Abstract
Fusion protein tags are widely used to capture and track proteins in research and industrial bioreactor processes. Quantifying fusion-tagged proteins normally requires several purification steps coupled with classical protein assays. Here, we developed a broadly-applicable nanosensor platform that quantifies glutathione-S-transferase (GST) fusion proteins in real-time. We synthesized a glutathione-DNA-carbon nanotube system to investigate glutathione-GST interactions via semiconducting single-walled carbon nanotube (SWCNT) photoluminescence. We found that SWCNT fluorescence wavelength and intensity modulation occurred specifically in response to GST and GST-fusions. The sensor response was dependent on SWCNT structure, wherein mod(n-m, 3) = 1 nanotube wavelength and intensity responses correlated with nanotube diameter distinctly from mod(n-m, 3) = 2 SWCNT responses. We also found broad functionality of this sensor to diverse GST-tagged proteins. This work comprises the first label-free optical sensor for GST and has implications for the assessment of protein expression in situ, including in imaging and industrial bioreactor settings.
Keywords: Fluorescent sensor, nanocarbon, solvatochromism, affinity tag
Graphical Abstract

Introduction
Fusion protein tags are co-expressed proteins or peptides used to capture and track a recombinant protein of interest, in protein expression systems or in biological investigations1–3. Common tags include the poly-histidine tag (his tag)4, the FLAG tag5, and the glutathione-S-transferase (GST) tag6, among many others1–3. Although essential for the purification and labeling of many proteins, fusion protein tags present unrealized opportunities for real-time, rapid analysis methods that function in situ.
Glutathione-S-transferase (GST) refers to a family of proteins across the phylogenetic spectrum6–7. These enzymes function by conjugating the thiol of glutathione to organic compounds for cell detoxification processes8–9, giving them a generality that was developed into a fusion tag, wherein glutathione is typically conjugated to a substrate, allowing binding to GST co-expressed proteins. The glutathione-GST system is a commonly-used protein tag6 for bacterial, yeast, insect, or mammalian expression systems10, consisting of a 26 kDa/211 amino acid sequence that promotes enhanced expression and solubility of recombinant proteins due to its size, tertiary structure, and hydrophilicity7, 11. Expressed fusion proteins are purified by GST binding to glutathione, which is preparatively immobilized through a sulfhydryl group in its reduced form (GSH) to a solid matrix, often in a spin-column format3, 7. In addition to this affinity tag application, GST tags are also useful in molecular or cell biology experiments where in vitro and in vivo protein tracking and visualization are applicable12–13.
GST-tagged proteins secreted from cells in industrial expression systems are primarily quantified following steps to collect and purify them from the production media. These systems typically produce up to 50 mg/L of the intended protein14. Common protein quantification techniques used include the Bradford assay, the Lowry assay, the bicinchoninic acid assay, and the ultraviolet absorption assay, along with semi-quantitative Western blotting15. These traditional methods require significant preparatory effort and multiple hours to yield results. GST was used as the recognition element to detect captan16 and glutathione17 in prior studies. Others used non-GSH capture molecules to detect GST protein and its function18–19. Electrochemical and aggregate-induction sensor platforms were used in these studies17, 20. More broadly, optical biosensors for other enzymes have been demonstrated to detect renal and hepatic injury, including for caspases, glucosaminidase, and galactosidase21–23. We believe that a rapid, label-free optical sensor for fusion protein tags, which can access the media of industrial protein expression systems, would confer a significant benefit to biotechnology and molecular biology fields.
Single-walled carbon nanotubes (SWCNT) consist of single tubes of graphene with diameters typically in the 0.5–1.5 nm range. Semiconducting SWCNTs emit near-infrared (NIR) fluorescence/photoluminescence24–30 which exhibits unique photostability31, enabling continuous monitoring and quantitative detection. The optical properties of SWCNTs can be made exquisitely sensitive to their local environment, enabling modulation for sensing applications32. They have been applied to detect small molecules, nucleic acids, and proteins33–35, in live cells32, 36, and in living organisms29, 35, 37–38. Nanotube-based sensors utilizing a variety of biological recognition elements have been developed, including antibodies29–30, 39, oligonucleotide aptamers40–41, complementary oligonucleotides35, and peptides42–43, among others including polymer-based molecular recognition screens44–46. None, however, have been developed using small molecule biological recognition elements.
In this work, we designed a label-free optical nanosensor for GST-tagged proteins wherein analyte recognition occurs via the small molecule glutathione at the surface of the transduction element. This sensor, comprised of glutathione-bound DNA-SWCNT complexes, exhibited a low-nanomolar limit of detection and strong selectivity for GST-tagged proteins, as well as generality to multiple classes of GST-tagged proteins, including cell cycle proteins, RNA-binding proteins, and ovarian cancer protein biomarkers. We identified and investigated a SWCNT species/chirality-dependent mechanistic response to GST chiral species, finding a distinct relationship to SWCNT structure, wherein mod(n-m, 3) = 1 nanotube responses negatively correlated with nanotube diameter and strongly correlated with wavelength shifting, while mod(n-m, 3) = 2 responses positively correlated with diameter and strongly correlated with intensity responses. We expect this class of nanosensor to have broad applications for protein quantification in the protein bioproduction industry and as research tools in molecular biology processes.
Results and Discussion
We first covalently conjugated single-stranded DNA with the sequence (TAT)6, and functionalized with 3’ acrydite, to reduced glutathione (GSH) (Figure 1a). The DNA-glutathione conjugate was purified and used to suspend HiPco single-walled carbon nanotubes via ultrasonication. Single-stranded DNA is known to separate and homogenously disperse single nanotube suspensions in solution, allowing for a stable hybrid construct and strong photoluminescence47–48. The glutathione-DNA-nanotube complexes (GSH-DNA-SWCNTs) were subjected to several purification steps before stability and optical properties were assessed using UV-Vis-NIR absorbance (Figure 1b) and NIR fluorescence spectroscopy (Figure. 1c).
Figure 1. Synthesis and characterization of GSH-DNA-SWCNT complexes.

(a) Schematic of the synthesis scheme. (b) Absorption spectrum of the GSH-DNA-SWCNT complexes. (c) Two-dimensional photoluminescence (PL) excitation-emission plot of the GSH-DNA-SWCNT complexes. Scale represents relative fluorescence from minimum (0) to maximum (1).
We sought to assess the sensitivity and selectivity of the optical behavior of the GSH-DNA-SWCNT complexes in the presence of GST via NIR fluorescence spectroscopy. Using a custom-built NIR plate reader, we investigated the dose-response to GST across six orders of magnitude. Near-infrared photoluminescence spectra (900 – 1400 nm) were obtained after 5, 30, and 60 minutes of incubation, via excitation with either a 660 nm or 730 nm laser diode, to allow the visualization of multiple SWCNT chiralities using a 2-second exposure time. The change in wavelength and intensity of each discernable SWCNT band, compared to the complexes in the absence of GST, was obtained in triplicate for each concentration. From these measurements, we observed gross differences in wavelength and intensity for multiple SWCNT chiralities compared to the control (Figure 2a). These changes occurred in a dose-dependent manner for the nanotube species observed upon 730 nm (Figure S1a) or 640 nm (Figure S1b) illumination.
Figure 2. Response of GSH-DNA-SWCNT complexes to GST.
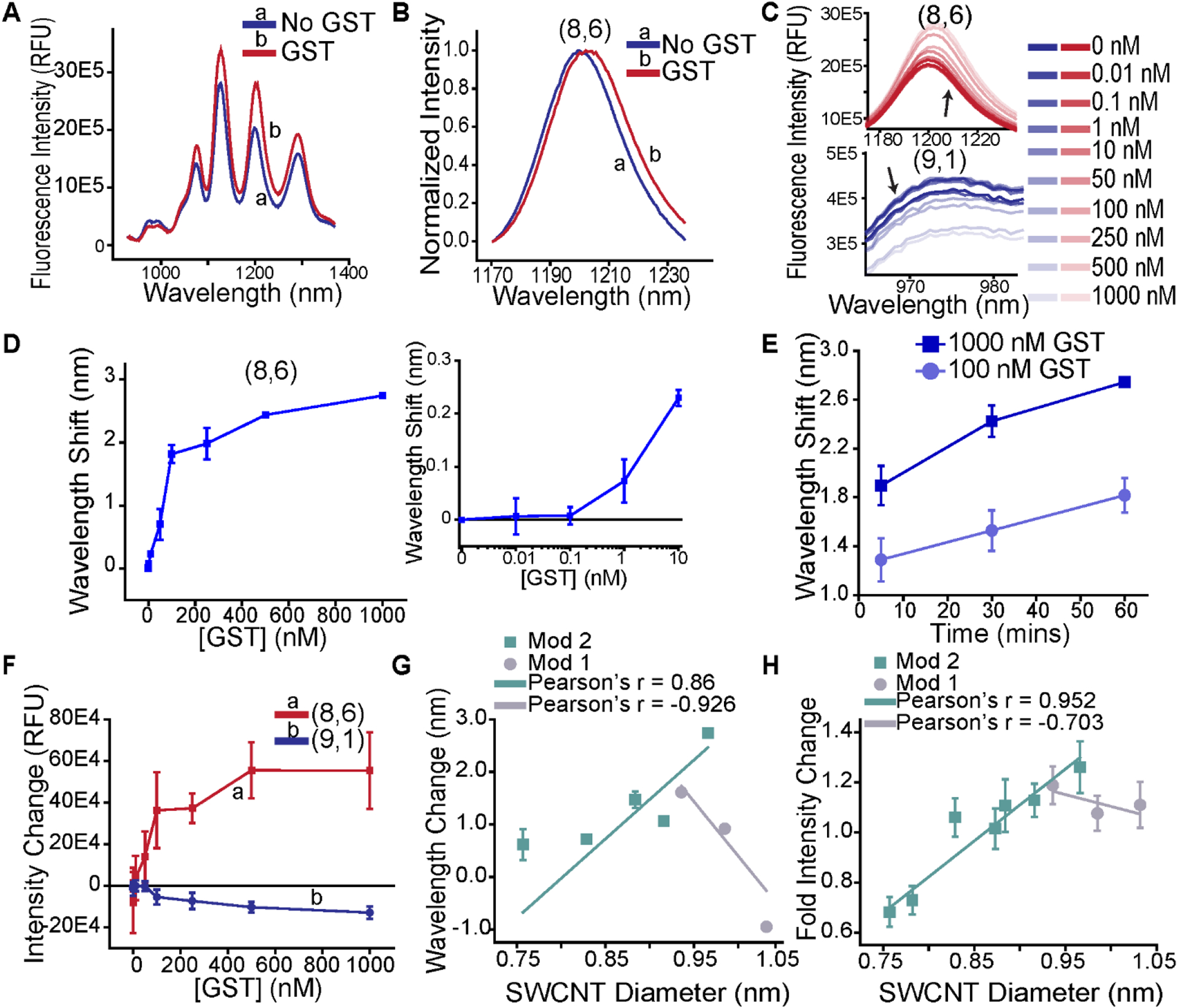
(a) Representative photoluminescence spectrum of the GSH-DNA-SWCNT complexes in the presence of 1000 nM GST, upon excitation at 730 nm. (b) Representative normalized intensity spectra (0 = minimum, 1 = maximum) of the (8,6) nanotube chirality without and with 1000 nM GST upon excitation at 730 nm to illuminate the bathochromic shift. (c) Representative emission spectra of the (8,6) chirality (top) and (9,1) chirality (bottom) for each concentration of added GST. (d) Change in emission center wavelength of the (8,6) chirality as a function of GST concentration. Inset (right): Magnified low-concentration regime of (8,6) response to GST to illuminate the lower range of sensitivity. (e) Temporal response in (8,6) center wavelength in response to maximum (1000 nM) or plateau (100 nM) concentrations of GST. All points d-e represent mean (n = 3 separate samples) ± SD. (f) Photoluminescence intensity change for the (8,6) and (9,1) chiralities as a function of added GST concentration. (g) Wavelength change for each nanotube chirality as a function of chiral diameter. (h) Intensity change for each chirality as a function of chiral diameter. Each point is mean (n = 3 separate samples) ± SD.
We found distinct differences in emission behavior among the SWCNT chiralities in response to GST. The (8,6) chirality exhibited the most pronounced wavelength-shifting response, reaching a maximum bathochromic (red) shift of 2.74 ± 0.04 nm at 60 minutes upon interrogation with 1 μM GST (Figure 2b). The emission intensity responses of the GSH-DNA-SWCNT complexes also varied widely. We found that the intensity of the (8,6) chirality increased most prominently, but the intensity of the (9,1) chirality decreased with increasing GST concentration (Figure 2c).
Investigating the concentration-dependence of the response, the (8,6) wavelength shift appeared to plateau at a concentration of 100 nM GST (Figure 2d). The limit of detection was approximately 1 nM GST, compared to controls (Figure 2d inset). We found that the dynamic range was between 1 nM and 100 nM GST, after incubation of as little as 5 minutes (Figure S2a–d) and was consistent at later timepoints (Figure 2e). The respective intensity changes, whether decrease or increase, appeared monotonic in response to increasing GST concentration (Figure 2f). We also found that the interaction of GST with the sensor construct yielded highly consistent photoluminescence emission from 1 hour to 24 hours, signifying rapid equilibration of the sensor response and a stable complex formation with no aggregation or quenching (Figure S2).
We observed differences in the sensitivities, kinetics, and dynamic ranges of optical response of the complexes among the nanotube chiralities. As expected, most nanotube species exhibited a bathrochromic shift beginning 5 minutes after exposure to GST, measured under laser excitation at 730 nm (Figure S3a–d) or 640 nm (Figure S3e–h). Interestingly, the response of some nanotubes, such as the (8,7), (9,4), and (10,2) chiralities appeared to reach a maximum shift by 5 minutes (Figure S3d), whereas others, such as the (10,3) chirality, appeared to undergo shifts from the 5 minute timepoint to the 30 minute timepoint (Figure S3h). The temporal changes for all nanotube chiralities at 100 nM followed a similar pattern compared to the maximum GST concentration of 1 μM (Figure S4). While all chiralities plateaued at 100 nM GST concentrations (Figure S3), detection limits were between 1 nM and 100 nM (Figure S5). Interestingly, the (8,7) chirality exhibited a hypsochromic instead of bathochromic shift (Figure S3a–d; S4a), warranting further investigations of the SWCNT chirality-dependence of the responses.
We further investigated the SWCNT chirality- and diameter-dependence of the sensor responses to GST. We first observed a correspondence in the solvatochromic response as a function of nanotube diameter of different mod. Semiconducting SWCNT chiral mods are defined by their (n,m) chiral index vector, when mod(n - m, 3) = 1 or 2 for mod1 and mod2, respectively49–50. We discovered a strong negative correlation (r = −0.926) of the wavelength shifting response and SWCNT diameter for mod2 nanotube chiralities and a positive correlation (r = 0.86) for mod1 nanotube chiralities. We further investigated the intensity response of the sensor (Figure 2h). We found a strong positive correlation (r = 0.952) for mod1 nanotubes, and a negative correlation for mod2 nanotubes (r = −0.703) with nanotube diameter. It is established that the differences in nanotube chiral geometry contribute to mod-dependent photoluminescence responses50–52. We therefore conclude that the intrinsic response of each nanotube to GST is largely defined by the chirality of SWCNT species.
To account for potential artifacts in measurements of sensor intensity, such as variabilities in GSH-DNA-SWCNT concentration or stability, we investigated whether ratiometric intensity changes of two chiralities could improve the robustness of the response. We found that the ratiometric response of the (8,6) chirality to the (9,1) chirality was monotonic, appearing to nearly plateau at 100 nM GST, similar to the wavelength responses (Figure 3a). This metric exhibited a 10 nM minimum sensitivity (Figure 3a inset). The kinetics of this response were slower than the solvatochromic response, suggesting a different mechanism of sensing (Figure 3b). Investigating other chiralities, we found that the (7,6) species also exhibited an increasing intensity response relative to the (9,1) species upon GST addition and illumination at 640 nm (Figure S6a). We found similar characteristics to the (8,6)/(9,1) response, with slightly greater variation at certain concentrations (Figure S6b) and dampened maximum responses at all time points (Figure S7). The (7,6)/(9,1) metric exhibited a higher limit of detection of 50 nM GST compared to the (8,6)/(9,1) (Figure S8).
Figure 3. Sensor response evaluation.
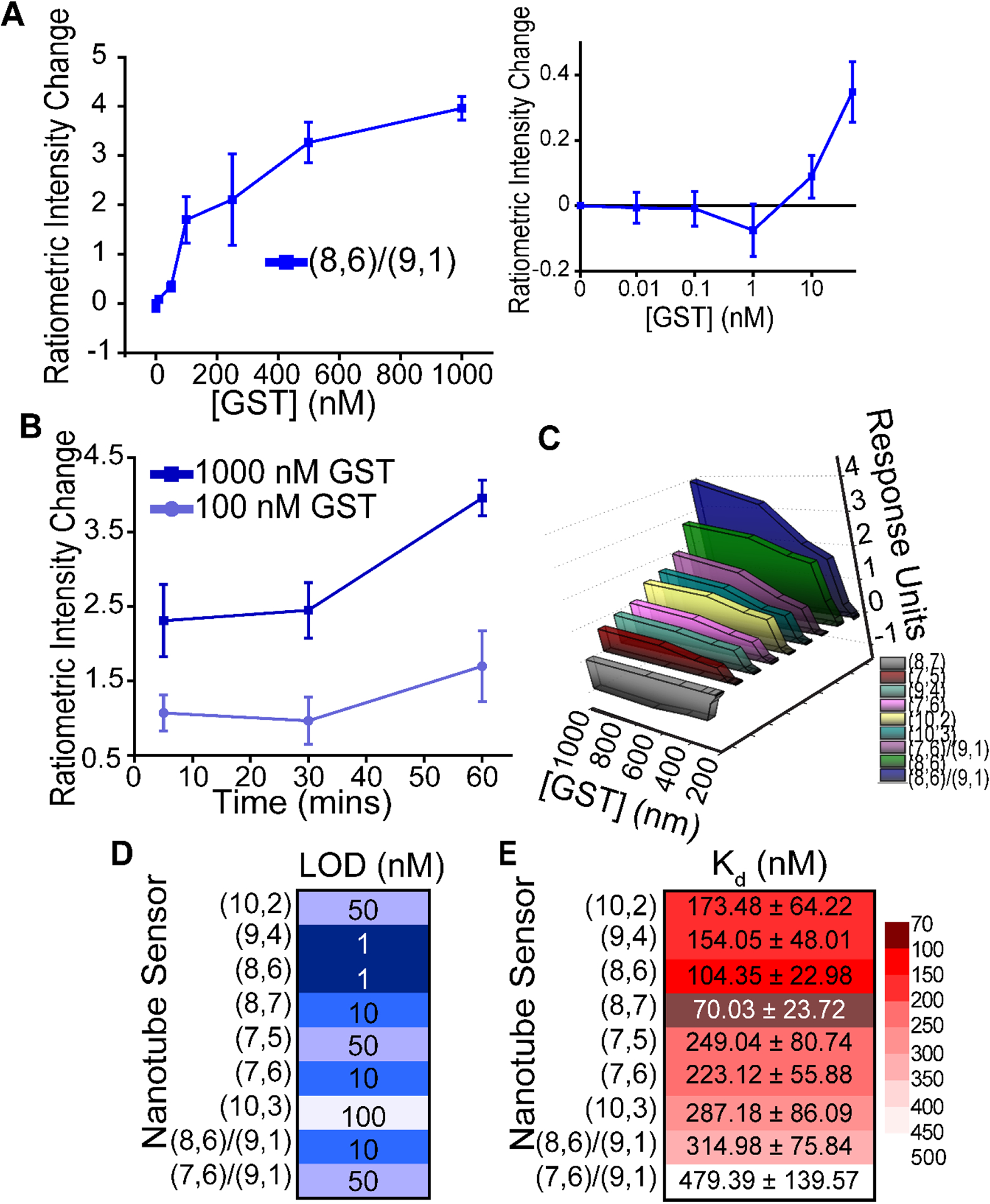
(a) Ratiometric intensity change, defined as a change in (8,6) intensity divided by change in (9,1) intensity, as a function of GST concentration. Inset (right): Magnified low-concentration regime of (8,6)/(9,1) ratiometric intensity response to GST to illuminate the lower range of sensitivity. (b) Temporal response of (8,6)/(9,1) ratiometric intensity change in response to maximum (1000 nM) or plateau (100 nM) concentrations of GST. All points a-b represent mean (n = 3 separate samples) ± SD. (c) Three-dimensional representation of the solvatochromic (7 chiralities) or ratiometric intensity responses (2 sets of chiralities) viewed from smallest to largest maximum response. Response units are defined as wavelength change or intensity ratio. Each point represents the mean of 3 separate samples. (d) Limit of detection (in nM) of each sensor output response, with darker squares corresponding to lower LOD and lighter squares corresponding to higher LOD. LOD is determined for the mean response of 3 separate samples. (e) Dissociation constant (in nM) of each sensor output response, with darker squares corresponding to lower Kd and light squares corresponding to higher Kd. Kd is determined for the mean response of 3 separate samples.
Given the possibility of sensing GST through either solvatochromic shifts or ratiometric intensity changes, we next compared the overall robustness of each of the sensor responses we investigated. The largest magnitude of response appeared to be the (8,6)/(9,1) ratio, followed closely by the bathochromic shift of the (8,6) chirality alone (Figure 3c). The limits of detection of the various sensor responses ranged from 1 nM to 100 nM GST, with both the (9,4) and (8,6) bathochromic shifts at the 1 nM level (Figure 3d). The (8,6)/(9,1) ratiometric intensity change, however, was among the next most sensitive.
For a more straightforward comparison of sensor responsivity to GST, we obtained the dissociation constant of the GST-glutathione interaction as measured by each type of sensor output. To do so, we fit each set of responsivity values to a standard model of noncooperative saturation single-site specific binding kinetics with the equation (Figure S9 Non-Linear Curve Fit)30: Y = Bmax * X / (Kd + X); wherein Bmax is the maximum specific response unit and Kd is the equilibrium binding constant in nM GST. Goodness of fit was determined by R2 values (Figure S9j), with all non-linear fit GST values greater than 0.947. For comparison, these data were also fit to linear equations, though in all cases the goodness of fit was less than the single-site saturation curve (Figure S9j), strongly suggesting site-specific binding between the GST and glutathione molecules instead of non-specific adsorption. The lowest dissociation constant measurable was 70.03 ± 23.72 nM, as recorded by the bathochromic shift of the (8,7) chirality (Figure 4e). The (8,6) bathochromic response exhibited the next lowest Kd, however, interestingly, the ratiometric intensity responses exhibited high dissociation constants of 315 – 479 nM GST. We therefore conclude that SWCNT wavelength changes have lower limits of detection and sharper dynamic ranges, while intensity changes may be useful over a wider range of protein concentrations.
Figure 4. Sensor selectivity and response to GST-tagged proteins.
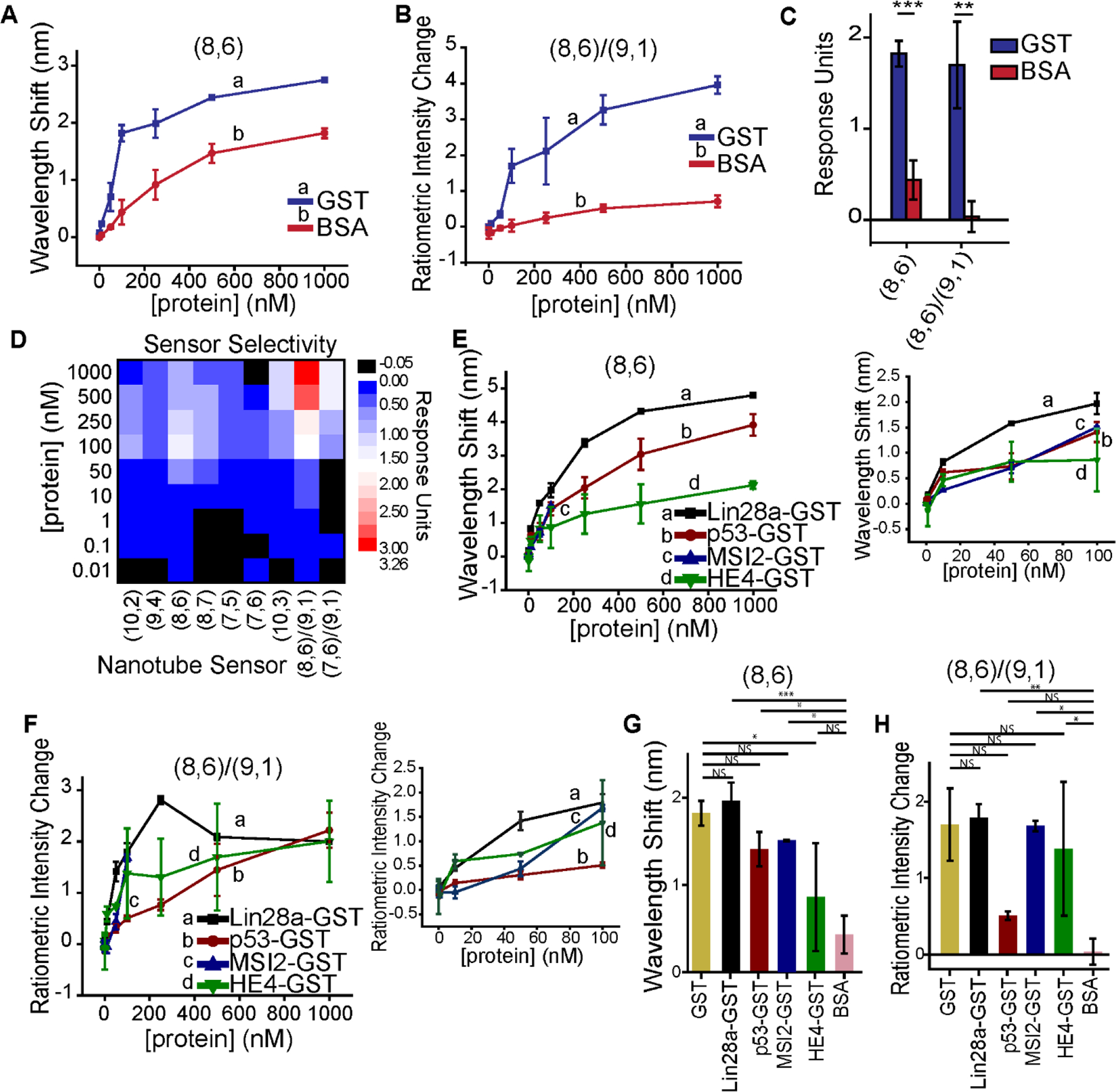
(a) Wavelength response of the (8,6) chirality as a function of GST or BSA concentration. (b) Ratiometric intensity response of the (8,6)/(9,1) chiralities as a function of GST or BSA concentration. (c) Direct comparison of sensor response to GST or BSA at the plateau concentration (100 nM) for the (8,6) wavelength response (left; *** = p = 7.5E−4) or (8,6)/(9,1) intensity response (right; ** = p = 4.7E−3); two-sided t-test. (d) Sensor selectivity for each solvatochromic (7) nanotube response and each ratiometric intensity response (2) at each concentration of protein added. Response units are defined as the mean (n = 3 separate samples) GST change minus mean BSA change in each type of measurement. (e) Ratiometric intensity change of the (8,6)/(9,1) chiralities for four GST-tagged proteins: Lin28a, p53, MSI2 (note: only extends to 100 nM), and HE4. Inset (right): magnification of the same data displayed to 100 nM. (f) Bathochromic response of the (8,6) chirality to the same four proteins. Inset (right): magnification of the same data displayed to 100 nM. (g) Comparative (8,6)/(9,1) ratiometric intensity change at 100 nM protein for the same four proteins as (e) with GST and BSA responses from above. Two-sided one-way ANOVA with Tukey post-hoc analysis; GST:Lin28a-GST NS = p = 0.999; GST:p53GST NS = p = 0.758; GST:MSI2GST NS = p = 1.00; GST:HE4GST NS = p = 0.936; BSA:Lin28a-GST ** = p = 3.29E−3; BSA:p53GST NS = p = 0.758; BSA:MSI2GST * = p = 0.0105; BSA:HE4GST * = p = 0.0346. (h) Comparative (8,6) bathochromic change at 100 nM protein for the same proteins as (e) with GST and BSA responses from above. Two-sided one-way ANOVA with Tukey post-hoc analysis; GST:Lin28a-GST NS = p = 0.991; GST:p53GST NS = p = 0.581; GST:MSI2GST NS = p = 0.790; GST:HE4GST * = p = 0.0293; BSA:Lin28a-GST *** = p = 5.77E−4; BSA:p53GST * = p = 0.0254; BSA:MSI2GST * = p = 0.0147; BSA:HE4GST NS = p = 0.525. All points a-h represent mean (n = 3 separate samples) ± SD; NS = not significant.
We sought to investigate the selectivity of the sensor. We interrogated the sensor with a very high concentration of powdered milk (Blotto), often used as a protein blocking agent in protein staining. We added 333 μg/mL powdered milk to simulate the protein-rich environment possible in certain applications, compared to the maximum of 26 μg/mL (1 μM) of GST tested above. We found that the (8,6) nanotube exhibited a 2 nm hypsochromic shift in response to the milk, compared to the maximal 2.7 bathochromic shift with GST, a difference of 4.7 nm (Figure S10). The powdered milk effected a change in the ratiometric intensity of (8,6)/(9,1) chiralities of just 0.47, which is 12% of the maximal GST response (Figure S10).
To more thoroughly evaluate the selectivity of the sensor, we performed experiments similar to those described above for GST interrogation, adding globular protein bovine serum albumin (BSA) as a model of potential interferent proteins in bioreactor systems. In the same context as with GST, we found there was indeed some bathochromic wavelength shifting in response to BSA when observing the (8,6) chirality (Figure 4a). Above 50 nM concentrations, this response was between 2 – 5-fold less for BSA compared to GST. Below 50 nM concentrations, the response to GST was up to 100-fold greater than BSA. The ratiometric sensing mechanism exhibited between a 5 – 50-fold greater response to GST than BSA (Figure 4b). The differences in each sensor response at the plateau value of 100 nM were functionally and statistically significant ((8,6) bathochromic p = 7.5E−4; (8,6)/(9,1) intensity p = 4.7E−3) (Figure 4c). This trend held true when evaluating all solvatochromic and ratiometric intensity responses (Figure S11a–c). The responses to BSA at maximum concentration were higher in solvatochromic responses than in ratiometric intensity responses (Figure S11d–j) and were consistent whether measured at the 5, 30, or 60-minute timepoint (Figure S12). Upon global evaluation of all sensor response selectivity values, it is clear that in the GST plateau region (100 nM – 1000 nM), ratiometric intensity outputs were much more selective for GST than BSA, whereas within the dynamic range of the sensor most outputs were comparable (Figure 4d; Figure S13). It should be noted, however, that the bathochromic (8,6) response and the ratiometric (8,6)/(9,1) intensity response were the most selective for GST over BSA. We therefore concluded that these two responses would have the greatest potential in more complex sensing environments.
To further investigate the specific response of each sensor output, we tested whether the response to BSA was a non-specific interaction. We hypothesized this to be the case, as prior work has found that hydrophobic regions of BSA adsorb to hydrophobic surfaces such as the carbon nanotube sidewall30, 53–55. To test this, we investigated whether the response to BSA was diffusion-controlled. On measuring sensor response to a BSA titration, we found that the data was successfully fit by a linear trendline, with all R2 > 0.897 (Figure S14). The trends were strongly suggestive of a non-specific, diffusion-controlled adsorptive linear response56–57. This data leads us to conclude that BSA binding is non-specific adsorption onto the nanotube, unlike the site-specific kinetics exhibited upon interrogation with GST.
We investigated the functionality of this sensor in the context of GST-tagged proteins. We measured the response of sensor to four different GST fusion proteins: human epididymis protein 4 (HE4), an FDA-approved serum-based biomarker for ovarian cancer58; Lin28 homolog a (Lin28a), a RNA-binding intracellular translation suppressor59; p53, a critical tumor suppressor and DNA damage repair protein60; and Musashi RNA binding protein 2 (MSI2), an RNA-binding protein involved in tumorigenesis61. Each protein was produced by bacteria and purified and chosen due to their important functional biological role potentially of interest for in vitro or in vivo quantification or tracking. The protein fusions further served as models for sensor function investigations, as they range widely in structure, function, molecular weight, from 23 kDa (Lin28a) – 53 kDa (p53), and isoelectric point, from 4.5 (MSI2) – 8.4 (Lin28A). We found that the sensor exhibited similar wavelength shifting response patterns to all four GST-tagged proteins (Figure 4e), although there were differences in dynamic range. The ratiometric intensity response of the sensors behaved similarly (Figure 4f). We then compared these sensor outputs to their responses to either GST or BSA at the concentration plateau value of 100 nM. On investigating the solvatochromic output, we found that the values ranged between 78% and 108% of the response to GST for Lin28a-GST, p53-GST, and MSI2-GST (Figure 4g). The response to HE4-GST was the only response significantly different than GST alone, and it was not significantly different than BSA alone. We found that the ratiometric intensity output was between 81% and 105% of the response to GST for Lin28a-GST, MSI2-GST, and HE4-GST. These values were not statistically different from the response to GST alone (Figure 4h). All response values, except the p53-GST, were statistically different from the response to BSA alone.
From these experiments with various GST-tagged fusion proteins, we conclude that the sensor is responsive to diverse GST fusions. However, we do note that each sensor output exhibited some differences in response to individual proteins. While the sensor exhibited an appropriate response to each GST-tagged protein, they were not identical to GST protein alone. We therefore conclude that the intrinsic properties of each protein have some effect on the sensor response. Those controlling intrinsic properties do not appear to include molecular weight or isoelectric point of each protein, as there is no clear trend, however it may be affected by protein hydrophobicity, tertiary structure, or a combination of several factors. Thus, while we believe this is a widely generalizable sensor to most GST-tagged fusion proteins, it may be necessary, though facile, to establish a standard curve for each different GST-tagged fusion protein prior to application and to determine which response (intensity/wavelength) is the most appropriate benchmark.
Conclusions
Here, we demonstrated the first broad application of a fusion-tagged protein sensor and the first label-free sensor for GST-tagged proteins. The sensor designed in this work was thoroughly evaluated for high sensitivity and specificity, while we performed experiments to understand the mechanisms of both specific and non-specific sensor responses. The sensor emission exhibited robust and reproducible solvatochromic and intensity responses, resulting in approximately 40-fold greater sensitivity than previously-described methods to detect GST17. Industrial protein expression systems typically produce maximally 0.18 – 1.9 μM GST concentrations (10 – 50 mg/L) in solution14, which are well above the detection limit of the sensor and overlap with the greater part of its dynamic range. We found interesting patterns between the sensor response and SWCNT species, in which we found mod(n-m, 3) - dependent changes that suggest a strong dependence on nanotube chirality. Additionally, this work demonstrates the first sensor based on SWCNTs involving protein molecular recognition using a small molecule recognition element. This method potentially introduces a large change in the local environment of the nanotube by decreasing the distance from the SWCNT surface to a large (protein) analyte. A small molecule recognition element, such as glutathione, presents advantages for improved nanosensor devices. A similar strategy may be applied to detect other fusion proteins such as poly-histidine (his-tag)4 or FLAG-tag5, among others. This method is applicable as a research tool that could include facile labeling of tagged proteins for live cell or in vivo imaging. Primarily, however, we envision this class of sensor as allowing for rapid and transient/real-time quantification of proteins secreted from cells in industrial bioreactor processes.
Supplementary Material
Acknowledgements
This work was supported in part by the NIH New Innovator Award (DP2-HD075698), NCI (R01-CA215719, P30-CA008748), NIDDK (R01-DK114321), CTSC (UL1-TR002384), the National Science Foundation CAREER Award (1752506), the American Cancer Society Research Scholar Grant (GC230452), the Pershing Square Sohn Cancer Research Alliance, the Honorable Tina Brozman Foundation for Ovarian Cancer Research, the Expect Miracles Foundation - Financial Services Against Cancer, the Anna Fuller Fund, the Louis V. Gerstner Jr. Young Investigator’s Fund, the Frank A. Howard Scholars Program, Cycle for Survival, the Alan and Sandra Gerry Metastasis Research Initiative, Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center, the Imaging & Radiation Sciences Program, and the Center for Molecular Imaging and Nanotechnology of Memorial Sloan Kettering Cancer. R.M.W. was supported by the Ovarian Cancer Research Fund [Ann Schreiber Mentored Investigator Award 370463], the American Heart Association Postdoctoral Fellowship (17POST33650043), and the City College of New York Grove School of Engineering. J.B. was supported by the Tow Foundation Postdoctoral Fellowship, Center for Molecular Imaging and Nanotechnology at MSKCC and the Wilson College of Textiles, North Carolina State University. The authors would like to thank P. Jena and D. Roxbury for instrument automation and MATLAB code, as well as helpful discussions, T. Galassi, H. Reed, S. Albanese, M. Shandell, and D. Deep for preliminary experimentation and helpful discussions, and M. Kharas and G. Dinares who provided GST-tagged proteins. All materials/methods and data needed to evaluate the conclusions in the paper are present in the paper and/or the supplementary materials. Additional raw data and code related to this paper may be requested from the authors.
Footnotes
Conflicts of Interest
D.A.H. is a cofounder and officer with equity interest in LipidSense, Inc., Nirova Biosense, Inc., and Goldilocks Therapeutics, Inc. D.A.H. is a member of the scientific advisory board of Concarlo Holdings, LLC and Nanorobotics, Inc. R.M.W. is a scientific advisor with equity interest in Goldilocks Therapeutics, Inc.
Supporting Information
Supporting Information is available online. This document contains a full Materials and Methods for this Letter. It also includes Supplementary Figures S1 – S14.
References
- 1.Terpe K, Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Applied microbiology and biotechnology 2003, 60 (5), 523–533. [DOI] [PubMed] [Google Scholar]
- 2.Kimple ME; Brill AL; Pasker RL, Overview of affinity tags for protein purification. Current Protocols in Protein Science 2013, 9.9. 1–9.9. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lichty JJ; Malecki JL; Agnew HD; Michelson-Horowitz DJ; Tan S, Comparison of affinity tags for protein purification. Protein expression and purification 2005, 41 (1), 98–105. [DOI] [PubMed] [Google Scholar]
- 4.Porath J; Carlsson J; Olsson I; Belfrage G, Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 1975, 258, 598–599. [DOI] [PubMed] [Google Scholar]
- 5.Hopp TP; Prickett KS; Price VL; Libby RT; March CJ; Cerretti DP; Urdal DL; Conlon PJ, A short polypeptide marker sequence useful for recombinant protein identification and purification. Biotechnology 1988, 6 (10), 1204–1210. [Google Scholar]
- 6.Smith DB; Johnson KS, Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 1988, 67 (1), 31–40. [DOI] [PubMed] [Google Scholar]
- 7.Harper S; Speicher DW, Purification of proteins fused to glutathione S-transferase. Protein Chromatography: Methods and Protocols 2011, 259–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayes JD; Pulford DJ, The glut athione S-transferase supergene family: regulation of GST and the contribution of the lsoenzymes to cancer chemoprotection and drug resistance part I. Critical reviews in biochemistry and molecular biology 1995, 30 (6), 445–520. [DOI] [PubMed] [Google Scholar]
- 9.Boyer TD; Vessey D; Holcomb C; Saley N, Studies of the relationship between the catalytic activity and binding of non-substrate ligands by the glutathione S-transferases. Biochemical Journal 1984, 217 (1), 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith DB; Corcoran LM, Expression and Purification of Glutathione‐S‐Transferase Fusion Proteins. Current protocols in molecular biology 2001, 16.7. 1–16.7. 7. [DOI] [PubMed] [Google Scholar]
- 11.Esposito D; Chatterjee DK, Enhancement of soluble protein expression through the use of fusion tags. Current opinion in biotechnology 2006, 17 (4), 353–358. [DOI] [PubMed] [Google Scholar]
- 12.Goda N; Tenno T; Inomata K; Iwaya N; Sasaki Y; Shirakawa M; Hiroaki H, LBT/PTD dual tagged vector for purification, cellular protein delivery and visualization in living cells. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 2007, 1773 (2), 141–146. [DOI] [PubMed] [Google Scholar]
- 13.Bell MR; Engleka MJ; Malik A; Strickler JE, To fuse or not to fuse: What is your purpose? Protein Science 2013, 22 (11), 1466–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harper S; Speicher DW, Purification of proteins fused to glutathione S-transferase. In Protein chromatography, Springer: 2011; pp 259–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldring JD, Protein quantification methods to determine protein concentration prior to electrophoresis. Protein Electrophoresis: Methods and Protocols 2012, 869, 29–35. [DOI] [PubMed] [Google Scholar]
- 16.Choi J-W; Kim Y-K; Song S-Y; Lee I.-h.; Lee WH, Optical biosensor consisting of glutathione-S-transferase for detection of captan. Biosensors and Bioelectronics 2003, 18 (12), 1461–1466. [DOI] [PubMed] [Google Scholar]
- 17.Barman U; Mukhopadhyay G; Goswami N; Ghosh SS; Paily RP, Detection of Glutathione by Glutathione-S-Transferase-Nanoconjugate Ensemble Electrochemical Device. IEEE transactions on nanobioscience 2017, 16 (4), 271–279. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J; Jin Z; Hu X-X; Meng H-M; Li J; Zhang X-B; Liu H-W; Deng T; Yao S; Feng L, Efficient two-photon fluorescent probe for glutathione S-transferase detection and imaging in drug-induced liver injury sample. Analytical chemistry 2017, 89 (15), 8097–8103. [DOI] [PubMed] [Google Scholar]
- 19.Borah H; Dutta RR; Gogoi S; Medhi T; Puzari P, Glutathione-S-transferase-catalyzed reaction of glutathione for electrochemical biosensing of temephos, fenobucarb and dimethoate. Analytical Methods 2017, 9 (27), 4044–4051. [Google Scholar]
- 20.Qin L; He X; Chen L; Zhang Y, Turn-on fluorescent sensing of glutathione s-transferase at near-infrared region based on FRET between gold nanoclusters and gold nanorods. ACS applied materials & interfaces 2015, 7 (10), 5965–5971. [DOI] [PubMed] [Google Scholar]
- 21.Huang J; Li J; Lyu Y; Miao Q; Pu K, Molecular optical imaging probes for early diagnosis of drug-induced acute kidney injury. Nat Mater 2019, 18, 1133–1143. [DOI] [PubMed] [Google Scholar]
- 22.Cheng P; Miao Q; Li J; Huang J; Xie C; Pu K, Unimolecular chemo-fluoro-luminescent reporter for crosstalk-free duplex imaging of hepatotoxicity. J. Am. Chem. Soc 2019, 141 (27), 10581–10584. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J; Cheng P; Pu K, Recent advances of molecular optical probes in imaging of β-galactosidase. Bioconjugate Chemistry 2019, 30 (8), 2089–2101. [DOI] [PubMed] [Google Scholar]
- 24.Welsher K; Sherlock SP; Dai H, Deep-tissue anatomical imaging of mice using carbon nanotube fluorophores in the second near-infrared window. Proceedings of the National Academy of Sciences 2011, 108 (22), 8943–8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi H; Ghosh D; Ham M-H; Qi J; Barone PW; Strano MS; Belcher AM, M13 phage-functionalized single-walled carbon nanotubes as nanoprobes for second near-infrared window fluorescence imaging of targeted tumors. Nano letters 2012, 12 (3), 1176–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong G; Lee JC; Robinson JT; Raaz U; Xie L; Huang NF; Cooke JP; Dai H, Multifunctional in vivo vascular imaging using near-infrared II fluorescence. Nat Med 2012, 18 (12), 1841–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Connell MJ; Bachilo SM; Huffman CB; Moore VC; Strano MS; Haroz EH; Rialon KL; Boul PJ; Noon WH; Kittrell C; Ma JP; Hauge RH; Weisman RB; Smalley RE, Band gap fluorescence from individual single-walled carbon nanotubes. Science 2002, 297 (5581), 593–596. [DOI] [PubMed] [Google Scholar]
- 28.Bachilo SM; Strano MS; Kittrell C; Hauge RH; Smalley RE; Weisman RB, Structure-assigned optical spectra of single-walled carbon nanotubes. Science 2002, 298 (5602), 2361–2366. [DOI] [PubMed] [Google Scholar]
- 29.Williams RM; Lee C; Galassi TV; Harvey JD; Leicher R; Sirenko M; Dorso MA; Shah J; Olvera N; Dao F, Noninvasive ovarian cancer biomarker detection via an optical nanosensor implant. Science advances 2018, 4 (4), eaaq1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams RM; Lee C; Heller DA, A Fluorescent Carbon Nanotube Sensor Detects the Metastatic Prostate Cancer Biomarker uPA. ACS Sensors 2018, 3 (9), 1838–1845. [DOI] [PubMed] [Google Scholar]
- 31.Heller DA; Baik S; Eurell TE; Strano MS, Single‐walled carbon nanotube spectroscopy in live cells: towards long‐term labels and optical sensors. Advanced Materials 2005, 17 (23), 2793–2799. [Google Scholar]
- 32.Heller DA; Jin H; Martinez BM; Patel D; Miller BM; Yeung T-K; Jena PV; Höbartner C; Ha T; Silverman SK, Multimodal optical sensing and analyte specificity using single-walled carbon nanotubes. Nat Nanotechnol 2008, 4 (2), 114–120. [DOI] [PubMed] [Google Scholar]
- 33.Roxbury D; Jena PV; Shamay Y; Horoszko CP; Heller DA, Cell Membrane Proteins Modulate the Carbon Nanotube Optical Bandgap via Surface Charge Accumulation. ACS nano 2015, 10 (1), 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J; Landry MP; Barone PW; Kim J-H; Lin S; Ulissi ZW; Lin D; Mu B; Boghossian AA; Hilmer AJ, Molecular recognition using corona phase complexes made of synthetic polymers adsorbed on carbon nanotubes. Nature nanotechnology 2013, 8 (12), 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harvey JD; Jena PV; Baker HA; Zerze GH; Williams RM; Galassi TV; Roxbury D; Mittal J; Heller DA, A carbon nanotube reporter of microRNA hybridization events in vivo. Nature Biomedical Engineering 2017, 1, 0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jena PV; Roxbury D; Galassi TV; Akkari L; Horoszko CP; Iaea DB; Budhathoki-Uprety J; Pipalia NH; Haka AS; Harvey JD; Mittal J; Maxfield FR; Joyce JA; Heller DA, A Carbon Nanotube Optical Reporter Maps Endolysosomal Lipid Flux. ACS Nano 2017, 11 (11), 10689–10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iverson NM; Barone PW; Shandell M; Trudel LJ; Sen S; Sen F; Ivanov V; Atolia E; Farias E; McNicholas TP, In vivo biosensing via tissue-localizable near-infrared-fluorescent single-walled carbon nanotubes. Nature nanotechnology 2013, 8 (11), 873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galassi TV; Jena PV; Shah J; Ao G; Molitor E; Bram Y; Frankel A; Park J; Jessurun J; Ory DS, An optical nanoreporter of endolysosomal lipid accumulation reveals enduring effects of diet on hepatic macrophages in vivo. Science translational medicine 2018, 10 (461), eaar2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J; Kruss S; Hilmer AJ; Shimizu S; Schmois Z; De La Cruz F; Barone PW; Reuel NF; Heller DA; Strano MS, A Rapid, Direct, Quantitative, and Label‐Free Detector of Cardiac Biomarker Troponin T Using Near‐Infrared Fluorescent Single‐Walled Carbon Nanotube Sensors. Advanced healthcare materials 2014, 3 (3), 412–423. [DOI] [PubMed] [Google Scholar]
- 40.So H-M; Won K; Kim YH; Kim B-K; Ryu BH; Na PS; Kim H; Lee J-O, Single-walled carbon nanotube biosensors using aptamers as molecular recognition elements. J. Am. Chem. Soc 2005, 127 (34), 11906–11907. [DOI] [PubMed] [Google Scholar]
- 41.Yang R; Tang Z; Yan J; Kang H; Kim Y; Zhu Z; Tan W, Noncovalent assembly of carbon nanotubes and single-stranded DNA: an effective sensing platform for probing biomolecular interactions. Analytical chemistry 2008, 80 (19), 7408–7413. [DOI] [PubMed] [Google Scholar]
- 42.Tung NT; Tue PT; Lien TTN; Ohno Y; Maehashi K; Matsumoto K; Nishigaki K; Biyani M; Takamura Y, Peptide aptamer-modified single-walled carbon nanotube-based transistors for high-performance biosensors. Scientific reports 2017, 7 (1), 17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heller DA; Pratt GW; Zhang J; Nair N; Hansborough AJ; Boghossian AA; Reuel NF; Barone PW; Strano MS, Peptide secondary structure modulates single-walled carbon nanotube fluorescence as a chaperone sensor for nitroaromatics. Proceedings of the National Academy of Sciences 2011, 108 (21), 8544–8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bisker G; Dong J; Park HD; Iverson NM; Ahn J; Nelson JT; Landry MP; Kruss S; Strano MS, Protein-targeted corona phase molecular recognition. Nat Commun 2016, 7, 10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bisker G; Ahn J; Kruss S; Ulissi ZW; Salem DP; Strano MS, A mathematical formulation and solution of the CoPhMoRe inverse problem for helically wrapping polymer corona phases on cylindrical substrates. The Journal of Physical Chemistry C 2015, 119 (24), 13876–13886. [Google Scholar]
- 46.Bisker G; Bakh NA; Lee MA; Ahn J; Park M; O’Connell EB; Iverson NM; Strano MS, Insulin detection using a corona phase molecular recognition site on single-walled carbon nanotubes. ACS sensors 2018, 3 (2), 367–377. [DOI] [PubMed] [Google Scholar]
- 47.Zheng M; Jagota A; Semke ED; Diner BA; McLean RS; Lustig SR; Richardson RE; Tassi N. G. J. N. m., DNA-assisted dispersion and separation of carbon nanotubes. 2003, 2 (5), 338–342. [DOI] [PubMed] [Google Scholar]
- 48.Roxbury D; Mittal J; Jagota A. J. N. l., Molecular-basis of single-walled carbon nanotube recognition by single-stranded DNA. 2012, 12 (3), 1464–1469. [DOI] [PubMed] [Google Scholar]
- 49.Hamada N; Sawada S.-i.; Oshiyama A, New one-dimensional conductors: graphitic microtubules. Physical review letters 1992, 68 (10), 1579–1581. [DOI] [PubMed] [Google Scholar]
- 50.Sfeir MY; Beetz T; Wang F; Huang L; Huang XH; Huang M; Hone J; O’Brien S; Misewich J; Heinz TF, Optical spectroscopy of individual single-walled carbon nanotubes of defined chiral structure. Science 2006, 312 (5773), 554–556. [DOI] [PubMed] [Google Scholar]
- 51.Budhathoki-Uprety J; Jena PV; Roxbury D; Heller DA, Helical polycarbodiimide cloaking of carbon nanotubes enables inter-nanotube exciton energy transfer modulation. J. Am. Chem. Soc 2014, 136 (44), 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim M; Wu X; Ao G; He X; Kwon H; Hartmann NF; Zheng M; Doorn SK; Wang Y, Mapping Structure-Property Relationships of Organic Color Centers. Chem 2018, 4 (9), 2180–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haskard CA; Li-Chan EC, Hydrophobicity of bovine serum albumin and ovalbumin determined using uncharged (PRODAN) and anionic (ANS-) fluorescent probes. Journal of Agricultural and Food Chemistry 1998, 46 (7), 2671–2677. [Google Scholar]
- 54.Jeyachandran Y; Mielczarski E; Rai B; Mielczarski J, Quantitative and qualitative evaluation of adsorption/desorption of bovine serum albumin on hydrophilic and hydrophobic surfaces. Langmuir 2009, 25 (19), 11614–11620. [DOI] [PubMed] [Google Scholar]
- 55.Budhathoki-Uprety J; Harvey JD; Isaac E; Williams RM; Galassi TV; Langenbacher RE; Heller DA, Polymer cloaking modulates the carbon nanotube protein corona and delivery into cancer cells. Journal of Materials Chemistry B 2017, 5 (32), 6637–6644. [DOI] [PubMed] [Google Scholar]
- 56.Song Q, Protein Adsorption in Microengraving Immunoassays. Sensors 2015, 15 (10), 26236–26250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang G; Lu Y; Hou H; Liu Y, Probing the binding behavior and kinetics of silver nanoparticles with bovine serum albumin. RSC advances 2017, 7 (15), 9393–9401. [Google Scholar]
- 58.Moore RG; Hill EK; Horan T; Yano N; Kim K; MacLaughlan S; Lambert-Messerlian G; Tseng YD; Padbury JF; Miller MC, HE4 (WFDC2) gene overexpression promotes ovarian tumor growth. Scientific reports 2014, 4, 3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piskounova E; Polytarchou C; Thornton JE; LaPierre RJ; Pothoulakis C; Hagan JP; Iliopoulos D; Gregory RI, Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 2011, 147 (5), 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harris CC; Hollstein M, Clinical implications of the p53 tumor-suppressor gene. New England Journal of Medicine 1993, 329 (18), 1318–1327. [DOI] [PubMed] [Google Scholar]
- 61.Kharas MG; Lengner CJ; Al-Shahrour F; Bullinger L; Ball B; Zaidi S; Morgan K; Tam W; Paktinat M; Okabe R, Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med 2010, 16 (8), 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


