Abstract
Chemotherapy has been reported to upregulate sphingomylinases and increase cellular ceramide, often linked to the induction to cell death. In this work, we show that sublethal doses of doxorubicin and vorinostat still increased cellular ceramide, which was located predominantly at the plasma membrane. To interrogate possible functions of this specific pool of ceramide, we used recombinant enzymes to mimic physiological levels of ceramide at the plasma membrane upon chemotherapy treatment. Using mass spectrometry and network analysis, followed by experimental confirmation, the results revealed that this pool of ceramide acutely regulates cell adhesion and cell migration pathways with weak connections to commonly stablished ceramide functions (e.g. cell death). Neutral sphingomyelinase 2 (nSMase2) was identified as responsible for the generation of plasma membrane ceramide upon chemotherapy treatment, and both ceramide at the plasma membrane and nSMase2 were necessary and sufficient to mediate these “side” effects of chemotherapy on cell adhesion and migration. This is the first time a specific pool of ceramide is interrogated for acute signaling functions and the results define plasma membrane ceramide as an acute signaling effector necessary and sufficient for regulation of cell adhesion and cell migration under chemotherapeutical stress.
Keywords: Sphingolipids, plasma membrane, sphingomyelinase, doxorubicin, vorinostat
Introduction
Studies spanning three decades have shown the bioactive sphingolipid ceramide to be involved in a myriad of biologies, the most prominent of which are the induction of growth arrest and cell death (1). However, ceramide species have been found to also mediate/regulate cytokine secretion, vesicle transport, and cell cycle. Ceramide is also involved in mechanisms employed by some pathogenic bacteria and viruses in order to engage and penetrate mammalian cells (2). The molecular mechanisms on how ceramide mediates these biologies are still not fully determined, although a few direct targets have been advanced, including atypical PKC, cathepsin D, protein phosphatases 1 and 2A, and VDAC (3, 4). Moreover, the specific subcellular (i.e., organelle) location for ceramide function is still not fully understood. Birbes et al. reported that targeting bacterial sphingomyelinase (bSMase) to mitochondria--but not to other compartments--in the cell induced apoptosis (5), and this finding has been supported and extended by more recent experimental studies on regulating mitochondrial ceramide by specifically transferring it from the ER to mitochondria (6). On the other hand, in earlier studies, our group and other investigators had found that generating plasma membrane ceramide through exogenously applied sphingomyelinases (SMases) did not induce cell death in many cell lines although it induced hemolysis in erythrocytes (7, 8).
These studies and several other lines of evidence prompted the development of the “Many Ceramides” hypothesis (9), which posits that different species of ceramide(s) at different cellular membranes are involved in different biological phenomena. These compartment-specific pathways would presumably operate in response to distinct stimuli that putatively activate specific enzymes of ceramide generation. Surprisingly, barely any of the previous studies hypothesized where ceramide at the onset of a specific cell biologic response is generated and localized. On a further level of complexity, ceramide can also be metabolized to other lipids, and these byproducts may potentially be responsible for the observed cell signaling and cellular responses (10). For example, actions of tumor necrosis factor (TNF) on SMases in fibroblasts results in subsequent formation of sphingosine 1-phosphate (S1P), which in turn promotes cell proliferation (11). In this context, it is becoming increasingly clear that ceramide-mediated cellular responses have to be investigated at the level of specific enzymes and specific subcellular compartments, while also taking into consideration the likelihood of further metabolism.
One of the challenges in dissecting and/or studying ceramide signaling is the lack of biochemical pathways that are promptly activated to induce ceramide generation. Indeed, the vast majority of studies on ceramide signaling use either exogenously applied ceramides (12, 13) or ceramide inducers, and responses often occur many hours or even days after induction (14). The localization of this generated or applied ceramide and their associated metabolism are not well defined. Moreover, many of the inducers used to generate endogenous ceramide may regulate more than a single pathway of ceramide generation (15); thus, greatly complicating the dissection of specific ceramide-mediated cell signaling pathways.
To address such challenges, a few analytical approaches have been used in order to identify direct targets that might propagate the subsequent steps in signaling after initial ceramide generation. The most direct biochemical approaches have utilized mostly short chain ceramide species bound to agarose beads for either affinity chromatography columns or in tandem pull-down/mass spectrometry studies (16). Other assays use functionalized ceramides that may be crosslinked with docking proteins to functionalized beads (17, 18). Together, these classic studies have found many associated proteins that bind ceramide and may be activated or inhibited, but still lacking the context on cellular topology.
In our own research group, we have long studied protein phosphatases that not only have been found to bind ceramide, but that are also directly activated in vitro by natural ceramide but not unnatural stereoisomers, thereby showing stereospecificity for the natural ceramide (19, 20). Indeed, in previous work, we have found that the PP1 family of phosphatases can be activated in cellulo by ceramide within just a few minutes after its generation in situ (21). Although the site of action was not defined, this might be the most robust effector for ceramide so far reported, and in this work, we used changes in protein (de)phosphorylation to screen for most direct ceramide actions.
Here, in order to advance studies on compartment-specific functions, we have focused on plasma-membrane ceramide to determine if there are specific biological attributes to that subcellular pool. Towards this goal, we screened for compounds that could potentially generate ceramide in the plasma membrane. We found doxorubicin and vorinostat generated ceramide at the plasma membrane, even at sublethal doses. Using recombinant tools, we acutely generated ceramide at the plasma membrane and defined the experimental conditions in which this particular pool of ceramide remained in the plasma membrane. Under these conditions, we screened which proteins underwent changes in the corresponding phosphorylation status. The results showed that plasma membrane-generated ceramide induces acute dephosphorylation of a large set of substrates regulating cell adhesion and migration. Interestingly, it has been reported that some chemotherapeutic treatments, including those from doxorubicin and vorinostat, present side effects including modulation of cell adhesion resulting in increasing the risk in cell invasion and metastasis (22), suggesting that these biologies are mediated by plasma membrane ceramide under the control of chemotherapy.
In summary, this work defines three novel key points about the mechanism on how sublethal doses of chemotherapy lead to increase cancer cell migration. First, chemotherapy increases expression of nSMase2. Elevation of nSMase2 is sufficient to drive cell migration, and suppression or inhibition of nSMase2 shows it is necessary for the chemotherapy effects on migration. Second, nSMase2 acts on the plasma membrane, generating a previously uncharacterized pool of ceramide, which our data show to mediate the effects on cell migration. Third, the mechanism on how ceramide acts on a biological pathway by acutely inducing protein (de)phosphorylation, leading to regulation of adhesion and migration.
Materials and Methods
Cell culture
All cell culture media (DMEM, OPTI-MEM, FBS), DPBS, Human Fibronectin MW 440KDa and BCA Protein Assay Kit (cat# 23225) were purchased from Thermo Fisher (Waltham, MS, USA). Fluorescent conjugated antibodies were from Invitrogen (Waltham, MS, USA) (donkey/Goat anti-mouse/rabbit Alexa Fluor 488/555). Xtremegene 9 DNA Transfection reagent, fatty acid free bovine serum albumin (BSA) lyophilized powder (cat#126609), Optiprep density gradient medium Doxorubicin and vorinostat were purchased from Sigma (St. Louis, MO, USA). The nuclear staining by using DAPI with mounting medium (H-1200) was from Vector Laboratories (Burlingame, CA, USA). The mouse hybridoma clone 2A2 was generated and its cognate anti-ceramide IgM monoclonal antibodies purified as previously reported (23). HeLa human cervical carcinoma cells (ATCC, Manassas, VA, USA), were maintained in DMEM supplemented with 10% FBS. Cells were incubated and imaged in a humidified environment at 37 °C with 5% CO2. Cells were monthly evaluated for mycoplasma and annually typed.
Cell Viability Assay
Cell viability was measured using the oxidation of the water soluble 3-(4,5- dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) to the insoluble formazan (Sigma, St. Louis, MO, USA), quantified by absorbance at 570 nm (SpectraMax M5, Molecular Devices). HeLa cells were seeded in 6 well plate at 10^5 cells/ml/well. At the end of the treatment period, medium was removed and replaced with fresh medium. MTT (5 mg/ml) in fresh DMEM medium was added to each well at 37 °C for 2 h. Formazan product was solubilized in DMSO and absorbance was measured.
Lactate Dehydrogenase Release assay
Lactate dehydrogenase (LDH) release was determined using a colorimetric assay kit (Biovision, Milpitas, CA) following the manufacturer's instructions.
Protein expression and purification
His-Venus-Lysenin (24), bSMase from Bacillus cereus (7), and pCDase from Pseudomonas aeruginosa (25) were expressed in E.coli BL21 DE3 and purified as previously described with minor modifications (7). Briefly, four liters of Terrific Broth was inoculated with bacteria, and bacteria were induced with 1mM of isopropyl β-D-1 thiogalactopyranoside (IPTG), followed by incubation at 25°C for 16 hours. Pelleted bacteria were kept at −80°C until purification. Pellets were thawed and sonicated with on/off cycles at 85% of amplitude (58W) during a 5-minute series (2 s on/ 2 s off); sonication was prolonged until the bacteria were visually cleared. Lysate was centrifuged and filtered through a 0.22 micrometer filter. Purification of His-tagged proteins was performed using a HisTrap FF column (GE Healthcare Life Sciences, Chicago, IL, USA) followed by size-exclusion chromatography (Superdex 200) in an ÄKTA purifier system. Fractions containing pure proteins were combined, the final volume was concentrated to 1 mg/mL, frozen directly in liquid nitrogen, and stored at −80°C until use.
Reverse-Transcriptase PCR
mRNA from HeLa cells was extracted by using the RNeasy kit (Qiagen, Venlo, Netherlands). RNA (0.5–1 μg) was used to synthesize cDNA with the Superscript III Kit (Invitrogen). Real-time PCR was performed using the Taqman assays on an ABI 7500 real-time system. Standard reaction volume was 20 μl containing 10 μl supermix, 4.67 μl dH2O, 0.33 μl 60× Taqman assay, and 5 μl cDNA template. The quantitative PCR protocol consisted of 2 min at 50°C followed by 10 min at 95°C, and 40 cycles consisting of a 15s at 95°C and 60s at 60°C. All reactions were in triplicates. Actin was used as a reference gene in all cases, and Ct values were converted to mean normalized expression using the ΔΔCt method. Taqman assays were purchased from Life Technologies (gene: Taqman assay number. actin: Hs99999903_m1, neutral SMase 2: Hs00920354_m1).
siRNA and plasmid DNA transfection
Cells were maintained at 37°C, 5% CO2 in a humidified atmosphere. For siRNA experiments, 300.000 HeLa cells were cultured in 100 mm dishes with media being changed 1h before transfection. Cells were transfected with 20 nM negative control (AllStar, Qiagen) or siRNA for nSMAse2 (gene symbol SMPD3, siRNAs Life technologies s30926 and results confirmed using s30927) using lipofectamine RNAimax (Life Technologies, Chicago, IL, USA) according to the manufacturer’s protocol. Ater 48 h, cells were incubated in fresh media for 1–2 h prior to stimulation as indicated. For overexpression experiments, HeLa cells were transfected at confluence in 60 mm dishes (500K cells/dish) with V5-nSMase2 (SMPD3) previously described (26) and using Xtremegene 9 standard protocol (SIGMA).
Wound-Healing (scratch) assays
Cell wound-healing (scratch) assay was adapted from Liang et al. (27). Briefly, 12-well plates were coated with fibronectin-1 (FN1) 5μg/ml for 16h (overnight). Wells were washed with PBS three times. Cells were plated at 200K cells/ well to generate a confluent monolayer of cells and was left overnight to attach. The appropriate amount of serum was determined at 2% FBS to prevent apoptosis and detachment. The scratch was made with a 200 pipet tip. The scratch area was measured from contrast-phase microscope images taken at 20X magnification. Pictures were taken at time zero and 20h (about 50% area was covered). Image analysis was performed with the Fiji software (28) extracting the area of the remaining wound area, and calculating the area covered by the cells in 20h. Automatization of image analysis (n=30 or otherwise as indicated) was performed using python OpenCV (OpenCV.org) and plotting and statistical analysis anova 1-way (Tukey posthoc test) were performed by using python-numpy-pandas.
Transwell cell migration assay
Directional migration of HeLa cells was measured using Fluoroblock Transwells 8μM (Costar, Cambridge, MA, USA) pre-coated with 5mM FN1for 16h, and allowing the cells to migrate in DMEM containing 1% FBS for 16h. Cells on the top and bottom of the fluoroblock transwell membrane were quantified by staining the cells with CellTracker Green CMFDA for 30 min and fluorescent measured using a plate reader Spectramax M5 (Molecular Devices, San Jose, CA, USA) at excitation/emission spectra (492/517 nm maxima). Each condition represented 5 independent experiments. Statistics one-way ANOVA Tukey posthoc vs control.
Adhesion assay
Cell adhesion assays were adapted from Piccolo et al. (29). Briefly, HeLa cells were plated in 100 mm diameter dishes at 1 million cells per condition to analyze. Next day, cell media were changed to DMEM serum free and left overnight. When required, cells were treated with drugs (doxorubicin, vorinostat, and GW-4869). Next day, cells were washed with serum free media and collected in PBS containing 10mM EDTA for 20 min using a cell lifter. Cells were pelleted at 600 x g for 5 min and resuspended in serum free media at 100.000 cells/ ml. Enzymatic treatments were applied at this step (bSMase, pCDase) for 3 min, and media were replaced by centrifugation. One ml of cell suspension was added to 35mm dishes containing 1ml of DMEM containing 1% FBS. Dishes were previously coated with fibronectin 5ug/ml for 16h and washed 3 times with PBS. Then cells were left for different amount of time (specified in each case) at 37°C for attachment. Each dish was then washed 3 times with PBS, and cells were fixed with 40°C warmed paraformaldehyde at 4% (w/v) in PBS. Cells were permeabilized with Triton X-100 0.1% (v/v) in PBS and stained for the actin cytoskeleton with rhodhamine-phalloidin and the nucleus DNA with DAPI. Attached cells were visualized using a Leica TCS SP8 laser scanning confocal microscope. Cell number was calculated by the number of nuclei, and cell area was determined by the area stained with phalloidin-rhodamine. Transfected cells were determined using the green channel for Venus-lysenin staining or tagged proteins (e.g. nSMase2 V5-tagged) by using anti-V5 mouse IgG labeled with Donkey anti-mouse conjugated to Alexa Fluor 488. Automatization of imaging processing (cell counting and area measurements and statistics) was performed using python-OpenCV software. Briefly, python-bioformats package was used to read raw data from Leica files and extract each image channel and metadata. RGB channels were filtered for noise and threshold was set using OTSU method. Individual nuclei were detected by using the cv2.connectedComponents algorithm. Cell area was measured using cv2.contourArea algorithm. When transfected cells were used, green channel was used as a mask on the blue and red channels to only count transfected cells. All data plotting was performed by using matplotlib and Seaborn python packages.
Lipid measurements
Lipid measurements were performed as previously described (30).
Western Blot
HeLa cells were plated in 6-well plates (80,000 cells/well). Culture media were aspirated and 1% (w/v) SDS was added to the cells. BCA protein assay test was used to assess the amount of protein, and 20 μg of protein was loaded in SDS-PAGE gradient gels (4-20% in Tris-HCl). Proteins were transferred onto nitrocellulose membranes and blocked in PBS-buffered non-fat milk containing 0.1% Tween-20. Primary antibodies were incubated overnight at 4°C using manufacturer-recommended dilutions.
Immunofluorescence analysis
HeLa cells were plated in 35 mm diameter uncoated glass bottom dishes (MatTek, Ashland MA) at 40,000 cells/dish (or 80,000 cells/dish when DNA cell transfection was not required) in DMEM-10% FBS and incubated overnight. Cells were transfected with plasmid DNA or V5 using Xtremegene 9 standard protocol. After 16h, cell culture media were changed to serum free media for 2 hours before treatments. For protein visualization, cells were fixed using fresh, pre-warmed (40°C) 4% paraformaldehyde (PFA) for 15 minutes. Samples were washed with phosphate-buffered saline (PBS) and permeabilized with 0.1% Triton X-100 in PBS for 20 minutes at room temperature. Permeabilized cells were blocked with 2% BSA for 20 minutes and washed with PBS. Primary antibodies were added in 2% BSA at recommended dilutions (1:500 for V5) and incubated overnight at 4°C. After incubation, samples were washed with PBS, and phalloidin and DAPI were added if required for 10 minutes at room temperature.
For lipid staining, cold serum-free media were added to samples before staining and after treatments. For sphingomyelin staining, samples were maintained at 4°C and freshly thawed, cold lysenin in BSA was added to the media, and protein-lipid interaction was allowed for 20 minutes. Samples were collected on ice and fixed using cold PFA for 30 minutes. Then samples were treated as previously described. For ceramide staining using the ceramide antibody, samples were treated, ceramide antibody was added to serum-free media, and samples were kept at 4°C for 20 minutes. Warmed PFA was added to samples and incubated at room temperature for 15 minutes. For dual protein and lipid staining, samples were fixed with warm followed by cold PFA and kept at 4°C; PFA was neutralized with glycine, samples were washed with cold PBS, ceramide antibody (1:80 in serum free DMEM media) was added and incubated for 20 minutes, and then fixed as described. Images were collected using a Multiphoton microscope Leica TCS SP8 using a HC PL APO 63X/1.40 oil objective CS2.
Cell fractionation.
Cells were plated in 4 x 15cm diameter plates at 5 million cells/dish and then starved for 24h in serum free media. Cells were then treated with bSMase 10milli-units for 2 min and washed in PBS. Cell fractionation was performed as recommended by Axis-Shield Optiprep density gradient media (SIGMA). Cells were collected in lysis buffer [HEPES 20mM, 2mM KCl, 2mM MgCl2, 0.25M sucrose + protease cocktail inhibitors from Sigma cat# P8340] using a cell lifter. Cells were disrupted by repeat passages through a needle syringe [gauge 25G]. Cell lysis was centrifuged at 2000xg for 10 min, and supernatant was centrifuged at 100.000xg 4°C for 60 min. The resultant pellet was resuspended in 200microliter lysis buffer and applied into an Iodixanol discontinuous gradient [0.5%, 2.5%, 10%, 15%, 20%, 30%. Final volume 13 ml] and centrifuged at 28.000 rpm (104.000 x g average point) in SW28 Beckman rotor for 20h. Following centrifugation, fractions were collected from the top to the bottom. Fractions were analyzed for western blot and for lipid contain using mass spectrometry.
Phosphoproteomics analysis
HeLa cells were growth in SILAC media (Fisher PI89985) with 10% dialyzed serum (Fisher PI88440) supplemented with heavy (13C6 15N4 L-arginine + 13C6 L-lysine) or light arginine and lysine. Hela cells (light or heavy labeled) were plated in 8 x 15 cm diameter dishes at 5 million cells/dish. Media were changed after 4h, and cells were incubated for 48h. Cells were treated with bSMase 10mU for 3 min, then cells were collected in 4 ml 2.5% (w/v) SDS, and cell lysate was frozen at −80C. For each sample, heavy and light cell lysates were mixed. Each sample was aliquoted in 4 ml. DTT (5mM, 10 min at 55°C) and Iodoacetamide (10mM 30 min room temperature, in the dark) were added to the samples. 4 volumes of methanol at −20°C were added followed by two volumes of −20°C chloroform and 3 volumes of water. Samples were vortexed and precipitated for 1h at −20°C. Samples were centrifuged at 4700 x g for 20 min at 4°C. The upper layer was removed without bothering the interphase, 3 volumes of MeOH were added and vortexed for 5 s, centrifuged at 4700 x g for 20 min, and the pellet was frozen until processing. 5ml Urea 8M in HEPES 20mM was added to each pellet. Samples were re-treated with DTT and Iodoacetamide. Urea was diluted to 2M with NH4CO3 and samples were digested with trypsin overnight. Next day samples were treated with TFA final concentration 0.5%. Desalting was performed in Waters WAT036810 Sep-Pak Plus tC18, and eluted in 10%, 20% and 50% TFA and lyophilized. Fractionation of samples was performed in Discovery DSC-SCX SPE 1g 52689-U eluted from 50-1000 mM KCl. Eluates were dried using a SpeedVac and desalted using OASIS 186000367 MAX 3cc columns. Phospho-enrichment was performed using TiO2 resin from GL Science, Japan.
Software
Fiji (ImageJ2) was used to measure the scratch area in scratch assay and single cell image tracking and quantification of fluorescent signal channels. OpenCV was used for image analysis for identification of single cells, measurement cell area, measurement of scratch area. Numpy (31), Pandas, Matplotlib, Seaborn were used for data processing, data representation, and data clustering. R language (32) package Graphite (33) and Cytoscape software (34) were used for the network generation, analysis and visualization. NetworkX was used to calculate degree of connectivity to filter the network for less significant interactions (35). GraphPad Prism (V4.05) and Seaborn (V0.9.0) were used for plotting and statistical analysis. Text mining analysis and Gene Ontology analysis were performed by using Biopython (36) and the Uniprot Rest API (37). Functional annotation in the network was extracted from Gene Ontology database considering only experimental evidences. Additional functional annotation was obtained via text mining of PubMed abstracts by using the gene2pubmed relationships files (38) and The Comparative Toxicogenomics Database (39).
Results
Vorinostat, and doxorubicin generate ceramide at the plasma membrane.
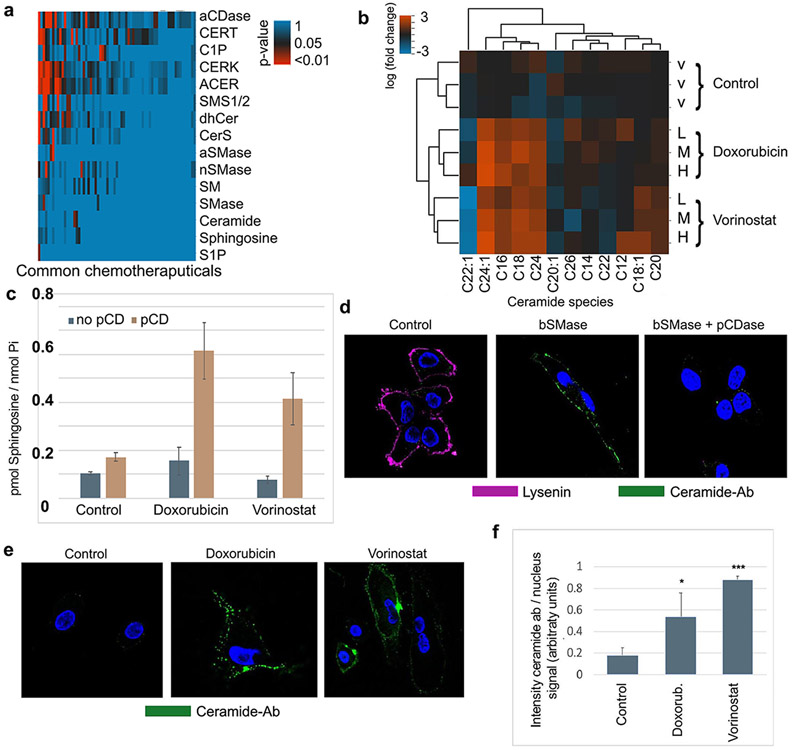
Many previous reports have shown that several chemotherapeutic treatments modulate the sphingolipid pathway (15, 40), including activation and/or induction of acid and neutral SMases (aSMase and nSMase, respectively), especially nSMase 2 (41-43). These enzymes localize in certain subcellular compartments, such as lysosomes and Golgi, as well as the plasma membrane (41, 44). In order to focus on the most clinically relevant chemotherapeutic drugs and targeted agents, we analyzed the available literature and performed cluster studies based on drugs commonly employed in contemporary cancer treatment, for the modulation of sphingolipids and their metabolizing enzymes (Figure 1a). From them, 68 drugs reported to have effects on the sphingolipid pathway were categorized based on their p-value using a Fisher t-test analysis (supplemental table s1). Ten of them, including etoposide, cisplatin, vorinostat, paclitaxel and some anthracyclines (doxorubicin, daunorubicin) presented the most significant p-values for ceramide. Doxorubicin was selected as it resulted in the most statistically significant increase in the expression of nSMase2, which is associated to the plasma membrane, and increases in cellular ceramide levels but less significant for other bioactive sphingolipids such as S1P and sphingosine. Vorinostat, was also more selective for ceramide and nSMase2, and it was also selected in our studies so as to provide representatives of two very much studied groups of compounds in cancer, albeit with markedly distinct mechanisms of action. Although reported in different cell lines, we examined these compounds in HeLa cells for ceramide generation. HeLa cells were treated with different sublethal doses of doxorubicin (0.6, 0.8 and 1 μM) and vorinostat (2.5, 5 and 10 μM), and the sphingolipid profile was measured at 24h after treatment (See supplemental figure 1). As previously reported by our group and others, both doxorubicin and vorinostat increased ceramide levels, including the most abundant species (C16-ceramide and C24:1 ceramide) by several fold (Figure 1b).
Fig 1. Selected chemotherapeutics induce plasma membrane ceramide.
(A) Text mining analysis using curated databases on NCBI literature (gene2pubmed and CTD database). Common chemotherapy compounds -x axis- were associated to sphingolipid terms -y axis-. Statistically significant relations between drugs and sphingolipid terms were plotted as adjusted p-values upon exact Fisher test analysis. P-values for doxorubicin/ceramide 0.00037, doxorubicin/nSMase2 0.040, vorinostat/ceramide 0.0443, vorinostat/nSMase2 0.0435 (B). Total cellular ceramide species were measured by mass spectrometry at 24h upon chemotherapeutic compounds treatment in HeLa cells. Natural logarithm of fold change versus control was plotted in a heatmap and clustered graphs. V-vehicle treatment (methanol). L-low doses (0.6 μM doxorubicin or 10 μM vorinostat), M-Medium doses (0.8 μM doxorubicin or 15 μM vorinostat), High doses (1 μM doxorubicin or 20 μM vorinostat). (C) Detection of ceramide at the plasma membrane by using recombinant pCDase to form sphingosine. Hydrolysis of plasma membrane ceramide to sphingosine is then measured by HPLC-mass spectrometry (see Methods). Doxorubicin and vorinostat did not induce sphingosine production, but when combined with pCDase, the levels of sphingosine dramatically increased, indicating formation of plasma membrane ceramide. (D) Validation of the anti-ceramide antibody. HeLa cells outer leaflet sphingomyelin and ceramide were stained with fluorescent-lysenin protein, which binds sphingomyelin (visualized in magenta), and with anti-ceramide antibody (visualized in green). Control cells (left panel) only showed lysenin but not ceramide staining. High doses of bSMase (100mU and 10 min) decreased lysenin staining and markedly increased cell staining with anti-ceramide antibody (middle panel). Treatment cells with 200mU pCDase reduced the ceramide staining (E) HeLa cells were treated with different chemotherapeutic compounds (doxorubicin 0.8uM and vorinostat 15uM) and stained using the anti-ceramide antibody. (F) Several images were quantified for anti-ceramide antibody showing statistical significance. Statistics one-way Anova Tukey post-test * p<0.05, *** p<0.001.
However, since the total cellular extract was used for the lipid measurements, the increase in ceramide species could occur in any sub-cellular membrane. Currently, there is a need for methods to quantify ceramide specifically at the plasma membrane. In order to quantitatively analyze the ceramide generated at the plasma membrane, we used an indirect analytical methodology, based on measuring the sphingosine produced by hydrolysis of ceramide at the plasma membrane. This method takes advantage that no basal or very little ceramide is detected in basal conditions, and very low levels of sphingosine are also detected basely. When unstimulated cells are treated with pCDase, no ceramide (or very few) is hydrolyzed to sphingosine. However, if cells are stimulated to generate ceramide in the plasma membrane, pCDase generates high levels of sphingosine, that can only come from ceramide in the plasma membrane. Then the produced sphingosine is in a 1:1 molar ratio with the hydrolyzed plasma membrane ceramide. At the 24h timepoint after treatment with the different drugs, ceramide was acutely hydrolyzed to sphingosine using exogenously added pCDase applied for 5 minutes. As shown in Figure 1c, significant levels of sphingosine were generated in response of the chemotherapeutic agents, thereby demonstrating the production of ceramide that is accessible to the exogenous pCDase. Collectively, these results establish that doxorubicin and vorinostat generate ceramide in the plasma membrane.
Next, we attempted to visualize this pool of ceramide in the plasma membrane using a ceramide antibody. To that end, we evaluated most of the available anti-ceramide antibodies in HeLa cells (3, 23, 45) even if their specificity to ceramide might not have been necessarily established. We ultimately chose to use the monoclonal antibody clone 2A2, a mouse IgM previously generated and validated against ceramide because it empirically stained the cells responding to exogenous recombinant sphingomyelinase (bSMase). To validate the antibody and monitor sphingomyelin hydrolysis, this last was visualized by using the binding probe lysenin conjugated to GFP protein as previously reported (7, 24). Thus, by using bSMase (100mU for 10 min), plasma membrane sphingomyelin was completely hydrolyzed (Figure 1d, left panel). Ceramide was barely detected in untreated cells, but dramatically increased in response to bSMase (Figure 1d, middle panel). To assure the ceramide-ligand specificity of the 2A2 monoclonal antibody, we employed soluble bacterial ceramidase cloned from Pseudomonas (pCDase) (7). The co-treatment of cells with pCDase and bSMase resulted in loss of binding of the ceramide antibody, thus verifying its specificity (Figure 1d, left panel).
Having experimentally confirmed the ligand specificity of the monoclonal antibody 2A2 against ceramide, we subsequently tested whether the increase in ceramide in response to the chemotherapeutical compounds could be detected in the plasma membrane. HeLa cells were treated with the ceramide inducers, and after 24h, a positive signal of anti-2A2 ceramide monoclonal antibody was detected at the plasma membrane, suggesting that at least part of the ceramide generated was located in the plasma membrane (Figure 1e shows representative images). Figure 1f plots the quantification of all images (as detailed in Materials and Methods). The anti-ceramide 2A2 monoclonal antibody in the immunofluorescent experiment provided a qualitative measurement of ceramide production in the plasma membrane. Based on these results, we conclude that treatment with these two chemotherapeutical compounds induced ceramide in the plasma membrane.
Regulating the generation of plasma-membrane ceramide at physiological levels.
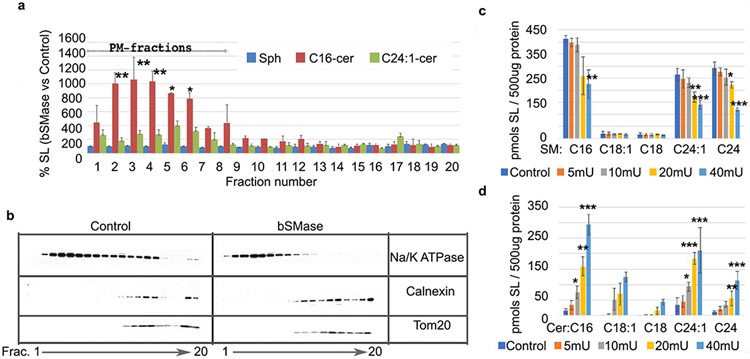
Besides generating ceramide at the plasma membrane, doxorubicin and vorinostat are known to trigger many cellular pathways (46). To determine if the formation of plasma membrane ceramide had a specific function, we attempted to isolate this putative plasma membrane signaling event from other compartments by utilizing recombinant bSMase which hydrolyzes plasma membrane sphingomyelin, with two caveats: 1) it has been used in large amounts removing most/all plasma membrane sphingomyelin and possibly acting on other phospholipids, and 2) the subsequent re-localization of ceramide has not been ascertained. In previous work, we showed that bSMase-generated ceramide is not metabolized to other known metabolites such as sphingosine, glycolipids, or ceramide 1-phosphate within a short (few minutes) timeframe (7). To establish the localization of this ceramide in the early phases after its generation, we measured the sphingolipid content in the different cellular membranes by liquid chromatography/tandem mass spectrometry (LC/MS/MS) upon cell fractionation (Figure 2a) and performed Western blotting to assign cellular compartment distribution (Figure 2b). Fractions containing Na-K ATPase but not the other cellular markers were considered plasma membrane fractions. Heavier membranes (e.g., ER-Golgi, mitochondria) were evaluated by using antibodies specific for calnexin and tom 20. The addition of bSMase for 10 minutes caused accumulation of ceramide in the plasma membrane fractions, and no detectable changes were found in other membranes. Other measured sphingolipids such as sphingosine (Figure 2a), along with GlcCer, ceramide-1-phosphate, and sphingosine-1-phospate did not show any detectable changes. Therefore, bSMase not only acts on the plasma membrane generating plasma membrane ceramide, but within the following 10 minutes, this ceramide is not metabolized to other sphingolipids, and it is not mobilized to other cellular membranes, but remained, at the plasma membrane. These conditions offer a time-window of at least 10 minutes after bSMase that can be used as a tool to study ceramide signaling at the plasma membrane isolated from more long term ceramide metabolism or after being translocated to other compartments.
Figure 2. Generation of ceramide at the plasma membrane in physiological conditions.
(A) Percentage in ceramide and sphingosine content measured upon bSMase treatment 100mU for 10 min. After treatment, cellular membranes were separated using gradient centrifugation optimized to separate plasma membrane from heavier membranes. Frc1-20 refers to the first twenty fractions collected. Sphingosine (Sph); ceramide with C16-acyl chain (C16Cer); ceramide with C24:1 acyl chain (C24:1Cer). (B) The identification of the different cellular membranes was carried out using stablished marker proteins by Western Blot. For plasma membrane, the Na/K ATPase was used. For heavier membranes, calnexin and Tom20 were used as markers for endoplasmic reticulum and mitochondria, respectively. (C) Mass spectrometry analysis of sphingomyelin content per 500ug cellular protein after treatment with increasing amounts of bSMase (in milliunits). (D) Same samples were also processed for ceramide quantification by mass spectrometry analysis. Statistics one–way Anova Tukey post test vs control. * p<0.05, ** p<0.01, *** p<0.001
Next, we limited the amount of hydrolysis of SM in the plasma membrane in order to mimic more physiologic and pharmacologic situations of activation of endogenous SMases, which have been shown to hydrolyze up to 20% of total sphingomyelin (47). At 10mU bSMase, 10-20% of sphingomyelin was hydrolyzed, and statistically significant levels of ceramide were generated (Figures 2c and 2d). These results demonstrate that physiological increases in ceramide species and physiological decreases of sphingomyelin species can be mimicked by using bSMase and by isolating this signaling event from other possible downstream targets and compartments. Thus, for subsequent experiments, we utilized 10mU bSMase applied for 3 minutes to generate bioactive ceramide at the plasma membrane.
Phosphoproteomic analysis reveals plasma membrane ceramide is involved in several biological phenomena with cell adhesion proteins constituting a dominant functional group.
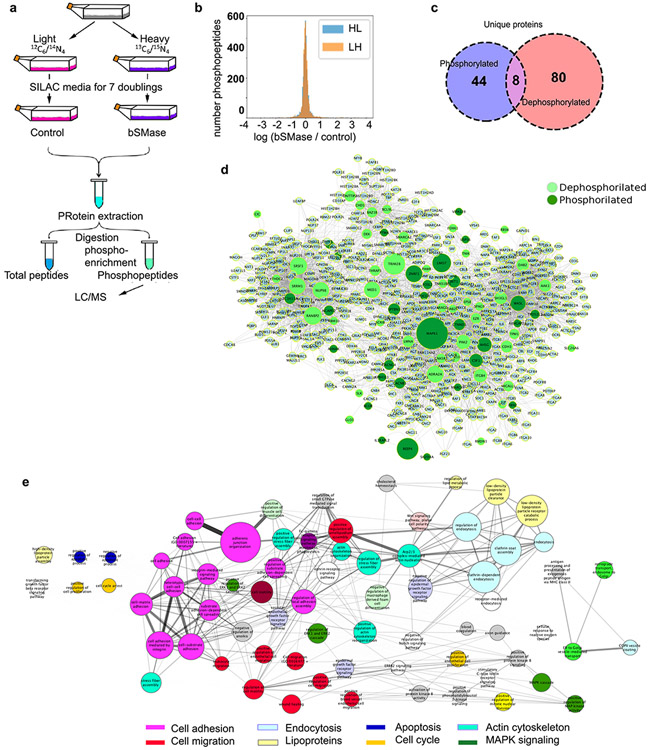
Ceramide activated protein phosphatases have emerged as mediators of many of the cell responses triggered by ceramide (1, 48). Moreover, protein kinases, especially PKCz, have also been advanced as molecular targets for ceramide action. Therefore, we evaluated the consequences of ceramide generation in the plasma membrane on the phosphorylation status of proteins utilizing phosphoproteomics. HeLa cells were cultured in SILAC media (as detailed in Materials and Methods) and treated with bSMase (Figure 3a). Around 4,000 phosphopeptides were detected in both control and bSMase-treated samples. From this experimental dataset, most phosphopeptides did not show changes in their phosphorylation levels in response to bSMase, but a significant fraction showed a decrease in phosphorylation, and another subset showed an increase in their phosphorylation level. For this study, phosphopeptides were selected with the following statistical criteria: assuming a normal distribution (49) for the changes in phosphorylation levels (control/ treated, in log2 scale), ratios between Z-score<−1.6 and Z-Score>1.6 reject the null hypothesis with a significance of α=0.05 (Figure 3b). This selection yielded 114 phosphopeptides that were dephosphorylated, corresponding to 88 unique proteins; 117 phosphopeptides showed increased phosphorylation corresponding to 52 unique proteins. Both ‘dephosphorylated’ and ‘phosphorylated’ groups had 8 proteins in common, meaning that these proteins were dephosphorylated at least in one residue, but phosphorylated on at least another distinct residue (Figure 3c). Supplemental table 2 lists the proteins dephosphorylated and phosphorylated in response to ceramide generation.
Figure 3. Interrogating ceramide function at the plasma membrane.
(A) Scheme of the phosphoproteomics experiment. HeLa cells were cultured in SILAC media containing (isotopically) heavy or light amino acids as described in Materials and Methods for at least 7 doubling to ensure incorporation in all cellular proteins. Cells fed in light conditions constituted the untreated (control) group, while cells fed with heavy amino acids were treated with bSMase 10mU for 2 min (LH experiment). A replicate experiment was also performed treating cells fed in light conditions with bSMase while cells fed with heavy amino acids were untreated (HL experiment). Cells were collected in lysis solution as indicated in Materials and Methods, and light and heavy protein lysates were mixed, proteins were digested, phospho-enriched and phospho-peptides were quantified by mass spectrometry analysis. (B) The distribution of both experiments on the natural logarithm of the ratio (treated/ control) of the detected abundance of each phospho-peptide detected followed a normal distribution. (C) Statistically significant ratios were assigned to phosphorylation or dephosphorylation events in response of treatment (p-value 0.05). The Venn diagram shows the amount of proteins for each group. Note that 8 proteins contained both events but on different phospho-sites. (D). Network analysis for proteins dephosphorylated (light green) and phosphorylated (dark green) enriched with interactions in Reactome+ String + KEGG + innateDB. To reduce less significant edges, the network was filtered for 3 or more degree of connectivity using NetworkX software. (E) Pathway analysis of the build network was performed using Reactome database and more statistically significant pathways (Fisher test) were selected. Pathways cluster node size are proportional to the intra-connectivity value and edges represent connections between proteins from within pathway clusters. Pathway cluster under a general GO term within the same hierarchy biology are colored with the same color (e.g. cell adhesion pathways are colored in magenta).
In order to analyze if some of these ceramide-regulated proteins were part of common pathways, we performed an enrichment analysis using only curated information from different interactome databases. We combined information from STRING (score > 700)(50) + Reactome (51) + KEGG (52) + innateDB (53) databases to build a connectivity network. The network was filtered removing nodes with less than 2 degree of connectivity (54) (Figure 3d). The network was then clustered based on biological pathways (Figure 3e). Interestingly, the resulting network showed pathways classically related to ceramide such as apoptosis (55), cell cycle regulation (56), endocytosis (57), and regulation of MAPK pathways (58, 59). However, three sets of pathways: ‘actin cytoskeleton, ‘cell migration’ and ‘cell adhesion’ showed the highest scores in intra-pathway connectivity and inter-pathway connectivity. We decided to focus on these biologies, since these were the most significant biologies in response to ceramide, they were highly inter-connected, and they are biologies were not commonly attributed to ceramide.
Plasma membrane ceramide regulates cell adhesion.
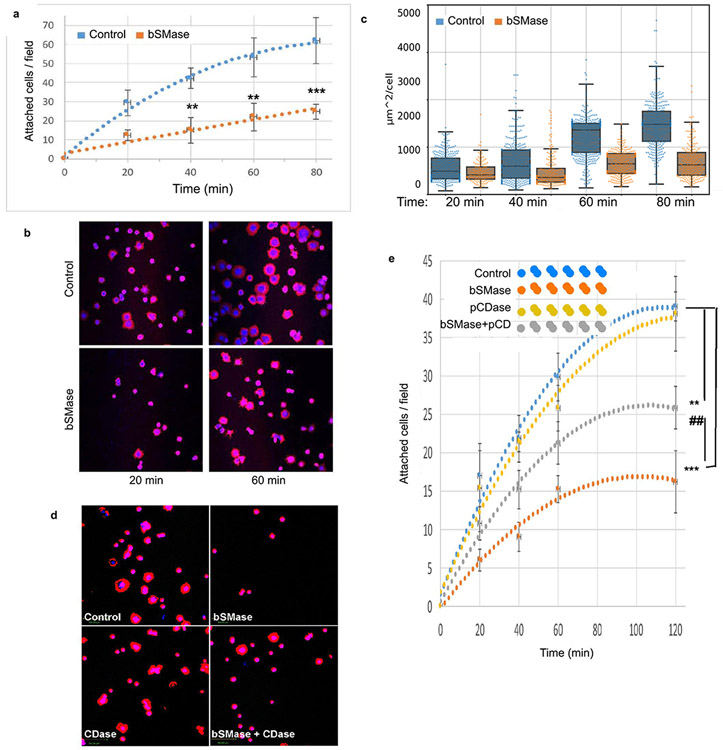
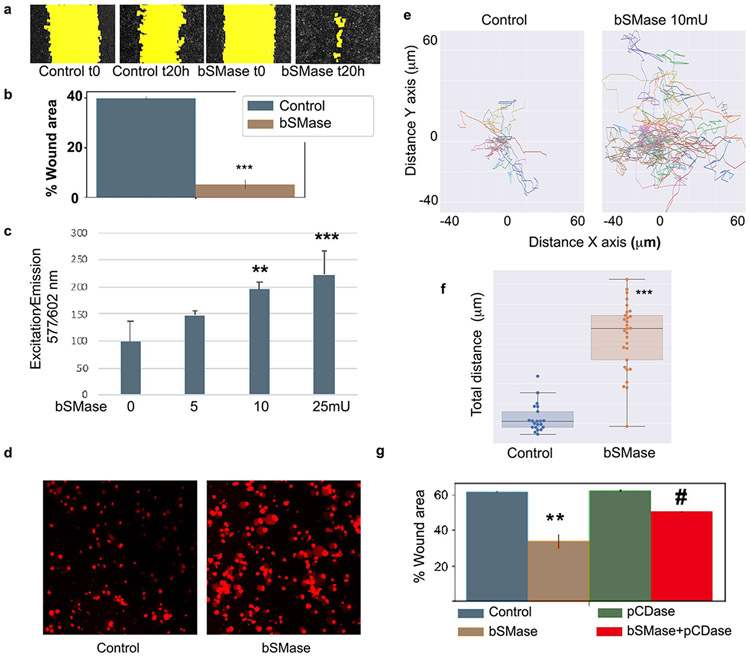
Since cell adhesion was the main functional class found for the largest number of identified phosphoproteins, we aimed to explore whether this prediction translates into specific cellular activities. Thus, HeLa cells treated with bSMase were tested for cell adhesion as detailed in Materials and Methods. Treatment with bSMase dramatically reduced the ability of cells to attach on fibronectin (Figure 4a), suggesting a critical role played by ceramide in this setting.
Figure 4. Plasma membrane ceramide regulates cell adhesion.
(A) HeLa cells were scrapped from culture dish in 10mM EDTA, washed in PBS by centrifugation and resuspended in serum free DMEM media. Cell suspension was treated with bSMase 10mU or vehicle for 2 min and washed in PBS and resuspended in low FBS media (1%). Cells were then re-plated in fibronectin1-coated dishes as specified in methods section. At different time points after re-plating, the dishes were washed from floating cells and the cells fixed with paraformaldehyde. Attached cells were stained with phalloidin-rhodamine and DAPI and visualized using fluorescent microscopy (representative images in B). Images from independent experiments (n=15) were used for cell counting and (C) cell area quantification. Area quantification represents n>1000 cells and was performed by image processing software as described in methods. (D) Adhesion assay was performed treating cells in suspension prior to re-attachment with bSMase, pCDase or [bSMase + pCDase] together. Representative images shown, and (E) quantification. Statistics one–way Anova Tukey post test vs control (*) or vs pCDase-combined treatment (#). * or # p<0.05, ** or ## p<0.01, *** or ### p<0.001.
As an immediate consequence of cell adhesion, we also measured cell spreading which requires the deformation of the plasma membrane and the formation of cell–substrate attachments. The area of the attached cells was measured and monitored for each attached cell during the 2-hour period of the assay (Figures 4b and 4c). The results showed that plasma membrane sphingolipids not only regulate the initial cell adhesion, but also affected cell spreading.
Given the established reciprocal changes in sphingomyelin and ceramide, we reasoned that it would be important to determine whether the effects of bSMase were due to the gain of ceramide or to the loss of sphingomyelin. To differentiate between these possibilities, HeLa cells were co-treated with an excess of recombinant soluble pCDase to hydrolyze ceramide formed by the action of bSMase. Figures 4d and 4e show that exogenously added pCDase had no effect on cell adhesion per se. However, pCDase significantly countered the effect of SMase. These results suggest that the generation of ceramide, and not the loss of sphingomyelin, was the main mediator of effects on cell adhesion to the extracellular matrix. Also, considering that pCDase acts only on ceramides, the results further support that the phenotype is mediated by the ceramide product specifically and not any subsequent metabolite(s).
Plasma membrane ceramide enhances cell migration.
Functionally, cell adhesion has been implicated in both increased cell migration with loss of adhesion allowing cells to migrate and in impairment of cell migration through loss of the required mechanical support on the extracellular matrix during movement. Moreover, cell migration can be evaluated by using different assays that assess different types of cell migration. The wound-healing assay was first chosen due to its technical simplicity, which enables simultaneous multiplex testing of many different experimental conditions and because it evaluates collective cell migration (60). HeLa cells were seeded on fibronectin at confluent density. A scratch was made, and migration of cells was serially monitored at different times. After 20h, 40-60% of the scratch was covered, and this time point was chosen to evaluate the effects of plasma membrane ceramide manipulation. bSMase (10mU/ml) treatment dramatically increased cell migration, with cells practically covering the entire area by 20h (representative images are shown in Figure 5a). Quantification of independent experiments is plotted in Figure 5b (n=15, p<0.001). These results show that the generation of ceramide at the plasma membrane reduced adhesion and accelerated migration of cells.
Figure 5. Generation of plasma membrane ceramide increases cell migration.
(A) Scratch wound healing migration assay in HeLa cells treated with bSMase was performed for 20h. Random images at time zero and after 20h are shown. Yellow color highlights the wound area. (B) quantification of independent experiments (n=10) showed that bSMase induced cell migration. Statistics t-test *** p<0.001. (C) Cell migration transwell using Fluoroblock. Different amounts of bSMase were added into the upper well for 2 min, washed and cells were left to migrate for 16h. Cells were stained with cell tracker and fluorescence was read from the bottom using a plate reader. Statistics one-way ANOVA Tukey posthoc vs bSMase 0 mU. (D) Random images of migrated cells stained with Red cell tracker, and visualized in a fluorescent microscopy. (E) Live imaging cell migration was performed in confocal dishes coated with fibronectin 1. Cells were plated at low confluency and low serum media. bSMase was added to plates for 2 min and images were taken every 15 min for 16h. Individual cell position was tracked using imaging processing software as detailed in methods. Individual cells tracks were plotted for qualitative view and (F) total distance for each individual cell was measured as the sum of the travelled distance each 15 min up to 16h. Student t-test *** p<0.001 (G) Scratch assay on cells treated with bSMase, pCDase and with the combination of both [bSMase+pCDase]. One-Way ANOVA Tukey post hoc test * p<0.01. # One-way ANOVA Tukey posthoc vs bSMase
Next, single cell migration was measured in transwell assays on fibronectin 1-coated membranes. HeLa cells responded to increasing amounts of bSMase with increasing cell migration in a concentration-dependent manner (Figure 5c, and representative images of migrated cells on Figure 5d).
Finally, we aimed to visualize the increase in cell migration by using live imaging microscopy. The positions of individual cells were tracked for 16h. Cells stimulated with bSMase underwent a marked increase in cell migration (Figure 5e, quantified in 5f and video 1). These results demonstrate that hydrolysis of SM to produce ceramide at the plasma membrane is sufficient to trigger an increase in cell migration phenotype.
Moreover, cells co-treated with pCDase overcame the effects of bSMase on migration (Figure 5g), confirming that the effects on cell migration were mediated by the production of ceramide and not by loss of sphingomyelin or by further metabolism of ceramide (to sphingosine and then to S1P).
Collectively, these results strongly suggested that plasma membrane ceramide modulates cell adhesion, spreading, and migration.
Activation of endogenous programs for the generation of plasma membrane ceramide.
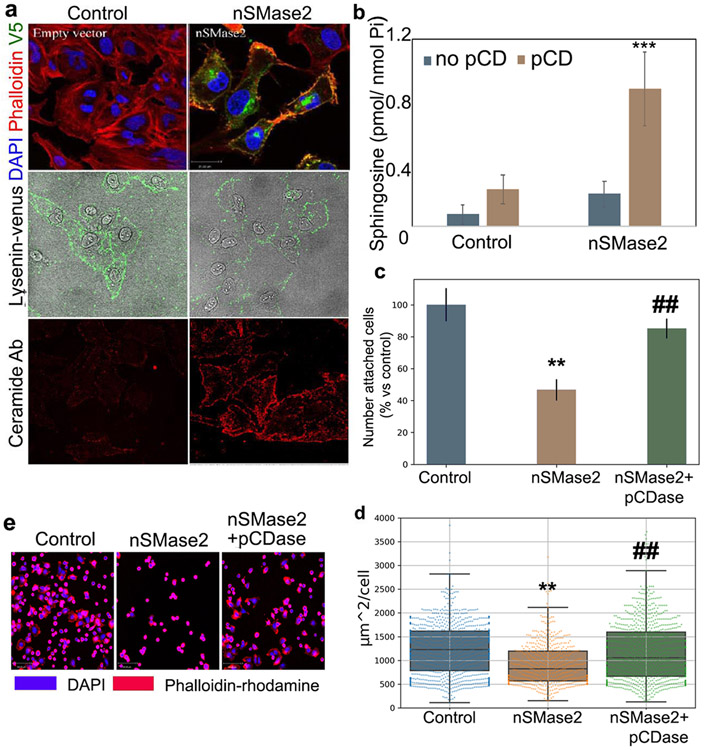
Next, it became important to determine whether endogenous enzymes associated with ceramide generation were able to induce ceramide at the plasma membrane and to regulate cell adhesion. We focused on nSMase2 because it has been reported to respond to doxorubicin and vorinostat increasing the mRNA and protein levels, and it has also been reported to be located at the plasma membrane (26, 61, 62). However, its ability to generate ceramide at the plasma membrane has not been documented. To test if expression of this enzyme (26) would be sufficient to generate ceramide at the plasma membrane and trigger the same biological phenotypes as seen with the bSMase treatment, we first expressed and confirmed by immunostaining that nSMase2 was localized at the plasma membrane in HeLa cells (Figure 6a upper panels). As shown with the 2A2 anti-ceramide monoclonal antibody, recombinant nSMase2 was capable of generating qualitative amounts of ceramide (lower panels) in the plasma membrane; however, the effects on changes in the plasma membrane sphingomyelin were only modest (Figure 6a middle panels). The presence of ceramide in the plasma membrane was also confirmed by using recombinant pCDase as previously shown. HeLa cells transfected with nSMase2 were treated with pCDase, and the sphingosine generated was measured. Control cells did not show a statistically significant increase in sphingosine levels whereas cells expressing nSMase2 showed an increase in sphingosine, concluding that overexpressing nSMase2 is sufficient to increase plasma membrane ceramide (Figure 6b).
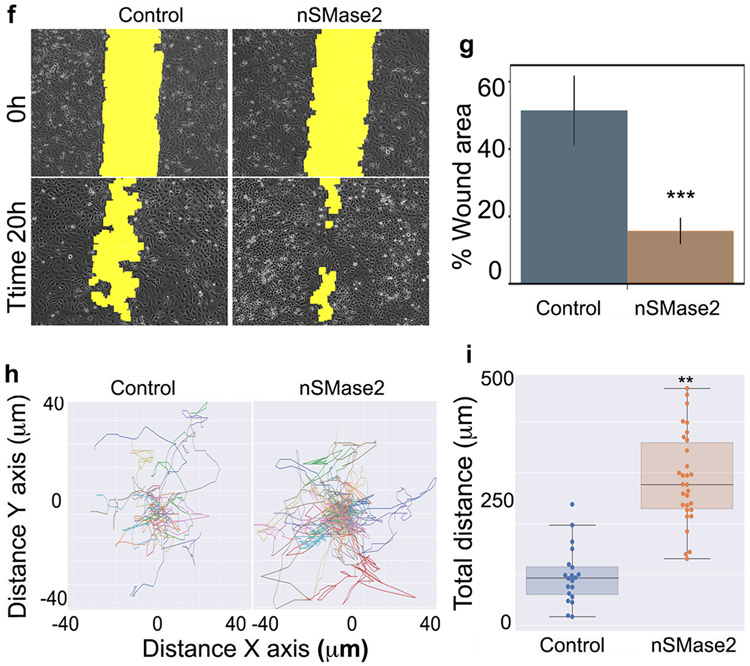
Figure 6. Expression of nSMase2 is active at the plasma membrane and recapitulates bSMase effects on cell adhesion.
(A) V5-tagged nSMase2 expressing HeLa cells and visualized (upper panels :V5 staining in green, phalloidin-rhodamine [F-actin cytoskeleton] in red, and DAPI [nucleus] in blue). The effects of the enzymes in the plasma membrane were also visualized by staining outer leaflet sphingomyelin (Middle panels using venus-lysenin, green) and ceramide (bottom panels, ceramide antibody in red). (B) Control cells and cells expressing recombinant nSMase2 for 24h were treated in serum free media with PBS (blue bars) or 200mU of pCDase (brown bars) for 5 min. Cells were collected in organic solvent and analyzed for sphingosine content by HPLC-MS. Samples were normalized based on inorganic phosphate (Pi) content. (C) Cells expressing the neutral nSMase2 were evaluated for cell adhesion. Cells were scraped from dish in EDTA and re-plated in fibronectin1-coated dishes. After 60 min, floating cells were washed, and attached cells were fixed with paraformaldehyde. Cells were stained for nuclei (DAPI) and F-actin cytoskeleton (rhodamine-phalloidin) and visualized in a fluorescent microscopy. Cells were counted in 10 independent experiments and (D) cell area was measured in >1000 cells using image processing software as detailed in methods. (E) Random images are shown. (F) Scratch wound healing migration assay in HeLa cells overexpressing nSMase2 was performed for 20h. Random images at time zero and after 20h are shown. Yellow color highlights the wound area.(G) Fifteen independent experiments were quantified. Statistics t-test *** p<0.001. Random images are displayed. (H) Control cells and cells expressing nSMase2 for 24 were seeded on fibronectin FN1. Cell migration was recorded for 16h, and individual cell tracks were were graphed on a dispersion plot. (I) Total distance for each individual track in (H) was calculated and total distance for 25 individual cells were plotted. Statistical test were t-Test (* p<0.05, ** p<0.01, *** p<0.0001) for G and I, one-way ANOVA with Tukey post hoc test for C and D. Two-way ANOVA for B. For C and D, * vs control, # vs nSMase.
Cell adhesion and spreading were next evaluated under these experimental conditions. Similar to the bSMase treatment, expression of nSMase2 impaired both functional activities when compared to non-transfected control cells. As observed for bSMase, the addition of pCDase during the cell adhesion assay was sufficient to revert the effects on loss of adhesion and cell spreading (Figure 6c and 6d, representative images in Figure 6e). These results indicated that the effects on cell adhesion and cell spreading by nSMase2 were mediated by plasma membrane ceramide.
Since the effects of ceramide on cell adhesion also interfered in migration when bSMase was used, it was predicted that overexpressing of nSMAse2 also should modulate cell migration. HeLa cells were transfected with nSMase2 and evaluated for changes in cell migration using the wound healing assay. Cells expressing nSMase2 showed accelerated rates in closing the wound, suggesting an effect on cell migration (examples of representative experiments in Figure 6f, quantified in Figure 6g). The effect on cell migration were extended by tracking cell motility on individual cells following cell motility on FN1 under the microscope taking pictures every 10 min for 20h, confirming the effect of nSMase2 on cell motility (examples of individual tracks in Figure 6h, quantified in Figure 6i)
From these experiments, we concluded that increased expression of nSMase2 generates ceramide at the plasma membrane and this level of ceramide was sufficient to induce loss of cell adhesion, loss of cell spreading increasing cell migration rate.
Doxorubicin and vorinostat induced loss of cell adhesion and increased cell migration mediated by nSMase2 and ceramide at the plasma membrane.
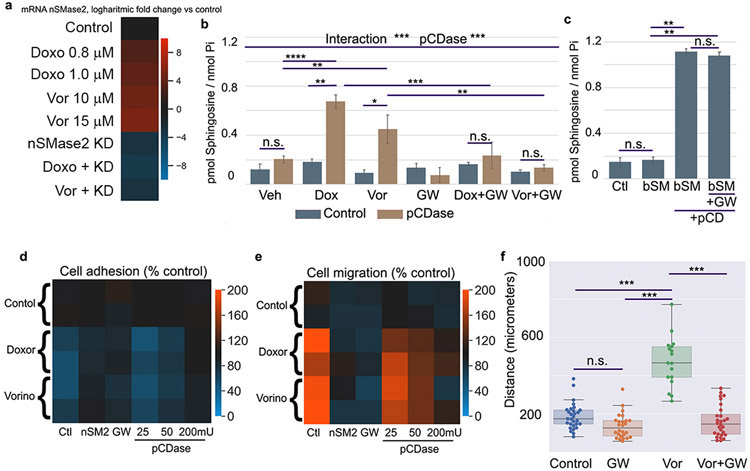
As previously reported (15), both doxorubicin and vorinostat induce cellular ceramide, and in figure 1 we showed this ceramide is at least partly located in the plasma membrane. We reasoned that generation of ceramide at the plasma membrane level may be sufficient to induce cell detachment and cell migration. Taking this experimental evidence into account, we predicted that vorinostat and doxorubicin may also induce loss of cell adhesion and stimulate migration. To our knowledge, there are no previous reports on doxorubicin or vorinostat modulating cellular adhesion. However, there are a few reports where a connection was found between cell adhesion molecules and doxorubicin. For instance, FAK protein was found to antagonize doxorubicin cardiotoxicity (63) and doxorubicin when combined with other drugs reduced adhesion of breast cancer cells (64). Moreover, these two compounds were selected in this study because they have been shown to induce transcription of nSMase2. We hypothesized that induction of nSMase2 by these compounds will generate ceramide in the plasma membrane that would trigger loss of cell adhesion and induce cell migration. In order to test this hypothesis, first we validated that the two compounds were also capable to induce nSMase2 in HeLa cells. As reported for other cell lines, both compounds at sublethal concentrations were capable to induce nSMase2 mRNA in a dose dependent manner in HeLa cells (Figure 7a). Next, we tested if the generation of plasma membrane ceramide induced by doxorubicin and vorinostat was dependent on nSMase2, and its generation could be blocked by inhibition of nSMase2. As shown in figure 1, we measured indirectly ceramide by using pCDase and measuring the generation of sphingosine, and as shown in figure 1, both compounds generated ceramide at the plasma membrane. Inhibition of nSMase2 was accomplished using the pharmacological inhibitor GW-4869. Inhibition of nSMase2 blocked the increase of plasma membrane ceramide in response to doxorubicin and vorinostat (Figure 7b). GW-4869 had no statistically significant effect in control conditions, but the mean of ceramide measured was decreased. In order to test if GW-4869 per se could affect the levels of ceramide independently of nSMase2 activation, GW-4869 was tested on plasma membrane ceramide generated using bSMase. In this scenario, GW-4869 failed to block bSMase and ceramide generation at the plasma membrane. This last result confirmed that the effects of GW-4869 were due to blockage of ceramide generation, and not secondary effects on plasma membrane sphingolipids (Figure 7c).
Figure 7. Chemotherapeutics induce loss of cell adhesion and induction of cell migration, mediated through plasma membrane nSMase2/ceramide axis.
(A) Doxorubicin and vorinostat at the indicated concentrations were evaluated for expression of nSMase2 measured by RT PCR. Messenger RNA was quantitated at 24h after induction. Validation of siRNA sequences on control (nSMAse KD) and induced samples (doxo +KD and Vor + KD) by RT-PCR. (B) Control cells and cells treated with doxorubicin 0.8 μM, vorinostat 10 μM, or co-treated with GW, were treated in serum free media with PBS (blue bars) or 200mU of pCDase (brown bars) for 5 min. Cells were collected in organic solvent and analyzed for sphingosine content by HPLC-MS. Samples were normalized based on inorganic phosphate (Pi) content. Statistics were two-way ANOVA with post hoc analysis. N.s – no significant. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. (C) Cells in serum free media were treated with PBS (control) or bSMase (bSM) 10mU for 3 min. Media was replaced for fresh serum free media and treated with pCDase (+pCD) for 20 min. Sphingosine was quantified as described in panel (B). Samples GW were pre-treated for 20h with GW inhibitor. (D) Heatmap summarizing the results on cell adhesion assays, being orange color a theorical gain of adhesion and blue loss in cell adhesion compared to untreated control cells (given value of 100% attachment). Chemotherapeutical compounds doxorubicin (Doxor) and vorinostat (Vorino) resulted in loss of cell adhesion. Inhibition of nSMase2 with siRNA or GW (nSMase inhibitor) blocked the effects of doxorubicin and vorinostat on cell adhesion. Cells treated with increasing doses of pCDase (25, 50 and 200mU) before re-attachment overcame the effects on loss of cell adhesion in a dose dependent manner. Two independent experiments are shown per each condition) (E) Heatmap summarizing scratch assays on cells treated with the two chemotherapeutical compounds. Orange color represents increase in cell migration (as % vs control untreated cells) and blue color a loss on cell migration vs control cells. Each condition is shown as two independent experiments. The two compounds, doxorubicin (Doxor) and vorinostat (Vorino) (F) Individual cell migration assay on cells treated with the inhibitors GW upon chemotherapeutical compound vorinostat.
The previous experiments allowed us to modulate the levels of plasma membrane ceramide generated in response to the chemotherapeutical compounds and use these tools to study if this specific pool of ceramide had the same biology that the one mimicked using bSMase. HeLa cells were treated with sub-lethal doses of vorinostat and doxorubicin, and the two compounds resulted in loss of cell adhesion. Inhibition of nSMase2 activity was evaluated using the inhibitor GW-4869, and it blocked the loss of cell adhesion induced by the two compounds (Figure 7d). Knock down of nSMase2 achieved the same results proving that the effects of GW-4869 were due to targeting nSMase2. The use of recombinant pCDase confirmed the role of plasma membrane ceramide on doxorubicin and vorinostat effects on cell adhesion (Figure 7d). Therefore, the loss of cell adhesion induced by the chemotherapeutical compounds was regulated by ceramide generated by induction of nSMase2, and this ceramide was acting at the plasma membrane level.
From the previous results, the generation of ceramide from doxorubicin and vorinostat is also expected to modulate cell migration. Again, we used the wound-healing assay to screen for several conditions and summarized as % cell migration versus untreated control cells (Figure 7e). First, we confirmed that both chemotherapeutical compounds induce cell migration in HeLa cells. Then we used both siRNA for nSMase2 and GW-4869 inhibitor to block nSMase2 effects on ceramide, and both methods completely abolished the increase in cell migration returning to basal levels, confirming the role of nSMase2 on allowing the induction of cell migration. To consolidate the role of the ceramide formation we used pCDase during the cell migration assay. Increasing concentration of pCDase resulted in more inhibition on cell migration. This last result also confirmed that ceramide produced by nSMase2 upon chemotherapy induction, was not only necessary to trigger cell migration and it was still located at the plasma membrane.
Interestingly, some chemotherapeutical compounds, including doxorubicin and vorinostat, have been reported to induce cell migration in some cell lines, contributing to the risk of cancer invasion to surrounding tissues and generating metastatic lesions. Since our data showed that loss of cell adhesion (which is shown to be related to a potential increase in cell invasion) and gain in cell migration were dependent on plasma membrane ceramide, we tested GW-4869 to block random cell migration on individually tracked cells upon stimulation. We chose vorinostat for this assay (unfortunately, doxorubicin-treated cells underwent cell death when they were exposed to the laser light in the microscope, rendering it technically unreliable to track them). Cells exposed to sublethal doses of vorinostat were stimulated for random migration. GW-4869, which had no effect on basal migration, blocked vorinostat- induced migration (Figure 7f). This assay not only confirmed the results on the wound-healing assay using an independent approach, but it also demonstrates that a small molecule inhibitor of nSMase2 can be used to block individual cell migration potentially induced after chemotherapy treatment.
Together, these results showed that some chemotherapeutical compounds, at sublethal doses, induce a loss in cell adhesion followed by an increase in both collective and individual cell migration. This gain of function can be blocked by specifically targeting plasma membrane ceramide, which can be achieved by inhibiting nSMase2 activity.
Discussion
In this study, we provide evidence of a specific pathway launched by nSMase2 acting on plasma membrane sphingomyelin to generate ceramide in the same compartment. This ceramide acutely triggers changes in the phosphorylation status of a set of proteins, which were predicted in-silico to be part of a network regulating cell adhesion and cell migration. Experimental results confirmed this prediction and defined these biologies as a cell anti-adhesion program resulting in enhancement of cell migration. Furthermore, this mechanism was found to fit the biological niche downstream of chemotherapeutical compounds inducing cell migration. Finally, enzymatic or pharmacological modulation of the cellular surface ceramide prevented the effects of these chemotherapeutics on enhancing cell migration, suggesting new avenues in optimizing cancer cell therapy.
A major conclusion from this study is in defining a mechanism-based specific action of sublethal chemotherapeutic agents on cell adhesion and migration. It has been suggested for many years that many chemotherapeutical treatments can affect cancer cells in a paradoxical way that might increase tumor cell spreading and metastatic dissemination (65, 66). Taxanes (65, 67), anthracyclines such as doxorubicin (65), and platinum salts such as cisplatin (67) are some of the drugs implicated in potentially increasing metastatic tumor cell spreading of cancers. Reported molecular mechanisms to explain these effects involve overexpression and or activation of adhesion proteins, such as FAK (68), ROCK (69), RhoA/MLC (22), ezrin (69, 70), src, (70) and EpCAM (71). However, the mechanism on how these proteins are regulated by chemotherapy or whether each reported adhesion protein works individually towards regulation of cell adhesion, or they actually work as part of a network has not being explored. The results in this work reveal that a pool of ceramide in the plasma membrane mediates the effects of at least two chemotherapeutical compounds on enhancing cell migration and loss of cell adhesion. This hypothesis is supported by several lines of evidence. First, Doxorubicin and vorinostat induced expression of nSMase2 which was necessary to trigger loss of cell adhesion and cell migration. Second, overexpression of recombinant nSMase2 was sufficient to trigger the same biology. Third, the use of bSMase to artificially generate ceramide at the plasma membrane, at physiologically and pharmacologically relevant levels, recapitulated nSMase2 effects on cell adhesion and migration. Moreover, plasma membrane ceramide generated using bSMase was sufficient to regulate a network of cell adhesion proteins within just 2-3 minutes after its generation. Fourth, Removal of this pool of ceramide using recombinant enzymes blocked the effects on adhesion and migration, independently from the source of this plasma membrane ceramide (bSMase, nSMase2, doxorubicin and vorinostat), and finally, pharmaceutical or genetic inactivation of nSMase blocked the effects on adhesion and migration upon doxorubicin and vorinostat treatment, but failed to block the effects upon bSMase treatment. Altogether, although we cannot rule out that other sphingolipids can originate from metabolism of this identified pool of ceramide and might be also involved in the studied biologies, these results strongly suggest that the mechanism by which chemotherapy induces cell migration requires ceramide at the plasma membrane, generated by increase expression of nSMase2.
These results are potentially very significant for cancer treatment. As mentioned above, sublethal doses of chemotherapy might increase the chances of cancer cell invasion and eventually cancer metastasis. Here, at sublethal concentrations of two chemotherapeutics, we still measure significant ceramide production, and we find it is necessary and sufficient to mediate the effects on cell adhesion and migration. Based on the present study, it could be speculated that reducing ceramide levels at the plasma membrane could be used combined with chemotherapy to reduce the potentially undesired effects on cell spreading. We showed that pharmacological inhibition of nSMase2 using GW significantly reduced the production of plasma membrane ceramide and completely blocks the effects of chemotherapy on cell migration and cell adhesion, and potentially reduce the risk of cancer spreading.
Another major conclusion from this study is that ceramide can act at independent subcellular compartments to couple to specific biologies. One of the conclusions of the ‘Many ceramides’ hypothesis was that different pools of ceramide might coexist in the cells, possibly regulating different biological processes (9). Changes in ceramide levels upon cell treatment or cell stress are routinely measured at the whole cell levels (30), thus, mixing all subcellular components and treating ceramide as one unique entity. This might represent a problem by ignoring or masking changes in some compartments compared to other compartments that may contribute more to the total mass of cellular ceramide, but with potentially dramatic effects on the cell fate. In that way, only changes in ceramide that are comparable to the whole cellular ceramide pool would be detected with the possibility to be related to certain biologies, for example as it has been done for cell death (55). There are just a few examples where a specific pool of ceramide have been suggested to be generated with functionality in one particular compartment (5, 72). However, this idea has not been further developed, probably because of the lack of tools that allow us to detect ceramide in specific compartments, and to be able to modulate ceramide levels in that specific compartment. Ceramide antibodies have been successfully used to detect ceramide production in individual cells and in tissues (45, 73). However, they have not been used to dissect the ceramide topology, and based on individual images, it is a qualitative more than a quantitative tool. We have developed a new assay to measure ceramide at the plasma membrane based on quantifying sphingosine as the result of the hydrolysis of the plasma membrane ceramide by adding exogenous recombinant and purified CDase and measuring the generated sphingosine by mass spectrometry. This method has allowed us to detect changes in ceramide levels in the plasma membrane, that otherwise would have been diluted by the whole cell ceramide. With respect to chemotherapeutics, we know from previous studies and this one that ceramide production upon chemotherapy is capable to mediate different biologies. For example, at lethal concentrations, doxorubicin and vorinostat are reported to increase ceramide, and this ceramide is necessary to induce cell death (74-76). This would be consistent with the ability of mitochondrial ceramide to induce cell death (5, 77). On the other hand, the current results not only demonstrate that plasma membrane ceramide has functions different from those classically assigned to cellular ceramide, but new functions not previously directly attributed to ceramide. Thus, compartment specific functions of ceramide are becoming indispensable for our understanding of the functions of various ceramides. Moreover, we can hypothesize that analogous tools working in different subcellular compartments would unveil new functions of ceramide, or at least assign known ceramide functions to specific pools of ceramide.
This work supports the idea of ceramide as a second messenger, where low or non-present ceramide levels in the plasma membrane increase in response of signaling events resulting in changes in the phosphorylation status of several proteins, many of them responsible to regulate biologically significant changes in cell adhesion and migration. Although we have identified the first ceramide responders towards regulation of these biologies, we have not identified the direct targets and mediators of ceramide. However, we suspect that since protein kinases and protein phosphatases have been reported to be directly modulated by ceramide in vitro, and the effects we found are in changes in phosphorylation levels, we suspect a direct o very first responder of ceramide would be a protein kinase or a protein phosphatase. Our group is currently working towards this direction.
In this work we have also advanced the understanding on nSMase2. The localization and place of actions of nSMase2 are still controversial, all works on localization are based on tagged recombinant forms, and not based the endogenous enzyme (26, 41). Recombinant forms of nSMase2 localize in the ER-Golgi and in the plasma membrane. Less is known about its substrate subcellular localization. Here, we shown that both physiologically induced and recombinant overexpression of nSMase2 are capable to form ceramide at the plasma membrane. This function in the plasma membrane was highly suspected, but not confirmed. In this work we were able to visualize with the ceramide antibody ceramide generation at the plasma membrane upon induction of endogenous nSMase2 or overexpression of nSMase2. Using recombinant pCDase coupled to detection of sphingosine, we were also capable to measure this ceramide generated at the plasma membrane. These results do not rule out that ceramide might be formed in other membranes in response to nSMase2, but they do demonstrate that ceramide can function at least at the plasma membrane. Exposed cells to bSMase or overexpressing nSMase2 resulted in the same phenotype on cell adhesion, cell spreading and cell migration, dependent on plasma membrane ceramide. Bacterial sphingomyelinase acts on the outer leaflet of the plasma membrane whereas nSMase2 is believed to act on the inner leaflet of the plasma membrane (61, 62). However, ceramide generated from nSMase2 can be detected in the outer leaflet as evidenced with the ceramide antibody and the use of recombinant pCDase, suggesting that ceramide can quickly equilibrate between two membrane bilayers as also shown by others (78).
The current results show that nSMase2 not only acts at the plasma membrane, but it is an essential component of a program regulating cell adhesion, spreading, and migration. NSMase2 is involved in the cell cycle, and it has also been found to mediate TNF and CD95 responses towards cell death (1). nSMase2 have been only indirectly shown to be related to cell adhesion, such as modulating some proteins with roles in cell adhesion such as ICAM (79), and integrins (80), in the regulation of chemokines (81, 82) and even more indirectly affecting somehow cytoskeletal proteins (40). Here, we have shown that nSMase2 is required for the physiological regulation of cell adhesion and cell migration. Knockdown of nSMase2 was sufficient to revert the chemotherapeutic drug effects on cell migration and adhesion, whereas overexpression of nSMase2 was sufficient to trigger these functions. Also, it is important to mention that probably other sphingolipid metabolizing enzymes capable to acutely modulate ceramide levels at the plasma membrane are part of this pathway. Thus, sphingomyelin synthase 2 and neutral ceramidase are two immediate candidates to play roles in cell adhesion and migration and should also be evaluated.
In summary, we have defined plasma membrane ceramide as an independent and as yet unappreciated second messenger that is triggered by some cytotoxic drugs, mediating loss of cell adhesion and spreading while enhancing migration. We have defined a previously unrecognized plasma membrane ceramide cell signaling pathway. Finally, one might speculate on inhibition of nSMase2 as a viable strategy to prevent or mitigate potentially undesirable effects of cancer chemotherapy.
Supplementary Material
Acknowledgements
We thank Dr Justin Snider and Izolda Mileva for lipidomics support. We acknowledge use of the Stony Brook Cancer Center Biomolecular Spectroscopy Shared Resource. We also thank Samia Mohammed and Dr. Chiara Luberto for their technical and intellectual support. We also thank all the students that have spent time in the lab during this work, including Mariam Elhawary, Zunair Ahmad and Laiqa Jalil. This work was supported by NIH grant CA218678.
Nonstandard Abbreviations:
- aSMase
acid sphingomyelinase
- BP
biological process
- bSMase
bacterial sphingomyelinase
- CC
cellular compartment
- GO
gene ontology
- MF
molecular function
- nSMase
neutral sphingomyelinase
- nSMase2
neutral sphingomyelinase2
- pCDase
Pseudomonas bacterial ceramidase
- S1P
sphingosine 1-phosphate
- SMase(s)
Sphingomyelinase(s)
Footnotes
Competing Interests Statement
Dr. Wadih Arap and Dr. Renata Pasqualini declare that they are inventors in filed patents on intellectual property related to the monoclonal antibody 2A2 and that they will be entitled to royalties if licensing or commercialization occurs. These arrangements are currently managed in accordance with institutional conflict-of-interest (COI) policies at Rutgers, the State University of new Jersey
References
- 1.Hannun YA, and Obeid LM (2008) Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 2.Seitz AP, Grassme H, Edwards MJ, Pewzner-Jung Y, and Gulbins E (2015) Ceramide and sphingosine in pulmonary infections. Biol Chem 396, 611–620 [DOI] [PubMed] [Google Scholar]
- 3.Canals D, Salamone S, and Hannun YA (2018) Visualizing bioactive ceramides. Chem Phys Lipids 216, 142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dadsena S, Bockelmann S, Mina JGM, Hassan DG, Korneev S, Razzera G, Jahn H, Niekamp P, Muller D, Schneider M, Tafesse FG, Marrink SJ, Melo MN, and Holthuis JCM (2019) Ceramides bind VDAC2 to trigger mitochondrial apoptosis. Nat Commun 10, 1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birbes H, El Bawab S, Hannun YA, and Obeid LM (2001) Selective hydrolysis of a mitochondrial pool of sphingomyelin induces apoptosis. FASEB J 15, 2669–2679 [DOI] [PubMed] [Google Scholar]
- 6.Bockelmann S, Mina JGM, Korneev S, Hassan DG, Muller D, Hilderink A, Vlieg HC, Raijmakers R, Heck AJR, Haberkant P, and Holthuis JCM (2018) A search for ceramide binding proteins using bifunctional lipid analogs yields CERT-related protein StarD7. J Lipid Res 59, 515–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canals D, Jenkins RW, Roddy P, Hernandez-Corbacho MJ, Obeid LM, and Hannun YA (2010) Differential effects of ceramide and sphingosine 1-phosphate on ERM phosphorylation: probing sphingolipid signaling at the outer plasma membrane. J Biol Chem 285, 32476–32485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walev I, Weller U, Strauch S, Foster T, and Bhakdi S (1996) Selective killing of human monocytes and cytokine release provoked by sphingomyelinase (beta-toxin) of Staphylococcus aureus. Infect Immun 64, 2974–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hannun YA, and Obeid LM (2011) Many ceramides. J Biol Chem 286, 27855–27862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannun YA, and Obeid LM (2018) Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Bio 19, 175–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tellier E, Negre-Salvayre A, Bocquet B, Itohara S, Hannun YA, Salvayre R, and Auge N (2007) Role for furin in tumor necrosis factor alpha-induced activation of the matrix metalloproteinase/sphingolipid mitogenic pathway. Mol Cell Biol 27, 2997–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang DI, Yeh CH, Chen S, Xu J, and Hsu CY (2004) Neutral sphingomyelinase activation in endothelial and glial cell death induced by amyloid beta-peptide. Neurobiol Dis 17, 99–107 [DOI] [PubMed] [Google Scholar]
- 13.Jaramillo-Gomez J, Nino A, Arboleda H, and Arboleda G (2015) Overexpression of DJ-1 protects against C2-ceramide-induced neuronal death through activation of the PI3K/AKT pathway and inhibition of autophagy. Neurosci Lett 603, 71–76 [DOI] [PubMed] [Google Scholar]
- 14.Mathias S, Dressler KA, and Kolesnick RN (1991) Characterization of a ceramide-activated protein kinase: stimulation by tumor necrosis factor alpha. Proc Natl Acad Sci U S A 88, 10009–10013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snider JM, Trayssac M, Clarke CJ, Schwartz N, Snider AJ, Obeid LM, Luberto C, and Hannun YA (2019) Multiple actions of doxorubicin on the sphingolipid network revealed by flux analysis. J Lipid Res 60, 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kota V, Szulc ZM, and Hama H (2012) Identification of C(6) -ceramide-interacting proteins in D6P2T Schwannoma cells. Proteomics 12, 2179–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulte-Zweckel J, Schneidewind T, Abad JL, Brockmeyer A, Janning P, and Triola G (2018) Azide-tagged sphingolipids for the proteome-wide identification of C16-ceramide-binding proteins. Chem Commun (Camb) 54, 13742–13745 [DOI] [PubMed] [Google Scholar]
- 18.Izquierdo E, and Delgado A (2018) Click chemistry in sphingolipid research. Chem Phys Lipids 215, 71–83 [DOI] [PubMed] [Google Scholar]
- 19.Dobrowsky RT, and Hannun YA (1992) Ceramide stimulates a cytosolic protein phosphatase. J Biol Chem 267, 5048–5051 [PubMed] [Google Scholar]
- 20.Dobrowsky RT, and Hannun YA (1993) Ceramide-activated protein phosphatase: partial purification and relationship to protein phosphatase 2A. Adv Lipid Res 25, 91–104 [PubMed] [Google Scholar]
- 21.Canals D, Roddy P, and Hannun YA (2012) Protein phosphatase 1alpha mediates ceramide-induced ERM protein dephosphorylation: a novel mechanism independent of phosphatidylinositol 4, 5-biphosphate (PIP2) and myosin/ERM phosphatase. J Biol Chem 287, 10145–10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu CL, Chen MJ, Lin JC, Lin CH, Huang WC, Cheng SP, Chen SN, and Chang YC (2019) Doxorubicin Promotes Migration and Invasion of Breast Cancer Cells through the Upregulation of the RhoA/MLC Pathway. J Breast Cancer 22, 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rotolo J, Stancevic B, Zhang J, Hua G, Fuller J, Yin X, Haimovitz-Friedman A, Kim K, Qian M, Cardo-Vila M, Fuks Z, Pasqualini R, Arap W, and Kolesnick R (2012) Anti-ceramide antibody prevents the radiation gastrointestinal syndrome in mice. J Clin Invest 122, 1786–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi T, Takahashi M, Nagatsuka Y, and Hirabayashi Y (2006) Lipid rafts: new tools and a new component. Biol Pharm Bull 29, 1526–1531 [DOI] [PubMed] [Google Scholar]
- 25.Wu BX, Snook CF, Tani M, Bullesbach EE, and Hannun YA (2007) Large-scale purification and characterization of recombinant Pseudomonas ceramidase: regulation by calcium. J Lipid Res 48, 600–608 [DOI] [PubMed] [Google Scholar]
- 26.Milhas D, Clarke CJ, Idkowiak-Baldys J, Canals D, and Hannun YA (2010) Anterograde and retrograde transport of neutral sphingomyelinase-2 between the Golgi and the plasma membrane. Biochim Biophys Acta 1801, 1361–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang CC, Park AY, and Guan JL (2007) In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2, 329–333 [DOI] [PubMed] [Google Scholar]
- 28.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, and Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piccolo SR, Hoffman LM, Conner T, Shrestha G, Cohen AL, Marks JR, Neumayer LA, Agarwal CA, Beckerle MC, Andrulis IL, Spira AE, Moos PJ, Buys SS, Johnson WE, and Bild AH (2016) Integrative analyses reveal signaling pathways underlying familial breast cancer susceptibility. Mol Syst Biol 12, 860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bielawski J, Szulc ZM, Hannun YA, and Bielawska A (2006) Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods 39, 82–91 [DOI] [PubMed] [Google Scholar]
- 31.van der Walt S, Colbert SC, and Varoquaux G (2011) The NumPy Array: A Structure for Efficient Numerical Computation. Comput Sci Eng 13, 22–30 [Google Scholar]
- 32.R Core Team. (2018) R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria [Google Scholar]
- 33.Sales G, Calura E, Cavalieri D, and Romualdi C (2012) graphite - a Bioconductor package to convert pathway topology to gene network. BMC Bioinformatics 13, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elliott A, Leicht E, Whitmore A, Reinert G, and Reed-Tsochas F (2018) A nonparametric significance test for sampled networks. Bioinformatics 34, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, and de Hoon MJ (2009) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.UniProt C (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47, D506–D515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zwick M, Kraemer O, and Carter AJ (2019) Dataset of the frequency patterns of publications annotated to human protein-coding genes, their protein products and genetic relevance. Data Brief 25, 104284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, Wiegers J, Wiegers TC, and Mattingly CJ (2017) The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res 45, D972–D978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeidan YH, Jenkins RW, and Hannun YA (2008) Remodeling of cellular cytoskeleton by the acid sphingomyelinase/ceramide pathway. J Cell Biol 181, 335–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shamseddine AA, Airola MV, and Hannun YA (2015) Roles and regulation of neutral sphingomyelinase-2 in cellular and pathological processes. Adv Biol Regul 57, 24–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grammatikos G, Teichgraber V, Carpinteiro A, Trarbach T, Weller M, Hengge UR, and Gulbins E (2007) Overexpression of acid sphingomyelinase sensitizes glioma cells to chemotherapy. Antioxid Redox Signal 9, 1449–1456 [DOI] [PubMed] [Google Scholar]
- 43.Airola MV, and Hannun YA (2013) Sphingolipid metabolism and neutral sphingomyelinases. Handb Exp Pharmacol, 57–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Canals D, Perry DM, Jenkins RW, and Hannun YA (2011) Drug targeting of sphingolipid metabolism: sphingomyelinases and ceramidases. Br J Pharmacol 163, 694–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishnamurthy K, Dasgupta S, and Bieberich E (2007) Development and characterization of a novel anti-ceramide antibody. J Lipid Res 48, 968–975 [DOI] [PubMed] [Google Scholar]
- 46.Dong X, Xiong L, Jiang X, and Wang Y (2010) Quantitative proteomic analysis reveals the perturbation of multiple cellular pathways in jurkat-T cells induced by doxorubicin. J Proteome Res 9, 5943–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrieu-Abadie N, and Levade T (2002) Sphingomyelin hydrolysis during apoptosis. Biochim Biophys Acta 1585, 126–134 [DOI] [PubMed] [Google Scholar]
- 48.Canals D, and Hannun YA (2013) Novel chemotherapeutic drugs in sphingolipid cancer research. Handb Exp Pharmacol, 211–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Key M (2012) A tutorial in displaying mass spectrometry-based proteomic data using heat maps. BMC Bioinformatics 13 Suppl 16, S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, and Mering CV (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47, D607–D613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, Milacic M, Roca CD, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Viteri G, Weiser J, Wu G, Stein L, Hermjakob H, and D'Eustachio P (2018) The Reactome Pathway Knowledgebase. Nucleic Acids Res 46, D649–D655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M, Sato Y, Kawashima M, Furumichi M, and Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44, D457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, Winsor GL, Hancock RE, Brinkman FS, and Lynn DJ (2013) InnateDB: systems biology of innate immunity and beyond--recent updates and continuing curation. Nucleic Acids Res 41, D1228–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mistry D, Wise RP, and Dickerson JA (2017) DiffSLC: A graph centrality method to detect essential proteins of a protein-protein interaction network. PLoS One 12, e0187091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Obeid LM, Linardic CM, Karolak LA, and Hannun YA (1993) Programmed cell death induced by ceramide. Science 259, 1769–1771 [DOI] [PubMed] [Google Scholar]
- 56.Marchesini N, Osta W, Bielawski J, Luberto C, Obeid LM, and Hannun YA (2004) Role for mammalian neutral sphingomyelinase 2 in confluence-induced growth arrest of MCF7 cells. J Biol Chem 279, 25101–25111 [DOI] [PubMed] [Google Scholar]
- 57.Chen CS, Rosenwald AG, and Pagano RE (1995) Ceramide as a modulator of endocytosis. J Biol Chem 270, 13291–13297 [DOI] [PubMed] [Google Scholar]
- 58.Kitatani K, Idkowiak-Baldys J, Bielawski J, Taha TA, Jenkins RW, Senkal CE, Ogretmen B, Obeid LM, and Hannun YA (2006) Protein kinase C-induced activation of a ceramide/protein phosphatase 1 pathway leading to dephosphorylation of p38 MAPK. J Biol Chem 281, 36793–36802 [DOI] [PubMed] [Google Scholar]
- 59.Oh HL, Seok JY, Kwon CH, Kang SK, and Kim YK (2006) Role of MAPK in ceramide-induced cell death in primary cultured astrocytes from mouse embryonic brain. Neurotoxicology 27, 31–38 [DOI] [PubMed] [Google Scholar]
- 60.Grada A, Otero-Vinas M, Prieto-Castrillo F, Obagi Z, and Falanga V (2017) Research Techniques Made Simple: Analysis of Collective Cell Migration Using the Wound Healing Assay. J Invest Dermatol 137, e11–e16 [DOI] [PubMed] [Google Scholar]
- 61.Tani M, and Hannun YA (2007) Analysis of membrane topology of neutral sphingomyelinase 2. FEBS Lett 581, 1323–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tani M, and Hannun YA (2007) Neutral sphingomyelinase 2 is palmitoylated on multiple cysteine residues. Role of palmitoylation in subcellular localization. J Biol Chem 282, 10047–10056 [DOI] [PubMed] [Google Scholar]
- 63.Cheng Z, DiMichele LA, Rojas M, Vaziri C, Mack CP, and Taylor JM (2014) Focal adhesion kinase antagonizes doxorubicin cardiotoxicity via p21(Cip1.). J Mol Cell Cardiol 67, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mustafa EH, Mahmoud HT, Al-Hudhud MY, Abdalla MY, Ahmad IM, Yasin SR, Elkarmi AZ, and Tahtamouni LH (2015) 2-deoxy-D-Glucose Synergizes with Doxorubicin or L-Buthionine Sulfoximine to Reduce Adhesion and Migration of Breast Cancer Cells. Asian Pac J Cancer Prev 16, 3213–3222 [DOI] [PubMed] [Google Scholar]
- 65.Karagiannis GS, Pastoriza JM, Wang Y, Harney AS, Entenberg D, Pignatelli J, Sharma VP, Xue EA, Cheng E, D'Alfonso TM, Jones JG, Anampa J, Rohan TE, Sparano JA, Condeelis JS, and Oktay MH (2017) Neoadjuvant chemotherapy induces breast cancer metastasis through a TMEM-mediated mechanism. Sci Transl Med 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Y, Campisi J, Higano C, Beer TM, Porter P, Coleman I, True L, and Nelson PS (2012) Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med 18, 1359–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roodhart JM, He H, Daenen LG, Monvoisin A, Barber CL, van Amersfoort M, Hofmann JJ, Radtke F, Lane TF, Voest EE, and Iruela-Arispe ML (2013) Notch1 regulates angio-supportive bone marrow-derived cells in mice: relevance to chemoresistance. Blood 122, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diaz Osterman CJ, Ozmadenci D, Kleinschmidt EG, Taylor KN, Barrie AM, Jiang S, Bean LM, Sulzmaier FJ, Jean C, Tancioni I, Anderson K, Uryu S, Cordasco EA, Li J, Chen XL, Fu G, Ojalill M, Rappu P, Heino J, Mark AM, Xu G, Fisch KM, Kolev VN, Weaver DT, Pachter JA, Gyorffy B, McHale MT, Connolly DC, Molinolo A, Stupack DG, and Schlaepfer DD (2019) FAK activity sustains intrinsic and acquired ovarian cancer resistance to platinum chemotherapy. Elife 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rebillard A, Jouan-Lanhouet S, Jouan E, Legembre P, Pizon M, Sergent O, Gilot D, Tekpli X, Lagadic-Gossmann D, and Dimanche-Boitrel MT (2010) Cisplatin-induced apoptosis involves a Fas-ROCK-ezrin-dependent actin remodelling in human colon cancer cells. Eur J Cancer 46, 1445–1455 [DOI] [PubMed] [Google Scholar]
- 70.Zhang S, Ma H, Zhang D, Xie S, Wang W, Li Q, Lin Z, and Wang Y (2018) LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis 9, 742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Y, Farmer RW, Yang Y, and Martin RC (2016) Epithelial cell adhesion molecule in human hepatocellular carcinoma cell lines: a target of chemoresistence. BMC Cancer 16, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Colombini M (2010) Ceramide channels and their role in mitochondria-mediated apoptosis. Biochim Biophys Acta 1797, 1239–1244 [DOI] [PubMed] [Google Scholar]
- 73.Wang G, Krishnamurthy K, Chiang YW, Dasgupta S, and Bieberich E (2008) Regulation of neural progenitor cell motility by ceramide and potential implications for mouse brain development. J Neurochem 106, 718–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu YY, Yu JY, Yin D, Patwardhan GA, Gupta V, Hirabayashi Y, Holleran WM, Giuliano AE, Jazwinski SM, Gouaze-Andersson V, Consoli DP, and Cabot MC (2008) A role for ceramide in driving cancer cell resistance to doxorubicin. FASEB J 22, 2541–2551 [DOI] [PubMed] [Google Scholar]
- 75.Dumitru CA, Carpinteiro A, Trarbach T, Hengge UR, and Gulbins E (2007) Doxorubicin enhances TRAIL-induced cell death via ceramide-enriched membrane platforms. Apoptosis 12, 1533–1541 [DOI] [PubMed] [Google Scholar]
- 76.Park MA, Mitchell C, Zhang G, Yacoub A, Allegood J, Haussinger D, Reinehr R, Larner A, Spiegel S, Fisher PB, Voelkel-Johnson C, Ogretmen B, Grant S, and Dent P (2010) Vorinostat and sorafenib increase CD95 activation in gastrointestinal tumor cells through a Ca(2+)-de novo ceramide-PP2A-reactive oxygen species-dependent signaling pathway. Cancer Res 70, 6313–6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dadsena S, Hassan DG, and Holthuis JCM (2019) Unraveling the molecular principles by which ceramides commit cells to death. Cell Stress 3, 280–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bennett WF, and Tieleman DP (2012) Molecular simulation of rapid translocation of cholesterol, diacylglycerol, and ceramide in model raft and nonraft membranes. J Lipid Res 53, 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lopes Pinheiro MA, Kroon J, Hoogenboezem M, Geerts D, van Het Hof B, van der Pol SM, van Buul JD, and de Vries HE (2016) Acid Sphingomyelinase-Derived Ceramide Regulates ICAM-1 Function during T Cell Transmigration across Brain Endothelial Cells. J Immunol 196, 72–79 [DOI] [PubMed] [Google Scholar]
- 80.Chudakova DA, Zeidan YH, Wheeler BW, Yu J, Novgorodov SA, Kindy MS, Hannun YA, and Gudz TI (2008) Integrin-associated Lyn kinase promotes cell survival by suppressing acid sphingomyelinase activity. J Biol Chem 283, 28806–28816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Newcomb B, Rhein C, Mileva I, Ahmad R, Clarke CJ, Snider J, Obeid LM, and Hannun YA (2018) Identification of an acid sphingomyelinase ceramide kinase pathway in the regulation of the chemokine CCL5. J Lipid Res 59, 1219–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jenkins RW, Clarke CJ, Canals D, Snider AJ, Gault CR, Heffernan-Stroud L, Wu BX, Simbari F, Roddy P, Kitatani K, Obeid LM, and Hannun YA (2011) Regulation of CC ligand 5/RANTES by acid sphingomyelinase and acid ceramidase. J Biol Chem 286, 13292–13303 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.