Abstract
Culturing skin cells outside of the body has been a cornerstone of dermatological investigation for many years; however, human skin equivalent systems typically lack the full complexity of native skin. Notably, skin appendages, such as hair follicles and sweat glands, remain a challenge to generate or maintain in cell cultures and reconstruct in damaged skin. Recent work from our lab has demonstrated methods for generating appendage-bearing skin tissue—known as skin organoids—from pluripotent stem cells. Here, we will summarize this work and other related works, and then discuss the potential future applications of skin organoids in dermatological research.
1 |. INTRODUCTION
For the past fifty years, human cell culture models have helped to minimize animal use and diminish the burden of skin diseases 1–7. However, the mouse remains the model of choice to study dynamic mechanisms of skin development and homeostasis. A key barrier to progress is that mouse models do not entirely mimic human biology and that human skin equivalent systems do not typically contain skin appendages, such as hair follicles (HFs) and sweat glands, or other skin-related cells, such as dermal fat cells and sensory neurons. Therefore, there has been a critical need for improved models of human skin generation in vitro for use as a research tool and, potentially, as a source of cells for skin reconstruction. Significant effort has been devoted to recreating hair-bearing skin from patient-derived epidermal and dermal cells. These studies have shown that, in particular, adult human dermal papilla cells retain some level of hair follicle inductive capacity and can be coaxed to initiate de novo follicle-like structures in 3D cultures 8–10. Nevertheless, these efforts have generally failed to fully reconstruct the entire skin architecture and its appendages outside of the body 11–14. Recent work from our laboratory shows that complex hair-bearing human skin tissue—referred to as skin organoids—can be generated from human pluripotent stem cells (hPSCs) over a months-long process of guided differentiation. In this Viewpoint, we will summarize our work on skin organoids and its evolution from our previous work on inner ear organogenesis and the hPSC work of other researchers in the field. Then, we will highlight key focus areas where the skin organoid model could accelerate progress.
2 |. THE CHALLENGE OF GROWING SKIN APPENDAGES IN A DISH
The challenge of de novo hair generation, in a wound or in a culture dish, has stumped researchers for years. The magnitude of the problem becomes apparent when one considers the complex integration of cellular, mechanical, and chemical inputs that lead to appendage induction during fetal development. The number of spatiotemporal variables that need to be controlled are staggering. In normal development, the dermis and epidermis, the major layers of the skin, undergo a series of structural and chemical changes in parallel—both tissues becoming stratified with multiple cell subtypes with distinct gene and protein expression programs. During the stratification process, the mechanical force at the interface of the epidermis and dermis is modulated by deposition of extracellular matrix proteins in the dermis and changes in cell organization in the epidermis. Moreover, a varying combination of secreted chemical cues (e.g. WNT, BMP, and FGF proteins) are exchanged between the epidermis and dermis over time, leading to development and maturation of skin. One major difficulty has been that we have an incomplete knowledge of the epidermal and dermal cellular players most critical to the establishment of hair placodes and germs. Recent work using single-cell genomics has made great strides toward elucidating these populations 15–17. Building off of the limited existing knowledge of how skin cells develop, many research teams have attempted to recreate the epidermis and dermis from hPSCs.
There are two types of hPSCs: embryonic stem cells (hESCs), derived from pre-implantation embryos, and induced pluripotent stem cells (hiPSCs), derived from reprogrammed adult cells 18. hPSCs can self-renew indefinitely and generate cells representing any tissue; thus, hPSCs could be an unlimited source of cells for experimentation or therapy 19,20. To guide hPSC differentiation in vitro, researchers can activate or inhibit cell signaling pathways known to pattern the vertebrate embryo. Prior to our study, an efficient means to generate most skin-related cell types from hPSCs remained elusive. It had been well established that epidermal keratinocytes could be generated from hPSCs using a combination of retinoic acid (RA) and bone morphogenetic protein-4 (BMP-4) 13,21. This combination treatment induces surface ectoderm progenitors that mature into various keratinocyte subtypes depending on the culture conditions. Wild-type or genetically engineered keratinocytes generated using this approach have been tested in pre-clinical studies for diseases, such as epidermolysis bullosa 20,22–24. In contrast, dermal cells have been difficult to generate from hPSCs in a targeted manner. Previous attempts to generate dermal fibroblasts have started by generating paraxial mesoderm or cranial neural crest (CNC) progenitors—the two developmental precursors of dermal fibroblasts in the body and head, respectively 13,25. The derived cells appear to contain a mixture of other related cell subtypes in these lineages and have failed to produce the entire dermal fibroblast lineage, which comprises the papillary, reticular, and hypodermal layers 26. A key limitation of existing methodologies is the use of undefined factors, such a fetal bovine serum (FBS), and the technical barrier of directing the co-differentiation of skin progenitor cells in parallel or in tandem in 2D monolayer cultures 27,28. Studies in which partial hair follicles have been generated from hPSC-derived cells have relied on a chimeric approach using human/mouse epidermal/dermal cells, xenografting onto nude mice to encourage folliculogenesis, or complicated bioengineering approaches 12,29.
3D organoid cultures have emerged in recent years that exquisitely mimic the structure and function of native organs, such as the brain and intestines 30,31. In the early days of organoid research, Mototsugu Eiraku, Yoshiki Sasai, and colleagues, published landmark papers on PSC-derived cerebral 32 and retinal 33 organoid models. Their general approach was to apply stepwise treatments of small molecules, recombinant proteins, and extracellular matrix proteins (from diluted Matrigel) to an aggregate of 3,000-10,000 PSCs to induce ectodermal epithelia. This approach was foundational for our later work on inner ear and skin organoids, because it allows the cells to dynamically self-organize during differentiation—a critical requirement for the emergence of higher-order cellular structures, such as otic or hair placodes 27,34,35. While optimizing our technique for generating inner ear organoids from mouse PSCs, we observed the spontaneous co-development of epidermis and dermis in 3D cultures resulting in the first hair-bearing skin organoids 27. Moving forward from mouse to human hPSCs, our strategy was to co-induce epithelial and mesenchymal cells and allow the two cell types to develop together as they do in the fetus. The resulting human skin organoid culture system proceeds in three stages (Fig. 1). In Stage-I, we co-induce surface ectoderm and CNC cells—two tissue types critical for cranial skin development—using an optimized sequence of TGFß, BMP, and FGF signaling modulators. In Stage-II, we place the cell aggregate in a floating rotational culture environment where it forms a cystic organoid, which contains CNC-derived mesenchymal, neural, and glial progenitor cells clustered around a sphere of keratinocytes. In Stage-III, the organoids form a keratinized epidermis surrounded by dermal fibroblasts, fat, cartilage, sensory neurons, Schwann cells, and melanocytes. The organoids then produce hair follicles that grow radially outward (Fig. 2) and receive innervation from sensory neurons. Thus, by directing the differentiation of a starting population of progenitors (epidermal, dermal, and neuro-glial), we can establish a self-organizing system that recapitulates nearly the entire skin developmental program, including appendage induction.
Figure 1.
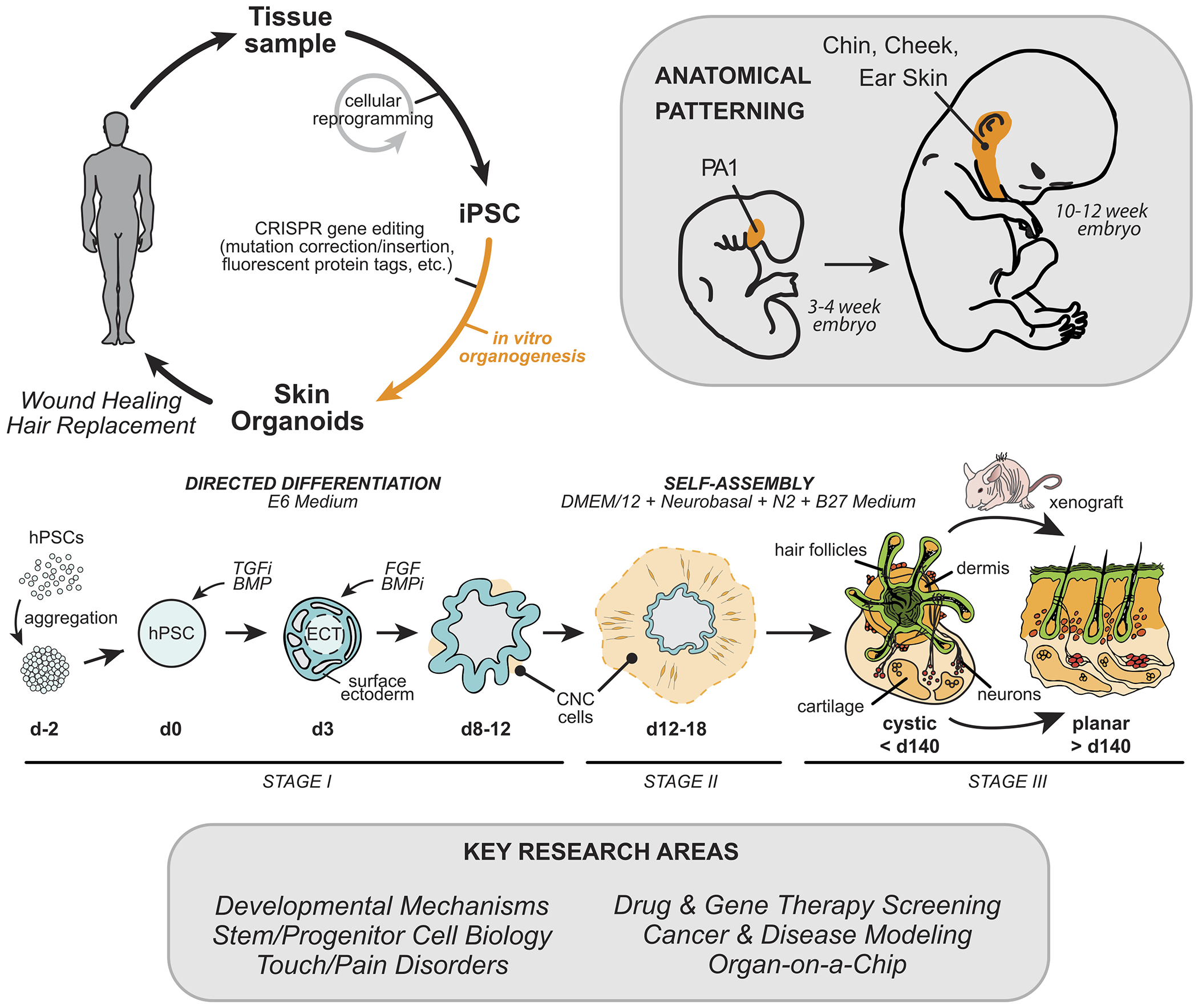
Overview of human skin organoid method and key areas of research that could benefit. Adapted from Lee et al Nature, 2020.
Figure 2.
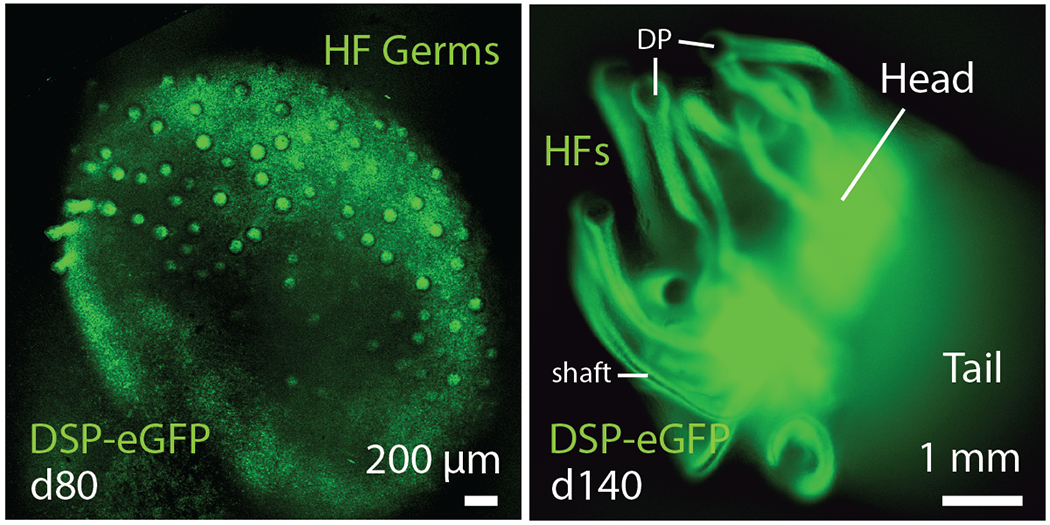
Emerging hair follicles in day-80 (left) and day-140 (right) skin organoids derived from a Desmoplakin (DSP)-eGFP hiPSC line.
3 |. PEERING INSIDE THE BLACK BOX OF HUMAN SKIN DEVELOPMENT
Skin organoids are particularly well suited for probing developmental biology research questions. Investigations into human skin development have been limited by the lack of availability of embryonic and fetal tissue specimens to work with for experimentation. In the United States, work with human fetal tissue has recently come under renewed scrutiny, which is predicted to have a chilling effect on future projects using fetal tissue 36. Nevertheless, there is much to be gained from expanding our understanding of human skin development. As mentioned, current research is heavily reliant on murine models, which do not fully mimic human biology. Consequently, there are critical gaps in our knowledge about how human skin develops. Our understanding of the early stages of human epidermis, dermis, melanocyte, and skin appendage fate specification are limited to basic histological studies 37–41. Now, with skin organoids, we have access to a nearly limitless source of fetal-like skin tissue for experimentation. A future focus of organoid-based research could be to determine how various signaling pathways (e.g. FGF, WNT, TGF, etc.) control early differentiation of the epidermal and neural crest cell lineages that lead to higher-order development, such as hair follicle induction. Although the role of developmental signaling cues have been extensively studied in mice, few of these studies have been directly investigated in developing human skin. In addition to investigations of chemical signaling messengers, future work could elucidate the role of mechanical forces, such as dermis induced constriction forces, on human skin development in the organoid model 15,42–45.
Positional patterning is another aspect of embryonic development reflected in skin organoid cultures. The human body contains a variety of skin types, specialized to anatomical locations (e.g. hairless glabrous skin on the palms). It is thought that the developmental origin and patterning of dermal fibroblast cells dictates the type of skin tissue 46–48. The dermis in skin organoids appears to arise from CNC cells, which produces facial dermis during development 49. Based on expression of a number of gene markers (e.g. DLX1-6, GSC, BARX1, and PITX1) in the developing dermis, we were able to estimate the anatomical location as similar to the first pharyngeal arch, which develops into cheek, chin, and ear skin 50. Interestingly, this appears to be the default anatomical location of skin organoids, which is undoubtedly influenced by the culture conditions we chose for the induction phase of the culture. A focus of future research could be to determine if skin can be generated by combining dermal progenitor cells from another lineage, the mesoderm, with surface ectoderm progenitors. We suspect that co-development of epidermal and dermal progenitors for an extended period of time (e.g. over 30 days) is critical for skin organoid formation; however, it is unclear whether skin organoids can be “built” by combining progenitors prior to co-development. If this approach is achievable, building organoids from separate epidermal and dermal progenitors could be one way to control the size, shape, and composition of skin of a particular anatomical site for precision reconstructive surgery in patients. This work could also lead to production of other skin appendages, such as mammary glands or teeth 29.
To gain additional insight into the endogenous mechanisms that shape and pattern skin organoids, more extensive single-cell RNA-sequencing (scRNA-seq) should be performed, encompassing time points throughout the entire ~150 days of organoid induction and maturation. In our recent study, we provided single-cell RNA-seq datasets for three timepoints of organoid induction. Anyone can readily access the datasets and query their genes of interest on our website at https://koehler-lab.org/resources. Our published data have provided insight into the endogenous mechanisms driving skin organoid self-patterning. In particular, WNT and FGF signalling modulators were differentially expressed between dermal and epidermal cell populations, consistent with known signaling factors present in developing mouse skin 51–53. As organoid generation is a dynamic process, we are currently expanding these analyses to additional timepoints encompassing the entire 4-5 month induction period. These datasets could be integrated with reference skin atlases like those under development by the Human Cell Atlas (HCA) project—an international consortium focused on molecular mapping of every cell in the human body 54,55. For example, Botting et al. recently generated a reference map of fetal liver hematopoiesis and establishment of immune cells in the skin and lung 56,57. A component of the HCA’s ongoing work on a Developmental Cell Atlas will use stem cell-derived organoids as a complementary cell source for difficult to obtain human fetal tissue 58,59. This work will provide a tremendous resource for future investigations into the stem/progenitor cells that arise during human skin formation.
4 |. IDENTIFYING NOVEL MECHANISMS OF HUMAN SKIN PLASTICITY AND REGENERATION
Skin organoids could be used to discover new regenerative drug or gene therapy approaches and for deriving cells to support wound healing. Although, a prerequisite for these uses of the model will be to better understand how the physical structure of organoids can be manipulated. Skin organoids are remarkably long-lived in vitro, developing over a period of 140 days or more. However, organoids develop as a cyst—a format that is not conducive to long-term maintenance of differentiated keratinocytes, which need to be periodically shed from the surface of the skin. We have shown that skin organoids integrate into nude mouse skin upon transplantation, creating a planar layer of in vivo-like epidermis with properly oriented hair follicles. These findings suggest that skin organoids could feasibly be used for skin reconstruction or for facilitating wound healing. Although we performed our xenografting experiments with whole organoids, previous work has shown that completely dissociated human fetal skin is capable of reconstituting hair-bearing skin in the nude mouse model as well 60. A major challenge will be to elucidate the process of planar skin reconstitution from cystic skin organoids in vivo. In a recent study on the hair-bearing potential of neonatal and adult mouse skin, Lei et al. suggest that sequential treatments of various signaling factors (e.g. IGF2, VEGF, Wnt3a, Wnt10b, and MMP14) can be used to guide the cystic-to-planar transition 28. Expanded studies like this using human primary skin cells will be insightful for future PSC-derived skin organoid studies 61. We do not yet know which cell populations are most critical for establishing planar skin. We also do not know whether nascent (hairless) skin organoids can develop hair after transplantation. Future studies could investigate unrecognized mechanisms that could be targeted to improve skin regeneration in vivo. This line of inquiry could be useful engineering planar skin organoids in vitro, which could extend the longevity of cultures (i.e. allowing shedding of squames) and be more suitable for investigating developmental, disease, or aging-related mechanisms that manifest in mature skin tissue.
Moving skin organoids to the clinic has significant challenges. Perhaps the largest hurdle will be managing the immune response. Most skin reconstruction, hair restoration, and wound healing applications for skin organoids will expose the cells to the patient’s immune system. Although various compartments of hair follicle maintain some degree of ‘immune privilege’ during the growth cycle under normal circumstances, this privilege would be broken in a wound or scar setting 62. Recent clinical trials on cell therapies for macular degeneration and Parkinson’s disease show promising early results of autologous iPSC-based therapies 63,64. However, these studies also reveal that the time required for iPSC derivation, validation, and differentiation can be over a year, which limits the patient population that could benefit from these therapies. The use of donor iPSCs may be an option in the future for producing allogeneic skin grafts—without immunosuppression. Several groups are working on gene-editing strategies to make iPSCs invisible to the immune system; thus, creating universal donor cells that will avoid rejection 65. However, this technology is nascent and has potential drawbacks, such as the unintended generation of transmissible cancers 66.
5 |. ADVANCED MODELS OF DISEASE AND CANCER
Skin organoids appear to contain the precursor cells to every major form of skin cancer: basal cell carcinoma, squamous cell carcinoma, melanoma, and Merkel cell carcinoma. Thus, the skin organoid model has the potential to provide excellent insight into cell growth and differentiation in disease modelling applications such as artificial cancer induction in an in vivo-like microenvironment. Recently, organoid cultures were shown to model induction of glioblastomas (in brain organoids) and lung cancer 67–69. Likewise, genetic skin disorders could be modelled using patient-derived or gene edited healthy donor hiPSCs. For example, this general approach has already been explored using hiPSC-derived keratinocytes from patients with epidermolysis bullosa 20,22,24,70. The ability to investigate human-specific molecular mechanisms will be highly advantageous for identifying novel therapeutic targets for human diseases or testing drug/gene therapies.
An obstacle to disease modeling and reproducibility between labs is the inherent variability of hPSC lines 71. Experimental results obtained using directed differentiation on one cell line may not entirely translate to other cell lines. In our recent report, we showed that skin organoids can be generated reproducibly from three stem cell lines, but noted one hESC line that performed poorly. Future studies will need to confirm key results with test sets of hiPSC lines from different male and female donors, ideally from racially diverse donors to better represent variation in the human population 72.
6 |. INCREASING COMPLEXITY: SKIN ORGANOIDS WITH VASCULATURE, IMMUNE CELLS, AND SWEAT GLANDS
Skin organoids are remarkably complex, but lack key cell populations, such as endothelial cells, pericytes, and immune cells (e.g. Langerhans cells and T cells). An often-cited weakness of organoid models is the lack of vasculature. Recent work has led to blood vessel organoids and incorporation of a blood-brain barrier (BBB) in cerebral organoids 73,74. Likewise, these novel platforms could be leveraged to seed skin organoids with endothelial cells and pericytes to investigate the role of vasculature in skin maturation or to derive skin grafts with built-in vasculature. Additionally, like many other organoid models, the utility of in vitro skin organoid systems may be expanded by incorporating key immune cell populations, perhaps generated from autologous iPSCs. A prime example of immune cell incorporation into organoid models has been the use of microglia in cerebral organoids 75,76. Microglia are the resident immune cells of the brain, and recently, researchers have successfully derived microglial cells by guiding hPSCs into the mesodermal lineage 75,77. In parallel, brain organoids were induced and then seeded with microglia using a simple co-culture approach 76. The timing of seeding was chosen to reflect the timing of microglial migration to the brain during normal development. Similarly, a parallel-induction and seeding approach of immune cells could be employed to incorporate myeloid and lymphoid lineage cells into skin organoids to set the stage for inflammation studies. These modifications to skin organoids would be critical for modeling skin disorders with an immune component, such as psoriasis; however, in the interim, researchers could follow the example of previous skin equivalent culture studies by treating skin organoids with various cytokines to mimic an inflammatory response 78,79.
Sweat glands are also missing from the skin organoids generated in our study 50. Sweat glands are a critical component of the skin for thermoregulation and, like hair follicles, poorly regenerate during wound healing. During development, sweat glands arise much later than the earliest hair follicles—at around weeks 18-20 of gestation. It is possible that our skin organoids are just reaching this equivalent time point of development in culture; thus, sweat glands do not have time to develop. Alternatively, the critical inductive cues for sweat glands may be missing in organoids 80. An exciting focus of research could be to further elucidate the mechanisms of sweat gland formation in the organoid model.
7 |. FUTURE OUTLOOK
In summary, the skin organoid model sets the stage for gaining a more in-depth understanding of how human skin forms and how fetal skin grows in culture that could be harnessed for cell therapy. However, there is much work to be done to improve and refine the skin organoid system to truly open the technology up for wide adoption in the field. Due to lingering caveats of using iPSCs for research, such as cell line heterogeneity, labor intensive cell cultures, and batch-to-batch variability, we suggest that skin organoids should not be viewed as a replacement for, but rather a complement to existing mouse models. The organoid model should be most useful for pursuing future directions focused on stem/progenitor cell biology in human hair follicle generation under healthy and pathophysiological conditions and exploring novel approaches to skin and hair restoration in patients with alopecia and extensive scarring. Although not discussed here, skin organoids could also be used for exploratory research on neurosensory (i.e. pain and touch) functions of the skin, microbiota-skin interactions, and viral/bacterial infections of the skin. We anticipate that skin organoids—much like cerebral, retinal, and gut organoids in their respective fields—will provide the Dermatology field with exciting new insights into human biology.
ACKNOWLEDGEMENTS
This work was supported by NIH grant R01 AR075018-01. We would like to thank members of the Koehler Lab for critical comments on the manuscript.
Footnotes
CONFLICT OF INTEREST
J.L. and K.R.K have submitted a patent application relating to the skin organoid system discussed in this manuscript (WO2017070506A1). The author declares no other conflicts of interest.
REFERENCES
- 1.Bell E, Ehrlich HP, Buttle DJ, Nakatsuji T. Living tissue formed in vitro and accepted as skin-equivalent tissue of full thickness. Science. 1981;211(4486):1052–1054. doi: 10.1126/science.7008197 [DOI] [PubMed] [Google Scholar]
- 2.Sun BK, Siprashvili Z, Khavari PA. Advances in skin grafting and treatment of cutaneous wounds. Science. 2014;346(6212):941–945. doi: 10.1126/science.1253836 [DOI] [PubMed] [Google Scholar]
- 3.Karimkhani C, Dellavalle RP, Coffeng LE, et al. Global skin disease morbidity and mortality: an update from the global burden of disease study 2013. JAMA Dermatol. 2017;153(5):406–412. doi: 10.1001/jamadermatol.2016.5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hay RJ, Johns NE, Williams HC, et al. The global burden of skin disease in 2010: an analysis of the prevalence and impact of skin conditions. J Invest Dermatol. 2014;134(6):1527–1534. doi: 10.1038/jid.2013.446 [DOI] [PubMed] [Google Scholar]
- 5.Lim HW, Collins SAB, Resneck JS, et al. The burden of skin disease in the United States. J Am Acad Dermatol. 2017;76(5):958–972.e2. doi: 10.1016/j.jaad.2016.12.043 [DOI] [PubMed] [Google Scholar]
- 6.Giesey RL, Mehrmal S, Uppal P, Delost ME, Delost GR. Dermatoses of the world: Burden of skin disease and associated socioeconomic status in the world. J Am Acad Dermatol. June 2020. doi: 10.1016/j.jaad.2020.05.157 [DOI] [PubMed] [Google Scholar]
- 7.Sen CK, Gordillo GM, Roy S, et al. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 2009;17(6):763–771. doi: 10.1111/j.1524-475X.2009.00543.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topouzi H, Logan NJ, Williams G, Higgins CA. Methods for the isolation and 3D culture of dermal papilla cells from human hair follicles. Exp Dermatol. 2017;26(6):491–496. doi: 10.1111/exd.13368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins CA, Chen JC, Cerise JE, Jahoda CAB, Christiano AM. Microenvironmental reprogramming by three-dimensional culture enables dermal papilla cells to induce de novo human hair-follicle growth. Proc Natl Acad Sci USA. 2013;110(49):19679–19688. doi: 10.1073/pnas.1309970110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Messenger AG. The culture of dermal papilla cells from human hair follicles. Br J Dermatol. 1984;110(6):685–689. doi: 10.1111/j.1365-2133.1984.tb04705.x [DOI] [PubMed] [Google Scholar]
- 11.Yang R, Zheng Y, Burrows M, et al. Generation of folliculogenic human epithelial stem cells from induced pluripotent stem cells. Nat Commun. 2014;5:3071. doi: 10.1038/ncomms4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abaci HE, Coffman A, Doucet Y, et al. Tissue engineering of human hair follicles using a biomimetic developmental approach. Nat Commun. 2018;9(1):5301. doi: 10.1038/s41467-018-07579-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh M, Umegaki-Arao N, Guo Z, Liu L, Higgins CA, Christiano AM. Generation of 3D skin equivalents fully reconstituted from human induced pluripotent stem cells (iPSCs). PLoS ONE. 2013;8(10):e77673. doi: 10.1371/journal.pone.0077673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gledhill K, Guo Z, Umegaki-Arao N, Higgins CA, Itoh M, Christiano AM. Melanin Transfer in Human 3D Skin Equivalents Generated Exclusively from Induced Pluripotent Stem Cells. PLoS ONE. 2015;10(8):e0136713. doi: 10.1371/journal.pone.0136713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mok K-W, Saxena N, Heitman N, et al. Dermal condensate niche fate specification occurs prior to formation and is placode progenitor dependent. Dev Cell. 2019;48(1):32–48.e5. doi: 10.1016/j.devcel.2018.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heitman N, Saxena N, Rendl M. Advancing insights into stem cell niche complexities with next-generation technologies. Curr Opin Cell Biol. 2018;55:87–95. doi: 10.1016/j.ceb.2018.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joost S, Zeisel A, Jacob T, et al. Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell Syst. 2016;3(3):221–237.e9. doi: 10.1016/j.cels.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 19.Kimbrel EA, Lanza R. Next-generation stem cells - ushering in a new era of cell-based therapies. Nat Rev Drug Discov. 2020;19(7):463–479. doi: 10.1038/s41573-020-0064-x [DOI] [PubMed] [Google Scholar]
- 20.Wenzel D, Bayerl J, Nyström A, Bruckner-Tuderman L, Meixner A, Penninger JM. Genetically corrected iPSCs as cell therapy for recessive dystrophic epidermolysis bullosa. Sci Transl Med. 2014;6(264):264ra165. doi: 10.1126/scitranslmed.3010083 [DOI] [PubMed] [Google Scholar]
- 21.Li L, Wang Y, Torkelson JL, et al. TFAP2C- and p63-Dependent Networks Sequentially Rearrange Chromatin Landscapes to Drive Human Epidermal Lineage Commitment. Cell Stem Cell. 2019;24(2):271–284.e8. doi: 10.1016/j.stem.2018.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tolar J, Xia L, Riddle MJ, et al. Induced pluripotent stem cells from individuals with recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2011;131(4):848–856. doi: 10.1038/jid.2010.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mavilio F, Pellegrini G, Ferrari S, et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat Med. 2006;12(12):1397–1402. doi: 10.1038/nm1504 [DOI] [PubMed] [Google Scholar]
- 24.Sebastiano V, Zhen HH, Haddad B, et al. Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci Transl Med. 2014;6(264):264ra163. doi: 10.1126/scitranslmed.3009540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gnedeva K, Vorotelyak E, Cimadamore F, et al. Derivation of hair-inducing cell from human pluripotent stem cells. PLoS ONE. 2015;10(1):e0116892. doi: 10.1371/journal.pone.0116892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Driskell RR, Lichtenberger BM, Hoste E, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504(7479):277–281. doi: 10.1038/nature12783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Böscke R, Tang P-C, Hartman BH, Heller S, Koehler KR. Hair Follicle Development in Mouse Pluripotent Stem Cell-Derived Skin Organoids. Cell Rep. 2018;22(1):242–254. doi: 10.1016/j.celrep.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei M, Schumacher LJ, Lai Y-C, et al. Self-organization process in newborn skin organoid formation inspires strategy to restore hair regeneration of adult cells. Proc Natl Acad Sci USA. 2017;114(34):E7101–E7110. doi: 10.1073/pnas.1700475114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeo M, Tsuji T. Organ regeneration based on developmental biology: past and future. Curr Opin Genet Dev. 2018;52:42–47. doi: 10.1016/j.gde.2018.05.008 [DOI] [PubMed] [Google Scholar]
- 30.Li M, Izpisua Belmonte JC. Organoids - Preclinical Models of Human Disease. N Engl J Med. 2019;380(6):569–579. doi: 10.1056/NEJMra1806175 [DOI] [PubMed] [Google Scholar]
- 31.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345(6194):1247125. doi: 10.1126/science.1247125 [DOI] [PubMed] [Google Scholar]
- 32.Eiraku M, Watanabe K, Matsuo-Takasaki M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3(5):519–532. doi: 10.1016/j.stem.2008.09.002 [DOI] [PubMed] [Google Scholar]
- 33.Eiraku M, Takata N, Ishibashi H, et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472(7341):51–56. doi: 10.1038/nature09941 [DOI] [PubMed] [Google Scholar]
- 34.Koehler KR, Hashino E. 3D mouse embryonic stem cell culture for generating inner ear organoids. Nat Protoc. 2014;9(6):1229–1244. doi: 10.1038/nprot.2014.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koehler KR, Mikosz AM, Molosh AI, Patel D, Hashino E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 2013;500(7461):217–221. doi: 10.1038/nature12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCune JM, Weissman IL. The Ban on US Government Funding Research Using Human Fetal Tissues: How Does This Fit with the NIH Mission to Advance Medical Science for the Benefit of the Citizenry? Stem Cell Reports. 2019;13(5):777–786. doi: 10.1016/j.stemcr.2019.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silva LMA, Hsieh R, Lourenço SV, Ottoni V, Valente N, Fernandes JD. Immunoexpression of adhesion molecules during human fetal hair development. Histol Histopathol. 2020;35(8):911–917. doi: 10.14670/HH-18-224 [DOI] [PubMed] [Google Scholar]
- 38.Arita K, Akiyama M, Tsuji Y, McMillan JR, Eady RAJ, Shimizu H. Changes in gap junction distribution and connexin expression pattern during human fetal skin development. J Histochem Cytochem. 2002;50(11):1493–1500. doi: 10.1177/002215540205001109 [DOI] [PubMed] [Google Scholar]
- 39.Hardman MJ, Moore L, Ferguson MW, Byrne C. Barrier formation in the human fetus is patterned. J Invest Dermatol. 1999;113(6):1106–1113. doi: 10.1046/j.1523-1747.1999.00800.x [DOI] [PubMed] [Google Scholar]
- 40.Akiyama M, Smith LT, Yoneda K, Holbrook KA, Hohl D, Shimizu H. Periderm cells form cornified cell envelope in their regression process during human epidermal development. J Invest Dermatol. 1999;112(6):903–909. doi: 10.1046/j.1523-1747.1999.00592.x [DOI] [PubMed] [Google Scholar]
- 41.Lee SC, Lee JB, Kook JP, et al. Expression of differentiation markers during fetal skin development in humans: immunohistochemical studies on the precursor proteins forming the cornified cell envelope. J Invest Dermatol. 1999;112(6):882–886. doi: 10.1046/j.1523-1747.1999.00602.x [DOI] [PubMed] [Google Scholar]
- 42.Guillot C, Lecuit T. Mechanics of epithelial tissue homeostasis and morphogenesis. Science. 2013;340(6137):1185–1189. doi: 10.1126/science.1235249 [DOI] [PubMed] [Google Scholar]
- 43.Biggs LC, Kim CS, Miroshnikova YA, Wickström SA. Mechanical forces in the skin: roles in tissue architecture, stability, and function. J Invest Dermatol. 2020;140(2):284–290. doi: 10.1016/j.jid.2019.06.137 [DOI] [PubMed] [Google Scholar]
- 44.Shyer AE, Rodrigues AR, Schroeder GG, Kassianidou E, Kumar S, Harland RM. Emergent cellular self-organization and mechanosensation initiate follicle pattern in the avian skin. Science. 2017;357(6353):811–815. doi: 10.1126/science.aai7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martino PA, Heitman N, Rendl M. The dermal sheath: an emerging component of the hair follicle stem cell niche. Exp Dermatol. October 2020. doi: 10.1111/exd.14204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rinn JL, Wang JK, Liu H, Montgomery K, van de Rijn M, Chang HY. A systems biology approach to anatomic diversity of skin. J Invest Dermatol. 2008;128(4):776–782. doi: 10.1038/sj.jid.5700986 [DOI] [PubMed] [Google Scholar]
- 47.Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2(7):e119. doi: 10.1371/journal.pgen.0020119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rinn JL, Wang JK, Allen N, et al. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 2008;22(3):303–307. doi: 10.1101/gad.1610508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137(16):2605–2621. doi: 10.1242/dev.040048 [DOI] [PubMed] [Google Scholar]
- 50.Lee J, Rabbani CC, Gao H, et al. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature. 2020;582(7812):399–404. doi: 10.1038/s41586-020-2352-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim X, Nusse R. Wnt signaling in skin development, homeostasis, and disease. Cold Spring Harb Perspect Biol. 2013;5(2). doi: 10.1101/cshperspect.a008029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu X-J, Liu Y, Dai Z-M, et al. BMP-FGF signaling axis mediates Wnt-induced epidermal stratification in developing mammalian skin. PLoS Genet. 2014;10(10):e1004687. doi: 10.1371/journal.pgen.1004687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richardson GD, Bazzi H, Fantauzzo KA, et al. KGF and EGF signalling block hair follicle induction and promote interfollicular epidermal fate in developing mouse skin. Development. 2009;136(13):2153–2164. doi: 10.1242/dev.031427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Regev A, Teichmann SA, Lander ES, et al. The human cell atlas. elife. 2017;6. doi: 10.7554/eLife.27041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reynolds G, Vegh P, Fletcher J, et al. Poised cell circuits in human skin are activated in disease. BioRxiv. November 2020. doi: 10.1101/2020.11.05.369363 [DOI] [Google Scholar]
- 56.Popescu D-M, Botting RA, Stephenson E, et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574(7778):365–371. doi: 10.1038/s41586-019-1652-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vento-Tormo R, Efremova M, Botting RA, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563(7731):347–353. doi: 10.1038/s41586-018-0698-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haniffa M, Taylor D, Linnarsson S, et al. Human Developmental Cell Atlas: milestones achieved and the roadmap ahead. October 2020. doi: 10.21203/rs.3.rs-73986/v1 [DOI] [Google Scholar]
- 59.Miller AJ, Yu Q, Czerwinski M, et al. In Vitro and In Vivo Development of the Human Airway at Single-Cell Resolution. Dev Cell. 2020;53(1):117–128.e6. doi: 10.1016/j.devcel.2020.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu X, Scott L, Washenik K, Stenn K. Full-thickness skin with mature hair follicles generated from tissue culture expanded human cells. Tissue Eng Part A. 2014;20(23-24):3314–3321. doi: 10.1089/ten.TEA.2013.0759 [DOI] [PubMed] [Google Scholar]
- 61.Weber EL, Woolley TE, Yeh C-Y, Ou K-L, Maini PK, Chuong C-M. Self-organizing hair peg-like structures from dissociated skin progenitor cells: New insights for human hair follicle organoid engineering and Turing patterning in an asymmetric morphogenetic field. Exp Dermatol. 2019;28(4):355–366. doi: 10.1111/exd.13891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rahmani W, Sinha S, Biernaskie J. Immune modulation of hair follicle regeneration. npj Regen Med. 2020;5:9. doi: 10.1038/s41536-020-0095-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schweitzer JS, Song B, Herrington TM, et al. Personalized iPSC-Derived Dopamine Progenitor Cells for Parkinson’s Disease. New Engl J Med. 2020;382(20):1926–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mandai M, Watanabe A, Kurimoto Y, et al. Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration. N Engl J Med. 2017;376(11):1038–1046. doi: 10.1056/NEJMoa1608368 [DOI] [PubMed] [Google Scholar]
- 65.Lanza R, Russell DW, Nagy A. Engineering universal cells that evade immune detection. Nat Rev Immunol. 2019;19(12):723–733. doi: 10.1038/s41577-019-0200-1 [DOI] [PubMed] [Google Scholar]
- 66.González BJ, Creusot RJ, Sykes M, Egli D. How safe are universal pluripotent stem cells? Cell Stem Cell. 2020;26(3):307–308. doi: 10.1016/j.stem.2020.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogawa J, Pao GM, Shokhirev MN, Verma IM. Glioblastoma model using human cerebral organoids. Cell Rep. 2018;23(4):1220–1229. doi: 10.1016/j.celrep.2018.03.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Linkous A, Balamatsias D, Snuderl M, et al. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep. 2019;26(12):3203–3211.e5. doi: 10.1016/j.celrep.2019.02.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dost AFM, Moye AL, Vedaie M, et al. Organoids model transcriptional hallmarks of oncogenic KRAS activation in lung epithelial progenitor cells. Cell Stem Cell. 2020;27(4):663–678.e8. doi: 10.1016/j.stem.2020.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Itoh M, Kiuru M, Cairo MS, Christiano AM. Generation of keratinocytes from normal and recessive dystrophic epidermolysis bullosa-induced pluripotent stem cells. Proc Natl Acad Sci USA. 2011;108(21):8797–8802. doi: 10.1073/pnas.1100332108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Volpato V, Webber C. Addressing variability in iPSC-derived models of human disease: guidelines to promote reproducibility. Dis Model Mech. 2020;13(1). doi: 10.1242/dmm.042317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vigilante A, Laddach A, Moens N, et al. Identifying Extrinsic versus Intrinsic Drivers of Variation in Cell Behavior in Human iPSC Lines from Healthy Donors. Cell Rep. 2019;26(8):2078–2087.e3. doi: 10.1016/j.celrep.2019.01.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cakir B, Xiang Y, Tanaka Y, et al. Engineering of human brain organoids with a functional vascular-like system. Nat Methods. 2019;16(11):1169–1175. doi: 10.1038/s41592-019-0586-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wimmer RA, Leopoldi A, Aichinger M, et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565(7740):505–510. doi: 10.1038/s41586-018-0858-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ormel PR, Vieira de Sá R, van Bodegraven EJ, et al. Microglia innately develop within cerebral organoids. Nat Commun. 2018;9(1):4167. doi: 10.1038/s41467-018-06684-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin Y-T, Seo J, Gao F, et al. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron. 2018;98(6):1141–1154.e7. doi: 10.1016/j.neuron.2018.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abud EM, Ramirez RN, Martinez ES, et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron. 2017;94(2):278–293.e9. doi: 10.1016/j.neuron.2017.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roy B, Simard M, Lorthois I, et al. In vitro models of psoriasis. In: Skin Tissue Models for Regenerative Medicine. Elsevier; 2018:103–128. doi: 10.1016/B978-0-12-810545-0.00005-X [DOI] [Google Scholar]
- 79.Naik S, Larsen SB, Gomez NC, et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature. 2017;550(7677):475–480. doi: 10.1038/nature24271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu CP, Polak L, Keyes BE, Fuchs E. Spatiotemporal antagonism in mesenchymal-epithelial signaling in sweat versus hair fate decision. Science. 2016;354(6319). doi: 10.1126/science.aah6102 [DOI] [PMC free article] [PubMed] [Google Scholar]


