Abstract
Background
The impact of HER2 somatic mutations in colorectal carcinoma (CRC) has not been well studied and its relationship with microsatellite instability‐high (MSI‐H) is yet to be fully elucidated.
Materials and Methods
From February 2017 to February 2020, the data of patients with CRC who underwent next‐generation sequencing and had detailed record of clinicopathological information were investigated. HER2 alteration and its relationship with MSI‐H were analyzed.
Results
Among 731 patients who underwent sequencing, 55 patients (7.5%) had HER2 alteration, including 29 (4.0%) with HER2 somatic mutations, 24 (3.3%) with HER2 gene amplification, and 2 patients (0.2%) with both HER2 mutations and amplification. R678Q was the most common mutated kinase domain, and no HER2 kinase domain in‐frame insertions/deletions were found in HER2 mutated cases. MSI‐H was found in 5.2% of our cohort and 36.8% of MSI‐H patients had HER2 mutation. For HER2 mutated cases, 48.3% were MSI‐H, whereas none of the HER2 amplification cases were MSI‐H. MSI‐H patients with HER2 mutation had significantly worse median progression‐free survival for programmed death‐1 (PD‐1) antibody than those without HER2 alteration (p = .036).
Conclusion
High MSI‐H rate was found in HER2 mutated cases, but no MSI‐H was found in HER2 amplification cases. MSI‐H patients with HER2 mutated had worse progression‐free survival for PD‐1 antibody than those without.
Implications for Practice
This study highlights the high microsatellite instability‐high (MSI‐H) rate in HER2 mutated cases but no MSI‐H in HER2 amplification cases. Moreover MSI‐H patients with HER2 mutated had worse progression‐free survival for programmed death‐1 antibody than those without. Further research to explore the internal relationship between HER2 alteration and MSI‐H is needed.
Keywords: HER2, Mutation, Amplification, Microsatellite instability‐high, Colorectal cancer
Short abstract
The clinical significance of HER2 somatic mutations and its relationship with high microsatellite instability in colorectal cancer is not fully understood. This study investigated the relationship by analyzing data of patients with colorectal cancer who underwent next‐generation sequencing.
Introduction
Colorectal cancer (CRC) was identified as the third most common malignancy worldwide in 2018 [1]. In China, CRC was the fourth most commonly diagnosed cancer and the fifth leading cause of cancer‐related mortality [2, 3]. The incidence of CRC can be largely attributed to cancer‐related genes, such as Kirsten rat sarcoma viral oncogene homolog (KRAS), v‐raf murine sarcoma viral oncogene homolog B (BRAF), microsatellite instability (MSI), Erythroblastic leukemia viral oncogene homolog 2 (ERBB2), and more [4].
ERBB2, also known as the human epidermal growth factor receptor 2 (HER2), is a confirmed therapeutic target for patients with breast and gastric cancers [5, 6, 7]. It has been reported to be amplified or overexpressed in 3%–5% of genetically unselected metastatic CRC (mCRC) [8, 9, 10, 11, 12] and could also serve as potential therapeutic targets in patients with mCRC [11, 13]. The HERACLES and MyPathway clinical trials assessed the feasibility of anti‐HER2 therapy in HER2 overexpressed/amplified patients with mCRC and observed an objective response rate (ORR) of 30% when using trastuzumab and lapatinib [11] or 38% with pertuzumab and trastuzumab [13]. MyPathway is a phase IIa, multiple basket study [13]. Patients with treatment‐refractory, histologically confirmed HER2‐amplified metastatic colorectal cancer received the combination therapy with pertuzumab and trastuzumab. In regard to treatment‐related resistance, HER2 amplifications/overexpressions have been shown to be predictive of resistance to epidermal growth factor receptor (EGFR)‐target therapy [14, 15]. It is therefore recommended to test HER2 amplification for patients with mCRC for better therapeutic selection and outcomes.
HER2 amplification can be assessed using a variety of tissue‐based approaches, including fluorescence in situ hybridization, polymerase chain reaction (PCR), or next‐generation sequencing (NGS). In the MyPathway study, HER2 amplification was defined as HER2 to chromosome 17 ratio > 2.0 or HER2 copy number > 6.0 based on NGS copy number gain [13].
HER2 somatic mutations are found in a wide range of solid tumors, including 9% of bladder cancer cases, 7% of glioblastoma cases, 5% of gastric cancer cases, 4% of lung adenocarcinoma, 3% of esophageal cancer, and 1.5%–2% of breast cancer cases [16]. Little is known about HER2 mutation in patients with CRC. The Cancer Genome Atlas (TCGA) project identified HER2 alterations in 7% (14/212) of CRC cases, including six HER2 somatic mutations (2.8%), five HER2 gene amplification (2.4%), and three cases with both HER2 mutations and HER2 amplification (1.4%) [17]. HER2 activating mutations demonstrated resistance to cetuximab and panitumumab by sustaining MAPK phosphorylation both in vitro and in patient‐derived xenograft model [18] but were responsive to HER2 target drugs [18].
Microsatellite instability‐high (MSI‐H) occurred for less than 5% in patients with metastatic CRC and it is found to be more common in patients with BRAFV600E mutation [19]. A previous study showed that MSI and PIK3CA mutations were associated with ERBB2/ERBB3 mutations [12]. Until now, the clinical significance of HER2 somatic mutations in CRC has not been fully studied and the relationship between HER2 mutation and MSI status is still undetermined.
In the present study, we retrospectively collected patients with CRC who had HER2 alteration or MSI‐H by NGS and analyzed their relationship with clinic‐pathologic features and response to treatment.
Materials and Methods
Patients
This protocol was approved by the Ethics Committee of Sun Yat‐Sen University Cancer Center (Guangzhou, China). Written informed consent was obtained from patients at their first visit.
All the patients with CRC whose tumor specimens underwent NGS from February 2017 to February 2020 were screened from our institution database using the following inclusion criteria: (a) had a known or confirmed diagnosis of colorectal adenocarcinoma, and (b) had complete records of clinicopathological data. In total, 731 patients with CRC were enrolled into this study. All the tumor samples were reviewed by pathologists prior to DNA extraction for sequencing. Paired nontumor tissues or peripheral blood white cells were used for exclusion of germline mutations.
DNA isolation and sequencing were processed using a customized analysis pipeline as described in our previous study [20, 21]. Sequencing of tumor tissues was performed with an average sequencing depth > 500×, with >50× of 99% exons. This method of sequencing allowed detections of copy number variations, gene rearrangement, and somatic mutations. For this study, the sequenced CRC tumors were queried for alterations of HER2, coaberrant genes, and MSI status. In brief, DNA from the tissues and peripheral blood cells were isolated using the QIAamp DNA FFPE Tissue Kit or QIAamp DNA Blood kit (Qiagen, Hilden, Germany). DNA concentration was detected in a Qubit Fluorometer 3.0 (Life Technologies, Carlsbad, CA). Two hundred nanograms of genomic DNA was used to perform hybridization, hybrid selection, and PCR amplification using commercial NGS assays targeting 295 or 1021 cancer‐related genes. The 295 and 1,021 cancer‐related genes were provided as supplementary tables. As of August 2019, the 1,021‐gene panel of cancer‐related genes replaced the 295‐gene panel. Both panels included HER2 and microsatellite stability (MSS) genes. Thirty‐three patients were assessed by the 295‐gene panel and 22 by the 1,021‐gene panel. A DNA library was then constructed using the Illumina NextSeq500 sequencer with pair‐end reads (Illumina, Inc., San Diego, CA), according to the commercial protocol, and the indexed samples were sequenced. A minimal median unique sequencing depth of 500× was necessary and sufficient to assess low‐frequency mutations for each tumor sample.
Analysis of Sequencing Data
Quality control checks of the sequence data were carried out using the FastQC v0.11.7 software (Babraham Bioinformatics, Cambridge, U.K.), on sequencing data in the FASTQ format. Low‐quality reads were filtered using the Trimmomatic‐0.36 software (http://www.usadellab.org/cms/?page = trimmomatic, Germany). The filtered data were mapped to the human genome (hg19) using Burrows‐Wheeler Aligner 0.7.10 (http://bio-bwa.sourceforge.net/). The alignments were processed using Samtools 0.1.19 (http://www.htslib.org/) and picard‐tools‐1.138 (https://sourceforge.net/projects/picard/). Local alignment optimization and variant (single nucleotide variant [SNV] and insertion‐deletion (INDEL)) calling were performed using the GATK 3.2 (https://software.broadinstitute.org/gatk/), VarDict (https://github.com/AstraZeneca-NGS/VarDict), and VarScan 2.4.3 software programs (http://varscan.sourceforge.net/), respectively. Genetic variations were filtered with the VarScan fpfilter pipeline. The remaining genetic variations were annotated using the ANNOVAR (http://annovar.openbioinformatics.org/), SnpEff v3.6 (http://snpeff.sourceforge.net/) and InterVar (https://github.com/WGLab/InterVar) software programs.
Gene‐level copy number variations (CNV) was assessed for significant changes compared with control using a t statistic after normalizing read depth in each region by the total read number and region size and correcting any GC‐bias using a LOESS algorithm as previously described [21, 22]. DNA translocation analysis for fusion genes was performed using both Tophat2 (http://ccb.jhu.edu/software/tophat/index.shtml) and Factera 1.4.3 (https://factera.stanford.edu/). The tumor mutation burden (TMB) value was calculated by dividing the total number of tissue SNVs and INDEL variations by the size of the 295‐gene panel (Burning Rock Biotech Ltd., Guangdong, China) or 1,021‐gene panel (Geneplus‐Beijing Institute, Beijing, China).
Statistical Analysis
All statistical analyses were performed using the R and Intercooled Stata 13.0 (Stata Corporation, College Station, TX). Significance was defined as p values <.05. Mutation profiles were performed using the maftools packages to explore the mutual relationships between genes such as co‐occurrence and exclusiveness [23]. TCGA Colon and Rectal Adenocarcinoma data were downloaded from the University of California Santa Cruz Xena [1] database exploration program (https://xena.ucsc.edu/). Kaplan‐Meier curves with log‐rank analysis were used for prognosis analyses. The last date of follow‐up was May 31, 2020.
Results
HER2 Alteration
There were a total of 55 (7.5%) patients with HER2 alteration, including 29 (4.0%) cases with HER2 somatic mutations, 24 (3.3%) with HER2 gene amplification, and 2 (0.2%) with both HER2 mutations and amplification. One patient had HER2 amplification and fusion (IKZF3‐ERBB2). The median copy number for HER2 amplification was 11.32 (mean ± SD, 33.9 ± 39.2), ranging from 2.93 to 134.
No HER2 kinase domain in‐frame insertions/deletions were found. Besides the common identification of previously reported mutation kinases in our cohort [18, 24, 25], we also identified 25 new HER2 mutations. Some HER2 mutations were seen in multiple patients, for example, R678Q was found in eight patients (Fig. 1). Some patients had multiple mutations. Two patients had three HER2 mutation domains and six patients had two HER2 mutation domains. The comparison of mutation locations of ERBB2 in the MSI‐H and MSS subgroups is listed in Figure 1.
Figure 1.
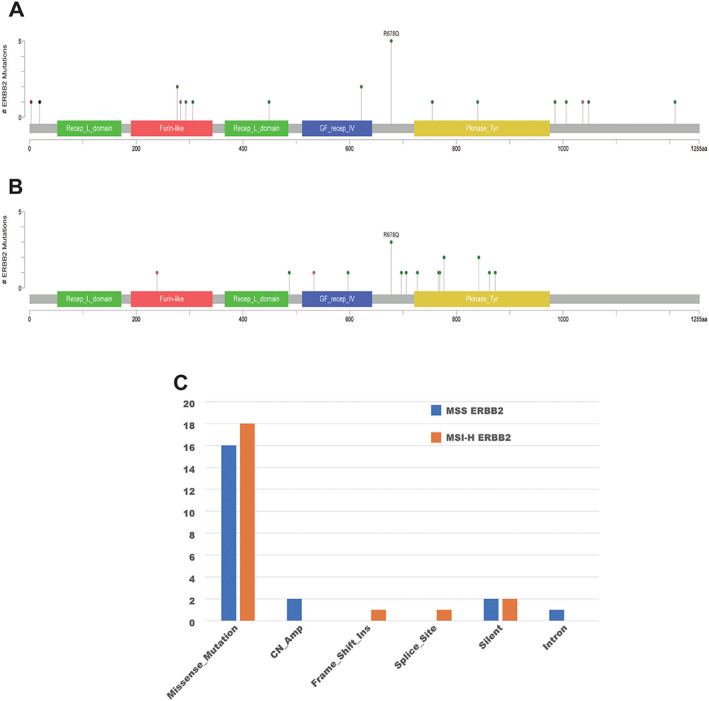
Mutation locations of ERBB2 in the MSI‐H (A), and MSS (B) subgroups. The comparison of ERBB2 alteration between the MSI‐H and MSS subgroups (C).
Abbreviations: MSI‐H, microsatellite instability‐high; MSS, microsatellite stability.
The comparison of clinicopathological features between HER2 mutated and amplification cases is shown in Table 1. Ratsarcoma viral oncogene homolog (RAS) mutation was more common in HER2 mutated cases than in HER2 amplification cases (48.3% vs. 23.1%; p = .052). The only co‐occurring BRAF mutation was found in HER2 mutated case with BRAF V600E. Left‐sided colon cases were more likely to have HER2 alteration than right‐sided colon and rectum. The MSS or MSI status of these cancers was reported and 48.3% HER2 mutated case were MSI‐H, whereas none of the HER2 amplification cases were MSI‐H. Patients with HER2 mutation had a significantly higher TMB than patients with HER2 amplification (p = .0098).
Table 1.
Comparison of clinic‐pathologic features between patients with colorectal cancer with HER2 mutation and amplification
| Characteristics | Whole cohort (n = 55), n (%) | HER2 mutation (n = 29), n (%) | HER2 amplification (n = 26), n (%) | p value |
|---|---|---|---|---|
| Sex | ||||
| Female | 23 (41.8) | 12 (41.4) | 11 (42.3) | |
| Male | 32 (58.2) | 17 (58.6) | 15 (57.7) | .94 |
| Age, yr | ||||
| Median | 54 | 52 | 55 | |
| Mean ± SD | 50.5 ± 14.5 | 50.4 ± 15.2 | 50.6 ± 13.9 | .97 |
| Family history | ||||
| Yes | 19 (34.6) | 11 (37.9) | 8 (30.8) | |
| No | 36 (65.4) | 18 (62.1) | 18 (69.2) | .58 |
| Location | ||||
| Right‐sided | 17 (30.9) | 10 (34.5) | 7 (26.9) | |
| Left‐sided | 25 (45.5) | 12 (41.4) | 13 (50.0) | |
| Rectum | 13 (23.6) | 7 (24.2) | 6 (23.1) | .79 |
| Grade | ||||
| Moderate + high differentiated | 36 (65.5) | 18 (62.1) | 20 (76.9) | |
| Poorly + undifferentiated | 19 (34.5) | 11 (38.9) | 6 (23.1) | .23 |
| HER2 expression | ||||
| Negative | 11 (45.8) | 28 (96.6) | 7 (26.9) | |
| Positive | 13 (54.2) | 1 (3.4) | 19 (73.1) | <.001 |
| RAS | ||||
| Wild type | 35 (63.6) | 15 (51.7) | 20 (76.9) | .052 |
| Mutation | 20 (36.4) | 14 (48.3) | 6 (23.1) | |
| MSS | ||||
| MSI‐H | 14 (25.5) | 14 (48.3) | 0 (0) | |
| MSS | 41 (74.5) | 15 (51.7) | 26 (100) | <.001 |
| TMB | ||||
| Median | 11.22 | 15.8 | 8.2 | |
| Mean ± SD | 31.0 ± 56.8 | 48.6 ± 71.0 | 8.0 ± 3.7 | .0098 |
| TNM stage | n = 54 a | n = 28 a | n = 26 | |
| I | 1 (1.8) | 1 (3.6) | 0 (0) | |
| II | 3 (5.6) | 2 (7.1) | 1 (3.8) | |
| III | 17 (31.5) | 9 (32.1) | 8 (30.8) | |
| IV | 33 (61.1) | 16 (57.2) | 17 (65.4) | .72 |
| Metastatic sites | n = 48 | n = 24 | n = 24 | |
| Liver | 26 (54.2) | 10 (41.7) | 16 (66.7) | .082 |
| Lung | 20 (41.7) | 6 (25.0) | 14 (58.3) | .019 |
| Distant lymph nodes | 20 (41.7) | 10 (41.7) | 10 (41.7) | .99 |
| Peritoneum | 17 (35.4) | 10 (41.7) | 7 (29.2) | .37 |
One MSI‐H patient got pathologic complete response after neoadjuvant chemotherapy with PD‐1 antibody. This patient is not included for TNM stage analysis.
Abbreviations: HER2, human epidermal growth factor receptor‐2; MSI‐H, microsatellite instability high; MSS, microsatellite stability; RAS, rat sarcoma viral oncogene homolog; TMB, tumor mutation burden; TNM, tumor‐node‐metastasis.
Approximately one third of the patients (37.9% with HER2 mutation and 30.8% with HER2 amplifications) had a family history of cancer. HER2 overexpression was found in 3.4% of the HER2 mutated cases and 73.1% of the HER2 amplification cases (p < .001). Sixty percent of the patients (33/55) were diagnosed at stage IV CRC at diagnosis and another 15 patients developed recurrence or distant metastasis. Patients with HER2 amplification had a significantly higher rate of lung metastasis than HER2 mutated cases (p = .019).
MSI‐H and HER2 Alteration
We found that HER2 mutated cases had a high percentage of MSI‐H, whereas no MSI‐H was found in HER2 amplification cases. Upon inspecting the 731 CRC cases who underwent NGS, we identified 38 (5.2%) MSI‐H cases including 14 HER2 mutated and 24 without HER2 alteration cases. Next, we compared the clinicopathological features of patients who were MSS‐HER2 mutated, MSI‐H HER2 mutated, and MSI‐H without HER2 alteration (Table 2).
Table 2.
Comparison of clinic‐pathologic features among patients with colorectal cancer with different MSS and HER2 mutation status
| Clinic‐pathologic features | HER2 mutation | MSI‐H without HER2 alteration (n = 2), n (%) | p value a | P value b | |
|---|---|---|---|---|---|
| MSS (n = 15), n (%) | MSI‐H (n = 14), n (%) | ||||
| Sex | |||||
| Female | 9 (60.0) | 3 (21.4) | 7 (29.2) | ||
| Male | 6 (40.0) | 11 (78.6) | 17 (70.8) | .035 | .60 |
| Age, yr | |||||
| Median | 56 | 47 | 42 | ||
| Mean ± SD | 52.5 ± 13.9 | 44.7 ± 14.9 | 42.8 ± 14.7 | .048 | .71 |
| Family history | |||||
| Yes | 3 (20.0) | 8 (57.1) | 12 (50.0) | ||
| No | 12 (80.0) | 6 (42.9) | 12 (50.0) | .039 | .67 |
| Location | |||||
| Right‐sided | 4 (26.7) | 6 (42.9) | 11 (45.8) | ||
| Left‐sided | 6 (40.0) | 6 (42.9) | 7 (29.2) | ||
| Rectum | 5 (33.3) | 2 (14.2) | 6 (25.0) | .44 | .61 |
| Grade | |||||
| Moderate + high differentiated | 10 (66.7) | 7 (50.0) | 11 (45.8) | ||
| Poorly + undifferentiated | 5 (33.3) | 7 (50.0) | 13 (54.2) | .36 | .80 |
| RAS | |||||
| Wild type | 8 (53.3) | 7 (50.0) | 11 (45.8) | ||
| Mutation | 7 (46.7) | 7 (50.0) | 13 (54.2) | .86 | .80 |
| TMB | |||||
| Median | 8.16 | 73.9 | 77.28 | ||
| Mean ± SD | 9.9 ± 4.7 | 99.3 ± 85.2 | 103.7 ± 114.8 | .0002 | .90 |
| TNM stage | |||||
| 0 | 0 (0) | 1 (7.1) | 5 (20.8) | ||
| I | 1 (6.7) | 0 (0) | 1 (4.2) | ||
| II | 0 (0) | 2 (14.3) | 6 (25.0) | ||
| III | 6 (40.0) | 3 (21.4) | 4 (16.7) | ||
| IV | 8 (53.3) | 8 (57.1) | 8 (33.3) | .29 | .49 |
| Metastatic sites | n = 14 | n = 10 | n = 11 | ||
| Liver | 6 (42.9) | 4 (40.0) | 4 (36.4) | .89 | .86 |
| Lung | 6 (42.9) | 0 (0) | 2 (18.2) | .017 | .16 |
| Distant lymph nodes | 5 (35.7) | 5 (50.0) | 4 (36.4) | .48 | .53 |
| Peritoneum | 7 (50.0) | 3 (30.0) | 6 (54.5) | .33 | .26 |
p comparison between MSI‐H group and MSS‐HER2 mutated group.
p comparison between MSS‐HER2 mutated group and HER2 amplification group.
Abbreviations: HER2, human epidermal growth factor receptor‐2; MSI‐H, microsatellite instability high; MSS, microsatellite stability; RAS, rat sarcoma viral oncogene homolog; TMB, tumor mutation burden; TNM, tumor‐node‐metastasis.
Among the HER2 mutated cases, patients with MSI‐H were predominantly men, were younger, were more likely to have a family history of cancer, and had a higher TMB. None of the MSI‐H patients developed lung metastasis, whereas MSS cases had a higher rate of peritoneum metastasis (64.3% vs. 30%).
Among the MSI‐H cases, no significant difference in clinicopathological features in Table 2 was found between patients with and without HER2 alteration, although patients without HER2 alteration comprised a numerically lower number of stage IV cases and higher rate of lung and peritoneum metastasis.
Mutation Type Analysis
HER2 amplification and intron mutation were only found in MSS cases, whereas Frame_Shift_Ins and Splice_Site were only seen in MSI‐H cases (Fig. 1C). The mutation domain of HER2 was quite different between MSI‐H and MSS cases. Only R678Q and D277Y were shared in these two groups (Fig. 2). The rate of A622T was double in MSI‐H patients and was not reported in previous HER2 mutated CRC studies [18, 24, 25]. MSI‐H cases had more mutation than MSS‐HER2 mutated cases and HER2 amplification cases, especially missense mutations, and the mutations were not concentrated in certain genes. For MSS‐HER2 mutated cases, except HER2 mutation, the more frequently mutated genes were APC and KRAS. We found that ERBB2 amplification was accompanied by amplification of CDK12 (57.7%) and RARA (34.6%), and most mutations of the APC gene were frameshift deletion or nonsense mutations (Fig. 3).
Figure 2.
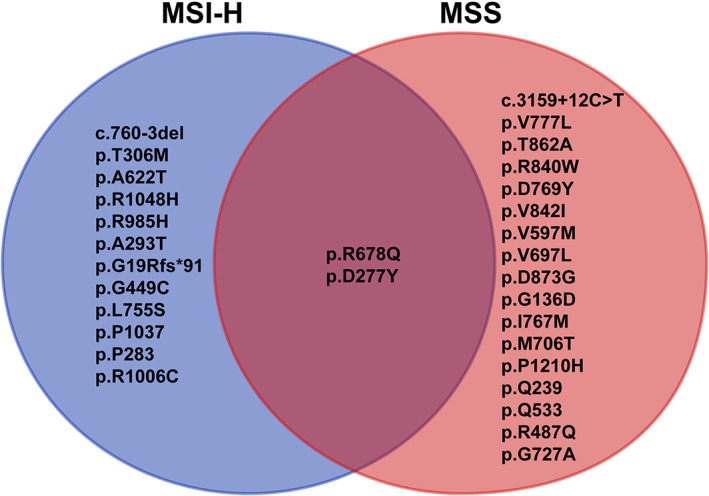
Association between MSI‐H cases and MSS cases in our present study and previous reports. Abbreviations: MSI‐H, microsatellite instability‐high; MSS, microsatellite stability.
Figure 3.
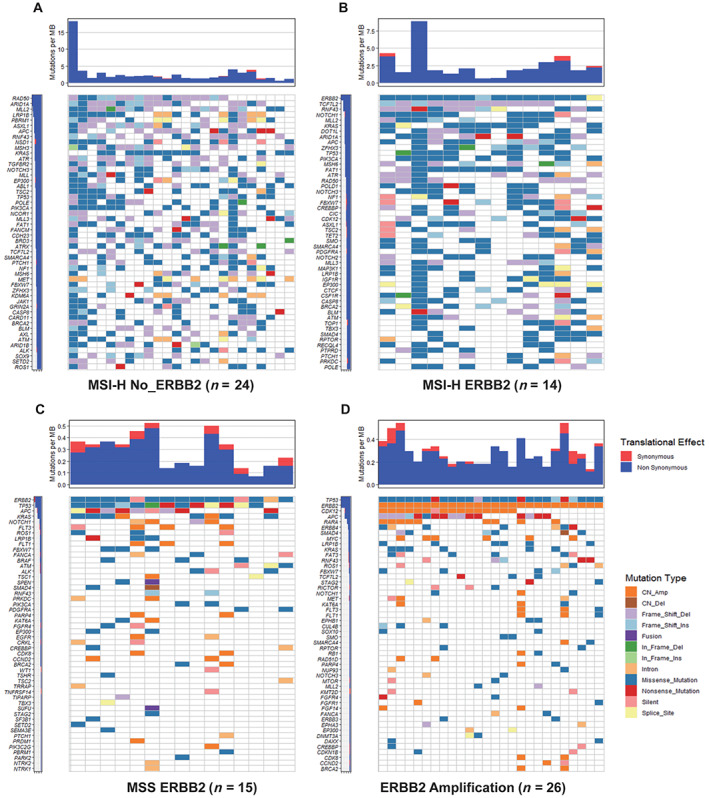
Type of mutations among patients with MSI‐H and without HER2 alteration (A), MSI‐H with HER2 mutation (B), MSS with HER2 mutation (C), and HER2 amplification (D).
Abbreviations: CN_Amp, copy number amplification; CN_Del, copy number deletion; Frame_Shift_Del, Frame_Shift_deletion; Frame_Shift_Ins, Frame_Shift_insertion; HER2, human epidermal growth receptor 2; In_Frame_Del, In_Frame_deletion; In_Frame_Ins, In_Frame_insertion MB, mutation burden; MSI‐H, microsatellite instability‐high; MSS, microsatellite stability.
Next, we compared MSI‐H patients with and without HER2 mutation. Patients with HER2 mutation had more missense mutations and SNV, whereas patients without HER2 mutation had more frameshift mutation (indel; p < .001). Except for ERBB2, there were another two genes whose mutation rates were significantly different between these two groups. CTNNA1 mutation was only found in patients without HER2 alteration, whereas PPM1D was more common in patients with HER2 mutation. HER2 mutation ranked 42nd among all the mutations genes in MSI‐H patients.
Treatment
The median disease‐free survival (mDFS) for patients with HER2 alteration was 17.6 months. For patients with HER2 mutation and amplification, the mDFS was 18.4 months and 13.3 months (p = .68). The mDFS for MSS‐HER2 alteration and MSI‐H patients was 16.2 months and 23.4 months (p = .95). The mDFS was not significantly different between MSI‐H patients with and without HER2 mutation, not reached and 23.4 months, respectively (p = .65; supplemental online Fig. 1).
Patients were treated with standard care based on the decision of physicians. The median progression‐free survival (mPFS) of first‐line therapy for patients with HER2 alteration was 10.7 months (95% confidence interval [CI], 6.4–13.8 months). The mPFS for first‐line therapy was not significantly different between patients with HER2 mutation and amplification, 10.7 months (95% CI, 4.1 months to not applicable) and 11.5 months (95% CI, 6.3–13.8 months; p = .83). No significant difference in mPFS using first‐line therapy was found between MSI‐H patients with and without HER2 mutation, 3.1 months (95% CI, 1.4–10.7 months) and 3.6 months (95% CI, 2.9–13.8 months; p = .66; supplemental online Fig. 2). Patients with MSI‐H using first‐line therapy had significantly shorter mPFS than those with MSS, 3.5 months (95% CI, 2.8–10.7 months) and 11.5 months (95% CI, 6.9–18.5 months; p = .032; Fig. 4A).
Figure 4.
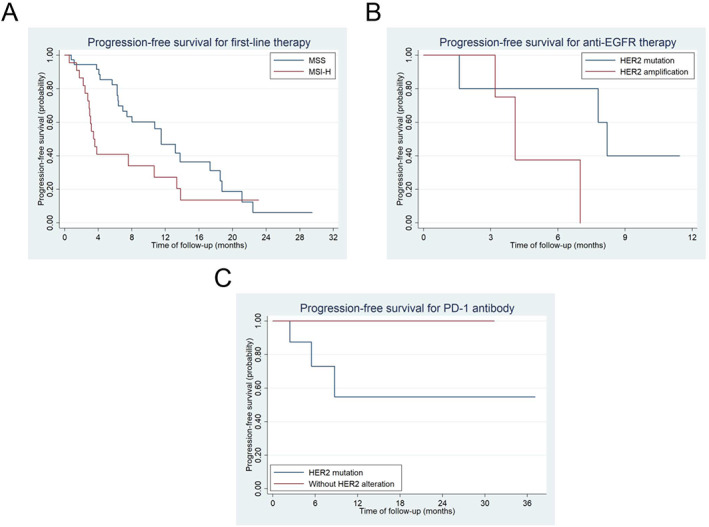
Progression‐free survival (PFS) using first‐line therapy in patients with MSS and MSI‐H (A), PFS using anti‐EGFR therapy between HER2 mutated and amplification cases (B), and PFS using PD‐1 antibody between MSI‐H with and without HER2 mutation (C). Abbreviations: EGFR, epidermal growth factor receptor; HER2, human epidermal growth receptor 2; MSI‐H, microsatellite instability‐high; MSS, microsatellite stability; PD‐1, programmed death‐1.
In total, 11 patients received anti‐EGFR therapy including 5 HER2 mutated cases (1 Partial response (PR), 3 stable disease (SD), and 1 progression disease (PD)) and 6 HER2 amplification cases (1 PR, 2 SD, and 2 PD). There were three patients in the first‐line setting, four in the second‐line setting, and four in the third‐line setting. The mPFS for anti‐EGFR therapy was 8.2 months and 4.1 months in HER2 mutated and amplification cases, respectively (p = .12; Fig. 4B).
Twenty‐five MSI‐H patients received PD‐1 antibody including 9 patients with HER2 mutation and 16 without HER2 alteration. The ORR and disease control rate (DCR) were 55.6% and 88.9% in HER2 mutated patients and 64.3% and 100% in patients (14 evaluable patients) without HER2 mutation (p = .68). Although mPFS to PD‐1 antibody was not reached in both groups, patients without HER2 alteration had significantly better mPFS than those with HER2 mutation (p = .036; Fig. 4C).
Discussion
The prevalence of HER2 somatic mutations and gene amplification in this study cohort was 7.5% in patients with colorectal cancer, similar to the reported prevalence of 7.0% in the TCGA CRC cohort [17]. Recently HER2 amplification and mutation were reported to be 3% and 6% in a Chinese CRC cohort [26]. We found that none of the patients with HER2 amplification were MSI‐H, whereas 48.3% of patients with HER2 mutation were MSI‐H and 36.8% (14/38) of patients who were MSI‐H had HER2 mutation. MSI‐H was found in approximately 15% of unselected patients with CRC [27]. To our knowledge, this is the first study to report the high MSI‐H rate in the HER2 mutated population. Using the TCGA CRC cohort, we found that 33.3% (2/6) of HER2 mutated cases were MSI‐H [17]. In the Chinese CRC cohort, 34.3% (12/35) HER2 mutated cases were MSI‐H, and 21.8% MSI‐H cases were HER2 mutated [26]. Kloth et al. found that 15% of patients with Lynch syndrome or Lynch‐like CRC had HER2 mutations [25]. The high MSI‐H rates in patients with CRC with HER2 mutations existed in previous studies but it was not pointed out. The potential reason could be that HER2 mutation is not paid enough attention. In the status of MSI‐H, all kinds of mutations may happen randomly. Is HER2 mutation just one of the high‐frequency events? However, we found that HER2 mutation was not in the top 40 mutated genes for all the MSI‐H patients. The internal relationship between MSI‐H and HER2 mutated deserves further study. Furthermore, no MSI‐H was found in the HER2 amplification subgroup. Until now, no study had reported the negative relationship between MSI‐H and HER2 amplification. Comparatively, we found that in the MyPathway study, 28 patients among 57 HER2‐amplified patients with mCRC had MSI status and none of them were MSI‐H [13]. In another study, none of the 11 HER2 amplification or overexpression patients with CRC were MSI‐H [28]. Similarly, none of the 16 HER2 amplification cases were MSI‐H [26]. Although it is too early to conclude that HER2 amplification and MSI‐H are exclusive, we observed that the prevalence of MSI‐H in patients with HER2 amplification was lower than that in the unselected CRC population. Larger sample size studies are required to validate these findings.
Among the HER2 mutated cases, compared with MSI‐H, MSS patients were predominately women, which was different from the sex distribution in the Chinese CRC population [2]. Furthermore, we found that the metastatic pattern was different between these two groups.
Until May 31, 2020, only four patients died and there were not enough events to analyze overall survival, and only DFS and PFS to first‐line therapy were analyzed. For first‐line therapy, MSS patients had significantly longer mPFS than MSI‐H patients. Although Kim et al. reported no significant difference in PFS between MSI‐H and MSS patients with CRC [29], more and more data support the concept that MSI‐H patients with colon cancer could benefit less from chemotherapy than MSS patients [30, 31].
Currently, for patients with CRC, MSI‐H is the only effective biomarker to predict response to PD‐1 antibody, with a response rate of 40% [32, 33] and a median PFS of 14.3 months [34]. In this present study, we found that MSI‐H patients with HER2 mutation had significantly worse mPFS for PD‐1 antibody than those without HER2 alteration. Further studies are merited to explore the association of HER2 mutation to PD‐1 antibody response in MSI‐H patients.
Several preclinical studies have suggested that HER2 amplification was a negative predictor of response to anti‐EGFR therapy [8, 15, 35]. To our knowledge, there is no report on the response of HER2 mutated mCRC cases to anti‐EGFR therapy. In our present study, we found that the mPFS for anti‐EGFR therapy was 8.2 months and 4.1 months in HER2 mutated and amplification cases. Although it is a retrospective study with a small sample size, it seems that patients with HER2 mutation with mCRC could benefit from anti‐EGFR therapy.
The limitations of the present study include the following: (a) not enough events for survival analysis, there were few cases followed, all stages of disease were mixed together, and different regimens were grouped together, so the analysis for DFS and PFS should be considered carefully; (b) a retrospective study with small sample sizes; and (c) potential selection bias. NGS was not a mandatory requirement for all the patients, and physicians are prone to use NGS in recurrent or metastatic patients. However, the alteration rate of HER2 was comparable to the TCGA data, meaning that our patients could be representative of patients with CRC.
Conclusion
To our knowledge, this is the first study to compare the clinicopathological features between HER2 mutated and amplification in CRC cases. We found a high MSI‐H rate in HER2 mutated cases but no MSI‐H in HER2 amplification cases. Furthermore, MSI‐H cases had less frequent lung metastasis. Our present study also showed that MSI‐H patients with HER2 mutation could have worse PFS for PD‐1 antibody than those without HER2 mutation. Studies with larger sample sizes are still warranted to confirm our findings.
Author Contributions
Conception/design: Miao‐Zhen Qiu, Da‐Jun Yang, Rui‐Hua Xu
Provision of study material or patients: Ying Jin, Yu‐Hong Li, Feng‐Hua Wang
Collection and/or assembly of data: Li‐Qiong Yang, Jun‐Zhong Lin, Wen‐Long Guan
Data analysis and interpretation: Xin‐Hua Yang, Da‐Lei Zhou, Ya‐Kang Long
Manuscript writing: Miao‐Zhen Qiu, Cai‐Yun He, Xin‐Hua Yang
Final approval of manuscript: Miao‐Zhen Qiu, Cai‐Yun He, Xin‐Hua Yang, Li‐Qiong Yang, Jun‐Zhong Lin, Da‐Lei Zhou, Ya‐Kang Long, Wen‐Long Guan, Ying Jin, Yu‐Hong Li, Feng‐Hua Wang, Da‐Jun Yang, Rui‐Hua Xu
Disclosures
The authors indicated no financial relationships.
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Supplementary figure 1 Disease free survival (DFS) in patients with HER2 mutation and amplification (A), patients with MSS and MSI‐H (B), MSI‐H patients with and without HER2 alteration (C).
Supplementary figure 2. Progression free survival (PFS) of first‐line therapy in patients with HER2 mutation and amplification (A), MSI‐H patients with and without HER2 alteration (B).
Table S1: Comparison of SNV and indel
Table S2: Gene list for 295 genes
Table S3: Gene list for 1021 genes
Acknowledgments
We thank all the patients for participating in the analysis. This study was supported by the National Natural Science Foundation of China (82073377, 81930065, 81772587), Science and Technology Program of Guangdong (2019B020227002), Science and Technology Program of Guangzhou (201904020046, 201803040019, 201704020228), and the third outstanding young talents training plan and Medical Scientist program of Sun Yat‐Sen University Cancer Center.
Disclosures of potential conflicts of interest may be found at the end of this article.
Contributor Information
Miao‐Zhen Qiu, Email: qiumzh@sysucc.org.cn.
Da‐Jun Yang, Email: xurh@sysucc.org.cn, Email: yangdj@sysucc.org.cn.
Rui‐Hua Xu, Email: xurh@sysucc.org.cn.
References
- 1. Bray F, Ferlay J, Soerjomataram I et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–132. [DOI] [PubMed] [Google Scholar]
- 3. Cao M, Li H, Sun D et al. Cancer burden of major cancers in China: A need for sustainable actions. Cancer Commun (Lond) 2020;40:205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berg M, Soreide K. EGFR and downstream genetic alterations in KRAS/BRAF and PI3K/AKT pathways in colorectal cancer: Implications for targeted therapy. Discov Med 2012;14:207–214. [PubMed] [Google Scholar]
- 5. Bang YJ, Van Cutsem E, Feyereislova A et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2‐positive advanced gastric or gastro‐oesophageal junction cancer (ToGA): A phase 3, open‐label, randomised controlled trial. Lancet 2010;376:687–697. [DOI] [PubMed] [Google Scholar]
- 6. Slamon DJ, Leyland‐Jones B, Shak S et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783–792. [DOI] [PubMed] [Google Scholar]
- 7. Liu T, Qin Y, Li J et al. Pertuzumab in combination with trastuzumab and chemotherapy for Chinese patients with HER2‐positive metastatic gastric or gastroesophageal junction cancer: A subpopulation analysis of the JACOB trial. Cancer Commun (Lond) 2019;39:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bertotti A, Migliardi G, Galimi F et al. A molecularly annotated platform of patient‐derived xenografts ("xenopatients") identifies HER2 as an effective therapeutic target in cetuximab‐resistant colorectal cancer. Cancer Discov 2011;1:508–523. [DOI] [PubMed] [Google Scholar]
- 9. Richman SD, Southward K, Chambers P et al. HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: Analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials. J Pathol 2016;238:562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu F, Ren C, Jin Y et al. Assessment of two different HER2 scoring systems and clinical relevance for colorectal cancer. Virchows Arch 2020;476:391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sartore‐Bianchi A, Trusolino L, Martino C et al. Dual‐targeted therapy with trastuzumab and lapatinib in treatment‐refractory, KRAS codon 12/13 wild‐type, HER2‐positive metastatic colorectal cancer (HERACLES): A proof‐of‐concept, multicentre, open‐label, phase 2 trial. Lancet Oncol 2016;17:738–746. [DOI] [PubMed] [Google Scholar]
- 12. Loree JM, Bailey AM, Johnson AM et al. Molecular landscape of ERBB2/ERBB3 mutated colorectal cancer. J Natl Cancer Inst 2018;110:1409–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meric‐Bernstam F, Hurwitz H, Raghav KPS et al. Pertuzumab plus trastuzumab for HER2‐amplified metastatic colorectal cancer (MyPathway): An updated report from a multicentre, open‐label, phase 2a, multiple basket study. Lancet Oncol 2019;20:518–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin V, Landi L, Molinari F et al. HER2 gene copy number status may influence clinical efficacy to anti‐EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer 2013;108:668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yonesaka K, Zejnullahu K, Okamoto I et al. Activation of ERBB2 signaling causes resistance to the EGFR‐directed therapeutic antibody cetuximab. Sci Transl Med 2011;3:99ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cerami E, Gao J, Dogrusoz U et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cancer Genome Atlas Network . Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kavuri SM, Jain N, Galimi F et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov 2015;5:832–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Noepel‐Duennebacke S, Juette H, Feder IS et al. High microsatellite instability (MSI‐H) is associated with distinct clinical and molecular characteristics and an improved survival in early colon cancer (CC); real world data from the AIO molecular registry Colopredict Plus. Z Gastroenterol 2020;58:533–541. [DOI] [PubMed] [Google Scholar]
- 20. Ye ZL, Qiu MZ, Tang T et al. Gene mutation profiling in Chinese colorectal cancer patients and its association with clinicopathological characteristics and prognosis. Cancer Med 2020;9:745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He CY, Qiu MZ, Yang XH et al. Classification of gastric cancer by EBV status combined with molecular profiling predicts patient prognosis. Clin Transl Med 2020;10:353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li A, Yang JJ, Zhang XC et al. Acquired MET Y1248H and D1246N mutations mediate resistance to MET inhibitors in non‐small cell lung cancer. Clin Cancer Res 2017;23:4929–4937. [DOI] [PubMed] [Google Scholar]
- 23. Mayakonda A, Lin DC, Assenov Y et al. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brannon AR, Vakiani E, Sylvester BE et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol 2014;15:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kloth M, Ruesseler V, Engel C et al. Activating ERBB2/HER2 mutations indicate susceptibility to pan‐HER inhibitors in Lynch and Lynch‐like colorectal cancer. Gut 2016;65:1296–1305. [DOI] [PubMed] [Google Scholar]
- 26. Guo Y, Guo XL, Wang S et al. Genomic alterations of NTRK, POLE, ERBB2 and MSI status in Chinese colorectal cancers. The Oncologist 2020;25:e1671–e1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Afrăsânie VA, Marinca MV, Alexa‐Stratulat T et al. KRAS, NRAS, BRAF, HER2 and microsatellite instability in metastatic colorectal cancer ‐ Practical implications for the clinician. Radiol Oncol 2019;53:265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park JS, Yoon G, Kim HJ et al. HER2 status in patients with residual rectal cancer after preoperative chemoradiotherapy: The relationship with molecular results and clinicopathologic features. Virchows Arch 2018;473:413–423. [DOI] [PubMed] [Google Scholar]
- 29. Kim ST, Kim HK, Lee J et al. The impact of microsatellite instability status and sidedness of the primary tumor on the effect of bevacizumab‐containing chemotherapy in patients with metastatic colorectal cancer. J Cancer 2018;9:1791–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cercek A, Dos Santos Fernandes G, Roxburgh CS et al. Mismatch repair‐deficient rectal cancer and resistance to neoadjuvant chemotherapy. Clin Cancer Res 2020;26:3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Rosa N, Rodriguez‐Bigas MA, Chang GJ, et al. DNA Mismatch Repair Deficiency in Rectal Cancer: Benchmarking Its Impact on Prognosis, Neoadjuvant Response Prediction, and Clinical Cancer Genetics. J Clin Oncol. 2016;34(25):3039–3046. doi: 10.1200/JCO.2016.66.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao P, Li L, Jiang X et al. Mismatch repair deficiency/microsatellite instability‐high as a predictor for anti‐PD‐1/PD‐L1 immunotherapy efficacy. J Hematol Oncol 2019;12:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Le DT, Uram JN, Wang H et al. PD‐1 Blockade in tumors with mismatch‐repair deficiency. N Engl J Med 2015;372:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Overman MJ, McDermott R, Leach JL et al. Nivolumab in patients with metastatic DNA mismatch repair‐deficient or microsatellite instability‐high colorectal cancer (CheckMate 142): An open‐label, multicentre, phase 2 study. Lancet Oncol 2017;18:1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Consonni D, Mensi C. Comment on the paper 'Boffetta et al. Validation of the diagnosis of mesothelioma and BAP1 protein expression in a cohort of asbestos textile workers from Northern Italy. Ann Oncol 2018; 29(2): 484‐489'. Ann Oncol 2019;30:340–341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Supplementary figure 1 Disease free survival (DFS) in patients with HER2 mutation and amplification (A), patients with MSS and MSI‐H (B), MSI‐H patients with and without HER2 alteration (C).
Supplementary figure 2. Progression free survival (PFS) of first‐line therapy in patients with HER2 mutation and amplification (A), MSI‐H patients with and without HER2 alteration (B).
Table S1: Comparison of SNV and indel
Table S2: Gene list for 295 genes
Table S3: Gene list for 1021 genes


