Abstract
Thousands of potential endocrine-disrupting chemicals present difficult regulatory challenges. Endocrine-disrupting chemicals can interfere with several nuclear hormone receptors associated with a variety of adverse health effects. The U.S. Environmental Protection Agency (U.S. EPA) has released its reviews of Tier 1 screening assay results for a set of pesticides in the Endocrine Disruptor Screening Program (EDSP), and recently, the Collaborative Estrogen Receptor Activity Prediction Project (CERAPP) data. In this study, the predictive ability of QSAR and docking approaches is evaluated using these data sets. This study also presents a computational systems biology approach using carbaryl (1-naphthyl methylcarbamate) as a case study. For estrogen receptor and androgen receptor binding predictions, two commercial and two open source QSAR tools were used, as was the publicly available docking tool Endocrine Disruptome. For estrogen receptor binding predictions, the ADMET Predictor, VEGA, and OCHEM models (specificity: 0.88, 0.88, and 0.86, and accuracy: 0.81, 0.84, and 0.88, respectively) were each more reliable than the MetaDrug™ model (specificity 0.81 and accuracy 0.77). For androgen receptor binding predictions, the Endocrine Disruptome and ADMET Predictor models (specificity: 0.94 and 0.8, and accuracy: 0.78 and 0.71, respectively) were more reliable than the MetaDrug™ model (specificity 0.33 and accuracy 0.4). A consensus approach is proposed that reaches general agreement among the models (specificity 0.94 and accuracy 0.89). This study integrates QSAR, docking, and systems biology approaches as a virtual screening tool for use in risk assessment. As such, this systems biology pathways and network analysis approach provides a means to more critically assess the potential effects of endocrine-disrupting chemicals.
Keywords: QSAR, Pesticides, Estrogen receptor, Androgen receptor, Developmental toxicity, In silico metabolism, Systems biology
1. Introduction
Endocrine-disrupting chemicals are exogenous compounds that alter the normal function of the endocrine system, potentially causing adverse health effects in humans and wildlife (McKinlay et al., 2008; Mnif et al., 2011). The endocrine-disrupting potential of many pesticides is a major health concern (Klotz et al., 1997; Bjorling-Poulsen et al., 2008; Mnif et al., 2011; Andersen et al., 2015; Ewence et al., 2015). Pesticide residues are found in the environment and in many food items as a result of the widespread use of pesticides. Humans are exposed to pesticides in many ways throughout life, knowingly and unknowingly. They might use pesticides at work, consume pesticide residue on foods they eat, or contact pesticides in air, soil, or water. Several studies have found pesticide residues and their metabolites in human tissues, demonstrating a global distribution (Wohlfahrt-Veje et al., 2011; Roca et al., 2014; Andersen et al., 2015).
Most endocrine-disrupting pesticides mimic estrogen function by acting as a ligand for the estrogen receptor, converting other steroids to active estrogen or increasing the expression of estrogen responsive genes, as shown by some organochlorines, organophosphates, carbamates, and pyrethroids. Antiandrogenic effects also have been reported for organochlorine and carbamate insecticides, and for triazines, a group of herbicides, through inhibition of binding natural ligand to androgen-binding receptors. Endocrine disruption also occurs through competitive inhibition of thyroid hormone receptors by organophosphates and inhibition of progesterone action by pyrethroids (McKinlay et al., 2008). The results of various transactivation assays using mammalian and yeast cells have indicated agonistic or antagonistic activity of pesticides toward aryl hydrocarbon receptors and some members of the nuclear receptor superfamily, including retinoic acid receptors, pregnane X receptors, and peroxisome proliferator-activated receptors.
Structure activity relationship (SAR) and quantitative structure-activity relationship (QSAR) approaches have been used by academia, pharmaceutical, agrochemical, food, and other industries, and by various advisory and regulatory government agencies as decision support tools to address data gaps and provide provisional data in the toxicity database of a chemical. These tools allow for the assessment of chemicals for which no data are available. SAR/QSAR approaches are increasingly being used as core prediction systems in toxicology and have proven to be powerful tools to increase our understanding of the potential harmful effects of chemicals on the environment and human health.
As the number and variety of potentially hazardous chemicals continues to increase, regulatory authorities have approached the challenge by applying these approaches as decision support to chemical risk-based approaches. Furthermore, the ever-increasing economic, social, and political call to reduce animal testing in toxicity evaluation has led to an expansion of the use of these tools. Many different SAR/QSAR models and platforms for predicting a wide range of toxicological endpoints have been documented. Many of these are commercially available (e.g., MultiCASE, DEREK, TOPKAT®, Leadscope), others are open source (e.g., OncoLogic™, ToxCast™, ECOSAR, OASIS, VEGA, CAESAR, TEST, OECD tool box), and some are proprietary in-house systems (e.g., U.S. Food and Drug Administration QSAR models). Although several SAR/QSAR computer-based models have been developed, only some have been routinely used in assessment of chemical toxicity. Their validation, reliability, and use protocols are crucial to further enhance their regulatory acceptance.
Government programs use structure–activity modeling to help protect human populations from exposure to environmental contaminants (Tong et al., 2004; Netzeva et al., 2005; Worth et al., 2007; Arvidson et al., 2010; Ruiz et al., 2011). Computational toxicology methods can identify chemicals that might pose an endocrin–disruption hazard and then prioritize those chemicals for additional in vitro or in vivo testing (Tong et al., 2004; Ruiz et al., 2013; Vuorinen et al., 2013; Zhang et al., 2013). Because of the diversity and complexity of endocrine disruption mechanisms and the limited data available for in silico modeling, most studies have focused on endocrine-disrupting chemicals that act via estrogen or androgen receptors (Tong et al., 2004; Li and Gramatica, 2010b, a; Ruiz et al., 2013; Zhang et al., 2013). These modeling approaches include QSAR modeling (Li and Gramatica, 2010b; Ruiz et al., 2013, 2016; Devillers et al., 2015; Ruiz et al., 2016a), molecular dynamics simulations (Celik et al., 2008; Tsakovska et al., 2011; Chen et al., 2014), and docking (Celik et al., 2008; Vedani et al., 2012; Kolsek et al., 2014; Plosnik et al., 2015). Regulators and industry stakeholders can select from several commercial and freely available computational tools to evaluate the potential toxicity of compounds. Comparative analysis of in silico methods and their interpretation on relevant data sets is of high interest to these stakeholders.
A lack of test data for many of the thousands (>84,000) of chemicals in the Toxic Substance Control Act (TSCA) inventory presents a challenge for regulatory decision making. One of the high-interest toxicity endpoints is endocrine-disrupting activity, which includes interference with several nuclear hormone receptors, such as estrogen, androgen, thyroid, glucocorticoid, and mineralocorticoid receptors that have been associated with cancers, diabetes, obesity, reproductive, or immune problems, and metabolic disorders. Recently, the U.S. Environmental Protection Agency (U.S. EPA) released its reviews of the ESDP Tier 1 screening assay results for a set of pesticides (U.S. EPA, 2015). Even more recently, the Collaborative Estrogen Receptor Activity Prediction Project (CERAPP) provided a training set data of 1677 chemicals derived from ToxCast and Tox21 programs (Mansouri et al., 2016).
The study provides an analysis into the scientific understanding and value of the practical application of these methods for classification and regulatory decision making. This work also aims to stimulate formation of new testable hypotheses to address some of the data gaps previously identified by in the U.S. EPA Endocrine Disruptor Screening Program (EDSP) Tier 1 assessment (U.S. EPA, 2015). Collaborations that focus on alternative methods and in silico studies with endocrine disruptors continue to evolve (e.g., CERAPP) (Mansouri et al., 2016). The data set from CERAPP is also used as a comparative reference with the defined regulatory Tier 1 data set. For estrogen and androgen receptor binding predictions, comparisons among two commercially available QSAR tools (ADMET Predictor™ and MetaDrug™), two publicly available QSAR tools (Virtual models for property Evaluation of chemicals within a Global Architecture [VEGA] and the Online Chemical Modeling Environment [OCHEM]), and an open source docking tool (Endocrine Disruptome) were performed (Roncaglioni et al., 2008; Sushko et al., 2011; Kolsek et al., 2014).
As part of the study, a computational systems biology approach was applied to assess the underlying mechanisms related to endocrine disruption; a case study using a well-study carbamate pesticide, carbaryl, which has been associated with endocrine-disrupting toxicity is presented. This carbaryl case study includes an in silico generation of metabolites and a biological pathway enrichment analysis, and the results of this analysis were compared with some known findings for carbaryl. The purpose of this study is not the development of a new in silico model. Instead, this work evaluates and integrates in silico methods and computational systems biology approaches to assess their usefulness in screening endocrine-disrupting chemicals.
2. Material and methods
2.1. Dataset
U.S. EPA recently released the Tier 1 screening results for 52 chemicals included in its Endocrine Disruptor Screening Program (U.S. EPA, 2015). This screening is used to determine if chemicals have the potential to interact with the three hormonal pathways (estrogen, androgen, and thyroid) in the body’s endocrine system. Eleven assays — five in vitro (cell systems) and six in vivo (live animal) — are used to determine if the chemicals interact with these three hormone pathways, and whether or not they are potential developmental and carcinogenic toxicants. The results of the 52 chemicals studied in the EDSP Tier 1 were used as a reference for androgen binding (Table 1S). The recently available training data set described by CERAPP (Mansouri et al., 2016), which consist of 1677 chemicals, was used for the estrogenicity endpoint.
2.2. QSAR modeling
For this study, the ability of five QSAR modeling packages to estimate the potential of these pesticide chemicals to interact with the estrogen and androgen receptor hormonal pathways was evaluated using ADMET Predictor™ 5.0 (Simulations Plus Inc.), MetaDrug™ with System Toxicology Module, VEGA, OCHEM, and the Endocrine Disruptome docking program.
In silico predictions for determining potential human metabolite profiles were performed using MetaDrug™ and ADMET Predictor. Chemical structures for all compounds in the dataset were drawn in MOL file format using ChemBioDraw Ultra 12.0 (CambridgeSoft), from which they could be imported into the QSAR packages. Qualitative assessment of the in silico predictions of potential metabolite structures was established by comparison with human in vivo metabolic profiles characterized in available biomonitoring studies from the literature.
2.2.1. ADMET Predictor™
ADMET Predictor™ is a state-of-the-art computer program designed to estimate certain absorption, distribution, metabolism, elimination, and toxicity properties of a chemical from its 2- and 3-dimensional molecular structures. The program uses molecular descriptor values as inputs to independent mathematical models (generally, nonlinear machine learning techniques) to generate estimates for each of the ADMET properties. Numerous studies have been gathered in U.S. EPA’s Distributed Structure-Searchable Toxicity (DSSTox) database, which was used to train ADMET Predictor’s models of endocrine receptor binding.
ADMET Predictor™ estimates qualitative and quantitative models of several measures of toxicity. It includes a qualitative assessment of estrogen receptor toxicity in rats (TOX_ER_filter), together with a quantitative measure of estrogen receptor toxicity in rats (TOX_ER [IC50 (estrogen)]) that is applied only for compounds classified as “toxic” by the previous quantitative assessment model.
Two neural network ensemble models are used to assess a compound’s likelihood of binding to the estrogen receptor. The first is a straightforward prediction of whether the molecule will have a detectable affinity for the receptor. A result of “nontoxic” indicates that the compound is unlikely to cause endocrine toxicity by binding to the receptor. A result of “toxic” indicates only that the compound is likely to bind detectably to the receptor. The degree of toxicity is not specified.
The second model predicts the degree of binding for those compounds that are identified by the filter as “toxic.” This model displays the relative binding affinity (%RBA) of a molecule determined by a competitive binding assay. The %RBA is a dimensionless number expressed as the percent ratio: 100% * (50% of the maximum inhibitory concentration (IC50) for 17 β-estradiol/IC50 for the chemical in question). Higher values indicate greater binding affinity and likelihood for endocrine-related toxicity.
Likewise, two neural network ensemble models were built for the androgen receptor: qualitative TOX_AR_Filter (toxic/nontoxic) and quantitative TOX_AR. Unlike the corresponding estrogen receptor models referenced by a natural estrogen, the reference ligand here was 17R-methyl-[3H]-methyltrienolone (R1881), a synthetic substrate that binds more strongly than testosterone. The binary classification of TOX_AR_Filter was based on the LC25 value of R1881 displacement assay. Compounds with LC25 ≤ 10 μmol/L were labeled “toxic” (likely to bind). Those >10 μmol/L were considered “nontoxic.”
The TOX_AR model predicts the degree of binding for those compounds identified by the filter as “toxic.” In this case, IC50 measurements were used from R1881 displacement assays. The % RBA is defined in a way analogous to TOX_ER.
ADMET Predictor tool automatically determines whether a given compound is within the scope (applicability domain) of each of these models. The phrase “within the scope” is defined in terms of molecular descriptor space, not in terms of the relative value of an ADMET property. Let X denote an ADMET model with known training set and let be a descriptor set on which model X is based. For each descriptor, xi(i = 1, …, N), its minimum and maximum values, and, are determined over the training set of X. A new compound C is said to be within the scope of X if the value of each relevant descriptor ci(i = 1, …, N) calculated for C is contained within the corresponding interval 〈, 〉 with tolerance equal to 10% of the interval length. Such a compound has its X value typed in black bold font. Otherwise, the compound in question is outside the scope of X and its X value is marked by magenta font.
2.2.2. MetaDrug™
MetaDrug™ includes more than 70 QSAR models calculated with ChemTree™ (Golden Helix, Bozeman, MT, USA) using a recursive partitioning algorithm. Overall, these models cover the full spectrum of absorption, metabolism, distribution, excretion, and toxicology (ADMET) properties for a given chemical compound. QSAR models are built based on two types of training sets, those taken directly from the literature and those manually annotated from the MetaCore™/MetaDrug™ database. Every molecule in the training set is carefully analyzed for duplicates, stereoisomers, salts, etc.
A QSAR model built with ChemTree™ is passed through internal validation (Myshkin et al., 2012). The coefficient of determination (R2) and root mean squared error are calculated for real vs. predicted values of the training set. Activity predicted from the corresponding QSAR model for the uploaded compound constitutes its QSAR value. The QSAR value has to comply with two QSAR thresholds. One corresponds to the negative logarithm of activity value of the most active compound of the training set defining the predictability limit of the model. The other is the negative common logarithm of 50 μM (−1.7), because less potent compounds are considered to be non-active. If the QSAR value falls within two thresholds it is colored green and the compound is considered as active. Red indicates QSAR values outside the thresholds and non-active compounds. Blue relates to physicochemical properties.
MetaDrug™ estimates qualitative and quantitative models of several measures of toxicity. It includes a qualitative assessment of estrogen and androgen receptor toxicity in mammals. The MetaDrug™ tool for androgen binding (ADR-lig, prob) predicts the potential to bind to androgen receptor, range from 0 to 1. Cutoff is 0.5. Values higher than 0.5 indicate potential androgen receptor ligands. The compounds used in this QSAR model are from Fang et al. (2003).
The MetaDrug™ tool for estrogen binding (ESR-lig, prob) predicts the potential to bind to the estrogen receptor at 100 μM or less, range from 0 to 1. Cutoff is 0.5. Values higher than 0.5 indicate potential estrogen receptor ligands. The 219 compounds used in this QSAR model are based on U.S. EPA DSSTox KIERBL data. To establish the reliability of the QSAR to evaluate the chemical compound, the structure similarity is calculated for the most similar compound in the training set. This is done using the Tanimoto Prioritization score, which is calculated using the Accord Chemistry Cartridge™ (Accelrys, San Diego, CA, USA). Tanimoto prioritization scores are reported on a scale from 0 to 1.
2.2.3. The Online Chemical Modeling Environment (OCHEM)
OCHEM is a web-based platform that aims to automate and simplify the typical steps required for QSAR modeling. The platform consists of two major subsystems: the database of experimental measurements and the modeling framework (Sushko et al., 2011). OCHEM is widely used to perform QSPR/QSAR studies online and share those with other web users. The ultimate goal of OCHEM is to collect all possible chemoinformatics tools within one simple, reliable, and user-friendly resource. OCHEM is free to use and available online at http://www.ochem.eu.
For this work, consensus estrogen-binding class QSAR modeling was applied to the data set. This model was developed and applied under CERAPP. The descriptors were calculated using several program packages, which cover different representations of chemical structures from simple fingerprints and a count of chemical groups. Detailed information about chemicals in the data sets, descriptor calculations, model parameters, and statistics are provided on the OCHEM platform (Sushko et al., 2011).
2.2.4. VEGA-QSAR
VEGA is a freely available platform of QSAR models for regulatory purposes (Roncaglioni et al., 2008). It assesses the chemical space where the model predictions are reliable through the Applicability Domain Index (ADI), with scoring from 0 to 1 (VEGA v. 1.1.1. (http://www.vega-qsar.eu/)). This work uses the relative binding affinity (RBA) QSAR model for the qualitative prediction of RBA for screening estrogen mediated endocrine-disrupting activity. This is a classification model based on a classification and regression tree (CART) algorithm and data set of 806 chemicals from the Japanese Ministry of Economy, Trade, and Industry database, as reported by Roncaglioni et al. (2008).
2.2.5. Endocrine Disruptome
Endocrine Disruptome is a freely available program package for estimating binding activities toward the following14 human nuclear receptors: androgen receptor (AR), estrogen receptors (ER α and ER β), glucocorticoid receptor (GR), liver X receptors (LXR α and LXR β), mineralocorticoid receptor (MR), peroxisome proliferator activated-receptors (PPAR α, PPAR β, and PPAR γ), progesterone receptor (PR), retinoid X receptor (RXR α), and thyroid receptors (TR α and TR β). (Kolsek et al., 2014) provide details of the docking protocol and development of the models, and data sets of compounds. Endocrine Disruptome’s docking simulation, which is an assessment of a ligand’s position within a target receptor, is performed by the Docking interface for Target Systems (DoTS) platform using the AutoDock Vina software package (Vedani et al., 2012; Kolsek et al., 2014).
The 18 proteins (14 agonists and 4 antagonists) in Endocrine Disruptome were carefully selected from a pool of 103 proteins extracted from Protein Data Bank, with consideration being given to the quality of data and docking results on reference compounds. The final results of the predictions are the binding affinities given numerically as binding free energies (in kcal mol−1) and, alternatively, presented as four colored classes at the tool output (Kolsek et al., 2014). For this classification, the program uses three threshold values, defined by calculations of sensitivity (SE) and the validation experiments. The threshold values are SE < 0.25 (red) for high binding affinity, 0.25 < SE < 0.5 (orange) and 0.5 < SE < 0.75 (yellow) for medium binding affinity (orange), and SE > 0.75 (green) for low binding affinity. The absolute values for the thresholds are also reported by (Plosnik et al., 2015). Thus, in this study, for classification of endocrine-disrupting chemicals as potential androgen or estrogen disruptors, if the receptor binding energy for agonists or antagonists is predicted as high or medium high, the chemical is classified as a potential disrupter (active).
2.3. QSAR modeling evaluations
A common way to evaluate the performance of classification models is to use a confusion matrix (Fjodorova et al., 2010). Such a matrix was used to show the number of correct and incorrect predictions made by each classification model compared with the actual (experimental value) in the data. The matrix is N × N, where N is the number of actual values (positive and negative). Performance of such models is commonly evaluated using the data in the matrix. The following illustration shows a 2 × 2 confusion matrix for two classes (positive and negative).
Several statistical parameters were used:
Sensitivity or recall: the proportion of actual positive cases correctly identified
Specificity: the proportion of actual negative cases correctly identified
Accuracy: the proportion of the total number of predictions that were correct
Balanced accuracy: (sensitivity + specificity)/2
Positive predictive value or precision (PPV): the proportion of positive cases correctly identified
Negative predictive value (NPV): the proportion of negative cases correctly identified
Matthews correlation coefficient (MCC): a coefficient of +1 represents a perfect prediction, 0 an average random prediction, and −1 an inverse prediction
2.4. Chemoinformatics and ontology enrichment analysis: carbaryl case study
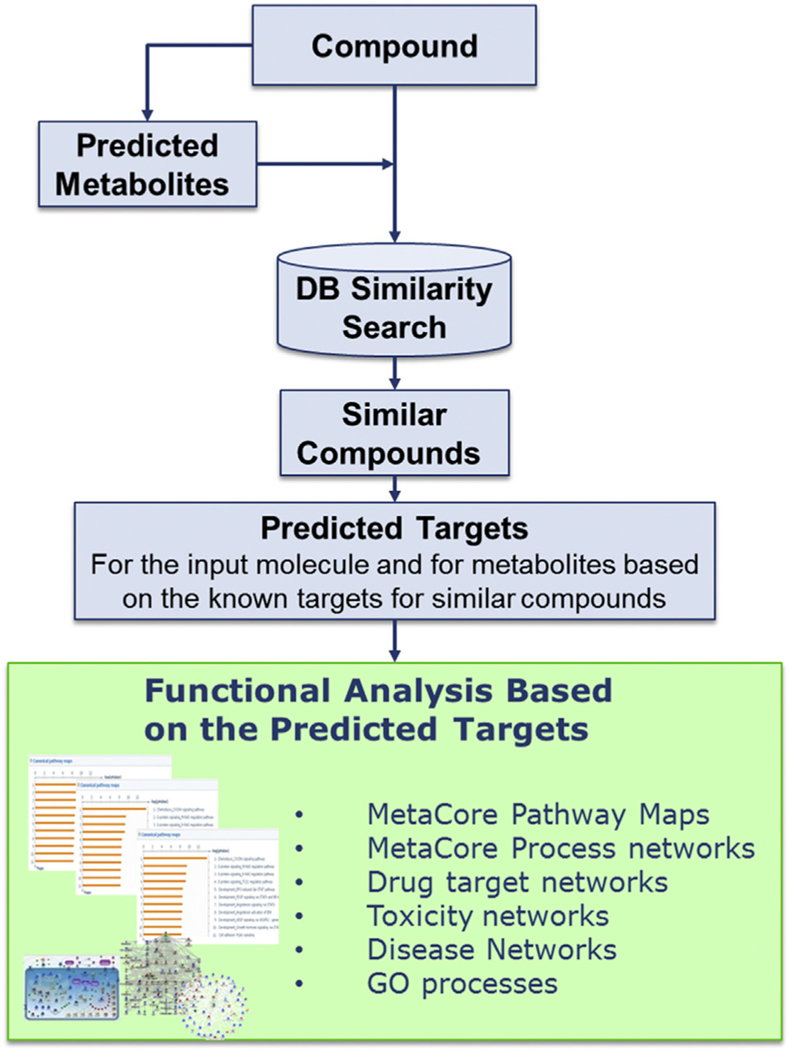
The chemoinformatics tools in MetaDrug™ identify structurally exact or similar compounds annotated in the MetaDrug™ database and report information on proteins with which these exact or similar compounds interact. This allows users to predict biological effects using the workflow scheme shown in Fig.1. As a case study, a chemical similarity search was conducted for carbaryl, a carbamate insecticide used on a variety of crops, in a database of some 700,000 manually annotated compounds linked via physical compound-protein interactions with some 4500 proteins known as targets for at least one small molecule xenobiotic. Accord Chemistry Cartridge™ (Accelrys, San Diego, CA, USA) fragment-based fingerprints were applied to perform a similarity search. A list of compounds was generated using the chosen similarity of exact match or 100% similarity score (Tanimoto coefficient = 1), which will search the information related to this specific chemical to validate the system biology analysis. An exact match analysis was used to mine this database for curated information about carbaryl. In this way, only targets for carbaryl reported in the literature and those predicted from QSAR models would be used for further pathway analysis. From this list, protein targets were retrieved using the collection of protein–target interactions annotated and stored in the MetaDrug™ database.
Fig. 1.
Analysis methods. The workflow for predicting biological effects based on a 2-dimensional chemical structure. Structures of the chemical was uploaded into MetaDrug™. A similarity search was performed based on the structure of input compound (Tanimoto coefficient 1) against the underlying database (MetaBase™) that contains nearly 700,000 annotated compound structures. The known targets of the chemical plus the targets added via the MetaDrug™ similarity search were added to the final target list by the MetaDrug™ software. These target lists were then used for further functional enrichment analysis that showed overrepresented pathway maps, as well as network construction and analysis, which displays annotated connections between targets, other nearby nodes, and the relevant diseases.
A list of potential protein targets can be used for enrichment analysis in Canonical Pathway Maps, one of the proprietary Thomson Reuters ontologies, to identify and rank cellular pathways and processes most influenced by the uploaded chemical. Every map consists of molecular entities (genes, proteins, microRNAs, or compounds) participating in a pathway and linked by well-established interactions. The significance of enrichment was defined by p-values calculated using a hypergeometric distribution (applied Fisher exact test). As a result, the chemical was associated with a quantitatively ranked list of maps summarizing its possibly toxic effects at a systems-biology level.
In the next step, the protein target list for carbaryl was computationally expanded by the nearest neighbors in the process of network building (i.e., by first-step protein–protein interactions) and intersected to identify unique targets for carbaryl compounds. The network is based on valid protein–protein and compound–protein interactions identified through manual curation using full text articles within MetaDrug™. Individual chemicals and proteins are mapped to functional ontologies such as gene-disease associations, biological processes, and mechanisms of toxicity, and later used to reconstruct biological networks linking the component nodes into biologically meaningful clusters.
3. Results and discussion
The performance and predictive ability of in silico methods, such as QSAR and docking, were investigated and compared with the U.S. EPA EDSP Tier 1 androgen assay results and with the CERAPP training set estrogen results.
The U.S. EPA Tier 1 androgen screening assay assessed the potential binding with the androgen receptor for 52 chemicals (Table 1S). Of these, 33 chemicals displayed no evidence of binding, 10 chemicals showed binding, and the remaining 9 chemicals showed an equivocal result. The CERAPP collected concentration-response results from 18 in vitro high-throughput screening (HTS) assays that explore several sites in the mammalian estrogen receptor pathway. (Judson et al., 2015) reported a mathematical model that integrates the in vitro data and calculates an area under the curve score, and proposed an activity consensus score across the estrogen receptor assays. The CERAPP training set data contain 1677 chemicals, of which 1440 were classified as inactive and 237 as active.
High priority issues in risk assessment include prioritizing chemicals for review, filling data gaps, and reducing the use of animals for experimental testing. HTS aids the investigation of thousands of chemical and provides a means to rapidly predict potential chemical toxicities or identify key receptors that regulate specific toxicity pathways (Judson et al., 2015).
In silico methods such as QSAR and docking approaches could support the identification of key characteristics in chemical structures responsible for a toxicity endpoint. These can be used to provide predictions about the possible activity of a chemical in virtual screening settings for regulatory purposes. The quality of in silico model development and assessment of commercial or open source models, based on chemical data sets from HTS experiments, varies depending on the efficiency of the assays, experimental design, data analysis, validation, access to the data, and quantity. However, the accuracy is usually high enough to support prioritizing chemicals identified as worth being subjected to experimental testing.
When using the U.S. EPA EDSP Tier 1 and CERAPP data set, differences were identified between the reported activities for an endpoint and a need for a balanced data set. CERAPP and Judson et al. (2015) reported the development of a classification method that integrates several HTS assays for estrogenic activity. This proposed classification for estrogen-binding activity was used to develop and evaluate QSAR models under CERAPP, led by U.S. EPA (Judson et al., 2015; Mansouri et al., 2016). Use of these data, which covers several different levels of the estrogen signaling pathway, is not equivalent to an individual-specific assay result.
3.1. Model performance evaluations
3.1.1. Androgen receptor and U.S. EPA tier 1 data set
For QSAR androgen receptor binding predictions, the predictive ability of the ADMET Predictor™ models were more reliable than those obtained with MetaDrug™. The Endocrine Disruptome docking results had similar reliability when compared with the ADMET Predictor QSAR model. The Endocrine Disruptome androgen receptor model was able to correctly predict the binding activity for more than 70% of the chemicals, with an overall prediction accuracy of 70% for the androgen receptor binding activity. This model showed good specificity (79%) and a low sensitivity (40%) (Table 1). Two molecules were outside of the applicability domain of the model: abamectin was predicted toxic and fenbutatin oxide was predicted negative.
Table 1.
Models predictions for the androgen binding receptor Tier 1 EDSP data.
| MD | AP | Disruptome | |
|---|---|---|---|
| TP | 4 | 3 | 4 |
| TN | 11 | 29 | 26 |
| FN | 6 | 7 | 6 |
| FP | 22 | 2 | 7 |
| Total Predicted | 43 | 41 | 43 |
| Sn | 0.40 | 0.30 | 0.40 |
| Sp | 0.33 | 0.94 | 0.79 |
| Accuracy | 0.35 | 0.78 | 0.70 |
| Balanced accuracy | 0.37 | 0.62 | 0.60 |
| PPV | 0.15 | 0.60 | 0.36 |
| NPV | 0.65 | 0.81 | 0.81 |
| MCC | −0.23 | 0.31 | 0.18 |
TP:true positives, TN:true negatives, FN:false negatives, FP:false positives, Sn:sensitivity, Sp:specificity, PPV:positive predictive value, NPV:negative predictive value, MCC:Matthews correlation coefficient, MD:MetaDrug, AP:ADMET Predictor.
For the androgen receptor MetaDrug™ model, overall prediction accuracy for binding activity was 35%, with low specificity (33%) and low sensitivity (40%) (Table 1). The applicability domain of the MetaDrug™ QSAR model for this particular data set of chemicals was poor; only 17 of the 52 chemicals showed a favorable Tanimoto score. Thus, it showed poor accuracy, sensitivity, and specificity, which could be because of the dissimilarity between the training (n = 162) and test set data (n = 32) used to develop and evaluate the QSAR model with a data set of pesticides. The model predicted the inactive chemicals better than the active chemicals. The ADMET Predictor docking model was able to correctly predict more than 70% of the chemicals, with an overall prediction accuracy of 78% for androgen receptor binding activity. The ADMET Predictor docking model also showed good specificity (94%), but low sensitivity (30%) (Table 1). The Endocrine Disruptome tool could not perform docking predictions for abamectin and fenbutatin oxide chemicals. The ADMET predictor QSAR model and the Endocrine Disruptome docking tools had a high chemical applicability domain for this data set; most chemicals were within the optimum prediction space, with high reliability of predictions. Both models show very good specificity, accuracy, and negative predictive value, which might be because these models were developed and evaluated on data sets that mainly consist of inactive chemicals.
3.1.2. Estrogen receptor and CERAPP data set
Table 2 shows the results of the four QSAR models predictions (ADMET predictor, MetaDrug™, VEGA, and OCHEM) for the CERAPP estrogen-binding data set. The dataset collects 1677 chemicals, of which 1440 are classified as inactives, and 237 as active. Because these data contain an unbalanced ratio of actives (positive) and inactive (negative) chemicals, balanced accuracy and MCC (which makes use of all the four calculated measures) are best taken into consideration. For balanced accuracy, the higher the value the better (the values range from 0 to 1). MCC ranges from −1 to 1, where −1 indicates perfect negative correlation, 0 predicts random distribution, and 1 predicts perfect correlation. Sensitivity, also called true positive rate, and specificity (true negative rate) show the ratio of the active and inactive chemicals correctly identified by the QSAR models. Positive predictive value and negative predictive value are conditional probabilities. MetaDrug™ and ADMET predictor estrogen receptor QSAR models showed low MCC, compared with VEGA and OCHEM estrogen receptor QSAR models. The balanced accuracy values are higher for VEGA and OCHEM.
Table 2.
Models predictions for the estrogen binding receptor CERAPP data set.
| MD | AP | VEGA | OCHEM | |
|---|---|---|---|---|
| TP | 128 | 103 | 131 | 203 |
| TN | 1170 | 1263 | 1273 | 1228 |
| FN | 109 | 134 | 104 | 32 |
| FP | 270 | 177 | 167 | 200 |
| Total Predicted | 1677 | 1677 | 1675 | 1665 |
| Sn | 0.54 | 0.43 | 0.56 | 0.86 |
| Sp | 0.81 | 0.88 | 0.88 | 0.86 |
| Accuracy | 0.77 | 0.81 | 0.84 | 0.88 |
| Balanced Accuracy | 0.68 | 0.66 | 0.72 | 0.86 |
| PPV | 0.32 | 0.37 | 0.44 | 0.50 |
| NPV | 0.91 | 0.90 | 0.92 | 0.97 |
| MCC | 0.28 | 0.29 | 0.40 | 0.56 |
TP:true positives, TN:true negatives, FN:false negatives, FP:false positives, Sn:sensitivity, Sp:specificity, PPV:positive predictive value, NPV:negative predictive value, MCC:Matthews correlation coefficient, MD:MetaDrug, AP:ADMET Predictor.
The best performance was observed with the OCHEM model, which was expected, because the model was developed using the CERAPP data set. The other models have a good balanced accuracy, but low MCC value compared with OCHEM. This could be because of the size and the distribution of the imbalanced data set used to develop the QSAR models. The models have good random distribution but less than perfect predictions. In general, all these models have high negative predictive value, most likely because they have a low number of active chemicals in their training sets, and positive predictive value is also poor for most of these models. These models provide relatively reliable predictions for inactive chemicals, but many incorrect active chemical predictions.
In terms of sensitivity (Sn), the ADMET predictor shows the lowest value (0.43) and also the highest number of false negatives (FN) (134), compared with the other models. Specificity (Sp) was very good for all models, with values ranging from 0.81 to 0.88. ADMET Predictor and OCHEM had higher chemical applicability domains for this data set compared with VEGA. The Tanimoto prioritization score calculated by the MetaDrug™ model for some chemical predictions was low, which brings into question the applicability of the QSAR results for those compounds.
3.1.3. QSAR consensus approaches
The consensus approach combines the predictions of several QSAR models to maximize their strengths, minimize their weaknesses, and increase their global prediction accuracy. Estrogen receptor and androgen receptor QSAR and docking models were selected. These are based on different molecular descriptors and statistical and modeling techniques. They were merged in two types of consensus approaches. In the first (consensus I), all the model predictions agreed on the predicted class. In the second (consensus II), based on the majority agreement approach, the compound was classified according to the most frequently predicted class.
For androgen binding model predictions, a 2-model or 3-model consensus using both approaches did not increase prediction reliability, especially for the active compounds (Sn ranging from 0.3 to 0.4). When using a 2-model consensus, a combination of the ADMET Predictor or MetaDrug™ model with the docking model, results are strongly influenced by the docking predictions, showing good accuracy and specificity. However, the consensus model using all three models shows higher specificity, accuracy, and MCC (Table 3).
Table 3.
Consensus model results for the androgen binding receptor Tier 1 EDSP data set.
| Consensus II 3-models | Consensus II 2-models combinations | |||
|---|---|---|---|---|
| MD+D | AP+D | MD+AP | ||
| TP | 3 | 2 | 3 | 3 |
| TN | 26 | 12 | 24 | 9 |
| FN | 7 | 8 | 7 | 7 |
| FP | 5 | 19 | 7 | 24 |
| Sn | 0.30 | 0.20 | 0.30 | 0.30 |
| Sp | 0.84 | 0.39 | 0.77 | 0.27 |
| Accuracy | 0.71 | 0.34 | 0.66 | 0.28 |
| Balanced accuracy | 0.57 | 0.30 | 0.54 | 0.29 |
| PPV | 0.38 | 0.10 | 0.30 | 0.11 |
| NPV | 0.79 | 0.60 | 0.77 | 0.56 |
| MCC | 0.15 | −0.35 | 0.07 | −0.37 |
TP:true positives, TN:true negatives, FN:false negatives, FP:false positives, Sn:sensitivity, Sp:specificity, PPV:positive predictive value, NPV:negative predictive value, MCC:Matthews correlation coefficient, MD:MetaDrug, AP:ADMET Predictor, D:Endocrine Disruptome.
For estrogen-binding model predictions, the consensus II approach was used to obtain a 3- and 4-models consensus. A 4-model consensus, (164 chemicals have a tied call [2 and 2]), provided an increase in predictions reliability and a substantial decrease in false positives, with a resultant increase in specificity and accuracy. When using a 3-model consensus, the OCHEM model was excluded because the training set data used to develop the model were the CERAPP data, so the OCHEM model would have strongly influenced the predictions. The 4-model consensus performed better when compared with the 3-model consensus, as shown in Table 4.
Table 4.
Consensus model results for the estrogen binding receptor CERAPP data set.
| Consensus II 4-models | Consensus II 3-models (excluded OCHEM) | |
|---|---|---|
| TP | 108 | 109 |
| TN | 1232 | 1299 |
| FN | 85 | 141 |
| FP | 88 | 128 |
| Total predicted | 1513 | 1677 |
| Sn | 0.55 | 0.46 |
| Sp | 0.94 | 0.90 |
| Accuracy | 0.89 | 0.84 |
| Balanced accuracy | 0.75 | 0.68 |
| PPV | 0.56 | 0.44 |
| NPV | 0.93 | 0.84 |
| MCC | 0.49 | 0.35 |
TP:true positives, TN:true negatives, FN:false negatives, FP:false positives, Sn:sensitivity, Sp:specificity, PPV:positive predictive value, NPV:negative predictive value, MCC:Matthews’s correlation coefficient.
The strengths and weaknesses of each model were considered, knowing that none of them was a perfect model. If the strength and advantage of a model is clearly known, the output of such a model can be used with greater certainty. The applicability domain of the estrogen or androgen binding QSAR models for the pesticide data set could be poor or strong (i.e., the chemical could be outside or inside the optimum prediction space). Estrogen and androgen receptor ADMET predictor models had higher chemical applicability domain for this data set when compared with the MetaDrug™ models. Both commercially available QSAR models were internally and externally validated. However, the ADMET predictor models had superior performance compared with the MetaDrug™ models, which might be because of the size and heterogeneity of the training data sets. A comparative QSAR analysis approach when using multiple models (commercially and open sources) could compensate for the limitations of the individual models that use different descriptors and statistical methods to model different aspects of the toxicological effects. Thus, the use of a consensus model approach could be more beneficial than using individual models. Such a consensus model approach could be used to confirm model predictions for the same endpoint or as a weight of evidence when supporting available data with related endpoints describing various modes of action.
When using multiple models with varying modeling techniques (molecular descriptors, statistical methods, and validation), judging the output from each can be particularly challenging when their performance is comparable but slightly different (Li and Gramatica, 2010a; Novic and Vracko, 2010; Ruiz et al., 2012, 2013; Plosnik et al., 2015). In such cases, conventional wisdom and expert knowledge could be used to examine how the QSAR models follow the development, validation, and application principles described by OECD.
3.2. Chemoinformatics and ontology enrichment analysis: carbaryl case study
In silico predictions for the estrogen-binding potential of carbaryl were negative across the QSAR models, and for androgen receptor binding, positive predictions were in agreement with the experimental data (U.S. EPA, 2010, 2011, 2015; Mansouri et al., 2016).
In this case study, Metadrug™ was used to predict the most likely metabolites of carbaryl, to allow a qualitative prediction for the metabolism of carbaryl, and to compare results with available published information. In silico metabolism approaches could be used as part of the in vitro testing as a means to improve metabolite identification strategies, particularly those mediated by CYP450 family enzymes. These approaches have been used to predict potential profile metabolites from a chemical structure, and to highlight the involvement of enzymatic reactions with the cytochrome P450 superfamily of enzymes.
Prediction of first pass and sequential metabolites, both phase I and phase II reactions, was selected to evaluate metabolite predictions for carbaryl, using MetaDrug™. Prediction of first pass and sequential metabolites, both phase I and phase II reactions, identified 59 possible phase I, and 2 phase II reactions. These include aromatic hydroxylation, N-dealkylation, epoxidation, epoxidation and hydrolysis, N-hydroxylation, N-/O-acetyl transfer reactions, N-/O-glucuronide transfer reactions, N-/O-methyl transfer reactions, N-/O-sulfate transfer reactions.
The prioritization of metabolites in MetaDrug™ is based on a score representing the occurrence rate. Occurrence rate (OC) is the ratio of the occurrence of a particular metabolic reaction to the total number of metabolic reactions in the MetaCore™/MetaDrug™ database. This occurrence frequency is then assigned to predicted metabolites as a negative log value (logOC). The larger the score, the higher the frequency of the metabolic reaction in the database. Metabolites are prioritized into major metabolites and minor metabolites on the basis of the logOC. Ranking the metabolites by the logOC values indicated three possible aromatic hydroxylation reactions, each with logOC = −1.08, as being the most likely to occur (Table 2S), and N-dealkylation was predicted to be only slightly less prevalent (logOC = −1.21). Nevertheless, numerous other metabolites were identified, confirming the diversity and relative ranking of metabolites predicted by MetaDrug™.
Carbaryl metabolite predictions using MetaDrug™ agreed very well with available published data (Tang et al., 2002). Carbaryl can be hydrolyzed by esterases and oxidized by cytochrome P450-mediated monooxygenases (CYP) to form hydrolysis and hydroxylation products, respectively. These are subject to further conjugation, such as sulfate and glucuronic acid conjugates of 1-naphthol and 4-hydroxycarbaryl. The major hydroxylation products include 5-hydroxycarbaryl (5-hydroxy 1-naphthyl N-methylcarbamate), 4-hydroxycarbaryl (4-hydroxy 1-naphthyl N-methylcarbamate), and carbaryl methylol (1-naphthyl N-[hydroxymethyl] carbamate). Although the contributions of hydrolysis and hydroxylation toward total metabolism of carbaryl have yet to be elucidated, hydroxylation by CYP is thought to be the more important route of carbaryl metabolism (Tang et al., 2002).
Assessment of the in silico prediction of potential carbaryl metabolites from a qualitative standpoint can substantially help in analyzing the available metabolism data. Fast and reliable in silico predictions could accelerate the in vitro/in vivo characterization of carbaryl metabolites. In silico tools could be continually used to explore or develop analysis of the potential endocrine disrupting effects of chemicals, the potential interferences with the metabolism of endogenous hormones, and the prediction of metabolism of chemicals by phase I and II enzymes. One of the most frequently cited limitations of in vitro assays without metabolic capacity concerns the qualitative and quantitative deficiencies in the metabolism of test chemicals. The case study presents the use of in silico metabolism prediction as a potential tool to integrate with endocrine-disrupting chemical HTS to be used for screening chemicals before endocrine disrupting chemical testing.
Based on QSAR predictions for carbaryl from the underlying database (MetaBase™), MetaDrug™ software generates a list of known and possible targets. Of these targets, three were cytochromes P450, including CYP1A2, CYP3A4, and CYP2D6, which were identified as possible targets by QSAR models (Tang et al., 2002; Myshkin et al., 2012). The androgen receptor was also identified as a potential target in this search, again supporting the results of the QSAR prediction (U.S. EPA, 2010, 2011, 2015). Another potential target included the fatty acid amide hydrolase (FAAH), with which carbaryl was previously reported to interact (Tarzia et al., 2003). This protein degrades bioactive fatty acid amides, such as oleamide, the endogenous cannabinoid, anandamide, and myristic amide to their corresponding acids, thereby serving to end the signaling functions of these molecules. This suggested that carbaryl has the potential to strongly affect metabolism and clearance.
For this study, the biological functions of the predicted protein targets for carbaryl were investigated in several ways, beginning with enrichment of the target list across three different ontological categories. The hypergeometric distribution p-value of Gene Ontology (GO) biological processes, GeneGo canonical pathway maps, and GeneGo toxicity networks, were calculated with respect to each category. GO processes and toxicity networks both confirmed the initial suggestion of a strong effect. Metabolic process and catabolic process were the two most affected GO processes. Metabolism_CYPs and Fanconi anemia group proteins and protein folding_HDAC, nucleophosmin were the two most affected GO toxicity networks (Fig. 1S).
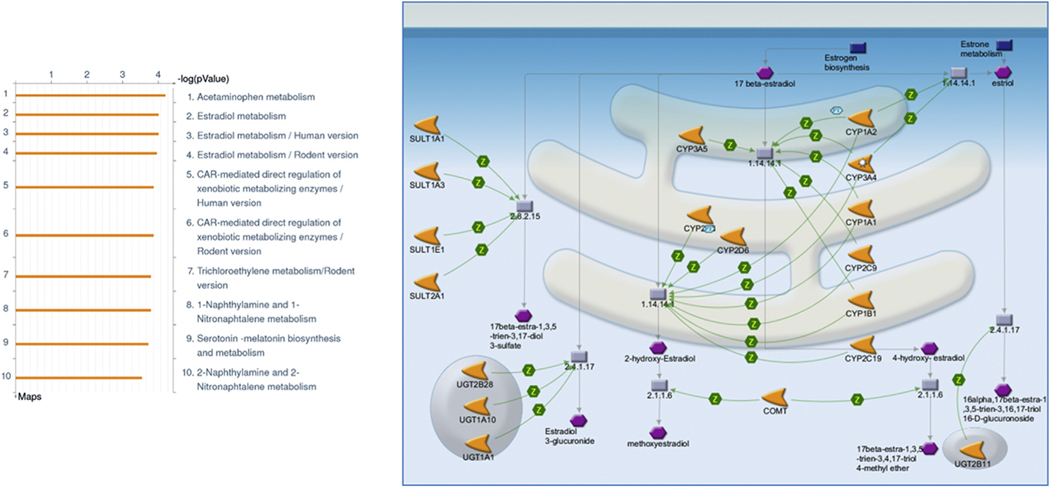
Enrichment in canonical pathway maps gave a slightly different picture, with the highest enrichment in pathways representing metabolism of endogenous hormones, particularly estrogenic and androgenic steroid hormones. Fig. 2 shows the map for estradiol metabolism, the pathway with the highest enrichment in the dataset. Disruption of steroid hormone homeostasis, as is suggested by the enrichment in these pathways for the predicted targets of carbaryl, is a plausible mechanism for the developmental defects associated with exposure to this chemical and its metabolites. Indeed, disruption of steroid hormone homeostasis as a result of altered metabolism after activation of nuclear receptors such as pregnane X receptor (PXR), androgen receptor, estrogen receptor, and aryl hydrocarbon receptor (AhR) has recently been hypothesized as a possible mechanism of the observed developmental and reproductive effects.
Fig. 2.
Representation of canonical pathway map of predicted targets of carbaryl and the estradiol metabolism canonical map at the bottom. Blue hexagon shows the CYPs regulated targets. Green arrows = activating interactions. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
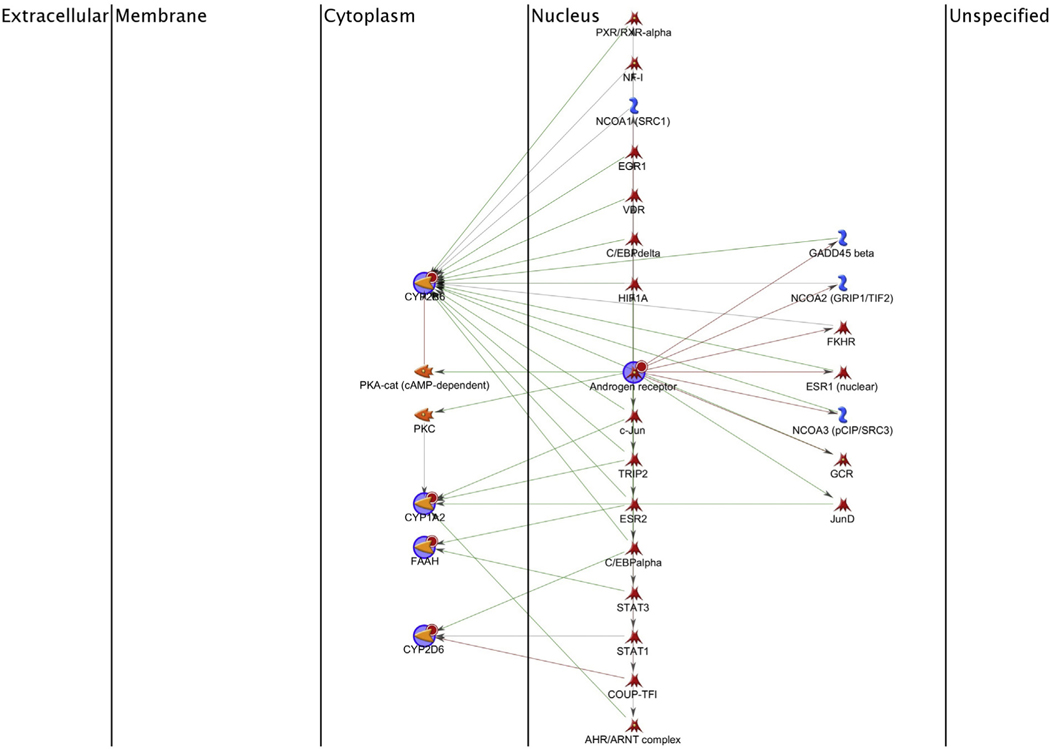
This list of potential protein targets was used to build a biological network to further study pathway interactions and potential downstream effects. The carbaryl network was further refined using the shortest path algorithm network option on MetaDrug™. A Dijkstra’s shortest paths algorithm calculates the shortest directed paths between selected objects. Thus, a complete network of interactions was generated for carbaryl targets (Fig. 3). The advanced search function, a Java application tool within the MetaDrug™ software, was also used to look for combined information. The advanced search made it possible to create Boolean queries and retrieve the results. A detailed methodological description of the systems biology procedures and protocols for using the software are available at http://lsresearch.thomsonreuters.com/. The advanced search tool was used to find all transcription targets of the androgen receptor that are part of the GO process “reproductive/development structure” and its sub-folders then used this list to build a network representation (Fig. 2S).
Fig. 3.
Proposed shortest paths network for potential genes associated to carbaryl. Green arrows = activating interactions; red arrows = inhibiting interactions. Large blue circles represent the genes that are the seed nodes the shortest paths algorithm was required to go through. Small red circles next to the genes indicate the targets of carbaryl. Catalytic factors = yellow; transcription factors = red; cytokines and lipoproteins = green; receptors and adaptor proteins = blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Computational systems biology was applied to carbaryl as a case study, for which few experimental developmental effects have been reported. Carbaryl is a carbamate pesticide still in use today. Animal studies on young and adult rodents have been the basis for risk assessment. Those studies have indicated that neonate animals are more sensitive to carbamates than are adult animals. The goal of the case study was to understand carbaryl’s potential toxicity pathways and biological network interactions related to endocrine disruption. The results were compared with published findings (Klotz et al.,1997; Bjorling-Poulsen et al., 2008; U.S. EPA, 2010, 2011, 2015; Mansouri et al., 2016).
In toxicology, computational systems biology facilitates the identification of important pathways and molecules from large data sets. These tasks can be extremely laborious when performed through a classical literature search. Computational systems biology offers more advantages than just providing a high-throughput literature search engine. These tools might provide the basis for establishing hypotheses on potential links between environmental chemicals and human diseases. Comprehensive databases containing information describing networks of human protein–protein interactions and protein–disease associations make this possible. Experimentally determined targets of the specific chemical of interest can be fed into these networks to obtain additional information that can be used to establish hypothetical links between the chemical and human diseases. Such information can also be applied for designing animal and cell-based laboratory experiments that can test the established hypotheses more intelligently (Ruiz et al., 2016b).
By using the in silico metabolite prediction and the QSAR tools in MetaDrug™ for this study, modes of toxic action were inferred and a major mechanism of toxicity of carbaryl was identified. QSAR models and targets of structurally similar compounds in the software database, when analyzed using the systems biology molecular interaction data and network analysis tools in the product, pointed toward profound effects on metabolic enzymes, including cytochromes P450, conjugating phase II metabolic enzymes, and xenobiotic transporters. Pathway and network analysis suggested that the androgen receptor was a key regulator of these processes and, through its activation, carbaryl has the potential to affect xenobiotic metabolism and homeostasis of endogenous steroid hormones.
The pathway and network for carbaryl presents a novel computational systems biology and in silico models of different molecular mechanisms of endocrine disruption action that point to potential disease outcomes. Future experimental evaluation of this model might lead to the development of new predictive markers of endocrine-disrupting chemical effects that could translate into new disease prevention and clinical use strategies. A major objective of the present study is to stimulate experimental laboratory research by identifying good biological pathways and chemical candidates for investigation. Specific avenues of laboratory research might include in vitro and in vivo studies that should be conducted using exposure to selected endocrine-disrupting chemicals on an individual or mixture basis using a factorial design approach. Specific receptors or pathway nodes of interest identified using these combined in silico and computational systems biology model approaches could be technically evaluated by application of genomic, proteomic, or metabolomic methods.
4. Conclusions
This study integrated QSAR, docking, and computational system biology tools to develop a novel approach for evaluation of the estrogen and androgen binding activity of pesticides. The predictions of two commercially available (ADMET Predictor and MetaDrug™) and two open source QSAR tools (VEGA and OCHEM) were evaluated. The study used the publicly available Endocrine Disruptome docking tool using the U.S. EPA Tier 1 screening assay results for a set of pesticides in the Endocrine Disruptor screening program, and CERAPP data set. For estrogen receptor binding predictions, the predictive ability of the ADMET Predictor, VEGA, and OCHEM models (specificity: 0.88, 0.88, and 0.86, and accuracy: 0.81, 0.84, and 0.88, respectively) were more reliable than those obtained by MetaDrug™ (specificity 0.81 and accuracy 0.77). In general, all models have high negative predictive value, most likely because they have a low number of active chemicals in their training sets; thus, the positive predictive value is also poor for most of the models. The models have shown more accurate predictions for inactive chemicals than for active chemicals. On the other hand, for androgen receptor binding predictions, the predictive ability of the Endocrine Disruptome and ADMET Predictor models (specificity: 0.94 and 0.8, and accuracy: 0.78 and 0.71, respectively) were more reliable to those obtained by MetaDrug™ (specificity 0.33 and accuracy 0.4).
It is proposed that the consensus approach for the estrogen receptor prediction reduces the limitations related to an individual in silico prediction by reaching a general agreement among a collection of models (specificity 0.94 and accuracy 0.89). This systems biology pathways and network analysis shows insightful potential effects on metabolic enzymes (CYP450), androgen receptors, and xenobiotic transporters. The results provided here show that application or use of these tools as an integrated approach could support chemical safety assessment and guide further experimental testing. It could also be extended to evaluate other environmental chemicals classes because these tools allow screening of large libraries of molecules for potential endocrine-disrupting activity. This approach could also help circumvent the resource-intensive laboratory work currently used to evaluate the rapidly increasing number of chemicals detected in humans and the environment.
Key molecular networks, toxicity pathways, potential modes of action, and potential biomarkers of diseases can potentially be identified early during the process of chemical safety assessment. In silico tools such as QSAR and docking can be used for hazard identification and prioritization of chemicals for further experimental testing (Jensen et al., 2008; Mays et al., 2012; Nendza et al., 2013; Rybacka et al., 2015). Later, when empirical computational systems biology data have been generated for a chemical, (e.g., omics data, HTS, in vitro assays) or when specific toxicogenomic, metabolic, or toxicologic pathways are known, all these data can be analyzed and integrated. Using virtual screening to generate new hypotheses will expand and improve the comprehensive systems evaluation of a chemical’s effects on biological systems.
Supplementary Material
HIGHLIGHTS.
An integrative virtual screening tool for risk assessment of EDCs is proposed.
Contemporary estrogen and androgen models show agreement with experimental data.
QSAR consensus modeling presented and evaluated for transparency and reliability.
Approach applied to Carbaryl show usefulness in screening EDCs and further testing.
Acknowledgment
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry. Mention of trade names is not an endorsement of any commercial product. We thank Kamel Mansouri, U.S. EPA for assistance with the CERAPP data, providing insight and expertise that greatly assisted this project. We are also grateful to Michael Lawless for his comments on an earlier version of the manuscript. We appreciate Donald Meadows for his editorial technical support.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.chemosphere.2017.03.026.
References
- Andersen HR, Debes F, Wohlfahrt-Veje C, Murata K, Grandjean P, 2015. Occupational pesticide exposure in early pregnancy associated with sex-specific neurobehavioral deficits in the children at school age. Neurotoxicol. Teratol. 47, 1–9. [DOI] [PubMed] [Google Scholar]
- Arvidson KB, Chanderbhan R, Muldoon-Jacobs K, Mayer J, Ogungbesan A, 2010. Regulatory use of computational toxicology tools and databases at the United States food and Drug Administration’s office of food additive safety. Expert Opin. Drug. Met. 6, 793–796. [DOI] [PubMed] [Google Scholar]
- Bjorling-Poulsen M, Andersen HR, Grandjean P, 2008. Potential developmental neurotoxicity of pesticides used in Europe. Environ. Health 7, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik L, Davey J, Lund D, Schiott B, 2008. Exploring interactions of endocrine-disrupting compounds with different conformations of the human estrogen receptor alpha ligand binding domain: a molecular docking study. Chem. Res. Toxicol. 21, 2195–2206. [DOI] [PubMed] [Google Scholar]
- Chen Y, Cheng F, Sun L, Li W, Liu G, Tang Y, 2014. Computational models to predict endocrine-disrupting chemical binding with androgen or oestrogen receptors. Ecotox. Environ. Safe 110, 280–287. [DOI] [PubMed] [Google Scholar]
- Devillers J, Bro E, Millot F, 2015. Prediction of the endocrine disruption profile of pesticides. Sar. QSAR Environ. Res. 26, 831–852. [DOI] [PubMed] [Google Scholar]
- Ewence A, Brescia S, Johnson I, Rumsby PC, 2015. An approach to the identification and regulation of endocrine disrupting pesticides. Food Chem. Toxicol. 78, 214–220. [DOI] [PubMed] [Google Scholar]
- Fang H, Tong W, Branham WS, Moland CL, Dial SL, Hong H, Xie Q, Perkins R, Owens W, Sheehan DM, 2003. Study of 202 natural, synthetic, and environmental chemicals for binding to the androgen receptor. Chem. Res. Toxicol. 16, 1338–1358. [DOI] [PubMed] [Google Scholar]
- Fjodorova N, Vracko M, Novic M, Roncaglioni A, Benfenati E, 2010. New public QSAR model for carcinogenicity. Chem. Cent. J. 4 (Suppl. 1), S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen GE, Niemelä JR, Wedebye EB, Nikolov NG, 2008. QSAR models for reproductive toxicity and endocrine disruption in regulatory use – a preliminary investigation. Sar. QSAR Environ. Res. 19, 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Magpantay FM, Chickarmane V, Haskell C, Tania N, Taylor J, Xia M, Huang R, Rotroff DM, Filer DL, Houck KA, Martin MT, Sipes N, Richard AM, Mansouri K, Setzer RW, Knudsen TB, Crofton KM, Thomas RS, 2015. Integrated model of chemical perturbations of a biological pathway using 18 in vitro high-throughput screening assays for the estrogen receptor. Toxicol. Sci. 148, 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz DM, Arnold SF, McLachlan JA, 1997. Inhibition of 17 beta-estradiol and progesterone activity in human breast and endometrial cancer cells by carbamate insecticides. Life Sci. 60, 1467–1475. [DOI] [PubMed] [Google Scholar]
- Kolsek K, Mavri J, Sollner Dolenc M, Gobec S, Turk S, 2014. Endocrine disruptome–an open source prediction tool for assessing endocrine disruption potential through nuclear receptor binding. J. Chem. Inf. Model. 54, 1254–1267. [DOI] [PubMed] [Google Scholar]
- Li J, Gramatica P, 2010a. Classification and virtual screening of androgen receptor antagonists. J. Chem. Inf. Model. 50, 861–874. [DOI] [PubMed] [Google Scholar]
- Li J, Gramatica P, 2010b. QSAR classification of estrogen receptor binders and prescreening of potential pleiotropic EDCs. Sar. QSAR Environ. Res. 21, 657–669. [DOI] [PubMed] [Google Scholar]
- Mansouri K, Abdelaziz A, Rybacka A, Roncaglioni A, Tropsha A, Varnek A, Zakharov A, Worth A, Richard AM, Grulke CM, Trisciuzzi D, Fourches D, Horvath D, Benfenati E, Muratov E, Wedebye EB, Grisoni F, Mangiatordi GF, Incisivo GM, Hong H, Ng HW, Tetko IV, Balabin I, Kancherla J, Shen J, Burton J, Nicklaus M, Cassotti M, Nikolov NG, Nicolotti O, Andersson PL, Zang Q, Politi R, Beger RD, Todeschini R, Huang R, Farag S, Rosenberg SA, Slavov S, Hu X, Judson RS, 2016. CERAPP: collaborative estrogen receptor activity prediction Project. Environ. Health Persp 124, 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays C, Benfenati E, Pardoe S, 2012. Use and perceived benefits and barriers of QSAR models for REACH: findings from a questionnaire to stakeholders. Chem. Cent. J. 6, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinlay R, Plant JA, Bell JN, Voulvoulis N, 2008. Endocrine disrupting pesticides: implications for risk assessment. Environ. Intern 34, 168–183. [DOI] [PubMed] [Google Scholar]
- Mnif W, Hassine AI, Bouaziz A, Bartegi A, Thomas O, Roig B, 2011. Effect of endocrine disruptor pesticides: a review. Int. J. Env. Res. Pub. He 8, 2265–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myshkin E, Brennan R, Khasanova T, Sitnik T, Serebriyskaya T, Litvinova E, Guryanov A, Nikolsky Y, Nikolskaya T, Bureeva S, 2012. Prediction of organ toxicity endpoints by QSAR modeling based on precise chemical-histopathology annotations. Chem. Biol. Drug Des. 80, 406–416. [DOI] [PubMed] [Google Scholar]
- Nendza M, Gabbert S, Kühne R, Lombardo A, Roncaglioni A, Benfenati E, Benigni R, Bossa C, Strempel S, Scheringer M, Fernández A, Rallo R, Giralt F, Dimitrov S, Mekenyan O, Bringezu F, Schüürmann G, 2013. A comparative survey of chemistry-driven in silico methods to identify hazardous substances under REACH. Regul. Toxicol. Pharm. 66, 301–314. [DOI] [PubMed] [Google Scholar]
- Netzeva TI, Worth A, Aldenberg T, Benigni R, Cronin MT, Gramatica P, Jaworska JS, Kahn S, Klopman G, Marchant CA, Myatt G, Nikolova-Jeliazkova N, Patlewicz GY, Perkins R, Roberts D, Schultz T, Stanton DW, van de Sandt JJ, Tong W, Veith G, Yang C, 2005. Current status of methods for defining the applicability domain of (quantitative) structure-activity relationships. The report and recommendations of ECVAM Workshop 52. ATLA 33, 155–173. [DOI] [PubMed] [Google Scholar]
- Novic M, Vracko M, 2010. QSAR models for reproductive toxicity and endocrine disruption activity. Mol. (Basel, Switzerland) 15, 1987–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plosnik A, Vracko M, Mavri J, 2015. Computational study of binding affinity to nuclear receptors for some cosmetic ingredients. Chemosphere 135, 325–334. [DOI] [PubMed] [Google Scholar]
- Roca M, Miralles-Marco A, Ferre J, Perez R, Yusa V, 2014. Biomonitoring exposure assessment to contemporary pesticides in a school children population of Spain. Environ. Res. 131, 77–85. [DOI] [PubMed] [Google Scholar]
- Roncaglioni A, Piclin N, Pintore M, Benfenati E, 2008. Binary classification models for endocrine disrupter effects mediated through the estrogen receptor. Sar. QSAR Environ. Res. 19, 697–733. [DOI] [PubMed] [Google Scholar]
- Ruiz P, Mumtaz M, Gombar V, 2011. Assessing the toxic effects of ethylene glycol ethers using Quantitative Structure Toxicity Relationship models. Toxicol. Appl. Pharm. 254, 198–205. [DOI] [PubMed] [Google Scholar]
- Ruiz P, Begluitti G, Tincher T, Wheeler J, Mumtaz M, 2012. Prediction of acute mammalian toxicity using QSAR methods: a case study of sulfur mustard and its breakdown products. Mol. (Basel, Switzerland) 17, 8982–9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz P, Myshkin E, Quigley P, Faroon O, Wheeler JS, Mumtaz MM, Brennan RJ, 2013. Assessment of hydroxylated metabolites of polychlorinated biphenyls as potential xenoestrogens: a QSAR comparative analysis. Sar. QSAR Environ. Res. 24, 393–416. [DOI] [PubMed] [Google Scholar]
- Ruiz P, Ingale K, Wheeler JS, Mumtaz M, 2016a. 3D QSAR studies of hydroxylated polychlorinated biphenyls as potential xenoestrogens. Chemosphere 144, 2238–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz P, Perlina A, Mumtaz M, Fowler BA, 2016b. A systems biology approach reveals converging molecular mechanisms that link different POPs to common metabolic diseases. Environ. Health Persp 124, 1034–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybacka A, Ruden C, Tetko IV, Andersson PL, 2015. Identifying potential endocrine disruptors among industrial chemicals and their metabolites – development and evaluation of in silico tools. Chemosphere 139, 372–378. [DOI] [PubMed] [Google Scholar]
- Sushko I, Novotarskyi S, Körner R, Pandey AK, Rupp M, Teetz W, Brandmaier S, Abdelaziz A, Prokopenko VV, Tanchuk VY, Todeschini R, Varnek A, Marcou G, Ertl P, Potemkin V, Grishina M, Gasteiger J, Schwab C, Baskin II, Palyulin VA, Radchenko EV, Welsh WJ, Kholodovych V, Chekmarev D, Cherkasov A, Aires-de-Sousa J, Zhang Q-Y, Bender A, Nigsch F, Patiny L, Williams A, Tkachenko V, Tetko IV, 2011. Online chemical modeling environment (OCHEM): web platform for data storage, model development and publishing of chemical information. J. Comp. Aid. Mol. Des. 25, 533–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cao Y, Rose RL, Hodgson E, 2002. In vitro metabolism of carbaryl by human cytochrome P450 and its inhibition by chlorpyrifos. Chem. Biol. Interact. 141, 229–241. [DOI] [PubMed] [Google Scholar]
- Tarzia G, Duranti A, Tontini A, Piersanti G, Mor M, Rivara S, Plazzi PV, Park C, Kathuria S, Piomelli D, 2003. Design, synthesis, and Structure–Activity relationships of alkylcarbamic acid aryl esters, a new class of fatty acid amide hydrolase inhibitors. J. Med. Chem. 46, 2352–2360. [DOI] [PubMed] [Google Scholar]
- Tong W, Xie Q, Hong H, Shi L, Fang H, Perkins R, 2004. Assessment of prediction confidence and domain extrapolation of two structure-activity relationship models for predicting estrogen receptor binding activity. Environ. Health Persp 112, 1249–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakovska I, Pajeva I, Alov P, Worth A, 2011. Recent advances in the molecular modeling of estrogen receptor-mediated toxicity. Adv. Protein Chem. Str. 85, 217–251. [DOI] [PubMed] [Google Scholar]
- U.S. EPA, 2010. Carbaryl: Report of the Endocrine Disruptor Review Team. Test Order #: EDSP-056801–57. US Environmental Protection Agency, Washington, DC. https://www.epa.gov/endocrine-disruption/endocrine-disruptor-screening-program-tier-1-screening-determinations-and (Accessed May 2016). [Google Scholar]
- U.S. EPA, 2011. Endocrine Disruptor Screening Program – Weight of Evidence: Evaluating Results of EDSP Tier 1 Screening to Identify the Need for Tier 2 Testing. US Environmental Protection Agency. [Google Scholar]
- U.S. EPA, 2015. US Environmental Protection Agency. Endocrine Disruptor Screening Program Tier 1 Assessments. https://www.epa.gov/ingredients-used-pesticide-products/endocrine-disruptor-screening-program-tier-1-assessments (Accessed May 2016). [Google Scholar]
- Vedani A, Dobler M, Smiesko M, 2012. VirtualToxLab - a platform for estimating the toxic potential of drugs, chemicals and natural products. Toxicol. Appl. Pharm. 261, 142–153. [DOI] [PubMed] [Google Scholar]
- Vuorinen A, Odermatt A, Schuster D, 2013. In silico methods in the discovery of endocrine disrupting chemicals. J. Steroid Biochem. 137, 18–26. [DOI] [PubMed] [Google Scholar]
- Wohlfahrt-Veje C, Main KM, Schmidt IM, Boas M, Jensen TK, Grandjean P, Skakkebaek NE, Andersen HR, 2011. Lower birth weight and increased body fat at school age in children prenatally exposed to modern pesticides: a prospective study. Environ. Health 10, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth AP, Bassan A, De Bruijn J, Gallegos Saliner A, Netzeva T, Patlewicz G, Pavan M, Tsakovska I, Eisenreich S, 2007. The role of the European Chemicals Bureau in promoting the regulatory use of (Q)SAR methods. Sar. QSAR Environ. Res. 18, 111–125. [DOI] [PubMed] [Google Scholar]
- Zhang L, Sedykh A, Tripathi A, Zhu H, Afantitis A, Mouchlis VD, Melagraki G, Rusyn I, Tropsha A, 2013. Identification of putative estrogen receptor-mediated endocrine disrupting chemicals using QSAR- and structure-based virtual screening approaches. Toxicol. Appl. Pharm. 272, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.