Abstract
Objective
The purpose of our research is to systematically explore the multiple mechanisms of Hemerocallis fulva Flowers (HF) on depressive disorder (DD).
Methods
The components of HF were searched from the literature. The targets of components were obtained from PharmMapper. After that, Cytoscape software was used to build a component-target network. The targets of DD were collected from DisGeNET, PharmGKB, TTD, and OMIM. Protein-protein interactions (PPIs) among the DD targets were executed to screen the key targets. Afterward, the GO and KEGG pathway enrichment analysis were performed by the KOBAS database. A compound-target-KEGG pathway network was built to analyze the key compounds and targets. Finally, the potential active substances and targets were validated by molecular docking.
Results
A total of 55 active compounds in HF, 646 compound-related targets, and 527 DD-related targets were identified from public databases. After treated with PPI, 219 key targets of DD were acquired. The gene enrichment analysis suggested that HF probably benefits DD patients by modulating pathways related to the nervous system, endocrine system, amino acid metabolism, and signal transduction. The network analysis showed the critical components and targets of HF on DD. Results of molecular docking increased the reliability of this study.
Conclusions
It predicted and verified the pharmacological and molecular mechanism of HF against DD from a holistic perspective, which will also lay a foundation for further experimental research and rational clinical application of DD.
1. Introduction
Depressive disorder (DD) is a severe and occasionally fatal mental disorder that occurs in 4.4% to 20% of the general population. It happens at any time peaking in older adulthood, and it is more prevalent in women than in men. According to the world health organization (WHO) report, depression will be the second most burdensome disease in terms of treatment and care costs by 2020 [1, 2]. Treatment for depression is necessary because depression can interfere with one's daily life, which can significantly affect relationships with family and friends. However, the specific pathophysiology basis of the development of depression remains unclear [3]. As a critical component of complementary and alternative medicine, Traditional Chinese Medicine (TCM) plays an essential role in treating depression.
TCM is a multicomponent, multitarget, and multipathway therapy, which can achieve its unique therapeutic effect by adjusting the biological network of the human body system. Therefore, it is difficult to detect the exact mechanism of TCM only through traditional experimental methods [4]. With the rapid development of bioinformatics, network pharmacology has become a new and efficient method to systematically reveal the molecular and pharmacological mechanisms of TCM [5]. Network pharmacology can reflect and elucidate the relationship among multiple components, multiple targets, and multiple diseases. At the same time, it abstracts the relationship into a network model and illustrates the action of drugs on the human biological network from a systematic perspective [6].
Hemerocallis fulva L. is a perennial herb of the Liliaceae family [7], which is indigenous to Asia, and its flowers are used as ornamental flowers, as well as for food and medicine. It was first recorded in the publication Supplement to Compendium of Materia Medica (Bencao Gangmu Shiyi). The root, seeding, and flower of Hemerocallis fulva L. are considered to have sweet, cool, and nontoxic properties and to be associated with the spleen, liver, and bladder meridians [8]. Previous investigations revealed its predominant chemical constituents including alkaloids [9], flavones [10], terpenes [11], steroidal saponins [12], and phenolic glycosides [13]. According to previous studies, Hemerocallis fulva Flowers (HF) have been used to treat various diseases including depression [14, 15], inflammation [16], insomnia [17], hepatosis [18], and cancer [19].
However, there were few research studies on the network pharmacology of HF, and the mechanism of HF in treating depression was not very clear. In this study, we proposed a network pharmacology method aiming at uncovering the active substances and mechanisms of HF in the treatment of DD. Firstly, the components from HF were searched from the literature. The components were filtered by the metrics of oral bioavailability and drug-likeness. The targets were predicted by PharmMapper. Secondly, protein-protein interactions (PPIs) among the targets associated with DD searched from the DisGeNET, PharmGKB, TTD, and OMIM databases were predicted by the STRING database, and the key targets of DD were acquired. Then, the intersection of the compound-compound target network and PPI network of DD targets was taken to find overlapping targets. After that, the GO and KEGG pathway enrichment analysis were performed by KOBAS. The compound-target-pathway (C-T-P) network was analyzed to obtain the key potential active substances and targets. Lastly, the key targets were further validated by molecular docking. The whole framework is shown in Figure 1.
Figure 1.

Workflow of the network pharmacology analysis of HF constituents against DD.
Based on network pharmacology technology, this study aimed to explore the overall regulatory role of multicomponent, multitarget antidepressants in the molecular-level system of HF and to provide a theoretical basis for further experimental study and rational clinical application of HF.
2. Materials and Methods
2.1. Chemical Database Collection and Active Components Screening
To collect the compounds of HF, we searched the TCM Integrated Database [20] (TCMID, http://www.megabionet.org/tcmid/), which records a great deal of information about herbal ingredients, and the Traditional Chinese Medicine Systems Pharmacology Database [21] (TCMSP, http://lsp.nwu.edu.cn/), a unique systemic pharmacology platform for Chinese herbal medicine. Unfortunately, few compounds were collected, except for some simple amino acid compounds. So, we consulted a lot of studies [9, 10, 22–28] to collect the compounds of HF. The chemical structural formulas of HF compounds were drawn with ChemBioDraw Ultra 14.0.
Absorption, distribution, metabolism, and excretion (ADME) are used in drug discovery. Appropriate ADME screening can ensure that these candidate compounds have suitable pharmacokinetic properties. In this study, two ADME-related parameters were employed to screen out the active compounds in HF, including gastrointestinal (GI) absorption and drug-likeness evaluated by Lipinski's rule. GI absorption and drug-likeness were obtained from SwissADME (http://www.swissadme.ch/index.php) [29]. GI absorption is a pharmacokinetic behavior that is critical to evaluate the various stages of the drug discovery process. It can be calculated using an accurate prediction model, the Brain Or IntestinaL EstimateD permeation method (BOILED-Egg) [30]. Lipinski's rule of five is a rule of thumb to evaluate if a chemical compound with certain pharmacological or biological activities could be a likely orally active drug in humans [31, 32]. All screen-out compounds should follow Lipinski's rule, and the GI absorption value of compounds should be high.
2.2. Predicted Compound Targets for HF
The chemical structural formulas of HF active compounds were saved as “mol2.” formatted files and treated with the function of MM2 to optimize the 3D molecular structures by ChemBio3D Ultra 14.0. The 3D molecular structure files of HF were imported into PharmMapper [33] (http://lilab.ecust.edu.cn/pharmmapper/), which was an online server that utilized pharmacophore mapping to approach for potential drug target identification. In this study, the targets of each compound obtained from PharmMapper were selected as potential targets. The details about the selected targets are described in Table S1.
2.3. Depressive Disorder Targets
The genes associated with targets of DD were collected from DisGeNET [34] (http://www.disgenet.org/), PharmGKB [35] (https://www.pharmgkb.org/), TTD [36] (http://db.idrblab.net/ttd/), and OMIM (https://omim.org/). We searched these platforms with the keywords “depressive disorder,” “depression,” or “major depressive disorder.” Humans' protein targets were selected in this study. The details about the selected targets are described in Table S2.
2.4. Protein-Protein Interaction
STRING is a database, which can predict protein-protein interactions. The interactions include direct (physical) and indirect (functional) associations. They stem from computational prediction, from knowledge transfer between organisms, and from interactions aggregated from other (primary) databases. The data of protein-protein interaction (PPI) were obtained from the STRING database [37] (https://string-db.org/, ver. 11.0), with the species limited to “Homo sapiens.” PPIs with comprehensive scores >0.7 were reserved in this study.
2.5. Network Construction
Network construction was executed as follows: (1) compound-compound target network was structured by connecting active compounds and corresponding targets; (2) PPI network of DD targets was built by linking DD targets; (3) compound-DD target-KEGG pathway network was established by connecting compounds, overlapping targets between compound targets and core DD targets, and top 15 KEGG pathways. In network interactions, nodes represent compounds, targets, and KEGG pathways, while edges represent the interaction of each other. The network visualization software Cytoscape [38] (http://cytoscape.org/, ver. 3.7.2) was used to show all the above networks. The software is well suited for visualizing molecular interactions in networks. Besides, the tool of NetworkAnalyzer [39] provides a powerful set of data integration, analysis, and visualization capabilities for analyzing complex networks. For each node in the interactive network, three metrics are calculated to evaluate its topology characteristics. “Degree” is defined as the number of edges of node i. “Node betweenness” represents the number of shortest paths between pairs of nodes passing through node i. “Closeness” is the reciprocal of the sum of distances between node i and the other nodes.
2.6. Gene Ontology and KEGG Pathway Enrichment
Gene Ontology (GO) is a bioinformatics project aimed to unify the characteristics of genes and genetic products in all species. Semantic lexical criteria that define and describe the function of genes and proteins can be updated for further study. GO has three classifications, including molecular function (MF), biological process (BP), and cell component (CC). MF describes the activity of gene products in molecular biology, such as catalytic or binding activity. BP is a multistep process composed of ordered molecular functions. CC refers to organelles or gene product groups and represents the role of gene products. Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism, and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies.
KOBAS is a widely used gene set enrichment (GSE) analysis tool [40]. The current version is KOBAS 3.0, which was released at the end of 2019, covering 5,945 species with incorporated knowledge. The enrichment module can accept the gene list or gene expression data as input and generate the enriched gene set, corresponding name, p value, enrichment probability, and enrichment score according to the results of various methods. In this study, KOBAS was applied to perform GO and KEGG pathway enrichment analysis. Enriched GO terms and pathways were defined as those p value <0.05. The pvalue was corrected using the method introduced by Benjamini and Hochberg [41]. It controlled the false discovery rate, which was the expected percentage of rejected assumptions. Therefore, this method is more effective than others. The horizontal bar of GO enrichment and bubble chart of KEGG pathway enrichment were plotted by using the bioinformatic tool (http://www.bioinformatics.com.cn/), which was a free online data analysis platform.
2.7. Molecular Docking Verification
Molecular docking plays an important role in rational drug design. It is often used to predict the binding sites and binding postures between candidate drugs and targets, as well as to estimate the binding affinity of molecules [42, 43]. Surflex-Dock is an accurate docking program based on a protomol that can be automatically generated or user-defined. In this study, the Surflex-Dock plug-in included in the Sybyl-X (version 2.0, TRIPOS Inc.) was used to perform molecular docking. The binding ability was evaluated using a scoring function analysis, and the higher docking score represented better binding ability. The files of protein molecular structures were obtained from the PDB database (https://www.rcsb.org/). For docking studies with Surflex-Dock, the ligand-binding site protomol was generated using the ligand from the PDB file. The visualization of intermolecular forces between the candidate compound and their potential target was performed on Discovery Studio 2020 program.
3. Results
3.1. Compounds in HF and Pharmacokinetic Evaluation
After consulting literature, 81 herbal compounds with structural information were gathered. After the GI absorption and drug-likeness process, some compounds that did not meet the ADME criteria were added back into the database because of their high bioactivity proved in previous studies [14, 44–52]. Eventually, 55 compounds were reserved, which meant that these compounds might be the active compounds of HF. The detailed information is shown in Table 1.
Table 1.
The information of active components in Hemerocallis fulva flowers (HF).
| ID | Name | Molecular formula | Molecular weight | Structure | GI absorption | Conform to rules of Lipinski |
|---|---|---|---|---|---|---|
| HF01 | (-)-(1S,3S)-1-Methyl-1, 2, 3, 4-tetrahydro-β-carboline-3-carboxylic acid | C13H14N2O2 | 230.3 |

|
High | Yes |
|
| ||||||
| HF02 | (+)-Dehydrovomifoliol | C13H18O3 | 222.3 |

|
High | Yes |
|
| ||||||
| HF03 | (3S,5 R)-Butyl-3-hydroxy-2-oxopyrrolidine-5-carboxylate | C9H15NO4 | 201.2 |

|
High | Yes |
|
| ||||||
| HF04 | (S)-2,4-Dibutoxy-3-(hydroxymethyl) cyclopent-2-en-1-one | C14H24O4 | 256.3 |

|
High | Yes |
|
| ||||||
| HF05 | (S)-Abscisic acid | C15H20O4 | 264.3 |

|
High | Yes |
|
| ||||||
| HF06 | 2′-Deoxyadenosine | C10H13N5O3 | 251.1 |

|
High | Yes |
|
| ||||||
| HF07 | 5, 6-Epoxy-3-hydroxy-7-megastigmen-9-ene | C13H20O3 | 224.3 |

|
High | Yes |
|
| ||||||
| HF08 | 5-O-p-Coumaroyl-1,5-quinide lactone | C16H16O7 | 320.3 |

|
High | Yes |
|
| ||||||
| HF09 | 5-O-p-Coumaroylquinic acid butyl ester | C20H26O8 | 394.4 |

|
High | Yes |
|
| ||||||
| HF10 | 5-O-p-Coumaroylquinic acid methyl ester | C17H20O8 | 352.3 |

|
High | Yes |
|
| ||||||
| HF11 | Apigenin | C15H10O5 | 270.3 |

|
High | Yes |
|
| ||||||
| HF12 | Catechin | C15H14O6 | 290.1 |

|
High | Yes |
|
| ||||||
| HF13 | Chlorogenic acid | C16H18O9 | 354.3 |

|
Low | No |
|
| ||||||
| HF14 | Chrysin | C15H10O4 | 254.3 |

|
High | Yes |
|
| ||||||
| HF15 | Daidzein | C15H10O4 | 254.3 |

|
High | Yes |
|
| ||||||
| HF16 | Dehydrololiolide | C11H14O3 | 194.2 |

|
High | Yes |
|
| ||||||
| HF17 | Ellagic acid | C14H6O8 | 302.2 |

|
High | Yes |
|
| ||||||
| HF18 | (-)-Epicatechin | C15H14O6 | 290.1 |

|
High | Yes |
|
| ||||||
| HF19 | Epigallocatechin gallate | C22H18O11 | 458.4 |

|
Low | No |
|
| ||||||
| HF20 | Fulvanine D | C10H15NO5 | 229.2 |

|
High | Yes |
|
| ||||||
| HF21 | Galangin | C15H10O5 | 270.3 |

|
High | Yes |
|
| ||||||
| HF22 | Hemerocallisamine I | C13H18N2O6 | 298.3 |

|
High | Yes |
|
| ||||||
| HF23 | Hemerocallisamine II | C10H15NO2 | 181.2 |

|
High | Yes |
|
| ||||||
| HF24 | Hemerocallisamine III | C10H15NO4 | 213.2 |

|
High | Yes |
|
| ||||||
| HF25 | Hemerocallisamine IV | C10H15NO4 | 213.2 |

|
High | Yes |
|
| ||||||
| HF26 | Hemerocallisamine V | C9H13NO4 | 199.2 |

|
High | Yes |
|
| ||||||
| HF27 | Hemerocallisamine VI | C9H11NO4 | 197.2 |

|
High | Yes |
|
| ||||||
| HF28 | Hemerocallisamine VII | C11H17NO5 | 243.2 |
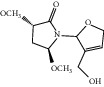
|
High | Yes |
|
| ||||||
| HF29 | Hesperidin | C28H34O15 | 610.6 |

|
Low | No |
|
| ||||||
| HF30 | Hydroxydihydrobovolide | C11H18O3 | 198.3 |

|
High | Yes |
|
| ||||||
| HF31 | Isololiolide | C11H16O3 | 196.3 |

|
High | Yes |
|
| ||||||
| HF32 | Kaempherol | C15H10O6 | 286.25 |

|
High | Yes |
|
| ||||||
| HF33 | Linoleic acid | C18H32O2 | 280.5 |

|
High | No |
|
| ||||||
| HF34 | Loliolide | C11H16O3 | 196.3 |

|
High | Yes |
|
| ||||||
| HF35 | Longitubanine A | C10H16N2O5 | 244.3 |

|
High | Yes |
|
| ||||||
| HF36 | Longitubanine B | C10H16N2O4 | 228.3 |

|
High | Yes |
|
| ||||||
| HF37 | Luteolin | C15H10O6 | 286.0 |

|
High | Yes |
|
| ||||||
| HF38 | Lycoperodine-1 | C12H12N2O2 | 216.2 |

|
High | Yes |
|
| ||||||
| HF39 | Methyl 2,4-dihydroxy-6-(4-hydroxyphenethyl)-3-methylbenzoate | C17H18O5 | 302.3 |

|
High | Yes |
|
| ||||||
| HF40 | Morin | C15H10O7 | 302.3 |

|
High | Yes |
|
| ||||||
| HF41 | Myricetin | C15H10O8 | 318.3 |

|
Low | No |
|
| ||||||
| HF42 | Naringenin | C15H12O5 | 272.3 |

|
High | Yes |
|
| ||||||
| HF43 | Naringin | C27H32O14 | 580.6 |

|
Low | No |
|
| ||||||
| HF44 | Pinocembrin | C15H12O4 | 256.3 |

|
High | Yes |
|
| ||||||
| HF45 | Prunasin | C14H17NO6 | 295.3 |

|
High | Yes |
|
| ||||||
| HF46 | Pseudolaroside C | C14H18O8 | 314.3 |

|
High | Yes |
|
| ||||||
| HF47 | Quercetin | C15H10O7 | 302.3 |

|
High | Yes |
|
| ||||||
| HF48 | Quercetin-3-O-rutinoside | C27H30O16 | 610.5 |

|
Low | No |
|
| ||||||
| HF49 | Quercetin-3-O-β-D-galactopyranoside | C21H20O12 | 464.4 |

|
Low | No |
|
| ||||||
| HF50 | Quercetin-3-O-β-D-glucopyranoside | C21H20O12 | 464.4 |
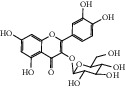
|
Low | No |
|
| ||||||
| HF51 | Rosin | C15H20O6 | 296.3 |

|
High | Yes |
|
| ||||||
| HF52 | Salidroside | C14H20O7 | 300.3 |

|
High | Yes |
|
| ||||||
| HF53 | Vitexin | C21H20O10 | 432.4 |

|
Low | No |
|
| ||||||
| HF54 | Wogonin | C16H12O5 | 284.3 |

|
High | Yes |
|
| ||||||
| HF55 | α-Linolenic acid | C18H30O2 | 278.4 |

|
High | No |
3.2. Compound-Compound Target Network Analysis
The compound-compound target network is depicted in Figure 2, including 701 nodes (55 active compound nodes and 646 compound target nodes) and 3112 edges. In this network, the rectangle represented the target, and the oval represented the compound. We found that many targets were hit by multiple compounds; for example, vascular endothelial growth factor A (VEGFA) and albumin (ALB) were together modulated by multiple ingredients, including hesperidin and quercetin-3-O-β-D-glucopyranoside. The average number of targets per component is 56.6, and the mean degree of components per target is 4.8. It clearly showed that HF fit the multicomponent and multitarget characteristics of TCM. Consequently, we not only obtained an approximate observation of the relationship between bioactive compounds and compound targets, but also discovered the potential pharmacological effects of HF from this network.
Figure 2.

Compound-compound target network (red ellipses represent compounds contained in HF, blue rectangles represent compound targets).
3.3. Disease PPI Network Construction
Based on the gene database DisGeNET, PharmGKB, TTD, and OMIM, there were a total of 527 candidate targets relevant to depressive disorder. After PPI was acquired, there were 446 nodes and 3905 edges in the DD target PPI network (Figure 3). The results of network analysis showed that there were 219 nodes whose degree value was higher than the median value, which meant that these targets were likely to be the key targets in the development of DD. The targets in the interior circle showed more interactions with targets than those in the exterior. After the intersection process, we found that there were 18 overlapping targets between compound targets of HF and key targets of DD. They are ALB, amyloid-beta precursor protein (APP), androgen receptor (AR), voltage-dependent L-type calcium channel subunit alpha-1C (CACNA1C), calcium-dependent protein kinase II alpha (CAMK2A), corticoliberin (CRH), discs large MAGUK scaffold protein 3 (DLG3), FK506 binding protein 4 (FKBP4), glutamate decarboxylase 2 (GAD2), monoamine oxidase A (MAOA), monoamine oxidase B (MAOB), neural cell adhesion molecule 1 (NCAM1), nitric oxide synthase 2 (NOS2), nuclear receptor subfamily 3 group C member 2 (NR3C2), Rac family small GTPase 1 (RAC1), superoxide dismutase 2 (SOD2), transthyretin (TTR), and VEGFA, which meant that these targets might be the key targets for HF treating DD.
Figure 3.

PPI network of DD targets (yellow circles represent nonkey targets of DD, orange circles represent key targets of DD, red circles represent overlapping targets between the compound target of HF and key targets of DD).
3.4. Potential Synergistic Mechanisms Analysis of HF Target-DD Target Network
3.4.1. GO Enrichment Analysis
After GO enrichment analysis of 18 overlapping targets, a total of 42 GO entries were found with the corrected pvalue <0.05. Figure 4 lists the top10 entries of each category, namely, BP, CC, and MF. BP included regulation of circadian sleep/wake cycle (GO: 0042749), programmed cell death (GO: 0012501), head development (GO: 0060322), metabolic process (GO: 0008152), and positive regulation of calcium ion transport (GO: 0051928). CC included main axon (GO: 0044304), vesicle lumen (GO: 0031983), cation channel complex (GO: 0034703), synapse (GO: 0045202), and vesicle (GO: 0031982). MF included protein binding (GO: 0005515), cation binding (GO: 0043169), and hydrolase activity (GO: 0016787). The GO entries mentioned above were strongly associated with the central nervous system and mental diseases. This demonstrated that HF probably worked by engaging in the above biological processes, cellular components, and molecular functions.
Figure 4.

GO enrichment analysis for 18 overlapping targets.
3.4.2. KEGG Pathway Analysis to Explore the Therapeutic Mechanisms of HF on DD
The 18 overlapping targets were further mapped to 34 pathways with p < 0.05. The 34 pathways belonged to four categories: human diseases (9/34), organismal systems (13/34), environmental information processing (4/34), and metabolism (8/34). Thus, our findings showed that HF integrated multiple signaling pathways to the nervous system, endocrine system, amino acid metabolism, signal transduction, and substance dependence. Based on the results of pathway analysis, it was found that these high-degree pathways were closely related to the DD, such as serotonergic synapse and dopaminergic synapse. The top 15 KEGG pathways are shown in Figure 5.
Figure 5.

KEGG enrichment analysis for 18 overlapping targets.
3.4.3. Compound-Target-KEGG Pathway Network Analysis
The details of KEGG pathways are described in Table 2, and the compound-target-pathway (C-T-P) network is shown in Figure 6, which contained 52 nodes including top 15 KEGG pathways, 10 targets related to top 15 KEGG pathways and 27 compounds, and 178 edges. The network was calculated to find the major nodes of compounds and targets. Finally, 12 compound nodes with their values of degree more than average were selected as crucial compound nodes, namely, naringin, (S)-2,4-dibutoxy-3-(hydroxymethyl)cyclopent-2-en-1-one, 5-O-p-coumaroylquinic acid butyl ester, hesperidin, lycoperodine-1, (-)-(1S,3S)-1-methyl-1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid, hydroxydihydrobovolide, epigallocatechin gallate (EGCG), prunasin, quercetin-3-O-β-D-galactopyranoside, vitexin, and α-linolenic acid. It indicated that these compounds might play a more important role than others in the treatment of depressive disorder with HF. Five target nodes with values of degree, betweenness, and closeness more than average were selected as key target nodes, namely, MAOA, MAOB, AR, CAMK2A, and GAD2. It indicated that these targets may be crucial to the treatment of depressive disorder with HF.
Table 2.
Pathways associated with 18 overlapping targets according to enrichment analysis base on KEGG.
| ID | Pathway | pvalue | Count | Gene IDs |
|---|---|---|---|---|
| hsa05031 | Amphetamine addiction | 7.39E-09 | 4 | MAOB, MAOA, CACNA1C, CAMK2A |
| hsa04726 | Serotonergic synapse | 5.65E-08 | 4 | MAOB, APP, CACNA1C, MAOA |
| hsa04728 | Dopaminergic synapse | 9.39E-08 | 4 | MAOB, MAOA, CACNA1C, CAMK2A |
| hsa00330 | Arginine and proline metabolism | 6.34E-07 | 3 | MAOB, NOS2, MAOA |
| hsa04066 | HIF-1 signaling pathway | 6.10E-06 | 3 | NOS2, VEGFA, CAMK2A |
| hsa00360 | Phenylalanine metabolism | 1.64E-05 | 2 | MAOB, MAOA |
| hsa04934 | Cushing syndrome | 1.71E-05 | 3 | CRH, CACNA1C, CAMK2A |
| hsa05034 | Alcoholism | 2.65E-05 | 3 | MAOB, CRH, MAOA |
| hsa00340 | Histidine metabolism | 2.88E-05 | 2 | MAOB, MAOA |
| hsa04020 | Calcium signaling pathway | 3.25E-05 | 3 | NOS2, CACNA1C, CAMK2A |
| hsa00350 | Tyrosine metabolism | 6.73E-05 | 2 | MAOB, MAOA |
| hsa01100 | Metabolic pathways | 6.98E-05 | 5 | MAOB, NOS2, AR, MAOA, GAD2 |
| hsa00260 | Glycine, serine, and threonine metabolism | 8.23E-05 | 2 | MAOB, MAOA |
| hsa00380 | Tryptophan metabolism | 9.04E-05 | 2 | MAOB, MAOA |
| hsa05030 | Cocaine addiction | 1.22E-04 | 2 | MAOB, MAOA |
Figure 6.
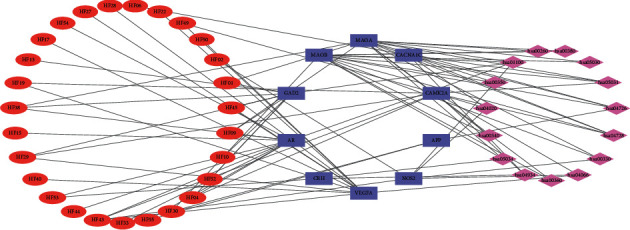
Compound-target-KEGG pathway network (red ellipses represent compounds contained in HF, blue rectangles represent compound targets, purple rhombuses represent KEGG pathway).
3.5. Molecular Docking Results and Analysis
To verify the top 5 key targets and their interacting compounds, the molecular docking simulation was carried out by the Surflex-Dock method. As the result in Table 3illustrated, most compounds had strong interactions with their targets. The average docking scores of compounds were more than 6. The results confirmed that most of these potential active compounds had strong binding affinities with the key targets, and the present network pharmacology method was reasonable. We take lycoperodine-1, (-)-(1s,3s)-1-methyl-1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid, and 5-O-p-coumaroylquinic acid butyl ester as examples to show the visualization of intermolecular forces between compounds and MAOA target, which are displayed in Figure 7.
Table 3.
Results of molecular docking of 5 key targets.
| ID | Name | Target | PDB ID | Total score |
|---|---|---|---|---|
| HF01 | (-)-(1s, 3s)-1-Methyl-1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid | MAOA | 2bxr | 6.764 |
| HF09 | 5-O-p-Coumaroylquinic acid butyl ester | MAOA | 2bxr | 9.395 |
| HF38 | Lycoperodine-1 | MAOA | 2bxr | 6.035 |
| HF30 | Hydroxydihydrobovolide | MAOB | 2bk3 | 6.579 |
| HF04 | (s)-2,4-Dibutoxy-3-(hydroxymethyl)cyclopent-2-en-1-one | AR | 3b68 | 8.112 |
| HF09 | 5-o-p-Coumaroylquinic acid butyl ester | AR | 3b68 | 7.554 |
| HF38 | Lycoperodine-1 | AR | 3b68 | 4.619 |
| HF43 | Naringin | AR | 3b68 | -0.167 |
| HF53 | Vitexin | AR | 3b68 | 4.508 |
| HF55 | α-Linolenic acid | AR | 3b68 | 10.392 |
| HF04 | (s)-2,4-Dibutoxy-3-(hydroxymethyl)cyclopent-2-en-1-one | CAMK2A | 2vz6 | 7.689 |
| HF19 | Epigallocatechin gallate | CAMK2A | 2vz6 | 8.921 |
| HF29 | Hesperidin | CAMK2A | 2vz6 | 7.035 |
| HF43 | Naringin | CAMK2A | 2vz6 | 4.630 |
| HF49 | Quercetin-3-o-β-d-galactopyranoside | CAMK2A | 2vz6 | 5.367 |
| HF01 | (-)-(1s,3s)-1-Methyl-1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid | GAD2 | 2okk | 5.580 |
| HF04 | (s)-2,4-Dibutoxy-3-(hydroxymethyl)cyclopent-2-en-1-one | GAD2 | 2okk | 6.110 |
| HF09 | 5-O-p-Coumaroylquinic acid butyl ester | GAD2 | 2okk | 5.097 |
| HF29 | Hesperidin | GAD2 | 2okk | 4.159 |
| HF38 | Lycoperodine-1 | GAD2 | 2okk | 4.555 |
| HF45 | Prunasin | GAD2 | 2okk | 4.916 |
Figure 7.

2D diagram of docking between compounds of HF and MAOA. (a) (-)-(1S,3S)-1-Methyl-1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid. (b) 5-O-p-Coumaroylquinic acid butyl ester. (c) Lycoperodine-1.
4. Discussion
Researchers have conducted a large number of studies to explore the pathogenesis of depression, but its exact pathogenesis is still unclear. The supposed pathogenesis of depression is the epigenetic hypothesis, neurotransmitter hypothesis, neurotrophic regeneration hypothesis, neurokinin hypothesis, neuroendocrine dysfunction hypothesis, and immune system abnormality hypothesis [53]. The research strategy of network pharmacology provides a unique and innovative pathway to study the mechanism of action of multicomponent and multitarget. HF is not only a nutrient food but also a drug with antidepressant activity. To elucidate the beneficial effects of HF on DD, the putative active ingredients and underlying mechanisms were comprehensively investigated using network pharmacology.
In the present study, a total of 55 active compounds in HF were screened by pharmacokinetic analysis, 646 compound-related targets were predicted by PharmMapper, and 527 DD-related targets were identified from public databases. After treated with PPI, 219 key targets in the development of DD were acquired. Among these targets, 18 targets were shared between compound-related and DD-related targets. Most entries from GO enrichment play important parts in the central nervous system, which affect different steps of synthesis or transportation of neurotransmitters. The KEGG pathway analysis proved that bioactive compounds from HF exerted a synergistic effect on the treatment of DD through numerous pathways and brain amino acid metabolism, such as amphetamine addiction, serotonergic synapse, dopaminergic synapse, arginine and proline metabolism, HIF-1 signaling pathway, phenylalanine metabolism, alcoholism, calcium signaling pathway, tyrosine metabolism, and tryptophan metabolism.
Furthermore, the top 5 key targets and top 12 key compounds were screened from the C-T-P network. These five screening targets have been shown to be highly correlated with depression. Monoamine oxidases (MAOs) play a crucial role during the development of various mental diseases [8]. There are two MAO isozymes, MAOA and MAOB. They are flavoenzymes, which bind to the outer mitochondrial membrane and catalyze the oxidative transformations of neurotransmitters like serotonin, norepinephrine, and dopamine [54]. However, deficiency of serotonin can lead to depression, so MAOA and MAOB were two important targets for DD. Hung et al. [55] found that loss of AR accelerated the development of depressive-like behaviors in mice under chronic mild stress (CMS), and mice with low androgen were more prone to depression-like behaviors [56]. The gene of CAMK2A has been reported to be associated with depression, and the expression of CAMK2A was significantly elevated in the depression tissues by 29% [57]. The role of CAMK2A as a signaling molecule inside neurons could influence the function of the brain in learning and memory. The translation regulation of the protein encoded by the gene could prove the long-term regulation of potentiation and depression [58]. Glutamate decarboxylase (GAD) is a rate-limiting enzyme for the conversion of glutamic acid to gamma-aminobutyric acid (GABA). GAD2 is an important enzyme that regulates depression-related neurotransmitters, which shows enhanced availability in situations of stress, responding to short-term demands for GABA [59]. GABAergic dysfunction in schizophrenia and mood disorders was reflected by decreased levels of GAD2 [60]. Therefore, HF may improve depression symptoms by regulating including but not limited to MAOA, MAOB, AR, CAMK2A, and GAD2.
Meanwhile, some of the screened compounds had been shown to antidepressant activity. It reported that naringin was a neuroactive flavonoid. It possessed functional beneficial neurobehavioral effects including anxiolytic, antidepressant, and memory enhancing [46]. Hesperidin, a well-known flavanone glycoside mostly found in citrus fruits, showed neuroprotective and antidepressant activity [61]. EGCG could attenuate the depressive status of mice, and the underlying mechanism may be related to the reduction of serum cortisol (CORT) and adrenocorticotropic hormone (ACTH), downregulation of malonyldialdehyde (MDA), interleukin-1 beta (IL-1β), interleukin 6 (IL-6), indoleamine 2,3-dioxygenase (IDO), and upregulation of superoxide dismutase (SOD) and glutathione peroxidase (GSH-PX) in the hippocampus [62]. Quercetin-3-O-β-D-galactopyranoside may improve depression-like effects by regulating the hypothalamic-pituitary-adrenal (HPA) axis and reducing the level of oxidative stress in the hippocampus [63]. Can et al. found that the antidepressant-like effect of vitexin was mediated through an increase in catecholamine levels in the synaptic cleft [64]. In a study of 150 elderly males from Crete, omega-3 α-linolenic acid adipose tissue stores were negatively correlated with depression [65]. The antidepressant-like effect of Hemerocallis extract was mainly related to flavonoids [14], which were similar to our findings. Our study suggested that, along with these reported compounds, some other putative active ingredients in HF might also possess antidepressant-like effects with diverse underlying mechanisms. Besides that, we also found some potential antidepressant compounds that have not been reported in the existing literature. They were (-)-(1S,3S)-1-methyl-1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid, (S)-2,4-dibutoxy-3-(hydroxymethyl)cyclopent-2-en-1-one, 5-O-p-coumaroylquinic acid butyl ester, lycoperodine-1, hydroxydihydrobovolide, and prunasin. Subsequent experiments could verify their activity.
Molecular docking experiment is a new method that uses computer simulation of compound structure and related disease targets to execute molecular docking, calculate and analyze the bioactivity of the compound, and screen the material basis of pharmacodynamics. This method could quickly and efficiently discover some new bioactive lead compounds from the database. In this study, molecular docking results showed that targets predicted by PharmMapper were reasonable. Most of the screened compounds and their corresponding targets scored well. The structures of lycoperodine-1 and (-)-(1S,3S)-1-methyl-1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid are similar to that of harmine, which was a carboline alkaloid and also was a MAOA inhibitor [66]. The similar scores of these compounds are also due to their structural similarity, which can prove the reliability of the molecular docking results.
5. Conclusion
In summary, the present study is the first one that combines active components, target prediction, network analysis, and gene enrichment analysis by a network pharmacology method to elucidate the molecular and pharmacological mechanism of HF against DD from a systematic perspective. In this research, we showed multiple targets of HF against DD for the first time. Based on this neural network model, active components, targets, and pathways in depressive disorder were initially explored to provide a preliminary theoretical basis for the design of subsequently targeted drugs. The results of molecular docking confirmed that the present network pharmacology method was reasonable. Nonetheless, more experiments should be implemented to verify the validity of our findings in further pharmacological and molecular research. Moreover, we hope that our study will be useful for antidepression drug discovery.
Acknowledgments
This work was supported by the Qiqihar Academy of Medical Sciences Program (QMSI2019M-10 and QMSI2019B-03), Key Laboratory of Polysaccharide Bioactivity Evaluation of TCM of Liaoning Province, Key Techniques Study of Consistency Evaluation of Drug Quality and Therapeutic Effect (18-400-4-08), and Liaoning Distinguished Professor Project for Ying Jia (2017).
Contributor Information
Yu Sun, Email: zsy5811321@126.com.
Ying Jia, Email: jiayingsyphu@126.com.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Tiancheng Ma and Ying Jia designed the study and guided the experiment. Yu Sun and Tiancheng Ma conducted experiments and wrote the manuscript. The data were analyzed by Tingxu Yan and Bo Wu. Chang Jiang and Weilin Xiong revised the paper. All authors read and approved the final manuscript.
Supplementary Materials
Table S1: the results of compound targets predicted by PharmMapper. Table S2: targets of depressive disorder obtained from the database.
References
- 1.Majdi A., Hosseini S. H., Roozbeh M., Mohammadi A. Antidepressant and anxiolytic effects of geraniol in mice: the possible role of oxidative stress and apoptosis. Iranian Red Crescent Medical Journal. 2019;21(6) doi: 10.5812/ircmj.91593.e91593 [DOI] [Google Scholar]
- 2.Murray C. J., Lopez A. D. Evidence-based health policy--lessons from the global burden of disease study. Science. 1996;274(5288):740–743. doi: 10.1126/science.274.5288.740. [DOI] [PubMed] [Google Scholar]
- 3.Zhang K., He M., Su D., et al. Quantitative proteomics reveal antidepressant potential protein targets of xiaochaihutang in corticosterone induced model of depression. Journal of Ethnopharmacology. 2019;231:438–445. doi: 10.1016/j.jep.2018.11.020. [DOI] [PubMed] [Google Scholar]
- 4.Liu X. K., Wu J. R., Zhang D., et al. A network pharmacology approach to uncover the multiple mechanisms of hedyotis diffusa willd. On colorectal cancer. Evidence-Based Complementary and Alternative Medicine. 2018;2018 doi: 10.1155/2018/6517034.6517034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hopkins A. L. Network pharmacology: the next paradigm in drug discovery. Nature Chemical Biology. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 6.Zeng L. T., Yang K. L. Exploring the pharmacological mechanism of Yanghe decoction on HER2-positive breast cancer by a network pharmacology approach. Journal of Ethnopharmacology. 2017;199:68–85. doi: 10.1016/j.jep.2017.01.045. [DOI] [PubMed] [Google Scholar]
- 7.Wang J., Hu D. M., Hou J., et al. Ethyl Acetate Fraction of Hemerocallis citrina Baroni decreases tert-butyl hydroperoxide-induced oxidative stress damage in BRL-3A cells. Oxidative Medicine and Cellular Longevity. 2018;2018 doi: 10.1155/2018/1526125.1526125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin H. Y., Tsai J. C., Wu L. Y., et al. Reveals of new candidate active components in Hemerocallis Radix and its anti-depression action of mechanism based on network pharmacology approach. International Journal of Molecular Sciences. 2020;21(5) doi: 10.3390/ijms21051868.1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto T., Nakamura S., Nakashima S., et al. Gamma-Lactam alkaloids from the flower buds of daylily. Journal of Natural Medicines. 2016;70(3):376–383. doi: 10.1007/s11418-015-0963-z. [DOI] [PubMed] [Google Scholar]
- 10.Liu L. Y., Chang L. Y., Chou S. S., et al. Studies on the antioxidant components and activities of the methanol extracts of commercially grown Hemerocallis fulva l. (daylily) in Taiwan. Journal of Food Biochemistry. 2010;34:90–104. [Google Scholar]
- 11.Yang Z.-d., Chen H., Li Y.-c. A new glycoside and a novel-type diterpene from Hemerocallis fulva (L.) L. Helvetica Chimica Acta. 2003;86(10):3305–3309. doi: 10.1002/hlca.200390271. [DOI] [Google Scholar]
- 12.Konishi T., Fujiwara Y., Konoshima T., et al. Steroidal saponins from Hemerocallis fulva var. kwanso. Chemical & Pharmaceutical Bulletin. 2001;49(3):318–320. doi: 10.1248/cpb.49.318. [DOI] [PubMed] [Google Scholar]
- 13.Cichewicz R. H., Nair M. G. Isolation and characterization of stelladerol, a new antioxidant naphthalene glycoside, and other antioxidant glycosides from edible daylily (Hemerocallis) flowers. Journal of Agricultural and Food Chemistry. 2002;50(1):87–91. doi: 10.1021/jf010914k. [DOI] [PubMed] [Google Scholar]
- 14.Du B., Tang X., Liu F., et al. Antidepressant-like effects of the hydroalcoholic extracts of Hemerocallis citrina and its potential active components. BMC Complementary and Alternative Medicine. 2014;14:p. 326. doi: 10.1186/1472-6882-14-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin S.-H., Chang H.-C., Chen P.-J., Hsieh C.-L., Su K.-P., Sheen L.-Y. The antidepressant-like effect of ethanol extract of daylily flowers (金針花 Jīn Zhēn huā) in rats. Journal of Traditional and Complementary Medicine. 2013;3(1):53–61. doi: 10.4103/2225-4110.106548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X. L., Luo L., Liu B. B., et al. Ethanol extracts from Hemerocallis citrina attenuate the upregulation of proinflammatory cytokines and indoleamine 2,3-dioxygenase in rats. Journal of Ethnopharmacology. 2014;153(2):484–490. doi: 10.1016/j.jep.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Szewczyk K., Kalemba D., Miazga-Karska M., et al. The essential oil composition of selected Hemerocallis cultivars and their biological activity. Open Chemistry. 2019;17(1):1412–1422. [Google Scholar]
- 18.Shen N., Huang X. d., Li Z. w., et al. Effects of Hemerocallis citrine baroni flavonids on CCl4-induced liver fibrosis of rats. Acta Pharmaceutica Sinica. 2015;50(5):547–551. [PubMed] [Google Scholar]
- 19.Cichewicz R. H., Zhang Y. J., Seeram N. P., et al. Inhibition of human tumor cell proliferation by novel anthraquinones from daylilies. Life Sciences. 2004;74(14):1791–1799. doi: 10.1016/j.lfs.2003.08.034. [DOI] [PubMed] [Google Scholar]
- 20.Xue R. C., Fang Z., Zhang M. X., et al. TCMID: traditional Chinese medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Research. 2013;41(D1):D1089–D1095. doi: 10.1093/nar/gks1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ru J. L., Li P., Wang J. N., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6 doi: 10.1186/1758-2946-6-13.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X., Guo Y., Zhang Y., et al. Monoterpene derivatives from the flowers of the Hemerocallis minor Mill. Phytochemistry Letters. 2017;21:134–138. [Google Scholar]
- 23.Kao F. J., Chiang W. D., Liu H. M. Inhibitory effect of daylily buds at various stages of maturity on nitric oxide production and the involved phenolic compounds. Lwt-Food Science and Technology. 2015;61(1):130–137. [Google Scholar]
- 24.Yoshikawa K., Nagai M., Arihara S. Amino-acid amides from hemerocallis-longituba. Phytochemistry. 1994;35(4):1057–1058. [Google Scholar]
- 25.Lin Y. L., Lu C. K., Huang Y. J., Chen H. J. Antioxidative caffeoylquinic acids and flavonoids from Hemerocallis fulva flowers. Journal of Agricultural and Food Chemistry. 2011;59(16):8789–8795. doi: 10.1021/jf201166b. [DOI] [PubMed] [Google Scholar]
- 26.Zhao X. C., Du J. L., Xie Y. G., et al. Chemical constituents of the flowers of Hemerocallis minor. Chemistry of Natural Compounds. 2018;54(3):556–558. [Google Scholar]
- 27.Matsumoto T., Nakamura S., Ohta T., et al. A rare glutamine derivative from the flower buds of daylily. Organic Letters. 2014;16(11):3076–3078. doi: 10.1021/ol501182v. [DOI] [PubMed] [Google Scholar]
- 28.Sun J., Liu W., Zhang M., et al. The analysis of phenolic compounds in daylily using UHPLC-HRMSn and evaluation of drying processing method by fingerprinting and metabolomic approaches. Journal of Food Processing and Preservation. 2018;42(1) doi: 10.1111/jfpp.13325.e13325 [DOI] [Google Scholar]
- 29.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports. 2017;7 doi: 10.1038/srep42717.42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daina A., Zoete V. A BOILED-Egg to predict gastrointestinal absorption and brain penetration of small molecules. Chemmedchem. 2016;11(11):1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipinski C. A., Lombardo F., Dominy B. W., et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews. 2012;64:4–17. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 32.Lipinski C. A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discovery Today Technologies. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Wang X., Shen Y. H., Wang S. W., et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Research. 2017;45(W1):W356–W360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piñero J., Ramírez-Anguita J. M., Saüch-Pitarch J., et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Research. 2020;48(D1):D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whirl-Carrillo M., McDonagh E. M., Hebert J. M., et al. Pharmacogenomics knowledge for personalized medicine. Clinical Pharmacology & Therapeutics. 2012;92(4):414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y. X., Zhang S., Li F. C., et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Research. 2020;48(D1):D1031–D1041. doi: 10.1093/nar/gkz981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szklarczyk D., Gable A. L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Assenov Y., Ramirez F., Schelhorn S. E., et al. Computing topological parameters of biological networks. Bioinformatics. 2008;24(2):282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 40.Xie C., Mao X. Z., Huang J. J., et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 42.Zhang Y. Y., Chen J. J., Li D. Q., et al. Network pharmacology uncovers anti-cancer activity of vibsane-type diterpenes from Viburnum odoratissimum. Natural Product Research. 2019;34(4):1–4. doi: 10.1080/14786419.2019.1582047. [DOI] [PubMed] [Google Scholar]
- 43.Tan X. J., Wang D., Hei X. M., et al. Synthesis, crystal structures, antiproliferative activities and reverse docking studies of eight novel Schiff bases derived from benzil. Acta Crystallographica Section C-Structural Chemistry. 2020;76:p. 44. doi: 10.1107/S2053229619015687. [DOI] [PubMed] [Google Scholar]
- 44.Wang T. D., Niu K., Fan A., et al. Dietary intake of polyunsaturated fatty acids alleviates cognition deficits and depression-like behaviour via cannabinoid system in sleep deprivation rats. Behavioural Brain Research. 2020;384 doi: 10.1016/j.bbr.2020.112545.112545 [DOI] [PubMed] [Google Scholar]
- 45.Ma Z. G., Wang G. L., Cui L., et al. Myricetin attenuates depressant-like behavior in mice subjected to repeated restraint stress. International Journal of Molecular Sciences. 2015;16(12):28377–28385. doi: 10.3390/ijms161226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ben-Azu B., Nwoke E. E., Aderibigbe A. O., et al. Possible neuroprotective mechanisms of action involved in the neurobehavioral property of naringin in mice. Biomedicine & Pharmacotherapy. 2019;109:536–546. doi: 10.1016/j.biopha.2018.10.055. [DOI] [PubMed] [Google Scholar]
- 47.Zheng M. Z., Liu C. M., Pan F. G., et al. Antidepressant-like effect of hyperoside isolated from Apocynum venetum leaves: possible cellular mechanisms. Phytomedicine. 2012;19(2):145–149. doi: 10.1016/j.phymed.2011.06.029. [DOI] [PubMed] [Google Scholar]
- 48.Barauna S. C., Delwing-Dal Magro D., Brueckheimer M. B., et al. Antioxidant and antidepressant-like effects of Eugenia catharinensis D. Legrand in an animal model of depression induced by corticosterone. Metabolic Brain Disease. 2018;33(6):1985–1994. doi: 10.1007/s11011-018-0306-3. [DOI] [PubMed] [Google Scholar]
- 49.Mao L. N., Zhu Q., Li J. X. Research progress on neuroprotective effects of vitexin and its mechanisms. Chinese Pharmacological Bulletin. 2016;32(10):1353–1356. [Google Scholar]
- 50.Nishizaki T., Ikeuchi Y., Matsuoka T., et al. Short-term depression and long-term enhancement of ACh-gated channel currents induced by linoleic and linolenic acid. Brain Research. 1997;751(2):253–258. doi: 10.1016/s0006-8993(96)01405-9. [DOI] [PubMed] [Google Scholar]
- 51.Jing S., Nian Z., Weini M., et al. Modulation of gut microbiota by chlorogenic acid pretreatment on rats with adrenocorticotropic hormone induced depression-like behavior. Food & Function. 2019;10(5) doi: 10.1039/c8fo02599a. [DOI] [PubMed] [Google Scholar]
- 52.Marilyn H. Effect of epigallocatechin gallate supplementation in schizophrenia and bipolar disorder: an 8-week, randomized, double-blind, placebo-controlled study. Therapeutic Advances in Psychopharmacology. 2013;3(1):21–27. doi: 10.1177/2045125312464103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H. Q., Wang Z. Z., Chen N. H. Advance in relationship between receptor gene abnormality and depression. Acta Pharmaceutica Sinica. 2020;55(03):384–391. [Google Scholar]
- 54.Rehuman N. A., Mathew B., Jat R. K., et al. A comprehensive review of monoamine oxidase-a inhibitors in their syntheses and potencies. Combinatorial Chemistry & High Throughput Screening. 2020;23(9) doi: 10.2174/1386207323666200428091306. [DOI] [PubMed] [Google Scholar]
- 55.Hung Y. Y., Huang Y. L., Chang C., et al. Deficiency in androgen receptor aggravates the depressive-like behaviors in chronic mild stress model of depression. Cells. 2019;8(9) doi: 10.3390/cells8091021.1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shao S., Cui Y., Chen Z. B., et al. Androgen deficit changes the response to antidepressant drugs in tail suspension test in mice. The Aging Male. 2020;23(5):1–7. doi: 10.1080/13685538.2020.1762074. [DOI] [PubMed] [Google Scholar]
- 57.Jian X. Q., Wang K. S., Wu T. J., et al. Association of ADAM10 and CAMK2A polymorphisms with conduct disorder: evidence from family-based studies. Journal of Abnormal Child Psychology. 2011;39(6):773–782. doi: 10.1007/s10802-011-9524-4. [DOI] [PubMed] [Google Scholar]
- 58.Athira K. S., Muddashetty R. Molecular cloning of CAMK2A gene. International Journal of Advanced Research in Engineering and Applied Sciences. 2016;5(1):7–19. [Google Scholar]
- 59.Unschuld P. G., Ising M., Specht M., et al. Polymorphisms in the GAD2 gene-region are associated with susceptibility for unipolar depression and with a risk factor for anxiety disorders. American Journal of Medical Genetics Part B-Neuropsychiatric Genetics. 2009;150B(8):1100–1109. doi: 10.1002/ajmg.b.30938. [DOI] [PubMed] [Google Scholar]
- 60.Fatemi S. H., Stary J. M., Earle J. A., et al. GABAergic dysfunction in schizophrenia and mood disorders as reflected by decreased levels of glutamic acid decarboxylase 65 and 67 kDa and reelin proteins in cerebellum. Schizophrenia Research. 2005;74(2-3):p. 287. doi: 10.1016/j.schres.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 61.Nadar J. S., Kale P. P., Kadu P. K., et al. Potentiation of antidepressant effects of agomelatine and bupropion by hesperidin in mice. Neurology Research International. 2018;2018 doi: 10.1155/2018/9828639.9828639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen J., Lin X. D., Liu L. Y., et al. Protective effects of epigallocatechin gallate on depressive mice induced by chronic unpredictable mild stress. Chinese Traditional Herbal Drugs. 2018;49(06):1351–1357. [Google Scholar]
- 63.Zhang Y., Zhu C. C. Effect of hyperin on depressive behavior of rats induced by chronic unpredicted mild stress. Chinese Journal of New Drugs and Clinical Remedies. 2017;36(03):150–156. [Google Scholar]
- 64.Can O. D., Ozkay U. D., Ucel U. I. Anti-depressant-like effect of vitexin in BALB/c mice and evidence for the involvement of monoaminergic mechanisms. European Journal of Pharmacology. 2013;699(1-3):250–257. doi: 10.1016/j.ejphar.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 65.Mamalakis G., Tornaritis M., Kafatos A. Depression and adipose essential polyunsaturated fatty acids. Prostaglandins Leukotrienes and Essential Fatty Acids. 2002;67(5):311–318. doi: 10.1054/plef.2002.0435. [DOI] [PubMed] [Google Scholar]
- 66.Ramsay R. R., Hunter D. J. B. Inhibitors alter the spectrum and redox properties of monoamine oxidase A. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2002;1601(2):178–184. doi: 10.1016/s1570-9639(02)00466-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: the results of compound targets predicted by PharmMapper. Table S2: targets of depressive disorder obtained from the database.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


