KDM5A is required for transcription elongation of Myc target genes in myeloma, and a small-molecule selective inhibitor shows activity in myeloma models.
Abstract
Lysine demethylase 5A (KDM5A) is a negative regulator of histone H3 lysine 4 trimethylation (H3K4me3), a histone mark associated with activate gene transcription. We identify that KDM5A interacts with the P-TEFb complex and cooperates with MYC to control MYC-targeted genes in multiple myeloma cells. We develop a cell-permeable and selective KDM5 inhibitor, JQKD82, that increases H3K4me3 but paradoxically inhibits downstream MYC-driven transcriptional output in vitro and in vivo. Using genetic ablation together with our inhibitor, we establish that KDM5A supports MYC target gene transcription independent of MYC itself by supporting TFIIH (CDK7)- and P-TEFb (CDK9)–mediated phosphorylation of RNAPII. These data identify KDM5A as a unique vulnerability in multiple myeloma functioning through regulation of MYC target gene transcription and establish JQKD82 as a tool compound to block KDM5A function as a potential therapeutic strategy for multiple myeloma.
Significance:
We delineate the function of KDM5A in activating the MYC-driven transcriptional landscape. We develop a cell-permeable KDM5 inhibitor to define the activating role of KDM5A on MYC target gene expression and implicate the therapeutic potential of this compound in mouse models and multiple myeloma patient samples.
See related video from the AACR Annual Meeting 2021: https://vimeo.com/554896826
Introduction
Multiple myeloma is a malignant plasma cell disorder accounting for 10% of hematologic malignancies (1). Although high-dose chemotherapy and targeted agents have improved patient outcomes, multiple myeloma remains an incurable disorder (1). Thus, there is an urgent need to develop novel therapeutic strategies. One of the most crucial molecular events in the development of multiple myeloma is c-MYC (MYC) dysregulation resulting from translocation or amplification of the MYC locus, loss of MYC checkpoints such as p53, or activation of upstream signaling (2,3,4). MYC is an oncogenic transcription factor that controls gene expression and is broadly dysregulated in a variety of human cancers (5, 6). High expression of MYC proteins in tumor cells results in transcriptional amplification, leading to selective dependence on transcriptional processes (5, 7, 8). Importantly, MYC plays a key role in the progression from monoclonal gammopathy of undermined significance (MGUS) to multiple myeloma, which is evidenced by the Vk*MYC mouse model (9). In this mouse model, transgenic MYC is sporadically activated in B cells undergoing somatic hypermutation in an activation-induced deaminase-dependent manner, resulting in the development of indolent multiple myeloma; however, additional oncogenic events are required for developing aggressive multiple myeloma (10). In agreement with this finding, MYC pathway genes are activated in multiple myeloma patient samples, but not in MGUS (9, 11). MYC is also required for the maintenance of multiple myeloma cells. Knockdown or pharmacologic inhibition of MYC critically impairs the growth of multiple myeloma cells (12,13,14). Thus, MYC is a central target of interest in multiple myeloma. Unfortunately, therapeutic targeting of MYC proteins has been challenging to accomplish (15). Furthermore, baseline MYC function is required for survival and differentiation of a variety of untransformed cells (16). As a result, interest has developed in identifying highly expressed, lineage-specific targets that cooperate with MYC function, to restrict the scope of inhibition to a particular cell of interest.
Targeting epigenetic-modifying enzymes represents one method of inhibiting MYC function (15). Our previous studies have demonstrated that inhibition of epigenetic proteins involved in the regulation of posttranslational modifications (PTM) of histones results in antitumor effects by modifying chromatin dynamics and gene expression programs (17, 18). Importantly, inhibition of these epigenetic proteins is tolerated in vivo, indicating a potential therapeutic window for their use (17, 18). These PTMs, including methylation and acetylation, are modulated by “writer” and “eraser” enzymes that may be context-specific targets and result in potent transcriptional effects (15, 19, 20). Jumonji C domain–containing proteins are “eraser” enzymes that remove histone methylation marks (21). One Jumonji C domain–containing protein is Lysine demethylase 5A (KDM5A; also known as JARID1A/RBP2), which, along with its subfamily members (KDM5B-5D), functions to remove histone H3 lysine 4 dimethylation and trimethylation (H3K4me2 and H3K4me3) marks. H3K4me3 is a key epigenetic mark associated with transcriptional gene activation (22, 23), and involved in development and differentiation (23,24,25). KDM5A promotes tumorigenesis, metastasis, and drug resistance in various cancers, including acute myeloid leukemia, lung cancer, and breast cancer (26,27,28,29). MYC binding to gene promoters is exquisitely sensitive to H3K4me3 (30), and loss of KDM proteins results in altered differentiation patterns and cell-cycle arrest across normal cells (31, 32), suggesting that KDM proteins may represent a potential cooperating oncogene with MYC. To investigate the function of KDM5, multiple KDM5 inhibitors have been developed (33,34,35). These compounds display a high degree of selectivity for KDM5 proteins but have poor cell permeability and modest cellular activity. Despite these issues, inhibition of KDM5 proteins in MM.1S multiple myeloma cells results in cell-cycle arrest and H3K4me3 hypermethylation, indicating that KDM5 proteins represent tractable candidates for therapeutic targeting in multiple myeloma (35).
Building on this evidence, here we demonstrate that KDM5A cooperates with MYC to regulate MYC target gene transcription in multiple myeloma. We develop a novel prodrug-type KDM5 inhibitor, JQKD82, which delivers the active binding molecule KDM5-C49 to potently block KDM5 function in multiple myeloma cells both in vitro and in vivo. We use this inhibitor, in combination with genetic knockdown studies, to demonstrate that KDM5A cobinds with MYC at target gene loci across the genome and activates MYC target genes by physically interacting with the positive transcription elongation factor b (P-TEFb) complex. KDM5A promotes transcriptional pause release at MYC target genes by demethylating H3K4me3, resulting in release of the transcription factor IID (TFIID) subunit TATA-box binding protein associated factor 3 (TAF3) and phosphorylation of the carboxy-terminal domain (CTD) of RNA polymerase II (RNAPII) subunit B1 (POLR2A). These findings identify a novel mechanism by which KDM5A promotes multiple myeloma growth through cooperation with MYC to regulate target gene transcription and provide a novel pharmacologic agent for interrogation of KDM5 protein function.
Results
KDM5 Mediates Multiple Myeloma Cell Growth
The oncogenic roles of the KDM5 family of H3K4 demethylases have been studied in multiple cancers (36). Although KDM5B has previously been implicated in multiple myeloma (33, 35), the biological impact of other KDM5 members in multiple myeloma remains unknown. To identify the individual relevance of the KDM5 familiy, we first examined gene expression of KDM5 isoforms in multiple myeloma using a dataset of 559 newly diagnosed patients (37). Each KDM5 member (KDM5A, KDM5B, and KDM5C) was highly expressed in primary multiple myeloma samples (Fig. 1A), and higher KDM5A expression was associated with poor overall survival, whereas KDM5B and KDM5C expression was not related to overall survival in this dataset (Supplementary Fig. S1A–S1C). Evaluation of the Cancer Cell Line Encyclopedia (CCLE) RNA-sequencing (RNA-seq) dataset (38) also indicated that KDM5A–C were expressed across multiple myeloma cell lines (n = 20; Fig. 1B). We further confirmed KDM5 protein expression in multiple myeloma cell lines and primary multiple myeloma patient samples (Fig. 1C). We did not obtain enough normal plasma cell samples and thus did not assess KDM5 protein expression in the normal counterpart of multiple myeloma cells, but KDM5 protein was not detected in healthy volunteer–derived B cells (Fig. 1C).
Figure 1.
KDM5 mediates multiple myeloma (MM) cell growth. A and B, Violin plots depicting KDM5A (blue), KDM5B (red), and KDM5C (green) expression in primary multiple myeloma samples (GSE2658, n = 559; A) and multiple myeloma cell lines (CCLE, n = 20; B). Center dashed lines represent the median, and lower and upper dashed lines represent the interquartile range. TPM, transcripts per million. C, Immunoblot analysis for KDM5A, KDM5B, and KDM5C in multiple myeloma cell lines, normal B cells, and multiple myeloma patient samples. MCF7 cells served as positive control for KDM5A and KDM5B expression levels. GAPDH served as loading control. D, Multiple myeloma cell lines were transduced with shKDM5A, shKDM5B, shKDM5C, or control shRNA targeting luciferase (shLuc). Three days after infection is designated as day 0. On day 0, whole-cell lysates were extracted and subjected to immunoblot analysis for KDM5A, KDM5B, and KDM5C. GAPDH served as loading control (top). Viable cells were measured by MTT assay on days 0 and 5. The cell growth rate (day 5/day 0) relative to shLuc is shown (bottom). Data represent mean ± SD of quadruplicate cultures. ***, P < 0.001 compared with shLuc; unpaired Student t test.
Because KDM5 proteins were expressed in multiple myeloma cells, we sought to identify the selective effects of KDM5 on multiple myeloma growth. To do so, we transduced multiple myeloma cell lines with lentivirus expressing shRNAs targeting KDM5A, KDM5B, and KDM5C (shKDM5A, shKDM5B, shKDM5C) or a control luciferase (shLuc), and measured the response of multiple myeloma cell lines in cell growth assays. Knockdown of either KDM isoform resulted in specific loss of each targeted protein without reduction of other isoforms (Fig. 1D). Furthermore, loss of any KDM5 impaired the growth of three c-MYC–expressing multiple myeloma cell lines, although the magnitude of effect was strongest for KDM5A loss, and reduction of KDM5C did not decrease the growth of MOLP-8 cells (Fig. 1D). These data indicate that KDM5A and to a lesser extent, KDM5B and KDM5C play a functional role in driving multiple myeloma cell growth.
Development of a Novel Cell-Permeable KDM5-Selective Inhibitor
Because multiple myeloma displayed reliance on KDM5 isoforms for growth, we sought to develop a high-potency, KDM5-selective inhibitor to study KDM5 inhibition in cancer cells and animal models. To do so, we took advantage of a prior KDM5-binding molecule, KDM5-C49, that displays excellent inhibitory activity in biochemical assays though poor activity in cell culture models due to the lack of cell permeability (33). A derivative compound, KDM5-C70, is metabolized to KDM5-C49 in cells, and while KDM5-C70 has cellular activity for KDM5 inhibition, it still suffers from poor delivery efficiency in cells and has limited activity in vivo (33). To overcome this obstacle and study the effects of KDM5 inhbition in cell and animal models, we used a prodrug approach, and designed a compound called JQKD82. JQKD82 is a more stable ester of KDM5-C49 that is able to deliver the active molecule KDM5-C49 to cells more efficiently (Fig. 2A). We first assessed the ability of JQKD82 to inhibit KDM5 in biochemical assays. JQKD82 showed similar enzymatic inhibitory activity as the prior report of ester KDM5-C70 in biochemical assays of KDM5 function (Supplementary Fig. S2A; ref. 33). As with KDM5-C49, JQKD82 did not show activity toward other KDMs, such as KDM3 (Supplementary Fig. S2B), and demonstrated some selectivity for KDM5A over other KDM5 isoforms (Supplementary Fig. S2B). Cocrystallization of JQKD82 with KDM5B revealed that JQKD82 hydrolyzed under cocrystallization conditions and produced the product KDM5-C49 bound to KDM5B (Fig. 2B) to give matched cocrystal structure as the previously reported KDM5-C49 crystal structure (33).
Figure 2.
JQKD82 is a cell-permeable KDM5-selective inhibitor. A, Chemical structures of the prodrug esters JQKD82 and KDM5-C70, and their active metabolite KDM5-C49. B, The crystal structure of JQKD82 bound to KDM5B was resolved after soaking the prodrug ester JQKD82 with KDM5B. This resulted in a crystal resolved for the acid derivative KDM5-C49 derived from JQKD82 under crystallization conditions, demonstrating that KDM5-C49 interacts with Glu501 to chelate with metal and with Lysine 517 on the backbone for selectivity against KDM5 in the demethylase catalytic domain.C, Intracellular concentrations of compounds (both parent compounds and metabolite KDM5-C49) were determined using LC/MS after treatment withKDM5-C70 or JQKD82 at 10 μmol/L for 2 hours in Caco2 cells. Data are shown as average ± SD of duplicate testing. *, P < 0.05; unpaired Student t test.D, Immunoblot analysis for H3K4me3 using H3 as loading control after treatment with KDM5-C49, KDM5-C70, or JQKD82 at 0.3 μmol/L or DMSO for 24 hours in MM.1S cells. E, Immunoblot analysis for H3K4me3, H3K9me3, H3K27me3, H3K36me3, and H3K79me3 using H3 as loading control after treatment with JQKD82 (0.3 or 1 μmol/L) or DMSO for 24 hours in MM.1S and MOLP-8 cells. Densitometry analysis displayed on the right (numbers are levels of methylation as a percentage of DMSO control).
Next, we assessed whether the phenol ester introduced in JQKD82 improved drug delivery into cells. To this end, we performed cell permeability assays using Caco2 monolayer cells treated with JQKD82 for 2 hours, after which we measured JQKD82 and KDM5-C49 concentration in cell lysates by liquid chromatography/mass spectrometry (LC/MS). JQKD82-treated Caco2 cells displayed a higher intracellular concentration of both JQKD82 and its metabolite KDM5-C49 than did Caco2 cells treated with KDM5-C70, indicating that JQKD82 has improved cell permeability and more efficiently delivers the active metabolite KDM5-C49 in vitro (Fig. 2C). As expected, even at low concentrations, JQKD82 caused an increase in the global H3K4me3 level of MM.1S cells, which was greater than that induced by equimolar concentrations of KDM5-C70 or KDM5-C49 (Fig. 2D). Assessment of multiple other histone lysine methylation marks indicated that JQKD82 induced specific increases of H3K4me3, but has no effect with either other histone methylation marks, such as H3K9me3 and H3K27me3, or H3K4me1, which is in agreement with the KDM5 methylation profile (Fig. 2E; Supplementary Fig. S2C; ref. 33). Together, these data suggest that JQKD82 represents a specific and cell-permeable KDM5 inhibitor.
KDM5 Inhibition with JQKD82 Inhibits Multiple Myeloma Cell Growth
Because KDM5 expression is required for multiple myeloma cell growth, we sought to examine the effects of KDM5 inhibition with JQKD82 on multiple myeloma cells. JQKD82 inhibited the growth of MM.1S cells in a dose- and time-dependent manner (Supplementary Fig. S3A). JQKD82 was 7-fold more potent than KDM5-C70 and more than 20-fold more potent than KDM5-C49 at eliciting growth suppression in MM.1S cells (JQKD82 IC50 = 0.42 μmol/L, KDM5-C49 IC50 > 10 μmol/L, KDM5-C70 IC50 = 3.1 μmol/L; Fig. 3A). Furthermore, JQKD82 treatment resulted in suppressed growth in a panel of multiple myeloma cell lines (Fig. 3B). These results were validated by treatment of five primary multiple myeloma patient samples with JQKD82, which demonstrated a 40% to 50% reduction in cell viability after 5 days of treatment (Fig. 3C). As a control, we isolated normal B cells from healthy individuals and treated these cells with JQKD82. CD40 antibody– and IL4-stimulated B cells were insensitive to the effects of JQKD82 (Fig. 3D), indicating that KDM5 inhibition may have a favorable in vivo therapeutic index. To further establish whether JQKD82 was effective at suppressing growth across multiple distinct lineages, we performed a screen of 367 distinct cancer cell lines, which were barcoded and pooled (39). JQKD82 was effective at reducing the growth of multiple tumor cell lineages, including multiple myeloma cells, indicating potential cross-cancer utility (Supplementary Fig. S3B).
Figure 3.
JQKD82 suppresses multiple myeloma (MM) cell growth. A, MM.1S cells were cultured with the indicated concentrations of KDM5-C49, KDM5-C70, or JQKD82 for 5 days. Viable cells were determined by MTT assay, and the cell growth relative to untreated control cells is shown. Data represent mean ± SD of triplicate cultures. B, Multiple myeloma cell lines were cultured with the indicated concentrations of JQKD82 for 5 days. Viable cells were determined by MTT assay, and the cell growth relative to untreated control cells is shown. Data represent mean ± SD of triplicate cultures.C, CD138-positive primary multiple myeloma cells were treated with 3 μmol/L of JQKD82 or DMSO for 5 days. The cell viability relative to untreated control was assessed by Cell TiterGlo assay. Data represent mean ± SD of duplicate or triplicate cultures. D, B cells from healthy donors were stimulated by 10 μg/mL of CD40 antibody and 100 U/ml of IL4, and were then treated with JQKD82 for 5 days. The cell growth relative to JQKD82-untreated control was assessed by Cell TiterGlo assay. Data represent mean ± SD of triplicate cultures. E, MM.1S and MOLP-8 cells were incubated with 1 μmol/L of JQKD82 for 48 hours. Cells were fixed, stained with propidium iodide, and analyzed for cell-cycle distribution using flow cytometry. F, MM.1S and MOLP-8 cells were incubated with 1 μmol/L of JQKD82 for 48 to 96 hours. Cells were stained with Annexin V and analyzed for apoptosis using flow cytometry. Data represent mean ± SD of triplicate samples (E and F). *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with control; unpaired Student t test. G, Schema of in vivo study using a disseminated model. MOLP-8 TurboGFP-Luc cells were intravenously inoculated into NSG mice. After disease establishment confirmed by BLI, the mice were randomized to JQKD82 or vehicle group, and treated i.p. with JQKD82 at 50 mg/kg or vehicle twice a day (BID), respectively, for 3 weeks, and followed for survival. H, Representative BLI of MOLP-8 TurboGFP-Luc cell xenografted mice after treatment with JQKD82 or vehicle control at a dose of 50 mg/kg i.p. twice daily. Images were obtained on day 17 after treatment initiation. Data are representative of nine mice per group. I, Tumor burden was serially evaluated by BLI. Data represent mean ± SEM. n = 9 mice per group. P = 0.028 by comparing treatment group against control group by unpaired Student t test. J, Kaplan–Meier curves showing the survival of JQKD82-treated mice or vehicle control mice. P < 0.0001 by the log-rank test. K, IHC analysis for H3K4me3, Ki67, and MYC in the subcutaneous tumor samples from the MOLP-8 TurboGFP-Luc–injected mice treated with JQKD82 or control. Scale bars, 50 μm. Data are representative of three independent tumors per treatment group.
Because JQKD82 suppressed growth of multiple myeloma cells in vitro, we sought to identify whether this was due to loss of proliferation, induction of apoptosis, or both. Importantly, treatment with JQKD82 induced G1 cell-cycle arrest by 48 hours (Fig. 3E). This arrest was also observed at 96 hours (Supplementary Fig. S3C). JQKD82 modestly elicited apoptosis after 72 hours, as evidenced by annexin V staining (Fig. 3F). These results indicate that the growth inhibitory effect of JQKD82 is primarily due to cell-cycle arrest rather than apoptosis. Consistent with JQKD82's effect, G1 cell-cycle arrest was also observed after KDM5A knockdown (Supplementary Fig. S3D).
JQKD82 Has Anti–Multiple Myeloma Activity In Vivo
Because JQKD82 has induced anti–multiple myeloma activity in vitro with no effects on normal B cells, we sought to identify whether JQKD82 was effective in vivo. To begin, we performed pharmacokinetic analysis of JQKD82 on CD1 mice treated at 50 mg/kg by intraperitoneal (i.p.) injection. Pharmacokinetic analysis demonstrated that the active component, KDM5-C49, was detectable in murine serum with a 6-hour half-life (Supplementary Fig. S3E) and a maximum concentration (Cmax) of 330 μmol/L. Mice treated with JQKD82 twice daily at 50 or 75 mg/kg, i.p., maintained stable body weights, indicating that compound treatment was tolerable (Supplementary Fig. S3F and S3G). Next, we examined the effects of JQKD82 on multiple myeloma in vivo, using a disseminated tumor model. To do so, we engineered MOLP-8 cells to express luciferase and then intravenously injected these MOLP-8 TurboGFP-Luc cells into NSG mice. Once these cells had systemically engrafted, as confirmed by sequential bioluminescence imaging (BLI), we randomized mice to receive JQKD82 or vehicle (n = 9 for each group) via intraperitoneal injection (Fig. 3G). Treatment with JQKD82 significantly reduced tumor burden, as detected by sequential BLI (Fig. 3H and I), and improved overall survival when compared with the vehicle-treated control group (Fig. 3J).
To assess the pharmacodynamic (PD) effects and efficacy of JQKD82 at higher doses, we evaluated the JQKD82 using a plasmacytoma model of multiple myeloma. We subcutaneously injected MOLP-8 TurboGFP-Luc cells into NSG mice and then treated them with JQKD82 (75 mg/kg twice a day) or vehicle control after tumor engraftment. Three mice for each group, treated with either JQKD82 or control vehicle for 7 days, were sacrificed for a PD study. Tumors were collected from treated or control mice and subjected to immunohistochemical (IHC) analysis. Consistent with our in vitro observations, JQKD82-treated tumors displayed an increase in H3K4me3 levels and reduction in Ki67 staining, indicative of a slower growth rate (Fig. 3K). These data indicated that the cellular mechanism of JQKD82 activity remains on target in vivo (Fig. 3K). In addition to this, we sought to determine whether JQKD82 treatment affected primary drivers of multiple myeloma in vivo. Thus, we performed IHC to MYC in JQKD82- and vehicle-treated tumors, demonstrating that JQKD82 resulted in a dramatic reduction of MYC immunostaining in vivo (Fig. 3K). In parallel to this, two groups of mice were treated with either JQKD82 or vehicle (n = 10 for each group) once tumor engraftment was confirmed by sequential BLI. As in our intravenous model, JQKD82 treatment for 14 days also significantly inhibited tumor growth, evidenced both by BLI and tumor size measurement (Supplementary Fig. S3H and S3I). These results indicate that JQKD82 is effective and well tolerated in vivo in two models of multiple myeloma, and suggests a link between KDM5 function and MYC expression.
KDM5 Inhibition Downregulates Expression of MYC Target Genes
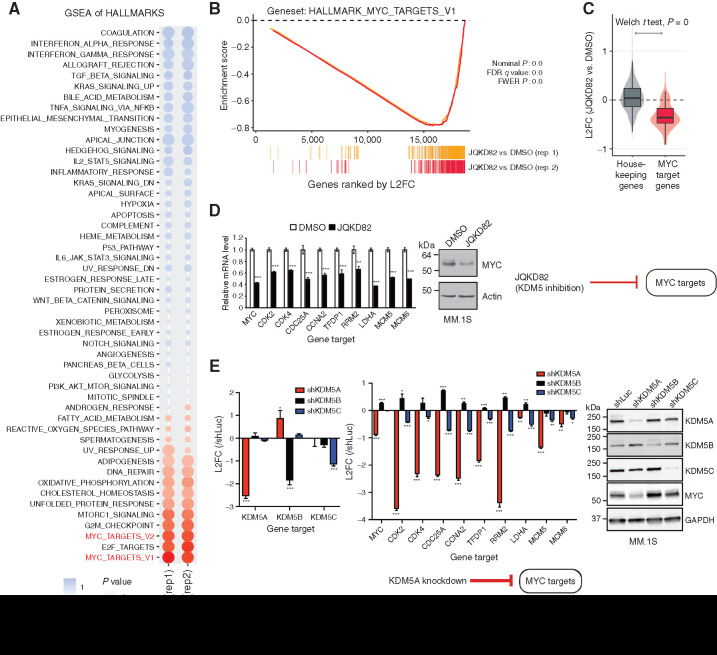
To further elucidate the molecular mechanism of KDM5 function and JQKD82 activity, we next examined the effects of compound treatment for 48 hours on gene expression using RNA-seq. At this time point, a total of 1,450 genes were upregulated, while 1,253 genes were downregulated, compared with DMSO-treated control cells (Padj < 0.05; Supplementary Table S1). These data are in agreement with previous reports that the KDM5 demethylase functions not only as a transcriptional repressor but also as a transcriptional activator (40,41,42,43,44). Gene-set enrichment analysis (GSEA) showed that expression of G1–S cell-cycle checkpoint genes were reduced by JQKD82 treatment, consistent with our cell-cycle analysis (Supplementary Fig. S4A). Importantly, GSEA across the hallmark gene sets from the Molecular Signatures Database (MSigDB) showed MYC gene sets were the most strongly downregulated class (Fig. 4A and B), paralleling the downregulation of MYC in our in vivo xenograft results. Because disruption of MYC may result in loss of transcriptional amplification (5, 8), we next examined the expression of housekeeping genes, which were relatively unchanged at this time point (Fig. 4C). Quantitative real-time PCR confirmed reduced mRNA expression of both MYC and MYC target genes after JQKD82 treatment in MM.1S and MOLP-8 cells (Fig. 4D; Supplementary Fig. S4B). We next asked whether KDM5A, KDM5B, or KDM5C mediated the transcriptional activation of MYC target genes. The expression of MYC target genes was largely diminished by KDM5A knockdown in MM.1S and MOLP-8 cells, whereas these genes were only modestly diminished by KDM5C knockdown and not at all by KDM5B knockdown (Fig. 4E; Supplementary Fig. S4C). MYC protein expression was also consistently reduced by depletion of KDM5A (Fig. 4E; Supplementary Fig. S4C), again paralleling our in vivo analysis. These results suggest that KDM5A, but not KDM5B and KDM5C, mainly functions to control expression of MYC and MYC target genes.
Figure 4.
Inhibition or knockdown of KDM5A downregulates expression of MYC-regulated genes. A, RNA-seq was performed on RNA extracted from MM.1S cells treated with 1 μmol/L of JQKD82 or DMSO control for 48 hours. GSEA for transcriptional hallmarks (Broad) summarized by a bubble plot is shown. Size of the bubbles indicates significance from nominal P value, and the normalized enrichment score (NES) indicates the strength and direction of the enrichment. n = 2 independent treatments, RNA extractions, and RNA-seq reactions per group. B, GSEA plots for MYC target genes after treatment with JQKD82 in MM.1S cells, across two biological replicates. FWER, family-wise error rate. C, Direct comparison of log2 fold change (L2FC) in RNA expression from samples in A, for housekeeping genes (left) and MYC target genes (right), summarized by boxplots and violin distributions. Comparison by Welch t test resulted in highest significance, P = 0. D, Expression levels of representative MYC target genes were assessed by quantitative real-time PCR after treatment with 1 μmol/L of JQKD82 or DMSO control for 48 hours in MM.1S cells (left). Data are normalized against the housekeeping control gene RPLP0. Theexpression relative to DMSO (control) is shown as mean ± SD of triplicate measurements. **, P < 0.01; ***, P < 0.001 compared with control; unpaired Student t test. Immunoblot analysis for MYC and actin (loading control) after treatment with JQKD82 at 1 μmol/L or DMSO for 48 hours in MM.1S cells (right).E, Expression levels of representative MYC target genes were assessed by quantitative real-time PCR after transduction of shKDM5A, shKDM5B, or shKDM5C in MM.1S cells (left). Data are normalized against the housekeeping control gene RPLP0. The expression relative to shLuc (log2 fold change) is shown as mean ± SD of triplicate measurements. *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with shLuc (control); unpaired Student t test. Immunoblot analysis for KDM5A, KDM5B, KDM5C, MYC, and GAPDH (loading control) after transduction of shKDM5A, shKDM5B, shKDM5C, or shLuc in MM.1S cells (right).
KDM5A and MYC Co-occupy and Activate Their Target Genes
MYC is key to multiple myeloma pathogenesis and maintenance, and our data demonstrated its mRNA expression was under KDM5A regulation. Thus, we sought to further investigate the function of KDM5A in multiple myeloma. To begin, we performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) analysis of the KDM5A protein in MM.1S cells (Supplementary Fig. S5A). KDM5A was noted to bind preferentially to intergenic (49%) and upstream/promoter (29%) loci in MM.1S cells (Supplementary Fig. S5B). Indeed, substantial KDM5A enrichment was observed around the transcriptional start sites (TSS; Supplementary Fig. S5C). Motif analysis of the top 200 KDM5A binding peaks revealed enrichment for the previously reported KDM5A consensus motif (Supplementary Fig. S5D; ref. 45). Satisfied that KDM5A was binding to KDM5A sites, we next compared KDM5A-bound sites with activated promoter (H3K4me3), promoter/enhancer (H3K27ac), and repressed promoter (H3K27me3) histone marks. KDM5A colocalized genome wide to activated promoter and promoter/enhancer marks, indicating the presence of KDM5A at transcriptionally competent loci (Supplementary Fig. S5C). To identify proteins that cobound along with KDM5A across the genome, we compared these KDM5A ChIP-seq results with publicly available ChIP-seq data in MM.1S cells by ChIP-Atlas (46). By this analysis, KDM5A colocalized with MYC across the genome in MM.1S cells (Fig. 5A; Supplementary Table S2). Furthermore, we found that KDM5A-bound sites significantly overlapped with ChIP peaks of transcriptional machinery components, including CDK7, CDK9, and RNAPII (Fig. 5A; Supplementary Table S2).
Figure 5.
KDM5A and MYC co-occupy and activate the genes downregulated by JQKD82. A, Heatmap showing KDM5A enrichment at regions bound by MYC, CDK7, CDK9, and RNAPII in MM.1S cells, resolved by ChIP-sequencing (± 5 kb from each of the protein-bound regions are shown). B, Venn diagram depicting the overlap of genes co-occupied by KDM5A and MYC at TSS and genes downregulated by JQKD82 in MM.1S cells (top). Metagene plots showing occupancy of MYC and KDM5A at genes downregulated by JQKD82 (right) and those not downregulated by JQKD82 (left) in MM.1S cells (bottom). RPM, reads per million mapped reads. C, Representative gene tracks demonstrating enrichment of KDM5A, H3K4me3, MYC, CDK7, CDK9, and RNAPII at MYC target gene loci (CDK4 and NOLC1) in MM.1S cells. D, 293T cells were cotransfected with 50 ng of human CDK4 regulatory element-luciferase reporter and 300 ng of KDM5A expression plasmid or empty vector. After 8 hours of transfection, the cells were treated with 10 μmol/L of JQKD82 or DMSO for 16 hours and assayed for luciferase activity. RLU, relative luminescence unit. E, 293T cells were transfected with 50 ng of human CDK4 regulatory element-luciferase reporter together with 200 ng of KDM5A and/or MYC expression plasmids. After 24 hours of transfection, luciferase activities were measured. Data represent mean ± SD of triplicate samples. **, P < 0.01; ***, P < 0.001; unpaired Student t test (D and E). F, 293T cells expressing FLAG-tagged KDM5A were harvested; cell lysates were then immunoprecipitated (IP) with anti-FLAG (mouse monoclonal), and subjected to immunoblot analysis with anti-FLAG, anti-CCNT2 (rabbit polyclonal), and anti-CDK9 (rabbit monoclonal). G, 293T cells were transfected with eGFP-KDM5A together with mCherry-CCNT2. After fixation, nuclei were stained with DAPI. Localizations of KDM5A and CCNT2 were observed using confocal laser scanning microscopy. Scale bars, 10 μm. H, Cell lysates from MM.1S cells were immunoprecipitated with anti-KDM5A, anti-CCNT2, or normal rabbit IgG (control) and subjected to immunoblot analysis with anti-KDM5A or anti-CCNT2. I, Cell lysates from MM.1S cells were incubated with KDM5-C49-Biotin, free biotin, or KDM5-C49 at 100 μmol/L for 24 hours. The chemical probes were pull down with streptavidin agarose resin, and proteins associated with probes were subjected to immunoblot analysis with anti-KDM5A, anti-CDK9, or anti-CCNT2.
Next, we sought to examine the locus-specific effects of KDM5A by integrating our RNA-seq and ChIP-seq results. KDM5 inhibition activated or repressed KDM5A-bound genes in a target-dependent manner (Supplementary Fig. S5E). Notably, most of the genes downregulated by JQKD82 (84%, 1,048 out of 1,253) were co-occupied by KDM5A and MYC at the TSS (Fig. 5B), in addition to CDK7, CDK9, and RNAPII. The gene list included the reprentative MYC target genes, including CDK4 and NOLC1 (Fig. 5C; Supplementary Fig. S5F; refs. 47, 48). Downregulated genes had greater occupancy of MYC around the TSS compared with other KDM5A and MYC co-occupied genes that were not downregulated by JQKD82 (Fig. 5B). Thus, based on these analyses, we hypothesized that KDM5A may directly activate MYC target genes downregulated by JQKD82. To test this possibility, we performed reporter assays in 293T cells using a reporter construct containing the regulatory element (between base pairs −563 and +574 relative to the TSS) of human CDK4, a known representative MYC target in B cells (48). Our ChIP-seq analysis demonstrated that this element is marked by binding of KDM5A, MYC, CDK7, CDK9, and RNAPII in multiple myeloma cells (Fig. 5C). Expression of KDM5A increased CDK4 promoter activity, which could be partially blunted by JQKD82 (Fig. 5D). Moreover, KDM5A and MYC cooperatively increased CDK4 promoter activity (Fig. 5E). These results indicate that KDM5A directly enables MYC target gene expression and this activation can be partly blocked by catalytic inhibition of KDM5A.
To further characterize the mechanism of transcriptional regulation by KDM5A, we next identified the proteins that physically interact with KDM5A. We expressed Flag-tagged KDM5A in 293T cells, and then performed coimmunoprecipitation followed by mass spectrometry. This analysis identified 201 potential KDM5A binding proteins, with a cutoff of >2-fold enrichment compared with empty vector–transduced control (Supplementary Table S3), and this list included several previously known KDM5A binding partners, such as SIN3B, EMSY, PHF12, GATAD1, ZMYND8, ZNF687, and ZNF592 (49,50,51). Consistent with our ChIP-seq analysis, we also identified a physical interaction of KDM5A with CDK9 in addition to the P-TEFb complex member CCNT2. P-TEFb is a CCNT2- and CDK9-containing protein complex that promotes transcriptional elongation (52). We then validated that Flag-tagged KDM5A physically interacts with CCNT2 and CDK9 by coimmunoprecipitation assays in 293T cells (Fig. 5F). Supporting this interaction, we identified that KDM5A and CCNT2 could be colocalized in the nucleus of 293T cells by immunofluorescent assay (Fig. 5G). To further establish this interaction, we then performed coimmunoprecipitation assays with antibodies recognizing endogenous KDM5A and CCNT2 in MM.1S cells. Immunoprecipitates of either KDM5A or CCNT2 demonstrated the presence of the other protein, indicating that this interaction is conserved in multiple myeloma cells (Fig. 5H). To further explore these interactions in the setting of KDM5A inhibition, we then developed a biotinylated chemical probe, KDM5-C49-Biotin, based on the structure guidance by linking the biotin on KDM5-C49 at the position that doesn't interfere with the binding to KDM5 (synthesis described in Supplementary Methods). Using KDM5-C49-Biotin, we performed chemical pull-down experiments using cell lysates from MM.1S cells. We used free biotin or the parental compound KDM5-C49 as control. The streptavidin agarose resin was then used to pull down the KDM5-C49-Biotin and proteins associated with this probe from cell lysates after 24-hour incubation of KDM5-C49-Biotin with MM.1S cell lysate. Our results showed that KDM5-C49-Biotin can pull down KDM5A, together with CCNT2 and CDK9, while the parental compound KDM5-C49 was not pulled down by the beads and biotin alone did not associate with these proteins (Fig. 5I). This experiment provides additional evidence that KDM5A physically interacts with CCNT2 and CDK9 in the P-TEFb complex. Together, these data suggest that KDM5A physically interacts with components of P-TEFb in multiple myeloma cells and may play a role in the regulation of transcriptional elongation in multiple myeloma cells.
JQKD82 Inhibits KDM5A Transcriptional Activating Function via Suppressing Phosphorylation of RNAPII
We next examined the effects of JQKD82 on H3K4me3 genome wide in MM.1S cells. We found that H3K4me3 levels were globally increased after JQKD82 treatment, evidenced by ChIP with reference exogenous genome (ChIP-Rx), where Drosophila chromatin is spiked in as a control for normalization. This strategy is needed because increases in H3K4me3 were expected to be global in nature, and would therefore confound an analysis using conventional reads per million mapped reads ChIP-seq normalization (53). With this approach, we observed that the absolute increase in H3K4me3 levels was more prominent at TSS proximal sites (promoters) compared with TSS distal sites (enhancers; Supplementary Fig. S6A and S6B). We then analyzed H3K4me3 levels at the TSS of KDM5A and MYC co-occupied genes, splitting into genes that were downregulated or not. The baseline H3K4me3 level at the TSS of genes downregulated by JQKD82 treatment was higher than in nondownregulated genes even after the increase, suggesting that genes with the highest H3K4me3 levels are the most modulated by KDM5A enzymatic activity (Fig. 6A). Knockdown of KDM5A phenocopied the effects of JQKD82, increasing H3K4me3 levels at MYC target genes (Supplementary Fig. S6C). Because H3K4me3 is an important modification for active transcription and assembly of RNAPII into the transcriptional preinitiation complex (54), we next examined how excess H3K4me3 induced by KDM5A inhibition impacted active genes. The H3K4me3-decorated +1 nucleosome is physically located at the boundary between the RNAPII pause site and the rest of the gene body. Strong, fixed positioning of the +1 nucleosome results in RNAPII stalling (55), and the H3K4me3 mark anchors the basal RNAPII-bound transcription factor TFIID (54). Thus, since KDM5 inhibition resulted in hypermethylation preferentially at proximal TSS, we hypothesized that KDM5A activity demethylates the H3K4me3 anchor, enabling RNAPII phosphorylation and pause release. As predicted, treatment with JQKD82 followed by ChIP-seq using phospho-specific antibodies to Ser2 and Ser5 of RNAPII demonstrated diminished phosphorylation levels at KDM5A and MYC co-occupied genes whose expression was downregulated by JQKD82 (Fig. 6B), which is exemplified by MYC and the MYC target, NOLC1, loci (Fig. 6C). Furthermore, JQKD82 only modestly hindered recruitment of CDK7 and CDK9, the RNAPII Ser2/5 kinases, to these loci (Supplementary Fig. S6D), suggesting that hyper-H3K4me3 primarily interferes with the function, not the localization, of these kinases.
Figure 6.
JQKD82 increases H3K4me3 and TFIID anchoring via TAF3, resulting in reduced RNAPII phosphorylation. A, Metagene plot of H3K4me3 level at the genes co-occupied by KDM5A and MYC at the TSS in MM.1S cells, and separated into groups either downregulated by JQKD82 (red) or not downregulated by JQKD82 (blue). The gene set defined in Fig. 5B was used for analysis. MM.1S cells were treated with 1 μmol/L of JQKD82 or DMSO for 48 hours. H3K4me3 level was measured by ChIP-seq normalized to reference exogenous spiked-in Drosophila chromatin (ChIP-Rx). RRPM, reference-normalized reads per million. B, Metagene plot of RNAPII phosphorylated at Ser5 (Ser5p) and Ser2 (Ser2p) ChIP-seq reads mapped to the gene loci co-occupied by KDM5A and MYC at the TSS and downregulated by JQKD82 in MM.1S cells. MM.1S cells were treated with 1 μmol/L of JQKD82 or DMSO for 48 hours. RPM, reads per million mapped reads. TES, transcription end site. C, ChIP-seq tracks of RNAPII (Ser5p), RNAPII (Ser2p), and RNAPII at representative MYC target gene loci (MYC and NOLC1) after treatment with JQKD82 in MM.1S cells. D, JQKD82 induced changes in the pausing index [the ratio of RNAPII ChIP-seq signal in the TSS region (TSSR) to the signal in the gene body] at CDK7 inhibitor (THZ1)–sensitive or –insensitive genes, and JQKD82-sensitive or -insensitive genes in MM.1S cells. Gene sensitivity status was defined by gene expression changes genome wide in MM.1S cells exposed to THZ1 (4 hours) or JQKD82 (48 hours). P values were calculated by unpaired Student t test comparing genes of different sensitivity status.E, JQKD82-induced changes in ChIP-seq signal of RNAPII (Ser5p) in the body of genes classified as sensitive or insensitive to CDK7 or JQKD82 in MM.1S cells as in D. P values were calculated by unpaired Student t test comparing genes of different sensitivity status. F, TAF3 (the subunit of TFIID with a H3K4me3 recognizing PHD domain) binding at JQKD82-downregulated genes was measured by ChIP-seq normalized to reference exogenous (ChIP-Rx) spiked-in Drosophila chromatin in MM.1S cells. Comparison is made to H3K4me3 ChIP-Rx data in MM.1S cells treated with 1 μmol/L of JQKD82 or DMSO for 48 hours. G, ChIP-seq signal of H3K4me3, CDK7, TAF3, and KDM5A at the TSS of genes either sensitive to CDK7 inhibition by THZ1 (black line) compared with THZ1-insensitive genes (gray line). H, Proposed model of KDM5A functions and the mechanism of action of JQKD82. KDM5A interacts with P-TEFb (CCNT2 and CDK9) and transactivates MYC target genes. JQKD82 or knockdown of KDM5A induces hypermethylation of H3K4me3 (hyper-H3K4me3), leading to anchoring of TFIID via TAF3 binding, which may function as a barrier to productive RNAPII phosphorylation by TFIIH (CDK7) and P-TEFb (CDK9), thereby dampening pause release and reducing MYC target gene transcription (indicated by faded RNAPII elongation complex).
Next, to interrogate the contribution of KDM5A to RNAPII pause release, we analyzed the traveling ratio (TR, or the pausing index) at genes co-occupied by KDM5A and MYC and whose expression was downregulated after JQKD82 treatment, and found an increase in pausing both with JQKD82 treatment and by shKDM5A knockdown (Supplementary Fig. S6E and S6F). Pausing, just as the H3K4me3 levels, was increased globally for genes sensitive or insensitive to KDM5A inhibition (Fig. 6D). Then we separated the genes according to CDK7 inhibitor (THZ1; ref. 56)–sensitive and –insensitive groups to determine if transcription pausing correlated with CDK7 sensitivity. Again we saw similar increase in pausing index in both THZ1-sensitive and -insensitive genes (Fig. 6D). This indicated that failed pause release is not the primary determinant of the transcription decrease elicited by JQKD82. We then reasoned that because KDM5A and CDK7 are positively correlated (Fig. 5A), that the genes most sensitive to CDK7 inhibition would also lose RNAPII Ser5 phosphorylation to a greater extent than insensitive genes. Global analysis showed most genes are mildly pausing and also losing RNAPII Ser5 phosphorylation in the gene body (Supplementary Fig. S6G). However, while the global pausing trend was not different among these gene sets, loss of RNAPII Ser5p was significantly greater at CDK7- and KDM5A-sensitive genes (Fig. 6E).
We then looked at the assembly of the transcriptional preinitiation complex. TAF3, one of the TFIID subunits, anchors TFIID to H3K4me3 at TSS-proximal nucleosomes (54, 57). Furthermore, evidence indicates that other components of TFIID, such as the subunit TAF7, are able to inhibit TFIIH (CDK7) and P-TEFb (CDK9) kinase activities, in a context-dependent manner (58). These data suggested a potential link between binding of TAF3 to H3K4me3 and TFIIH and P-TEFb function. Thus, we hypothesized that hypermethylation of H3K4me3 at TSS-proximal nucleosomes would result in anchored TAF3 and abnormal TFIID retainment, resulting in hindered TFIIH and P-TEFb activities. To test this hypothesis, we measured the genome-wide binding changes of TAF3 in MM.1S cells treated with either JQKD82 or vehicle control. To ensure our detection of global changes in TAF3 were appropriately normalized, we again performed spike-in normalization with ChIP-Rx. In contrast with vehicle-treated controls, JQKD82-treated MM.1S cells displayed increased H3K4me3 and TAF3 binding at the TSS of genes that were downregulated by JQKD82 (Fig. 6F). Indeed, TAF3 is bound to CDK7-sensitive genes at a much higher level, suggesting that there is an H3K4me3/TAF3/CDK7 axis that KDM5A is recruited to modulate (Fig. 6G).
Collectively, these results support a model that KDM5A functions to control expression of MYC target genes by reducing TSS H3K4m3 levels at these gene loci, resulting in TFIID (TAF3) release, and phosphorylation of serine 5 residue on RNAPII by CDK7. In contrast, inhibition or knockdown of KDM5A induces TSS-proximal hyper-H3K4me3, resulting in aberrant TFIID (TAF3) anchoring that inhibits productive RNAPII phosphorylation by TFIIH and P-TEFb, thereby reducing MYC target gene transcription (Fig. 6H).
MYC Target Genes Corequire KDM5A
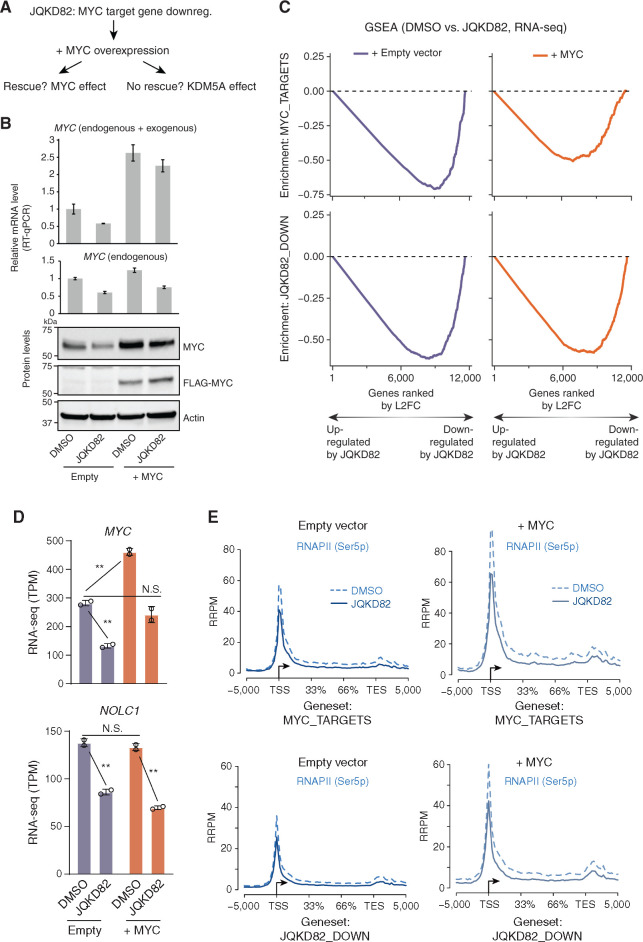
Our data support a model of inhibition of phosphorylation of RNAPII by hypermethylation of H3K4me3, demonstrated in Fig. 6H. However, because JQKD82 results in hypermethylation of H3K4me3, downregulation of MYC and, indeed, downregulation of MYC target genes, we could not formally identify whether the dominant effect of KDM5A inhibition is mediated by MYC itself or whether KDM5A inhibition has effects on gene expression independent of MYC. To address this question, we hypothesized that if the dominant effect of KDM5A inhibition was mediated by MYC itself, then exogenous overexpression of MYC in a manner insensitive to KDM5A inhibition should result in rescue of MYC target gene expression. In contrast, if KDM5A directly regulates MYC target genes in multiple myeloma cells, then we hypothesized that JQKD82 would result in their downregulation even in the setting of exogenously expressed MYC protein (Fig. 7A).
Figure 7.
Dissecting the requirement of MYC and KDM5A for transcription and RNAPII phosphorylation of JQKD82-sensitive genes. A, Framing the question: Is the effect of JQKD82 on downregulating MYC targets an indirect effect that can be rescued by MYC overexpression, or do MYC target genes require KDM5A even in the presence of supraphysiologic levels of MYC? B, FLAG-tagged MYC or empty vector was overexpressed in MM.1S cells. Expression level of MYC measured by quantitative real-time PCR (RT-qPCR; top) and by immunoblot (bottom) compared to cells with empty vector after treated with DMSO or 1 μmol/L of JQKD82 for 48 hours. MYC quantitative real-time PCR results are normalized against RPLP0 as a housekeeping control. For detecting both endogenous and exogenous MYC, the primers target only the CDS region (top). For detecting only endogenous MYC, the primers target the region including 5′UTR because exogenous MYC does not include 5′ UTR of the MYC gene (bottom). The expression relative to DMSO-treated empty vector control is shown as mean ± SD of triplicate measurements. C, MM.1S cells were treated as in B, and RNA was extracted for RNA-seq analysis. This was then used to perform GSEA of MYC targets (top) and JQKD82-sensitive genes (bottom). Gene-level quantification was ERCC spike-in normalized on a per-cell basis to adjust for changes in total RNA per cell. Changes in expression upon JQKD82 treatment were quantified and ranked by log2 fold change (L2FC, of transcripts per million), and enrichment scores were generated using the GSEA desktop software package. Gene expression changes were evaluated in this way from MM.1S cells with either empty vector (purple) or MYC overexpression vector (orange). n = 2 independent biological replicates per condition. D, RNA-seq from MM.1S cells for MYC and NOLC1 genes in TPM (transcripts per million) comparing DMSO versus JQKD82 treatment in the cellular context of either empty vector versus MYC overexpression. Error bars represent the SD across two independent biological replicates. N.S., P > 0.1; **, P < 0.005, unpaired Student t test. E, RNAPII (Ser5p) ChIP-seq with exogenous reference genomes (ChIP-Rx) from MM.1S cells with either empty vector (left two panels) or MYC overexpression vector (right two panels), viewed at both MYC target genes (top two panels) and JQKD82-sensitive genes (bottom two panels). Dotted lines indicate metagene signal from DMSO-treated cells, and solid lines indicate metagene signal from JQKD82-treated cells.
To test these hypotheses, we derived MM.1S stably expressing either exogenous Flag-tagged MYC or empty vector as a control (Fig. 7B). Flag-tagged MYC was identified in stable cells by quantitative real-time PCR and immunoblotting (Fig. 7B). Treatment of empty vector cells with JQKD82 resulted in downregulation of MYC mRNA and protein, which could also be observed in MYC-overexpressing cells because endogenous MYC level in these cells is similarly dependent on KDM5A, although exogenous MYC is not affected (Fig. 7B). Next, we treated these empty vector and Flag-tagged MYC MM.1S cells with either vehicle or JQKD82 for 48 hours and performed RNA-seq, correcting for global effects on transcriptional amplification by using exogenous RNA spike-in controls (ERCC) on a per-cell level (ref. 59; Fig. 7C). Increased MYC levels were moderate in MYC-overexpressing cells but enough to rescue MYC (during JQKD82 exposure) back to a level indistinguishable from empty vector–transduced MM.1S cells without JQKD82 treatment (Fig. 7D, top). In this condition, JQKD82 resulted in loss of expression of canonical (pan-cancer) MYC targets and the MM.1S cell–specific MYC/KDM5A cobound genes sensitive to JQKD82, similar to empty vector control (Fig. 7C and D, shown is the MYC target gene NOLC1 as a representative example). Overexpression of MYC alone did not induce further NOLC1 expression, suggesting that NOLC1 expression is upregulated to the maximum level by endogenous MYC in MM.1S cells (Fig. 7D). On the basis of our model (presented in Fig. 6H) and given that exogenous MYC could not rescue the expression of MYC target genes downregulated in response to JQKD82, we then hypothesized that overexpression of MYC would similarly be unable to rescue the defect in RNAPII phosphorylation. To test this hypothesis, we treated empty vector and Flag-tagged MYC MM.1S cells with either vehicle or JQKD82 for 48 hours and performed ChIP-Rx to RNAP II Ser5 phosphorylation (Fig. 7E). Consistent with our model and RNA-seq results, overexpression of MYC resulted in increased Ser5 phosphorylation, which was reduced by JQKD82 treatment (Fig. 7E). Together, these data indicated that KDM5A is directly required for the transcription of MYC target genes, and that inhibition of KDM5A using JQKD82 results in loss of RNAPII phosphorylation mediated by hypermethylation of H3K4me3 and anchoring of TFIID.
Discussion
In the context of epigenetic regulation, recent studies have suggested the pathologic relevance of KDM5B in multiple myeloma (33, 35). Here, we demonstrate that the distinct KDM5 subfamily member KDM5A plays a critical role in multiple myeloma cell growth through a unique link to MYC-driven transcriptional programs. KDM5A, but not KDM5B, regulates both MYC and the MYC-driven downstream transcriptional network, suggesting distinct functions of these two H3K4 demethylases in multiple myeloma. KDM5A and MYC co-occupy target genes on a genome-wide scale to orchestrate their regulation of oncogenic target gene transcription in multiple myeloma cells. Most of these KDM5A and MYC co-occupied regions, including the MYC locus itself, are located proximally to the TSS of various genes, suggesting that KDM5A controls MYC target gene expression at promoter regions.
KDM5 family members remove the active transcription mark H3K4me3. They are components of repressor complexes, including the Sin3B/HDAC and NuRD complexes, and are therefore transcriptional corepressors (22, 23, 50, 51). Indeed, KDM5s repress target gene expression by H3K4 demethylation to induce various cellular and biological processes, including DNA repair, differentiation, and transformation (60,61,62,63). Despite these findings, accumulating evidence indicates that KDM5 proteins also function as context-specific transcriptional activators (40,41,42,43,44). KDM5 proteins activate target genes via several distinct mechanisms, including inhibition of HDAC activity or enhancement of nuclear receptor–mediated transcription (41, 42, 64). Here, using a novel prodrug KDM5 inhibitor, we identify a subset of genes that are bound and positively regulated by KDM5A. Importantly, the dominant phenotype induced by KDM5A inhibition is cell-cycle arrest, with a gene expression pattern consistent with inhibition of MYC target gene expression. These data indicate that the dominant effect of KDM5A inhibition is loss of MYC target gene oncogenic transcriptional output. We show that KDM5A colocalizes with CDK7, CDK9, MYC, and RNAPII. Furthermore, the KDM5 inhibitor JQKD82 induces hypermethylation of H3K4me3 and reduces phosphorylation of RNAPII, accompanied by RNAPII pausing at the MYC target loci. Our data support a model by which hypermethylation of H3K4me3 results in ectopic anchoring of TAF3, forcing reduced phosphorylation of RNAPII. In multiple myeloma cells, this occurs predominantly at MYC target gene loci and intriguingly, is partially independent of MYC expression itself. These data suggest that KDM5A may promote phosphorylation of the C-terminal domain of RNAPII with TFIIH and P-TEFb by promoting the release of TFIID. Indeed, inhibition of KDM5A results in anchoring of TFIID (Fig. 6F), which includes TAF7 that may inhibit CDK7 and CDK9 kinase activities (58). Consistent with this model, overexpression of MYC is both insufficient to restore the JQKD82-induced downregulation of MYC targets and RNAPII phosphorylation, indicating that KDM5A plays a functional role in demethylating H3K4me3, resulting in the phosphorylation of RNAPII at MYC target gene loci, partly independent of MYC expression. While the precise mechanism of how KDM5A facilitates the phosphorylation of RNAPII and RNAPII pause release is undetermined, it is likely to involve protein–protein interactions with members of TFIID that suppress CDK7 and CDK9 activities, and remains an active area of interest.
With the design of the prodrug JQKD82, we have successfully delivered a previously validated KDM5-selective inhibitor KDM5-C49 (33) to cancer cells both in vitro and in vivo. JQKD82 significantly prolongs the survival of tumor-bearing mice, and no apparent toxicity was observed during 3-week treatment, demonstrating its tolerability. In combination with evidence for broad anticancer activity and limited toxicity in vitro and in vivo, these data imply a promising therapeutic index for KDM5 inhibition. Ongoing studies are directed at understanding the mechanism of selectivity for this apparent therapeutic window, prior to clinical translation.
In summary, here we have revealed the biological significance and molecular functions of KDM5A in multiple myeloma. Our data identify a novel function of KDM5A that is required to sustain and reinforce the MYC oncogenic transcriptional program in multiple myeloma. Catalytic inhibition of KDM5A results in hypermethylation of proximal TSS H3K4me3, resulting in failed pause release at MYC-regulated genes. As MYC plays critical roles in driving tumorigenesis, these data indicate potential clinical implications for KDM5 inhibition using in vivo active selective KDM5 inhibitors, and a mechanism for identifying on-target behavior of these compounds. Our results therefore provide the rationale for further evaluation and development of KDM5 inhibitors as novel therapeutic strategies in multiple myeloma and other KDM5A-dependent malignancies.
Methods
Primary Cells (Multiple Myeloma Cells and Normal B Cells)
Bone marrow or peripheral blood samples were obtained from patients with multiple myeloma or healthy donors with written informed consent after approval of the Institutional Review Board of the Dana-Farber Cancer Institute or Kumamoto University in accordance with the Declaration of Helsinki. Mononuclear cells were isolated from samples by Ficoll-Paque PLUS (GE Healthcare). Multiple myeloma cells were enriched by anti-CD138 magnetic activated cell separation microbeads (Miltenyi Biotec). Normal B cells from healthy volunteers' peripheral blood were enriched by negative selection methods using EasySep Human B Cell Isolation Kit (STEMCELL Technologies). For B-cell proliferation, isolated B cells were stimulated by 10 μg/mL of human CD40 antibody (R&D systems) in the presence of 100 U/mL of recombinant human IL4 (R&D Systems).
Cell Lines
Human multiple myeloma cell lines MM.1S, U266, and NCI-H929 were obtained from ATCC. The human multiple myeloma cell line MOLP-8 was purchased from DSMZ. Human multiple myeloma cell lines KMS-11 and KMS-20 were purchased from the JCRB Cell Bank. Human multiple myeloma cell line OPM-1 is a gift from Edward Thompson (University of Texas, Galveston, TX). The identities of MM.1S, U266, OPM-1, NCI-H929, and KMS-11 were validated by STR profiling (GenePrint10 System, Promega). MOLP-8 cells expressing TurboGFP and luciferase (MOLP-8 TurboGFP-Luc) were generated by retrovirally transducing TurboGFP-IRES-luciferase bicistronic expression vector into MOLP-8 cells. Human embryonic kidney cell line 293T, human breast cancer cell line MCF7, and human colon cancer cell line Caco2 were otatined from ATCC. Cell lines were used within 3 months after thawing. Mycoplasma contamination was excluded using the MycoAlert Mycoplasma Detection Kit (Lonza). All multiple myeloma cell lines were maintained in RPMI1640 containing 100 U/mL penicillin and 100 μg/mL streptomycin, supplemented with 10% (v/v) FBS, and 2 μmol/L l-glutamine in 5% CO2 at 37°C. 293T and MCF7 cells were maintained in DMEM supplemented with 10% (v/v) FBS. Caco2 cells were maintained in EMEM supplemented with 20% (v/v) FBS.
Chemicals
KDM5-C49 and KDM5-C70 were synthesized in the Qi Lab based on literature-reported procedures (33). Compound JQKD82 and KDM5-C49-Biotin were designed and synthesized based on the scheme listed in Supplementary Methods. The structure and purity of these compounds were further confirmed by nuclear magnetic resonance and LC/MS, with detailed descriptions in Supplementary Methods.
In Vivo Xenograft Models
Six-week-old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (from The Jackson Laboratory, catalog no: 0005557) were used in this study.
For the disseminated model, 1 × 106 MOLP-8 TurboGFP-Luc cells were injected intravenously into NSG mice. Tumor burden was serially monitored by BLI using IVIS Imaging System and Living Image Software (PerkinElmer). After tumor engraftment, the mice were randomly divided into two groups (JQKD82 treatment or vehicle control group), and then i.p. treated with JQKD82 at 50 mg/kg as prepared in the same manner in the pharmacokinetics study described in Supplementary Methods or vehicle (DMSO in 10% of hydroxypropyl beta cyclodextrin) twice a day for 3 weeks.
For the plasmacytoma model, 1 × 106 MOLP-8 TurboGFP-Luc cells were injected subcutaneously into the right flank of NSG mice within PBS mixed with 30% of Matrigel. Mice were serially imaged after inoculation. In parallel, tumor size was measured using an electronic caliper, and tumor volume was determined using the formula: (length × width+) × 2−1, where length is greater than width. After tumor engraftment confirmed by BLI signal and tumor size measurement, the mice were randomly divided into two groups (JQKD82 treatment or vehicle control group), and then i.p. treated with JQKD82 at 75 mg/kg or vehicle twice a day for 2 weeks. Animal studies were performed under a protocol approved by the Dana-Farber Institutional Animal Care and Use Committee.
RNA-seq
Total RNA was extracted from MM.1S cells using RNeasy Mini Kit (Qiagen) after 48 hours of treatment with 1 μmol/L of JQKD82 or DMSO in biological duplicate. For RNA-seq using MYC-overexpressed MM.1S cells, external control spike-ins were used to allow normalization to cell number. Briefly, 8 μL of a 1:100 dilution of ERCC RNA Spike-In Mix#1 (Thermo Fisher Scientific) was added to the lysate from 5 × 105 MM.1S-MYC or MM.1S-empty cells before RNA extraction. RNAs were then treated with TURBO DNA-free reagents to remove contaminating DNA (Thermo Fisher Scientific). The libraries were prepared using NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs) or TruSeq Stranded mRNA Library Prep Kit (Illumina), and subjected to 75 base-pair single-read sequencing on an Illumina HiSeq 2000 or NextSeq 500. Detailed RNA-seq analysis is descrbed in Supplementary Methods. RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE148047.
ChIP-seq
ChIP procedure is described in Supplementary Methods. The libraries were constructed from 3 to 10 ng of ChIP samples or 50 ng of input samples using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs), and quantified using GenNext NGS Library Quantification kit (Toyobo). The fragment length of libraries was assessed using 2200 TapeStation (Agilent). Seventy-five base-pair single-read sequencing was performed on an Illumina NextSeq 500. The MM.1S ChIP-seq data for H3K27Ac (GSM894083), H3K27me3 (GSM1252088), MYC (GSM894108), CDK7 (GSM1121098), CDK9 (GSM1085735), and RNAPII (GSM1070127) were downloaded from NCBI (8, 65, 66). Detailed ChIP-seq analysis is descrbed in Supplementary Methods. The ChIP-seq data in this study have been deposited in the GEO database under accession number GSE148046.
Statistical Analysis
For statistical comparison, two-tailed Student t test was performed between two groups, assuming normal distribution. IC50 values were determined by nonlinear regression using GraphPad Prism8 software. Log-rank test was carried out to assess significance of survival differences using GraphPad Prism8 software. A value ofP < 0.05 was considered statistically significant.
Material Availability
The chemical probes generated in this study will be available for research purposes through proper material transfer agreement.
Data Availability
RNA-seq and ChIP-seq data have been deposited in the GEO database under SuperSeries accession number GSE148048, which is comprised of SubSeries accession numbers GSE148046 and GSE148047.
Supplementary Material
Acknowledgments
The authors thank Hitoshi Takizawa, Goro Sashida, Bob Roader, and Toshio Suda for helpful suggestions and discussions. They thank the Liaison Laboratory Research Promotion Center, Institute of Molecular Embryology and Genetics, Kumamoto University for assistance with ChIP-seq and RNA-seq analyses; the International Research Center for Medical Sciences, Kumamoto University for help with MOLP-8 TurboGFP-Luc cell sorting; the Center for Cancer Computational Biology, Dana-Farber Cancer Institute for assistance with RNA-seq analysis; the Animal Resources Facility, Dana-Farber Cancer Institute for support with animal studies; and the PRISM program at Broad Institute for support on profiling JQKD82 in a panel of 767 cancer cell lines. The authors also thank Charles Lin and Jošt Vrabič Koren for the ERCC spike-in per cell normalization algorithm. They thank Vineela Gangalapudi and Sivasish Sindiri for assistance with constructing the ERCC spike-in RNA-seq pipeline. They thank David Milewski and Young Song for assistance with ChIP-Rx. This research was supported by the Japan Society for the Promotion of Science grants KAKENHI (1 8 H 0 6 1 6 7 and 1 9 K 2 1 2 7 6, to H. Ohguchi), grants from Mochida Memorial Foundation for Medical and Pharmaceutical Research (to H. Ohguchi), the Shinnihon Foundation of Advanced Medical Treatment Research (to H. Ohguchi), the Princess Takamatsu Cancer Research Fund (18–25002; to H. Ohguchi), Kobayashi Foundation for Cancer Research (to H. Ohguchi), the Ichiro Kanehara Foundation for the Promotion of Medical Sciences and Medical Care (to H. Ohguchi), Japanese Society of Myeloma Research Award (to H. Ohguchi), Japanese Society of Hematology Research Grant (to H. Ohguchi), the program of the Joint Usage/Research Center for Developmental Medicine, Institute of Molecular Embryology and Genetics, Kumamoto University (to H. Ohguchi), International Research Center for Medical Sciences grant for international collaborative research (to H. Ohguchi), Dana-Farber/Harvard Cancer Center SPORE in Multiple Myeloma Career Enhancement Award (to H. Ohguchi), NIH training grant 5T32CA236754 (to X. Zhang), Cancer Research UK grant C41580/A23900 (to U. Oppermann), the LEAN program grant from the Leducq Foundation (to U. Oppermann), NIH grant 5P50CA100707 (to K.C. Anderson), grants from the Paula and Rodger Riney Foundation (to K.C. Anderson), NIH grant R01CA233601 (to J. Qi), and a Leukemia & Lymphoma Society TRP grant (to J. Qi). A.D. Durbin is the recipient of a Damon Runyon-Sohn Fellowship and funding from the Alex's Lemonade Stand Foundation, CureSearch for Children's Cancer foundation, and the American Lebanese Syrian Associated Charities.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
#H. Ohguchi, P.M.C. Park, T. Wang, B.E. Gryder contributed equally to this article.
Blood Cancer Discov 2021;2:370–87
Authors' Disclosures
H. Ohguchi reports grants from Japan Society for the Promotion of Science KAKENHI (1 8 H 0 6 1 6 7 and 1 9 K 2 1 2 7 6), Mochida Memorial Foundation for Medical and Pharmaceutical Research, Shinnihon Foundation of Advanced Medical Treatment Research, Princess Takamatsu Cancer Research Fund (18–25002), Kobayashi Foundation for Cancer Research, Ichiro Kanehara Foundation for the Promotion of Medical Sciences and Medical Care, Japanese Society of Myeloma Research Award, Japanese Society of Hematology Research Grant, and the program of the Joint Usage/Research Center for Developmental Medicine, Institute of Molecular Embryology and Genetics, Kumamoto University during the conduct of the study. Y. Kawano reports personal fees from Janssen Pharmaceuticals, Sanofi, Takeda Pharmaceutical Company Limited, and Ono Pharmaceutical CO., LTD. outside the submitted work. N.C. Munshi reports other support from Bristol Myers Squibb, Janssen, Amgen, Takeda, Oncopep, AbbVie, Karyopharm, Novartis, and Legend outside the submitted work. M. Nakao reports grants from Japan Society for the Promotion of Science and Japan Agency for Medical Research and Development during the conduct of the study. U. Oppermann reports nonfinancial support from Oxford NIHR BRC during the conduct of the study, as well as grants from Leducq Foundation, Cancer Research UK, Arthritis Research UK, Bristol Myers Squibb, Bayer Healthcare, and Innovate UK outside the submitted work. A.D. Durbin reports grants from NCI, Damon Runyon Cancer Research Foundation, Rally Foundation for Childhood Cancer Research, Alex's Lemonade Stand Foundation, and CureSearch for Childhood Cancer Research during the conduct of the study. K.C. Anderson reports personal fees from Pfizer, AstraZeneca, Precision Biosciences, Windmill, Janssen, Starton, C4 Therapeutics, Oncopep, Raqia, and Mana Therapeutics outside the submitted work. J. Qi reports grants from Leukemia & Lymphoma Society and NIH during the conduct of the study; other support from Epiphanes outside the submitted work; and a patent for PCT/US2019/045259 pending. No disclosures were reported by the other authors.
One of the Editors-in-Chief is an author on this article. In keeping with the AACR's editorial policy, the peer review of this submission was managed by a member of Blood Cancer Discovery's Board of Scientific Editors, who rendered the final decision concerning acceptability.
Authors' Contributions
H. Ohguchi: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. P.M.C. Park: Investigation, methodology, writing–review and editing. T. Wang: Investigation, methodology, writing–review and editing. B.E. Gryder: Formal analysis, investigation. D. Ogiya: Investigation. K. Kurata: Investigation. X. Zhang: Investigation.D. Li: Investigation. C. Pei: Investigation. T. Masuda: Investigation. C. Johansson: Investigation. V.K. Wimalasena: Investigation.Y. Kim: Investigation. S. Hino: Resources, investigation. S. Usuki: Investigation. Y. Kawano: Investigation. M.K. Samur: Investigation. Y.-T. Tai: Resources. N.C. Munshi: Resources. M. Matsuoka: Investigation. S. Ohtsuki: Investigation. M. Nakao: Investigation.T. Minami: Investigation. S. Lauberth: Resources. J. Khan: Resources. U. Oppermann: Supervision, investigation. A.D. Durbin: Formal analysis, investigation, writing–review and editing. K.C. Anderson: Conceptualization, supervision, funding acquisition, investigation, writing–review and editing. T. Hideshima: Conceptualization, formal analysis, supervision, validation, investigation, writing–review and editing. J. Qi: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1.Kumar SK, Rajkumar V, Kyle RA, van Duin M, Sonneveld P, Mateos MV, et al. Multiple myeloma. Nat Rev Dis Primers 2017;3:17046. [DOI] [PubMed] [Google Scholar]
- 2.Dang CV.MYC on the path to cancer. Cell 2012;149:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV.MYC, metabolism, and cancer. Cancer Discov 2015;5:1024–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jovanovic KK, Roche-Lestienne C, Ghobrial IM, Facon T, Quesnel B, Manier S.Targeting MYC in multiple myeloma. Leukemia 2018;32:1295–306. [DOI] [PubMed] [Google Scholar]
- 5.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012;151:68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeid R, Lawlor MA, Poon E, Reyes JM, Fulciniti M, Lopez MA, et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat Genet 2018;50:515–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradner JE, Hnisz D, Young RA.Transcriptional addiction in cancer. Cell 2017;168:629–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012;151:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chesi M, Robbiani DF, Sebag M, Chng WJ, Affer M, Tiedemann R, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell 2008;13:167–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chesi M, Stein CK, Garbitt VM, Sharik ME, Asmann YW, Bergsagel M, et al. Monosomic loss of MIR15A/MIR16–1 is a driver of multiple myeloma proliferation and disease progression. Blood Cancer Discov 2020;1:68–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T, et al. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia 2011;25:1026–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, et al. IRF4 addiction in multiple myeloma. Nature 2008;454:226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holien T, Vatsveen TK, Hella H, Waage A, Sundan A.Addiction to c-MYC in multiple myeloma. Blood 2012;120:2450–3. [DOI] [PubMed] [Google Scholar]
- 14.Harada T, Ohguchi H, Grondin Y, Kikuchi S, Sagawa M, Tai YT, et al. HDAC3 regulates DNMT1 expression in multiple myeloma: therapeutic implications. Leukemia 2017;31:2670–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wimalasena VK, Wang T, Sigua LH, Durbin AD, Qi J.Using chemical epigenetics to target cancer. Mol Cell 2020;78:1086–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev 2004;18:2747–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011;146:904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durbin AD, Zimmerman MW, Dharia NV, Abraham BJ, Iniguez AB, Weichert-Leahey N, et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet 2018;50:1240–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson MA, Kouzarides T.Cancer epigenetics: from mechanism to therapy. Cell 2012;150:12–27. [DOI] [PubMed] [Google Scholar]
- 20.Ohguchi H, Hideshima T, Anderson KC.The biological significance of histone modifiers in multiple myeloma: clinical applications. Blood Cancer J 2018;8:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klose RJ, Kallin EM, Zhang Y.JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 2006;7:715–27. [DOI] [PubMed] [Google Scholar]
- 22.Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, et al. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 2007;128:889–900. [DOI] [PubMed] [Google Scholar]
- 23.Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 2007;128:1063–76. [DOI] [PubMed] [Google Scholar]
- 24.Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG, Jr.Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell 2005;18:623–35. [DOI] [PubMed] [Google Scholar]
- 25.Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016;537:548–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 2009;459:847–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng YC, Lee CF, Li YS, Chen YR, Hsiao PW, Chan MY, et al. Histone demethylase RBP2 promotes lung tumorigenesis and cancer metastasis. Cancer Res 2013;73:4711–21. [DOI] [PubMed] [Google Scholar]
- 28.Cao J, Liu Z, Cheung WK, Zhao M, Chen SY, Chan SW, et al. Histone demethylase RBP2 is critical for breast cancer progression and metastasis. Cell Rep 2014;6:868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010;141:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol 2006;8:764–70. [DOI] [PubMed] [Google Scholar]
- 31.Kidder BL, Hu G, Yu ZX, Liu C, Zhao K.Extended self-renewal and accelerated reprogramming in the absence of Kdm5b. Mol Cell Biol 2013;33:4793–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong PP, Miranda F, Chan KV, Berlato C, Hurst HC, Scibetta AG.Histone demethylase KDM5B collaborates with TFAP2C and Myc to repress the cell cycle inhibitor p21(cip) (CDKN1A). Mol Cell Biol 2012;32:1633–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johansson C, Velupillai S, Tumber A, Szykowska A, Hookway ES, Nowak RP, et al. Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol 2016;12:539–45. [DOI] [PubMed] [Google Scholar]
- 34.Vinogradova M, Gehling VS, Gustafson A, Arora S, Tindell CA, Wilson C, et al. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat Chem Biol 2016;12:531–8. [DOI] [PubMed] [Google Scholar]
- 35.Tumber A, Nuzzi A, Hookway ES, Hatch SB, Velupillai S, Johansson C, et al. Potent and selective KDM5 inhibitor stops cellular demethylation of H3K4me3 at transcription start sites and proliferation of MM1S myeloma cells. Cell Chem Biol 2017;24:371–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harmeyer KM, Facompre ND, Herlyn M, Basu D.JARID1 histone demethylases: emerging targets in cancer. Trends Cancer 2017;3:713–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood 2006;108:2020–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER, 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corsello SM, Nagari RT, Spangler RD, Rossen J, Kocak M, Bryan JG, et al. Discovering the anticancer potential of non-oncology drugs by systematic viability profiling. Nat Cancer 2020;1:235–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brier AB, Loft A, Madsen JGS, Rosengren T, Nielsen R, Schmidt SF, et al. The KDM5 family is required for activation of pro-proliferative cell cycle genes during adipocyte differentiation. Nucleic Acids Res 2017;45:1743–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, Secombe J, et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 2011;333:1881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee N, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y.The H3K4 demethylase lid associates with and inhibits histone deacetylase Rpd3. Mol Cell Biol 2009;29:1401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Secombe J.The histone demethylase KDM5 activates gene expression by recognizing chromatin context through its PHD reader motif. Cell Rep 2015;13:2219–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lloret-Llinares M, Perez-Lluch S, Rossell D, Moran T, Ponsa-Cobas J, Auer H, et al. dKDM5/LID regulates H3K4me3 dynamics at the transcription-start site (TSS) of actively transcribed developmental genes. Nucleic Acids Res 2012;40:9493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu S, Teng YC, Yuan C, Wu YT, Chan MY, Cheng AN, et al. The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat Struct Mol Biol 2008;15:419–21. [DOI] [PubMed] [Google Scholar]
- 46.Oki S, Ohta T, Shioi G, Hatanaka H, Ogasawara O, Okuda Y, et al. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep 2018;19:e46255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lafita-Navarro MC, Kim M, Borenstein-Auerbach N, Venkateswaran N, Hao YH, Ray R, et al. The aryl hydrocarbon receptor regulates nucleolar activity and protein synthesis in MYC-expressing cells. Genes Dev 2018;32:1303–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A 2006;103:17834–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, et al. Analysis of the human endogenous coregulator complexome. Cell 2011;145:787–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishibuchi G, Shibata Y, Hayakawa T, Hayakawa N, Ohtani Y, Sinmyozu K, et al. Physical and functional interactions between the histone H3K4 demethylase KDM5A and the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem 2014;289:28956–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varier RA, Carrillo de Santa Pau E, van der Groep P, Lindeboom RG, Matarese F, Mensinga A, et al. Recruitment of the mammalian histone-modifying EMSY complex to target genes is regulated by ZNF131. J Biol Chem 2016;291:7313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schier AC, Taatjes DJ.Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev 2020;34:465–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER, et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 2014;9:1163–70. [DOI] [PubMed] [Google Scholar]
- 54.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 2007;131:58–69. [DOI] [PubMed] [Google Scholar]
- 55.Jimeno-Gonzalez S, Ceballos-Chavez M, Reyes JC.A positioned +1 nucleosome enhances promoter-proximal pausing. Nucleic Acids Res 2015;43:3068–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014;511:616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 2013;152:1021–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gegonne A, Weissman JD, Lu H, Zhou M, Dasgupta A, Ribble R, et al. TFIID component TAF7 functionally interacts with both TFIIH and P-TEFb. Proc Natl Acad Sci U S A 2008;105:5367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, et al. Revisiting global gene expression analysis. Cell 2012;151:476–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gong F, Clouaire T, Aguirrebengoa M, Legube G, Miller KM.Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J Cell Biol 2017;216:1959–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beshiri ML, Holmes KB, Richter WF, Hess S, Islam AB, Yan Q, et al. Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proc Natl Acad Sci U S A 2012;109:18499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oser MG, Sabet AH, Gao W, Chakraborty AA, Schinzel AC, Jennings RB, et al. The KDM5A/RBP2 histone demethylase represses NOTCH signaling to sustain neuroendocrine differentiation and promote small cell lung cancer tumorigenesis. Genes Dev 2019;33:1718–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell 2007;25:801–12. [DOI] [PubMed] [Google Scholar]
- 64.Chan SW, Hong W.Retinoblastoma-binding protein 2 (Rbp2) potentiates nuclear hormone receptor-mediated transcription. J Biol Chem 2001;276:28402–12. [DOI] [PubMed] [Google Scholar]
- 65.Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, et al. Genome-wide localization of small molecules. Nat Biotechnol 2014;32:92–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013;153:320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ChIP-seq data have been deposited in the GEO database under SuperSeries accession number GSE148048, which is comprised of SubSeries accession numbers GSE148046 and GSE148047.