ABSTRACT
The emergence of carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates in Egyptian hospitals has been reported. However, the genetic basis and analysis of the plasmids associated with carbapenem-resistant hypervirulent K. pneumoniae (CR-HvKP) in Egypt have not been presented. Therefore, we attempted to decipher the plasmid sequences that are responsible for transferring the determinants of carbapenem resistance, particularly blaNDM-1 and blaKPC-2. Out of 34 K. pneumoniae isolates collected from two tertiary hospitals in Egypt, 31 were CRKP. Whole-genome sequencing revealed that our isolates were related to 13 different sequence types (STs). The most prevalent ST was ST101, followed by ST383 and ST11. Among the CRKP isolates, one isolate named EBSI036 has been reassessed by Nanopore sequencing. Genetic environment analysis showed that EBSI036 carried 20 antibiotic resistance genes and was identified as a CR-HvKP strain: it harbored four plasmids, namely, pEBSI036-1-NDM-VIR, pEBSI036-2-KPC, pEBSI036-3, and pEBSI036-4. The two carbapenemase genes blaNDM-1 and blaKPC-2 were located on plasmids pEBSI036-1-NDM-VIR and pEBSI036-2-KPC, respectively. The IncFIB:IncHI1B hybrid plasmid pEBSI036-1-NDM-VIR also carried some virulence factors, including the regulator of the mucoid phenotype (rmpA), the regulator of mucoid phenotype 2 (rmpA2), and aerobactin (iucABCD and iutA). Thus, we set out in this study to analyze in depth the genetic basis of the pEBSI036-1-NDM-VIR and pEBSI036-2-KPC plasmids. We report a high-risk clone ST11 KL47 serotype of a CR-HvKP strain isolated from the blood of a 60-year-old hospitalized female patient from the intensive care unit (ICU) in a tertiary care hospital in Egypt, which showed the cohabitation of a novel hybrid plasmid coharboring the blaNDM-1 and virulence genes and a blaKPC-2-carrying plasmid.
IMPORTANCE CRKP has been registered in the critical priority tier by the World Health Organization and has become a significant menace to public health. The emergence of CR-HvKP is of great concern in terms of both disease and treatment. In-depth analysis of the carbapenemase-encoding and virulence plasmids may provide insight into ongoing recombination and evolution of virulence and multidrug resistance in K. pneumoniae. Thus, this study serves to alert contagious disease clinicians to the presence of hypervirulence in CRKP isolates in Egyptian hospitals.
KEYWORDS: Klebsiella pneumoniae, NDM-1, KPC-2, hybrid plasmid, virulent plasmid, Egypt
OBSERVATION
Several studies have reported the emergence of carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates in Egyptian hospitals (1–4); however, to the best of our knowledge, the genetic basis and analysis of the plasmids associated with CR-hypervirulent K. pneumoniae (CR-HvKP) in Egypt have not been presented. Furthermore, carbapenem resistance has been reported to be associated with increased length of hospital stay and mortality of bloodstream infection (BSI) patients in low- and middle-income countries (5). Therefore, we sought to analyze in depth the genetic basis of pEBSI036-1-NDM-VIR (a novel hybrid plasmid harboring blaNDM-1 and virulence genes) and pEBSI036-2-KPC (a blaKPC-2-carrying plasmid), which have been identified in a clinical K. pneumoniae strain from a blood sample of a patient in Egypt.
A total of 34 nonduplicate K. pneumoniae isolates were recovered from the blood of hospitalized patients in two tertiary care hospitals, namely, El-Demerdash Hospital (Cairo, Egypt) and the National Cancer Institute (Cairo, Egypt), in the period between June 2017 and March 2018 as a part of a study for the monitoring of antimicrobial resistance. Our isolates were selected based on their clinical characteristics, where all of them were primarily identified by Vitek 2 and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) as K. pneumoniae causing bloodstream infections (BSIs), among which 31 were confirmed phenotypically and genotypically as CRKP isolates. Overall, the 34 isolates were isolated from the blood of 55.9% (19/34) female and 44.1% (15/34) male hospitalized patients from 9 days to 75 years of age. MICs of all 34 isolates were determined for 17 antibiotics using the agar microdilution method according to CLSI (6), except for tigecycline and colistin, for which MICs were obtained by the broth microdilution method according to EUCAST (7). Out of 34 isolates, 91.2% (31/34) were resistant to ertapenem, while 73.5% (25/34) and 61.8% (21/34) were resistant to imipenem and meropenem, respectively. However, all isolates were susceptible to colistin.
All isolates were assessed by whole-genome sequencing (WGS) using an Illumina HiSeq 2000 platform. In silico multilocus sequence typing showed that our isolates belong to 13 different sequence types (STs). The most prevalent ST was ST101 (13/34 [38.2%]), followed by ST383 (5/34 [14.7%]). One isolate, EBSI036, belongs to ST11: ST11 is the dominant ST clone responsible for the prevalence of CRKP worldwide and is considered an emerging high-risk clone (1, 8–10). Among 31 CRKP isolates, the prevalence of carbapenemase genes blaNDM-1 and blaOXA-48 was 45.2% (14/31)—in addition, four strains carried both genes. Moreover, strain EBSI036 coharbors blaNDM-1 and blaKPC-2. According to the clinical data, the K. pneumoniae strain EBSI036 was isolated from the blood of a 60-year-old female patient 2 days after admission to the Gastroenterology Department of El-Demerdash Hospital with symptoms of pneumonia, diarrhea, and fever. The patient’s symptoms improved following the administration of intravenous ceftriaxone and colistin, and she was discharged from the hospital 8 days posthospitalization. With the further genome analysis of EBSI036, the plasmid-associated virulence determinants rmpA/rmpA2, iucABCD, and iutA in this strain were predicted using the Virulence Factor Database (VFDB [http://www.mgc.ac.cn/VFs/main.htm]). EBSI036 was determined as a KL47 capsular serotype by using Kaptive software (https://github.com/katholt/Kaptive). The serotype KL47 was the most reported type among CRKP infections in Asia (11–13). The virulence level of EBSI036 was confirmed using the Galleria mellonella larva model as previously described (11, 14) (see Fig. S1 in the supplemental material). These results revealed that EBSI036 is a CR-HvKP strain.
Virulence potential of K. pneumoniae strain EBSI036 as depicted in a Galleria mellonella infection model with an inoculum of 1 × 104 CFU. Download FIG S1, TIF file, 0.3 MB (296.4KB, tif) .
Copyright © 2021 Ahmed et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
As EBSI036 coharbors two carbapenem genes besides the plasmid-mediated virulence genes, we have further analyzed the characteristics of the related fully sequenced plasmids by using a long-read MinION sequencer (Oxford Nanopore Technologies, Oxford, United Kingdom). Genomic analysis showed that EBSI036 included a 5,513,124-bp chromosome and four plasmids, namely, pEBSI036-1-NDM-VIR (347,365 bp), pEBSI036-2-KPC (129,869 bp), pEBSI036-3 (10,060 bp), and pEBSI036-4 (5,596 bp) (see Table S1 in the supplemental material). Twenty antimicrobial resistance genes, including six β-lactamase-encoding genes, were identified in EBSI036 by using ABRicate version 0.5 (https://github.com/tseemann/abricate) and aligning genome sequences to the ResFinder database. Of these, blaSHV-11, oqxB, oqxA, and fosA6, were identified in the EBSI036 chromosome. The two carbapenemase genes blaNDM-1 and blaKPC-2 were located on plasmids pEBSI036-1-NDM-VIR and pEBSI036-2-KPC, respectively. In addition, 86 putative virulence genes were annotated in the genome of EBSI036, including genes coding for fimbriae, capsule, yersiniabactin, iron-enterobactin, mucoid, and aerobactin (see Table S2 in the supplemental material).
Overall features of the genome and minimum inhibitory concentrations (MICs) for K. pneumoniae EBSI036. Download Table S1, DOCX file, 0.03 MB (28.1KB, docx) .
Copyright © 2021 Ahmed et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Putative virulence genes detected on the K. pneumoniae EBSI036 chromosome. Download Table S2, XLS file, 0.04 MB (37.5KB, xls) .
Copyright © 2021 Ahmed et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
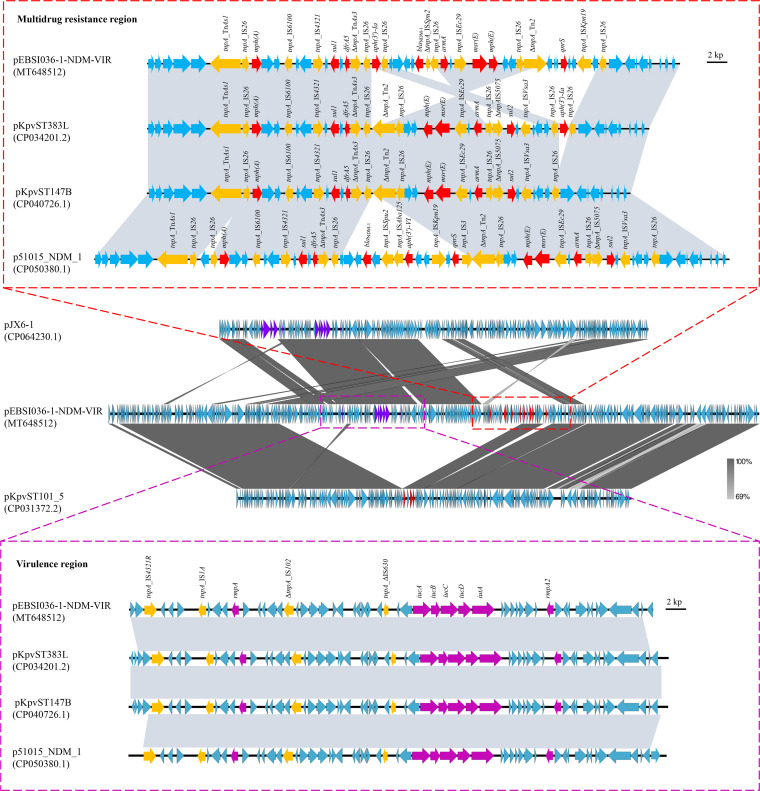
Hybrid plasmids that harbor resistance and virulence genes in a single genetic environment have been reported recently in various K. pneumoniae isolates, including the high risk of virulence clone ST23 and multidrug resistance (MDR) clone ST11 (15–17). Herein, the largest pEBSI036-1-NDM-VIR plasmid belongs to an IncFIB:IncHI1B hybrid plasmid. BLASTn showed that pEBSI036-1-NDM-VIR shared >99% identity with plasmids pKpvST383L (CP034201.2), pKpvST147B_virulence (CP040726.1), and p51015_NDM_1 (CP050380.1), with query coverages of 97 to 99% (see Fig. S2 in the supplemental material). The backbone region of pEBSI036-1-NDM-VIR almost covered the complete sequence of the MDR plasmid pKpvST101_5, with a length of 210,661 bp (CP031372.2) (Fig. 1). Most of the remaining sequences (∼130 kb) of pEBSI036-1-NDM-VIR were similar to those of the virulence plasmid pJX6-1, with a length of 228,974 bp (CP064230.1) (Fig. 1).
FIG 1.
Structure analysis of pEBSI036-1-NDM-VIR. Major structural features of plasmid pEBSI036-1-NDM-VIR were compared with those of plasmids pKpvST101_5 (GenBank accession no. CP031372.2) and pJX6-1 (GenBank accession no. CP064230.1). The comparative schematic diagrams of resistance regions and virulence regions in plasmids pEBSI036-1-NDM-VIR, pKpvST383L (GenBank accession no. CP034201.2), pKpvST147B_virulence (GenBank accession no. CP040726.1), and p51015_NDM_1 (GenBank accession no. CP050380.1) are shown. Gray shading indicates shared regions with a high degree of homology. Red and purple represent the antibiotic resistance and virulence genes, respectively, and yellow represents the insertion sequences and transposons.
Sequence alignment analysis among plasmids pEBSI036-1-NDM-VIR and pKpvST383L (GenBank accession no. CP034201.2), pKpvST147B_virulence (GenBank accession no. CP040726.1), and p51015_NDM_1 (GenBank accession no. CP050380.1). Download FIG S2, TIF file, 2.3 MB (2.4MB, tif) .
Copyright © 2021 Ahmed et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
An ∼38-kb MDR region in pEBSI036-1-NDM-VIR harbored carbapenemase-encoding gene blaNDM-1 and another eight resistance genes: mph(A), sul1, dfrA5, aph(3′)-Ia, armA, msr(E), mph(E), and qnrS. A truncated transposon, ΔTnAs1 (Tn3 family [6,694 bp]), and IS26 elements (IS6 family [820 bp]) were located upstream of mph(A). The mph(A) gene and the downstream complete IS6100 sequence (family IS6 [880 bp]) were separated by two open reading frames (ORFs). sul1 and dfrA5 were surrounded by IS4321 (family IS110 [1,327 bp]), ΔTnAs3 (Tn3 family [18,375 bp]), and IS26 elements. This fragment with 15,448 bp containing the resistance genes mentioned above was similar to plasmid pKpvST383L (Fig. 1). The aph(3′)-Ia gene was flanked by IS26 elements; a similar structure was also found downstream of the resistance region in pKpvST383L. The segment IS26-armA-IS26-msr(E)-mph(E)-ORF-ORF-IS26-ΔTn2 in pEBSI036-1-NDM-VIR was also found to be identical to an inverted sequence in pKpvST383L. In addition, the blaNDM-1 and qnrS genes were on either side of this fragment, while they were absent in pKpvST383L. By comparing the complete sequences of pEBSI036-1-NDM-VIR and pKpvST383L, it was found that pKpvST383L had another resistance region (26,683 bp) carrying blaNDM-5 and blaOXA-9. Compared with plasmid p51015_NDM_1, the resistance region of plasmid pEBSI036-1-NDM-VIR lacked the aph(3′)-VI and sul2 genes (Fig. 1). In pEBSI036-1-NDM-VIR, the MDR region contained six IS26 elements and other transposon elements. Some studies demonstrated that the resistance loci containing IS26 can be hot spots for the capture of further resistance genes to constitute a novel MDR region (18).
A set of virulence genes was detected in pEBSI036-1-NDM-VIR with increased colonization and infection-producing capabilities, including rmpA and rmpA2 for the hypermucoviscous phenotype and iucABCD and iutA, associated with virulence (19). The ∼39-kb region harboring virulence genes exhibited high similarity (99.9% identity and 98% query coverage) to pKpvST383L, pKpvST147B_virulence, and p51015_NDM_1 (Fig. 1).
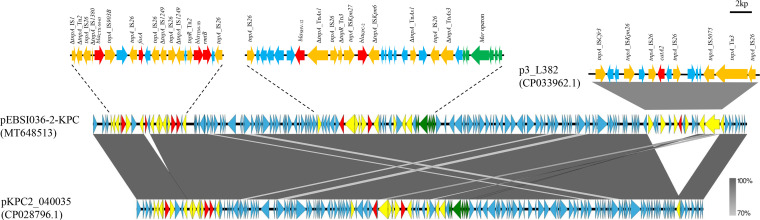
Comparative analysis showed that the IncR:IncFII-type plasmid pEBSI036-2-KPC had 98 to 99% query coverages and 99.9% nucleotide identity to the following plasmids: pKP19-2029-KPC2 (CP047161.1), p69-2 (CP025458.1), and p16HN-263_KPC (CP045264.1). The pEBSI036-2-KPC plasmid carried the carbapenemase-encoding gene blaKPC-2 and three β-lactamase-encoding genes: blaCTX-M-65, blaTEM-1B, and blaSHV-12. The pEBSI036-2-KPC plasmid had additional resistance genes: catA2, fosA3, and rmtB. These resistance genes were located in two regions (Fig. 2). The blaKPC-2 and blaSHV-12 genes were separated by sequence ΔTnAs1-IS26-ΔTn3-ISKpn27, and ISKpn6 was located downstream of blaKPC-2. Sequence downstream of blaKPC-2 contained a mer operon responsible for mercuric resistance and transposon elements (ΔTnAs1-IS26-ΔTnAs3). This segment carrying blaKPC-2, blaSHV-12, and a mer operon, was highly similar to those of other plasmids, such as pKP1034 (20). There was another MDR region (15,254 bp) that consisted of blaCTX-M-65, fosA3, blaTEM-1B, and rmtB genes and five IS26 fragments (Fig. 2). The basic structure of pEBSI036-2-KPC was similar to that of plasmid pKPC2_040035 (CP028796.1) (99.98% identity and 88% query coverage), except for two regions. One locus (10,379 to 10,795 bp) carried fosA3, which was flanked by IS26. The other region contained a ΔISCfr3-ISKpn26-IS26-catA2-IS26-IS5075-ΔTn3-IS26 structure with a length of 15,042 bp, which was the same as plasmid p3_L382 (CP033962.1), with 100% query coverage and 99.99% nucleotide identity (Fig. 2). Both fosA3 and catA2 were flanked by IS26, as previously reported (20, 21). That evidence emphasizes the role of insertion elements such as IS26 in regulating insertion and deletion of resistance genes again.
FIG 2.
Sequence alignment analysis among plasmids pEBSI036-2-KPC, pKPC2_040035 (GenBank accession no. CP028796.1), and p3_L382 (GenBank accession no. CP033962.1). Red and green represent the antibiotic resistance and heavy metal resistance genes, respectively, and yellow represents the insertion sequences and transposons.
In conclusion, we have reported a high-risk clone of ST11 KL47 of a CR-HvKP strain isolated from the blood of a patient from an ICU in Egypt, which coharbors two plasmids: one is a novel hybrid plasmid harboring the carbapenemase gene blaNDM-1 and virulence genes, and the other carries blaKPC-2. Further countrywide surveillance studies are needed to elucidate the rate of prevalence of this high-risk clone in Egypt and its burden on hospital-acquired infections.
Accession numbers.
The sequences of the plasmids pEBSI036-1-NDM-VIR and pEBSI036-2-KPC have been deposited in GenBank under accession no. MT648512 and MT648513.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 82061128001, 81722030, 81830103, and 81902123), the National Key Research and Development Program (grant no. 2017ZX10302301), the Guangdong Natural Science Foundation (grant no. 2017A030306012), the Project of High-Level Health Teams of Zhuhai at 2018 (the Innovation Team for Antimicrobial Resistance and Clinical Infection), the 111 Project (grant no. B12003), the Open Project of the Key Laboratory of Tropical Disease Control (Sun Yat-sen University), Ministry of Education (grant no. 2020kfkt04 and 2020kfkt07) and China Postdoctoral Science Foundation (grant no. 2019M653192), the Science, Technology & Innovation Commission of Shenzhen Municipality (grant no. JCYJ20190807151601699), the Science and Technology Planning Project of Guangdong (grant no. 2017A020215017), the Qingyuan People's Hospital Medical Scientific Research Fund Project (grant no. 20190209), and the Guangdong Provincial Bureau of Traditional Chinese Medicine Research Fund (grant no. 20201407).
The authors report no conflicts of interest in this work. All authors have read and approved the manuscript.
Contributor Information
Fan Yang, Email: yangf77@xxmu.edu.cn.
Lingqing Xu, Email: lingqing_xu@126.com.
Guo-Bao Tian, Email: tiangb@mail.sysu.edu.cn.
Patricia A. Bradford, Antimicrobial Development Specialists, LLC
REFERENCES
- 1.Abdulall AK, Tawfick MM, El Manakhly AR, El Kholy A. 2018. Carbapenem-resistant Gram-negative bacteria associated with catheter-related bloodstream infections in three intensive care units in Egypt. Eur J Clin Microbiol Infect Dis 37:1647–1652. doi: 10.1007/s10096-018-3294-7. [DOI] [PubMed] [Google Scholar]
- 2.Gamal D, Fernández-Martínez M, Salem D, El-Defrawy I, Montes L, Ocampo-Sosa AA, Martínez-Martínez L. 2016. Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtF. Int J Infect Dis 43:17–20. doi: 10.1016/j.ijid.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Hamza E, Dorgham SM, Hamza DA. 2016. Carbapenemase-producing Klebsiella pneumoniae in broiler poultry farming in Egypt. J Glob Antimicrob Resist 7:8–10. doi: 10.1016/j.jgar.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Khairy RMM, Mahmoud MS, Shady RR, Esmail MAM. 2020. Multidrug-resistant Klebsiella pneumoniae in hospital-acquired infections: concomitant analysis of antimicrobial resistant strains. Int J Clin Pract 74:e13463. doi: 10.1111/ijcp.13463. [DOI] [PubMed] [Google Scholar]
- 5.Stewardson AJ, Marimuthu K, Sengupta S, Allignol A, El-Bouseary M, Carvalho MJ, Hassan B, Delgado-Ramirez MA, Arora A, Bagga R, Owusu-Ofori AK, Ovosi JO, Aliyu S, Saad H, Kanj SS, Khanal B, Bhattarai B, Saha SK, Uddin J, Barman P, Sharma L, El-Banna T, Zahra R, Saleemi MA, Kaur A, Iregbu K, Uwaezuoke NSC, Abi Hanna P, Feghali R, Correa AL, Munera MI, Le TAT, Tran TTN, Phukan C, Phukan C, Valderrama-Beltrán SL, Alvarez-Moreno C, Walsh TR, Harbarth S. 2019. Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study. Lancet Infect Dis 19:601–610. doi: 10.1016/S1473-3099(18)30792-8. [DOI] [PubMed] [Google Scholar]
- 6.CLSI. 2018. Performance standards for antimicrobial susceptibility testing. M100-S28. CLSI, Wayne, PA. [Google Scholar]
- 7.Osei Sekyere J, Govinden U, Bester LA, Essack SY. 2016. Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: emerging resistance mechanisms and detection methods. J Appl Microbiol 121:601–617. doi: 10.1111/jam.13169. [DOI] [PubMed] [Google Scholar]
- 8.Dong N, Lin D, Zhang R, Chan EW, Chen S. 2018. Carriage of blaKPC-2 by a virulence plasmid in hypervirulent Klebsiella pneumoniae. J Antimicrob Chemother 73:3317–3321. doi: 10.1093/jac/dky358. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Long D, Xiang TX, Du FL, Wei DD, Wan LG, Deng Q, Cao XW, Zhang W. 2019. Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J Antimicrob Chemother 74:1233–1240. doi: 10.1093/jac/dkz023. [DOI] [PubMed] [Google Scholar]
- 10.Wang YC, Tang HL, Liao YC, Chiou CS, Chen YT, Chiang MK, Lu MC, Lai YC. 2019. Cocarriage of distinct blaKPC-2 and blaOXA-48 plasmids in a single sequence type 11 carbapenem-resistant Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 63:e02282-18. doi: 10.1128/AAC.02282-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Leung SSY, Huang Y, Guo Y, Jiang N, Li P, Chen J, Wang R, Bai C, Mi Z, Gao Z. 2020. Identification of two depolymerases from phage IME205 and their antivirulent functions on K47 capsule of Klebsiella pneumoniae. Front Microbiol 11:218. doi: 10.3389/fmicb.2020.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Gu Y, Li X, Liu Y, Ye Y, Guan S, Li J. 2019. Identification and characterization of NDM-1-producing hypervirulent (hypermucoviscous) Klebsiella pneumoniae in China. Ann Lab Med 39:167–175. doi: 10.3343/alm.2019.39.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLaughlin MM, Advincula MR, Malczynski M, Barajas G, Qi C, Scheetz MH. 2014. Quantifying the clinical virulence of Klebsiella pneumoniae producing carbapenemase Klebsiella pneumoniae with a Galleria mellonella model and a pilot study to translate to patient outcomes. BMC Infect Dis 14:31. doi: 10.1186/1471-2334-14-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang YH, Chou SH, Liang SW, Ni CE, Lin YT, Huang YW, Yang TC. 2018. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother 73:2039–2046. doi: 10.1093/jac/dky164. [DOI] [PubMed] [Google Scholar]
- 16.Li R, Cheng J, Dong H, Li L, Liu W, Zhang C, Feng X, Qin S. 2020. Emergence of a novel conjugative hybrid virulence multidrug-resistant plasmid in extensively drug-resistant Klebsiella pneumoniae ST15. Int J Antimicrob Agents 55:105952. doi: 10.1016/j.ijantimicag.2020.105952. [DOI] [PubMed] [Google Scholar]
- 17.Turton J, Davies F, Turton J, Perry C, Payne Z, Pike R. 2019. Hybrid resistance and virulence plasmids in “high-risk” clones of Klebsiella pneumoniae, including those carrying blaNDM-5. Microorganisms 7:326. doi: 10.3390/microorganisms7090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyrsch ER, Roy Chowdhury P, Chapman TA, Charles IG, Hammond JM, Djordjevic SP. 2016. Genomic microbial epidemiology is needed to comprehend the global problem of antibiotic resistance and to improve pathogen diagnosis. Front Microbiol 7:843. doi: 10.3389/fmicb.2016.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang HL, Chiang MK, Liou WJ, Chen YT, Peng HL, Chiou CS, Liu KS, Lu MC, Tung KC, Lai YC. 2010. Correlation between Klebsiella pneumoniae carrying pLVPK-derived loci and abscess formation. Eur J Clin Microbiol Infect Dis 29:689–698. doi: 10.1007/s10096-010-0915-1. [DOI] [PubMed] [Google Scholar]
- 20.Xiang D-R, Li J-J, Sheng Z-K, Yu H-Y, Deng M, Bi S, Hu F-S, Chen W, Xue X-W, Zhou Z-B, Doi Y, Sheng J-F, Li L-J. 2015. Complete sequence of a novel IncR-F33:A−:B− plasmid, pKP1034, harboring fosA3, blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB from an epidemic Klebsiella pneumoniae sequence type 11 strain in China. Antimicrob Agents Chemother 60:1343–1348. doi: 10.1128/AAC.01488-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sennati S, Riccobono E, Di Pilato V, Villagran AL, Pallecchi L, Bartoloni A, Rossolini GM. 2016. pHN7A8-related multiresistance plasmids (blaCTX-M-65, fosA3 and rmtB) detected in clinical isolates of Klebsiella pneumoniae from Bolivia: intercontinental plasmid dissemination? J Antimicrob Chemother 71:1732–1734. doi: 10.1093/jac/dkv506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Virulence potential of K. pneumoniae strain EBSI036 as depicted in a Galleria mellonella infection model with an inoculum of 1 × 104 CFU. Download FIG S1, TIF file, 0.3 MB (296.4KB, tif) .
Copyright © 2021 Ahmed et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overall features of the genome and minimum inhibitory concentrations (MICs) for K. pneumoniae EBSI036. Download Table S1, DOCX file, 0.03 MB (28.1KB, docx) .
Copyright © 2021 Ahmed et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Putative virulence genes detected on the K. pneumoniae EBSI036 chromosome. Download Table S2, XLS file, 0.04 MB (37.5KB, xls) .
Copyright © 2021 Ahmed et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence alignment analysis among plasmids pEBSI036-1-NDM-VIR and pKpvST383L (GenBank accession no. CP034201.2), pKpvST147B_virulence (GenBank accession no. CP040726.1), and p51015_NDM_1 (GenBank accession no. CP050380.1). Download FIG S2, TIF file, 2.3 MB (2.4MB, tif) .
Copyright © 2021 Ahmed et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.