Abstract
Eukaryotic ribosome biogenesis is an elaborate process during which ribosomal proteins assemble with the pre-rRNA while it is being processed and folded. Hundreds of assembly factors (AF) are required and transiently recruited to assist the sequential remodeling events. One of the most intricate ones is the stepwise removal of the internal transcribed spacer 2 (ITS2), between the 5.8S and 25S rRNAs, that constitutes together with five AFs the pre-60S ‘foot’. In the transition from nucleolus to nucleoplasm, Nop53 replaces Erb1 at the basis of the foot and recruits the RNA exosome for the ITS2 cleavage and foot disassembly. Here we comprehensively analyze the impact of Nop53 recruitment on the pre-60S compositional changes. We show that depletion of Nop53, different from nop53 mutants lacking the exosome-interacting motif, not only causes retention of the unprocessed foot in late pre-60S intermediates but also affects the transition from nucleolar state E particle to subsequent nuclear stages. Additionally, we reveal that Nop53 depletion causes the impairment of late maturation events such as Yvh1 recruitment. In light of recently described pre-60S cryo-EM structures, our results provide biochemical evidence for the structural role of Nop53 rearranging and stabilizing the foot interface to assist the Nog2 particle formation.
INTRODUCTION
Ribosomes are the ribonucleoprotein complexes selected through evolution to translate the nucleotide-based genetic code into polypeptide chains in the three Domains of life (1,2). To this end, the eukaryotic small (40S) ribosomal subunit harbors the decoding site, whereas the large (60S) subunit accounts for several prominent architectural features including the peptidyl transferase center (PTC), the polypeptide exit tunnel (PET), the L1 stalk, the 5S RNP-containing central protuberance (CP) and the GTPase-associated center between the sarcin-ricin loop (SRL) and the P-stalk (3–5). Both translation fidelity and proteostasis are highly dependent on the accurate ribosome synthesis to ensure the correct assembly of these functional centers out of 79 ribosomal proteins (r-proteins) and four non-coding ribosomal RNAs (rRNAs) (6–8).
The eukaryotic ribosome biogenesis is energetically demanding, hierarchically organized, and a highly regulated multi-step process that takes place across different subcellular compartments (9–11). In the yeast Saccharomyces cerevisiae, it starts in the nucleolus with the independent transcription of the preribosomal RNAs (pre-rRNAs) 5S and 35S, which is a polycistronic transcript, comprising the 40S component 18S rRNA, and the 60S components 5.8S and 25S rRNAs. In the 35S pre-rRNA, these rRNAs are separated by internal transcribed spacers (ITS1 and ITS2) and flanked by external spacers (5′ETS and 3′ETS) (11), which are removed through an ordered sequence of site-specific endo- and exonucleolytic cleavages. The pre-rRNAs undergo progressive structural compaction, as they are co- and post-transcriptionally processed, covalently modified, and folded (10–14).
During this process, not only r-proteins are hierarchically recruited, becoming stably bound (15,16), but also more than 200 assembly factors (AF) transiently participate, spatiotemporally coordinating the conformational and compositional changes of the preribosomal intermediates. Several preribosomal particles have been characterized (Supplementary Table S1), and their specific pre-rRNA and AF compositions provided the basis for our understanding of the sequence of maturation events (9–11).
The small and large ribosomal subunits follow independent maturation routes after the endonucleolytic cleavage at the A2 site within ITS1, separating the 20S and 27SA2 pre-rRNAs (13,14). Although the very first nucleolar pre-60S intermediates remain poorly characterized, the 5S rRNA in association with the r-proteins uL18 and uL5 (5S RNP) appears to be already flexibly bound to early pre-60S particles (17–20). At this stage, 12 AFs, called A3-factors, are required for the removal of the remaining ITS1 sequence at the 5′-end of the 27S pre-rRNA (11,21).
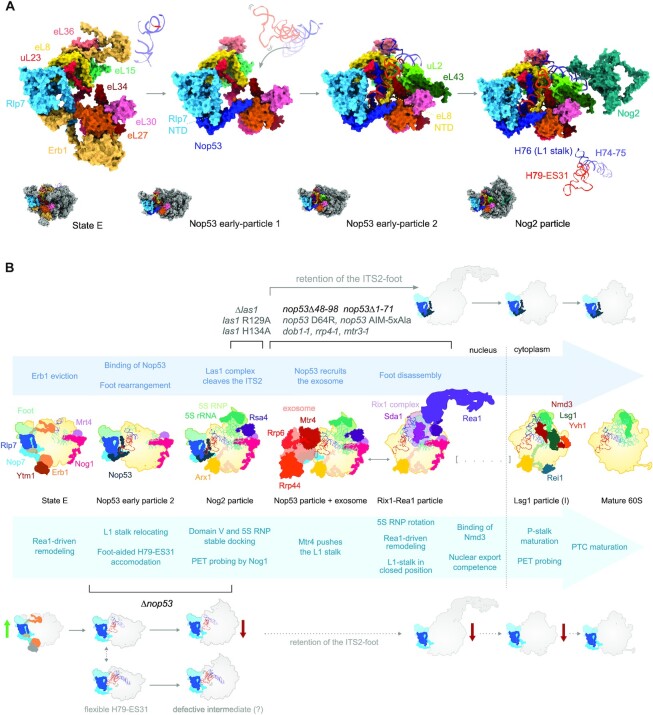
Remarkably, five of the A3-factors (Cic1, Nop15, Rlp7, Nop7 and Erb1) assemble around the ITS2, forming one of the first regions to stably fold into the pre-60S core particle (17–19), a structural hallmark called pre-60S ‘foot’. Cic1, Nop15 and Rlp7 directly interact with the ITS2 (22,23) and assemble in a mutually interdependent manner together with Nop7 and the Erb1 N-terminus, chaperoning and stabilizing the foot structure (17,21,24). The protein composition of the foot remains mostly unchanged in nucleolar pre-60S intermediates from state A up to state E. During the transition from the nucleolar state E particle to the nucleoplasmic Nog2 particle, the dissociation of Erb1 (25–27) enables the recruitment of Nop53 to the basis of the foot. It happens with the nearly concurrent release of 11 AFs (17,22,23,28).
Important pre-60S remodeling events take place when the foot is still present, including the L1 stalk repositioning, the recruitment of 5 AFs (Bud20, Arx1, Nog2, Rsa4, Cgr1), and 3 r-proteins (uL2, eL43, eL39), the Nog1-assisted PET maturation, the folding of the 25S rRNA Domains IV and V, and the 5S RNP stable docking to form the CP of the Nog2 particle (22,28–30). After that, the foot disassembles as the ITS2 is removed in a complex multistep process, during which Nop53 plays a fundamental role. The underlying mechanism of the ITS2 processing has recently been unveiled (31,32), showing that it starts with an endonucleolytic cleavage by the Las1-Gcr3 complex at the C2 site within ITS2, that generates the 7S and 26S pre-rRNAs (33–35). This cleavage requires the recruitment in previous steps of 14 AFs and 11 r-proteins, called B-factors (36,37). The 5′-end of the 26S pre-rRNA is then trimmed by Rat1-Rai1, whereas the 3′-end of the 7S pre-rRNA is targeted to the RNA exosome complex (Exo14). Nop53 is the 60S AF responsible for recruiting and positioning the exosome on the pre-60S by directly interacting, through its conserved N-terminal arch-interacting motif (AIM), with the exosome-associated DExH RNA helicase Mtr4 (38–41). As the 7S pre-rRNA is threaded through Mtr4 into the exosome hexameric channel up to the catalytic subunit Rrp44, the foot AFs dissociate (40,42). Nop53 and Nop7 are the last foot AFs to disassemble as the remaining 5.8S+30 pre-rRNA is handed over to be trimmed by Rrp6, the other exosome catalytic subunit (32,43,44).
In the nucleoplasm, the pre-60S intermediates acquire nuclear export competence through several remodeling events, including Rea1-driven removal of Rsa4, 5S RNP 180° rotation, and multiple compositional changes, such as the eviction of Nog2, allowing the binding of the export factor Nmd3 (22,28,45–47). Interestingly, the removal of the foot was shown not to be a pre-requisite for the pre-60S nuclear export, as las1, mtr4 and nop53 point mutants concentrate foot AFs, 7S and 27S pre-rRNAs in the cytoplasm (37,48,49). Considering that foot-containing 60S subunits have been captured engaged in translation, it has been proposed that the foot removal does not strongly affect late 60S maturation events (37,48).
In the cytoplasm, the last r-proteins assemble into the pre-60S, whilst the remaining AFs dissociate in a process dependent on some exclusively cytoplasmic AFs (e.g. Lsg1, Drg1, Rei1, Reh1) (50–52). The AAA-ATPase Drg1 starts a compositional remodeling that enables Nog1 release (52–54). In parallel, PET and P-stalk undergo independent final maturation (55). In the first case, as Arx1 dissociates, the tunnel is sequentially probed by Rei1 and Reh1 (50,51,56,57). In the second case, also observed in late nuclear stages, Yvh1 triggers Mrt4 eviction, licensing the P-stalk r-proteins to assemble, (50,58–62). The PTC acquires its mature conformation only in the last maturation steps as Nmd3 is released with Lsg1, enabling the dissociation of the last AF - eIF6 (50,51,63).
Important contributions have been made to detail the mechanisms by which the 60S is assembled, mostly on account of the increasing number of high-resolution cryo-EM structures of pre-60S particles (9,10,64–66). More than answers, these structures have provided a source of new biological questions. The cryo-EM structure of Nog2-particles, for instance, revealed that Nop53 is positioned close to the ITS2, exhibiting a tetrahedral architecture embedded at the basis of the pre-60S foot with multiple points of interaction. Indeed, among all the foot AFs present in the Nog2-particle, Nop53 interacts with the largest number of different proteins (Nop7, Rlp7, eL8, uL29 and uL23) (22,23). This observation raised the hypothesis that Nop53 could also have a structural role, acting as a scaffold capable of connecting different regions of the pre-60S (23). To further address this possibility, we evaluated the impact of Nop53 depletion on several pre-60S intermediates.
Using MS/MS-based quantitative proteomics, we reveal the importance of Nop53 for an efficient transition from the state E particle to the Nog2 particle. Our results show that upon depletion of Nop53, not only the unprocessed ITS2-foot is retained, but specific late maturation events such as the recruitment of Yvh1 to the pre-60S are impaired. By comparing Nop53 depletion with nop53 truncation mutants incapable of recruiting the exosome, we demonstrate that the 60S assembly is differentially affected, strongly supporting an expanded role of Nop53 in this process, in addition to the exosome-driven ITS2 processing. Taking into account recently described pre-60S intermediates (28), we propose that our findings reflect the structural role of Nop53 in organizing and stabilizing the docking site of the H79-ES31. We postulate that in the absence of Nop53 the pre-60S foot structure is not an efficient scaffold to assist the remodeling of the 25S rRNA Domain V, impairing the L1 stalk accommodation, and thereby affecting the Nog2 particle assembly.
MATERIALS AND METHODS
Yeast strains and plasmids
Maintenance, growth and genetic manipulation of yeast cultures were performed according to standard procedures. The full list of Saccharomyces cerevisiae strains used in this study is described in Supplementary Table S2. They were grown at 30°C in minimal medium (YNB) supplemented with 2% glucose or galactose as indicated, in addition to the essential amino acids and nitrogenous bases. To evaluate the effect of Nop53 depletion, the conditional strain Δnop53 expressing either the plasmid-borne ptet-Off-GFP-NOP53 or pGAL1-ProtA-NOP53 was employed as previously described (41,67). The transcriptional repression of NOP53 expression was achieved by shifting cells to doxycycline-supplemented (1.5 μg/ml) medium or from galactose to glucose-containing medium, respectively, for 18h before harvesting. The strains used for fluorescence microscopy (FBY2138, FBY2139 FBY2141 and FBY2142) derive from Δnop53/GAL1-ProtA-NOP53 and were obtained by homologous recombination using PCR cassettes, containing HIS3 selectable marker, amplified from genomic DNA extracted from specific strains of the yeast GFP Clone Collection (68). Plasmids used in this study were constructed by restriction cloning or with the InFusion HD (Clontech) recombination cloning system using either competent E. coli DH5α or Stellar (Clontech) cells. All the plasmids used in this study are listed in Supplementary Table S3.
Growth complementation assay
Precultures of the conditional strain Δnop53/tet-Off-GFP-NOP53 expressing the plasmid-borne nop53 truncation mutants (FBY2086-2091; FBY2130-2133) were shifted to fresh DOX-free or DOX-containing (1.5 μg/ml) minimal medium and grown to an OD600nm = 1.0. Aliquots of each yeast culture were harvested for analysis by SDS-PAGE and western blot. 10-fold serial dilutions, starting from the same OD 600nm, were spotted onto glucose-containing solid minimal medium and incubated at different growth conditions [temperature: 16°C or 30°C; doxycycline (1.5 μg/ml), rapamycin (400 nM) or hygromycin B (200 μg/ml)] for a period from 2 to 9 days. Growth curve measurements were performed in microplates incubated over 24h at 30°C using a SpectraMax ie3 Microplate Detection Platform (Molecular Devices) set to read OD600 nm every 60 min.
SDS-PAGE and immunoblot
Yeast whole-cell lysates, the fractions from the glycerol gradient sedimentation, and the coimmunoprecipitated proteins were resuspended in SDS sample buffer, resolved on 8 or 10% SDS-PAGE (Tris-glycine running buffer), transferred to nitrocellulose membranes (Ambion) and incubated with anti-CBP (Millipore), anti-GFP (Sigma-Aldrich), anti-uL18 (gift from Dr Cleslei F. Zanelli, UNESP), anti-Nog1 (gift from Dr John L. Woolford Jr., Carnegie Mellon University) and anti-Pgk1 (Abcam). Secondary antibodies conjugated to IR700dye (anti-rabbit IgG, LI-COR) or IR800dye (anti-mouse IgG, LI-COR) were employed, and near-infrared Western blot detection was carried out using ChemiDoc MP Imaging System (BioRad) or Odyssey equipment (LI-COR). Images were processed using Image Studio™ Lite (ver. 5.2) software (LI-COR).
Fluorescence microscopy
The strains Δnop53/GAL1-ProtA-NOP53 carrying GFP C-terminal fusions chromosomally integrated and expressing the plasmid-borne Nop1-RFP as a nucleolar control were used to survey the subcellular localization of different 60S AFs upon depletion of Nop53. Exponentially growing cells cultured for 18 h in glucose or galactose-containing minimal media were evaluated by fluorescence microscopy. An aliquot of each yeast culture was also analyzed by SDS-PAGE and western blot to confirm Nop53 depletion and evaluate the expression levels of the GFP fusions. Live yeast cells were mounted on agarose pads and imaged using a Nikon Eclipse Ti microscope fitted with a Plan Apo VC 100× Oil DIC N2 objective and an EM-CCD camera (DU-885, Andor) controlled with NIS Elements AR software (version 4.11, Nikon). Images were captured at exposure times ranging from 1 to 3 s, at 13 MHz readout speed, using filters for green (GFP-3035B-000-ZERO, Semrock) and red fluorescence (Texas Red BrightLine set, TXRED4040-B, Semrock) as previously described (69). Images were processed and analyzed using FIJI (Fiji Is Just ImageJ) 2.0.0-rc-69/1.53c software (National Institutes of Health, Bethesda, MD, USA). More than 150 cells were analyzed in both conditions, in replicate experiments (Supplementary Table S4). The RGB profile plots were analyzed using the open-source RGB_Profiler plugin.
Coimmunoprecipitation of pre-60S intermediates
Preribosomes were affinity-purified from the conditional strain Δnop53/tet-Off-GFP-NOP53 expressing plasmid-borne TAP-tagged 60S AFs (Nop7, Rsa4, Arx1, Nmd3, Erb1, Yvh1, Lsg1, and Mrt4). These yeast strains were cultured in 2L glucose containing-medium both with and without supplementation with doxycycline at a final concentration of 1.5 μg/ml. Biological triplicates were harvested at mid-log phase by centrifugation. The cell pellets were resuspended in R buffer (20 mM Tris–HCl pH 8.0, 200 mM KCl, 5 mM MgOAc, 5% glycerol, 1 mM PMSF, 1:50 cOmplete EDTA-free Protease Inhibitor (Roche)) and flash-frozen in liquid N2. The coimmunoprecipitation assays were carried out as previously described (41,70), with minor modifications. Briefly, the whole-cell lysates were obtained by cryogenic milling on a Ball Mill device (Retsch PM 100) and cleared by centrifugation at 17 000 rpm (F12-6×500 LEX Rotor, Sorvall R6 C Plus) for 30 min at 4°C. The lysates were incubated for 2 h at 4°C with 200 μl IgG-Sepharose 6 Fast Flow (GE Healthcare) previously equilibrated with R buffer. For RNA analysis, after washing six times with 1 ml of buffer R supplemented with 0.04 U/ml RNAsin (Promega), RNA was directly extracted from the beads. For proteomic analyses, beads were washed four times with 1mL of R buffer and twice with 100 mM NH4OAc pH 7.4, 0.1 mM MgCl2, before elution under denaturing conditions (500 mM NH4OH). Eluates were lyophilized and further processed before LC–MS/MS analysis. Both input and eluate fractions were separated for analysis by SDS-PAGE and western blot.
Sample preparation for LC–MS/MS
The lyophilized eluates were subjected to in-solution digestion for LC–MS/MS analysis as previously described (41,71). Briefly, proteins were resuspended in 50 mM NH4HCO3, 8 M urea and reduced with DTT at a final concentration of 5 mM at 56°C for 25 min. Samples were then alkylated with 14 mM iodoacetamide for 30 min at RT under protection from light. After dilution with 50 mM NH4HCO3, samples were digested for 16 h with sequencing-grade modified trypsin (Promega) at a 1:50 (E:S) ratio at 37°C. The proteolysis was stopped by adding TFA to a final concentration of 0.4% (v/v) and the resultant peptides were desalted using SepPak tC18 cartridges (Waters).
LC–MS/MS data acquisition
Digested peptides were resuspended in 0.1% formic acid and fractionated on an in-house reversed-phase capillary emitter column (10 cm × 75 μm, filled with 5 μm particle diameter C18 Aqua resins-Phenomenex) coupled to a nano-HPLC (Thermo). The fractionation was carried out through a 60 min gradient of 2–80% acetonitrile in 0.1% formic acid (buffer B) (0–28% of buffer B for 45 min, 28–80% for 13 min; 80–5% for 2 min) at a flow rate of 300 nl min−1. The eluted peptides were analyzed by an LTQ-Orbitrap Velos (Thermo Scientific) (source voltage of 1.9 kV, capillary temperature of 200°C). The mass spectrometer was operated in DDA mode with dynamic exclusion enabled (exclusion duration of 45 seconds), MS1 resolution of 30 000, and MS2 normalized collision energy of 30. For each cycle, one full MS1 scan range of 200–2000 m/z was followed by ten MS2 scans (for the most intense ions) using an isolation window size of 1.0 m/z and collision-induced dissociation (CID). The 10 most intense ions were selected for fragmentation by CID in DDA. Data from the LTQ Velos-Orbitrap were analyzed with the MaxQuant (version 1.5.3.12) software using the UniProt database (taxonomy Saccharomyces cerevisiae—strain ATCC 204508 / S288c), setting tolerance for the MS of 20 ppm and the MS/MS of 0.5 Da; carbamidomethylation of cysteine as a fixed modification; oxidation of methionine and acetylation of protein N-termini as variable modifications; and 1% False Discovery Rate (FDR). The match between runs option was used to increase the number of trusted IDs. The normalized LFQ Intensity values of the proteingroups.txt output were used for the quantitative analysis.
Label-free quantitative proteomics - data analysis
Proteomic data analysis was performed using Perseus software (version 1.6.10.50, Max Planck Institute of Biochemistry) as previously described (72), with minor changes. The affinity purification assays using different baits were analyzed separately. The MaxQuant output data was initially filtered removing contaminants, hits to the reverse database, and proteins identified only by a modification site. The label-free quantification (LFQ) intensities were log2-transformed and the biological triplicates were grouped by categorical annotation in the two conditions assessed: depletion [Nop53-] and non-depletion [Nop53+] of Nop53. A list of all 60S r-proteins and AFs classified according to the 60S maturation stage (10) was added as an annotation column. The identified proteins were filtered for at least two valid LFQ intensity values in at least one replicate group. From this list, proteins that were identified in only one replicate group were considered uniquely identified in one condition (either [Nop53-] or [Nop53+]). LFQ-Intensity correlation multi-scatterplots were used to evaluate reproducibility among replicates, showing the Pearson correlation coefficient for each comparison. Heatmaps of the log2-transformed LFQ-Intensities of all the 60S AFs identified in at least one group and at least two biological replicates were generated hierarchically clustering the replicates and the 60S AFs (Euclidian distance; average-linkage; preprocess with k-means; number of clusters set at 300; number of iterations set at 10). Before the statistical analysis, missing LFQ-Intensity values were imputed from normal distribution around the detection limit (width set at 0.3, downshift of 2.0 standard deviations) and the LFQ-Intensities histogram was verified to confirm that the distribution of these imputed values fitted the normal distribution. A two-tailed Student's t-test (S0 = 0, threshold P-value = 0.05) was performed comparing [Nop53–] with [Nop53+]. Using the GraphPad 8 (Prism) software the –log10-transformed P-values were plotted against the log2-transformed fold change ([Nop53–]/[Nop53+]) in a volcano plot. Statistically significant differences are above the horizontal cut-off line (P-value < 0.05) either on the left of the cut-off line x = –1 (fold change < 0.5) or on the right of the cut-off line x = 1 (fold change > 2). These fold change values were also depicted in a bar graph against the corresponding 60S AFs orderly plotted according to their stepwise association with the pre-60S from early nucleolar to late cytoplasmic stages. The 60S AFs exclusively identified in one condition (either [Nop53–] or [Nop53+]) were depicted as dashed columns. For additional analysis, nine clusters of 60S AFs, representative of specific pre-60S intermediates, were unambiguously defined based on a state-of-the-art model of the 60S assembly pathway (Supplementary Table S1), excluding components present in almost all maturation stages. Violin plots were created using the GraphPad 8 (Prism) software, showing for each cluster the distribution of log2-transformed LFQ Intensity values of the identified 60S AFs in both conditions. For each violin plot: the height is delimited by the highest (upper bound) and the lowest (lower bound) LFQ Intensity values found in the cluster; the median is depicted as a dark gray line; and the shape stems from a rotated and smoothed kernel density plot reflecting the frequency distribution of the log2-transformed LFQ Intensity values observed in the cluster. The 60S AFs exclusively identified in one condition were depicted as gray triangles. The non-parametric two-tailed Mann–Whitney U-test (confidence level: 95%) was performed to compare the 60S AFs distributions in both conditions ([Nop53–] and [Nop53+]). To compare nop53 truncated mutants with Nop53 depletion, an untargeted principal component analysis (PCA) was performed using Perseus software with Benjamini–Hochberg corrected FDR = 0.05 and number of components set at 5.
RNA extraction and Northern blotting
A modified hot phenol RNA extraction method was used to extract total RNA from exponentially growing yeast cells, and affinity-purified RNAs from the beads as previously described (41,73). After quantification using NanoDrop 2000 spectrophotometer (Thermo Scientific), equal amounts of RNA were denatured and resolved either on a 1.5% agarose gel or on a denaturing TBE-urea (7.5 M) 8% polyacrylamide gel. RNA was transferred to Hybond N+ membranes (GE Healthcare) and Northern blotting analyses were carried out using oligonucleotides listed in Supplementary Table S5, previously 3′-biotinylated using Biotin 3′End DNA Labeling Kit according to manufacturer's protocol (Thermo Scientific). Detection of biotin-labeled probes was performed using Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific).
In vivo RNA structure analysis by SHAPE
In vivo rRNA structure was probed by selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) as previously described (37,74,75), with minor changes. In brief, after a gentle wash with PBS, yeast cells at mid-log phase were incubated with the acylating reagent 2-methylnicotinic acid imidazolide (NAI) at a final concentration of 100 mM for 20 min at 30°C. As a control, the same protocol was performed for each yeast culture replacing NAI by DMSO. Cells were harvested and total RNA was immediately extracted by a modified hot phenol method (67). Aliquots of each yeast culture were separated for further analysis by SDS-PAGE and western blot. To detect the 2′-O-adducts formed, 5 μg of total RNA were subjected to primer extensions with SuperScript III Reverse Transcriptase (Invitrogen). The IR700dye-labelled oligonucleotides (Alpha DNA, Quebec), which were used to probe specific regions of the 25S rRNA, are listed in Supplementary Table S5. The nucleotide position of each modification site was identified using a DNA sequencing ladder obtained with the plasmid pMS2-ITS2 as a template. The sequencing reactions were performed with the IR700dye-labeled UC2 probe and the Sequenase Version 2.0 DNA Sequencing Kit (Affymetrix). The IR700dye-labelled cDNAs and the four ddNTP termination reactions were resolved in parallel on a denaturing TBE-urea (7 M) 6% polyacrylamide gel under protection from light (76). Near-infrared detection was carried out using Odyssey Imaging System (LI-COR) and images were processed using Image Studio™ Lite (ver. 5.2) software (LI-COR).
Structure visualization with UCSF ChimeraX
The UCSF ChimeraX (1.0-rc-2020.05.22) software was employed for molecular visualization of pre-60S particles, highlighting the position and the protein-protein interactions of the 60S AFs affinity-purified or used as baits in our proteomic analyses. The following Protein Data Bank (PDB) files were used: 4V88 (5), 3JCT (22), 6ELZ (17), 6C0F (18), 6FT6, 6FSZ (40), 6N8K, 6N8O (50), 6RZZ, 6RI5 (51), 6YLX, 6YLY and 6YLH (28).
Glycerol density gradient sedimentation
Whole yeast cell extracts obtained by cryogenic milling on a Ball Mill device (Retsch PM 100) from the same strains used in the affinity purification assays were fractionated after density gradient sedimentation as previously described (41,77). Briefly, pre-cleared lysates − obtained from Nop53 depleted (−) or non-depleted (+) cells − corresponding to 5 mg of total protein were loaded on the top of a 12 ml linear 10–30% glycerol gradient (20 mM Tris–HCl pH 8.0, 200 mM KCl, 5 mM MgOAc, 5% glycerol, 1 mM PMSF), previously obtained using a Gradient Master device (BioComp), and were centrifuged at 23 000 rpm for 12.5 h at 4°C in a P40ST rotor (Hitachi). Each gradient was fractionated from the top in 24 aliquots (500 μl) using an EconoSystem (Bio-Rad) fitted with a UV monitor plotting the protein/nucleic acid sedimentation profile. All the gradient fractions were flash-frozen in liquid N2 and later precipitated with ice-cold 10% trichloroacetic acid (TCA) before analysis by SDS-PAGE and western blot. The 48 gradient fractions (24 Nop53+ and 24 Nop53–) of each strain were processed and analyzed together. The same amount of Input was loaded in each gel. All the western blot membranes (Nop53+ and Nop53–) of the same strain were scanned together under the same parameters using the Odyssey Imaging System (LI-COR) with near-infrared fluorescence detection.
RESULTS
Nop53 depletion impairs the transition from the nucleolar state E to the nuclear Nog2 particle, affecting late pre-60S intermediates
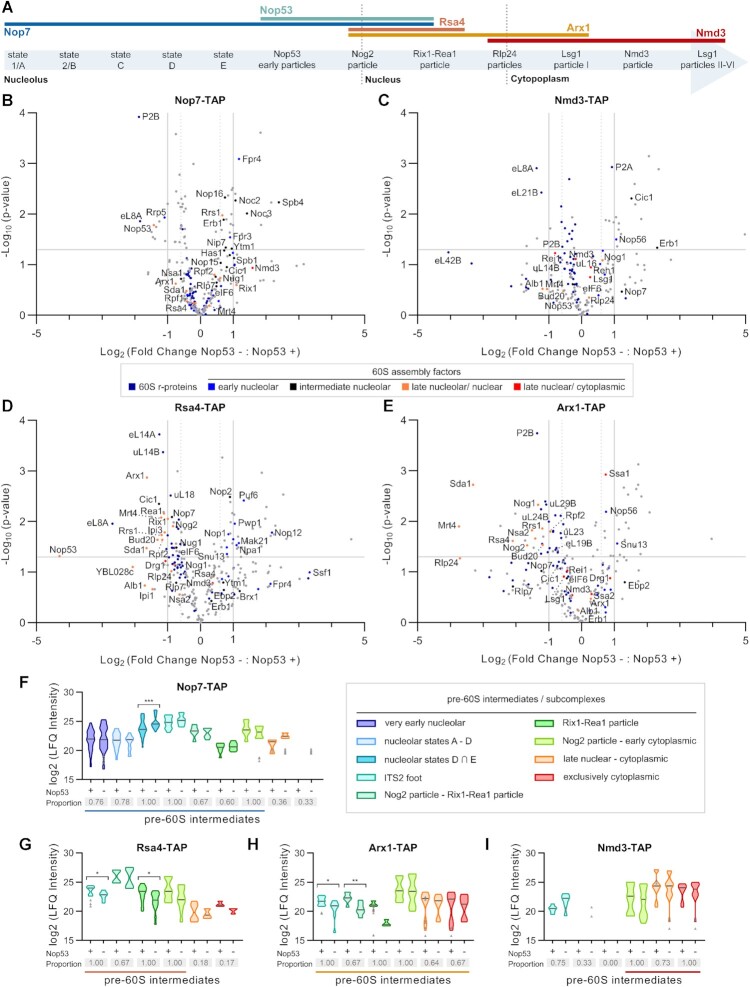
To evaluate how the recruitment of Nop53 to the pre-60S affects the compositional changes during the large ribosomal subunit assembly, four specific 60S AFs representative of different maturation stages (Figure 1A) were selected to be TAP-tagged. Three of them, namely Nop7, Rsa4 and Arx1, are present in Nop53-containing particles, whereas Nmd3 is recruited after Nop53 release (Supplementary Figures S1A, E and S2A, E). Nop7 is an early-binding AF, component of the pre-60S foot, that directly interacts with both Erb1 and Nop53 in a mutually exclusive manner. The replacement of Erb1 by Nop53 takes place in the transition from the nucleolar state E particle to the nucleoplasmic Nog2 particle and is essential for Nop7 release, as it requires the exosome-driven ITS2 removal (17,22,32,40,78,79). Following Nop53 recruitment, Rsa4 and Arx1 are incorporated to form the Nog2 particle (28). Whereas Rsa4 is released in the nucleoplasm by the action of the Rix1–Rea1 complex (22,28,46,80,81), Arx1 remains bound to the pre-60S even after the nuclear export (22,28,81–83). At late nuclear stages, after the foot disassembly, Nmd3 is recruited to the pre-60S, binding on top of the maturing intersubunit surface, where it contributes to the PTC maturation (50,51,84,85). Both Arx1 and Nmd3 are nuclear export AFs that assist the pre-60S nucleocytoplasmic shuttling (86,87) and dissociate from it only in the cytoplasm (50,51,55–57,88).
Figure 1.
Pre-60S protein compositional changes caused by Nop53 depletion. The 60S AFs Nop7, Nmd3, Rsa4 and Arx1 were selected to probe the effect of Nop53 depletion on the 60S biogenesis. (A) The hierarchical order of their recruitment and release is depicted on top of a schematic representation of the 60S assembly pathway listing pre-60S intermediates, whose cryo-EM structures have already been described. (B–I) Mass spectrometry-based label-free quantitative proteomic analysis of preribosomes affinity-purified in biological triplicates with the indicated TAP-tagged AFs. Volcano plots show the comparison between Nop53 depleted (−) and non-depleted (+) conditions for (B) Nop7-TAP, (C) Nmd3-TAP, (D) Rsa4-TAP and (E) Arx1-TAP particles. –Log10-transformed P-values were plotted against log2-transformed fold change values. 60S r-proteins and AFs are highlighted in different colors, the latter being classified according to the 60S maturation stage they participate in. (F–I) Analysis of clusters of 60S AFs representative of specific pre-60S intermediates. Violin plots show the distribution of the copurified 60S AFs based on their log2-transformed LFQ Intensity values (states D ∩ E designates intersection—common AFs between these particles); height: delimited by the highest and lowest LFQ Intensity values of the cluster; width: frequency (high: wide; low: narrow) of the LFQ Intensity values observed in the cluster. The 60S AFs identified in only one condition (Nop53+ or Nop53–) are depicted as gray triangles. The median is indicated by a dark gray line. Increased (higher median) or decreased (lower median) association with components of the indicated pre-60S intermediates is revealed by comparing Nop53 depleted (−) and non-depleted (+) conditions. Statistically significant (P < 0.05) differences are highlighted in the graph (Nop7-TAP, ***P = 0.0007; Rsa4-TAP, *P = 0.0188, *P = 0.0186; Arx1-TAP, *P = 0.0315, **P = 0.0087). Gray boxes show the proportion of each cluster that was affinity-purified (number of affinity-purified 60S AFs components of the cluster/ total number of 60S AFs in the cluster). A horizontal bar indicates the pre-60S intermediates with which the bait is expected to be associated.
These TAP-tagged 60S AFs were independently expressed in the conditional strain Δnop53 with GFP-NOP53 under control of a tetracycline repressible promoter (tet-OFF). Upon doxycycline treatment, GFP-NOP53 was downregulated (Supplementary Figures S1B,F, S2B,F) partially and sufficiently depleting Nop53 to affect both cell growth and pre-rRNA processing as previously described (41,89–91). Preribosomes affinity-purified in biological triplicates both in the presence and upon repression of Nop53 expression were analyzed by mass spectrometry-based label-free quantitative proteomics (Figure 1 and Supplementary Figures S1 and S2). Upon Nop53 depletion, the copurified preribosomes exhibited significant and specific changes in composition (Figure 1B-I and Supplementary Figures S1C–D, G–I and S2C–D, G–I). To circumvent any possible contamination with mature ribosomal particles, we focused our analysis only on the 60S AFs, inferring the precise maturation steps that were affected by comparing the distribution of their log2-transformed LFQ Intensity values. The identified 60S AFs were assigned and analyzed in clusters representative of different pre-60S intermediates (Figure 1F–I), which were unambiguously defined based on a state-of-the-art model of the 60S assembly pathway (Supplementary Table S1). Changes in the shape and height of the violin plots upon Nop53 depletion directly reflect the increase (higher median) or decrease (lower median) in the levels of copurified components of specific pre-60S intermediates. Statistically significant differences were determined using the Mann-Whitney U-test.
Given the role of Nop53 in recruiting the RNA exosome to trim the 7S pre-rRNA (38,41), we first asked what would be the effect of Nop53 depletion on the ITS2-foot structure. We observed in this condition that whereas the foot AF Nop7 copurified few late-binding AFs (Reh1 and Lsg1), the late Nmd3 particles coenriched some foot AFs (Cic1 and Rlp7) (Figure 1C, F, I and Supplementary Figure S1I), indicating that, upon Nop53 depletion, the foot structure, whose disassembly was blocked, could be found in late nuclear and cytoplasmic pre-60S intermediates. Previous studies have shown that nop53, mtr4 and las1 point mutants impair the ITS2 processing and cause the misplacement of foot AFs to cytoplasmic particles (37,48,49). We would have expected, therefore, under repression of Nop53 expression, a strong and significant enrichment of several late-binding AFs copurified with Nop7. Instead, Nop7 particles coenriched mostly nucleolar 60S AFs (Figure 1B,F), suggesting an effect of Nop53 depletion on early 60S maturation stages, which might indirectly affect the formation of late pre-60S intermediates.
Among the nucleolar 60S AFs identified upon Nop53 depletion, Nop7 significantly and highly coenriched constituents of the states D and E particles (e.g. Spb4, Noc3, Nip7) (Figure 1B,F and Supplementary Figure S1I), which correspond to the pre-60S intermediates preceding Nop53 recruitment (Figure 1A and Supplementary Table S1). It was, therefore, conceivable to hypothesize that in this condition the transition from nucleolar stages to the Nop53-containing Nog2 particle was impaired. This observation was confirmed when we analyzed the effect of Nop53 depletion on the Nog2 particle AFs Rsa4 and Arx1, which are incorporated only after Nop53 (Figure 1D-E,G-H and Supplementary Figure S2I). In both cases, the level of several Nog2 particle components (Bud20, Nog2, Rsa4, Arx1) are significantly reduced upon Nop53 depletion, including foot AFs and those (Rrs1, Rpf2, Nog1, eIF6, Nug1, Nsa2, Mrt4, YBL028c) remaining from earlier particles (22,28). Similarly, the Rix1-Rea1 particle AFs (Sda1, Rix1, Ipi1, Ipi3, Rea1) that are subsequently recruited to the Nog2-particle to trigger the removal of Rsa4 and the CP remodeling (22,28,46), were also found at decreased levels. All these findings strongly indicate that depletion of Nop53 causes an accumulation of nucleolar pre-60S intermediates, which is in agreement with the previously reported accumulation of 27S pre-rRNA in this condition (41,89–91).
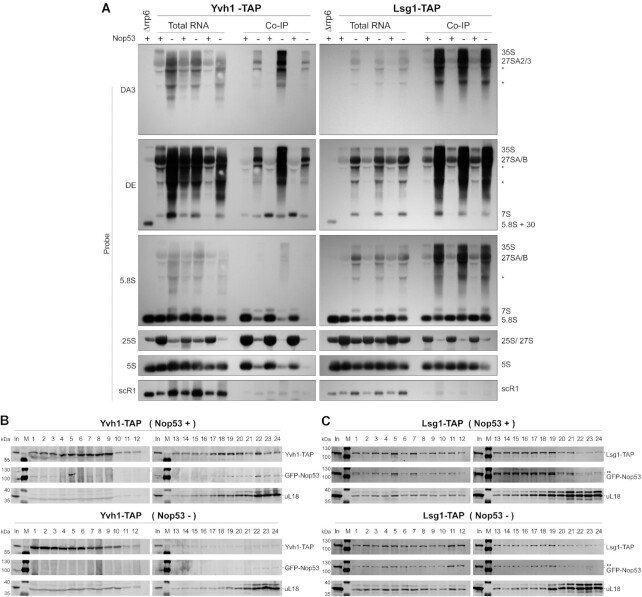
Analysis of preribosomes copurified with Nmd3 and Arx1 allowed us to evaluate late nuclear and cytoplasmic particles, revealing an effect of Nop53 depletion on late-binding AFs (Figure 1H-I and Supplementary Figures S1H–I and S2H–I). Of note, under repression of Nop53 expression, both Arx1 and Nmd3 particles were depleted of the late-binding AF Yvh1, which is involved in the P-stalk maturation (58,59,62). From these results, we observe that late nuclear and cytoplasmic pre-60S intermediates are still formed upon Nop53 depletion, although to a lesser extent and with specific late 60S maturation events affected.
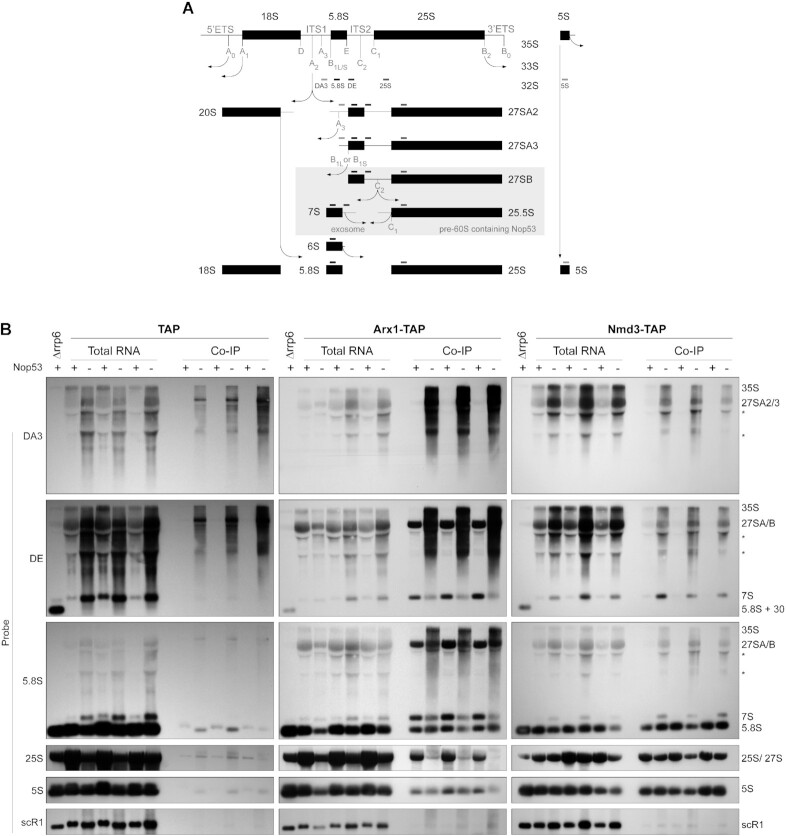
To further characterize the effect of Nop53 depletion on late pre-60S intermediates, complementing the proteomic analyses, the pre-rRNA content of Arx1 and Nmd3 preribosomes was assessed in biological triplicates by Northern blot (Figure 2). As expected, depletion of Nop53 caused a significant accumulation of both 7S and 27S pre-rRNAs, visible in the total extract lanes (Figure 2B). The negative control (Figure 2B, left panels) was important to reveal background amounts of 35S and 27SA pre-rRNAs, which are not part of Arx1 or Nmd3 particles. Importantly, the results confirmed the proteomic analysis, showing lower levels of 7S pre-rRNA, 5.8S, and 25S rRNAs copurified with Arx1 upon depletion of Nop53, even though 7S pre-rRNA accumulated in this condition (Figure 2B, middle panels). Comparatively, Rsa4 showed a profile similar to Arx1, whereas Nop7 coenriched 27S and 7S pre-rRNAs (Supplementary Figure S3A), consonant with the proteomic data and evidencing less efficient recruitment of both Arx1 and Rsa4 to nucleoplasmic pre-60S intermediates. Of note, this effect seemed slightly more pronounced on Arx1. As the formation of Nog2 particles was impaired, but not blocked when Nop53 is lacking, we show by glycerol density gradient analysis, that both Arx1 and Rsa4, which exhibit similar sedimentation profiles, are, as Nmd3, still associated with pre-60S intermediates upon Nop53 depletion (Supplementary Figure S3B–D). Consistent with this result and with the above-mentioned retention of the foot structure, Nmd3, which normally copurifies only mature rRNAs, coenriched the 7S pre-rRNA with a slight decrease in the levels of 25S rRNA upon Nop53 depletion (Figure 2B, right panel).
Figure 2.
Nop53 depletion differentially affects the association of the nucleocytoplasmic AFs Arx1 and Nmd3 with pre-60S intermediates. (A) Schematic representation of the pre-rRNA processing in S. cerevisiae, highlighting the stages during which Nop53 is associated with the pre-60S and the hybridization position of the Northern blot probes. (B) The pre-rRNAs copurified (IP) with Arx1-TAP and Nmd3-TAP in biological triplicates both in Nop53 depleted (−) and non-depleted (+) conditions were assayed along with the Input (total RNA) by Northern blot using the indicated probes. The empty TAP tag was used as a negative control and the Δrrp6 strain as a control for the accumulation of 5.8S+30 pre-rRNA. *non-specific band.
We then wondered if the observed effects of Nop53 would also extend to an altered subcellular localization of nucleocytoplasmic or foot AFs. For this purpose, we analyzed GFP-tagged Nmd3, Arx1 and Nop7 both in the presence and upon repression of Nop53 expression (Supplementary Figure S4 and Supplementary Table S4). Consistent with the findings described above, Nmd3 subcellular localization was affected by Nop53 depletion (Supplementary Figure S4A). Although still present in the cytoplasm, Nmd3 was found highly concentrated in the nucleus upon Nop53 depletion, indicating partial retention of Nmd3 in association with nuclear intermediates. Interestingly, the foot AF Nop7, which has been shown to mislocalize to the cytoplasm in mutant backgrounds that block ITS2 processing, such as nop53 AIM mutants (48), was still mostly retained in the nucleus under Nop53-depleted condition (Supplementary Figure S4B). As expected, in agreement with the above-mentioned analyses, the subcellular localization of Arx1 was not affected by Nop53 depletion, remaining mainly nuclear (Supplementary Figure S4C). Taken together, these observations indicate that Nop53 depletion, in addition to the effect on nucleolar intermediates, also causes partial retention of nucleocytoplasmic AFs in the nucleus, reducing the formation of cytoplasmic pre-60S intermediates.
Nop53 depletion leads to the accumulation of state E particles and the association of Erb1 with late 60S assembly intermediates
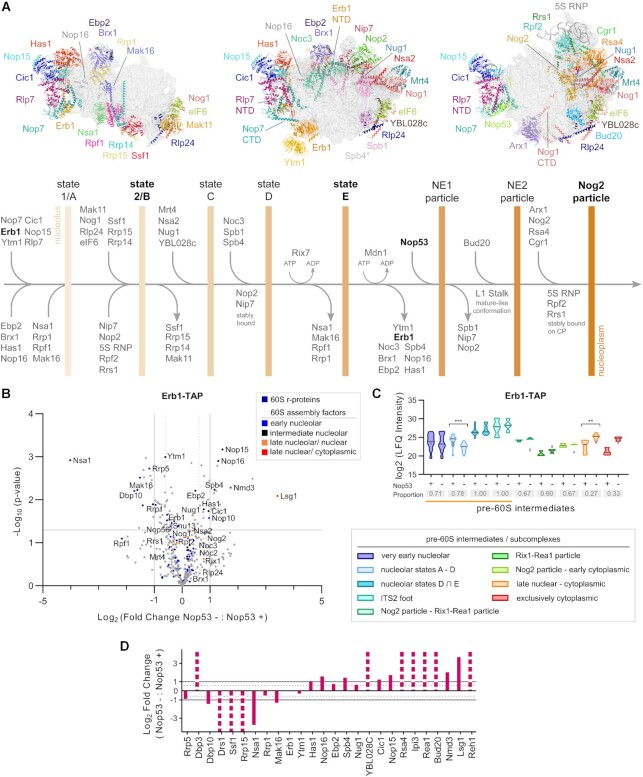
To better characterize the impact of Nop53 depletion on early nucleolar 60S maturation stages, and particularly on the state E particle, we set out to analyze the protein and pre-rRNA content of preribosomes affinity-purified with the TAP-tagged nucleolar AF Erb1 both in the presence and upon repression of Nop53 expression (Figure 3 and Supplementary Figure S5). Erb1 associates very early with nascent pre-60S intermediates (17–19,21,92,93) interacting with several AFs (Supplementary Figure S5A), including foot components such as Nop7 (17). However, different from Nop7, which dissociates only after the recruitment of Nop53 and the Nog2-particle formation, Erb1 remains uniquely bound to nucleolar intermediates up to the state E particle (Figure 3A and Supplementary Table S1) (17,22). It was, therefore, chosen as an adequate bait to probe nucleolar maturation stages that precede Nop53 binding. In fact, the dissociation of Erb1 is necessary for the recruitment of Nop53 to the pre-60S (17,22,28).
Figure 3.
Erb1 is mostly associated with state E and late pre-60S particles upon depletion of Nop53. (A) Schematic representation of early nucleolar and nuclear 60S maturation steps showing the sequential changes in protein composition that leads to the pre-60S intermediates, indicated by vertical bars. Structures correspond to PDB IDs: 6C0F, 6ELZand 3JCT. *Spb4 is present in state E but not shown. NE: Nop53 early particle, NTD: N-terminal domain, CTD: C-terminal domain. (B–D) Mass spectrometry-based label-free quantitative proteomic analysis of preribosomes affinity-purified in biological triplicates with Erb1-TAP. (B) Comparison between Nop53 depleted (−) and non-depleted (+) conditions highlighting the identified 60S r-proteins and AFs. The latter were classified according to the 60S maturation stage they participate in. (C) Analysis of clusters of 60S AFs representative of specific pre-60S intermediates. Violin plots show the distribution of the copurified 60S AFs based on their log2-transformed LFQ Intensity values (states D ∩ E designates intersection—common AFs between these particles); height: delimited by the highest and lowest LFQ Intensity values of the cluster; width: frequency (high: wide; low: narrow) of the LFQ Intensity values observed in the cluster. The 60S AFs identified in only one condition (Nop53+ or Nop53–) are depicted as gray triangles. The median is indicated by a dark gray line. Increased (higher median) or decreased (lower median) association with components of the indicated pre-60S intermediates is revealed by comparing Nop53 depleted (−) and non-depleted (+) conditions. Statistically significant (P < 0.05) differences are highlighted in the graph (***P = 0.0001; **P = 0.0043). Gray boxes show the proportion of each cluster that was affinity-purified (number of affinity-purified 60S AFs components of the cluster/ total number of 60S AFs in the cluster). A horizontal bar indicates the pre-60S intermediates with which Erb1 is expected to be associated. (D) The log2-transformed fold change value of each identified 60S AF was normalized to the bait and orderly plotted according to their sequential engagement in the 60S assembly. Dashed columns indicate the 60S AFs exclusively identified in one condition.
Upon Nop53 depletion, Erb1 copurified higher levels of state E AFs (e.g. Spb4, Has1, Nop16, Ebp2), some of which remaining from earlier particles. Its association with AFs specific to nucleolar state A to D particles (e.g. Nsa1, Rrp1, Mak16, Rrp15 and Ssf1), however, significantly decreased (Figure 3B–D), suggesting a reduced association of Erb1 with these early nucleolar particles. In line with this, nucleolar AFs that constitute the earliest pre-60S intermediates (Rrp5, Drs1, Dbp10) were also copurified with Erb1 at decreased levels upon Nop53 depletion, strongly indicating that in this condition most of the nucleolar Erb1 particles correspond to state E intermediates. This is coherent with the observation that under Nop53-depleted condition, Erb1 affinity-purifies less 27SA2/A3 pre-rRNAs, but is still associated with 27SB pre-rRNA, which constitutes state E particles (Supplementary Figure S5F). Furthermore, no significant changes were observed in the Erb1 sedimentation profile after Nop53 depletion (Supplementary Figure S5B), confirming that it was still bound to preribosomal subunits.
Interestingly, Nog2 particle components (e.g. Rsa4, Bud20) and particularly late nuclear/ cytoplasmic AFs (e.g. Nmd3, Lsg1, Reh1) were also coenriched with Erb1 upon Nop53 depletion, in agreement with the higher levels of 7S pre-rRNA copurified with Erb1 in this condition (Supplementary Figure S5F). On the other hand, upon Nop53 depletion, the late AF Nmd3 affinity-purified higher levels of Erb1 (Figure 1C), which, congruently, has also been reported upon depletion of the ITS2 endonuclease Las1 (37). Altogether, these findings indicate that ITS2-foot-containing late pre-60S particles prompt the otherwise unusual Erb1 binding to late assembly intermediates, which is particularly favored in this condition given that the Erb1 binding site on the foot is vacant when Nop53 is lacking.
Nop53 depletion affects late maturation events impairing the Yvh1 recruitment to the pre-60S
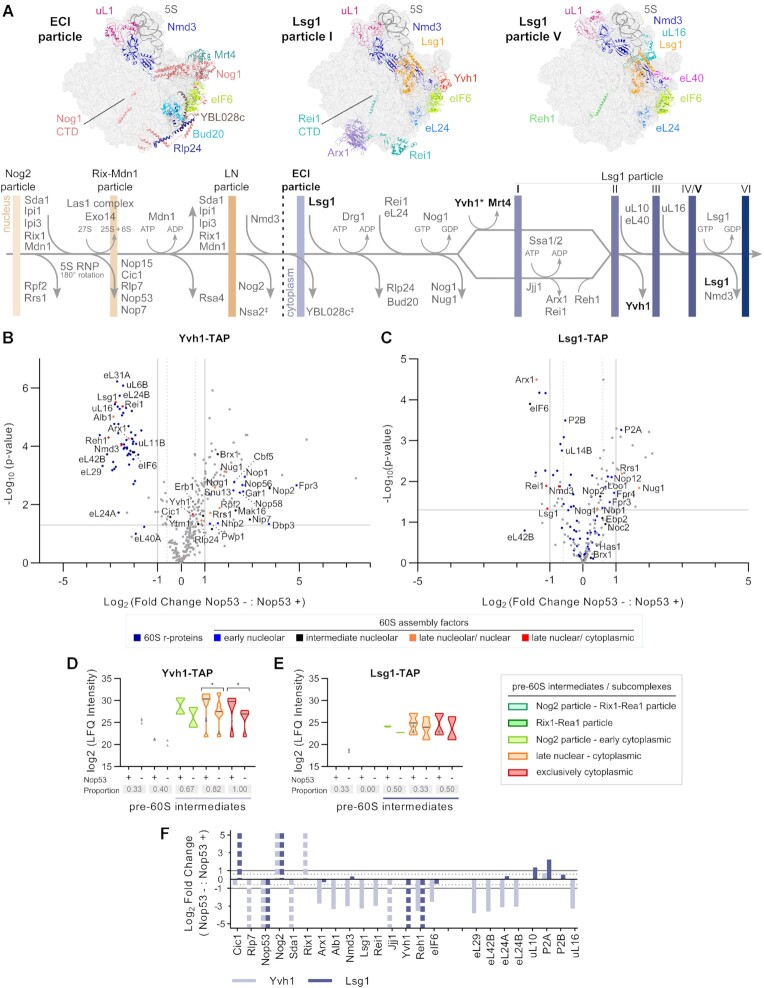
As described above, both Nmd3 and Arx1 particles were depleted of the late-binding nucleocytoplasmic AF Yvh1 upon Nop53 depletion (Supplementary Figures S1H-I and S2H-I). This prompted us to further evaluate the interplay between Nop53 recruitment and Yvh1, which is required for the late nuclear/cytoplasmic maturation of the P-stalk (Figure 4A) (58,59,62). To verify in more detail whether Nop53 depletion could affect the recruitment of Yvh1 to late pre-60S particles, we analyzed pre-60S intermediates copurified with Yvh1-TAP both in the presence and upon repression of Nop53 expression by mass-spectrometry based label-free quantitative proteomics (Figure 4, and Supplementary Figure S6A–D). As a control, the exclusively cytoplasmic AF Lsg1-TAP was used as bait (Figure 4, and Supplementary Figure S6E–H). Upon Nop53 depletion, both Yvh1 and Lsg1, copurified some nucleolar AFs and early pre-rRNAs, reflecting their accumulation already shown in this condition (Figure 2B, left panels). Focusing our analysis on late-binding AFs, our data revealed a striking effect of Nop53 depletion on the Yvh1 interactome (Figure 4B, D, and Supplementary Figure S6D). Both nucleocytoplasmic (e.g. Arx1, Alb1, Nmd3) and cytoplasmic AFs (e.g. Lsg1, Rei1, Jjj1, Reh1), components of late nuclear and cytoplasmic particles, were significantly less affinity-purified with Yvh1-TAP under Nop53-depleted condition, as well as several r-proteins including the late-associating ones incorporated only at cytoplasmic stages (eL29, eL42, eL24, uL16). Consistent with this observation, the levels of 7S pre-rRNA and the mature 5.8S, 25S and 5S rRNAs copurified with Yvh1 were all significantly decreased (Figure 5A, left panels). Indeed, compared to all the baits used in this study, Yvh1 was the only one for which the association with all mature rRNAs was drastically reduced upon Nop53 depletion.
Figure 4.
Nop53 depletion has different impacts on the interactome of the late-binding 60S AFs Yvh1 and Lsg1. (A) Schematic representation of late nuclear and cytoplasmic 60S maturation events highlighting the protein compositional changes and the resulting pre-60S intermediates, indicated by vertical bars (Structures correspond to PDB IDs: 6N8K, 6RZZ and 6RI5). LN: late nuclear Rlp24 particle; ECI: early cytoplasmic-immediate Rlp24 particle; CTD: C-terminal domain; ‡ uncertain dissociation stage; * late nuclear/cytoplasmic recruitment. (B–F) Effect of Nop53 depletion on late 60S maturation stages. Preribosomes affinity-purified in biological triplicates with the late-binding AFs (B) Yvh1 and (C) Lsg1 were analyzed by mass spectrometry-based label-free quantitative proteomics. Volcano plots show the comparison between Nop53 depleted (−) and non-depleted (+) conditions. The identified 60S r-proteins and AFs are highlighted in colors according to the 60S maturation stages they participate in. (D, E) Analysis of clusters of 60S AFs representative of specific pre-60S intermediates. Violin plots show the distribution of the copurified 60S AFs based on their log2-transformed LFQ Intensity values; height: delimited by the highest and lowest LFQ Intensity values of the cluster; width: frequency (high: wide; low: narrow) of the LFQ Intensity values observed in the cluster. The 60S AFs identified in only one condition (Nop53+ or Nop53-) are depicted as gray triangles. The median is indicated by a dark gray line. Increased (higher median) or decreased (lower median) association with components of the indicated pre-60S intermediates is revealed by comparing Nop53 depleted (−) and non-depleted (+) conditions. Statistically significant (P < 0.05) differences are highlighted in the graph (Yvh1-TAP * P = 0.03150; * P = 0.00205). Gray boxes show the proportion of each cluster that was affinity-purified (number of affinity-purified 60S AFs components of the cluster/ total number of 60S AFs in the cluster). A horizontal bar indicates the pre-60S intermediates with which Yvh1 or Lsg1 are expected to be associated. (F) log2-transformed fold change values of intermediate/late 60S AFs and cytoplasmic 60S r-proteins, normalized to the bait, were plotted following their order of association with the pre-60S. Dashed columns indicate the 60S AFs exclusively identified in one condition.
Figure 5.
Yvh1 recruitment to the pre-60S is impaired upon depletion of Nop53. (A) Northern blot analysis of the pre-rRNAs affinity-purified (IP) in biological triplicates with Yvh1-TAP and Lsg1-TAP both in Nop53 depleted (−) and non-depleted (+) conditions, together with the Input (total RNA), using the indicated probes. The Δrrp6 strain is a control for the accumulation of 5.8S+30 pre-rRNA. *non-specific band. (B, C) Glycerol density gradient sedimentation of the TAP-tagged AFs (B) Yvh1 and (C) Lsg1 in Nop53 depleted (−) and non-depleted (+) conditions. Whole-cell lysates were loaded on a 10–30% linear glycerol gradient. The 24 fractions collected were analyzed by western blot with anti-CBP, anti-GFP and anti-uL18. ** indicates the Lsg1-TAP band close to GFP-Nop53 (In: input - whole cell lysate; M: protein MW marker).
In the same condition, Lsg1 exhibited a decrease of only 25S rRNA levels (Figure 5A, right panels), and copurified lower levels of some late-binding AFs (e.g. Arx1, Rei1, Reh1), including Yvh1 (Figure 4C, E–F and Supplementary Figure S6H), indicating a reduced formation of cytoplasmic pre-60S intermediates upon Nop53 depletion. However, Lsg1 did not show a significant decrease in the levels of most r-proteins, co-enriched 7S pre-rRNA, and exhibited almost no effect on the levels of 5S rRNA under Nop53-depleted condition (Figure 5A, right panels). Taken together, these observations strongly suggest a higher impact of Nop53 depletion on Yvh1 recruitment than on Lsg1 association with pre-60S intermediates. This hypothesis was further substantiated by the analysis of the sedimentation profiles of Yvh1 and Lsg1 (Figure 5B, C). Whereas Lsg1 still co-sediments, with the r-protein uL18 (fractions 18–24) upon Nop53 depletion, Yvh1 becomes mostly concentrated in the top fractions [1-10] of the gradient, with almost no signal detected in the bottom fractions corresponding to pre-60S intermediates. The depletion of Nop53, therefore, affects not only nucleolar steps of the 60S maturation, as shown above, but also impairs the late nuclear/cytoplasmic recruitment of Yvh1.
As Yvh1 is required for Mrt4 release (Figure 4A) (58,59), we wondered if Nop53 depletion would also affect the association of this latter AF with pre-60S intermediates. Mrt4 is an early-binding AF recruited in the nucleolus (state C – Figure 4A and Supplementary Table S1) (17–19), which dissociates from the pre-60S only in late nuclear/cytoplasmic stages upon recruitment of Yvh1 (50,52,58–62). The results show that upon Nop53 depletion, Mrt4 is still associated with pre-60S intermediates in bottom fractions [18-24] of the density gradient, but also considerably enriched in the free fractions [1-10] (Supplementary Figure S7A), indicating that in this condition Mrt4 is not trapped in cytoplasmic pre-60S intermediates, as previously reported in Δyvh1 cells (58,59). Consistent with this interpretation, Nop53 depletion led to an increased association of Mrt4 with 7S and 27SA/B pre-rRNAs containing particles, and decreased association with mature rRNAs (Supplementary Figure S7B). In this condition, most of the Mrt4 particles might correspond to nucleolar and nuclear intermediates. Accordingly, the subcellular localization of Mrt4 was not significantly affected upon depletion of Nop53, remaining mostly nucleolar (Supplementary Figure S7C, D).
Effect of nop53 truncated mutants on cellular growth and late 60S maturation events
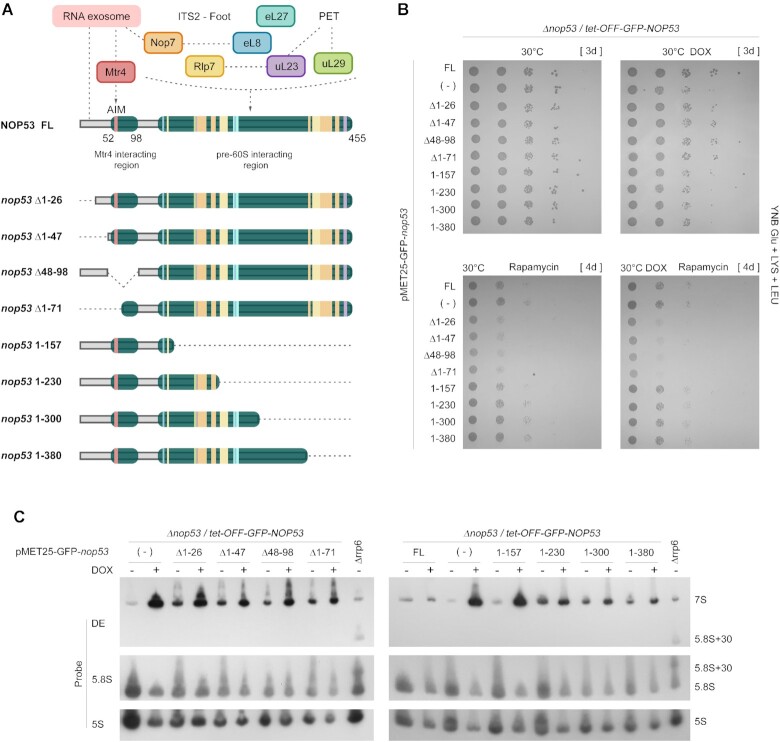
To further characterize the role of different regions of Nop53 during ribosome assembly, we constructed N- and C-terminally truncated mutants of Nop53 fused to GFP (Figure 6A). Nop53 N-terminal half covers a highly conserved region, which harbors between the amino acid residues 60 and 64 the motif (AIM) of interaction with the RNA helicase Mtr4, that interacting with the RNA exosome complex, assists the 3′-5′ processing of the 7S pre-rRNA (38,39,94). Four different N-terminal mutants were created to assess the role of the region surrounding the AIM: nop53Δ1–26 and nop53Δ1–47, which still contain the AIM, as well as nop53Δ48–98 and nop53Δ1–71, which are both devoid of the AIM. In parallel, four nop53 C-terminal mutants were constructed by progressively removing 50–80 amino acids from this end (nop53 1–157, nop53 1–230, nop53 1–300 and nop53 1–380). Nop53 C-terminus is mostly engaged in interactions with r-proteins, 25S rRNA, and AFs such as Rlp7 and Nop7. Plasmids expressing the mutants were transformed into the strain Δnop53/tet-GFP-NOP53, allowing the exclusive transcriptional repression of the full-length NOP53 upon treatment with doxycycline (DOX), but not that of the tested mutant. GFP was used as a negative control (−), and the full-length GFP-Nop53 (FL) as a positive control (Supplementary Fig S8A).
Figure 6.
Effect of nop53 truncation mutants on cell growth and pre-rRNA processing. (A) Schematic representation of the nop53 N- and C-terminal truncated mutants (Δ1–26, Δ1–47, Δ48–98, Δ1–71, 1–157, 1–230, 1–300, 1–380) indicating the regions involved in protein-protein interactions. The exosome and the RNA helicase Mtr4 interact with Nop53 N-terminus where the conserved AIM is found. 60S AFs (Rlp7, Nop7) and r-proteins (eL8, uL23, eL27, and uL29) which are associated with the ITS2-containing foot or PET directly interact with Nop53 at different points along the middle region and the C-terminus. (B) Spot growth assays performed with the conditional strain Δnop53/tet-OFF-GFP-NOP53, expressing the positive control GFP-Nop53 full length (FL), the negative control empty vector (−), or the indicated GFP-tagged nop53 N- and C-terminal truncated mutants. These strains were spotted in 10-fold serial dilution on to YNB Glu +LYS+LEU solid medium supplemented or not with doxycycline (DOX – 1.5μg/ml) and rapamycin (400 nM). They were incubated at 30°C for the period indicated in square brackets. (C) Northern blot analysis of total RNA extracted from the same strains grown at 30°C with or without doxycycline (DOX – 1.5 μg/ml) to evaluate the effect of the nop53 truncated mutants on the pre-rRNA processing. The RNA resolved through 8% polyacrylamide denaturing gel electrophoresis was analyzed using the indicated probes. The Δrrp6 strain was employed as a control for the accumulation of 5.8S + 30 pre-rRNA. All tested nop53 truncated mutants accumulated 7S pre-rRNA, although at different levels. The quantification of the bands is shown in Supplementary Figure S8D.
The growth complementation assay shows that the tested truncation mutants partially complemented the growth of Δnop53 at 30°C (Figure 6B and Supplementary Figure S8B-C). Consistent with this phenotype, all the truncation mutants accumulated 7S pre-rRNA although at different levels (Figure 6C and Supplementary Figure S8D). The N-terminal deletions and the mutant nop53 1–157 exhibited the highest increases of 7S pre-rRNA levels, similar but inferior to Nop53 depletion. These observations not only confirm the essential role of the AIM, and the important function of the central region of Nop53 (amino acids 157–300) stabilizing its association with the pre-60S, but also indicate that the region immediately preceding the AIM, comprising the first 47 amino acid residues, is important to ensure adequate 7S pre-rRNA processing.
To better differentiate the effect of the nop53 deletion mutants, we performed the complementation assay at different growth conditions. The C-terminally truncated mutants seemed slightly more cold-sensitive than the N-terminal deletion mutants (Supplementary Figure S8C). Both N- and C-terminal mutants exhibited, as previously shown for nop53 AIM mutants (48), comparable sensitivity to hygromycin B, a translation inhibitor that affects the decoding fidelity (95) (Supplementary Figure S8C). However, upon treatment with the TOR inhibitor, rapamycin, which triggers a decrease of protein synthesis and ribosome biogenesis (96–99), the N-terminal truncated mutants were significantly more sensitive than the C-terminal deletion mutants, and remarkably impaired the cellular growth more than the depletion of Nop53 (Figure 6B). This effect was also detected without doxycycline treatment (in the presence of full-length Nop53), characterizing a dominant-negative phenotype.
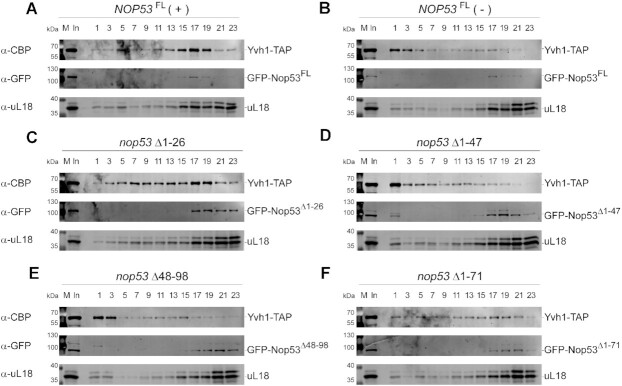
As the nop53 N-terminal truncated mutants mostly affect the interaction of Nop53 with the exosome, without impairing the association of Nop53 with the pre-60S, we compared their effect to the depletion of Nop53. Although in both cases the 7S pre-rRNA accumulates with consequent retention of the foot in late pre-60S intermediates, in the first case the nop53-truncated mutants would be found embedded at the basis of the foot, whereas in the second case Nop53 binding sites would be vacant. To assess whether the recruitment of Yvh1 to the pre-60S could be affected by the different nop53 N-terminal mutants, we performed density gradient fractionation (Figure 7). Importantly, all tested nop53 N-terminal truncated mutants co-sedimented with uL18 in bottom fractions of the gradient, confirming that the N-terminus is not necessary for the stable association of Nop53 with the pre-60S (Figure 7C–F). As expected, and shown above, depletion of Nop53 decreased the association of Yvh1 with pre-60S intermediates, displacing it from fractions 13–19 to the free fractions 1–5 (Figure 7A, B). Interestingly, the N-terminal mutants exhibited different impacts on the Yvh1 sedimentation profile. Yvh1 was still mainly associated with pre-60S in the nop53Δ1–26 mutant, which caused only a slight increase of the free fractions. On the contrary, the mutants nop53Δ1–47, nop53Δ48–98, and nop53Δ1–71 significantly affected the association of Yvh1 with the pre-60S, similar to Nop53 depletion. Together, these results pointed to the importance of the Nop53 N-terminus and the ITS2-foot disassembly for the Yvh1 recruitment.
Figure 7.
nop53 N-terminal truncated mutants impair the recruitment of Yvh1 to the pre-60S. Sedimentation profile analysis of Yvh1-TAP expressed in the conditional strain Δnop53 carrying (A) the positive control GFP-NOP53FL full length (+), (B) the negative control empty vector (−), or the GFP-tagged N-terminal truncated mutants (C) nop53Δ1–26, (D) nop53Δ1–47, (E) nop53Δ48–98 and (F) nop53Δ1–71. Whole-cell lysates were subjected to 10–30% glycerol density gradient sedimentation followed by top to bottom fractionation. All the odd fractions collected were analyzed by western blot with anti-CBP, anti-GFP, and anti-uL18. (In: input – whole cell lysate; M: protein MW marker).
Complementary to these observations, we compared the global effect of Nop53 depletion and its N-terminal mutants on the preribosomal and mature 60S subunits by chemically probing the RNA structure of the 25S rRNA using in vivo SHAPE assay (Supplementary Figure S9) (75). We focused on pre-60 binding sites of Nop53 (H16–17 in Domain I (38)) and Yvh1 (P stalk and SRL in Domains II and VI, respectively (50)). If the effect of Nop53 depletion and the N-terminal mutations on the 60S subunits were the same, we would have observed a very similar chemical modification pattern. Instead, the chemical reactivity of several regions of the 25S rRNA were differentially affected. Interestingly, the nucleotide accessibility of the Nop53 interacting site was increased only upon depletion of Nop53 (Supplementary Figure S9A). The mutant nop53 Δ1–26, which had almost no effect on the Yvh1 recruitment (Figure 7C), exhibited a modification pattern very similar to NOP53 full-length with all tested probes. Coherently, the other N-terminal mutants (nop53Δ1–47, nop53Δ48–98 and nop53Δ1–71) exhibited very similar modification patterns among them, as observed in the P-stalk and SRL regions, but different from Nop53 depletion (Supplementary Figure S9B, C). These changes in the modification patterns might correlate with the accumulation of different pre-60S intermediates, as well as of defective preribosomal and mature 60S particles. Regardless of the underlying cause, the observed differences attest to the involvement of different impacts on the 60S subunits, strengthening the hypothesis that the role of Nop53 during the pre-60S maturation exceeds that of its N-terminus.
Nop53 depletion has different impacts on the pre-60S maturation compared to truncation mutants lacking the exosome recruiting motif
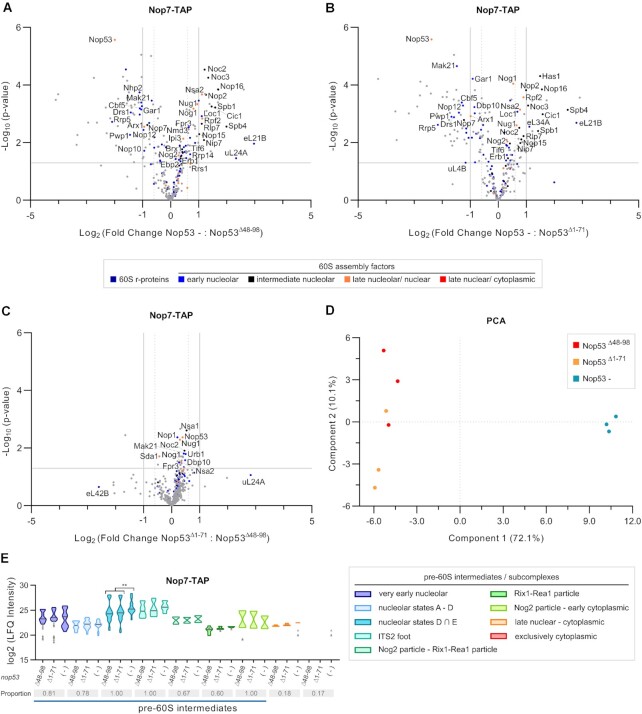
To more thoroughly evaluate whether the nop53 N-terminal truncation mutants differentially affect the 60S biogenesis pathway in comparison to Nop53 depletion, we set out to analyze the preribosomal intermediates affinity-purified with Nop7-TAP in these different conditions by mass-spectrometry based label-free quantitative proteomics (Figure 8 and Supplementary Figure S10). Since Nop7 remains associated with the pre-60S from an early nucleolar stage up to Nop53 release, this AF was chosen for probing not only all Nop53-containing intermediates but also earlier maturation stages. We focused our analysis on the nop53Δ48–98 and nop53Δ1–71 mutants, both devoid of the Mtr4 interacting motif (AIM), and therefore incapable of recruiting the exosome for the 7S pre-rRNA trimming and the foot disassembly. Interestingly, the pre-60S intermediates co-purified with Nop7 upon Nop53 depletion showed significant differences compared to both nop53Δ48–98 and nop53Δ1–71, whereas these mutants showed similar profiles between them (Figure 8). Upon depletion of Nop53, Nop7 particles exhibited decreased levels of some very early nucleolar AFs (e.g. Rrp5, Pwp1, Mak21), when comparing with both nop53 mutant strains, but statistically significant enrichment of several state D-E particle components (e.g. Noc3, Spb4, Nop2, Nip7, Spb1), strongly suggesting a predominant association of Nop7 with these nucleolar pre-60S intermediates when Nop53 is lacking (Figure 8E). These observations are not only in agreement with the above-mentioned results showing that Nop53 depletion impairs the transition from state E to Nog2 particle, but also indicate that this process is more dependent on the recruitment of Nop53 to the pre-60S than on the ability of Nop53 to recruit the RNA exosome. Consistent with this interpretation, the hierarchical clustering of samples, based on the LFQ intensity values of all identified 60S AFs, clustered the nop53 N-terminal mutants together, separated from the Nop53 depletion replicates (Supplementary Figure S10C), which is also shown in the Principal Component Analysis (PCA) (Figure 8D). Collectively, these findings corroborate recently revealed structural details of the transition from state E to Nog2 particle (Figure 9A) (28), showing that the recruitment of Nop53 to the foot helps to reposition the L1 stalk at a near mature conformation, which is essential for the subsequent formation of the Nog2 particle.
Figure 8.
Impact of nop53 N-terminal truncated mutants on the 60S subunit assembly compared to depletion of Nop53. Nop7-TAP was expressed in the conditional strain Δnop53/ptet-Off-GFP-NOP53 carrying the empty vector, or expressing one of the mutants GFP-nop53Δ48–98 and GFP-nop53Δ1–71. Strains were grown at the same conditions, under repression of NOP53 full-length expression. Preribosomes copurified with Nop7-TAP in biological triplicates upon depletion of Nop53 or in the presence of nop53 N-terminal mutants were analyzed by mass spectrometry-based label-free quantitative proteomics. Volcano plots show the comparison of Nop53 depletion (Nop53-) with (A) nop53Δ48–98 and with (B) nop53Δ1–71, as well as between the (C) nop53 mutants (nop53Δ1–71 versus nop53Δ48–98). 60S r-proteins and AFs are highlighted in colors, the latter being classified according to the 60S maturation stage they participate in. (D) PCA plot (Benjamini-Hochberg cut-off FDR 0.05) compares the proteomic profiles of the biological replicates. (E) Analysis of clusters of 60S AFs representative of specific pre-60S intermediates. Violin plots show the distribution of the copurified 60S AFs based on their log2-transformed LFQ Intensity values (states D ∩ E designates intersection – common AFs between these particles); height: delimited by the highest and lowest LFQ Intensity values of the cluster; width: frequency (high: wide; low: narrow) of the LFQ Intensity values observed in the cluster. The 60S AFs identified in only one or two conditions are depicted as gray triangles. The median is indicated by a dark gray line. The three conditions were pairwise compared and statistically significant (P < 0.05) differences are highlighted in the graph (Nop53 – versus nop53Δ48–98 ** P = 0.0015; Nop53 – versus nop53Δ1–71 **P = 0.0065). Gray boxes show the proportion of each cluster that was affinity-purified (number of affinity-purified 60S AFs components of the cluster/ total number of 60S AFs in the cluster). A horizontal bar indicates the pre-60S intermediates with which Nop7 is expected to be associated.
Figure 9.
Nop53 plays an essential structural role during the transition from nucleolar state E to the Nog2 particle. (A) Structural and compositional changes undergone by the ITS2-foot, which allow the stable accommodation of the H79-ES31 during the 25S rRNA Domain V anchoring to the pre-60S. The transition from the state E particle to the Nog2 particle is depicted by the cryo-EM structures PDB IDs 6ELZ, 6YLX, 6YLY and 3JCT, highlighting only the 60S AFs and r-proteins that interact directly with the L1-stalk basis (H74-H76) and the H79-ES31. For simplicity, Nip7 and Nop2 are not shown in the state E particle. Upon Erb1 eviction and Nop53 binding, Rlp7 N-terminus (NTD) is displaced. Nop53 extensions interact directly with the H79-ES31 and with 4 (Rlp7, eL8, uL23, and eL27) out of the 8 proteins that comprise the H79-ES31 docking site. (B) Schematic representation of the 60S assembly pathway based on the PDB IDs 6ELZ, 6YLY, 3JCT, 6FT6, 6FSZ, 6YLH, 6RZZ and 4V88. The 25S rRNA Domain V is depicted in green, whereas the L1-stalk basis and the H79-ES31 are colored as in (A). Important remodeling events are summarized below the structures, whereas foot-associated events above. Our proposed model indicates that due to its structural role, Nop53 depletion directly affects the formation of Nog2 particles impairing later steps of pre-60S maturation. This effect was neither observed upon removal (nop53Δ48–98, nop53Δ1–71) or mutation (nop53 D64R, nop53 AIM 5xAla, (48)) of Nop53 exosome-interacting region, nor upon mutation of Las1 (las1 R129A, las1 H134A, (37,48)) or of the exosome subunits and cofactors (dob1-1, rrp4-1, mtr3-1, (49)). These mutants, by impairing ITS2 processing, cause the retention of the foot in late pre-60S intermediates, without significant impacts on the export competence or cytoplasmic maturation events.
DISCUSSION
Nop53 depletion affects nucleolar and late 60S maturation stages
The first studies characterizing the role of Nop53 in yeast S. cerevisiae showed that the depletion of this essential 60S AF caused accumulation of 7S and 27S pre-rRNAs (89–91). The underlying molecular mechanism, however, has only recently been described, revealing that Nop53 directly recruits the Mtr4-associated exosome complex for the 7S pre-rRNA 3′-5′ processing (32,38,39,41). Excepting its role in the ITS2 removal and foot disassembly (32,40), little is known about the global impact of Nop53 recruitment on the compositional changes of the maturing large ribosomal subunit. To address this question, this study has evaluated how Nop53 depletion affects different stages of 60S assembly, probing it with 8 different AFs used as baits to affinity-purify preribosomal intermediates.
Nop7-TAP and Erb1-TAP allowed us to evaluate the earliest nucleolar 60S maturation stages, whereas the Nog2 particle components, Arx1-TAP and Rsa4-TAP, provided a window spanning the nucleoplasmic stages. Our quantitative proteomic analyses showed that upon depletion of Nop53, pre-60S subunits copurified with both A3-factors Nop7 and Erb1 coenriched components of the nucleolar state E particle and decreased association with earlier AFs. Of note, the Nsa1 module, which is removed by the AAA-ATPase Rix7 in the transition from nucleolar state D to E (17–20), was significantly decreased in Erb1 particles upon Nop53 depletion. As the transition from the nucleolar state E to the nuclear Nog2 particle coincides with the replacement of Erb1 by Nop53 at the basis of the foot (17,22,25), these observations strongly suggested an impaired formation of Nog2 particles. Consistent with this hypothesis, both Rsa4-TAP and Arx1-TAP copurified significantly reduced levels of Nog2 particle components in the Nop53-depleted condition.
Comparing the observed phenotype with already described mutants of AFs that constitute the state E or Nog2 particles, we noticed that depletion of Nop53 differs from ytm1 point mutants, which completely blocks the pre-60S maturation at state E particles, avoiding the Rea1-driven Erb1 release (17,25,27). Nop53 depletion is also not comparable to the depletion of most of the B-factors (required for the C2 cleavage), that block early nucleolar stages, impairing the transition from state C to D particles (36,37). Nog2 depletion, on the contrary, exhibited a similar profile to the Nop53-depleted condition, disrupting Rsa4 and Arx1 association with pre-60S particles (100,101). Interestingly, however, it caused almost no effect on state E components (36,37,100), suggesting a critical role for Nop53 binding on the transition from state E to Nog2 particle.
The disassembly of the foot structure, which is dependent on the ITS2 processing, has been shown nonessential for most of the late nuclear remodeling events, including nuclear export, so that late pre-60S subunits still carrying the foot can escape to the cytoplasm and engage in translation (37,48,49). Here we show that depletion of Nop53 also causes retention of the foot structure in late pre-60S intermediates, similar to mutant strains impaired in the recruitment or activity of the exosome (nop53 D64R, nop53 AIM-5xAla, dob1-1, rrp4-1, mtr3-1) or the Las1-complex (las1Δ, las1 R129A, las1 H134A).
However, different from the above-mentioned ITS2-processing mutants, the effect of Nop53 depletion reverberates in the following steps of the 60S maturation pathway. Both Rsa4 and Arx1 exhibited reduced association with components of the nucleoplasmic Rix1-Rea1 particle, reflecting the delayed formation of Nog2 particles. This effect was slightly more pronounced on the interaction of Arx1 with pre-60S intermediates. As a non-essential nuclear export AF, however, it would not be sufficient to halt the pre-60S maturation or cause a significant nuclear export defect (86). Indeed, the analysis of preribosomal particles affinity purified with the late-binding AFs Nmd3 and Lsg1 showed that late nuclear and early cytoplasmic intermediates were still formed under Nop53-depleted condition, although at decreased levels. In line with that, the nucleocytoplasmic AF Nmd3 exhibited a partial nuclear enrichment upon Nop53 depletion, reminiscent of the phenotype of depletion of export factors (102,103). Likewise, the nucleolar foot AF Nop7 was not significantly misplaced to the cytoplasm, as previously reported for nop53 AIM-5xAla (48), but found mostly in the nucleoplasm, similar to Arx1, whose subcellular localization remained unaltered.
Remarkably, the late-acting nucleocytoplasmic AF Yvh1 was absent on preribosomes isolated with all tested late-binding AFs (Arx1, Nmd3, Lsg1) upon depletion of Nop53. This observation was further confirmed using Yvh1 itself as a bait, showing a prominent decrease of copurified late AFs, r-proteins, as well as precursor and mature rRNAs. In the same condition, cytoplasmic Lsg1 particles did not exhibit the same compositional change, suggesting that Nop53 depletion affects more intensely particular late maturation events, specifically impairing the Yvh1 recruitment. The retention of the ITS2-foot seems to contribute to this effect, as nop53 N-terminal truncation mutants lacking the exosome-interaction motif (AIM) also caused a similar effect on the Yvh1 recruitment. The main cause, however, seems to be related to the less efficient formation of late pre-60S intermediates to which Yvh1 binds, given that earlier steps are affected upon Nop53 depletion, similar to what has been reported in Arx1 particles upon Cgr1 depletion (104).
Consistent with a delayed formation of late pre-60S intermediates, depletion of Nop53 did not phenocopy the Δyvh1 effect on cytoplasmic pre-60S intermediates. The binding of Yvh1, which is a dual-specificity phosphatase and zinc-binding protein, allosterically induces the dissociation of Mrt4 from the P-stalk (50,58–62). Upon depletion of Nop53, Mrt4 was still associated with foot-containing pre-60S particles but did not accumulate in the cytoplasm as in Δyvh1 (58,59), evidencing that the pre-60S maturation was not specifically blocked at the Yvh1-dependent release of Mrt4, but at an earlier step. The implications of the increased free pool of Yvh1 remain to be tested, considering the numerous cellular processes to which it has been linked (105–108).
Evidence of additional roles of Nop53 during the 60S subunit assembly
Comparing the overall effect of Nop53 depletion with previous reports on the retention of the ITS2-foot in late pre-60S subunits, interesting differences arise. Whereas depletion of Nop53 causes nuclear enrichment of Nmd3, point mutations in nop53 N-terminal AIM are sufficient to cause noticeable cytoplasmic mislocalization of Nop7 and Cic1 (48), so as depletion or mutation of the endoribonuclease Las1, generating translation competent foot-containing 60S subunits (37,48). The early pre-60S maturation steps are likewise differentially affected. In contrast to Nop53 depletion, Nop7 particles affinity-purified from cells expressing the catalytic-dead mutant las1 H134A did not exhibit enrichment of early-binding AFs, but instead, decreased levels of Erb1, Ytm1, and other components of state E particles (37). Taken together, these observations illustrate how Nop53 depletion has a different effect on the 60S assembly pathway compared to other conditions that exclusively block the ITS2 processing, strengthening the hypothesis of an additional exosome-independent structural role played by Nop53, which was not hindered through AIM point mutations nor upon Las1 depletion.
This prompted us to evaluate the effect of new nop53 truncation mutants. Interestingly, we observed that the Nop53 N-terminal region upstream the AIM is also important for the ITS2 processing, as nop53 N-terminal mutants lacking the first 47 aa, although still carrying the AIM, accumulated 7S pre-rRNA. Testing both nop53 N- and C-terminal mutants in different growth conditions, we showed that only the N-terminal mutants were strongly affected upon treatment with the TOR-inhibitor rapamycin, yielding a dominant-negative phenotype. Upon TOR inhibition, several cellular processes are affected including translation and ribosome biogenesis, decreasing RNA Pol I and Pol III activities, and downregulating the expression of r-proteins and AFs (96–99). In this condition, we speculate that the nop53 C-terminal mutants, being devoid of large anchoring extensions (426–455 aa) involved in interactions with Nop7, Rlp7, and uL23 (22,23), might be less stably associated with the pre-60S, causing a similar or milder effect than depletion of Nop53. The nop53 N-terminal mutants, on the other hand, may compete with the full-length copy and cause, once bound to the pre-60S, the formation of foot-containing 60S subunits, which trigger not only translational stress, but retention of A3 AFs in the cytoplasm synergistically affecting the ribosome biogenesis with TOR inhibition (37,48).
Preribosomes affinity-purified with Nop7 upon depletion of Nop53 exhibited significant compositional changes in comparison to those isolated from cells expressing nop53 N-terminal mutants (nop53Δ48–98 and nop53Δ1–71), that bind to the pre-60S but cannot recruit the exosome. These observations are consistent with the in vivo SHAPE analysis of the 25S rRNA, revealing strikingly different modification patterns between Nop53 deletion and N-terminal mutants. If the only function of Nop53 during the 60S assembly were associated with the recruitment of the exosome, we would expect rather similar profiles. Instead, compared to both N-terminal mutants, Nop7 particles isolated upon Nop53 depletion were highly enriched for components of the state E particle, which is not only consistent with the other results but also shows that an efficient formation of Nog2 particles requires the association of Nop53 with the pre-60S regardless of its capacity of recruiting the exosome.
Structural basis of the expanded role of Nop53 in the 60S subunit assembly
Taking into account recently described cryo-EM structures, we can explain the molecular basis of the structural role played by Nop53 (Figure 9A), whose implications we have here discussed. During the assembly of the 60S subunit, the ITS2-containing foot plays an important role as a scaffold (9) in early nucleolar steps affecting the ITS1 processing (11,21), and later, assisting the L1 stalk (H74-79-ES31) repositioning (17,28). In the first case with Erb1, and in the second, after the recruitment of Nop53. In the transition from state E to Nog2 particle, several AFs are released, including those that stabilize L1 stalk in a premature conformation (17–19,22,28). After the recruitment of Nop53, the 25S rRNA Domain V, which contains the PTC, undergoes a significant remodeling with accommodation of the flexible L1 stalk (H74-H79-ES31) in a mature-like position and binding of the r-proteins uL2 and eL43 (Figure 9A) (28). Nop53 plays an important role during this process, allowing the repositioning of the N-terminus of Rlp7, which enables the accommodation of the H79-ES31 (Figure 9A) (17,28). In Figure 9A we highlight that Nop53 not only directly interacts with the H79-ES31, but also with AFs and r-proteins (Rlp7, eL8, uL23 and eL27) that comprise the H79-ES31 docking site (22,23,28). At this position, Nop53 seems to play a central role in organizing and stabilizing the interface of the foot with the r-proteins, impacting on eL8 NTD (Figure 9A, Nop53 early-particle 2), making room for the stable accommodation of H79-ES31 and assisting the L1 stalk repositioning. In light of this interpretation, we propose that in the absence of Nop53, the foot structure is likely more flexible or not properly oriented to scaffold the 25S rRNA Domain V remodeling, which would affect but not completely hinder L1 stalk rearrangement, explaining the observed delay in the transition from state E particle to Nog2 particle.
Remarkably, a similar effect has been reported upon depletion of uL2 and eL43, which bind to H79-ES31 just after the L1 stalk repositioning (Figure 9A). The absence of these r-proteins causes enrichment of early-binding AFs and decrease of Nog2-particle components affinity-purified with Nog1 (70). Likewise, truncation of the eL8 N-terminal extension, which directly interacts with H79-ES31 (Figure 9A), although not sufficient to avoid the formation of the Nog2 particle, impairs the recruitment of Sda1 and blocks the following 60S maturation steps (109), conceivably due to misfolding of the L1-stalk and H68-69, which are Sda1 binding sites on the pre-60S. These r-proteins share with Nop53 the interaction with the H79-ES31, highlighting the importance of an adequate 25S rRNA Domain V docking, in the absence of which Nog2 particles might not be properly assembled. It remains to be tested if upon Nop53 depletion part of the Nog2 particles formed are also defective, impairing later remodeling events (Figure 9B).
The scaffolding role of the foot may be the reason why Nop53 recruitment at NE1 stage is not sufficient to trigger the ITS2 removal. Stable anchoring of Domain V and proper formation of the Nog2 particle must happen before the ITS2 processing starts. Interestingly, it has been recently shown that failure to recruit the AF Sda1, which interacts with the L1 stalk and the Rix1 complex (28), generates defective Nog2 particles, which are devoid of the ITS2-foot, with unstructured H75-H79-ES31, and exhibiting L1 stalk in an inward unusual position (29). As a possible interpretation, we propose that removal of the foot before stable binding of Sda1 or the Rix1 complex may destabilize the anchoring of H79-ES31 and induce the incorrect positioning of the L1 stalk, since during the ITS2 trimming by the exosome, the RNA helicase Mtr4 directly contacts both the H79-ES31 and the L1-stalk, pushing it towards a closed position (40). In the absence of Sda1 or the Rix1 complex, the exosome-induced L1 stalk repositioning would yield a deflected position halting later maturation steps. We postulate, therefore, that the ITS2 removal may act as a quality-control mechanism ensuring the proper assembly of Nog2 and Rix1-Rea1 particles, both sensitive to a premature foot disassembly. The interconnection of the foot with other functional centers such as PET, through uL23, and PTC, through H79-ES31, may provide cues for the exact timing of disassembly (64,65,109) and Nop53 seems to play a central role as sensor interacting with both uL23 and H79-ES31. Whereas Las1 cleavage depends on 5S RNP docking and binding of B-factors to Domain V (36,37), the activity of the exosome seems to be affected by, but not to depend on the 5S RNP rotation and association of Sda1 and Rix1 complex, which might provide mechanical information to the foot through Nop53. This functional connection between Domain V and the ITS2 remains to be further investigated.
Altogether, our results provide biochemical evidence to an expanded role of Nop53, revealing that its depletion affects both nucleolar and late maturation events, provoking different defects in the 60S biogenesis compared to N-terminal deletion mutants. These findings support the important structural role played by Nop53 during the transition from nucleolar state E to Nog2 particle, remodeling and stabilizing the foot structure to perform its last scaffolding function: to assist the Domain V anchoring by accommodating the H79-ES31 (Figure 9B). It is interesting to note that Nop53 association with the pre-60S is flanked by two major remodeling events triggered by the same AAA-ATPase, Rea1. It is tempting therefore to consider that by coordinating the exact moment of ITS2 removal and assisting Domain V anchoring, Nop53 plays a central role in the quality control of the first remodeling event before the second can take place. We postulate that in contrast to the retention of the foot, which does not impair most of the following pre-60S maturation events, the premature removal of the foot in the transition from state E to Rix1-Rea1 particle may lead to defective particles with L1 stalk incorrectly positioned and unstructured Domain V (29,30). The role of Nop53 during the 60S biogenesis is, therefore, not restricted to the recruitment of the RNA exosome, affecting essential rRNA remodeling events for the formation of late pre-60S particles.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD023383.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr John L. Woolford Jr. (Carnegie Mellon University) and Dr Cleslei F. Zanelli (UNESP, Araraquara, SP, Brazil) for the kind gifts of anti-Nog1 and anti-uL18 sera, respectively, and GFP Clone Collection strains. We also thank the members of our Department Dr Frederico Gueiros Filho for the use of the fluorescence microscope, Dr Mauricio S. Baptista for the use of the SpectraMax ie3 Microplate Detection Platform, and Dr Fabio L. Forti for the gift of hygromycin B. We thank all the members of the Oliveira lab for their support and critical discussions.
Contributor Information
Felipe F M Bagatelli, Department of Biochemistry, Institute of Chemistry, University of São Paulo, São Paulo, SP 05508-000, Brazil.
Francisca N de Luna Vitorino, Laboratory of Cell Cycle, Butantan Institute, São Paulo, SP 05503-900, Brazil; Center of Toxins, Immune-Response and Cell Signaling, Butantan Institute, São Paulo, SP 05503-900, Brazil.
Julia P C da Cunha, Laboratory of Cell Cycle, Butantan Institute, São Paulo, SP 05503-900, Brazil; Center of Toxins, Immune-Response and Cell Signaling, Butantan Institute, São Paulo, SP 05503-900, Brazil.
Carla C Oliveira, Department of Biochemistry, Institute of Chemistry, University of São Paulo, São Paulo, SP 05508-000, Brazil.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [15/06477-9 to C.C.O., 13/07467-1 to J.P.C.C. - CeTICS]; CAPES and FAPESP fellowships [2015/05776-2 to F.F.M.B., 2017-18344-9 to F.N.L.V.]. Funding for open access charge: FAPESP [15/06477-9 to C.C.O.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Timsit Y., Sergeant-Perthuis G., Bennequin D.. Evolution of ribosomal protein network architectures. Sci. Rep. 2021; 11:625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Petrov A.S., Gulen B., Norris A.M., Kovacs N.A., Bernier C.R., Lanier K.A., Fox G.E., Harvey S.C., Wartell R.M., Hud N.V.et al.. History of the ribosome and the origin of translation. Proc. Natl Acad. Sci. U.S.A. 2015; 112:15396–15401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klinge S., Voigts-Hoffmann F., Leibundgut M., Ban N.. Atomic structures of the eukaryotic ribosome. Trends Biochem. Sci. 2012; 37:189–198. [DOI] [PubMed] [Google Scholar]
- 4. Melnikov S., Ben-Shem A., Garreau de Loubresse N., Jenner L., Yusupova G., Yusupov M.. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012; 19:560–567. [DOI] [PubMed] [Google Scholar]
- 5. Ben-Shem A., Loubresse N.G., Melnikov S., Jenner L., Yusupova G., Yusupov M.. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011; 334:1524–1529. [DOI] [PubMed] [Google Scholar]
- 6. Tye B.W., Commins N., Ryazanova L.V., Wühr M., Springer M., Pincus D., Churchman L.S.. Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness. eLife. 2019; 8:e43002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang H., Ghalei H., Karbstein K.. Quality control of 40S ribosome head assembly ensures scanning competence. J. Cell Biol. 2020; 219:e202004161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collins J.C., Ghalei H., Doherty J.R., Huang H., Culver R.N., Karbstein K.. Ribosome biogenesis factor Ltv1 chaperones the assembly of the small subunit head. J. Cell Biol. 2018; 217:4141–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baßler J., Hurt E.. Eukaryotic ribosome assembly. Annu. Rev. Biochem. 2019; 88:281–306. [DOI] [PubMed] [Google Scholar]
- 10. Klinge S., Woolford J.L.. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019; 20:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Woolford J.L., Baserga S.J.. Ribosome biogenesis in the yeast saccharomyces cerevisiae. Genetics. 2013; 195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koš M., Tollervey D.. Yeast Pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010; 37:809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tomecki R., Sikorski P.J., Zakrzewska-Placzek M.. Comparison of preribosomal RNA processing pathways in yeast, plant and human cells – focus on coordinated action of endo- and exoribonucleases. FEBS Lett. 2017; 591:1801–1850. [DOI] [PubMed] [Google Scholar]
- 14. Fernández-Pevida A., Kressler D., de la Cruz J.. Processing of preribosomal RNA in Saccharomyces cerevisiae. WIREs RNA. 2015; 6:191–209. [DOI] [PubMed] [Google Scholar]
- 15. de la Cruz J., Karbstein K., Woolford J.L.. Functions of ribosomal proteins in assembly of eukaryotic ribosomes In Vivo. Annu. Rev. Biochem. 2015; 84:93–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gamalinda M., Ohmayer U., Jakovljevic J., Kumcuoglu B., Woolford J., Mbom B., Lin L., Woolford J.L.. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev. 2014; 28:198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kater L., Thoms M., Barrio-Garcia C., Cheng J., Ismail S., Ahmed Y.L., Bange G., Kressler D., Berninghausen O., Sinning I.et al.. Visualizing the assembly pathway of nucleolar Pre-60S ribosomes. Cell. 2017; 171:1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sanghai Z.A., Miller L., Molloy K.R., Barandun J., Hunziker M., Chaker-Margot M., Wang J., Chait B.T., Klinge S.. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature. 2018; 556:126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou D., Zhu X., Zheng S., Tan D., Dong M.-Q., Ye K.. Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate. Protein Cell. 2019; 10:120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kressler D., Roser D., Pertschy B., Hurt E.. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J. Cell Biol. 2008; 181:935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sahasranaman A., Dembowski J., Strahler J., Andrews P., Maddock J., Woolford J.L. Jr. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J. 2011; 30:4020–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu S., Tutuncuoglu B., Yan K., Brown H., Zhang Y., Tan D., Gamalinda M., Yuan Y., Li Z., Jakovljevic J.et al.. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature. 2016; 534:133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu S., Tan D., Woolford J.L., Dong M.-Q., Gao N.. Atomic modeling of the ITS2 ribosome assembly subcomplex from cryo-EM together with mass spectrometry-identified protein–protein crosslinks. Protein Sci. 2017; 26:103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Granneman S., Petfalski E., Tollervey D.. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 2011; 30:4006–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baßler J., Kallas M., Pertschy B., Ulbrich C., Thoms M., Hurt E.. The AAA-ATPase rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol. Cell. 2010; 38:712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wegrecki M., Rodríguez-Galán O., de la Cruz J., Bravo J.. The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two β-propellers during the assembly of large ribosomal subunits in eukaryotes. Nucleic Acids Res. 2015; 43:11017–11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thoms M., Ahmed Y.L., Maddi K., Hurt E., Sinning I.. Concerted removal of the Erb1–Ytm1 complex in ribosome biogenesis relies on an elaborate interface. Nucleic Acids Res. 2016; 44:926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kater L., Mitterer V., Thoms M., Cheng J., Berninghausen O., Beckmann R., Hurt E.. Construction of the central protuberance and L1 stalk during 60S subunit biogenesis. Mol. Cell. 2020; 79:615–628. [DOI] [PubMed] [Google Scholar]
- 29. Micic J., Li Y., Wu S., Wilson D., Tutuncuoglu B., Gao N., Woolford J.L.. Coupling of 5S RNP rotation with maturation of functional centers during large ribosomal subunit assembly. Nat. Commun. 2020; 11:3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilson D.M., Li Y., LaPeruta A., Gamalinda M., Gao N., Woolford J.L.. Structural insights into assembly of the ribosomal nascent polypeptide exit tunnel. Nat. Commun. 2020; 11:5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pillon M.C., Lo Y.-H., Stanley R.E.. IT’S 2 for the price of 1: multifaceted ITS2 processing machines in RNA and DNA maintenance. DNA Repair (Amst.). 2019; 81:102653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fromm L., Falk S., Flemming D., Schuller J.M., Thoms M., Conti E., Hurt E.. Reconstitution of the complete pathway of ITS2 processing at the pre-ribosome. Nat. Commun. 2017; 8:1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gasse L., Flemming D., Hurt E.. Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol. Cell. 2015; 60:808–815. [DOI] [PubMed] [Google Scholar]
- 34. Pillon M.C., Sobhany M., Borgnia M.J., Williams J.G., Stanley R.E.. Grc3 programs the essential endoribonuclease Las1 for specific RNA cleavage. Proc. Natl Acad. Sci. U.S.A. 2017; 114:E5530–E5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pillon M.C., Hsu A.L., Krahn J.M., Williams J.G., Goslen K.H., Sobhany M., Borgnia M.J., Stanley R.E.. Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex. Nat. Struct. Mol. Biol. 2019; 26:830–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Talkish J., Zhang J., Jakovljevic J., Horsey E.W., Woolford J.L. Jr. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2012; 40:8646–8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Biedka S., Micic J., Wilson D., Brown H., Diorio-Toth L., Woolford J.L. Jr. Hierarchical recruitment of ribosomal proteins and assembly factors remodels nucleolar pre-60S ribosomes. J. Cell Biol. 2018; 217:2503–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thoms M., Thomson E., Baßler J., Gnädig M., Griesel S., Hurt E.. The exosome is recruited to RNA substrates through specific adaptor proteins. Cell. 2015; 162:1029–1038. [DOI] [PubMed] [Google Scholar]
- 39. Falk S., Tants J.-N., Basquin J., Thoms M., Hurt E., Sattler M., Conti E.. Structural insights into the interaction of the nuclear exosome helicase Mtr4 with the preribosomal protein Nop53. RNA. 2017; 23:1780–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schuller J.M., Falk S., Fromm L., Hurt E., Conti E.. Structure of the nuclear exosome captured on a maturing preribosome. Science. 2018; 360:219–222. [DOI] [PubMed] [Google Scholar]
- 41. Cepeda L.P.P., Bagatelli F.F.M., Santos R.M., Santos M.D.M., Nogueira F.C.S., Oliveira C.C.. The ribosome assembly factor Nop53 controls association of the RNA exosome with pre-60S particles in yeast. J. Biol. Chem. 2019; 294:19365–19380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lingaraju M., Schuller J.M., Falk S., Gerlach P., Bonneau F., Basquin J., Benda C., Conti E.. To process or to decay: A mechanistic view of the nuclear RNA exosome. Cold Spring Harb. Symp. Quant. Biol. 2019; 84:155–163. [DOI] [PubMed] [Google Scholar]
- 43. Briggs M.W., Burkard K.T.D., Butler J.S.. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ End Formation*. J. Biol. Chem. 1998; 273:13255–13263. [DOI] [PubMed] [Google Scholar]
- 44. Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D.. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999; 18:5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barrio-Garcia C., Thoms M., Flemming D., Kater L., Berninghausen O., Baßler J., Beckmann R., Hurt E.. Architecture of the Rix1–Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nat. Struct. Mol. Biol. 2016; 23:37–44. [DOI] [PubMed] [Google Scholar]
- 46. Ulbrich C., Diepholz M., Bassler J., Kressler D., Pertschy B., Galani K., Böttcher B., Hurt E.. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell. 2009; 138:911–922. [DOI] [PubMed] [Google Scholar]
- 47. Matsuo Y., Granneman S., Thoms M., Manikas R.-G., Tollervey D., Hurt E.. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature. 2014; 505:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sarkar A., Thoms M., Barrio-Garcia C., Thomson E., Flemming D., Beckmann R., Hurt E.. Preribosomes escaping from the nucleus are caught during translation by cytoplasmic quality control. Nat. Struct. Mol. Biol. 2017; 24:1107–1115. [DOI] [PubMed] [Google Scholar]
- 49. Rodríguez-Galán O., García-Gómez J.J., Kressler D., de la Cruz J.. Immature large ribosomal subunits containing the 7S pre-rRNA can engage in translation in Saccharomyces cerevisiae. RNA Biology. 2015; 12:838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou Y., Musalgaonkar S., Johnson A.W., Taylor D.W.. Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome. Nat. Commun. 2019; 10:958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kargas V., Castro-Hartmann P., Escudero-Urquijo N., Dent K., Hilcenko C., Sailer C., Zisser G., Marques-Carvalho M.J., Pellegrino S., Wawiórka L.et al.. Mechanism of completion of peptidyltransferase centre assembly in eukaryotes. eLife. 2019; 8:e44904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Klingauf-Nerurkar P., Gillet L.C., Portugal-Calisto D., Oborská-Oplová M., Jäger M., Schubert O.T., Pisano A., Peña C., Rao S., Altvater M.et al.. The GTPase Nog1 co-ordinates the assembly, maturation and quality control of distant ribosomal functional centers. eLife. 2020; 9:e52474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kappel L., Loibl M., Zisser G., Klein I., Fruhmann G., Gruber C., Unterweger S., Rechberger G., Pertschy B., Bergler H.. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J. Cell Biol. 2012; 199:771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pertschy B., Saveanu C., Zisser G., Lebreton A., Tengg M., Jacquier A., Liebminger E., Nobis B., Kappel L., van der Klei I.et al.. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol. 2007; 27:6581–6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lo K.-Y., Li Z., Bussiere C., Bresson S., Marcotte E.M., Johnson A.W.. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell. 2010; 39:196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Meyer A.E., Hoover L.A., Craig E.A.. The cytosolic J-protein, Jjj1, and Rei1 function in the removal of the Pre-60 S subunit factor Arx1. J. Biol. Chem. 2010; 285:961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Greber B.J., Gerhardy S., Leitner A., Leibundgut M., Salem M., Boehringer D., Leulliot N., Aebersold R., Panse V.G., Ban N.. Insertion of the biogenesis factor Rei1 probes the ribosomal tunnel during 60S maturation. Cell. 2016; 164:91–102. [DOI] [PubMed] [Google Scholar]
- 58. Lo K.-Y., Li Z., Wang F., Marcotte E.M., Johnson A.W.. Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J. Cell Biol. 2009; 186:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kemmler S., Occhipinti L., Veisu M., Panse V.G.. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J. Cell Biol. 2009; 186:863–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rodríguez-Mateos M., Abia D., García-Gómez J.J., Morreale A., de la Cruz J., Santos C., Remacha M., Ballesta J.P.G.. The amino terminal domain from Mrt4 protein can functionally replace the RNA binding domain of the ribosomal P0 protein. Nucleic Acids Res. 2009; 37:3514–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rodríguez-Mateos M., García-Gómez J.J., Francisco-Velilla R., Remacha M., de la Cruz J., Ballesta J.P.G.. Role and dynamics of the ribosomal protein P0 and its related trans -acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2009; 37:7519–7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sarkar A., Pech M., Thoms M., Beckmann R., Hurt E.. Ribosome-stalk biogenesis is coupled with recruitment of nuclear-export factor to the nascent 60S subunit. Nat. Struct. Mol. Biol. 2016; 23:1074–1082. [DOI] [PubMed] [Google Scholar]
- 63. Weis F., Giudice E., Churcher M., Jin L., Hilcenko C., Wong C.C., Traynor D., Kay R.R., Warren A.J.. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2015; 22:914–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Konikkat S., Woolford J.L. Jr. Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast. Biochem. J. 2017; 474:195–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Biedka S., Wu S., LaPeruta A.J., Gao N., Woolford J.L. Jr. Insights into remodeling events during eukaryotic large ribosomal subunit assembly provided by high resolution cryo-EM structures. RNA Biology. 2017; 14:1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Greber B.J. Mechanistic insight into eukaryotic 60S ribosomal subunit biogenesis by cryo-electron microscopy. RNA. 2016; 22:1643–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Granato D.C., Machado-Santelli G.M., Oliveira C.C.. Nop53p interacts with 5.8S rRNA co-transcriptionally, and regulates processing of pre-rRNA by the exosome. FEBS J. 2008; 275:4164–4178. [DOI] [PubMed] [Google Scholar]
- 68. Huh W.-K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K.. Global analysis of protein localization in budding yeast. Nature. 2003; 425:686–691. [DOI] [PubMed] [Google Scholar]
- 69. Okuda E.K., Gonzales-Zubiate F.A., Gadal O., Oliveira C.C.. Nucleolar localization of the yeast RNA exosome subunit Rrp44 hints at early pre-rRNA processing as its main function. J. Biol. Chem. 2020; 295:11195–11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ohmayer U., Gamalinda M., Sauert M., Ossowski J., Pöll G., Linnemann J., Hierlmeier T., Perez-Fernandez J., Kumcuoglu B., Leger-Silvestre I.et al.. Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae. PLoS One. 2013; 8:e68412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Villén J., Gygi S.P.. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 2008; 3:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J.. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016; 13:731–740. [DOI] [PubMed] [Google Scholar]
- 73. Linnemann J., Pöll G., Jakob S., Ferreira-Cerca S., Griesenbeck J., Tschochner H., Milkereit P.. Impact of two neighbouring ribosomal protein clusters on biogenesis factor binding and assembly of yeast late small ribosomal subunit precursors. PLoS One. 2019; 14:e0203415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Konikkat S., Biedka S., Woolford J.L. Jr. The assembly factor Erb1 functions in multiple remodeling events during 60S ribosomal subunit assembly in S. cerevisiae. Nucleic Acids Res. 2017; 45:4853–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Spitale R.C., Crisalli P., Flynn R.A., Torre E.A., Kool E.T., Chang H.Y.. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 2013; 9:18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ying B.-W., Fourmy D., Yoshizawa S.. Substitution of the use of radioactivity by fluorescence for biochemical studies of RNA. RNA. 2007; 13:2042–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dez C., Houseley J., Tollervey D.. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006; 25:1534–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tang L., Sahasranaman A., Jakovljevic J., Schleifman E., Woolford J.L.. Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-Subcomplex in yeast preribosomes. MBoC. 2008; 19:2844–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Adams C.C., Jakovljevic J., Roman J., Harnpicharnchai P., Woolford J.L.. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA. 2002; 8:150–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ahmed Y.L., Thoms M., Mitterer V., Sinning I., Hurt E.. Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin–ligand-type complexes. Nat. Commun. 2019; 10:3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Leidig C., Thoms M., Holdermann I., Bradatsch B., Berninghausen O., Bange G., Sinning I., Hurt E., Beckmann R.. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat. Commun. 2014; 5:3491. [DOI] [PubMed] [Google Scholar]
- 82. Bradatsch B., Leidig C., Granneman S., Gnädig M., Tollervey D., Böttcher B., Beckmann R., Hurt E.. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat. Struct. Mol. Biol. 2012; 19:1234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Greber B.J., Boehringer D., Montellese C., Ban N.. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2012; 19:1228–1233. [DOI] [PubMed] [Google Scholar]
- 84. Ma C., Wu S., Li N., Chen Y., Yan K., Li Z., Zheng L., Lei J., Woolford J.L., Gao N.. Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat. Struct. Mol. Biol. 2017; 24:214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Malyutin A.G., Musalgaonkar S., Patchett S., Frank J., Johnson A.W.. Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis. EMBO J. 2017; 36:854–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bradatsch B., Katahira J., Kowalinski E., Bange G., Yao W., Sekimoto T., Baumgärtel V., Boese G., Bassler J., Wild K.et al.. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell. 2007; 27:767–779. [DOI] [PubMed] [Google Scholar]
- 87. Ho J.H.-N., Kallstrom G., Johnson A.W.. Nmd3p is a Crm1p-Dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 2000; 151:1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hedges J., West M., Johnson A.W.. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005; 24:567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Granato D.C., Gonzales F.A., Luz J.S., Cassiola F., Machado-Santelli G.M., Oliveira C.C.. Nop53p, an essential nucleolar protein that interacts with Nop17p and Nip7p, is required for pre-rRNA processing in Saccharomyces cerevisiae. FEBS J. 2005; 272:4450–4463. [DOI] [PubMed] [Google Scholar]
- 90. Thomson E., Tollervey D.. Nop53p is required for late 60S ribosome subunit maturation and nuclear export in yeast. RNA. 2005; 11:1215–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sydorskyy Y., Dilworth D.J., Halloran B., Yi E.C., Makhnevych T., Wozniak R.W., Aitchison J.D.. Nop53p is a novel nucleolar 60S ribosomal subunit biogenesis protein. Biochem. J. 2005; 388:819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chaker-Margot M., Klinge S.. Assembly and early maturation of large subunit precursors. RNA. 2019; 25:465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chen W., Xie Z., Yang F., Ye K.. Stepwise assembly of the earliest precursors of large ribosomal subunits in yeast. Nucleic Acids Res. 2017; 45:6837–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Schuch B., Feigenbutz M., Makino D.L., Falk S., Basquin C., Mitchell P., Conti E.. The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase. EMBO J. 2014; 33:2829–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Borovinskaya M.A., Shoji S., Fredrick K., Cate J.H.D.. Structural basis for hygromycin B inhibition of protein biosynthesis. RNA. 2008; 14:1590–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zencir S., Dilg D., Rueda M.P., Shore D., Albert B.. Mechanisms coordinating ribosomal protein gene transcription in response to stress. Nucleic Acids Res. 2020; 48:11408–11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Albert B., Knight B., Merwin J., Martin V., Ottoz D., Gloor Y., Bruzzone M.J., Rudner A., Shore D.. A molecular titration system coordinates ribosomal protein gene transcription with ribosomal RNA synthesis. Mol. Cell. 2016; 64:720–733. [DOI] [PubMed] [Google Scholar]
- 98. Loewith R., Hall M.N.. Target of Rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011; 189:1177–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jorgensen P., Rupeš I., Sharom J.R., Schneper L., Broach J.R., Tyers M.. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004; 18:2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lebreton A., Rousselle J.-C., Lenormand P., Namane A., Jacquier A., Fromont-Racine M., Saveanu C.. 60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes. Nucleic Acids Res. 2008; 36:4988–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Saveanu C., Bienvenu D., Namane A., Gleizes P.-E., Gas N., Jacquier A., Fromont-Racine M.. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001; 20:6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yao W., Roser D., Köhler A., Bradatsch B., Baßler J., Hurt E.. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell. 2007; 26:51–62. [DOI] [PubMed] [Google Scholar]
- 103. Altvater M., Chang Y., Melnik A., Occhipinti L., Schütz S., Rothenbusch U., Picotti P., Panse V.G.. Targeted proteomics reveals compositional dynamics of 60S pre-ribosomes after nuclear export. Mol. Syst. Biol. 2012; 8:628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Thoms M., Mitterer V., Kater L., Falquet L., Beckmann R., Kressler D., Hurt E.. Suppressor mutations in Rpf2–Rrs1 or Rpl5 bypass the Cgr1 function for pre-ribosomal 5S RNP-rotation. Nat. Commun. 2018; 9:4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Geng Q., Xhabija B., Knuckle C., Bonham C.A., Vacratsis P.O.. The atypical dual specificity phosphatase hYVH1 associates with multiple ribonucleoprotein particles. J. Biol. Chem. 2017; 292:539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sugiyama M., Nugroho S., Iida N., Sakai T., Kaneko Y., Harashima S.. Genetic interactions of ribosome maturation factors Yvh1 and Mrt4 influence mRNA decay, glycogen accumulation, and the expression of early meiotic genes in Saccharomyces cerevisiae. J. Biochem. 2011; 150:103–111. [DOI] [PubMed] [Google Scholar]
- 107. Liu Y., Chang A.. A mutant plasma membrane protein is stabilized upon loss of Yvh1, a novel ribosome assembly factor. Genetics. 2009; 181:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sakumoto N., Yamashita H., Mukai Y., Kaneko Y., Harashima S.. Dual-Specificity protein phosphatase Yvh1p, which is required for vegetative growth and sporulation, interacts with yeast pescadillo homolog in saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2001; 289:608–615. [DOI] [PubMed] [Google Scholar]
- 109. Tutuncuoglu B., Jakovljevic J., Wu S., Gao N., Woolford J.L.. The N-terminal extension of yeast ribosomal protein L8 is involved in two major remodeling events during late nuclear stages of 60S ribosomal subunit assembly. RNA. 2016; 22:1386–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD023383.