Abstract
Objective
Our study investigated the presence of regional differences in hexanucleotide repeat number in postmortem brain tissues of 2 patients with X-linked dystonia-parkinsonism (XDP), a combined dystonia-parkinsonism syndrome modified by a (CCCTCT)n repeat within the causal SINE-VNTR-Alu retrotransposon insertion in the TAF1 gene.
Methods
Genomic DNA was extracted from blood and postmortem brain samples, including the basal ganglia and cortex from both patients and from the cerebellum, midbrain, and pituitary gland from 1 patient. Repeat sizing was performed using fragment analysis, small-pool PCR-based Southern blotting, and Oxford nanopore sequencing.
Results
The basal ganglia (p < 0.001) and cerebellum (p < 0.001) showed higher median repeat numbers and higher degrees of repeat instability compared with blood.
Conclusions
Somatic repeat instability may predominate in brain regions selectively affected in XDP, thereby hinting at its potential role in disease manifestation and modification.
X-linked dystonia-parkinsonism (XDP) is an adult-onset neurodegenerative disorder characterized by rapidly progressive dystonia and parkinsonism.1 Unique to individuals of Filipino descent, XDP is caused by a single founder mutation: a SINE-VNTR-Alu (SVA) retrotransposon insertion in the TAF1 gene.2,3 This change downregulates the expression of TAF1,4,5 likely leading to basal ganglia degeneration.6 Consequently, XDP has long been considered a model disorder to study how basal ganglia pathology leads to dystonia and parkinsonism.6 However, the notion of exclusive striatal atrophy in this movement disorder has been recently challenged by neuroimaging studies proposing that other brain regions such as the cortex and the cerebellum may be affected as well.7 Of note, patients with XDP exhibit a range of nonmotor features, including cognitive impairment, anxiety, and depression.8
Recent studies suggest that XDP shares genetic similarities with microsatellite repeat expansion disorders such as Huntington disease (HD), several forms of spinocerebellar ataxia, and more than 20 other human diseases.9,10 Namely, it has been reported that the length of the polymorphic (CCCTCT)n domain within the TAF1 SVA retrotransposon acts as a genetic modifier of disease expressivity in XDP. The longer the (CCCTCT)n repeat, the more severe the disease, and the earlier the age at onset.9,10 The hexanucleotide repeat also showed somatic repeat variability,10 raising the possibility that higher RNs in specific brain tissues may contribute to selective neurodegeneration. Hence, this study was undertaken to investigate in greater detail whether regional differences in (CCCTCT)n RN reflect the pattern of neuronal loss seen in XDP.
Methods
Standard Protocol Approvals, Registrations, and Patient Consents
This study was approved by the Ethics Committee of the University of Lübeck, Germany. Postmortem brain samples from the abovementioned patients were used in this investigation in accordance with the “Gesetz über das Leichen-, Bestattungs- und Friedhofswesen (Bestattungsgesetz) des Landes Schleswig-Holstein vom 04.02.2005, Abschnitt II, § 9 (Leichenöffnung, anatomisch).” In this case, it is allowed to dissect the bodies of the patients (Körperspender/in) for scientific and/or educational purposes without explicit prior approval from patients or their legal representatives. The first patient (L-10322) died in the Philippines, and ethics approval was obtained at the Philippine Children's Medical Center, Department of Pathology. The second patient (L-7995) died in Lübeck, Germany, and 2 family members (1 sister and 1 brother) provided written and signed consent with Ethics Committee approval.
Genetic Analyses
To further investigate the link between RN differences and neurodegeneration in XDP, we extracted genomic DNA from blood and postmortem brain samples from 2 affected individuals and performed hexanucleotide repeat sizing with 4 different methods: (1) fragment analysis,10 (2) small-pool PCR (SP-PCR) coupled with Southern blotting, (3) Cas9-targeted nanopore sequencing, and (4) deep nanopore sequencing of the SVA amplicon. Genomic DNA was extracted using the QIAamp DNA Mini Kit and Blood and Cell Culture DNA Midi Kit (Qiagen, Hilden, Germany) for SP-PCR-based Southern blotting and nanopore sequencing, respectively, following the manufacturer's prescribed procedure. Oxford nanopore sequencing permits the detection of single molecules of DNA without the need for PCR amplification.11 We thus performed long-read sequencing by enriching the SVA insertion using the Nanopore Cas9-targeted sequencing protocol.12 Long-read sequencing was applied only to basal ganglia (striatum) and cerebellum samples from L-7995.
SP-PCR–Based Southern Blotting
SP-PCR is an efficient and sensitive method for accurately measuring the heterogeneity of RNs in specific tissues in the absence of artifacts.13 In brief, 30-mg tissue fragments derived from 2 (cortex and basal ganglia) and 5 (cortex, basal ganglia, cerebellum, pituitary gland, and midbrain) brain regions were dissected from L-10322 and L-7995, respectively. For patient L-10322, DNA from the basal ganglia was isolated from the caudate, anterior putamen, posterior putamen, globus pallidus internus, and globus pallidus externus. For the cortex, DNA from the frontal and temporal cortices was used. Similarly, for patient L-7995, DNA from the basal ganglia was extracted from the striatum, globus pallidus internus, and globus pallidus externus. For the cortex, DNA from the frontal and oculomotor cortices was used.
Brain tissues from these individuals were previously observed to exhibit somatic mosaicism.10 Hence, for each brain region, 75 pg of genomic DNA derived from ∼10 cells was used as template for a previously established long-range PCR reaction amplifying the TAF1 SVA insertion (eTable 1, links.lww.com/NXG/A435).14 The said concentration was used to rapidly assess the diversity of RNs per tissue while still avoiding the drawbacks of bulk PCR.13 Then, PCR products were digested with the restriction enzymes BsaI, HindIII, HaeIII, and MseI (New England Biolabs, Ipswich, MA) to generate a 458 + (n × 6) bp fragment (n = number of repeats) containing the hexanucleotide repeat region and adjacent base pairs. This digestion mixture was electrophoresed on a denaturing 7% polyacrylamide gel and then transferred to a positively charged nylon membrane (GE Healthcare, Buckinghamshire, United Kingdom) by overnight capillary blotting. After prehybridization, (AGAGGG)6 probes, 3′ labeled with digoxigenin (DIG) using the DIG Oligonucleotide 3′-End Labeling Kit (Roche Applied Science, Mannheim, Germany), were hybridized overnight at 42°C.
All hybridization, washing, and detection procedures were performed following the DIG Application Manual (Roche Applied Science, Mannheim, Germany). The ChemiDoc MP Imaging System (Bio-Rad Laboratories, Hercules, CA) was used for visualizing the chemiluminescent signal. Fragments amplified were sized using custom-made markers containing 30, 35, 40, 45, 50, and 55 hexanucleotide repeats. The marker bands were generated using the aforementioned procedures except that bulk genomic DNA (10 ng) from the previously genotyped patients was used as template. The use of such markers ensured that the hybridization probes were specific to the repeat-bearing fragments and the observed bands with different sizes were not a result of PCR artifacts or DNA degradation.
Oxford Nanopore Sequencing
Customizable crRNAs guided the Cas9 ribonucleoprotein complex to cleave out the region of interest. The resulting blunt ends with 5′ phosphates were used for the specific ligation of the sequencing adapters to the region of interest.12 Hence, for library preparation, 4 CRISPR–Cas9 crRNAs were designed with the ChopChop online tool15 (chopchop.cbu.uib.no): 2 upstream (genomic location GRCh38, [target sequence] chrX:70,657,572 [AAACTTCCCCCGATCCTGCTTGG], chrX:70,658,413 [GTCCAGTCTACCAAGTAAACAGG]) and 2 downstream (genomic location [target sequence] chrX:70,660,444 [GACGTAAGTGTGACGACATGGG], chrX:70,660,445 [TGACGTAAGTGTGACGACA TTGG]) of the SVA insertion (eFigure 1, links.lww.com/NXG/A435). For the Cas9-targeted nanopore sequencing, 4 flow cells (R9.4.1) loaded with 5 libraries (4 × 5 µg and 1 × 1 µg of DNA) were used for blood. Three flow cells (R9.4.1) loaded with 5 libraries (4 × 5 µg and 1 × 10 µg of DNA) were used for the cerebellum. Four flow cells (R9.4.1) loaded with 5 libraries (3 × 3 µg, 1 × 2 µg, and 1 × 1 µg of DNA) were used for the basal ganglia (striatum). In addition, 1 flow cell with 1 µg of barcoded PCR product was run for each tissue. The locations of the guide RNAs used for Cas9-targeted enrichment and the PCR primer for deep nanopore sequencing are shown in eFigure 1. The Cas9 excision resulted in a 5.5-kb product, including the 2.6-kb SVA insertion. The LSK109 ligation sequencing kit (Oxford Nanopore Technologies, Oxford, United Kingdom) was used for library preparation without barcoding, and subsequent sequencing was performed on the R9.4.1 flow cell on the MinION. Base calling was performed with integrated Guppy algorithm in MinKNOW (version 2.2.3) on the MinIT. For Cas9-targeted enrichment, we obtained 148,975 reads on average per sample (blood-derived DNA: 107,138; basal ganglia–derived DNA: 178,449; and cerebellum-derived DNA: 161,284) and 1,630 reads that were mapped to the target with at least 1 kb of alignment length (blood-derived DNA: 1,081; basal ganglia–derived DNA: 1,133; and cerebellum-derived DNA: 2,677). Thus, the proportion of reads that mapped to the target was on average 1.1% per sample.
Separately, a 3.1-kb amplicon product of the SVA was generated with long-range PCR with the use of primers, as previously published.14 For this 3.1-kb PCR amplicon sequencing, 1 µg of DNA per product was used, and all samples were multiplexed together on 1 flow cell (R9.4.1) with the native barcoding kit (EXP-NBD104). After multiplex barcoding the PCR products, the LSK109 ligation sequencing kit was used for library preparation, and nanopore sequencing was performed with the MinION and the R9.4.1 flow cell. Base calling was performed with integrated Guppy algorithm in MinKNOW (version 2.2.3) on the MinIT.
For the PCR amplicon, the proportion of reads that mapped to the region of interest was larger for the sequenced amplicons that were generated for the deep nanopore sequencing. We obtained 98,883 reads on average per sample (blood-derived DNA: 129,463; basal ganglia–derived DNA: 49,704; and cerebellum-derived DNA: 117,482) and 56,695 reads that were mapped to the target with at least 1 kb of alignment length (blood-derived DNA: 78,438; basal ganglia–derived DNA: 29,725; and cerebellum-derived DNA: 61,923) resulting in a proportion of 57.7%.
To analyze all sequence data with NCRF, the FASTQ files were aligned to the reference (hg19 including the SVA insertion) FASTA file with minimap2 (version 2.17). To extract those reads that were on target, a BAM file was generated that only contained reads that had an alignment length of more than 1 kb with samtools (version 1.3.1). Subsequently, the BAM file was converted to FASTQ format with samtools as well. As NCRF requires the FASTA format as an input, the FASTQ file was converted. To optimize the NCRF analysis, we set the maximum mismatch ratio to 20% and the minimum query length to 108 bp, which equals a repeat count of 18. The script used in this study is available on GitHub (github.com/nanopol/xdp_sva/). Further filtering with the NCRF software tool resulted in 1,137 reads per sample (blood-derived DNA: 721; basal ganglia–derived DNA: 759; and cerebellum-derived DNA: 1,932) for the Cas9-targeted enrichment. After the performed NCRF filtering, 60,589 reads remained (blood-derived DNA: 83,470; basal ganglia–derived DNA: 31,544; and cerebellum-derived DNA: 66,754).
Statistical Analyses
Both Southern blotting and long-read sequencing were used to assess the RNs. We compared RNs derived from different tissues from the respective same individual by statistical tests. RNs per tissue showed deviations from a symmetric normal distribution and had different variances. As a result, the Mood's median test was applied to test whether the median RNs differ between tissues. To account for multiple comparisons of varying brain tissues with blood, the overall significance level of α = 0.05 per patient was split up to 0.05/2 = 0.025 for patient L-10322 and 0.05/5 = 0.01 for patient L-7995. Boxplots were used to show the distribution of RNs per tissue, whereas quartile coefficients of dispersions (QCDs) and corresponding 95% confidence intervals were calculated to describe the interquartile range relative to the distribution's median.
Data Availability
The scripts for the Cas9 enrichment and amplicon runs on the nanopore, FASTA files are available on a GitHub repository (github.com/nanopol/xdp_sva/).
Results
Clinical Description of the Patients
The first patient (L-10322), a former seaman, developed first symptoms at age 50 years. At age 56 years, he presented with a parkinsonian phenotype, including drooling, dysphagia, resting tremor of the right hand, and loss of balance when walking. He was treated with clonazepam and biperiden. In addition to XDP, he was diagnosed with pulmonary fibrosis at age 50 years. He died at age 57 years.
The second patient (L-7995), a former customs employee, initially manifested with blepharospasm at age 31 years. Dystonia rapidly progressed in this patient and became generalized within a few years. On neurologic examination, he showed severe generalized dystonia, mostly pronounced in the upper half of the body (the Burke-Fahn-Marsden Dystonia Rating Scale score: 69/120). In addition, he had parkinsonian signs, including limb rigidity, but dystonia was clearly prominent. At age 35 years, he died in a cachexic state from pneumogenic sepsis due to bacterial aspiration pneumonia and concurrent pulmonary tuberculosis.
Genetic Findings
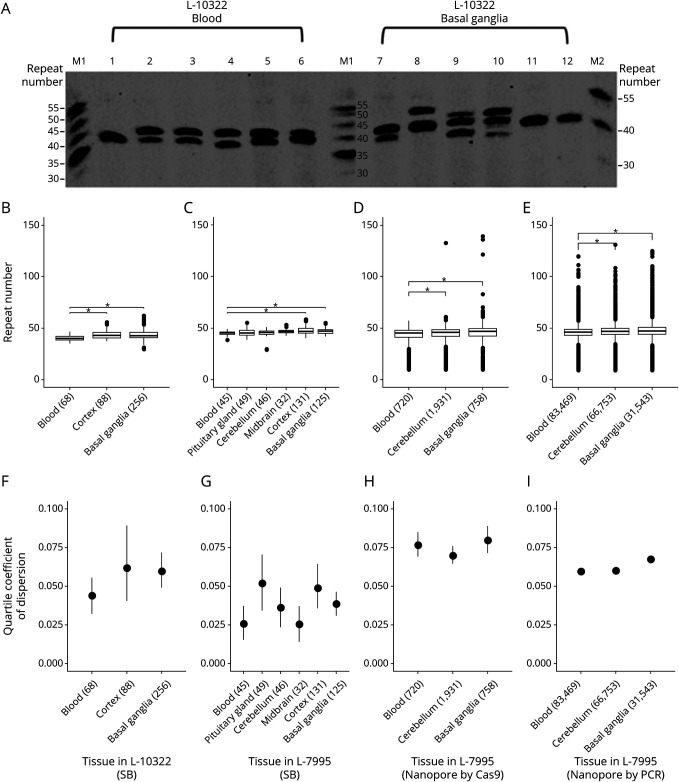
Our standard fragment analysis identified 41 and 45 (CCCTCT)n repeats in the blood-derived DNA from L-10322 and L-7995, respectively. However, the SP-PCR–based Southern blot analysis revealed heterogeneity across RNs in all tissues examined from both individuals (Figure 1A). In particular, median RNs were significantly higher in the basal ganglia (RN = 42.56, p = 4 × 10−7) and cortex (RN = 42.85, p = 1.83 × 10−4) compared with blood (RN = 39.94) from L-10322 (Figure 1B). This observation was consistent with RN counts in L-7995, who also exhibited higher median RNs in the aforementioned brain regions (RN = 46.98, p = 4.31 × 10−4 and RN = 46.78, p = 4.67 × 10−4 for the basal ganglia and cortex, respectively) compared with blood (RN = 45.30) (Figure 1C). Furthermore, although no significant difference in median RNs was observed between the other brain areas and blood from L-7995, a slight tendency toward higher median RNs was found in the cerebellum (RN = 45.67) and midbrain (RN = 46.54). These findings were consistent with the results of nanopore sequencing after Cas9 enrichment for L-7995. With Cas9-targeted enrichment and nanopore sequencing, we obtained >700 single-molecule reads for each sample type. This also showed higher median RNs in the basal ganglia (RN = 46.8, p < 0.0001) and cerebellum (RN = 45.8, p = 0.0002) compared with blood (RN = 45.2) for L-7995 (Figure 1D). In addition, long-read deep sequencing of the amplicon encompassing the SVA detected n > 31,000 single-molecule reads for each tissue type and also showed higher median RNs in the basal ganglia (RN = 49.3, p < 0.0001) and cerebellum (RN = 49.0, p < 0.0001) compared with blood (RN = 48.2) (Figure 1E).
Figure 1. Number and Instability of (CCCTCT)n Repeats in Blood and Different Brain Regions.
(A) Representative Southern blot image illustrating repeat number (RN) heterogeneity in the blood (lanes 1–6) and basal ganglia (lanes 7–12) of L-10322. Each lane represents 1 small-pool PCR reaction using DNA from the corresponding tissue as template. Custom-made size markers in lanes M1 and M2 show the sizes of alleles in RNs. Boxplots showing median RNs for (B) L-10322 and (C) L-7995 measured using Southern blotting (SB) and (D) L-7995 with Oxford Nanopore long-read sequencing after Cas9 enrichment (E) L-7995 measured with PCR amplification of the SVA and subsequent Oxford Nanopore deep long-read sequencing. Forest plots presenting repeat instability measured as the quartile coefficient of dispersion (QCD) in (F) L-10322 and (G) L-7995 based on SB and (H) L-7995 with Nanopore sequencing with Cas9 enrichment (I) L-7995 with Nanopore sequencing of PCR product. Parentheses indicate the number of bands or alleles measured for each tissue. Asterisks indicate significant differences between samples.
Correspondingly, based on Southern blotting, the basal ganglia and cortex showed higher QCDs (QCD = 0.060 and QCD = 0.062, respectively) compared with blood in L-10322 (QCD = 0.044) (Figure 1F), whereas all brain regions, except the midbrain (QCD = 0.026), exhibited higher QCDs compared with blood (QCD = 0.026) in L-7995 (Figure 1G). Cas9-targeted sequencing also showed higher QCD for the basal ganglia (QCD = 0.080) compared with blood in L-7995 (QCD = 0.077) (Figure 1H). However, the cerebellum had a lower QCD (QCD = 0.070) compared with blood. The PCR amplicons and deep sequencing showed higher QCD for the basal ganglia and cerebellum compared with blood (QCD = 0.070, 0.063, and 0.062, respectively) (Figure 1I).
Discussion
In addition to significant striatal volume loss in the basal ganglia,6 patients with XDP show gray matter volume loss in the cerebellum and a mild reduction in cortical thickness that has been linked to cognitive dysfunction.7 Thus, it is remarkable that in our study, both the basal ganglia and the cortex showed higher median RNs and higher degrees of repeat instability compared with blood in the 2 patients based on SP-PCR-based Southern blotting. Nanopore-based long-read sequencing also detected a similar tendency of the repeats to expand in the basal ganglia and the cerebellum of 1 patient. In HD, the brain regions that exhibited the most evident neuropathology were also reported to have the most pronounced mosaicism.16 Thus, our finding of the presence of higher median RNs in the basal ganglia, cortex, and cerebellum may suggest a role for somatic mosaicism in disease manifestation and modification. In keeping with this notion, somatic repeat instability in the cortex was found to be an age at onset modifier in HD.17 Furthermore, genetic and pharmacologic inhibition of somatic repeat expansions significantly delays disease onset and rescues progressive motor dysfunction in a transgenic mouse model of HD.18
Our approach to analyze somatic repeat instability in different tissues entailed a number of technical limitations. First, 75 pg of genomic DNA was used as template to rapidly evaluate the diversity of RNs per tissue.13 However, in the Southern blots, the empirical determination of average number of amplifiable molecules for each sample (i.e., calculation of genome equivalents) was not performed, which affects the estimation of degree of somatic instability. Nevertheless, we used long-read sequencing to detect single molecules.12 From this analysis, we obtained the same median RN as Southern blot analysis with the Cas9-targeted enrichment. Deep amplicon-based sequencing was performed to further validate the repeat expansion in the brain tissue, while considering the caveat of preferential amplification during PCR. Still, the range in RNs was striking and more variable than Southern blot data (eFigure 2, links.lww.com/NXG/A435), likely due to noise from the raw squiggles.19–21 The accuracy of nanopore sequencing can be hampered by errors from indels and homopolymers.19 As better bioinformatic tools and training mechanisms are developed over time, the accuracy of long-read sequencing technologies will improve. In fact, nanopore sequencing has already been used to investigate the size and methylation status of unstable microsatellite repeat expansions.20,22,23 The robustness of our findings is strengthened by the simultaneous comparison between Southern blotting and nanopore sequencing on the TAF1 SVA hexanucleotide repeat in XDP patients.
In conclusion, our work extends the utility of long-read sequencing to repeat instability, helping us provide further evidence of brain region–specific instability of the (CCCTCT)n repeat in XDP. Our study of 2 independently analyzed patients is currently unable to determine the exact contribution of somatic repeat expansions to the characteristic pattern of neuronal death seen in XDP. Nevertheless, our work indicates that somatic mosaicism may predominate in brain regions selectively affected in the disease, thereby hinting at its vital role in modifying the yet-unresolved, repeat-mediated pathogenic process leading to neurodegeneration. This finding holds possible translational significance as it highlights the potential of somatic repeat instability as a viable therapeutic target for this emerging repeat expansion disorder.
Acknowledgment
The authors acknowledge Dr. Lillian V. Lee, Dr. Madelyn Pascual, Dr. Mark Angelo C. Ang, Dr. Aloysius Domingo, Dr. Ivy Lique, Dr. Edwin Muñoz, Atty. Geraldine Acuña-Sunshine, and the Sunshine Care Foundation for referral of patients and clinical information. They also thank Ms. Heike Pawlack for the technical assistance.
Glossary
- DIG
digoxigenin
- HD
Huntington disease
- QCD
quartile coefficients of dispersion
- RN
repeat number
- SP-PCR
small-pool PCR
- SVA
SINE-VNTR-Alu
- XDP
X-linked dystonia-parkinsonism
Appendix. Authors
Contributor Information
Charles Jourdan Reyes, Email: charlesreyes24@gmail.com.
Björn-Hergen Laabs, Email: laabs@imbs.uni-luebeck.de.
Susen Schaake, Email: susen.schaake@neuro.uni-luebeck.de.
Theresa Lüth, Email: theresa.lueth@gmx.de.
Raphaela Ardicoglu, Email: rapha.ardico@gmail.com.
Aleksandar Rakovic, Email: aleksandar.rakovic@neuro.uni-luebeck.de.
Karen Grütz, Email: karen.gruetz@neuro.uni-luebeck.de.
Daniel Alvarez-Fischer, Email: daniel.alvarez@neuro.uni-luebeck.de.
Roland Dominic Jamora, Email: rgjamora@up.edu.ph.
Raymond L. Rosales, Email: rlrosalesmd88@gmail.com.
Imke Weyers, Email: weyers@anat.uni-luebeck.de.
Inke R. König, Email: inke.koenig@imbs.uni-luebeck.de.
Norbert Brüggemann, Email: norbert.brueggemann@neuro.uni-luebeck.de.
Christine Klein, Email: christine.klein@neuro.uni-luebeck.de.
Valerija Dobricic, Email: valerija.dobricic@uni-luebeck.de.
Ana Westenberger, Email: ana.westenberger@neuro.uni-luebeck.de.
Study Funding
The study was supported by the Deutsche Forschungsgemeinschaft (DFG FOR2488; to A.R., I.R.K., N.B., A.W., C.K., and J.T.), by funding by the Collaborative Center for X-linked Dystonia-Parkinsonism at Massachusetts General Hospital (to N.B. and C.K.), the Else-Kröner Fresenius Foundation (to N.B. and J.T.), by intramural funds from the University of Lübeck (to C.K. and J.T.), career development award from Peter Engelhorn Foundation (to J.T.), and by a career development award from the Hermann and Lilly Schilling Foundation (to C.K.). C.J. Reyes is supported by a PhD scholarship from the Katholischer Akademischer Ausländer-Dienst.
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org/NG for full disclosures.
References
- 1.Lee LV, Rivera C, Teleg RA, et al. The unique phenomenology of sex-linked dystonia parkinsonism (XDP, DYT3, ‘Lubag’). Int J Neurosci. 2011;121(suppl 1):3-11. [DOI] [PubMed] [Google Scholar]
- 2.Domingo A, Westenberger A, Lee LV, et al. New insights into the genetics of X-linked dystonia-parkinsonism (XDP, DYT3). Eur J Hum Genet. 2015;23(10):1334-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makino S, Kaji R, Ando S, et al. Reduced neuron-specific expression of the TAF1 gene is associated with X-linked dystonia-parkinsonism. Am J Hum Genet. 2007;80(3):393-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aneichyk T, Hendriks WT, Yadav R, et al. Dissecting the causal mechanism of X-linked dystonia-parkinsonism by integrating genome and transcriptome assembly. Cell. 2018;172(5):897-909.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakovic A, Domingo A, Grütz K, et al. Genome editing in induced pluripotent stem cells rescues TAF1 levels in X-linked dystonia-parkinsonism. Mov Disord. 2018;33(7):1108-1118. [DOI] [PubMed] [Google Scholar]
- 6.Goto S, Lee LV, Munoz EL, et al. Functional anatomy of the basal ganglia in X-linked recessive dystonia-parkinsonism. Ann Neurol. 2005;58(1):7-17. [DOI] [PubMed] [Google Scholar]
- 7.Hanssen H, Heldmann M, Prasuhn J, et al. Basal ganglia and cerebellar pathology in X-linked dystonia-parkinsonism. Brain. 2018;141(10):2995-3008. [DOI] [PubMed] [Google Scholar]
- 8.Jamora RDG, Ledesma LK, Domingo A, Cenina ARF, Lee LV. Nonmotor features in sex-linked dystonia parkinsonism. Neurodegener Dis Manag. 2014;4(3):283-289. [DOI] [PubMed] [Google Scholar]
- 9.Bragg DC, Mangkalaphiban K, Vaine CA, et al. Disease onset in X-linked dystonia-parkinsonism correlates with expansion of a hexameric repeat within an SVA retrotransposon in TAF1. Proc Natl Acad Sci USA. 2017;114(51):E11020-E11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westenberger A, Reyes CJ, Saranza G, et al. A hexanucleotide repeat modifies expressivity of X‐linked dystonia parkinsonism. Ann Neurol. 2019;85(6):812-822. [DOI] [PubMed] [Google Scholar]
- 11.Ameur A, Kloosterman WP, Hestand MS. Single-molecule sequencing: towards clinical applications. Trends Biotechnol. 2019;37(1):72-85. [DOI] [PubMed] [Google Scholar]
- 12.Gilpatrick T, Lee I, Graham JE, et al. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat Biotechnol. 2020;38(4):433-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes-Pereira M, Bidichandani SI, Monckton DG. Analysis of unstable triplet repeats using small-pool polymerase chain reaction. Methods Mol Biol. 2004;277:61-76. [DOI] [PubMed] [Google Scholar]
- 14.Kawarai T, Pasco PMD, Teleg RA, et al. Application of long-range polymerase chain reaction in the diagnosis of X-linked dystonia-parkinsonism. Neurogenetics. 2013;14(2):167-169. [DOI] [PubMed] [Google Scholar]
- 15.Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47(W1):W171-W174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Telenius H, Kremer B, Goldberg YP, et al. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet. 1994;6(4):409-414. [DOI] [PubMed] [Google Scholar]
- 17.Swami M, Hendricks AE, Gillis T, et al. Somatic expansion of the Huntington's disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum Mol Genet. 2009;18(16):3039-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budworth H, Harris FR, Williams P, et al. Suppression of somatic expansion delays the onset of pathophysiology in a mouse model of Huntington's disease. PLoS Genet. 2015;11(8):1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris RS, Cechova M, Makova KD. Noise-cancelling repeat finder: uncovering tandem repeats in error-prone long-read sequencing data. Bioinformatics. 2019;35(22):4809-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giesselmann P, Brändl B, Raimondeau E, et al. Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nat Biotechnol. 2019;37(12):1478-1481. [DOI] [PubMed] [Google Scholar]
- 21.Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev Genet. 2020;21(10):597-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebbert MTW, Farrugia SL, Sens JP, et al. Long-read sequencing across the C9orf72 ‘GGGGCC’ repeat expansion: implications for clinical use and genetic discovery efforts in human disease. Mol Neurodegener. 2018;13(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Roeck A, Duchateau L, Van Dongen J, et al. An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer's disease. Acta Neuropathol. 2018;135(6):827-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The scripts for the Cas9 enrichment and amplicon runs on the nanopore, FASTA files are available on a GitHub repository (github.com/nanopol/xdp_sva/).