ABSTRACT
Tuberculosis continues to be an important public health problem. Particularly considering Beijing-family strains of Mycobacterium tuberculosis, which have been associated with drug-resistance and hypervirulence. The Beijing-like SIT190 (BL) is the most prevalent Beijing strain in Colombia. The pathogenic mechanism and immune response against this pathogen is unknown. Thus, we compared the course of pulmonary TB in BALB/c mice infected with Classical-Beijing strain 391 and BL strain 323. The disease course was different among infected animals with Classical-Beijing and BL strain. Mice infected with BL had a 100% mortality at 45 days post-infection (dpi), with high bacillary loads and massive pneumonia, whereas infected animals with Classical-Beijing survived until 60 dpi and showed extensive pneumonia and necrosis. Lung RNA extraction was carried out at early (day 3 dpi), intermediate (day 14 dpi), and late (days 28 and 60 dpi) time points of infection. Transcriptional analysis of infected mice with Classical-Beijing showed several over-expressed genes, associated with a pro-inflammatory profile, including those for coding for CCL3 and CCL4 chemokines, both biomarkers of disease severity. Conversely, mice infected with BL displayed a profile which included the over-expression of several genes associated with immune-suppression, including Nkiras, Dleu2, and Sphk2, highlighting an anti-inflammatory milieu which would allow high bacterial replication followed by an intense inflammatory response. In summary, both Beijing strains induced a non-protective immune response which induced extensive tissue damage, BL strain induced rapidly extensive pneumonia and death, whereas Classical-Beijing strain produced slower extensive pneumonia later associated with extensive necrosis.
KEYWORDS: M. tuberculosis, Beijing, Beijing-like, virulence, transcriptomics, immune response, Colombia
Introduction
Although in recent years the World Health Organization (WHO) has reported a small but sustained decrease in the incidence of tuberculosis (TB) worldwide, TB remains a serious public health problem and Mycobacterium tuberculosis (MtB) is considered one of the main infectious agents in terms of single-pathogen mortality. In 2019, the WHO estimated that about 10 million people suffered active TB and 1.2 million died [1].
Colombia is ranked as having a medium-high TB burden, with approximately 14,684 cases in 2019 [2]. Studies carried out in Valle del Cauca reported that the Mtb Beijing genotype (recently renamed lineage 2) has been circulating in Colombia since 1997, specifically in the port of Buenaventura (Principal commercial port of the country) in the pacific coast, whose inhabitants are mainly African–Colombian [3]. Beijing's genotype is the most prevalent in this region and the third most common genotype in the entire country [4]. The BL SIT 190 genotype, was isolated and identified in 2010 [5] and is also prevalent in Colombia. This subtype of Beijing strain is characterized by the absence of spacers 1–34 and 40 of the direct repeats (DR) polymorphic locus. Beijing isolates are frequently drug resistant and patients suffer severe disease with a fatal outcome in most cases. Since its identification, the Beijing genotype has been studied by comparative molecular genomics in order to elucidate its virulence disease mechanisms [6,7]. However, there is still a considerable knowledge gap that needs to be addressed. In order to contribute to this significant matter, the present study assessed the in-vivo and in-vitro virulence of Colombian Classical-Beijing and BL genotypes and the host immune response.
For the in vivo study, we used a well-established murine model of progressive pulmonary TB in BALB/c mice infected by the intratracheal route with a high bacterial dose. Since mice are not a natural host of Mtb, a high bacterial infecting dose is required to produce progressive disease [8,9]. In this model, when mice are infected with the laboratory mild virulence strain H37Rv there is an initial cell-mediated immune response characterized by high expression of Th1 cytokines, peaking at 21-days post infection (dpi) [9,10]. High production of interferon gamma (IFN-γ) and interleukin 12 (IL-12) efficiently control bacterial replication by inducing macrophage activation, which consequently increases the production of TNF-α. This response is associated with granuloma formation and temporal control of bacillary growth. Four weeks after the infection, there is a progressive immunomodulation mediated by the emergence of Th-2 cells and high production of anti-inflammatory cytokines, including IL-10 and TGF-β, that in coexistence with a decrease in the production of Th1 cytokines, progressive pneumonia, and high bacillary load, produce animal’s death [11]. We hypothesized that although both Beijing strains are highly similar genetically, the disease phenotype would vary significantly in terms of animal mortality, lung bacillary load, and histopathological damage, in association with a differential mice lung gene expression profile during the early, intermediate, and late phase of the disease determined by RNASeq.
Methods
Ethics statement
All animal experiments were performed in accordance with the ethical criteria of the Medicine Faculty at Universidad Nacional de Colombia committee, Assessment Act No. 011–104-15 and the Mexican Law NOM 061-Z00-1999 and approved by the Internal Committee for the Care and Use of Laboratory Animals (CICUAL) of the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán (Protocol number PAT-1846-16/20).
Bacterial strains:
The Mtb strains used in this study belong to the Beijing family lineage 2. BL 323 SIT 190 strain and Classical-Beijing 391 SIT 1 strain. BL 323 strain is an Extensively Drug-Resistant (XDR) strain and was isolated from a patient from the Port of Buenaventura on the Colombian west coast. This patient died of pulmonary and extrapulmonary TB and was the first clinical isolate described as SIT 190. The Classical-Beijing 391 strain (also a multi-drug-resistant MDR strain) was kindly donated by the Mycobacteria Laboratory from the National Laboratory Network of the National Institute of Health in Colombia. To confirm the bacteria genotype, the Spoligotyping [12] and MIRU-VNTR 24-loci [13] tests were performed. The typing results were analyzed by the web application MIRU-VNTRPlus (https://www.miru-vntrplus.org/) using the default parameters. To confirm the antibiotic susceptibility profile, liquid culture was performed in BACTEC MGIT 960® automated 7H9 medium, according to the manufacturer’s instructions.
Bacterial culture
The stored bacterial strains in glycerol were thawed and cultivated in Lowenstein Jensen (LJ) solid culture medium. For the infection of animals and cells, a bacterial massive culture was carried out in 60 ml of Middlebrook 7H9 medium supplemented with 5% glycerol, 10% OADC, and 0.02% of Tyloxapol Premium (SIGMA/T8761) to avoid clump formation, without antibiotics. Bacteria were recovered in the mid logarithmic growth phase (DO: 0,013 for BL and DO: 0,0325 for Classical-Beijing) and were centrifuged and recovered free of growth medium and were stored in aliquots of 1-ml. The aliquots were standardized to a concentration of 250.000 bacteria in 100 µl of sterile saline and stored at −80°C. The aliquots were thawed and prepared on the day of infection and after animal infection, the remnant of the bacterial inoculum was plated to confirm the number of CFU administered to the animals.
Cell culture for the in-vitro infection assay
The mouse alveolar macrophage cell-line MH-S (ATCC® CRL-2019 ™) was used for the in vitro assays. Cells were thawed at 37°C in RPMI medium (Caisson Labs, USA, Cat. RPL03) and supplemented with 10% fetal bovine serum (GIBCO, USA); a mass culture was generated until obtaining 70% of confluence for in vitro infection with the Mtb strains.
In vitro assays of macrophage infection
The MH-S cells were adjusted until obtaining 1 × 105 cells per well in 96-wells plates with 200 µl of RPMIc medium. Following that, the cells were incubated for 24 hours at 37°C with 5% CO2. From bacterial aliquots, the bacterial concentration was adjusted to a final concentration of 5 × 105 bacteria/well to obtain a multiplicity of infection (MOI) of 1:5. Cells were infected with 100 µl of each strain (three wells per strain and a control of cells without infection), and as a control we used Mtb reference strain H37Rv (ATCC 25,618). Bacteria were sonicated for 30 seconds at 20 kHz and added to each well for posterior incubation for 3 hours at 37°C and 5% of CO2. The supernatants from each well were collected in a pool for cytokine detection and cells were washed with RPMI at three different time-points: 3 hours (day 0), 3 and 4 days after incubation. Thereafter, cells were lysed with 1% SDS for 10 min and 20% bovine serum albumin was added to stop the reaction. Subsequently, serial dilutions were made in the liquid culture medium Middlebrook 7H9 USA and were sown in 7H10 Middlebrook plates. The viability of intracellular bacteria was determined by counting colony forming units (CFUs) [14]. The cytokines TNF-α, IFN-γ, IL-12P70 and IL-10 present in the supernatant of infected cells and in the supernatant of non-infected macrophages were determined in a single measure using ELISA kits (BioLegend Company, CA, USA) according to manufacturer´s instructions. The culture supernatants of non-infected and infected macrophages with either strain were collected in one pool for each condition.
Experimental model of progressive pulmonary TB in BALB/c mice:
The model of progressive pulmonary TB has been extensively described [9]. Briefly, bacterial aliquots were thawed and sonicated for 30 seconds at 20 kHz to disaggregate the bacterial clumps. Male BALB/c mice of 6–8 weeks old and ~22 gr weight were used. Mice were anesthetized with 100 µl of Sevofluorane at 100% in a gas chamber, and infected with either strain (Classical-Beijing or BL) intratracheally with approximately ~2.5×105 (232.500 bacteria/100 µl for BL and 215.000/100 µl in Classical-Beijing) bacteria using a cannula. After spontaneously recovering, groups of six animals were housed in microisolators connected to equipment with negative air pressure in a bioterium with level III safety facilities. Animals were monitored daily in order to follow disease progression. In case of severe signs of respiratory insufficiency or significant weight loss, humanitarian euthanasia was performed under pentobarbital anesthesia. Groups of six mice were euthanized by exsanguination after anesthesia with 210 mg/kg of intraperitoneal pentobarbital on day 1, 3, 7, 14, 21, 28 and 60 dpi, lungs and spleens were immediately recovered. The right lungs of the six animals were immediately frozen in liquid nitrogen, stored at −80°C and used to determine the bacillary load (CFUs). The left lungs of three animals were perfused with absolute ethanol, fixed for at least 72 hours and used for histopathological studies. The other three remaining lungs were immediately stored in liquid nitrogen at −80°C to perform RNA extraction and quantitative PCR assessment of transcripts (qRT-PCR). Two independent experiments were performed.
For the histopathologic study of lungs, the left lungs of three animals from the two different groups infected with either Beijing strain for each day dpi was perfused intratracheally with absolute ethanol. Parasagittal sections were dehydrated and embedded in paraffin (Oxford Labware, St Louis, MO, EE. UU). Sections of 5 µM were obtained with a microtome and stained using Hematoxylin-Eosin (HE). Using an automated histology system (Leica Microsystems, QWin Leica, Milton Keynes, United Kingdom), the size and number of granulomas per lung area was measured. The percentage of lung surface affected by pneumonia was also determined.
After CFUs quantification and histopathology analysis, the representative time-points for early, intermediate, and advanced disease were determined. These were set at day 3 (early), 14 (intermediate), and 28 (late) for both strains and at day 60 (very late) in animals infected with Classical-Beijing. Three lungs were used for each time point, considered as biological replicates to perform the RNA-seq.
RNA Extraction, library construction and RNA-Seq:
Lungs stored at −80°C were removed from the freezer and kept in liquid nitrogen until processed. In a sterile mortar homogenizer, lungs were pulverized using liquid nitrogen and aliquots of lung powder were stored in Eppendorf tubes; subsequently, Buffer RLT Plus (Cat No./ID: 1,053,393 Qiagen) supplemented with 10% β-mercaptoethanol was added to preserve RNA integrity. Then, RNA extraction was performed using the Quick-RNA Miniprep Kit (Zymo Research cat. R1054/R1055), following the manufacturer’s instructions. Once the RNA was extracted, its quality was evaluated at a 260/280 ratio using the EpochTM Microplate Spectrophotometer, and the integrity of ribosomal subunits was evaluated in 2% agarose gel.
RNA with quality 260/280 ratio higher than 0.8 with well-preserved ribosomal subunits was confirmed in gel, evaluating the integrity of ribosomal subunits (RIN) with the microchip bioanalyzer Agilent, following the manufacturer’s instructions (Agilent RNA 6000 picoKit, cat. No. 5067–1513). Samples with RIN higher than 8 were used for the production of DNA libraries and sequencing. The quantification of RNA for libraries construction was carried out with Qubit RNA HS (ThermoFisher cat., Q32852).
Libraries for mouse transcriptome sequencing were prepared using the TrueSeq® Illumina Kit, following the manufacturer’s instructions. Then, libraries concentration was determined by fluorometry using the qubit dsDNA HS (ThermoFisher cat. Q32851), and size fragments of libraries were evaluated with a bioanalyzer (High sensitivity DNA Kit Agilent, Cat 50,674,626). Finally, the equimolar mixing of the samples was carried out, and they were sequenced in an Illumina HiSeq instrument.
Bioinformatics:
Libraries were sequenced in paired-end versions (101 bases) in an Illumina HiSeq2500 equipment at the RAI (Red de Apoyo a la Investigación, México City, México). Raw reads were deposited under the bioproject PRJNA635984 (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA635984) and in the Gene Expression Omnibus database (GSE169541) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169541). The quality control was checked and trimmed (>20Q) using fastqc and Trim_Galore, respectively [15]. Next, filtered reads were aligned with the Mus musculus from NCBI (assembly GRCm38.p6) as the reference genome using BWA_MEM [16] with default parameters. Read counts were obtained using htseq-count [17]. The differentially expressed genes (DEG) were identified considering DEG those with a p-value of <0.01 by DESeq2 (version 3.7) [18]. The GO (Gene Ontology) and KEGG enrichment were evaluated using WebGestalt [19]. GO and pathway Over-Representation Analysis (ORA) were also conducted using the InnateDB platform [20]. For the KEGG and GO enrichment p-value of 0.01 was used as the cutoff. Protein–protein interactions and network analysis were performed with the same platform, applying the NetworkAnalyst program [21].
Comparative transcriptome analysis was carried out at representative time-points of the infection with each strain, as mentioned before: day 3, 14 and 28 dpi in the case of infection with the BL 323 strain; and day 3, 14, 28 and 60 in the case of infection with strain Classical-Beijing 391. A sample from day 3 was used as a reference for all comparisons. Additionally, we look for a comparative transcriptome analysis between strains at day 3, 14 and 28 dpi. We performed RNA-Seq studies on three biological and independent experiments for each condition. The lungs infected with either strain was at early infection (3 dpi), after 2 weeks (14 dpi) when the formation of granulomas began, at 28 dpi when different disease pattern was produced by either strain, and 60 dpi when extensive pneumonia with necrosis was observed. For the lungs infected with the BL strain, we obtained an average of 19,018,945 (+/−1,112,382.23) of reads for the reference condition (3 dpi), 21,177,772 (+/−2,616,700.08) for the 14 dpi, and 19,591,732 (+/−2,174,103.73) for the 28 dpi. Regarding lungs infected with the Classical-Beijing, we obtained an average of 18,178,599 (+/−3,520,162.36) of reads for the reference condition (3 dpi), 21,438,481 (+/−909,426.19) for the 14 dpi, 19,977,170 (+/−2,420,729.24) for the 28 dpi and 18,634,158 (+/−2,462,101.56) for 60 dpi.
Statistical Analysis: Statistical significance between the experimental groups was determined by two-way ANOVA tests.
An overview of the methodology is shown in Figure 1.
Figure 1.
Methodological scheme. Graphical view of the methodological approach. A. In-vitro approach of the study. B. In-vivo approach of the study
Results
Molecular typing of strains and drug susceptibility determination.
The strains genotype were confirmed by Spoligotyping [12] and MIRUs VNTR [13]. Each strain profile was analyzed using the web application MIRU-VNTRPlus https://www.miru-vntrplus.org/ [22,23] with the typing results. A phylogenetic tree using the algorithm Neighbor-Joining was constructed to determine the closeness of strains used in this study and is shown in (Supplementary Figure 1.) The 323 strain was classified as BL genotype; this genotype has a characteristic Spoligotype in which the spacers 1 to 34 are absent, the spacers 35 to 43 are present and have additional absence of spacer 40; the majority of the Colombian Beijing strains have this Spoligotype [5]. On the other hand, strain 391 has all the spacers from 35 to 43 and was considered in international databases as SIT1 Classical-Beijing genotype.
Regarding drug susceptibility, using the BACTEC MGIT 960® automated 7H9 medium, strain 323 BL is resistant to all first-line antibiotics. In a previous report on the whole genome of BL strain, the analysis show SNPs in genes associated with second line antibiotic resistance. So, this is an extensively drug-resistant strain (XDR) [24]. Classical-Beijing 391 is an MDR strain, with resistance to isoniazid, rifampin and streptomycin.
BL genotype shows higher replicative ability and induce more production of pro-inflammatory cytokines than Classical-Beijing genotype in infected murine alveolar macrophages
For the in vitro infection study, we used alveolar macrophages MH-S transformed by SV40 virus from BALB/c mouse. With this in vitro infection model, the macrophage ability of bacterial killing and cytokine secretion was studied, comparing the reference H37Rv strain with Classical-Beijing and BL strains. Previously we determined the growth curves of both strains in-vitro (Supplementary Figure 2). Classical-Beijing showed higher replicative activity (g = 31.36 h) than BL (g = 41.8 h) in liquid culture 7H9 broth. Within the macrophages, BL strain showed significantly higher replicative activity than Classical-Beijing and H37Rv strains at 3 and 4 dpi (Figure 2a); in order to understand the expression of pro- and anti-inflammatory cytokines by infected macrophages, a qualitative measurement was performed. IL-1-β production was undetectable in macrophages infected with either of the three strains (data not shown). TNF-α production increased at day 4 dpi by macrophages infected with strain BL and was very low with the other strains. IFN-γ at the beginning was induced higher by H37Rv, which decreased on day 3 and on day 4 was equal to day 0, but higher production was induced by those infected with BL on day 3. IL-12P70 was induced in very similar amounts, but macrophages infected with either of the two Beijing strains did not produce anything on day 3, and on day 4 produced similar levels. IL-10 was not detected on day 0 in any of the strains, on day 3 was more induced by H37rv, while Classical-Beijing strain induced very low IL-10 production after 4 days of incubation (Figure 2 B-E). Thus, during late in vitro infection of alveolar macrophages (3 and 4 days), BL strain induced higher production of pro-inflammatory cytokines, while macrophages infected with Classical-Beijing were arrested in the production of the anti-inflammatory cytokine IL-10.
Figure 2.
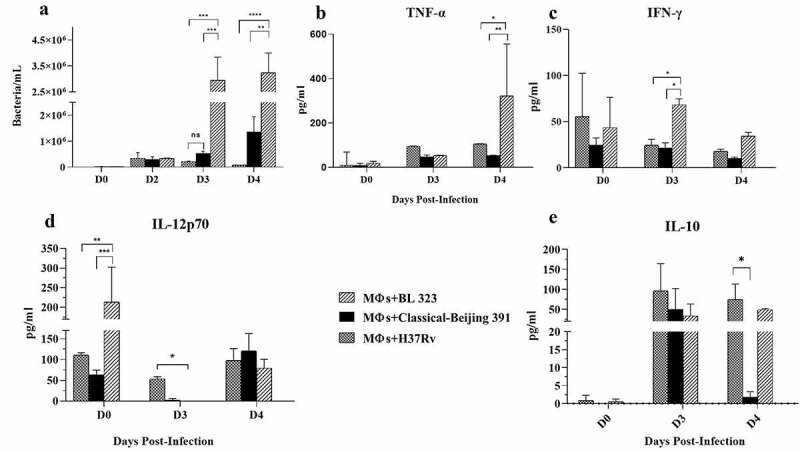
Determination in-vitro of the intracellular proliferative activity of Beijing strains and cytokine production in infected alveolar macrophages. A) Alveolar macrophages cell line MH-S were infected n = 3 with the indicated strain (MOI 5/1), and at different time points infected cells of the three replicates were collected and prepared to determine bacterial viability by colony forming units quantification. BL showed significant higher proliferative activity after 3 and 4 days of incubation. B-F) The supernatants were collected in pools from the infected macrophages after each day PI, and the indicated cytokines were measured by ELISA (asterisk indicate statistical significance * = P < 0.05, ** = P < 0.005, *** = P < 0.0005, **** = P < 0.0001
Survival, Bacillary load and histopathology produced by Beijing strains infection in BALB/c mice
In order to compare the level of virulence between Beijing strains, groups of BALB/c mice (40 per group for 2 separate experiments) were infected intratracheally with 2.5 × 105 Mtb-CFUs of BL or Classical-Beijing. Virulence was determined by survival rates, quantification of CFU from lungs and spleen at different time points, and the extension of tissue damage was determined by evaluating the percentage of the lung surface affected by pneumonia and granuloma size.
BL showed higher virulence, animals infected with this strain started to die after 28 dpi, and at 45 dpi all the animals were dead (Figure 3a); whereas animals infected with strain Classical-Beijing lived up to 60 dpi. In comparison with mice infected with Classical-Beijing strain, the lungs of animals infected with strain BL showed significantly higher bacillary burden from 2 weeks of infection (Figure 3b), and this was more striking in the spleen where 20 times more CFUs were counted, suggesting that strain BL was highly efficient to disseminate. This result was in concordance with the clinical evolution of the patient from which this strain was isolated (5).
Figure 3.
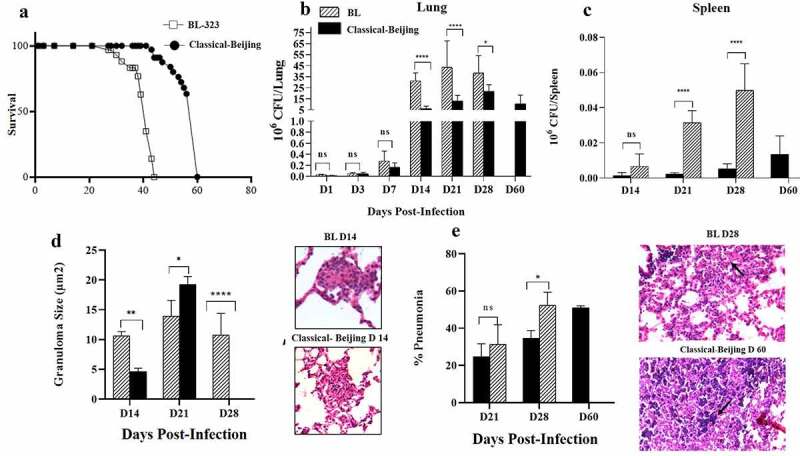
Survival, bacillary loads and histological changes in the lungs and spleen of BALB/c mice infected with Beijing strains. A). Survival of BALB/c mice infected by intratracheal injection of M. tuberculosis strains BL or Classical Beijing. B) Kinetics of bacillary loads in lungs infected with Beijing Strains. C). Bacterial burdens in the spleen, which indicate dissemination from lungs causing extrapulmonary TB. D) Granuloma size determined by automated morphometry and representative micrographs of granulomas (Black arrows) after two weeks of infection with the indicated strain. E) Percentage of lung surface affected by pneumonia (Black arrows strain BL day 28) determined by automated morphometry, and representative low-power micrographs of the lung from mouse infected after one month with strain BL, showing extensive areas of lung consolidation, and extensive pneumonia with areas of necrosis (Black arrows strain Classical Beijing day 60) in the lung of mouse after 2 months of infection with strain Classical Beijing. Multiple comparisons were performed using two-way ANOVA. Asterisks represent statistical significance * = P < 0.05, ** = P < 0.005, *** = P < 0.0005, **** = P < 0.0001 and n.s. Non-significant differences
Regarding histopathology, granuloma formation started with both strains at 14 dpi (Figure 3d). At 21 dpi, mice infected with Classical-Beijing showed more and larger granulomas, while those infected with BL strain exhibited some patches of pneumonia and scarce disorganized granulomas, pneumonia increased 20% after 1 week at 28 dpi (Figure 3e) when the animals infected with this strain started to die, and all the animals were dead at 45 dpi. Whereas animals infected with Classic-Beijing strain showed lesser pneumonia at 28 dpi but after 2 months of infection there were extensive pneumonia and necrosis (Figure 3e).
To obtain general information about the immune response produced in the infected animals, pro-inflammatory and anti-inflammatory cytokines, as well as inducible nitric oxide synthase (iNOS) were quantified by qRT-PCR. Both strains induced similar expression during early infection (3 dpi), whereas after 2 weeks’ post-infection (14 dpi)BL induced higher expression of the anti-inflammatory cytokines IL-10 and TGFβ. While both strains induced low expression of TNF α; however, mice infected with Classical-Beijing exhibited very high TNF expression at late disease (60 dpi), these results show that mice infected with these Beijing strains do not produce a protective immune response, allowing the progression of an aggressive pathology in short time (Figure Supplementary 3).
Transcription profile of the lungs from mice infected with strain BL
In comparison with 3 dpi, DE analysis using DEseq2 showed 447 and 487 over-expressed genes at 14 and 28 dpi for BL strain, respectively (Figure Supplementary 4A, Supplementary File 1: Tables supplementary 1 and 2). Of these, 349 were shared (Figure supplementary 4A). We found 602 down-regulated genes at 14 dpi and 620 at 28 dpi. Of these, 473 down-regulated genes were shared at 14 dpi and 147 at 28 dpi. (Figure Supplementary 4A and B, Supplementary File 1: Table supplementary 3).
KEGG enrichment pathway analysis (EPA) on the down-regulated genes at 14 dpi showed genes involved in antigen presentation and processing, phagosome, response to Staphylococcus aureus, asthma, and allograft rejection, as the most significant pathways (Figure 4a). Additionally, Gene ontology over-representation analysis (ORA) (Significance threshold 1,3) also indicated genes related with immune response, including MHC class II protein complex, cellular response to IFNγ and IFNβ, among others (Figure Supplementary 5). Genes involved in response to Stapylococcus aureus included MHC class II genes, such as H2-Eb1 and H2-Ab1, also are included the integrin alpha M and complement cascade genes such as C3, C2, Cfb, Clqa and Clqc [25]. Additionally, the protein–protein interactions showed high nodes of interaction involved in the immune response, particularly the pro-inflammatory responses like the Ikbkg gene and the Hsp90ab1 (Figure Supplementary 6). The most repressed gene at 14 dpi with BL strain was the Tnfrsf8 gene (Fold change: −7.79;) (Supplementary File 1: Supplementary Table 3), which is a member of the TNF-receptor superfamily. The second-most repressed gene was the CXCL9 chemokine (Fold change: −7.7,) (Supplementary File 1: Supplementary Table 3), which contributes to the growth, movement, or activation of immune and inflammatory cells that participate in the granuloma formation to mycobacterial infection [26]. The down-regulated genes that are involved in pro-inflammatory responses were CCL3 or macrophage inflammatory protein 1 (MIP-1α), CCL4, CXCl16, interleukin 7 (IL-7) and SHARPIN (Fold change: −2.19), whose product plays a major role in pro-inflammatory cytokine induction in response to TLR activation in macrophages [27] and it is required for optimal NLRP inflammasome activation [28]. Moreover, we found similar results in the EPA and ORA analyses at 28 dpi (Supplementary figure 7).
Figure 4.
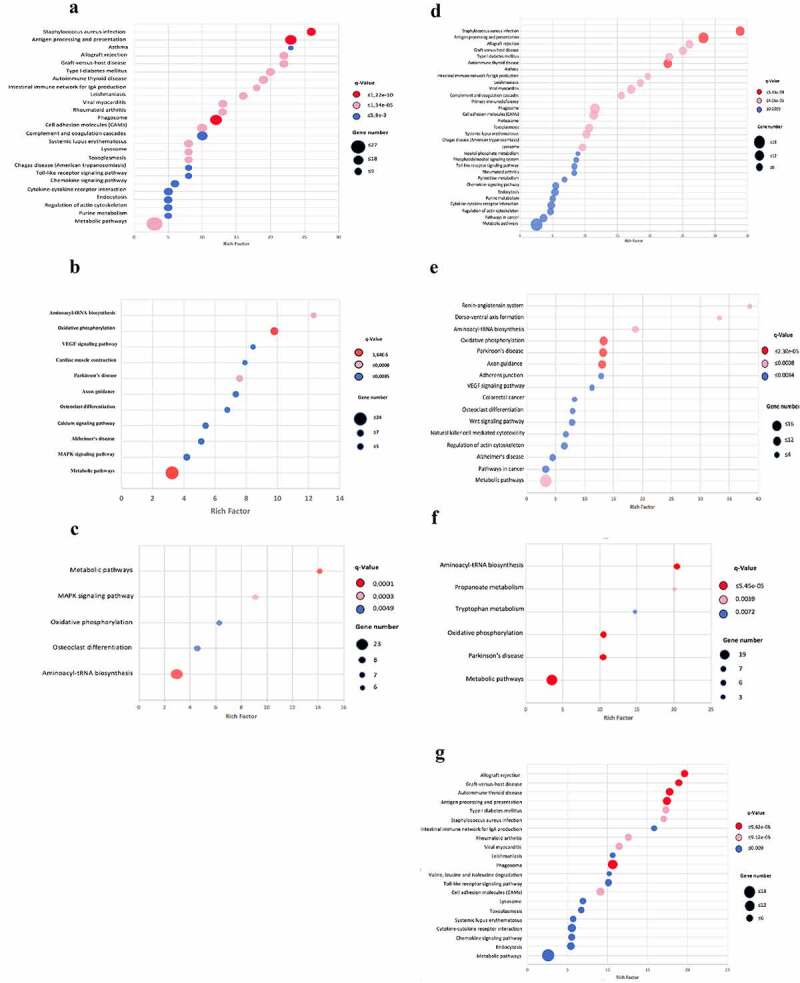
Scatter plot of the KEGG pathway enrichment analysis for DEGs during infection with Beijing strains. Each circle in the graph represent a KEGG pathway, with its name in the Y-axis and the enrichment factor indicated in the X-axis. Rich factor is the ratio of the numbers of DEGs annotated in each pathway related to the proportion of all genes mentioned in the same pathway. Higher enrichment factor means a more significant enrichment of the DEGs in a given pathway. The color of the circle represented the q-value, which was the adjusted P after multiple hypothesis testing, ranging from 0–1. A smaller q-value indicated a higher reliability of the significance of the enrichment of DEGs in each pathway. The sizes of the circles represent the number of enriched genes. Left panel: mice infected with BL strain; A) Down regulated genes at 14 dpi. B) Over-expression genes at 14 dpi. C) Over-expression genes at 28 dpi. Right panel: mice infected with Classical Beijing strain; D) Down regulated genes at 14 dpi. E) Over-expression genes at 14 dpi. F) Over-expression genes at 28 dpi. G) Over-expression genes at 60 dpi
On the other hand, we found that the most enriched pathways of the over-expression genes at 14 dpi with BL strain were as follows: aminoacyl-tRNA biosynthesis, oxidative phosphorylation, VEGF, and calcium signaling pathways and MAPK signaling pathway (Figure 4b). Results obtained by ORA showed genes related to protein binding, mitochondrial activity, blood vessel remodeling, and sphingosine kinase activity (Supplementary Figure 8). Network analysis of protein–protein interactions displayed genes involved in immune response as important nodes of interaction, like the Fyn gene (Fold change: 1.09, supp. material) that encodes for a membrane-associated tyrosine kinase implicated in the control of cell growth and the pro-inflammatory IL16 (Figure supplementary 9). Another important hub of interaction is the Smarca2 gene, which is involved in transcriptional activation and repression of selected genes by chromatin remodeling (alteration of DNA-nucleosome topology) [29].
Moreover, the Fyn gene with a fold change of 1.09 (Supplementary File 1: Supplementary Table 1) is a member of the protein-tyrosine kinase oncogene family that encodes a membrane-associated tyrosine kinase, which has been implicated in the control of cell growth and is related to diverse significant immune activities, such as IFN-γ pathway, IL-2-mediated signaling events, T-cell receptor, TGF-β receptor signaling, and TRAIL (TNF superfamily, member 10). The over-expression of the Spkh2 gene was also found (Fold change: 1.3; Supplementary File 1: Supplementary Table 1) which encodes for a sphingosine kinase, and its responsible for suppressing the inflammation [30] and macrophage activation [31]. The over-expression of the Cacna1c and Rras2 genes which are related to calcium signaling and MAPK pathways is striking, as it is associated with pro-inflammatory silencing during Mtb infection [32,33]. Thus, it seems that the expression profile at 14 dpi with BL strain shows a bias toward an anti-inflammatory response.
After 1 month of BL strain infection (28 dpi), there are lesser-activated pathways than at 14 dpi, but several are more common at both points (Supplementary Figure 4A), indicating a slight change in the host response (Figure 4c). One of the activated pathways was the MAPK signaling pathway, over-expressing the immune-suppressing MAPK7 and MAPK12 that suggest a maintained anti-inflammatory response. ORA analysis revealed similar gene profile at 14 and 28 dpi, which belong to pathways related to protein binding, mitochondrial activity and blood vessel remodeling. In this case, the sphingosine kinase activity was not active, indicating a pro-inflammatory environment (Supplementary Figure 10). The most interconnected gene network was Fyn and Smarca2, followed by Bmpr2 gene, which has been involved in pulmonary arterial hypertension (Supplementary Figure 11).
Transcription profile of the lungs from mice infected with strain Classical-Beijing
Mice infected with the strain Classical-Beijing 391 showed longer survival; therefore, pulmonary RNA was isolated at 3, 14, 28 and 60 dpi. In comparison with day 3dpi, we found 489, 441 and 395 down-regulated genes at 14, 28 and 60 dpi, respectively. Of these, most of the genes were shared at 14 and 28 dpi (339 genes) (Supplementary Figure 12 B). Regarding over-expressed genes, we found 285 genes at 14 dpi and after one (28 dpi) and two (60 dpi) months were 325 and 468, respectively (Supplementary Figure 12 A, Supplementary File 1: Supplementary Tables 4, 5, 6, and 7). Of these, 188 over-expressed genes were shared only at 14 and 28 dpi.
The EPA analysis on the down-regulated genes at 14 dpi revealed that the enriched genes are those involved in antigen presentation and processing, response to Staphylococcus aureus, asthma, and allograft rejection. These results were similar to those observed in mice infected with the BL strain at the same point (Figure 4d). Moreover, ORA results also indicated a very akin profile of genes related to immune response between both strains under this timeframe (Supplementary Figure 5) such as TLRs. Network analysis displayed similar down-regulated genes in the top of the interactions, being the Ikbkg and Hsp90ab1 significant genes involved in the immune response (Supplementary Figure 6). It is important to note that we found repression of SHARPIN gene in both strains, which has a major role in pro-inflammatory cytokine induction in response to TLR activation in macrophages [34] and optimal NLRP inflammasome activation [28]. Additionally, the Nos2 gene (Nitric Oxide Synthase 2) is also one of the most interconnected genes and suggests that the pro-inflammatory response is active at early infection. The chemokine repression profile (CXCLl9, CCl4, and IL7) observed in Classical-Beijing strain was also similar to BL (Supplementary File 1: Table supplementary 4).
In addition, the EPA on over-expressed genes at 14 dpi showed enriched pathways associated with renin-angiotensin systems, aminoacyl tRNA biosynthesis, oxidative phosphorylation, and VEGF signaling, among others. Pathways linked to the immunology process include the natural killer cell (NK) mediated cytotoxicity pathway (Figure 4e). Additionally, the ORA analysis revealed genes involved in oxidative phosphorylation, mitochondrial activity, renin–angiotensin system, VEGF signaling pathway and CD4 T cell receptor signaling, among others (Supplementary Figure 8). Overexpressed genes that belong to the NK cells mediated cytotoxicity pathways, such as Itgb2, RAC3, Kras, and Ppp3cc, were also detected under this timeframe. The Itgb2 gene encoded an integrin that contributes to NK cell cytotoxicity [35] and it is also involved in leukocyte adhesion and transmigration of leukocytes, including T-cells and neutrophils [36,37].
Protein–protein interactions showed the genes Aph1, Dynclil, Notch3, and Gnb1 as the most interconnected in this network (Supplementary Figure 9). Cytoplasmic dynein 1 coded by the gene Dynclil contributes to the intracellular retrograde motility of vesicles and organelles along microtubules, and Gnb1 is a heterotrimeric guanine nucleotide-binding protein (G proteins), which integrates signals between receptors and effector proteins.
The EPA, at 28 dpi with Classical-Beijing strain, showed lesser activated pathways than at 14 dpi (figure 4f). However, some genes are expressed at both points (Figure 4e), and new pathways were over-expressed, such as the propanoate and tryptophan metabolism. Similar ORA results were obtained when compared with 14 dpi, which included mitochondrial activity and angiogenesis. Some new pathways include regulation of lipid metabolic processes. (Supplementary Figure 10). Analysis of the protein–protein interactions showed Smarca2, Gata2, Rbbp4, and Aph1 genes to be the most interconnected genes in this network (Supplementary Figure 11).
Finally, the over-expression genes at 60 dpi with the strain Classical-Beijing, showed enriched pathways involved in the immune response similar to those observed in the down-regulated genes at 14 dpi (Figures 4d and 4g); some of these pathways were antigen presentation and processing, response to Staphylococcus aureus, asthma and allograft rejection. ORA and network analysis confirmed these results (Supplementary Figure 13 and 14). Interestingly, we found that the over-expressed genes CCL4, IL-7 and SHARPIN were down-regulated in BL at 14 dpi [38].
Comparative transcriptome analysis between strain BL and Classical-Beijing
Comparisons were only made at 3, 14 and 28 days, as the mice infected with the BL strain died at 45 days. The DEseq2 analysis did not find significant differences between the transcriptomes of the mice infected with either strain at 3 and 28, only at 14 days were found differences. At day 14, compared with mice infected with strain Classical-Beijing, only 21 genes were over-expressed in animals infected with BL strain, and nine of these were non-coding RNA (42%) (Table 1A). Among these overexpressed genes induced by BL strain were Sphk2 whose product is Sphingosine kinase 2 a negative regulator of inflammatory macrophage activation [31] and of IL-2 pathways; Dleu2 that is a long noncoding RNA that controls B cell differentiation [39]. Nkiras1 acts as a potent regulator of NF-kappa-B activity by preventing the degradation of NF-kappa-B inhibitor beta (NFKBIB) [40]. Network analysis showed interactions between Dleu2, Sphk2, and Nkiras1 (Supplementary Figure 15) This profile suggests an anti-inflammatory response.
Table 1.
Comparison of overexpressed genes between Beijing strains at 14 dpi
| A. Overexpressed genes of BL at 14 dpi | |||||
|---|---|---|---|---|---|
| Entrez ID | Name | Biotype | Description | Log FC | p-value |
| 17,739 | TrnP | tRNA | tRNA proline,mitochondrial | 0,66 | 0,0008 |
| 17,977 | Ncoa1 | Protein | Nuclear receptor coactivator | 0,44 | 0,0199 |
| 23,874 | Farsb | tRNA | Phenylalanyl-tRNA synthetase, beta subunit | 0,48 | 0,0077 |
| 55,936 | Ctps2 | Protein | Cytidine 5´-triphosphate synthase | 1,27 | 0,0000 |
| 56,632 | Sphk2 | Protein | Sphingosine kinase 2 | 0,88 | 0,0000 |
| 69,721 | Nkiras1 | Protein | NFKB inhibitor interactions | 0,46 | 0,0022 |
| 71,056 | 4933405E24Rik | ncRNA | Non coding RNA | 0,40 | 0,0077 |
| 74,453 | Cfap53 | Protein | Cilia and flagella associated protein 53 | 0,52 | 0,0004 |
| 75,138 | 4930526L06Rik | ncRNA | Non coding RNA | 0,84 | 0,0000 |
| 75,388 | Boll | Protein | Boule homolog, RNA binding protein | 0,52 | 0,0102 |
| 75,784 | Cfap299 | Protein | Cilia and flagella associated protein 299 | 0,55 | 0,0054 |
| 75,888 | 4930573H18Rik | ncRNA | RIKEN cDNA 4930573H18 | 0,74 | 0,0000 |
| 76,933 | Ifi27l2a | Protein | Interferon alpha-inducible protein 27 like 2A | 0,55 | 0,0004 |
| 330,401 | Tmcc1 | Protein | Transmembrane and coiled coil domains 1 | 0,43 | 0,0030 |
| 664,942 | Gm13858 | Predicted gene | Cancer susceptibility candidate 4 pseudogene | 0,48 | 0,0040 |
| 668,253 | Dleu2 | lncRNA | Long non coding RNA | 0,42 | 0,0069 |
| 100,504,464 | E230016K23Rik | lncRNA | Long non coding RNA | 0,72 | 0,0000 |
| 102,632,404 | Gm15396 | lncRNA | Long non coding RNA | 0,30 | 0,0062 |
| 102,635,296 | Gm32673 | lncRNA | Long non coding RNA | 0,60 | 0,0027 |
| 102,636,584 | Gm32673 | lncRNA | Long non coding RNA | 0,42 | 0,0061 |
| 108,167,382 | LOC108167382 | lncRNA | Long non coding RNA | 0,85 | 0,0093 |
| B. Overexpressed genes of Classical-Beijing at 14 dpi | |||||
| Entrez ID | Name | Biotype | Description | Log FC | p-value |
| 15,516 | Hsp90ab1 | Protein | Heat shock protein 90 alpha (cytosolic)2 C class B member 1 | 0,44 | 0,0172 |
| 16,468 | Jarid2 | Protein | Jumonji, AT rich interactive domain 2 | 0,64 | 0,0003 |
| 16,651 | Sspn | Protein | Sarcospan | 0,78 | 0,0000 |
| 21,577 | Tcrb | Protein | T cell receptor beta chain | 0,56 | 0,0001 |
| 27,204 | Syn3 | Protein | Synapsin III | 0,41 | 0,0042 |
| 27,660 | 1700088E04Rik | cDNA | RIKEN cDNA 1700088E04 gene | 0,40 | 0,0102 |
| 76,392 | Slfn5os | lncRNA | Schlafen 5 opposite strand | 0,46 | 0,0028 |
| 78,749 | Filip1 | Protein | Filamin A interacting protein 1-like | 0,68 | 0,0001 |
| 106,205 | Zc3h7a | Protein | Zinc finger CCCH type containing 7 | 0,65 | 0,0001 |
| 106,585 | Ankrd12 | Protein | Ankyrin repeat domain 12 | 0,48 | 0,0038 |
| 109,979 | Art3 | Protein | ADP -ribosyltransferase 3 | 0,37 | 0,0180 |
| 225,182 | Rbbp8 | Protein | Retinoblastoma binding protein | 0,88 | 0,0001 |
| 232,414 | Clec9a | Protein | C -type lectin domain family 9, member a | 0,45 | 0,0168 |
| 278,279 | Tmtc2 | Protein | Tansmembrane and tetratricopeptide repeat containing | 0,62 | 0,0008 |
| 319,371 | D030028A08Rik | lncRNA | Long non coding RNA | 0,37 | 0,0180 |
| 320,249 | D130009I18Rik | lncRNA | Long non coding RNA | 0,71 | 0,0000 |
| 667,373 | Ifit1bl1 | Protein | Interferon induced protein with tetratricopeptide repeats | 0,30 | 0,0066 |
| 101,056,014 | LOC101056014 | misc_RNA | Long non coding RNA | 0,47 | 0,0011 |
| 102,465,200 | Mir6381 | Micro RNA | Micro RNA 6381 | 0,44 | 0,0005 |
| 102,632,231 | LOC102632231 | lncRNA | Long non coding RNA | 0,49 | 0,0038 |
| 102,634,124 | Gm31785 | lncRNA | Long non coding RNA | 0,52 | 0,0003 |
| 102,635,265 | Gm32650 | lncRNA | Long non coding RNA | 0,42 | 0,0036 |
| 102,636,152 | Gm28068 | lncRNA | Long non coding RNA | 0,28 | 0,0151 |
| 102,639,821 | Gm36043 | lncRNA | Long non coding RNA | 0,46 | 0,0006 |
| 102,639,982 | LOC102639982 | lncRNA | Long non coding RNA | 0,41 | 0,0119 |
| 102,640,929 | Gm36876 | lncRNA | Long non coding RNA | 0,66 | 0,0002 |
| 105,243,537 | Gm39429 | lncRNA | Long non coding RNA | 0,57 | 0,0013 |
| 108,167,336 | 108,167,336 | lncRNA | Long non coding RNA | 0,52 | 0,0001 |
| 108,168,339 | 108,168,339 | lncRNA | Long non coding RNA | 0,69 | 0,0002 |
Likewise, DE analysis showed 29 genes with high expression with Classical-Beijing strain. Interestingly, we also found a high proportion of non-coding RNAs (48%), this suggests that the post-transcriptional regulation at 14 dpi could play an important role in the response to different strains (specific host response) (Table 1B). Among the interesting genes, Hsp90ab1 is a molecular chaperone that promotes the maturation, structural maintenance, and proper regulation of specific target proteins involved for instance in cell cycle control and signal transduction [41]. The Alpha-beta T cell receptors (Tcrb) are antigen-specific receptors which are essential to the immune response and are present on the cell surface of T lymphocytes [42]. Network interactions shown the Hsp90ab1 the principal hub in this network (Supplementary Figure 16) These genes and the others in the list, suggest a pro-inflammatory response.
Discussion
The Beijing family or lineage 2 is one of the most prevalent and virulent Mtb genotypes worldwide [43]. This has been shown as its initial description with strains collected from Asia [7]. The Beijing genotype is frequently associated with antibiotic resistance and immune evasion, allowing high bacterial replication, dissemination, and transmission [44–46]. Interestingly, the Beijing genotype is not frequent in Latin America (incidence less than 5%) [47]. The Beijing lineage in Colombia has been circulating in the southwest of the country since 1998 [48]. Two Beijing variants have been identified, SIT190 BL and SIT1 Classical-Beijing, being the former the most prevalent and cause of the highest mortality [49]. At the genomic level, there are 18 single nucleotide polymorphisms (SNPs) between these two Beijing strains, which could be partially related to differences in virulence [49]. Thus, the principal objective of the present work was to study and compare the immune response induced by BL strain 323 (SIT 190) and Classical-Beijing strain 391 (SIT 1), using a murine model of progressive TB and transcriptomics Our results are in agreement with the epidemiological results, showing that BL is highly virulent, which is particularly worrisome considering that it is an XDR strain.
Our in-vitro results infecting murine alveolar macrophages showed higher intracellular proliferation of strain BL at four dpi. BL strain also induced high production of TNFα, while Classical-Beijing strain induced very low production of IL-10 at the same point of infection. These results contrast with a previous study with macrophages derived from monocytes infected with seven Beijing clinical isolates, which induced early production of IL-1β and TNF-α [50]. Thus, it seems that the variations in the response of infected macrophages depend on not only the Mtb strain, but also the type of infected cells [51–53]. In another in vitro study with classical Beijing strains, they induced low production of IL-1β and TNF-α at early times of infection [54], which contrast with the high production of this cytokine after 4 days of incubation with our strain BL, suggesting that the time of infection is also a significant factor related to the production of cytokines.
Regarding murine models, we showed that Beijing strains collected from Asia were highly virulent and induced a non-protective immune response [10]. Then, with isolates from South Africa, we demonstrated that typical highly transmissible Beijing strains produced severe disease. Whereas non-transmissible atypical strains Beijing were attenuated, but both highly virulent and attenuated strains may exist in the same sublineage [55]. In the present study now using Beijing strains from Colombia, we confirmed the high virulence of this genotype, but the comparison between two predominant strains showed that BL is more virulent than Classical Beijing. BL induced rapid exacerbation of the disease from day 21, causing rapid animal’s death due to extensive pneumonia in coexistence with higher bacillary burden. However, the Classical-Beijing strain is also virulent, and although it exhibited slower production of pneumonia and delayed death, there was necrosis at day 60 dpi with high bacillary loads. Thus, both strains are highly virulent and they showed different immune responses that was studied in more detail using transcriptomics in different moments of the disease.
No significant differences were observed between transcriptomes of infected mice with either strain at days 3 and 28, the expression profile was essentially pro-inflammatory, in which over-expression of SHARPIN seems to play an important role, as it activates the inflammasome [28] and participates in cytokine induction in response to TLR activation in macrophages [34]. Although well-formed granulomas were observed after 14 dpi, it seems that at day 3 dpi started the cell recruitment for their formation because several genes involved in cell recruitment were over-expressed, such as those that encode the chemokines CXCL9, CCL3, and CCL4. Gene Hsp90ab showed higher expression (two-fold more) in mice infected with Classical-Beijing strain (Fold change: 1,54), the product of this gene antagonizes CHIP-mediated ubiquitination and degradation of Smad3, allowing TGF-β mediated anti-inflammatory and tissue regenerative signaling.
More significant changes were observed at 14 dpi. At this time point, infection with BL strain showed only 21 up-regulated genes. Interestingly, nine of these were non-coding RNA (42%). Likewise, infected mice with Classical-Beijing strain only showed 29 up-regulated genes; again, a remarkable observation was the high proportion of non-coding RNAs (48%) (Table 1B). Interestingly, the lower virulent strain showed more non-coding RNAs and none of these were shared, indicating a specific host response to Mtb strain. Over-expression of gene ZC3H7A (Zinc Finger CCCH-Type Containing 7A) could be related to this process, because it codes a protein that is a specific regulator of miRNA biogenesis [56]. After 14 dpi, it seems that a partially suppressed immune response was induced by the strain BL, because the genes Nkiras1, Dleu2, and Sphk2 were over-expressed. Nkiras1 is an atypical Ras-like gene that codifies a protein, that prevents the degradation of NF-kappa-B inhibitor beta (NFKBIB), and mediate cytoplasmic retention of p65/RELA NF-kappa-B subunit [57]. The over-expression of the long non-coding gene Dleu2 is important in the humoral immune response because it controls B cell proliferation, while the protein codified by gene Sphk2 should be important in the response of cell-mediated immunity because it regulates IL-2 pathways in T cells suppressing the inflammatory response [30]. Moreover, the over-expression of this gene that codifies sphingosine kinase 2 is a negative regulator of macrophage activation [31], inhibits cell growth and enhances apoptosis [58]. Additionally, the gene Ifi27l2a that is involved in type-I interferon-induced lymphocyte apoptosis should contribute to immune modulation [59,60].
Other epigenomic events could be important after 2 weeks of infection with strain BL as suggested by the network analysis that informs expression of the nuclear receptor coactivator 1 (NCOA1 or SRC-1), which is a transcriptional co-regulatory protein that shows several nuclear receptor interacting domains and an intrinsic histone acetyltransferase activity that modulate gene expression [61]. Another significant cellular activity was cellular proliferation, considering the over-expression of the gene that codifies the protein Ctps2, which is the rate-limiting enzyme in the synthesis of cytosine nucleotides related to cellular replication [62]. Thus, BL strain produced a change in the expression profile after 2 weeks of infection that decreased the pro-inflammatory response, but at day 28 dpi the strong pro-inflammatory response returned as suggested by the lack of the sphingosine kinase gene expression, that probably contributed to the development of rapid and extensive pneumonia producing the animal’s death by respiratory insufficiency [63].
A different transcription profile was seen at 2 weeks dpi with Classical-Beijing strain, which was characterized by strong signaling pathway of NK cells and overexpressed genes related to pro-inflammatory responses, such as Tcrb (T cell receptor b), CLEC9A group V C-type lectin-like receptor (CTLR), which is a receptor whose activation induces pro-inflammatory cytokine production [64], Rbbp8 gene (retinoblastoma-binding protein) that is important for immunological genes recombination and Syn3 which is a member of the synapsin gene family and is important for phagosome maturation. Over-expression of gene Art3, ADP-ribosyltransferase 3 (ADPRT) is important for cell-mediated immune regulation because it encodes a protein that catalyzes a reversible reaction that modifies proteins by the addition or removal of ADP-ribose to an arginine residue, regulating the function of the modified protein; mature T cells, but not B cells or macrophages, express ADPRT and are able to ADP-ribosylate cell-surface proteins that are essential for T cells functions, such as LFA-1, CD8, CD27, CD43, CD44, and CD45 [65].
The communication between cytoskeleton and extracellular matrix is also important at this point of infection with Classical-Beijing strain because there was over-expression of Sspn gene, a member of the dystrophin-glycoprotein complex (DGC) that provides a structural link between the cytoskeleton and the extracellular matrix [66]; Filip1 which encodes a filamin A binding protein that promotes its degradation [67], and Ankrd12 that encodes a member of the ankyrin repeats-containing cofactor family. These proteins may inhibit the transcriptional activity of nuclear receptors through the recruitment of histone deacetylases and in plasma from active TB patients is detected circRNA Hsa_circ_102296 that aligns with ANKRD12 [68]. Another important activity is the function of the endoplasmic rough reticulum (ER) as indicated by the over-expression of Tmtc2, whose encoded protein plays a role in calcium homeostasis in the ER [69]. Finally, the over-expression of the Ifit1bl1 gene (Interferon-induced protein with tetratricopeptide repeats 1B-like 1) is striking because this protein inhibits viral replication and translational initiation [70]. Considering that Mtb is an intracellular pathogen, similar to viruses, it is possible that they share similar intracellular defense mechanisms.
Mice infected with the strain Classical-Beijing showed higher survival; some of them were alive up to 60 dpi. Interestingly, at this point, the expression profile was pro-inflammatory, very similar to day 3 of infection, including some considered as markers of disease severity, such as CCL3 and CCL4, which well correlated with high bacillary burdens and extensive tissue damage (pneumonia with necrosis).
These Beijing strains are highly similar at the genomic level, as reported before they only differ in 18 SNPs and one spacer in the DR locus [49]. However, they showed different virulence and host gene expression profiles, suggesting that small changes at the bacterial genomic level could have a high impact on the phenotype. In comparison with strain Classical-Beijing, the SNPs of the strain BL are located in genes related to the synthesis of ptiocerol dimicoserosate (PDIM) and the transcription factor WhiB6 [49]. These could partially explain the differences described in the present study, but other studies are required to determine more precisely the differences, such as the characterization of the Mtb transcriptome with bacterial RNA isolated from the infected lung tissue, a novel analysis that is now in course in our laboratory.
In conclusion and although our study has several limitations, such as intratracheal high infecting bacterial dose in BALB/c mice that could produce a different profile than obtained with the most common TB model in C57BL/6 mice infected with low bacterial dose by aerosols, we show that two Beijing strains prevalent in Colombia induced a non-protective immune response with extensive tissue damage, BL strain produced rapidly extensive pneumonia and death, while Classical-Beijing strain produced slower extensive pneumonia later associated with necrosis. The transcriptional analysis of infected mice with Classical-Beijing showed several over-expressed genes associated with a pro-inflammatory profile, while mice infected with BL displayed an over-expression of several genes associated with immune-suppression, which would allow high bacterial replication followed by an intense inflammatory response with lung consolidation and necrosis. Further investigations must explore the bacterial mechanisms that regulate these different inflammatory and immune responses.
Supplementary Material
Acknowledgments
We thank the following entities for the financial support of this work:
- Universidad Nacional de Colombia
- Departamento Administrativo de Ciencia, Tecnología e Innovación (Colciencias)-Colombia, Contract Number: CT-731-2018
- Consejo Nacional de Ciencia y Tecnología (CONACyT)-Mexico, Contract Number: 223279
- Sistema de Información de la Investigación, Extensión y Laboratorios de la Universidad Nacional de Colombia (Hermes), Project code: 42665.
- Juan Manuel Hurtado-Ramírez for the technical support, and the installation of programs in the server.
Funding Statement
This work was supported by the Departamento Administrativo de Ciencia, Tecnología e Innovación (CO) [CT-731-2018]; Universidad Nacional de Colombia, Proyecto Hermes [42665]; Facultad de Medicina, Universidad Nacional de Colombia, Proyecto Hermes (53688). We acknowledge the support by CONACyT grant CB-2013-223279 to RHP.
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].WHO | Global tuberculosis report 2020. In: WHO [Internet]. World Health Organization; [cited 2020 Oct 19]. Available: http://www.who.int/tb/publications/global_report/en/
- [2].2019_Boletin_epidemiologico_semana_52.pdf. [22-28 December 2019]. Available: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2019_Boletin_epidemiologico_semana_52.pdf
- [3].Acquired multidrug-resistant tuberculosis—Buenaventura, Colombia, 1998. JAMA. 1998;280(19):1653. doi: 10.1001/jama.280.19.1653-JWR1118-3-1. [DOI] [PubMed] [Google Scholar]
- [4].Moreira CA, Hernández HL, Arias NL, et al. Resistencia inicial a drogas antituberculosas en Buenaventura, Colombia. biomedica. 2004;24:73. [PubMed] [Google Scholar]
- [5].Murcia MI, Manotas M, Jiménez YJ, et al. First case of multidrug-resistant tuberculosis caused by a rare “Beijing-like” genotype of Mycobacterium tuberculosis in Bogotá, Colombia. Infect Genet Evol. 2010;10(5):678–681. [DOI] [PubMed] [Google Scholar]
- [6].Parwati I, Van Crevel R, Van Soolingen D.. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis. 2010;10:103–111. [DOI] [PubMed] [Google Scholar]
- [7].Van Soolingen D, Qian L, de Haas PE, et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol. 1995;33:3234–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hernandez-Pando R, Orozco H, Honour J, et al. Adrenal changes in murine pulmonary tuberculosis; a clue to pathogenesis? FEMS Immunol Med Microbiol. 1995;12:63–72. [DOI] [PubMed] [Google Scholar]
- [9].Hernández-Pando R, Orozcoe H, Sampieri A, et al. Correlation between the kinetics of Th1, Th2 cells and pathology in a murine model of experimental pulmonary tuberculosis. Immunology. 1996;89:26–33. [PMC free article] [PubMed] [Google Scholar]
- [10].López B, Aguilar D, Orozco H, et al. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin Exp Immunol. 2003;133:30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Moreno JR, García IE, de la Luz García Hernández M, et al. The role of prostaglandin E2 in the immunopathogenesis of experimental pulmonary tuberculosis. Immunology. 2002;106:257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kamerbeek J, Schouls L, Kolk A, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Supply P, Allix C, Lesjean S, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zetter M, Barrios-Payán J, Mata-Espinosa D, et al. Involvement of Vasopressin in the pathogenesis of pulmonary tuberculosis: a new therapeutic target? Front Endocrinol (Lausanne). 2019;10:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Babraham bioinformatics - Trim Galore! [cited 2020 Nov 18]. Available: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- [16].Fast and accurate short read alignment with burrows-wheeler transform - PubMed. [cited 2020 Nov 18]. Available: https://pubmed.ncbi.nlm.nih.gov/19451168/ [DOI] [PMC free article] [PubMed]
- [17].Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15. DOI: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang J, Vasaikar S, Shi Z, et al. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45(W1):W130–W137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Breuer K, Foroushani AK, Laird MR, et al. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 2013;41(D1):D1228–D1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhou G, Soufan O, Ewald J, et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234–W241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Weniger T, Krawczyk J, Supply P, et al. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38(Web Server):W326–W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Allix-Béguec C, Harmsen D, Weniger T, et al. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2008;46(8):2692–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rodríguez JG, Pino C, Tauch A, et al. Complete genome sequence of the clinical Beijing-like strain Mycobacterium tuberculosis 323 using the PacBio real-time sequencing platform. Genome Announc. 2015;3(2):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lowy FD. Staphylococcus aureus Infections. N Engl J Med. 1998;339(8):520–532. [DOI] [PubMed] [Google Scholar]
- [26].KEISER TL, PURDY GE. Killing Mycobacterium tuberculosis in vitro: what model systems can teach us. Microbiol Spectr. 2017;5(3):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Algood HMS, Chan J, Flynn JL. Chemokines and tuberculosis. Cytokine Growth Factor Rev. 2003;14(6):467–477. [DOI] [PubMed] [Google Scholar]
- [28].Gurung P, Li B, Subbarao Malireddi RK, et al. Chronic TLR stimulation controls NLRP3 inflammasome activation through IL-10 mediated regulation of NLRP3 expression and caspase-8 activation. Sci Rep. 2015;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Euskirchen G, Auerbach RK, Snyder M. SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J Biol Chem. 2012;287:30897–30905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Samy ET, Meyer CA, Caplazi P, et al. Cutting edge: modulation of intestinal autoimmunity and IL-2 signaling by sphingosine kinase 2 independent of sphingosine 1-phosphate. J Immunol. 2007;179:5644–5648. [DOI] [PubMed] [Google Scholar]
- [31].Weigert A, von Knethen A, Thomas D, et al. Sphingosine kinase 2 is a negative regulator of inflammatory macrophage activation. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:1235–1246. [DOI] [PubMed] [Google Scholar]
- [32].Mehra S, Pahar B, Dutta NK, et al. Transcriptional Reprogramming in Nonhuman Primate (Rhesus Macaque) Tuberculosis Granulomas. PLoS One. 2010;5. DOI: 10.1371/journal.pone.0012266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang X, Wu Y, Jiao J, et al. Mycobacterium tuberculosis infection induces IL-10 gene expression by disturbing histone deacetylase 6 and histone deacetylase 11 equilibrium in macrophages. Tuberculosis (Edinb). 2018;108:118–123. [DOI] [PubMed] [Google Scholar]
- [34].Zak DE, Tam VC, Aderem A. Systems-level analysis of innate immunity. Annu Rev Immunol. 2014;32:547–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Barber DF, Faure M, Long EO. LFA-1 contributes an early signal for NK cell cytotoxicity. J Immunol. 2004;173:3653–3659. [DOI] [PubMed] [Google Scholar]
- [36].Ostermann G, Weber KSC, Zernecke A, et al. is a ligand of the beta(2) integrin LFA-1 involved in transendothelial migration of leukocytes. Nat Immunol. 2002;3:151–158. [DOI] [PubMed] [Google Scholar]
- [37].Bai M, Grieshaber-Bouyer R, Wang J, et al. CD177 modulates human neutrophil migration through activation-mediated integrin and chemoreceptor regulation. Blood. 2017;130(19):2092–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Xu J, Gao X-P, Ramchandran R, et al. Nonmuscle myosin light-chain kinase mediates neutrophil transmigration in sepsis-induced lung inflammation by activating β2 integrins. Nat Immunol. 2008;9:880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Klein U, Lia M, Crespo M, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. [DOI] [PubMed] [Google Scholar]
- [40].Chen Y, Vallee S, Wu J, et al. Inhibition of NF-κB activity by IκBβ in association with κB-Ras. Mol Cell Biol. 2004;24:3048–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Horgan CP, Hanscom SR, Jolly RS, et al. Rab11-FIP3 binds dynein light intermediate chain 2 and its overexpression fragments the Golgi complex. Biochem Biophys Res Commun. 2010;394:387–392. [DOI] [PubMed] [Google Scholar]
- [42].Rossjohn J, Gras S, Miles JJ, et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015;33:169–200. [DOI] [PubMed] [Google Scholar]
- [43].Glynn JR, Whiteley J, Bifani PJ, et al. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg Infect Dis. 2002;8(8):843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Phyu S, Stavrum R, Lwin T, et al. Predominance of Mycobacterium tuberculosis EAI and Beijing Lineages in Yangon, Myanmar. J Clin Microbiol. 2009;47:335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg Infect Dis. 2006 May;12(5):736-743. doi: 10.3201/eid1205.050400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chihota VN, Niehaus A, Streicher EM, et al. Geospatial distribution of Mycobacterium tuberculosis genotypes in Africa. PLoS One. 2018;13. DOI: 10.1371/journal.pone.0200632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cerezo-Cortés MI, Rodríguez-Castillo JG, Hernández-Pando R, et al. Circulation of M. tuberculosis Beijing genotype in Latin America and the Caribbean. Pathog Glob Health. 2019;113:336–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Laserson KF, Osorio L, Sheppard JD, et al. Clinical and programmatic mismanagement rather than community outbreak as the cause of chronic, drug-resistant tuberculosis in Buenaventura, Colombia, 1998. Int J Tuberc Lung Dis. 2000;4(7):673–683. [PubMed] [Google Scholar]
- [49].Rodríguez-Castillo JG, Pino C, Niño LF, et al. Comparative genomic analysis of Mycobacterium tuberculosis Beijing-like strains revealed specific genetic variations associated with virulence and drug resistance. Infect Genet Evol. 2017;54:314–323. [DOI] [PubMed] [Google Scholar]
- [50].Krishnan N, Malaga W, Constant P, et al. Mycobacterium tuberculosis lineage influences innate immune response and virulence and is associated with distinct cell envelope lipid profiles. PLoS One. 2011;6(9):e23870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chen -Y-Y, Chang J-R, Huang W-F, et al. The pattern of cytokine production in vitro induced by ancient and modern Beijing Mycobacterium tuberculosis strains. PLoS One. 2014;9. DOI: 10.1371/journal.pone.0094296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Romagnoli A, Petruccioli E, Palucci I, et al. Clinical isolates of the modern Mycobacterium tuberculosis lineage 4 evade host defense in human macrophages through eluding IL-1β-induced autophagy. Cell Death Dis. 2018;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Portevin D, Gagneux S, Comas I, et al. Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 2011;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].van Laarhoven A, Mandemakers JJ, Kleinnijenhuis J, et al. Low induction of proinflammatory cytokines parallels evolutionary success of modern strains within the Mycobacterium tuberculosis Beijing genotype. Infect Immun. 2013;81:3750–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ates LS, Dippenaar A, Ummels R, et al. Mutations in ppe38 block PE_PGRS secretion and increase virulence of Mycobacterium tuberculosis. Nat Microbiol. 2018;3:181–188. [DOI] [PubMed] [Google Scholar]
- [56].Liang J, Song W, Tromp G, et al. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS One. 2008;3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Fenwick C, Na SY, Voll RE, et al. A subclass of Ras proteins that regulate the degradation of IkappaB. Science. 2000;287:869–873. [DOI] [PubMed] [Google Scholar]
- [58].Maceyka M, Sankala H, Hait NC, et al. SphK1 and SphK2, sphingosine kinase isoenzymes with opposing functions in sphingolipid metabolism. J Biol Chem. 2005;280:37118–37129. [DOI] [PubMed] [Google Scholar]
- [59].Rosebeck S, Leaman DW. Mitochondrial localization and pro-apoptotic effects of the interferon-inducible protein ISG12a. Apoptosis. 2008;13:562–572. [DOI] [PubMed] [Google Scholar]
- [60].Gytz H, Hansen MF, Skovbjerg S, et al. Apoptotic properties of the type 1 interferon induced family of human mitochondrial membrane ISG12 proteins. Biol Cell. 2017;109:94–112. [DOI] [PubMed] [Google Scholar]
- [61].Onate SA, Boonyaratanakornkit V, Spencer TE, et al. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–12108. [DOI] [PubMed] [Google Scholar]
- [62].Qin L, Gibson PG, Simpson JL, et al. Dysregulation of sputum columnar epithelial cells and products in distinct asthma phenotypes. Clin Exp Allergy. 2019;49:1418–1428. [DOI] [PubMed] [Google Scholar]
- [63].Martin E, Palmic N, Sanquer S, et al, CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation. Nature. 2014. Jun 12;510(7504):288–292. doi: 10.1038/nature13386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Huang LS, Sudhadevi T, Fu P, et al. Sphingosine Kinase 1/S1P Signaling Contributes to Pulmonary Fibrosis by Activating Hippo/YAP Pathway and Mitochondrial Reactive Oxygen Species in Lung Fibroblasts. Int J Mol Sci. 2020;21:2064.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Okamoto S, Azhipa O, Yu Y, et al. Expression of ADP-ribosyltransferase on normal T lymphocytes and effects of nicotinamide adenine dinucleotide on their function. J Immunol. 1998;160:4190–4198. [PubMed] [Google Scholar]
- [66].Crosbie RH, Heighway J, Venzke DP, et al. Sarcospan, the 25-kDa transmembrane component of the dystrophin-glycoprotein complex. J Biol Chem. 1997;272:31221–31224. [DOI] [PubMed] [Google Scholar]
- [67].Nagano T, Yoneda T, Hatanaka Y, et al. A-interacting protein (FILIP) regulates cortical cell migration out of the ventricular zone. Nat Cell Biol. 2002;4:495–501. [DOI] [PubMed] [Google Scholar]
- [68].Yi Z, Gao K, Li R, et al. Dysregulated circRNAs in plasma from active tuberculosis patients. J Cell Mol Med. 2018;22:4076–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sunryd JC, Cheon B, Graham JB, et al. TMTC1 and TMTC2 are novel endoplasmic reticulum tetratricopeptide repeat-containing adapter proteins involved in calcium homeostasis. J Biol Chem. 2014;289:16085–16099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Fensterl V, Sen GC. Interferon-induced ifit proteins: their role in viral pathogenesis. J Virol. 2014;89:2462–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


