Shi, Li, et al. report the development of label-retention expansion microscopy (LR-ExM) with a set of trifunctional anchors that not only prevent fluorescent signal loss but also enable high-efficiency labeling using SNAP and CLIP tags.
Abstract
Expansion microscopy (ExM) increases the effective resolving power of any microscope by expanding the sample with swellable hydrogel. Since its invention, ExM has been successfully applied to a wide range of cell, tissue, and animal samples. Still, fluorescence signal loss during polymerization and digestion limits molecular-scale imaging using ExM. Here, we report the development of label-retention ExM (LR-ExM) with a set of trifunctional anchors that not only prevent signal loss but also enable high-efficiency labeling using SNAP and CLIP tags. We have demonstrated multicolor LR-ExM for a variety of subcellular structures. Combining LR-ExM with superresolution stochastic optical reconstruction microscopy (STORM), we have achieved molecular resolution in the visualization of polyhedral lattice of clathrin-coated pits in situ.
Introduction
By physically expanding the sample before image acquisition, expansion microscopy (ExM) has enabled the use of a conventional confocal microscope to achieve ∼70-nm lateral spatial resolution (Chen et al., 2015; Chozinski et al., 2016; Ku et al., 2016; Tillberg et al., 2016). Recent efforts have further enhanced the resolution of ExM either by increasing the volume expansion ratio (Chang et al., 2017; M’Saad and Bewersdorf, 2020; Truckenbrodt et al., 2018) or by combining ExM with superresolution microscopy, such as structured illumination microscopy (SIM; Cahoon et al., 2017; Halpern et al., 2017; Wang et al., 2018) and stimulated emission depletion (STED) microscopy (Gambarotto et al., 2019; Gao et al., 2018; Li et al., 2018; Xu et al., 2019). In all of these cases, the homogenization of the specimen through either protease digestion (Chen et al., 2015; Chozinski et al., 2016; Tillberg et al., 2016) or protein denaturation (Ku et al., 2016; Tillberg et al., 2016) is essential to achieve isotropic expansion without structural distortion. To retain the spatial information of the target structures, the biomolecules of interest, such as protein (Chozinski et al., 2016; Ku et al., 2016; Tillberg et al., 2016) or RNA (Chen et al., 2016), and/or labels, such as dye-labeled DNA (Chen et al., 2015), dye-labeled antibodies (Chozinski et al., 2016; Tillberg et al., 2016), or fluorescent proteins (Ku et al., 2016; Tillberg et al., 2016), are anchored to the hydrogel matrix before digestion or denaturation. Nevertheless, digestion and denaturation cause incompletely anchored proteins or protein fragments to be washed out (Chozinski et al., 2016; Chung et al., 2013; Yang et al., 2014), the polymerization to make the hydrogel produces free radicals that readily destroy fluorophores (Chen et al., 2015; Tillberg et al., 2016; Truckenbrodt et al., 2019), and both factors can damage fluorescent proteins. Consequently, >50% of the target molecules can lose labeling after expansion (Tillberg et al., 2016; Truckenbrodt et al., 2019), which is a major issue of current ExM methods. The label-loss problem is exacerbated when aiming for higher spatial resolution. First, ensuring isotropic expansion at the nanometer scale requires more thorough proteinase digestion or denaturation; hence, more labeled antibodies are washed out (Chen et al., 2015; Chozinski et al., 2016; Tillberg et al., 2016). Second, superresolution microscopy often requires certain dyes that do not survive hydrogel polymerization. For example, Alexa Fluor 647 (AF647), one of the best fluorophores for superresolution stochastic optical reconstruction microscopy (STORM) and photoactivated localization microscopy, suffers >90% loss of the fluorescent molecules after polymerization and digestion (Chozinski et al., 2016; Tillberg et al., 2016; Xu et al., 2019).
When the spatial resolution approaches the molecular scale, high-efficiency labeling of target molecules becomes a distinctly new requirement, as previously recognized in the development and application of superresolution microscopy (Hell et al., 2015; Kamiyama and Huang, 2012; Thevathasan et al., 2019). This requirement is because information from unlabeled target molecules is permanently lost. Although amplifying retained labels by amplifying DNA-barcoded antibody, such as immunosignal hybridization chain reaction (Lin et al., 2018) and immunostaining with signal amplification by exchange reaction using hybridization chain reaction (Saka et al., 2019), or using biotin (Chozinski et al., 2016; Xu et al., 2019) can significantly enhance the brightness, it cannot recover the lost positional information from washed-out antibodies. These methods result in bright but still incompletely labeled structures. Moreover, the bulky labels will introduce localization displacement that is detectable with the molecular resolution. On the other hand, postexpansion immunostaining can avoid dye loss during the gelation and digestion steps, such as magnified analysis of proteome (Ku et al., 2016), CUBIC-X (clear, unobstructed brain/body imaging cocktails and computational analysis-X; Murakami et al., 2018), and SHIELD (stabilization under harsh conditions via intramolecular epoxide linkages to prevent degradation; Park et al., 2018). Nevertheless, post-gel immunostaining can be problematic for certain targets such as neuroligin and GAP, because these proteins are damaged by denaturation and cannot be recognized by antibodies after expansion (Ku et al., 2016). The damage and loss of target proteins will cause underlabeling. In fact, the density of labeled targets fundamentally sets the lower limit of effective spatial resolution (Nieuwenhuizen et al., 2013; Shroff et al., 2008; Thevathasan et al., 2019).
With the combination of ExM and superresolution microscopy, fluorophores can theoretically be localized with the target proteins with molecular resolution. However, in practice, most ExM methods use indirect immunostaining to label target proteins. Although convenient, immunostaining ExM has two practical problems. First, high-quality antibodies are not always available. Second, bulky primary and secondary antibodies can add up to 20-nm distance between the target protein and the dye molecule or barcoding DNA strand (Chang et al., 2017), which is larger than the resolution of ExSTED and ExSTORM. Therefore, immunostaining is insufficient to reveal biological structures with molecular resolution. An effective solution is to express enzymatic protein tags, such as SNAP and CLIP tags, which can be recognized by highly specific and efficient fluorescent ligands (Gautier et al., 2008; Keppler et al., 2004; Thevathasan et al., 2019). These two tags are ∼2 nm long, much smaller than antibodies and even smaller than fluorescent proteins.
Herein, we report label-retention ExM (LR-ExM), a method that solves the signal loss problem with a set of small molecule trifunctional anchors that are inert to polymerization, digestion, and denaturation and that can be fluorescently labeled after expansion. We have developed this method not only for immunofluorescence labeling but also for SNAP and CLIP tags, which are particularly advantageous because of their small size and high labeling efficiency by organic dyes with tag-recognizing ligands.
Results
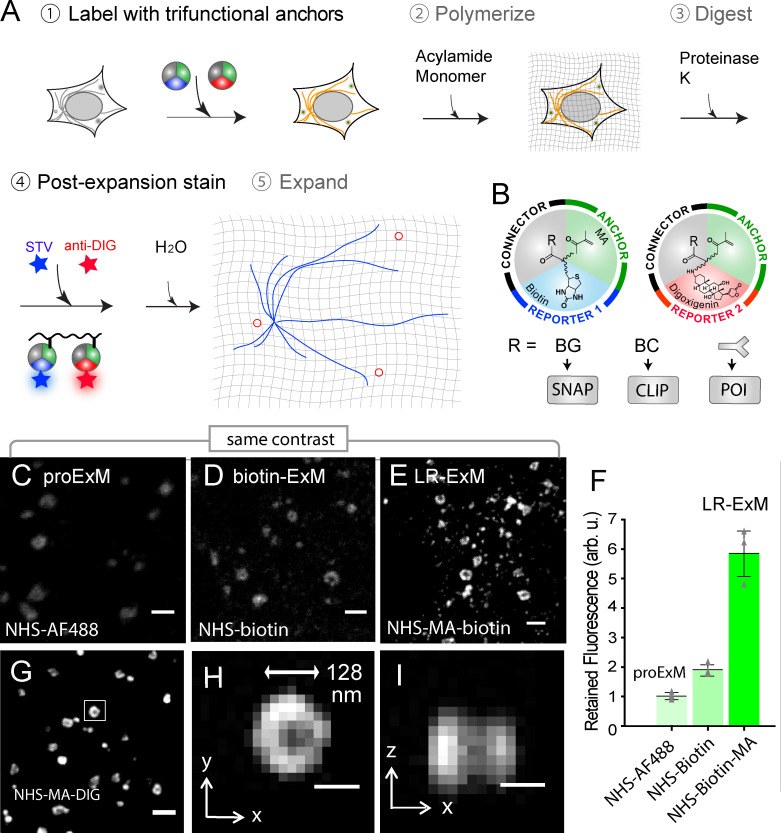
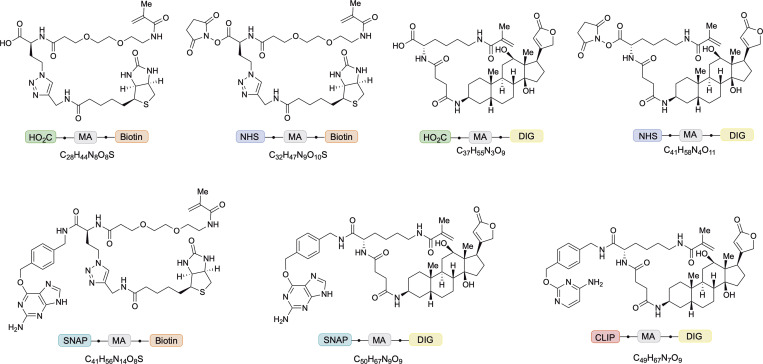
The workflow of LR-ExM includes five steps: labeling target molecules with trifunctional anchors, forming in situ hydrogel with cells or tissues, proteinase digestion, postexpansion fluorescence staining, and expansion (Fig. 1 A). Specifically, we designed and synthesized trifunctional anchors with three arms (Fig. 1 B): (1) N-hydroxysuccinimide (NHS) for connection to antibodies, benzylguanine (BG) for SNAP tag recognition, or benzylcytosine (BC) for CLIP tag recognition; (2) methacrylamide (MA) group for anchoring to the polymer matrix; (3) biotin or digoxigenin (DIG) as two orthogonal reporter handles for conjugation to an organic dye after expansion. We have chosen these functional groups and the molecular scaffold so that the anchors are resistant to both major causes of fluorescence loss, proteinase digestion and acrylamide gel polymerization. Therefore, target molecules labeled with the trifunctional anchors retain high labeling efficiency. The two orthogonal reporter handles enable two-color labeling and imaging. The structures and syntheses of the anchors are described in detail in Figs. 2 and S1 and Data S1.
Figure 1.
Workflow and characterization of LR-ExM.(A) Workflow of LR-ExM. (B) Schematic of trifunctional anchors. (C–E) ExM confocal images of CCPs in U2OS cells indirectly immunostained for clathrin heavy-chain (POI). (C) ProExM using AF488-conjugated secondary antibodies. (D) ExM with postexpansion labeling using biotin-conjugated antibodies. (E) LR-ExM using antibodies conjugated with NHS-MA-biotin trifunctional anchor. Samples in D and E were postexpansion stained with streptavidin-AF488. (F) Intensity quantification of C–E. Error bars represent SD. n = 3 for each case. (G) LR-ExM confocal image of CCPs in U2OS cells immunostained indirectly with secondary antibodies conjugated with NHS-MA-DIG anchor, postexpansion stained with anti-DIG antibody. (H and I) Cross sections of the CCP in the boxed area of G. The length expansion ratios for images in C, D,E, and G–I are 4.3, 4.5, 4.6, and 4.3, respectively. The length expansion ratio for the samples used in plot F is 4.5 ± 0.2. Scale bars, 500 nm (C–E and G) and 100 nm (H and I). All scale bars are in preexpansion units. arb. u., arbitrary units; STV, streptavidin.
Figure 2.
Structures of trifunctional anchors. HOOC/NHS-MA-biotin, HOOC/NHS-MA-DIG, BG-MA-biotin, BG-MA-DIG, and BC-MA-DIG.
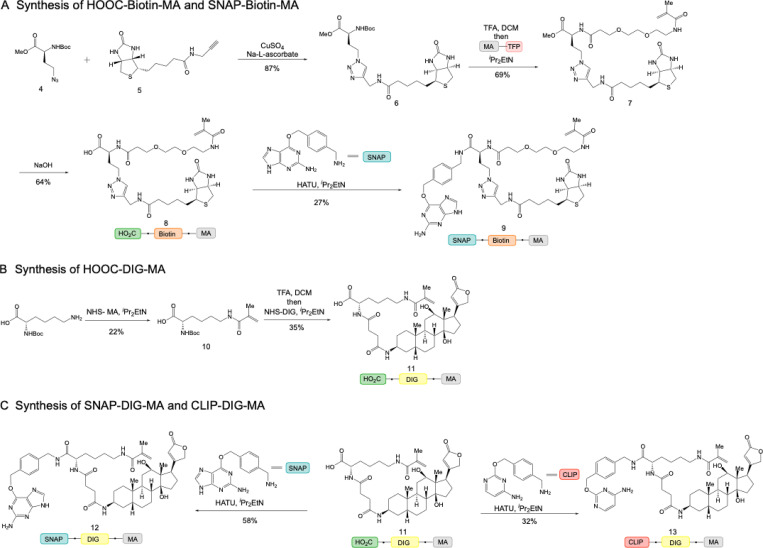
Figure S1.
Synthetic schemes of trifunctional anchors. (A) Synthetic routes of HOOC-biotin-MA and SNAP-biotin-MA. (B) Synthetic route of HOOC-DIG-MA. (C) Synthetic routes of SNAP-DIG-MA and CLIP-DIG-MA.
With different trifunctional anchors, LR-ExM is compatible with both immunofluorescent and enzymatic protein tags (Fig. 1 A). For immunofluorescence, we stained fixed cells with antibodies conjugated to NHS-MA-biotin or NHS-MA-DIG, proceeded with the standard ExM procedure of gel polymerization and proteinase K digestion, and then stained the gel with fluorescently labeled streptavidin for biotin anchor and/or anti-DIG antibody for DIG anchor before expanding the gel in water and imaging. For labeling with SNAP or CLIP tag, the procedure was nearly identical, except that fixed cells were directly treated with BG or BC trifunctional anchors.
To demonstrate label retention, we compared ExM of U2OS cells immunostained using secondary antibodies conjugated to AF488 dye (following the protocol in Tillberg et al., 2016), biotin (ExM followed by postexpansion staining with AF488-streptavidin; Chozinski et al., 2016; Xu et al., 2019), or NHS-MA-biotin (our LR-ExM). Fig. 1, C–E showcased the clathrin-coated pit (CCP) images (Fig. 1, C–E) processed with these three kinds of secondary antibodies, with the same contrast. We conjugated streptavidin with an average dye/protein ratio of 1:1 so that the fluorescence level in the three cases can be directly compared. On average, the LR-ExM generated a fluorescence signal 5.8 ± 0.8 (mean ± SD, n = 3) times higher than that generated by AF488 antibody (Fig. 1 F; for quantification methods, see Data S1 and Fig. S2). The biotin-antibody sample generated a fluorescence signal 1.9 ± 0.2 (mean ± SD, n = 3) times higher than that generated by protein-retention ExM (proExM). Taking these values together, we concluded that out of the ∼83% label loss by proExM, ∼15% can be attributed to polymerization and ∼68% to digestion. These numbers may be rescaled since the biotin may not be completely conserved. The fraction of fluorescence loss caused by digestion is dependent on the labeling tags such as antibody, streptavidin, and GFP, and is affected by the digestion condition such as digestion duration, buffer, and temperature. Differences between the protocols may lead to contradictory results. In this evaluation, we focused on IgG antibodies and followed the digestion condition of proExM (Tillberg et al., 2016). See Materials and methods and Fig. S2 for details of the procedure.
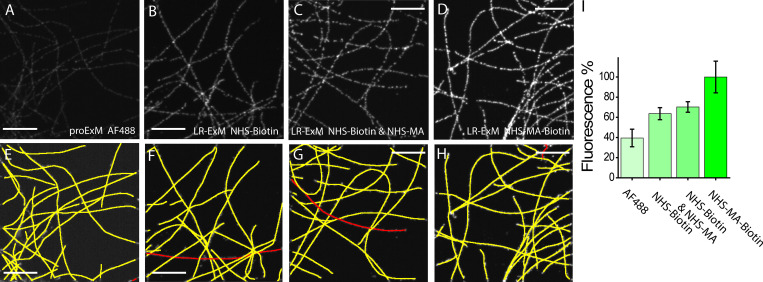
Figure S2.
Comparison of fluorescence intensities resulting from different ExM methods.(A–D) Images of microtubules prepared with proExM with AF488-labeled secondary antibody (A), biotin-ExM with the biotin-NHS–labeled secondary antibody (B), LR-ExM with biotin-NHS and MA-NHS–colabeled secondary antibody (C), and LR-ExM with biotin-MA-NHS–labeled secondary antibody (D). Images A–D have the same contrast to show the relative brightness of the stain achieved in each case. Samples were processed side by side with the same immunostaining, digestion, and imaging conditions. (E–H) The microtubules in A–D are tracked and marked in yellow and red in E–H by a Fiji plugin JFilament, respectively. (I) Histogram of retained fluorescence represented in A–D. n = 3 for each case. The retained fluorescence was normalized by the total length of microtubules in each image and the ratios of AF488/Ab, biotin/Ab, and AF488/streptavidin. All samples were digested by incubating in 8 U/ml proteinase K for 16 h at room temperature. Scale bars, 2 µm.
We calibrated the length expansion ratio of our LR-ExM protocol to range from 4.3 to 4.7. Fitting the cross-section profiles of microtubules to a Gaussian peak then gave a full width at half maximum (FWHM) of 84 ± 1.3 nm (mean ± SD, n = 3 independent experiments) after rescaling by the expansion ratio. Taking the size of immunostained microtubules (Dempsey et al., 2011) into the cross-section profile for fitting (Chen et al., 2015; Olivier et al., 2013), we obtained an effective resolution of 71 ± 1.6 nm. At this effective resolution, we could resolve the hollow shape of CCPs (Fig. 1, H and I). Using an anti-DIG antibody with a high dye/protein ratio (10:1) produces a brighter signal (Fig. 1 G), though we do not expect obvious differences in the actual labeling efficiency.
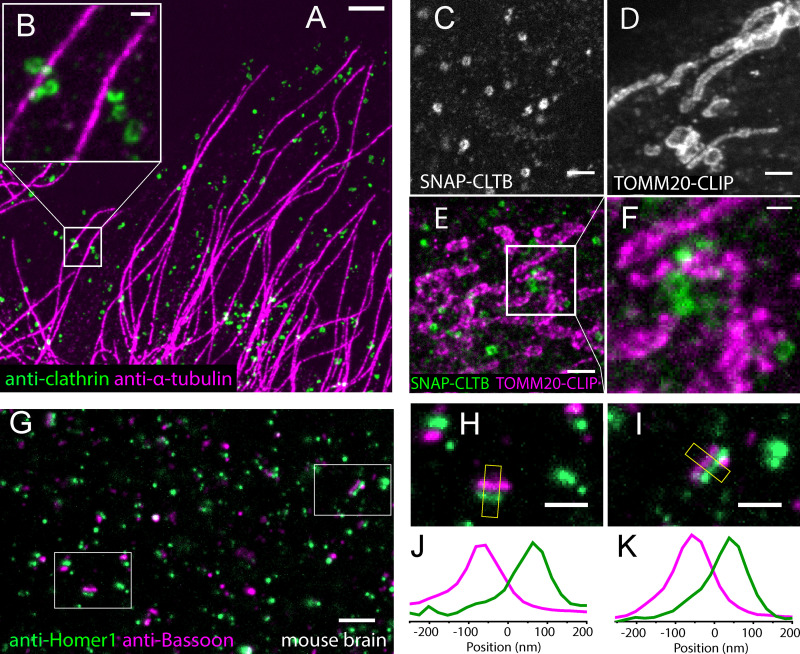
We demonstrated two-color LR-ExM by coimmunostaining CCPs and microtubules with antibodies conjugated with NHS-MA-DIG and NHS-MA-biotin, respectively (Fig. 3, A and B). Similarly, we demonstrated LR-ExM for SNAP tag and CLIP tag with BG-MA-biotin and BC-MA-DIG trifunctional anchors, respectively (see Fig. 3 C for CCPs by overexpressing SNAP-clathrin and Fig. 3 D for mitochondria by TOMM20-CLIP), including two-color imaging using both enzymatic protein tags owing to their orthogonality (Fig. 3, E and F). The above methods are compatible with brain tissue. We immunostained mouse brain slices for the presynaptic and postsynaptic markers Bassoon and Homer1 using secondary antibodies conjugated with NHS-MA-biotin and NHS-MA-DIG and then treated these slices with digestion, postexpansion staining, and expansion (Fig. 3 G). Presynaptic and postsynaptic densities were well separated, and junctions between them were clearly observable with a confocal microscope (Fig. 3, H–K).
Figure 3.
Two-color LR-ExM images using immunostaining and protein tag approaches.(A) Two-color LR-ExM confocal image of microtubules labeled with NHS-MA-biotin–conjugated secondary antibodies (magenta) and CCPs labeled with NHS-MA-DIG–conjugated secondary antibodies (green) in a U2OS cell. (B) Magnified view. (C–E) LR-ExM confocal images of CCPs and/or mitochondria in HeLa cells labeled using SNAP tag–labeled clathrin (C), CLIP tag–labeled TOMM20 (D), and two-color imaging (E). (F) Magnified view. (G) LR-ExM confocal image of mouse brain slice indirectly immunostained for the presynaptic marker Bassoon (magenta) and the postsynaptic marker Homer1 (green). (H and I) Zoomed-in images of synapses. (J and K) Transverse intensity profiles along the yellow box long axes. Bassoon is labeled with NHS-MA-DIG–conjugated secondary antibodies, and Homer1 is labeled with NHS-MA-biotin–conjugated secondary antibodies. All samples are postexpansion stained with streptavindin-AF488 and or anti-Digoxin-AF594. The length expansion ratios for images in A and B; C; D; E and F; and G–I are 4.7, 4.4, 4.4, 4.5, and 4.2, respectively. Scale bars, 1 µm (A and G), 200 nm (B, F, H, and I), and 500 nm (C–E). All scale bars are in preexpansion units.
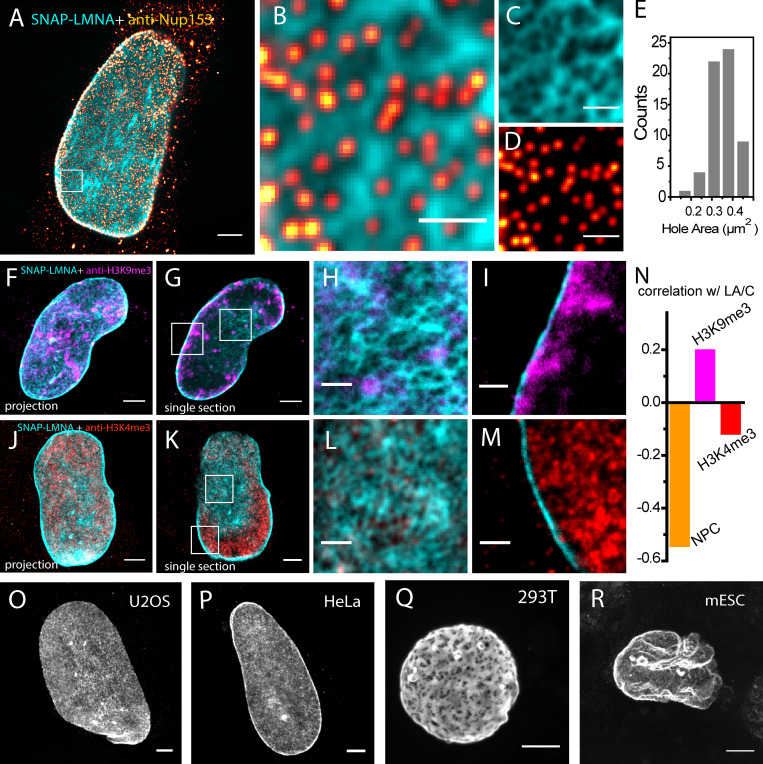
The enzymatic and immunostaining LR-ExM approaches can be combined easily. As a demonstration, we imaged nuclear lamina with SNAP tag–labeled lamin A/C and antibody-stained nuclear pore complex (NPC; Fig. 4, A–D). Nuclear lamina is a dense fibrillar network bridging the nuclear envelope and chromatin. It positions the NPCs (Turgay et al., 2017) and participates in chromatin organization (Shumaker et al., 2003; Zheng et al., 2018). Two-color LR-ExM cleanly resolved the holes in the nuclear lamina where NPCs are located. The high labeling efficiency of enzymatic protein tags was the key to achieving superior image quality compared with previous superresolution microscopy results using antibody staining (Shimi et al., 2015). The area of the holes in the lamin A/C network varies from 0.1 to 0.5 µm2, with an average of 0.35 µm2 (Fig. 4 E), which is in agreement with previous SIM studies (Shimi et al., 2015) and EM (Turgay et al., 2017). We observed a strong anticorrelation between lamin A/C and NPC signal (Fig. 4, A and B).
Figure 4.
LR-ExM reveals subcellular protein organizations.(A) Two-color confocal LR-ExM of SNAP-tagged lamin A/C (cyan) and immunostained NPC (red hot) of a HeLa cell. (B) Magnified view. (C and D) Views of individual channels of B. Note the cytoplasmic background in A is caused by the anti-NUP153 antibody. (E) Histogram of lamin hole area in the boxed region. (F–I) Two-color confocal LR-ExM of SNAP-tagged lamin A/C (cyan) and immunostained H3K9me3 (magenta) of a HeLa cell, with a maximum intensity project of a z stack covering the bottom half of the nucleus (F), a single section of the nucleus (G), and magnified views of the boxed regions in G (H and I). (J–M) Two-color confocal LR-ExM of SNAP-tagged lamin A/C (cyan) and immunostained H3K4me3 (red), with a maximum intensity projection of a z stack covering the bottom half of the nucleus (J), a single section of the nucleus (K), and magnified views (L and M) of the boxed regions in K. (N) Correlation coefficients of NPC with lamin A/C, H3K9me3 with lamin A/C, and H3K4me3 with lamin A/C. (O–R) Confocal LR-ExM of lamin A/C in U2OS (O), HeLa (P), HEK 293T (Q), and mouse embryonic stem cells (mESC; R) showing maximum intensity projections over the bottom half of a nucleus. The length expansion ratios for images in A–D, F–I, J–M, O, P, Q, and R are 4.5, 4.5, 4.3, 4.2, 4.3, 4.6, and 4.4, respectively. Scale bars, 2 µm (A, F, G, J, K, and O–R) and 500 nm (B–D, H, I, L, and M). All scale bars are in preexpansion units.
We further characterized the spatial relationship between the lamin A/C network and two different chromatin markers, H3K9me3 for repressed chromatin (Fig. 4, C–I; Nakayama et al., 2001) and H3K4me3 for active chromatin (Fig. 4, D–M; Liang et al., 2004). With the enhanced spatial resolution, two-color LR-ExM images clearly showed that near the nuclear envelope, H3K9me3 was concentrated near lamin A/C–rich regions, whereas H3K4me3 had an anticorrelation with lamin A/C signal in a similar manner as NPC (Fig. 4 N). This result agrees with the model for lamin’s association with heterochromatin and the NPC’s association with euchromatin at the nuclear periphery. We were also able to resolve the distinct fine network features of lamin A/C in a variety of cell lines including mouse embryonic stem cells (Fig. 4, O–R). All of these results illustrate the application of LR-ExM (potentially in conjugation with fluorescence in situ hybridization to visualize DNA) in studying chromatin organization and subdiffraction limit-sized chromatin domains such as lamina-associated domains.
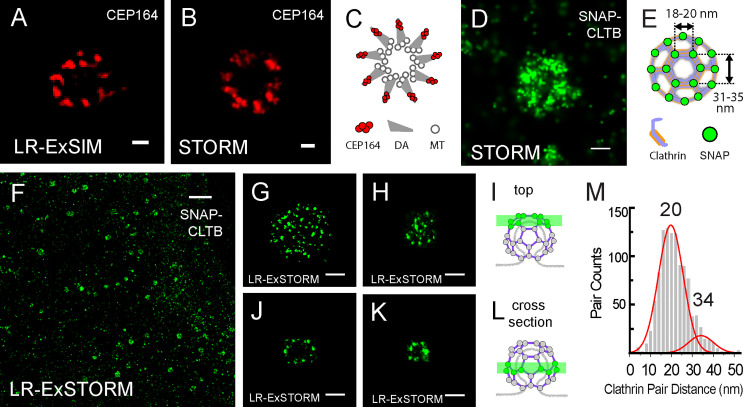
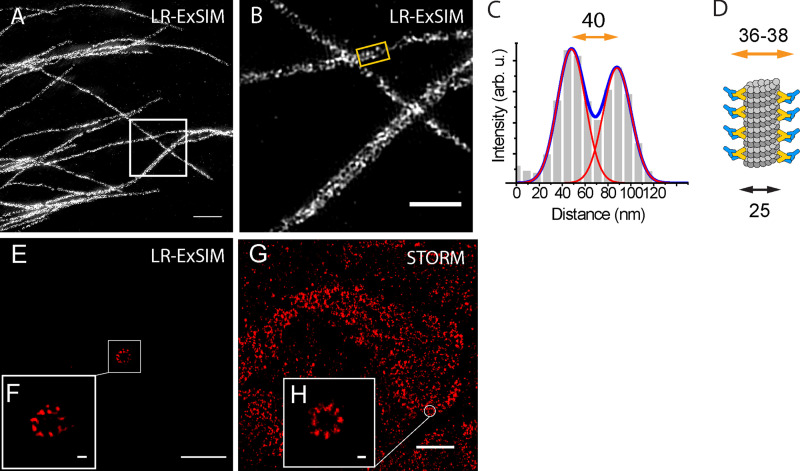
The high label retention of trifunctional anchors and high labeling efficiency of enzymatic tags in LR-ExM enhances its combination with superresolution microscopy. We first demonstrated this application by imaging antibody-stained distal appendages in primary cilia of retinal pigment epithelium cells using LR-ExM combined with SIM (LR-ExSIM; Fig. 5 A). The LR-ExSIM image clearly showed nine clusters of distal appendage marker CEP164, achieving a resolution and image quality similar to the STORM image of the same target protein (Fig. 5 B). The symmetry and size of distal appendages measured using LR-ExM agreed with the model based on STED (Lau et al., 2012) and STORM studies (Shi et al., 2017; Fig. 3 C). We calculated the resolution (FWHM) of LR-ExSIM to be 34 nm.
Figure 5.
LR-ExSIM and LR-ExSTORM reveal subcellular protein organization.(A) LR-ExSIM image of Cep164 in distal appendages of a primary cilium indirectly immunostained with NHS-MA-biotin secondary antibodies. (B) STORM image of Cep164 distal appendages of unexpanded cilium. (C) Schematic of the structure of distal appendages (DA) of the primary cilium. MT, microtubule. (D) STORM image of an unexpanded HeLa cell overexpressing SNAP-CLTB and stained with BG-AF647. (E) Schematic of the structure of a CCP with SNAP tag–labeled CLTB. (F) LR-ExSTORM image of a HeLa cell overexpressing SNAP-CLTB, stained with BG-MA-biotin, and postexpansion labeled with streptavidin-AF647. (G and H) Images of x-y cross sections at the top of single CCPs as illustrated in I. (J–L) Images of x-y cross sections in the middle of single CCPs (J and K) as illustrated in L. Images in G–K are different CCPs. (M) Nearest cluster distance analysis of 134 CCPs imaged with LR-STORM. The length expansion ratios for images in A, F, G, H, J, and K are 4.2, 3.3, 3.3, 3.1, 3.1, and 3.1, respectively. The length expansion ratio for samples used in plot M is 3.2 ± 0.2. Scale bars, 100 nm (A, B, and G–K), 200 nm (D), and 2 µm (F). All scale bars in LR-ExM images are in preexpansion units.
Finally, we pushed forward the resolution to the molecular level by combining LR-ExM with STORM (LR-ExSTORM). We have examined commonly used photoswitchable dyes, including Cy5, Cy5.5, and AF647, all of which show no loss in either molecular brightness or photoswitching kinetics compared with nonexpansion STORM conditions (Dempsey et al., 2011). The length expansion ratio for LR-ExSTORM ranges from 3 to 3.3. It is smaller than four because the STORM mounting medium for dye photoswitching has a relatively high ionic strength. In U2OS cells expressing SNAP tag–labeled clathrin light chain B (CLTB), we demonstrated that LR-ExSTORM revealed far more details than STORM alone (Fig. 5, D–M). While STORM showed the hollow structure of the CCP (Fig. 5 D), LR-ExSTORM was able to resolve the lattice vertices of CCPs as clusters of localization points, each cluster resulting from repetitive photoswitching of one AF647 label (Fig. 5, G–K). The effective localization precision of LR-ExSTORM was measured to be ∼4 nm (FHWM of clusters rescaled by the expansion ratio), which is comparable to the size of a typical protein molecule.
At such a small scale, localization precision is not the only parameter to determine image resolution. The distortion of hydrogel at nanoscale needs to be considered. In addition, the size and orientation of the label also add uncertainty to the measurement. To evaluate this uncertainty, we compared the unit length of clathrin lattice measured by ExSTORM and EM. In ExSTORM images focused on the top of CCPs, where the lattice plane is horizontal (e.g., Fig. 5, G and H), we measured distances from the centroid of one cluster to the centroid of its nearest neighbor. The histogram of these nearest-neighbor distances (1,102 pairs from 134 CCPs) showed a main peak at 20 nm and a small shoulder peak at 34 nm (Fig. 5 M). The main peak matched the distance between adjacent vertices of clathrin lattice, as previously measured by EM (Jin and Nossal, 2000; Kirchhausen et al., 2014), while the shoulder peak matched the distance between every other vertex (Fig. 5 E). This agreement confirmed the ability of LR-ExSTORM to faithfully expand protein complexes at the 10–20-nm scale, possibly attributed to the high degree of isotropic expansion. The 6-nm standard deviation of the histogram is contributed by polyacrylamide gel distortion, SNAP tag, and heterogeneity in the clathrin lattice. Moreover, the histogram also indicates that our labeling efficiency has resulted in a majority of vertices containing at least one labeled CLTB, noting that not all clathrin light chains had SNAP tag in our case because of the presence of endogenous CLTB and the other clathrin light chain isoform, CLTA. More ExSIM images of microtubules and ciliary distal appendages are shown in Fig. S4.
Figure S4.
LR-ExSIM of microtubules.(A) LR-ExSIM image of microtubules in a U2OS cell stained with antibody conjugated with NHS-MA-DIG anchors. (B) Magnification of A. (C) The transverse profile of the microtubule in the gold box in B. (D) Schematic of the structure of an immunostained microtubule. By fitting the peaks to Gaussian functions, we calculated the resolution (FWHM) of LR-ExSIM to be 34 nm. (E) LR-ExSIM image of Cep164 in distal appendages of a primary cilium of an expanded mouse embryonic fibroblast indirectly immunostained with NHS-MA-biotin secondary antibodies. The length expansion ratio is 4.2. (F) Magnified view of E. (G) STORM image of Cep164 in distal appendages of motile cilia of an unexpanded multiciliated mouse tracheal epithelial cell. (H) Magnified view of G. LR-ExSIM (E and F) and STORM (G and H) reveal structure of distal appendages with similar super resolution. The same primary antibody was used for both images. Scale bars, 1 µm (A), 500 nm (B), 2 µm (E and G), and 100 nm (F and H).
Discussion
We developed LR-ExM using trifunctional anchors, which eliminate the fluorescence loss caused by both polymerization and digestion and therefore improve the fluorescence intensity severalfold. Paring the trifunctional anchors with SNAP and CLIP tags, LR-ExM achieved high labeling efficiency. It paves the way for the combination of ExM and superresolution microscopy, which elevates the resolution of optical microscopy to the molecular scale. For such high resolution (<10 nm), high labeling efficiency of individual target proteins rather than overall brightness is required to obtain complete molecular architecture of protein complexes. This requirement is much more stringent compared with diffraction limited imaging. The high labeling efficiency is a major advantage of LR-ExM in combination with superresolution microscopy, such as STORM, photoactivated localization microscopy, STED microscopy, and SIM.
There are other methods that can efficiently recover the fluorescent signal lost in polymerization reactions. For example, immunosignal hybridization chain reaction (Lin et al., 2018) and immunostaining with signal amplification by exchange reaction using hybridization chain reaction (Saka et al., 2019) increase fluorescence by two to three orders of magnitude, which remarkably boosts the detection sensitivity of tissue imaging. However, these methods are unable to rescue the information lost from antibodies that are washed away during the digestion step, resulting in incompletely but still brightly labeled structures. Moreover, the bulky labels introduce localization displacement that is detectable with the molecular resolution. Compared with these label amplification methods, LR-ExM not only avoids polymerization-induced fluorophore damage by postexpansion labeling but also eliminates digestion-induced fluorophore loss by directly cross-linking the reporter to the hydrogel. Wen and coworkers presented a similar chemical linking approach that enables direct grafting of a targeting molecule and fluorophore to the hydrogel and demonstrated that the design principle of trifunctional anchors also works for antibody-free staining of small biomolecules like lipid and actin in ExM (Wen et al., 2020).
To address the limitations of immunostaining, we developed the enzymatic approach of LR-ExM using SNAP tag, CLIP tag, and trifunctional anchors bearing BG or BC as the connector group. This approach is ideal for molecular-resolution microscopy with the advantages of the high ligand-binding efficiency of SNAP tag or CLIP tag and their much smaller size compared with antibodies (19 kD for SNAP vs. 150 kD for IgG). For the same reasons, cell lines with SNAP-tagged nuclear pore protein are used to benchmark the quality of superresolution microscopes (Thevathasan et al., 2019). Compared with diffraction limited imaging, molecular-resolution imaging has more stringent requirements for small labels. LR-ExM minimizes the size of the label by using not only small enzymatic tags but also short trifunctional anchors only ∼1 nm long. We demonstrated the resolving power of LR-ExSTORM by visualizing the polyhedral lattice of CCPs in situ (Fig. 5, F–M), which is not resolvable using STORM alone (Fig. 5 D; Bates et al., 2008; Huang et al., 2008). While using SNAP and CLIP tags can improve resolution by reducing the tag size and provide high labeling efficiency, it does require genetic approaches to fuse the tags to the targeted proteins. Extra attention needs to be paid to the expression level of enzymatic tags, as overexpression may cause structural or functional artifacts. We recommend keeping the expression level similar to the endogenous level of the target protein.
Another advantage of LR-ExM is robustness. Most ExM protocols consist of nonspecific protein-hydrogel anchoring with chemical cross-linkers (e.g., MA-NHS and glutaraldehyde) and proteinase digestion (Cahoon et al., 2017; Chang et al., 2017; Chozinski et al., 2016; Gambarotto et al., 2019; Gao et al., 2018; Halpern et al., 2017; Li et al., 2018; Lin et al., 2018; Saka et al., 2019; Tillberg et al., 2016; Truckenbrodt et al., 2018; Wang et al., 2018; Xu et al., 2019). How much fluorescence is retained depends on the balance of anchoring density and digestion efficiency, resulting in largely variable fluorescence intensity across experiments (Chozinski et al., 2016; Ku et al., 2016; Truckenbrodt et al., 2019). Higher anchoring density increases fluorescence retention, while stronger digestion duration decreases fluorescence retention. However, the fluorescence retention rate of LR-ExM is independent from digestion conditions, since the reporter is directly anchored to the hydrogel, not through antibodies. Consequently, the LR-ExM protocol is robust and reproducible across experiments.
LR-ExM is a versatile method that can be integrated with ExM protocols targeting nucleic acids (e.g., expansion fish fluorescent in situ hybridization; Chen et al., 2016), lipids (e.g., membrane ExM; Karagiannis et al., 2019 Preprint; or trivalent anchoring; Wen et al., 2020), or context proteins (e.g., fluorescent labeling of abundant reactive entities; Mao et al., 2020; or pan-ExM; M'Saad and Bewersdorf, 2020). Although only two-color LR-ExM using biotin- and DIG-bearing trifunctional anchors was demonstrated, additional color channels were imaged by postexpansion labeling DNA with DAPI and postexpansion labeling telomere with locked nucleic acid oligonucleotides (data not shown). The multiplexity can be further extended by developing new trifunctional anchors with more small chemical reporters, such as alkyne (pairing with azide) and chloroalkane (pairing with HaloTag).
In summary, LR-ExM is an effective, robust, and versatile method to enhance the signal and labeling efficiency of expansion microscopy. Our trifunctional anchors can be applied to both antibody and enzymatic labeling. They can also be integrated into most existing ExM protocols, greatly increasing their signals and multiplexity. Overcoming the bottleneck of label loss, the currently achieved postexpansion resolutions of ∼70 nm with confocal microscopy, ∼30 nm with SIM, and localization precision of ∼4 nm with STORM are suitable to cover a wide range of biological questions at various scales.
Materials and methods
Trifunctional anchors
We synthesized five trifunctional anchors, including HOOC-MA-biotin, HOOC-MA-DIG, BG-MA-biotin, BG-MA-DIG, and BC-MA-DIG (Fig. 2). HOOC-MA-biotin and HOOC-MA-DIG anchors were converted to NHS-MA-biotin and NHS-MA-DIG, respectively, to conjugate antibodies for the immunostaining approach of LR-ExM. The BG-MA-biotin, BG-MA-DIG, and BC-MA-DIG anchors were directly used for the protein tag approach of LR-ExM. The synthetic schemes are shown in Fig. S1.
All reactions were performed in flame- or oven-dried glassware fitted with rubber septa under a positive pressure of nitrogen, unless otherwise noted. All reaction mixtures were stirred throughout the course of each procedure using Teflon-coated magnetic stir bars. Air- and moisture-sensitive liquids were transferred via syringe. Solutions were concentrated by rotary evaporation <30°C. Analytical TLC was performed using glass plates precoated with silica gel (Silicycle; 0.25 mm, 60-Å pore size, 230−400 mesh) impregnated with a fluorescent indicator (254 nm). TLC plates were visualized by exposure to UV light and then stained by submersion in a basic aqueous solution of potassium permanganate or with an acidic ethanolic solution of anisaldehyde, followed by brief heating. For synthetic procedures and characterization data (TLC, nuclear magnetic resonance [NMR], and mass spectroscopy), see “Reagents for trifunctional anchor synthesis” at the end of Materials and methods. Samples of trifunctional anchors described in this paper are available upon request.
Antibodies
The following primary antibodies were used for the immunostaining approach of LR-ExM: rabbit anti-clathrin heavy-chain antibody (Abcam; ab21679); rat anti–α-tubulin antibody, tyrosinated, clone YL1/2 (Millipore Sigma; MAB1864-I); monoclonal mouse anti-Nup153 antibody (Abcam; ab24700); anti-H3K9me3 (Abcam; ab176916); and anti-H3K4me3 (Abcam; ab8580). The secondary antibodies used for trifunctional anchor conjugation were unconjugated AffiniPure Donkey Anti-Rabbit IgG (Jackson ImmunoResearch; H+L, 711–005-152) and unconjugated AffiniPure Donkey Anti-Rat IgG (Jackson ImmunoResearch; H+L, 712–005-153).
Antibody conjugation
Secondary antibodies were labeled with the amine-reactive trifunctional anchor NHS-MA-biotin or NHS-MA-DIG. Amine-reactive trifunctional anchors were freshly made from stock solutions of synthesized trifunctional anchors HOOC-MA-biotin or HOOC-MA-DIG (26 mM in DMSO, stored at −20°C). 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide in solution (20 µl, 20 mg/ml in DMSO), NHS solution (20 µl, 15 mg/ml in DMSO), and N,N-Diisopropylethylamine solution (10 µl, 70 mg/ml in DMSO) were sequentially added into the solution of HOOC-MA-biotin or HOOC-MA-DIG (50 µl, 26 mM). The mixture was gently shaken at room temperature for 16 h while shielding from light with aluminum foil. Using the aforementioned volumes, the final concentration of in situ–prepared NHS-MA-biotin or NHS-MA-DIG is 13 mM.
To conjugate the secondary antibodies with the amine-reactive trifunctional anchor, the following procedure was performed: 10 µl aqueous NaHCO3 solution (1 M) was added to an Eppendorf tube containing 80 µl unconjugated antibody solution (1 mg/ml). A solution of the amine-reactive trifunctional anchor (NHS-MA-biotin or NHS-MA-DIG, 13 mM, 24 µl) was then added to the NaHCO3-buffered antibody solution. The labeling reaction mixture was gently rocked for 20 min at room temperature. During the reaction, a Sephadex G-25 column (GE Healthcare; NAP-5) was equilibrated with PBS (pH 7.4). The labeling reaction mixture was loaded onto the column, followed by flowing with 650 µl PBS. The antibodies conjugated with trifunctional anchors were collected by eluting the column with another 250 µl PBS, and stored at 4°C.
The procedure used for antibody conjugation with commercially available bifunctional linker NHS-biotin was the same as that used for conjugation with trifunctional anchors, except that a solution of NHS-biotin (26 mM, 4 µl) instead of the trifunctional anchor was added to the NaHCO3-buffered antibody solution.
Quantification of the biotin to antibody ratio
Antibody concentration was characterized by measuring the absorption at 280 nm with a UV-Vis spectrophotometer. The concentration of biotin was measured using a HABA/Avidin reagent kit, following the protocol provided by the supplier (Thermo Fisher Scientific/Pierce; Biotin Quantitation Kit #28005). The biotin to antibody ratios of the antibody conjugated with NHS-biotin in Fig. 1 D (Fig. S2 B), the antibody conjugated with NHS-MA-biotin in Fig. 1 E (Fig. S2 D), and the antibody conjugated with NHS-biotin and NHS-MA in Fig. S2 C are 12.1, 8.6, and 9.9, respectively. The dye to antibody ratio of the antibody conjugated with AF488 in Fig. 1 C (Fig. S2 A) is 8.9. These biotin to antibody ratios and the dye to antibody ratio are used to normalize the label-retention efficiency of proExM, biotin-ExM, and LR-ExM (Figs. 1 F and S2 I).
Cell culture
U2OS cells (ATCC; HTB-96) were cultured in McCoy's 5a (ATCC; 30–2007) supplemented with 10% FBS at 37°C in 5% CO2. HeLa (ATCC; CCL-2) and HEK 293T (CRL-3216) cells were cultured in DMEM-Glutamax (Thermo Fisher Scientific) supplemented with 10% FBS at 37°C in 5% CO2. U2OS, HeLa, and HEK 293T cells were seeded at 104 cells/cm2 in 16-well chambers (Grace Bio-Labs; 112358) and grown to 75% confluency.
Human retinal epithelial (ATCC; RPE1-hTERT) cells were cultured in DMEM/F12 supplemented with 10% FBS at 37°C in 5% CO2. Cells were plated on 16-well chambered slides coated with 0.1% gelatin (Sigma-Aldrich; G1393) at 104 cells/cm2 per well and serum starved in OptiMEM reduced serum media for 24 h to induce ciliation.
Cell lines were not authenticated. No commonly misidentified cell lines were used. All growing cell lines were routinely tested for mycoplasma.
Molecular cloning
To generate the pTOMM20-N-10-CLIPf mammalian expression plasmid, the DNA of CLIP tag was PCR amplified from pCLIPf vector (plasmid source: the Michael Davidson Fluorescent Protein Collection at the University of California, San Francisco (UCSF) Nikon Imaging Center) using primers (forward: 5′-GCGGGGATCCACCGGTCGCCACCATGGACAAAGACTGCGAAATGAAGC-3′; reverse: 5′-TCTAGAGTCGCGGCCGCTTAACCCAGCCCAGGCTTGCCC-3′). We then performed restriction enzyme digestion on vector pmEmerald-TOMM20-N-10 (plasmid source: the Michael Davidson Fluorescent Protein Collection at the UCSF Nikon Imaging Center), cutting out the mEmerald sequence between BamHI and NotI. The PCR-amplified CLIP tag was then ligated with the digested vectors using the In-Fusion HD Cloning kit (Clontech). The plasmids pSNAPf-Clathrin-15 and pSNAPf-LMNA were directly purchased from UCSF Nikon Imaging Center. For constructing the lentiviral production vectors, DNAs of TOMM20-N-10-CLIPf and SNAPf-Clathrin-15 were directly PCR amplified from mammalian expression constructs and subcloned into lentiviral pHR-SFFV vector (BamHI/NotI) using the In-Fusion HD Cloning kit (Clontech).
Immunostaining
For microtubule immunostaining and microtubule-clathrin coimmunostaining, cells were fixed with 3.2% PFA in PEM buffer (100 mM Pipes, 1 mM EGTA, and 1 mM MgCl2, pH 6.9) at room temperature for 10 min. The fixation was reduced with 0.1% sodium borohydride in PBS for 7 min. Sodium borohydride was removed by washing with PBS three times with 5-min incubation between washes. The fixed cells were incubated in blocking buffer (PBS, 3% BSA, and 0.1% Triton X-100) for 30 min at room temperature. Primary antibodies at a concentration of 2 µg/ml were added to the fixed cells in blocking buffer for 16 h at 4°C. The primary antibodies used for this paper are listed in Data S1. After, primary antibody incubation, the cells were washed with PBS three times with 5 min of incubation between washes. Secondary antibodies conjugated with trifunctional anchors were added at a concentration of 3 µg/ml and incubated for 1 h in blocking buffer on an orbital shaker. The secondary antibodies were removed by three washes with PBS buffer.
For CEP164 immunostaining, cells were fixed with 100% cold methanol for 3 min, incubated in blocking buffer (2.5% BSA and 0.1% Triton X-100 in PBS) for 1 h at room temperature, and then incubated with primary antibody in blocking buffer (Santa Cruz Biotechnology; 1:100 dilution of goat anti-CEP164, sc-240226) overnight at 4°C. Cells were washed with PBS four times with 5 min of incubation between washes and then incubated with donkey anti-goat secondary antibody conjugated with NHS-MA-biotin anchors for 1 h at room temperature. Secondary antibodies were removed by three washes with PBS buffer.
For mouse brain tissue immunostaining, wild-type adult mouse brain was fixed in 4% PFA overnight, before transferring to 30% sucrose and then optimal cutting temperature compound for cryoprotection. Tissue block was sectioned coronally at 20 µm thick and incubated in blocking buffer (10% goat serum, 3% BSA, 1% glycine, 0.4% Triton X-100 in TBS) for 1 h at room temperature. Tissue slices were then stained with primary antibodies (1:500 dilution of mouse anti-Bassoon, VAM-PS003 from StressGene and 1:200 dilution of rabbit anti-Homer1, 160003 from Synaptic Systems) in blocking buffer overnight at room temperature. Slices were washed with TBS three times for 5 min each time and then incubated with donkey anti-mouse (conjugated with NHS-MA-DIG) and donkey anti-rabbit (conjugated with NHS-MA-biotin) secondary antibodies (both at 1:100 dilution in blocking buffer) for 2 h at room temperature.
SNAP and CLIP tag labeling
The cells expressing SNAP tag and or CLIP tag were fixed for 10 min with 4% PFA in PBS buffer. The PFA was removed by a PBS wash. Fixed cells were incubated in blocking buffer (PBS, 3% BSA, and 0.1% Triton X-100) for 30 min at room temperature. Cells were then incubated in 3 µM trifunctional anchor SNAP-MA-biotin and/or 5 µM CLIP-MA-DIG for 1 h.
Polymerization, proteinase digestion, postexpansion labeling, and expansion
The polymerization and proteinase digestion steps are similar to the proExM protocol (Chozinski et al., 2016; Tillberg et al., 2016), with two exceptions: (1) we also treated the sample with DNase I before polymerization to fragment the genomic DNA, with the intention to reduce potential distortions inside and around the nucleus; and (2) protein anchoring with MA NHS ester or glutaraldehyde is not necessary but optional.
Specifically, fixed cells were incubated in DNase I buffer (New England Biolabs; M0303S, 1:100 dilution in PBS buffer) for 30 min at 37°C and then were polymerized in a mixture of monomer solution (8.6 g sodium acrylate, 2.5 g acrylamide, 0.15 g N,N′-methylenebisacrylamide, 11.7 g sodium chloride per 100 ml PBS buffer), N,N,N′,N′-Tetramethylethylenediamine (final concentration 0.2% [wt/wt]), and ammonium persulfate (final concentration 0.2% [wt/wt]) for 1 h at 37°C. The gel was then digested with proteinase K (Sigma-Aldrich; P4850-5ML) at a final concentration of 8 U/ml in digestion buffer (50 mM Tris, pH 8.0, 1 mM EDTA, 0.5% Triton X-100, and 0.8 M guanidine HCl) for 18 h at room temperature or 4 h at 37°C. After digestion, proteinase K was removed by four washes with excessive water (30 min each time). To introduce fluorophores to the trifunctional anchors on the target cellular structures, we incubated the hydrogel in 2 µg/ml AF488-labeled streptavidin (Jackson ImmunoResearch Laboratories; 0165400084) and/or DyLight 594–labeled anti-DIG/Digoxin (DIG; Vector Laboratories; DI-7594) in Hepes buffer (10 mM Hepes and 15 mM NaCl, pH 7.5) for 24 h. For LR-ExSTORM and LR-SIM, AF647 streptavidin (Jackson ImmunoResearch Laboratories; 0160600084) was used for postexpansion staining. The postexpansion labeled hydrogel was then washed and expanded by four washes with excessive water (at least 30 min each time). It is optional to treat the cells with 25 mM methacrylic acid NHS ester for 1 h before polymerization.
The length expansion ratio of the LR-ExM protocol is determined by measuring the diameters of the gel before and after expansion with calipers.
STORM image acquisition and analysis
STORM was performed on a custom-built microscope based on a Nikon Ti-U inverted microscope. Two lasers (Coherent CUBE 405 and CUBE 642) were combined using dichroic mirrors, aligned, expanded and focused to the back focal plane of the objective (Nikon Plan Apo 100× oil NA 1.45). The lasers were controlled directly by the computer. A quad band dichroic mirror (zt405/488/561/640rpc, Chroma) and a band-pass filter (ET705/70m, Chroma) separated the fluorescence emission from the excitation light. During image acquisition, the focusing of the sample was stabilized by a closed-loop system that monitored the back reflection from the sample coverglass via an infrared laser beam sent through the edge of the microscope objective. A low-end piezoelectric deformable mirror (DM; Thorlabs; DMP40-P01) was added in the emission path at the conjugate plane of the objective pupil plane (Shi et al., 2017). By first flattening the mirror and then manually adjusting key Zernike polynomials, this DM corrected aberrations induced by both the optical system and the glass–water refractive index mismatch when the sample was several micrometers away from the coverglass. The fluorescence was recorded at a frame rate of 57 Hz on an electron-multiplying charge-coupled device camera (Andor; iXon+ DU897E-CS0-BV).
The mounting medium used for STORM imaging was water with the addition of 10 mM mercaptoethylamine at pH 8.5, 5% glucose (wt/vol) and oxygen-scavenging enzymes 0.5 mg/ml glucose oxidase (Sigma-Aldrich), and 40 mg/ml catalase (Roche Applied Science). The pH of the final solution was adjusted to 8.4. The mounting medium remained suitable for imaging for 1–2 h. The photoswitchable dye AF647 was conjugated on streptavidin and used for imaging with a ratio of 0.8:1 of dye/streptavidin. The stained hydrogel was incubated in the mounting medium for 15 min before mounting. The hydrogel was them transferred to laser-cut sample chamber with a polylysine-coated coverglass that we devised to mechanically stabilize the expanded gel during image acquisition (Fig. S3).
Figure S3.
3D-printed chamber for drift reduction of the hydrogel.
AF647 was excited with a 642-nm imaging laser with typically 1 kW/cm2 laser intensity at the focal plane. Analysis of STORM raw data was performed in the Insight3 software (Shi et al., 2017), which identified and fitted single-molecule spots in each camera frame to determine their x and y coordinates as well as photon numbers.
Confocal and SIM image acquisition and analysis
Confocal imaging was performed on a spinning-disk confocal microscope (Nikon CSU-W1) with a 60× water-immersion objective (Nikon CFI SR Plan Apo IR 60× WI NA 1.27) in the UCSF Nikon Imaging Center. SIM imaging was performed on a SIM microscope (GE HealthCare DeltaVision OMX) with a 100× objective (CFI Plan Apo Lambda 100× oil NA 1.45) in the UCSF Nikon Imaging Center. The SIM reconstruction was done using the reconstruction software associated with the OMX microscope. The fluorescence intensity of confocal and SIM images was analyzed using the open-source software Fiji (ImageJ). No deconvolution was applied to any images in this work. The resolution of confocal LR-ExM and LR-ExSIM is qualified using microtubules, as demonstrated in Figs. S5 and S4, respectively.
Figure S5.
Resolution measurement for LR-ExM confocal images. The transverse profiles of the microtubule cross sections marked in yellow were used to measure the resolution of LR-ExM using a confocal microscope. Scale bar, 2 µm.
Drift reduction and correction
We minimized the sample drift during data acquisition by mounting the hydrogel in a 3D-printed chamber (Fig. S3). The bottom of the chamber is a coverglass modified with poly-L-lysine, which creates a strong adhesion to the negatively charged hydrogel. The drift during data acquisition was further corrected using imaging correlation analysis. The drift-corrected coordinates, photon number, and the frame of appearance of each identified molecule were saved in a molecule list for further analysis.
Storage and reimaging
The imaged hydrogel can be stored in PBS buffer. Fluorescence will be retained for at least 1 mo for multiple rounds of imaging. Note that in water, the fluorescence signal will completely fade away in 1 wk.
Quantification and comparison of fluorescence retention efficiencies
To compare the fluorescence retention efficiencies of different ExM methods, we took the images of immunostained microtubules in U2OS cells prepared with different ExM methods in the same imaging condition. We calculated the retained fluorescence by dividing the total fluorescence intensity of all microtubules by their total length. The total length of all microtubules in each image was quantified using the Fiji plugin JFilament. The quantification process and results are shown in Fig. S2.
Quantification of LR-ExSTORM images of CCPs
We LR-ExSTORM imaged CCPs in U2OS cells expressing SNAP tag–labeled CLTB stained with BG-MA-biotin anchor before expansion and biotin-AF488 after expansion. CCPs focused at the top were selected for the quantification. We measured the distances from the centroid of one cluster to the centroid of its nearest neighbor in the central area of each CCP and excluded the clusters at the CCP edge to avoid off-focus localizations. We plotted the histogram of these nearest-neighbor distances (1,102 pairs from 134 CCPs) and fitted the distance distribution with Gaussian functions. The position of the center and the SD of the Gaussian peak are respectively used as the distance between neighboring vertices of the polyhedral CCPs and the SD of the distance (Fig. 3). The effective localization precision of LR-ExSTORM was calculated by dividing the mean of measured FHWM of clusters in the STORM images by the length expansion ratio. Each cluster is the superimposition of fitted Gaussian peaks of repetitive photoswitching of one AF647 label.
Reagents for trifunctional anchor synthesis
The bifunctional linkers NHS-functionalized MA, biotin, and digoxin were obtained from Lumiprobe. 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide, NHS, 4-dimethylaminopyridine, trifluoroacetic acid (TFA), and all of the organic solvents were purchased from Sigma-Aldrich. The HABA/Avidin reagent kit was obtained from Thermo Fisher Scientific. Nap-5 Sephadex G-25 size exclusion columns were ordered from GE Healthcare (17085301). PBS, pH 7.4 (catalog number: 10010–023), was ordered from Thermo Fisher Scientific. Acetonitrile, dichlormethane, dimethylformamide, ether, and tetrahydrofuran (to be used in anhydrous reaction mixtures) were dried by passage through activated alumina columns immediately before use. Hexanes used were ≥85% n-hexane. Other commercial solvents and reagents were used as received, unless otherwise noted.
Synthesis and qualitative analysis of trifunctional anchors
General
All reactions were performed in flame- or oven-dried glassware fitted with rubber septa under a positive pressure of nitrogen, unless otherwise noted. All reaction mixtures were stirred throughout the course of each procedure using Teflon-coated magnetic stir bars. Air- and moisture-sensitive liquids were transferred via syringe. Solutions were concentrated by rotary evaporation below 30°C. Analytical TLC was performed using glass plates precoated with silica gel (0.25-mm, 60-Å pore size, 230−400 mesh; Silicycle) impregnated with a fluorescent indicator (254 nm). TLC plates were visualized by exposure to UV light and then stained by submersion in a basic aqueous solution of potassium permanganate or with an acidic ethanolic solution of anisaldehyde, followed by brief heating.
Materials
Dichloromethane (DCM), N,N-dimethylformamide (DMF), tetrahydrofuran, ethyl ether, and acetonitrile (used in anhydrous reaction mixtures) were dried by passage through activated alumina columns immediately before use. Hexanes used were ≥85% n-hexane. Other commercial solvents and reagents were used as received, unless otherwise noted.
Instrumentation
Unless otherwise noted, proton NMR (1H-NMR) spectra and carbon NMR (13C-NMR) spectra were recorded on a 400-MHz Bruker Avance III HD 2-channel instrument NMR spectrometer at 23°C. Proton chemical shifts are expressed in parts per million (δ scale) and are referenced to residual protium in the NMR solvent (CHC13: δ 7.26, DMSO-d5: δ 2.50, CHD2OD: δ 3.31, and HDO: δ 4.79). Carbon chemical shifts are expressed in parts per million (δ scale) and are referenced to the carbon resonance of the NMR solvent (CDC13: δ 77.16, DMSO-d6: δ 39.52, and CD3OD: δ 49.0). Data are represented as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, dd = doublet of doublets, dt = doublet of triplets, sxt = sextet, m = multiplet, br = broad, and app = apparent), integration, and coupling constant (J) in hertz (Hz). Reverse-phase preparative HPLC was performed on a Waters Delta Prep 4000 preparative chromatography system containing solvents A (0.1% TFA in Milli-Q water) and B (0.1% TFA in acetonitrile), Gemini-NX, 5 µm, C18, 110 Å, 30.00 mm i.d. × 100 mm, wavelength 220 nm with a gradient of 5–60% B over 17 min followed by 60–95% B over 2 min. High-resolution mass spectra were obtained on a Waters Acquity UPLC/Xevo G2-XS QTOF mass spectrometer with electrospray ionization (ESI; special thanks to Dr. Ziyang Zhang in the Shokat Laboratory for assistance). HPLC traces were obtained on an HP 1100 analytical HPLC containing solvents A (0.1% TFA in Milli-Q water) and B (0.1% TFA, 1% Milli-Q water in acetonitrile), Jupiter, 5 µm, C4, 300 Å, 4.6 mm i.d. × 250 mm, wavelength 220 nm with a gradient of 5–100% B over 45 min (special thanks to Dr. Kui Zhang in the DeGrado Laboratory for assistance).
Scheme 1.
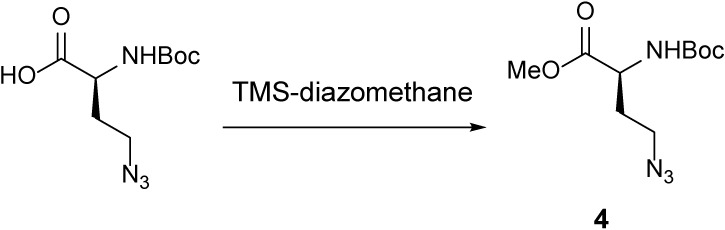
Azide 4 ( McLaughlin et al., 2003 ).
A 50-ml round-bottom flask containing (S)-4-azido-2-((tert-butoxycarbonyl)amino)butanoic acid (500 mg, 2.05 mmol, 1 equivalent [equiv]) was evacuated and flushed with nitrogen (this process was repeated a total of three times). A solution of MeOH and DCM (1:3, 22.8 ml) was added, resulting in a colorless solution. TMS-diazomethane (2 ml, 4.09 mmol, 2 equiv) was added dropwise. After 2 h, the mixture was concentrated, and azide 4 (theoretical: 529 mg, 2.05 mmol) was used without further purification. The 1H-NMR spectral data for azide 4 is in agreement with the tabulated data published in reference (McLaughlin et al., 2003).
Scheme 2.
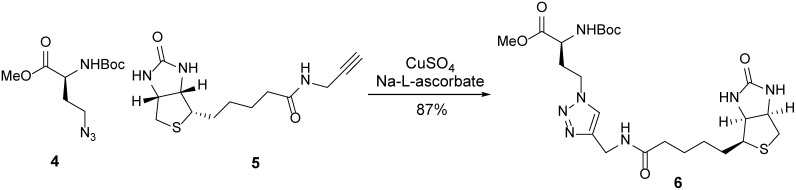
Click product 6.
A 100-ml round-bottom flask containing azide 4 (529 mg, 2.05 mmol, 1.05 equiv) and alkyne 5 (Nainar et al., 2017; 549 mg, 1.95 mmol, 1 equiv) was evacuated and flushed with nitrogen (this process was repeated a total of three times). DCM (21 ml) was added, followed by adding a solution of copper (II) sulfate (31.1 mg, 195 μmol, 0.1 equiv) and sodium L-ascorbate (155 mg, 780 μmol, 0.4 equiv) in t-BuOH and H2O solvent (1:1, 19.5 ml). The resulting suspension was sonicated, resulting in a light-yellow solution. After 2 h, the solution was concentrated to reveal a yellow foam. The resulting crude residue was purified by flash chromatography (silica gel, eluent:MeOH:DCM = 1:9) to afford click product 6 (920 mg, 87%) as an off-white solid.
TLC (MeOH/DCM = 1:9): Rf = 0.27 (anisaldehyde, KMnO4).
1H NMR (400 MHz, DMSO-d6): δ 8.28 (t, J = 5.7 Hz, 1H), 7.86 (s, 1H), 7.44 (d, J = 7.9 Hz, 1H), 6.41 (d, J = 1.9 Hz, 1H), 6.36 (s, 1H), 4.51–4.21 (m, 5H), 4.16–4.04 (m, 1H), 4.00–3.89 (m, 1H), 3.61 (s, 3H), 3.09 (ddd, J = 8.5, 6.2, 4.4 Hz, 1H), 2.82 (dd, J = 12.4, 5.1 Hz, 1H), 2.57 (d, J = 12.4 Hz, 1H), 2.31–2.16 (m, 1H), 2.14–2.00 (m, 3H), 1.70–1.11 (m, 15H).
13C-NMR (100 MHz, DMSO-d6): δ 172.3, 172.0, 162.7, 155.6, 145.1, 122.9, 78.6, 61.0, 59.2, 55.4, 52.0, 51.0, 46.2, 39.9, 35.0, 34.1, 31.3, 28.2, 28.2, 28.0, 25.2.
HRMS-ESI m/z calculated [calcd] for C23H37N7O6S [M+H]+ 540.2606, found 540.2612.
Scheme 3.
Methyl ester 7.
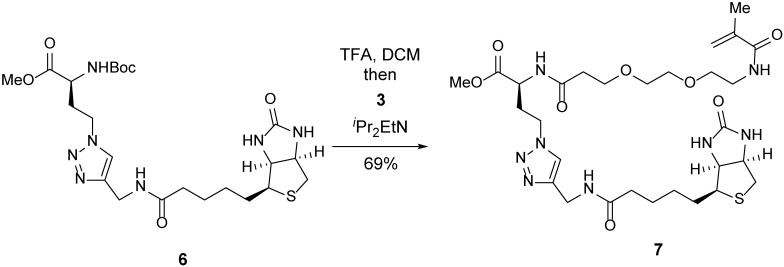
A 25-ml round-bottom flask containing click product 6 (200 mg, 371 μmol, 1 equiv) was evacuated and flushed with nitrogen (this process was repeated three times). DCM (4 ml) was added, followed by TFA (4 ml). After 30 min, the mixture was concentrated. Toluene (10 ml) was added to the resulting crude residue, and the mixture was concentrated to remove residual TFA (this process was repeated three times). The residue was dissolved in DMF (8 ml), and a solution of TFP-ester 3 (160 mg, 408 μmol, 1.1 equiv) in DMF (4 ml) was added. iPr2EtN (0.32 ml, 1.85 mmol, 5 equiv) was then added dropwise. After 5 h, the mixture was concentrated. The resulting crude residue was purified by flash chromatography (silica gel, eluent: MeOH/DCM = 1:19 to 1:9 with 1% ammonium hydroxide) to afford methyl ester 7 (170 mg, 69%) as a light-pink oil.
TLC (MeOH:DCM = 1:9): Rf =0.17 (UV, anisaldehyde).
1H-NMR (400 MHz, DMSO-d6): δ 8.44 (d, J = 7.8 Hz, 1H), 8.27 (t, J = 5.7 Hz, 1H), 7.91 (t, J = 5.7 Hz, 1H), 7.85 (s, 1H), 6.41 (s, 1H), 6.36 (s, 1H), 5.64 (s, 1H), 5.31 (t, J = 1.6 Hz, 1H), 4.42–4.33 (m, 2H), 4.34–4.20 (m, 4H), 4.12 (ddd, J = 7.8, 4.4, 1.9 Hz, 1H), 3.66–3.56 (m, 5H), 3.49 (s, 3H), 3.41 (t, J = 6.1 Hz, 3H), 3.23 (q, J = 6.0 Hz, 2H), 3.09 (ddd, J = 8.5, 6.2, 4.4 Hz, 1H), 2.82 (dd, J = 12.4, 5.0 Hz, 1H), 2.57 (d, J = 12.4 Hz, 1H), 2.48–2.22 (m, 3H), 2.10 (q, J = 7.2 Hz, 3H), 1.83 (s, 3H), 1.67–1.39 (m, 4H), 1.37–1.20 (m, 2H).
13C-NMR (100 MHz, DMSO-d6): δ 172.0, 171.8, 170.6, 167.5, 162.7, 145.0, 139.8, 123.0, 119.1, 69.5, 69.5, 68.8, 66.7, 61.0, 59.2, 55.4, 52.1, 49.3, 46.1, 39.9, 38.8, 35.9, 35.0, 34.1, 31.5, 28.2, 28.0, 25.2, 18.6.
HRMS-ESI m/z calcd for C29H46N8O8S [M+H]+ 667.3239, found 667.3251.
Scheme 4.
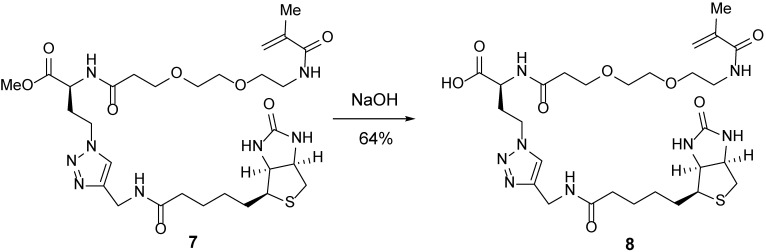
Carboxylic acid 8.
Water (5 ml) was added to a 20-ml scintillation vial containing methyl ester 7 (166 mg, 371 μmol, 1 equiv). Sodium hydroxide (49.8 mg, 1.24 mmol, 5 equiv) was added. After 1 h, the reaction was quenched with 1N HCl (1.6 ml) and concentrated. The resulting crude residue was purified by reverse-phase HPLC to afford carboxylic acid 8 (103 mg, 64%) as a white powder.
TLC (MeOH:DCM = 1:1): Rf =0.17 (UV, anisaldehyde).
1H-NMR (400 MHz, D2O): δ 7.90 (s, 1H), 5.67 (s, 1H), 5.43 (s, 1H), 4.63–4.56 (m, 1H), 4.57–4.48 (m, 2H), 4.46 (s, 2H), 4.38 (dd, J = 7.9, 4.5 Hz, 1H), 4.31 (dd, J = 9.7, 4.6 Hz, 1H), 3.79 (t, J = 6.1 Hz, 2H), 3.68 (s, 4H), 3.63 (t, J = 5.4 Hz, 2H), 3.43 (t, J = 5.4 Hz, 2H), 3.28 (p, J = 5.6 Hz, 1H), 2.98 (dd, J = 13.0, 5.0 Hz, 1H), 2.77 (d, J = 13.0 Hz, 1H), 2.63–2.50 (m, 3H), 2.40–2.25 (m, 3H), 1.91 (s, 3H), 1.77–1.43 (m, 4H), 1.43–1.24 (m, 2H).
13C-NMR (100 MHz, D2O): δ 176.7, 174.4, 174.0, 171.9, 165.3, 144.7, 139.0, 124.1, 121.0, 69.6, 69.4, 68.8, 66.6, 62.0, 60.2, 55.4, 40.0, 47.0, 39.7, 39.1, 35.7, 35.2, 34.2, 30.8, 27.7, 27.6, 25.0, 17.7.
HRMS-ESI m/z calcd for C28H44N8O8S [M+H]+ 653.3083, found 653.3079.
Scheme 5.
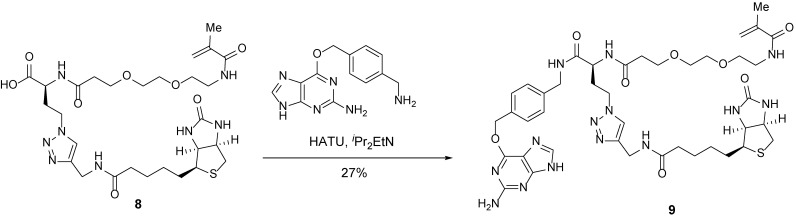
Trifunctional anchor 9.
A 10-ml round-bottom flask containing carboxylic acid 8 (20 mg, 30.6 μmol, 1 equiv) and 6-((4-(aminomethyl)benzyl)oxy)-9H-purin-2-amine (12.4 mg, 46.0 μmol, 1.5 equiv) was evacuated and flushed with nitrogen (this process was repeated three times). DMF (2 ml) was added. iPr2EtN (13 µl, 76.6 μmol, 2.5 equiv) was then added dropwise. After the solid reactants dissolved, HATU (hexafluorophosphate azabenzotriazole tetramethyl uronium; 23.3 mg, 61.3 μmol, 2 equiv) was added, resulting in a yellow solution. After 2 h, the mixture was concentrated to reveal a yellow oil. The resulting crude residue was purified by reverse-phase high-performance liquid chromatography to afford trifunctional anchor 9 (7.5 mg, 27%) as a white powder.
TLC (MeOH:DCM = 1:4): Rf =0.18 (UV, anisaldehyde).
1H-NMR (400 MHz, MeOD): δ 8.38 (s, 1H), 7.83 (s, 1H), 7.50 (d, J = 8.1 Hz, 2H), 7.32 (d, J = 8.1 Hz, 2H), 5.68 (s, 1H), 5.64 (s, 2H), 5.38–5.34 (m, 1H), 4.52–4.32 (m, 8H), 4.27 (dd, J = 7.9, 4.5 Hz, 1H), 3.84–3.68 (m, 2H), 3.60–3.53 (m, 4H), 3.50 (t, J = 5.8 Hz, 2H), 3.35 (t, J = 5.8 Hz, 2H), 3.21–3.13 (m, 1H), 2.90 (ddd, J = 12.8, 5.0, 2.7 Hz, 1H), 2.69 (dd, J = 12.8, 3.0 Hz, 1H), 2.61–2.37 (m, 3H), 2.31–2.15 (m, 3H), 1.91 (s, 3H), 1.80–1.51 (m, 4H), 1.47–1.33 (m, 2H).
13C-NMR (100 MHz, MeOD): δ 176.0, 174.3, 171.3, 166.1, 161.1, 153.4, 146.3, 143.9, 141.2, 140.7, 135.3, 130.3, 128.7, 124.7, 120.6, 107.7, 71.3, 71.2, 71.0, 70.4, 68.3, 63.3, 61.6, 57.0, 52.2, 49.7, 49.5, 49.3, 49.1, 48.1, 43.8, 41.0, 40.4, 37.6, 36.5, 35.6, 33.4, 29.7, 29.4, 26.7, 18.8.
HRMS-ESI m/z calcd for C41H56N14O8S [M+H]+ 905.4206, found 905.4189.
Scheme 6.
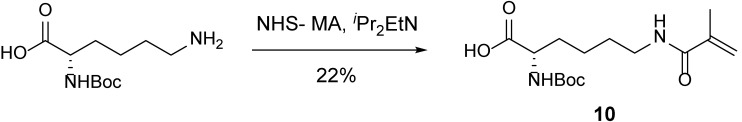
MA 10.
A 10-ml round-bottom flask containing (tert-butoxycarbonyl)-L-lysine (200 mg, 812 μmol, 1 equiv) and NHS-methacrylate (178 mg, 974 μmol, 1.2 equiv) was evacuated and flushed with nitrogen (this process was repeated a total of three times). DMF (8 ml) was added. iPr2EtN (0.28 ml, 1.62 mmol, 2 equiv) was then added dropwise, resulting in a white suspension. After stirring overnight, the mixture was concentrated to reveal a yellow oil. The resulting crude residue was purified by flash chromatography (silica gel, eluent: MeOH/DCM = 1:19), to yield an oil contaminated with n-hydroxysuccinimide. The oil was dissolved in DCM (10 ml) and then washed with water (3 × 10 ml) and brine (10 ml). The washed solution was dried (Na2SO4). The dried solution was filtered, and the filtrate was concentrated to yield MA 10 (71.4 mg, 22%) as a clear oil.
TLC (MeOH:DCM = 1:9): Rf =0.19 (UV, anisaldehyde).
1H-NMR (400 MHz, CDCl3): δ 8.70 (brs, 1H), 6.31 (brs, 1H), 5.69 (s, 1H), 5.38 (brs, 1H), 5.31 (s, 1H), 4.23 (brs, 1H), 3.38–3.18 (m, 2H), 1.93 (s, 3H), 1.84 (bs, 1H), 1.76–1.49 (m, 4H), 1.42 (s, 9H), 1.30–1.19 (m, 1H).
13C-NMR (100 MHz, CDCl3): δ 176.0, 169.2, 156.0, 139.9, 120.1, 80.1, 53.6, 39.5, 32.2, 29.0, 28.5, 22.6, 18.8.
HRMS-ESI m/z calcd for C15H26N2O5 [M+Na]+ 337.1739, found 337.1774.
Scheme 7.
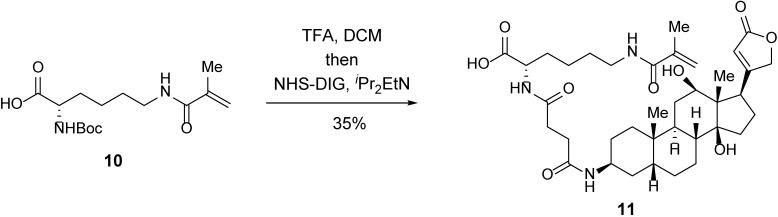
Carboxylic acid 11.
A 5-ml round-bottom flask containing MA 10 (2.6 mg, 8.3 μmol, 1 equiv) was evacuated and flushed with nitrogen (this process was repeated a total of three times). DCM (1 ml) was added. TFA (1 ml) was then added dropwise. After 30 min, the mixture was concentrated. Toluene was added to the resulting crude residue, and the mixture was concentrated to remove residual TFA (this process for repeated a total of three times). The residue was dissolved in DCM, and 3-amino-3-deoxydigoxigenin hemisuccinamide, succinimidyl ester (5.0 mg, 8.5 μmol, 1.0 equiv) was added in one portion. iPr2EtN (7.2 µl, 41 μmol, 5.0 equiv) was then added dropwise. After stirring overnight, the mixture was concentrated. The resulting crude residue was purified by reverse-phase HPLC to afford carboxylic acid 11 (2.0 mg, 35%) as a white powder.
TLC (MeOH:DCM = 1:4): Rf =0.12 (UV, anisaldehyde).
1H-NMR (400 MHz, MeOD): δ 5.91 (brt, J = 1.8 Hz, 1H), 5.67 (brt, J = 1.0 Hz, 1H), 5.37–5.34 (m, 1H), 5.03–4.87 (m, 2H), 4.37 (dd, J = 8.9, 4.9 Hz, 1H), 4.07 (bs, 1H), 3.44–3.32 (m, 2H), 3.24 (t, J = 6.9 Hz, 2H), 2.67–2.36 (m, 4H), 2.22–1.07 (m, 25H), 1.93 (s, 3H), 1.00 (s, 3H), 0.79 (s, 3H).
HRMS-ESI m/z calcd for C37H55N3O9 [M+H]+ 686.4018, found 686.4000.
Scheme 8.
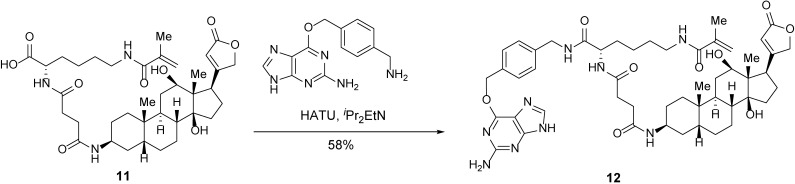
Trifunctional anchor 12.
A 5-ml round-bottom flask containing carboxylic acid 11 (1.5 mg, 2.2 μmol, 1 equiv) was evacuated and flushed with nitrogen (this process was repeated a total of three times). DMF (0.3 ml) was added. iPr2EtN (0.95 µl, 5.5 μmol, 2.5 equiv) was then added dropwise. After stirring overnight, 6-((4-(aminomethyl)benzyl)oxy)-7H-purin-2-amine (0.89 mg, 3.3 μmol, 1.5 equiv) and HATU (1.7 mg, 4.4 μmol, 2 equiv) were added in succession. After 2 h, the mixture was concentrated. The resulting crude was purified by reverse-phase HPLC to afford trifunctional anchor 12 (1.2 mg, 58%) as a white powder.
TLC (MeOH:DCM = 1:4): Rf =0.53 (UV, anisaldehyde).
1H-NMR (400 MHz, MeOD) δ 8.66 (t, J = 6.0 Hz, 1H), 8.28 (s, 1H), 7.49 (d, J = 8.0 Hz, 2H), 7.32 (d, J = 8.0 Hz, 2H), 5.90 (s, 1H), 5.67 (s, 1H), 5.62 (s, 2H), 5.37–5.33 (m, 1H), 5.03–4.87 (m, 2H), 4.46–4.34 (m, 2H), 4.30 (dd, J = 9.3, 4.7 Hz, 1H), 3.97 (bs, 1H), 3.44–3.32 (m, 2H), 3.23 (t, J = 7.0 Hz, 2H), 2.92–2.85 (m, 1H), 2.71–2.35 (m, 5H), 2.24–1.06 (m, 25H), 1.93 (s, 3H), 0.97 (s, 3H), 0.78 (s, 3H).
HRMS-ESI m/z calcd for C50H67N9O9 [M+H]+ 938.5142, found 938.5134.
Scheme 9.
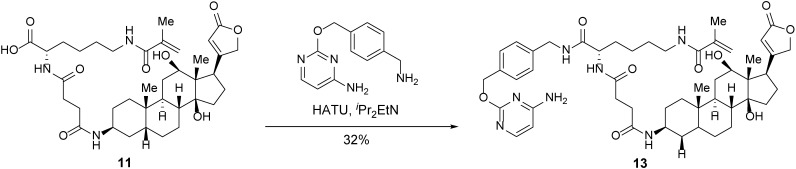
Trifunctional anchor 13.
A 5-ml round-bottom flask containing carboxylic acid 11 (1.5 mg, 2.2 μmol, 1 equiv) was evacuated and flushed with nitrogen (this process was repeated a total of three times). DMF (0.3 ml) was added. iPr2EtN (0.95 µl, 5.5 μmol, 2.5 equiv) was then added dropwise. After stirring overnight, 2-((4-(aminomethyl)benzyl)oxy)pyrimidin-4-amine (0.76 mg, 3.3 μmol, 1.5 equiv) and HATU (1.7 mg, 4.4 μmol, 2 equiv) were added in succession. After 2 h, the mixture was concentrated. The resulting crude was purified by reverse-phase HPLC to afford trifunctional anchor 13 (0.6 mg, 32%) as a white powder.
TLC (MeOH:DCM = 1:4): Rf =0.53 (UV, anisaldehyde).
1H-NMR (400 MHz, MeOD) δ 8.67 (t, J = 6.0 Hz, 1H), 7.87 (d, J = 7.1 Hz, 1H), 7.45 (d, J = 8.0 Hz, 2H), 7.33 (d, J = 8.0 Hz, 2H), 6.39 (d, J = 7.1 Hz, 1H), 5.91 (s, 1H), 5.67 (s, 1H), 5.51 (s, 2H), 5.37–5.33 (m, 1H), 5.03–4.87 (m, 2H), 4.45–4.37 (m, 2H), 4.30 (dd, J = 9.3, 4.6 Hz, 1H), 3.99 (bs, 1H), 3.44–3.32 (m, 2H), 3.23 (t, J = 7.0 Hz, 2H), 2.89 (d, J = 11.0 Hz, 1H), 2.69 –.39 (m, 5H), 2.20–1.15 (m, 25H), 1.93 (s, 3H), 0.98 (s, 3H), 0.78 (s, 3H).
HRMS-ESI m/z calcd for C49H67N7O9 [M+H]+ 898.5008, found 898.5031.
Online supplemental material
Fig. S1 shows the synthetic scheme of trifunctional anchors. Fig. S2 shows a comparison of fluorescence intensities resulting from different ExM methods. Fig. S3 shows a 3D-printed chamber for drift reduction of hydrogel. Fig. S4 shows LR-ExSIM of microtubules and distal appendages of cilia; LR-ExSIM and STORM reveal the structure of distal appendages with similar superresolution. Fig. S5 shows a resolution measurement for LR-ExM confocal images. Data S1 shows 1H- and 13C-NMR spectra of compounds 6–10 and 1H-NMR and HPLC analysis of compounds 11–13 (y axes are in arbitrary units).
Supplementary Material
shows the 1H- and 13C-NMR spectra of compounds 6–10 and 1H-NMR and HPLC analysis of compounds 11–13.
Acknowledgments
We thank Drs. Ed Boyden and Fei Chen for their help with the proExM protocol and Drs. Joshua C. Vaughan and Aaron Halpern for help with the ExM protocol. We are grateful to Dr. Keith DeLuca from the Jenifer DeLuca Lab for validating the expansion factor of compact structures in the nucleus, Stefan Niekamp from the Ron Vale Lab for measuring the labeling efficiency, and Dr. Dan Xie for optimizing the DM and 3D printing slide adapters for the STORM microscope. We appreciate Dr. Luke Lavis for his consultation on protein tags, Dr. Xiangpeng Li from the Adam Abate Lab, and Dr. Xiao Huang from the Tejal Desai Lab for their consultation on biochemistry and microfluidics. We thank Eric Simental for transfecting mouse embryonic stem cells. This work was also highly inspired by personal conversations with Drs. David Brown and Juan Guan and discussions with all Huang laboratory members.
This project was supported by the National Institutes of Health (Director’s New Innovator Awards DP2OD008479 and R01GM124334 to B. Huang; NIH Pathway to Independence Award K99GM126136/R00GM126136 to X. Shi) the University of California, San Francisco Mary Anne Koda-Kimble Seed Award for Innovation (to X. Shi), the American Heart Association (predoctoral fellowship 19PRE3480616 to J. Chen), the National Science Foundation (graduate research fellowship 1650113 to A.A. Tran), the David and Lucile Packard Foundation (I.B. Seiple), and the Damon Runyon Cancer Research Foundation (postdoctoral fellowship DRG2168-13 to T.T. Chow). B. Huang and J.F. Reiter are Chan Zuckerberg Biohub Investigators.
The authors declare no competing financial interests.
Author contributions: X. Shi, Q. Li, Z. Dai, I.B. Seiple, and B. Huang designed the experiments and interpreted the results. X. Shi designed LR-ExM protocols. X. Shi, Z. Lin, X. Wang, T.T. Chow, and A.R. McColloch imaged samples. Q. Li and A.A. Tran synthesized the trifunctional anchors, and Z. Dai performed image quantification and antibody conjugation. S. Feng and A.D. Ramirez designed and made plasmids. J. Chen prepared and stained the mouse brain. D. Kumar, X. Shi, J.F. Reiter, and B. Huang designed the experiments on imaging primary cilia with ExSIM. D. Kumar and X. Shi took the ExSIM images. X. Shi drafted the manuscript. B. Huang, I.B. Seiple, A.A. Tran, Z. Dai, S. Feng, J. Chen, E.J. Huang, J.F. Reiter, and X. Shi edited the manuscript.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the online supplemental material. All other data are available upon request. Protocols are described in Materials and methods in detail. We are seeking commercial channels for distribution of the trifunctional anchors described in this paper. Before they are commercially available, samples will be provided upon request.
References
- Bates, M., Huang B., and Zhuang X.. 2008. Super-resolution microscopy by nanoscale localization of photo-switchable fluorescent probes. Curr. Opin. Chem. Biol. 12:505–514. 10.1016/j.cbpa.2008.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon, C.K., Yu Z., Wang Y., Guo F., Unruh J.R., Slaughter B.D., and Hawley R.S.. 2017. Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. USA. 114:E6857–E6866. 10.1073/pnas.1705623114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J.B., Chen F., Yoon Y.G., Jung E.E., Babcock H., Kang J.S., Asano S., Suk H.J., Pak N., Tillberg P.W., et al. 2017. Iterative expansion microscopy. Nat. Methods. 14:593–599. 10.1038/nmeth.4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., Tillberg P.W., and Boyden E.S.. 2015. Optical imaging. Expansion microscopy. Science. 347:543–548. 10.1126/science.1260088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., Wassie A.T., Cote A.J., Sinha A., Alon S., Asano S., Daugharthy E.R., Chang J.B., Marblestone A., Church G.M., et al. 2016. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods. 13:679–684. 10.1038/nmeth.3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chozinski, T.J., Halpern A.R., Okawa H., Kim H.J., Tremel G.J., Wong R.O., and Vaughan J.C.. 2016. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods. 13:485–488. 10.1038/nmeth.3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, K., Wallace J., Kim S.Y., Kalyanasundaram S., Andalman A.S., Davidson T.J., Mirzabekov J.J., Zalocusky K.A., Mattis J., Denisin A.K., et al. 2013. Structural and molecular interrogation of intact biological systems. Nature. 497:332–337. 10.1038/nature12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey, G.T., Vaughan J.C., Chen K.H., Bates M., and Zhuang X.. 2011. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods. 8:1027–1036. 10.1038/nmeth.1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambarotto, D., Zwettler F.U., Le Guennec M., Schmidt-Cernohorska M., Fortun D., Borgers S., Heine J., Schloetel J.G., Reuss M., Unser M., et al. 2019. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods. 16:71–74. 10.1038/s41592-018-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., Maraspini R., Beutel O., Zehtabian A., Eickholt B., Honigmann A., and Ewers H.. 2018. Expansion Stimulated Emission Depletion Microscopy (ExSTED). ACS Nano. 12:4178–4185. 10.1021/acsnano.8b00776 [DOI] [PubMed] [Google Scholar]
- Gautier, A., Juillerat A., Heinis C., Corrêa I.R. Jr., Kindermann M., Beaufils F., and Johnsson K.. 2008. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 15:128–136. 10.1016/j.chembiol.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Halpern, A.R., Alas G.C.M., Chozinski T.J., Paredez A.R., and Vaughan J.C.. 2017. Hybrid Structured Illumination Expansion Microscopy Reveals Microbial Cytoskeleton Organization. ACS Nano. 11:12677–12686. 10.1021/acsnano.7b07200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell, S.W., Sahl S.J., Bates M., Zhuang X., Heintzmann R., Booth M.J., Bewersdorf J., Shtengel G., Hess H., Tinnefeld P., et al. 2015. The 2015 super-resolution microscopy roadmap. J. Phys. D Appl. Phys. 48:443001. 10.1088/0022-3727/48/44/443001 [DOI] [Google Scholar]
- Huang, B., Jones S.A., Brandenburg B., and Zhuang X.. 2008. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat. Methods. 5:1047–1052. 10.1038/nmeth.1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, A.J., and Nossal R.. 2000. Rigidity of triskelion arms and clathrin nets. Biophys. J. 78:1183–1194. 10.1016/S0006-3495(00)76676-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyama, D., and Huang B.. 2012. Development in the STORM. Dev. Cell. 23:1103–1110. 10.1016/j.devcel.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis, E.D., Kang J.S., Shin T.W., Emenari A., Asano S., Lin L., Costa E.K., Marblestone A.H., Kasthuri N., and Boyden E.S.. 2019. Expansion Microscopy of Lipid Membranes. bioRxiv. (Preprint posted November 4, 2019) 10.1101/829903 [DOI]
- Keppler, A., Pick H., Arrivoli C., Vogel H., and Johnsson K.. 2004. Labeling of fusion proteins with synthetic fluorophores in live cells. Proc. Natl. Acad. Sci. USA. 101:9955–9959. 10.1073/pnas.0401923101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen, T., Owen D., and Harrison S.C.. 2014. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb. Perspect. Biol. 6:a016725. 10.1101/cshperspect.a016725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, T., Swaney J., Park J.Y., Albanese A., Murray E., Cho J.H., Park Y.G., Mangena V., Chen J., and Chung K.. 2016. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 34:973–981. 10.1038/nbt.3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, L., Lee Y.L., Sahl S.J., Stearns T., and Moerner W.E.. 2012. STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophys. J. 102:2926–2935. 10.1016/j.bpj.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., Chen X., Lin Z., Wang Y., and Sun Y.. 2018. Expansion enhanced nanoscopy. Nanoscale. 10:17552–17556. 10.1039/C8NR04267E [DOI] [PubMed] [Google Scholar]
- Liang, G., Lin J.C., Wei V., Yoo C., Cheng J.C., Nguyen C.T., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., and Jones P.A.. 2004. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. USA. 101:7357–7362. 10.1073/pnas.0401866101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, R., Feng Q., Li P., Zhou P., Wang R., Liu Z., Wang Z., Qi X., Tang N., Shao F., and Luo M.. 2018. A hybridization-chain-reaction-based method for amplifying immunosignals. Nat. Methods. 15:275–278. 10.1038/nmeth.4611 [DOI] [PubMed] [Google Scholar]
- M’Saad, O., and Bewersdorf J.. 2020. Light microscopy of proteins in their ultrastructural context. Nat. Commun. 11:3850. 10.1038/s41467-020-17523-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, C., Lee M.Y., Jhan J.R., Halpern A.R., Woodworth M.A., Glaser A.K., Chozinski T.J., Shin L., Pippin J.W., Shankland S.J., et al. 2020. Feature-rich covalent stains for super-resolution and cleared tissue fluorescence microscopy. Sci. Adv. 6:eaba4542. 10.1126/sciadv.aba4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin, M., Mohareb R.M., and Rapoport H.. 2003. An efficient procedure for the preparation of 4-substituted 5-aminoimidazoles. J. Org. Chem. 68:50–54. 10.1021/jo026257s [DOI] [PubMed] [Google Scholar]
- Murakami, T.C., Mano T., Saikawa S., Horiguchi S.A., Shigeta D., Baba K., Sekiya H., Shimizu Y., Tanaka K.F., Kiyonari H., et al. 2018. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 21:625–637. 10.1038/s41593-018-0109-1 [DOI] [PubMed] [Google Scholar]
- Nainar, S., Kubota M., McNitt C., Tran C., Popik V.V., and Spitale R.C.. 2017. Temporal Labeling of Nascent RNA Using Photoclick Chemistry in Live Cells. J. Am. Chem. Soc. 139:8090–8093. 10.1021/jacs.7b03121 [DOI] [PubMed] [Google Scholar]
- Nakayama, J., Rice J.C., Strahl B.D., Allis C.D., and Grewal S.I.. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 292:110–113. 10.1126/science.1060118 [DOI] [PubMed] [Google Scholar]
- Nieuwenhuizen, R.P.J., Lidke K.A., Bates M., Puig D.L., Grünwald D., Stallinga S., and Rieger B.. 2013. Measuring image resolution in optical nanoscopy. Nat. Methods. 10:557–562. 10.1038/nmeth.2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier, N., Keller D., Gönczy P., and Manley S.. 2013. Resolution doubling in 3D-STORM imaging through improved buffers. PLoS One. 8:e69004. 10.1371/journal.pone.0069004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y.-G., Sohn C.H., Chen R., McCue M., Yun D.H., Drummond G.T., Ku T., Evans N.B., Oak H.C., Trieu W., et al. 2018. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat. Biotechnol. 37:73–83. 10.1038/nbt.4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka, S.K., Wang Y., Kishi J.Y., Zhu A., Zeng Y., Xie W., Kirli K., Yapp C., Cicconet M., Beliveau B.J., et al. 2019. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 37:1080–1090. 10.1038/s41587-019-0207-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X., Garcia G. III, Van De Weghe J.C., McGorty R., Pazour G.J., Doherty D., Huang B., and Reiter J.F.. 2017. Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nat. Cell Biol. 19:1178–1188. 10.1038/ncb3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi, T., Kittisopikul M., Tran J., Goldman A.E., Adam S.A., Zheng Y., Jaqaman K., and Goldman R.D.. 2015. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell. 26:4075–4086. 10.1091/mbc.E15-07-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff, H., Galbraith C.G., Galbraith J.A., and Betzig E.. 2008. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods. 5:417–423. 10.1038/nmeth.1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker, D.K., Kuczmarski E.R., and Goldman R.D.. 2003. The nucleoskeleton: lamins and actin are major players in essential nuclear functions. Curr. Opin. Cell Biol. 15:358–366. 10.1016/S0955-0674(03)00050-4 [DOI] [PubMed] [Google Scholar]
- Thevathasan, J.V., Kahnwald M., Cieśliński K., Hoess P., Peneti S.K., Reitberger M., Heid D., Kasuba K.C., Hoerner S.J., Li Y., et al. 2019. Nuclear pores as versatile reference standards for quantitative superresolution microscopy. Nat. Methods. 16:1045–1053. 10.1038/s41592-019-0574-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillberg, P.W., Chen F., Piatkevich K.D., Zhao Y., Yu C.C., English B.P., Gao L., Martorell A., Suk H.J., Yoshida F., et al. 2016. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34:987–992. 10.1038/nbt.3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truckenbrodt, S., Maidorn M., Crzan D., Wildhagen H., Kabatas S., and Rizzoli S.O.. 2018. X10 expansion microscopy enables 25-nm resolution on conventional microscopes. EMBO Rep. 19:19. 10.15252/embr.201845836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truckenbrodt, S., Sommer C., Rizzoli S.O., and Danzl J.G.. 2019. A practical guide to optimization in X10 expansion microscopy. Nat. Protoc. 14:832–863. 10.1038/s41596-018-0117-3 [DOI] [PubMed] [Google Scholar]
- Turgay, Y., Eibauer M., Goldman A.E., Shimi T., Khayat M., Ben-Harush K., Dubrovsky-Gaupp A., Sapra K.T., Goldman R.D., and Medalia O.. 2017. The molecular architecture of lamins in somatic cells. Nature. 543:261–264. 10.1038/nature21382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Yu Z., Cahoon C.K., Parmely T., Thomas N., Unruh J.R., Slaughter B.D., and Hawley R.S.. 2018. Combined expansion microscopy with structured illumination microscopy for analyzing protein complexes. Nat. Protoc. 13:1869–1895. 10.1038/s41596-018-0023-8 [DOI] [PubMed] [Google Scholar]
- Wen, G., Vanheusden M., Acke A., Valli D., Neely R.K., Leen V., and Hofkens J.. 2020. Evaluation of Direct Grafting Strategies via Trivalent Anchoring for Enabling Lipid Membrane and Cytoskeleton Staining in Expansion Microscopy. ACS Nano. 14:7860–7867. 10.1021/acsnano.9b09259 [DOI] [PubMed] [Google Scholar]
- Xu, H., Tong Z., Ye Q., Sun T., Hong Z., Zhang L., Bortnick A., Cho S., Beuzer P., Axelrod J., et al. 2019. Molecular organization of mammalian meiotic chromosome axis revealed by expansion STORM microscopy. Proc. Natl. Acad. Sci. USA. 116:18423–18428. 10.1073/pnas.1902440116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B., Treweek J.B., Kulkarni R.P., Deverman B.E., Chen C.K., Lubeck E., Shah S., Cai L., and Gradinaru V.. 2014. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 158:945–958. 10.1016/j.cell.2014.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X., Hu J., Yue S., Kristiani L., Kim M., Sauria M., Taylor J., Kim Y., and Zheng Y.. 2018. Lamins Organize the Global Three-Dimensional Genome from the Nuclear Periphery. Mol. Cell. 71:802–815.e7. 10.1016/j.molcel.2018.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the 1H- and 13C-NMR spectra of compounds 6–10 and 1H-NMR and HPLC analysis of compounds 11–13.
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the online supplemental material. All other data are available upon request. Protocols are described in Materials and methods in detail. We are seeking commercial channels for distribution of the trifunctional anchors described in this paper. Before they are commercially available, samples will be provided upon request.