Abstract
Over the past decade, we have witnessed significant advances in the molecular characterization of systemic mastocytosis (SM). This has provided important information for a better understanding of the pathogenesis of the disease but has also practically impacted the way we diagnose and manage it. Advances in molecular testing have run in parallel with advances in therapeutic targeting of constitutive active KIT, the major driver of the disease. Therefore, assessing the molecular landscape in each SM patient is essential for diagnosis, prognosis, treatment, and therapeutic efficacy monitoring. This is facilitated by the routine availability of novel technologies like digital PCR and NGS. This review aims to summarize the pathogenesis of the disease, discuss the value of molecular diagnostic testing and how it should be performed, and provide an overview of present and future therapeutic concepts based on fine molecular characterization of SM patients.
Keywords: Systemic mastocytosis, KIT, Allele burden, Tyrosine kinase inhibitor
Introduction
Mastocytosis is a rare neoplasm characterized by the expansion and accumulation in different organs/tissues of clonal mast cells (MCs) that can be recognized as morphologically and immunophenotypically abnormal.1 The clinical presentation of mastocytosis is heterogeneous, with various manifestations ranging from skin-limited disease (cutaneous mastocytosis, CM) to extra-cutaneous involvement (systemic mastocytosis, SM). CM is frequent in the pediatric age but it can spontaneously regress during puberty.2,3 In contrast, SM is generally seen in adult patients and may be associated with multiorgan dysfunction and shortened survival.4 The 2016 World Health Organization (WHO) defined major categories and variants of SM. Based on histological criteria, clinical parameters, and organ involvement, SM is divided into indolent SM (ISM), smoldering SM (SSM), and advanced SM variants (AdvSM), including SM with an associated clonal hematopoietic non-MC disease (SM-AHN), aggressive SM (ASM) and MC leukemia (MCL).5 ISM is the most common subtype and has a relatively benign prognosis,6 although 5 to 10% of ISM patients progress to SSM or AdvSM.7,8 SSM is associated with an increased symptom burden, inferior survival compared to ISM, and a much higher transformation rate (15–20% for transformation to AdvSM or acute leukemia).9 In addition, patients with AdvSM have a reduced life expectancy.10
Whereas treatment of ISM is generally palliative and primarily directed towards preventing anaphylaxis and the control of symptoms, treatment of AdvSM needs cytoreductive therapy to ameliorate disease-related organ dysfunction. For a long time, high-dose chemotherapy possibly followed by transplant when feasible, interferon or cladribine have represented the main options for AdvSM patients. The knowledge of the driver lesion underlying the molecular pathogenesis of SM has ultimately enabled the development of targeted therapies. Despite the great hurdle represented by the rarity of AdvSM patients, the tyrosine kinase inhibitor midostaurin has recently been approved based on the results of phase 2 clinical trial that lasted almost 10 years.11 Other investigational agents, like avapritinib and ripretinib, are being evaluated. In addition, several additional compounds with the same or even different targets have been tested at the preclinical level.
In this review, we summarize the molecular studies and acquisitions that have led to a better understanding of the pathogenesis of SM, paving the way to the development of targeted therapies. Moreover, we discuss how the implementation of advanced molecular technologies has recently refined the diagnostic and prognostic algorithms and how molecular testing is acquiring a pivotal role in treatment individualization. Finally, we also provide an overview of a series of new putative therapeutic targets, some of whom might find a clinical translation, primarily via repurposing of already approved agents.
Activating KIT Mutations Drive the Pathogenesis of SM
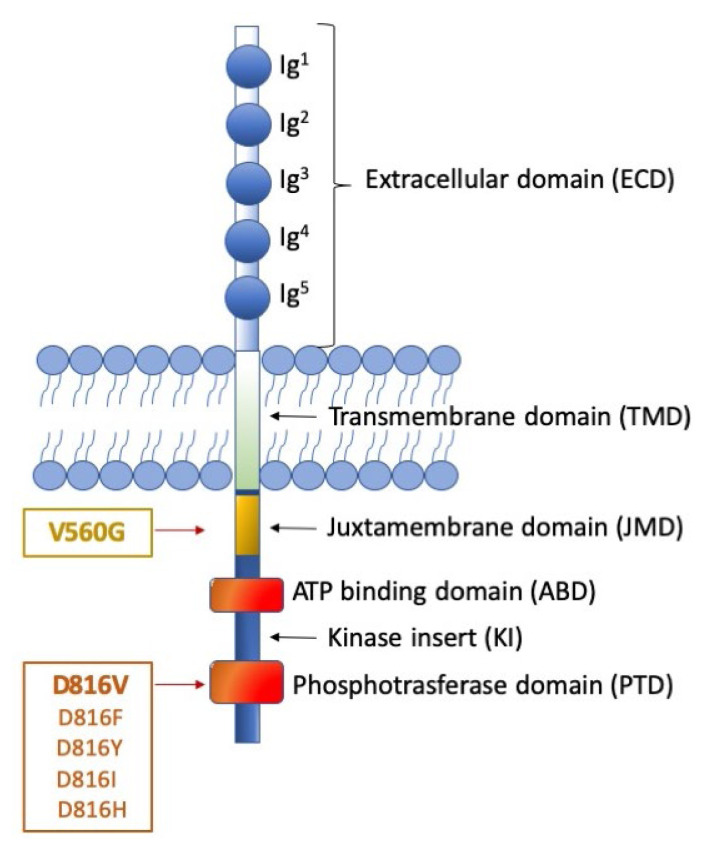
SM is virtually always associated with somatic gain-of-function mutations of the proto-oncogene c-KIT, which encodes a transmembrane protein belonging to type III tyrosine kinase receptors. KIT is expressed by MCs, hematopoietic progenitor cells, germ cells, melanocytes, and interstitial cells of Cajal in the gastrointestinal tract.12 KIT expression is down-regulated upon differentiation of hematopoietic progenitors into mature cells of all lineages, except in MCs that retain high cell surface KIT expression levels. The KIT receptor explicitly binds the stem cell factor (SCF) cytokine, also known as the KIT ligand, steel factor, or MC growth factor. The interaction between KIT and its ligand leads to dimerization of the receptor and activation of its intrinsic tyrosine kinase activity and drives a variety of downstream signal transduction pathways.13 The transduction process involves different players such as PI3 kinase, Src family kinases, the Ras-Erk pathway, and JAK/STAT, resulting in cell proliferation, survival, and migration. Aberrant activation of the KIT receptor, caused by gain-of-function mutations of the gene, is a frequent event in SM and results in uncontrolled production and proliferation of MCs. The first identification of somatic gain-of-function mutations in the KIT gene occurred in the HMC-1 human MCL cell line. Two heterozygous point mutations of KIT were revealed: a substitution of aspartic acid with valine at residue 816 (KIT D816V) and a substitution of valine with glycine at residue 560 (KIT V560G). D816V and V560G are the prototypes of two distinct categories of activating mutations, displaying different oncogenic properties and sensitivity to tyrosine kinase inhibitors (TKIs) (Figure 1).14 The first can indeed be classified as an ‘enzymatic pocket’ mutation since it falls in the activation loop at the entrance of the enzymatic pocket. The activation loop serves as a sort of molecular switch for kinase activity, and D816V turns it permanently on. Given its strategic position with respect to the enzymatic pocket, this mutation is an obstacle to the binding of some TKIs, like imatinib. The second, in contrast, belongs to the wider category of ‘regulatory type’ mutations. It is located in the intracellular juxta-membrane domain of the receptor, which has a critical auto-inhibitory function; thus, it results in constitutive kinase activation.
Figure 1. Representation of KIT receptor structure and position of the major mutations.
The figure shows the receptor under its monomeric form. The extracellular domain (ECD) (light blue) is characterized by 5 Immunoglobulin (Ig)-like domains that contain a ligand binding site for stem cell factor (SCF) and a dimerization site. The cytoplasmic region contains a transmembrane domain (light green) made by a single helix. The intracellular portion (dark blue) contains an auto-inhibitory juxta-membrane domain (yellow) and a kinase domain (orange) which is split into two parts: an ATP-binding domain and a phosphotransferase domain, linked by a kinase insert. On the left, two distinct categories of KIT activating mutations are reported: the ‘regulatory type’ mutations (with the V560G being the most frequent), that are located in the juxta-membrane domain; and the ‘enzymatic pocket’ mutations (best exemplified by the D816 mutations), that fall in the phosphotransferase domain. Though much rarer, the first category of mutations can be addressed by a wider spectrum of inhibitors, including imatinib. The second category of mutations, in contrast, impacts on kinase conformation, thus can be addressed, at present, only by midostaurin (approved) avapritinib and ripretinib (investigational).
‘Regulatory type’ mutations may also affect the binding of substrates or signal transducing or regulatory molecules, or induce ligand-independent dimerization with subsequent autophosphorylation and activation of the kinase. The greatest majority of SM patients, irrespective of the WHO sub-type, display the D816V mutation.4,15 More rare variants like D816Y, D816F, D816H, and D816I have occasionally been detected. Less common (<5%) KIT mutations at other codons (mostly belonging to the ‘regulatory type’ class) have been reported.16,17 Rare germline KIT mutations associated with familial mastocytosis have also been reported, including F522C, A533D and K509I. For these forms, the tyrosine kinase inhibitor imatinib offers a valuable therapeutic option.18 Moreover, several non-D816V mutations have anecdotally been reported in MCL and mast cell sarcomas. Mital et al. reported one case of MCL with a p.A502_Y503dup mutation in exon 9.19 Other studies conducted in patients with MCL and mast cell sarcomas described KIT mutations at exon 10, exon 11, and exon 13.20–22 Finally, children with CM display missense KIT mutations targeting exon 17 at codon 816 but also several alterations in exons 8 and 9 involving the fifth Ig loop of the KIT extracellular domain and leading to constitutive activation of the receptor.23
The abnormal kinase activity of KIT has been documented in other human malignancies such as germ cell tumors, melanoma, GIST, and acute myeloid leukemia (AML).24–27
The Role of KIT Mutation Detection in the Diagnostic Workout of SM
The current WHO classification of mastocytosis defines one major and four minor diagnostic criteria for SM.5 The major SM criterion is the presence of multifocal dense aggregates of MCs in bone marrow (BM) or other visceral organs (at least 15 MCs/cluster). Minor SM criteria include: i) abnormal morphology of MCs (>25%), ii) expression of CD2 and/or CD25 in MCs, iii) persistent serum tryptase concentration >20 ng/mL, and iv) presence of the KIT D816V point mutation in the BM or another extracutaneous organ. If at least one major and one minor or three minor SM criteria are fulfilled, the diagnosis of SM can be established.
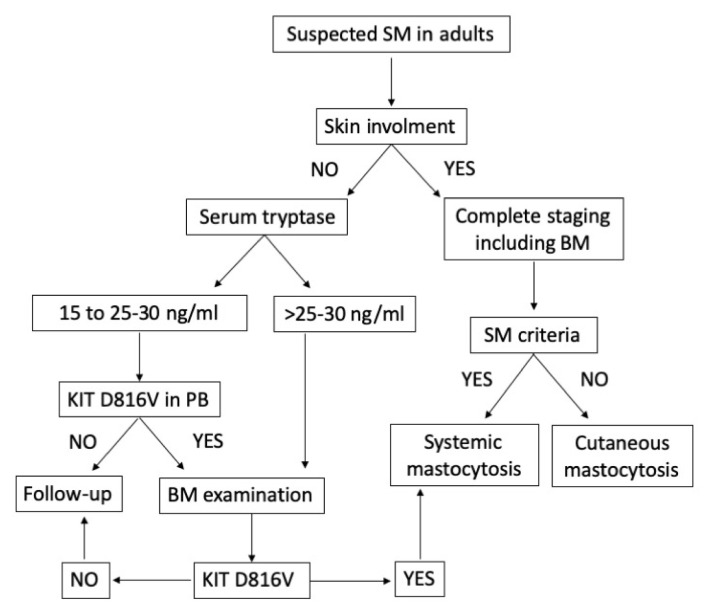
The European Competence Network on Mastocytosis (ECNM) suggests an algorithm for patients with suspected (systemic) mastocytosis and provides essential technical guidelines for KIT mutation detection.28 In The case of suspected mastocytosis in an adult patient without skin lesions and average or slightly increased tryptase level, the presence of KIT D816V mutation may be investigated at first in peripheral blood (PB). If the D816V mutation is detected in PB cells, a BM examination is recommended.29 In adult patients with suspected mastocytosis and documented skin involvement, a BM investigation is required. In the non-availability of fresh BM aspirate sample, KIT mutational analyses can be performed on cells detached from BM smears or a paraffin-embedded biopsy sample (Figure 2).30
Figure 2. Flowchart for adult patients with suspected systemic mastocytosis (SM).
In adult patients with suspected SM without skin involvement, the basal serum tryptase level is an important initial screen parameter. In conditions of slightly increased tryptase level, the presence of the KIT D816V mutation may be investigated at first in peripheral blood (PB) provided that a highly sensitive test is warranted. If the D816V mutation is detected in PB cells, a BM examination is recommended. If the KIT D816V mutant is not detectable in the BM and symptoms are non-specific, the patient will be examined in the follow up. By contrast, if the D816V mutation is detected in the BM and SM criteria are fulfilled, the diagnosis of SM can be established. In adult patients with suspected mastocytosis and documented skin involvement, a BM investigation is required. Based on the presence or absence of criteria for SM, the final diagnosis will be SM or cutaneous mastocytosis (CM).
In most SM patients, particularly ISM patients, the MC burden, that is, the number of neoplastic MCs infiltrating the BM, is very low. Moreover, mononuclear cells (MNCs) from PB are almost invariably KIT D816V negative when investigated by low sensitivity techniques. This means that sensitivity is crucial to avoid false-negative results and misdiagnosis. Over the past few years, substantial advances have been made to develop more and more sensitive and accurate diagnostic assays. Chronologically, the following techniques have been proposed for KIT D816V mutation testing. They all have advantages as well as disadvantages and differ in terms of lower detection limit, target mutation(s), and requirement of dedicated instrumentations: i) RT-PCR plus restriction fragment length polymorphism (RFLP), ii) nested RT-PCR followed by denaturing high-performance liquid chromatography (D-HPLC) of amplicons, iii) peptide nucleic acid (PNA)-mediated PCR, iv) ASO-qPCR on DNA or cDNA, v) droplet digital PCR (ddPCR) and vi) Next-generation sequencing (NGS).
RT-PCR plus RFLP: it is a simple, cheap, and fast technique that allows identifying the KIT D816V mutation with a sensitivity of 0,05%. No dedicated instrumentation is needed except for a thermal cycler and an electrophoretic system. However, it is not quantitative, and it does not allow the detection of D816 variants and the identification of mutations at other codons.31
Nested RT-PCR followed by D-HPLC of amplicons: this assay allows detecting different KIT mutations in the same reaction.32 Nevertheless, it has a relatively low sensitivity (0,5–1%), and it is not quantitative. Moreover, it is time-consuming and needs expensive and not widely available instrumentation (D-HLPC).
PNA-mediated PCR: it is the recommended method for FFPE BM trephine biopsies.31 It allows detection of KIT mutations at position 816 or in adjacent codons, but it is not a quantitative assay, and it has an intermediate sensitivity (0,1%).33
ASO-qPCR: it is a simple, fast, cheap, reliable, and highly sensitive method (0,01%) that allows to detect and quantitate KIT D816V in different tissues and specimens (BM, PB, and organ biopsies).34,35 However, it requires standard curves for the quantitative assessment, thorough validation, and possibly multi-laboratory standardization.
Droplet Digital PCR: it is a promising new method for sensitive and accurate quantification of KIT D816V with a sensitivity of 0,01%.36 Moreover, ddPCR has been shown to sensitively and reproducibly detect and quantify KIT D816V in FFPE material.37 In contrast to ASO-qPCR, absolute quantification does not need a standard curve; thus, it is more straightforward. Commercial kits are available.
Next-generation sequencing (NGS): it has the lowest sensitivity (1–5%), but it is the best remaining option to study those patients who have no evidence of mutations at codon 816. Commercial myeloid panels are available that include several KIT hotspot exons among their target genes. Such panels may also highlight the presence of mutations in genes other than KIT, providing additional prognostic information (see below).
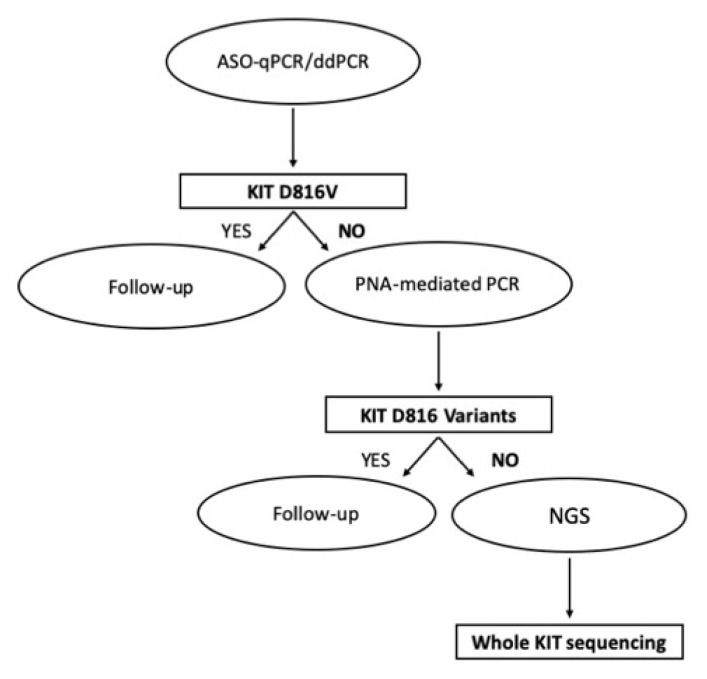
Being by far the most common, the presence of a KIT D816V needs to be investigated first. However, in the case of D816V negativity, variants like D816Y, D816H, etc., should be ruled out. If no mutation at all can be identified at codon 816, more rare mutations in other KIT exons (like those mentioned in the previous section) should be investigated. A stepwise approach is thus needed. For D816V detection and quantitation, ASO-qPCR or ddPCR are the most frequent option. D816 variants in confirmed D816V-negative patients may be investigated using PNA-mediated PCR. Mutations in other hotspot regions of the KIT gene may then be assessed by NGS (Figure 3).
Figure 3. Proposed flowchart for KIT mutation analysis in SM, taking advantage of all the currently available technologies.
Allele-specific oligonucleotide (ASO)-qPCR or droplet digital PCR (ddPCR) need to be performed in first instance for KIT D816V detection and quantitation. In confirmed D816V-negative patients, D816 variants may be investigated using peptide nucleic acid (PNA)-mediated PCR. If no mutation at all can be identified at codon 816, mutations in other hotspot regions of the KIT gene may then be assessed by Next Generation Sequencing (NGS).
Importantly, ASO-qPCR and ddPCR have also shown the capability to detect the D816V mutation non-invasively in the PB.32,37–39 Their potential for the study of cell-free DNA is under investigation.
Prognostic Implications of KIT D816v Allele Burden Quantification and of Multilineage Involvement
For the assessment of KIT D816V, quantitative methods are desirable for several reasons. The measurement of KIT D816V mutation percentage (the so-called ‘allele burden’) has prognostic significance and can also be employed for monitoring treatment response and disease progression. Recent studies show that the KIT D816V allele burden is indicative of the WHO SM subtype and correlated with the severity of the disease.36,40 Indeed, patients with AdvSM show a significantly higher KIT D816V mutation percentage compared to ISM and CM patients, the latter showing the lowest KIT D816V allele burden. The higher KIT D816V allele burden in AdvSM may be associated with the presence of non-MC-lineage cells harboring the KIT D816V mutation and could explain why it is not always correlated with the number of MC in BM biopsies, and it is only moderately associated with serum tryptase levels. It is now well known that some ISM and all AdvSM patients display a multilineage disorder whose characteristics are closely related to the risk of disease progression.41 In fact, the KIT D816V mutation is detected not only in bone marrow MCs but also in myeloid and lymphoid hematopoietic cell lineages, supporting the stem cell origin of this disease starting from a hematopoietic precursor with the ability for multilineage differentiation.42–44 The greater or smaller involvement of non-MC-lineage cells harboring the KIT D816V mutation reflects the risk of evolving to a more aggressive subtype of the disease. Therefore, SM patients with multilineage KIT D816V mutation have been shown to have a higher probability of progression compared to patients with MC-restricted KIT D816V.41
The assessment of the KIT D816V allele burden also has a remarkable potential in monitoring treatment response. Hoermann et al. observed that in SM patients with stable clinical course, there were not variations in the KIT D816V allele burden, while advSM patients with disease progression showed a marked increase.40 In line with this, Jawhar et al. used the measurement of KIT D816V allele burden for monitoring midostaurin-treated AdvSM patients. They showed that at month 6 of therapy, a significant reduction ≥25% of the allele burden was the strongest on-treatment predictor for improved survival.45
The KIT D816V allele burden was also identified as a prognostic factor regarding overall survival (OS), and to the purpose, a cut-off of ≥ or <2% mutant alleles was used to define two distinct SM groups with different survival times.36,40 The group under the cut-off level had a significantly longer life expectancy compared to patients over the 2% cut-off.
These findings are still under validation, but the wider implementation of KIT D816V allele burden assessment in clinical practice holds the potential to improve treatment and monitoring of patients with SM.
Molecular Alterations Additional to KIT Mutations
The somatic KIT D816V mutation is undoubtedly the major driver of SM pathogenesis. However, it is detectable both in patients with ISM, who have a relatively benign prognosis and in patients with AdvSM, who are frequently characterized by rapidly deteriorating clinical courses and poor outcomes. Furthermore, while childhood-onset and adult-onset mastocytosis are both associated with activating KIT mutations, the natural history of the two conditions is quite different. Together with experimental shreds of evidence suggesting that KIT D816V alone is not a fully transforming oncoprotein,46–49 this emphasized the importance of a better understanding of SM pathogenesis. Prognostically relevant mutations additional to KIT have now been identified in AdvSM. These have been found to affect genes encoding for epigenetic regulators (ASXL1, DNMT3A, EZH2, TET2), transcription factors (RUNX1), signaling molecules (CBL, JAK2, KRAS, NRAS), or splicing factors (SRSF2, SF3B1, U2AF1). These mutations are not specific for SM but are frequently detected in myeloid neoplasm, including myelodysplastic syndromes (MDS), myeloproliferative neoplasms (MPN), chronic myelomonocytic leukemia (CMML), and acute myeloid leukemia (AML) where they are often associated with poor prognosis and shorter survival.50–52 TET2 mutations are detected in 20–40% of AdvSM patients.47,48 Evidence in mice suggests that the cooperation between KIT D816V and the loss of function of TET2 induces a transformation to a more aggressive disease phenotype.53 Moreover, when TET2 mutations are associated with DNMT3A mutations, the prognosis of SM patients is severely impaired compared to those with wild-type genes.48 Analogously to TET2, the presence of other mutations additional to KIT may confer an adverse prognosis compared with patients without such abnormalities. Jawhar et al. were the first to observe that in SM-AHN, the presence and number of mutated genes within the SRSF2/ASXL1/RUNX1 (S/A/R) panel was associated with poor outcome and adverse clinical characteristics.54 In particular, mutations in SRSF2 and ASXL1 were the most predictive adverse indicators concerning OS. As expected, these mutations were observed at a higher frequency in ASM-AHN and MCL-AHN and at a lower frequency in ISM-AHN. Muñoz-González et al. suggested that, in addition to S/A/R gene panel, somatic EZH2 gene mutations can be detected in ISM patients who show a higher risk of disease progression.55
Moreover, the overall response rate (ORR) and OS were significantly higher in patients S/A/Rneg than in patients S/A/Rpos. The Mayo Clinic group evaluated the impact of ASXL1 and/or CBL mutations and the occurrence of ≥3 non-KIT mutations on the clinical outcome of AdvSM patients and showed that they were independently associated with inferior OS.56 Moreover, based on clinical and molecular parameters (including ASXL1 mutations), Pardanani et al. proposed a “mutation-augmented” prognostic scoring system to stratify advSM patients into 3 subgroups: low-, intermediate- and high-risk with significantly different OS.57 Alternative prognostic scoring systems that include information about S/A/R profiling and other molecular markers have been proposed, and others are still under investigation.58–61
KIT as Therapeutic Target
The clinical spectrum of this disease is highly heterogeneous. The treatment should be individualized and related to symptoms and diagnosis of each patient. Therapy of CM is mainly directed towards skin lesions.62 Anti-mediator drug therapy for MC activation symptoms represents the mainstay of treatment for symptomatic CM, ISM and SSM however, it should be considered in all SM patients.63 The goal is to regulate mediator secretion, keep allergies under control, and counteract osteopenia/osteoporosis. AdvSM is frequently associated with organ damage, so cytoreductive therapy has long been considered the best therapeutic strategy. For a long time, interferon64–66 and cladribine67–69 have represented the main options for AdvSM patients. However, the introduction of tyrosine kinase inhibitors (TKIs) has revolutionized the prognosis and the outcome of patients with AdvSM.70 Several TKIs, such as imatinib, nilotinib, and masitinib, have shown activity against wild-type KIT. They belong to type II TKI inhibitors, so they recognize only the inactive conformation of the receptor and stabilize it. The presence of mutations in the activation loop (like KIT D816V) that result in a constitutively active conformation of the receptor renders it inaccessible to type II inhibitors. Imatinib (IM) is currently approved by the FDA for the treatment of ASM with WT KIT or with unknown KIT mutational status. It demonstrated activity against certain trans-membrane (F522C) and juxta-membrane (V560G) KIT mutants.71–73 IM efficacy has also been reported in a patient with familiar SM with KIT K509I mutation in the germinal line.18 However, since the major of SM patients harbor the KIT D816V mutation, it has a limited role in the treatment of this disease. Nilotinib was evaluated in phase II multicenter study of SM patients with or without KIT D816V mutation. However, it showed modest clinical benefit, observed only in patients with WT KIT.74 Masitinib showed a great in vitro activity in ISM and SSM patients with WT KIT or with KIT mutations outside the phospho-transferase domain,75,76 but it has yet to be tested in AdvSM. Type I TKIs (able to bind both the active and the inactive form of kinases) tested in SM include dasatinib, midostaurin, and avapritinib. Dasatinib is a multikinase inhibitor that showed in vitro activity against both WT KIT and KIT activation loop mutants (KIT D816V/Y/F).77 It was also shown to re-localize KIT D816V from the cytoplasm to the cell surface. Interestingly, the extent of this re-localization correlates with SM severity (AdvSM > ISM).78 However, given its short half-life, it showed poor in vivo effectiveness.79 Midostaurin is another multikinase inhibitor; currently, the only TKI approved for the treatment of AdvSM. The encouraging results related to the use of midostaurin in a patient with MCL80 led to a multicenter phase II study (CPKC412D2213) of midostaurin in 26 AdvSM patients. Results consisted in an unprecedented overall response rate (ORR) of 69% regardless of KIT mutational status, with 38% of Major Responses (MR)(complete resolution of at least one clinical(C)-finding such as cytopenias, osteolysis with or without pathologic fractures, hepatosplenomegaly and/or with impaired liver function and/or ascites, and malabsorption).81 Starting from these data, a global phase II trial (CPKC412D2201) was initiated to evaluate the efficacy and safety of midostaurin in 89 AdvSM patients.82 The ORR was 60% and was independent of KIT mutation status and the number of previous therapies. Almost half of the patients (45%) had an MR, 15% had a partial response. After the closure of this study, a compassionate use program of midostaurin was approved in France. ORR was 71%, with a median response duration of 17 months.83 The most frequents adverse effects of midostaurin are low grade (nausea, vomiting, diarrhea), but also new or worsening grade 3 or 4 (neutropenia, anemia, and thrombocytopenia) adverse events can be observed.
Finally, avapritinib is a type I kinase inhibitor that selectively inhibits the activation-loop mutants of KIT (exon 11/17, including KIT D816V). It is approved in the USA for PDGFRA exon 18 mutant gastrointestinal stromal tumors (GIST), and it is also in clinical development for SM treatment. The phase I study (Explorer; NCT02561988) in adult patients with AdvSM has demonstrated potent antineoplastic activity of avapritinib across all advSM subtypes with complete (disappearance of all target lesions) and durable responses.84 The ORR was 77%; 74% of patients maintained the response for at least 12 months. Avapritinib was also showed to improve overall symptoms of SM. Avapritinib is currently being evaluated in an ongoing, double-blind, placebo-controlled, phase II study (Pioneer), where ISM patients with moderate or severe symptoms, refractory to ≥2 best supportive care drugs, are enrolled.85 Most adverse reactions of this drug are of grade 1 or 2.
Another agent under investigation is ripretinib (DCC-2618). It is a type II kinase inhibitor designed to inhibit the full spectrum of mutant KIT and PDGFRA. In addition, it shows strong activity against activation loop mutants, previously thought to be only achievable with type I inhibitors.86 Its efficacy and safety are being tested in a Phase I open-label dose-escalation study (NCT02571036) to treat AdvSM, GIST, and advanced cancer.
Other Non-KIT Targets and Novel Agents
Several studies have shed light on novel altered pathways involved in the pathogenesis of SM. Under normal conditions, SCF binds KIT receptor leading to its dimerization and activation. This binding activates several downstream signaling molecules involved in the proliferation and survival of MCs, such as the phosphatidylinositol triphosphate kinase (PI3K), the Janus kinase (JAK)/signal transducers and activators of transcription (STAT), and the rat sarcoma (RAS)/extracellular signal-regulated kinases (ERK). The presence of the KIT D816V mutation contributes to divert these pathways leading to a malignant transformation of MCs.
Baumgartner et al. showed that the SM-related oncoprotein KIT D816V promotes STAT5 activation and phosphorylation and that pSTAT5 contributes to the growth of neoplastic MCs.87 Moreover, STAT5 was found to localize to the cytoplasm and to form a signaling complex with PI3K,88 whose activation has been described to be constitutively associated with D816V KIT mutant.89 Once activated, PI3K activates its downstream effector AKT promoting MCs growth and survival. This evidence suggests that the STAT5-PI3K-AKT pathway plays an important role in the pathogenesis of SM. Indeed, the knock-down of either STAT5 or AKT activity resulted in growth inhibition of KIT D816V+ neoplastic MCs.88 Therefore, drugs targeting the STAT5- PI3K-AKT axis should be considered a potentially novel therapeutic approach in SM patients. Another interesting strategy may be targeting the JAK/STAT signaling pathway. Starting from the evidence that there is often an increased expression of JAK/STAT pathway components in SM, Lasho et al. showed KIT D816V-mediated cell growth decrease in MC cell lines treated with the JAK2 inhibitor TG101348.90
One of the major pathways downstream PI3K is represented by rapamycin (mTOR). It is a serine/threonine kinase found in two protein complexes called mTOR complex 1 (mTORC1) and complex 2 (mTORC2). PI3K is able to regulate the mTORC1 pathway by the activation of AKT. Once activated, AKT directly phosphorylates the negative regulator of mTOR, leading to mTOR activation and phosphorylation of two effector molecules: p70 ribosomal S6 kinase (p70S6K) and eukaryotic initiation factor 4E-binding protein1 (4E-BP1).91 The increased activation of the mTORC1 pathway, reported in neoplastic MCs lines and immature MCs,92 seems to contribute to MCs dysregulated proliferation and survival. Therefore, the inhibition of mTORC1 by rapamycin should be considered to contrast this event. Indeed, it has been shown to block mTORC1-dependent p70S6K phosphorylation.91 Moreover, the dual PI3K/mTOR blocker NVP-BEZ235 showed profound inhibitory effects on the growth of primary and neoplastic MCs in vitro.93
In addition to KIT downstream effectors, other non-KIT targets have been described as therapeutically interesting. For example, MCL-1, a BCL-2 family member with antiapoptotic properties, was expressed constitutively both in neoplastic MCs of all SM variants and MC leukemia cell lines HMC-1.1 (D816V negative) and HMC-1.2 (D816V positive); furthermore, MCL-1 knock-down mediated by antisense oligonucleotides or specific siRNA resulted in increased apoptosis and a decreased survival of neoplastic MCs, showing synergistic effects in combination with midostaurin and other TKIs including nilotinib and imatinib.94 These data indicate that MCL-1 may be a novel, potential target in neoplastic MCs.
KIT D816V was also found to downregulate BIM expression.95 BIM is a proapoptotic member of the BCL-2 family that acts as a tumor suppressor in various myeloid neoplasms.96–98 The KIT-targeting drug midostaurin was able to induce BIM re-expression in neoplastic HMC-1.1 and HMC-1.2 cells and to promote growth inhibition in both subclones.95 Moreover, in the HMC-1 cell line, the use of the proteasome inhibitor bortezomib was associated with a substantial increase in BIM expression and induction of apoptosis. Indeed, several studies have shown that phosphorylated BIM degradation in neoplastic myeloid cells is usually mediated by the proteasome.97,99 In addition to midostaurin and bortezomib, Aichberger et al. showed that the pan Bcl-2 blocker obatoclax contributed to inducing growth arrest and promoting apoptosis in HMC-1 cells and had synergic effects with midostaurin.95 In summary, BIM acts as a regulator of the growth and survival of neoplastic human MCs. Therefore, promoting BIM re-expression or targeting antiapoptotic members of the Bcl-2 family, combined with a KIT inhibitor, may be a novel, interesting approach in AdvSM patients.
Finally, the loss of function of the SETD2 tumor suppressor gene has recently been reported in AdvSM.100 It was already observed in various solid tumors and hematologic malignancies of both myeloid and lymphoid origin.101 The SETD2 tumor suppressor gene encodes the only methyltransferase able to tri-methylate histone H3 on lysine 36 (H3K36Me3).102 H3K36Me3 by SETD2 is critical for the maintenance of chromatin structure and transcriptional fidelity. Moreover, SETD2 plays pivotal roles in RNA alternative splicing regulation, DNA damage repair, and cytoskeleton protein methylation.101,103 SETD2 loss of function may be due to biallelic missense or truncating mutations, as seen in acute leukemias, or associated with monoallelic deletions at chromosome 3p and mutations of the remaining allele, as reported in clear cell renal cell carcinoma (ccRCC). However, other mechanisms acting at the transcript or protein level can be observed. In SM, a new post-translational mechanism of SETD2 loss of function has been described: both subclones of the HMC-1 cell line and up to 80% of patients with AdvSM displayed H3K36Me3 deficiency as a result of non-genomic loss of function of SETD2, in the absence of mutations or structural aberrations.100 The extent of H3K36Me3/SETD2 downmodulation was correlated with disease aggressiveness (AdvSM>ISM). Proteasome inhibition restored H3K36Me3 and SETD2 protein expression, suggesting that a functional protein is produced but rapidly degraded. Moreover, the treatment with the proteasome inhibitor bortezomib resulted in induction of apoptosis and reduced colony growth both in the HMC-1 cell line and in primary neoplastic MCs from patients with AdvSM. SETD2 loss of function in acute leukemias is often associated with chromosomal aberrations and activated gene expression in the mTOR and Jak/Stat signaling pathways, contributing directly to leukemogenesis in acute leukemias and SM. All these data suggest that reverting SETD2 loss of function, in combination with KIT-targeting drugs, could be a promising therapeutic strategy to improve the prognosis of AdvSM patients who do not respond to or relapse on midostaurin. Further investigations into the mechanisms leading to SETD2 altered turnover in SM and the cooperative effects of KIT constitutive activation and SETD2 loss of function in AdvSM are warranted.
Conclusions and Future Perspectives
Over the past decade, we have witnessed major advances in the molecular characterization of SM. This has provided important information for a better understanding of the pathogenesis of the disease but has also practically impacted the way we diagnose and manage it.
Sensitive and accurate assessment of KIT mutation status is critical for appropriate diagnostic and therapy. Moreover, quantitation of KIT D816V allele burden has prognostic significance and can be employed for monitoring treatment response and disease progression. Cooperating mutations in genes other than KIT contribute to the greater aggressiveness of the disease and may provide additional prognostic information. Over the past few years, substantial advances have been made in developing more sensitive and accurate diagnostic assays that have shown the capability to detect the KIT D816V mutation percentage at very low levels (down to 0,001%), and non-invasively in the PB (ASO-qPCR, ddPCR). Both these features are important, considering that SM is generally underdiagnosed.
Moreover, a role for targeted NGS in identifying KIT non-D816V mutants or mutations in other key myeloid genes has been recently established. The same myeloid panels commonly interrogated in diagnostic laboratories for acute myeloid leukemias, myelodysplasias, Philadelphia chromosome-negative myeloproliferative neoplasms etc are suitable, facilitating the application of relatively expensive and high-throughput technology to such a rare setting like SM patients. However, molecular testing will require standardization, and although the ECNM has already provided some useful technical recommendations in the past29 updated and more detailed guidelines and greater harmonization will be needed.
The recent approval of the pan-inhibitor midostaurin (effective against both wild-type and mutant KIT) for the treatment of AdvSM patients has represented a breakthrough and has paved the way for targeted therapy of SM. Additional KIT inhibitors, like avapritinib and ripretinib, are currently being evaluated in clinical trials. As more and more options may become available, comprehensive molecular profiling can assist in individualizing therapy, including treatment intensification of ISM and SSM cases at higher risk of progression. Other novel drug targets (STAT5-PI3K-AKT pathway, mTORC1, MCL-1, BIM, SETD2) have been validated at the preclinical level, but more studies would need to be undertaken before the respective inhibitors can advance to clinical use. Patients with AdvSM failing or not tolerating approved therapies and unfit for transplant might indeed benefit from additional therapeutic options chosen on the basis of their molecular characterization. In this specific, very small setting, repurposing of compounds already tested and approved for other conditions could be more straightforward.
Thus, biological and clinical research move forward in close synergy in SM to offer these rare and long-neglected patients the best quality of life and outcome expectations.
Footnotes
Competing interests: The authors declare no conflict of Interest.
References
- 1.Horny HP, Sotlar K, Valent P. Mastocytosis: state of the art. Pathobiology. 2007;74(2):121–32. doi: 10.1159/000101711. [DOI] [PubMed] [Google Scholar]
- 2.Wiechers T, Rabenhorst A, Schick T, Preussner LM, Förster A, Valent P, Horny HP, Sotlar K, Hartmann K. Large maculopapular cutaneous lesions are associated with favorable outcome in childhood-onset mastocytosis. J Allergy Clin Immunol. 2015;136(6):1581–1590. doi: 10.1016/j.jaci.2015.05.034. [DOI] [PubMed] [Google Scholar]
- 3.Hartmann K, Metcalfe DD. Pediatric mastocytosis. Hematol Oncol Clin North Am. 2000;14(3):625–40. doi: 10.1016/S0889-8588(05)70299-9. [DOI] [PubMed] [Google Scholar]
- 4.Lim KH, Tefferi A, Lasho TL, Finke C, Patnaik M, Butterfield JH, McClure RF, Li CY, Pardanani A. Systemic mastocytosis in 342 consecutive adults: survival studies and prognostic factors. Blood. 2009;113(23):5727–36. doi: 10.1182/blood-2009-02-205237. [DOI] [PubMed] [Google Scholar]
- 5.Valent P, Akin C, Metcalfe DD. Mastocytosis: 2016 updated WHO classification and novel emerging treatment concepts. Blood. 2017;129(11):1420–1427. doi: 10.1182/blood-2016-09-731893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pieri L, Bonadonna P, Elena C, Papayannidis C, Grifoni FI, Rondoni M, Girlanda S, Mauro M, Magliacane D, Elli EM, Iorno ML, Almerigogna F, Scarfì F, Salerno R, Fanelli T, Gesullo F, Corbizi Fattori G, Bonifacio M, Perbellini O, Artuso A, Soverini S, De Benedittis C, Muratori S, Pravettoni V, Cova V, Cortellini G, Ciceri F, Cortelezzi A, Martinelli G, Triggiani M, Merante S, Vannucchi AM, Zanotti R. Clinical presentation and management practice of systemic mastocytosis. A survey on 460 Italian patients. Am J Hematol. 2016;91(7):692–9. doi: 10.1002/ajh.24382. [DOI] [PubMed] [Google Scholar]
- 7.Trizuljak J, Sperr WR, Nekvindová L, Elberink HO, Gleixner KV, Gorska A, Lange M, Hartmann K, Illerhaus A, Bonifacio M, Perkins C, Elena C, Malcovati L, Fortina AB, Shoumariyeh K, Jawhar M, Zanotti R, Bonadonna P, Caroppo F, Zink A, Triggiani M, Parente R, von Bubnoff N, Yavuz AS, Hägglund H, Mattsson M, Panse J, Jäkel N, Kilbertus A, Hermine O, Arock M, Fuchs D, Sabato V, Brockow K, Bretterklieber A, Niedoszytko M, van Anrooij B, Reiter A, Gotlib J, Kluin-Nelemans HC, Mayer J, Doubek M, Valent P. Clinical features and survival of patients with indolent systemic mastocytosis defined by the updated WHO classification. Allergy. 2020;75(8):1927–1938. doi: 10.1111/all.14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardanani A, Tefferi A. Systemic mastocytosis in adults: a review on prognosis and treatment based on 342 Mayo Clinic patients and current literature. Curr Opin Hematol. 2010 Mar;17(2):125–32. doi: 10.1097/MOH.0b013e3283366c59. [DOI] [PubMed] [Google Scholar]
- 9.Tefferi A, Shah S, Reichard KK, Hanson CA, Pardanani A. Smoldering mastocytosis: Survival comparisons with indolent and aggressive mastocytosis. Am J Hematol. 2019;94(1):E1–E2. doi: 10.1002/ajh.25302. [DOI] [PubMed] [Google Scholar]
- 10.Ustun C, Arock M, Kluin-Nelemans HC, Reiter A, Sperr WR, George T, Horny HP, Hartmann K, Sotlar K, Damaj G, Hermine O, Verstovsek S, Metcalfe DD, Gotlib J, Akin C, Valent P. Advanced systemic mastocytosis: from molecular and genetic progress to clinical practice. Haematologica. 2016;101(10):1133–1143. doi: 10.3324/haematol.2016.146563. [DOI] [PubMed] [Google Scholar]
- 11.Gilreath JA, Tchertanov L, Deininger MW. Novel approaches to treating advanced systemic mastocytosis. Clin Pharmacol. 2019;11:77–92. doi: 10.2147/CPAA.S206615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miettinen M, Lasota J. KIT (CD117): a review on expression in normal and neoplastic tissues, and mutations and their clinicopathologic correlation. Appl Immunohistochem Mol Morphol. 2005;13(3):205–20. doi: 10.1097/01.pai.0000173054.83414.22. [DOI] [PubMed] [Google Scholar]
- 13.Valent P, Spanblöchl E, Sperr WR, Sillaber C, Zsebo KM, Agis H, Strobl H, Geissler K, Bettelheim P, Lechner K. Induction of differentiation of human mast cells from bone marrow and peripheral blood mononuclear cells by recombinant human stem cell factor/kit-ligand in long-term culture. Blood. 1992;80(9):2237–45. doi: 10.1182/blood.V80.9.2237.2237. [DOI] [PubMed] [Google Scholar]
- 14.Longley BJ, Reguera MJ, Ma Y. Classes of c-KIT activating mutations: proposed mechanisms of action and implications for disease classification and therapy. Leuk Res. 2001 Jul;25(7):571–6. doi: 10.1016/S0145-2126(01)00028-5. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Montero AC, Jara-Acevedo M, Teodosio C, Sanchez ML, Nunez R, Prados A, Aldanondo I, Sanchez L, Dominguez M, Botana LM, Sanchez-Jimenez F, Sotlar K, Almeida J, Escribano L, Orfao A. KIT mutation in mast cells and other bone marrow hematopoietic cell lineages in systemic mast cell disorders: a prospective study of the Spanish Network on Mastocytosis (REMA) in a series of 113 patients. Blood. 2006;108(7):2366–72. doi: 10.1182/blood-2006-04-015545. [DOI] [PubMed] [Google Scholar]
- 16.Tang X, Boxer M, Drummond A, Ogston P, Hodgins M, Burden AD. A germline mutation in KIT in familial diffuse cutaneous mastocytosis. J Med Genet. 2004;41(6):e88. doi: 10.1136/jmg.2003.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgin-Lavialle S, Lhermitte L, Dubreuil P, Chandesris MO, Hermine O, Damaj G. Mast cell leukemia. Blood. 2013;121(8):1285–95. doi: 10.1182/blood-2012-07-442400. [DOI] [PubMed] [Google Scholar]
- 18.Zhang LY, Smith ML, Schultheis B, Fitzgibbon J, Lister TA, Melo JV, Cross NC, Cavenagh JD. A novel K509I mutation of KIT identified in familial mastocytosis-in vitro and in vivo responsiveness to imatinib therapy. Leuk Res. 2006;30(4):373–8. doi: 10.1016/j.leukres.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Mital A, Piskorz A, Lewandowski K, Wasąg B, Limon J, Hellmann A. A case of mast cell leukaemia with exon 9 KIT mutation and good response to imatinib. Eur J Haematol. 2011;86(6):531–5. doi: 10.1111/j.1600-0609.2011.01598.x. [DOI] [PubMed] [Google Scholar]
- 20.Akin C, Fumo G, Yavuz AS, Lipsky PE, Neckers L, Metcalfe DD. A novel form of mastocytosis associated with a transmembrane c-kit mutation and response to imatinib. Blood. 2004;103(8):3222–5. doi: 10.1182/blood-2003-11-3816. [DOI] [PubMed] [Google Scholar]
- 21.Georgin-Lavialle S, Aguilar C, Guieze R, Lhermitte L, Bruneau J, Fraitag S, Canioni D, Chandesris MO, Suarez F, Grandpeix-Guyodo C, Damaj G, Barete S, Aouba A, Fite C, Robert C, Gaulard P, Lortholary O, Tournilhac O, Dubreuil P, Hermine O. Mast cell sarcoma: a rare and aggressive entity--report of two cases and review of the literature. J Clin Oncol. 2013;31(6):e90–7. doi: 10.1200/JCO.2012.41.9549. [DOI] [PubMed] [Google Scholar]
- 22.Spector MS, Iossifov I, Kritharis A, He C, Kolitz JE, Lowe SW, Allen SL. Mast-cell leukemia exome sequencing reveals a mutation in the IgE mast-cell receptor β chain and KIT V654A. Leukemia. 2012;26(6):1422–5. doi: 10.1038/leu.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bodemer C, Hermine O, Palmérini F, Yang Y, Grandpeix-Guyodo C, Leventhal PS, Hadj-Rabia S, Nasca L, Georgin-Lavialle S, Cohen-Akenine A, Launay JM, Barete S, Feger F, Arock M, Catteau B, Sans B, Stalder JF, Skowron F, Thomas L, Lorette G, Plantin P, Bordigoni P, Lortholary O, de Prost Y, Moussy A, Sobol H, Dubreuil P. Pediatric mastocytosis is a clonal disease associated with D816V and other activating c-KIT mutations. J Invest Dermatol. 2010 Mar;130(3):804–15. doi: 10.1038/jid.2009.281. [DOI] [PubMed] [Google Scholar]
- 24.Abbaspour Babaei M, Kamalidehghan B, Saleem M, Huri HZ, Ahmadipour F. Receptor tyrosine kinase (c-Kit) inhibitors: a potential therapeutic target in cancer cells. Drug Des Devel Ther. 2016;10:2443–59. doi: 10.2147/DDDT.S89114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartmann K, Wardelmann E, Ma Y, Merkelbach-Bruse S, Preussner LM, Woolery C, Baldus SE, Heinicke T, Thiele J, Buettner R, Longley BJ. Novel germline mutation of KIT associated with familial gastrointestinal stromal tumors and mastocytosis. Gastroenterology. 2005;129(3):1042–6. doi: 10.1053/j.gastro.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 26.Gari M, Goodeve A, Wilson G, Winship P, Langabeer S, Linch D, Vandenberghe E, Peake I, Reilly J. c-kit proto-oncogene exon 8 in-frame deletion plus insertion mutations in acute myeloid leukaemia. Br J Haematol. 1999;105(4):894–900. doi: 10.1046/j.1365-2141.1999.01449.x. [DOI] [PubMed] [Google Scholar]
- 27.Goemans BF, Zwaan CM, Miller M, Zimmermann M, Harlow A, Meshinchi S, Loonen AH, Hählen K, Reinhardt D, Creutzig U, Kaspers GJ, Heinrich MC. Mutations in KIT and RAS are frequent events in pediatric core-binding factor acute myeloid leukemia. Leukemia. 2005;19(9):1536–42. doi: 10.1038/sj.leu.2403870. [DOI] [PubMed] [Google Scholar]
- 28.Valent P, Escribano L, Broesby-Olsen S, Hartmann K, Grattan C, Brockow K, Niedoszytko M, Nedoszytko B, Oude Elberink JN, Kristensen T, Butterfield JH, Triggiani M, Alvarez-Twose I, Reiter A, Sperr WR, Sotlar K, Yavuz S, Kluin-Nelemans HC, Hermine O, Radia D, van Doormaal JJ, Gotlib J, Orfao A, Siebenhaar F, Schwartz LB, Castells M, Maurer M, Horny HP, Akin C, Metcalfe DD, Arock M European Competence Network on Mastocytosis. Proposed diagnostic algorithm for patients with suspected mastocytosis: a proposal of the European Competence Network on Mastocytosis. Allergy. 2014;69(10):1267–74. doi: 10.1111/all.12436. [DOI] [PubMed] [Google Scholar]
- 29.Arock M, Sotlar K, Akin C, Broesby-Olsen S, Hoermann G, Escribano L, Kristensen TK, Kluin-Nelemans HC, Hermine O, Dubreuil P, Sperr WR, Hartmann K, Gotlib J, Cross NC, Haferlach T, Garcia-Montero A, Orfao A, Schwaab J, Triggiani M, Horny HP, Metcalfe DD, Reiter A, Valent P. KIT mutation analysis in mast cell neoplasms: recommendations of the European Competence Network on Mastocytosis. Leukemia. 2015;29(6):1223–32. doi: 10.1038/leu.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sotlar K. c-kit mutational analysis in paraffin material. Methods Mol Biol. 2013;999:59–78. doi: 10.1007/978-1-62703-357-2_4. [DOI] [PubMed] [Google Scholar]
- 31.Fritsche-Polanz R, Jordan JH, Feix A, Sperr WR, Sunder-Plassmann G, Valent P, Födinger M. Mutation analysis of C-KIT in patients with myelodysplastic syndromes without mastocytosis and cases of systemic mastocytosis. Br J Haematol. 2001;113(2):357–64. doi: 10.1046/j.1365-2141.2001.02783.x. [DOI] [PubMed] [Google Scholar]
- 32.Erben P, Schwaab J, Metzgeroth G, Horny HP, Jawhar M, Sotlar K, Fabarius A, Teichmann M, Schneider S, Ernst T, Müller MC, Giehl M, Marx A, Hartmann K, Hochhaus A, Hofmann WK, Cross NC, Reiter A. The KIT D816V expressed allele burden for diagnosis and disease monitoring of systemic mastocytosis. Ann Hematol. 2014;93(1):81–8. doi: 10.1007/s00277-013-1964-1. [DOI] [PubMed] [Google Scholar]
- 33.Sotlar K, Escribano L, Landt O, Möhrle S, Herrero S, Torrelo A, Lass U, Horny HP, Bültmann B. One-step detection of c-kit point mutations using peptide nucleic acid-mediated polymerase chain reaction clamping and hybridization probes. Am J Pathol. 2003;162(3):737–46. doi: 10.1016/S0002-9440(10)63870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schumacher JA, Elenitoba-Johnson KS, Lim MS. Detection of the c-kit D816V mutation in systemic mastocytosis by allele-specific PCR. J Clin Pathol. 2008;61(1):109–14. doi: 10.1136/jcp.2007.047928. [DOI] [PubMed] [Google Scholar]
- 35.Kristensen T, Broesby-Olsen S, Vestergaard H, Bindslev-Jensen C, Møller MB Mastocytosis Centre Odense University Hospital (MastOUH) Circulating KIT D816V mutation-positive non-mast cells in peripheral blood are characteristic of indolent systemic mastocytosis. Eur J Haematol. 2012;89(1):42–6. doi: 10.1111/j.1600-0609.2012.01789.x. [DOI] [PubMed] [Google Scholar]
- 36.Greiner G, Gurbisz M, Ratzinger F, Witzeneder N, Simonitsch-Klupp I, Mitterbauer-Hohendanner G, Mayerhofer M, Müllauer L, Sperr WR, Valent P, Hoermann G. Digital PCR: A Sensitive and Precise Method for KIT D816V Quantification in Mastocytosis. Clin Chem. 2018;64(3):547–555. doi: 10.1373/clinchem.2017.277897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greiner G, Gurbisz M, Ratzinger F, Witzeneder N, Class SV, Eisenwort G, Simonitsch-Klupp I, Esterbauer H, Mayerhofer M, Müllauer L, Sperr WR, Valent P, Hoermann G. Molecular quantification of tissue disease burden is a new biomarker and independent predictor of survival in mastocytosis. Haematologica. 2020;105(2):366–374. doi: 10.3324/haematol.2019.217950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jara-Acevedo M, Teodosio C, Sanchez-Muñoz L, Álvarez-Twose I, Mayado A, Caldas C, Matito A, Morgado JM, Muñoz-González JI, Escribano L, Garcia-Montero AC, Orfao A. Detection of the KIT D816V mutation in peripheral blood of systemic mastocytosis: diagnostic implications. Mod Pathol. 2015;28(8):1138–49. doi: 10.1038/modpathol.2015.72. [DOI] [PubMed] [Google Scholar]
- 39.Broesby-Olsen S, Oropeza AR, Bindslev-Jensen C, Vestergaard H, Møller MB, Siebenhaar F, Kristensen T, Mortz CG Mastocytosis Centre Odense University Hospital (MastOUH); Odense Research Centre for Anaphylaxis. Recognizing mastocytosis in patients with anaphylaxis: value of KIT D816V mutation analysis of peripheral blood. J Allergy Clin Immunol. 2015;135(1):262–4. doi: 10.1016/j.jaci.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 40.Hoermann G, Gleixner KV, Dinu GE, Kundi M, Greiner G, Wimazal F, Hadzijusufovic E, Mitterbauer G, Mannhalter C, Valent P, Sperr WR. The KIT D816V allele burden predicts survival in patients with mastocytosis and correlates with the WHO type of the disease. Allergy. 2014;69(6):810–3. doi: 10.1111/all.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Escribano L, Alvarez-Twose I, Sánchez-Muñoz L, Garcia-Montero A, Núñez R, Almeida J, Jara-Acevedo M, Teodósio C, García-Cosío M, Bellas C, Orfao A. Prognosis in adult indolent systemic mastocytosis: a long-term study of the Spanish Network on Mastocytosis in a series of 145 patients. J Allergy Clin Immunol. 2009;124(3):514–21. doi: 10.1016/j.jaci.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Akin C, Kirshenbaum AS, Semere T, Worobec AS, Scott LM, Metcalfe DD. Analysis of the surface expression of c-kit and occurrence of the c-kit Asp816Val activating mutation in T cells, B cells, and myelomonocytic cells in patients with mastocytosis. Exp Hematol. 2000;28(2):140–7. doi: 10.1016/S0301-472X(99)00145-9. [DOI] [PubMed] [Google Scholar]
- 43.Yavuz AS, Lipsky PE, Yavuz S, Metcalfe DD, Akin C. Evidence for the involvement of a hematopoietic progenitor cell in systemic mastocytosis from single-cell analysis of mutations in the c-kit gene. Blood. 2002 Jul 15;100(2):661–5. doi: 10.1182/blood-2002-01-0203. [DOI] [PubMed] [Google Scholar]
- 44.Kocabas CN, Yavuz AS, Lipsky PE, Metcalfe DD, Akin C. Analysis of the lineage relationship between mast cells and basophils using the c-kit D816V mutation as a biologic signature. J Allergy Clin Immunol. 2005;115(6):1155–61. doi: 10.1016/j.jaci.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 45.Jawhar M, Schwaab J, Naumann N, Horny HP, Sotlar K, Haferlach T, Metzgeroth G, Fabarius A, Valent P, Hofmann WK, Cross NCP, Meggendorfer M, Reiter A. Response and progression on midostaurin in advanced systemic mastocytosis: KIT D816V and other molecular markers. Blood. 2017;130(2):137–145. doi: 10.1182/blood-2017-01-764423. [DOI] [PubMed] [Google Scholar]
- 46.Schwaab J, Schnittger S, Sotlar K, Walz C, Fabarius A, Pfirrmann M, Kohlmann A, Grossmann V, Meggendorfer M, Horny HP, Valent P, Jawhar M, Teichmann M, Metzgeroth G, Erben P, Ernst T, Hochhaus A, Haferlach T, Hofmann WK, Cross NC, Reiter A. Comprehensive mutational profiling in advanced systemic mastocytosis. Blood. 2013;122(14):2460–6. doi: 10.1182/blood-2013-04-496448. [DOI] [PubMed] [Google Scholar]
- 47.Tefferi A, Levine RL, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, Finke CM, Mullally A, Li CY, Pardanani A, Gilliland DG. Frequent TET2 mutations in systemic mastocytosis: clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia. 2009;23(5):900–4. doi: 10.1038/leu.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Traina F, Visconte V, Jankowska AM, Makishima H, O’Keefe CL, Elson P, Han Y, Hsieh FH, Sekeres MA, Mali RS, Kalaycio M, Lichtin AE, Advani AS, Duong HK, Copelan E, Kapur R, Olalla Saad ST, Maciejewski JP, Tiu RV. Single nucleotide polymorphism array lesions, TET2, DNMT3A, ASXL1 and CBL mutations are present in systemic mastocytosis. PLoS One. 2012;7(8):e43090. doi: 10.1371/journal.pone.0043090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jawhar M, Schwaab J, Schnittger S, Sotlar K, Horny HP, Metzgeroth G, Müller N, Schneider S, Naumann N, Walz C, Haferlach T, Valent P, Hofmann WK, Cross NC, Fabarius A, Reiter A. Molecular profiling of myeloid progenitor cells in multi-mutated advanced systemic mastocytosis identifies KIT D816V as a distinct and late event. Leukemia. 2015;29(5):1115–22. doi: 10.1038/leu.2015.4. [DOI] [PubMed] [Google Scholar]
- 50.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T, Yoshida K, Roller A, Nadarajah N, Shiraishi Y, Shiozawa Y, Chiba K, Tanaka H, Koeffler HP, Klein HU, Dugas M, Aburatani H, Kohlmann A, Miyano S, Haferlach C, Kern W, Ogawa S. Landscape of genetic lesi ons in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–7. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B, Dicker F, Schnittger S, Dugas M, Kern W, Haferlach C, Haferlach T. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010;28(24):3858–65. doi: 10.1200/JCO.2009.27.1361. [DOI] [PubMed] [Google Scholar]
- 52.Jalili M, Yaghmaie M, Ahmadvand M, Alimoghaddam K, Mousavi SA, Vaezi M, Ghavamzadeh A. Prognostic Value of RUNX1 Mutations in AML: A Meta-Analysis. Asian Pac J Cancer Prev. 2018;19(2):325–329. doi: 10.22034/APJCP.2018.19.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Vita S, Schneider RK, Garcia M, Wood J, Gavillet M, Ebert BL, Gerbaulet A, Roers A, Levine RL, Mullally A, Williams DA. Loss of function of TET2 cooperates with constitutively active KIT in murine and human models of mastocytosis. PLoS One. 2014;9(5):e96209. doi: 10.1371/journal.pone.0096209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jawhar M, Schwaab J, Schnittger S, Meggendorfer M, Pfirrmann M, Sotlar K, Horny HP, Metzgeroth G, Kluger S, Naumann N, Haferlach C, Haferlach T, Valent P, Hofmann WK, Fabarius A, Cross NC, Reiter A. Additional mutations in SRSF2, ASXL1 and/or RUNX1 identify a high-risk group of patients with KIT D816V(+) advanced systemic mastocytosis. Leukemia. 2016;30(1):136–43. doi: 10.1038/leu.2015.284. [DOI] [PubMed] [Google Scholar]
- 55.Muñoz-González JI, Jara-Acevedo M, Alvarez-Twose I, Merker JD, Teodosio C, Hou Y, Henriques A, Roskin KM, Sanchez-Muñoz L, Tsai AG, Caldas C, Matito A, Sánchez-Gallego JI, Mayado A, Dasilva-Freire N, Gotlib JR, Escribano L, Orfao A, García-Montero AC. Impact of somatic and germline mutations on the outcome of systemic mastocytosis. Blood Adv. 2018;2(21):2814–2828. doi: 10.1182/bloodadvances.2018020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pardanani AD, Lasho TL, Finke C, Zblewski DL, Abdelrahman RA, Wassie EA, Gangat N, Hanson CA, Ketterling RP, Tefferi A. ASXL1 and CBL mutations are independently predictive of inferior survival in advanced systemic mastocytosis. Br J Haematol. 2016;175(3):534–536. doi: 10.1111/bjh.13865. [DOI] [PubMed] [Google Scholar]
- 57.Pardanani A, Lasho T, Elala Y, Wassie E, Finke C, Reichard KK, Chen D, Hanson CA, Ketterling RP, Tefferi A. Next-generation sequencing in systemic mastocytosis: Derivation of a mutation-augmented clinical prognostic model for survival. Am J Hematol. 2016;91(9):888–93. doi: 10.1002/ajh.24426. [DOI] [PubMed] [Google Scholar]
- 58.Jawhar M, Schwaab J, Álvarez-Twose I, Shoumariyeh K, Naumann N, Lübke J, Perkins C, Muñoz-González JI, Meggendorfer M, Kennedy V, Metzgeroth G, Fabarius A, Pfeifer D, Sotlar K, Horny HP, von Bubnoff N, Haferlach T, Cross NCP, Hofmann WK, Sperr WR, García-Montero AC, Valent P, Gotlib J, Orfao A, Reiter A. MARS: Mutation-Adjusted Risk Score for Advanced Systemic Mastocytosis. J Clin Oncol. 2019;37(31):2846–2856. doi: 10.1200/JCO.19.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muñoz-González JI, Álvarez-Twose I, Jara-Acevedo M, Henriques A, Viñas E, Prieto C, Sánchez-Muñoz L, Caldas C, Mayado A, Matito A, Dasilva-Freire N, Orfao A, García-Montero AC. Frequency and prognostic impact of KIT and other genetic variants in indolent systemic mastocytosis. Blood. 2019;134(5):456–468. doi: 10.1182/blood.2018886507. [DOI] [PubMed] [Google Scholar]
- 60.Pardanani A, Shah S, Mannelli F, Elala YC, Guglielmelli P, Lasho TL, Patnaik MM, Gangat N, Ketterling RP, Reichard KK, Hanson CA, Vannucchi AM, Tefferi A. Mayo alliance prognostic system for mastocytosis: clinical and hybrid clinical-molecular models. Blood Adv. 2018;2(21):2964–2972. doi: 10.1182/bloodadvances.2018026245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sperr WR, Kundi M, Alvarez-Twose I, van Anrooij B, Oude Elberink JNG, Gorska A, Niedoszytko M, Gleixner KV, Hadzijusufovic E, Zanotti R, Bonadonna P, Bonifacio M, Perkins C, Illerhaus A, Elena C, Merante S, Shoumariyeh K, von Bubnoff N, Parente R, Jawhar M, Belloni Fortina A, Caroppo F, Brockow K, Zink A, Fuchs D, Kilbertus AJ, Yavuz AS, Doubek M, Hägglund H, Panse J, Sabato V, Bretterklieber A, Niederwieser D, Breynaert C, Hartmann K, Triggiani M, Nedoszytko B, Reiter A, Orfao A, Hermine O, Gotlib J, Arock M, Kluin-Nelemans HC, Valent P. International prognostic scoring system for mastocytosis (IPSM): a retrospective cohort study. Lancet Haematol. 2019;6(12):e638–e649. doi: 10.1016/S2352-3026(19)30166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scherber RM, Borate U. How we diagnose and treat systemic mastocytosis in adults. Br J Haematol. 2018;180(1):11–23. doi: 10.1111/bjh.14967. [DOI] [PubMed] [Google Scholar]
- 63.Pardanani A. Systemic mastocytosis in adults: 2019 update on diagnosis, risk stratification and management. Am J Hematol. 2019;94(3):363–377. doi: 10.1002/ajh.25371. [DOI] [PubMed] [Google Scholar]
- 64.Butterfield JH. Response of severe systemic mastocytosis to interferon alpha. Br J Dermatol. 1998;138(3):489–95. doi: 10.1046/j.1365-2133.1998.02131.x. [DOI] [PubMed] [Google Scholar]
- 65.Butterfield JH, Tefferi A, Kozuh GF. Successful treatment of systemic mastocytosis with high-dose interferon-alfa: long-term follow-up of a case. Leuk Res. 2005;29(2):131–4. doi: 10.1016/j.leukres.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 66.Hauswirth AW, Simonitsch-Klupp I, Uffmann M, Koller E, Sperr WR, Lechner K, Valent P. Response to therapy with interferon alpha-2b and prednisolone in aggressive systemic mastocytosis: report of five cases and review of the literature. Leuk Res. 2004;28(3):249–57. doi: 10.1016/S0145-2126(03)00259-5. [DOI] [PubMed] [Google Scholar]
- 67.Pardanani A, Hoffbrand AV, Butterfield JH, Tefferi A. Treatment of systemic mast cell disease with 2-chlorodeoxyadenosine. Leuk Res. 2004;28(2):127–31. doi: 10.1016/S0145-2126(03)00185-1. [DOI] [PubMed] [Google Scholar]
- 68.Böhm A, Sonneck K, Gleixner KV, Schuch K, Pickl WF, Blatt K, Peter B, Herrmann H, Schernthaner GH, Pehamberger H, Rabitsch W, Sperr WR, Valent P. In vitro and in vivo growth-inhibitory effects of cladribine on neoplastic mast cells exhibiting the imatinib-resistant KIT mutation D816V. Exp Hematol. 2010;38(9):744–55. doi: 10.1016/j.exphem.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 69.Kluin-Nelemans HC, Oldhoff JM, Van Doormaal JJ, Van ‘t Wout JW, Verhoef G, Gerrits WB, van Dobbenburgh OA, Pasmans SG, Fijnheer R. Cladribine therapy for systemic mastocytosis. Blood. 2003;102(13):4270–6. doi: 10.1182/blood-2003-05-1699. [DOI] [PubMed] [Google Scholar]
- 70.Gilreath JA, Tchertanov L, Deininger MW. Novel approaches to treating advanced systemic mastocytosis. Clin Pharmacol. 2019;11:77–92. doi: 10.2147/CPAA.S206615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akin C, Fumo G, Yavuz AS, Lipsky PE, Neckers L, Metcalfe DD. A novel form of mastocytosis associated with a transmembrane c-kit mutation and response to imatinib. Blood. 2004;103(8):3222–5. doi: 10.1182/blood-2003-11-3816. [DOI] [PubMed] [Google Scholar]
- 72.Zermati Y, De Sepulveda P, Féger F, Létard S, Kersual J, Castéran N, Gorochov G, Dy M, Ribadeau Dumas A, Dorgham K, Parizot C, Bieche Y, Vidaud M, Lortholary O, Arock M, Hermine O, Dubreuil P. Effect of tyrosine kinase inhibitor STI571 on the kinase activity of wild-type and various mutated c-kit receptors found in mast cell neoplasms. Oncogene. 2003;22(5):660–4. doi: 10.1038/sj.onc.1206120. [DOI] [PubMed] [Google Scholar]
- 73.Ma Y, Zeng S, Metcalfe DD, Akin C, Dimitrijevic S, Butterfield JH, McMahon G, Longley BJ. The c-KIT mutation causing human mastocytosis is resistant to STI571 and other KIT kinase inhibitors; kinases with enzymatic site mutations show different inhibitor sensitivity profiles than wild-type kinases and those with regulatory-type mutations. Blood. 2002;99(5):1741–4. doi: 10.1182/blood.V99.5.1741. [DOI] [PubMed] [Google Scholar]
- 74.Hochhaus A, Baccarani M, Giles FJ, le Coutre PD, Müller MC, Reiter A, Santanastasio H, Leung M, Novick S, Kantarjian HM. Nilotinib in patients with systemic mastocytosis: analysis of the phase 2, open-label, single-arm nilotinib registration study. J Cancer Res Clin Oncol. 2015;141(11):2047–60. doi: 10.1007/s00432-015-1988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dubreuil P, Letard S, Ciufolini M, Gros L, Humbert M, Castéran N, Borge L, Hajem B, Lermet A, Sippl W, Voisset E, Arock M, Auclair C, Leventhal PS, Mansfield CD, Moussy A, Hermine O. Masitinib (AB1010), a potent and selective tyrosine kinase inhibitor targeting KIT. PLoS One. 2009;4(9):e7258. doi: 10.1371/journal.pone.0007258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lortholary O, Chandesris MO, Bulai Livideanu C, Paul C, Guillet G, Jassem E, Niedoszytko M, Barete S, Verstovsek S, Grattan C, Damaj G, Canioni D, Fraitag S, Lhermitte L, Georgin Lavialle S, Frenzel L, Afrin LB, Hanssens K, Agopian J, Gaillard R, Kinet JP, Auclair C, Mansfield C, Moussy A, Dubreuil P, Hermine O. Masitinib for treatment of severely symptomatic indolent systemic mastocytosis: a randomised, placebo-controlled, phase 3 study. Lancet. 2017;389(10069):612–620. doi: 10.1016/S0140-6736(16)31403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schittenhelm MM, Shiraga S, Schroeder A, Corbin AS, Griffith D, Lee FY, Bokemeyer C, Deininger MW, Druker BJ, Heinrich MC. Dasatinib (BMS-354825), a dual SRC/ABL kinase inhibitor, inhibits the kinase activity of wild-type, juxtamembrane, and activation loop mutant KIT isoforms associated with human malignancies. Cancer Res. 2006;66(1):473–81. doi: 10.1158/0008-5472.CAN-05-2050. [DOI] [PubMed] [Google Scholar]
- 78.Bougherara H, Georgin-Lavialle S, Damaj G, Launay JM, Lhermitte L, Auclair C, Arock M, Dubreuil P, Hermine O, Poul MA. Relocalization of KIT D816V to cell surface after dasatinib treatment: potential clinical implications. Clin Lymphoma Myeloma Leuk. 2013;13(1):62–9. doi: 10.1016/j.clml.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 79.Verstovsek S, Tefferi A, Cortes J, O’Brien S, Garcia-Manero G, Pardanani A, Akin C, Faderl S, Manshouri T, Thomas D, Kantarjian H. Phase II study of dasatinib in Philadelphia chromosome-negative acute and chronic myeloid diseases, including systemic mastocytosis. Clin Cancer Res. 2008;14(12):3906–15. doi: 10.1158/1078-0432.CCR-08-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gotlib J, Berubé C, Growney JD, Chen CC, George TI, Williams C, Kajiguchi T, Ruan J, Lilleberg SL, Durocher JA, Lichy JH, Wang Y, Cohen PS, Arber DA, Heinrich MC, Neckers L, Galli SJ, Gilliland DG, Coutré SE. Activity of the tyrosine kinase inhibitor PKC412 in a patient with mast cell leukemia with the D816V KIT mutation. Blood. 2005;106(8):2865–70. doi: 10.1182/blood-2005-04-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DeAngelo DJ, George TI, Linder A, Langford C, Perkins C, Ma J, Westervelt P, Merker JD, Berube C, Coutre S, Liedtke M, Medeiros B, Sternberg D, Dutreix C, Ruffie PA, Corless C, Graubert TJ, Gotlib J. Efficacy and safety of midostaurin in patients with advanced systemic mastocytosis: 10-year median follow-up of a phase II trial. Leukemia. 2018;32(2):470–478. doi: 10.1038/leu.2017.234. [DOI] [PubMed] [Google Scholar]
- 82.Gotlib J, Kluin-Nelemans HC, George TI, Akin C, Sotlar K, Hermine O, Awan FT, Hexner E, Mauro MJ, Sternberg DW, Villeneuve M, Huntsman Labed A, Stanek EJ, Hartmann K, Horny HP, Valent P, Reiter A. Efficacy and Safety of Midostaurin in Advanced Systemic Mastocytosis. N Engl J Med. 2016;374(26):2530–41. doi: 10.1056/NEJMoa1513098. [DOI] [PubMed] [Google Scholar]
- 83.Chandesris MO, Damaj G, Canioni D, Brouzes C, Lhermitte L, Hanssens K, Frenzel L, Cherquaoui Z, Durieu I, Durupt S, Gyan E, Beyne-Rauzy O, Launay D, Faure C, Hamidou M, Besnard S, Diouf M, Schiffmann A, Niault M, Jeandel PY, Ranta D, Gressin R, Chantepie S, Barete S, Dubreuil P, Bourget P, Lortholary O, Hermine O CEREMAST Study Group. Midostaurin in Advanced Systemic Mastocytosis. N Engl J Med. 2016;374(26):2605–7. doi: 10.1056/NEJMc1515403. [DOI] [PubMed] [Google Scholar]
- 84.Radia D, Deininger MW, Gotlib J, Bose P, Drummond MW, Hexner E, Robinson W, Quiery A, Winton E, George T, Horny HP, Oren R, Shi H, Schmidt-Kittler O, Mar B, DeAngelo D. Avapritinib, a potent and selective inhibitor of Kit D816V, induces complete and durable responses in patients (pts) with advanced systemic mastocytosis (AdvSM) (abstract no. S830 and oral presentation) HemaSphere. 2019. [DOI]
- 85.Maurer M, Elberink HO, Gotlib J, Sabato V, Hartmann K, Broesby-Olsen S, Castells M, Deininger M, Heaney M, George T, Siebenhaar F, Radia D, Triggiani M, Daele P, DeAngelo D, Schmidt-Kittler O, Lin H, Morrison A, Mar B, Maurer M. Results from PIONEER: a randomized, double-blind, placebo-controlled, phase 2 study of avapritinib in patients with indolent systemic mastocytosis (ISM) Oncol Res Treat. 2020;43:77–77. [Google Scholar]
- 86.Smith BD, Kaufman MD, Lu WP, Gupta A, Leary CB, Wise SC, Rutkoski TJ, Ahn YM, Al-Ani G, Bulfer SL, Caldwell TM, Chun L, Ensinger CL, Hood MM, McKinley A, Patt WC, Ruiz-Soto R, Su Y, Telikepalli H, Town A, Turner BA, Vogeti L, Vogeti S, Yates K, Janku F, Abdul Razak AR, Rosen O, Heinrich MC, Flynn DL. Ripretinib (DCC-2618) Is a Switch Control Kinase Inhibitor of a Broad Spectrum of Oncogenic and Drug-Resistant KIT and PDGFRA Variants. Cancer Cell. 2019;35(5):738–751.e9. doi: 10.1016/j.ccell.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 87.Baumgartner C, Cerny-Reiterer S, Sonneck K, Mayerhofer M, Gleixner KV, Fritz R, Kerenyi M, Boudot C, Gouilleux F, Kornfeld JW, Sillaber C, Moriggl R, Valent P. Expression of activated STAT5 in neoplastic mast cells in systemic mastocytosis: subcellular distribution and role of the transforming oncoprotein KIT D816V. Am J Pathol. 2009;175(6):2416–29. doi: 10.2353/ajpath.2009.080953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harir N, Boudot C, Friedbichler K, Sonneck K, Kondo R, Martin-Lannerée S, Kenner L, Kerenyi M, Yahiaoui S, Gouilleux-Gruart V, Gondry J, Bénit L, Dusanter-Fourt I, Lassoued K, Valent P, Moriggl R, Gouilleux F. Oncogenic Kit controls neoplastic mast cell growth through a Stat5/PI3-kinase signaling cascade. Blood. 2008;112(6):2463–73. doi: 10.1182/blood-2007-09-115477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sundström M, Vliagoftis H, Karlberg P, Butterfield JH, Nilsson K, Metcalfe DD, Nilsson G. Functional and phenotypic studies of two variants of a human mast cell line with a distinct set of mutations in the c-kit proto-oncogene. Immunology. 2003;108(1):89–97. doi: 10.1046/j.1365-2567.2003.01559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lasho T, Tefferi A, Pardanani A. Inhibition of JAK-STAT signaling by TG101348: a novel mechanism for inhibition of KITD816V-dependent growth in mast cell leukemia cells. Leukemia. 2010;24(7):1378–80. doi: 10.1038/leu.2010.109. [DOI] [PubMed] [Google Scholar]
- 91.Kim MS, Kuehn HS, Metcalfe DD, Gilfillan AM. Activation and function of the mTORC1 pathway in mast cells. J Immunol. 2008;180(7):4586–95. doi: 10.4049/jimmunol.180.7.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smrz D, Kim MS, Zhang S, Mock BA, Smrzová S, DuBois W, Simakova O, Maric I, Wilson TM, Metcalfe DD, Gilfillan AM. mTORC1 and mTORC2 differentially regulate homeostasis of neoplastic and non-neoplastic human mast cells. Blood. 2011;118(26):6803–13. doi: 10.1182/blood-2011-06-359984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blatt K, Herrmann H, Mirkina I, Hadzijusufovic E, Peter B, Strommer S, Hoermann G, Mayerhofer M, Hoetzenecker K, Klepetko W, Ghanim V, Marth K, Füreder T, Wacheck V, Valenta R, Valent P. The PI3-kinase/mTOR-targeting drug NVP-BEZ235 inhibits growth and IgE-dependent activation of human mast cells and basophils. PLoS One. 2012;7(1):e29925. doi: 10.1371/journal.pone.0029925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aichberger KJ, Mayerhofer M, Gleixner KV, Krauth MT, Gruze A, Pickl WF, Wacheck V, Selzer E, Müllauer L, Agis H, Sillaber C, Valent P. Identification of MCL1 as a novel target in neoplastic mast cells in systemic mastocytosis: inhibition of mast cell survival by MCL1 antisense oligonucleotides and synergism with PKC412. Blood. 2007;109(7):3031–41. doi: 10.1182/blood-2006-07-032714. [DOI] [PubMed] [Google Scholar]
- 95.Aichberger KJ, Gleixner KV, Mirkina I, Cerny-Reiterer S, Peter B, Ferenc V, Kneidinger M, Baumgartner C, Mayerhofer M, Gruze A, Pickl WF, Sillaber C, Valent P. Identification of proapoptotic Bim as a tumor suppressor in neoplastic mast cells: role of KIT D816V and effects of various targeted drugs. Blood. 2009;114(26):5342–51. doi: 10.1182/blood-2008-08-175190. [DOI] [PubMed] [Google Scholar]
- 96.Shinjyo T, Kuribara R, Inukai T, Hosoi H, Kinoshita T, Miyajima A, Houghton PJ, Look AT, Ozawa K, Inaba T. Downregulation of Bim, a proapoptotic relative of Bcl-2, is a pivotal step in cytokine-initiated survival signaling in murine hematopoietic progenitors. Mol Cell Biol. 2001;21(3):854–64. doi: 10.1128/MCB.21.3.854-864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luciano F, Jacquel A, Colosetti P, Herrant M, Cagnol S, Pages G, Auberger P. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene. 2003;22(43):6785–93. doi: 10.1038/sj.onc.1206792. [DOI] [PubMed] [Google Scholar]
- 98.Kuribara R, Honda H, Matsui H, Shinjyo T, Inukai T, Sugita K, Nakazawa S, Hirai H, Ozawa K, Inaba T. Roles of Bim in apoptosis of normal and Bcr-Abl-expressing hematopoietic progenitors. Mol Cell Biol. 2004;24(14):6172–83. doi: 10.1128/MCB.24.14.6172-6183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aichberger KJ, Mayerhofer M, Krauth MT, Vales A, Kondo R, Derdak S, Pickl WF, Selzer E, Deininger M, Druker BJ, Sillaber C, Esterbauer H, Valent P. Low-level expression of proapoptotic Bcl-2-interacting mediator in leukemic cells in patients with chronic myeloid leukemia: role of BCR/ABL, characterization of underlying signaling pathways, and re-expression by novel pharmacologic compounds. Cancer Res. 2005;65(20):9436–44. doi: 10.1158/0008-5472.CAN-05-0972. [DOI] [PubMed] [Google Scholar]
- 100.Martinelli G, Mancini M, De Benedittis C, Rondoni M, Papayannidis C, Manfrini M, Meggendorfer M, Calogero R, Guadagnuolo V, Fontana MC, Bavaro L, Padella A, Zago E, Pagano L, Zanotti R, Scaffidi L, Specchia G, Albano F, Merante S, Elena C, Savini P, Gangemi D, Tosi P, Ciceri F, Poletti G, Riccioni L, Morigi F, Delledonne M, Haferlach T, Cavo M, Valent P, Soverini S. SETD2 and histone H3 lysine 36 methylation deficiency in advanced systemic mastocytosis. Leukemia. 2018;32(1):139–148. doi: 10.1038/leu.2017.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li J, Duns G, Westers H, Sijmons R, van den Berg A, Kok K. SETD2: an epigenetic modifier with tumor suppressor functionality. Oncotarget. 2016;7(31):50719–50734. doi: 10.18632/oncotarget.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27(2):406–20. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park IY, Powell RT, Tripathi DN, Dere R, Ho TH, Blasius TL, Chiang YC, Davis IJ, Fahey CC, Hacker KE, Verhey KJ, Bedford MT, Jonasch E, Rathmell WK, Walker CL. Dual Chromatin and Cytoskeletal Remodeling by SETD2. Cell. 2016;166(4):950–962. doi: 10.1016/j.cell.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]