PURPOSE
Using a candidate gene approach, we tested the hypothesis that individual single nucleotide polymorphisms (SNPs) and gene-level variants are associated with cognitive impairment in patients with hematologic malignancies treated with blood or marrow transplantation (BMT) and that inclusion of these SNPs improves risk prediction beyond that offered by clinical and demographic characteristics.
PATIENTS AND METHODS
In the discovery cohort, BMT recipients underwent a standardized battery of neuropsychological tests pre-BMT and at 6 months, 1 year, 2 years, and 3 years post-BMT. Associations between 68 candidate genes and cognitive impairment were assessed using generalized estimating equation models. Elastic-Net regression was used to build Base (sociodemographic), Clinical, and Combined (Base plus Clinical plus genetic) risk prediction models of post-BMT impairment. An independent nonoverlapping cohort from the BMT Survivor Study with self-report of learning/memory problems (as identified by their health care provider) was used for model replication.
RESULTS
The discovery cohort included 277 participants (58.5% males; 68.6% non-Hispanic whites; and 46.6% allogeneic BMT recipients). Adjusting for BMT type, age at BMT, sex, race/ethnicity, and cognitive reserve, SNPs in the blood-brain barrier, telomere homeostasis, and DNA repair genes were significantly associated with cognitive impairment. Compared with the Clinical Model, the Combined Model had higher predictive power in both the discovery cohort (mean area under the receiver operating characteristic curve [AUC], 0.89; 95% CI, 0.85 to 0.93 v 0.77; 95% CI, 0.71 to 0.83; P = 1.24 × 10−9) and the replication cohort (AUC, 0.71; 95% CI, 0.66 to 0.76 v 0.63; 95% CI, 0.57 to 0.68; P = .004).
CONCLUSION
Inclusion of candidate genetic variants enhanced the prediction of risk of post-BMT cognitive impairment beyond that offered by demographic/clinical characteristics and represents a step toward a personalized approach to managing patients at high risk for cognitive impairment after BMT.
INTRODUCTION
Blood or marrow transplantation (BMT), a potentially curative option for hematologic malignancies,1,2 is unfortunately accompanied by unintended adverse consequences.3,4 We5 and others6-9 have demonstrated that cognitive impairment is prevalent and persists in patients with hematologic malignancies treated with BMT, albeit with significant interindividual variability in risk, suggesting a role for genetic susceptibility.10,11 Although the impact of genetic factors on cognitive ability is well recognized in nononcology populations,12,13 the limited information on patients with cancer is essentially restricted to breast cancer survivors, implicating APOE, COMT, DNA repair, and oxidative stress genes.14-17 Genetic susceptibility to cognitive impairment in patients with hematologic malignancy treated with BMT and use of this information to identify those at highest risk remain unstudied and were addressed in this study.
Single nucleotide polymorphisms (SNPs) are the most common type of genetic variation at the population level that may help explain interindividual variability in susceptibility to disease. We used a candidate gene approach to curate a list of biologically plausible SNPs.18-21 A common practice is to examine the association between the outcome of interest and SNPs individually, one at a time. Joint modeling of interaction effects of multiple SNPs within a gene, however, may have more power to detect genetic variants, as well as to potentially improve risk prediction.22 Accordingly, we aimed to identify individual SNP and gene-level associations with risk of post-BMT cognitive impairment. We then evaluated the ability of identified SNPs to enhance prediction of cognitive impairment in BMT survivors beyond demographic and clinical characteristics alone.23-28 We used a prospective, longitudinally followed cohort of BMT survivors for discovery to (1) identify candidate SNP associations with cognitive impairment and (2) build 3 risk prediction models using demographic, clinical, and genetic predictors. A nonoverlapping cohort of BMT survivors3,29 was used as an independent replication cohort to validate the performance of the models.
PATIENTS AND METHODS
Study Cohort, Risk Predictors, and Outcome
Discovery cohort.
The discovery cohort was a prospective, longitudinally followed cohort of patients undergoing autologous or allogeneic BMT for hematologic malignancies at City of Hope (COH) between 2005 and 2011.5 Eligible participants were 18 years of age or older at the time of BMT and fluent in English; for this study, we restricted the cohort to those with availability of pre-BMT DNA. Patients with a history of preexisting neurologic or major psychiatric disorders, significant auditory/visual/motor impairments, and/or neuropsychological intervention within the preceding 6 months were excluded. Comprehensive assessment of cognitive function was performed using a 2-hour battery of 14 standardized neurocognitive tests in 8 cognitive domains at 5 time points: pre-BMT and 6 months, 1 year, 2 years, and 3 years post-BMT. Demographic variables (age, sex, race/ethnicity, education, and income) were self-reported, and clinical variables (primary cancer diagnosis, conditioning regimens and intensity, type of BMT, risk of relapse at BMT, disease status post-BMT, stem cell source, and chronic graft versus host disease [GvHD]) were abstracted from medical records. Intelligence quotient was assessed pre-BMT as a measure of cognitive reserve, and level of fatigue was assessed at each time point.30,31 We used the Global Deficit Score (GDS) to represent overall cognitive impairment.32,33 GDS is a well-recognized summary score of overall cognitive performance that has been used in previous studies of both patients with cancer6,34,35 and patients without cancer.36,37 Individual T scores were converted to deficit scores (range, 0-5) and averaged across the 14 tests to estimate the GDS.5 We used a cutoff of GDS ≥ 0.5 to create a binary indicator for cognitive impairment.32 The neuropsychological tests, GDS calculation, cohort diagram, and average test scores over time are described in the Data Supplement. The study was approved by the institutional review board at COH; informed consent was provided according to the Declaration of Helsinki.
Replication cohort.
An independent nonoverlapping cohort from the BMT Survivor Study (BMTSS)3,29 was used to replicate the predictive models from the discovery cohort. BMTSS is a collaborative study between the University of Alabama at Birmingham, COH, and University of Minnesota examining outcomes in patients who received BMT between 1974 and 2014 and survived ≥ 2 years after BMT. Study participants completed a 255-item questionnaire that included sociodemographic factors and medical outcomes, including self-report of learning/memory problems (as diagnosed by their health care provider; Data Supplement). A measure of cognitive reserve was not available. BMT survivors were included as cases if they reported learning/memory problems after BMT (n = 192) and controls (n = 354) if they did not report any cognitive problems, matched (up to 2 per case) on race/ethnicity, primary hematologic malignancy, and time from BMT to cognitive problems for cases and from BMT to questionnaire completion for controls.
SNP Selection and Genotyping
Discovery cohort.
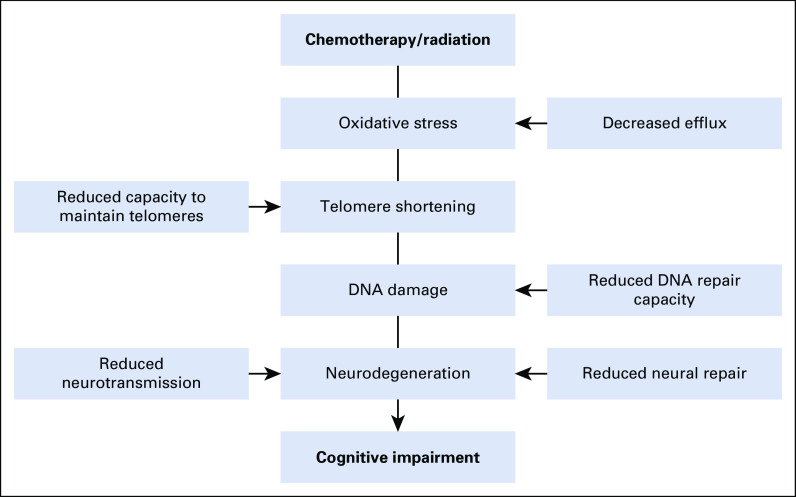
We hypothesized that chemotherapy and/or radiation induce oxidative stress resulting in DNA damage and telomere shortening, which could result in neurodegeneration and present as cognitive impairment. Impaired capacity to repair damaged DNA, to effectively pump genotoxic agents out of cells, and to maintain telomere homeostasis could further affect cognitive functioning. Finally, reduced capacity for neural repair and neurotransmitter activity could exacerbate and/or have independent effects on cognitive impairment.18-21,38-41 We selected candidate SNPs involving 5 mechanisms: blood-brain barrier (BBB) transport, telomere homeostasis, neural repair, neurotransmission, and DNA repair (Fig 1). The Data Supplement lists the candidate genes.
FIG 1.
Hypothesized model of candidate mechanisms for cognitive impairment after blood or marrow transplantation.
Germline DNA was collected pre-BMT (94% blood; 6% saliva) and genotyped using Illumina Infinium HumanExome BeadChip (Illumina, San Diego, CA). Quality control was performed using PLINK.42 Of the 278 study participants with available genetic data, we excluded 1 with > 10% missing genotype data. Of a total of 1,503 successfully genotyped SNPs, we removed 35 SNPs with > 5% missing genotype, 9 SNPs because of deviation from Hardy-Weinberg equilibrium (P < .001), and 474 SNPs with minor allele frequency < 5%. The genetic association analysis included 985 SNPs in 68 candidate genes, with a total genotyping call rate of 99% in 277 BMT recipients.
Replication cohort.
We genotyped 69 SNPs identified from the discovery cohort. We used the Fluidigm SNP genotyping assay (Fluidigm, South San Francisco, CA) in 546 BMTSS samples (19% blood, 80% saliva, and 1% nail samples); 68 SNPs were successfully genotyped.
Statistical Analysis
The complexity of genetic data calls for the adaptation of data-intense methodologic techniques, such as machine learning, a computer science field that has emerged as a primary technique for analysis and representation of highly complex data in genetics43 and cancer research.44 We based gene selection on biologic plausibility and applied previously tested machine learning techniques to identify genetic markers and construct risk prediction models.
Single-SNP analysis.
Repeated measurements of post-BMT GDS in the discovery cohort were analyzed using generalized estimating equation models. Primary independent variables were the candidate SNPs; other covariables considered included BMT type, age, sex, race/ethnicity, education, income, fatigue, cognitive reserve, conditioning regimen, risk of relapse at BMT, and chronic GvHD. By default, the minor allele was considered the “risk” allele.45 Linkage disequilibrium-based pruning yielded 326 independent tests, with an overall P value threshold of 1.53 × 10−4 using Bonferroni correction for multiple testing (Data Supplement).42 We followed the genomic control method to adjust for population structure and estimated corrected P values after adjusting the test statistics at individual loci by the genomic inflation factor (quantile-quantile plots in the Data Supplement).
Gene-level analysis.
We included genes with ≥ 2 measured SNPs (n = 63) in the discovery cohort, excluding 5 genes with only 1 measured SNP: APOE, APEX1, FEN1, PNKP, and XRCC6. We used the machine-learning approach of logic regression to search for SNP-SNP interactions at the gene level, referred to as logic trees (Data Supplement).46-48
Risk prediction modeling.
Significant genetic signals identified from the single-SNP and gene-level analyses were used in the discovery and replication cohorts to predict and replicate risk of post-BMT cognitive impairment. Logistic regression models were built using Elastic-Net regression, which employs a hybrid of 2 regularization techniques, lasso and ridge regression, to address feature selection and overfitting.49,50 Elastic-Net regression overcomes the limitations of traditional methods for risk prediction, such as cross-validation and step-wise regression, when a set of predictors is large and bigger than the number of observations, such as the case with high-dimensional genetic data. Elastic-Net penalty encourages a grouping effect where strongly correlated predictors tend to be in or out of the model together addressing the limitations of overfitting and feature selection, thus creating a parsimonious model. Models for predicting cognitive impairment were developed for the discovery cohort (6 months post-BMT using GDS) and for the replication cohort (using self-report of post-BMT learning/memory problems as identified by their health care provider).
We built 3 risk prediction models: Base Model (sociodemographic), Clinical Model (Base Model plus clinical characteristics, therapeutic exposures, and baseline cognitive reserve), and Combined Model (Base plus Clinical plus significant SNPs). Models were fit with predetermined tuning parameters, and predictions of the binary outcome were used to calculate the C-statistic equal to the area under the curve (AUC) of the receiver operating characteristic curve.51 Differences in AUC comparing Clinical versus Base and Combined versus Clinical models were estimated52; empirical 95% CIs, not including 0 and P value < 5%, were considered statistically significant (Data Supplement). The Data Supplement provides a complete list of variables included in the 3 models. All analyses were performed using R and Stata 14 (Stata, College Station, TX).
RESULTS
Discovery Cohort
Patient characteristics.
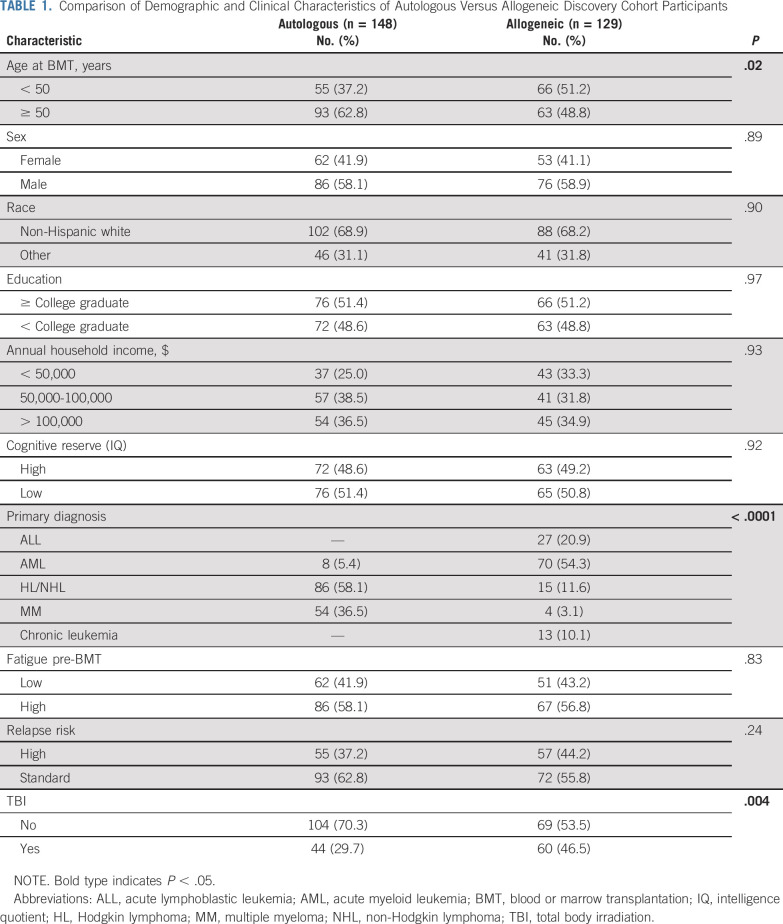
Of the 277 participants in the discovery cohort, 129 (46.6%) were allogeneic BMT recipients (Table 1). Sociodemographic characteristics were comparable between autologous and allogeneic BMT recipients, except for younger age at transplantation for allogeneic BMT recipients (mean age, 48 v 51.5 years; P = .03) and higher proportion with total body irradiation (TBI)-based conditioning (46.5% v 29.7%; P = .004). The most common primary hematologic malignancy diagnoses were acute myeloid leukemia (54.3%) for allogeneic BMT and Hodgkin/non-Hodgkin lymphoma (58.1%) for autologous BMT.
TABLE 1.
Comparison of Demographic and Clinical Characteristics of Autologous Versus Allogeneic Discovery Cohort Participants
Demographic and clinical predictors of cognitive impairment.
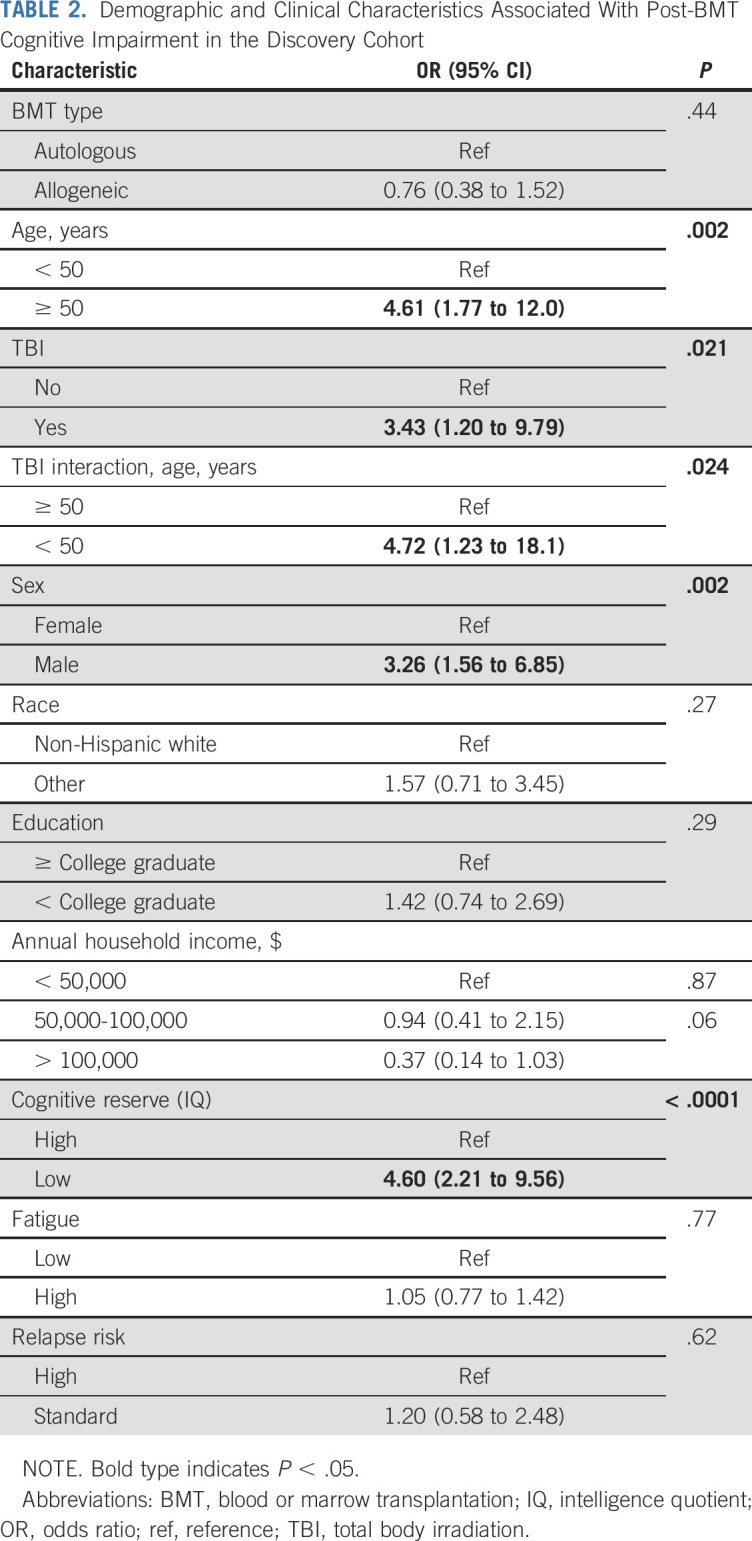
Older age (odds ratio [OR], 4.6; 95% CI, 1.8 to 12.0; P = .002), male sex (OR, 3.3; 95% CI, 1.6 to 6.9; P = .002), and lower baseline cognitive reserve (OR, 4.6; 95% CI, 2.2 to 9.6; P < .0001) were associated with cognitive impairment (Table 2). Significant interaction was identified between younger age at BMT (< 50 years) and receipt of TBI (interaction OR, 4.7; 95% CI, 1.2 to 18.1; P = .024).
TABLE 2.
Demographic and Clinical Characteristics Associated With Post-BMT Cognitive Impairment in the Discovery Cohort
Genetic Predictors
Single-SNP analysis.
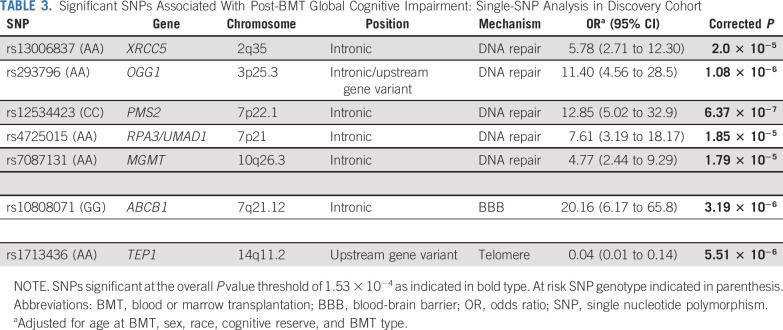
Post-BMT global cognitive impairment was associated with 5 SNPs on DNA repair genes (rs13006837 [XRCC5], P = 2.0 × 10−5; rs293796 [OGG1], P = 1.1 × 10−6; rs12534423 [PMS2], P = 6.4 × 10−7; rs4725015 [RPA3], P = 1.9 × 10−5; and rs7087131 [MGMT], P = 1.8 × 10−5), 1 SNP on the BBB gene (rs10808071 [ABCB1], P = 3.2 × 10−6), and 1 SNP on the telomere homeostasis gene (rs1713436[TEP1]: P = 5.5 × 10−6; Table 3). At the individual cognitive-domain level, we found significant associations of SNPs in DNA repair genes with processing speed and working memory (Data Supplement).
TABLE 3.
Significant SNPs Associated With Post-BMT Global Cognitive Impairment: Single-SNP Analysis in Discovery Cohort
Gene-level analysis.
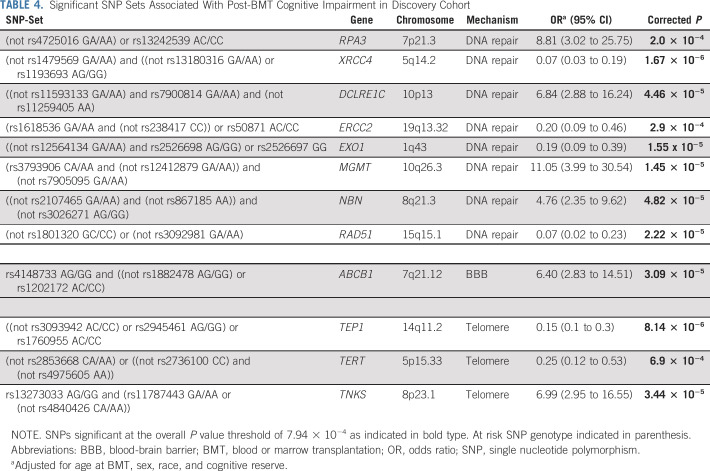
Post-BMT global cognitive impairment was associated with the BBB gene (ABCB1, P = 3.1 × 10−5), telomere homeostasis genes (TEP1, P = 8.1 × 10−6; TERT, P = 6.9 × 10−4; TNKS, P = 3.4 × 10−5), and DNA repair genes (RPA3, P = 2.0 × 10−4; XRCC4, P = 1.7 × 10−6; DCLRE1C, P = 4.5 × 10−5; ERCC2, P = 2.9 × 10−4; EXO1, P = 1.6 × 10−5; MGMT, P = 1.5 × 10−5; NBN, P = 4.8 × 10−5; RAD51, P = 2.2 × 10−5; Table 4). All models were adjusted for age at BMT, sex, race/ethnicity, and baseline cognitive reserve.
TABLE 4.
Significant SNP Sets Associated With Post-BMT Cognitive Impairment in Discovery Cohort
Risk Prediction Analysis
The mean AUCs for the 3 models were as follows: Base Model, 0.69 (95% CI, 0.63 to 0.76), Clinical Model, 0.77 (95% CI, 0.71 to 0.83), and Combined Model, 0.89 (95% CI, 0.85 to 0.93). The Clinical Model performed significantly better than the Base Model (P = .003; cross-validated difference = .08 [.03, .14]), and the Combined Model performed significantly better than the Clinical Model (P = 1.24 × 10−9; cross-validated difference = .12 [.08, .17] Fig 2).
FIG 2.
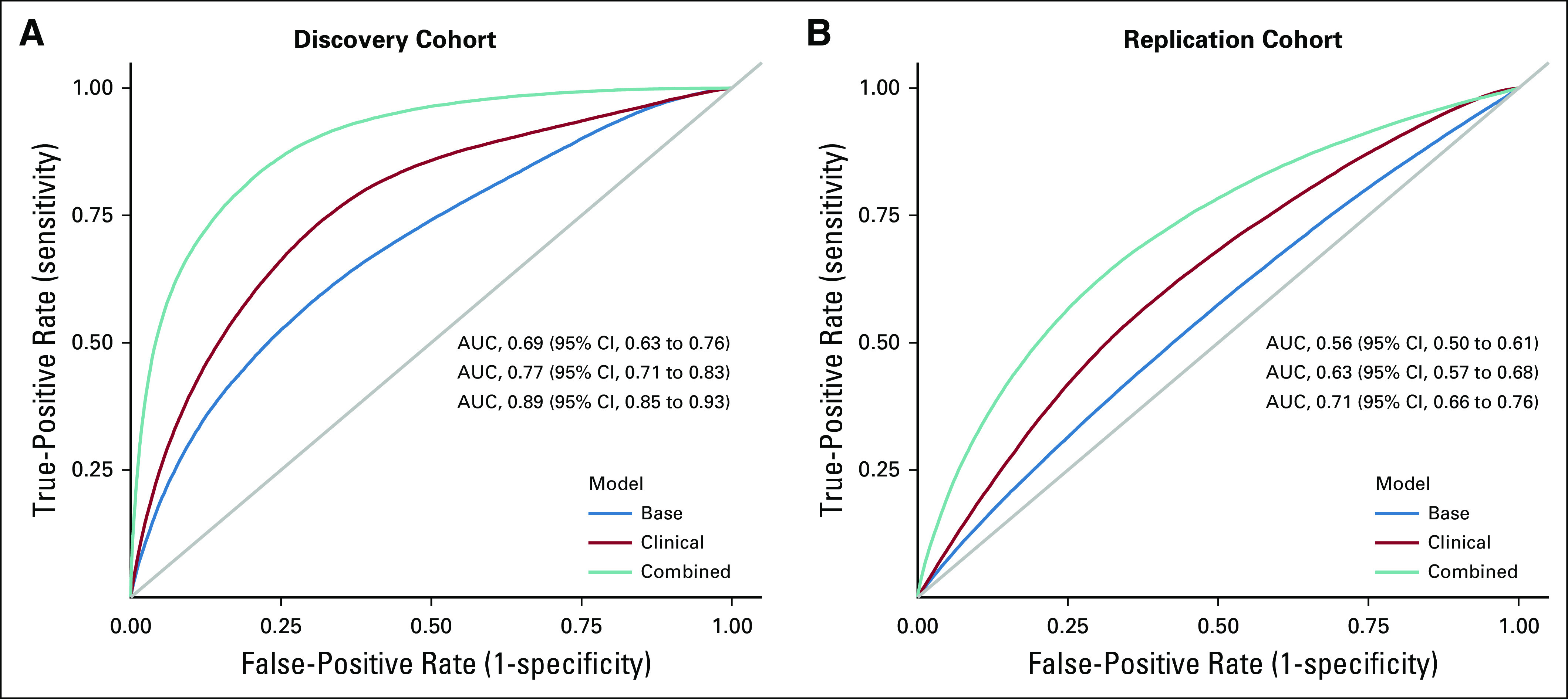
Area under receiver operating characteristic curves (AUCs) for prediction of global cognitive impairment after blood or marrow transplantation. (A) Discovery cohort. (B) Replication cohort.
Replication Cohort
Cases and controls were comparable with respect to age at BMT, sex, race/ethnicity, education, and annual household income (Data Supplement). Median time from BMT to development of memory problems was 2 years (interquartile range, 1-7 years). No demographic or clinical characteristics were independently associated with learning/memory severity (Data Supplement). Compared with the discovery cohort, the replication cohort was younger at BMT and had an over-representation of autologous BMT recipients, as well as females and nonwhite racial/ethnic groups (Data Supplement). There were no significant differences in level of education, income, marital status, or receipt of TBI. The mean AUCs of the 3 replicated models were as follows: Base Model, 0.56,(95% CI, 0.50 to 0.61); Clinical Model, 0.63 (95% CI, 0.57 to 0.68); and Combined Model, 0.71 (95% CI, 0.66 to 0.76). The Clinical Model performed significantly better than the Base Model (P = .001; cross-validated difference = .07 [.02, .15]) and the Combined Model performed significantly better than the Clinical Model (P = .004; cross-validated difference = .08 [.03, .14] Fig 2). Variables retained in the models for both cohorts are listed in the Data Supplement.
DISCUSSION
To our knowledge, this is the first study to examine the role of genetic susceptibility in the development of post-BMT cognitive impairment in patients with hematologic malignancies using a candidate gene approach12,18,19 and the first to develop a comprehensive risk prediction model to identify BMT recipients at risk for cognitive impairment. We identified individual SNP and gene-level variants in DNA repair, BBB transport, and telomere homeostasis genes. Similar to previous reports, we found significant associations between post-BMT cognitive impairment and older age, male sex, and lower baseline cognitive reserve.5,9,53 On the basis of previous reports on associations of fatigue with cognitive decline in allogeneic BMT survivors54 and with self-reported cognitive problems,55,56 we a priori included fatigue as a variable to be accounted for when predicting risk of cognitive impairment. However, fatigue was not independently associated with cognitive impairment in either the discovery or replication cohorts in the current study.
Among patients with Alzheimer’s disease, several DNA repair mechanisms are associated with mild cognitive impairment and increased oxidative DNA damage.57-59 In patients with cancer, chemotherapy and radiation induce oxidative stress resulting in DNA damage; hence, DNA repair variants can potentially lead to cognitive impairment. We identified novel SNPs on 5 DNA repair genes that span several DNA repair mechanisms: XRCC5 (nonhomologous end-joining mechanism), OGG1 (base excision repair), PMS2 (mismatch excision repair), RPA3 (nucleotide excision repair), and MGMT (direct reversal), in addition to 2 SNPs on ABCB1 and TEP1 genes. ABCB1 encodes for P-glycoprotein (P-gp) expressed in the brain capillary endothelial cells protecting brain cells from toxic substances such as chemotherapeutic agents, which are mostly P-gp substrates. ABCB1 is an important amyloid-beta (Aβ) exporter, and variants on ABC drug transporter genes, including ABCB1, are linked to Alzheimer’s disease.60-62 Pathogenesis could be explained by decreased P-gp expression leading to accumulation of Aβ in the brain63-66 and impeding efflux of toxic agents at the BBB,67 further exacerbated by downregulation of P-gp expression.68 The TEP1 gene encodes a component of the ribonucleoprotein complex essential for the addition of new telomeres on chromosome ends and binds to the telomerase RNA component (TERC). TERC and the telomerase reverse transcription (TERT) together constitute the 2 components of human telomerase. Decreased expression of TERT was found to affect cognitive function in animal studies,69 whereas common variants on TERT and TERC are associated with susceptibility to Alzheimer’s disease.70
The gene-level analyses identified additional signals along the same 3 candidate gene mechanisms, providing additional support to the biologic plausibility of these findings. Our results showed that incorporating identified genetic factors significantly enhanced the risk prediction of cognitive impairment and improved the accuracy, as assessed by the C-statistic, in both the discovery and replication cohorts. In applied psychology, when considering factors that can influence behavioral outcomes, an AUC of 0.5 indicates that a model performed no better than chance alone, and an AUC approaching 1 indicates perfect discrimination and prediction; AUC values ≥ 0.7 are generally considered good, and values ≥ 0.80 are considered excellent.71 In our study, the Combined Model that incorporated genetic markers had an AUC of 0.89; in comparison, the model without the genetic variants had an AUC of 0.77. These findings indicate that a model including genetic information was superior to a model including only demographic and clinical information.
Our study provides useful insights regarding the utility of collecting genetic data in clinical practice to predict post-BMT cognitive impairment. For example, patients undergoing allogeneic BMT nearly always have host germline DNA banked in the HLA laboratory after completion of HLA typing. Thus, there is no extra cost in collecting and banking germline DNA from these patients. We showed that incorporating the discovered SNPs enhanced classification of cognitive impairment risk in BMT survivors as estimated by AUC curves, because such SNP testing can serve to complement traditional demographic and clinical risk factors when assessed in a clinical setting.72 Furthermore, this analysis is a step toward risk stratification to separate those at high risk from those at low risk.73,74 In light of the steady decline in the cost of SNP testing technologies, this supports the incorporation of SNPs in a risk prediction model to guide risk stratification and hence an informed clinical decision-making process.75 The cost of a select few candidate SNPs using a customized array is inexpensive, and the cost is decreasing steadily.76,77 In fact, 2 SNPs in our analysis, rs7087131 [MGMT] and rs12534423 [PMS2], are part of a custom chip independently designed by the Neuro Consortium collaborative research group for the investigation of genetic variation in various neurodegenerative diseases, including Alzheimer's disease and dementia.76-78 These observations speak to the clinical utility of our study.
Our study needs to be placed in the context of its limitations. We used a candidate gene approach, which may limit identifying novel associations. SNP-set refinement may be needed in future studies. Another limitation is the difference in the measurement of cognitive impairment between the discovery (objective assessment) and replication (self-report of learning/memory problems identified by their health care provider) cohorts. Previous reports indicate that self-endorsed cognitive problems do not always correlate with objectively measured cognitive impairment.79 However, the current study did not rely on self-endorsed cognitive problems in the replication set; instead, the patients were asked to report on learning/memory problems identified by their health care providers. In a previous report, we have shown that BMT survivors are able to report previously diagnosed health conditions with a fair degree of accuracy.80 An ideal replication would have used an objective measure of cognition to ensure comparability. However, such a population was not available to us. Nonetheless, successful replication of the risk prediction model despite these differences speaks to the robustness of the model. The Combined Models included SNPs identified from both single-SNP and gene-level analyses, which may compromise precision where a significant signal points to multiple potential effectors; nonetheless, findings overall would likely be of more potential clinical/public health relevance than examining single factors.22 A strength of our study lies in the longitudinal design of the discovery cohort and the repeated measurement analysis of cognitive impairment that helped reduce random error and increase statistical power to identify significant genetic associations.81
In summary, we found associations between global cognitive impairment in patients with hematologic malignancies treated with BMT and genetic mechanisms that influence DNA repair, BBB, and telomere homeostasis. Significant SNPs enhanced risk prediction model performance beyond standard demographic and clinical factors. This study represents a first step toward identification of BMT survivors at high risk for cognitive impairment, informing personalized management of cognitive outcomes in patients undergoing BMT.
PRIOR PRESENTATION
Early findings presented at the 58th American Society of Hematology (ASH) Annual Meeting and Exposition, San Diego, CA, December 3-6, 2016; 66th American Society of Human Genetics Annual Meeting, Vancouver, Canada, October 18-22, 2016; and 60th ASH Annual Meeting and Exposition, San Diego, CA, December 1-4, 2018; and as a poster discussion at the 2019 American Society of Clinical Oncology Annual Meeting, Chicago, IL, May 31-June 4.
SUPPORT
Supported in part by Leukemia and Lymphoma Society (LLS) (62771-11, S.B.) and LLS Career Development Award (3386-19, N.S.).
AUTHOR CONTRIBUTIONS
Conception and design: Noha Sharafeldin, Sunita K. Patel, Smita Bhatia
Provision of study materials or patients: Stephen J. Forman, Smita Bhatia
Collection and assembly of data: Noha Sharafeldin, Alysia Bosworth, Purnima Singh, Liton Francisco, Smita Bhatia
Data analysis and interpretation: Noha Sharafeldin, Joshua Richman, Yanjun Chen, Purnima Singh, Xuexia Wang, F. Lennie Wong, Smita Bhatia
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Clinical and Genetic Risk Prediction of Cognitive Impairment After Blood or Marrow Transplantation for Hematologic Malignancy
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/journal/jco/site/ifc.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Stephen J. Forman
Consulting or Advisory Role: Allogene, Lixte
Patents, Royalties, Other Intellectual Property: Mustang Bio
No other potential conflicts of interest were reported.
REFERENCES
- 1. Horowitz MM: Uses and growth of hematopoietic cell transplantation, in Blume KG, Forman SJ, Appelbaum ER (eds): Thomas’ Hematopoietic Cell Transplantation. Malden, Blackwell Publishing Ltd., 2004, pp 9-15. [Google Scholar]
- 2.Devine H, DeMeyer E: Hematopoietic cell transplantation in the treatment of leukemia. Semin Oncol Nurs 19:118-132, 2003 [DOI] [PubMed] [Google Scholar]
- 3.Sun CL, Francisco L, Kawashima T, et al. : Prevalence and predictors of chronic health conditions after hematopoietic cell transplantation: A report from the Bone Marrow Transplant Survivor Study. Blood 116:3129-3139, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark CA, Savani M, Mohty M, et al. : What do we need to know about allogeneic hematopoietic stem cell transplant survivors? Bone Marrow Transplant 51:1025-1031, 2016 [DOI] [PubMed] [Google Scholar]
- 5.Sharafeldin N, Bosworth A, Patel SK, et al. : Cognitive functioning after hematopoietic cell transplantation for hematologic malignancy: Results from a prospective longitudinal study. J Clin Oncol 36:463-475, 2018 [DOI] [PubMed] [Google Scholar]
- 6.Syrjala KL, Artherholt SB, Kurland BF, et al. : Prospective neurocognitive function over 5 years after allogeneic hematopoietic cell transplantation for cancer survivors compared with matched controls at 5 years. J Clin Oncol 29:2397-2404, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Syrjala KL, Dikmen S, Langer SL, et al. : Neuropsychologic changes from before transplantation to 1 year in patients receiving myeloablative allogeneic hematopoietic cell transplant. Blood 104:3386-3392, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Booth-Jones M, Jacobsen PB, Ransom S, et al. : Characteristics and correlates of cognitive functioning following bone marrow transplantation. Bone Marrow Transplant 36:695-702, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Scherwath A, Schirmer L, Kruse M, et al. : Cognitive functioning in allogeneic hematopoietic stem cell transplantation recipients and its medical correlates: A prospective multicenter study. Psychooncology 22:1509-1516, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Harris SE, Deary IJ: The genetics of cognitive ability and cognitive ageing in healthy older people. Trends Cogn Sci 15:388-394, 2011 [DOI] [PubMed] [Google Scholar]
- 11.Kremen WS, Panizzon MS, Cannon TD: Genetics and neuropsychology: A merger whose time has come. Neuropsychology 30:1-5, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahles TA, Root JC, Ryan EL: Cancer- and cancer treatment-associated cognitive change: An update on the state of the science. J Clin Oncol 30:3675-3686, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janelsins MC, Kohli S, Mohile SG, et al. : An update on cancer- and chemotherapy-related cognitive dysfunction: Current status. Semin Oncol 38:431-438, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahles TA, Saykin AJ, Noll WW, et al. : The relationship of APOE genotype to neuropsychological performance in long-term cancer survivors treated with standard dose chemotherapy. Psychooncology 12:612-619, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Small BJ, Rawson KS, Walsh E, et al. : Catechol-O-methyltransferase genotype modulates cancer treatment-related cognitive deficits in breast cancer survivors. Cancer 117:1369-1376, 2011 [DOI] [PubMed] [Google Scholar]
- 16.Koleck TA, Bender CM, Sereika SM, et al. : Polymorphisms in DNA repair and oxidative stress genes associated with pre-treatment cognitive function in breast cancer survivors: An exploratory study. Springerplus 5:422, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janelsins MC, Kesler SR, Ahles TA, et al. : Prevalence, mechanisms, and management of cancer-related cognitive impairment. Int Rev Psychiatry 26:102-113, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahles TA, Saykin AJ: Candidate mechanisms for chemotherapy-induced cognitive changes. Nat Rev Cancer 7:192-201, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morley KI, Montgomery GW: The genetics of cognitive processes: Candidate genes in humans and animals. Behav Genet 31:511-531, 2001 [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Zhou R, Sulman EP, et al. : Genetic modulation of neurocognitive function in glioma patients. Clin Cancer Res 21:3340-3346, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wefel JS, Noll KR, Scheurer ME: Neurocognitive functioning and genetic variation in patients with primary brain tumours. Lancet Oncol 17:e97-e108, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aschard H: A perspective on interaction effects in genetic association studies. Genet Epidemiol 40:678-688, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altman DG, Vergouwe Y, Royston P, et al. : Prognosis and prognostic research: Validating a prognostic model. BMJ 338:b605, 2009 [DOI] [PubMed] [Google Scholar]
- 24.Moons KG, Altman DG, Vergouwe Y, et al. : Prognosis and prognostic research: Application and impact of prognostic models in clinical practice. BMJ 338:b606, 2009 [DOI] [PubMed] [Google Scholar]
- 25.Moons KG, Royston P, Vergouwe Y, et al. : Prognosis and prognostic research: What, why, and how? BMJ 338:b375, 2009 [DOI] [PubMed] [Google Scholar]
- 26.Riley RD, Hayden JA, Steyerberg EW, et al. : Prognosis Research Strategy (PROGRESS) 2: Prognostic factor research. PLoS Med 10:e1001380, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Royston P, Moons KG, Altman DG, et al. : Prognosis and prognostic research: Developing a prognostic model. BMJ 338:b604, 2009 [DOI] [PubMed] [Google Scholar]
- 28.Steyerberg EW, Moons KG, van der Windt DA, et al. : Prognosis Research Strategy (PROGRESS) 3: Prognostic model research. PLoS Med 10:e1001381, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker KS, Gurney JG, Ness KK, et al. : Late effects in survivors of chronic myeloid leukemia treated with hematopoietic cell transplantation: Results from the Bone Marrow Transplant Survivor Study. Blood 104:1898-1906, 2004 [DOI] [PubMed] [Google Scholar]
- 30.Hann DM, Jacobsen PB, Azzarello LM, et al. : Measurement of fatigue in cancer patients: Development and validation of the Fatigue Symptom Inventory. Qual Life Res 7:301-310, 1998 [DOI] [PubMed] [Google Scholar]
- 31. McNair DM, Heuchert JWP: Profile of Mood States: Technical Update. Toronto, Canada, Multi-Health Systems, 2005. [Google Scholar]
- 32.Carey CL, Woods SP, Gonzalez R, et al. : Predictive validity of global deficit scores in detecting neuropsychological impairment in HIV infection. J Clin Exp Neuropsychol 26:307-319, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Blackstone K, Moore DJ, Franklin DR, et al. : Defining neurocognitive impairment in HIV: Deficit scores versus clinical ratings. Clin Neuropsychol 26:894-908, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vardy JL, Dhillon HM, Pond GR, et al. : Cognitive function in patients with colorectal cancer who do and do not receive chemotherapy: A prospective, longitudinal, controlled study. J Clin Oncol 33:4085-4092, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vardy JL, Stouten-Kemperman MM, Pond G, et al. : A mechanistic cohort study evaluating cognitive impairment in women treated for breast cancer. Brain Imaging Behav 13:15-26, 2019 [DOI] [PubMed] [Google Scholar]
- 36.Hulgan T, Kallianpur AR, Guo Y, et al. : Peripheral blood mitochondrial DNA copy number obtained from genome-wide genotype data is associated with neurocognitive impairment in persons with chronic HIV infection. J Acquir Immune Defic Syndr 80:e95-e102, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dampier W, Antell GC, Aiamkitsumrit B, et al. : Specific amino acids in HIV-1 Vpr are significantly associated with differences in patient neurocognitive status. J Neurovirol 23:113-124, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seigers R, Fardell JE: Neurobiological basis of chemotherapy-induced cognitive impairment: A review of rodent research. Neurosci Biobehav Rev 35:729-741, 2011 [DOI] [PubMed] [Google Scholar]
- 39.Ren X, Boriero D, Chaiswing L, et al. : Plausible biochemical mechanisms of chemotherapy-induced cognitive impairment (“chemobrain”), a condition that significantly impairs the quality of life of many cancer survivors. Biochim Biophys Acta Mol Basis Dis; 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Correa DD, Satagopan J, Baser RE, et al. : APOE polymorphisms and cognitive functions in patients with brain tumors. Neurology 83:320-327, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Correa DD, Satagopan J, Cheung K, et al. : COMT, BDNF, and DTNBP1 polymorphisms and cognitive functions in patients with brain tumors. Neuro-oncol 18:1425-1433, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell S, Neale B, Todd-Brown K, et al. : PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559-575, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Libbrecht MW, Noble WS: Machine learning applications in genetics and genomics. Nat Rev Genet 16:321-332, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran BX, Latkin CA, Sharafeldin N, et al. : Characterizing artificial intelligence applications in cancer research: A Latent Dirichlet Allocation Analysis. JMIR Med Inform 7:e14401, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park JH, Gail MH, Weinberg CR, et al. : Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc Natl Acad Sci USA 108:18026-18031, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ruczinski I, Kooperberg C, LeBlanc M: Logic regression. J Comp Graph Stat. 12:475-511, 2003.
- 47.Schwender H, Ruczinski I: Logic regression and its extensions. Adv Genet 72:25-45, 2010 [DOI] [PubMed] [Google Scholar]
- 48.Schwender H, Ickstadt K: Identification of SNP interactions using logic regression. Biostatistics 9:187-198, 2008 [DOI] [PubMed] [Google Scholar]
- 49.Tibshirani R: Regression shrinkage and selection via the lasso. J R Stat Soc Ser A Stat Soc 58:267-288, 1996 [Google Scholar]
- 50.Zou F, Chai HS, Younkin CS, et al. : Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet 8:e1002707, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeLong ER, DeLong DM, Clarke-Pearson DL: Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 44:837-845, 1988 [PubMed] [Google Scholar]
- 52.Demler OV, Pencina MJ, Cook NR, et al. : Asymptotic distribution of ∆AUC, NRIs, and IDI based on theory of U-statistics. Stat Med 36:3334-3360, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beglinger LJ, Duff K, Van Der Heiden S, et al. : Neuropsychological and psychiatric functioning pre- and posthematopoietic stem cell transplantation in adult cancer patients: A preliminary study. J Int Neuropsychol Soc 13:172-177, 2007 [DOI] [PubMed] [Google Scholar]
- 54.Harder H, Cornelissen JJ, Van Gool AR, et al. : Cognitive functioning and quality of life in long-term adult survivors of bone marrow transplantation. Cancer 95:183-192, 2002 [DOI] [PubMed] [Google Scholar]
- 55.Wefel JS, Kesler SR, Noll KR, et al. : Clinical characteristics, pathophysiology, and management of noncentral nervous system cancer-related cognitive impairment in adults. CA Cancer J Clin 65:123-138, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ganz PA, Kwan L, Castellon SA, et al. : Cognitive complaints after breast cancer treatments: Examining the relationship with neuropsychological test performance. J Natl Cancer Inst 105:791-801, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bucholtz N, Demuth I: DNA-repair in mild cognitive impairment and Alzheimer’s disease. DNA Repair (Amst) 12:811-816, 2013 [DOI] [PubMed] [Google Scholar]
- 58.Kwiatkowski D, Czarny P, Toma M, et al. : Associations between DNA damage, DNA base excision repair gene variability and Alzheimer’s disease risk. Dement Geriatr Cogn Disord 41:152-171, 2016 [DOI] [PubMed] [Google Scholar]
- 59.Kwiatkowski D, Czarny P, Toma M, et al. : Association between single-nucleotide polymorphisms of the hOGG1,NEIL1,APEX1, FEN1,LIG1, and LIG3 genes, and Alzheimer’s disease risk. Neuropsychobiology 73:98-107, 2016 [DOI] [PubMed] [Google Scholar]
- 60. Pahnke J, Langer O, Krohn M: Alzheimer's and ABC transporters–new opportunities for diagnostics and treatment. Neurobiol Dis 72:54-60, 2014. [DOI] [PMC free article] [PubMed]
- 61.Cascorbi I, Flüh C, Remmler C, et al. : Association of ATP-binding cassette transporter variants with the risk of Alzheimer’s disease. Pharmacogenomics 14:485-494, 2013 [DOI] [PubMed] [Google Scholar]
- 62.Fehér Á, Juhász A, Pákáski M, et al. : ABCB1 C3435T polymorphism influences the risk for Alzheimer’s disease. J Mol Neurosci 54:826-829, 2014 [DOI] [PubMed] [Google Scholar]
- 63.Chiu C, Miller MC, Monahan R, et al. : P-glycoprotein expression and amyloid accumulation in human aging and Alzheimer’s disease: Preliminary observations. Neurobiol Aging 36:2475-2482, 2015 [DOI] [PubMed] [Google Scholar]
- 64.Wang W, Bodles-Brakhop AM, Barger SW: A role for P-glycoprotein in clearance of Alzheimer amyloid β-peptide from the brain. Curr Alzheimer Res 13:615-620, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jedlitschky G, Vogelgesang S, Kroemer HK: MDR1-P-glycoprotein (ABCB1)-mediated disposition of amyloid-β peptides: Implications for the pathogenesis and therapy of Alzheimer’s disease. Clin Pharmacol Ther 88:441-443, 2010 [DOI] [PubMed] [Google Scholar]
- 66.Jeynes B, Provias J: An investigation into the role of P-glycoprotein in Alzheimer’s disease lesion pathogenesis. Neurosci Lett 487:389-393, 2011 [DOI] [PubMed] [Google Scholar]
- 67.Jeynes B, Provias J: The case for blood-brain barrier dysfunction in the pathogenesis of Alzheimer’s disease. J Neurosci Res 89:22-28, 2011 [DOI] [PubMed] [Google Scholar]
- 68.Brenn A, Grube M, Peters M, et al. : Beta-amyloid downregulates MDR1-P-glycoprotein (Abcb1) Expression at the blood-brain barrier in mice. Int J Alzheimers Dis 2011:690121, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang F, Chang G, Geng X: NGF and TERT co-transfected BMSCs improve the restoration of cognitive impairment in vascular dementia rats. PLoS One 9:e98774, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scarabino D, Broggio E, Gambina G, et al. : Common variants of human TERT and TERC genes and susceptibility to sporadic Alzheimers disease. Exp Gerontol 88:19-24, 2017 [DOI] [PubMed] [Google Scholar]
- 71.Rice ME, Harris GT: Comparing effect sizes in follow-up studies: ROC area, Cohen’s d, and r. Law Hum Behav 29:615-620, 2005 [DOI] [PubMed] [Google Scholar]
- 72.Torkamani A, Wineinger NE, Topol EJ: The personal and clinical utility of polygenic risk scores. Nat Rev Genet 19:581-590, 2018 [DOI] [PubMed] [Google Scholar]
- 73.Howe R, Miron-Shatz T, Hanoch Y, et al. : Personalized medicine through SNP testing for breast cancer risk: Clinical implementation. J Genet Couns 24:744-751, 2015 [DOI] [PubMed] [Google Scholar]
- 74.Torkamani A, Andersen KG, Steinhubl SR, et al. : High-definition medicine. Cell 170:828-843, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katki HA: Quantifying risk stratification provided by diagnostic tests and risk predictions: Comparison to AUC and decision curve analysis. Stat Med 38:2943-2955, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Blauwendraat C, Faghri F, Pihlstrom L, et al: NeuroChip, an updated version of the NeuroX genotyping platform to rapidly screen for variants associated with neurological diseases. Neurobiol Aging 57:247.e9-247.e13, 2017. [DOI] [PMC free article] [PubMed]
- 77. Nalls MA, Bras J, Hernandez DG, et al: NeuroX, a fast and efficient genotyping platform for investigation of neurodegenerative diseases. Neurobiol Aging 36:1605.e7-1605.12, 2015. [DOI] [PMC free article] [PubMed]
- 78. illumina.com: Advancing genomic discoveries in neurodegenerative diseases [Internet], 2019 Available from: https://www.illumina.com/science/consortia/human-consortia/neuro-consortium.html.
- 79.Costa DSJ, Fardell JE: Why are objective and perceived cognitive function weakly correlated in patients with cancer? J Clin Oncol 37:1154-1158, 2019 [DOI] [PubMed] [Google Scholar]
- 80.Louie AD, Robison LL, Bogue M, et al. : Validation of self-reported complications by bone marrow transplantation survivors. Bone Marrow Transplant 25:1191-1196, 2000 [DOI] [PubMed] [Google Scholar]
- 81.Sung YJ, Simino J, Kume R, et al. : Comparison of two methods for analysis of gene-environment interactions in longitudinal family data: The Framingham Heart Study. Front Genet 5:9, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]