Abstract
Biofilms formed by Staphylococcus aureus are one of the predominant causes of implant-associated infections (IAIs). Previous studies have found that S. aureus nucleases nuc1 and nuc2 modulate biofilm formation. In this study, we found low nuc1/nuc2 expression and high biofilm-forming ability among IAI isolates. Furthermore, in a mouse model of exogenous IAIs, Δnuc1/2 exhibited higher bacterial load on the surface of the implant than that exhibited by the other groups (WT, Δnuc1, and Δnuc2). Survival analysis of the hematogenous IAI mouse model indicated that nuc1 is a virulence factor related to mortality. We then detected the influence of nuc1 and nuc2 on biofilm formation and immune evasion in vitro. Observation of in vitro biofilm structures with scanning electron microscopy and evaluation of bacterial aggregation with flow cytometry revealed that both nuc1 and nuc2 are involved in biofilm structuring and bacterial aggregation. Unlike nuc1, which is reported to participate in immune evasion, nuc2 cannot degrade neutrophil extracellular traps. Moreover, we found that nuc1/nuc2 transcription is negatively correlated during S. aureus growth, and a possible complementary relationship has been proposed. In conclusion, nuc1/nuc2 are complementary genes involved in biofilm formation in exogenous IAIs. However, nuc2 contributes less to virulence and is not involved in immune evasion.
Keywords: Staphylococcus aureus, biofilm, thermonuclease, implant associated infections, periprosthetic joint infection
Introduction
Orthopedic implants are mainly used for bone fixation and joint replacement. Owing to locally compromised host defense, implanted foreign structures are highly susceptible to microbial colonization (Zimmerli and Moser, 2012; Zimmerli, 2014). As a devastating complication after arthroplasty or internal fixation, implant-associated infections (IAIs) frequently lead to the failure of the prosthetic device or requirement of implant replacement and are associated with substantial patient morbidity (Kapadia et al., 2016; Depypere et al., 2020). Orthopedic IAIs are often caused by Staphylococcus aureus, although many other pathogens can lead to such infections (Arciola et al., 2005; Pulido et al., 2008). IAIs can be classified as exogenous or hematogenous (Zimmerli, 2014; Wang et al., 2017; Arciola et al., 2018). Exogenous infections, which are the most common type, occur as a consequence of direct seeding from external contaminants or contiguous spread during the perioperative period. Hematogenous infections involve bacterial seeding on implants through the bloodstream. Although hematogenous infections occur less frequently, they represent up to 20% of prosthetic joint infections (PJIs) (Sendi et al., 2011; Konigsberg et al., 2014; Tande et al., 2016).
In contrast to other infections such as bacteremia and skin abscess, microbes in IAIs generally form biofilms, which are aggregated structured bacterial communities encased in an extracellular matrix. Biofilms are responsible for the recalcitrance of implant infection to therapy and serve as a source of bacterial dissemination (Arciola et al., 2018). Biofilms are characterized by the production of extracellular polymeric substances (EPSs), which commonly comprise lipids, extracellular proteins, extracellular DNA (eDNA), and exopolysaccharides (Hobley et al., 2015; Schilcher and Horswill, 2020). EPSs typically account for 90% or more of the biofilm dry weight and perform various functions for the inhabitants, such as providing structural rigidity or protecting them from external environmental stress (Flemming and Wingender, 2010; Flemming, 2016). Researchers found that methicillin-sensitive S. aureus (MSSA) strains commonly produce polysaccharide intercellular adhesin (PIA)-dependent biofilms. In contrast, the release of eDNA and cell surface expression of a number of sortase-anchored proteins have been implicated in the biofilm phenotype of methicillin-resistant S. aureus (MRSA) (Mccarthy et al., 2015).
Extracellular DNA has been recognized as a component of the EPS matrix for a long time. However, its role in the EPS was underestimated until the discovery that it is an essential component in Pseudomonas aeruginosa biofilms (Whitchurch et al., 2002). Further investigation revealed that eDNA stabilizes the biofilm matrix and promotes antimicrobial resistance (Hall-Stoodley et al., 2012). In addition, two clinical studies have recently reported a relationship between the presence of eDNA in the biofilm and the outcome of orthopedic IAIs (Zatorska et al., 2017, 2018).
Extracellular DNA is released through bacterial autolysis and digested by nucleases (Okshevsky et al., 2015). According to previous reports, S. aureus secretes thermonuclease enzymes to regulate biofilm formation by modulating eDNA (Kiedrowski et al., 2011; Tang et al., 2011). To our knowledge, the chromosome of S. aureus encodes two thermonucleases, nuc1 and nuc2 (Tang et al., 2008; Hu et al., 2012). nuc1, also called micrococcal nuclease, was the first documented thermonuclease, and it is a secreted virulence factor controlled by the SaeRS two-component system (Olson et al., 2013). nuc2 is a cell surface-binding protein with functional nuclease activity (Kiedrowski et al., 2014). Previous studies have reported that S. aureus secretes nuc1 to degrade neutrophil extracellular traps (NETs) and kill phagocytes (Berends et al., 2010; Thammavongsa et al., 2013; Sultan et al., 2019). Interestingly, the two abovementioned phenotypes (biofilm formation and immune evasion) seem incompatible because nuc1 upregulation contributes to immune evasion, whereas nuc1 downregulation leads to biofilm formation. The mechanism by which nucleases regulate the survival of S. aureus in the IAI microenvironment remains unknown. In addition, the contribution of nuc2 to S. aureus pathogenesis in biofilm-related infections and whether nuc2 contributes to immune evasion are particularly unclear because this nuclease was more recently discovered than nuc1 and has received limited attention.
In this study, we evaluated the activity of thermonucleases in IAI isolates. We also examined the impact of nuc1 and nuc2 on biofilm formation and immune evasion under in vitro and in vivo conditions. Finally, we discussed the relationship between the two nucleases and their function in S. aureus survival and adaptation in the IAI microenvironment.
Results
Low Thermonuclease Expression and High Biofilm-Forming Ability in IAI Strains
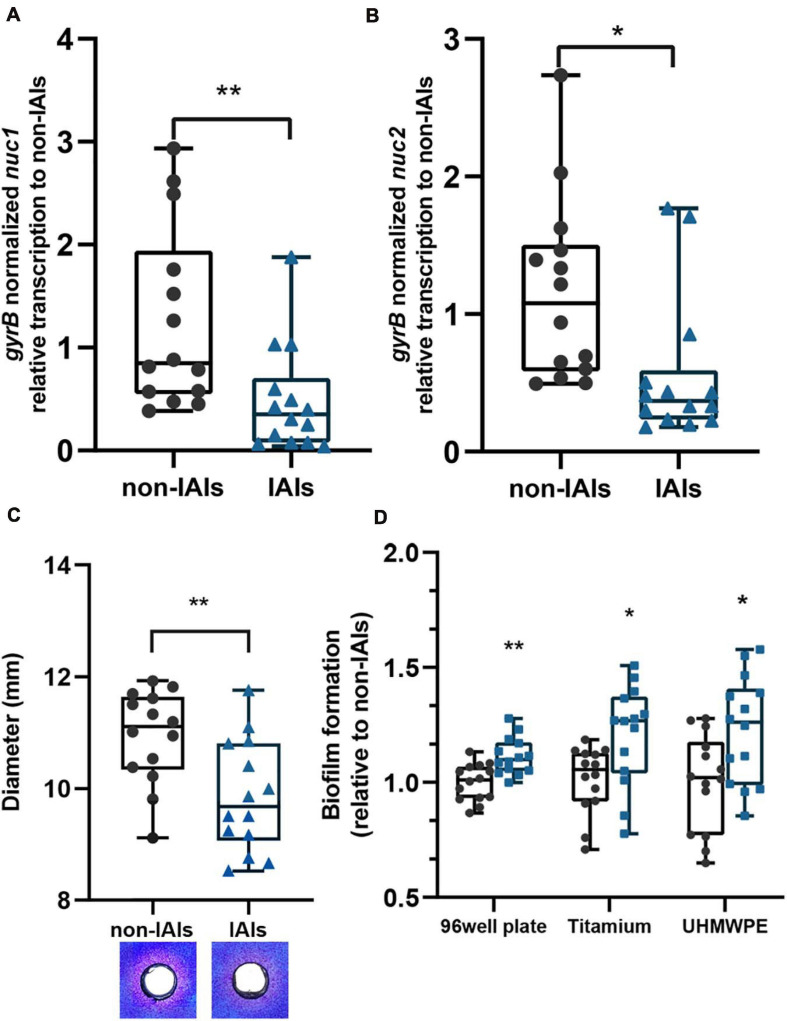
We analyzed the transcription levels of S. aureus thermonucleases among 28 clinical isolates using quantitative PCR (qPCR; Figures 1A,B). Significantly lower transcription levels of both nuc1 (p < 0.01) and nuc2 (p < 0.05) were observed in IAI strains (n = 14) than in non-IAI strains (n = 14). nuc1 and nuc2 transcription levels in the IAI group were 2.57- and 2.47-fold lower than those in the non-IAI groups, respectively. Since gene expression is highly dependent on the involved environment, we included human synovial fluid to mimic the environment encountered by the bacteria in the host, and the results were similar to isolates grown in TSB (Supplementary Figure 1).
FIGURE 1.
The expression levels of thermonucleases and biofilm-forming ability in clinical isolates. Expression of nuc1 (A) and nuc2 (B) in IAI and non-IAI isolates (n = 14/group) determined by qPCR. (C) Nuclease activity in toluidine blue DNA agar represented in the diameter of red zones. Representative images are also presented. Red zones indicate nuclease activity. (D) Biofilm formation of IAI (n = 14) and non-IAI isolates (n = 14) on different materials including polypropylene 96-well plates (non-IAIs = 1.334 ± 0.103), titanium disk (non-IAIs = 1.664 ± 0.229), and UHMWPE (non-IAI = 1.407 ± 0.284). Biofilm biomass was stained with crystal violet. Statistical significance was calculated using two-tail Student’s t-test in panels (A–C); the multiple t-test (Bonferroni–Dunn’s test) was used in panel (D). *p < 0.05; **p < 0.01 vs. non-IAI strains.
To determine whether there were differences in nuclease enzyme activity between the two groups, thermonuclease activity was measured directly using toluidine blue DNA agar, and the zones of clearing were measured. The majority of strains from the IAI group had smaller zones of clearing than those of the strains from the non-IAI group (p < 0.01), which indicated lower thermonuclease activity. Representative images for each group are shown in Figure 1C.
Previous reports demonstrated that S. aureus nuclease (nuc1) could affect biofilm formation by modulating eDNA (Kiedrowski et al., 2011). Here, we noted an increased biofilm eDNA in the IAI group (Supplementary Figure 2A). By relating the eDNA amount to nuc1 and nuc2 expression levels, we found a moderate correlation between nuc1 and eDNA (Pearson R = −0.4592; Supplementary Figure 2B), but no significant correlation was found between nuc2 and eDNA (Pearson R = −0.2983, p > 0.05). Then, we wonder if the IAI isolates with low thermonuclease activity also have higher biofilm-forming ability. Static microtiter biofilm assay found that IAI isolates had higher biofilm-forming ability (Figure 1D). Considering that the most used materials in orthopedic implants are titanium alloy and ultra-high-molecular-weight polyethene (UHMWPE), we further performed biofilm formation assay on titanium disk and UHMWPE, and similar results were obtained (Figure 1D). These data together showed that IAI strains are more prone to form biofilms on the surface of various materials than their non-IAI counterparts.
Construction and Characterization of nuc1/nuc2 Mutant Strains
To study the pathogenesis of S. aureus nuc1 and nuc2 in IAIs, we constructed nuc1 and/or nuc2 mutants using the clinical IAI isolate ST1792, which we termed Δnuc1, Δnuc2, and Δnuc1/2. After in-frame mutation, the strain genotypes were validated by Sanger sequencing (Supplementary Figure 3A). Interestingly, the colony formed by Δnuc1/2 was much stickier (Supplementary Figure 3B) than that formed by wild type (WT), Δnuc1, and Δnuc2. The same phenomenon was also observed in the USA300 nuc1/nuc2 isogenic mutant (BD1281).
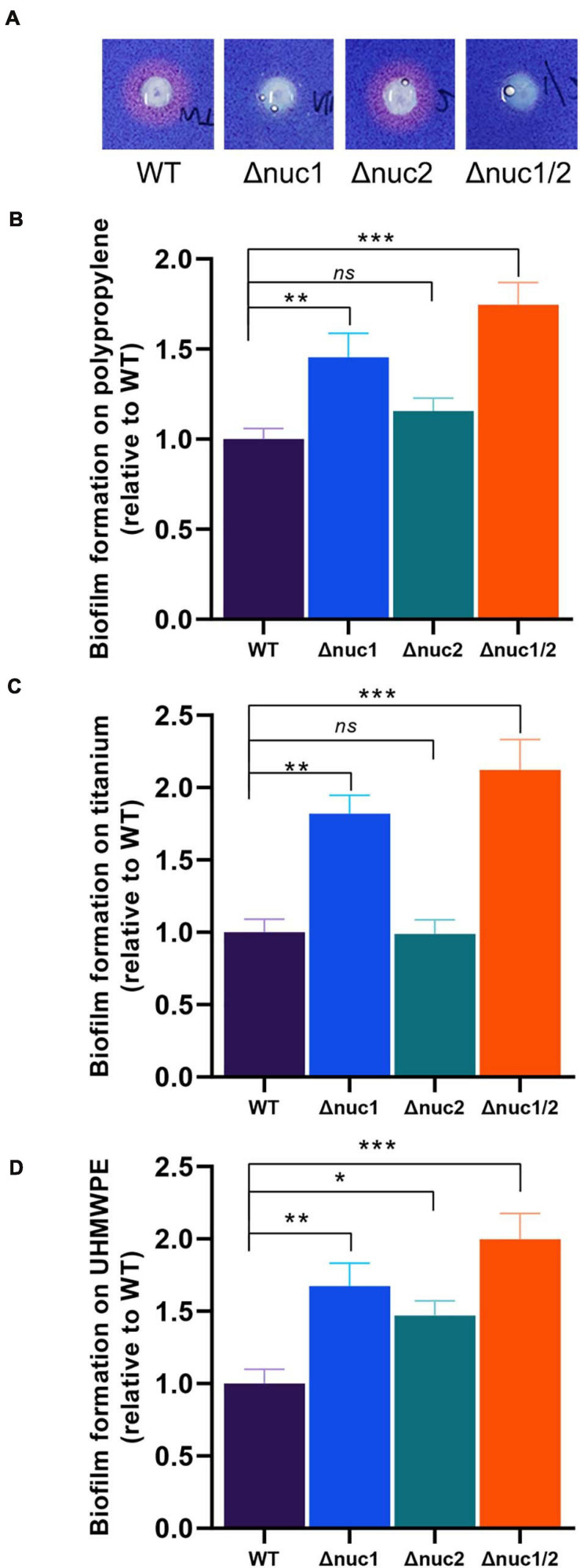
Nuclease activity was then compared among the four strains (Δnuc1, Δnuc2, Δnuc1/2, and WT) using toluidine blue DNA agar (Figure 2A). No observable difference was found between Δnuc2 and WT with a wide area of the clearing zone. In contrast, no detectable nuclease activity was observed for Δnuc1 and Δnuc1/2. We also quantified the biofilm-forming capacity of these strains. Following crystal violet staining, we observed that the biomass of Δnuc1 and Δnuc1/2 increased significantly (Figures 2B–D, Δnuc1: p < 0.01, Δnuc1/2: p < 0.001) in various materials, including titanium, UHMWPE, and polypropylene 96-well plates. However, the biomass of the Δnuc2 biofilm varied with the materials. For example, when grown in UHMWPE, Δnuc2 bacteria developed a more robust biofilm than developed by the WT bacteria. However, biofilms grown on titanium disks and 96-well plates were comparable to the WT biofilms. In addition to quantifying biofilm biomass, the number of culturable cells was also assessed. The results showed that Δnuc1/2 biofilms contained more bacterial cells than the other genotypes (Supplementary Figure 4).
FIGURE 2.
In vitro nuclease activity and biofilm-forming ability of ST1792 and its isogenic mutant strains. (A) Toluidine blue DNA agar test with red zones representing nuclease activity. (B–D) Biofilm-forming ability of tested strains on polypropylene 96-well plates (B, WT = 1.135 ± 0.055), titanium disk (C, WT = 1.237 ± 0.091), and UHMWPE disk (D, WT = 1.457 ± 0.119). Biofilm biomass was stained with crystal violet. Statistical significance was calculated using ANOVA with Dunnett multiple column comparisons. n = 3/group for each experiment. *p < 0.05; **p < 0.01; ***p < 0.001 vs. WT.
Δnuc1/2 Has Higher Biofilm-Forming Capacity in the Exogenous IAI Mouse Model
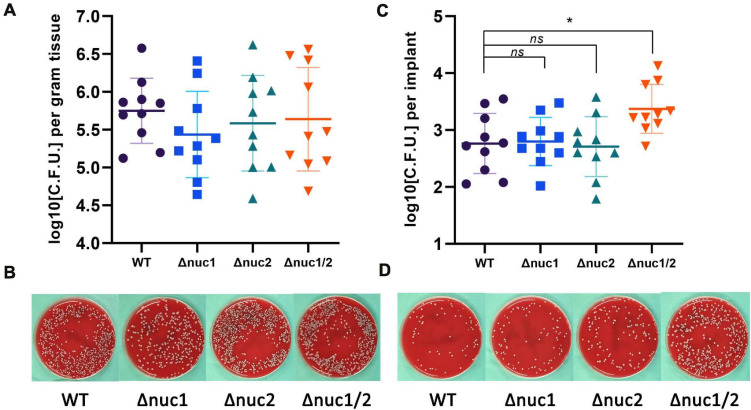
By inoculating bacteria around the implant locally, we constructed an exogenous IAI mouse model. All mice were euthanized 7 days after infection. No significant differences were found among the groups (WT, Δnuc1, Δnuc2, and Δnuc1/2) when evaluating the bacterial load in the peri-implant tissues (Figures 3A,B). However, when quantifying adherent bacteria on the implant, a higher bacterial load was exhibited by Δnuc1/2 than by the WT (Figures 3C,D). Interestingly, the adherent bacterial load showed no statistical difference among the Δnuc1, Δnuc2, and WT groups.
FIGURE 3.
Bacterial burden in an exogenous IAI mouse model. CFU for peri-implant tissue and biofilms on implant was determined 7 days after infection. (A) Bacterial count for peri-implant tissues and representative photos (B). (C) Bacterial count for biofilms on implant and representative photos (D). Statistical significance was calculated using ANOVA with Dunnett multiple column comparisons. n = 10/group. *p < 0.05 vs. WT.
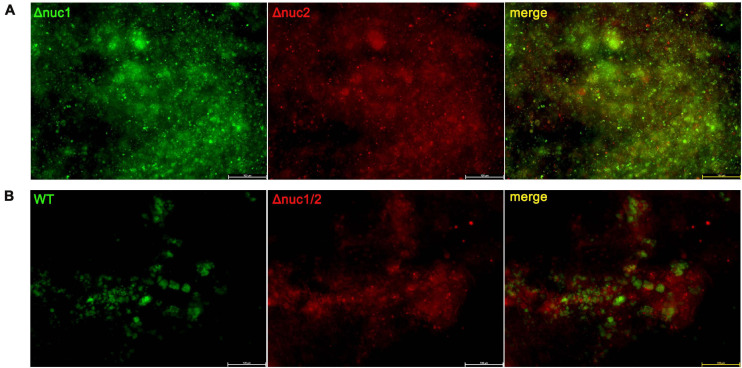
To investigate the effect of nucleases on environmental adaptations in vivo, a competitive assay was conducted. Bacteria with different genotypes and fluorescent labels were mixed and inoculated in vivo. The implants were harvested on day 7 and observed under a fluorescence microscope. Groups infected with a mixture of Δnuc1 and Δnuc2 presented overlapping red and green fluorescence, and no difference was detected between them (Figure 4). The Pearson correlation test showed a strong correlation (R = 0.83) between Δnuc1 (green) and Δnuc2 (red) signals (Supplementary Figure 5A). However, in groups infected with a mixture of WT and Δnuc1/2, a difference in bacterial distribution (Figure 4B) and a low Pearson correlation (Supplementary Figure 5B) were observed. Specifically, Δnuc1/2 strains labeled with mCherry exhibited broad and even distribution, whereas WT strain labeled with superfolder GFP (sfGFP) was distributed in clusters with less area covered.
FIGURE 4.
Implant from a competition infection mouse model observed with a fluorescent microscope. Implants were harvested on the seventh day since infection and observed with a fluorescence microscope. (A) Implant from mice infected with a ∼1:1 mixture of Δnuc1 (green) and Δnuc2 (red). (B) Implant from mice infected with a ∼1:1 mixture of WT (green) and Δnuc1/2 (red). Scale bar = 100 μm, n = 3/group.
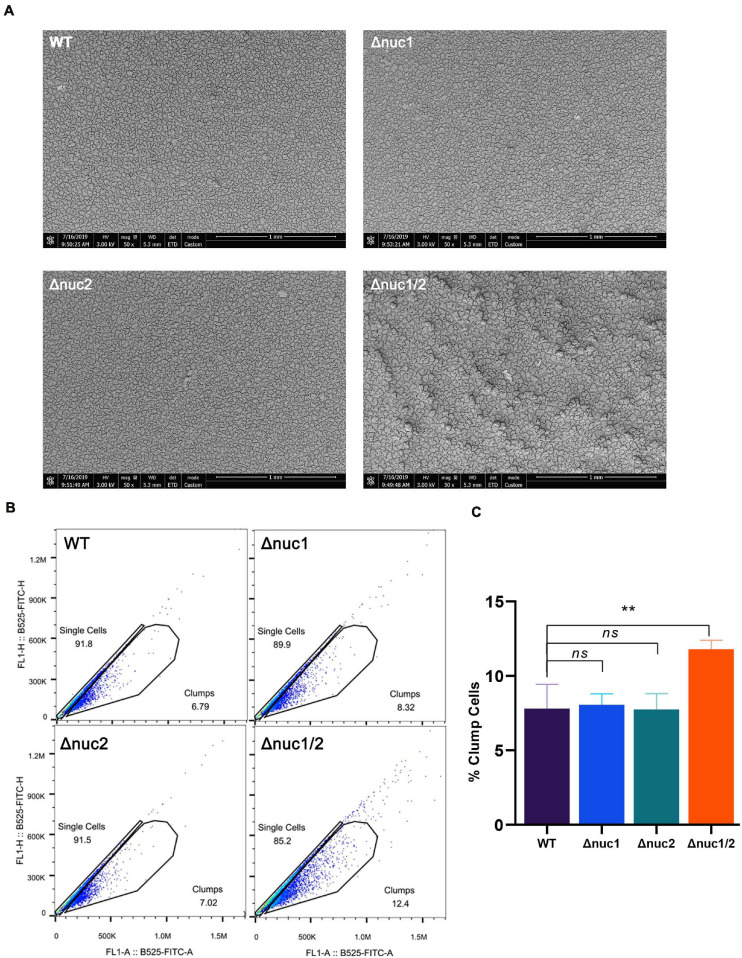
Δnuc1/2 Affects Bacterial Aggregation and Biofilm Structure in vitro
Next, we examined biofilm structure, in vitro, using scanning electronic microscopy (SEM). The biofilm structure of Δnuc1/2 was different from that of the other three genotypes (Figure 5A). Δnuc1/2 bacteria developed “valley and mountain-like” structures, whereas the other bacterial strains did not. However, this difference was only observed at ×50 magnification. When the biofilm was observed at ×2,000 magnification, no difference was detected (Supplementary Figure 6). In order to observe the extracellular matrix, we used a confocal microscope. We noticed that the biofilms formed by Δnuc1/2 were thicker and had higher PI signals, which represent both eDNA and dead cells.
FIGURE 5.
Role of nuc1 and nuc2 on bacterial aggregation and biofilm structure in vitro. (A) Biofilm formed by ST1792 and its isogenic mutant on titanium disk observed with SEM (scale bar = 1 mm). (B) Flow cytometry was used to determine bacterial aggregation. (C) Quantification result for the flow cytometry experiment. Statistical significance was calculated using ANOVA with Dunnett multiple column comparisons. n = 3/group. **p < 0.01 vs. WT.
We also measured the percentage of bacterial aggregation using flow cytometry (Figures 5B,C). Δnuc1/2 was more likely to aggregate between bacterial cells (p < 0.01). No statistical difference was observed among the remaining groups (Δnuc1, Δnuc2, and WT).
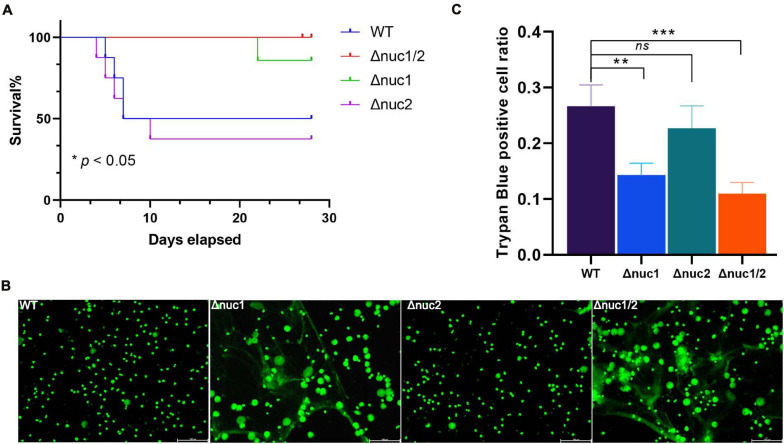
nuc2 Is Not a Virulence Factor Like nuc1 in a Hematogenous Mice Model
The work mentioned above was based on an IAI mouse model induced by surgical site contamination. However, hematogenous infections represent up to 20% of IAIs (Wang et al., 2017). Therefore, we investigated the pathogenesis of Δnuc1 and/or Δnuc2 strains in a hematogenous IAI mouse model. Based on our observations, the group infected with WT and its isogenic nuc2 mutant had significantly higher mortality rates (p < 0.05, Figure 6A). Mutual comparisons of survival curves among the four groups are presented in Supplementary Figure 8. Interestingly, most of the death events occurred within 7 days of infection. However, no difference was detected in the bacterial load in peri-implant tissue and on the implant among the four groups (Supplementary Figure 9).
FIGURE 6.
nuc1 and nuc2 involvement in immune evasion. (A) Survival curve for a hematogenous IAI mouse model infected with ST1792 WT, Δnuc2 (n = 8 each), and Δnuc1 and Δnuc1/2 (n = 7 each). (B) Representative fluorescent images for NET degradation assay. S. aureus were incubated with PMA-stimulated neutrophils (n = 3/group). A green signal represents NETs and cell nucleus, scale bar = 100 μm. (C) THP-1 macrophages were incubated with DNA and S. aureus. Cell viability was determined by trypan blue staining (n = 3/group). The survival curve was analyzed with a log-rank (Mantel–Cox) test. ANOVA with Dunnett multiple column comparisons was used in panel (C). **p < 0.01; ***p < 0.001 vs. WT.
According to previous reports, nuc1 is involved in immune evasion (Tang et al., 2011; Thammavongsa et al., 2013). Hence, we speculated whether nuc2 had the same function. In the NET degradation assay, in which WT and Δnuc1/2 were considered positive and negative controls, respectively, we did not detect any difference when comparing Δnuc1 with Δnuc1/2 (Figure 6B). A previous study reported that nuc1 could lead to immune cell death (Thammavongsa et al., 2013). In line with this result, the trypan blue staining assay (Figure 6C) showed increased THP-1 cell viability in the Δnuc1 and Δnuc1/2 groups when compared with the WT. However, there was no difference in cell viability between the WT and Δnuc2 groups or between the Δnuc1 and Δnuc1/2 groups.
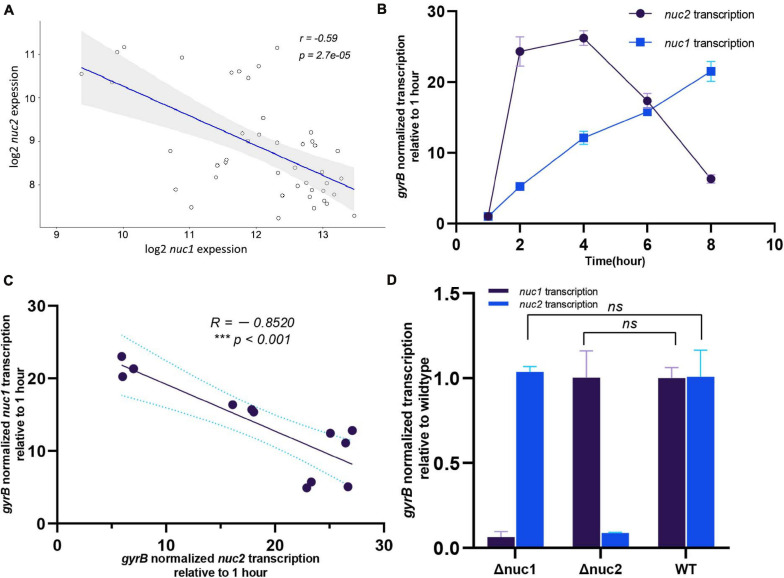
Staphylococcus aureus Sequentially Expresses nuc1 and nuc2 for Environmental Adaptation
Our study indicates that nuc1 and nuc2 are both essential for biofilm formation. However, the redundancy of thermonucleases in the S. aureus chromosome prompted us to investigate the underlying mechanism. By analyzing the public microarray dataset GSE25454, we found that nuc1 and nuc2 were negatively correlated (Figure 7A, R = −0.59, p < 0.001). Our qPCR results confirmed this phenomenon: during S. aureus growth in tryptic soy broth (TSB), nuc2 was upregulated in 2–4 h and then decreased. In contrast, nuc1 transcription peaked in the post-exponential stage (Figure 7B). Also, the correlation between nuc1 and nuc2 in our study was similar to what we found in the dataset GSE25454 (Figure 7C, R = −0.8520, p < 0.001). To exclude the possible regulation between the two genes, we also investigated nuc1/nuc2 transcription in Δnuc2/Δnuc1 (Figure 7D), and the data obtained showed no regulation between nuc1 and nuc2. Such a negative correlation led to our hypothesis that nuc1 and nuc2 are complementary. Therefore, it is possible that nuc2 functions in the early growth phase and that nuc1 plays its role during the later phase.
FIGURE 7.
Transcription relationship between nuc1 and nuc2. (A) nuc1 and nuc2 correlation found by analyzing the publicly available data GSE25454. (B) Transcription levels of nuc1 and nuc2 in WT ST1792 at various time points determined through qPCR (n = 3/time point). (C) Transcription levels of nuc2 at various time points (2, 4, 6, and 8 h) were related to nuc1. (D) nuc1 and nuc2 transcription levels in ST1792 WT, Δnuc1, and Δnuc2 determined using qPCR (n = 3/group). A Pearson correlation test was performed in panels (A,C), and unpaired Student’s t test was performed in panel (D).
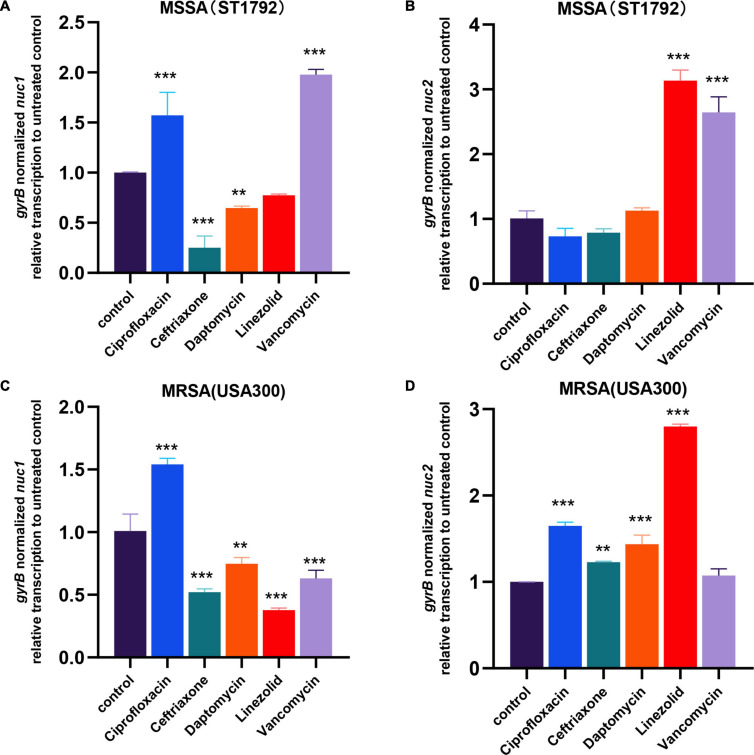
Staphylococcus aureus Modulates Nuclease Transcription When Exposed to Antibiotics
In the first part of our study, we found low expression of nucleases in IAI isolates. Considering patients with IAIs generally require long-term antibiotic administration, we further speculated whether antibiotics would affect nuc1/2 expression. We then exposed MRSA (USA300) and MSSA (ST1792) to several of the most commonly used antibiotics at sub-minimum inhibitory concentration (MIC) levels. First, we determined the MIC and sub-MIC of both strains, and the results are listed in Table 1. Following sub-MIC exposure, qPCR was used to measure nuc1 and nuc2 transcriptions (Figure 8).
TABLE 1.
Minimum Inhibitory Concentration and sub-MIC for strains tested#.
| Strain | (μ g/ml) | Ciprofloxacin | Ceftriaxone | Daptomycin | Linezolid | Vancomycin |
| ST1792 | MIC | 0.25 | 2 | >16 | 1 | 2 |
| USA300 | MIC | 0.25 | >8 | >16 | 1 | 2 |
| ST1792 | sub-MIC* | 0.125 | 1 | 8 | 0.5 | 1 |
| USA300 | sub-MIC* | 0.125 | 4 | 8 | 0.5 | 1 |
#MIC level was determined based on three technical replicates. *Sub-MIC was equal to 1/2 MIC.
FIGURE 8.
nuc1 and nuc2 transcription changes when exposed to antibiotics in MRSA (USA300) and MSSA (ST1792). (A,B) nuc1 (A) and nuc2 (B) transcription changes in MSSA (ST1792). (C,D) nuc1 (C) and nuc2 (D) transcriptional changes in MRSA (USA300). Statistical significance was calculated using ANOVA with Dunnett multiple column comparisons. **p < 0.01; ***p < 0.001 vs. non-treated control.
We found that MSSA and MRSA had different responses to antibiotics. Fewer differences in expression patterns were seen for nuc1 in the two strains with only a major shift seen for vancomycin (decreased in MRSA but increased in MSSA). In both ST1792 and USA300, nuc1 was upregulated when exposed to ciprofloxacin and downregulated after exposure to ceftriaxone and daptomycin. However, for nuc2 expression, an increase was observed in both MRSA and MSSA strains after linezolid. Ciprofloxacin, ceftriaxone, and daptomycin increased nuc2 expression only in the MRSA strain. Finally, nuc2 expression increased in response to vancomycin for the MSSA strain, which is similar to nuc1.
Discussion
To our knowledge, this is the first study reporting that S. aureus IAI isolates have low nuclease (nuc1 and nuc2) expression levels, which may be relevant for the high biofilm-forming capacity of IAI isolates. By constructing nuclease mutant strains, we found that Δnuc1/2 exhibited higher biofilm-forming capacity in an exogenous IAI mouse model. However, a previous study reported that nuc1 and nuc2 had no significant impact on biofilm formation using a murine model of catheter-associated biofilm formation (Beenken et al., 2012). Although the reason underlying this discrepancy with our results is unclear, we noticed that the strain used is different, which could explain, in part, the discrepancies observed.
According to previous reports, nuc1 regulates biofilm formation by modulating eDNA in the biofilm matrix (Kiedrowski et al., 2011; Tang et al., 2011). However, the impact of nuc2 on biofilms has received limited attention. In this study, we noticed that Δnuc1/2 strains formed sticky colonies and distinct biofilm morphology. One possible explanation for this phenomenon is that the mutation of both nuc1 and nuc2 leads to the accumulation of eDNA, which in turn may increase colony and biofilm viscosity. This hypothesis is partially corroborated by a previous study (Kaito et al., 2011), where it was reported that the presence of eDNA increases extracellular matrix viscosity. Also, biofilms observed with confocal microscopy showed that Δnuc1/2 formed thicker biofilms with higher PI signal. According to a previous report, eDNA degradation is involved in the “exodus” and “dispersal” steps during biofilm maturation (Moormeier et al., 2014). Their study prompted us to speculate that a high eDNA content in the biofilm matrix may make both live and dead bacteria unable to egress from the biofilm and get trapped, thus resulting in a thicker biofilm with a higher PI signal. Nevertheless, we cannot exclude other possible mechanisms accounting for the observed phenotypes. Since previous studies have not determined if nuc1 and nuc2 have an influence on other biofilm-related genes, the detected phenomenon could also be caused by the regulation between nucleases and other genes.
By analyzing nuc1 and nuc2 transcription levels at different time points, we found that nuc1 and nuc2 transcription levels were temporally regulated during S. aureus growth, and this was also reported in a previous study (Hu et al., 2012). It seems nuc2 was expressed when cell density was low, and nuc1 was prone to be expressed at high cell density. Such temporal gene regulation was most likely dependent on a quorum-sensing system. However, agr, the most well-studied quorum sensing, is not directly associated with nuc1 or nuc2 regulation (Olson et al., 2013; Kiedrowski et al., 2014). Hence, other quorum-sensing systems (e.g., LuxS) and stimuli rather than population density might be involved in the temporal regulation of thermonucleases. Meanwhile, it should be noted that biofilms are not synchronized in terms of growth phase and that it would lead to special difference in gene expression. Previous studies found that nuc1 and nuc2 are highly expressed in the peripheral colony, highlighting the need to study in more detail the spatial regulation of nuc1 and nuc2 in biofilms (Kaito et al., 2011).
The redundancy of nuc1 and nuc2 does not mean equal contribution to S. aureus virulence. For instance, nuc1 in hematogenous IAIs contributed to the mortality rate of infection while nuc2 did not. Interestingly, we observed that most deaths, in the hematogenous IAI model, occurred during the first 7 days of infection. Based on the knowledge that adaptive immunity takes 4–7 days to mount a response (Iwasaki and Medzhitov, 2010), we speculated that nuc1 may be relevant for innate immune evasion instead of adaptive immune evasion. This was consistent with published researches which demonstrated that S. aureus escapes innate immune defense through NET degradation and phagocyte apoptosis (Berends et al., 2010; Thammavongsa et al., 2013; Winstel et al., 2018). However, concerning nuc2, no observable immune evasion function was detected in our study. It may be due to its reported low enzymic activity (Kiedrowski et al., 2014), and thus, NETs could not be sufficiently degraded with nuc2.
Considering patients with IAIs require long-term administration of antibiotics, our study also examined the impact of antibiotics on nuclease expression. Although MSSA and MRSA had different responses to antibiotics, ceftriaxone and daptomycin reduced nuc1 levels in both MSSA and MRSA. Because MRSA strains are resistant to ceftriaxone, which seems to downregulate nuc1 expression, the use of this antibiotic could lead to increased biofilm formation. As such, caution should be taken when adopting ceftriaxone for IAI treatment before the antibiotic sensitivity testing is clear.
Our study has some limitations. First, the higher biofilm-forming capacity of IAI groups cannot be fully explained by the low expression of thermonucleases, as other factors such as sarA, clfA/B, srtA, and agr loci also contribute to biofilm formation regulation (Paharik and Horswill, 2016; Otto, 2018). Second, the regulation mechanism of nuc2 transcription in S. aureus remains unknown. Finally, in our hematogenous IAI mouse model, we did not check the bacterial load in other systemic organs, which could help us understand the capacity of these strains to disperse to distant sites. Also, the bacterial load in the hematogenous IAI mouse model showed no significant differences even in the Δnuc1/2 group. Although the mechanism underlying the discrepancy observed between the two mouse models remains unclear, we speculated that the virulence adopted by S. aureus to colonize bone implants and develop biofilms might vary depending on how they invade the human body.
In summary, we identified temporal regulation for nuc1 and nuc2. The pathogenesis of both nucleases was explored using two types of infection models. Low expression of both nuc1 and nuc2 is essential in S. aureus IAIs caused by surgical site contamination. However, in hematogenous IAIs, upregulation of nuc1, rather than nuc2, contributes to S. aureus pathogenesis. Our study may provide new insights into the prevention and treatment of IAIs.
Materials and Methods
Bacterial Strains and Growth Conditions
Staphylococcus aureus strains used in this study were either strains that were maintained in our laboratory or clinically isolated. To construct a fluorescence-labeled strain, pRN11 and pCM29 plasmids (Pang et al., 2010; de Jong et al., 2017) expressing mCherry and sfGFP, respectively, were introduced into S. aureus competent cells RN4220 via electroporation using a MicroPulser (Bio-Rad, United States). After adding 0.5 μg of plasmid into 50 μl of RN4220 competent cells, the default Staph program was performed (2-mm gap, 1.8 kV, 2.5 ms). Then, the cells were immediately resuspended in 1 ml of TSB (Haibo, Qingdao, China) and cultured on a shaking incubator at 200 rpm for 1 h at 37°C. A total of 100 μl of the recovery culture was grown overnight at 37°C on a TSB-chloramphenicol (10 μg/ml) agar plate. Next morning, a single chloramphenicol-resistant colony harboring pRN11 or pCM29 plasmids was selected and grown overnight in 4 ml of TSB with chloramphenicol (10 μg/ml). Next, according to a previously described bacteriophage transformation method (Olson, 2014), the plasmid was transformed into S. aureus ST1792, which was isolated from an infected prosthesis, with bacteriophage 11.
Thermonuclease Activity Detection
Bacterial cultures were grown in TSB on a shaking incubator at 37°C and 200 rpm for 6 h and then heat-inactivated at 100°C for 10 min. Toluidine blue DNase agar was used to detect thermonuclease activity according to the manufacturer’s instructions (Haibo, Qingdao, China). A total of 80 μl of inactivated culture was added into a 5-mm-diameter hole in the agar plate made with a sterile pellet tip. The plate was incubated for 6 h at 37°C, and the diameter of the clearing zone was measured.
Construction of Thermonuclease Mutants
In-frame deletion of nuc1 and nuc2 genes in clinical isolate ST1792 was performed by allelic replacement using the plasmid pKOR1 as previously described elsewhere (Bae and Schneewind, 2006). The primers used are listed in Supplementary Table 1. Briefly, after amplifying the upstream and downstream regions of the target gene, we used SOE-PCR to ligate the upstream and downstream fragments. The PCR product was cloned into pKOR1, and the resulting recombinant plasmids pKOR1-nuc1 and pKOR1-nuc2 were further transformed into S. aureus competent cells RN4220 via electroporation and maintained using chloramphenicol (10 μg/ml). Next, the plasmid was transformed into S. aureus ST1792 using bacteriophage 11. S. aureus ST1792 containing the plasmid constructed was used for construction of mutants by allele replacement with temperature shifting as described previously (Bae and Schneewind, 2006). Candidate mutant strains were validated by Sanger sequencing.
In vitro Static Biofilm Assays
All bacterial strains involved in this experiment were cultured overnight at 37°C in TSB supplemented with 0.25% glucose (TSBG). The overnight culture was serially diluted to a concentration of ∼1 × 106 CFU/ml, and then, 100 μl of the culture was inoculated into a 96-well plate with a flat bottom (BIOFIL, Guangzhou, China). UHMWPE disks (5-mm diameter) were sterilized and placed on a 96-well plate with a round bottom (BIOFIL) and then inoculated with 100 μl of the bacterial culture followed by incubation. Titanium disks (10-mm diameter) were sterilized and placed on a 24-well plate with a flat bottom (BIOFIL) and then inoculated with 1 ml of bacterial culture followed by incubation. After incubation at 37°C for 24 h, the culture medium was aspirated from each well, and wells were washed three times with either 200 μl of PBS in case of the 96-well plate or 1 ml of PBS in case of the 24-well plate. After fixation with methanol, the plate was air-dried, and the biofilm was stained with 200 μl of crystal violet. The crystal violet bound at the bottom of the well was dissolved in 200 μl of 33% acetic acid, and 100 μl aliquots from each well were transferred into a new 96-well plate with a flat bottom. Optical absorbance was measured at 590 nm using a microplate reader (BioTek Instruments, Inc., United States) to quantify the biofilm biomass.
Biofilm eDNA Content Measurement
Staphylococcus aureus biofilms were grown as described above in a six-well plate (1.5 ml TSBG per well). After gently removing the supernatant, biofilm cells were resuspended with 1 ml of PBS and then filtered using a 0.2-μm filter. To measure eDNA content, 100 μl of filtered resuspension was mixed with 100 μl of 2 μM SYTOX Green (Invitrogen, United States). Fluorescence was measured by using a plate reader (BIO-TEK, ELX 800, United States) with excitation and emission wavelengths of 485 and 520 nm.
Staphylococcus aureus RNA Isolation and Quantitative PCR
To investigate the transcription levels of thermonucleases between an IAI strain and a non-IAI strain, S. aureus was cultured in TSB (or TSB supplied with 20% human synovial fluid) in a shaking incubator at 37°C and 200 rpm for 6 h. To investigate thermonuclease expression at different time points, ST1792 was cultured in TSB (37°C/200 rpm) and collected at several time points (1, 2, 4, 6, and 8 h). To determine nuc1/2 expression in ST1792 and its isogenic mutants (Δnuc1 and Δnuc2), bacteria were cultured in TSB and incubated at 37°C and 200 rpm for 6 h. To explore nuc1/nuc2 transcription changes after exposure to antibiotic, ST1792 and USA300 were cultured in TSB supplied with sub-MIC antibiotics for 6 h (37°C/200 rpm). The abovementioned bacteria were harvested and transferred to a tissue lyser (ScientzTM, Ningbo, China), and the cell wall was physically disrupted for 30 s at a frequency of 50 Hz. RNA was isolated using the EZ-press RNA Purification Kit (EZBioscience, United States) according to the manufacturer’s instructions. The quality of the RNA was measured using a Nanodrop device (Thermo Fisher Scientific, United States), and RNA samples with absorbance ratios of 260 nm/280 nm and 260 nm/230 nm higher than 2.0 were selected for reverse transcription. After adding DNase to remove gDNA, 1 μg fresh RNA was immediately reverse-transcribed into cDNA using an RT-PCR kit (EZBioscience, United States). cDNA was diluted with ddH2O (1:5 dilution) and subsequently used as the DNA template for qPCR, performed with the kit 2 × SYBR Green qPCR Master Mix (EZBioscience). DNA amplification was performed by thermal cycling: initial denaturation at 95°C for 5 min, followed by 40 amplification cycles at 95°C for 10 s and at 60°C for 30 s using a Roche LightCycler 480 (Roche, Switzerland). The primers used in this study and other related information such as product size, primer efficiency, and cycling parameters are provided in Supplementary Table 1. Relative gene expression levels were quantified using the 2–ΔΔCT method, with the expression levels of gyrB as the internal reference.
Biofilm Structure Observation Using SEM and Confocal Microscopy
Overnight-grown ST1792 biofilms were formed on a titanium disk as described above. After gently being washed with PBS, the samples were fixed with 2.5% glutaraldehyde at 4°C for 4 h, dehydrated using a graded ethanol series (50, 70, 80, 90, 95, and 100% v/v) for 10 min, freeze-dried, coated with platinum, and visualized using SEM (Magellan 400, FEI, United States).
For confocal microscopy observation, biofilms grown on titanium disk were stained with a Live/Dead BacLight bacterial viability kit (Invitrogen, United States) according to the instructions of the manufacturer. Stained biofilms were observed immediately using a confocal microscope (Leica TCS SP8, Germany). The optimal exposure time and laser intensity for both channels (excitation: 488 and 555 nm) were manually set to ensure no overexposure among groups. Then, all of the images were acquired with the same setting for comparability among groups. Raw data were imported into Imaris 9.0.1 for biovolume and biofilm thickness calculation.
Detection of Bacterial Clumps Through Flow Cytometry
sfGFP-labeled ST1792 and its isogenic mutants were incubated in TSB at 37°C and 200 rpm overnight in the presence of chloramphenicol. Then, overnight bacterial cultures were used for flow cytometry (Beckman Coulter CytoFLEX, United States). The sample flow rate was 10 μl/min, and 20,000 events were recorded for further analysis. First, the GFP+ cell population, comprising the bacteria, was selected. Then, single/clumping populations were labeled using the correlation between FITC-H and FITC-A.
Neutrophil Extracellular Traps Degradation Assay
Neutrophils were isolated from the blood of healthy donors. The anti-coagulated whole blood (5 ml) was carefully layered over 5.0 ml of PolymorphprepTM (Alere Technologies, Norway) in a 15-ml centrifuge tube. Tubes were centrifuged at 500× g for 30 min at 18–22°C. PMNs were gently separated, and 3 ml of RPMI medium was added to restore normal osmolality. Samples were centrifuged at 400× g for 10 min to collect cells. Finally, the cells were resuspended in the RPMI medium containing 10% fetal bovine serum (FBS). For NET induction, phorbol-12-myristate-13-acetate (PMA, Sigma, United States) was added to the culture medium to reach a final concentration of 90 nM, and samples were incubated at 37°C with 5% CO2 for 4 h. Then, heat-inactivated bacterial culture (ST1792 and its isogenic mutants) was added (MOI = 100) and incubated for 2 h to degrade the NETs. After fixation with 4% paraformaldehyde, NETs were stained with SYTOX Green (Invitrogen, United States) and observed using a Leica DMI8 microscope (Leica, Germany).
Cellular Cytotoxicity Assays Using Trypan Blue Staining
Human monocytic THP-1 cells were obtained from the Institute of Biochemistry and Cell Biology (Shanghai, China). A total of 1 × 105 THP-1 cells were cultured, overnight, in an RPMI medium containing 10% FBS and penicillin/streptomycin together with heat-inactivated WT, Δnuc1, Δnuc2, or Δnuc1/2 bacterial suspensions (MOI = 10) and DNA (final concentration of 100 ng/μl). Subsequently, cells were stained with trypan blue and visualized using light microscopy.
Bacterial MIC Assay
We adopted a macrodilution method to determine the MIC of different antibiotics for MRSA and MSSA, and 1 ml of twofold serial dilutions of antibiotics dissolved in TSB was added to 1 ml of TSB, which contained nearly 106 CFU/ml, in separate tubes. After overnight incubation at 37°C, the turbidity of the test tubes was visually inspected, as turbid test tubes were indicative of bacterial growth, whereas tubes that remained clear indicated no growth. The MIC of the antibiotics tested was considered to be the lowest concentration that inhibited growth. Gentamycin, linezolid, daptomycin, ciprofloxacin, and ceftriaxone were purchased from Aladdin (Shanghai, China).
GEO Microarray Data Analysis
Public microarray dataset GSE25454 including 74 samples was selected. First, we downloaded the raw data (.CEL files) from the GEO database (GSE25454). Then, the following process was performed in R (4.0.1). We used the readAffy function (limma package; Ritchie et al., 2015) to import the .CEL files and performed background correction with the gcrma function (GCRMA package; Lim et al., 2007). Note that the resulting expression data were represented as intensity and transformed into log-2 scale according to descriptions in the GCRMA package manual. After that, we used the “normalizeWithinArray” function (from the limma package; Ritchie et al., 2015) to normalize the data among samples to remove batch effects. Finally, we extracted the nuc1 and nuc2 expression values for each sample at various time points and performed a Pearson correlation test. The scatter plot was presented with the ggscatter function (ggpubr package).
Implant-Associated Infections Mouse Model
Mice Model of Exogenous IAIs
Twenty BALB/c mice (6 weeks old) were randomly divided into four groups (WT, Δnuc1, Δnuc2, and Δnuc1/2). Mice were intraperitoneally anesthetized with 1% pelltobarbitalum natricum (provided by the animal center), and both knees were shaved and disinfected. Then, the distal femur was exposed through a medial parapatellar incision, and a narrow channel was created at the femoral end using a 25G needle. Subsequently, the prepared sterile titanium wires (0.5-mm diameter) were inserted in a retrograde direction into the intramedullary canal. The overlying subcutaneous tissue and skin were closed using absorbable subcuticular sutures. Finally, 25 μl of the corresponding bacterial inoculum (ST1792 and its isogenic mutants, ∼1 × 107/ml) was injected intraarticularly into the knee joint space. All mice were anesthetized and euthanized by cervical dislocation 7 days after the infection. Peri-implant tissues were harvested and homogenized in 1 ml of sterile saline before CFU counting. The biofilm on the titanium wires together with 1 ml of sterile saline was subjected to sonication (30 kHz, 10 min) in an ultrasound bath (CQ-200B-DST, Yuejin, China), and the resulting sonicated fluid was used for further bacterial load quantification.
Competition Infection Model
To observe the in vivo biofilm structure and explore the adaptability of different S. aureus genotypes, a titanium disk was implanted subcutaneously into the dorsal area of the mice, and ∼1 × 106 CFU of the strain mixture was inoculated around the implant. We labeled Δnuc1/WT with sfGFP and Δnuc2/Δnuc1/2 with mCherry. Mice were infected with either a WT/Δnuc1/2 mixture or a Δnuc1/Δnuc2 mixture. All mice were euthanized by cervical dislocation and anesthetized 7 days after the infection. The biofilm on the titanium disks was observed using a fluorescence microscope (Leica DMI8, Germany).
Mouse Model of Hematogenous IAIs
Thirty-two adult BALB/c mice were randomly divided into four groups (WT, Δnuc1, Δnuc2, and Δnuc1/2). After the implantation surgery performed as described previously, mice were infected via tail vein injection (ST1792 and its isogenic mutants, 1 × 107 CFU/100 μl). Survival was recorded daily. All mice were anesthetized and euthanized by cervical dislocation 28 days after the infection. Peri-implant tissues and biofilm cells on the implant were prepared as described above. Bacterial load enumeration was performed according to the protocol listed in Section “Bacterial Load Enumeration.”
Bacterial Load Enumeration
The bacterial suspension or sonicated fluid was serially diluted tenfold. A total of 100 μl of the diluted suspension was spread on sheep blood agar plates. After incubation at 37°C overnight, CFU counts were performed according to the National Standard of China GB/T 4789.2 protocol. The resulting bacterial load for peri-implant tissues was normalized to tissue weight, and all bacterial loads were presented on a log10 scale.
Statistical Analysis
The GEO microarray was analyzed with R 4.0.1. The fluorescence distribution pattern in Figure 4 was quantified with a Pearson correlation test using Coloc 2, a program in ImageJ (1.53c, Fiji). The remaining data analysis was performed using GraphPad Prism 8.3.0. Statistical significance was indicated as a two-sided p < 0.05. Results are represented as mean ± SD unless stated otherwise.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
Neutrophils were isolated from the blood of healthy donors. Human synovial fluid was collected from osteoarthritis patients before they received intra-articular injection of hyaluronic acid. The procedures were approved by the Ethics Committee of the Shanghai Sixth Peoples Hospital. The handling of mice and related procedures in this study were approved by the Animal Care and Experiment Committee of the Medical College of Shanghai Jiao Tong University affiliated Sixth People’s Hospital.
Author Contributions
JY contributed to the concept of the study and wrote the manuscript. JD and FZ contributed to the qPCR experiment. YM and JY performed S. aureus mutant construction. JT and MH collected clinical strains and performed antibiotic susceptibility testing. JY, FJ, and FZ performed the in vivo experiment. QW and PH contributed to data analysis and data interpretation. HS contributed to the study design, manuscript editing and revision. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Diep BA for the gift of USA300 and its isogenic mutants (BD1276, BD1280, and BD1281).
Abbreviations
- MRSA/MSSA
methicillin-resistant/methicillin-sensitive Staphylococcus aureus
- EPS
extracellular polymeric substances
- SEM
scanning electronic microscope
- IAIs
implant-associated infections
- MIC
minimum inhibitory concentration
- TSB
tryptic soy broth.
Footnotes
Funding. This study was supported by the National Natural Science Foundation of China (Grant No. 81772364) and Medical Guidance Scientific Research Support Project of Shanghai Science and Technology Commission (Grant No. 19411962600).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.687888/full#supplementary-material
Transcription levels of thermonucleases in clinical isolates cultured in TSB (supplied with 20% human synovial fluid). Expression of nuc1 (A) and nuc2 (B) in IAI and non-IAI isolates (n = 14/group) determined by qPCR. Statistical significance was calculated using two-tailed Student’s t-test; ∗p < 0.05; ∗∗p < 0.01 vs. non-IAI strains.
Biofilm eDNA measurement for clinical isolates and its relation to nuc1 or nuc2 transcription. (A) Biofilm eDNA content in IAI and non-IAI biofilms was measured by with SYTOX green staining and presented as fluorescence signals (n = 14/group). Two-tailed Student’s t-test was adopted; ∗p < 0.05 vs. the non-IAI group. (B) Biofilm eDNA content was related to nuc1 or nuc2 expression using the Pearson correlation test.
(A) Sanger sequence results of nuc1 and nuc2 mutations with alignment to WT ST1792. (B) ST1792 nuc1 and nuc2 double mutation leads to the sticky characteristic of the bacterial colony.
Bacterial load enumeration for biofilms formed in vitro by ST1792 and its isogenic mutants. Statistical significance was calculated using ANOVA with Dunnett multiple column comparisons. n = 3/group. ∗∗p < 0.01 vs. WT.
Fluorescence co-localization test using ImageJ. (A) Δnuc1 and Δnuc2 co-localization 2D intensity plot. (B) WT and Δnuc1/2 co-localization 2D intensity plot. A Pearson correlation test was performed in this study.
Biofilms formed on titanium disk in vitro by ST1792 and its isogenic mutant strains observed using SEM with ×2,000 magnification. Scale bar=10 μm.
Biofilms formed by ST1792 and its isogenic mutant strains observed using a confocal microscope. Green represents live cells, and red represents eDNA and dead cells. Scale bar=50 μm.
Mutual comparison of survival curves among four groups. (A) Survival analysis between WT (n = 8) and Δnuc1 (n = 7); (B) survival analysis between WT (n = 8) and Δnuc2 (n = 8); (C) survival analysis between WT (n = 8) and Δnuc1/2 (n = 7); (D) survival analysis between Δnuc1 (n = 7) and Δnuc2 (n = 8); (E) survival analysis between Δnuc1/2 (n = 7) and Δnuc2 (n = 8). Statistical significance was analyzed with a log-rank (Mantel–Cox) test.
Bacterial count for implant (right) and peri-implant tissues (left) in a hematogenous IAI mouse model (n = 8, 12, 6, and 16 h for WT, Δnuc1, Δnuc2, and Δnuc1/2, respectively). Statistical significance was calculated using ANOVA with Dunnett multiple column comparisons.
References
- Arciola C. R., An Y. H., Campoccia D., Donati M. E., Montanaro L. (2005). Etiology of implant orthopedic infections: a survey on 1027 clinical isolates. Int. J. Artif. Organs 28 1091–1100. 10.1177/039139880502801106 [DOI] [PubMed] [Google Scholar]
- Arciola C. R., Campoccia D., Montanaro L. (2018). Implant infections: adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 16 397–409. 10.1038/s41579-018-0019-y [DOI] [PubMed] [Google Scholar]
- Bae T., Schneewind O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55 58–63. 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Beenken K. E., Spencer H., Griffin L. M., Smeltzer M. S. (2012). Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under in vitro and in vivo conditions. Infect. Immun. 80 1634–1638. 10.1128/IAI.06134-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berends E. T., Horswill A. R., Haste N. M., Monestier M., Nizet V., von Kockritz-Blickwede M. (2010). Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J. Innate Immun. 2 576–586. 10.1159/000319909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong N. W., van der Horst T., van Strijp J. A., Nijland R. (2017). Fluorescent reporters for markerless genomic integration in Staphylococcus aureus. Sci. Rep. 7:43889. 10.1038/srep43889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depypere M., Morgenstern M., Kuehl R., Senneville E., Moriarty T. F., Obremskey W. T., et al. (2020). Pathogenesis and management of fracture-related infection. Clin. Microbiol. Infect. 26 572–578. 10.1016/j.cmi.2019.08.006 [DOI] [PubMed] [Google Scholar]
- Flemming H. C. (2016). EPS-then and now. Microorganisms 4:41. 10.3390/microorganisms4040041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H. C., Wingender J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8 623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L., Stoodley P., Kathju S., Hoiby N., Moser C., Costerton J. W., et al. (2012). Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol. Med. Microbiol. 65 127–145. 10.1111/j.1574-695X.2012.00968.x [DOI] [PubMed] [Google Scholar]
- Hobley L., Harkins C., MacPhee C. E., Stanley-Wall N. R. (2015). Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol. Rev. 39 649–669. 10.1093/femsre/fuv015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Xie Y., Tang J., Shi X. (2012). Comparative expression analysis of two thermostable nuclease genes in Staphylococcus aureus. Foodborne Pathog. Dis. 9 265–271. 10.1089/fpd.2011.1033 [DOI] [PubMed] [Google Scholar]
- Iwasaki A., Medzhitov R. (2010). Regulation of adaptive immunity by the innate immune system. Science 327 291–295. 10.1126/science.1183021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaito C., Hirano T., Omae Y., Sekimizu K. (2011). Digestion of extracellular DNA is required for giant colony formation of Staphylococcus aureus. Microb. Pathog. 51 142–148. 10.1016/j.micpath.2011.04.007 [DOI] [PubMed] [Google Scholar]
- Kapadia B. H., Berg R. A., Daley J. A., Fritz J., Bhave A., Mont M. A. (2016). Periprosthetic joint infection. Lancet 387 386–394. 10.1016/S0140-6736(14)61798-0 [DOI] [PubMed] [Google Scholar]
- Kiedrowski M. R., Crosby H. A., Hernandez F. J., Malone C. L., McNamara J. O., II, Horswill A. R. (2014). Staphylococcus aureus Nuc2 is a functional, surface-attached extracellular nuclease. PLoS One 9:e95574. 10.1371/journal.pone.0095574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski M. R., Kavanaugh J. S., Malone C. L., Mootz J. M., Voyich J. M., Smeltzer M. S., et al. (2011). Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One 6:e26714. 10.1371/journal.pone.0026714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konigsberg B. S., Della Valle C. J., Ting N. T., Qiu F., Sporer S. M. (2014). Acute hematogenous infection following total hip and knee arthroplasty. J. Arthroplasty. 29 469–472. 10.1016/j.arth.2013.07.021 [DOI] [PubMed] [Google Scholar]
- Lim W. K., Wang K., Lefebvre C., Califano A. (2007). Comparative analysis of microarray normalization procedures: effects on reverse engineering gene networks. Bioinformatics 23 i282–i288. 10.1093/bioinformatics/btm201 [DOI] [PubMed] [Google Scholar]
- Mccarthy H., Rudkin J. K., Black N. S., Gallagher L., O’Neill E., O’Gara J. P. (2015). Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 5:1. 10.3389/fcimb.2015.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moormeier D. E., Bose J. L., Horswill A. R., Bayles K. W. (2014). Temporal and stochastic control of Staphylococcus aureus biofilm development. mBio 5:e01341–14. 10.1128/mBio.01341-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okshevsky M., Regina V. R., Meyer R. L. (2015). Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 33 73–80. 10.1016/j.copbio.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Olson M. E. (2014). “Bacteriophage transduction in Staphylococcus aureus,” in Methods in Molecular Biology, ed. Bose J. L. (New York, NY: Springer; ), 69–74. 10.1007/7651_2014_186 [DOI] [PubMed] [Google Scholar]
- Olson M. E., Nygaard T. K., Ackermann L., Watkins R. L., Zurek O. W., Pallister K. B., et al. (2013). Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect. Immun. 81 1316–1324. 10.1128/IAI.01242-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2018). Staphylococcal biofilms. Microbiol. Spectr. 6:6.4.27. 10.1128/microbiolspec.GPP3-0023-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paharik A. E., Horswill A. R. (2016). The Staphylococcal Biofilm: adhesins, regulation, and host response. Microbiol. Spectr. 4:4.2.06. 10.1128/microbiolspec.VMBF-0022-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y. Y., Schwartz J., Thoendel M., Ackermann L. W., Horswill A. R., Nauseef W. M. (2010). agr-Dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J. Innate Immun. 2 546–559. 10.1159/000319855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido L., Ghanem E., Joshi A., Purtill J. J., Parvizi J. (2008). Periprosthetic joint infection: the incidence, timing, and predisposing factors. Clin. Orthop. Relat. Res. 466 1710–1715. 10.1007/s11999-008-0209-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilcher K., Horswill A. R. (2020). Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 84:e00026–19. 10.1128/MMBR.00026-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendi P., Banderet F., Graber P., Zimmerli W. (2011). Periprosthetic joint infection following Staphylococcus aureus bacteremia. J. Infect. 63 17–22. 10.1016/j.jinf.2011.05.005 [DOI] [PubMed] [Google Scholar]
- Sultan A. R., Hoppenbrouwers T., Lemmens-den Toom N. A., Snijders S. V., van Neck J. W., Verbon A., et al. (2019). During the early stages of Staphylococcus aureus biofilm formation, induced neutrophil extracellular traps are degraded by autologous thermonuclease. Infect. Immun. 87 e00605–19. 10.1128/IAI.00605-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tande A. J., Palraj B. R., Osmon D. R., Berbari E. F., Baddour L. M., Lohse C. M., et al. (2016). Clinical presentation, risk factors, and outcomes of hematogenous prosthetic joint infection in patients with Staphylococcus aureus bacteremia. Am. J. Med. 129:221 e211–e220. 10.1016/j.amjmed.2015.09.006 [DOI] [PubMed] [Google Scholar]
- Tang J., Kang M., Chen H., Shi X., Zhou R., Chen J., et al. (2011). The staphylococcal nuclease prevents biofilm formation in Staphylococcus aureus and other biofilm-forming bacteria. Sci. China Life Sci. 54 863–869. 10.1007/s11427-011-4195-5 [DOI] [PubMed] [Google Scholar]
- Tang J., Zhou R., Shi X., Kang M., Wang H., Chen H. (2008). Two thermostable nucleases coexisted in Staphylococcus aureus: evidence from mutagenesis and in vitro expression. FEMS Microbiol. Lett. 284 176–183. 10.1111/j.1574-6968.2008.01194.x [DOI] [PubMed] [Google Scholar]
- Thammavongsa V., Missiakas D. M., Schneewind O. (2013). Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science 342 863–866. 10.1126/science.1242255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cheng L. I., Helfer D. R., Ashbaugh A. G., Miller R. J., Tzomides A. J., et al. (2017). Mouse model of hematogenous implant-related Staphylococcus aureus biofilm infection reveals therapeutic targets. Proc. Natl. Acad. Sci. U.S.A. 114 E5094–E5102. 10.1073/pnas.1703427114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch C. B., Tolker-Nielsen T., Ragas P. C., Mattick J. S. (2002). Extracellular DNA required for bacterial biofilm formation. Science 295:1487. 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- Winstel V., Missiakas D., Schneewind O. (2018). Staphylococcus aureus targets the purine salvage pathway to kill phagocytes. Proc. Natl. Acad. Sci. U.S.A. 115 6846–6851. 10.1073/pnas.1805622115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatorska B., Arciola C. R., Haffner N., Segagni Lusignani L., Presterl E., Diab-Elschahawi M. (2018). Bacterial extracellular DNA production is associated with outcome of prosthetic joint infections. Biomed. Res. Int. 2018:1067413. 10.1155/2018/1067413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatorska B., Groger M., Moser D., Diab-Elschahawi M., Lusignani L. S., Presterl E. (2017). Does extracellular DNA production vary in staphylococcal biofilms isolated from infected implants versus controls? Clin. Orthop. Relat. Res. 475 2105–2113. 10.1007/s11999-017-5266-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli W. (2014). Clinical presentation and treatment of orthopaedic implant-associated infection. J. Intern. Med. 276 111–119. 10.1111/joim.12233 [DOI] [PubMed] [Google Scholar]
- Zimmerli W., Moser C. (2012). Pathogenesis and treatment concepts of orthopaedic biofilm infections. FEMS Immunol. Med. Microbiol. 65 158–168. 10.1111/j.1574-695X.2012.00938.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcription levels of thermonucleases in clinical isolates cultured in TSB (supplied with 20% human synovial fluid). Expression of nuc1 (A) and nuc2 (B) in IAI and non-IAI isolates (n = 14/group) determined by qPCR. Statistical significance was calculated using two-tailed Student’s t-test; ∗p < 0.05; ∗∗p < 0.01 vs. non-IAI strains.
Biofilm eDNA measurement for clinical isolates and its relation to nuc1 or nuc2 transcription. (A) Biofilm eDNA content in IAI and non-IAI biofilms was measured by with SYTOX green staining and presented as fluorescence signals (n = 14/group). Two-tailed Student’s t-test was adopted; ∗p < 0.05 vs. the non-IAI group. (B) Biofilm eDNA content was related to nuc1 or nuc2 expression using the Pearson correlation test.
(A) Sanger sequence results of nuc1 and nuc2 mutations with alignment to WT ST1792. (B) ST1792 nuc1 and nuc2 double mutation leads to the sticky characteristic of the bacterial colony.
Bacterial load enumeration for biofilms formed in vitro by ST1792 and its isogenic mutants. Statistical significance was calculated using ANOVA with Dunnett multiple column comparisons. n = 3/group. ∗∗p < 0.01 vs. WT.
Fluorescence co-localization test using ImageJ. (A) Δnuc1 and Δnuc2 co-localization 2D intensity plot. (B) WT and Δnuc1/2 co-localization 2D intensity plot. A Pearson correlation test was performed in this study.
Biofilms formed on titanium disk in vitro by ST1792 and its isogenic mutant strains observed using SEM with ×2,000 magnification. Scale bar=10 μm.
Biofilms formed by ST1792 and its isogenic mutant strains observed using a confocal microscope. Green represents live cells, and red represents eDNA and dead cells. Scale bar=50 μm.
Mutual comparison of survival curves among four groups. (A) Survival analysis between WT (n = 8) and Δnuc1 (n = 7); (B) survival analysis between WT (n = 8) and Δnuc2 (n = 8); (C) survival analysis between WT (n = 8) and Δnuc1/2 (n = 7); (D) survival analysis between Δnuc1 (n = 7) and Δnuc2 (n = 8); (E) survival analysis between Δnuc1/2 (n = 7) and Δnuc2 (n = 8). Statistical significance was analyzed with a log-rank (Mantel–Cox) test.
Bacterial count for implant (right) and peri-implant tissues (left) in a hematogenous IAI mouse model (n = 8, 12, 6, and 16 h for WT, Δnuc1, Δnuc2, and Δnuc1/2, respectively). Statistical significance was calculated using ANOVA with Dunnett multiple column comparisons.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.