Abstract
Context
Although primary adrenal insufficiency (PAI) in children and young people is often due to congenital adrenal hyperplasia (CAH) or autoimmunity, other genetic causes occur. The relative prevalence of these conditions is poorly understood.
Objective
We investigated genetic causes of PAI in children and young people over a 25 year period.
Design, Setting and Participants
Unpublished and published data were reviewed for 155 young people in the United Kingdom who underwent genetic analysis for PAI of unknown etiology in three major research centers between 1993 and 2018. We pre-excluded those with CAH, autoimmune, or metabolic causes. We obtained additional data from NR0B1 (DAX-1) clinical testing centers.
Intervention and Outcome Measurements
Genetic analysis involved a candidate gene approach (1993 onward) or next generation sequencing (NGS; targeted panels, exomes) (2013-2018).
Results
A genetic diagnosis was reached in 103/155 (66.5%) individuals. In 5 children the adrenal insufficiency resolved and no genetic cause was found. Pathogenic variants occurred in 11 genes: MC2R (adrenocorticotropin receptor; 30/155, 19.4%), NR0B1 (DAX-1; 7.7%), CYP11A1 (7.7%), AAAS (7.1%), NNT (6.5%), MRAP (4.5%), TXNRD2 (4.5%), STAR (3.9%), SAMD9 (3.2%), CDKN1C (1.3%), and NR5A1/steroidogenic factor-1 (SF-1; 0.6%). Additionally, 51 boys had NR0B1 variants identified through clinical testing. Although age at presentation, treatment, ancestral background, and birthweight can provide diagnostic clues, genetic testing was often needed to define the cause.
Conclusions
PAI in children and young people often has a genetic basis. Establishing the specific etiology can influence management of this lifelong condition. NGS approaches improve the diagnostic yield when many potential candidate genes are involved.
Keywords: adrenal, adrenal insufficiency, Addison disease, genetics, NGS
Primary adrenal insufficiency (PAI) is a potentially life-threatening condition that can be difficult to diagnose and that requires urgent treatment with glucocorticoid and sometimes mineralocorticoid replacement therapy [1]. PAI can occur across the lifespan and has many well established causes, such as congenital adrenal hyperplasia (CAH), autoimmune disorders (Addison disease) or physical damage of the adrenals (eg, hemorrhage, tumor) [1-4]. However, in many situations, a specific diagnosis is not readily reached.
The past 25 years have seen significant progress in our understanding of the genetic causes of PAI, especially for conditions presenting in children or in young people [4-8]. More than 30 different single gene disorders have now been reported with a diverse range of genetic inheritance patterns (recessive, dominant/de novo, X-linked, or even imprinted).
Establishing a specific diagnosis is important for understanding the natural history of a condition and how it might affect an individual, personalizing therapy (eg, the need for lifelong mineralocorticoid treatment), monitoring associated features (eg, puberty and gonadal function, renal), counseling about inheritance patterns and fertility, and identifying presymptomatic family members at risk. Many forms of PAI do not have diagnostic phenotypic or biochemical features, so genetic analysis is the only way to establish a precise diagnosis.
Genetic analysis traditionally required each candidate gene to be sequenced individually. This approach was time-consuming and expensive. More recently, next-generation sequencing (NGS) technologies have allowed parallel sequencing of many genes at the same time, either as a targeted panel of adrenal-related genes or as whole exome or genome sequencing. Recently, we have used a target-based NGS approach in a cohort study in Turkey of 95 children with PAI of unknown etiology (ie, CAH, autoimmune, metabolic causes excluded). With this approach, a genetic diagnosis was reached in 78/95 (82%) young people, involving just 9 genes [eg, MC2R, NR0B1 (DAX-1), STAR, CYP11A1, MRAP, NNT, ABCD1, NR5A1, AAAS] [9]. However, the population studied had a high degree of consanguinity and marked genetic founder effects, so it is unclear if the findings can be translated to other populations in the world.
Here, we report our findings on the genetic analysis of children and young people with PAI of unknown etiology referred from within the United Kingdom (UK) to 3 major research centers and clinical testing centers for NR0B1 (DAX-1) and provide a combined analysis of our published and unpublished experiences of diagnosing these conditions over 25 years.
Materials and Methods
Patient Cohort
The total cohort studied consisted of 155 children and young people (102 males, 48 females, five 46,XY girls) with PAI of unknown etiology living in the UK who were referred to our research groups (UCL GOS Institute of Child Health, London, UK; Queen Mary University, London, UK and Technische Universität Dresden, Dresden, Germany) for genetic analysis from several pediatric endocrine centers across the UK and where a genetic diagnosis was made between 1993 and 2018. Within the cohort there were 16 sibling pairs or familial pairs, 3 trios, and an extended family with 7 affected members.
All individuals included in the analysis had a provisional diagnosis of adrenal hypoplasia, familial glucocorticoid deficiency (FGD)/adrenocorticotropin (ACTH)-resistance, triple A syndrome/Allgrove syndrome (adrenal insufficiency, achalasia, alacrima), or just PAI with an unknown cause.
Subjects had undergone standard clinical and biochemical analysis to try to reach a diagnosis and were excluded if a specific cause was established [eg, CAH (21-hydroxylase deficiency, 11-hydroxylase deficiency, 3β-hydroxysteroid dehydrogenase deficiency type 2, 17α-hydroxylase deficiency, P450 oxidoreductase deficiency, aldosterone synthase deficiency), autoimmune adrenal disorders, metabolic dysfunction (eg, adrenoleukodystrophy), or physical causes]. Children with syndromic features, fetal growth restriction (intrauterine growth restriction), or genital anomalies (eg, hypospadias) were included.
Informed consent was obtained from affected individuals and/or their parents. Ethical approval for more recent studies was obtained from the London-Bloomsbury National Research Ethics Service committee (reference number 07/Q0508/24) and Outer North East London Research Ethics committee (reference number 09/H0701/12). Historical approvals for published data are detailed in the relevant publications (see Table 1).
Table 1.
Overview of conditions, associated features, common presenting features and genetic variants identified
| Gene (inheritance) | Potential associated features | N | Pathogenic variants (n) | Refs.a |
|---|---|---|---|---|
| MC2R (AR) | Possible tall stature, hyperpigmentation | 30 | p.S74I (20); p.S74I/p.R128C; p.S74I/p.L224R; p.N81Kfs*3; p.D107N (3); p.R146H; p.R146H/p.V187Afs*29 (2); p.I154fs*248 | [14-18] |
| NR0B1 (X-linked) | Hypogonadotropic hypogonadism, infertility, possible early puberty | 12 | NR0B1 deletion (3); contiguous gene deletion b (2); p.W171*; p.L262Q; T265R; p.Y399*; p.H419Dfs*7 (2);p.F448S | [20-22,59] |
| CYP11A1 (AR) | Possible gonadal steroid insufficiency | 12 | p.R120Q/p.Q395K; p.R120*/c.940G > A (2); c.426-2A > G/c.940G > A; c.790-802del/c.940G > A (3); p.A277Dfs*11/c.940G > A; p.I279Yfs*10/c.940G > A; p.S391 = /c.940G > A; p.R424*/c.940G > A; p.R451W | [9,11,25] |
| AAAS (AR) | Triple A syndrome (alacrima, achalasia, neurological features, dermatological features) | 11 | p.S382Rfs*33/p.W474*; p.H71Ifs*23/p.R230* (2); p.R286*/ c.525_545 + 4dupCCGTGTGTATAATGCCAGCAGGTGT ; p.R286*/p.R478*; p.Q145*/p.S263P; p.R230*/p.Q456*; p.G14Vfs*45 (2); c.689 + 1G > C (2) | [60-62] |
| NNT (AR) | Possible early puberty | 10 | p.R71*; p.Y201Kfs*1 (2); c.599 + 2T > C; p.G255E (2); p.G664R/p.T689Lfs*32 (2); p.G678R; p.L977P | [28,29] |
| MRAP (AR) | Hyperpigmentation | 7 | p.M1? (2); p.E28K/c.106 + 2dupT; c.106 + 2dupT (2); c.106 + 1G > C (2) | [31] |
| TXNRD2 (AD) | Cardiac defects | 7 | p.Y447* (7) | [33] |
| STAR (AR) | Possible gonadal steroid insufficiency | 6 | p.A10T; p.V187M; p.G201D/p.G221S (2); p.G221D; p.T223Lfs*98 | [35,37,38] |
| SAMD9 c (AD/de novo) | MIRAGE syndrome (myelodysplasia, infection, restriction of growth, adrenal hypoplasia, genital phenotypes, enteropathy) | 5 | p.R459Q; p.R982C; p.R982H; p.R1293Q; p.K1569N | [39] |
| CDKN1C (AD, imprinted) | IMAGe syndrome (intrauterine growth restriction, metaphyseal dysplasia, adrenal hypoplasia, genitourinary anomalies) | 2 | p.D274N; p.K278E | [40] |
| NR5A1 (AD/de novo) | Gonadal dysgenesis/steroid insufficiency | 1 | p.G35E | [41] |
Italics indicates heterozygous (monoallelic) changes either as a dominant or compound heterozygous condition; bold indicates novel changes not reported previously by our groups or others; numbers in parentheses show the total number of individuals with this specific variant.
Abbreviations: AD, autosomal dominant; AR, autosomal recessive.
a References indicate our previous reports including these findings or other groups’ reports of these genetic variants (see supplementary table in [13]).
b One girl with a Xp21 contiguous gene deletion syndrome expressed a phenotype due to skewed X-inactivation.
c For SAMD9 the primary genetic change is shown, although several children had developed somatic revertant events such as monosomy 7 or second loss-of-function variants in SAMD9.
An additional cohort of children were identified (n = 51) who had been diagnosed with X-linked adrenal hypoplasia due to pathogenic variants in NR0B1 (DAX-1) through clinical genetic testing pathways. This supplementary group was important to consider as clinical testing became more widespread in the 2000s, and we did not want the NR0B1 group to be underrepresented in the cohort, as it is an important diagnosis to make. Data were obtained primarily from Wessex Regional Genetics Laboratory and from the Northern Genetics Service, Newcastle. Individuals with NR0B1 mutations in the research cohort and the clinical testing group were independent.
Genetic Analysis: Sanger Sequencing
In the first 20 years of the study (1993-2012), genetic analysis was mostly undertaken by Sanger sequencing using a candidate gene approach. The main genes analyzed using this method at the time were MC2R, MRAP, AAAS, NNT, NR0B1 (DAX-1), NR5A1 (SF-1), CDKN1C, STAR, and CYP11A1, and key findings and sequencing methods were published where appropriate. Subsequently, a more refined Sanger sequencing approach was used for initial analysis (MC2R, MRAP exon 3, CYP11A1 exon 5, STAR exons 4-5, NR0B1).
Genetic Analysis: HaloPlex-Targeted Capture and NGS
Since 2013, a NGS strategy was used to try to reach a specific genetic diagnosis in 75 patients in whom no diagnosis had been reached, either using a HaloPlex-targeted panel or exome sequencing.
HaloPlex targeted capture
A HaloPlex DNA target enrichment panel was designed using SureDesign (Agilent Technologies Inc., Santa Clara, CA, USA) to include all known genes involved in PAI at the time (2013) [9,10].
Patient DNA was extracted from leukocytes using standard methods. Batches of 24 samples were run together, including 23 patient samples and one enhanced control DNA for quality control. Genomic DNA samples (225 ng) were processed for Illumina Sequencing according to the “HaloPlex Target Enrichment System” protocol (Version D.5, Agilent Technologies Inc.) and as described previously [9]. Resultant libraries were then subjected to NGS using an Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA). Raw FASTQ files were analyzed using SureCall (v3.0.1.4) software (Agilent Technologies, Inc.) and Ingenuity Pathway Analysis (Qiagen Bioinformatics, Inc., Redwood City, CA, USA). Aligned BAM files were also visualized manually to review for potential whole gene or single exon deletions.
Whole exome sequencing
Exome sequencing was undertaken using the Agilent SureSelect all exon V4 capture and paired-end (2 × 100) sequencing on an Illumina HiSeq 2000 at Otogenetics (Norcross, GA, USA). Alignment, variant calling, and annotation were performed by DNAnexus (DNAnexus Inc., Mountain View, CA, USA) using the DNA nexus Classic platform or Ingenuity Variant Analysis. Raw data were also reanalyzed by BWA-MEM FastQ Readmapper, variants called using Vendor Human Exome GATK-Lite (Unified Genotyper) and analyzed in Ingenuity Variant Analysis using filtering criteria reported previously [11].
Variant Validation
Sanger sequencing was used to confirm the presence of key variants identified by NGS. Regions of interest were polymerase chain reaction amplified and sequenced using a Big Dye Terminator v.1.1 Cycle Sequencing Kit (Applied Biosystems) and ABI3130 sequencer (Applied Biosystems), and visualized using Sequencher v5.2.4 (Gene Codes Corporation). Population genomic variation control data were obtained from Genome Aggregation Database (gnomAD, Cambridge, MA, USA; https://gnomad.broadinstitute.org) [12].
Functional Prediction of Genetic Variants
The predicted functional effects of specific variants were performed using Ensembl Variant Effect Predictor, SIFT, PolyPhen2, and MutationTaster algorithms (https://www.ensembl.org/info/docs/tools/vep/index.html; http://sift.jcvi.org/; http://genetics.bwh.harvard.edu/pph2/; http://www.mutationtaster.org/, respectively).
Statistical Analysis
Chi-square tests for sex differences were performed using GraphPad Prism version 8.4.3 for Windows (GraphPad Software, San Diego, CA, USA; www.graphpad.com).
Results
Cohort Results
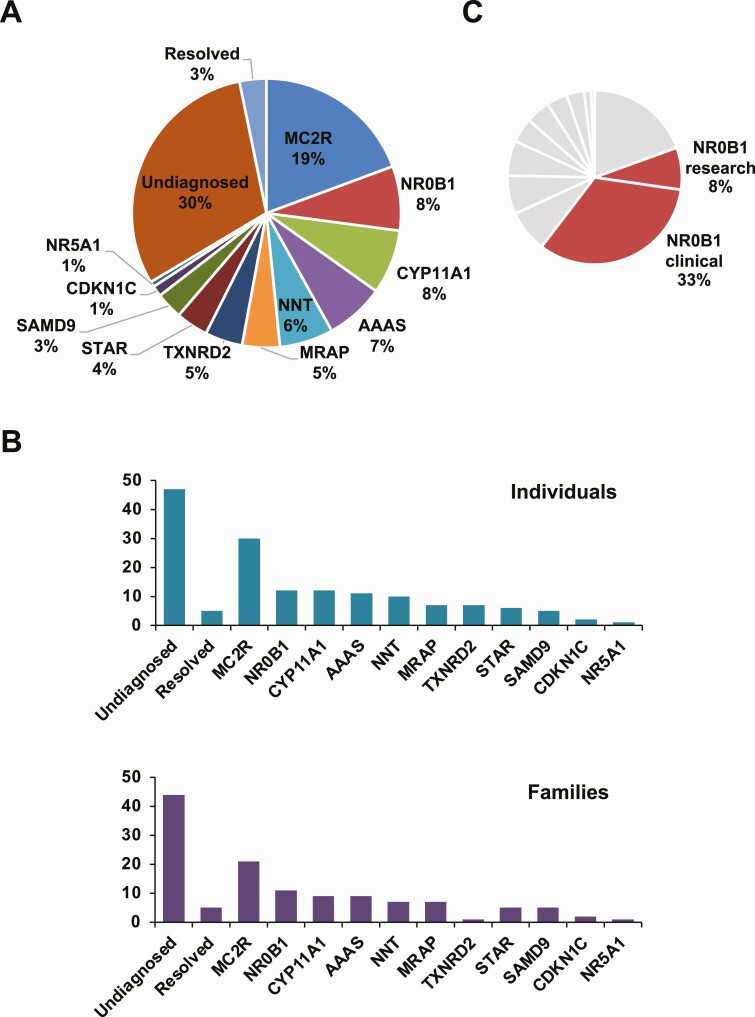
Within the entire cohort, a specific genetic diagnosis was reached in 103/155 (66.5%) individuals (Fig. 1A). No specific genetic diagnosis was found in 47/155 (30.3%) individuals (29 had NGS; 18 had candidate gene sequencing). The adrenal insufficiency resolved and steroid replacement treatment was gradually withdrawn during the course of this study in 5/155 (3.2%) children, including in 1 boy 13 years after initial treatment. Of note, no genetic cause was found in this group.
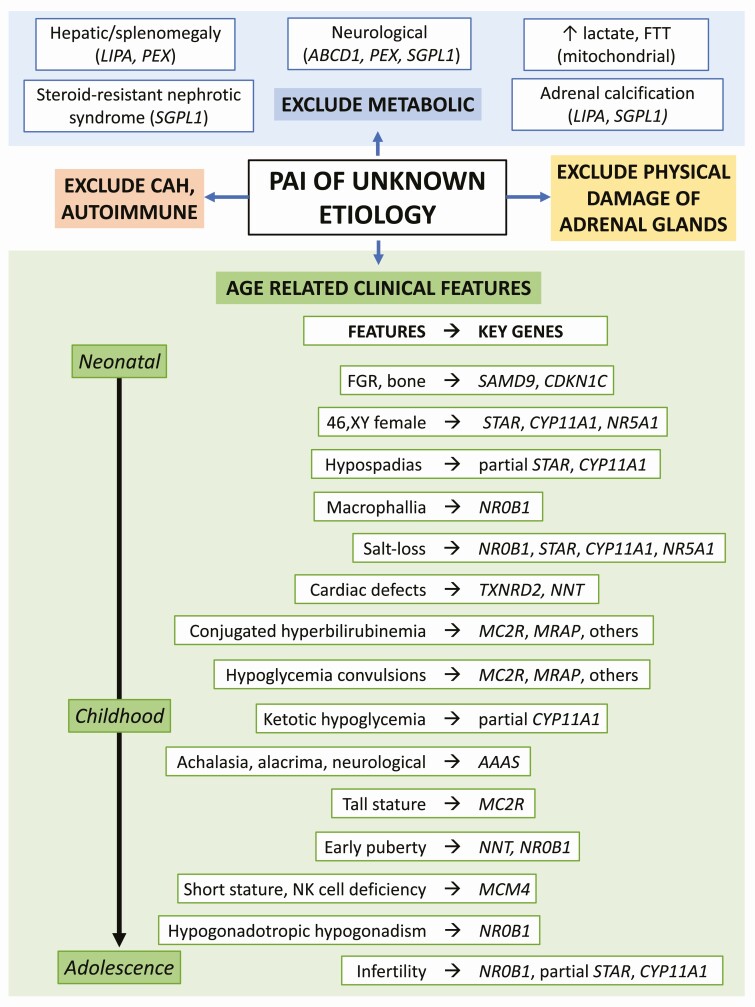
Figure 1.
Overview of the study cohort and genetic diagnoses reached. (A) Relative percentages of each diagnosis for the research cohort in relation to undiagnosed and resolved categories (n = 155). (B) Numbers of individuals (n = 155) with each genetic diagnosis (upper panel) and number of different affected families (n = 127) (lower panel). Note: data for X-linked adrenal hypoplasia (DAX-1/NR0B1) identified through clinical services not included here. (C) Relative percentages of NR0B1 diagnoses (research and clinical) (combined n = 63) in relation to all diagnosed individuals (n = 154). Data represent the number of individuals, not families.
Pathogenic variants were found in 11 different genes (Fig. 1A and B; Tables 1 and 2; also see supplementary table in [13]). Related characteristics for each genetic diagnosis (eg, treatment, family history, fetal growth restriction, ancestry) are shown in Fig. 2A-2D, and age at presentation of the adrenal insufficiency in each subgroup is shown in Figure 3.
Table 2.
Novel pathogenic variants identified and their predicted pathogenicity
| Gene | Sequence variation | gnomAD Allele Frequency | VEP | SIFT | PolyPhen-2 | Mutation Taster | ||
|---|---|---|---|---|---|---|---|---|
| cDNA | Protein | SIFT | PolyPhen | |||||
| MC2R | c.243delT | p.N81Kfs*3 | nd | Frameshift | na | na | Disease causing | |
| c.671T > G | p.L224R | 4/282270 | 0 | 0.945 | 0 | 0.995 | Disease causing | |
| NR0B1 | c.1343T > C | p.F448S | nd | 0 | 0.8 | 0.001 | 0.981 | Disease causing |
| AAAS | c.525_545 + 4dupCCGTGTGTA TAATGCCAGCAGGTGT | na | 3/281760 | Splice defect | na | na | Disease causing | |
| c.689 + 1G > C | na | nd | Splice donor | na | na | Disease causing | ||
| NNT | c.599 + 2T > C | na | nd | Splice donor | na | na | Disease causing | |
| c.764G > A | p.G255E | nd | 0 | 0.988 | 0.001 | 0.999 | Disease causing | |
| MRAP | c.82G > A | p.E28K | 3/251322 | 0.04 | 0.998 | 0.071 | 1 | Disease causing |
| STAR | c.28G > A | p.A10T | nd | 0.01 | 0.503 | 0.014 | 0.95 | Disease causing |
| c.602G > A | p.G201D | 3/282724 | 0 | 0.944 | 0 | 0.992 | Disease causing | |
| c.666delC | p.T223Lfs*98 | nd | Frameshift | na | na | Disease causing |
Abbreviations: na, not applicable; nd, not detected; VEP, Ensembl Variant Effect Predictor.
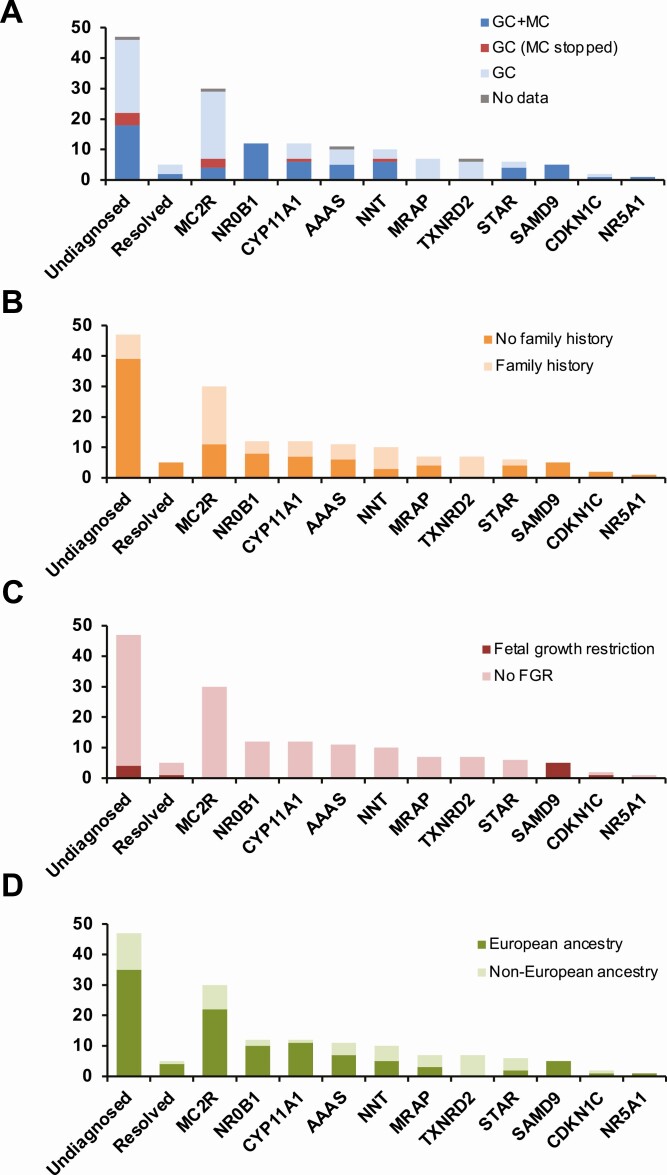
Figure 2.
Key features of each diagnostic group. (A) Steroid replacement received. (B) Presence or absence of a positive family history of adrenal insufficiency. (C) Presence or absence of fetal growth restriction (FGR) (<2.5 kg at term). (D) Ancestry. European was defined as of a White European background compared to other backgrounds such as Asian, Asian British, African, or Black British. Data for X-linked adrenal hypoplasia (DAX-1/NR0B1) identified through clinical services not included here or in subsequent figures. Abbreviations: GC, glucocorticoids; MC, mineralocorticoids.
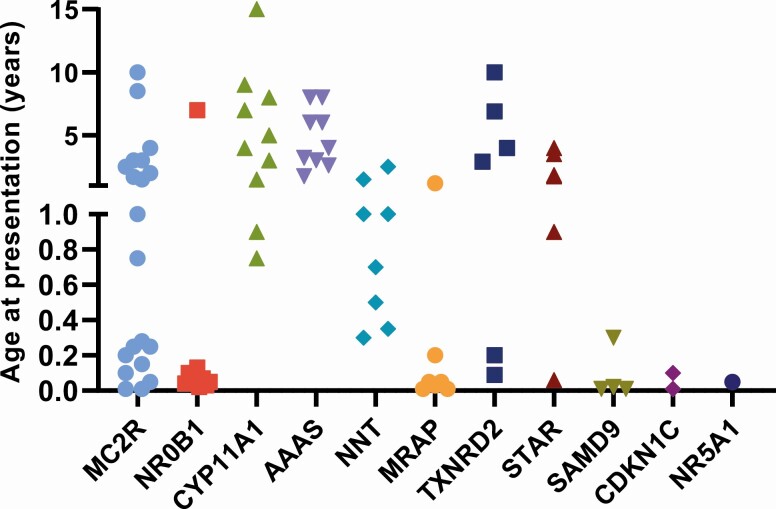
Figure 3.
Age of main presentation of PAI in the different groups with a genetic diagnosis. Note the nonlinear age scale.
An additional 51 boys were identified with pathogenic variants in NR0B1 (DAX-1) through clinical testing pathways (Fig. 1C).
MC2R
One of the most prevalent conditions diagnosed was FGD type 1 due to disruption of the ACTH receptor melanocortin receptor-2 (MC2R) [14-18]. Pathogenic variants were identified in 19.4% (30/155) of the entire cohort. This condition was most often (20/30) due to the p.S74I variant found in individuals of European ancestry from Ireland and Scotland [19]. Two siblings had a p.V187Afs*29 (c.560delT) variant, which is now being seen in Turkey and neighboring regions [9,17]. Most young people were treated with glucocorticoids alone. Seven children were treated with additional mineralocorticoids, but 3 of these were able to stop the mineralocorticoid treatment once the diagnosis had been made (Fig. 2A).
NR0B1 (DAX-1)
Disruption of NR0B1 (DAX-1), causing X-linked adrenal hypoplasia congenita (AHC) was found in 7.7% (12/155) of children in the research cohort [20,21]. Presentation was mostly in the newborn period with salt-losing adrenal insufficiency in boys, but sometimes more insidiously in later childhood (Fig. 3). Analysis showed that 7 boys had missense, frameshift, or stop gain mutations, whereas 3 had localized deletions of the NR0B1 gene itself. An Xp21 contiguous gene deletion syndrome including glycerol kinase deficiency and Duchenne muscular dystrophy was also diagnosed in 2 children; surprisingly, 1 of these was a girl who had a large Xp21 deletion and expressed the phenotype due to skewed X inactivation [22]. All boys of a pubertal age have shown evidence of hypogonadotropic hypogonadism and required pubertal induction.
An additional 51 boys from 42 families were identified with pathogenic NR0B1 variants through clinical testing pathways. Taken together with the research cohort (NR0B1, n = 12), 63 children had a diagnosis of X-linked AHC out of a total of 154 children with a positive genetic diagnosis (103 research cohort, 51 NR0B1 clinical cohort). These combined data suggest that X-linked AHC represented the largest diagnostic group overall (63/154, 40.9%) (Fig. 1C).
CYP11A1
CYP11A1 mutations were found in 7.7% (12/155) of the cohort. CYP11A1 encodes the enzyme P450 side-chain cleavage enzyme (P450scc), which converts cholesterol to pregnenolone. P450scc is needed for the synthesis of all adrenal and gonadal steroids and severe disruption of this protein causes early-onset salt-losing adrenal insufficiency and impaired virilization/46,XY DSD [23,24]. Most of the patients in our cohort had partial loss of CYP11A1 activity and often presented in childhood with signs of glucocorticoid insufficiency, such as ketotic hypoglycemia (Fig. 3). Approximately half of them had evidence of mildly impaired zona glomerulosa function and were treated with mineralocorticoids (Fig. 2A). Ten of the 12 individuals were from a UK background and had the c.940G > A variant in compound heterozygosity with another severely disruptive change [11,25,26]. One child was compound heterozygous for p.R120Q and p.Q395K and 1 individual, whose family were from central Turkey, was homozygous for p.R451W, a variant that is prevalent in that region [9].
AAAS
Triple A syndrome (Allgrove syndrome) is a rare autosomal recessive multisystem disease characterized by adrenal insufficiency, alacrima, and achalasia caused by pathogenic variants in AAAS [27]. In this cohort, 7.1% (11/155) of children had AAAS variants. Most of these were nonsense or frameshift changes, with 1 novel, homozygous splice site disruption (c.689 + 1G > C) found in 2 cousins and a second novel splice site mutation (c.525_545 + 4dupCCGTGTGTATAATGCCAGCAGGTGT) with predicted loss of protein function. Children with triple A usually developed adrenal insufficiency in mid-childhood (age range 1.75-8 years) (Fig. 3) and often had other features such as alacrima, achalasia, and neurological symptoms at this time. Approximately half required treatment with both glucocorticoids and mineralocorticoids, whereas half received glucocorticoids alone.
NNT
Defects in nicotinamide nucleotide transhydrogenase (NNT) were found in 10/155 (6.5%) children who presented between 4 months and 2.5 years of age [28,29]. This protein is involved in regulating mitochondrial oxidative stress. Three children experienced early puberty, which has been reported recently as a potential associated feature of this condition [30]. One child of Pakistani ancestry was homozygous for a stop gain (p.R71*); this variant has been previously identified in a child living in Australia whose mother originated from this region [29].
MRAP
Disruption of the melanocortin 2 receptor-associated protein (MRAP) causes FGD2 and was found in 7/155 (4.5%) of individuals [31]. Most of these changes affected translation initiation (p.M1?) or splicing events, as reported previously, and usually occurred in children who presented in the neonatal period or first 3 months of life [32].
TXNRD2
A similar number of individuals (4.5%) were found to have adrenal insufficiency due to a homozygous stop gain mutation (p.Y447*) in thioredoxin reductase 2 (TXNRD2) (Fig. 1B) [33]. However, individuals were all from a single Kashmiri kindred (Fig. 1C). No pathogenic variants in TXNRD2 were found in other patients.
STAR
Similar to CYP11A1, defects in steroidogenic acute regulatory protein (STAR) can affect both adrenal and gonadal steroid synthesis. Severe disruption of STAR causes classic congenital lipoid adrenal hyperplasia (CLAH) [34]. This condition was diagnosed in a 46,XY girl with early-onset salt-losing adrenal insufficiency due to a homozygous frameshift mutation. Five others who presented between 9 months of age and 4 years with predominant glucocorticoid insufficiency had nonclassic CLAH, although mineralocorticoids were given to most of them [35-38]. Nonclassic CLAH was not associated with obvious genital anomalies.
SAMD9 and CDKN1C
Five children (5/155, 3.2%) had de novo, gain of function mutations in sterile alpha motif domain-containing 9 (SAMD9) [39]. They were all born preterm and had fetal growth restriction, together with other features of MIRAGE syndrome (myelodysplasia, infection, restriction of growth, adrenal hypoplasia, genital phenotypes, enteropathy). Two other children in the cohort had gain of function changes in the proliferating cell nuclear antigen–binding domain of cyclin dependent kinase inhibitor 1C (CDKN1C) associated with IMAGe syndrome (intrauterine growth restriction, metaphyseal dysplasia, adrenal hypoplasia, genitourinary anomalies) [40]. CDKN1C is an imprinted gene. In 1 child, there was no marked growth restriction and features were mild.
NR5A1 (SF-1)
Only 1 individual was found to have a pathogenic variant in NR5A1, encoding steroidogenic factor-1 (SF-1) [41]. This 46,XY child presented with salt-loss and a female phenotype. No variants in NR5A1 were found to cause PAI alone.
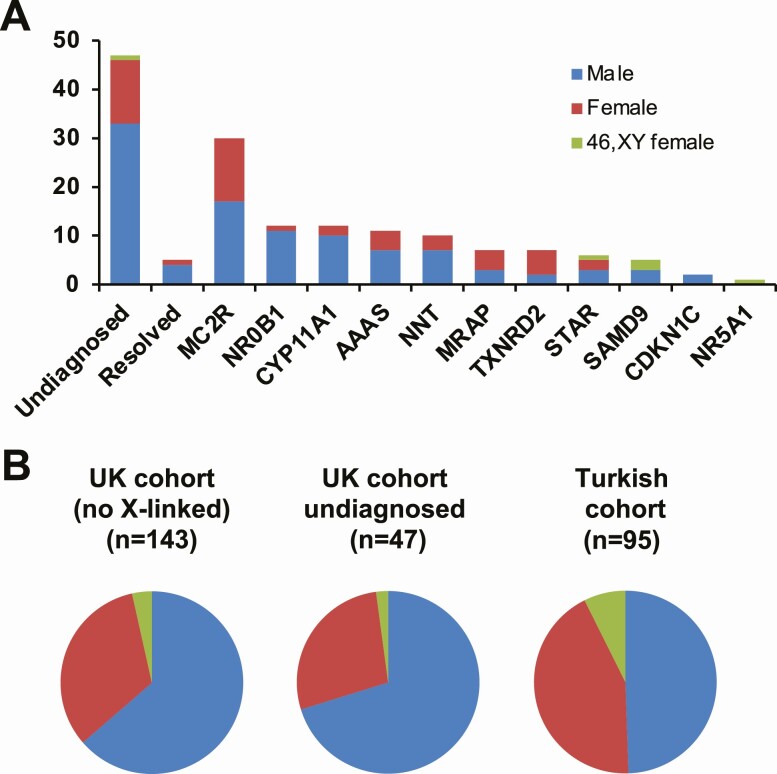
Sex Differences
Analysis of sex differences within the cohort revealed that there was an excess of boys in the UK cohort (102/155, 65.8%; χ 2 = 22.17, P < 0.001) (Fig. 4A). This observation persisted even when individuals with X-linked adrenal hypoplasia (NR0B1) were removed from analysis (91/143, 63.6%; χ 2 = 16.20, P < 0.001) (Fig. 4B). Indeed, the sex-differences were even more marked in the “undiagnosed” group (33/47, 70.2%; χ 2 = 8.7, P = 0.0032) and in the group where PAI had resolved (4/5) (Fig. 4B). This observation contrasts with data from the Turkish cohort where sex-differences were not seen (47/95, 49.5%; χ 2 = 0.4, P = 0.5) [9].
Figure 4.
Sex differences in the cohort. (A) Number of males, females and 46,XY females in each group. (B) Relative proportions of males, females and 46,XY females in the total UK cohort with X-linked conditions removed (left chart), the “undiagnosed” UK cohort (center chart) and previously published Turkish cohort (right chart).
Discussion
PAI is an extremely important diagnosis to make and establishing a specific genetic diagnosis can have important implications. The most common cause of PAI in infants and children is CAH, accounting for approximately 80% to 85% of PAI in studies to date [2,42-44]. CAH occurs most often due to 21-hydroxylase deficiency, and the diagnosis can usually be confirmed by biochemical and genetic analysis. Several “hotspots” of other forms of CAH exist (eg, 11β-hydroxylase deficiency in Jewish populations from Morocco); indeed, congenital lipoid adrenal hyperplasia due to disruption of STAR is particularly prevalent in South Korea and Japan [2,6]. Other well-recognized causes of PAI include autoimmune disorders (eg, autoimmune polyglandular syndrome type 1/AIRE; autoimmune Addison disease in teenagers) and established but rare metabolic disorders (eg, Wolman disease, Zellweger spectrum, mitochondrial disorders, adrenoleukodystrophy) [4]. Of these metabolic conditions, X-linked adrenoleukodystrophy is an important diagnosis to make in boys, and very-long change fatty acid analysis should be considered. However, in many children a specific cause of PAI is not obvious and additional genetic investigation is needed.
Here, we have combined published and unpublished data over a 25-year period from 3 major research centers investigating childhood PAI in the UK to provide insight into the genetic basis of some of the rarer forms of PAI in children and young people, where CAH, autoimmune and metabolic causes had been excluded. A diagnosis was reached in 66.5% of the cohort. This figure is lower than in the recent Turkish study (82%) where there was higher consanguinity but still represents a significant diagnostic yield for monogenic disorders, especially as CAH and other metabolic conditions had been excluded [9]. Furthermore, 51 boys from 42 independently ascertained families had NR0B1 variants identified through clinical testing pathways, so the UK diagnostic yield is likely to be even higher.
The proposed benefits of achieving a specific genetic diagnosis include being able to counsel the family appropriately and identifying at risk relatives before the onset of an adrenal crisis; understanding the natural history of different conditions and monitoring for the development of known associated features; and, in some situations, being able to modify treatment strategies. Within this cohort there are several examples of each of these situations.
First, in several children with familial forms of X-linked AHC (NR0B1/DAX-1), MC2R defects or TXNRD2 disruption, we were able to make a diagnosis of adrenal insufficiency early on in the presentation or whilst the child was presymptomatic. In 1 family (NR0B1), steroid treatment was commenced as sodium started to fall in the first week of life, but before the onset of a severe salt-losing state or extreme hyperkalemia. In another family (TXNRD2), symptoms such as malaise were linked to PAI rather than being overlooked. PAI can be a difficult diagnosis to make in any child presenting for the first time as features can be nonspecific. We have previously shown in families where there are 2 brothers with X-linked AHC due to NR0B1/DAX-1 mutations that the second boy is typically diagnosed at a younger age than the first, often because of heightened awareness [45]. By combining genetic testing with biochemistry, a presymptomatic diagnosis should be possible. Indeed, in some situations a child’s biochemistry will be within normal limits early on and genetic analysis may be the only diagnostic test.
Second, detecting potential associated features is important (Fig. 5). For example, boys with X-linked adrenal hypoplasia (NR0B1/DAX-1) are at risk of developing hypogonadotropic hypogonadism in adolescence, whereas there is a potential risk of long-term sex steroid insufficiency or hypofertility with partial defects in STAR or CYP11A1/P450scc [7,25,35,36,46]. Appropriate monitoring and counseling can be provided, and cryopreservation of sperm might be considered. There are reports of testicular adrenal rest tumors with poor control in some of these conditions too [46,47]. Patients with triple A syndrome often present with additional features such as alacrima (93% of all patients, 64% of them within the first 36 months of life), achalasia (86% of all patients with an average age of 6.4 years), dermatological symptoms (71% of all patients), and neurological symptom (72% of all patients) [48]. Peripheral muscle weakness, nasal speech, and hyperreflexia without other evidence of spasticity are pathognomonic signs of triple A syndrome. In addition, growth-restricted infants diagnosed with MIRAGE syndrome or IMAGe syndrome should have appropriate monitoring of associated features. In some situations children with MIRAGE syndrome may require stem cell transplant if monosomy 7-related myelodysplastic syndrome and subsequent hematopoietic changes develop [39,49]. Finally, in conditions with no strong associated features (eg, MC2R, MRAP) reassurance can be given.
Figure 5.
Selected clinical features that might be associated with different genetic etiologies of PAI. Abbreviations: CAH, congenital adrenal hyperplasia; FGR, fetal growth restriction; FTT, failure to thrive.
Several examples also emerged where treatment was modified or personalized based on genetic analysis. Several children with ACTH resistance (MC2R mutations) were started on mineralocorticoid treatment at diagnosis, but this was withdrawn later in life when the diagnosis was made and mineralocorticoid requirements are less [17]. Adequate salt and fluid intake are still recommended during heat and exertion though. Also, there can be a tendency to try to suppress ACTH concentrations in some forms of ACTH resistance, which can lead to overtreatment with replacement glucocorticoids, and an awareness of the diagnosis can help in this regard. Of particular interest is the small group of children who were able to stop steroid hormone replacement and recover adrenal function during the course of this work. None of them was found subsequently to have a genetic diagnosis in these key genes. Diagnosing adrenal insufficiency can be difficult especially in a sick child with hyponatremia or hypoglycemia, and if a full diagnostic work-up including ACTH measurements is not available, children may have been started on treatment at the time, erring on the side of caution. Readdressing the diagnosis can be difficult once established on steroid treatment, as the hypothalamo-pituitary-adrenal axis may be suppressed for years, even if relatively low-maintenance doses have been used [50]. While we would not support generalized withdrawal of treatment when a genetic cause is not found, especially as this is research rather than diagnostic testing, it could be envisaged that a lower threshold for a trial off treatment would be considered in some situations, especially where evidence for initial diagnosis is not robust, and is based on incomplete biochemical data. Being able to make any genuine change in treatment for children and young people has major implications, as this is a lifelong condition.
With the emergence of NGS approaches, either as panels of genes or whole exomes, it is now often cheaper and quicker to adopt this approach rather than a candidate gene approach. In the UK, clinical testing with panels has now been introduced. However, in some settings, resources are limited and more focused genetic analysis may be needed. By assimilating data over a 25-year period, some useful insights have emerged. For example, X-linked adrenal hypoplasia usually affects boys and often presents with salt-loss in the first 2 months of life, whereas glucocorticoid insufficiency often presents with prolonged neonatal jaundice or hypoglycemic convulsions. Defects in MRAP tend to present in early life, whereas presentation with NNT mutations is usually between 6 months and 4 years of age, and triple A syndrome more often in mid-childhood (Fig. 3). Partial loss of CYP11A1 or STAR can present in childhood with ketotic hypoglycemia. These data are largely supportive of our findings in the Turkish cohort study [9].
Furthermore, ancestral background is emerging as an important predictor of genetic etiology in some children. For example, the p.S74I variant in MC2R is by far the most common change seen in our UK population with FGD, which likely arises from an Irish/Scottish founder effect [19]. The c.940G > A variant in CYP11A1 causing misplicing and a partial phenotype was found in 10 patients of UK ancestry and has been reported in families from France, Spain, the United States, Canada, and Australia recently [25,26,46]. A p.R451W variant in CYP11A1 was found in a child born to a family originating from central Turkey; reports have also described this change in Germany in a family of central Turkish origin [9,51]. The c.560delT change in MC2R also has a founder effect from Turkey and neighboring regions [9,17]. Finally, the p.R71* stop gain in NNT in a family of Pakistani origin living in Scotland has also been recently reported by us in a woman living in Australia who also originated from this region [29]. Therefore, a careful family history is always important when assessing a child with PAI.
This study has several limitations. First, this cohort represents those children and families sent to us for genetic analysis and does not represent a cross-sectional cohort or means of calculating incidence or prevalence of these conditions. We were likely to be biased in receiving more familial cases rather than sporadic, and in recent years clinical genetic testing for conditions such as X-linked adrenal hypoplasia (NR0B1/DAX-1) has become more widely available, meaning that this group in particular may be underrepresented. We addressed this to some extent by identifying clinically diagnosed children with pathogenic variants in NR0B1, but it was not possible to combine these data with the research cohort as we lacked details about specific characteristics for this group, as well as for those children who tested negative for DAX-1. We did not find any pathogenic variants in MCM4 (adrenal insufficiency, short stature, natural killer cell deficiency), which, to date, has only been reported in the Irish Traveller population, nor in SGPL1 (adrenal insufficiency, steroid-resistant nephrotic syndrome, ichthyosis, other features) in this UK resident population, although not all children had full analysis with panels or exomes [52,53]. Very recently described causes of adrenal insufficiency (such as POLE1 deficiency) were not considered, although none of our cohort had clear phenotypic features of this syndrome [54]. Finally, adrenal insufficiency can also arise from nongenetic causes in some situations, such a bilateral adrenal hemorrhage [55]. Adrenal imaging is not always undertaken or features may resolve, so potentially this is underdiagnosed.
Within our undiagnosed cohort, it is possible there are new genetic causes of adrenal insufficiency to be discovered or we may have overlooked deep intronic changes that are emerging from our work as a more common cause of gene disruption than initially thought [29,56,57]. It is also interesting that we see a sex-bias in the UK-based population, with more boys being diagnosed with PAI than girls, especially in the undiagnosed group (Fig. 4). This suggests that an X-linked phenomenon might exist that we are overlooking, such as a new genetic cause or alteration in a regulatory region of a key gene such as NR0B1 [58]. We hypothesize that there could also be sex differences in adrenal function in males and females, which could manifest as a susceptibility to transient adrenal hypofunction in the newborn period in boys and diagnosis with PAI. Although numbers are small, 4/5 children where PAI resolved were boys. This hypothesis requires further investigation, particularly linked to developmental sex differences [10]. We have recently shown how the phenotype associated with MIRAGE syndrome can be influenced by developmental events such as somatic reversion [39]. It is also possible that genetic plasticity and sex differences can influence adrenal function across the life course in other conditions.
In summary, this retrospective observational study shows that genetic analysis can help reach a specific diagnosis in almost 70% of children and young people with PAI of unknown etiology, with potential implications for long-term counseling and management. Taken together with CAH and other established causes of adrenal dysfunction, it is now likely that a specific diagnosis can be reached in the vast majority of children and young people with PAI. The adoption of PAI into clinical genetic testing platforms in countries such as the UK should help considerably in improving the speed and rate of diagnosis for these conditions in the future.
Acknowledgments
We are grateful to the participating individuals, and to the following clinicians for their contributions to these studies: Faisal Ahmed, Indi Banerjee, Kausik Banerjee, Ritwik Banerjee, John Barton, Angela Brady, Caroline Brain, Nicola Bridges, Gary Butler, Gerard Conway, Mohammed Didi, Paul Dmitri, Malcolm Donaldson, William Drake, David Dunger, Sarah Ehtisham, Sherif El-Refee, James Greening, Ashley Grossman, Tessa Homfray, Ieuan Hughes, Shelagh Joss, Avril Mason, Shane McKee, Talat Mushtaq, Nisha Nathwani, Evelien Pease-Gevers, Catherine Peters, Annie Procter, Richard Ross, Nick Shaw, Debbie Shears, Savitha Shenoy, Helen Simpson, Helen Spoudeas, Jerry Wales, and John Wass.
Financial Support: This research was funded in whole, or in part, by the Wellcome Trust (grants 098513/Z/12/Z and 209328/Z/17/Z). For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission. JCA also has research support from Great Ormond Street Hospital Children’s Charity (grant V2518) and the National Institute for Health Research, Great Ormond Street Hospital Biomedical Research Centre (grant IS-BRC-1215–20012). LAM is supported by funding from Barts Charity (grant MGU0438) and the Medical Research Council (MRC; Project Grant MR/K020455/1). LFC received funding from the MRC (G0802796), IFCAH, BSPED, and Barts Charity (MGU0458). RP is supported by funding from the MRC (Project Grant MR/T02402X/1). The views expressed are those of the authors and not necessarily those of the National Health Service, National Institute for Health Research, or Department of Health. KK and AH are supported by funding from the German Research Foundation (grants KO 3588/2-1, HU 895/3-3, 3–4, 3–5, 4-1, 5-1, 5-2 and 314061271 CRC-TRR205).
Additional Information
Disclosures: The authors have nothing to declare.
Data Availability
Some or all data sets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
- 1. Bornstein SR, Allolio B, Arlt W, et al. . Diagnosis and treatment of primary adrenal insufficiency: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab. 2016;101(2):364-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Perry R, Kecha O, Paquette J, Huot C, Van Vliet G, Deal C. Primary adrenal insufficiency in children: twenty years experience at the Sainte-Justine Hospital, Montreal. J Clin Endocrinol Metab. 2005;90(6):3243-3250. [DOI] [PubMed] [Google Scholar]
- 3. Buonocore F, Achermann JC. Primary adrenal insufficiency: new genetic causes and their long-term consequences. Clin Endocrinol (Oxf). 2020;92(1):11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buonocore F, McGlacken-Byrne SM, Del Valle I, Achermann JC. Current insights into adrenal insufficiency in the newborn and young infant. Front Pediatr. 2020;8:619041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Flück CE. Mechanism in endocrinology: update on pathogenesis of primary adrenal insufficiency: beyond steroid enzyme deficiency and autoimmune adrenal destruction. Eur J Endocrinol. 2017;177(3):R99-R111. [DOI] [PubMed] [Google Scholar]
- 6. Amano N, Narumi S, Hayashi M, et al. . Genetic defects in pediatric-onset adrenal insufficiency in Japan. Eur J Endocrinol. 2017;177(2):187-194. [DOI] [PubMed] [Google Scholar]
- 7. Suntharalingham JP, Buonocore F, Duncan AJ, Achermann JC. DAX-1 (NR0B1) and steroidogenic factor-1 (SF-1, NR5A1) in human disease. Best Pract Res Clin Endocrinol Metab. 2015;29(4):607-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maharaj A, Maudhoo A, Chan LF, et al. . Isolated glucocorticoid deficiency: genetic causes and animal models. J Steroid Biochem Mol Biol. 2019;189:73-80. [DOI] [PubMed] [Google Scholar]
- 9. Guran T, Buonocore F, Saka N, et al. . Rare causes of primary adrenal insufficiency: genetic and clinical characterization of a large nationwide cohort. J Clin Endocrinol Metab. 2016;101(1):284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Del Valle I, Buonocore F, Duncan AJ, et al. . A genomic atlas of human adrenal and gonad development. Wellcome Open Res. 2017;2:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan LF, Campbell DC, Novoselova TV, Clark AJ, Metherell LA. Whole-exome sequencing in the differential diagnosis of primary adrenal insufficiency in children. Front Endocrinol (Lausanne). 2015;6:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium. Variation across 141 456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. Published online January 30, 2019. doi: 10.1101/531210 [DOI]
- 13. Buonocore F, Maharaj A,Qamar Y,. Data from: genetic analysis of pediatric primary adrenal insufficiency (PAI) of unknown etiology: 25 years experience in the United Kingdom. Open Science Framework. Deposited March 3, 2021. doi: 10.17605/OSF.IO/Z7UVW [DOI] [PMC free article] [PubMed]
- 14. Clark AJL, Grossman A, McLoughlin L. Familial glucocorticoid deficiency associated with point mutation in the adrenocorticotropin receptor. Lancet. 1993;341(8843):461-462. [DOI] [PubMed] [Google Scholar]
- 15. Weber A, Toppari J, Harvey RD, et al. . Adrenocorticotropin receptor gene mutations in familial glucocorticoid deficiency: relationships with clinical features in four families. J Clin Endocrinol Metab. 1995;80(1):65-71. [DOI] [PubMed] [Google Scholar]
- 16. Naville D, Barjhoux L, Jaillard C, et al. . Demonstration by transfection studies that mutations in the adrenocorticotropin receptor gene are one cause of the hereditary syndrome of glucocorticoid deficiency. J Clin Endocrinol Metab. 1996;81(4):1442-1448. [DOI] [PubMed] [Google Scholar]
- 17. Lin L, Hindmarsh PC, Metherell LA, et al. . Severe loss-of-function mutations in the adrenocorticotropin receptor (ACTHR, MC2R) can be found in patients diagnosed with salt-losing adrenal hypoplasia. Clin Endocrinol (Oxf). 2007;66(2):205-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan LF, Metherell LA, Krude H, et al. . Homozygous nonsense and frameshift mutations of the ACTH receptor in children with familial glucocorticoid deficiency (FGD) are not associated with long-term mineralocorticoid deficiency. Clin Endocrinol (Oxf). 2009;71(2):171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clark AJL. Adrenal insufficiency. In: Jameson JL, ed. Hormone Resistance Syndromes. Humana Press; 1999:245-257. doi: 10.1007/978-1-59259-698-0_13 [DOI] [Google Scholar]
- 20. Reutens AT, Achermann JC, Ito M, et al. . Clinical and functional effects of mutations in the DAX-1 gene in patients with adrenal hypoplasia congenita. J Clin Endocrinol Metab. 1999;84(2):504-511. [DOI] [PubMed] [Google Scholar]
- 21. Lin L, Gu WX, Ozisik G, et al. . Analysis of DAX1 (NR0B1) and steroidogenic factor-1 (NR5A1) in children and adults with primary adrenal failure: ten years’ experience. J Clin Endocrinol Metab. 2006;91(8):3048-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shaikh MG, Boyes L, Kingston H, et al. . Skewed X inactivation is associated with phenotype in a female with adrenal hypoplasia congenita. J Med Genet. 2008;45(9):e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hiort O, Holterhus PM, Werner R, et al. . Homozygous disruption of P450 side-chain cleavage (CYP11A1) is associated with prematurity, complete 46,XY sex reversal, and severe adrenal failure. J Clin Endocrinol Metab. 2005;90(1):538-541. [DOI] [PubMed] [Google Scholar]
- 24. Kim CJ, Lin L, Huang N, et al. . Severe combined adrenal and gonadal deficiency caused by novel mutations in the cholesterol side chain cleavage enzyme, P450scc. J Clin Endocrinol Metab. 2008;93(3):696-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maharaj A, Buonocore F, Meimaridou E, et al. . Predicted benign and synonymous variants in CYP11A1 cause primary adrenal insufficiency through missplicing. J Endocr Soc. 2019;3(1):201-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goursaud C, Mallet D, Janin A, et al. . Aberrant splicing is the pathogenicity mechanism of the p.Glu314Lys variant in CYP11A1 gene. Front Endocrinol (Lausanne). 2018;9:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Handschug K, Sperling S, Yoon SJ, Hennig S, Clark AJ, Huebner A. Triple A syndrome is caused by mutations in AAAS, a new WD-repeat protein gene. Hum Mol Genet. 2001;10(3):283-290. [DOI] [PubMed] [Google Scholar]
- 28. Meimaridou E, Kowalczyk J, Guasti L, et al. . Mutations in NNT encoding nicotinamide nucleotide transhydrogenase cause familial glucocorticoid deficiency. Nat Genet. 2012;44(7):740-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Novoselova TV, Rath SR, Carpenter K, et al. . NNT pseudoexon activation as a novel mechanism for disease in two siblings with familial glucocorticoid deficiency. J Clin Endocrinol Metab. 2015;100(2):E350-E354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roucher-Boulez F, Mallet-Motak D, Samara-Boustani D, et al. . NNT mutations: a cause of primary adrenal insufficiency, oxidative stress and extra-adrenal defects. Eur J Endocrinol. 2016;175(1):73-84. [DOI] [PubMed] [Google Scholar]
- 31. Metherell LA, Chapple JP, Cooray S, et al. . Mutations in MRAP, encoding a new interacting partner of the ACTH receptor, cause familial glucocorticoid deficiency type 2. Nat Genet. 2005;37(2):166-170. [DOI] [PubMed] [Google Scholar]
- 32. Chung TT, Chan LF, Metherell LA, Clark AJ. Phenotypic characteristics of familial glucocorticoid deficiency (FGD) type 1 and 2. Clin Endocrinol (Oxf). 2010;72(5):589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prasad R, Chan LF, Hughes CR, et al. . Thioredoxin Reductase 2 (TXNRD2) mutation associated with familial glucocorticoid deficiency (FGD). J Clin Endocrinol Metab. 2014;99(8):E1556-E1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bose HS, Sugawara T, Strauss JF, Miller WL. The pathophysiology and genetics of congenital lipoid adrenal hyperplasia. N Engl J Med. 2002;335(25):1870-1879. [DOI] [PubMed] [Google Scholar]
- 35. Baker BY, Lin L, Kim CJ, et al. . Nonclassic congenital lipoid adrenal hyperplasia: a new disorder of the steroidogenic acute regulatory protein with very late presentation and normal male genitalia. J Clin Endocrinol Metab. 2006;91(12):4781-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Metherell LA, Naville D, Halaby G, et al. . Nonclassic lipoid congenital adrenal hyperplasia masquerading as familial glucocorticoid deficiency. J Clin Endocrinol Metab. 2009;94(10):3865-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sahakitrungruang T, Soccio RE, Lang-Muritano M, Walker JM, Achermann JC, Miller WL. Clinical, genetic, and functional characterization of four patients carrying partial loss-of-function mutations in the steroidogenic acute regulatory protein (StAR). J Clin Endocrinol Metab. 2010;95(7):3352-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Flück CE, Pandey AV, Dick B, et al. . Characterization of novel StAR (steroidogenic acute regulatory protein) mutations causing non-classic lipoid adrenal hyperplasia. PloS One. 2011;6(5):e20178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Buonocore F, Kühnen P, Suntharalingham JP, et al. . Somatic mutations and progressive monosomy modify SAMD9-related phenotypes in humans. J Clin Invest. 2017;8(4):315-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arboleda VA, Lee H, Parnaik R, et al. . Mutations in the PCNA-binding domain of CDKN1C cause IMAGe syndrome. Nat Genet. 2012;44(7):788-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Achermann JC, Ito M, Ito M, Hindmarsh PC, Jameson JL. A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat Genet. 1999;22(2):125-126. [DOI] [PubMed] [Google Scholar]
- 42. Chang Z, Lu W, Zhao Z-H, et al. . Genetic aetiology of primary adrenal insufficiency in Chinese children. Published online July 22, 2020. doi: 10.21203/rs.3.rs-45355/v1 [DOI] [PMC free article] [PubMed]
- 43. Capalbo D, Moracas C, Cappa M, et al. . Primary adrenal insufficiency in childhood: data from a large nationwide cohort. J Clin Endocrinol Metab. 2020;106(3):762-773. [DOI] [PubMed] [Google Scholar]
- 44. Osuwannaratana P, Nimkarn S, Santiprabhob J, Likitmaskul S, Sawathiparnich P. The etiologies of adrenal insufficiency in 73 Thai children: 20 years experience. J Med Assoc Thai. 2008;91(10):1544-1550. [PubMed] [Google Scholar]
- 45. Achermann JC, Silverman BL, Habiby RL, Jameson JL. Presymptomatic diagnosis of X-linked adrenal hypoplasia congenita by analysis of DAX1. J Pediatr. 2000;137(6):878-881. [DOI] [PubMed] [Google Scholar]
- 46. Kolli V, Kim H, Torky A, et al. . Characterization of the CYP11A1 nonsynonymous variant p.E314K in children presenting with adrenal insufficiency. J Clin Endocrinol Metab. 2019;104(2):269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kallali W, Gray E, Mehdi MZ, et al. . Long-term outcome of partial P450 side-chain cleavage enzyme deficiency in three brothers: the importance of early diagnosis. Eur J Endocrinol. 2020;182(3):K15-K24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klaus D. Genotype phenotype relationship in triple A syndrome. [MD thesis]. Medizinische Fakultät, Technische Universität Dresden; 2015. [Google Scholar]
- 49. Narumi S, Amano N, Ishii T, et al. . SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7. Nat Genet. 2016;48(7):792-797. [DOI] [PubMed] [Google Scholar]
- 50. Tan TSE, Manfredonia C, Kumar R, et al. . Retrospective review of Synacthen testing in infants. Arch Dis Child. 2018;103(10):984-986. [DOI] [PubMed] [Google Scholar]
- 51. Parajes S, Kamrath C, Rose IT, et al. . A novel entity of clinically isolated adrenal insufficiency caused by a partially inactivating mutation of the gene encoding for P450 side chain cleavage enzyme (CYP11A1). J Clin Endocrinol Metab. 2011;96(11):E1798-E1806. [DOI] [PubMed] [Google Scholar]
- 52. Hughes CR, Guasti L, Meimaridou E, et al. . MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J Clin Invest. 2012;122(3):814-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Prasad R, Hadjidemetriou I, Maharaj A, et al. . Sphingosine-1-phosphate lyase mutations cause primary adrenal insufficiency and steroid-resistant nephrotic syndrome. J Clin Invest. 2017;127(3):942-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Logan CV, Murray JE, Parry DA, et al. . DNA polymerase epsilon deficiency causes image syndrome with variable immunodeficiency. Am J Hum Genet. 2018;103(6):1038-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Toti MS, Ghirri P, Bartoli A, et al. . Adrenal hemorrhage in newborn: how, when and why—from case report to literature review. Ital J Pediatr. 2019;45(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. David A, Camacho-Hübner C, Bhangoo A, et al. . An intronic growth hormone receptor mutation causing activation of a pseudoexon is associated with a broad spectrum of growth hormone insensitivity phenotypes. J Clin Endocrinol Metab. 2007;92(2):655-659. [DOI] [PubMed] [Google Scholar]
- 57. Chatterjee S, Shapiro L, Rose SJ, et al. . Phenotypic spectrum and responses to recombinant human IGF1 (rhIGF1) therapy in patients with homozygous intronic pseudoexon growth hormone receptor mutation. Eur J Endocrinol. 2018;178(5):481-489. [DOI] [PubMed] [Google Scholar]
- 58. Salvi R, Gomez F, Fiaux M, et al. . Clinical case seminar: progressive onset of adrenal insufficiency and hypogonadism of pituitary origin caused by a complex genetic rearrangement within DAX-1. J Clin Endocrinol Metab. 2002;87:4094–4100. [DOI] [PubMed] [Google Scholar]
- 59. Ahmad I,, Paterson WF,, Lin L,, et al. A novel missense mutation in Dax-1 with an unusual presentation of X-linked adrenal hypoplasia congenita. Horm Res Paediatr. 2007;68(1):32-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Allgrove J, Clayden GS, Grant DB, Macaulay JC. Familial glucocorticoid deficiency with achalasia of the cardia and deficient tear production. Lancet. 1978;311(8077):1284-1286. [DOI] [PubMed] [Google Scholar]
- 61.Houlden H. Clinical and genetic characterization of families with triple A (Allgrove) syndrome. Brain. 2002;125(12):2681-2690. [DOI] [PubMed] [Google Scholar]
- 62.Reshmi-Skarja S, Huebner A, Handschug K, Finegold DN, Clark AJL, Gollin SM. Chromosomal fragility in patients with triple A syndrome. Am J Med Genet. 2003;117A(1):30-36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data sets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.