Abstract
Exon skipping using antisense oligonucleotides (ASOs) has recently proven to be a powerful tool for mRNA splicing modulation. Several exon-skipping ASOs have been approved to treat genetic diseases worldwide. However, a significant challenge is the difficulty in selecting an optimal sequence for exon skipping. The efficacy of ASOs is often unpredictable, because of the numerous factors involved in exon skipping. To address this gap, we have developed a computational method using machine-learning algorithms that factors in many parameters as well as experimental data to design highly effective ASOs for exon skipping. eSkip-Finder (https://eskip-finder.org) is the first web-based resource for helping researchers identify effective exon skipping ASOs. eSkip-Finder features two sections: (i) a predictor of the exon skipping efficacy of novel ASOs and (ii) a database of exon skipping ASOs. The predictor facilitates rapid analysis of a given set of exon/intron sequences and ASO lengths to identify effective ASOs for exon skipping based on a machine learning model trained by experimental data. We confirmed that predictions correlated well with in vitro skipping efficacy of sequences that were not included in the training data. The database enables users to search for ASOs using queries such as gene name, species, and exon number.
Graphical Abstract
Graphical Abstract.
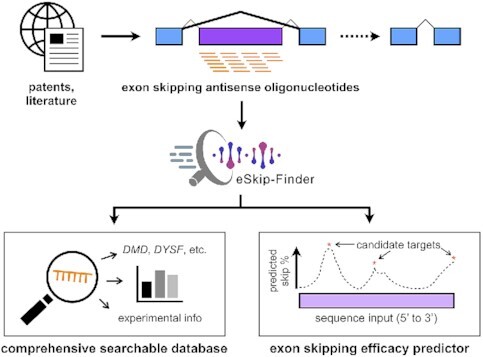
eSkip-Finder uses information on exon skipping antisense oligonucleotides from the literature to produce a database and a skipping efficacy predictive tool to aid researchers in designing effective exon skipping therapies.
INTRODUCTION
Exon skipping is a strategy that uses antisense oligonucleotides (ASOs) to exclude specific exons from the mature mRNA transcript of a given gene. ASOs are short nucleic acid analogs of diverse chemistry that recognize target mRNA sequences by base pairing. Once hybridized to their targets, ASOs act as steric blockers that prevent splicing factors and other critical proteins from accessing these sequences (1). It is through this mechanism that ASOs could be designed to modulate splicing, for example, by targeting exonic splice enhancer sequences. Given its simplicity and versatility, exon skipping has evolved to become a promising treatment for various genetic disorders, particularly muscular dystrophies (2,3).
Exon skipping is showing promise as a therapy to treat Duchenne muscular dystrophy (DMD) and other genetic diseases (1). Most cases of DMD are caused by large, out-of-frame deletions in the DMD gene, leading to an absence of the sarcolemma-stabilizing dystrophin protein in muscle cells (4–6). Exon skipping was adapted to make out-of-frame DMD mutations in-frame by removing incompatible exons from the final transcript. In this manner, exon skipping facilitates the production of shorter but partially functional dystrophin protein in muscle, ameliorating DMD pathology. Recent years have seen the approval of four exon-skipping ASOs for DMD therapy by the U.S. Food and Drug Administration (FDA): eteplirsen (2016, Sarepta), golodirsen (2019, Sarepta), viltolarsen (2020, NS and NS Pharma), and casimersen (2021, Sarepta) (7–9). In addition, the FDA approved the first n-of-1 clinical trial with an exon-skipping ASO named milasen to treat a single patient with Batten's disease in 2018 (10).
While these support the outlook of exon skipping as a viable therapeutic strategy for genetic diseases, there is much to improve especially regarding efficacy. For instance, eteplirsen could only restore up to about 1% dystrophin of healthy levels after 180 weeks of treatment in DMD patients (7). Previous studies from our group demonstrate the utility of in silico methods to design more effective ASOs (11–14). In one study, we developed an ASO with 12-fold higher in vitro exon skipping efficacy than eteplirsen using an in silico predictive tool based on statistical modelling (12). Such work and others have since uncovered numerous factors that could influence the exon skipping efficacy of an ASO including length, proximity to splice sites, target mRNA secondary structure, chemistry, and binding energy, among others (13,15–19)—all of which would be useful considerations in ASO design. However, previously developed online tools lack the capacity to simultaneously integrate many parameters critical to ASO design.
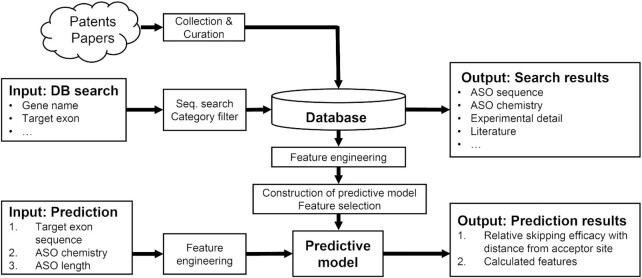
To address this gap, we previously developed a computational method using a mathematical model based on 60 descriptor candidates as well as experimental data to design highly effective ASOs for exon skipping (13). Here, we improved this framework further using machine-learning algorithms and have developed eSkip-Finder, a web server to aid the design of effective ASOs for exon skipping. The overview of the webserver is presented in Figure 1. One part of eSkip-Finder is a first-of-its-kind comprehensive database of exon skipping ASOs for DMD and other genes. This database was populated using published scientific literature and patents as sources, and contains information such as ASO chemistry, ASO sequence, and experimentally obtained skipping efficacies. The second part is a first-of-its-kind machine learning-based application to predict highly effective ASO sequences for exon skipping, based on a training set of 566 skipping values from 209 unique ASOs extracted from the database above. Here, we describe the features of eSkip-Finder in-depth and outline the ways by which it can be used for the design of exon skipping ASOs.
Figure 1.
Overview of eSkip-Finder.
RESULTS
Construction of database
A database of exon-skipping ASOs and their skipping efficacy was built by manually collecting and curating research papers and patents written in English. The database compiles data on exon-skipping ASOs for various genes, including their sequence, target exon, chemistry, literature information, and experimental information such as the ASO concentration, the cell type used for testing, and the target species. The database statistics as of 15 April 2021, are shown in Supplementary Table S1. The complete dataset extracted for each ASO in the database is provided in the web server.
Predictive model of exon-skipping efficacy
We extracted skipping data that met the following criteria from the database to prepare our training and test datasets: (i) an absolute skipping efficacy was given by a numerical value; (ii) ASO concentration used in the experiment was given; (iii) rhabdomyosarcoma (RD) cells were used in the experiment to normalize experimental conditions; (iv) the skipping efficacy was not given as an EC50 value; (v) an ASO sequence that was sequential (not dual-targeting) in the pre-mRNA of dystrophin was used. After filtering the database, 426 skipping values from 109 unique ASO sequences and 228 skipping values from 124 unique ASO sequences were obtained for ASOs with phosphorodiamidate morpholino oligomers (PMO) and 2′-O-methyl oligonucleotides (2OMe), respectively. Predictive models were built for the PMO and 2OMe separately. We split the filtered data into a training set (90%) and a test set (10%), as shown in Supplementary Table S2, under two conditions, that is, training and test sets reproduced a similar distribution of skipping efficacy, and they did not share identical sequences, as shown in Supplementary Figure S1.
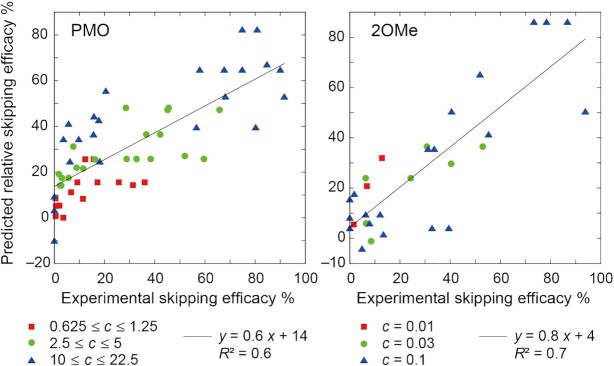
We built a predictive model for the relative skipping efficacy of a target exon of dystrophin mRNA using the support vector regressor (SVR) implemented in scikit-learn version 0.23.2 (20). First, 32 features, tabulated in Table 1 and Supplementary Table S3, were prepared by feature engineering of ASO and/or its target exon sequences such as predicted binding score between the ASO and its target exon (21), predicted local RNA structure at the target site (22), and GC contents of the ASO and target exon. We also included the ASO concentration used in experimental studies as a feature. More details on the features used are provided elsewhere (13). Each feature was standardized before fitting the model. To select fewer important features, we built all possible combinations of the SVR model that used fewer than seven features, where the experimental ASO concentration was always included as a selected feature. The upper limit number of features, six, was chosen according to the available computational resources. For each model, the hyper-parameter optimization by a grid search for C, gamma, and epsilon was conducted with 100-time repeated splitting of the training data into 80% used to build a model and 20% used to validate the built model under the condition that they did not share identical sequences. Finally, we selected the SVR model that yielded the highest average R2 of the validation sets as shown in Supplementary Figure S2, the features of which are given in Table 1. The selected models for PMO and 2OMe were applied to the test set, yielding R2 values of 0.6 and 0.7, as shown in Figure 2. The correlation between experimental and predicted skipping efficacy was confirmed for various concentrations. The contributions of each feature to predictive performance (feature importance) were estimated by permutation importance (23). The importance of each feature was defined by decrease of the R2 value when the feature in the test set was permutated randomly. The feature importance calculation was repeated 100 times and the averaged values are shown in Table 1. The current model is focused on the prediction of the relative skipping efficacy of ASOs. However, other parameters should be also considered when designing ASOs, one of which is the off-target effect. Other bioinformatics tools such as SKIP-E (https://skip-e.geneticsandbioinformatics.eu/) could complement it.
Table 1.
Selected features
| Selected features for PMO | Selected features for 2OMe | ||||
|---|---|---|---|---|---|
| Name | Description | FIa | Name | Description | FIa |
| ASO concentrationb | Concentration of oligomer used in the experiment | 0.64±0.14 | ASO concentrationb | Concentration of oligomer used in the experiment | 0.11±0.05 |
| Exon v intron %GC after blocking by oligo | %GC in exon when blocked by oligo / %GC 5′ intron 200 bases upstream | 0.68±0.15 | GCs (number of) | Total GCs in ASO sequence | 0.67±0.20 |
| dG (50BaseFlanksAroundTarget) | Predicted binding energy (21) of ASO to the target sequence plus 50-base flanks (13) | 0.66±0.16 | ACP | Distance in bases from the splice acceptor site to the center of the target site (17) | 0.49±0.21 |
| ACC_LAST15 | Predicted accessibility scores (22) of the 3′ end of the target (last 15 bases) | 0.32±0.09 | %GC of exon when blocked by oligo | Total remaining %GCs of target exon sequence when blocked by ASOs | 0.46±0.11 |
| niscore_per_base | Cumulative NI score (24) divided by the number of exon bases. | 0.18±0.09 | |||
| ACC_LAST8 | Predicted accessibility scores of the 3′ end of the target (last 8 bases) | 0.12±0.07 | |||
aThe feature importance (FI) was calculated by the permutation importance method (23).
bThe ASO concentration used in the experiment is always included as one of the features of the predictive model.
Figure 2.
Predictive performance of SVR models for PMO and 2OMe. Symbols represent oligomer concentration (c) given in μM used in the experiment. The coefficient of determination, R2, was calculated by linear regression (black lines).
Implementation
The selected predictive models (Figure 2 and Table 1) are implemented on the web server with scikit-learn (20). Features of local accessibility scores of target exon sequences and binding scores between ASOs and their target exons were calculated with the ViennaRNA Package (22) and RNAstructure (21). The dictionary of NI scores was retrieved from Ref. (24). The concentrations of ASOs were set to typical values, that is, 3 μM for PMO and 0.1 μM for 2OMe. The database was built using PostgreSQL.
Case study
Database search
The web server provides an intuitive search interface of relevant information on exon skipping efficacy with search queries, such as gene name, species, and exon number.
Prediction of the efficacy of exon-skipping ASOs
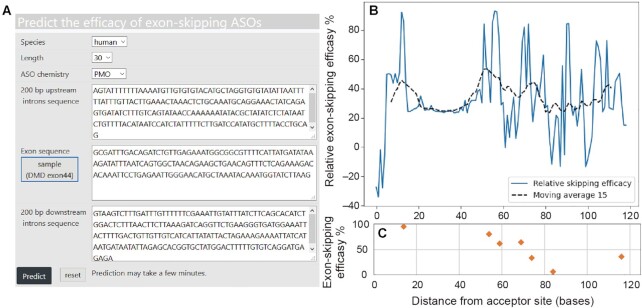
The web server provides a prediction of the relative exon-skipping efficacy of a target exon specified by a user as shown in Figure 3 under the following conditions: 3 μM of PMO or 0.1 μM of 2OMe introduced into cultured cells.
Figure 3.
Case study on predicting skipping ASOs for exon 44 of the dystrophin pre-mRNA. (A) Input image of the predictive model. A user specifies the length of ASO and its chemistry (PMO or 2OMe). The upstream (200 bases) and downstream (200 bases) intron sequences of the target exon are required in addition to the target exon sequence, which are used to calculate features. (B) Output image. The relative exon-skipping efficacy is predicted by scanning the target exon sequence with a window size of the length specified by the user. Moving averages with 15 bases are plotted with a dashed line. (C) Efficacy of dystrophin exon 44 skipping observed under identical experimental conditions (cell type used = healthy primary human myotubes, ASO chemistry = PMO, ASO length = 30, ASO concentration = 0.5 μM) as previously reported (15), which is not included in the training dataset. The correlation between predicted and experimental skipping efficacies R2 was 0.7 as shown in Supplementary Figure S3.
In this case study, we targeted exon 44 of the dystrophin pre-mRNA using a single ASO, the chemistry and length of which were PMO and 30-mer, respectively. A user needs to input 200 bp of upstream and 200 bp of downstream intron sequences in addition to the target exon sequence, as this sequence information is required to calculate the features. The prediction took 79 s. We obtained the promising regions for exon 44 skipping, that is, the regions between 10–20 and 50–80. We found that these regions were indeed included in experimentally observed effective ASOs (15).
As a validation of predicting exon skipping efficiency beyond DMD, we present a test case of PMO-mediated exon 73 skipping of collagen type VII alpha 1 chain (COL7A1) (Supplementary Table S4). (25). Although the experimental conditions (e.g. ASO concentration) were different, we found that predicted and experimental values correlated well with each other, and the model correctly ranked the efficacy of the three PMOs, indicating a potential predictive ability of the current model for other genes. Currently, the amount of available experimental data of exon skipping for other genes is limited. To examine the applicability of our model to other genes, we plan to further validate with various genes when sufficient data become available. We expect that adding various genes and oligo chemistries to the database will help expand the applicability of the predictive model further.
DATA AVAILABILITY
The authors confirm that the data supporting the findings of this study are available within the article and/or in the supplementary material.
Supplementary Material
Contributor Information
Shuntaro Chiba, HPC- and AI-driven Drug Development Platform Division, RIKEN Center for Computational Science, Yokohama 230-0045, Japan.
Kenji Rowel Q Lim, Department of Medical Genetics, University of Alberta Faculty of Medicine and Dentistry, 8613-114 St, Edmonton, AB, Canada.
Narin Sheri, Department of Medical Genetics, University of Alberta Faculty of Medicine and Dentistry, 8613-114 St, Edmonton, AB, Canada.
Saeed Anwar, Department of Medical Genetics, University of Alberta Faculty of Medicine and Dentistry, 8613-114 St, Edmonton, AB, Canada.
Esra Erkut, Department of Medical Genetics, University of Alberta Faculty of Medicine and Dentistry, 8613-114 St, Edmonton, AB, Canada.
Md Nur Ahad Shah, Department of Medical Genetics, University of Alberta Faculty of Medicine and Dentistry, 8613-114 St, Edmonton, AB, Canada.
Tejal Aslesh, Department of Medical Genetics, University of Alberta Faculty of Medicine and Dentistry, 8613-114 St, Edmonton, AB, Canada.
Stanley Woo, Department of Medical Genetics, University of Alberta Faculty of Medicine and Dentistry, 8613-114 St, Edmonton, AB, Canada.
Omar Sheikh, Department of Medical Genetics, University of Alberta Faculty of Medicine and Dentistry, 8613-114 St, Edmonton, AB, Canada.
Rika Maruyama, Department of Medical Genetics, University of Alberta Faculty of Medicine and Dentistry, 8613-114 St, Edmonton, AB, Canada.
Hiroaki Takano, HPC- and AI-driven Drug Development Platform Division, RIKEN Center for Computational Science, Yokohama 230-0045, Japan.
Katsuhiko Kunitake, Department of Molecular Therapy, National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP), Kodaira, Tokyo 187-8551, Japan.
William Duddy, Northern Ireland Center for Stratified Medicine, Biomedical Sciences Research Institute, C-TRIC, Altnagelvin Hospital Campus, Ulster University, Londonderry BT47 6SB, UK.
Yasushi Okuno, HPC- and AI-driven Drug Development Platform Division, RIKEN Center for Computational Science, Yokohama 230-0045, Japan; Department of Biomedical Data Intelligence, Graduate School of Medicine, Kyoto University, Kyoto 606-8507, Japan.
Yoshitsugu Aoki, Department of Molecular Therapy, National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP), Kodaira, Tokyo 187-8551, Japan.
Toshifumi Yokota, Department of Medical Genetics, University of Alberta Faculty of Medicine and Dentistry, 8613-114 St, Edmonton, AB, Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid for Research on Nervous and Mental Disorders [2–6 to Y.A. and S.C.]; T.Y. is supported by Muscular Dystrophy Canada; Friends of Garrett Cumming Research Fund, HM Toupin Neurological Science Research Fund; Canadian Institutes of Health Research (CIHR); Alberta Innovates: Health Solutions (AIHS), Jesse's Journey; Women and Children's Health Research Institute (WCHRI). S.C. is supported by HOKUSAI BigWaterfall system. Funding for open access charge: Women and Children’s Health Research Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lim K.R.Q., Yokota T.. Invention and early history of exon skipping and splice modulation. Methods Mol. Biol. 2018; 1828:3–30. [DOI] [PubMed] [Google Scholar]
- 2. Rodrigues M., Yokota T.. Yokota T., Maruyama R.. Exon Skipping and Inclusion Therapies: Methods and Protocols. 2018; NY: Springer; 31–55. [Google Scholar]
- 3. Siva K., Covello G., Denti M.A.. Exon-skipping antisense oligonucleotides to correct missplicing in neurogenetic diseases. Nucleic Acid Ther. 2014; 24:69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Petrof B.J., Shrager J.B., Stedman H.H., Kelly A.M., Sweeney H.L.. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:3710–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bladen C.L., Salgado D., Monges S., Foncuberta M.E., Kekou K., Kosma K., Dawkins H., Lamont L., Roy A.J., Chamova T.et al.. The TREAT-NMD DMD global database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum. Mutat. 2015; 36:395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tuffery-Giraud S., Béroud C., Leturcq F., Yaou R.B., Hamroun D., Michel-Calemard L., Moizard M.-P., Bernard R., Cossée M., Boisseau P.et al.. Genotype–phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD–DMD database: a model of nationwide knowledgebase. Hum. Mutat. 2009; 30:934–945. [DOI] [PubMed] [Google Scholar]
- 7. Lim K.R., Maruyama R., Yokota T.. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des. Dev. Ther. 2017; 11:533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anwar S., Yokota T.. Golodirsen for Duchenne muscular dystrophy. Drugs Today (Barc.). 2020; 56:491–504. [DOI] [PubMed] [Google Scholar]
- 9. Roshmi R.R., Yokota T.. Viltolarsen for the treatment of Duchenne muscular dystrophy. Drugs Today (Barc.). 2019; 55:627–639. [DOI] [PubMed] [Google Scholar]
- 10. Kim J., Hu C., Moufawad El Achkar C., Black L.E., Douville J., Larson A., Pendergast M.K., Goldkind S.F., Lee E.A., Kuniholm A.et al.. Patient-customized oligonucleotide therapy for a rare genetic disease. N. Engl. J. Med. 2019; 381:1644–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Echigoya Y., Lim K.R.Q., Melo D., Bao B., Trieu N., Mizobe Y., Maruyama R., Mamchaoui K., Tanihata J., Aoki Y.et al.. Exons 45–55 skipping using mutation-tailored cocktails of antisense morpholinos in the DMD gene. Mol. Ther. 2019; 27:2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Echigoya Y., Lim K.R.Q., Trieu N., Bao B., Miskew Nichols B., Vila M.C., Novak J.S., Hara Y., Lee J., Touznik A.et al.. Quantitative antisense screening and optimization for Exon 51 skipping in Duchenne muscular dystrophy. Mol. Ther. 2017; 25:2561–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Echigoya Y., Mouly V., Garcia L., Yokota T., Duddy W.. In silico screening based on predictive algorithms as a design tool for exon skipping oligonucleotides in Duchenne muscular dystrophy. PLoS One. 2015; 10:e0120058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee J.J.A., Maruyama R., Duddy W., Sakurai H., Yokota T.. Identification of novel antisense-mediated exon skipping targets in DYSF for therapeutic treatment of dysferlinopathy. Mol. Ther. - Nucleic Acids. 2018; 13:596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Popplewell L.J., Trollet C., Dickson G., Graham I.R.. Design of phosphorodiamidate morpholino oligomers (PMOs) for the induction of exon skipping of the human DMD gene. Mol. Ther. 2009; 17:554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harding P.L., Fall A.M., Honeyman K., Fletcher S., Wilton S.D.. The influence of antisense oligonucleotide length on dystrophin exon skipping. Mol. Ther. 2007; 15:157–166. [DOI] [PubMed] [Google Scholar]
- 17. Pramono Z.A.D., Wee K.B., Wang J.L., Chen Y.J., Xiong Q.B., Lai P.S., Yee W.C.. A prospective study in the rational design of efficient antisense oligonucleotides for exon skipping in the DMD gene. Hum. Gene Ther. 2012; 23:781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wee K.B., Pramono Z.A.D., Wang J.L., MacDorman K.F., Lai P.S., Yee W.C.. Dynamics of co-transcriptional pre-mRNA folding influences the induction of dystrophin exon skipping by antisense oligonucleotides. PLoS One. 2008; 3:e1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aartsma-Rus A., Houlleberghs H., van Deutekom J.C.T., van Ommen G.-J.B., t Hoen P.A.C.. Exonic sequences provide better targets for antisense oligonucleotides than splice site sequences in the modulation of Duchenne muscular dystrophy splicing. Oligonucleotides. 2010; 20:69–77. [DOI] [PubMed] [Google Scholar]
- 20. Pedregosa F., Varoquaux G.e., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V.et al.. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011; 12:2825–2830. [Google Scholar]
- 21. Reuter J.S., Mathews D.H.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010; 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lorenz R., Bernhart S.H., Höner zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorith. Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Altmann A., Tolosi L., Sander O., Lengauer T.. Permutation importance: a corrected feature importance measure. Bioinformatics. 2010; 26:1340–1347. [DOI] [PubMed] [Google Scholar]
- 24. Stadler M.B., Shomron N., Yeo G.W., Schneider A., Xiao X., Burge C.B.. Inference of splicing regulatory activities by sequence neighborhood analysis. PLoS Genet. 2006; 2:e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ham K.A., Aung-Htut M.T., Fletcher S., Wilton S.D.. Nonsequential splicing events alter antisense-mediated exon skipping outcome in COL7A1. Int. J. Mol. Sci. 2020; 21:7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or in the supplementary material.