Summary
Background
Mutations accrued by SARS-CoV-2 lineage P.1—first detected in Brazil in early January, 2021—include amino acid changes in the receptor-binding domain of the viral spike protein that also are reported in other variants of concern, including B.1.1.7 and B.1.351. We aimed to investigate whether isolates of wild-type P.1 lineage SARS-CoV-2 can escape from neutralising antibodies generated by a polyclonal immune response.
Methods
We did an immunological study to assess the neutralising effects of antibodies on lineage P.1 and lineage B isolates of SARS-CoV-2, using plasma samples from patients previously infected with or vaccinated against SARS-CoV-2. Two specimens (P.1/28 and P.1/30) containing SARS-CoV-2 lineage P.1 (as confirmed by viral genome sequencing) were obtained from nasopharyngeal and bronchoalveolar lavage samples collected from patients in Manaus, Brazil, and compared against an isolate of SARS-CoV-2 lineage B (SARS.CoV2/SP02.2020) recovered from a patient in Brazil in February, 2020. Isolates were incubated with plasma samples from 21 blood donors who had previously had COVID-19 and from a total of 53 recipients of the chemically inactivated SARS-CoV-2 vaccine CoronaVac: 18 individuals after receipt of a single dose and an additional 20 individuals (38 in total) after receipt of two doses (collected 17–38 days after the most recent dose); and 15 individuals who received two doses during the phase 3 trial of the vaccine (collected 134–230 days after the second dose). Antibody neutralisation of P.1/28, P.1/30, and B isolates by plasma samples were compared in terms of median virus neutralisation titre (VNT50, defined as the reciprocal value of the sample dilution that showed 50% protection against cytopathic effects).
Findings
In terms of VNT50, plasma from individuals previously infected with SARS-CoV-2 had an 8·6 times lower neutralising capacity against the P.1 isolates (median VNT50 30 [IQR <20–45] for P.1/28 and 30 [<20–40] for P.1/30) than against the lineage B isolate (260 [160–400]), with a binominal model showing significant reductions in lineage P.1 isolates compared with the lineage B isolate (p≤0·0001). Efficient neutralisation of P.1 isolates was not seen with plasma samples collected from individuals vaccinated with a first dose of CoronaVac 20–23 days earlier (VNT50s below the limit of detection [<20] for most plasma samples), a second dose 17–38 days earlier (median VNT50 24 [IQR <20–25] for P.1/28 and 28 [<20–25] for P.1/30), or a second dose 134–260 days earlier (all VNT50s below limit of detection). Median VNT50s against the lineage B isolate were 20 (IQR 20–30) after a first dose of CoronaVac 20–23 days earlier, 75 (<20–263) after a second dose 17–38 days earlier, and 20 (<20–30) after a second dose 134–260 days earlier. In plasma collected 17–38 days after a second dose of CoronaVac, neutralising capacity against both P.1 isolates was significantly decreased (p=0·0051 for P.1/28 and p=0·0336 for P.1/30) compared with that against the lineage B isolate. All data were corroborated by results obtained through plaque reduction neutralisation tests.
Interpretation
SARS-CoV-2 lineage P.1 might escape neutralisation by antibodies generated in response to polyclonal stimulation against previously circulating variants of SARS-CoV-2. Continuous genomic surveillance of SARS-CoV-2 combined with antibody neutralisation assays could help to guide national immunisation programmes.
Funding
São Paulo Research Foundation, Brazilian Ministry of Science, Technology and Innovation and Funding Authority for Studies, Medical Research Council, National Council for Scientific and Technological Development, National Institutes of Health.
Translation
For the Portuguese translation of the abstract see Supplementary Materials section.
Introduction
SARS-CoV-2 is a betacoronavirus (in the Coronaviridae family) that was first reported in Wuhan, China, in December, 2019.1 As of May 7, 2021, SARS-CoV-2 has caused more than 155 million cases and 3·2 million deaths globally.2 More than 1·45 million SARS-CoV-2 genome sequences have been classified in over 900 lineages.3 The appearance and spread of some mutations in the spike protein, such as Asp614Gly, have resulted in more transmissible SARS-CoV-2 variants.4 The spike protein's receptor-binding domain (RBD) and N-terminal domain (NTD) are the primary targets of neutralising antibodies in the SARS-CoV-2 response;5, 6 however, the RBD is a highly variable region, and circulating SARS-CoV-2 might be under antibody-mediated selective pressure.7 Consequently, the emergence of SARS-CoV-2 variants with mutations in the RBD has raised concerns that neutralising antibody responses, and the effectiveness of vaccination programmes, could be compromised.8
In late 2020, the B.1.1.7 lineage was detected in the UK and the B.1.351 lineage detected in South Africa.9, 10 As of May 7, 2021, B.1.1.7 has been identified in 114 countries and B.1.351 in 68 countries.3 Both these lineages have enhanced transmissibility compared with previously circulating SARS-CoV-2 lineages, and carry unique constellations of spike protein mutations. Wild-type SARS-CoV-2 isolates or pseudoviruses carrying the same mutations described in these lineages showed reduced neutralisation by immune sera from individuals who had received an mRNA vaccine (eg, BNT162b2 [tozinameran; developed by Pfizer–BioNTech] or mRNA-1273 [Moderna]) or adenoviral-vectored vaccine (eg, ChAdOx1 nCoV-19 [Oxford University–AstraZeneca]), suggesting that these lineages might be inhibited by vaccine-mediated humoral immunity.11, 12, 13 A new SARS-CoV-2 lineage P.1 was discovered in Manaus, Brazil, in early January, 2021.14 P.1 has a signature set of 15 unique amino acid changes, including a trio of mutations (Lys417Thr, Glu484Lys, and Asn501Tyr) in the RBD that also are present in the B.1.351 lineage.14 In this study, we investigated whether the full set of mutations found in the spike protein of the P.1 lineage can escape from neutralising antibodies generated by patients with a history of COVID-19 or by individuals previously immunised with the inactivated SARS-CoV-2 vaccine CoronaVac.15
Methods
Overview
We did an immunological study to assess the neutralising effects of antibodies on lineage P.1 and lineage B isolates of SARS-CoV-2, using plasma samples from patients previously infected with or vaccinated against SARS-CoV-2.
All procedures followed the ethical standards of the responsible committee on human experimentation and approved by the ethics committees from the University of Campinas (Campinas, Brazil; approval numbers CONEP 4.021.484 [for plasma collection of blood donors], and CAEE 32078620.4.0000.5404 and 30227920.9.0000.5404 [for the sampling of vaccinated and viral genome sequencing, respectively]). All patient data were anonymised before use in the study, and patients gave informed consent for inclusion in the study.
Research in context.
Evidence before this study
The emergence of variants of concerns of SARS-CoV-2 with multiple mutations in the spike protein has caused concern about the potential for the virus to evade antibody responses elicited by previous SARS-CoV-2 infections or vaccinations. We searched PubMed without language restriction until May 5, 2021, for studies published with the terms “SARS-CoV-2 lineage P.1” or “escape neutralization” and “CoronaVac vaccine”, and found evidence that P.1 can escape neutralisation by antibodies generated through natural SARS-CoV-2 infection or through vaccination with mRNA or adenoviral-vectored SARS-CoV-2 vaccines. However, most of these studies have used pseudoviruses or an infectious clone system with chimeric viruses. To our knowledge, there is scarce information about the ability of wild-type P.1 isolates of SARS-CoV-2, containing the complete set of signature mutations, to escape from neutralising antibodies present in individuals previously infected with SARS-CoV-2 or vaccinated with an inactivated vaccine.
Added value of this study
This study showed that antibodies present in the plasma of blood donors with a history of COVID-19 or individuals vaccinated with CoronaVac were less efficient at neutralising SARS-CoV-2 lineage P.1 isolates than a lineage B isolate.
Implications of all the available evidence
Neutralising antibodies are an important correlate of protection of the human immune response against SARS-CoV-2. Therefore, the capacity of the P.1 lineage to evade antibodies present in the plasma of naturally infected or CoronaVac-immunised individuals suggests that variants of concern might escape population immunity and continue to circulate in populations, even with high vaccination coverage with inactivated vaccines. Consequently, continued surveillance of neutralising antibody levels and genetic diversity of SARS-CoV-2 strains will be needed to monitor potential viral mutants with increased ability to escape immune responses in vaccinated individuals and to guide potential updates of national and international immunisation programmes.
SARS-CoV-2 isolates
Samples of SARS-CoV-2 P.1 were obtained from residual nasopharyngeal lavage specimens from two patients with COVID-19, obtained from the Hematology and Hemotherapy Hospital Foundation of Amazonas (Fundação Hospitalar de Hematologia e Hemoterapia do Amazonas, Manaus, Brazil). The samples were confirmed to be positive for SARS-CoV-2 infection by real-time quantitative RT-PCR (RT-qPCR) with cycle threshold (Ct) values of 22 and 17. All nasopharyngeal samples used in this study were collected from patients on Dec 21, 2020, and had been previously classified as being infected with SARS-CoV-2 of the P.1 lineage by viral genome sequencing.14
Nasopharyngeal lavage samples were inoculated into Vero cells (CCL-81; ATCC, Manassas, VA, USA) for virus isolation, using previously described methods.16 Briefly, Vero cells were plated in a T25 cell culture flask at a concentration of 5 × 105 cells per mL with Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin solution (10 000 U/mL penicillin and 10 mg/mL streptomycin; Sigma-Aldrich, St Louis, MO, USA). Samples were thawed on ice, diluted 1:10 in DMEM, and centrifuged at 12 000 × g for 5 min at 4°C. Samples were filtered with 0·22 μm syringe filters and incubated on ice for 1 h with penicillin-streptomycin solution and amphotericin B (250 μg/mL, 1:1; Sigma-Aldrich) in a final dilution of 1:10. After incubation at 37°C for 1 h for adsorption, the inoculum was removed from the culture and replaced with fresh culture medium. Cells were incubated at 37°C and observed for cytopathic effects daily, to 72 h. Subsequently, the cell culture supernatant was collected and viral replication was confirmed through RT-qPCR. Viral RNA was extracted from the supernatant cells with a Quick-RNA Viral Kit (Zymo Research, Irvine, CA, USA) following the manufacturer-recommended procedures. RT-qPCR was used to confirm viral isolation through a decrease in the Ct value.17 All experiments related to cell culture and viral replication were done in the biosafety level 3 laboratory of the University of Campinas Emerging Viruses Laboratory (Campinas, Brazil). We also sequenced SARS-CoV-2 genomes from two isolates using the ARTIC version 3 protocol with MinION sequencing (Oxford Nanopore Technologies, Oxford, UK), as described elsewhere.14
For comparison, we also used a B lineage isolate (SARS.CoV2/SP02.2020, GenBank accession number MT126808) recovered from a sample collected on Feb 28, 2020, in Brazil.18
Immunofluorescent staining
Infection by isolates was confirmed by an immunofluorescent staining. Vero cells were plated onto silanised glass slides, fixed, and stained as previously described.19 Briefly, after fixation with 4% paraformaldehyde, cells were washed with phosphate-buffered saline containing Tween 20 (0·1 M, pH 7·4), then incubated for 10 min with glycine (0·1 M) and treated for 30 min with 1% bovine serum albumin (BSA) solution (Sigma-Aldrich). Cells then were incubated overnight at 4°C with a recombinant human IgG1 antibody against SARS-CoV-2 spike protein S1 subunit RBD (clone HC2001, catalogue number A02038; GenScript, Piscataway, NJ, USA) or a rabbit IgG antibody against SARS-CoV-2 nucleocapsid protein (clone 001, catalogue number 40143-R001; Sino Biological, Wayne, PA, USA) diluted 1:100 in 1% BSA. The slides were washed and incubated for 2 h with secondary antibodies (Alexa Fluor 488-conjugated goat anti-human IgG, catalogue number A11013, Thermo Fisher Scientific, Waltham, MA, USA; or Alexa Fluor 594-conjugated donkey anti-rabbit IgG, catalogue number A21207, Thermo Fisher Scientific) diluted 1:500 in 1% BSA. Cells then were washed and stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; catalogue number SC3598, Santa Cruz Biotechnology, Dallas, TX, USA) for nuclear labelling and Alexa Fluor 647 phalloidin (catalogue number A22287, Thermo Fisher Scientific) to visualise F-actin. Microscopic images were acquired with a Zeiss LSM 880 with Airyscan on an Axio Observer 7 inverted microscope (Carl Zeiss, Jena, Germany) with a C Plan Apochromat 63 × /1·4 oil differential interference contrast objective lens with 4 × optical zoom. Before visual inspection of the images, raw .czi files were automatically processed into deconvoluted Airyscan images using Zen Black 2.3 software. DAPI staining was visualised with conventional confocal imaging using a 405 nm laser line for excitation and pinhole set to 1 Airy unit.
Plasma specimens
To evaluate the capacity of P.1 isolates to be neutralised by antibodies generated against other, previously circulating SARS-CoV-2 virus lineages, we collected plasma from 21 blood donors who had a history of SARS-CoV-2 infection confirmed by laboratory methods between May and August, 2020 (months before the first recorded infections associated with the P.1 lineage). These patients had donated blood to a plasmapheresis programme being run at the University of Campinas (Campinas, Brazil). Plasma samples were obtained using the Amicus automated blood cell separator (Fresenius Kabi, Bad Homburg, Germany) at the Hematology and Hemotherapy Center in the University of Campinas. Clinical data for these patients were obtained from electronic health records and included age, sex, method of SARS-CoV-2 diagnosis (eg, laboratory confirmed), duration of COVID-19 symptoms, time between COVID-19 symptom onset and sample collection, and whether hospitalisation was required during SARS-CoV-2 infection (appendix 2 p 8). As a negative control, we also obtained plasma from a person who had not been previously infected with SARS-CoV-2.
Plasma samples from vaccinated individuals were acquired from 15 participants in the Sinovac phase 3 trial (NCT04456595), and from 18 individuals who received a single dose and 38 who received two doses of CoronaVac during the Brazilian vaccination programme against SARS-CoV-2. Samples were collected by venipuncture at the Clinical Hospital of the University of Campinas (Campinas, Brazil). The clinical trial was done in Brazil following the Declaration of Helsinki and Good Clinical Practice Guidelines and with approval by the Brazilian Health Regulatory Agency. Full information was collected for each individual and included age, sex, and dates of first and second vaccine doses (appendix 2 pp 9–11).
IgM and IgG antibodies against SARS-CoV-2 spike and nucleocapsid proteins in the plasma samples were measured with Abbott SARS-CoV-2 chemiluminescence microparticle immunoassays (catalogue numbers 6R87 and 6R86, Abbott, Chicago, IL, USA) using the Architect instrument, in accordance with the manufacturer's instructions.
SARS-CoV-2 neutralisation tests
SARS-CoV-2 neutralisation tests were done as previously described.20 In brief, plasma samples from previously infected and vaccinated participants were inactivated at 56°C for 30 min. 100 μL aliquots of two-fold serial dilutions (final dilution range 1:20 to 1:2560) of heat-inactivated plasma were then incubated for 1 h at 37°C with 100 μL of solution containing 103 plaque-forming units (PFU) per mL of SARS-CoV-2 lineage P.1 (isolates P.1/28 and P.1/30) or lineage B (isolate SARS.CoV2/SP02.2020).18 After incubation, 120 μL solution was transferred to 5 × 103 Vero cell monolayers (multiplicity of infection 0·01) for 1 h at 37°C with 5% CO2. After virus removal, cells were incubated for 72 h with DMEM containing 10% FBS. The median virus neutralisation titre (VNT50) for a plasma sample was defined as the reciprocal value of the sample dilution that showed 50% protection against cytopathic effects based on the number of wells in which cytopathic effects were observed, similar to a median tissue culture infectious dose analysis (appendix 2 p 2). Each plasma sample was tested in duplicate using two independent assays.
Plaque reduction neutralisation test
An in-house plaque reduction neutralisation test (PRNT) was done as previously described.16 Serial dilutions of each serum sample from both previously infected and vaccinated participants were incubated with a solution containing 2 × 103 PFU/mL of SARS-CoV-2 lineages P.1 or B for 1 h at 37°C. The virus–serum mixtures were added to pre-formed Vero cell monolayers and incubated for 1 h at 37°C in 5% CO2. Subsequently, 1 mL of DMEM containing 1% carboxymethylcelluloses and 5% FBS was gently added to each well, and the plates were incubated at 37°C in a 5% CO2 atmosphere for 3 days. Finally, cells were fixed with 2 mL of 8% formaldehyde solution overnight and stained with 1% methylene blue (Sigma-Aldrich) for 5 min. Plaque reduction was calculated for each plasma sample-containing well relative to the number of plaques in wells inoculated with SARS-CoV-2 alone (positive control; appendix 2 p 2). PRNT50 was defined as the sample dilution that showed a 50% reduction in number of plaques formed compared with that in the positive control well.
Statistical analysis
To assess whether plasma VNT50s against isolates of SARS-CoV-2 lineage P.1 were reduced compared with those against SARS-CoV-2 lineage B in individuals with a history of SARS-CoV-2 infection, we used a binomial distribution model based on the proportion of samples in each of two categories (differences >0 and differences ≤0). The analysis was done in R Studio version 1.3.1073. The statistical method is described in appendix 2 (p 1).
For analysis of differences in plasma neutralising antibody titres, median PRNT (PRNT50) for each sample was calculated as a mean of two technical duplicates, and statistical significance of the differences between group medians was determined by one-way ANOVA followed by Tukey's test of non-linear regression curves using GraphPad Prism software (version 8.2.1).
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
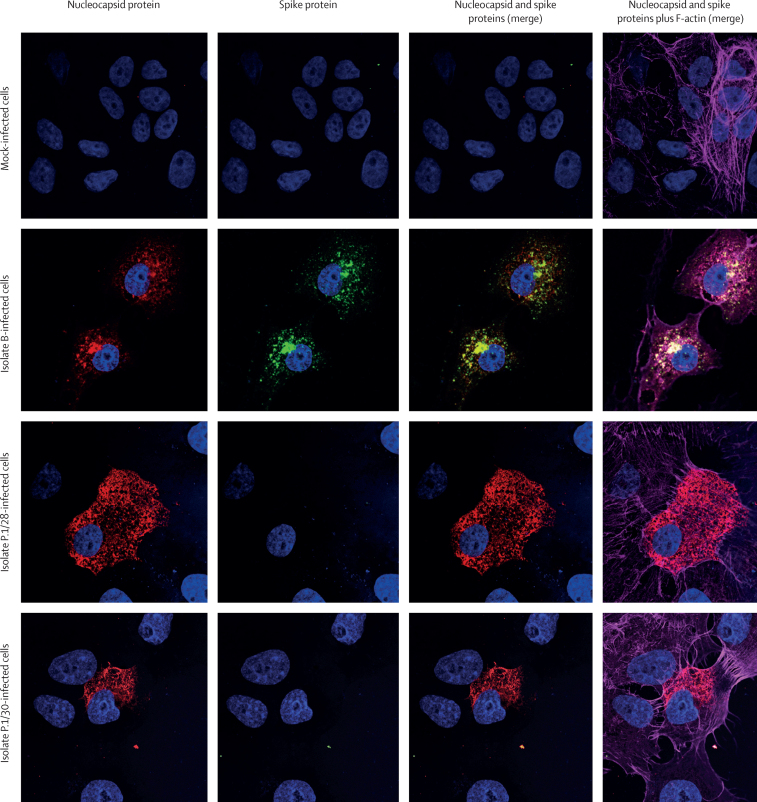
The two SARS-CoV-2 isolates obtained from nasopharyngeal samples (P.1/28 and P.1/30) were confirmed as belonging to the P.1 lineage by genome sequencing. We obtained near-complete sequences, with genome coverage of 83·7% for isolate P.1/28 and 99·6% for isolate P.1/30. We confirmed all signature mutations of the P.1 lineage on isolate P.1/30, and 13 of 15 amino acid signature mutations3 in isolate P.1/28 (appendix 2 p 12). The sequences were deposited in GISAID (EPI_ISL_1708317 and EPI_ISL_1708318). The full mutation set of the P.1 signature mutations in the original samples was previously confirmed.14 Cytopathic effects in a Vero cell monolayer were observed 3 days post-inoculation and cell culture supernatant was harvested 4 days post-inoculation (appendix 2 p 3). Viral titres were 7·2 × 104 for P.1/28 and 1·2 × 105 PFU/mL for P.1/30. The presence of SARS-CoV-2 RNA was confirmed using RT-qPCR targeting the envelope gene (appendix 2 p 3). The immunofluorescence assay using commercial monoclonal antibodies against SARS-CoV-2 spike protein subunit S1 RDB and nucleocapsid protein showed robust binding to the nucleocapsid protein but reduced binding to the spike protein in SARS-CoV-2 lineage P.1-infected Vero cells compared with binding in cells infected with the lineage B isolate (figure 1).
Figure 1.
Immunofluorescent staining with commercial antibodies against SARS-CoV-2 proteins
Images show staining of mock-infected Vero cells and of Vero cells inoculated with a SARS-CoV-2 lineage B isolate or a SARS-CoV-2 lineage P.1 isolate (P.1/28 or P.1/30). Cells were stained with antibodies against SARS-CoV-2 nucleocapsid protein (red fluorescence, columns 1, 3, and 4) and spike protein (green fluorescence, columns 2–4) and phalloidin for visualisation of F-actin filaments (pink fluorescence, column 4). Nuclei were labelled with 4′,6-diamidino-2-phenylindole dihydrochloride (blue fluorescence). Slides were analysed by confocal microscopy and images were merged with ImageJ.
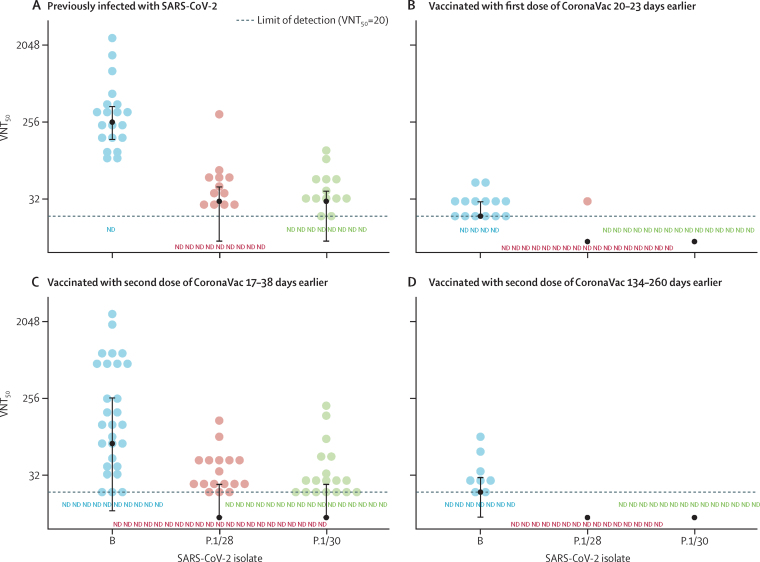
Plasma samples were collected from 21 blood donors (nine [43%] men and 12 [57%] women; median age 34 years [IQR 31–43]) with a history of laboratory-confirmed SARS-CoV-2 infection. The median time between onset of symptoms and blood sample collection was 65 days (45–69), and all samples had high concentrations of SARS-CoV-2-specific IgM or IgG antibodies (appendix 2 p 8). The negative control sample, obtained from a person who had not previously been infected with SARS-CoV-2, had undetectable levels of anti-SARS-CoV-2 IgM and IgG antibodies. We tested these plasma samples for neutralisation of the isolates P.1/28 and P.1/30 and a Brazilian SARS-CoV-2 lineage B reference isolate (SARS.CoV-2/SP02.2020) on the basis of VNT50. A binominal model showed that VNT50s of plasma from previously SARS-CoV-2-infected patients against the lineage P.1 isolates were significantly reduced compared with those against the lineage B isolate (p≤0·0001; figure 2A). Plasma from previously SARS-CoV-2-infected individuals had median VNT50s of 30 for isolate P.1/28 (IQR <20–45) and 30 for isolate P.1/30 (<20–40), compared with a VNT50 of 260 (160–400) for the B lineage isolate, indicating that the capacity of neutralisation antibodies was around 8·6 times lower for P.1 lineage isolates compared with the B lineage isolate (figure 2A; appendix 2 p 8). Additionally, higher paired differences in VNT50s between P.1/28 and B lineage isolates corresponded to higher paired differences (data not shown) between P.1/30 and B lineage isolates (Pearson's correlation 0·997), indicating consistency between results for both isolates of P.1.
Figure 2.
Neutralisation of SARS-CoV-2 lineages B and P.1 by plasma from previously infected or vaccinated individuals, according to VNT50
Plasma samples were incubated with Vero cells infected with a SARS-CoV-2 lineage B isolate (SARS.CoV2/SP02.2020) or one of two lineage P.1 isolates (P.1/28 and P.1/30) to assess VNT50 (defined as the reciprocal value of the plasma sample dilution that showed 50% protection against cytopathic effects). (A) Plasma from blood donors previously infected with SARS-CoV-2 (n=21). (B) Plasma from individuals vaccinated with a single dose of CoronaVac vaccine during the Brazilian vaccination programme (n=18), collected at a median 21 days (IQR 21–21; range 20–23) after receipt of the first dose. (C) Plasma from individuals vaccinated with two doses of CoronaVac vaccine during the Brazilian vaccination programme (n=38), collected at a median 21 days (IQR 20–23; range 17–38) after receipt of the second dose. (D) Plasma from individuals vaccinated with two doses of CoronaVac vaccine in the Sinovac phase 3 trial (n=15), collected at a median 158 days (IQR 156–170; range 134–260) after receipt of the second dose. Dashed lines indicate the lower LOD of the VNT50 assay for samples with low or absent virus neutralisation capacity. Black circles indicate group medians for each isolate, with bars showing IQRs. Each data point is the average of a duplicate assay for each plasma sample, and two independent assays were done for all groups (except the group of participants who received a single dose of vaccine, panel B). LOD=limit of detection (VNT50 titre <20). ND=not detected (below LOD). VNT50=median virus neutralisation titre.
To investigate whether the P.1 lineage might also escape neutralisation by antibodies induced by the CoronaVac vaccine, a set of plasma samples was obtained from 18 individuals (six [33%] men and 12 [67%] women); median age 29 years [IQR 26–36]) who had received a first dose of the vaccine 20–23 days earlier (median 21 days [21–21]), and from an additional 20 individuals (38 in total; ten [26%] men and 28 [74%] women; 30 years [26–40]) who had received a second dose of vaccine 17–38 days earlier (median 21 days [20–23]; appendix 2 pp 9–10). The median VNT50 of plasma samples from participants after a single dose of CoronaVac was 20 (IQR 20–30) against the lineage B isolate, and almost all plasma samples against both P.1 isolates had VNT50s below the limit of detection (<20; figure 2B; appendix 2 pp 9–10). The median VNT50s of plasma samples from individuals who had received two doses of CoronaVac were 75 (<20–263) against the lineage B isolate, 24 (<20–25) against isolate P.1/28, and 28 (<20–25) against isolate P.1/30 (figure 2C; appendix 2 pp 9–10), showing significant reductions with both lineage P.1 isolates compared with the lineage B isolate (p=0·0051 for P.1/28 and p=0·0336 for P.1/30).
Subsequently, we evaluated a set of plasma samples from 15 participants who had received the vaccine, not placebo, in the previously reported Sinovac phase 3 trial in Brazil in August, 2020. CoronaVac-immunised individuals (five [33%] men and ten [67%] women; median age 34 years [32–39]) had received their CoronaVac booster vaccination 134–260 days before sample collection (median 158 days [156–170]). These individuals had antibody titres (measured as relative binding signals, compared with the cutoff value of each assay) between 0·03 and 1·09 for IgM and between 0·06 and 0·38 for IgG antibodies (appendix 2 p 11). Plasma samples from all trial participants had VNT50s below the limit of detection against both P.1 isolates, and a median VNT50 of 20 (<20–30) against the lineage B isolate (figure 2D; appendix 2 p 11). Because these neutralisation titres were near or below the limit of detection, statistical significance could not be reached in the comparison of the lineage B isolate with the P.1 isolates (figure 2B, D).
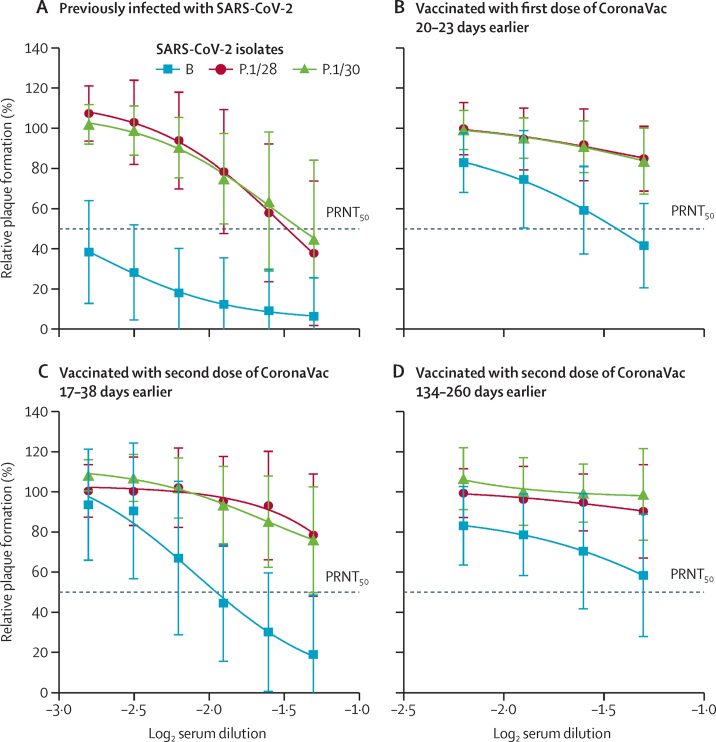
Plasma samples from previously SARS-CoV-2-infected individuals and CoronaVac-vaccinated individuals (including those who had received one or two doses in the Brazilian vaccination programme and those who received two doses in the Sinovac trial) were tested by PRNT against both P.1 isolates and the B isolate. Plasma samples from previously infected individuals had PRNT50 values of 1:25 against P.1/28 and 1:23 against P.1/30, whereas the PRNT50against the lineage B isolate more than 1:640—a neutralising antibody capacity more than 25 times higher (p<0·0001; figure 3A). In plasma samples from individuals who had received a first dose of CoronaVac 20–23 days earlier or a second dose of CoronaVac 134–260 days earlier, we found no detectable neutralising activity against the P.1 isolates (limit of detection 1:20), whereas the PRNT50 value against the B isolate was around 1:20 (figure 3B, D). By comparison, plasma collected 17–38 days after a second dose of CoronaVac (in those participants immunised in the national vaccination programme) had a PRNT50value around 1:80 for B lineage and less than 1:20 for both P.1 lineage isolates, indicating a neutralising antibody capacity of around a quarter against P.1 isolates compared with those against the lineage B isolate (p=0·0033; figure 3C).
Figure 3.
Neutralisation of SARS-CoV-2 lineages B and P.1 by plasma from previously infected or vaccinated individuals, according to PRNT50
Plasma samples were tested by PRNT in Vero cells after incubation with 100 plaque-forming units of different isolates of SARS-CoV-2 (B and two isolates of P.1 lineages). PRNT50 represents the sample dilution that showed a 50% reduction in plaque formation compared with a control well inoculated with SARS-CoV-2 alone (without plasma), after linear regression analysis. Each data point represents the mean of all plasma samples for each group at each dilution level (shown as log2 serum dilution) and error bars represent SD. (A) Plasma from blood donors previously infected with SARS-CoV-2 (n=21). (B) Plasma from individuals vaccinated with a single dose of CoronaVac vaccine during the Brazilian vaccination programme (n=18), collected at a median 21 days (IQR 21–21; range 20–23) after receipt of the first dose. (C) Plasma from individuals vaccinated with two doses of CoronaVac vaccine during the Brazilian vaccination programme (n=38), collected at a median 21 days (IQR 20–23; range 17–38) after receipt of the second dose. (D) Plasma from individuals vaccinated with two doses of CoronaVac vaccine in the Sinovac phase 3 trial (n=15), collected at a median 158 days (IQR 156–170; range 134–260) after receipt of the second dose. All PRNT curves for each sample used in the study are shown in appendix 2 (pp 4–7). PRNT=plaque reduction neutralisation test.
Discussion
Our data suggest that P.1 lineage SARS-CoV-2 can escape from neutralising antibody responses generated by previous SARS-CoV-2 infection and, thus, that reinfection might be possible with antigenically distinct variants with mutations in spike protein. Reinfection with P.1 lineage SARS-CoV-2 has been detected in Manaus,21 where high seroprevalence rates have previously been observed.22 Previous studies using serum or plasma from people with a history of COVID-19 have shown neutralising titres between 2·2 and 13·3 times lower against isolates of wild-type virus or pseudoviruses featuring the B.1.1.7, B.1.351, and P.1 constellations of spike protein mutations, compared with other lineages not containing these mutations.12, 13
The neutralisation potencies of plasma samples from individuals vaccinated with the BNT162b2, mRNA-1273, and ChAdOx1 nCoV-19 vaccines were up to 13·3 times lower (but remained neutralising) against the wild-type P.1 isolate12 and SARS-CoV-2 pseudoviruses containing spike protein mutations (Glu484Lys, Lys417Thr, and Asn501Tyr) present in the B.1.1.7, B.1.351, and P.1 lineages, in comparison with their neutralisation ability against lineages not containing those mutation.11, 23 To date, the CoronaVac vaccine has been approved for emergency use in Brazil, China, Colombia, Indonesia, Mexico, and Turkey, and was shown to be well tolerated and to induce humoral responses against SARS-CoV-2 in phase 1/2 and phase 3 clinical trials.15, 24 Additionally, its efficacy against symptomatic COVID-19 was estimated to be 50·7%.25 In Brazil, where the P.1 lineage first emerged,14 CoronaVac has been the main vaccine used in the national immunisation programme against SARS-CoV-2, accounting for around 41 million (76%) of the total 53 million doses administered in the country. Our data show that plasma collected 20–23 days after a first dose of CoronaVac or 134–260 days after a second dose had low levels of neutralising antibodies against the SARS-CoV-2 B lineage, and many of these samples showed no inhibitory activity against two wild-type P.1 isolates at the limit of detection of the VNT or PRNT assays. The low levels of neutralising antibodies induced by this vaccine are consistent with the phase 1/2 and phase 3 trial data.15, 24 By contrast, neutralising titres in individuals who had received two doses of CoronaVac 21 days before sample collection was near 1:80 for B.1 and 1:20 for P.1 isolates. Collectively, the data suggest that P.1 lineage virus might escape from neutralising antibodies induced by an inactivated SARS-CoV-2 vaccine, especially at 5 months after vaccination as immunity wanes.
Human influenza virus vaccines are typically updated when a more than four times reduction of neutralising activity (based on haemagglutination-inhibition titres) is detected against the new strains of the virus versus the previously circulating strains.26 Our results showed that neutralising capacity of plasma from patients previously infected with SARS-CoV-2 was at least 8·6 times lower against two isolates of lineage P.1 than against a lineage B isolate recovered from a virus previously circulating in Brazil in late February, 2020.18 Sera from individuals infected with common cold coronavirus HCoV-229E in the 1980s and 1990s had a neutralising antibody capacity at least four times lower than those against contemporary HCoV-229E strains,27 an observation that is comparable to the differences we observed in neutralising activity between the B and P.1 SARS-CoV-2 strains. Another study has shown that SARS-CoV-2 antibody levels start decreasing within 5 months after initial infection,28 similar to our findings in the plasma of CoronaVac-immunised individuals. Monitoring the reduction of neutralisation activity of vaccinated individuals over time and boosting immunity with updated vaccine versions might be required to halt transmission of new variants.
Lower neutralising capacity of SARS-CoV-2 antibodies and partial immunity against new variants could suggest that reinfection might occur in convalescent or even vaccinated individuals. Additionally, the delayed production of neutralising antibodies has been associated with impaired viral control and fatal COVID-19.29 However, a phase 3 clinical trial showed that CoronaVac can protect against severe COVID-19 and death.25 Therefore, neutralising antibodies might not be the only contributing factor, and memory T cell or B cell responses might reduce disease severity. Further clinical and epidemiological studies are needed to clarify how previous exposure through natural infection and vaccination could protect against reinfection and primary infection with existing and newly emerging variants of concern.
In conclusion, our exploratory data suggest that SARS-CoV-2 lineage P.1 can escape from neutralising antibodies elicited during infection or immunisation with previously circulating viral variants. Continued and enhanced genetic surveillance of SARS-CoV-2 variants worldwide, paired with plasma neutralising antibody assays, could help to guide updates of immunisation programmes.
Data sharing
All metadata generated in this analysis are available in appendix 2.
Declaration of interests
MSD is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, and Carnival Corporation, and on the Scientific Advisory Boards of Moderna and Immunome. MSD is the principal investigator of a laboratory that has received funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions.
Acknowledgments
Acknowledgments
This study was supported by grants from the São Paulo Research Foundation (FAPESP; 2016/00194-8 and 2020/04558-0) and Fundo de Apoio ao Ensino, Pesquisa e Extensão (FAEPEX) of UNICAMP (2266/20); by the Brazilian Ministry of Science, Technology, and Innovation (MCTI) through the Rede Corona-ômica Br-MCTI/Financier of Studies and Projects (01.20.0003.00 to REM, affiliated to RedeVírus/MCTI [01.20.0029.000462/20 and 404096/2020-4]); and by the UK Medical Research Council and FAPESP–Brazil–UK Centre for (Arbo)virus Discovery, Diagnosis, Genomics and Epidemiology partnership award (MR/S0195/1 and FAPESP 2018/14389-0). WMS is supported by FAPESP (2017/13981-0 and 2019/24251-9) and the National Council for Scientific and Technological Development (CNPq; 408338/2018-0). HM-S is supported by FAEPEX (2005/20, 2319/20, and 2432/20). SPM is supported by FAPESP (#18/13645-3). GFdS is supported by FAPESP (#18/10224-7). NRF is supported by a Wellcome Trust and Royal Society Sir Henry Dale Fellowship (204311/Z/16/Z). BDB is supported by CNPq (401977/2020). KB-d-S, CLS, and PLP were supported by FAPESP fellowships (2020/02159-0, 2020/02448-2, and 2017/26908-0). MRA and PPB were supported by Coordination for the Improvement of Higher Education Personnel (88887.356527/2019-00) fellowships, and DAT-T and LSM were supported by CNPq fellowships (141844/2019-1 and 382206/2020-7). MSD was supported by a grant from the National Institutes of Health (U01 grant AI151810). We thank Wendy Barclay for helpful discussions; all blood donors, vaccinated individuals, and SARS-CoV-2 patients who provided clinical specimens for this study; Thermo Fisher Scientific for provision of an EVOS inverted microscope for the biosafety level 3 facility at the Emerging Viruses Laboratory; the National Institute of Science and Technology of Photonics Applied to Cell Biology for confocal microscopy analysis; UNICAMP-Task-Force against COVID-19, which facilitated this study; Elzira E Saviani for technical support; all individuals involved in the diagnosis and generation of SARS-CoV-2 sequences as part of the CADDE-Genomic-Network; and the MCTI and all members of the Corona-ômica network for support.
Contributors
WMS, MRA, RS-C, ECS, FG, NRF, and JLP-M conceptualised the study. WMS, MRA, RS-C, LDC, DAT-T, SPM, GFdS, PLP, PPB, KB-d-S, LSM, CLS, NSB, IMC, ASSD, TMC, ABZ, CC-L, ABSPG, LIB, FCS, VAC, LAMF, DSC, JGdJ, CAMS, MSR, GMF, MCP, LMS, ECR, PSA, MAEC, GCM, ERM, MNNS, CCC, RNA, CWA, NG, MLM, HM-S, FG, and JLP-M contributed to acquisition of data. WMS, MRA, RS-C, DAT-T, LSM, ECS, FG, and JLP-M contributed to data analysis. WMS, MRA, RS-C, LDC, DAT-T, LMS, NRF, ECS, FG, OGP, CD, and JLP-M contributed to data interpretation. WMS, RS-C, and JLP-M drafted the manuscript. WMS, RS-C, MRA, RNA, MLM, OGP, CD, MA-C, BDB, MARV, MASM, HM-S, REM, ASF, ECS, FG, NRF, MSD, and JLP-M revised the manuscript. WMS, FRS, MASM, REM, ASF, NRF, ECS, NRD, and JLP-M acquired funding for the study. All authors read and approved the final version of the manuscript and had access to all the data in the study. WMS, RS-C, and JLP-M accessed and verified the data underlying the study.
Supplementary Materials
References
- 1.Wu F, Zhao S, Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO WHO coronavirus (COVID-19) dashboard. https://covid19.who.int2021
- 3.Rambaut A, Holmes EC, O'Toole Á. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou YJ, Chiba S, Halfmann P. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chi X, Yan R, Zhang J. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piccoli L, Park YJ, Tortorici MA. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024. doi: 10.1016/j.cell.2020.09.037. 42.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomson EC, Rosen LE, Shepherd JG. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171. doi: 10.1016/j.cell.2021.01.037. 87.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Schmidt F, Weisblum Y. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tegally H, Wilkinson E, Giovanetti M. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 10.Davies NG, Abbott S, Barnard RC. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Beltran WF, Lam EC, St Denis K. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372. doi: 10.1016/j.cell.2021.03.013. 83.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dejnirattisai W, Zhou D, Supasa P. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021 doi: 10.1016/j.cell.2021.03.055. published online March 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen RE, Zhang X, Case JB. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faria NR, Mellan TA, Whittaker C. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Zeng G, Pan H. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18-59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect Dis. 2021;21:181–192. doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wölfel R, Corman VM, Guggemos W. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 17.Corman VM, Landt O, Kaiser M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davanzo GG, Codo AC, Brunetti NS. SARS-CoV-2 uses CD4 to infect T helper lymphocytes. medRxiv. 2020 doi: 10.1101/2020.09.25.20200329. published online Sept 28. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perera RA, Mok CK, Tsang OT. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.16.2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Araujo DB, Machado RRG, Amgarten DE. SARS-CoV-2 isolation from the first reported patients in Brazil and establishment of a coordinated task network. Mem Inst Oswaldo Cruz. 2020;115 doi: 10.1590/0074-02760200342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naveca F, da Costa C, Nascimento V. SARS-CoV-2 reinfection by the new variant of concern (VOC) P.1 in Amazonas, Brazil. https://virological.org/t/sars-cov-2-reinfection-by-the-new-variant-of-concern-voc-p-1-in-amazonas-brazil/596
- 22.Buss LF, Prete CA, Jr, Abrahim CMM. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science. 2021;371:288–292. doi: 10.1126/science.abe9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann M, Arora P, Groß R. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384. doi: 10.1016/j.cell.2021.03.036. 93.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bueno SM, Abarca K, González PA. Interim report: safety and immunogenicity of an inactivated vaccine against SARS-CoV-2 in healthy Chilean adults in a phase 3 clinical trial. medRxiv. 2021 doi: 10.1101/2021.03.31.21254494. published online April 1. (preprint). [DOI] [Google Scholar]
- 25.Palacios R, Batista AP, Albuquerque CSN. Efficacy and safety of a COVID-19 inactivated vaccine in healthcare professionals in Brazil: the PROFISCOV study. SSRN. 2021 doi: 10.2139/ssrn.3822780. published online April 14. (preprint). [DOI] [Google Scholar]
- 26.Smith DJ, Lapedes AS, de Jong JC. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 27.Eguia RT, Crawford KHD, Stevens-Ayers T. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Röltgen K, Powell AE, Wirz OF. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucas C, Klein J, Sundaram ME. Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nat Med. 2021 doi: 10.1038/s41591-021-01355-0. published online May 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All metadata generated in this analysis are available in appendix 2.