SUMMARY
The suprachiasmatic nucleus (SCN) acts as a master pacemaker driving circadian behavior and physiology. Although the SCN is small, it is composed of many cell types, making it difficult to study the roles of particular cells. Here we have developed bioluminescent circadian reporter mice that are Cre-dependent, allowing the circadian properties of genetically-defined populations of cells to be studied in real time. Using a Color-Switch PER2::LUCIFERASE reporter that switches from red PER2::LUCIFERASE to green PER2::LUCIFERASE upon Cre recombination, we assessed circadian rhythms in two of the major classes of peptidergic neurons in the SCN: AVP (arginine vasopressin) and VIP (vasoactive intestinal polypeptide). Surprisingly, we find that circadian function in AVP neurons, not VIP neurons, is essential for autonomous network synchrony of the SCN and stability of circadian rhythmicity.
Keywords: Circadian rhythms, suprachiasmatic nucleus (SCN), bioluminescence imaging, Per2 gene, vasopressin, luciferase, neuronal network, neuronal coupling
INTRODUCTION
In mammals, the hypothalamic suprachiasmatic nucleus (SCN) acts as a master pacemaker controlling circadian rhythms in behavioral states (sleep-wake), feeding behavior, body temperature, and endocrine physiology (Hastings et al., 2018; Saper, 2013; Welsh et al., 2010). The SCN, like other nuclei within the hypothalamus, contains a heterogenous population of ~10,000 neurons that secrete more than 100 identified neurotransmitters, neuropeptides, cytokines, and growth factors (Abrahamson and Moore, 2001; Lee et al., 2013; Morin, 2013; Welsh et al., 2010). Some of these signaling molecules play important roles in cell-cell coupling, a process that synchronizes period length and phase relationships among SCN neurons (Bedont and Blackshaw, 2015; Colwell, 2011; Evans et al., 2013; Hastings et al., 2018; Herzog, 2007; Mohawk and Takahashi, 2011). However, the roles of specific SCN cell types and the intercellular signaling mechanisms engaged in the generation of behavioral circadian rhythms are not fully understood.
The study of circadian rhythms requires very long times scales (days to weeks) and very sensitive detection methods. Pioneered by Steve Kay and colleagues (Millar et al., 1995), in vivo bioluminescence imaging has long been used to observe circadian rhythms in real time, due to its extremely low background and high signal-to-noise ratio (Troy et al., 2004; Yamazaki and Takahashi, 2005). The dynamics of circadian gene expression in the SCN have been studied using real-time imaging of Per1-luciferase and PER2::LUCIFERASE, as well as other transgenic mouse models (Cheng et al., 2009; Kuhlman et al., 2000; Mei et al., 2018; Ono et al., 2016; Welsh and Kay, 2005; Wilsbacher et al., 2002; Yamaguchi et al., 2003; Yamazaki and Takahashi, 2005; Yoo et al., 2004). However, circadian genes are ubiquitously expressed making it difficult to study the circadian properties of specific SCN cell types. This gap in our knowledge motivated us to develop circadian reporter mice that allow for conditional genetic manipulation to selectively interrogate subpopulations of SCN neurons. Our novel Color-Switch PER2::LUCIFERASE mouse line makes use of Cre-lox dependent reporters that allow the circadian properties of genetically-defined cell populations to be studied separately, but simultaneously.
Two major classes of neuropeptide-containing neurons, AVP (arginine vasopressin) and VIP (vasoactive intestinal polypeptide) are enriched in the “shell” and the “core” regions of the SCN, respectively. There is ample evidence that VIP/VPAC2R signaling is important for maintaining the coherence of circadian oscillators both within the SCN itself, as well as in vivo for the generation of circadian behavioral rhythms (Aton et al., 2005; Colwell et al., 2003; Harmar et al., 2002; Jones et al., 2018; Maywood et al., 2011; Maywood et al., 2006). AVP signaling may also mediate interneuronal communication within the SCN, as loss-of-function of both AVP V1a and V1b receptors profoundly alters the resetting response of mice to shifted light cycles (Yamaguchi et al., 2013). In addition, deletion of the core clock component, Bmal1, in AVP neurons lengthens free-running period and enhances re-entrainment to light (Mieda et al., 2015), suggesting that cell-autonomous circadian rhythms in AVP neurons are required to regulate circadian behavioral rhythms (Mieda et al., 2015). Deletion of Bmal1 in AVP neurons also reduces the amplitude of circadian rhythms in the dorsal SCN (Mieda et al., 2015); however, it is unclear how robust these effects are or which cell types are specifically affected. Interestingly, Bmal1 deletion from the liver has no effect on Per2 rhythms, suggesting that external signals (coordinated by the SCN) can compensate for loss of a functioning clock in some tissues (Kornmann et al., 2007; Lamia et al., 2008). In the current study, we sought to delineate cell-type-specific functions of the circadian clock in SCN neurons utilizing our novel Color-Switch PER2::LUCIFERASE mouse line. Our results reveal surprising observations about the circadian function of VIP and AVP neurons in SCN network synchrony.
RESULTS
Generation of Color-Switch PER2::LUCIFERASE reporter mice allows for single-cell analysis of rhythmicity in SCN neurons
To allow for cell-type-specific analysis of circadian rhytthms, we generated Color-Switch mice with click beetle red (CBR) luciferase fused to the C-terminus of PER2 that switches to click beetle green (CBG) luciferase fusion upon Cre-lox recombination (Gammon et al., 2006; Villalobos et al., 2010; Viviani et al., 2008). The targeting construct used to create the Cre-dependent click beetle luciferase reporter mice is shown in Figure S1. CB luciferase was used for red/green discrimination because firefly luciferase is pH sensitive and shifts to longer wavelengths so that red/green separation is compromised. The CB luciferases are also about 2–5X brighter than firefly luciferase (Miloud et al., 2007). In order to visualize the red and green bioluminescence signals from PER2::CBR and PER2::CBG, we also developed a dual-color imaging device that is capable of separating and capturing dual-color bioluminescence signals concurrently on the same image-capture device. The bioluminescence signal from the sample was separated with a beam splitter into long (>565nm) and short wavelengths (<565nm). The long- and short-wavelength signals were filtered with a long-pass filter (>625 nm) and a short-pass filter (<550 nm), respectively, to separate the green and red bioluminescence signals. The green- and red-channel images were then focused onto the right and left halves of a cooled CCD camera detector so that the red and green channels could be collected simultaneously frame-by-frame (Figure 1A).
Figure 1: Dual-color bioluminescence imaging of VIP-Cre and AVP-Cre SCNs.
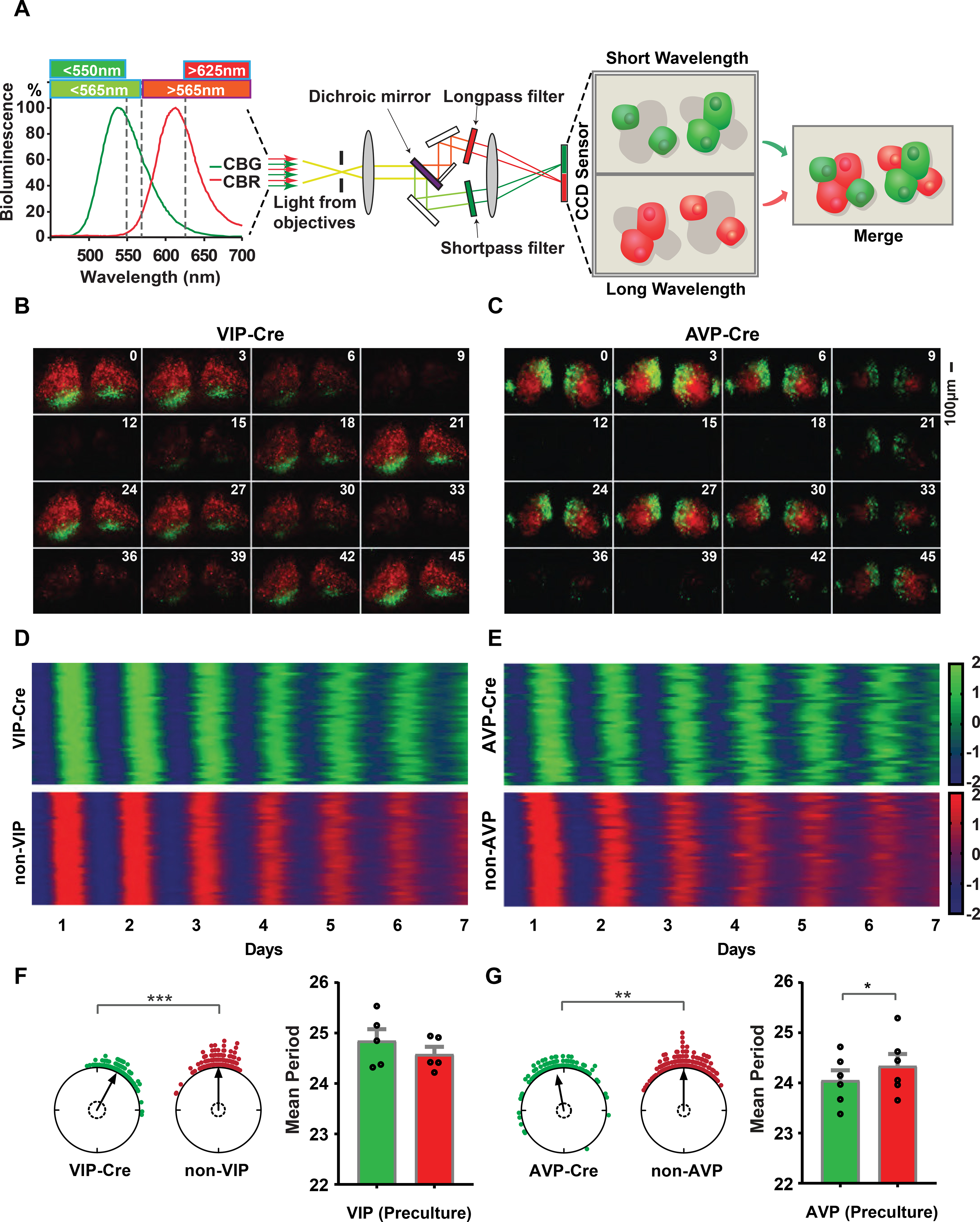
(A) Light-path of the dual-color imaging device. (B, C) Example time-lapse images for 48 hr from VIP-Cre and AVP-Cre SCN, respectively. Time interval between frames is three hours. SCN images were positioned with the ventral-dorsal axis vertically, optic chiasma at the bottom. Scalebar represents 100 μm. (D, E) Heat map representations of single-cell real-time imaging of two SCN slices for seven days. Each panel contains 40 single cells of each color in raster plots. The Z-score of detrended and denoised bioluminescence time series were color-coded between −2 to +2 as indicated by the color bar. Time series were ordered by phase with earlier phases placed at the top. (F, G) Rayleigh plots show the phases of single cells on day 6 and phases of green cells are shown relative to the normalized mean phase of red cells. Arrows are the R vector and dashed circles indicate the Rayleigh critical values. Individual data points are scattered as the circular plots. Asterisks indicate significant directional mean phase differences by Watson-Williams test (VIP, p value <0.001; AVP, p value=0.008). Mean +/− SEM period values from multiple slices and experiments (24.85±0.23 hrs, VIP; 24.58±0.15 hrs non-VIP; n=5 for VIP-Cre and 24.05±0.20hrs, AVP; 24.34±0.24, non-AVP; n=6 for AVP-Cre). Asterisks indicate the significance of t-test between green and red (* p ≤0.05; ** p ≤ 0.01; *** p ≤ 0.001). Detailed quantification results are included in Supplemental Data 1.
We next crossed Color-Switch PER2::LUCIFERASE mice with VIP-ires-Cre (Taniguchi et al., 2011) or AVP-ires-Cre (Pei et al., 2014) mouse lines and ex vivo cultures of SCN explants were prepared (Yamazaki and Takahashi, 2005). As expected, VIP cells with PER2::CBG expression were localized in the ventral/core region of the SCN close to the optic chiasm (Figure 1B; Video S1); whereas, AVP cells were localized in the dorsal/shell region of the SCN (Figure 1C; Video S2). Heatmap representations of single-cell rhythms from VIP (Figure 1D) and AVP (Figure 1E) neurons show that circadian rhythmicity was coherent in SCN explants, similar to that seen previously in pan-SCN PER2::LUC recordings (Buhr et al., 2010; Ko et al., 2010; Liu et al., 2007). Importantly, the SCN slice is not in steady state when the tissue slice is first prepared. Initially, the SCN slice reflects a state of the oscillator system that is similar to what it was previously in vivo. As documented in many studies, the relative phase coherence of SCN oscillators is regulated by photoperiod and this can be seen subsequently in SCN slices as the coherence of phases of the population of cells in the SCN (Buijink et al., 2016; Schaap et al., 2003; VanderLeest et al., 2007). With time in vitro, the SCN slices “relax” to a new steady state with respect to mutual coupling of the population of oscillators. During this first week in culture, the phase relationships among SCN neurons gradually change and SCN neurons can exhibit different period lengths as the slice goes from one state (from in vivo) to a new steady state (in vitro). With our methods, we can directly measure these subtle period changes during this transition interval.
To evaluate the periodicity of genetically-identified single cells from dual-color imaging, bioluminescence time series were analyzed with a custom made pipeline (Figure S2, and see Supplemental Methods). VIP neurons had a delayed average phase and longer average single-cell period compared to non-VIP cells (Figure 1F; Figure S3). In contrast, AVP neurons had shorter average single-cell period length, but no average phase difference, compared to non-AVP cells (Figure 1G; Figure S3). Because both VIP and AVP cell types used PER2::CBG in comparison to PER2::CBR, the red/green reporters cannot account for the average period differences seen between VIP and AVP SCN neurons. These results suggest that our new mouse line allows for the accurate measurement of circadian PER2::LUC rhythms from any genetically accessible cell type given an appropriate Cre driver line. As with the original PER2::LUC reporter mouse (Yoo et al., 2004), this conditional Color-Switch reporter mouse should become a valuable reagent for the field.
Intercellular coupling compensates for loss of functional clocks in VIP neurons
The SCN is a neuronal network and intercellular coupling mutually synchronizes the periods of individual cell-autonomous oscillators (Abel et al., 2016; Gonze et al., 2005; Herzog, 2007; Low-Zeddies and Takahashi, 2001; Schmal et al., 2018). Intercellular coupling of SCN neurons confers robustness to the network and can rescue cell-autonomous genetic defects observed in isolated SCN neurons (Ko et al., 2010; Liu et al., 2007; Tokuda et al., 2015). To study the neuropeptide-specific roles of neurons within this network, we applied tetrodotoxin (TTX), a sodium ion channel blocker, to inhibit the firing of action potentials in order to disrupt the coupling of SCN neurons (Yamaguchi et al., 2003). Neuronal firing results in the release of AVP and VIP in the SCN (Mazuski et al., 2018; Pennartz et al., 1998); therefore, blocking neuronal firing via TTX prevents intercellular communication within the SCN. Washout of TTX reverses the decoupling of the network, resulting in restored synchrony and high-amplitude circadian rhythmicity (Abel et al., 2016; Yamaguchi et al., 2003).
To examine the respective roles of VIP and AVP neurons in maintaining synchrony within the SCN network, we also compared the effects of TTX on VIP or AVP neurons in a Cre-only or Bmal1 conditional deletion genetic background. The VIP-Cre and AVP-Cre mouse lines were crossed to Color-Switch mice and a Cre-lox conditional Bmal1 knock-out mouse line (Johnson et al., 2014) to produce VIP-Cre; Bmal1fx/fx; Per2iLuc/iLuc (referred to as VIP-Bmal1−/−, Supplemental Table 1), and AVP-Cre; Bmal1fx/fx; Per2iLuc/iLuc (referred to as AVP-Bmal1−/−, Supplemental Table 1) mice. VIP-Cre and AVP-Cre lines were used as heterozygous drivers in order to mitigate potential hypomorphic expression of their peptide gene products. In these mice, cells with green bioluminescence signals also indicate the excision of Bmal1 and loss of cell-autonomous circadian function. We first compared TTX-mediated synchronization between the VIP-Cre labeled SCN, and the VIP-Bmal1−/− SCN, because VIP is known to play an important role in SCN synchrony (Aton et al., 2005). In both VIP-Cre and VIP-Bmal1−/−, individual cells exhibited high-amplitude oscillations with moderate damping. When 1 μM TTX was added, the amplitude of single cells and the overall SCN damped quickly, and the phase coherence was also disrupted. (Figure 2 A–F; Figure S3), as seen previously (Abel et al., 2016; Buhr et al., 2010; Yamaguchi et al., 2003). TTX was washed out at the end of second week, and the damped and desynchronized neurons recoupled when neuronal firing was restored following TTX washout. Surprisingly, this recoupling during washout resulted in sustained synchronous oscillations in both the VIP-Cre SCN and the VIP-Bmal1−/− SCN, despite the known importance of VIP neurons in SCN synchrony. Of note, previous work showing the importance of VIP signaling for SCN synchrony and rhythmicity employed either Vip or Vipr2 knockout mice, which are distinct from Bmal1 knockout within VIP neurons. High-amplitude oscillations (as quantified by LSP peak) were restored even in VIP-Bmal1−/− neurons following TTX washout and there were no effects of genotype on phase variance (Figure 2 G–H; Figure S3), indicating that input from the remaining SCN network is sufficiently strong to drive circadian oscillations in VIP-Bmal1−/− SCN neurons.
Figure 2: Cell-type-specific effects of Bmal loss of function in VIP neurons.
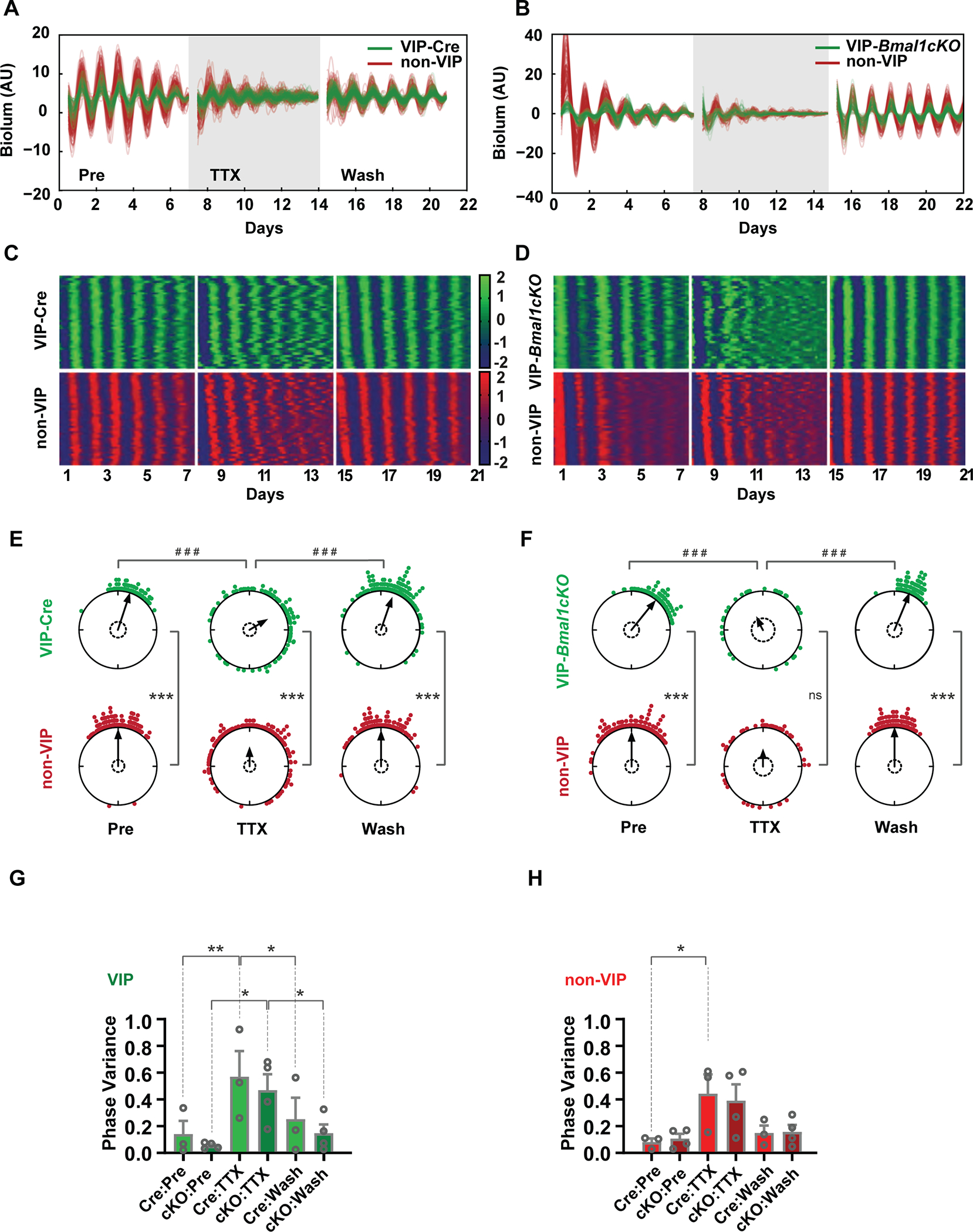
(A, B) Single-cell trajectories from VIP-Cre and VIP-Bmal1−/− (cKO) SCN through three conditions: pretreatment, 1μM TTX treatment and washout of TTX. (C, D) Heatmap representation of single-cell trajectories from VIP-Cre and VIP-Bmal1−/− SCN through three conditions. (E, F) Single-cell phase distributions of a VIP SCN (E) and a VIP-Bmal1−/− SCN (F). Sinusoid curve fitting was performed only on rhythmic cells, and phases on day 4 (midpoint) are presented in Rayleigh plots. Asterisks indicate significant directional mean phase differences by Watson-Williams test and pound signs indicate significant phase homogeneity or variance differences by Watson-Wheeler test. (G-H) Homogeneity of single-cell phase from multiple replicate SCN slices (VIP, n=3 and VIP-Bmal1−/−, n=4), were evaluated with Mean +/− SEM circular variances. Two-way ANOVA found no effect of genotype across pretreatment, TTX treatment, and washout. One-way repeated measures ANOVA with Tukey’s multiple comparisons was used to assess the effects of pretreatment, TTX treatment, and washout in each cell type (VIP-Cre, non-VIP, VIP-Bmal1−/−, and non-VIP-Bmal1−/−). Asterisks and pound signs indicate significant differences (* or #, p ≤0.05; ** or ##, p ≤ 0.01; *** or ###, p v≤ 0.001). Detailed quantification results are included in Supplemental Data 1.
Next, we examined the effects of TTX on AVP-Cre labeled SCN and AVP-Bmal1−/− SCN neurons. As expected, the AVP-Cre labeled SCN neurons were rhythmic and coherent (Figures 3A, C, and E; Figure S3). In contrast, AVP neurons within the AVP-Bmal1−/− SCN exhibited abnormal oscillations even before the application of TTX (Figures 3B, D, and F; Figure S3) and had increased phase variance as compared to AVP-Cre neurons (Figure 3G). At the same time, non-AVP neurons within the AVP-Bmal1−/− SCN had high amplitude and coherent rhythms despite the loss of rhythmicity in AVP neurons. TTX application led to a further loss of circadian amplitude of AVP neurons in AVP-Bmal1−/− SCN (Figures 3B and D; Figure S3). Surprisingly, removal of TTX failed to restore rhythmicity to AVP neurons in the AVP-Bmal1−/− SCN as compared to AVP-Cre neurons (Figure 3B and D; Figure S3). The absence of normal rhythmicity in AVP-Bmal1 −/− neurons, even when coherence remains in non-AVP cells, suggests that AVP neurons do not receive adequate coupling inputs from the rest of the SCN network to rescue aberrant cell-intrinsic rhythms.
Figure 3: Cell-type-specific effects of Bmal loss of function in AVP neurons.
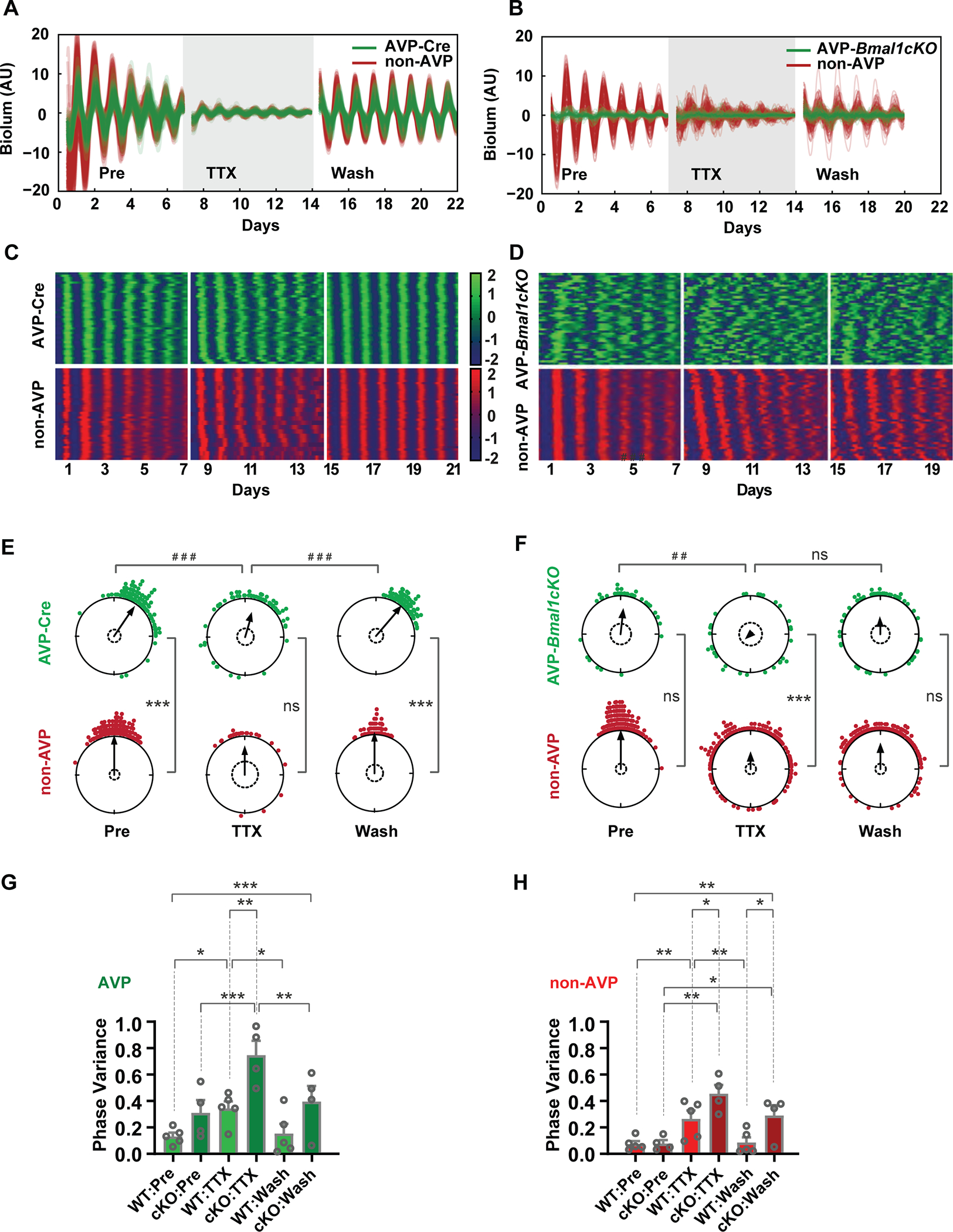
(A, B) Single-cell trajectories from AVP-Cre and AVP-Bmal1−/− (cKO) SCN through three conditions: pretreatment, 1μM TTX treatment and washout of TTX. (C, D) Heatmap representation of single-cell trajectories from AVP-Cre and an AVP-Bmal1−/− SCN through three conditions. (E, F) Single-cell phase distributions of an AVP SCN (E) and an AVP-Bmal1−/− SCN, (F). Sinusoid curve fitting was performed only on rhythmic cells, and phases on day 4 (midpoint) are presented in Rayleigh plots. Asterisks indicate significant directional mean phase differences by Watson-Williams test and pound signs indicate significant phase homogeneity or variance differences by Watson-Wheeler test. (G-H) Homogeneity of single-cell phase from multiple replicate SCN slices (AVP, n=5 and AVP-Bmal1−/−, n=4), were evaluated with Mean +/− SEM circular variances. Two-way ANOVA revealed a main effect of genotype, and asterisks indicate the significant Holm-Sidak’s multiple comparisons between AVP-Cre and AVP-Bmal1−/− across pretreatment, TTX treatment, and washout. One-way repeated measures ANOVA with Tukey’s multiple comparisons was used to assess the effects of pretreatment, TTX treatment and washout in each cell type (AVP-Cre, non-AVP, AVP-Bmal1−/−, and non-AVP Bmal1−/−). Asterisks and pound signs indicate significant differences (* or #, p ≤0.05; ** or ##, p ≤ 0.01; *** or ###, p ≤ 0.001). Detailed quantification results are included in Supplemental Data 1.
To test if loss of Bmal1 affected rhythmicity by altering PER2 expression, we normalized the green bioluminescence of Cre-positive cells to red bioluminescence of non-Cre cells, which have normal Bmal1 expression. Compared to Cre-only controls, loss of Bmal1 had no obvious effect on average PER2:LUC levels in VIP-Bmal1−/− or AVP-Bmal1−/− cells, except for slight upregulation in VIP-Bmal1−/− neurons during precultue (Figure S3). The lack of downregulation of PER2::LUC in Bmal1−/− neurons indicates that loss of circadian amplitude is not due to a reduction in PER2::LUC levels.
To examine the joint roles of VIP and AVP neurotransmission, we also generated combined VIP-Cre; AVP-Cre; Bmal1fx/fx; Per2iLuc/iLuc mice in which Bmal1 is excised in both AVP and VIP neurons. The VIP/AVP-Bmal1−/− SCN displayed a phenotype similar to an AVP-Bmal1−/− SCN, both in terms of rhythmicity and identified network structure (Figure S4). Although the VIP/AVP-Bmal1−/− SCN had a higher ratio of neurons with Bmal1 deletion, there was no obvious additional loss of rhythmicity or synchrony. These results suggest that it is not just the total number of SCN neurons with Bmal1 deletion that is important (Low-Zeddies and Takahashi, 2001), but rather that there is something specific to Bmal1 loss AVP neurons. Thus, our findings confirm previous suggestions that Bmal1 is necessary for circadian function in AVP neurons (Mieda et al., 2015) and that a functional circadian clock within AVP neurons is essential both for sustaining endogenous circadian rhythms and maintaining synchrony with the rest of the SCN network.
A functional circadian clock in AVP neurons is critical for network synchrony in the SCN
In previous studies, the coupling of circadian rhythms between SCN neurons was estimated with either period or phase distributions (Abraham et al., 2010; Evans et al., 2013; Gonze et al., 2005; Mieda et al., 2015; Schmal et al., 2018). In our dual-color imaging results from the AVP-Bmal1−/− SCN, the disrupted bioluminescence patterns are not sinusoidal (with poor phase estimation (Supplemental Data 1), and thus should not be parameterized as such (Figures 3B and 3D). Therefore, in addition to using phase distribution (Figures 3G and 3H) to assess coupling, we used the maximal information coefficient (MIC), a nonparametric pairwise correlation of noisy signals (Reshef et al., 2011), which has been used previously for this purpose (Abel et al., 2016), and combined this with LSP rhythmicity to assess intercellular communication between SCN neuronal subpopulations. Communication between SCN neurons was categorized into three types: red with red (no Cre with no Cre) red with green (no Cre with Cre only or Cre recombined Bmal1−/−), and green with green (Cre only with Cre only, or Cre recombined Bmal1−/− with Cre recombined Bmal1−/−). This analysis was performed for the four SCN types listed previously: VIP-Cre, VIP-Bmal1−/− (Figure 2), AVP-Cre, and AVP-Bmal1−/− (Figure 3). Intercellular coupling was significantly reduced during TTX treatment across the SCN and restored upon TTX washout. The connections between SCN neurons, either distant between the two lobes or adjacent inside a lobe, displayed similar MIC scores (Figure S3), indicating the high correlation between the two lobes.
We found that in both VIP- and AVP-Cre SCNs, the correlation within and between classes of SCN neurons decreased during TTX and rose to approximately the pre-TTX level following washout (Figure 4A and 4D, Supplemental Table 2). This is consistent with loss of intercellular coupling during TTX application, even among non-oscillatory cells. For example, in VIP-Cre SCN, there was strong coupling within and between all three classes of cells as seen in the correlation matrices and as indicated by the black lines connecting SCN cells in the cartoon representative of the anatomical position of the cell within the SCN (Figure 4A). For the VIP-Bmal1−/− SCN, the correlation throughout the experiment was similar to that seen with the VIP-Cre SCN, with a confirmed loss and restoration of oscillation and synchrony due to intercellular communication (Figure 4B). Furthermore, inter-class MIC was high following washout, indicating that VIP and non-VIP neurons exchange information to establish synchrony across the network, rather than confining communication within a single neuronal subtype (Figure 4C, Supplemental Table 2).
Figure 4: Comparison of the effects of loss of Bmal1 on Coupling in VIP and AVP neurons in the SCN.
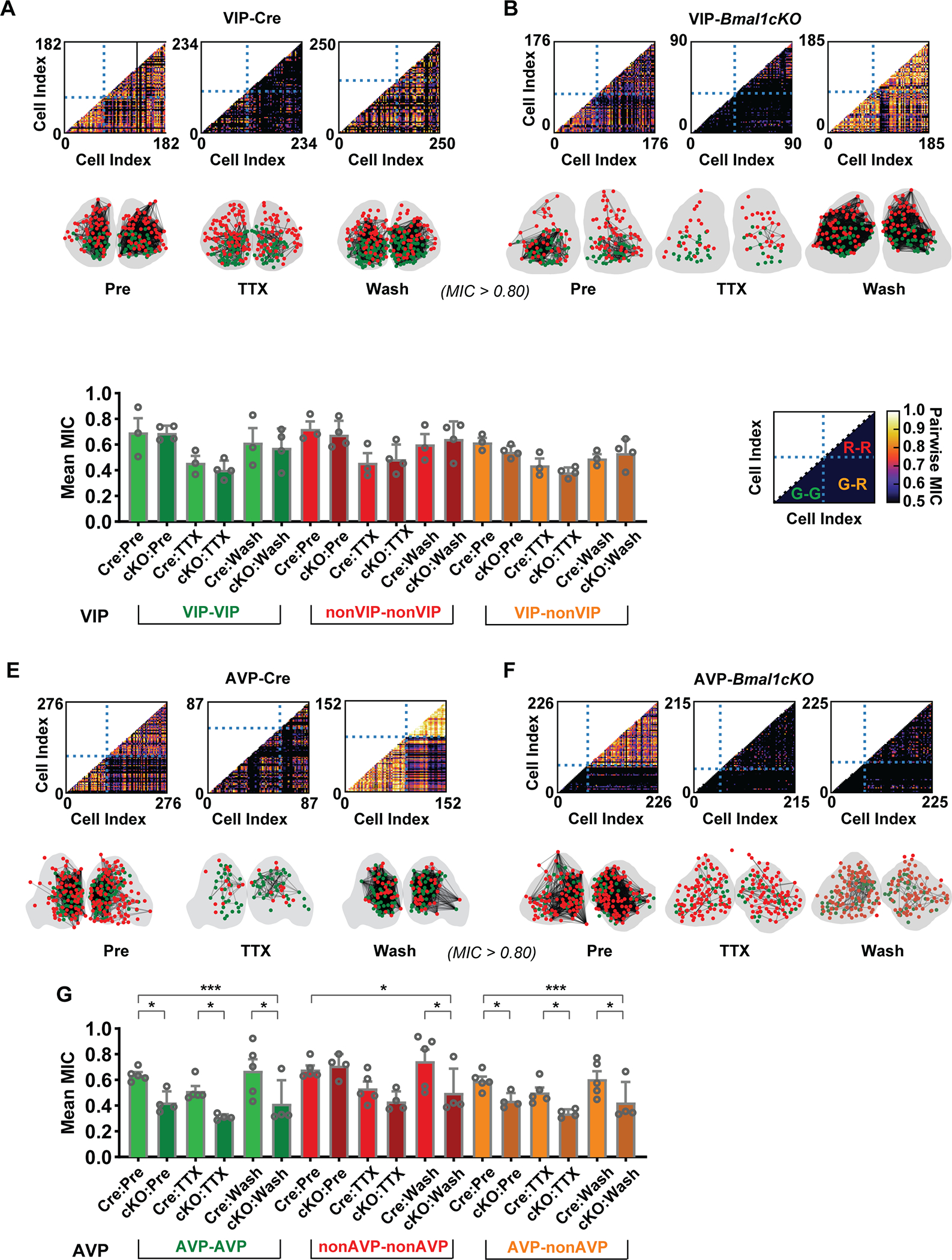
(A, B) Pairwise MIC score plots of neurons in the VIP SCN versus the VIP-Bmal1−/− (cKO) SCN, during pretreatment, TTX treatment, and washout. As annotated in D, horizontal and vertical dashed lines split the plot into four parts, including the lower-left part showing the pair-wise MIC value among green cells, lower-right part is MIC values of green-red pairs and higher-right part is red-red pairs. Correlation network was constructed from pairwise MIC analysis: A connection line was drawn when the MIC value of the cell pair is larger than 0.8. Right-left SCN associations are not shown. (C) Mean +/− SEM MIC scores are shown to compare three types of associations between VIP-Cre and VIP-Bmal1−/− SCNs. The associations were categorized as green-green (or KO-KO), red-red (or Cre-Cre) and green-red (or KO-Cre). Each mean MIC value from a replicate is represented by a point in the overlaid scatter plot (VIP, n=3; VIP-Bmal1−/−, n=4). Higher asterisks represent the significance of genotype factor in two-way ANOVA, and lower asterisks represent significance of Holm-Sidak’s multiple comparisons. (E, F) Pairwise MIC score plots of neurons in the AVP SCN versus the AVP-Bmal1−/− SCN, during pretreatment, TTX treatment, and washout. (G ) Mean +/− SEM of MIC scores are shown to compare three types of associations (as in C) between AVP-Cre and AVP-Bmal1−/− SCNs (AVP, n=5; AVP-Bmal1−/−, n=4). Asterisks indiciate significant differences (* p ≤0.05; ** p ≤ 0.01; *** p ≤ 0.001). Detailed quantification results are included in Supplemental Data 1.
In sharp contrast, the AVP-Bmal1−/− SCN exhibited low MIC scores during TTX and following washout, indicating little transmission of information between neurons of either population within the tissue (Figure 4F). Specifically, in AVP-Bmal1−/− SCN, there was a complete loss of coupling from AVP to AVP (G-G) and AVP to non-AVP (G-R) cells, but a persistence of non-AVP to non-AVP (R-R) coupling in the control pretreatment (Pre) interval, reminiscent of a status of in vivo entrainment. TTX treatment then almost completely abolished coupling among all classes of cells in the SCN, and surprisingly, the network was not capable of full recovery of synchrony after TTX washout (Figure 4G, Supplemental Table 2). The VIP/AVP-Bmal1−/− SCN displayed a phenotype similar to an AVP-Bmal1−/− SCN. Interestingly, although the average single-cell rhythmicity and overall synchrony of green Bmal1−/− cells was low in the VIP/AVP-Bmal1−/− SCN, a subgroup of Bmal1−/− cells in the ventral area displayed relatively higher rhythmicity and stronger coupling during pretreatment (Figure S4, Supplemental Table 2). These findings suggest that the VIP-Bmal1−/− cells are still able to acquire rhythms from other cells expressing Bmal1 that have been entrained in vivo. Thus, we conclude that Bmal1 and cell autonomous circadian clock function is necessary in AVP neurons for intercellular communication both among AVP neurons and between AVP and non-AVP cells in the SCN.
Conditional Bmal1 deletion from AVP neurons alters circadian behavior
To examine the in vivo effects of cell-type-specific deletions of Bmal1 on circadian activity rhythms of mice, we compared VIP-Cre and AVP-Cre controls, VIP-Bmal1−/−; AVP-Bmal1−/−; and VIP/AVP-Bmal1−/− mice. Exposure to constant light (LL) desynchronizes SCN neurons (Ohta et al., 2005), therefore mice were first subjected to an LD12:12 light-dark cycle for at least two weeks (designated LD1), then constant darkness (DD1) for three weeks, followed by exposure to constant light (LL) as a disruptive episode, a second interval in constant darkness (DD2), re-entrainment to LD12:12 (LD2) and a third interval in constant darkness (DD3). VIP-Cre and VIP-Bmal1−/− mice expressed normal circadian rhythms under all of these conditions (Figure S5), suggesting that conditional deletion of Bmal1 in VIP neurons has no effect on circadian activity rhythms. In stark contrast, AVP-Bmal1−/− mice exhibited lengthened period in DD2 following the exposure to constant light (LL) (Figure S5), although the effects were variable, possibly as a result of variance in Bmal1 loss between animals or due to individual differences in circadian periodicity (Evans et al., 2015). Nonetheless, these results corroborate previously reported results (Mieda et al., 2015). AVP-Bmal1−/− mice also had marked period instability in the three DD intervals with significantly increased period variance of each animal (Figure S5), although this was not as severe as that seen in NMS-Bmal1−/− mice (Lee et al., 2015). The labile circadian period phenotype, which can be modulated strongly by different lighting conditions, is consistent with the reduction in network coupling observed in AVP-Bmal1−/− SCN explants and also with previous reports of faster re-entrainment after a jet lag (Mieda et al., 2015; Yamaguchi et al., 2003). Furthermore, the VIP/AVP-Bmal1−/− mice displayed phenotypes similar to AVP-Bmal1−/− mice, with slightly milder period lengthening or instability (Figure S5), reiterating the importance of autonomous rhythms in AVP neurons. The lack of entrainment or circadian periodicity defects in VIP-Bmal1−/− mice further indicates that Bmal1 (and cell-autonomous circadian function) is dispensable in VIP neurons.
Disparities in coupling strength between VIP and AVP neurons, not cell numbers, underlie differential roles in network synchrony
AVP and VIP are thought to function in a complementary fashion (Maywood et al., 2011) in the SCN, and both AVP receptor V1a, and VIP receptor VPAC2R (G-protein coupled receptors with Gq or Gs alpha subunits, respectively) are expressed in the SCN. Thus, AVP and VIP increase calcium and cAMP signaling within a postsynaptic neuron, which ultimately increases the activity of cAMP response element-binding protein (CREB) and promotes Per transcription (Tischkau et al., 2003). Furthermore, because AVP and VIP are released in response to neuronal firing, and Gs or Gq pathways increase excitation, VIP release (e.g., in response to light) or AVP release may trigger the release of the complementary neurotransmitter, thus increasing the strength of such a signal. This has recently been suggested by increases in VIP neuron activity leading to increased cFOS expression in the dorsal SCN, far from the VIP-expressing ventral SCN (Mazuski et al., 2018). Finally, Avp transcription is regulated by an E-box, and AVP-Bmal1−/− therefore reduces overall AVP expression as well as other CLOCK:BMAL1 target genes such as Prok2 and Rgs16 in addition to suppressing circadian oscillations (Jin et al., 1999; Mieda et al., 2015). Indeed, immunostaining and quantification of the expression of VIP and AVP proteins in our mouse models revealed that AVP and AVP receptor (V1a) expression is decreased in AVP-Bmal1−/− SCN, consistent with previous findings of reduced AVP gene expression in the SCN of AVP-Bmal1−/− mice (Mieda et al., 2015), while VIP and VIP receptor (VPAC2) expression is not decreased (Figure 5). AVP and V1a expression is also decreased in VIP-Bmal1−/− SCN. Because CLOCK:BMAL1 regulates AVP and AVP receptor (V1a), it is to be expected that BMAL1 target genes in VIP-Bmal1−/− cells would be down-regulated. In this case, VIP neurons may have a reduction in AVP signaling but this is not sufficient to prevent VIP neurons from receiving network coupling inputs. Thus, it is unlikely that the effects of AVP-Bmal1−/− conditional deletion on network synchrony are caused by non-cell-autonomous changes in VIP or VIP receptor expression in the SCN, and intact VIP signaling may be the responsible for the unaltered light entrainment in AVP-Bmal1−/− animals.
Figure 5: Immunofluorescent staining of key components of VIP and AVP signaling in VIP-Bmal1−/−, AVP-Bmal1−/− and VIP/AVP-Bmal1−/− SCNs.
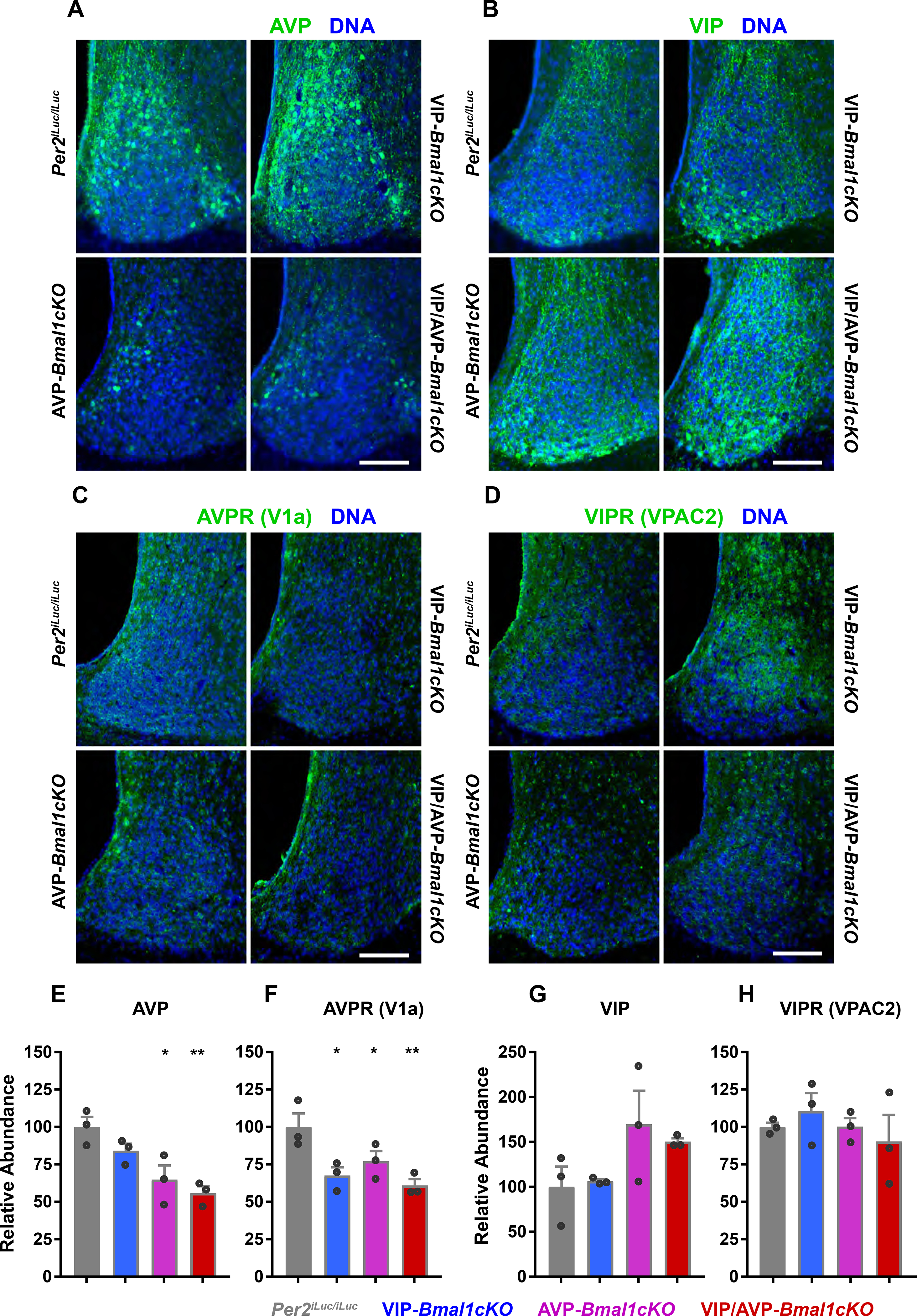
(A - D) Immunofluorescent images from 50μm thick SCN slices from wildtype control, VIP-Bmal1−/− (cKO), AVP-Bmal1−/−(cKO) and VIP/AVP-Bmal1−/− (cKO) SCNs, stained with AVP, VIP, AVP receptor V1a and VIP receptor VPAC2 antibodies. Green signals from the neuropeptides or receptors are merged with the blue staining of nuclear DNA. Scale bars represent 100μm. (E-H) Mean +/− SEM plots of fluorescence intensity normalized to wildtype, VIP-Cre and AVP-Cre controls (one of each) are shown as the relative abundance. Each replicate is shown as a point in the overlaid scatter plots (all n=3). Asterisks indicate the significance of Holm-Sidak’s comparisons to controls and ANOVA adjusted p-value (* p ≤0.05; ** p ≤ 0.01; *** p ≤ 0.001).
We next constructed a mathematical model to test the origin of the difference in phenotype between AVP-Bmal1−/− and VIP-Bmal1-/−. There are approximately 1100 VIP, 2100 AVP and 6800 other cells in the SCN (Welsh et al., 2010), so one possibility is that functional AVP cells are sufficient to drive VIP cells simply due to the larger number of cells. To test this hypothesis, we modified a simple, mechanistic and stochastic model of the circadian oscillator (Gonze et al., 2005) to reflect the interactions of the three SCN compartments. We first tested if relative AVP cell count was sufficient to explain the observed phenotypes. We parameterized AVP and VIP pathways identically and varied AVP:VIP cell count ratios from 9:1 to 1:9 (Figure S6). We found that only a relative abundance of at least 9:1 AVP:VIP was sufficient to drive oscillation and synchrony in VIP-Bmal1−/− while leaving AVP-Bmal1−/− arrhythmic and asynchronous, which is much higher than the AVP:VIP ratio that is observed physiologically. However, another possibility is that the coupling strength of an AVP cell on a postsynaptic cell in the absence of VIP is stronger than that of a VIP cell on a postsynaptic cell in the absence of AVP. To test this, we varied the relative coupling strength of the AVP and VIP pathways from 10:1 to 1:10 AVP:VIP with equal AVP and VIP cell counts. We found that a relative coupling strength of 5:1 AVP:VIP was sufficient to drive oscillation in VIP-Bmal1−/−, while AVP-Bmal1−/− remained arrhythmic and asynchronous (Figure S6, details and model parameterization in Methods). Thus, our mathematical findings support the hypothesis that differences observed experimentally reflect stronger AVP coupling than VIP coupling.
We next sought to model a typical SCN with physiologically relevant cell ratios (Figure 6A; VIP:nonVIP/AVP:AVP set to 27:170:53; for a total of 250 cells). Given that there are 2X more AVP than VIP cells, we chose a 2.5:1 AVP:VIP coupling strength as it yields a total power of 5:1 that we found was sufficient to drive oscillations in VIP-Bmal1−/− neurons. Using these parameters, stronger AVP signaling to the VIP-Bmal1−/− compartment complemented by a higher fraction of AVP cells was sufficient to drive synchrony, whereas weaker VIP input to the AVP-Bmal1−/− compartment was insufficient to restore synchrony and resulted in greatly diminished oscillation (Figures 6B and Figure S6). Furthermore, we found that the pairwise MIC in this model follows a pattern similar to that seen in the experiments presented here (Figures 2 and 3; Figures 6C and Figure S6). This result predicts that in vivo entrainment in the AVP-Bmal1−/− is due to an increase in VIP signal strength in response to light, as VIP neurons are thought to be the primary mediators of light entrainment (Jones et al., 2018; Mazuski et al., 2018). Moreover, our immunohistochemistry results suggest that decreased coupling strength in the SCN of AVP-Bmal1−/− mice may be due to a reduction in AVP and AVP receptor expression.
Figure 6: A stochastic circadian computation model to explain experimental observations.
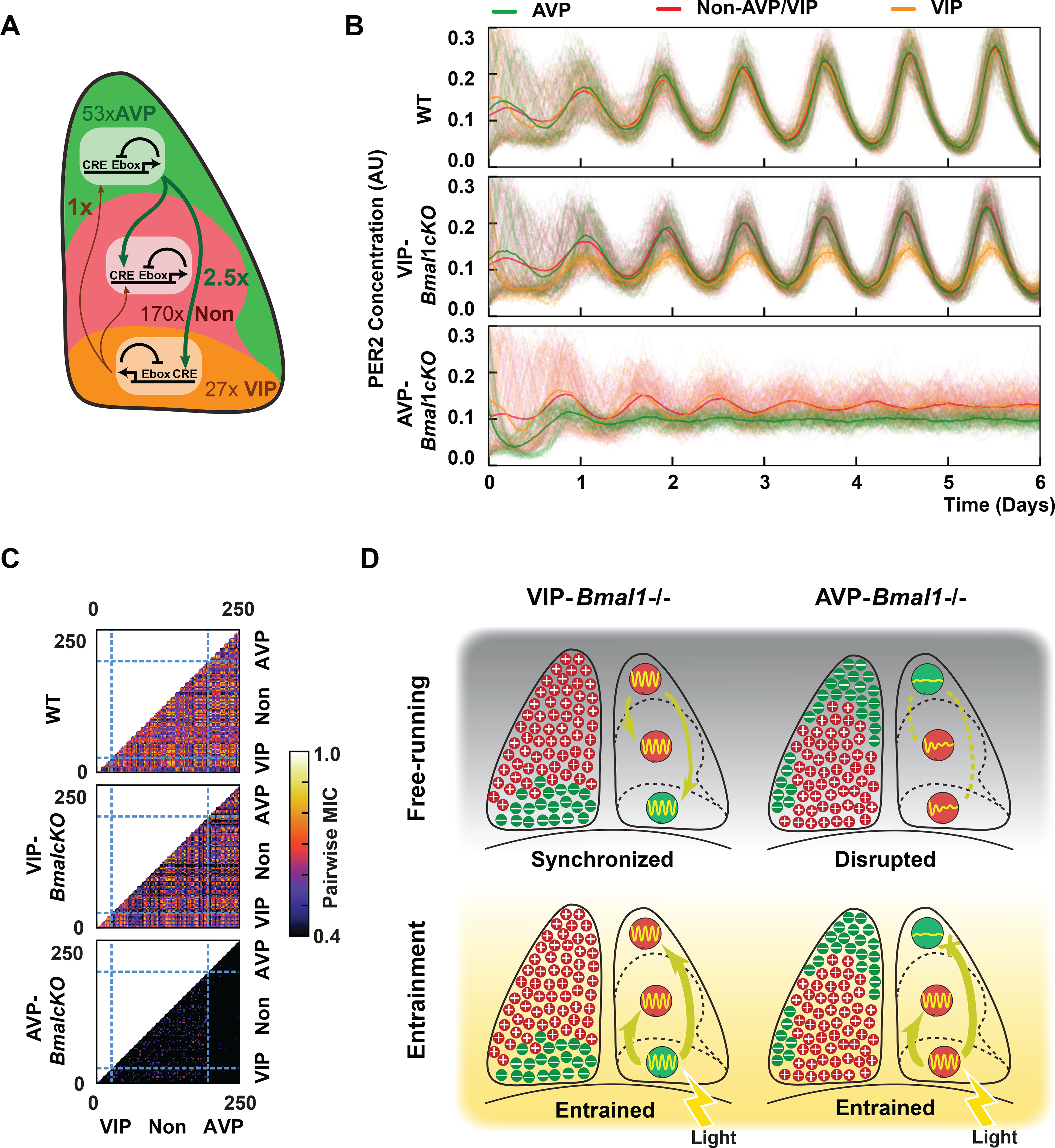
(A) Computational model as the outcome of parameterization of the circadian circuits based on approximated ratios of SCN neurons (53 AVP cells, 27 VIP cells, and 170 non-AVP/non-VIP cells). The AVP:VIP coupling strength was set to 2.5:1. (B) Single-cell trajectories of each cell type. The signals represent relative expression level. Three models representing a wildtype, a VIP-Bmal1−/− (cKO) or an AVP-Bmal1−/− (cKO) SCN are shown. (C) Pairwise MIC scores represent six inter- or intra-class connections and correlations. X axis and Y axis represent the cell classes, and the pairing pattern of the classes is divided by dashed lines. (D) Proposed mechanism of the differential roles of VIP and AVP neurons in the SCN. Entrainment occurs under daylight and switches to free-running under constant darkness. Green color and minus signs indicate the excision of Bmal1 and ablation of autonomous circadian rhythms, while red color and plus signs indicate intact cells.
DISCUSSION
The dynamics of circadian rhythms of the SCN have been studied extensively using real-time imaging of Per1-luciferase and PER2::LUC as well as other transgenic mouse models (Cheng et al., 2009; Kuhlman et al., 2000; Mei et al., 2018; Ono et al., 2016; Wilsbacher et al., 2002; Yamaguchi et al., 2003; Yoo et al., 2004). However, because circadian genes are expressed ubiquitously, it has been difficult to study the circadian properties of specific cell types in the SCN in order to understand their functions within the SCN network. Previous studies have indicated that VIP and AVP neurons may serve important non-redundant functions for circadian rhythm generation in SCN (Aton et al., 2005; Brancaccio et al., 2017; Jones et al., 2018; Maywood et al., 2011; Mieda et al., 2015; Yamaguchi et al., 2013). To delineate these functions we utilized our novel mouse model to examine the effects of conditional deletion of Bmal1 in either VIP or AVP neurons. We were surprised to see that Bmal1-dependent intrinsic rhythms in AVP, not VIP, neurons were essential for synchrony of the SCN. Even with the rest of the SCN network intact, intercellular signaling to AVP-Bmal1−/− neurons was insufficient to restore synchronous oscillation. In contrast, VIP-Bmal1−/− neurons were rhythmic and synchronized, indicating that coupling signals from the rest of the SCN network can rescue circadian rhythms in VIP-Bmal1−/− neurons. When network coupling was inhibited by TTX treatment, AVP-Bmal1−/− neurons displayed an even stronger phenotype in which synchrony of the entire SCN network was disrupted after TTX washout. Moreover, at the behavioral level, conditional deletion of Bmal1 in VIP neurons had no obvious effects on circadian activity rhythms; while conditional deletion of Bmal1 in AVP neurons lead to a modest lengthening of period and labile circadian activity rhythms that can be modulated strongly by different lighting conditions, consistent with previous reports (Mieda et al., 2015). These results demonstrate that Bmal1 (and presumably circadian function in AVP neurons) is necessary for network synchrony in the SCN and validate previously suggested functions of AVP neurons in SCN intercellular communication (Mieda et al., 2015; Yamaguchi et al., 2013).
To be clear, our findings do not argue against an important role for VIP signaling (VIP ligand and VIP receptors) per se, but rather the results presented here demonstrate that circadian function derived from Bmal1-dependent autonomous rhythms in AVP neurons is more impactful than that seen in VIP neurons. Our findings are consistent with a recent report that also suggests that circadian function in VIP neurons is not essential for SCN or behavioral rhythmicity (Mazuski et al., 2020). Ultimately, further study of the electrical interaction of these neurotransmitters is needed to understand communication in the SCN (Hastings et al., 2018; Honma, 2018). Moreover, neuronal firing is responsible for the timing of the release of these signaling molecules, and so oscillations in transcription alone do not fully address communication dynamics. Because the Avp gene is a direct target of CLOCK:BMAL1 transcriptional activation (Jin et al., 1999), the loss of Bmal1 in AVP neurons reduced AVP gene expression, as well as the expression of other genes, such Rgs16 and Prok2 (Mieda et al., 2015), which regulate additional SCN outputs (Cheng et al., 2002; Doi et al., 2011). In addition, recent work has shown that astrocytes in the SCN may be sufficient to regulate circadian behavior (Brancaccio et al., 2019). Future studies using our colorswitch model to probe various other SCN neuronal and glial populations will help to further clarify the complicated dynamics of the SCN.
In order to understand the different roles of VIP and AVP neurons within the SCN, we also developed a simple stochastic model of the SCN network composed of three classes of neurons. By imposing a difference in the coupling strength and abundance of VIP and AVP neurons, the model recapitulates the effects of Bmal1 loss-of-function in VIP or AVP neurons on the SCN network. Although the model predicts that a mechanistic difference in AVP and VIP signaling strength is essential to recapitulate observations, it cannot predict precisely where this difference lies. Two possible differences between AVP and VIP that are compatible with the model are: (i) differences in neurotransmitter abundance resulting from Bmal−/−, or (ii) differences in effect on membrane potential. Importantly, we did not attempt to model the interaction between AVP and VIP pathways or the membrane potential directly. Such a model would become quite complicated, as it would necessitate capturing electrical effects, which then modulate AVP or VIP release in the postsynaptic neuron. Furthermore, modeling other SCN neurotransmitters, e.g. GABA, (DeWoskin et al., 2015), and astrocyte contributions to neurotransmission (Brancaccio et al., 2017; Tso et al., 2017), may be necessary to fully understand the interactions of SCN neurons.
Together, our ex vivo, in vivo, and in silico results support a model in which circadian rhythmicity in AVP neurons is essential for SCN network synchrony and can even compensate for the loss of intrinsic rhythms in VIP neurons. One possible mechanism for the stronger AVP signal is that rhythmic excitatory AVP input may trigger action potentials and VIP release. Thus, VIP-Bmal1−/− neurons may still function effectively within the SCN network because of rhythmic AVP (or other) inputs driving rhythmic VIP release, despite a lack of autonomous circadian oscillation. Meanwhile, the AVP signal is weaker in AVP-Bmal1−/− mice because AVP production is reduced as it is regulated by an E-box. Similarly, VIP release is known to trigger shell neuron activity, and so entrainment in response to light may be augmented by VIP neurons stimulating AVP release. In conjunction with recent studies of the role of VIP neurons in light response (Jones et al., 2018) and the role of electrical activity in VIP release (Mazuski et al., 2018), our results help to clarify the hierarchy within the SCN (Figure 6D): VIP neurons receive photic input from the retina and entrain the SCN network to light and establish synchrony, but intrinsic circadian function is dispensable. In contrast, AVP neurons support the spontaneous synchronization of SCN network and maintain network oscillation in the absence of photic entrainment, and autonomous circadian function is required for AVP neurons to execute their role.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Joseph S. Takahashi (joseph.takahashi@utsouthwestern.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
The Color-Switch PER2::LUC mouse knock-in mouse line was created and maintained in a C57BL/6N background. As described in Supplemental Table 1, the Color-Switch PER2::LUC reporter was always used in the homozygous condition. VIP-ires-Cre and AVP-ires-Cre and the Bmal1fx/fx conditional knock-out mouse lines were described previously (Izumo et al., 2014; Johnson et al., 2014; Pei et al., 2014; Taniguchi et al., 2011). Note that this Bmal1fx/fx allele (gene symbol Arntl) (Izumo et al., 2014; Johnson et al., 2014) was made independently from the Arntltm1Weit (JAX #007668) line. The VIP-Cre line was obtained directly from Josh Huang before the Taniguchi et al. publication, and we made this line congenic on C57BL/6 in our laboratory, therefore it is similar to this Vip-IRES-Cre (C57BL/6J) line (JAX #031628). We have characterized and validated this line previously (Fan et al., 2015; Lee et al., 2015). Note that two recent studies report hypomorphic expression of VIP and/or AVP in Avp-IRES2-Cre (JAX #023530) and Vip-IRES-Cre (JAX #010908) mice (Cheng et al., 2019; Joye et al., 2020). The Viptm1(cre)Zjh/J line (JAX #010908) originates from a C57BL/6 X 129S4/SvJae F1 hybrid background. The AVP-Cre JAX #023530 line is also different from the AVP-Cre line used in our work (Pei et al., 2014). All Cre mouse driver lines reported here were used in the heterozygous condition, which did not affect circadian rhythm phenotypes, consistent with (Cheng et al., 2019; Joye et al., 2020). The mouse lines used in this work were backcrossed and maintained in a C57BL/6N background in order to maintain an isogenic background with the ES cells used for gene targeting (described below). All mice were maintained under an LD12:12 light dark cycle. Animals for SCN tissue culture and wheel-running experiments were used at two to six months of age. Wheel-running experiments were performed as previously reported (Siepka and Takahashi, 2005) and locomotor activity was recorded using ClockLab (Actimetrics, Inc., Wilmette, IL). Temperature and humidity were monitored, and the mice were housed under an LD12:12 cycle (green LEDs, ~100 lux at the level of the cage floor). Food and water were provided ad libitum throughout the study. All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Texas Southwestern Medical Center (APN 2015–100925). Periodicity of locomotor activity was analyzed with the Chi-square periodogram function in ClockLab Analysis (Actimetrics),
METHOD DETAILS
Knock-in Construct
The Per2::CBRiCBG knock-in targeting vector was derived from the backbone of the Per2::Luc targeting vector (Yoo et al., 2004). Firefly luciferase was replaced with a color-switchable reporter cassette, consisting of two click beetle luciferases, CBG99 and CBR (Gammon et al., 2006). A modified CBR cDNA, with C-terminal fragment substituted with C-terminal CBG (Villalobos et al., 2010) and its ATG start codon removed, was fused to the read through frame of loxP (11 aa), and placed in conjunction with the 3’ end of the Per2 open reading frame (ORF) in exon 23. CBG luciferase cDNA was fused to the second loxP site and placed next to the neomycin resistance cassette. To minimize non-specific expression of the CBG before Cre-lox recombination, the ATG start codon was removed and three stop codons were placed immediately upstream the loxP-CBG ORF. The 24 amino-acid (72bp) truncation in the previous version of the PER2::LUC reporter was repaired to make a full-length PER2 ORF (Yoo et al., 2004).
ES Cell Culture and gene targeting
JM8.F6 ES cells (passage #9) derived from the C56BL/6N background (Pettitt et al., 2009) were maintained in DMEM (Gibco) supplemented with 4 mM L-glutamine, 15% ES qualified Fetal Bovine Serum (Gemini), 55 μM β-mercaptoethanol (Gibco), 1x MEM non-essential amino acid (Gibco), 1x Penicillin-Streptomycin, 10,000 U/ml purified mLIF. A layer of SNL7 fibroblasts (neomycin resistant) was treated with 150 μg/ml mitomycin C and re-plated as the feeder layer. ES cells were transfected by electroporation (Bio-Rad), maintained in regular medium overnight before resistance selection with G418 (200 μg/ml) for two weeks for homologous recombinants. Colonies were picked and propagated in 96-well plates in duplicate. One of the duplicate plates was transfected with a lentiviral vector carrying a Cre recombinase cassette. Cells were imaged with a low-light camera and CCTV lens to select for Cre-dependent bioluminescence signal. Genomic DNA from the screened-out ES cells was prepared, then long-fragment PCR (LA Taq, Takara) and Southern blotting (Yoo et al., 2004) was carried out to verify the integration of non-genomic DNA sequence at both 5’ and 3’ arms of the Per2 locus. Targeted ES cell cells were injected into B6(Cg)-Tyrc−2J/J blastocysts and transferred to pseudopregnant B6(Cg)-Tyrc−2J/J female recipients. Resulting male chimeras were bred with C57BL/6N females. Germ-line transmission was confirmed by coat color, bioluminescence, and Southern blotting.
Ex vivo culture of SCN
Following procedures reported previously (Buhr et al., 2010; Izumo et al., 2014; Lee et al., 2015), the mouse brain was collected following cervical dislocation, before the onset of darkness. Peripheral tissues including liver were also collected. Coronal brain slices of 250–300 μm were made with a Vibratome in cold HBSS buffer. The SCN region was trimmed into a 1 mm × 1 mm square with optic chiasm attached and placed on top of a culture insert with pore size of 0.4 μm (Millipore). A 1 mm × 2 mm piece of liver was cultured in the same way as a control sample. All tissue slices were cultured in phenol red-free DMEM (Corning) supplemented with high glucose, glutamine, pyruvate, HEPES, and B27 supplement (Gibco). Culture medium was changed once a week. TTX treatment followed a previous report (Yamaguchi et al., 2003), except that 1 μM was applied for a period of a week. TTX was added by changing pre-mixed fresh medium, and wash-out of TTX was carried out by moving the culture insert to a new culture dish with pre-warmed fresh culture medium, four times, repeatedly.
Bioluminescence imaging
SCN explants were cultured as described above and imaging was carried out as described previously (Buhr et al., 2010; Welsh et al., 2004; Yoo et al., 2004) with modifications. Briefly, an explant culture was placed on an inverted custom-made microscope within an environmentally controlled, light-tight, heated chamber maintained at 36.5°C. The temperature sensor was attached to the sample stage of the microscope, close to the sample, and the sensor readout was maintained at 36.5 ± 0.2°C throughout the experiment. The chamber and microscope were mounted on an anti-vibration table (TMC).
A DV2 (Photometrics, Tucson, AZ) or OptoSplit II (Cairn Research, Kent, England) dual emission image splitter was equipped with Chroma T565lpxr-UF2 to split the short and long wavelengths. A 550 nm shortpass filter and a 625 nm longpass filter (Techspec OD 4, Edmund Optics, Barrington, NJ) were put into the light paths of the short- and long-wavelengths, respectively, to filter the red and green channels. The image splitter was attached to a 20X 0.75 NA Nikon objective or a 20X 1.0 NA water immersion Olympus objective, a zoom-focus tube, and an achromatic tube lens. An 850S cooled CCD camera operating at −90 °C (Spectral Instruments, Tucson, AZ) was mounted with the optics for image capture. In some experiments, an XR/MEGA-10Z dual intensified CCD camera (Stanford Photonics, Palo Alto, CA) was used.
The dual-color bioluminescence split red and green channels were focused onto the right and left halves of the CCD chip. Each exposure/readout cycle was 15 min, and 32-bit images were collected and stored. Image stacks were then analyzed using ImageJ (Rueden et al., 2017; Schindelin et al., 2012) equipped with a Trackmate plugin. The image series were processed and analyzed as follows: The cosmic ray noise was removed by using a top-hat filter with a radius of 2-pixel and threshold of 50. Local drifting of samples was corrected and aligned by rigid transformation alignment using landmark correspondence of identified cells. The filtered and aligned image sequences were then used to measure the bioluminescence intensity of single cells. The bioluminescence value of each single cell, outlined with a region of interest (ROI), was tracked along an image sequence with the Trackmate plugin (Tinevez et al., 2017). The cells in green and red channels were tracked separately and all recorded cells were included for rhythmicity analysis and quantification. The contour of an SCN slice was outlined from an average of all frames and both channels. For video presentation, the 32-bit image sequence was transformed to 16 bits and smoothed with a Kalman filter, and the brightness/contrast optimized. The image sequence was exported to Quicktime (Apple) for conversion to H264 videos.
Immunocytochemistry
Immunofluorescence was performed as described previously (An et al., 2012; Izumo et al., 2014; Lee et al., 2015). Mice were maintained in LD 12:12 and brain tissues collected at ZT16. Animals were anesthetized and euthanized with euthasol (phenytoin/pentobarbital) at 1ml/kg body weight. Intracardial perfusion was performed with 25ml 4% paraformaldehyde in PHEM buffer (pH6.9, 60mM PIPES, 25mM HEPES, 10mM EGTA, and 4mM MgSO4). Brains were removed and post-fixed for 10 hr at 4°C in 4% paraformaldehyde in PHEM buffer. Brains were sectioned into 50μm coronal sections with a vibratome (Leica VT1200S), and slices containing the rostral SCN were collected and processed free-floating. Brain slices were permeabilized with 0.1% Tween 20 for staining VIP and AVP receptors, or 0.3% Triton for staining neuropeptides. 1% BSA and 10% normal goat serum (Abcam, ab7481) were used as blocking agents. The permeabilized slices were then incubated with rabbit polyclonal antibodies to VIP/ Vasoactive Intestinal Peptide (ImmunoStar, 20077, 1:1000), AVP/ Vasopressin (ImmunoStar, 20069, 1:1000), AVPR1A/AVP Receptor V1a (Bioss, bs-11598R,1:500) and VIPR2/ VIP Receptor VPAC2 (Abcam, ab28624,1:1000), overnight. The secondary antibody was Alexa Fluor 488 conjugated goat anti rabbit IgG (Life tech, A-11034, 1:1000). DNA was stained with Hoechst 33258 (Molecular Probes). The sections were mounted on ultra-clean glass slides with antifade mountant (Prolong Glass, ThermoFisher) and imaged with a Leica LSM710 confocal microscope equipped with a 10x objective. Specificities of antibodies were verified with immunofluorescent staining patterns of AVP, VIP, V1a and VPAC2 in comparison with non-specific antibodies and are further supported by their use in other studies: VIP and AVP (Lee et al., 2015), AVP receptor V1a (Albee et al., 2018), and VIP receptor VPAC2 (An et al., 2012). Fluorescent images were analyzed with cellprofiler (Carpenter et al., 2006). Fluorescence intensity of background subtracted images was measured within the entire SCN region and then normalized to the counts of nuclei in the SCN. The percentage of relative abundance to Cre-only and wild-type control was acquired for each target from three replicate experiments.
Quantification and statistical analysis
Phase analysis and Rayleigh plots were generated using Oriana (Kovach Computing Services, Pentraeth, United Kingdom), with a Rayleigh critical value of p=0.05. Phases of Cre or green cells were normalized to the circular mean phase of non-Cre cells at CT0 to normalize variation between SCN slices or between treatments. Circular statistics were performed using Rayleigh test and Watson-Williams F-test in Oriana, with the circular package in R or with the Astropy package in Python. Watson-Wheeler test was performed to analyze the uniformity of phase distribution, between pretreatment and TTX treatment, between TTX and washout, or between genotypes (Wheeler and Watson, 1964). Phase estimates from experiments were normalized (zeroed) to the circular mean, to rule out the difference of mean direction. p-values of Watson-Wheeler test are shown with # (<0.05), ## (<0.01), ### (<0.001) to indicate the significance of the difference in the variance of phase distributions. Additional analyses and plots were generated using Prism 8 (GraphPad). Data points are presented as boxplots or as mean ± SEM unless indicated otherwise. One-way ANOVA was performed to compare experimental conditions with control conditions using Tukey’s multiple comparisons. Two-way ANOVA was performed to compare the main effects of TTX conditions and cell types, along with the interaction effect of the two variables. ANOVA analysis was performed with Prism 8 and R. Tests with p<0.05 were deemed statistically significant, with statistical results presented as: * p<0.05, ** p < 0.01 and *** p<0.001. Nonsignificant differences are not shown in the graphs.
Supplementary Material
Figure S1: Targeting of Per2 gene with a color-switching luciferase cassette.
(A) Diagram of the knock-in construct targeted to the Per2 locus. Southern blotting probes are indicated with 5’Probe and 3’Probe. 5’F and 5’R are the primer pair used in 5’ long fragment PCR. 3’F and 3’R are used in 3’ long fragment PCR. EcoRI and EcoRV sites indicated in the diagram are the restriction sites for Southern blotting. Neo is the neomycin resistance cassette, and DT is the diphtheria toxin cassette on the 5’ arm of the construct. (B) Southern blotting and long-fragment PCR (C) to verify the gene targeting in the ES cell clones. (D) Homozygous and heterozygous germline transduction in mice were tested by Southern blot. (E) Split channels of the dual-color imaging from the VIP-Cre and AVP-Cre SCN slices shown in Figure 1, compared with the merged images.
Figure S2. Analysis pipeline for single-cell rhythmicity.
Raw signals from single-cell trajectories were filtered by removing outliers and then detrended with a Hodrick-Prescott (HP) filter. (A) shows the green HP baseline estimated from the black raw signal and (B) shows the baseline-subtracted data. Detrended data carries high-frequency low-amplitude read noise, and an eigen decomposition of the autocorrelation matrix and an eigenvector-based signal reconstruction are performed to remove the low-amplitude noise, as shown in (C), with X axis as the eigenvalue index and Y axis as the eigenvalue. Denoised data is reconstructed and shown as the red trajectory in (D). To analyze the rhythmicity of detrended and denoised signals, Lomb-Scargle spectral analysis is performed and the resulting LS periodogram is shown in (E), with X axis as the period range and Y axis as the normalized power value. The power peak between 18–30 hr is used to estimate of period of the signal, and a sinusoid curve-fit, with a constraint of ±1 hr of the LS peak period, is performed to estimate the rhythmicity including fitted period, phase, amplitude and damping rate. The fitted cosine trajectory is shown in blue in (F). (G-L) shows the same analysis procedure as in (A-F), where (A-F) is a typical rhythmic cell from the AVP-Cre SCN, while (G-L) shows a noisy cell from the AVP-Bmal1−/− SCN that does not fit to a cosine model.
Figure S3: Single-cell quantification of Cre-only and Bmal1−/− SCN slices.
(A) Rayleigh plots of circular mean of the 4th-day phases from VIP-Cre and AVP-Cre SCN slices during pre-culture (VIP, n=5; AVP, n=6). Each replicate is represented by a point in the overlaid circular scatter plot. Mean phases of green, Cre positive cells are zeroed to red control cells for statistical analysis between cell types. (B) The average green signal of Cre-positive cells across the recording during pre-culture, TTX treatment or washout, was background subtracted and normalized the to average red signal of non-Cre cells in each SCN (AVP-Cre, n=5; AVP-Bmal1−/−, n=4; VIP-Cre, n=3; VIP-Bmal1−/−, n=4). * Significant difference (p<0.05) from one-way ANOVA with multiple comparisons is shown. (C, D) Mean +/− SEM of MIC scores are shown to compare the associations between or inside the two lobes of an SCN. The between lobe associations were categorized as green-green-inter, red-red inter and green-red inter and the within lobe associations were categorized as green-green-intra, red-red intra and green-red intra. Each mean MIC value from a replicate is represented by a point in the overlaid scatter plot (VIP-Cre, n=3; VIP-Bmal1−/−, n=4; AVP-Cre, n=5; AVP-Bmal1−/−, n=4). Nonsignificant differences between the inter- and intra-lobe comparison of each condition are not shown. (E-L) Cell trajectories were classified as rhythmic (p-value of peak <0.05) or arrhythmic (p-value of peak ≥ 0.05) with Lomb-Scargle periodogram between 18–30 hr, as described in Supplemental Methods. (E-F) Mean +/− SEM periods of VIP-Cre (n=3) and VIP-Bmal1−/− SCN slices (n=4). Green represents the VIP-cre cells and red represents non-VIP cells. Lower asterisks indicate significance of t-tests between green and red cells, and upper asterisks indicate significance of Holm-Sidak’s multiple comparisons test within green cells. (G-H) Mean +/− SEM LSP rhythmicities of VIP-Cre and VIP-Bmal1−/− SCN slices. Lower asterisks indicate significance of t-tests between green and red cells, and upper asterisks indicate significance of Holm-Sidak’s multiple comparisons test within green cells. (I-J) Mean +/− SEM periods of AVP-Cre and AVP-Bmal1−/− SCN slices (AVP-Cre n=5; AVP-Bmal1−/− n=4). Green represents the AVP-Cre cells and red represents non-AVP cells. (K-L) Mean +/− SEM LSP rhythmicities of AVP-Cre and AVP-Bmal1−/− SCN slices (* p ≤0.05; ** p ≤ 0.01; *** p ≤ 0.001). Detailed quantification results and ANOVA analyses are included in Supplemental Data 1.
Figure S4: Single-cell rhythmicity and network analysis of SCN explants with genetic excision of Bmal1 in both VIP and AVP cells.
(A) Single-cell trajectories from a VIP/AVP-Bmal1−/− SCN through pretreatment, TTX treatment and washout of TTX. (B) Heat map representations of the single-cells from the same SCN slices. (C-D) LSP rhythmicity values shown by anatomical location in the SCN. The histogram shows LSP Rhythmicity distributions of Bmal1−/− cells versus VIP/AVP-Cre cells. (E) Pairwise MIC score plot of neurons in the VIP/AVP-Bmal1−/− SCN. (F) Correlation network constructed from pair-wise MIC analysis. (G) Mean +/− SEM plots of MIC scores from the VIP/AVP-Bmal1−/− SCN slices (n=4) * p value ≤0.05; ** p value ≤ 0.01; *** p value ≤ 0.001. Detailed quantification results are included in Supplemental Data 1.
Figure S5: Circadian activity rhythms in Cre-only Control, VIP-Bmal1−/−, AVP-Bmal1−/−, and VIP/AVP-Bmal1−/− mice.
Circadian wheel-running activity of mice with light cycle treatments as follows: LD1, animals were maintained in regular LD12:12 cycles for at least two weeks; DD1, animals were released into constant darkness for three weeks; LL, constant light for 2–3 weeks; DD2, constant darkness for three weeks; LD2, LD12:12 light dark cycle for three weeks for re-entrainment; DD3, constant darkness for three weeks. (A - D) Actogram of the circadian behavioral activity of a Cre-only control, a VIP-Bmal1−/−, an AVP-Bmal1−/− and a VIP/AVP-Bmal1−/− mouse. (E) Constant darkness epochs DD1, DD2 and DD3 were used to calculate the period length of circadian rhythms. Scatter plots of the three period values are overlaid with the box plot showing median line, Turkey whiskers and the point of mean (DD2: Cre-only control, 23.73hrs, n=18; VIP-Bmal1−/−, 23.66hrs, n=22; AVP-Bmal1−/−, 24.05hrs, n=15; VIP/AVP-Bmal1−/−, 23.91hrs, n=13). Asterisks indicate the significance of Holm-Sidak’s multiple comparison between DD2, with ANOVA adjusted P value from the four DD2 groups. (F) Average variance in period for each animal, calculated from the six DD periods from G is compared among wildtype, VIP-Bmal1−/−, AVP-Bmal1−/− and VIP/AVP-Bmal1-/−. Detailed quantification results and are included in Table S4. (G) Period lengths calculated from two 10-day sub-intervals in each of the three constant darkness: DD1–1, DD1–2, DD2–1, DD2–2, DD3–1 and DD3–2. Each line connecting six data points are derived from the same animal. * p ≤0.05; ** p ≤ 0.01; *** p ≤ 0.001. Detailed quantification results are included in Supplemental Data 2.
Figure S6: Testing the role of neurotransmission strength and cell types in SCN synchrony.
A mini-SCN model was created and neurotransmission strength (A-B) and cell type ratios (C-D) were tested to determine if these parameters could account for the observed experimental phenotypes. Details are included in methods. (A) Diagram of the mini-SCN model for varying coupling strength. Three cellular populations (AVP, VIP, nonAVP nonVIP) of 20 cells each were connected via mean-field coupling, with the strength of the signals from each population varied.
(B) To test the hypothesis that difference in neurotransmission strength is sufficient to explain the observed phenotypes, we varied the relative strength of coupling between 1:10 and 10:1 AVP:VIP. Resulting whole-SCN mean MIC scores for 100 simulations each of control, VIP-Bmal1−/− (cKO), and AVP-Bmal1−/− (cKO) models are shown. We considered a model to replicate the phenotype if control and VIP-Bmal1−/− showed significantly higher (p<0.05) mean MIC scores than AVP-Bmal1−/−, and showed no significant difference (p>0.05) between each other (two-sided Mann-Whitney U-test with Bonferroni correction for 21 individual tests). This was true in two cases (denoted by asterisks), 5:1 and 10:1 AVP:VIP. Thus, stronger neurotransmission are sufficient to recapitulate the observed experimental phenotypes. (C) Diagram of the mini-SCN model where cell counts of VIP and AVP cells were varied. Neurotransmission strength was fixed at 1:1 AVP:VIP. (D) To test the hypothesis that the experimental phenotype is mediated by relative fractions of AVP and VIP cells, we varied the 40 AVP:VIP cell counts from 4:36 to 36:4 AVP:VIP. As with coupling strength, the phenotype (described above) was recapitulated when AVP cells greatly outnumber VIP cells (9:1). Thus, coupling strength, as mediated by a stronger AVP neurotransmission pathway or an abundance of AVP cells, is sufficient to explain the observed experimental differences between AVP and VIP coupling, and implicates AVP as of importance in SCN synchrony. (E) Distributions of pairwise MIC score of the three SCN genotypes during one simulation of resynchronization. (F) Heat map plots of z-score and Rayleigh plots of phases of single-cell trajectories are shown in the same color modes as in A and B. Each heat map includes 20 example cells of the corresponding cell type. Phases for the Rayleigh plot were estimated at the end of the sixth day.
Supplemental Table 1: Genotypes of mouse lines used in experiments.
Supplemental Table 2: Rhythmicity and synchrony of Bmal1−/− SCN during the decoupling and recoupling processes. Results of Bmal1−/− SCNs are presented in comparison to wildtype controls (one-way ANOVA of mean from multiple slices). VIP/AVP-Bmal1−/− is compared to AVP-Cre controls. The synchrony of red cells is not affected during pretreatment, which is likely to represent an in vivo entrainment by light-dark cycles. MIC quantification during TTX treatment is excluded because the circuit and coupling was turned off with TTX. * Synchrony between red cells with normal Bmal1, in the AVP-Bmal1−/− or VIP/AVP-Bmal1−/− SCN, are not affected during pretreatment, which is likely to represent an in vivo entrainment by light-dark cycles. # Some Bmal1−/− cells in the ventral part of VIP/AVP-Bmal1−/− SCN are not affected during pretreatment, which are possibly the VIP cells driven by the entrained network during pretreatment.
Supplemental Video 1: Dual-color, time-lapse imaging of a VIP-Cre SCN
Supplemental Video 2: Dual-color, time-lapse imaging of an AVP-Cre SCN
Supplemental Data 1: Single-cell rhythmicity, statistical analysis of significance and variance of rhythmicity, and MIC network quantification and statistics of all the SCN explants
Supplemental Data 2: Quantification of circadian activity rhythms.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical Commercial Assays | ||
| Standard polycarbonate mouse cages with running wheels | Actimetrics | http://actimetrics.com/products/clocklab/ running-wheel-cages/ |
| Deposited Data | ||
| PER2::LUC signal and location data of single-cell SCN imaging | This paper | To be done. |
| Raw data files for wheel-running activity | This paper | To be done. |
| Experimental Models: Cell lines | ||
| JM8.F6, ES cell line (Strain: C57BL/6N) | UT Southwestern Medical Center Transgenic Core | |
| SNL 76/7 fibroblast, feeder | UT Southwestern Medical Center Transgenic Core | |
| Experimental Models: Organisms/Strain | ||
| Mouse: C57BL/6N | NIH, NCI-Frederick maintained in lab |
Stock 01C55 |
| Mouse: B6N-Per2::iLuc | This paper | |
| Mouse: B6J-Bmal1fx | (Johnson et al., 2014) | |
| Mouse: Viptm1(cre)Zjh/J, congenic, Vip-Cre | Josh Huang, similar to #031628 | JAX: 031628 |
| Mouse: B6-AVP-iCre | (Pei et al., 2014) | AVP-iCre |
| Mouse: C57BL/6-Tg(Nms-icre)20Ywa/J | The Jackson Laboratory | JAX: 027205 |
| Recombinant DNA | ||
| Per2::iLuc targeting vector | This Paper | |
| Per2::Luc targeting vector | (Yoo et al., 2004) | |
| Puro.Cre empty vector | Addgene | Plasmid #17408 |
| DNA Plasmid | ||
| pGEX-2T-mLIF | (Mereau et al., 1993) | |
| Antibodies | ||
| VIP/ Vasoactive Intestinal Peptide | ImmunoStar | 20077 |
| AVP/ Vasopressin | ImmunoStar | 20069 |
| AVPR1A/AVP Receptor V1a | Bioss | bs-11598R |
| VIPR2/ VIP Receptor VPAC2 | Abcam | ab28624 |
| Alexa Fluor 488 conjugated goat anti rabbit IgG | Life tech | A-11034 |
| Experimental Models: Imaging Hardware | ||
| 20x 0.75 NA air microscope objective | Nikon | CFI Plan APO VC 20x |
| 20x 1.00 NA water microscope objective | Olympus | XLUMPLFLN-W 20x |
| Immersion medium for water objective | Zeiss | Immersol 2010 |
| Achromatic Doublet tube lens | Thorlabs | AC254–080-A |
| Non-rotating zoom barrel | Thorlabs | SM1NR1 |
| Beam-splitter units | Photometrics Cairn Research | DV2 OptoSplit II |
| Dichroic mirror | Chroma | T565lpxr-UF2 |
| 550nm short-pass filter | Edmundoptics | #84–708 |
| 625nm long-pass filter | Edmundoptics | #84–746 |
| Cooled CCD camera | Spectral Instruments | 850S |
| Intensified CCD camera | Stanford Photonics | XR/MEGA-10Z |
| Software and Algorithms | ||
| ClockLab USB Data Collection Program (v.3.15) | Actimetrics, Inc. | http://actimetrics.com/downloads/clocklab/ |
| ClockLab Analysis (v.6.0.34 – standalone) | Actimetrics, Inc. | http://actimetrics.com/downloads/clocklab/ |
| Prism (v.8.3) |
GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| SI Image SGL E | Spectral Instruments, Inc. | http://www.specinst.com |
| Piper Control (2.6) | StanfordPhotonics | http://www.stanfordphotonics.com/ |
| ImageJ2 (Fiji) with trackmate | NIH | https://imagej.net/ImageJ2 |
| Python, with minepy, networkx and astropy | Python | https://www.python.org/ |
| R, with multcomp and circular | R | https://www.r-project.org/ |
| Oriana (v.4.0.1) | Kovach Computing Services | https://www.kovcomp.co.uk/oriana/ |
ACKNOWLEDGMENTS
Research was supported by the Howard Hughes Medical Institute (J.S.T.), NIH/NINDS grant R01 NS106657 (J.S.T.), The Welch Foundation AU-1971–20180324 and R01 GM114424 (S.H.Y.), and NIH awards T32-HLO9701 and F32-AG064886 (J.H.A.). We would like to thank Lisa Thomas, Delali Bassowou, and Chryshanthi Joseph for assistance with animal care and maintenance, and Dr. Shin Yamazaki and Dr. Carla B. Green for helpful discussions. The constructs of click-beetle luciferases were generously donated by Dr. David Piwnica-Worms at MD Anderson Cancer Center. Blastocyst injection was performed at the transgenic core of UT Southwestern directed by Dr. Robert E. Hammer, and JM8.F6 ES cell line was also from the transgenic core. Puro.Cre empty vector was a gift from Tyler Jacks (Addgene plasmid # 17408). pGEX-2T-mLIF plasmid was a gift from Dr. Janet Rossant at University of Toronto. J.S.T. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abel JH, Meeker K, Granados-Fuentes D, St John PC, Wang TJ, Bales BB, Doyle FJ 3rd, Herzog ED, and Petzold LR (2016). Functional network inference of the suprachiasmatic nucleus. Proc Natl Acad Sci U S A 113, 4512–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham U, Granada AE, Westermark PO, Heine M, Kramer A, and Herzel H (2010). Coupling governs entrainment range of circadian clocks. Mol Syst Biol 6, 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamson EE, and Moore RY (2001). Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res 916, 172–191. [DOI] [PubMed] [Google Scholar]
- Albee LJ, LaPorte HM, Gao X, Eby JM, Cheng YH, Nevins AM, Volkman BF, Gaponenko V, and Majetschak M (2018). Identification and functional characterization of arginine vasopressin receptor 1A : atypical chemokine receptor 3 heteromers in vascular smooth muscle. Open Biol 8, 170207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Tsai C, Ronecker J, Bayly A, and Herzog ED (2012). Spatiotemporal distribution of vasoactive intestinal polypeptide receptor 2 in mouse suprachiasmatic nucleus. J Comp Neurol 520, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, and Herzog ED (2005). Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci 8, 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedont JL, and Blackshaw S (2015). Constructing the suprachiasmatic nucleus: a watchmaker’s perspective on the central clockworks. Front Syst Neurosci 9, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Edwards MD, Patton AP, Smyllie NJ, Chesham JE, Maywood ES, and Hastings MH (2019). Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science 363, 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Patton AP, Chesham JE, Maywood ES, and Hastings MH (2017). Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling. Neuron 93, 1420–1435 e1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, and Takahashi JS (2010). Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijink MR, Almog A, Wit CB, Roethler O, Olde Engberink AH, Meijer JH, Garlaschelli D, Rohling JH, and Michel S (2016). Evidence for Weakened Intercellular Coupling in the Mammalian Circadian Clock under Long Photoperiod. PLoS One 11, e0168954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AH, Fung SW, and Cheng HM (2019). Limitations of the Avp-IRES2-Cre (JAX #023530) and Vip-IRES-Cre (JAX #010908) Models for Chronobiological Investigations. J Biol Rhythms 34, 634–644. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Alvarez-Saavedra M, Dziema H, Choi YS, Li A, and Obrietan K (2009). Segregation of expression of mPeriod gene homologs in neurons and glia: possible divergent roles of mPeriod1 and mPeriod2 in the brain. Hum Mol Genet 18, 3110–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, Weaver DR, Leslie FM, and Zhou QY (2002). Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature 417, 405–410. [DOI] [PubMed] [Google Scholar]
- Colwell CS (2011). Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci 12, 553–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, and Waschek JA (2003). Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Regul Integr Comp Physiol 285, R939–949. [DOI] [PubMed] [Google Scholar]
- DeWoskin D, Myung J, Belle MD, Piggins HD, Takumi T, and Forger DB (2015). Distinct roles for GABA across multiple timescales in mammalian circadian timekeeping. Proc Natl Acad Sci U S A 112, E3911–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Ishida A, Miyake A, Sato M, Komatsu R, Yamazaki F, Kimura I, Tsuchiya S, Kori H, Seo K, et al. (2011). Circadian regulation of intracellular G-protein signalling mediates intercellular synchrony and rhythmicity in the suprachiasmatic nucleus. Nat Commun 2, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JA, Leise TL, Castanon-Cervantes O, and Davidson AJ (2013). Dynamic interactions mediated by nonredundant signaling mechanisms couple circadian clock neurons. Neuron 80, 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JA, Leise TL, Castanon-Cervantes O, and Davidson AJ (2015). Neural correlates of individual differences in circadian behaviour. Proc Biol Sci 282, 20150769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Zeng H, Olson DP, Huber KM, Gibson JR, and Takahashi JS (2015). Vasoactive intestinal polypeptide (VIP)-expressing neurons in the suprachiasmatic nucleus provide sparse GABAergic outputs to local neurons with circadian regulation occurring distal to the opening of postsynaptic GABAA ionotropic receptors. J Neurosci 35, 1905–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammon ST, Leevy WM, Gross S, Gokel GW, and Piwnica-Worms D (2006). Spectral unmixing of multicolored bioluminescence emitted from heterogeneous biological sources. Anal Chem 78, 1520–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonze D, Bernard S, Waltermann C, Kramer A, and Herzel H (2005). Spontaneous synchronization of coupled circadian oscillators. Biophys J 89, 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, et al. (2002). The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 109, 497–508. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Maywood ES, and Brancaccio M (2018). Generation of circadian rhythms in the suprachiasmatic nucleus. Nat Rev Neurosci 19, 453–469. [DOI] [PubMed] [Google Scholar]
- Herzog ED (2007). Neurons and networks in daily rhythms. Nat Rev Neurosci 8, 790–802. [DOI] [PubMed] [Google Scholar]
- Honma S (2018). The mammalian circadian system: a hierarchical multi-oscillator structure for generating circadian rhythm. J Physiol Sci 68, 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumo M, Pejchal M, Schook AC, Lange RP, Walisser JA, Sato TR, Wang X, Bradfield CA, and Takahashi JS (2014). Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. eLife 3, e04617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, and Reppert SM (1999). A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96, 57–68. [DOI] [PubMed] [Google Scholar]
- Johnson BP, Walisser JA, Liu Y, Shen AL, McDearmon EL, Moran SM, McIntosh BE, Vollrath AL, Schook AC, Takahashi JS, and Bradfield CA (2014). Hepatocyte circadian clock controls acetaminophen bioactivation through NADPH-cytochrome P450 oxidoreductase. Proc Natl Acad Sci U S A 111, 18757–18762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JR, Simon T, Lones L, and Herzog ED (2018). SCN VIP Neurons Are Essential for Normal Light-Mediated Resetting of the Circadian System. J Neurosci 38, 7986–7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joye DAM, Rohr KE, Keller D, Inda T, Telega A, Pancholi H, Carmona-Alcocer V, and Evans JA (2020). Reduced VIP Expression Affects Circadian Clock Function in VIP-IRES-CRE Mice (JAX 010908). J Biol Rhythms, DOI: 10.1177/0748730420925573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, Zhang EE, Ralph MR, Kay SA, Forger DB, and Takahashi JS (2010). Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol 8, e1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, and Schibler U (2007). System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, Quintero JE, and McMahon DG (2000). GFP fluorescence reports Period 1 circadian gene regulation in the mammalian biological clock. Neuroreport 11, 1479–1482. [PubMed] [Google Scholar]
- Lamia KA, Storch KF, and Weitz CJ (2008). Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A 105, 15172–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IT, Chang AS, Manandhar M, Shan Y, Fan J, Izumo M, Ikeda Y, Motoike T, Dixon S, Seinfeld JE, et al. (2015). Neuromedin S-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 85, 1086–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Zamdborg L, Southey BR, Atkins N Jr., Mitchell JW, Li M, Gillette MU, Kelleher NL, and Sweedler JV (2013). Quantitative peptidomics for discovery of circadian-related peptides from the rat suprachiasmatic nucleus. J Proteome Res 12, 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. (2007). Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low-Zeddies SS, and Takahashi JS (2001). Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell 105, 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, O’Brien JA, and Hastings MH (2011). A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci U S A 108, 14306–14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H, and Hastings MH (2006). Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol 16, 599–605. [DOI] [PubMed] [Google Scholar]
- Mazuski C, Abel JH, Chen SP, Hermanstyne TO, Jones JR, Simon T, Doyle FJ 3rd, and Herzog ED (2018). Entrainment of Circadian Rhythms Depends on Firing Rates and Neuropeptide Release of VIP SCN Neurons. Neuron 99, 555–563 e555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazuski C, Chen SP, and Herzog ED (2020). Different Roles for VIP Neurons in the Neonatal and Adult Suprachiasmatic Nucleus. J Biol Rhythms, DOI: 10.1177/0748730420932073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L, Fan Y, Lv X, Welsh DK, Zhan C, and Zhang EE (2018). Long-term in vivo recording of circadian rhythms in brains of freely moving mice. Proc Natl Acad Sci U S A 115, 4276–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M, Ono D, Hasegawa E, Okamoto H, Honma K, Honma S, and Sakurai T (2015). Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 85, 1103–1116. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carre IA, Strayer CA, Chua NH, and Kay SA (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267, 1161–1163. [DOI] [PubMed] [Google Scholar]
- Miloud T, Henrich C, and Hammerling GJ (2007). Quantitative comparison of click beetle and firefly luciferases for in vivo bioluminescence imaging. J Biomed Opt 12, 054018. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, and Takahashi JS (2011). Cell autonomy and synchrony of suprachiasmatic nucleus circadian oscillators. Trends Neurosci 34, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin LP (2013). Neuroanatomy of the extended circadian rhythm system. Exp Neurol 243, 4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H, Yamazaki S, and McMahon DG (2005). Constant light desynchronizes mammalian clock neurons. Nat Neurosci 8, 267–269. [DOI] [PubMed] [Google Scholar]
- Ono D, Honma S, and Honma K (2016). Differential roles of AVP and VIP signaling in the postnatal changes of neural networks for coherent circadian rhythms in the SCN. Sci Adv 2, e1600960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Sutton AK, Burnett KH, Fuller PM, and Olson DP (2014). AVP neurons in the paraventricular nucleus of the hypothalamus regulate feeding. Mol Metab 3, 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennartz CM, Bos NP, Jeu MT, Geurtsen AM, Mirmiran M, Sluiter AA, and Buijs RM (1998). Membrane properties and morphology of vasopressin neurons in slices of rat suprachiasmatic nucleus. J Neurophysiol 80, 2710–2717. [DOI] [PubMed] [Google Scholar]
- Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, and Skarnes WC (2009). Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nature Methods 6, 493–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, Lander ES, Mitzenmacher M, and Sabeti PC (2011). Detecting novel associations in large data sets. Science 334, 1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, and Eliceiri KW (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper CB (2013). The central circadian timing system. Curr Opin Neurobiol 23, 747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap J, Albus H, VanderLeest HT, Eilers PH, Detari L, and Meijer JH (2003). Heterogeneity of rhythmic suprachiasmatic nucleus neurons: Implications for circadian waveform and photoperiodic encoding. Proc Natl Acad Sci U S A 100, 15994–15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmal C, Herzog ED, and Herzel H (2018). Measuring Relative Coupling Strength in Circadian Systems. J Biol Rhythms 33, 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepka SM, and Takahashi JS (2005). Methods to record circadian rhythm wheel running activity in mice. Methods Enzymol 393, 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, and Eliceiri KW (2017). TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90. [DOI] [PubMed] [Google Scholar]
- Tischkau SA, Mitchell JW, Tyan SH, Buchanan GF, and Gillette MU (2003). Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J Biol Chem 278, 718–723. [DOI] [PubMed] [Google Scholar]
- Tokuda IT, Ono D, Ananthasubramaniam B, Honma S, Honma K, and Herzel H (2015). Coupling Controls the Synchrony of Clock Cells in Development and Knockouts. Biophys J 109, 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy T, Jekic-McMullen D, Sambucetti L, and Rice B (2004). Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models. Mol Imaging 3, 9–23. [DOI] [PubMed] [Google Scholar]
- Tso CF, Simon T, Greenlaw AC, Puri T, Mieda M, and Herzog ED (2017). Astrocytes Regulate Daily Rhythms in the Suprachiasmatic Nucleus and Behavior. Curr Biol 27, 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderLeest HT, Houben T, Michel S, Deboer T, Albus H, Vansteensel MJ, Block GD, and Meijer JH (2007). Seasonal encoding by the circadian pacemaker of the SCN. Curr Biol 17, 468–473. [DOI] [PubMed] [Google Scholar]
- Villalobos V, Naik S, Bruinsma M, Dothager RS, Pan MH, Samrakandi M, Moss B, Elhammali A, and Piwnica-Worms D (2010). Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chem Biol 17, 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viviani VR, Silva Neto AJ, Arnoldi FG, Barbosa JA, and Ohmiya Y (2008). The influence of the loop between residues 223–235 in beetle luciferase bioluminescence spectra: a solvent gate for the active site of pH-sensitive luciferases. Photochem Photobiol 84, 138–144. [DOI] [PubMed] [Google Scholar]
- Welsh DK, and Kay SA (2005). Bioluminescence imaging in living organisms. Curr Opin Biotechnol 16, 73–78. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, and Kay SA (2010). Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol 72, 551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Yoo SH, Liu AC, Takahashi JS, and Kay SA (2004). Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14, 2289–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler S, and Watson GS (1964). A distribution-free two-sample test on a circle. Biometrika 51, 256–257. [Google Scholar]
- Wilsbacher LD, Yamazaki S, Herzog ED, Song EJ, Radcliffe LA, Abe M, Block G, Spitznagel E, Menaker M, and Takahashi JS (2002). Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice invivo. Proc Natl Acad Sci U S A 99, 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, and Okamura H (2003). Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302, 1408–1412. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, Okada K, Chen Y, Fustin JM, Yamazaki F, Mizuguchi N, Zhang J, et al. (2013). Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 342, 85–90. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, and Takahashi JS (2005). Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol 393, 288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. (2004). PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101, 5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Targeting of Per2 gene with a color-switching luciferase cassette.
(A) Diagram of the knock-in construct targeted to the Per2 locus. Southern blotting probes are indicated with 5’Probe and 3’Probe. 5’F and 5’R are the primer pair used in 5’ long fragment PCR. 3’F and 3’R are used in 3’ long fragment PCR. EcoRI and EcoRV sites indicated in the diagram are the restriction sites for Southern blotting. Neo is the neomycin resistance cassette, and DT is the diphtheria toxin cassette on the 5’ arm of the construct. (B) Southern blotting and long-fragment PCR (C) to verify the gene targeting in the ES cell clones. (D) Homozygous and heterozygous germline transduction in mice were tested by Southern blot. (E) Split channels of the dual-color imaging from the VIP-Cre and AVP-Cre SCN slices shown in Figure 1, compared with the merged images.
Figure S2. Analysis pipeline for single-cell rhythmicity.
Raw signals from single-cell trajectories were filtered by removing outliers and then detrended with a Hodrick-Prescott (HP) filter. (A) shows the green HP baseline estimated from the black raw signal and (B) shows the baseline-subtracted data. Detrended data carries high-frequency low-amplitude read noise, and an eigen decomposition of the autocorrelation matrix and an eigenvector-based signal reconstruction are performed to remove the low-amplitude noise, as shown in (C), with X axis as the eigenvalue index and Y axis as the eigenvalue. Denoised data is reconstructed and shown as the red trajectory in (D). To analyze the rhythmicity of detrended and denoised signals, Lomb-Scargle spectral analysis is performed and the resulting LS periodogram is shown in (E), with X axis as the period range and Y axis as the normalized power value. The power peak between 18–30 hr is used to estimate of period of the signal, and a sinusoid curve-fit, with a constraint of ±1 hr of the LS peak period, is performed to estimate the rhythmicity including fitted period, phase, amplitude and damping rate. The fitted cosine trajectory is shown in blue in (F). (G-L) shows the same analysis procedure as in (A-F), where (A-F) is a typical rhythmic cell from the AVP-Cre SCN, while (G-L) shows a noisy cell from the AVP-Bmal1−/− SCN that does not fit to a cosine model.
Figure S3: Single-cell quantification of Cre-only and Bmal1−/− SCN slices.
(A) Rayleigh plots of circular mean of the 4th-day phases from VIP-Cre and AVP-Cre SCN slices during pre-culture (VIP, n=5; AVP, n=6). Each replicate is represented by a point in the overlaid circular scatter plot. Mean phases of green, Cre positive cells are zeroed to red control cells for statistical analysis between cell types. (B) The average green signal of Cre-positive cells across the recording during pre-culture, TTX treatment or washout, was background subtracted and normalized the to average red signal of non-Cre cells in each SCN (AVP-Cre, n=5; AVP-Bmal1−/−, n=4; VIP-Cre, n=3; VIP-Bmal1−/−, n=4). * Significant difference (p<0.05) from one-way ANOVA with multiple comparisons is shown. (C, D) Mean +/− SEM of MIC scores are shown to compare the associations between or inside the two lobes of an SCN. The between lobe associations were categorized as green-green-inter, red-red inter and green-red inter and the within lobe associations were categorized as green-green-intra, red-red intra and green-red intra. Each mean MIC value from a replicate is represented by a point in the overlaid scatter plot (VIP-Cre, n=3; VIP-Bmal1−/−, n=4; AVP-Cre, n=5; AVP-Bmal1−/−, n=4). Nonsignificant differences between the inter- and intra-lobe comparison of each condition are not shown. (E-L) Cell trajectories were classified as rhythmic (p-value of peak <0.05) or arrhythmic (p-value of peak ≥ 0.05) with Lomb-Scargle periodogram between 18–30 hr, as described in Supplemental Methods. (E-F) Mean +/− SEM periods of VIP-Cre (n=3) and VIP-Bmal1−/− SCN slices (n=4). Green represents the VIP-cre cells and red represents non-VIP cells. Lower asterisks indicate significance of t-tests between green and red cells, and upper asterisks indicate significance of Holm-Sidak’s multiple comparisons test within green cells. (G-H) Mean +/− SEM LSP rhythmicities of VIP-Cre and VIP-Bmal1−/− SCN slices. Lower asterisks indicate significance of t-tests between green and red cells, and upper asterisks indicate significance of Holm-Sidak’s multiple comparisons test within green cells. (I-J) Mean +/− SEM periods of AVP-Cre and AVP-Bmal1−/− SCN slices (AVP-Cre n=5; AVP-Bmal1−/− n=4). Green represents the AVP-Cre cells and red represents non-AVP cells. (K-L) Mean +/− SEM LSP rhythmicities of AVP-Cre and AVP-Bmal1−/− SCN slices (* p ≤0.05; ** p ≤ 0.01; *** p ≤ 0.001). Detailed quantification results and ANOVA analyses are included in Supplemental Data 1.
Figure S4: Single-cell rhythmicity and network analysis of SCN explants with genetic excision of Bmal1 in both VIP and AVP cells.
(A) Single-cell trajectories from a VIP/AVP-Bmal1−/− SCN through pretreatment, TTX treatment and washout of TTX. (B) Heat map representations of the single-cells from the same SCN slices. (C-D) LSP rhythmicity values shown by anatomical location in the SCN. The histogram shows LSP Rhythmicity distributions of Bmal1−/− cells versus VIP/AVP-Cre cells. (E) Pairwise MIC score plot of neurons in the VIP/AVP-Bmal1−/− SCN. (F) Correlation network constructed from pair-wise MIC analysis. (G) Mean +/− SEM plots of MIC scores from the VIP/AVP-Bmal1−/− SCN slices (n=4) * p value ≤0.05; ** p value ≤ 0.01; *** p value ≤ 0.001. Detailed quantification results are included in Supplemental Data 1.
Figure S5: Circadian activity rhythms in Cre-only Control, VIP-Bmal1−/−, AVP-Bmal1−/−, and VIP/AVP-Bmal1−/− mice.
Circadian wheel-running activity of mice with light cycle treatments as follows: LD1, animals were maintained in regular LD12:12 cycles for at least two weeks; DD1, animals were released into constant darkness for three weeks; LL, constant light for 2–3 weeks; DD2, constant darkness for three weeks; LD2, LD12:12 light dark cycle for three weeks for re-entrainment; DD3, constant darkness for three weeks. (A - D) Actogram of the circadian behavioral activity of a Cre-only control, a VIP-Bmal1−/−, an AVP-Bmal1−/− and a VIP/AVP-Bmal1−/− mouse. (E) Constant darkness epochs DD1, DD2 and DD3 were used to calculate the period length of circadian rhythms. Scatter plots of the three period values are overlaid with the box plot showing median line, Turkey whiskers and the point of mean (DD2: Cre-only control, 23.73hrs, n=18; VIP-Bmal1−/−, 23.66hrs, n=22; AVP-Bmal1−/−, 24.05hrs, n=15; VIP/AVP-Bmal1−/−, 23.91hrs, n=13). Asterisks indicate the significance of Holm-Sidak’s multiple comparison between DD2, with ANOVA adjusted P value from the four DD2 groups. (F) Average variance in period for each animal, calculated from the six DD periods from G is compared among wildtype, VIP-Bmal1−/−, AVP-Bmal1−/− and VIP/AVP-Bmal1-/−. Detailed quantification results and are included in Table S4. (G) Period lengths calculated from two 10-day sub-intervals in each of the three constant darkness: DD1–1, DD1–2, DD2–1, DD2–2, DD3–1 and DD3–2. Each line connecting six data points are derived from the same animal. * p ≤0.05; ** p ≤ 0.01; *** p ≤ 0.001. Detailed quantification results are included in Supplemental Data 2.
Figure S6: Testing the role of neurotransmission strength and cell types in SCN synchrony.
A mini-SCN model was created and neurotransmission strength (A-B) and cell type ratios (C-D) were tested to determine if these parameters could account for the observed experimental phenotypes. Details are included in methods. (A) Diagram of the mini-SCN model for varying coupling strength. Three cellular populations (AVP, VIP, nonAVP nonVIP) of 20 cells each were connected via mean-field coupling, with the strength of the signals from each population varied.
(B) To test the hypothesis that difference in neurotransmission strength is sufficient to explain the observed phenotypes, we varied the relative strength of coupling between 1:10 and 10:1 AVP:VIP. Resulting whole-SCN mean MIC scores for 100 simulations each of control, VIP-Bmal1−/− (cKO), and AVP-Bmal1−/− (cKO) models are shown. We considered a model to replicate the phenotype if control and VIP-Bmal1−/− showed significantly higher (p<0.05) mean MIC scores than AVP-Bmal1−/−, and showed no significant difference (p>0.05) between each other (two-sided Mann-Whitney U-test with Bonferroni correction for 21 individual tests). This was true in two cases (denoted by asterisks), 5:1 and 10:1 AVP:VIP. Thus, stronger neurotransmission are sufficient to recapitulate the observed experimental phenotypes. (C) Diagram of the mini-SCN model where cell counts of VIP and AVP cells were varied. Neurotransmission strength was fixed at 1:1 AVP:VIP. (D) To test the hypothesis that the experimental phenotype is mediated by relative fractions of AVP and VIP cells, we varied the 40 AVP:VIP cell counts from 4:36 to 36:4 AVP:VIP. As with coupling strength, the phenotype (described above) was recapitulated when AVP cells greatly outnumber VIP cells (9:1). Thus, coupling strength, as mediated by a stronger AVP neurotransmission pathway or an abundance of AVP cells, is sufficient to explain the observed experimental differences between AVP and VIP coupling, and implicates AVP as of importance in SCN synchrony. (E) Distributions of pairwise MIC score of the three SCN genotypes during one simulation of resynchronization. (F) Heat map plots of z-score and Rayleigh plots of phases of single-cell trajectories are shown in the same color modes as in A and B. Each heat map includes 20 example cells of the corresponding cell type. Phases for the Rayleigh plot were estimated at the end of the sixth day.
Supplemental Table 1: Genotypes of mouse lines used in experiments.
Supplemental Table 2: Rhythmicity and synchrony of Bmal1−/− SCN during the decoupling and recoupling processes. Results of Bmal1−/− SCNs are presented in comparison to wildtype controls (one-way ANOVA of mean from multiple slices). VIP/AVP-Bmal1−/− is compared to AVP-Cre controls. The synchrony of red cells is not affected during pretreatment, which is likely to represent an in vivo entrainment by light-dark cycles. MIC quantification during TTX treatment is excluded because the circuit and coupling was turned off with TTX. * Synchrony between red cells with normal Bmal1, in the AVP-Bmal1−/− or VIP/AVP-Bmal1−/− SCN, are not affected during pretreatment, which is likely to represent an in vivo entrainment by light-dark cycles. # Some Bmal1−/− cells in the ventral part of VIP/AVP-Bmal1−/− SCN are not affected during pretreatment, which are possibly the VIP cells driven by the entrained network during pretreatment.
Supplemental Video 1: Dual-color, time-lapse imaging of a VIP-Cre SCN
Supplemental Video 2: Dual-color, time-lapse imaging of an AVP-Cre SCN
Supplemental Data 1: Single-cell rhythmicity, statistical analysis of significance and variance of rhythmicity, and MIC network quantification and statistics of all the SCN explants
Supplemental Data 2: Quantification of circadian activity rhythms.


