Abstract
Background.
The translocase of the mitochondrial inner membrane (TIM) imports most of the nucleus-encoded proteins that are destined for the matrix, inner membrane (IM), and the intermembrane space (IMS). Trypanosoma brucei, the infectious agent for African trypanosomiasis, possesses a unique TIM complex consisting of several novel proteins in association with a relatively conserved protein TbTim17. Tandem affinity purification of the TbTim17 protein complex revealed TbTim54 as a potential component of this complex.
Results.
TbTim54, a trypanosome-specific IMS protein, is peripherally associated with the IM and is present in a protein complex slightly larger than the TbTim17 complex. TbTim54 knockdown (KD) reduced the import of TbTim17 and compromised the integrity of the TbTim17 complex. TbTim54 KD inhibited the in vitro mitochondrial import and assembly of the internal signal-containing mitochondrial carrier proteins MCP3, MCP5, and MCP11 to a greater extent than TbTim17 KD. Furthermore, TbTim54 KD, but not TbTim17 KD, significantly hampered the mitochondrial targeting of ectopically expressed MCP3 and MCP11. These observations along with our previous finding that the mitochondrial import of N-terminal signal-containing proteins like cytochrome oxidase subunit 4 and MRP2 was affected to a greater extent by TbTim17 KD than TbTim54 KD indicating a substrate-specificity of TbTim54 for internal-signal containing mitochondrial proteins. In other organisms, small Tim chaperones in the IMS are known to participate in the translocation of MCPs. We found that TbTim54 can directly interact with at least two of the six known small TbTim proteins, TbTim11 and TbTim13, as well as with the N-terminal domain of TbTim17.
Conclusion.
TbTim54 transiently interacts with TbTim17. It also plays a crucial role in the mitochondrial import and complex assembly of internal signal-containing IM proteins in T. brucei.
Significance.
We are the first to characterize TbTim54, a novel TbTim that is involved primarily in the mitochondrial import of MCPs and TbTim17 in T. brucei.
Keywords: Trypanosoma brucei, mitochondrial, translocase, TbTim17, TbTim54, MCPs
Introduction
Trypanosoma brucei belongs to a group of flagellated parasitic protozoa with a single mitochondrion in each cell. The mitochondrion contains a complex mitochondrial DNA structure known as the kinetoplast. For this reason, this group of microorganisms is known as kinetoplastida [Landfear and Zilberstein, 2019; Maslov et al., 2019]. Several species of kinetoplastids cause various vector-borne diseases that affect millions of people worldwide, such as African trypanosomiasis, Chagas disease, and leishmaniasis [Sternberg and Maclean, 2010; Echavarria at al., 2019; Ngere et al., 2020]. The kinetoplast in trypanosomes consists of hundreds of pieces of circular DNA concatenated to form a disc-like structure [Jensen and Englund, 2012]. Despite such complexity, this mitochondrial DNA encodes only a handful of mitochondrial proteins. Therefore, like other eukaryotes, trypanosomes need to import 99% of mitochondrial proteins from the cytosol [Lukes et al., 2005; Panigrahi et al., 2009].
The mitochondrial protein import machinery and mechanisms have been extensively studied in fungi, humans, and plants [Schmidt et al., 2010; Bauer et al., 1999; Lister et al., 2005]. Nuclear-encoded mitochondrial proteins differ from other cytosolic proteins due to the presence of mitochondrial targeting signals (MTSs) in these proteins [Schmidt et al., 2010]. There are three major types of MTSs: 1) cleavable N-terminal MTS, 2) cleavable N-terminal MTS associated with some sorting signal, and 3) internal MTS. The mitochondrial import pathway of a nuclear-encoded protein depends on the type of targeting signal it has and its destination in the mitochondria [Schmidt et al., 2010; Bauer et al., 1999; Lister et al., 2005]. The translocase of the mitochondrial outer membrane (TOM) imports virtually all nuclear-encoded mitochondrial proteins [Becker and Wagner, 2018]. Once the proteins cross the TOM complex, the N-terminal MTS-containing proteins targeted to the mitochondrial matrix are translocated via the translocase of the mitochondrial inner membrane 23 (TIM23) complex that also contains the presequence translocase-associated motor (PAM) components [Bomer et al., 1996; Neupert and Brunner, 2002]. Many inner membrane proteins those have a sorting signal along with an N-terminal MTS are also translocated via the TIM23 complex but do not require any PAM components [Herrmann and Neupert, 2003]. On the other hand, a large group of mitochondrial inner membrane proteins that have multiple transmembrane domains, like mitochondrial carrier proteins (MCPs), are recognized by their internal targeting signals and translocated by the TIM22 complex [Kolli et al., 2018; Koehler et al., 1998].
The internal signal-containing mitochondrial inner membrane (IM) proteins enter the TOM complex as a loop structure and are assisted by small Tim chaperone Tim9-Tim10 complex in the intermembrane space (IMS) in their release from the TOM complex [Baker et al., 2009]. The Tim9-Tim10 complex carrying the cargo protein then docks on Tim12 located in the TIM22 complex for further translocation [Gebert et al., 2008]. Tim54 and Tim18 are required for the stability of the TIM22 complex [Kerscher et al., 1997; Kerscher et al., 2000].
Although components of the TIM23 complex are relatively conserved, the components of the TIM22 complex vary between fungi and humans [Callegari et al., 2016]. There are no homologs of Tim54 and Tim18 in human. Instead, a novel protein, Tim29, is shown to be critical for the structure and function of the human TIM22 complex [Callegari et al., 2016; Kang et al., 2016]. Besides in fungi and mammals that belong to the eukaryotic super group opisthokont, the Tom and Tim components in other eukaryotic groups are also widely divergent in number, size, and homology [Pyrihova et al., 2018; Makki et al., 2019]. The most remarkable divergence has been found in excavata, a major supergroup of unicellular eukaryotes that includes Trypanosoma and Leishmania that diverge very early during evolution [Schneider et al., 2018; Eckers et al., 2012].
Interest in the T. brucei mitochondrial protein import machinery has recently emerged. T. brucei possesses an archaic TOM complex known as the ATOM [Harsman et al., 2012]. For the TIM complex, TbTim17 is the only member of the Tim17/Tim23/Tim22 protein family that was initially discovered by homology searches when the complete genomic sequence of T. brucei and information on its mitochondrial proteomes became available [www.genedb.org]. TbTim17 is essential to mitochondrial protein import at two major developmental stages of T. brucei [Singha et al., 2008]. TbTim17 exists in large protein complexes with molecular weights ranging from 300 to 1100 kDa [Singha et al., 2012]. Using tandem affinity purification (TAP) of the TbTim17 protein complex, we identified at least two novel components, TbTim62 and TbTim54 [Singha et al., 2012]. We showed that TbTim62 is an IM protein required for the stability of the TbTim17 protein complex [Singha et al., 2015]. We previously showed TbTim54 to be involved in mitochondrial protein import [Singha et al., 2012]. However, its role has not been characterized. Using reciprocal co-immunoprecipitation and SILAC proteomics, Harsman et al. also identified several other components of the TbTim17 complex, such as TbTim42, two rhomboid-like proteins, and several small TbTims [Harsman et al., 2016]. Interestingly, these immunoprecipitation techniques consistently pulled down metabolic enzyme acyl-CoA dehydrogenase (ACAD) with the TbTim17 complex [Singha et al., 2012; Harsman et al., 2016]. Its role in mitochondrial protein import warrants further investigation. On the other hand, TbTim54 has only been identified by the TAP-tag approach [Singha et al., 2012]. Here, we further characterize the function of TbTim54. We show that TbTim54 is peripherally associated with the IM, interacts with the N-terminal region of TbTim17, and is specifically involved in the mitochondrial import of internal targeting signal-containing proteins in T. brucei.
Results
TbTim54 is a novel peripherally associated IM protein
We previously identified TbTim54 (Tb927.6.2470) as a TbTim17-associated protein in T. brucei [Singha et al., 2012]. Protein sequence analysis of TbTim54 using Basic Local Alignment Search Tools (BLAST) [Altschul et al., 1990] revealed that homologs of TbTim54 are only present in trypanosomatids and not in other eukaryotes. Fungal Tim54, which is a component of the TIM22 complex, does not show any homology with TbTim54, indicating that the latter is a different protein. TbTim54 consists of 492 amino acids and possesses three predicted hydrophobic regions, but lacks a predicted N-terminal mitochondrial targeting signal (Fig. 1A). However, TbTim54 has been identified in the mitochondrial proteomes of kinetoplastids [Panigrahi et al., 2009]. Mitocarta analysis [Zhang et al., 2010] also predicted its localization in the T. brucei mitochondrion. Our BLAST analysis using different protein databases revealed TbTim54 possesses two structural motifs: 1) a lysine-rich motif (AAs 60–160) and 2) a Knr4-like repeat domain (AAs 198–351) (Fig 1A). The Knr4-like motif is an intrinsically disordered protein hub that interacts with many partners [Dagkessamanskaia et al., 2010]. None of these motifs have been found in any Tim proteins characterized so far, indicating that this trypanosome protein is unique.
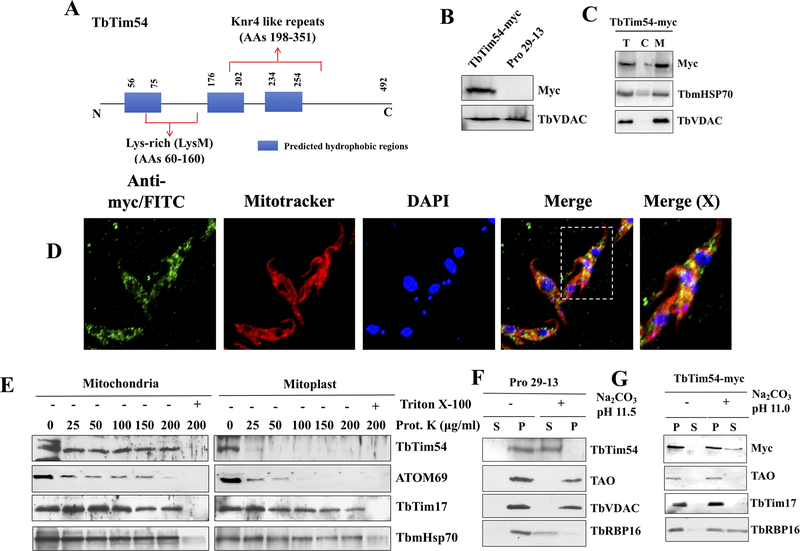
Figure 1|. A schematic of TbTim54 protein and its sub-cellular localization.
(A) The blue boxes represent the predicted hydrophobic regions. Lysine-rich site (AAs 60–160) and Knr4-like repeats (AAs 198–351) are shown. (B) T. brucei cell line where one TbTim54 locus was tagged in situ with 12X-Myc at the 3’ end of the coding region (TbTim54-Myc) and the parental control cell line (Pro29–13) were cultured. Equal number of cells were harvested from each cell line at the logarithmic phase. Total cellular proteins were analyzed by SDS-PAGE and immunoblotting using antibodies for Myc and T. brucei VDAC. (C) TbTim54-Myc cells were lysed and sub-cellular fractionation was performed as described in the materials and methods. Equal amounts of total (T), cytosolic (C), and mitochondrial (M) proteins from each sample were analyzed by SDS-PAGE and immunoblotting using antibodies for Myc, mitochondrial Hsp70, and VDAC. (D) In situ immunofluorescence analysis of the sub-cellular localization of TbTim54. The procyclic form of T. brucei cells expressing in situ-tagged TbTim54–12X-Myc were stained with anti-Myc monoclonal antibody and FITC-conjugated anti-mouse IgG. Before immunostaining, live cells were stained with MitoTracker Red to mark the mitochondria. DAPI was used to stain the nuclear and mitochondrial DNA. The merged picture shows the co-localization of TbTim54–12X-Myc with the mitochondria. The far-right panel shows the enlarged merged photo of one cell indicated in a dotted box. The correlation coefficients for colocalization ranged from 0.788 to 0.826 as calculated from 20 merged images. (E) Limited PK digestion of isolated mitochondria and mitoplasts to determine the sub-mitochondrial location of TbTim54. Concentrations of PK used are as shown. After digestion, mitochondria were re-isolated by centrifugation and proteins were analyzed by immunoblotting. Triton x-100 (1%) was added in one sample to show that these proteins are not resistant to PK. (F & G) Mitochondria from the parental (F) and TbTim54–12X-Myc (G) cells were treated with Na2CO3 (0.1 M) at pH 11.5 and 11.0, respectively. Soluble (S) and membrane (P) proteins were separated and analyzed by immunoblot using the indicated antibodies.
To confirm the subcellular localization of TbTim54, we tagged it in situ with 12X-Myc. Immunoblot analysis showed that TbTim54-Myc was of the expected size and enriched in the mitochondrial fraction (Fig. 1B & C). Immunostaining and confocal microscopy showed co-localization of TbTim54-Myc with the Mitotracker-stained mitochondrion in the procyclic form of T. brucei (Fig. 1D). To investigate the sub-mitochondrial localization of TbTim54, we treated isolated T. brucei mitochondria with various concentrations of proteinase K (PK) (up to 200 μg/ml) (Fig. 1E). We found that similar to IM proteins like Tim17 and matrix-localized mHsp70, TbTim54 was also protected from PK treatment, suggesting that TbTim54 is not an outer membrane protein. To verify that TbTim54 is localized at the IM, we exposed swollen mitochondria, where MOM has been disrupted (mitoplast fraction) to PK (up to 200 μg/ml). We observed that TbTim54 was completely digested even at the lowest PK concentration (25 μg/ml). Outer mitochondrial membrane protein ATOM69 was partially digested by treatment of mitochondria with PK at 25–100 μg/ml and was completely digested at PK concentrations above this range. In mitoplast, ATOM69 was digested at a much lower PK concentration, as expected (Fig. 1E). On the other hand, TbTim17 and mHsp70 were protected against PK digestion in the mitoplasts. These observations suggest that TbTim54 is exposed to the IMS. Furthermore, to check if TbTim54 is membrane-integrated, we perform an alkali extraction (pH 11.5) on the isolated mitochondria. Interestingly, we found that although TbTim54 has three predicted hydrophobic stretches, it was still enriched in the alkali-soluble supernatant, similar to T. brucei mitochondrial matrix protein RBP16 [Hayman and Read, 1999] (Fig. 1F). Other mitochondrial membrane proteins such as Tim17, TAO, and VDAC were present in the alkali-resistant pellets, as expected (Fig. 1F & 1G). Extraction of the mitochondrial fraction from TbTim54-Myc-expressing cells at pH 11.0 showed TbTim54-Myc to be distributed in the soluble and pellet fractions (Fig. 1G). Therefore, we conclude that TbTim54 is a peripherally associated IM protein that is exposed primarily to the IMS.
TbTim54 is required to preserve the integrity of the TbTim17 protein complex.
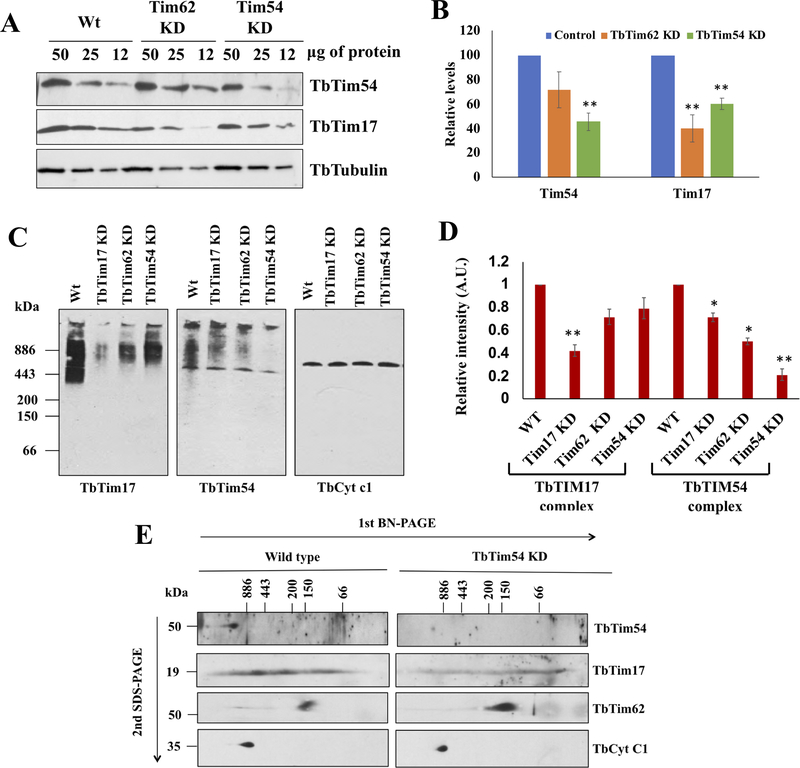
Next, we assessed the effect of TbTim54 depletion on the levels of TbTim17 and the TbTim17 protein complex. Comparison of TbTim54 protein levels in mitochondria isolated from wild type and TbTim54-knockdown (KD) cells revealed the TbTim54 KD to be about 50%. This KD also reduced the levels of TbTim17 protein by about 40%. TbTim17 levels were reduced to a greater extent in mitochondria from TbTim62-KD cells (Fig. 2A and B), as we have shown previously [Singha et al., 2012]. To investigate if TbTim54 exists in a protein complex similar to TbTim17, we performed Blue-native (BN)-PAGE analysis of mitochondrial protein complexes, followed by immunoblotting with TbTim54 and TbTim17 antibodies. The TbTim17 antibody recognized TbTim17 protein complexes with molecular weights ranging from 300 to 1100 kDa in the parental cells, as expected [Weems et al., 2015; Weems et al., 2017]. TbTim17 KD reduced the levels of these complexes by more than 60% (Fig. 2C first panel & 2D). Interestingly, TbTim54 KD reduced the levels of TbTim17 complexes about 20–30%. We also analyzed the mitochondria from TbTim62-KD cells in parallel. TbTim62 is a component of the TbTim17 complex. As shown earlier [Singha et al., 2015], TbTim62 KD reduced the levels of TbTim17 complexes (Fig. 2C first panel). TbTim54 antibody also recognized protein complexes of similar sizes in the mitochondria from the parental cells. The levels of these complexes were reduced about 80% in mitochondria from TbTim54-KD cells (Fig. 2C second panel & 2D). These observations suggest the presence of Tim54 in this protein complex. TbTim17 KD and TbTim62 KD also reduced the levels of the TbTim54 protein complex by about 20–30% (Fig. 2C and D). Levels of the cytochrome b-c1 complex were similar in all samples; hence, it served as the loading control for the BN-PAGE (Fig. 2C third panel).
Figure 2|. Effect of TbTim54 KD on the levels of mitochondrial proteins and protein complexes.
(A) Immunoblot analysis of the serially diluted mitochondrial proteins (50, 25 and 12 μg) from the wild type (Wt), TbTim62-KD, and TbTim54-KD T. brucei using antibodies for TbTim54 and TbTim17. (B) Densitometric analysis of the TbTim54 and TbTim17 protein bands. Their corresponding tubulin bands were used for normalization. Values shown are mean ± SEM from triplicate samples. ** P < 0.01, t-test. (C) Analysis of mitochondrial protein complexes containing TbTim17 and TbTim54. Mitochondria were isolated from wild type, TbTim17-KD, TbTim62-KD, and TbTim54-KD T. brucei cells four days post induction of RNAi with doxycycline. Mitochondria samples (100 μg) were solubilized with digitonin (1.0%). The solubilized supernatant was clarified by centrifugation at 100,000 × g and analyzed by BN-PAGE. Protein complexes were detected by immunoblotting using antibodies for TbTim17, TbTim54, and TbCyt c1. Molecular weight marker proteins apoferritin dimer (886 kDa), apoferritin monomer (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), and bovine serum albumin (66 kDa) were run on the gel and visualized by Coomassie Blue staining. Positions of the marker proteins are shown. (D) Intensities of the protein bands for TbTim17 and TbTim54 complexes were quantitated by densitometric analysis using Image J and normalized to the corresponding band intensities for the TbCyt c1 complex. Values shown are mean ± SEM from triplicate samples. ** P < 0.01, * < 0.05, t-test. (E) Gel strips from the first dimension BN-PAGE were excised and run on a second dimension SDS-PAGE. Proteins were transferred to nitrocellulose membranes and probed with the indicated antibodies.
To further analyze these protein complexes, we performed a two-dimensional BN-SDS-PAGE. For this purpose, gel strips from the first dimension BN-PAGE were used to separate proteins on a second dimension SDS-PAGE and immunoblotted with specific antibodies to detect TbTim17, TbTim54 and TbTim62. As shown in Fig. 2E, the TbTim54 antibody recognized a single spot at ~54-kDa in the mitochondrial sample from wild type cells on the second-dimension gel. The location of this protein on the first dimension BN-PAGE was above 886 kDa, which is slightly higher than the broad band observed for TbTim17. A similar analysis of the mitochondria from TbTim54-KD cells with the same antibody showed a much-reduced intensity of this 54-kDa spot (Fig. 2E). Previous analysis with two-dimensional BN-PAGE has shown protein complexes containing TbTim17 in wild type T. brucei mitochondria to be in a wide range of sizes (<886 to >150 kDa). The intensity of the TbTim17 in the second-dimension SDS-PAGE was also lower for the mitochondrial sample from TbTim54-KD cells. We also observed the presence of TbTim17 protein complexes with lower molecular weights in mitochondria from TbTim54-KD cells, suggesting that TbTim54 KD destabilized this complex or compromised the assembly of TbTim17 protein complexes with higher molecular weights. TbTim62 was primarily found in a ~120-kDa complex on BN-SDS-PAGE, as we reported earlier [Singha et al., 2015]. TbTim54 KD did not affect the levels of this complex. The levels of the bc1-reductase complex were similar in the mitochondria from wild type and TbTim54-KD cells, as expected. Taken together, these observations suggest that TbTim54 may exist in a protein complex slightly larger than the TbTim17 complex, and that TbTim54 depletion can disrupt the integrity of the TbTim17 complex. These observations prompted us to investigate if TbTim54 is required for the mitochondrial import of TbTim17 and the assembly of its complex.
TbTim54 is required for the mitochondrial import of TbTim17 and the assembly of its complex.
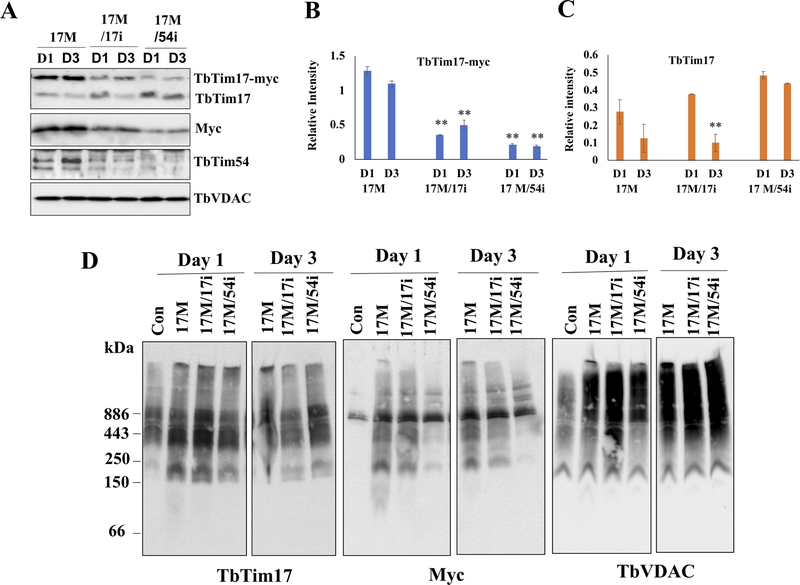
To determine the effect of TbTim54 KD on the mitochondrial import of TbTim17 and the assembly of its complex, we ectopically expressed TbTim17 with a C-terminal 2X-Myc tag in wild type, TbTim54-KD, and TbTim17-KD cells. We have shown previously that TbTim17-Myc can translocate to the mitochondria and be assembled into the TbTim17 protein complex in T. brucei [Singha et al., 2015; Smith et al., 2018]. We treated the KD cells with doxycycline to simultaneously induce TbTim17-Myc expression as well as TbTim54 KD and TbTim17 KD, respectively. We harvested the cells at day 1 and 3 post induction and determined the levels of TbTim17-Myc in the mitochondria by immunoblotting. As expected, the TbTim17 antibody recognized both TbTim17-Myc and endogenous TbTim17 (Fig. 3A). TbTim17-Myc levels were similar on day 1 and 3 post induction in the mitochondria from TbTim7-Myc (17M) cells. TbTim17-Myc levels were reduced in the mitochondria from TbTim17-KD cells due to perturbation by RNAi. We observed a slight increase in TbTim17-Myc in TbTim17-KD cells (17M/17i) at day 3 post induction, which we attribute to compensation by TbTim17-Myc. This is because we found the endogenous TbTim17 levels to be reduced when TbTim17-Myc was overexpressed in the 17M cells (Fig. 3A and B). We previously reported a similar observation [Singha et al., 2015]. In contrast, TbTim17-Myc levels were significantly reduced in mitochondria from TbTim54-KD cells (17M/54i) on both day 1 and 3 post-induction (Fig. 3A and B). TbTim54 KD did not affect the levels of endogenous TbTim17 within this time period (Fig. 3A and C), since there was a pre-existing pool of endogenous TbTim17 protein in the mitochondria before TbTim54 RNAi induction. The anti-Myc antibody recognized only the ectopically expressed TbTim17-Myc and revealed a similar pattern for TbTim17-Myc protein levels as that detected by the TbTim17 antibody (Fig. 3A). Taken together, these results showed that TbTim54 is critical for the mitochondrial import of TbTim17.
Figure 3|. Effect of TbTim54 KD on the import of TbTim17-Myc into mitochondria.
T. brucei cell lines TbTim17-Myc (17M), TbTim17-Myc/TbTim17-KD (17M/17i), and TbTim17-Myc/TbTim54-KD (17M/54i) were treated with doxycycline to simultaneously induce TbTim17-Myc expression and KD of TbTim17 and TbTim54, respectively. (A) On day 1 (D1) and day 3 (D3) post induction, cells were harvested to isolate their mitochondria. The mitochondrial samples were analyzed by immunoblot using antibodies for TbTim17, Myc, and TbTim54. VDAC served the loading control. Relative band intensity for TbTim17-Myc (B) and endogenous TbTim17 (C) were quantitated by densitometry and normalized to the intensities of the corresponding VDAC protein bands. Values shown are mean ± SEM from triplicate samples. ** P < 0.01, t-test in comparison to the 17M samples on day 1. (D) Analysis of mitochondrial protein complexes from wild type (con), 17M, 17M/17i, and 17M/54i cells. Mitochondrial proteins (100 μg) isolated at D1 and D3 post-induction were solubilized with digitonin (1.0%). The solubilized supernatant was clarified by centrifugation at 100,000 × g and analyzed by BN-PAGE. Protein complexes were detected by immunoblotting using antibodies for TbTim17, Myc and VDAC antibodies. Molecular size markers are shown. Blots are representatives of three independent experiments.
Next, we determined if TbTim54 KD can interfere with the assembly of TbTim17-Myc protein complexes. BN-PAGE analysis of mitochondria obtained from 17M, 17M/17i, and 17M/54i cells revealed that the assembly of TbTim17-Myc protein complexes was greatly hampered due to TbTim54 KD (Fig. 3D). The TbTim17 antibody that recognizes both TbTim17-Myc and endogenous TbTim17 detected similar levels of TbTim7 complexes in all samples except the wild type where TbTim17 was not overexpressed. However, immunoblotting with the anti-Myc antibody revealed a much lower level of TbTim17 complexes (from 200 to over 600 kDa) only in the 17M/54i samples collected on day 1 and day 3 post induction. These results show that TbTim17-myc was not assembled in these complexes in mitochondria from 17M/54i cells. The anti-Myc antibody also revealed a band of ~886 kDa in all samples, including control cells that do not have express TbTim17-Myc; thus, differences in the intensities of this band were not due to effects of RNAi. VDAC was used as the loading control. We found similar levels of the specific ~120 kDa band recognized by VDAC antibody in all samples.
TbTim54 KD reduces the mitochondrial import of MCP3, MCP5, and MCP11.
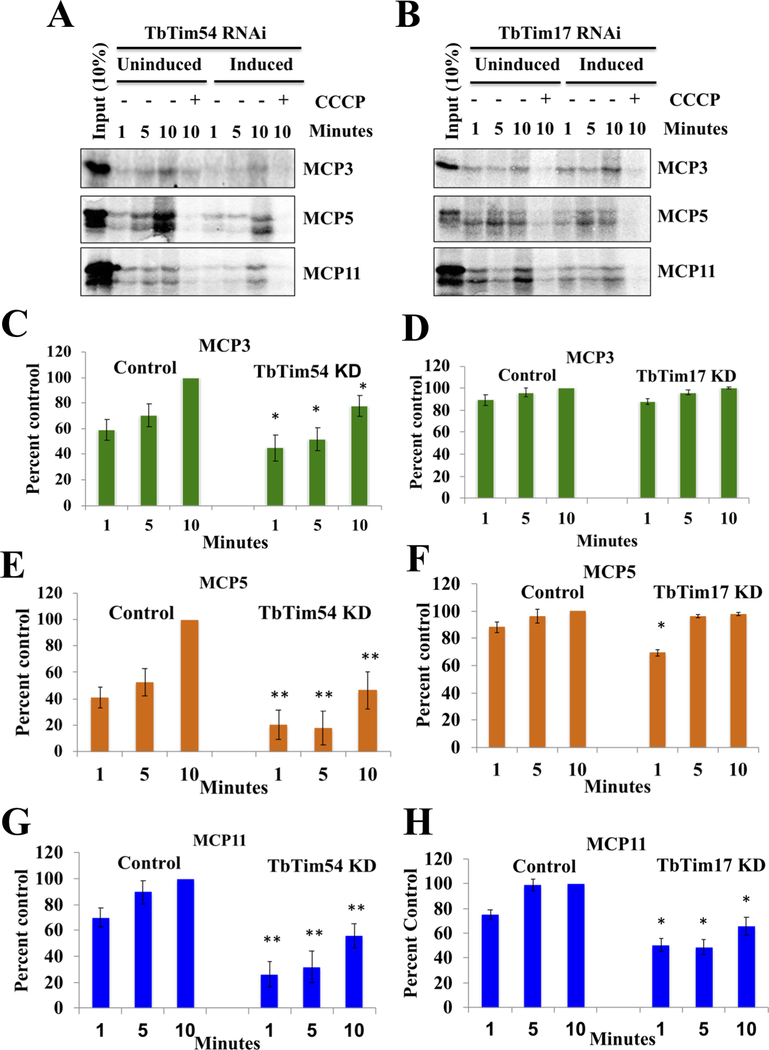
Next, we investigated the effect of TbTim54 KD on the mitochondrial import of other internal targeting signal-containing proteins in T. brucei. To do this, we performed an in vitro mitochondrial import assay using radiolabeled proteins and mitochondria isolated from induced and un-induced TbTim17-KD and TbTim54-KD T. brucei cells. We selected three mitochondrial carrier proteins—MCP3, MCP5, and MCP11—as substrates for this assay. Since these proteins do not have a cleavable N-terminal mitochondrial targeting signal, they resemble MCPs in other eukaryotes in that they likely depend on their internal targeting signals for translocation. To assess the extent of mitochondrial import, we treated the isolated mitochondria with PK after the import reaction and analyzed the radiolabeled proteins to determine if they were protected from PK digestion, as described in materials and methods. We found that TbTim54 KD reduced the mitochondrial import of MCP3, MCP5, and MCP11 by about 20%, 50%, and 40%, respectively (Fig. 4A, C, E, and G). Pretreatment of mitochondria with cyanide m-chloro-phenylhydrazone (CCCP) to disrupt mitochondrial membrane potential abolished the import, confirming the requirement of mitochondrial membrane potential for the import of these proteins. In contrast to TbTim54 KD, the effect of TbTim17 KD was less apparent on the import of MCP3 and MCP5, although both TbTim54 KD and TbTim17 KD affected the mitochondrial import of MCP11 to a similar extent (Fig. 4B, D, F, and H). Taken together, these results showed that inhibition of import of MCP3 and MCP5, two out of three MCPs tested, by TbTim54 KD is independent of TbTim17 function.
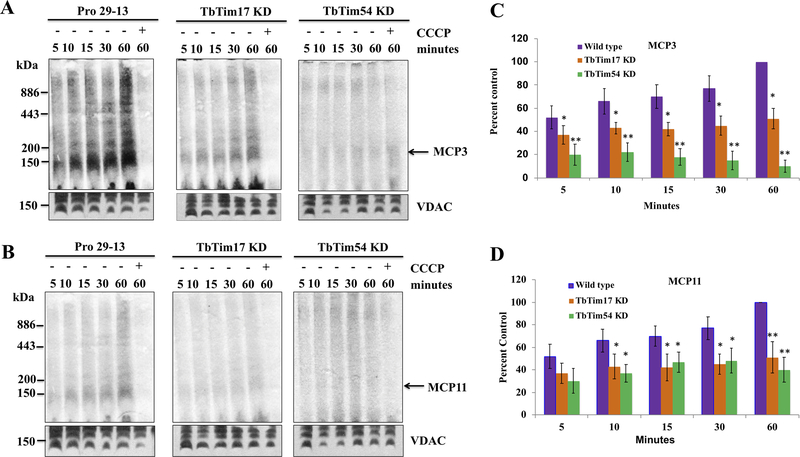
Figure 4|. Effect of TbTim54 KD on the mitochondrial import of MCPs.
(A & B) Radiolabeled MCP3, MCP5, and MCP11 served as substrates in an in vitro mitochondrial protein import assay using mitochondria isolated from TbTim54-KD and TbTim17-KD cells grown in the absence (Uninduced) and presence (Induced) of doxycycline for 96 h. Mitochondria were re-isolated at different time points during the assay. The proteins were analyzed by SDS-PAGE and autoradiography. Input lanes represent 10% of the radiolabeled substrates used for each reaction. One set of mitochondria was pretreated with CCCP to disrupt the mitochondrial membrane potential (ΔΨ). These pretreated mitochondria were used to show that import of MCPs was ΔΨ-dependent. (C–H) Intensities of protein bands protected from PK digestion were quantitated by densitometry using Image J. Band intensities of the substrates (MCP3, MCP5, and MCP11) in the uninduced control cells at the 10-min time point were set as 100% (control), and the band intensities at other time points for the uninduced and induced samples were calculated as percentages of these control values. Each experiment was repeated at least three times and standard errors are shown. ** P < 0.01, * < 0.05, t-test.
TbTim54 KD hampers the assembly of protein complexes containing MCP3 and MCP11 in the mitochondria of T. brucei.
Generally, MCPs have six trans-membrane domains. It has been shown in other eukaryotes that after entry through the TIM complex, MCPs assemble into dimeric or tetrameric complexes in the IMS. To investigate if TbMCP3 and TbMCP11 also form similar complexes, we performed an in vitro mitochondrial import assay using radiolabeled proteins and mitochondria from wild type cells. We then analyzed the mitochondrial membrane protein complexes by BN-PAGE and autoradiography. We observed that the imported MCPs assembled into a ~150-kDa complex in wild type cells and that its levels increased with time (Fig. 5A & 5B, first panel). Pretreatment of mitochondria with CCCP inhibited the formation of this complex due to the inhibition of the mitochondrial import of MCP3 and MCP11. Next, we assessed the effect of TbTim17 KD and TbTim54 KD on the assembly of this complex. We found that MCP inclusion in the ~150 kDa complex was reduced by 50% in the mitochondria from TbTim17-KD cells compared to the mitochondria from parental cells (Fig. 5A and C). The reduction of MCP3 inclusion was greater in the mitochondria from TbTim54-KD cells (~70%) compared to mitochondria from parental cells. In contrast to MCP3 inclusion, MCP11 inclusion was inhibited to a similar extent in mitochondria from TbTim17-KD and TbTim54-KD cells (Fig. 5B and D). These results echo our in vitro mitochondrial protein import data in Fig. 3.
Figure 5|. Effect of TbTim54 KD on the in vitro mitochondrial import and complex assembly of MCP3, MCP5, and MCP11.
Radiolabeled MCP3 (A) and MCP11 (B) proteins were incubated with mitochondria isolated from parental Pro29–13, TbTim17-KD, and TbTim54-KD cells for different periods (5–60 min). After the import reaction, the mitochondria were washed and treated with PK (20 μg/ml) for 30 min at 4°C. The mitochondria were then solubilized with 1% digitonin, and the solubilized supernatants were analyzed by BN-PAGE and autoradiography. One set of mitochondria was pretreated with CCCP to disrupt mitochondrial membrane potential. (C & D) Band intensities of the protein complexes at ~150 kDa were quantitated by densitometry using Image J and normalized to the corresponding VDAC bands. The values obtained for the Pro29–13 mitochondria sample at 60 min were set to 100% (control) and the band intensities at other time points in both sets of samples were calculated as percentages of the control values. The experiments were repeated at least three times and standard errors are shown. ** P < 0.01, * < 0.05, t-test.
TbTim54 is required for mitochondrial targeting of ectopically expressed MCP3 and MCP11 in T. brucei.
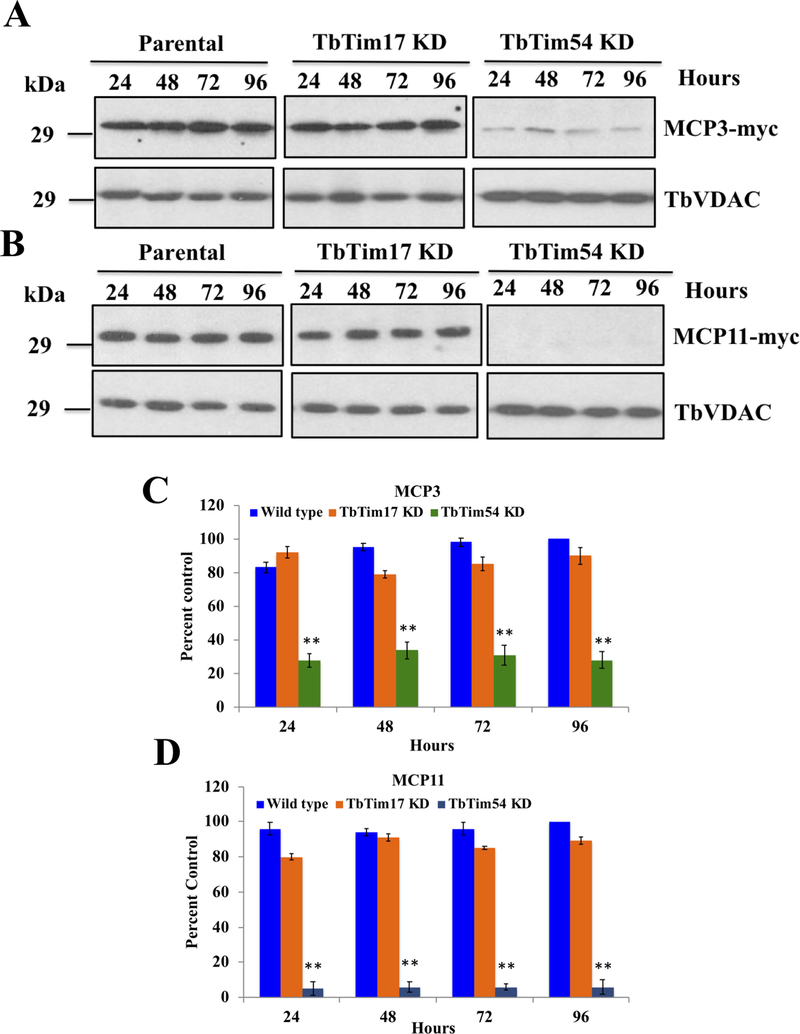
We have shown so far that TbTim54 KD inhibits the mitochondrial import and assembly of multiple MCPs into protein complexes in vitro. To verify these results in cells, we performed an in vivo mitochondrial protein import assay. To do this, we used doxycycline-inducible cassettes to ectopically express C-terminally 2X-Myc-tagged MCP3 and MCP11 in T. brucei wild type (MCP3- or MCP11–2X-Myc), TbTim54-KD, (MCP3- or MCP11–2X-Myc/TbTim54-KD), and TbTim17-KD (MCP3- or MCP11–2X-Myc/TbTim17-KD) cells. We simultaneously induced MCP expression and RNAi with doxycycline. We harvested the induced cells at different time points, isolated their mitochondria, and examined the levels of MCP3- and MCP11–2X-Myc by immunoblotting using the anti-Myc antibody (Fig. 6). We found the levels of MCP3–2X-Myc (~29 kDa) to be consistent throughout the entire experiment (0–96 hr) in the parental wild type T. brucei mitochondria (Fig. 6A). TbTim17 KD showed little effect on the mitochondrial targeting of MCP3–2X-Myc. However, TbTim54 KD drastically reduced the localization of MCP3–2X-Myc in mitochondria (Fig. 6A and C). We also observed similar results for MCP11–2X-Myc (Fig. 6B and D). We did not detect any accumulation of MCP3- or MCP11–2X-myc in the cytosolic fractions (not shown), probably because the unimported proteins were degraded in the cytosol. These results strongly suggest that TbTim54 is critical for the mitochondrial localization of these MCPs, and that TbTim54 is required upstream of TbTim17 in the import pathway.
Figure 6|. Effect of TbTim17 KD and TbTim54 KD on the mitochondrial localization of ectopically expressed MCP3 and MCP11 in T. brucei.
Double transfected TbMCP3-Myc/TbTim17-KD, TbMCP3-Myc/TbTim54-KD, TbMCP11-Myc/TbTim17-KD, and TbMCP11-Myc/TbTim54 KD cells as well as singly transfected TbMCP3-Myc and TbMCP11-Myc cells were grown in the presence of doxycycline for the indicated time periods. Crude mitochondrial fractions were isolated at each time point. An equal amount of proteins from each sample was analyzed by SDS-PAGE and immunoblotting using anti-Myc antibodies. VDAC served as the loading control. (C & D) Band intensities for MCP3-Myc (C) and MCP11-Myc (D) were quantitated by densitometry and normalized to the intensities of corresponding VDAC bands. The normalized values were plotted against time. Individual experiments were repeated three times and standard errors are shown. ** P < 0.01, * < 0.05, t-test.
TbTim54 interacts directly with small TbTims and the N-terminal region of TbTim17.
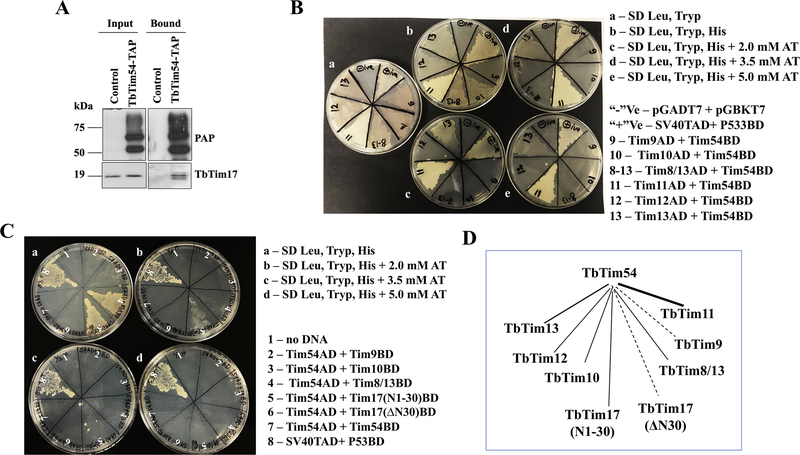
We initially identified TbTim54 by TAP of TbTim17. We further confirmed the association of these two proteins by co-immunoprecipitation [Singha et al., 2012]. Here, we repeated the co-immunoprecipitation analysis using mitochondrial lysates from cells expressing TAP-tagged TbTim54 (TbTim54-TAP) followed by immunoblotting using the anti-TbTim17 antibody. Our analysis showed TbTim17 to be associated with TbTim54-TAP (Fig. 7A). It has been shown in fungi and mammals that Tim9, Tim10, and Tim12 play critical roles in the translocation of MCPs, particularly that of mitochondrial ADP/ATP carrier (AAC), through the TIM22 complex. In these organisms, Tim9 and Tim10 form a 70-kDa hetero-hexameric complex in the IMS that are also associated with the larger TIM22 complex. T. brucei possesses five small TbTims with similar signature sequences—TbTim9, TbTim10, TbTim8/13, TbTim11, TbTim13—and one Tim-like protein, TbTim12. All these small TbTims are soluble IMS proteins that are stably associated with the TbTIM17 complex. However, the role of these proteins in the translocation of MCPs is unclear. Since TbTim54 is also a peripheral membrane-associated IMS protein and appears to play a role in the mitochondrial import of MCPs, we examined if TbTim54 can interact with any small TbTims using yeast two-hybrid analysis as described in the materials and methods. Our analysis showed TbTim54 to interact strongly with TbTim11 and also with TbTim13 (Table 1 & Fig. 7B). Yeast cells co-transformed with TbTim54-activation domain (AD)/TbTim11-DNA-binding domain (BD) plasmids grew on synthetic-defined (SD) plates even at 5.0 mM 3-amino-1,2,4-triazole (AT). Conversely, while yeast cells transformed with TbTim54-AD/TbTim13-BD grew on SD plates with up to 3.5 mM AT, their growth was inhibited at AT concentrations above 3.5 mM (Table 1 & Fig. 7B). TbTim54-AD/TbTim10-BD, TbTim54-AD/TbTim8/13-BD, and TbTim54-AD/TbTim12-BD grew on SD plates without AT. However, TbTim54-AD/TbTim9-BD did not grow on any histidine drop-out plates. Together, these results indicate that TbTim54 strongly interact with TbTim11 and weakly with TbTim13. TbTim54 showed weaker interaction pattern with TbTim10, TbTim8/13, and TbTim12, but it does not interact with TbTim9. Interestingly, we found the TbTim17 N-terminal region (AAs 1–30) that is exposed to the IMS to interact with TbTim54 (Table 1 & Fig. 7C). Yeast cells transformed with TbTim54-AD/Tim17(N1–30)-BD grew on SD plates with up to 2.0 mM AT. On the other hand, the rest of the protein (Δ30-TbTim17) failed to interact with TbTim54. These observations suggest that TbTim54 interacts with TbTim17 in the IMS. As expected, yeast cells co-transformed with empty pGADT7 and pGBKT7 vectors did not grow on SD –leu/–trp/–his plates with or without AT. Yeast cells expressing p53 and SV40-T antigen served as positive controls because these proteins are known to interact very strongly [Drayman et al., 2016]. As anticipated, transformants with P53-BD/SV40-T-AD grew even at 5.0 mM AT. Taken together, our findings indicate that TbTim54 interacts directly with the N-terminal region of TbTim17 that is exposed to the IMS. TbTim54 also interacts strongly with TbTim11 and weakly with some of the other small TbTims, except TbTim9 (Table 1 & Fig. 7D).
Figure 7|. Interaction of TbTim54 with other TbTims.
(A) Mitochondria were isolated from T. brucei cells expressing TbTim54-TAP and control parental cells. Mitochondrial extracts were subjected to immunoprecipitation using IgG-Sepharose. Input (10%) and bound (50%) fractions were analyzed by immunoblotting using peroxidase-anti-peroxidase (PAP) reagent and the TbTim17 antibody. (B) Yeast two-hybrid analysis of interaction between TbTim54 and small TbTims. Representative plates showing the growth of yeast cells co-transformed with a small TbTim (TbTim9, TbTim10, TbTim8/13, TbTim11, TbTim12, or TbTim13) in pGADT7 (AD) and TbTim54 in pGBKT7 (BD) plasmids on (a) SD medium lacking leucine and tryptophan (–leu/–trp), (b) SD medium lacking leucine, tryptophan, and histidine (–leu/–trp/–his), (c) –leu/–trp/–his SD medium containing 2.0 mM AT, (d) –leu/–trp/–his SD medium containing 3.5 mM AT, and (e) –leu/–trp/–his SD medium containing 5.0 mM AT. Yeast cells co-transformed with empty vectors pGADT7 and pGBKT7 served as negative (-ve) controls, and those co-transformed with SV40 T-antigen-AD and p53-BD served as positive (+ve) controls. (C) Interaction of the N-terminal fragment of TbTim17 (N1–30) and the rest of the protein (ΔN30) with TbTim54. Representative plates showing the growth of yeast cells co-transformed with TbTim54 in pGADT7 and one of the following TbTims: TbTim9, TbTim10, TbTim8/13, TbTim17(N1–30), TbTim17 ΔN30, TbTim54 in pGBKT7 on (a)–leu/–trp/–his SD medium, (b) –leu/–trp/–his SD medium containing 2.0 mM AT, (c) –leu/–trp/–his SD medium containing 3.5 mM AT, and (d) –leu/–trp/–his SD medium containing 5.0 mM AT. The plates shown here are from one of three independent experiments. (D) A schematic of interactions between TbTim54 with small TbTims and TbTim17 fragments. The thick solid line represents a strong interaction between TbTim54 and TbTim11, while the thinner solid lines indicate weaker interactions between TbTim54 with TbTim13, TbTim10, TbTim8/13, TbTim12 and between TbTim54 and TbTim17(N1–30). Dotted lines represent no significant interactions.
Table 1.
Yeast two-hybrid analysis of the interaction between TbTim54 and other TbTims.
| Interacting Partner | Growth at 0 mM AT | Growth at 2.0 mM AT | Growth at 3.5 mM AT | Growth at 5.0 mM AT |
|---|---|---|---|---|
| TbTim54AD + TbTim9 BD | − | − | − | − |
| TbTim54AD + TbTim10 BD | + | − | − | − |
| TbTim54AD + TbTim8/13 BD | ++ | − | − | − |
| TbTim54AD + TbTim11 BD | ++ | ++ | ++ | ++ |
| TbTim54AD + TbTim12 BD | + | − | − | − |
| TbTim54AD + TbTim13 BD | ++ | + | + | − |
| TbTim54AD + TbTim54 BD | − | − | − | − |
| TbTim54AD + TbTim17-N(1–30) | ++ | + | − | − |
| TbTim54AD + △30-TbTim17 | − | − | − | − |
| pGADT7 + pGBKT7 | − | − | − | − |
| SV40-T-AD + P53-BD | ++ | ++ | ++ | ++ |
Growth of yeast cells transformed with the respective pairs of AD and BD plasmids on SD medium lacking leucine, tryptophan, and histidine (−leu/−trp/−his) supplemented with various concentrations (up to 5 mM) of AT is indicated by ‘+’. ‘++’ indicates heavier growth and ‘−’ indicates no growth.
Discussion
We are the first to characterize TbTim54, a novel Tim protein in T. brucei that is involved primarily in the mitochondrial import of internal signal-containing proteins like MCPs and TbTim17. TbTim54 is peripherally associated with the IM and located in the IMS. We showed TbTim54 to be in a protein complex slightly larger than the TbTim17 complex and required to maintain the integrity of the latter. We also observed that TbTim54 is critical for the import of substrates containing internal mitochondrial targeting signals such as MCPs and TbTim17. Furthermore, TbTim54 plays an important role in the mitochondrial import of these proteins in a manner that is independent of TbTim17.
We identified TbTim54 by TAP of TbTim17 from cell/mitochondrial extracts. Reversibly, we also pulled down TbTim17 with TbTim54-TAP. These findings indicate that these two proteins interact in T. brucei. We showed TbTim54 to be an IMS protein that is peripherally associated with the IM and present in a protein complex slightly larger than the TbTim17 complex. Interestingly, we found the levels and integrity of the TbTim17 protein complex to be reduced in mitochondria from TbTim54-KD cells. Using mitochondrial import and assembly assays, we showed TbTim54 to be essential for the mitochondrial import of TbTim17 and assembly of the TbTim17 complex. In contrast, the reduction in TbTim17 levels minimally affect the mitochondrial import of the newly synthesized TbTim17 and the assembly of its complex, suggesting that TbTim54 participates in a step that does not require TbTim17. Based on these results, we speculate that TbTim54 first recognizes the imported TbTim17 in the IMS before its complex assembly. To support this, we further showed that TbTim54 interacts directly with the N-terminal domain of TbTim17 that is exposed to the IMS. Therefore, it is likely that while TbTim54 may not exist in the same complex as TbTim17, it may transiently interact with TbTim17 to facilitate the mitochondrial import of the latter and the assembly of the TbTim17 complex.
Both in vitro and in vivo import assays revealed that TbTim54 is required for the import and complex assembly of at least some MCPs. T. brucei expresses several MCPs [Colasante et al., 2009], of which we selected MCP3, MCP5, and MCP11 for analysis. MCP5 has been characterized as a mitochondrial AAC [Pena-Diaz et al., 2012; Gnipova et al., 2015]. MCP11 has recently been characterized as a mitochondrial phosphate carrier [Gao et al., 2020]. MCP3 has not been functionally characterized. Since TbTim54 KD affects the formation of the TbTim17 complex, we performed our import and import/assembly assays in mitochondria from both TbTim17-KD and TbTim54-KD cells. We consistently observed in the in vitro assays that TbTim54 KD inhibited the mitochondrial import of MCPs to a greater extent than TbTim17-KD, indicating that TbTim54 plays a role in the translocation of these proteins in a way that is independent of TbTim17. We also observed that the requirement of TbTim17 and TbTim54 varied for the mitochondrial import of different MCPs, indicating that the MCP import process is complex. In addition, we also observed in TbTim54-KD cells that the mitochondrial localization of MCP3 and MCP11 were greatly hampered. In contrast, the changes on mitochondrial targeting/localization of these two MCPs was minimal in TbTim17-KD cells. These findings further support the notion that TbTim54 is required at the earlier translocation step of these substrate proteins into the IM, whereas TbTim17 is needed for IM integration and assembly.
Mitochondrial carrier translocase in fungi, also known as the Tim22–54 complex, possesses Tim54P [Kurz et al., 1999]. However, we did not detect any sequence homology between the fungal Tim54P and TbTim54. Tim54P in fungi is an essential and integral IM protein facing the IMS that is tightly associated with Tim22, the central component of the carrier translocase [Kurz et al., 1999]. It has been shown that S. cerevisiae Tim54P is crucial for maintaining the integrity of the TIM22–54 complex; however, its role in MCP translocation is unclear [Wagner et al.,2008]. The mitochondrial translocation of AACs has been studied extensively in yeast [Kunji et al., 2016; Ruprecht et al., 2014]. It has been shown that the Tim9-Tim10 IMS chaperone complex carries AACs from the TOM complex and docks them on Tim12 attached to the TIM22 complex for further translocation. Experimental evidence showed that AACs directly interact with small Tims Tim9, Tim10, and Tim12, as well as the core component, Tim22 [Kunji et al., 2016; Ruprecht et al., 2014]. However, Tim54P does not show any direct interaction with AACs, indicating an indirect role of Tim54P in AAC translocation [Wagner et al., 2008].
Interestingly, it has been reported that yeast Tim54P not only plays a role in maintaining the integrity of the TIM22 complex, but it is also important for the assembly of the i-AAA protease complex known as the YMEL1 complex in mitochondria [Hwang et al., 2007]. YMEL1 is an ATPase-dependent metalloprotease that forms a large protein complex in the IM facing the IMS. This complex plays an important role in maintaining mitochondrial proteostasis by degrading unassembled and defective proteins in the mitochondria [Dunn et al., 2008]. Although YMEL1 enters the mitochondria via the TIM23 complex and not the TIM22 complex, its complex assembly and functional competence are greatly hampered in the absence of Tim54P [Hwang et al., 2007]. T. brucei possesses a homologue of Yme1 protease (Tb927.10.7620) that has not been fully characterized. Therefore, it would be interesting to examine whether TbTim54 also plays a role in the assembly of this protein complex in the IM.
Here, we show that TbTim54 is required for the mitochondrial import and complex assembly of several MCPs and TbTim17. T. brucei does not have any homologs of fungal Tim22 or Tim18; however, it possesses at least six small TbTims: TbTim9, TbTim10, TbTim8/13, TbTim11, TbTim12, and TbTim13 [Smith et al., 2018; Wenger et al., 2017]. Unlike small Tims in fungi and in other eukaryotes, all these small TbTims are associated with the TbTIM17 complex and are required for the stability of this complex [Smith et al., 2018; Wenger et al., 2017]. We also show that TbTim54 can interact directly with TbTim11 and TbTim13 and weekly with other small TbTims, except TbTim9. Whether these small TbTims act as chaperones for MCP translocation require further investigation. We also show that TbTim54 can interact with the N-terminal region of TbTim17, which is exposed to the IMS. Taken together, our findings indicate that TbTim54, in association with some small TbTims, facilitates the mitochondrial translocation of MCPs via the TbTIM17 complex.
Experimental Procedures
Cell culture and growth analysis.
The procyclic form of the T. brucei 427 double-resistant cell line (Pro 29–13) expressing a tetracycline repressor gene and T7 RNA polymerase was grown in SDM-79 medium supplemented with 10% fetal bovine serum, G418 (15 μg/mL), and hygromycin (50 μg/mL) [Wirtz et al., 1999]. For growth measurements, cells were seeded at 2×106 cells/mL in fresh medium containing the appropriate antibiotics. Cells were harvested at different time points (up to 10 days) and counted using a Neubauer hemocytometer. The log of cumulative cell numbers was plotted against time (in days) of incubation. T. brucei 29–13 cells served as wild type controls.
Generation of plasmids and T. brucei transgenic cell lines.
T. brucei cell lines expressing TbTim54 double-stranded RNA (TbTim54-KD) and overexpressing TbTim54 with a C-terminal TAP tag (TbTim54-TAP), respectively, were developed previously [Singha et al., 2012]. In situ epitope tagging of one of the two loci of TbTim54 was performed using a construct derived from the pNAT-12MYC vector [Alsford and Horn, 2008]. Briefly, the C-terminal coding fragment of TbTim54 (excluding the stop codon) was amplified by using primers TbTim54_1024 F and TbTim54_1476 R (Table 2) and cloned into the pNAT-12MYC vector in between HindIII and XbaI sites. The resulting construct was digested with MfeI and used to transform 427 procyclic form cells. The transformed cells were then selected with hygromycin (50 μg/ml). T. brucei cells expressing TbTim17–2X-myc via doxycycline induction were developed previously [Singha et al., 2015]. These transgenic cells were transfected with TbTim17RNAi and TbTim54RNAi constructs in the p2T7–177(Puro) vector and selected by puromycin (1 μg/ml). The coding regions for TbMCP3 (Tb927.9.11040), TbMCP5 (Tb927.9.10310), and TbMCP11 (Tb927.10.14820) were amplified by PCR using the appropriate primers (Table 2). The PCR products were cloned in between HindIII and BamHI sites in the pGem4z vector for in vitro transcription/translation and in the pLew-Myc vector for expression in T. brucei. The resulting constructs in pLew-Myc were linearized with NotI, transfected to T. brucei procyclic form cells, and selected by phleomycin to generate stable cell lines. These stable cells lines were further transfected with TbTim54RNAi and TbTim17RNAi constructs in the p2T7–177(Puro) vector. All these double-transfected cell lines were used in in vivo import assays as described below.
Table-2.
List of primers used in this study
| Name | Sequence (5’---3’) | Length (bp) |
|---|---|---|
| TbTim54_1024 F | GATCAAGCTTGGGCGAGGACCTTCATGAGTGTG | 32 |
| TbTim54_1476 R | GATCTCTAGAACACGCTGCTCGCATCCGAGAGTC | 34 |
| TbMCP3 F | GATCAAGCTTATGGATGTAATATGCTCCGC | 30 |
| TbMCP3 R | GATCTCTAGACGCCATATTACC | 22 |
| TbMCP11 F | GATCAAGCTTATGAGCGCAAAGAACAAAACTTGGGAC | 37 |
| TbMCP11 R | GATCTCTAGACTTCTTTCCGGAACCACC | 28 |
| TbMCP5 F | AGAAGCTTGCTATGACGGATAAAAAGCGGG | 30 |
| TbMCP5 R | AAGAATTCCTAATTCGATCTGGCGCC | 26 |
Sub-cellular fractionation.
Fractionation of T. brucei procyclic form cells was performed as described [Smith et al., 2018]. Briefly, 2×108 cells were pelleted and re-suspended in 500 μL of SMEP buffer (250 mM sucrose, 20 mM MOPS/KOH, pH 7.4, 2 mM EDTA, 1 mM PMSF) containing 0.03% digitonin and incubated on ice for 5 min. The cell suspension was then centrifuged for 5 min at 6,800 x g at 4°C. The resultant pellet was considered as the crude mitochondrial fraction, and the supernatant contained soluble cytosolic proteins.
Isolation and post-isolation treatment of mitochondria.
T. brucei cells were seeded at 2×106 cells/mL in a 2-L Erlenmeyer flask. The cells were constantly agitated at room temperature and allowed to grow for 2–4 days. Mitochondria were isolated from these parasites after lysis in an isotonic buffer using a nitrogen cavitation bomb followed by differential centrifugation, as previously described [Singha et al., 2008; Duncan et al., 2013]. The isolated mitochondria were stored at −70°C at a protein concentration of 10 mg/mL in SEMP buffer containing 50% glycerol. For sodium carbonate extraction, mitochondria (100 μg) were incubated with 100 μL of 100 mM sodium carbonate (pH 11.5 or pH 11.0) on ice for 30 minutes. The lysate was centrifuged at 14,000 x g and the supernatants and pellets were collected for further analysis. For limited PK digestion, mitochondria in SME buffer (1 mg/ml) were treated with various concentrations of PK (up to 200 μg/ml) for 30 min on ice. After incubation, PK was inhibited by phenyl-methyl-sulphonyl-fluoride (PMSF, 2 mM) and mitochondria were re-isolated by centrifugation at 10,000 x g at 4 °C for 10 min.
Two-dimensional BN/SDS-PAGE analysis.
Mitochondrial proteins (200 μg) were solubilized in 100 μL of ice-cold 1X native buffer (Life Technologies) containing 1% digitonin. The solubilized mitochondrial proteins were clarified by centrifugation at 100,000 x g for 30 min at 4°C. The supernatants were mixed with G250 sample additive (Invitrogen) and were electrophoresed on a precast 4–16% bis-tris polyacrylamide gel (Invitrogen), according to the manufacturer’s protocol. Protein complexes were detected by immunoblot analysis. Molecular size marker proteins apoferritin dimer (886 kDa), apoferritin monomer (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), and bovine serum albumin (66 kDa) were electrophoresed on the same gel and visualized by Coomassie staining. Strips representing individual lanes on the first-dimension native gel were excised. These gel strips were first incubated in a buffer containing 1% SDS and 5% β-mercaptoethanol for 30 minutes to denature proteins. The denatured proteins were then electrophoresed on the second dimension using Tricine-SDS-PAGE (12%). Finally, the proteins were transferred to a nitrocellulose membrane for western blot analysis.
SDS-PAGE and immunoblotting.
Proteins from whole cell lysates as well as cytosolic and mitochondrial extracts were separated on a 12% Tris-SDS polyacrylamide gel, transferred to nitrocellulose membranes, and probed with polyclonal antibodies for TbTim17 (Tb927.11.13920) [Singha et al., 2008], TbAAC (Tb927.10.14820), VDAC (Tb927.2.2510) [Singha et al., 2009], TbPP5 (Tb927.10.13670) [Chaudhuri 2001], cytochrome b-c1 (CytC1) (Tb927.8.1890) [Priest et al., 1993], RBP16 (Tb927.11.7900) [Hayman and Read, 1999], and mtHsp70 (Tb927.6.3740) [Effron et al., 1993]. Monoclonal antibodies for Myc (9E10 clone, ATCC), TAO (Tb927.10.7090), and T. brucei β-tubulin (Tb927.1.2350) were also used. The polyclonal antibodies for ATOM69 peptide (CTRGLQHFEQQEIQP) was custom synthesized by Bethyl laboraties. Blots were developed with the appropriate secondary antibodies and an enhanced chemiluminescence kit (Pierce).
In vitro import and assembly assay.
Radiolabeled (35S-L-methionine) TbMCP3, TbMCP5, and TbMCP11 were synthesized using a cell-free transcription-and-translation rabbit reticulocyte lysate (TNT®, Promega). Isolated T. brucei mitochondria (100 μg) were suspended in 90 μL of import buffer (250 mM sucrose, 80 mM potassium chloride, 5 mM magnesium chloride, 5 mM dithiothreitol, 1.0 mg/mL fatty acid-free bovine serum albumin, 10 mM 3-(N-morpholino)propanesulfonic acid [MOPS/KOH] pH 7.2, 2 mM adenosine triphosphate [ATP], 10 mM creatine phosphate, 0.1 mg/mL creatine kinase, 8 mM potassium ascorbate, 200 nM N,N,N’,N’-tetramethylphenylenediamine, and 5 mM nicotinamide adenine dinucleotide [NADH]). The mitochondrial suspension was mixed with 10 μL of TNT lysate containing the radiolabeled precursor proteins and incubated at 27°C for up to 15 min. Some mitochondria were pretreated with 50 μM carbonyl cyanide n-chlorophenyl hydrazone (CCCP) for 15 min to deplete mitochondrial membrane potential before the addition of the radiolabeled precursor proteins. The samples were washed twice with SEM buffer to remove excess radiolabeled precursor proteins. The washed mitochondria were resuspended in SME buffer (1 μg/μL) and treated with PK (20 μg/mL) for 10 min on ice. Proteins in the post-import fractions were separated by SDS-PAGE and transferred to nitrocellulose membranes. The radiolabeled proteins were detected by autoradiography. For in vitro assembly assay, the import reaction was carried out for up to 60 min. At different time points, an equal amount of reaction mixture from each sample was treated with PK as described above. The proteins were solubilized in 1X native buffer containing 1% digitonin and the soluble supernatants were analyzed by BN-PAGE. Radiolabeled protein complexes were detected by western blot analysis followed by autoradiography.
In vivo mitochondrial protein import assay.
The stable cell lines expressing either MCP3-Myc or MCP11-Myc and TbTim17-myc along with TbTim17TNAi or TbTim54RNAi constructs were treated with doxycycline (1.0 μg/mL) to simultaneously induce RNAi and the expression of the Myc-tagged proteins. Cells without the TbTim RNAi constructs served as controls. At different time points (up to 96 hr) after induction with doxycycline, cells were lysed, and the mitochondrial fractions were isolated. An equal amount of proteins from each mitochondrial fraction was analyzed by SDS-PAGE and immunoblotting to assess the level of MCP3-Myc or MCP11-Myc in the mitochondria. VDAC was used as a loading control.
MitoTracker staining and immunofluorescence microscopy.
Live T. brucei cells (5×106) expressing TbTim54–12X-myc (in situ-tagged) were used for MitoTracker staining as previously described [Singha et al., 2008]. Briefly, MitoTracker® Red CMXROS (Molecular Probe®) in DMSO (1 mM) was added to cells in culture medium to a final concentration of 0.5 μM. The mixture was incubated at 37 °C for 10 min. Cells were washed and incubated in fresh culture medium for an additional 30 min. Cells were then washed twice with PBS and spread evenly over poly-L-lysine (100 μg/ml in H2O)-coated slides. Once the cells had settled, the slides were washed with cold PBS to remove any unattached cells. The attached cells were fixed with 3.7% paraformaldehyde and permeabilized with 0.1% Triton X-100. After blocking with 5% non-fat milk for 30 min, the slides were washed with 1X PBS. Monoclonal anti-Myc antibody was used as the primary antibody and a fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG was used as a secondary antibody for visualization under a fluorescent microscope. DNA was stained with 1μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Cells were imaged using a Nikon TE2000E widefield microscope equipped with a 60× 1.4 NA Plan Apo VC oil immersion objective. Images were captured using a CoolSNAP HQ2 cooled CCD camera and the Nikon Elements Advanced Research software.
Yeast-two-hybrid analysis.
The ORF of TbTim54 was subcloned into yeast expression vector pGADT7 to generate the bait plasmid. The ORFs of small TbTims TbTim9, TbTim10, TbTim8/13, TbTim11, TbTim13, and TbTim12 were subcloned into yeast expression vector pGBKT7 to generate the prey plasmids. Fragments of the TbTim17 ORF from nucleotide 1–90 (AAs 1–30) and 91–459 (AAs 31–153 AAs) were also cloned into pGADT7 and pGBKT7 vectors. Approximately 2 μg each of bait and prey plasmids in different pairing combinations were used to transform S. cerevisiae Y2H Gold strain (Clontech, Cat. No. 630489) using the lithium acetate method. The transformed yeast cells were plated on SD medium lacking leu and trp, and the cells were allowed to grow for 3 days at 30 °C. Yeast clones that grew were then plated on SD medium lacking leucine, tryptophan, and histidine (–leu/–trp/–his) to select for protein-protein interactions between the bait and prey proteins. The SD –leu/–trp/–his plates were also supplemented with 2.0–5.0 mM AT, which inhibits growth due to leaky expression of the HIS3 gene [Smith et al., 2018], to limit the occurrence of false positives. Inoculated plates were incubated at 30 °C for 3–5 days. To confirm positive readouts, this process was repeated at least three times with multiple clones.
Densitometry and statistical analysis.
Image J (NIH) was used to perform densitometry of the western blots. The band intensities of the loading controls were used for normalization and values were plotted using MS Excel. Standard errors were calculated from three independent experiments for each blot. Statistical analysis was performed using t-test in PRISM (GraphPad). P-values are indicated by asterisks on bars as follows: ** P < 0.01, *** P < 0.001.
Supplementary Material
Acknowledgements
This work was supported by grants 1RO1AI125662 and 2SC1GM081146 from the National Institutes of Health to MC. The Core Facilities at Meharry Medical College is supported by NIH grant U54RR026140/U54MD007593. We thank George Cross for the T. brucei 427 (Pro 29-13) cell line and the pLew100 vectors, Sam Alsford for the pNAT vector, Laurie Read for the RBP16 antibody and p2T7Ti-177 RNAi(Puro) vector, the late Paul Englund for the mtHsp70 antibody, Steve Hajduk for the cytochrome c1 antibody, and Keith Gull for the p2T7Ti-177 RNAi (phleomycin resistance) vector. We thank the Meharry Office for Scientific Editing and Publications for scientific editing support (S21MD000104).
Footnotes
Conflict of Interest: The authors declare that no conflicts of interest exists.
References
- Alsford S, Horn D (2008). Single-locus targeting constructs for reliable regulated RNAi and transgene expression in Trypanosoma brucei. Mol. Biochem. Parasitol 161, 76–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990). Basic local alignment search tool. J. Mol. Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Baker MJ, Webb CT, Stroud DA, Palmer CS, Frazier AE, Guiard B, Chacinska A, Gulbis JM, Ryan MT (2009). Structural and functional requirements for activity of the Tim9-Tim10 complex in mitochondrial protein import. Mol. Biol. Cell 20, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer MF, Gempel K, Reichert AS, Rappold GA, Lichtner P, Gerbitz KD, Neupert W, Brunner M, Hofmann S (1999). Genetic and structural characterization of the human mitochondrial inner membrane translocase. J. Mol. Biol 289, 69–82. [DOI] [PubMed] [Google Scholar]
- Becker T, Wagner R (2018). Mitochondrial Outer Membrane Channels: Emerging Diversity in Transport Processes. Bioessays. 40, e1800013. [DOI] [PubMed] [Google Scholar]
- Bomer U, Rassow J, Zufall N, Pfanner N, Meijer M, Maarse AC (1996). The preprotein translocase of the inner mitochondrial membrane: evolutionary conservation of targeting and assembly of Tim17. J Mol. Biol 262:389–395. [DOI] [PubMed] [Google Scholar]
- Callegari S, Richter F, Chojnacka K, Jans DC, Lorenzi I, Pacheu-Grau D, Jakobs S, Lenz C, Urlaub H, Dudek J, Chacinska A, Rehling P (2016). TIM29 is a subunit of the human carrier translocase required for protein transport. FEBS Letter 590, 4147–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri M (2001). Cloning and characterization of a novel serine/threonine protein phosphatase type 5 from Trypanosoma brucei. Gene 266, 1–13. [DOI] [PubMed] [Google Scholar]
- Colasante C, Pena Diaz P, Clayton C, Voncken F (2009). Mitochondrial carrier family inventory of Trypanosoma brucei brucei: Identification, expression and subcellular localisation. Mol. Biochem. Parasitol 167, 104–117. [DOI] [PubMed] [Google Scholar]
- Dagkessamanskaia A, Durand F, Uversky VN, Binda M, Lopez F, El Azzouzi K, Francois JM, Martin-Yken H (2010). Functional dissection of an intrinsically disordered protein: understanding the roles of different domains of Knr4 protein in protein-protein interactions. Protein Sci 19, 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drayman N, Ben-Nun-Shaul O, Butin-Israeli V, Srivastava R, Rubinstein AM, Mock CS, Elyada E, Ben-Neriah Y, Lahav G, Oppenheim A (2016). P53 elevation in human cells halt SV40 infection by inhibiting T-ag expression. Oncotarget 7:5264333–52660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MR, Fullerton M, Chaudhuri M (2013). Tim50 in Trypanosoma brucei possesses a dual specificity phosphatase activity and is critical for mitochondrial protein import. J. Biol. Chem 288, 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CD, Tamura Y, Sesaki H, Jensen RE (2008). Mgr3p and Mgr1p are adaptors for the mitochondrial i-AAA protease complex. Mol. Biol Cell. 19, 5387–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echavarria NG, Echeverria LE, Stewart M, Gallego C, Saldarriaga C (2019). Chagas Disease: Chronic Chagas Cardiomyopathy. Curr. Probl. Cardiol 100507. [DOI] [PubMed] [Google Scholar]
- Eckers E, Cyrklaff M, Simpson L, Deponte M (2012) Mitochondrial protein import pathways are functionally conserved among eukaryotes despite compositional diversity of the import machineries. Biol. Chem 393, 513–524. [DOI] [PubMed] [Google Scholar]
- Effron PN, Torri AF, Engman DM, Donelson JE, Englund PT (1993). A mitochondrial heat shock protein from Crithidia fasciculata. Mol. Biochem. Parasitol 59, 191–200. [DOI] [PubMed] [Google Scholar]
- Gao F, Voncken F, Colasante C 2020. The mitochondrial phosphate carrier TbMCP11 is essential for mitochondrial function in the procyclic form of Trypanosoma brucei. Mol. Biochem. Parasitol 237, 111275. [DOI] [PubMed] [Google Scholar]
- Gebert N, Chacinska A, Wagner K, Guiard B, Koehler CM, Rehling P, Pfanner N, Wiedemann N (2008). Assembly of the three small Tim proteins precedes docking to the mitochondrial carrier translocase. EMBO Rep 9, 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnipova A, Subrtova K, Panicucci B, Horvath A, Lukes J, Zikova A (2015). The ADP/ATP carrier and its relationship to oxidative phosphorylation in ancestral protist Trypanosoma brucei. Eukaryot. Cell 14, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsman A, Niemann M, Pusnik M, Schmidt O, Burmann BM, Hiller S, Meisinger C, Schneider A, Wagner R (2012). Bacterial origin of a mitochondrial outer membrane protein translocase: new perspectives from comparative single channel electrophysiology. J. Biol. Chem 287, 31437–31445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsman A, Oeljeklaus S, Wenger C, Huot JL, Warscheid B, Schneider A (2016). The non-canonical mitochondrial inner membrane presequence translocase of trypanosomatids contains two essential rhomboid-like proteins. Nat. Commun 7, 13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman ML, Read LK (1999). Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem 274, 12067–12074. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Neupert W (2003). Protein insertion into the inner membrane of mitochondria. IUBMB Life 55, 219–225. [DOI] [PubMed] [Google Scholar]
- Hwang DK, Claypool SM, Leuenberger D, Tienson HL, Koehler CM (2007). Tim54p connects inner membrane assembly and proteolytic pathways in the mitochondrion. J. Cell Biol 178, 1161–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RE, Englund PT (2012). Network news: the replication of kinetoplast DNA. Annu Rev Microbiol 66, 473–491. [DOI] [PubMed] [Google Scholar]
- Kang Y, Baker MJ, Liem M, Louber J, McKenzie M, Atukorala I, Ang CS, Keerthikumar S, Mathivanan S, Stojanovski D (2016). Tim29 is a novel subunit of the human TIM22 translocase and is involved in complex assembly and stability. Elife 5. e17463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Holder J, Srinivasan M, Leung RS, Jensen RE (1997). The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J. Cell Biol 139, 1663–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Sepuri NB, Jensen RE (2000). Tim18p is a new component of the Tim54p-Tim22p translocon in the mitochondrial inner membrane. Mol. Biol. Cell 11, 103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler CM, Jarosch E, Tokatlidis K, Schmid K, Schweyen RJ, Schatz G (1998). Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279, 369–373. [DOI] [PubMed] [Google Scholar]
- Kolli R, Soll J, Carrie C (2018). Plant Mitochondrial Inner Membrane Protein Insertion. Int. J. Mol. Sci 19, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunji ER, Aleksandrova A, King MS, Majd H, Ashton VL, Cersonm E, Springett R, Kibalchenko M, Tavoulari S, Crichton PG, Ruprecht JJ (2016). The transport mechanism of the mitochondrial ADP/ATP carrier. Biochim. Biophys. Acta 1863, 2379–2393. [DOI] [PubMed] [Google Scholar]
- Kurz M, Martin H, Rassow J, Pfanner N, Ryan MT (1999). Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol. Biol. Cell 10, 2461–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landfear SM, Zilberstein D, (2019) Sensing What’s Out There - Kinetoplastid Parasites. Trends Parasitol 35, 274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Hulett JM, Lithgow T, Whelan J (2005). Protein import into mitochondria: origins and functions today (review). Mol. Membr. Biol 22, 87–100. [DOI] [PubMed] [Google Scholar]
- Lukes J, Hashimi H, Zikova A (2005). Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet 48, 277–299. [DOI] [PubMed] [Google Scholar]
- Makki A, Rada P, Zarsky V, Kereiche S, Kovacik L, Novotny M, Jores T, Rapaport D, Tachezy J (2019). Triplet-pore structure of a highly divergent TOM complex of hydrogenosomes in Trichomonas vaginalis. PLoS Biol 17, e3000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov DA, Opperdoes FR, Kostygov AY, Hashimi H, Lukes J, Yurchenko V (2019). Recent advances in trypanosomatid research: genome organization, expression, metabolism, taxonomy and evolution. Parasitology. 146, 1–27. [DOI] [PubMed] [Google Scholar]
- Ngere I, Gufu Boru W, Isack A, Muiruri J, Obonyo M, Matendechero S, Gura Z (2020). Burden and risk factors of cutaneous leishmaniasis in a peri-urban settlement in Kenya, 2016. PLoS One 15, e0227697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Brunner M (2002). The protein import motor of mitochondria. Nat Rev Mol Cell Biol. 3, 555–565. [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Ogata Y, Zikova A, Anupama A, Dalley RA, Acestor N, Myler PJ, Stuart K.D. (2009). A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9, 434–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Diaz P, Pelosi L, Ebikeme C, Colasante C, Gao F, Bringaud F, Voncken F (2012). Functional characterization of TbMCP5, a conserved and essential ADP/ATP carrier present in the mitochondrion of the human pathogen Trypanosoma brucei. J. Biol. Chem 287, 41861–41874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest JW, Wood ZA, Hajduk SL (1993). Cytochromes c1 of kinetoplastid protozoa lack mitochondrial targeting presequences. Biochim. Biophys. Acta 1144, 229–231. [DOI] [PubMed] [Google Scholar]
- Pyrihova E, Motyckova A, Voleman L, Wandyszewska N, Fiser R, Seydlova G, Roger A, Kolisko M, Dolezal P (2018). A Single Tim Translocase in the Mitosomes of Giardia intestinalis Illustrates Convergence of Protein Import Machines in Anaerobic Eukaryotes. Genome Biol. Evol 10, 2813–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht JJ, Hellawell AM, Harding M, Crichton PG, McCoy AJ, Kunji ER (2014). Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc. Natl. Acad. Sci. U S A 111, e426–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C (2010). Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol 11, 655–667. [DOI] [PubMed] [Google Scholar]
- Schneider A (2018). Mitochondrial protein import in trypanosomatids: Variations on a theme or fundamentally different? PLoS Pathog 14, e1007351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singha UK, Sharma S, Chaudhuri M (2009). Downregulation of mitochondrial porin inhibits cell growth and alters respiratory phenotype in Trypanosoma brucei. Eukaryot. Cell 8, 1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singha UK, Peprah E, Williams S, Walker R, Saha L, Chaudhuri M (2008). Characterization of the mitochondrial inner membrane protein translocator Tim17 from Trypanosoma brucei. Mol. Biochem. Parasitol 159, 30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singha UK, Hamilton V, Duncan MR, Weems E, Tripathi MK, Chaudhuri M (2012). Protein translocase of mitochondrial inner membrane in Trypanosoma brucei. J. Biol. Chem 287, 14480–14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singha UK, Hamilton V, Chaudhuri M (2015). Tim62, a Novel Mitochondrial Protein in Trypanosoma brucei, Is Essential for Assembly and Stability of the TbTim17 Protein Complex. J. Biol. Chem 290, 23226–23239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JT Jr., Singha UK, Misra S, Chaudhuri M (2018). Divergent Small Tim Homologues Are Associated with TbTim17 and Critical for the Biogenesis of TbTim17 Protein Complexes in Trypanosoma brucei. mSphere 3, e00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg JM, Maclean L (2010). A spectrum of disease in human African trypanosomiasis: the host and parasite genetics of virulence. Parasitology. 137, 2007–2015. [DOI] [PubMed] [Google Scholar]
- Wagner K, Gebert N, Guiard B, Brandner K, Truscott KN, Wiedemann N, Pfanner N, Rehling P (2008). The assembly pathway of the mitochondrial carrier translocase involves four preprotein translocases. Mol. Cell Biol 28, 4251–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger C, Oeljeklaus S, Warscheid B, Schneider A, Harsman A (2017). A trypanosomal orthologue of an intermembrane space chaperone has a non-canonical function in biogenesis of the single mitochondrial inner membrane protein translocase. PLoS Pathog 13, e1006550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weems E, Singha UK, Hamilton V, Smith JT Jr., Waegemann K, Mokranjac D, Chaudhuri M (2015). Functional complementation analyses reveal that the single PRAT family protein of Trypanosoma brucei is a divergent homolog of Tim17 in saccharomyces cerevisiae. Eukaryot. Cell. 14, 286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weems E, Singha UK, Smith JT Jr., Chaudhuri M (2017). The divergent N-terminal domain of Tim17 is critical for its assembly in the TIM complex in Trypanosoma brucei. Mol. Biochem. Parasitol 218, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA (1999). A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol 99, 89–101. [DOI] [PubMed] [Google Scholar]
- Zhang X, Cui J, Nilsson D, Gunasekera K, Chanfon A, Song X, Wang H, Xu Y, Ochsenreiter T (2010). The Trypanosoma brucei MitoCarta and its regulation and splicing pattern during development. Nucleic Acids Res 38, 7378–7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.