times. The Pearsons’s R between the values predicted by the fitted models and the measured values in the test sets is used as the metric for model evaluation, which we refer to as the cross-validated Pearson-R.
The special case, where k is the size of the dataset, is referred to as leave-one-out cross-validation, and we refer to the corresponding evaluation metric as leave-one-out cross-validated Pearson R. The machine learning models for the three biomarkers of interest were evaluated using the aforementioned metric and are reported in the Extended Data Figure 4b. These scores are similar to the cross-validated 5-fold scores in the main text.
Extended Data
Extended Data Figure 1.
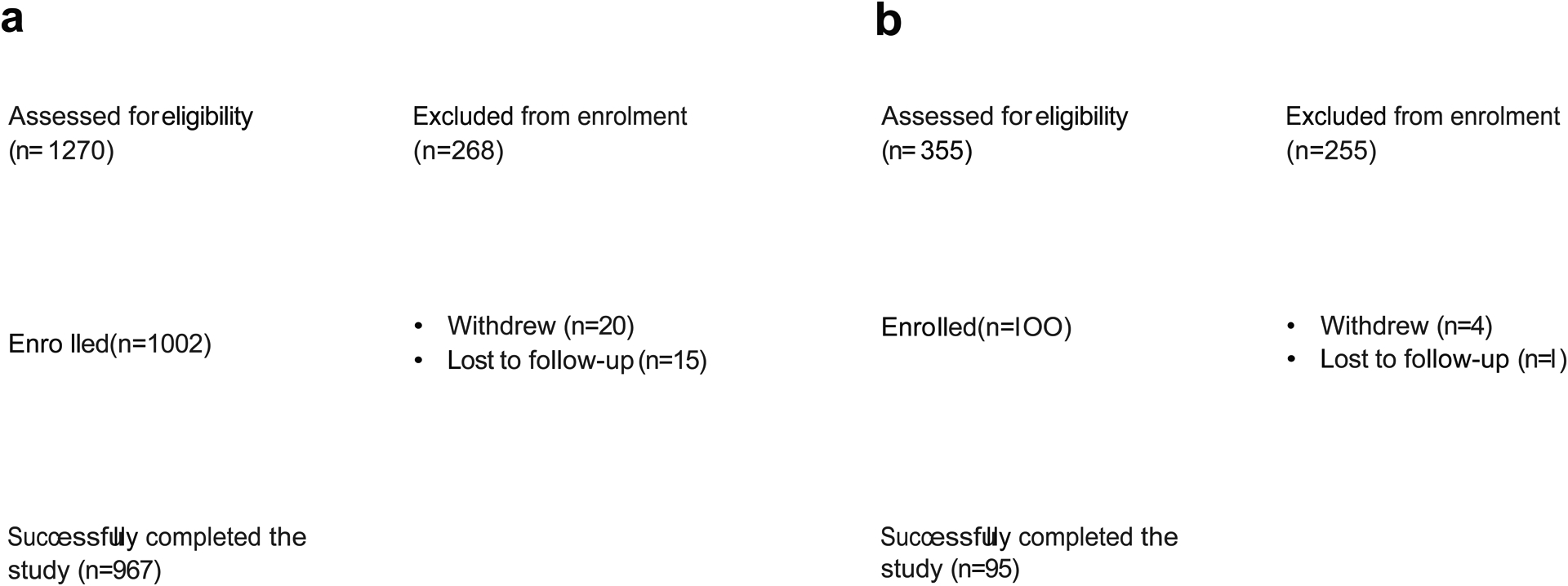
Consort Diagrams for (a) UK and (b) US populations in the PREDICT 1 study.
Extended Data Figure 2.
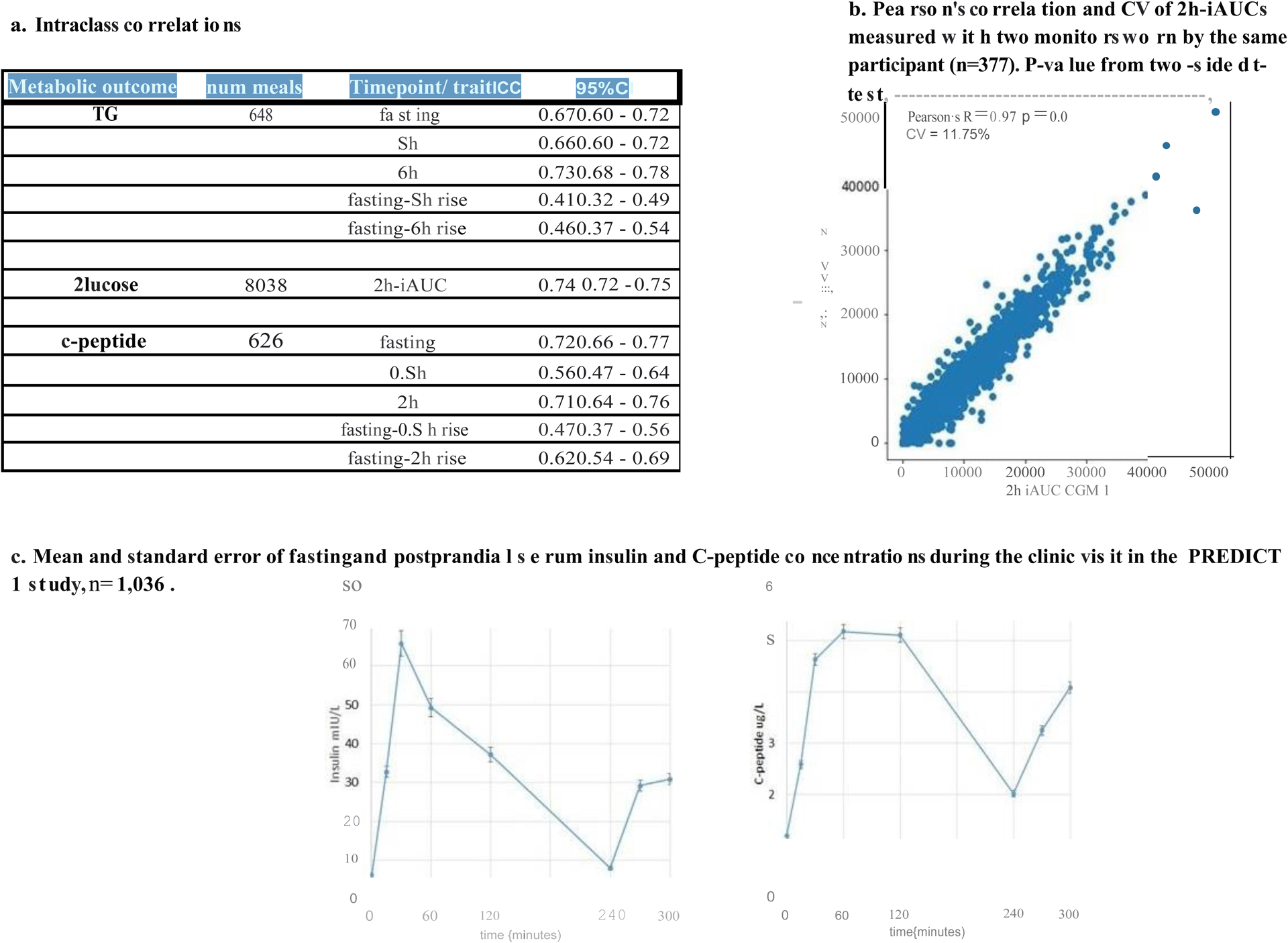
Repeatability in the PREDICT 1 study
Extended Data Figure 3.
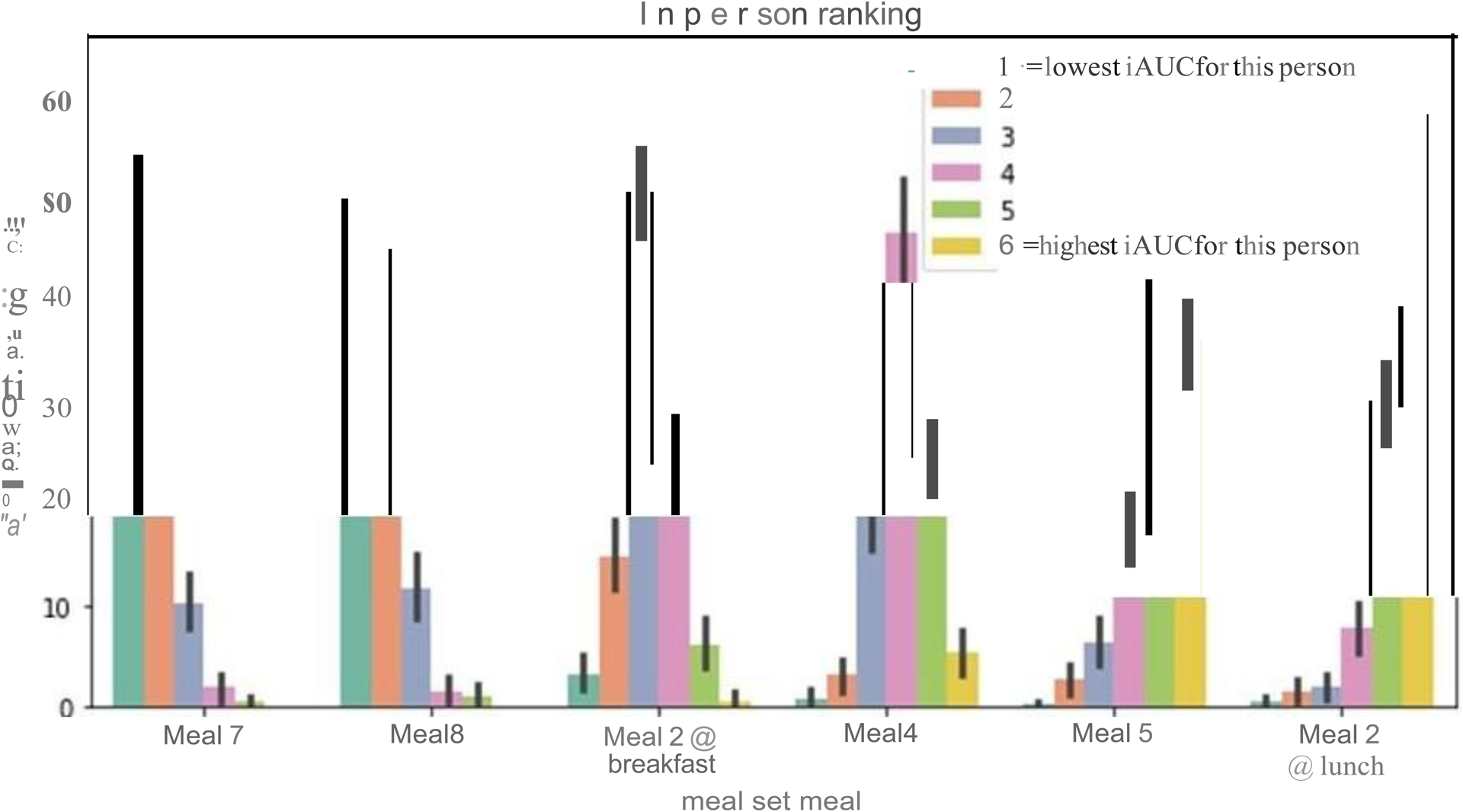
Frequency distribution of in-person ranking for 6 of meals shown in Figure 6a (High fat 40g = meal 7, High protein = meal 8, UK average = meal 2, High carb = meal 4, OGTT = meal 5, Uk avg at lunch = meal 2). n = 1102 participants
Extended Data Figure 4.
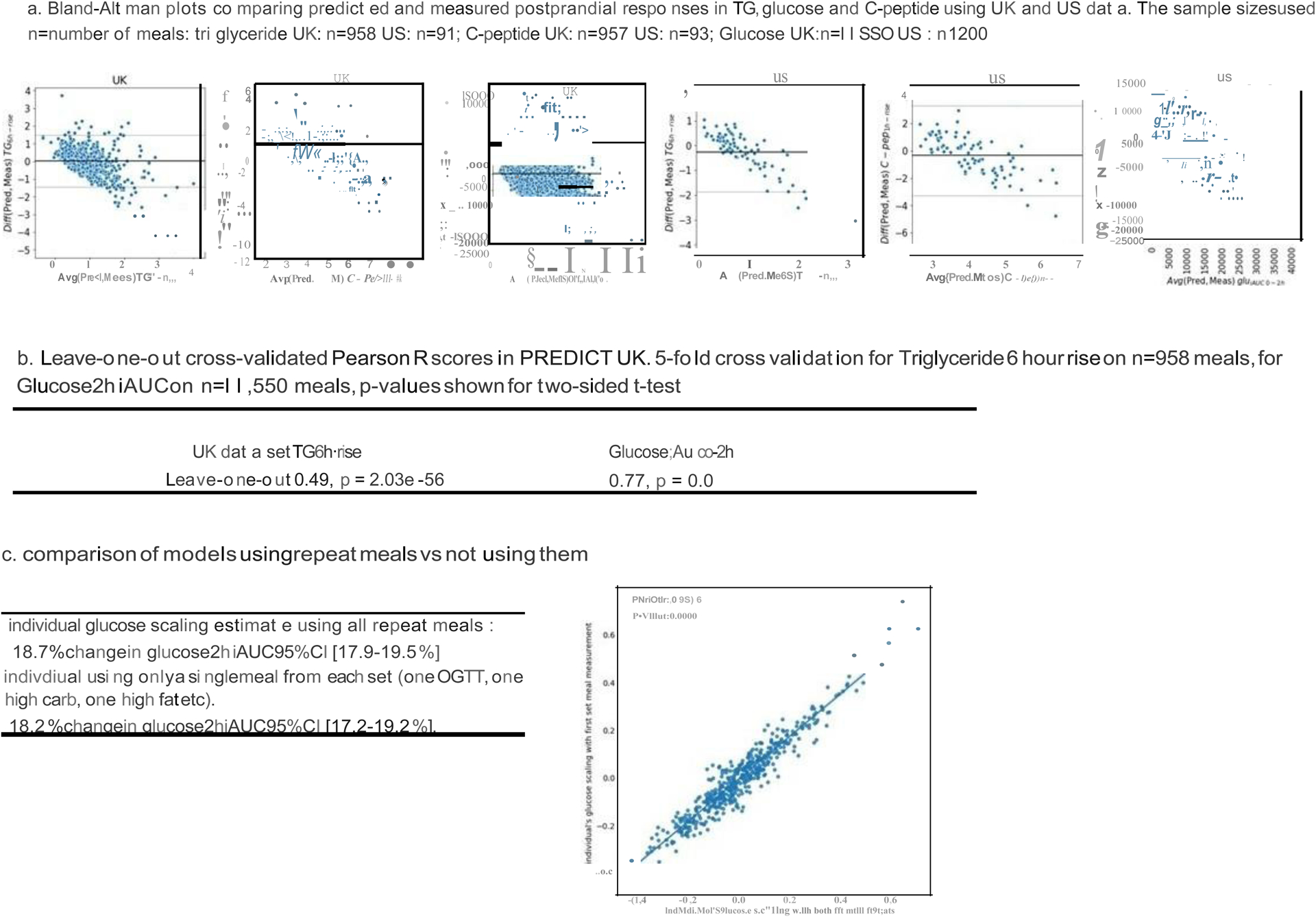
Machine Learning comparisons, cross validation and repeatability
Supplementary Material
Acknowledgements
We express our sincere thanks to the participants of the PREDICT 1 study. We thank Mark McCarthy (University of Oxford) and Leanne Hodson (University of Oxford) for valuable feedback on the manuscript. We thank the staff of Zoe Global, the Department of Twin Research and Massachusetts General Hospital for their tireless work in contributing to the running of the study and data collection. We thank Abbott for their support with using their CGMs. This work was supported by Zoe Global Ltd also received support from grants from the Wellcome Trust (212904/Z/18/Z) and the Medical Research Council (MRC)/British Heart Foundation Ancestry and Biological Informative Markers for Stratification of Hypertension (AIMHY; MR/M016560/1). PWF was supported in part by grants from the European Research Council (CoG-2015_681742_NASCENT), Swedish Research Council, Novo Nordisk Foundation and the Swedish Foundation for Strategic Research (IRC award). AMV was supported by the National Institute for Health Research Nottingham Biomedical Research Centre. TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union, Chronic Disease Research Foundation (CDRF), Zoe Global Ltd and the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London.
Footnotes
Conflict of interest statement: TD Spector, SE Berry, AM Valdes, F Asnicar, PW Franks, LM Delahanty, N Segata, are consultants to Zoe Global Ltd (“Zoe”). J Wolf, G Hadjigeorgiou, R Davies, H Al Khatib, J Capdevila, C Bonnett, S Ganesh, E Bakker, P Wyatt and I Linenberg are or have been employees of Zoe. Other authors have no conflict of interest to declare.
Data availability
The data used for analyzing this study are held by the department of Twin Research at Kings College London. The data can be released to bona fide researchers using our normal procedures overseen by the Wellcome Trust and its guidelines as part of our core funding. We receive around 100 requests per year for our datasets and have a meeting three times a month with independent members to assess proposals. Application is via https://twinsuk.ac.uk/resources-for-researchers/access-our-data/. This means that the data needs to be anonymized and conform to GDPR standards. Specifically for this paper, all the variables used in the models can be requested as well as the summary outcome measures for each person. The 16S microbiome data used here will be uploaded onto the EBI website (https://www.ebi.ac.uk/ with unlimited access.
Code availability
The scripts for statistical analysis are freely available upon request to the Department of Twins Research. Application is via https://twinsuk.ac.uk/resources-for-researchers/access-our-data/.
References
- 1.Collaborators, G.B.D.D. Health effects of dietary risks in 195 countries, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 393, 1958–1972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agriculture, U.S.D.o.H.a.H.S.a.U.S.D.o. 2015 – 2020 Dietary Guidelines for Americans. 8th Edition. December 2015. Available at https://health.gov/our-work/food-and-nutrition/2015-2020-dietary-guidelines/.(2015). [Google Scholar]
- 3.Karpyn A Food and public health : a practical introduction, xv, 368 pages (Oxford University Press, New York, 2018). [Google Scholar]
- 4.(EFSA), E.F.S.A. Dietary Reference Values for nutrients Summary report. 10.2903/sp.efsa.2017.e15121(2017). [DOI]
- 5.Zeevi D et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 163, 1079–1094 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Tebani A & Bekri S Paving the Way to Precision Nutrition Through Metabolomics. Front Nutr 6, 41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolovou GD et al. Assessment and clinical relevance of non-fasting and postprandial triglycerides: an expert panel statement. Curr Vasc Pharmacol 9, 258–70 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Astley CM et al. Genetic Evidence That Carbohydrate-Stimulated Insulin Secretion Leads to Obesity. Clin Chem 64, 192–200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaak EE et al. Impact of postprandial glycaemia on health and prevention of disease. Obes Rev 13, 923–84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decode Study Group, t.E.D.E.G. Glucose tolerance and cardiovascular mortality: comparison of fasting and 2-hour diagnostic criteria. Arch Intern Med 161, 397–405 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Ning F et al. Cardiovascular disease mortality in Europeans in relation to fasting and 2-h plasma glucose levels within a normoglycemic range. Diabetes Care 33, 2211–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bansal S et al. Fasting compared with nonfasting triglycerides and risk of cardiovascular events in women. JAMA 298, 309–16 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Lindman AS, Veierod MB, Tverdal A, Pedersen JI & Selmer R Nonfasting triglycerides and risk of cardiovascular death in men and women from the Norwegian Counties Study. Eur J Epidemiol 25, 789–98 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson KG, Poppitt SD & Minihane AM Postprandial lipemia and cardiovascular disease risk: Interrelationships between dietary, physiological and genetic determinants. Atherosclerosis 220, 22–33 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Stumvoll M, Goldstein BJ & van Haeften TW Type 2 diabetes: principles of pathogenesis and therapy. Lancet 365, 1333–46 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Moayyeri A, Hammond CJ, Hart DJ & Spector TD The UK Adult Twin Registry (TwinsUK Resource). Twin Res Hum Genet 16, 144–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prokopenko I et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet 10, e1004235 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxena R et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet 42, 142–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wojczynski MK et al. Genome-wide association study of triglyceride response to a high-fat meal among participants of the NHLBI Genetics of Lipid Lowering Drugs and Diet Network (GOLDN). Metabolism 64, 1359–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanna S et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet 51, 600–605 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almgren P et al. Genetic determinants of circulating GIP and GLP-1 concentrations. JCI Insight 2(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wojcikowski M, Ballester PJ & Siedlecki P Performance of machine-learning scoring functions in structure-based virtual screening. Sci Rep 7, 46710 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hastie T, Tibshirani R & Friedman J The Elements of Statistical Learning: Data Mining, Inference, and Prediction, (Springer; New York, 2013). [Google Scholar]
- 24.Russell WR et al. Impact of Diet Composition on Blood Glucose Regulation. Crit Rev Food Sci Nutr 56, 541–90 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Jeong Y et al. A Review of Recent Evidence from Meal-Based Diet Interventions and Clinical Biomarkers for Improvement of Glucose Regulation. Prev Nutr Food Sci 25, 9–24 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delahanty LM et al. Effects of weight loss, weight cycling, and weight loss maintenance on diabetes incidence and change in cardiometabolic traits in the Diabetes Prevention Program. Diabetes Care 37, 2738–45 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diabetes Prevention Program Research, G. et al. 10-year follow-up of diabetes incidence and weight loss in the Diabetes Prevention Program Outcomes Study. Lancet 374, 1677–86 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knowler WC et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 346, 393–403 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papandonatos GD et al. Genetic Predisposition to Weight Loss and Regain With Lifestyle Intervention: Analyses From the Diabetes Prevention Program and the Look AHEAD Randomized Controlled Trials. Diabetes 64, 4312–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor R Banting Memorial lecture 2012: reversing the twin cycles of type 2 diabetes. Diabet Med 30, 267–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell KJ et al. Impact of fat, protein, and glycemic index on postprandial glucose control in type 1 diabetes: implications for intensive diabetes management in the continuous glucose monitoring era. Diabetes Care 38, 1008–15 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Hansson P et al. Meals with Similar Fat Content from Different Dairy Products Induce Different Postprandial Triglyceride Responses in Healthy Adults: A Randomized Controlled Cross-Over Trial. J Nutr 149, 422–431 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atkinson FS, Hancock D, Petocz P & Brand-Miller JC The physiologic and phenotypic significance of variation in human amylase gene copy number. Am J Clin Nutr 108, 737–748 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Gardner CD et al. Effect of Low-Fat vs Low-Carbohydrate Diet on 12-Month Weight Loss in Overweight Adults and the Association With Genotype Pattern or Insulin Secretion: The DIETFITS Randomized Clinical Trial. JAMA 319, 667–679 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viitasalo A et al. Genetic predisposition to higher body fat yet lower cardiometabolic risk in children and adolescents. Int J Obes (Lond) 43, 2007–2016 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Hughes RL, Kable ME, Marco M & Keim NL The Role of the Gut Microbiome in Predicting Response to Diet and the Development of Precision Nutrition Models. Part II: Results. Adv Nutr (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JG et al. Deep Learning in Medical Imaging: General Overview. Korean J Radiol 18, 570–584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubois C et al. Effects of graded amounts (0–50 g) of dietary fat on postprandial lipemia and lipoproteins in normolipidemic adults. Am J Clin Nutr 67, 31–8 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Garcia-Perez I et al. Objective assessment of dietary patterns by use of metabolic phenotyping: a randomised, controlled, crossover trial. Lancet Diabetes Endocrinol 5, 184–195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zierer J et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet 50, 790–795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berry S, D. D, Linenberg I,Wolf J, Hadjigeorgiou G, Davies R, Al Khatib H, Hart D, Surdulescu G, Yarand D, Nessa A, Sheedy A, Vijay A, Asnicar, Segata N, Chan A, Franks P, Valdes A, Spector T. Personalised REsponses to DIetary Composition Trial (PREDICT): an intervention study to determine inter-individual differences in postprandial response to foods. Protocol Exchange (2020). [Google Scholar]
- 42.Verdi S et al. TwinsUK: The UK Adult Twin Registry Update. Twin Res Hum Genet, 1–7 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Mulligan AA et al. A new tool for converting food frequency questionnaire data into nutrient and food group values: FETA research methods and availability. BMJ Open 4, e004503 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flint A, Raben A, Blundell JE & Astrup A Reproducibility, power and validity of visual analogue scales in assessment of appetite sensations in single test meal studies. Int J Obes Relat Metab Disord 24, 38–48 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Callahan BJ et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gloor GB, Wu JR, Pawlowsky-Glahn V & Egozcue JJ It’s all relative: analyzing microbiome data as compositions. Ann Epidemiol 26, 322–9 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Thomas AM et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med 25, 667–678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soininen P, Kangas AJ, Wurtz P, Suna T & Ala-Korpela M Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet 8, 192–206 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Johansson J, Becker C, Persson NG, Fex M & Torn C C-peptide in dried blood spots. Scand J Clin Lab Invest 70, 404–9 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Samson CE, Galia AL, Llave KI, Zacarias MB & Mercado-Asis LB Postprandial Peaking and Plateauing of Triglycerides and VLDL in Patients with Underlying Cardiovascular Diseases Despite Treatment. Int J Endocrinol Metab 10, 587–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madhu S, Sinha B, Aslam M, Mehrotra G & Dwivedi S Postprandial triglyceride responses and endothelial function in prediabetic first-degree relatives of patients with diabetes. J Clin Lipidol 11, 1415–1420 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Nakamura A et al. Different postprandial lipid metabolism and insulin resistance between non-diabetic patients with and without coronary artery disease. J Cardiol 66, 435–44 (2015). [DOI] [PubMed] [Google Scholar]
- 53.van Hees VT et al. Estimating sleep parameters using an accelerometer without sleep diary. Sci Rep 8, 12975 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eitel I et al. Genome-wide association study in takotsubo syndrome - Preliminary results and future directions. Int J Cardiol 236, 335–339 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Slinker BK & Glantz SA Multiple regression for physiological data analysis: the problem of multicollinearity. Am J Physiol 249, R1–12 (1985). [DOI] [PubMed] [Google Scholar]
- 56.Purcell S Variance components models for gene-environment interaction in twin analysis. Twin Res 5, 554–71 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Gelman A & Hill J Data analysis using regression and multilevel/hierarchical models, xxii, 625 p. (Cambridge University Press, Cambridge; New York, 2007). [Google Scholar]
- 58.Salvatier J, Wiecki TV & Fonnesbeck C Probabilistic programming in Python using PyMC3. Peerj Computer Science (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goff DC Jr. et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 129, S49–73 (2014). [DOI] [PubMed] [Google Scholar]
- 60.American Diabetes, A. 2. Classification and Diagnosis of Diabetes. Diabetes Care 39 Suppl 1, S13–22 (2016).26696675 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


