Leukemia and nonmalignant clonal hematopoiesis (CH) are indistinguishable by bulk sequencing assays. Here, DNA probes tailored to detect patient-specific oncogenic lesions are combined with multiplex immunophenotyping to definitively resolve leukemia and CH at a single-cell level.
Abstract
Genetic mutations associated with acute myeloid leukemia (AML) also occur in age-related clonal hematopoiesis, often in the same individual. This makes confident assignment of detected variants to malignancy challenging. The issue is particularly crucial for AML posttreatment measurable residual disease monitoring, where results can be discordant between genetic sequencing and flow cytometry. We show here that it is possible to distinguish AML from clonal hematopoiesis and to resolve the immunophenotypic identity of clonal architecture. To achieve this, we first design patient-specific DNA probes based on patient's whole-genome sequencing and then use them for patient-personalized single-cell DNA sequencing with simultaneous single-cell antibody–oligonucleotide sequencing. Examples illustrate AML arising from DNMT3A- and TET2-mutated clones as well as independently. The ability to personalize single-cell proteogenomic assessment for individual patients based on leukemia-specific genomic features has implications for ongoing AML precision medicine efforts.
Significance:
This study offers a proof of principle of patient-personalized customized single-cell proteogenomics in AML including whole-genome sequencing–defined structural variants, currently unmeasurable by commercial “off-the-shelf” panels. This approach allows for the definition of genetic and immunophenotype features for an individual patient that would be best suited for measurable residual disease tracking.
Introduction
Age-related clonal hematopoiesis (also known as clonal hematopoiesis of indeterminate potential) is seen in those without hematologic malignancy and with a somatic mutation detectable in a clonal population of mature blood cells (1,2,3). These mutations, most commonly in epigenetic regulators DNMT3A and TET2, typically are found in older individuals and appear to be associated with an increased risk for death, predominately from inflammation-associated vascular events (4). These mutations are also common in patients with acute myeloid leukemia (AML; ref. 5) and can make genomic measurement of residual disease challenging (6, 7). Single-cell DNA sequencing (scDNA-seq) for AML has recently been described (8, 9) but has yet to be used for the detection of novel chromosomal fusions that are the key, and often only, defining feature for multiple subtypes of this disease. We performed deep laboratory interrogation of samples, using whole-genome and targeted error-corrected sequencing, scDNA-seq with antibody–oligonucleotide conjugates, and multiparameter flow cytometry, from three adult patients with relapsed AML who harbored mutations detected potentially attributable to either leukemia or clonal hematopoiesis.
Results
Patient 1 was a 74-year-old man with a second relapse of AML. Comorbidities included type 2 diabetes with neuropathy, retinopathy and peripheral vascular disease, hypertension, hyperlipidemia, coronary arterial disease, congestive heart failure, active tobacco use with chronic obstructive pulmonary disease, and obstructive sleep apnea. Prior chemotherapy included clofarabine and cytarabine. Clinical flow cytometry identified a 1% CD34-positive population of abnormal blasts in blood. Blood marrow metaphase cytogenetics revealed a 47,XY karyotype with inversion of chromosome 16 and trisomy of chromosome 8 in four metaphases. A mutation in DNMT3A was noted by clinical DNA sequencing (DNA-seq), with additional mutations in DNMT3A and TET2 also noted in error-corrected research DNA-seq (Table 1).
Table 1.
Patient genetic characteristics
| P | Age, y/sex | Cytogenetics | Gene | HGVSc/HGVSp | Chr | Position | Ref | Alt | Consequence | VAF |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 74/M | +8, inv(16)(p13.1q22), del(20)(q11.2q13.3) | DNMT3Aa | NM_022552.4:c.811delGNP_072046.2:p.Asp271ThrfsTer45 | 2 | 25470949 | TC | T | Frameshift | 0.22 |
| DNMT3A | NM_022552.4:c.2408+5G>A | 2 | 25461994 | C | T | Splicing | 0.03 | |||
| TET2 | NM_001127208.2:c.5037C>ANP_001120680.1:p.Tyr1679Ter | 4 | 106196704 | C | A | Nonsense | 0.01 | |||
| MYH11-CBFB | N/A | 1616 | 1581509467131781 | N/A | N/A | Fusion gene | N/A | |||
| 2 | 63/F | t(6;14)(q25;q22) | CUX1a | NM_181552.3:c.2195delNP_853530.2:p.Pro732HisfsTer33 | 7 | 101844770 | AC | A | Frameshift | 0.07 |
| DNMT3Aa | NM_022552.4:c.2645G>ANP_072046.2:p.Arg882His | 2 | 25457242 | C | T | Missense | 0.07 | |||
| NPM1a | NM_002520.6:c.860_863dupNP_002511.1:p.Trp288CysfsTer12 | 5 | 170837543 | C | CTCTG | Frameshift | 0.03 | |||
| U2AF1a | NM_006758.2:c.101C>ANP_006749.1:p.Ser34Tyr | 21 | 44524456 | G | T | Missense | 0.06 | |||
| TIAM2-PPP2R5E | N/A | 614 | 15515959063959402 | N/A | N/A | Fusion gene | N/A | |||
| 3 | 71/M | Normal | CEBPA | NM_004364.4:c.949_950insGTCNP_004355.2:p.Glu316_Leu317insArg | 19 | 33792371 | A | AGAC | Inframe insertion | 0.08 |
| CEBPA | NM_004364.4:c.68dupNP_004355.2:p.His24AlafsTer84 | 19 | 33793252 | C | CG | Frameshift | 0.07 | |||
| DNMT3Aa | NM_022552.4:c.1979A>GNP_072046.2:p.Tyr660Cys | 2 | 25464534 | T | C | Missense | 0.20 | |||
| GATA2 | NM_032638.4:c.1085G>ANP_116027.2:p.Arg362Gln | 3 | 128200720 | C | T | Missense | 0.06 | |||
| TET2a | NM_001127208.2:c.3954+1G>TN/A | 4 | 106180927 | G | T | Splicing | 0.17 |
Abbreviations: Alt, alternate; Chr, chromosome; F, female; HGVSc, human genome variation society cDNA nomenclature; HGVSp, HGVS protein nomenclature; M, male; N/A, not applicable; P, patient; Ref, reference; VAF, variant allele frequency; y, year.
aIdentified by clinical and research sequencing.
Patient 2 was a 63-year-old woman with a second relapse of AML. No vascular, diabetic, or other comorbidities were noted. Prior chemotherapy had included three rounds of intensive chemotherapy (idarubicin and cytarabine followed by cytarabine consolidation, and then salvage therapy with cytarabine, etoposide, and mitoxantrone). Clinical flow cytometry identified a 9% CD117-positive population of abnormal blasts in bone marrow (BM). CD117 immunohistochemistry was positive for 10% of cells. Translocation t(6;14)(q25;q22) was observed in 9 of 20 metaphases examined. Clinical DNA-seq noted mutations in DNMT3A, U2AF1, NPM1, and CUX1 (Table 1).
Patient 3 was a 71-year-old man with a first relapse of AML. Concurrent comorbidities included hypertension and hyperlipidemia. He had previously received intensive chemotherapy with idarubicin and cytarabine induction followed by cytarabine consolidation. Clinical flow cytometry identified a 4.4% CD117-positive population of abnormal blasts in BM. Clinical DNA-seq noted mutations in both DMNT3A and TET2, and research sequencing confirmed these and also identified mutations in GATA2 and CEBPA (Table 1).
In all three patients, the relationship between mutations discovered by targeted DNA-seq, leukemia-associated chromosomal abnormalities, and aberrant immunophenotype was unclear based on clinical evaluation.
In the first two patients, with known chromosomal structural abnormality from clinical metaphase cytogenetics, whole-genome sequencing (WGS) was performed for exact DNA breakpoint determination using BM aspirate sorted for either CD34 or CD117 positivity based on clinical immunophenotype (Fig. 1; Supplementary Fig. S1). Patient-specific primers for scDNA-seq (Mission Bio, Inc.) were then designed to detect these breakpoints and also mutations identified by clinical and research-targeted DNA-seq (Supplementary Table S1). Both chromosomal breakpoints and mutations found by bulk targeted DNA-seq were detected by scDNA-seq (Supplementary Fig. S2).
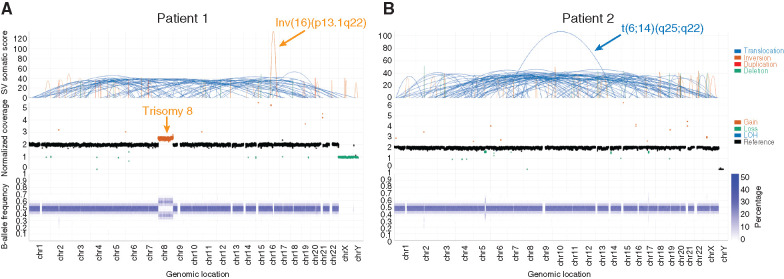
Figure 1.
WGS for identification of personalized structural variations. WGS of CD34+ BM aspirate from patient 1 (A) and CD117+ BM from patient 2 (B) identified structural aberrations consistent with cytogenetic studies (chromosomal locations displayed on the x-axis). Top: structural changes identified by the structural variant caller. Middle: Total normalized somatic copy-number estimates. Bottom: B-allele frequency at binned regions across the genome. Additional genetic material from chromosome 8 along with allelic imbalance was observed in patient 1, consistent with trisomy 8. Data visualized using Illumina Analysis Software. chr, chromosome; SV, structural variant.
Profiling cell-surface protein expression using antibody–oligo conjugates together with scDNA-seq has recently been described (10). The use of antibody–oligo conjugates in these scDNA-seq experiments could approximate the cell-surface immunophenotypic expression distributions as benchmarked by multiparameter flow cytometry (Supplementary Fig. S3). This ability to pair cell-surface immunophenotype with scDNA-seq genotyping for both chromosomal aberrations and sequence mutations allowed us to fully resolve the relationship between clonal hematopoiesis and AML in all three patients.
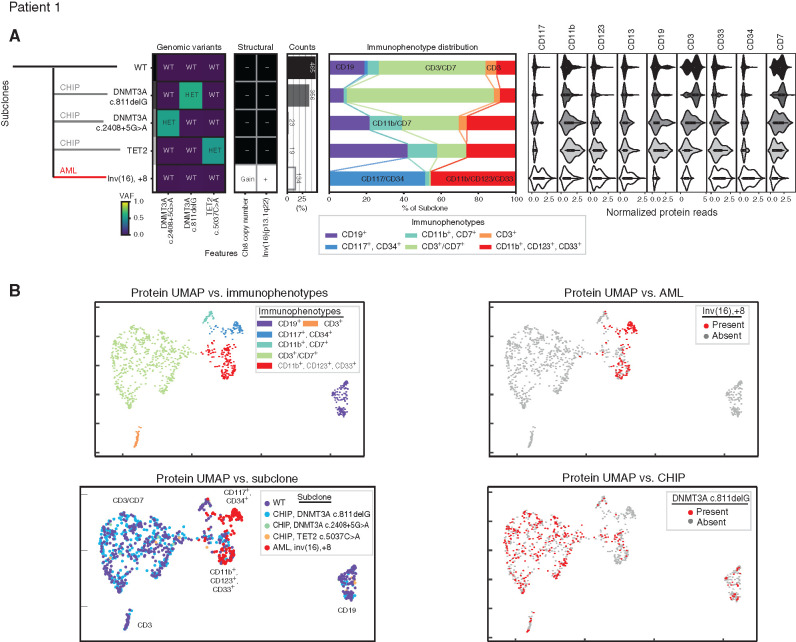
Five independent genetically defined clones were found in peripheral blood mononuclear cells (PBMC) of patient 1 (Fig. 2; Supplementary Fig. S4A). The pathognomonic inversion 16 breakpoint was detected in 13% of cells examined and was mutually exclusive from clones containing DNMT3A and TET2 mutations. Trisomy of chromosome 8 was found in cells containing inversion 16 but not in other clones. Although clinical flow cytometry described a 1% abnormal blast population positive for CD13, CD34, and CD117, this particular immunophenotype represented only half of the cells genotyped as containing inversion 16 and trisomy 8. This leukemic genotype was also found in cells with an immunophenotype consistent with more mature myeloid cell types (CD11b, CD33, and CD123 positive) but notably not in T (CD3) or B (CD19) lymphocytes. In contrast, the three subclones each containing one mutation in either DNMT3A or TET2 did not have any restriction to myeloid or lymphoid lineages.
Figure 2.
Development of leukemia independent from clonal hematopoiesis. A, Clonal architecture as determined by scDNA and antibody–oligonucleotide sequencing in patient 1 shows the leukemic (AML, red line) clone developed separately from three clonal hematopoiesis (CHIP, gray lines) clones. Left: Genomic subclones with wild-type (WT), heterozygous (HET), present (Gain/+), or absent (-) features. Center: Immunophenotype as a percentage of each subclone. Right: Cell-surface protein expression for each subclone. B, For patient 1, cells clustered by cell-surface protein expression. UMAP shown colored by immunophenotype (top left) and genotype, with all subclones (bottom left), the leukemic subclone (top right), and one clonal hematopoiesis subclone (bottom right) depicted. VAF, variant allele frequency.
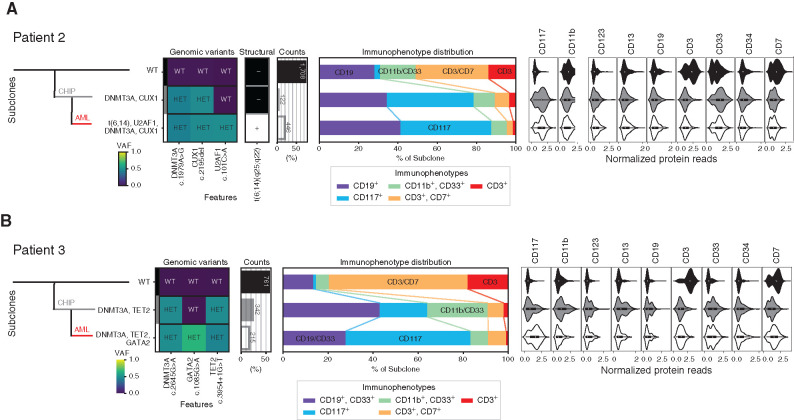
Unlike this first case, patients 2 and 3 both had AML that likely arose from a preceding clonal hematopoiesis cell population. ScDNA-seq of BM mononuclear cells (BMMC) from patient 2 demonstrated that 20% of cells contained the leukemia-defining features of t(6;14) translocation and mutated U2AF1, in addition to the DNMT3A and CUX1 mutations that were within but not specific to this clone (Fig. 3A; Supplementary Fig. S4B). The NPM1 mutation was incompletely genotyped (see Methods). This malignant clone had a diverse range of immunophenotypes including both CD117-positive and CD117-negative populations. Cells containing mutations in DNMT3A and CUX1 but not the t(6;14) translocation and U2AF1 mutation represented just 5% of cells.
Figure 3.
Sequential development of leukemia from clonal hematopoiesis. Clonal architecture as determined by scDNA and antibody–oligonucleotidesequencing in patient 2 (A) and patient 3 (B) shows the leukemic (AML, red line) clone developed from clonal hematopoiesis (CHIP, gray line). Left: Genomic subclones with wild-type (WT), heterozygous (HET), present (+), or absent (–) features. Center: Immunophenotype as a percentage of each subclone. Right: Cell-surface protein expression for each subclone. VAF, variant allele frequency.
ScDNA-seq of BMMCs from patient 3 identified a clone representing 16% of cells that contained DNMT3A, TET2, CEBPA, and GATA2 mutations and was markedly enriched for CD117 cell-surface protein expression (Fig. 3B; Supplementary Fig. S4C). CEBPA was incompletely genotyped but found only within cells with mutated GATA2 (see Methods). Cells containing mutations in DNMT3A and TET2 but not GATA2 or CEBPA, presumably representing the founder population of nonmalignant clonal hematopoiesis, represented 26% of all BM cells sequenced and had a diverse spectrum of immunophenotypes ranging across both myeloid and lymphoid lineages.
The individual patient clonal structures determined by scDNA-seq were confirmed by sorting on the predominant cell-surface protein followed by WGS. For patient 1, CD34-positive BM cells had enrichment of trisomy of chromosome 8 and inversion 16 relative to bulk BM sequencing, with DNMT3A and TET2 mutations not detected. For patients 2 and 3, WGS of CD117-positive cells confirmed the enrichment of all mutations, and, as predicted, unlike patient 1, the DNMT3A and TET2 mutations remained detectable (Supplementary Table S2).
Discussion
This is the first (to our knowledge) description of WGS-informed, patient-personalized scDNA-seq to accurately distinguish leukemic cells from clonal hematopoiesis. This is also the first direct comparison of antibody–oligonucleotide and such scDNA-seq with concurrent multiparameter flow cytometry. The ability to determine the optimum combinations of immunophenotypic markers necessary to identify malignant subclones may have utility in flow cytometric approaches for measurement of residual AML, which currently underrepresent total genetically defined leukemic burden. Although targeted DNA-seq cannot determine if mutations associated with clonal hematopoiesis are also represented within the malignant clone, detection of pathognomonic fusion genes by single-cell DNA and antibody–oligonucleotide sequencing can resolve, and determine the immunophenotype of, the relevant leukemic clonal architecture, allowing comprehensive and personalized assessment of AML. This integrated multimodal approach has implications for precision medicine in AML and other diseases.
Methods
Clinical Samples
BM aspirate and peripheral blood (PB) samples were collected from patients with relapsed or refractory AML after receipt of written informed consent on a National Heart, Lung, and Blood Institute (NHLBI) Institutional Review Board–approved protocol in accordance with the Declaration of Helsinki. BMMCs were purified from BM aspirate and PBMCs from PB using density centrifugation, cryopreserved, and stored in liquid nitrogen. Genomic DNA (gDNA) was isolated either from fresh heparinized BM aspirate using the QIAamp DNA Blood Midi Kit or from thawed PBMCs or BMMCs using the QIAamp DNA Micro Kit (QIAGEN).
Targeted DNA-seq
Mutations were identified in each patient by error-corrected DNA-seq of 50 ng of gDNA using a panel covering 75 genes commonly mutated in myeloid malignancies (Myeloid VariantPlex, ArcherDx). The resulting libraries were subjected to paired-end 150-bp sequencing on a Miseq (Illumina). Raw sequencing FASTQ files were analyzed using the Archer Analysis software version 6.0.2.3. AML-associated variants (Table 1), with a minimum variant allele frequency of 0.01, were identified using information regarding unique molecular identifier (UMI) read families, background error rate models, unique start sites, strand-specific priming, homopolymer runs, and predicted variant consequence.
WGS
WGS was performed on all patients to determine the DNA breakpoint locations of known cytogenetic rearrangements (patients 1 and 2) and to confirm immunophenotype:genotype relationships. gDNA isolated from buccal swabs, bulk BM aspirate, and microbead (Miltenyi Biotec) positively enriched CD34- or CD117-positive cells from BMMCs was used for WGS library preparation using the TruSeq DNA PCR-Free Kit with ligation of IDT for Illumina TruSeq DNA UD Indexes. Libraries were clustered in a single library per lane topography (with five lanes per AML library and one lane per germline library) on a cBot2 (Illumina) before sequencing on a HiSeq X platform (Illumina) to generate paired-end 150-bp reads. WGS raw data were converted from BCL to FASTQ format using Illumina's bcl2fastq2 v2.20. Samples were processed by the HAS2.2 (Illumina) resequencing analysis workflow. Reads were aligned to the hg38 reference genome plus decoy sequences using isaac (version Isaac-04.17.06.15). Then, matched tumor and normal results were analyzed by Illumina's tumor normal workflow, which included somatic variant detection by strelka2 (version 2.80), somatic structural variant detection by manta (version 1.1.1), and somatic copy-number detection by canvas (version 1.28.0.272). Breakpoint locations for fusion partners were chosen for further analysis (Table 1; Fig. 1; Supplementary Fig. S1).
Single-Cell DNA and Protein Sequencing
ScDNA-seq with antibody–oligonucleotide staining was performed using the Mission Bio Tapestri scDNA-seq V2 platform as per the manufacturer's instructions. In short, custom scDNA-seq panels targeting the patient-specific mutations and chromosomal structural abnormalities (Table 1; Supplementary Table S1) and oligonucleotide-conjugated antibodies (AOC) targeting cell-surface proteins of interest (Supplementary Table S3) were designed and manufactured by Mission Bio, Inc. Approximately 815,000 thawed BMMCs or PBMCs were stained for 30 minutes at room temperature with the AOC pool, followed by four rounds of washing with Dulbecco's phosphate-buffered saline (DPBS) containing 7% FBS. Stained cells were resuspended in cell buffer and subjected to microfluidic encapsulation, lysis, and cell barcoding on the Tapestri instrument. Amplification of the targeted DNA regions and antibody–oligonucleotide tags was performed by incubating the barcoded DNA emulsions in a thermocycler as follows: 98°C for 6 minutes (4°C/sec); 11 cycles of 95°C for 30 seconds, 72°C for 10 seconds, 61°C for 3 minutes, 72°C for 20 seconds (1°C/sec); 13 cycles of 95°C for 30 seconds, 72°C for 10 seconds, 48°C for 3 minutes, 72°C for 20 seconds (1°C/sec); and 72°C for 6 minutes (4°C/sec). Emulsions were broken and DNA digested and purified with 0.7X SPRISelect reagent (Beckman Coulter). The beads were pelleted, and the supernatant was retained for antibody library preparation, whereas the remaining beads were washed with 80% ethanol and the DNA targets were eluted in nuclease-free water. The supernatant containing the antibody tags was incubated with a biotinylated capture oligo (/5Biosg/CGAGATGACTACGCTACTCATGG/3C6/, Integrated DNA Technologies) at 96°C for 5 minutes, followed by ice for 5 minutes, and recovered with streptavidin beads (Dynabead MyOne Streptavidin C1, Thermo Fisher). Indexed Illumina libraries were generated by amplifying DNA libraries with Mission Bio V2 Index Primers and protein libraries bound to streptavidin beads with i5 and i7 index primers (Integrated DNA Technologies) in the thermocycler using the following program: 95°C for 3 minutes; 10 cycles (DNA library) or 20 cycles (protein library) of 98°C for 20 seconds, 62°C for 20 seconds, 72°C for 45 seconds; and 72°C for 2 minutes. Final libraries were purified with 0.69X (DNA) or 0.9X (protein) SPRISelect reagent. Libraries were pooled and subjected to paired-end 150-bp sequencing on a Novaseq 6000 (Illumina).
Raw FASTQ files were analyzed using the Tapestri pipeline (MissionBio) and custom python scripts. An average of 1,797 cells were sequenced per patient (Supplementary Table S4). Single-nucleotide variants and indels identified via GATK were filtered for quality score, read depth, and minimum frequency. In order to confidently delineate clonal structure, only variants genotyped in a high proportion of cells were taken forward for subclone analysis. Multiple variants exhibited characteristics of incomplete genotyping for various technical reasons. The amplicon covering the CEBPA c.68dup failed amplification in all patients. The NPM1 c.860_863dup variant is located just downstream from a homopolymer run region, which presented difficulty on the Illumina sequencer, resulting in poor quality reads and subsequent read trimming. In the custom panel used for patients 2 and 3 (CO-86), the reverse primer was situated such that the insertion site was not covered, resulting in allelic dropout across many cells. This technical issue was resolved in the newer custom panel used for patient 1 (CO-101), drastically improving coverage. The CEBPA c.949_950insGTC mutation is also located within a difficult-to-sequence region, which also resulted in poor read quality and subsequent allelic dropout across cells.
Next, UMAP dimension reduction and clustering were applied to identify subclone populations. The presence of inversions or translocations in those subclones was noted by counting amplicons spanning the splice junctions. Copy-number variation was calculated by normalizing read counts for each DNA panel target to the wild-type subpopulation and then observing a relative gain or loss of reads. Protein expression was calculated with a centered log-ratio transformation on the raw antibody counts to normalize for read depth variation between cells. From that, immunophenotypes were identified via UMAP dimension reduction and k-means clustering; the protein signature of each cluster was calculated by observing when the median expression for each target surpassed a fixed threshold across proteins.
Flow Cytometry
Thawed PBMCs or BMMCs were resuspended in Cell Staining Buffer (BioLegend) and blocked with Human TruStain FcX (Fc Receptor Blocking Solution; BioLegend) for 10 minutes. Cells were stained with a 1:100 dilution of Zombie UV Fixable Viability dye (BioLegend) for 10 minutes and then incubated with 5 μL of each antibody listed in Supplementary Table S5 for 20 minutes. All steps were performed at ambient temperatures. At least 100,000 events were acquired with a BD FACSymphony; data were analyzed with FlowJo Software version 10 (BD).
Data Availability
FASTQ files from scDNA and antibody sequencing and targeted DNA-seq are available in the NCBI SRA (PRJNA718560). FCS files for flow cytometry are available in FlowRepository (FR-FCM-Z3LT).
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (NIH) and by the Department of Defense award HU0001-18-2-0038 to C.L. Dalgard.
This work utilized the NHLBI Flow Cytometry Core, the NHLBI Sequencing and Genomics Core, and the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
#L.W. Dillon, J. Ghannam, and C. Nosiri contributed equally to this article.
Blood Cancer Discov 2021;2:319–25
Authors' Disclosures
M.D. Wilkerson reports grants from National Heart, Lung, and Blood Institute and Defense Health Agency during the conduct of the study. C.L. Dalgard report grants from National Heart, Lung, and Blood Institute and Defense Health Agency during the conduct of the study. A. Sciambi reports other support from Mission Bio, Inc. outside the submitted work, as well as a patent for US App. No. 16/839,055 pending. A. Llanso reports other support from Mission Bio, Inc. during the conduct of the study; other support from Mission Bio, Inc. outside the submitted work; and a patent for US App. No. 16/839,055 pending. S. Gulati reports personal fees from Mission Bio, Inc. during the conduct of the study, as well as personal fees from Mission Bio, Inc. outside the submitted work. S. Wang reports other support from Mission Bio, Inc. during the conduct of the study. A. Ooi reports grants from National Cancer Institute during the conduct of the study; other support from Mission Bio, Inc. outside the submitted work; and a patent for US App. No. 16/839,055 pending. C.S. Hourigan reports other support from Merck and Sellas outside the submitted work. No disclosures were reported by the other authors.
Disclaimer
The opinions and assertions expressed herein are those of the author(s) and do not necessarily reflect the official policy or position of the Uniformed Services University or the Department of Defense, the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., or the National Institutes of Health.
Authors' Contributions
L.W. Dillon: Conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. J. Ghannam: Conceptualization, data curation, formal analysis, writing–review and editing. C. Nosiri: Conceptualization, data curation, formal analysis, writing–review and editing. G. Gui: Data curation, formal analysis, visualization, writing–review and editing. M. Goswami: Data curation, formal analysis, writing–review and editing. K.R. Calvo: Data curation, writing–review and editing. K.E. Lindblad: Data curation, writing–review and editing. K.A. Oetjen: Data curation, writing–review and editing. M. Wilkerson: Data curation, writing–review and editing. A.R. Soltis: Data curation, writing–review and editing. G. Sukumar: Data curation, writing–review and editing. C.L. Dalgard: Data curation, writing–review and editing. J. Thompson: Data curation, writing–review and editing. J. Valdez: Data curation, writing–review and editing. C.B. DeStefano: Data curation, writing–review and editing. C. Lai: Data curation, writing–review and editing A. Sciambi: Formal analysis, visualization, writing–review and editing.R. Durruthy-Durruthy: Formal analysis, visualization, writing–review and editing. A. Llanso: Resources, writing–review and editing. S. Gulati: Resources, writing–review and editing. S. Wang: Resources, writing–review and editing. A. Ooi: Resources, writing–review and editing. P.K. Dagur: Data curation, formal analysis, writing–review and editing. J.P. McCoy: Formal analysis, writing–review and editing. P. Burr: Data curation, writing–review and editing. Y. Li: Resources, data curation, writing–review and editing. C.S. Hourigan: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014;20:1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 2012;44:1179–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young AL, Challen GA, Birmann BM, Druley TE.Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 2016;7:12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017;377:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jongen-Lavrencic M, Grob T, Hanekamp D, Kavelaars FG, Al Hinai A, Zeilemaker A, et al. Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med 2018;378:1189–99. [DOI] [PubMed] [Google Scholar]
- 7.Hourigan CS, Dillon LW, Gui G, Logan BR, Fei M, Ghannam J, et al. Impact of conditioning intensity of allogeneic transplantation for acute myeloid leukemia with genomic evidence of residual disease.J Clin Oncol 2020;38:1273–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy R, et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 2020;587:477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita K, Wang F, Jahn K, Hu T, Tanaka T, Sasaki Y, et al. Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat Commun 2020;11:5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demaree B, Delley CL, Vasudevan HN, Peretz CAC, Ruff D, Smith CC, et al. Joint profiling of DNA and proteins in single cells to dissect genotype-phenotype associations in leukemia. Nat Commun 2021;12:1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
FASTQ files from scDNA and antibody sequencing and targeted DNA-seq are available in the NCBI SRA (PRJNA718560). FCS files for flow cytometry are available in FlowRepository (FR-FCM-Z3LT).