Crossley et al. demonstrate that GFP-tagged, catalytically inactive RNase H1 protein is a versatile tool for imaging cellular R-loops. They also show that it is significantly more specific than the commonly used S9.6 antibody, which detects considerable nonspecific signal.
Abstract
R-loops are three-stranded nucleic acid structures with both physiological and pathological roles in cells. R-loop imaging generally relies on detection of the RNA–DNA hybrid component of these structures using the S9.6 antibody. We show that the use of this antibody for imaging can be problematic because it readily binds to double-stranded RNA (dsRNA) in vitro and in vivo, giving rise to nonspecific signal. In contrast, purified, catalytically inactive human RNase H1 tagged with GFP (GFP-dRNH1) is a more specific reagent for imaging RNA–DNA hybrids. GFP-dRNH1 binds strongly to RNA–DNA hybrids but not to dsRNA oligonucleotides in fixed human cells and is not susceptible to binding endogenous RNA. Furthermore, we demonstrate that purified GFP-dRNH1 can be applied to fixed cells to detect hybrids after their induction, thereby bypassing the need for cell line engineering. GFP-dRNH1 therefore promises to be a versatile tool for imaging and quantifying RNA–DNA hybrids under a wide range of conditions.
Introduction
R-loops are three-stranded nucleic acid structures, typically formed during transcription, consisting of an RNA–DNA hybrid and a displaced strand of single-stranded DNA (ssDNA). R-loops have been linked to the regulation of several cellular processes, but they can also promote genome instability in certain contexts (Crossley et al., 2019; García-Muse and Aguilera, 2019). Moreover, perturbations in R-loop levels have been linked to several human diseases (Crossley et al., 2019; Richard and Manley, 2017). Therefore, there is an imperative for accurate R-loop quantification in various experimental and disease contexts.
The RNA–DNA hybrid portion of the R-loop is typically detected as a marker for R-loop levels. Historically, the S9.6 mAb has been the main reagent used, having been developed to recognize RNA–DNA hybrids (Boguslawski et al., 1986). Indeed, the S9.6 antibody has been used extensively in biochemical, next-generation sequencing, and imaging experiments in different organisms, cell types, and disease states to probe RNA–DNA hybrid levels (Ginno et al., 2012; Wahba et al., 2016; Barroso et al., 2019; García-Rubio et al., 2015; Castellano-Pozo et al., 2012; Marabitti et al., 2019). However, the S9.6 antibody also binds to double-stranded RNA (dsRNA; Hartono et al., 2018; Kinney et al., 1989; Smolka et al., 2021) with a modestly weaker affinity than that of RNA–DNA hybrids (Kd 0.6 nM for RNA–DNA hybrids and Kd 2.7 nM for dsRNA; Phillips et al., 2013). For this reason, quantification of RNA–DNA hybrids with S9.6 has been shown to be problematic in some contexts, unless bulk RNA is removed before S9.6 immunoprecipitation (Zhang et al., 2015; Hartono et al., 2018; Smolka et al., 2021).
The RNase H enzymes are a conserved family of endonucleases that hydrolyze the phosphodiester backbone of the RNA moiety in RNA–DNA hybrids (Cerritelli and Crouch, 2009). In human cells, there are two RNase H proteins: H1 and H2. RNase H1 cleaves RNA in hybrids that are at least 4 bp in size, whereas RNase H2 is able to cleave RNA in RNA–DNA hybrids and to excise single ribonucleotides incorporated into DNA (Cerritelli and Crouch, 2009; Williams et al., 2016). As a monomer, RNase H1 is typically easier to manipulate experimentally than the RNase H2 trimer. RNase H1 harbors a hybrid binding domain and a catalytic domain that can be rendered inactive by mutating a single amino acid in the active site (Cerritelli and Crouch, 2009; Wu et al., 2001; Nowotny et al., 2007). Both the hybrid binding and catalytic domains of RNase H1 bind RNA–DNA hybrids with an affinity 25–30-fold over that of dsRNA (Nowotny et al., 2007, 2008), making RNase H1 an attractive alternative to the S9.6 antibody for hybrid detection.
R-loop detection by imaging is a rapid and efficient way to monitor RNA–DNA hybrid levels. It can provide information on a single-cell level and on populations of cells, and it can also be used to measure subcellular hybrid localization (Teloni et al., 2019; Silva et al., 2018; Koo et al., 2015). The S9.6 antibody has been widely used for hybrid quantification by indirect immunofluorescence in many different cell types. However, the binding of S9.6 to RNA makes it difficult to assess the nature of this readout. Recently, catalytically inactive RNase H has been employed as an alternative reagent for hybrid detection by being stably expressed in cells and used for imaging, fluorescence-activated cell sorting, and next-generation sequencing approaches (Nguyen et al., 2017; Teloni et al., 2019; Chappidi et al., 2020; Chen et al., 2017; Tan-Wong et al., 2019; Bhatia et al., 2014; Makharashvili et al., 2018). However, this approach is labor intensive because each cell line needs to be individually engineered and may require extensive optimization before use.
Here, we employed recombinant GFP-tagged, human RNase H1 protein harboring the inactivating D210N mutation within its catalytic domain (GFP-dRNH1) for in situ imaging of fixed cells. We present it as a versatile tool for RNA–DNA hybrid detection that can be used in place of the S9.6 antibody. Importantly, by directly comparing the binding of GFP-dRNH1 and S9.6 to RNA–DNA hybrids and dsRNA, we show that GFP-dRNH1 is a more specific reagent than the S9.6 antibody for hybrid detection in fixed cells.
Results
Purified, catalytically inactive GFP-dRNH1 is specific toward RNA–DNA hybrids in vitro
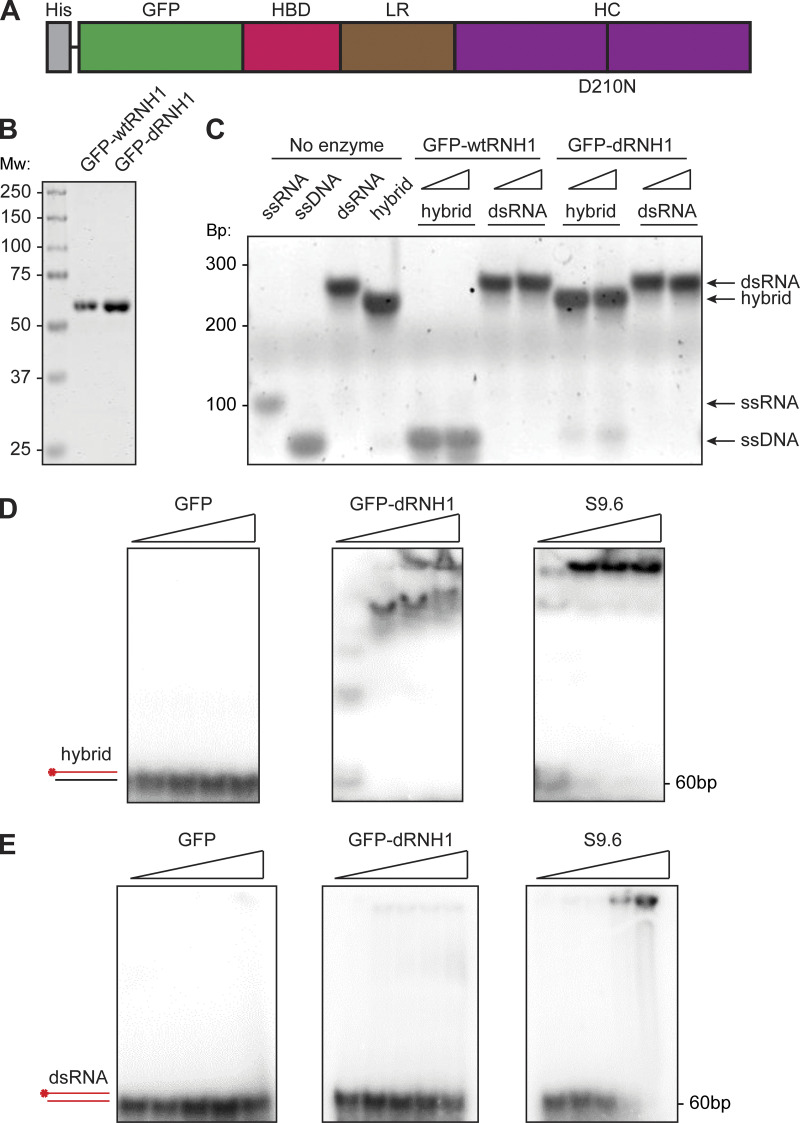
We engineered a construct of GFP-dRNH1 (Fig. 1 A; Nowotny et al., 2007; Wu et al., 2001). To ensure that protein integrity was maintained during purification, we also purified WT RNase H1 protein (GFP-wtRNH1) in parallel. We expressed these 59 kD fusion proteins in bacterial cells and purified them to homogeneity using affinity and ion-exchange columns (Fig. 1 B and Fig. S1). GFP-wtRNH1 was able to degrade the RNA strand of synthetic RNA–DNA hybrids but was not active on dsRNA, whereas GFP-dRNH1 had negligible activity on RNA–DNA hybrids, even at high concentrations (Fig. 1 C). We therefore conclude that GFP-dRNH1 is catalytically inactive and free from contaminating RNase activity.
Figure 1.
Purified GFP-dRNH1 is catalytically inactive and specific for RNA–DNA hybrids in vitro. (A) Schematic illustration of GFP-dRNH1. HBD, hybrid binding domain; HC, hybrid catalytic domain; His, His tag; LR, linker region. (B) Purified GFP-RNaseH1, both GFP-wtRNH1 and GFP-dRNH1, were resolved on a 4–12% Bis-Tris SDS-PAGE gel and stained with Coomassie G-250. Mw, molecular weight in kDa. (C) Products from enzymatic reactions following incubations of oligonucleotide substrates with GFP-wtRNH1 or GFP-dRNH1 were resolved on a polyacrylamide gel and stained with SYBR Gold. Bp, DNA size in base pairs. (D) RNA–DNA hybrids 60 bp in length were labeled with 32P, and 1 nM of labeled substrate was incubated with increasing concentrations of GFP (left), GFP-dRNH1 (middle), or S9.6 antibody (right; 1, 10, 20, 40 nM). The resulting complexes were resolved on a native polyacrylamide gel. Unbound RNA–DNA hybrids are indicated at the bottom of the gel. (E) dsRNA 60 bp in length was labeled with 32P, and 1 nM of substrate was incubated with increasing concentrations of GFP (left), GFP-dRNH1 (middle), or S9.6 antibody (right; 1, 10, 20, 40, 80 nM). The resulting complexes were resolved on a native polyacrylamide gel. Unbound dsRNA is indicated at the bottom of the gel.
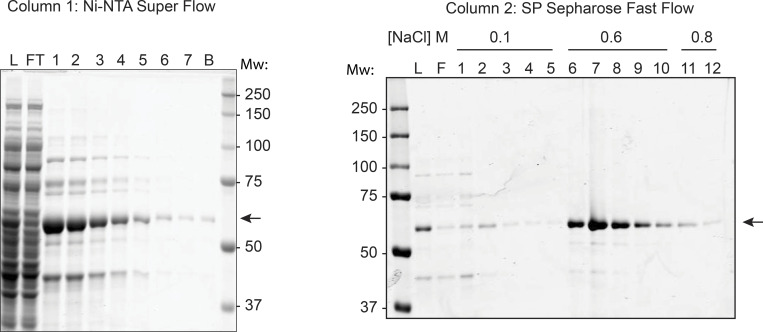
Figure S1.
Purification of GFP-RNH1. GFP-RNH1 was purified by Ni–nitrilotriacetic acid (Ni-NTA) superflow (left), followed by SP sepharose fast flow (right), and 5 μl of the indicated fractions were resolved on a 10% Tris-glycine SDS-PAGE gel and stained with Coomassie G-250 stain. Purification fractions analyzed included L, column load; FT, flow through; 1–7 or 1–12, eluted fractions; and B, leftover beads. The GFP-RNH1 protein is indicated by arrows. Mw, molecular weight in kDa.
Given that previous reports have shown that the S9.6 antibody can recognize both RNA–DNA hybrids and dsRNA (Phillips et al., 2013; Kinney et al., 1989; Hartono et al., 2018), we compared the recognition of S9.6 or GFP-dRNH1 for these nucleic acids by in vitro electrophoretic mobility shift assays. We found that both S9.6 and GFP-dRNH1 bind to RNA–DNA hybrids, even at low protein concentrations (Fig. 1 D). We further found that S9.6 bound robustly to dsRNA, whereas GFP-dRNH1 exhibited negligible binding at high concentrations (Fig. 1 E). A GFP control did not exhibit binding toward either substrate (Fig. 1, D and E). Thus, whereas S9.6 and GFP-dRNH1 both bind robustly to RNA–DNA hybrids, only S9.6 exhibits significant affinity for dsRNA, suggesting that GFP-dRNH1 could serve as a more specific reagent for imaging.
S9.6 antibody, but not GFP-dRNH1, binds to transfected dsRNA oligonucleotides
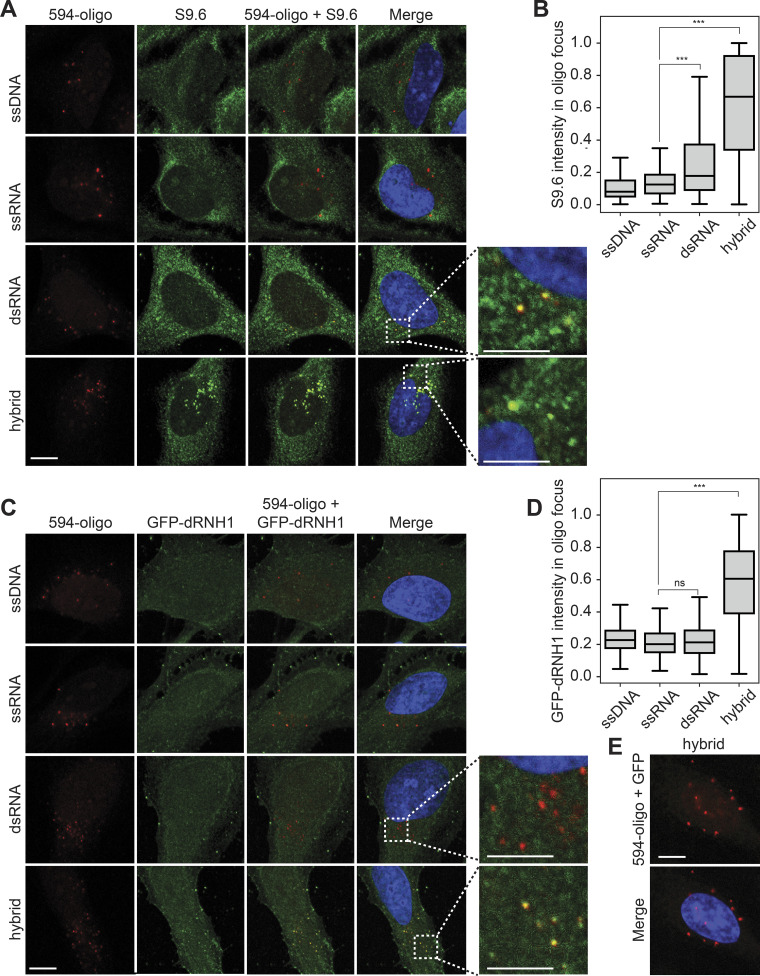
We then assessed whether GFP-dRNH1 can be used as a specific probe to image RNA–DNA hybrids in fixed human cells, comparing it with the current standard approach using the S9.6 antibody. To this end, we compared the binding specificities of GFP-dRNH1 and S9.6 to RNA–DNA hybrids and other nucleic acids in fixed cells. We transfected fluorescently (ATTO 594) labeled 60-mer oligonucleotides of ssDNA, single-stranded RNA (ssRNA), dsRNA, or RNA–DNA hybrids (hybrids) into cells and, after methanol fixation, used GFP-dRNH1 or S9.6 as a probe for imaging. S9.6 signal was amplified using a secondary antibody by standard indirect immunofluorescence, whereas GFP-dRNH1 was detected directly by measuring GFP intensity. All oligonucleotides formed distinct foci in transfected cells (Fig. 2, A and C). Cellular staining patterns showed that S9.6 exhibits a strong cytoplasmic signal, and, within the nucleus, binding was mostly enriched within the nucleoli. By contrast, GFP-dRNH1 exhibited pan-cellular staining, and, in the nucleus, the signal was not strongly enriched over nucleoli. As expected, both S9.6 and GFP-dRNH1 bound robustly to transfected hybrids, as seen by strong overlapping signal within hybrid foci, but not to ssDNA or ssRNA (Fig. 2, A–D). S9.6 also exhibited moderate binding to dsRNA oligonucleotides (Fig. 2, A and B). We detected similar binding properties in a different batch of S9.6 sourced commercially, indicating that S9.6 binding to dsRNA substrates is an inherent property of the antibody (Fig. S2). In contrast, GFP-dRNH1 did not bind to dsRNA above the background levels detected for ssDNA and ssRNA (Fig. 2, C and D). Importantly, GFP did not bind to cells or to transfected RNA–DNA hybrids (Fig. 2 E), indicating that dRNase H1 protein and not GFP determines substrate binding.
Figure 2.
S9.6 antibody but not GFP-dRNH1 binds to transfected dsRNA in human cells. (A) Representative confocal images showing HeLa cells transfected with ATTO-594 (red)-labeled ssDNA, ssRNA, dsRNA, or RNA–DNA hybrids and stained with S9.6 antibody (green). Inset images are magnified to show overlap of S9.6 signal and ATTO-594 foci. (B) Quantification of mean S9.6 intensities within individual oligonucleotide foci shown in A. (C) Same as in A, but with GFP-dRNH1 staining (green). (D) Quantification of mean GFP-dRNH1 intensities within individual oligonucleotide foci shown in C. (E) Representative confocal images of HeLa cells transfected with ATTO-594 (red) RNA–DNA hybrids and stained with GFP (green). Box plots show median (box central line), 25% and 75% percentiles (box edges), and minimum and maximum values (whiskers). Data are combined from three biological replicates (n = 3), with at least 60 oligonucleotide foci scored per condition per experiment. ***, P ≤ 0.001 by Mann-Whitney U test; ns, P > 0.05. Scale bars are 10 microns; 5 microns for inset images. Inset images are magnified to show overlap of S9.6 or GFP-dRNH1 signal with ATTO-594 foci.
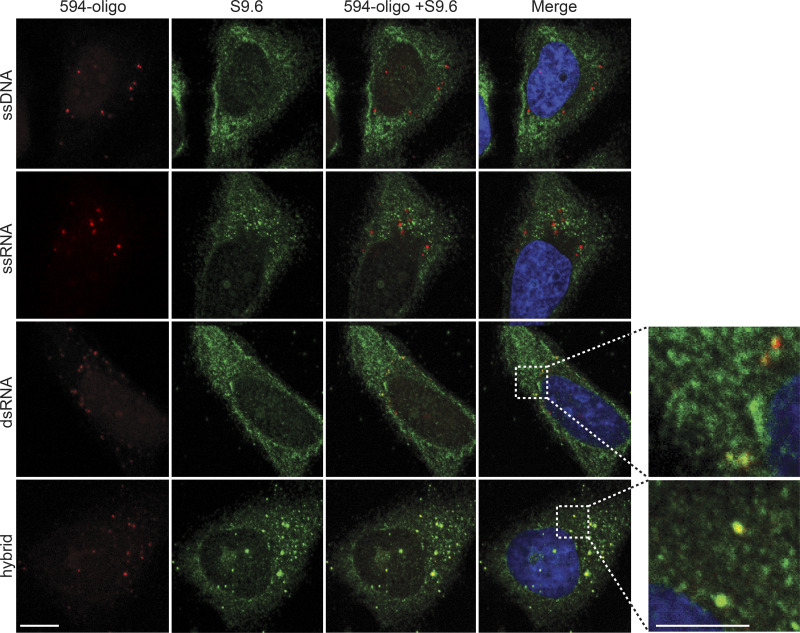
Figure S2.
Multiple batches of S9.6 antibody bind to transfected RNA–DNA hybrids and dsRNA in human cells. Representative confocal images of HeLa cells transfected with ATTO-594 (red)-labeled ssDNA, ssRNA, dsRNA, or RNA–DNA hybrids, followed by immunostaining with S9.6 antibody sourced from Kerafast (green). Inset images are magnified to show overlap of S9.6 signal and ATTO-594 foci. Scale bar is 10 microns; 5 microns for inset images.
S9.6 immunofluorescence signal in fixed cells arises predominantly from RNA binding
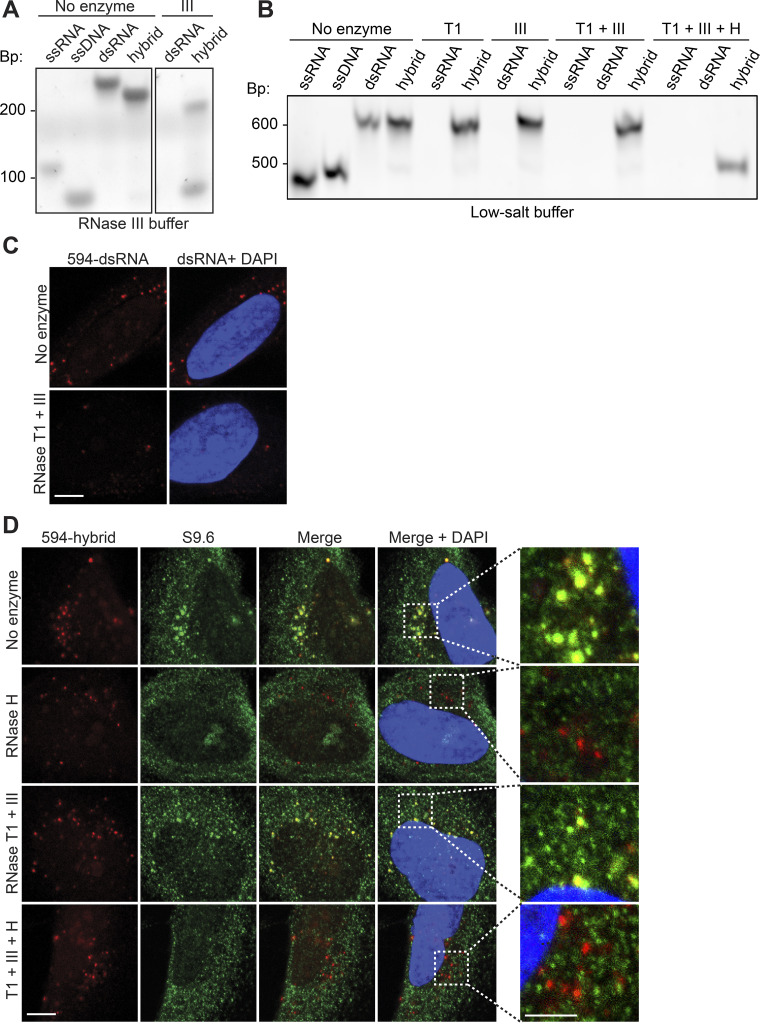
Given the observation that S9.6 bound to transfected dsRNA oligonucleotides, we next sought to compare the binding specificities of S9.6 and GFP-dRNH1 to endogenous RNA and RNA–DNA hybrids in fixed cells. To specifically degrade structured and dsRNAs without digesting RNA–DNA hybrids, we employed a combination of commercially sourced endoribonucleases (Ginno et al., 2012; Crossley et al., 2019; Chédin et al., 2021). RNase T1 acts on ssRNA and cleaves the phosphodiester backbone at guanine residues, whereas RNase III cleaves dsRNA into short fragments. We first established the specificities of these enzymes in vitro. We found that RNase III acted on RNA–DNA hybrids when incubated with its accompanying commercial buffer supplemented with manganese (Fig. S3 A). Moreover, this buffer removed cells from coverslips, making it inappropriate for our purposes. However, we found that in a low-salt, magnesium-containing buffer, RNases T1 and III specifically degraded ssRNA and dsRNA while leaving RNA–DNA hybrids intact (Fig. S3 B). A combination of these RNases, together with bacterial RNase H, effectively degraded all three nucleic acid substrates (Fig. S3 B). Treatment with RNases T1/III also reduced the fluorescence intensity of dsRNA oligonucleotides following transfection into cells (Fig. S3 C). Furthermore, treatment with RNase H, but not RNases T1 and III, reduced S9.6 signal intensity within RNA–DNA hybrid oligonucleotide foci (Fig. S3 D). Taken together, these data indicate that these RNases exhibit specific activity toward their intended substrates on fixed cells.
Figure S3.
Treatment of coverslips with RNases can selectively remove RNA and RNA–DNA hybrids. (A) Products from enzymatic reactions of oligonucleotide substrates with RNase III, incubated in commercially sourced ShortCut RNase III buffer supplemented with manganese. Hybrid, RNA–DNA hybrid. Products were resolved on a polyacrylamide gel and stained with SYBR Gold. (B) Same as in A, but oligonucleotides were digested with RNase T1 (T1); RNase III (III); RNases T1 and III combined (T1+ III); and RNases T1, III, and H combined (T1 + III + H) and incubated in low-salt, magnesium-containing buffer. (C) Representative confocal images of HeLa cells transfected with ATTO-594 (red)-labeled dsRNA. After fixation, coverslips were either mock treated or treated with a combination of RNases T1 and III. (D) Same as in C, but HeLa cells were transfected with ATTO-594 (red)-labeled RNA–DNA hybrids. After fixation, coverslips were treated with the following enzymes: none (No enzyme); RNase H; RNase T1 and RNase III combined (RNase T1 + III); or RNase H, RNase T1, and RNase III combined (T1 + III + H). S9.6 signal is shown in green and DAPI in blue. Scale bars are 10 microns; 5 microns for inset images. Inset images are magnified to show overlap of S9.6 signal and ATTO-594 foci. Bp, DNA size in base pairs.
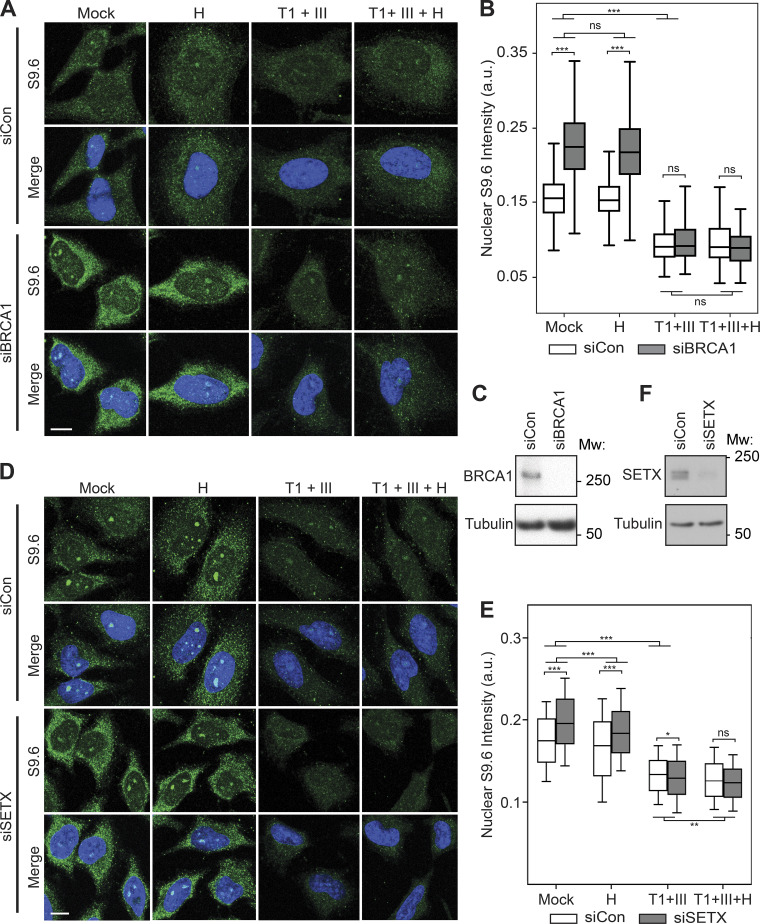
Next, we sought to determine the specificity of endogenous hybrid signal in fixed cells when R-loop levels are perturbed. To this end, we depleted cells of the R-loop resolution factor BRCA1 (Hatchi et al., 2015; Zhang et al., 2017; Chiang et al., 2019) or the RNA–DNA hybrid helicase senataxin (SETX; Hatchi et al., 2015; Cohen et al., 2018) using siRNA transfection, and we treated control (siCon), BRCA1-depleted (siBRCA1), and SETX-depleted (siSETX) cells with combinations of RNases before immunostaining with the S9.6 antibody. Without enzyme treatment, both siBRCA1 and siSETX cells showed a significant increase in S9.6 nuclear signal compared with siCon cells (Fig. 3, A, C, D, and F). Pretreatment of all samples with RNases T1 and III significantly reduced this signal. Importantly, following pretreatment of RNases T1 and III, the siCon, siBRCA1, and siSETX S9.6 signal intensities were reduced to parity (Fig. 3, B and E). This indicates that the majority of S9.6 signal under these conditions is derived from S9.6 binding to RNA. Conversely, pretreatment of fixed cells with RNase H minimally reduced S9.6 signal in siBRCA1 or siSETX cells and did not reduce signal to parity, further suggesting that the increased S9.6 signal is due to an increase in nonspecific binding of S9.6 to RNAs. Moreover, the triple combination of RNases H, T1, and III did not significantly further reduce S9.6 signal below that of RNases T1 and III combined. Taken together, these results indicate that the vast majority of S9.6 staining in fixed cells is derived from RNA and not from RNA–DNA hybrids, even upon perturbation of a factor that acts on R-loops in cells.
Figure 3.
Increased S9.6 immunofluorescent signal upon BRCA1 and SETX depletion is due to binding to dsRNA. (A) Representative confocal images of HeLa cells transfected with control or BRCA1-targeting siRNAs. After fixation, coverslips were treated with the following enzymes: none (Mock); RNase H (H); RNase T1 and RNase III combined (T1 + III); RNase H, RNase T1, and RNase III combined (T1 + III + H). S9.6 signal is shown in green, DAPI in blue. (B) Quantification of mean nuclear S9.6 intensities for the conditions shown in B. Box plots show median (box central line), 25% and 75% percentiles (box edges), and minimum and maximum values (whiskers). (C) Levels of BRCA1 analyzed by Western blotting. Tubulin serves as a loading control. (D) Same as in A, but with transfection of control or SETX-targeting siRNAs. (E) Quantification of mean nuclear S9.6 intensities for the conditions shown in D. Box plots show median (box central line), 25% and 75% percentiles (box edges), and 10% and 90% percentiles (whiskers). (F) Protein levels of SETX as analyzed by Western blotting. Tubulin serves as a loading control. Data are combined from three biological replicates (n = 3), with at least 120 nuclei scored per condition per experiment. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (by Mann-Whitney U test); ns, P > 0.05. Scale bars are 10 microns. Mw, molecular weight in kDa.
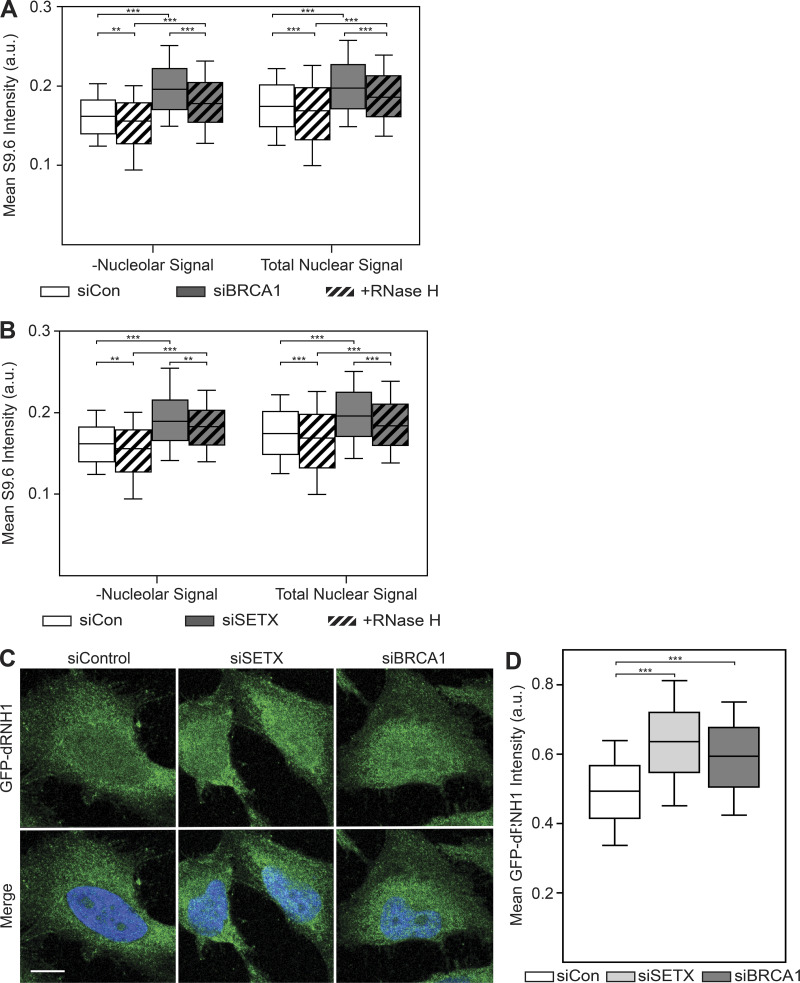
Because previous work has shown that the S9.6 antibody recognizes ribosomal RNAs (rRNAs) and that this likely accounts for the majority of its staining pattern, particularly in the nucleoli (Smolka et al., 2021), we sought to address whether the strong nucleolar staining of S9.6 was masking nuclear RNase H sensitivity. To this end, we excluded nucleolar regions, as identified by the nucleolin antibody, from quantification. With these parameters, RNase H pretreatment only modestly decreased the S9.6 signal and did not reduce it to parity with the siCon cells (Fig. S4, A and B). Importantly, reductions in S9.6 signal after RNase H treatment were comparable between intensity measurements that either excluded nucleolar regions or included them (compare left and right of Fig. S4, A and B). Taking these data together, we conclude that the majority of S9.6 signal is derived from dsRNAs even when nucleolar regions are excluded from measurement.
Figure S4.
Exclusion of nucleolar regions does not resensitize S9.6 immunostaining to RNase H treatment, whereas GFP-dRNH1 is compatible with PFA fixation.(A) Quantification of mean nuclear S9.6 signal in control or siBRCA1 cells after excluding the nucleolar regions (left). Total mean nuclear S9.6 signal after BRCA1 depletion (Fig. 3 B) is shown on the right for reference. (B) Same as in A but with SETX depletion. Nucleolar regions were excluded (left), whereas total mean nuclear S9.6 signal after SETX depletion (Fig. 3 E) is shown on the right for reference. (C) Representative confocal images of 4% PFA-fixed HeLa cells. After fixation, coverslips were incubated with GFP-dRNH1. GFP-dRNH1 signal is shown in green and DAPI in blue. (D) Quantification of mean nuclear GFP-dRNH1 intensities for the conditions shown in C. Box plots show median (box central line), 25% and 75% percentiles (box edges), and 10% and 90% percentiles (whiskers). Data are combined from three biological replicates (n = 3), with at least 100 nuclei scored per condition per experiment. **, P ≤ 0.01; ***, P ≤ 0.001 (Mann-Whitney U test). Scale bars are 10 microns.
GFP-dRNH1 is a more specific probe than S9.6 for imaging RNA–DNA hybrids in fixed cells
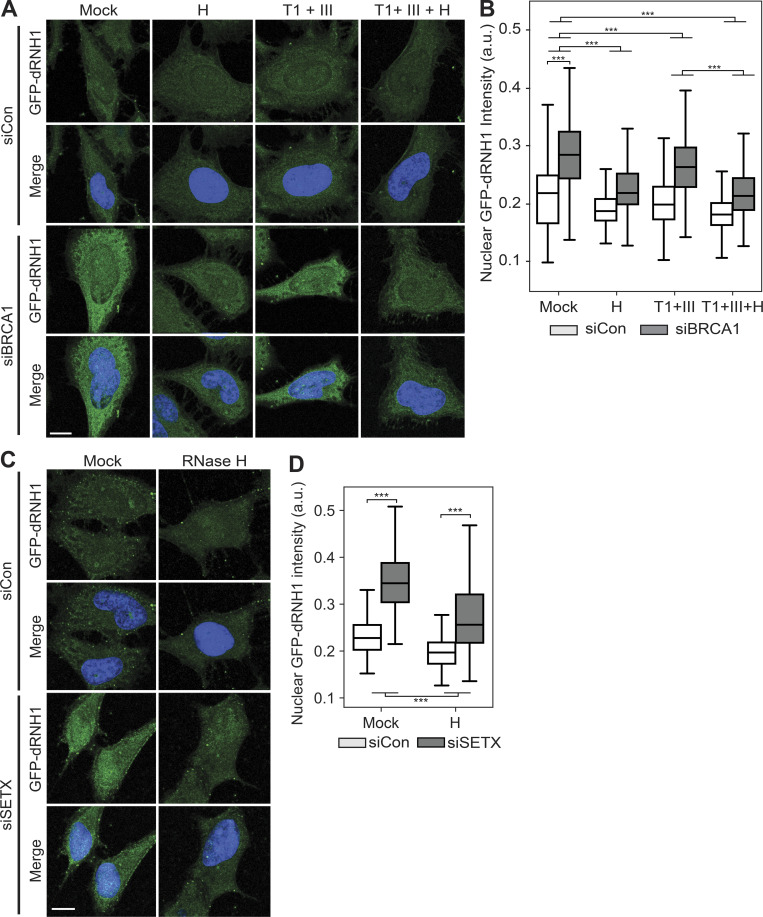
Next, we assessed the specificity of GFP-dRNH1 binding in fixed cells upon depletion of BRCA1. In mock-treated conditions, siBRCA1 cells showed a robust increase in GFP-dRNH1 signal compared with siCon cells (Fig. 4, A and B). Upon quantifying the nuclear signal, we found that pretreatment of siCon and siBRCA1 samples with RNase T1 and III resulted in a modest but significant reduction in signal, indicating that a minority of the signal was due to GFP-dRNH1 binding to RNA. Importantly, and in contrast with S9.6 staining, pretreatment with RNase H removed the majority of the GFP-dRNH1 signal in both siCon- and siBRCA1-treated cells. Furthermore, the triple combination of RNases H, T1, and III significantly reduced GFP-dRNH1 signal below that of RNases T1 and III combined, although a small amount of background signal still remained above that of siCon cells. Depletion of SETX also induced an increase in GFP-dRNH1 signal in cells, and this signal was also sensitive to pretreatment of coverslips with RNase H (Fig. 4, C and D). Notably, siBRCA1- and siSETX-treated cells also showed a robust increase in GFP-dRNH1 signal when cells were fixed with PFA (Fig. S4, C and D), suggesting that this probe is compatible with different cell fixation conditions. Taken together, these data indicate that GFP-dRNH1 is a more specific probe for RNA–DNA hybrids than S9.6 in fixed cells and is sensitive to changes in cellular levels of RNA–DNA hybrids upon perturbation.
Figure 4.
BRCA1 and SETX depletion leads to increases in RNA–DNA hybrids, detectable by GFP-dRNH1. (A) Representative confocal images of HeLa cells transfected with control or BRCA1-targeting siRNAs. After fixation, coverslips were treated with the following enzymes: none (Mock); RNase H (H); RNase T1 and RNase III combined (T1 + III); RNase H, RNase T1, and RNase III combined (T1 + III + H). GFP-dRNH1 signal is shown in green, DAPI in blue. (B) Quantification of mean nuclear GFP-dRNH1 intensities for the conditions shown in A. (C) Same as in A but with transfection of control or SETX-targeting siRNAs. (D) Quantification of mean nuclear GFP-dRNH1 intensities for the conditions shown in C. Box plots show median (box central line), 25% and 75% percentiles (box edges), and minimum and maximum values (whiskers). Data are combined from three biological replicates (n = 3), with at least 80 nuclei scored per condition per experiment. ***, P ≤ 0.001 (Mann-Whitney U test). Scale bars are 10 microns.
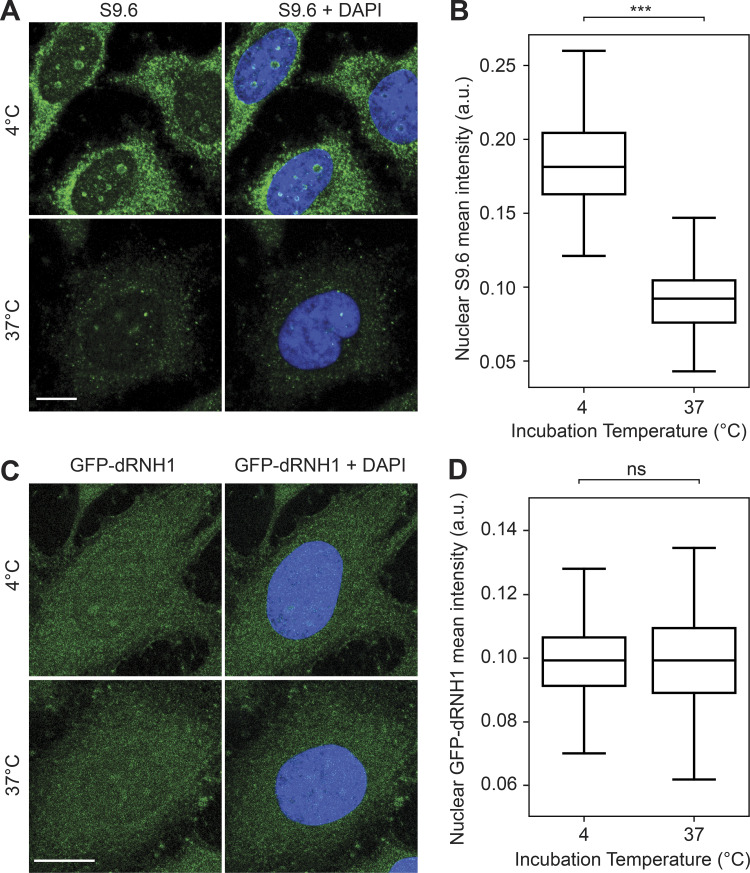
Somewhat surprisingly, we also found that S9.6 binding on coverslips is highly sensitive to incubation temperature, with a strong reduction in S9.6 fluorescence intensity if coverslips were kept at 37°C as compared with 4°C (Fig. S5, A and B). Conversely, we did not observe a similar decrease in GFP-dRNH1 fluorescence intensity when incubated at 37°C as compared with 4°C (Fig. S5, C and D). This finding highlights that GFP-dRNH1 is a more stable probe that is suitable for diverse experimental conditions.
Figure S5.
S9.6 immunostaining is sensitive to incubation temperature. (A) Representative confocal images of methanol-fixed HeLa cells. After fixation, coverslips were incubated in PBS and kept at 4°C or 37°C overnight, followed by S9.6 immunostaining. S9.6 signal is shown in green and DAPI in blue. (B) Quantification of mean nuclear S9.6 intensities for the conditions shown in A. (C) Same as in A but for GFP-dRNH1 staining. (D) Same as for B but for GFP-RNH1 staining. Scale bars are 10 microns. Box plots show median (box central line), 25% and 75% percentiles (box edges), and minimum and maximum values (whiskers). Data are combined from two biological replicates (n = 2), with at least 45 nuclei scored per condition per experiment. ***, P ≤ 0.001 (Mann-Whitney U test); ns, P > 0.05.
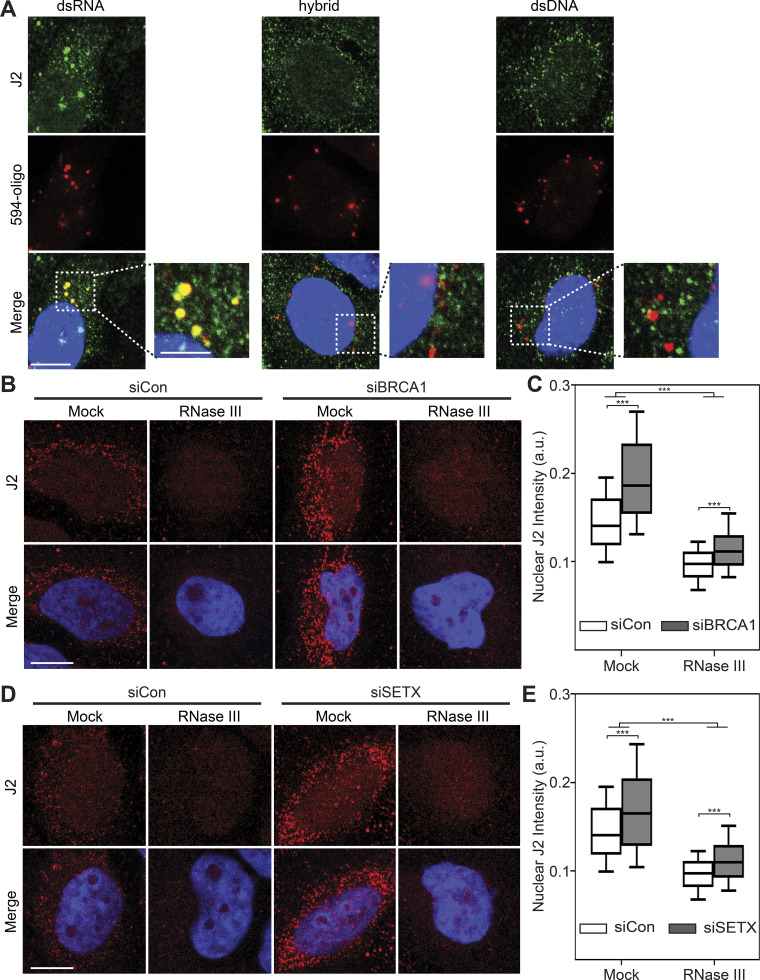
BRCA1 and SETX depletion results in increased production of dsRNA species
Although our results suggest that S9.6 signal predominantly arises from nonspecific binding to cellular RNAs, we consistently observed a reproducible and robust increase in S9.6 signal upon BRCA1 or SETX depletion (Fig. 3), suggesting that RNA levels or processing may be disrupted so that dsRNAs accumulate. To directly test this idea, we monitored dsRNA levels after siBRCA1 or siSETX treatment using the dsRNA-specific antibody J2. We first tested the specificity of this antibody by transfecting cells with fluorescently labeled dsDNA, dsRNA, or hybrid oligonucleotides and assessing colocalization with J2. The J2 antibody demonstrated overlapping signal only with dsRNA and not a hybrid or dsDNA oligonucleotides (Fig. 5 A), suggesting that this antibody is indeed specific for dsRNAs and can be used as a reliable readout for cellular dsRNA levels. We then monitored dsRNA levels after BRCA1 or SETX depletion using immunofluorescence with the J2 antibody. Loss of both BRCA1 and SETX resulted in a robust increase in J2 signal compared with siCon cells (Fig. 5, B–E). Importantly, pretreatment of samples with RNase III dramatically reduced the J2 signal and brought the signal to near parity with siCon cells. Taken together, our data suggest that, in addition to increasing RNA–DNA hybrid levels, dsRNA species accumulate in cells upon BRCA1 and SETX loss and identify a novel cellular phenomenon upon disruption of both these proteins.
Figure 5.
Loss of BRCA1 and SETX increases cellular dsRNA. (A) Representative confocal images showing HeLa cells transfected with ATTO-594 (red)-labeled dsRNA or RNA–DNA hybrids and stained with the J2 antibody (green). Inset images are magnified to show overlap of J2 signal and ATTO-594 foci. (B) Representative confocal images of HeLa cells transfected with control or BRCA1-targeting siRNAs. After fixation, coverslips were either mock treated or treated with RNase III. J2 signal is shown in red, DAPI in blue. (C) Quantification of mean nuclear J2 intensities shown in B. (D) Same as in B but following SETX depletion. (E) Quantification of mean nuclear J2 intensities shown in D. All box plots show median (box central line), 25% and 75% percentiles (box edges), and 10% and 90% percentiles (whiskers). Data are combined from three biological replicates (n = 3), with at least 75 nuclei scored per condition per experiment. ***, P ≤ 0.001 (by Mann-Whitney U test); ns, P > 0.05. Scale bar is 10 microns; 5 microns for inset images.
Discussion
We have compared the widely used S9.6 antibody with recombinant GFP-dRNH1 protein as tools for detecting RNA–DNA hybrids by imaging in fixed cells. As expected, we found that while S9.6 binds strongly to RNA–DNA hybrids, it also exhibits robust binding to dsRNA, both in vitro and in cells (Figs. 1, 2, and S2). In the cellular context where RNA is much more abundant than RNA–DNA hybrids, S9.6 immunofluorescent signal is predominantly attributable to RNA and not RNA–DNA hybrid binding (Fig. 3). We also show that recombinant GFP-dRNH1 can be readily purified (Figs. 1 and S1) and used directly on fixed cells to image RNA–DNA hybrids (Figs. 4 and S4). Because GFP-dRNH1 has a relatively weak affinity for dsRNA (Figs. 1 and 2), in a cellular context, GFP-dRNH1 is able to specifically bind to RNA–DNA hybrids, with a much smaller portion of cellular signal being attributable to RNA binding compared with S9.6. We also establish important controls to test the specificity of RNA–DNA hybrid probes in imaging and show that buffer composition and temperature are important variables that must be kept consistent during sample processing (Figs. S3 and S5). Finally, we show that increased S9.6 staining following the loss of the R-loop resolution factors BRCA1 and SETX is in fact due to increases in dsRNA (Figs. 3 and 5), whereas GFP-dRNH1 is able to detect increases in nuclear RNA–DNA hybrids under these cellular perturbations (Fig. 4).
Taken together, our results and those of a recent study (Smolka et al., 2021) show that S9.6 should be used with caution to detect RNA–DNA hybrids by immunofluorescence, especially under conditions in which RNA is highly abundant and when experimental readouts cannot readily distinguish between RNA–DNA hybrids and RNA. Although it may be possible to include steps in such experiments to remove abundant cellular RNA, we found that the S9.6 signal in siBRCA1 and siSETX cells was similar to siCon cells when coverslips were treated with RNases T1 and III before S9.6 immunolabeling. Moreover, the remaining S9.6 signal was not readily attributable to RNA–DNA hybrids, because it was not sensitive to RNase H. It is possible that residual RNA species remain under these conditions and prevent binding to RNA–DNA hybrids that are present in cells. Certain RNAs also could be refractory to RNases T1 and III, or complete enzymatic digestion may require treatment conditions that could perturb RNA–DNA hybrids. It is nonetheless possible to detect transfected RNA–DNA hybrids with S9.6 immunofluorescence, and enzymatic removal of RNA has also been reported to facilitate hybrid detection by imaging upon depletion of some cellular proteins (Barroso et al., 2019).
Our results also indicate that depletion of BRCA1 and SETX alters dsRNA levels in cells, in addition to increasing RNA–DNA hybrid levels. Because both SETX and BRCA1 regulate many aspects of transcription and RNA processing, and interact with RNA polymerase II, their loss could disrupt many of these processes (Skourti-Stathaki et al., 2011; Suraweera et al., 2009; Monteiro, 2000; Hatchi et al., 2015; Zhang et al., 2017). In turn, this could affect the levels and distribution of dsRNAs in the nucleus and cytoplasm (Chédin et al., 2021). Additionally, transcription of ribosomal DNA could also be affected, giving rise to nucleolar stress and increased ribosomal biogenesis (Yang et al., 2018). Indeed, because S9.6 signal was found to primarily reflect rRNA binding (Smolka et al., 2021), the increased S9.6 staining observed upon BRCA1 and SETX depletion may be suggestive of altered ribosomal biogenesis. Importantly, increases in RNA–DNA hybrids can be detected with GFP-dRNH1 under these conditions, whereas the high abundance and increased load of dsRNA masks the real increase in hybrid levels when using S9.6.
Because many factors associated with regulating R-loop levels are involved in transcriptional and post-transcriptional processes, increased S9.6 immunofluorescent signal from dsRNA species, including rRNAs, may be a widespread phenomenon when these processes are disrupted. In any case, sensitivity to RNase H is necessary to demonstrate that the readouts are due to the presence of RNA–DNA hybrids and not RNA. Importantly, because the S9.6 signal may be temperature sensitive, mock- and enzyme-digested samples must also be treated equally. Interestingly, we also observed GFP-dRNH1 binding in the cytoplasm, and this signal was RNase H sensitive. Although some studies have indicated the presence of RNA–DNA hybrids in mitochondria (Silva et al., 2018; Holt, 2019), the nature of this signal is currently poorly characterized and warrants further investigation.
The nonspecific binding of S9.6 to dsRNA may also be a confounding factor in other methods that use S9.6 for hybrid detection and do not exclude the presence of RNA in the readout. Indeed, this problem has been observed in S9.6-based sequencing experiments when sequencing RNA molecules following DNA-RNA immunoprecipitation (Hartono et al., 2018; Chédin et al., 2021). Nevertheless, the S9.6 antibody remains a powerful tool for detecting RNA–DNA hybrids in a wide range of applications, such as in DNA-RNA immunoprecipitation–based sequencing approaches in which the DNA portion of the hybrid is sequenced (Ginno et al., 2012; Crossley et al., 2019; Chédin et al., 2021; Crossley et al., 2020).
Although GFP-dRNH1 is more specific than the S9.6 antibody, the use of GFP-dRNH1 or other forms of recombinant RNase H1 protein may have its own limitations. For example, RNase H1 can bind several human proteins (Nguyen et al., 2017; Teloni et al., 2019; Chappidi et al., 2020), and it may have a binding preference for certain types of RNA–DNA hybrids that are G-rich (Nguyen et al., 2017; Teloni et al., 2019; Chappidi et al., 2020; Chen et al., 2017; Tan-Wong et al., 2019). Therefore, the binding of RNase H1 may be influenced by the localization of other binding proteins. Further engineering of RNase H as an RNA–DNA hybrid probe may improve its sensitivity and specificity, as well as extend the range of its potential applications. In contrast to methods that use stable expression of catalytically inactive RNase H in human cells for RNA–DNA hybrid detection (Nguyen et al., 2017; Teloni et al., 2019; Chappidi et al., 2020; Chen et al., 2017; Tan-Wong et al., 2019; Bhatia et al., 2014; Makharashvili et al., 2018), the purified protein we characterize herein is more versatile and does not require the engineering of individual cell lines. Furthermore, using purified GFP-dRNH1 protein avoids any alteration or stabilization of RNA–DNA hybrids levels in vivo, which can occur upon expression of the protein in cells (Tan-Wong et al., 2019; Kabeche et al., 2018; Li et al., 2020; Chédin et al., 2021). Finally, analysis of subcellular localization is limited with stably expressed RNase H due to the presence of localization domains and sequences that restrict its localization to specific cellular organelles (Suzuki et al., 2010; Shen et al., 2017). Use of purified GFP-dRNH1 protein on fixed cells could thus be applied across many cell types and experimental conditions.
In sum, the accurate detection and quantification of RNA–DNA hybrids in cells by imaging is highly desirable because it could provide insight into many areas of cell biology, but care must be taken to ensure such approaches are specific for detecting RNA–DNA hybrids. This remains an important challenge, and efforts must continue to better characterize and improve the use of S9.6, RNase H, and possibly other probes for RNA–DNA hybrid detection. We demonstrate catalytically inactive human RNase H1 protein as a sensitive, versatile, and more specific reagent for imaging RNA–DNA hybrids than the current standard approach for imaging using S9.6 immunofluorescence.
Materials and methods
Expression and purification of GFP-RNase H1
A tobacco etch virus–cleavable, N-terminal tandem His-GFP–tagged RNase H1 (both WT and D210N, excluding residues 1–27 containing the mitochondrial localization signal) was amplified by PCR, subcloned to vector 1GFP (California Institute for Quantitative Biosciences, University of California, Berkeley MacroLab; Addgene) using NEBuilder HiFi assembly (New England Biolabs), and sequence verified using the University of California, Berkeley Sequencing Facility. Plasmid 1GFP/RNase H1 was transformed into Escherichia coli BL21(DE3)/Rosetta (Novagen/MilliporeSigma), cultured in lysogeny broth media supplemented with kanamycin (50 µg/ml) and chloramphenicol (25 µg/ml), and induced by IPTG (final concentration of 0.3 mM) at 16°C for 18 h when cell density reached A600 ∼0.4. All of the following steps were performed on ice or at 4°C. Induced bacterial cells (50 ml) were harvested and pelleted by centrifugation at 10,000 ×g for 10 min. Cells were resuspended in 5 ml Ni binding buffer (50 mM NaH2PO4, 5 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.3% NP-40, 10% glycerol, 10 mM imidazole, 1 mM β-mercaptoethanol, 1 mM PMSF, and EDTA-free protease inhibitor cocktail tablet [11836170001; Roche/MilliporeSigma]) and lysed by sonication. Total cell lysate was centrifuged at 20,000 ×g for 30 min. The cleared supernatant was transferred to a new 15-ml conical tube containing preequilibrated Ni–nitrilotriacetic acid Superflow beads (Qiagen; column volume [CV] 350 μl for WT and CV 500 μl for D210N) and set up for batch binding for 1 h with gentle rotation, followed by centrifugation at 700 ×g for 5 min. The pelleted beads were washed five times with 10 ml Ni wash buffer (50 mM NaH2PO4, 5 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.3% NP-40, 10% glycerol, 20 mM imidazole, 1 mM β-mercaptoethanol, 1 mM PMSF), then transferred to a 1.7-ml microtube and washed thrice with 1 ml Ni wash buffer. Samples were eluted eight times with 1 CV Ni elution buffer (50 mM NaH2PO4, 5 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.3% NP-40, 10% glycerol, 300 mM imidazole, 1 mM β-mercaptoethanol, 1 mM PMSF), each with 5-min incubation. Peak fractions containing GFP-RNase H1 were pooled in a Slide-A-Lyzer dialysis cassette with 10 kDa molecular weight cutoff (Thermo Fisher Scientific) and dialyzed overnight against sulphopropyl (SP) binding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.3% NP-40, 10% glycerol, 1 mM β-mercaptoethanol, 1 mM PMSF). Dialyzed samples were applied to an SP sepharose column (GE Healthcare; CV 125 μl), and the flow-through fraction was collected and reapplied to the column two more times. The samples were washed and eluted via a step salt gradient (40 CV of 0.1 M, 8 CV of 0.6 M, and 2 CV of 0.8 M NaCl), and peaked eluted fractions containing GFP-RNase H1 were pooled and dialyzed overnight into the storage buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 0.3% NP-40, 10% glycerol, 1 mM β-mercaptoethanol, 1 mM PMSF). The dialyzed samples were flash frozen and stored at −80°C. Purified GFP-RNase H1 was analyzed with BSA standards by Coomassie-stained 4–12% Bis-Tris SDS-PAGE (Thermo Fisher Scientific) to determine protein purity and concentrations. The protein stocks at 0.188 mg/ml were aliquoted, stored at −80°C, and freshly thawed for each experiment.
In vitro testing of RNases T1, III, and H and GFP-RNase H protein
DNA and RNA oligonucleotides were synthesized and HPLC purified by Integrated DNA Technologies. Lyophilized oligonucleotides were resuspended in UltraPure distilled water (Invitrogen) at 250 µM and diluted to 25 µM before use. For annealing to form RNA–DNA hybrid or dsRNA oligonucleotides, DNA and/or RNA oligonucleotides were diluted to 2.5 µM in annealing buffer (10 mM Tris-HCl, pH 7.4, 50 mM NaCl), incubated at 95°C for 5 min, and cooled to RT. ssDNA and ssRNA oligonucleotides were similarly diluted to 2.5 µM in annealing buffer. Nucleic acid substrates (2 μl) were added to make a 10-µl reaction containing enzymes (2 units of RNase III [M0245L; New England Biolabs], 1,000 units RNase T1 [EN0542; Thermo Fisher Scientific], a combination of both, or both plus 5 units of RNase H [M0297L; New England Biolabs] or 500 ng of purified GFP-RNase H1 protein) and enzyme digestion buffer: For RNases T1 and III, the enzyme digestion buffer used was 50 mM Tris-HCl pH 7.6, 75 mM KCl, 3 mM MgCl2, 0.1% BSA; for RNase III, additionally 1× ShortCut buffer (New England Biolabs) was used with manganese added per the manufacturer’s instructions; for reactions containing purified GFP-RNase H, the buffer used was 50 mM Tris-HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2, 10 mM DTT. Reactions were mixed and incubated at 37°C for 1 h. Novex Hi-Density Tris-Borate-EDTA Sample Buffer (LC6678; Thermo Fisher Scientific) was added to each sample, which was then resolved on a 13.3% nondenaturing acrylamide gel at 10 mA for 1.5 h. The gel was stained for 10 min in SYBR Gold Nucleic Acid Gel Stain (S-11494; Thermo Fisher Scientific) and imaged on an Alpha Innotech FluorHD2 imager. Oligonucleotide sequences used for enzyme reactions are as follows: RNA1: 5′-AUAUGGGAACCACUGAUCCC-3′; RNA2: 5′-GGGAUCAGUGGUUCCCAUAU-3′; DNA: 5′-GGGATCAGTGGTTCCCATAT-3′. For dsRNA annealing, RNA1/RNA2 were used, and for RNA–DNA hybrid annealing, RNA1/DNA were used.
Cell culture
HeLa cells were obtained from the American Type Culture Collection, where they were tested for mycoplasma and verified by short tandem repeat profiling. These cells were grown in DMEM (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin/glutamine and grown in 5% CO2 at 37°C. Cells were seeded onto 1.5-mm coverslips in 24-well plates. For siRNA transfections, reverse transfection was performed according to the manufacturer’s instructions using Lipofectamine RNAiMax (Thermo Fisher Scientific) and 20 nM of the indicated siRNA. Transfection reaction mixtures were incubated at RT for 20 min before adding them dropwise to the cell mixture onto coverslips. Culture media were replaced with fresh media 24 h after transfection. Transfections were performed for 42–48 h, and cells were grown to ∼50–70% confluency before harvesting. Sequences of siRNAs used were as follows: siCon (luciferase GL#3, D-001400-01-20; Dharmacon), siBRCA1 (SI02654575; Qiagen), and siSETX (5′-GCCAGAUCGUAUACAAUUA-3′; Dharmacon).
Oligonucleotide transfection and imaging
DNA and RNA oligonucleotides were synthesized and HPLC purified by Integrated DNA Technologies. Lyophilized oligonucleotides were resuspended in UltraPure distilled water to make a 40 µM stock solution. For annealing to form RNA–DNA hybrids or dsRNA oligonucleotides, DNA and/or RNA oligonucleotides were diluted to 10 µM in annealing buffer (10 mM Tris-HCl, pH 7.4, 50 mM NaCl), incubated at 95°C for 5 min, and cooled to RT. ssDNA and ssRNA oligonucleotides were similarly diluted to 10 µM in annealing buffer, incubated at 95°C for 5 min, and cooled to RT. Transfections were performed according to the manufacturer’s instructions using Lipofectamine RNAiMax and a 1:100 dilution of hybridized oligonucleotides in 100 µl Opti-MEM. Transfection mix was added dropwise to cells in 500 µl complete media and performed for 3 h. After 3 h, cells were washed once with warm media and incubated for an additional 3 h before fixation. For immunofluorescence, all incubations were performed at RT with rocking unless otherwise indicated. Cells were washed once with 1× PBS, fixed with ice-cold methanol for 5 min at −20°C, and subsequently washed twice briefly with 1× PBS. For enzymatic digestion, coverslips were incubated in low-salt buffer (50 mM Tris-HCl, pH 7.6, 75 mM KCl, 3 mM MgCl2, 0.1% BSA) with 1:150 of RNase T1 (EN0542; Thermo Scientific) or ShortCut RNase III (New England Biolabs), or both enzymes added together, for 1.5–2 h at 37°C. RNase H (New England Biolabs) was diluted 1:50 in 1× RNase H buffer (New England Biolabs) and incubated on coverslips for 3–5 h at 37°C. For a combination of all three enzymes, coverslips were first incubated with RNase H, washed in low-salt buffer, and then incubated with RNases T1 and III in low-salt buffer as described above. Mock-treated coverslips were incubated in parallel in the same enzymatic reaction buffers but without enzymes added. Following enzyme incubations, coverslips were washed twice in PBS with Tween 20 (PBST), then once in PBS for 5 min each, and blocked with staining buffer (3% BSA in PBS) for 30 min. Samples were incubated with a 1:200 dilution of S9.6 antibody (mouse, ENH001 [Kerafast] or 1 mg/ml stock [Antibodies Inc.]), a 1:2,000 dilution of GFP-dRNH1 (0.188 mg/ml stock), or a 1:300 dilution of GFP (Thermo Fisher Scientific, 88899; 0.46 mg/ml stock) in staining buffer overnight at 4°C. For J2 antibody staining, cells were fixed with 4% PFA for 10 min, washed briefly twice with 1× PBS, and then permeabilized with 0.25% Triton X-100 in PBS for 10 min. Samples were blocked and stained with a 1:200 dilution of dsRNA (J2) antibody (from a 1 mg/ml stock, mouse; 10010200, SCICONS). In all cases, after being washed thrice with PBST for 5 min, samples were incubated with a 1:2,000 dilution of anti-mouse Alexa Fluor 488–conjugated secondary antibody (A11001 goat; Invitrogen), 5 ng/ml DAPI (32670; MilliporeSigma), and 0.2 µl HCS Cellmask Deep Red (H32721; Thermo Fisher Scientific) in staining buffer for 1 h. Samples were washed thrice for 10 min with PBS, and coverslips were mounted onto glass slides using Prolong Glass antifade mountant (P36984; Invitrogen) and stored at 4°C in the dark until imaging.
Oligonucleotide sequences used for transfection were as follows: ATTO590-conjugated DNA: 5′-ATTO590N-GTACCGGGATCCTCTAGAGTCGAGCGTCGATCCGAACTTGGCACTGGCCGTCGTTACAAC-3′; ATTO590-conjugated RNA: 5′-ATTO590N-GUACCGGGAUCCUCUAGAGUCGAGCGUCGAUCCGAACUUGGCACUGGCCGUCGUUACAAC-3′; 60-mer RNA (RNA60): 5′-GUUGUAACGACGGCCAGUGCCAAGUUCGGAUCGACGCUCGACUCUAGAGGAUCCCGGUAC-3′.
Electrophoretic mobility shift assays
A 60-mer RNA oligonucleotide (RNA60) was labeled with [γ-32P]dATP (PerkinElmer) and annealed with either an unlabeled 60-mer DNA (DNA60) or its antisense RNA (asRNA60) oligonucleotide to form a 60-bp-long hybrid or dsRNA substrate. To start the reaction, GFP, S9.6 antibody, or GFP-dRNaseH1 protein was mixed with 1 nM labeled substrate at molar oligonucleotide/protein ratios ranging from 1:1 to 1:80. Samples were incubated for 30 min at 37°C in a total volume of 10 µl containing 25 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA, pH 8.0, 1 mM DTT, 6% glycerol, 0.1% NP-40, 0.1 mg/ml BSA, RNaseOUT, and 0.5 mM PMSF. After the binding reaction, the samples were resolved on a 6% polyacrylamide gel in 0.5× Tris-Borate-EDTA buffer at 4°C and visualized on a Typhoon Phosphorimager.
Oligonucleotide sequences used were as follows: DNA60: 5′-GTACCGGGATCCTCTAGAGTCGAGCGTCGATCCGAACTTGGCACTGGCCGTCGTTACAAC-3′; RNA60: 5′-GUUGUAACGACGGCCAGUGCCAAGUUCGGAUCGACGCUCGACUCUAGAGGAUCCCGGUAC-3′; asRNA60: 5′-GUACCGGGAUCCUCUAGAGUCGAGCGUCGAUCCGAACUUGGCACUGGCCGUCGUUACAAC-3′.
Immunofluorescence with S9.6 antibody
Cells were washed once with 1× PBS and fixed with ice-cold 100% methanol for 5 min at −20°C. Cells were washed briefly twice with 1× PBS. For enzymatic digestion, coverslips were incubated in low-salt buffer (50 mM Tris-HCl, pH 7.6, 75 mM KCl, 3 mM MgCl2, 0.1% BSA) with 1:150 of RNase T1 (EN0542; Thermo Scientific) or ShortCut RNase III (New England Biolabs), or both enzymes added together, for 1.5–2 h at 37°C. RNase H (New England Biolabs) was diluted 1:50 in 1× RNase H buffer (New England Biolabs) and incubated on coverslips for 3–5 h at 37°C. For a combination of all three enzymes, coverslips were first incubated with RNase H, washed in low-salt buffer, and then incubated with RNases T1 and III in low-salt buffer as described above. Mock-treated coverslips were incubated in parallel in the same enzyme reaction buffers but without enzymes added. Following enzyme incubations, coverslips were washed twice in PBST, then once in PBS for 5 min each, and blocked with staining buffer (3% BSA in PBS) for 30 min. Samples were incubated in staining buffer with a 1:200 dilution of S9.6 antibody (mouse, from a 1 mg/ml stock; Antibodies Inc.) and a 1:2,000 dilution of anti-nucleolin (ab22758 rabbit; Abcam) overnight at 4°C. After washing thrice with PBS for 5 min, samples were incubated with a 1:1,000 dilution of anti-mouse Alexa Fluor 488–conjugated and anti-rabbit Alexa Fluor 594 (A32742 goat; Invitrogen)–conjugated secondary antibodies, 5 ng/ml DAPI, and 0.2 µl HCS Cellmask Deep Red in staining buffer for 1 h. Samples were washed thrice for 10 min with PBS, and coverslips were mounted onto glass slides using Prolong Glass antifade mountant and stored at 4°C in the dark until imaging.
Imaging with GFP-dRNase H1
Cells were washed once with 1× PBS and either fixed with ice-cold methanol for 5 min at −20°C or fixed with 4% PFA for 10 min, washed twice with 1× PBS, and permeabilized with 0.25% Triton X-100 in PBS. Cells were subsequently washed briefly twice with 1× PBS and blocked with staining buffer (3% BSA in PBS) for 30 min. For enzymatic digestion, samples were treated and then washed as described in the previous paragraph. Cells were blocked with staining buffer (3% BSA in PBS) for 30 min. Samples were then incubated with a 1:2,000 dilution of GFP-dRNH1 at 0.188 mg/ml in staining buffer for 1.5 h at 37°C. After washing with PBST thrice for 5 min, samples were incubated with 5 ng/ml DAPI and 0.2 µl HCS Cellmask Deep Red in PBS for 20 min. Samples were washed thrice for 10 min with PBS, and coverslips were mounted onto glass slides using Prolong Glass antifade mountant and stored at 4°C in the dark and imaged within 1–2 d.
Immunofluorescence with J2 antibody
Unless otherwise indicated, all incubations were performed at RT with rocking. Cells were washed once with 1× PBS and fixed with 4% PFA for 10 min. Cells were then washed twice briefly with 1× PBS, permeabilized with 0.25% Triton X-100 for 10 min, and again washed twice briefly with 1× PBS. Coverslips were then incubated in low-salt buffer (50 mM Tris-HCl, pH 7.6, 75 mM KCl, 3 mM MgCl2, 0.1% BSA) with a 1:150 dilution ShortCut RNase III (New England Biolabs) for 90 min at 37°C. Mock-treated coverslips were incubated in parallel in the same enzymatic reaction buffers without enzyme. Following enzyme incubations, coverslips were washed twice in PBST, then once in PBS for 5 min each, and blocked with staining buffer (3% BSA in PBS) for 30 min. Samples were incubated in staining buffer with a 1:200 dilution of dsRNA (J2) antibody overnight at 4°C. After being washed twice with PBST and once with PBS for 5 min, samples were incubated with a 1:1,000 dilution of anti-mouse Alexa Fluor 594–conjugated secondary antibody, 5 ng/ml DAPI, and 0.2 µl HCS Cellmask Deep Red in staining buffer for 1 h. Samples were washed thrice for 10 min with PBS, and coverslips were mounted onto glass slides using Prolong Glass antifade mountant and stored at 4°C in the dark until imaging.
Image acquisition and analysis
Image analysis was performed using CellProfiler (version 3.1.8). The DAPI channel was used to identify nuclei using the IdentifyPrimaryObjects module with manual thresholding. The 594 channel was used to identify nucleoli using the IdentifyPrimaryObjects module with global thresholding using minimum cross entropy and a threshold correction factor of 10. To exclude nucleolar areas from nuclear signal, the nucleoli were then expanded by 4 pixels and inverted from the nuclei to create a mask using the MaskObjects module. The mean intensity of S9.6, GFP-dRNH1, and J2 for each nucleus was calculated, and data were exported. For quantification of S9.6 or GFP-dRNH1 within oligonucleotide foci, the 594 channel of each image was used to identify oligonucleotide foci using the IdentifyPrimaryObjects module with manual thresholding and a diameter range of 1–35 pixel units to calculate an appropriate filter size. The mean intensity of the S9.6 and GFP-dRNH1 within these regions was then calculated. All images were acquired at 25°C on a Zeiss LSM800 confocal microscope equipped with an AxioImager.Z2 microscope using a 63×/1.40 NA differential interference contrast oil objective (Zeiss) with oil immersion. Images were acquired using ZEN (ZEISS Efficient Navigation) 2.6 blue edition software (version 2.6.76.0000), exported as 16-bit, all cropped, and adjusted equally in ImageJ (version 2.0.0).
Western blotting
Cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% deoxycholate, 0.1% SDS, 1% NP-40) supplemented with protease inhibitors (11873580001; Roche/MilliporeSigma). Whole-cell extracts were separated by gel electrophoresis, transferred onto polyvinylidene difluoride membranes, and blocked in 5% skim milk dissolved in 0.1% Tween/TBS for 1 h. Membranes were incubated with primary antibodies (mouse anti–α-tubulin mAb [T9026; Thermo Fisher Scientific], rabbit polyclonal anti-SETX [A301-105A; Bethyl Laboratories], and mouse anti-BRCA1 mAb [OP92; EMD Millipore]) overnight at 4°C followed by washing in 0.1% Tween/TBS. Membranes were incubated with HRP-linked secondary antibodies at 25°C for 1 h and washed thrice before signal detection. Membranes were developed by chemiluminescence using ECL reagent.
Data visualization and statistical analysis
Data visualization was performed either in Python using Matplotlib (version 2.1.2) and Seaborn (version 0.8.1) or in GraphPad Prism 8. Mann-Whitney rank tests were performed in Python using the SciPy stats package (version 1.0.0) or as calculated by GraphPad Prism 8.
Online supplemental material
Fig. S1 supports Fig. 1 and shows purification gels for GFP-dRNH1. Figs. S2 and S3 support Fig. 2. Fig. S2 shows oligonucleotide recognition with a commercial S9.6 antibody, whereas Fig. S3 characterizes the enzymatic activities of nucleases used in this study, both in vitro and on transfected oligonucleotides. Fig. S4 supports Figs. 3 and 4 and shows S9.6 signal when excluding nucleolar regions as well as GFP-dRNH1 staining with PFA fixation. Fig. S5 also supports Figs. 3 and 4 and demonstrates the respective sensitivities of S9.6 and GFP-dRNH1 to incubation temperature.
Acknowledgments
We thank Melody Kao for preliminary experiments that supported further development of this tool. We also thank Robert Veres and Aye Chan Thwin for their work in cloning and optimizing the expression and purification of GFP-RNase H proteins.
This work was supported by the Leukemia and Lymphoma Society (5455-17 to M.P. Crossley), the Jane Coffin Childs Memorial Fund for Medical Research (61-1755 to J.R. Brickner), and the National Institutes of Health (R01 GM119334 and GM126600 to K.A. Cimprich; R01 GM120607 to F. Chédin; and P01 CA092584 to M.-S. Tsai, S.M.T. Zar, S.S. Maw). K.A. Cimprich is an American Cancer Society Research Professor and is also supported by the V Foundation for Cancer Research (D2018.017 to K.A. Cimprich).
The authors declare no competing financial interests.
Author contributions: M.P. Crossley, J.R. Brickner, C. Song, S.M.T. Zar, S.S. Maw, and M.-S. Tsai performed experiments and analyzed data. F. Chédin provided guidance regarding use of RNases T1 and III and shared preliminary data. M.P. Crossley, J.R. Brickner, and K.A. Cimprich wrote the manuscript with contributions from M.-S. Tsai.
References
- Barroso, S., Herrera-Moyano E., Muñoz S., García-Rubio M., Gómez-González B., and Aguilera A.. 2019. The DNA damage response acts as a safeguard against harmful DNA–RNA hybrids of different origins. EMBO Rep. 20:e47250. 10.15252/embr.201847250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia, V., Barroso S.I., García-Rubio M.L., Tumini E., Herrera-Moyano E., and Aguilera A.. 2014. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 511:362–365. 10.1038/nature13374 [DOI] [PubMed] [Google Scholar]
- Boguslawski, S.J., Smith D.E., Michalak M.A., Mickelson K.E., Yehle C.O., Patterson W.L., and Carrico R.J.. 1986. Characterization of monoclonal antibody to DNA⋅RNA and its application to immunodetection of hybrids. J. Immunol. Methods. 89:123–130. 10.1016/0022-1759(86)90040-2 [DOI] [PubMed] [Google Scholar]
- Castellano-Pozo, M., García-Muse T., and Aguilera A.. 2012. R-loops cause replication impairment and genome instability during meiosis. EMBO Rep. 13:923–929. 10.1038/embor.2012.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli, S.M., and Crouch R.J.. 2009. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276:1494–1505. 10.1111/j.1742-4658.2009.06908.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappidi, N., Nascakova Z., Boleslavska B., Zellweger R., Isik E., Andrs M., Menon S., Dobrovolna J., Balbo Pogliano C., Matos J., et al. 2020. Fork cleavage-religation cycle and active transcription mediate replication restart after fork stalling at co-transcriptional R-loops. Mol. Cell. 77:528–541.e8. 10.1016/j.molcel.2019.10.026 [DOI] [PubMed] [Google Scholar]
- Chédin, F., Hartono S.R., Sanz L.A., and Vanoosthuyse V.. 2021. Best practices for the visualization, mapping, and manipulation of R-loops. EMBO J. 40:e106394. 10.15252/embj.2020106394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Chen J.-Y., Zhang X., Gu Y., Xiao R., Shao C., Tang P., Qian H., Luo D., Li H., et al. 2017. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell. 68:745–757.e5. 10.1016/j.molcel.2017.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, H.-C., Zhang X., Li J., Zhao X., Chen J., Wang H.T.-H., Jatoi I., Brenner A., Hu Y., and Li R.. 2019. BRCA1-associated R-loop affects transcription and differentiation in breast luminal epithelial cells. Nucleic Acids Res. 47:5086–5099. 10.1093/nar/gkz262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S., Puget N., Lin Y.-L., Clouaire T., Aguirrebengoa M., Rocher V., Pasero P., Canitrot Y., and Legube G.. 2018. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun. 9:533. 10.1038/s41467-018-02894-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley, M.P., Bocek M., and Cimprich K.A.. 2019. R-loops as cellular regulators and genomic threats. Mol. Cell. 73:398–411. 10.1016/j.molcel.2019.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley, M.P., Bocek M.J., Hamperl S., Swigut T., and Cimprich K.A.. 2020. qDRIP: a method to quantitatively assess RNA–DNA hybrid formation genome-wide. Nucleic Acids Res. 48:e84. 10.1093/nar/gkaa500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Muse, T., and Aguilera A.. 2019. R loops: from physiological to pathological roles. Cell. 179:604–618. 10.1016/j.cell.2019.08.055 [DOI] [PubMed] [Google Scholar]
- García-Rubio, M.L., Pérez-Calero C., Barroso S.I., Tumini E., Herrera-Moyano E., Rosado I.V., and Aguilera A.. 2015. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 11:e1005674. 10.1371/journal.pgen.1005674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno, P.A., Lott P.L., Christensen H.C., Korf I., and Chédin F.. 2012. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 45:814–825. 10.1016/j.molcel.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartono, S.R., Malapert A., Legros P., Bernard P., Chédin F., and Vanoosthuyse V.. 2018. The affinity of the S9.6 antibody for double-stranded RNAs impacts the accurate mapping of R-loops in fission yeast. J. Mol. Biol. 430:272–284. 10.1016/j.jmb.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchi, E., Skourti-Stathaki K., Ventz S., Pinello L., Yen A., Kamieniarz-Gdula K., Dimitrov S., Pathania S., McKinney K.M., Eaton M.L., et al. 2015. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell. 57:636–647. 10.1016/j.molcel.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, I.J.2019. The mitochondrial R-loop. Nucleic Acids Res. 47:5480–5489. 10.1093/nar/gkz277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche, L., Nguyen H.D., Buisson R., and Zou L.. 2018. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science. 359:108–114. 10.1126/science.aan6490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney, J.S., Viscidi R.P., Vonderfecht S.L., Eiden J.J., and Yolken R.H.. 1989. Monoclonal antibody assay for detection of double-stranded RNA and application for detection of group A and non-group A rotaviruses. J. Clin. Microbiol. 27:6–12. 10.1128/jcm.27.1.6-12.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, C.X., Kobiyama K., Shen Y.J., LeBert N., Ahmad S., Khatoo M., Aoshi T., Gasser S., and Ishii K.J.. 2015. RNA polymerase III regulates cytosolic RNA:DNA hybrids and intracellular microRNA expression. J. Biol. Chem. 290:7463–7473. 10.1074/jbc.M115.636365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Song Y., Xu W., Li Q., Wang X., Li K., Wang J., Liu Z., Velychko S., Ye R., et al. 2020. R-loops coordinate with SOX2 in regulating reprogramming to pluripotency. Sci. Adv. 6:eaba0777. 10.1126/sciadv.aba0777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makharashvili, N., Arora S., Yin Y., Fu Q., Wen X., Lee J.-H., Kao C.-H., Leung J.W., Miller K.M., and Paull T.T.. 2018. Sae2/CtIP prevents R-loop accumulation in eukaryotic cells. eLife. 7:e42733. 10.7554/eLife.42733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabitti, V., Lillo G., Malacaria E., Palermo V., Sanchez M., Pichierri P., and Franchitto A.. 2019. ATM pathway activation limits R-loop-associated genomic instability in Werner syndrome cells. Nucleic Acids Res. 47:3485–3502. 10.1093/nar/gkz025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro, A.N.2000. BRCA1: exploring the links to transcription. Trends Biochem. Sci. 25:469–474. 10.1016/S0968-0004(00)01632-7 [DOI] [PubMed] [Google Scholar]
- Nguyen, H.D., Yadav T., Giri S., Saez B., Graubert T.A., and Zou L.. 2017. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell. 65:832–847.e4. 10.1016/j.molcel.2017.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny, M., Gaidamakov S.A., Ghirlando R., Cerritelli S.M., Crouch R.J., and Yang W.. 2007. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell. 28:264–276. 10.1016/j.molcel.2007.08.015 [DOI] [PubMed] [Google Scholar]
- Nowotny, M., Cerritelli S.M., Ghirlando R., Gaidamakov S.A., Crouch R.J., and Yang W.. 2008. Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD. EMBO J. 27:1172–1181. 10.1038/emboj.2008.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, D.D., Garboczi D.N., Singh K., Hu Z., Leppla S.H., and Leysath C.E.. 2013. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J. Mol. Recognit. 26:376–381. 10.1002/jmr.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard, P., and Manley J.L.. 2017. R loops and links to human disease. J. Mol. Biol. 429:3168–3180. 10.1016/j.jmb.2016.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W., Sun H., De Hoyos C.L., Bailey J.K., Liang X.-H., and Crooke S.T.. 2017. Dynamic nucleoplasmic and nucleolar localization of mammalian RNase H1 in response to RNAP I transcriptional R-loops. Nucleic Acids Res. 45:10672–10692. 10.1093/nar/gkx710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, S., Camino L.P., and Aguilera A.. 2018. Human mitochondrial degradosome prevents harmful mitochondrial R loops and mitochondrial genome instability. Proc. Natl. Acad. Sci. USA. 115:11024–11029. 10.1073/pnas.1807258115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki, K., Proudfoot N.J., and Gromak N.. 2011. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 42:794–805. 10.1016/j.molcel.2011.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka, J.A., Sanz L.A., Hartono S.R., and Chédin F.. 2021. Recognition of RNA by the S9.6 antibody creates pervasive artifacts when imaging RNA:DNA hybrids. J. Cell Biol. 220:e202004079. 10.1083/jcb.202004079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera, A., Lim Y., Woods R., Birrell G.W., Nasim T., Becherel O.J., and Lavin M.F.. 2009. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum. Mol. Genet. 18:3384–3396. 10.1093/hmg/ddp278 [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Holmes J.B., Cerritelli S.M., Sakhuja K., Minczuk M., Holt I.J., and Crouch R.J.. 2010. An upstream open reading frame and the context of the two AUG codons affect the abundance of mitochondrial and nuclear RNase H1. Mol. Cell. Biol. 30:5123–5134. 10.1128/MCB.00619-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong, S.M., Dhir S., and Proudfoot N.J.. 2019. R-loops promote antisense transcription across the mammalian genome. Mol. Cell. 76:600–616.e6. 10.1016/j.molcel.2019.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teloni, F., Michelena J., Lezaja A., Kilic S., Ambrosi C., Menon S., Dobrovolna J., Imhof R., Janscak P., Baubec T., et al. 2019. Efficient pre-mRNA cleavage prevents replication-stress-associated genome instability. Mol. Cell. 73:670–683.e12. 10.1016/j.molcel.2018.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba, L., Costantino L., Tan F.J., Zimmer A., and Koshland D.. 2016. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 30:1327–1338. 10.1101/gad.280834.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J.S., Lujan S.A., and Kunkel T.A.. 2016. Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat. Rev. Mol. Cell Biol. 17:350–363. 10.1038/nrm.2016.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Lima W.F., and Crooke S.T.. 2001. Investigating the structure of human RNase H1 by site-directed mutagenesis. J. Biol. Chem. 276:23547–23553. 10.1074/jbc.M009676200 [DOI] [PubMed] [Google Scholar]
- Yang, K., Yang J., and Yi J.. 2018. Nucleolar stress: hallmarks, sensing mechanism and diseases. Cell Stress. 2:125–140. 10.15698/cst2018.06.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z.Z., Pannunzio N.R., Hsieh C.-L., Yu K., and Lieber M.R.. 2015. Complexities due to single-stranded RNA during antibody detection of genomic rna:dna hybrids. BMC Res. Notes. 8:127. 10.1186/s13104-015-1092-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Chiang H.-C., Wang Y., Zhang C., Smith S., Zhao X., Nair S.J., Michalek J., Jatoi I., Lautner M., et al. 2017. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat. Commun. 8:15908. 10.1038/ncomms15908 [DOI] [PMC free article] [PubMed] [Google Scholar]