ABSTRACT
Metagenome-assembled genome sequences (MAGs) were generated from two wastewater treatment systems in two German cities (Göttingen and Greifswald), based on metagenomes derived from hospital effluent, different wastewater treatment stages, and adjacent water bodies. The MAGs mainly originated from bacterial members of Proteobacteria, Bacteroidota, Firmicutes, “Candidatus Patescibacteria,” Actinobacteriota, Chloroflexota, Desulfobacterota, and Verrucomicrobiota.
ANNOUNCEMENT
Municipal wastewater, university hospital wastewater, sludge, and adjacent water bodies at nine and eight locations affiliated with wastewater treatment plants (WWTP) in Göttingen and Greifswald (Germany), respectively, were sampled quarterly (2016 to 2018) (Table S1; https://doi.org/10.6084/m9.figshare.14601126). Three technical replicates from each sample location were collected and processed, and DNA was isolated as described previously (1). Briefly, the planktonic fraction was harvested by centrifugation; the pellets were stabilized with RNAprotect (Qiagen, Hilden, Germany) and stored at 4°C. The RNAprotect was removed and DNA was extracted using the PowerSoil DNA isolation kit (MoBio Laboratories, Inc., Carlsbad, CA, USA). DNA isolations from each sampling site were pooled in equimolar concentrations. The sequencing libraries were constructed and indexed using a Nextera DNA sample preparation kit and an index kit as recommended by the manufacturer (Illumina, San Diego, CA, USA). Paired-end sequencing was performed using a HiSeq 2500 instrument (rapid run mode, 500 cycles) as recommended by the manufacturer (Illumina). Library construction failed for two Bodden samples (March and July 2017), and the sludge was not sampled in 2016 in Greifswald, resulting in 131 metagenomes (Table S1; https://doi.org/10.6084/m9.figshare.14601126).
Default parameters were used for all software unless otherwise specified. R v4.0.2 (2) and RStudio v1.3.1056 (3) were used for data table processing and figure generation. The data processing included fastp v0.20.0 (4) with overlap correction, quality filtering (removal of reads of <Q20), read clipping with a sliding window of 4, the removal of reads shorter than 50 bp, and Illumina adapter removal. After quality filtering, the metagenome sequences consisted of 5.7 billion paired-end reads with an average read length of 207 bp (forward) and 206 bp (reverse), respectively (Table S1; https://doi.org/10.6084/m9.figshare.14601126).
The samples were merged by site and city, resulting in 17 data sets, which were assembled using metaSPAdes v3.13.0 (5) with defined kmers (-k 21,33,55,77,99,127) and without error correction (- -only-assembler). Contigs with lengths of <1,000 bp were discarded using USEARCH v9.2.64 (6). The assembly characteristics were calculated using BBMap’s statswrapper.sh (https://sourceforge.net/projects/bbmap/) and are summarized in Table S2 (https://doi.org/10.6084/m9.figshare.14601129).
The contig coverage information was determined using Bowtie2 v2.3.5.1 (7) and SAMtools v1.9 (8). Metagenome-assembled genome sequences (MAGs) were generated using MetaBAT2 v2.12.1 (9). The MAG quality and average coverage were determined using CheckM v1.1.2 (10). The MAG bins were classified as high, medium, and low quality according to minimum information MAGs (MIMAGs) (11). The rRNA and tRNA genes were annotated using Prokka v1.14.5 (12). In addition, MAG bins with <10-fold coverage, <500 kbp, <20% completeness, and >1,000 contigs were removed. The overall average sequencing depth was 44-fold. The MAGs were classified taxonomically using GTDB-Tk v1.3.0 (13) and the Genome Taxonomy Database (GTDB) r95 (14).
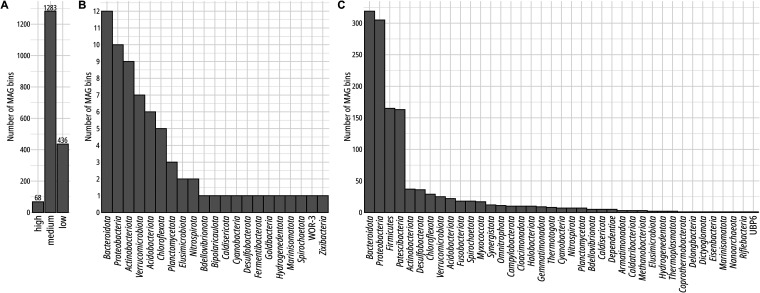
This resulted in 68 high-, 1,283 medium-, and 436 low-quality MAGs (Fig. 1), which belong to Archaea (21 MAGs) and Bacteria (1,766 MAGs). All high-quality MAGs were of bacterial origin and comprised Bacteroidota (Bacteroidia, 9), Proteobacteria (Alphaproteobacteria, 5; Gammaproteobacteria, 3), Verrucomicrobiota (Verrucomicrobiae, 5; Kiritimatiellae, 1), Acidobacteriota (Aminicenantia, 1; Thermoanaerobaculia, 2; “Candidatus UBA6911,” 1; Vicinamibacteria, 1), Actinobacteriota (Acidimicrobiia, 3; Actinomycetia, 1), Planctomycetota (Phycisphaerae, 2; “Candidatus UBA8108,” 1), Chloroflexota (Anaerolineae, 2), Elusimicrobiota (Elusimicrobia, 2), Bdellovibrionota (“Candidatus UBA2394,” 1), “Candidatus Bipolaricaulota” (Bipolaricaulia, 1), Caldisericota (Caldisericia, 1), Cyanobacteria (Vampirovibrionia, 1), Desulfobacterota (Syntrophia, 1), Fermentibacterota (“Candidatus Fermentibacteria,” 1), “Candidatus Goldbacteria” (“Candidatus PGYV01,” 1), Hydrogenedentota (Hydrogenedentia, 1), Marinisomatota (“Candidatus UBA2242,” 1), Nitrospirota (Nitrospiria, 1), Spirochaetota (“Candidatus UBA4802,” 1), “Candidatus WOR-3” (“Candidatus Hydrothermia,” 1), and “Candidatus Zixibacteria” (“Candidatus MSB-5A5,” 1). Details for all generated MAGs are provided in Table S3 (https://doi.org/10.6084/m9.figshare.14601141).
FIG 1.
Overview of all generated MAGs from two wastewater treatment systems in Göttingen and Greifswald. (A) Number of MAG bins with high, medium, and low quality according to MIMAG. (B and C) Phylum distribution of MAGs of high (B) and medium (C) quality.
Data availability.
The raw sequences of the metagenomes have been deposited in the NCBI Sequence Read Archive under the BioProject accession number PRJNA524094; details are listed in Table S1 (https://doi.org/10.6084/m9.figshare.14601126). The coassembled metagenomes (https://doi.org/10.6084/m9.figshare.14578308) and MAGs (https://doi.org/10.6084/m9.figshare.14578629) are also available.
ACKNOWLEDGMENTS
We thank Mechthild Bömeke and Melanie Heinemann for technical assistance and Sarah Zachmann, Maike Karnstedt, and Tim Böer for assistance during sampling and sample preparation.
We thank the University Hospital Göttingen and the Göttinger Entsorgungsbetriebe and the Stadtwerke Greifswald for permission to sample and assistance with the sample collection. This study was partly supported by the Bundesministerium für Bildung und Forschung (BMBF) within the ANTIRES project (FKZ 03ZZ0815B) in the framework of the InfectControl 2020 funding measure. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Rolf Daniel, Email: rdaniel@gwdg.de.
J. Cameron Thrash, University of Southern California.
REFERENCES
- 1.Schneider D, Aßmann N, Wicke D, Poehlein A, Daniel R. 2020. Metagenomes of wastewater at different treatment stages in central Germany. Microbiol Resour Announc 9:e00201-20. doi: 10.1128/MRA.00201-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 3.RStudio Team. 2020. RStudio: integrated development environment for R. RStudio Team, Boston, MA, USA.
- 4.Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 7.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, Tringe SG, Ivanova NN, Copeland A, Clum A, Becraft ED, Malmstrom RR, Birren B, Podar M, Bork P, Weinstock GM, Garrity GM, Dodsworth JA, Yooseph S, Sutton G, Glöckner FO, Gilbert JA, Nelson WC, Hallam SJ, Jungbluth SP, Ettema TJG, Tighe S, Konstantinidis KT, Liu W-T, Baker BJ, Rattei T, Eisen JA, Hedlund B, McMahon KD, Fierer N, Knight R, Finn R, Cochrane G, Karsch-Mizrachi I, Tyson GW, Rinke C, Lapidus A, Meyer F, Yilmaz P, Parks DH, Eren AM, Schriml L, Banfield JF, The Genome Standards Consortium , et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 13.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. 2020. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parks DH, Chuvochina M, Chaumeil P-A, Rinke C, Mussig AJ, Hugenholtz P. 2020. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol 38:1079–1086. doi: 10.1038/s41587-020-0501-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequences of the metagenomes have been deposited in the NCBI Sequence Read Archive under the BioProject accession number PRJNA524094; details are listed in Table S1 (https://doi.org/10.6084/m9.figshare.14601126). The coassembled metagenomes (https://doi.org/10.6084/m9.figshare.14578308) and MAGs (https://doi.org/10.6084/m9.figshare.14578629) are also available.