Figure 5.
Mutational analysis of zGDNF and zGFRα1 site 1 and 2 interactions with zRETECD
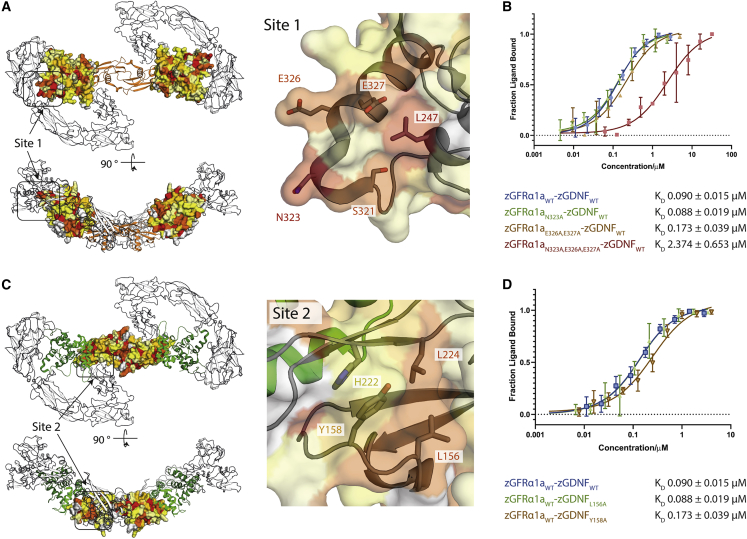
(A) Heatmap of the sequence conservation between hGFRα paralogs, and zGFRα1a mapped onto the structure of zGFRα1a D2-D3 domains reported here. Residues are colored by similarity (red highly similar to yellow through to white, least similar). Two orthogonal views are shown. Right panel, close-up of site 1 and conserved zGFRα1a residues.
(B) Binding curves and KD values obtained using MST for zGFRα1aD1-3 and mutations assessed in complex with zGDNFmat., with a minimum of n = 3 repeats for the WT and the mutations with the SE represented.
(C) Heatmap of the sequence similarity between GDNF paralogs depicted as a surface representation, mapped onto zGDNF138-235. Right panel, close-up of site 2 contact between RETCRD and zGDNF dimer.
(D) MST binding curves and KD values for zGDNF and mutations L156A and Y158A probed in complex with WT zGFRα1a binding to zRETECD.