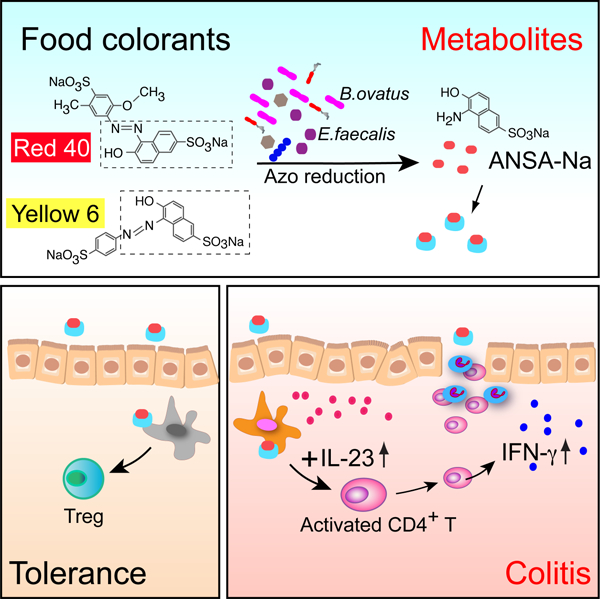
Summary
Both genetic predisposition and environmental factors appear to play a role in inflammatory bowel diseases (IBD) development. Genetic studies in humans have linked the interleukin (IL)-23 signaling pathway with IBD, but the environmental factors contributing to disease have remained elusive. Here we show that the azo dyes Red 40 and Yellow 6, the most abundant food colorants in the world, can trigger an IBD-like colitis in mice conditionally expressing IL-23, or in two additional animal models in which IL-23 expression was augmented. Increased IL-23 expression led to generation of activated CD4+ T cells that expressed interferon-γ and transferred disease to mice exposed to Red 40. Colitis induction was dependent on the commensal microbiota promoting the azo reduction of Red 40 and generation of a metabolite, 1-amino-2-naphthol-6-sulphonate sodium salt. Together these findings suggest that specific food colorants represent novel risk factors for development of colitis in mice with increased IL-23 signaling.
Keywords: Food colorants, food additives, CD4+ T cells, IFN-γ, IL-23, colitis, microbiota, azo bond, metabolites
Graphical Abstract
eTOC Blurb:
He et al. find that food colorants Red 40 and Yellow 6 induce colitis in mice with dysregulated expression of IL-23. Colitis development requires bacterial processing of the food colorants and is dependent on IL-23-induced pathogenic CD4+ T cells that produce IFN-γ.
INTRODUCTION
The dramatic changes in the concentration of air and water pollutants and the increased use of processed foods and food additives in the human diet in the last century correlate with an increase in the incidence of inflammatory and autoimmune diseases. These environmental changes are thought to contribute to development of these diseases, but relatively little is known about how they do so.
In the present study we focus on the contribution of a particular class of dietary products (food colorants) to development of inflammatory bowel disease (IBD), a condition that affects millions of people worldwide(Ng et al., 2018). Artificial food colorants were first introduced in the food chain at the end of the 19th century(Sharma et al., 2011), but despite being highly prevalent in world-wide diets, have not been studied in the context of IBD. Red 40, also known as Allura Red AC, is the most abundant food colorant in the world, with annual production exceeding 2.3 million kilograms(Sharma et al., 2011). Red 40 is a red azo dye that is approved as a food color additive in many countries and has been reported to have no adverse cytotoxic, carcinogenic or mutagenic effects on the host(Bastaki et al., 2017b; Honma, 2015; World Health Organization, 2017). The paucity of information of food colorants on IBD development stands in contrast with what is known about genetic and immune factors in the etiology of this condition. Over 200 loci and genes have been associated with IBD in humans(Liu et al., 2015) and several immune factors have been amply studied in mice and humans. Among the best-studied immune factors contributing to development of IBD is interleukin (IL)-23 (IL-23). Genome-wide association studies in humans link the IL-23 signaling pathway with IBD(Duerr et al., 2006) and recent clinical studies show that therapies targeting IL-23 are effective in patients with different forms of IBD, such as Crohn’s disease (CD) (Feagan et al., 2017; Sandborn et al., 2012; Sands et al., 2017) and ulcerative colitis (UC)(Sands et al., 2019).
IL-23 is a heterodimeric cytokine formed by the IL-23-specific p19 subunit and the p40 subunit that is also present on IL-12. Elevated production of IL-23 by myeloid cells has been noted in both patients with CD and UC(Kobayashi et al., 2008; Kvedaraite et al., 2016). IL-23 has been shown to contribute to colitis development in several studies using antibody blockade or gene-deficient mice(Bernshtein et al., 2019; Geremia and Jewell, 2012; Powrie et al., 1994). However, to our knowledge, no studies to date demonstrate that up-regulation of IL-23 expression is sufficient for induction of colitis in mice. A recent report suggests that up-regulation of IL-23 may contribute to colitis development (Bernshtein et al., 2019), but macrophage-derived IL-23 is unlikely to be the sole driver of pathology in that setting because such cells also lack IL-10 signaling (Bernshtein et al., 2019).
In our previous study, we found that overexpression of IL-23 by myeloid cells does not appear to be sufficient for induction of colitis in adult mice(Chen et al., 2018). The mice we developed conditionally express IL-23 in CX3CR1-positive myeloid cells (R23FR mice). R23FR mice were generated by crossing Rosa26-lox-STOP-lox-IL23 mice with CX3CR1CreER mice (FR mice)(Chen et al., 2018). Upon tamoxifen (TAM) treatment, R23FR mice express IL-23 in CX3CR1-positive cells but do not develop colitis when fed the standard diet used in our facility (Labdiet 5053). Colitis is only observed when R23FR mice are exposed to repeated cycles of custom diet 2019 (TD.160647, Envigo) (Chen et al., 2018). Histologically the colitis in R23FR mice induced by TAM plus diet 2019 resembles human ulcerative colitis, and it is apparent after the second cycle of TAM (d21)(Chen et al., 2018). This colitis is transient and the mice enter in remission after the diet 2019 is discontinued (Chen et al., 2018). Subsequent treatment of the R23FR mice in remission with diet 2019 without TAM causes a flare of colitis (d56)(Chen et al., 2018). Importantly, after the initial TAM treatment, IL-23 expression in CX3CR1-positive cells remains stable overtime, indicating that development of relapse is not caused by increased IL-23 expression(Chen et al., 2018).
Screening different components of the diets fed to R23FR mice we have now discovered that the food colorant Red 40, added to facilitate identification of one of the TAM-containing diets used in our experiments(Chen et al., 2018) is the colitogenic agent present in diet 2019. We show here that Red 40 alone does not induce colitis in control mice, but it can trigger severe IBD-like colitis in IL-23-overexpressing mice. Colitis development depends on processing of Red 40 by commensal bacteria and on CD4+ T cells that produced interferon (IFN)-γ. Animals given Red 40 prior to induction of IL-23 become unresponsive to IL-23 plus Red 40 challenge via induction of regulatory T cells (Treg), suggesting development of tolerance to this compound under normal conditions. We further show that Bacteroides ovatus (B.ovatus) and Enterococcus faecalis (E.faecalis) contribute to Red 40-induced colitis in R23FR mice using monocolonization of germ-free (GF) mice. Importantly, we show that 1-amino-2-naphthol-6-sulphonate sodium salt (ANSA-Na), metabolite derived from azo-reduction of both Red 40 and Yellow 6, is capable of inducing colitis in R23FR mice at remission. Finally, we also show that Red 40 functions as a colitogenic agent in non-transgenic wild-type mice in which expression of IL-23 is augmented. These findings provide experimental evidence to suggest that specific food colorants are environmental risk factors for colitis development in conditions where IL-23 expression is dysregulated.
RESULTS
Red 40 induces development of colitis in R23FR mice overexpressing IL-23.
To start identifying the major dietary components accounting for induction of colitis we solubilized the diet 2019 (TD 160647) with water and ethanol and tested the colitogenic activities of these fractions by feeding them to R23FR mice in remission (d48, after diet 2019 TAM treatment) (Fig. S1A). We found that the water-soluble fraction (aqueous extraction), but not the 95% ethanol-soluble fraction, induced colitis in R23FR mice as measured by fecal lipocalin-2 (Lcn-2) (Fig. S1B) and intestinal histology (Fig. S1C and S1D). These results suggested that the colitogenic component of diet 2019 was water-soluble. Further comparison of the water-soluble fraction with the ethanol-soluble fraction showed they differed in color, which was caused by the presence of the water-soluble food colorant Red 40 (Fig. 1A). Red 40 was added to the custom diet 2019 to indicate the presence of tamoxifen. To test if Red 40 contributed to induction of flares of colitis, we directly tested two versions of the diet 2019 (2019 grey, TD 130833 vs 2019 Red, TD 160647) (Fig. S1E). We found that the diet 2019 containing Red 40 (TD 160647), but not 2019 grey (TD 130833), caused flares of colitis in R23FR mice (Fig. S1F-S1H). In addition, administration of the diet 2019 grey (TD 130833) together with Red 40 in drinking water (Red water) induced severe disease similar to that induced by diet 2019 Red (TD 160647) (Fig. S1E-S1H).
Fig 1. Red 40 induces colitis in IL-23 overexpressing mice.
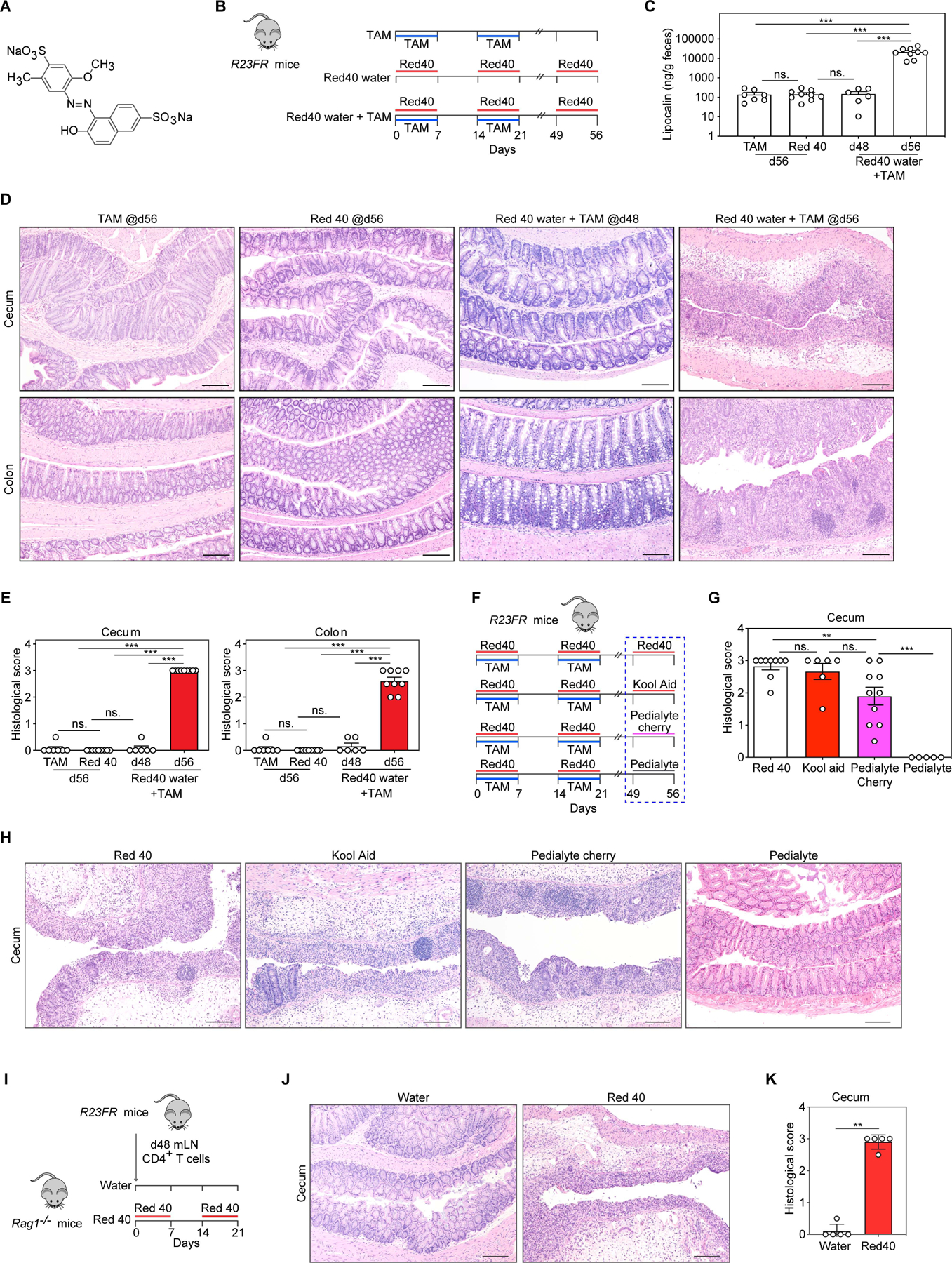
(A) Chemical structure of Red 40. (B) Experimental scheme. R23FR mice were treated with TAM and 0.025% Red 40 in drinking water (0.25g/L), or with TAM alone or with Red 40 in drinking water alone. (C) Fecal lipocalin-2 levels at day 56. (D,E) Representative H&E-stained sections (D) and histologic scores (E) of the cecum and colon of experimental mice described in Fig. 1B. (F) Experimental scheme. R23FR mice were treated with TAM + Red 40 for 2 cycles. For the final cycle, animals were treated with 0.25 g/L Red 40 in drinking water (positive control, 250mg/L), Kool aid (containing 200mg/L Red 40), Pedialyte AdvancedCare cherry punch flavor (containing 16 mg/L Red 40) or Pedialyte hydrating solution without Red 40 for 7 days. (G,H) Histologic scores (G) and representative H&E-stained sections (H) of the cecum of experimental mice described in Fig. 1F. (I) Experimental scheme. Unfractionated CD4+ T cells obtained from mLN of R23FR mice in remission (TAM + Red 40 treated group, d48) were injected into Rag1−/− mice (106 cells/mouse) fed with or without Red 40 in drinking water. (J,K) Representative H&E-stained sections (J) and histologic scores (K) of the cecum of experimental mice at day 21 described Fig. 1I.
Scale bars in (D) (H) and (J), 50 μm. In (C) (E) (G) and (K), each dot indicates an individual mouse. Error bars indicate SEM. ns p> 0.05, ** p<0.01, *** p<0.001, by nonparametric Mann-Whitney test.
To rule out any effect associated with the diet switch to the study, we tested if addition of TAM to our standard animal facility diet (LabDiet 5053) and administration of Red 40 in the drinking water would cause colitis. We fed the mice with the TAM diet (5053 TAM, TD.190129) without any colorants and added Red 40 in the drinking water (0.25g/L) at the same concentration found in the diet 2019 (Fig. 1B). Consistent with our previous results(Chen et al., 2018), TAM induced IL-23 expression in CX3CR1+ cells did not induce colitis when the mice drank water (Fig. 1C-1E). Treatment of mice with Red 40 alone did not cause colitis, but administration of Red 40 along with TAM (Red 40 water + TAM) caused a severe relapsing–remitting colitis in R23FR mice (Fig. 1C-1E). Mice treated with Red 40 water + TAM, had no inflammation in the large intestine at day 7, but had marked leukocytic infiltrates in the mucosa of the cecum at day 21. By day 48, these infiltrates were absent (Fig. 1C-1E). Notably, a relapse was induced with a third cycle of treatment (from d49 to d56) of Red 40 water without TAM (Fig. 1C-1E). At this point (day 56), both cecum and colon of R23FR mice (Red 40 water + TAM) showed a severe inflammation, with marked leukocytic infiltrates, crypt loss, epithelial damage, and ulceration (Fig. 1D). These findings are similar to those observed in R23FR mice treated with TAM and diet 2019 (0.25g/kg of Red 40) (Fig. S1H), indicating that Red 40 is the colitis-inducing agent present in diet 2019.
Red 40 is widely present in the food, drugs and beverages. Having shown that Red 40 in drinking water or in the diet can promote colitis in R23FR mice, we next tested if a Red 40-containing beverage or a Red 40-containing medicine given to humans could promote development of relapse in R23FR mice. Feeding R23FR mice at remission stage (d49, after TAM and Red 40 treatment) with the Red 40-containing beverage Kool-Aid (containing 200mg/L of Red 40) (Stevens et al., 2014) or the Red 40-containing hydrating solution Pedialyte cherry punch flavor (containing 16mg/L of Red 40), but not the Pedialyte hydrating solution without Red 40, promoted development of colitis (Fig. 1F-1H), further confirming that Red 40 can induce development of colitis in mice overexpressing IL-23 in myeloid cells.
Previously we have shown that unfractionated mesenteric lymph node (mLN) CD4+ T cells obtained from R23FR mice in remission transfer colitis to Rag1−/− mice exposed to diet switches (R23FR→Rag transfer model)(Chen et al., 2018). Based on the result shown above we hypothesized that Red 40 could contribute to disease development in the R23FR→Rag transfer model. As expected, unfractionated CD4+ T cells obtained from mLN of R23FR mice in remission stage (d48, after TAM and Red 40 treatment) transferred disease to Rag1−/− mice fed with Red 40 in drinking water, but not with regular water (Fig. 1I-1K). These results indicate that disease development in response to Red 40 and IL-23 is dependent on CD4+ T cells.
Red 40 exposure prior to IL-23 expression results in the generation Treg-mediated tolerance to Red 40
The colitis-inducing properties of Red 40 in R23FR mice are in contrast to its reported safety profile. Extensive safety testing has demonstrated that Red 40 has no adverse cytotoxic, carcinogenic, or mutagenic effects(World Health Organization, 2017). The fact that Red 40 consumption does not lead to development of colitis in the population at large suggests development of immunological tolerance to this compound (or to proteins that bind to it, or to its metabolites). Accordingly, we found that animals given Red 40 prior to induction of IL-23 became unresponsive to IL-23 plus Red 40 (Fig. 2A-2C), suggesting development of tolerance to this compound under normal conditions. We thus tested next whether regulatory T cells (Treg) derived from control mice exposed to Red 40 could inhibit or attenuate development of colitis (Fig. 2D). Colitis-inducing effector CD4+ T cells (CD45.2+) from mLN of R23FR mice in remission were transferred to Rag1−/− mice along with Treg cells isolated from wild-type mice (CD45.1) treated with Red 40 (Red 40 Treg) or water (water Treg) (Fig. 2D). Importantly, mice transferred with effector CD4+ T cells and Treg derived from water treated mice developed marked colitis (Fig. 2E-2F). In contrast, animals transferred with CD4+ effector cells and Treg isolated from Red 40 treated mice had significantly less colitis (Fig. 2E-2F). These results suggest that Red 40 exposure prior to an increase in IL-23 expression leads to development of tolerance mediated by Treg, and that elevated expression of IL-23 prevents development of tolerance to this compound.
Fig. 2. Red 40 induced Tregs protect from colitis development in R23FR mice.
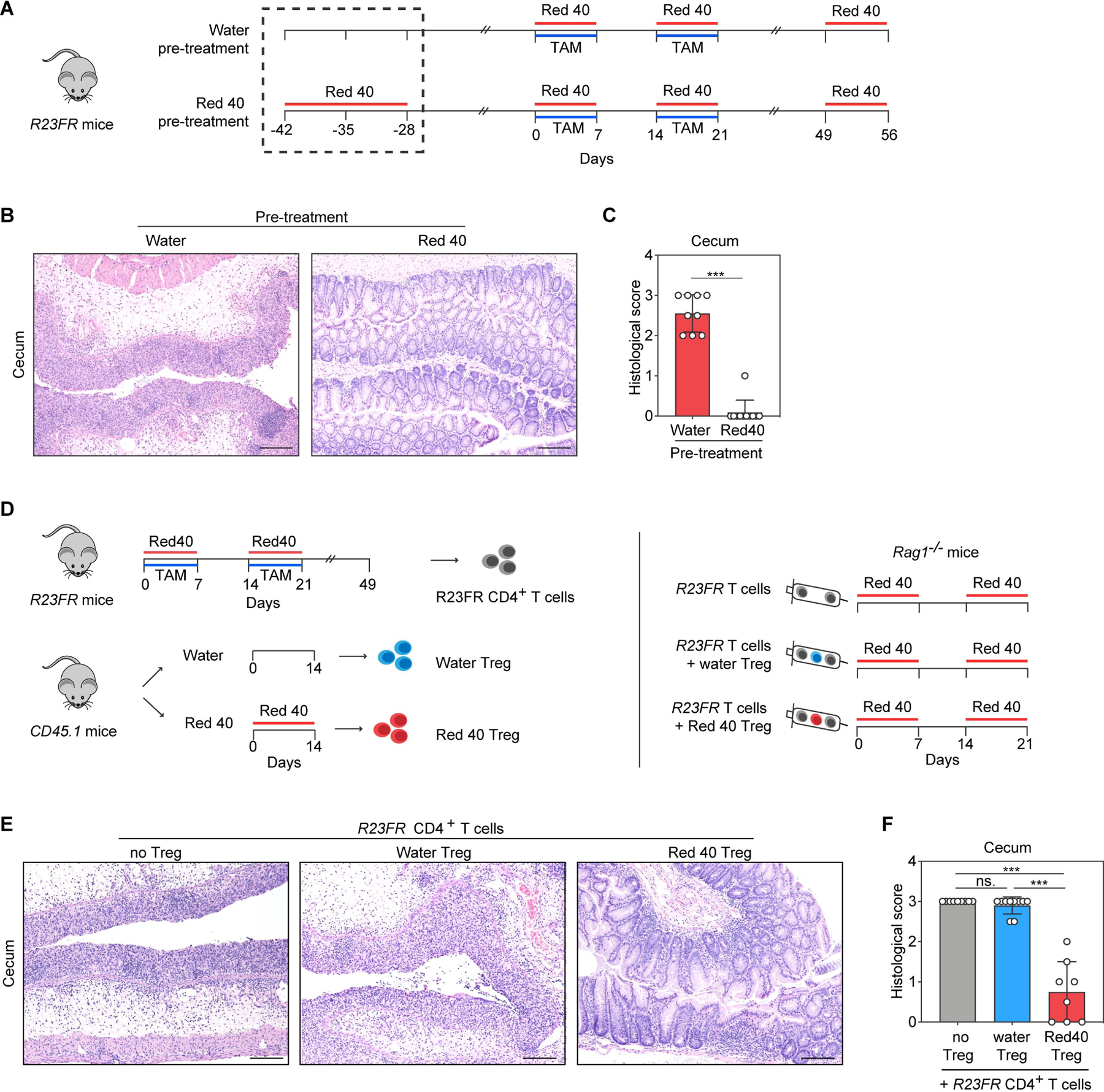
(A) Experimental scheme. R23FR mice were treated with Red 40 (0.25g/L) for 2 weeks before TAM and Red 40 treatment. (B,C) Representative H&E-stained sections (B) and histologic scores (C) of the cecum of experimental mice at day 56 described in Fig. 2A. (D) Experimental scheme. Unfractionated CD4+ T cells were isolated from the mLN of R23FR mice in remission (d48 after TAM and Red 40 treatment) (106 cells/ mouse) and co-injected with 4× 105 Treg, CD45.1+CD4+CD25+, isolated from WT mice (CD45.1+) treated with or without Red 40 into recipient Rag1−/− mice treated with Red 40 in the drinking water. (E,F) Representative H&E-stained sections (E) and histologic scores (F) of the cecum of experimental mice at d21 described in Fig. 2D. Scale bars in (B) and (E), 50 μm. In (C) and (F), each dot indicates an individual mouse. Error bars indicate SEM. *** p<0.001; ns, p >0.05 by nonparametric Mann-Whitney test.
Red 40 induced-colitis is dependent on IFN-γ, but not on IL-17A, IL-17F, TNF-α or IL-22
Having shown that Red 40 promotes disease via CD4+ T cells in R23FR mice, we sought next to define the immunopathogenic mechanisms responsible for development of colitis. IL-23 is known to regulate the production of cytokines such as IL-17, IL-22 and IFN-γ by CD4+ T cells. We tested next if these cytokines affected disease development using antibodies or gene-deficient mice. We found that blockade of TNF-α, IL-17A or IL-17F did not inhibit colitis development in the R23FR→Rag transfer model (Fig. 3A-3C). By using Il22−/− mice(Chen et al., 2019) intercrossed with R23FR mice, we found that IL-22 deficiency did not affect colitis development in R23FR mice primed with 2019 TAM (TD130968) and challenged with diet 2019 (Red, TD 160647)(Fig. 3D).
Fig 3. IFN-γ-producing CD4+ T cells are critical for the development of cecal inflammation.
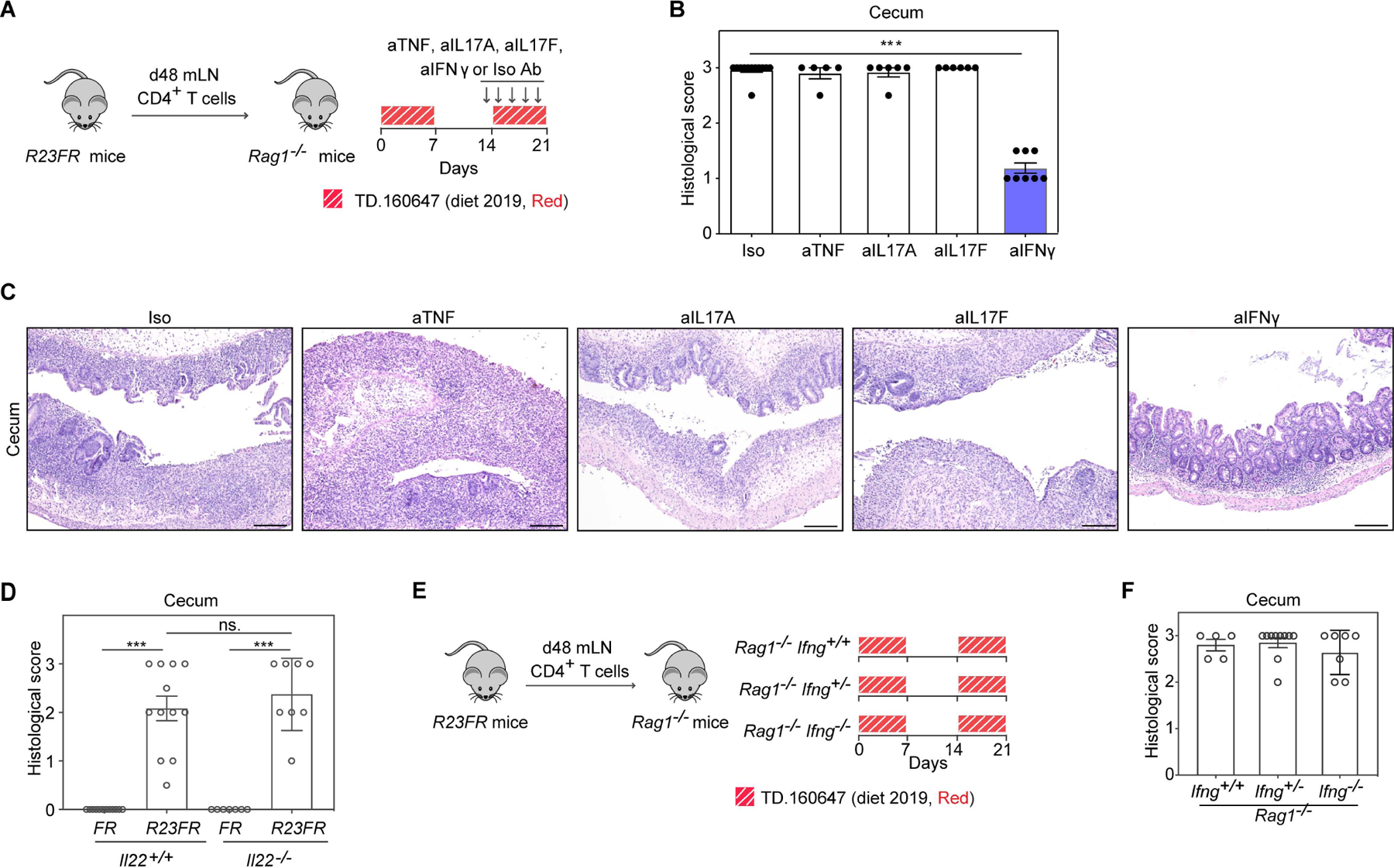
(A) Experimental scheme. (B,C) Histologic scores (B) and representative H&E-stained sections (C) of the cecum of adoptively transferred Rag1−/− mice at day 21 with different antibodies treatment described in Fig. 3A. (D) Histologic scores of the cecum of R23FR/Il22+/+ mice and R23FR/Il22−/− mice at day 56. Mice were primed with 2019 TAM (TD130968) and challenged with diet 2019 (Red, TD 160647) as described in Fig. S1A. (E) Experimental scheme. R23FR CD4+ T cells were transferred into littermate Ifng+/+Rag1−/− mice, Ifng+/−Rag1−/− mice and Ifng−/−Rag1−/− mice. Recipient mice were fed with two alternating cycles of diet 2019 (Red, TD 160647) for 21 days. (F) The histologic scores of the cecum of experimental mice at d21 described in Fig. 3E. Scale bars in (C), 50 μm. In (B) (D) and (F), each dot indicates an individual mouse. Error bars indicate SEM. *** p<0.001; ns, p >0.05 by nonparametric Mann-Whitney test.
In contrast to the results described above, we found that blockade of IFN-γ decreased colitis severity in the R23FR→Rag transfer model (Fig. 3A-3C). These results suggested that IFN-γ is a major cytokine contributing to CD4+ T cell effector function in this model. To further distinguish the cellular source of IFN-γ contributing to the disease, we sorted CD4+ T cells from donor R23FR mice in remission (d48) and transferred them to recipient Rag1−/− mice and Ifng−/− Rag1−/− double-knockout mice treated with diet 2019 (Red, TD 160647) (Fig. 3E). The Ifng−/− Rag1−/− double-knockout mice that received R23FR CD4+ T cells and were fed with 2 cycles of the Red 40-containing diet 2019 developed colitis comparable to that of control Rag−/− mice subjected to the same treatment (Fig. 3F). Given that the only source of IFN-γ in Ifng−/− Rag1−/− double-knockout mice were the donor CD4+ T cells (Fig. 3E), we conclude that the IFN-γ produced by CD4+ T cells is important for colitis development. In summary, the findings suggest that IFN-γ, but not IL-22, IL-17A, IL-17F nor TNF-α, is important for the effector function of CD4+ T cells in IL-23-overexpressing mice exposed to Red 40.
Red 40-induced colitis is dependent on the intestinal microbiota.
To probe into the colitogenic mechanisms associated with Red 40, we next tested if microbes were required for Red 40-induced colitis. We adoptively transferred mLN CD4+ T cells from d48 R23FR mice to germ-free (GF) Rag1−/− mice and fed them with Red 40 in drinking water (Fig. 4A). We did not observe colitis in GF conditions (Fig. 4B-4C). Colonization of these GF Rag1−/− mice with specific pathogen-free (SPF) commensal bacteria and treatment with Red 40 resulted in marked colitis (Fig. 4H-4J) indicating that the ability of Red 40 to cause colitis is dependent on the intestinal microbiota.
Fig 4. Red 40-induced colitis is dependent on the intestinal microbiota.
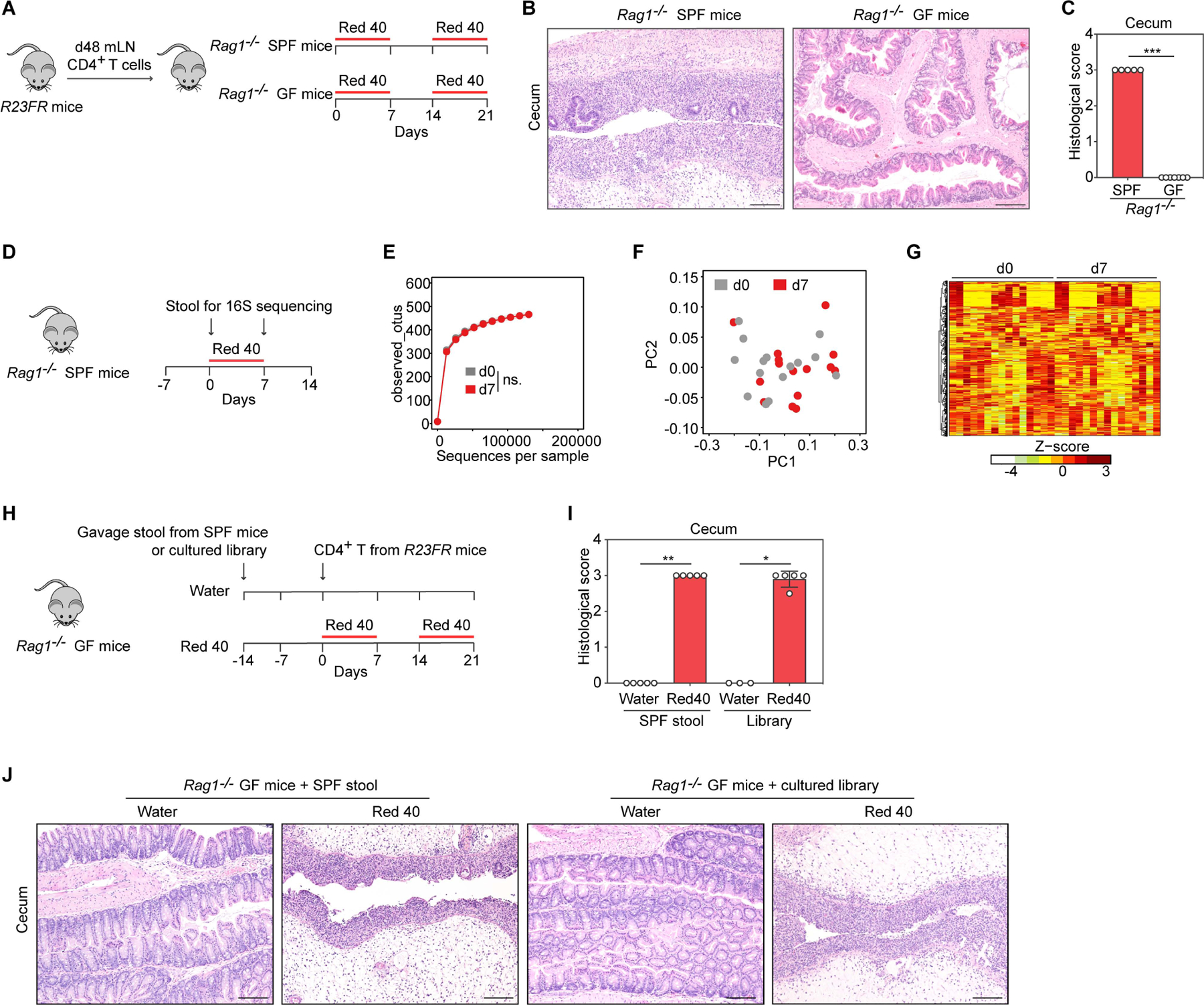
(A) Experimental scheme. (B,C) Representative H&E-stained sections (B) and histologic scores (C) of the cecum of experimental mice at day 21 described in Fig. 4A. (D) Experiments designed to test the impact of Red 40 on the intestinal bacterial community. Fecal samples were collected for 16S rRNA amplicon sequencing before (d0) and after (d7) Red 40 (0.25g/L) treatment in Rag1−/− SPF mice (n=15/time point). (E) Alpha diversity (estimated as number of observed OTUs) at a sequencing depth of 120,000 sequences/sample (p >0.05, by pairwise Wilcoxon rank sum test). (F) No clustering of d0 and d7 fecal samples in weighted Unifrac analysis (p > 0.05, by Adonis test). (G) Hierarchical clustering of the abundance profiles of all 620 OTUs detected after quality filtering. Rows in the heatmap represent individual OTUs. None of these OTUs showed a significant change in abundance between d0 and d7 (q < 0.05, fold > 1.5). (H) Experimental scheme. CD4+ T cells obtained from mLN of R23FR mice in remission (d48) were injected into SPF microbiota or defined commensal consortium (Table S1) reconstituted Rag1−/− GF (ex-GF) mice (106 cells/mouse) fed with or without Red 40 in drinking water. (I,J) Histologic scores (I) and representative H&E-stained sections (J) of the cecum of experimental mice at day 21 described in Fig. 4H. Scale bars in (B) and (J), 50 μm. In (C) and (I), each dot indicates an individual mouse. Error bars indicate SEM. * p<0.05, ** p<0.01, *** p<0.001, by nonparametric Mann-Whitney test.
To examine if Red 40 caused disease by changing the composition of the bacterial community we performed 16S rRNA amplicon sequencing. The administration of Red 40 in drinking water did not significantly change the fecal bacterial composition (Fig. 4D-4G), as indicated by no significant differences in alpha diversity (Fig. 4E), no clustering of d0 and d7 fecal samples in weighted Unifrac analyses (Fig. 4F) and no significant change in abundance of operational taxonomic units (OUT) between d0 and d7 (Fig.4G). These results suggested that Red 40-induced colitis was dependent on the microbiota but that it did not depend on marked shifts in its composition.
B.ovatus and E.faecalis contribute to Red 40-induced colitis.
To define if specific bacterial strains contributed to Red 40 induced-colitis, we pretreated SPF Rag1−/− mice with vancomycin (which preferentially targets Gram-positive bacteria) or polymyxin B (which preferentially targets Gram-negative bacteria) (Atarashi et al., 2011) for two weeks and injected them with CD4+ T cells as described above, and exposed them to Red 40 in the water (Fig. S2A). As expected, administration of vancomycin or polymyxin B led to a reduction of fecal microbiota density compared to non-antibiotic treated SPF mice (Fig. S2B). Importantly, treatment with vancomycin resulted in a substantial decrease in colitis severity (Fig. S2C and S2D). In contrast, mice treated with polymyxin B developed colitis comparable to that observed in non-antibiotic treated SPF Rag1−/− mice exposed to Red 40 (Fig. S2C and S2D). We thus hypothesized that disease-associated bacteria were enriched in the polymyxin B treated group. Analysis of the composition of cecal microbiota by 16S rRNA amplicon sequencing in mice after antibiotic treatment revealed that Bacteroides, a genus of gram-negative bacteria, was enriched in polymyxin B treated group (Fig. S2E), suggesting that Bacteroides could contribute to Red 40 induced colitis in mice.
To test if Bacteroides could be linked to disease, we gavaged a consortia of identified bacterial strains containing several Bacteroides (library number #1001136) (Table S1) (Britton et al., 2019) into GF Rag1−/− mice (ex-GF mice), injected them with CD4+ T cells from d48 R23FR mice in remission, and treated them with Red 40 in the drinking water (Fig. 4H-4J). Rag1−/− ex-GF mice colonized with the bacterial library treated with Red 40, but not treated with water, developed a marked colitis that was undistinguishable from that of Rag1−/− GF mice colonized with total stool of SPF C57BL/6 mice (Fig. 4H-4J). Analysis of the fecal microbiota from library #1001136 re-constituted mice by 16S rRNA amplicon sequencing showed that it contained most of the culturable species present in the bacterial library (Table S1). Bacteroides ovatus (B.ovatus) and Escherichia coli (E. coli) were among the most highly abundant strains detected in the Rag1−/− ex-GF mice (Table S1).
It is well established that the intestinal bacteria play a key role in the metabolism of Red 40 (Feng et al., 2012; Zou et al., 2020). Having defined that a consortium of identified bacterial strains (library number #1001136) could promote Red 40 induced colitis, we next tested whether specific bacteria present in library #1001136 (Britton et al., 2019) contribute to metabolize Red 40. To test this we cultured B.ovatus and E.coli, two highly abundant strains present in the stools of Rag1−/− ex-GF mice re-constituted with library #1001136 (Table S1), with Red 40 (Fig. S3). Consistent with previous observations(Feng et al., 2012; Walker et al., 1971), culture of B.ovatus, but not E. coli, in vitro with Red 40, resulted in metabolic processing of Red 40, rendering it colorless (Fig. S3A and S3C). Red 40 did not affect bacterial growth (by detection of OD600nm) (Fig. S3A and S3B), but completely disappeared from the culture media after incubation (as measured at OD504nm) (Fig. S3C).
Next we tested if these single bacterial species were sufficient to cause colitis development in vivo. We mono-colonized GF Rag1−/− mice with B.ovatus or with E. coli. We then injected mono-colonized mice with CD4+ T cells from R23FR mice in remission, and treated them with Red 40 in the water (Fig. 5A). We found that Red 40 treatment did not trigger development of colitis in E.coli-colonized Rag1−/− mice (Fig. 5B and 5C). In contrast, Red 40 treatment, but not water, triggered development of colitis in B. ovatus-colonized Rag1−/− mice (Fig. 5B and 5C). More importantly, this response was not observed when mice were repeatedly exposed to extracts of dead B. ovatus plus Red 40 treatment (Fig. 5D-5F), suggesting that live B. ovatus was required for Red 40 to promote colitis development.
Fig 5. B.ovatus and E.faecalis contribute to Red 40-induced colitis.
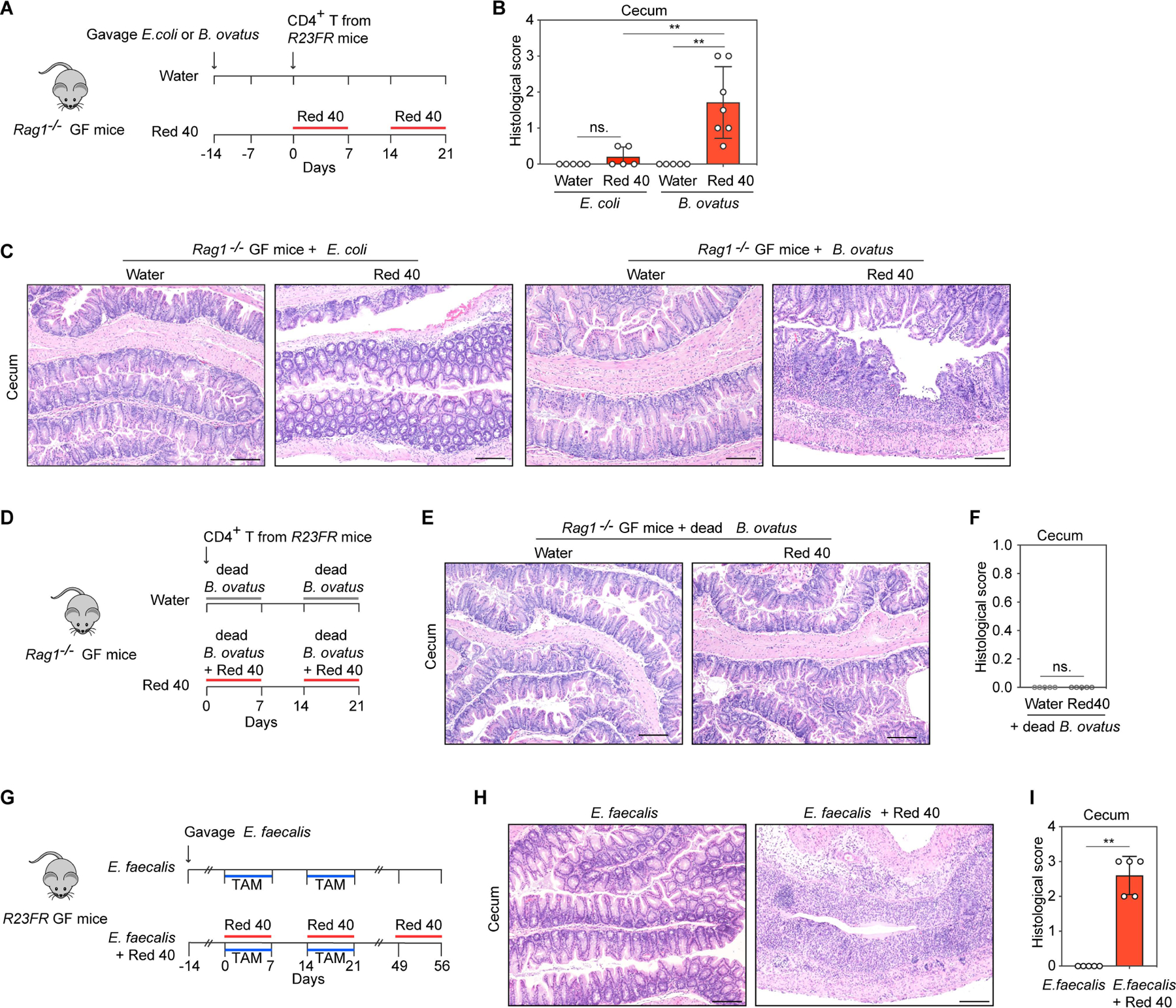
(A) Experimental scheme. CD4+ T cells from R23FR mice in remission were transferred into B. ovatus or E.coli monocolonized Rag1−/− germ-free (GF) mice (106 cells/mouse) treated with and without Red 40 in drinking water. (B,C) Histologic scores (B) and representative H&E-stained sections (C) of the cecum of experimental mice at day 21 described in Fig. 5A. (D) Experimental scheme. Rag1−/− GF mice adoptively transferred with CD4+ T cells from R23FR mice in remission (d48 after TAM + Red 40 treatment) (106 cells/mouse) were treated with dead B.ovatus in drinking water (10g/L) or dead B.ovatus (10g/L) plus Red 40 (0.25g/L) in drinking water. (E,F) Representative H&E-stained sections (E) and histologic scores (F) of the cecum of experimental mice at day 21 described in Fig. 5D. (G) Experimental scheme. E.faecalis monocolonized R23FR GF mice were treated with TAM in the food and treated with 0.025% Red 40 (0.25g/L) in drinking water. (H,I) Representative H&E-stained sections (H) and histologic scores (I) of the cecum of experimental mice at day 56 described in Fig. 5G. Scale bars (C), (E) and (H), 50 μm. In (B), (F) and (I) each dot indicates an individual mouse. Error bars indicate SEM. ns, p >0.05, ** p<0.01, by nonparametric Mann-Whitney test.
Enteroccocus faecalis (E.faecalis) has high capacity to metabolize Red 40 and a broader substrate spectrum compared to other mammalian intestinal bacteria(Feng et al., 2012; Walker et al., 1971). We hypothesized that E. faecalis could have a role similar to B. ovatus in our model. To test this hypothesis, we cultured E. faecalis with Red 40 for 5 hours, which rendered it colorless (Fig. S3A and S3C) and used this culture supernatant to treat R23FR mice in remission (d49) (Fig. S3D). We found that supernatants of the culture of E. faecalis with Red 40 but not those of the E. faecalis culture without Red 40 could induce colitis in R23FR mice pretreated with TAM and Red 40 (Fig. S3E and S3F). These results indicate that processing of Red 40 by commensal bacteria (E.faecalis) yields metabolites with colitogenic properties. In addition, we monocolonized germ-free R23FR mice with E.faecalis and treated them with TAM and Red 40 (Fig. 5G). Histological analysis of cecum of these animals at day 56 showed severe colitis, with marked leukocytic infiltrate, crypt loss, epithelial damage, and ulceration (Fig. 5H and 5I). In contrast, there was no colitis development in E. faecalis monocolonized germ-free R23FR mice treated with TAM alone (Fig. 5G-5I). These results indicate that monocolonization of R23FR mice with the commensal organism E. faecalis is sufficient to render R23FR mice responsive to Red 40 in the presence of IL-23.
Taken together, these findings suggested that processing of Red 40 by specific commensal bacteria was required for their colitis-inducing properties.
Inactivation of the azo bond significantly reduces the colitogenic properties of Red 40.
Red 40 is a red azo dye. Azo dyes are organic compounds that contain the functional azo group (–N=N–). Since both B.ovatus or E.faecalis contribute to Red 40-induced colitis (Fig. 5), we hypothesized that reduction of the Red 40 azo bond by these organisms could be important for generation of colitogenic agents. To test this hypothesis, we chemically synthesized a compound which lacked the azo bond, dihydro Red 40 (–HN-NH–) (Fig. S4A and Fig. S5) and tested its colitogenic activity in vivo (Fig. S4B-S4G). We found that dihydro Red 40 treatment could not induce colitis in the R23FR mice (Fig. S4B-S4D) or in Rag1−/− mice that received CD4+ T cells obtained from d48 R23FR mice (Fig. S4E-S4G). These results suggest that processing of the azo bond present in azo dyes by commensal bacteria is necessary for disease induction, and are consistent with our observation that Red 40 does not induce disease in GF Rag1−/− mice transferred with T cells.
A Red 40 metabolite is the colitis-inducing agent in primed mice.
Red 40 can be metabolized by azo-reduction in the gastrointestinal tract into two major metabolites: cresidine-4-sulfonate sodium salt (CSA-Na) and 1-amino-2-naphthol-6-sulphonate sodium salt (ANSA-Na) (Zou et al., 2020) (Fig. S3G). Analysis by LC-MS of the supernatants of E. faecalis cultures with Red 40, confirmed its processing, and revealed the presence of the two metabolites CSA-Na and ANSA-Na (Fig. S3H). Consistent with our previous data (Fig. S3C), Red 40, rather than its metabolites, was present in E. coli culture supernatants (Fig. S3H).
Having shown that supernatants from E. faecalis cultured with Red 40 could induce flares of colitis (Fig. S3D-S3F) we hypothesized that one or both of Red 40 metabolites could contribute to Red 40-induced colitis. To test this hypothesis we treated R23FR mice with Red 40 and TAM for two cycles and added CSA-Na or ANSA-Na to the water in the last cycle (Fig. 6A). We found that animals treated with ANSA-Na but not CSA-Na developed colitis (Fig. 6B and 6C), suggesting that this moiety is the colitis inducing metabolite of Red 40.
Fig 6. 1-amino-2-naphthol-6-sulphonate sodium salt (ANSA-Na) is the colitis-inducing agent in IL-23 expressing mice.
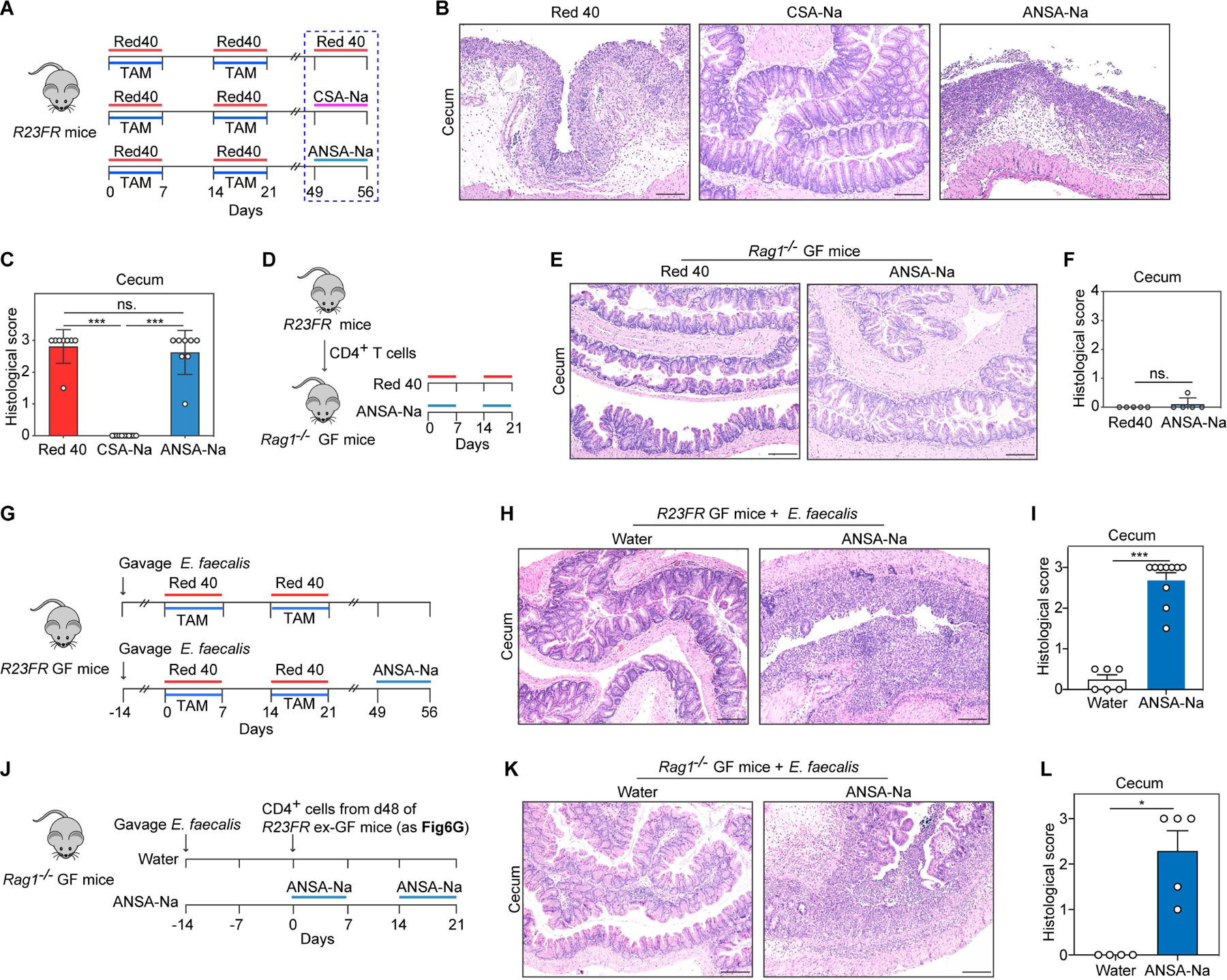
(A) Experimental scheme. R23FR remission mice were treated with Red 40 (0.5g/L), CSA-Na (0.5g/L) or ANSA-Na (0.5g/L) for 7 days. (B,C) Representative H&E-stained sections (B) and histologic scores (C) of the cecum of experimental mice described in Fig. 6A. (D) Experimental scheme. CD4+ T cells obtained from mLN of R23FR mice in remission (d48 after TAM + Red 40 treatment) were injected into Rag1−/− GF mice (106 cells/mouse) fed with Red 40 (0.5g/L) or ANSA-Na (0.5g/L) in drinking water. (E,F) Representative H&E-stained sections (E) and histologic scores (F) of the cecum of experimental mice at day 21 described in Fig. 6D. (G) Experimental scheme. E.faecalis monocolonized R23FR GF mice were treated with TAM in the food and treated with 0.025% Red 40 (0.25g/L) in drinking water for 2 cycles. From day 49, groups of remission animals were treated with or without ANSA-Na (0.5g/L) in drinking water for 7 days. (H,I) Representative H&E-stained sections (H) and histologic scores (I) of the cecum of experimental mice at day 56 described in Fig. 6G. (J) Experimental scheme. CD4+ T cells obtained from mLN of ex-GF R23FR mice in remission (d48 after TAM + Red 40 treatment) (Fig. 6G) were injected into E.faecalis monocolonized Rag1−/− GF mice (106 cells/mouse) fed with or without ANSA-Na (0.5g/L) in drinking water. (K,L) Representative H&E-stained sections (K) and histologic scores (L) of the cecum of experimental mice at day 21 described in Fig. 6J.
Scale bars in (B), (E), (H) and (K), 50μm. In (C), (F), (I) and (L) each dot indicates an individual mouse. Error bars indicate SEM. ns, p >0.05, * p<0.05, *** p<0.001, by nonparametric Mann-Whitney test.
To further confirm that ANSA-Na is the moiety important for colitis induction we tested Yellow 6 (Sunset Yellow FCF) (Fig. S6A), another abundantly used azo dye (Chung et al., 1992). Metabolism of Yellow 6 can also yield ANSA-Na via azo-reduction (Zou et al., 2020)(Fig. S6A). As expected, Yellow 6 promoted development of colitis in R23FR mice pretreated with TAM and Yellow 6 (Fig. S6B-S6D). Of interest, the non-azo dyes Red 3 and Blue 1, which do not contain the ANSA-Na moiety (Fig. S6A), did not elicit colitis in R23FR mice (Fig. S6B-S6D). We further hypothesized that the dyes Red 40 and Yellow 6 could induce similar immune reactions between them. Indeed, R23FR mice pretreated with TAM and Yellow 6 developed colitis when exposed to Red 40 (Fig. S6E-S6G) and vice-versa (Fig. S6E-S6G).
We next sought to test if microbes were required for ANSA-Na-induced colitis. We adoptively transferred mLN CD4+ T cells from d48 R23FR mice to GF Rag1−/− mice and fed them with ANSA-Na in the drinking water (Fig. 6D). Treatment of GF mice with this substance did not elicit colitis (Fig. 6E and 6F), indicating that it did not work as an irritant, and suggesting it required the presence of bacteria to cause disease. Accordingly, ANSA-Na promoted development of colitis in E.faecalis monocolonized R23FR GF mice pretreated with TAM and Red 40 (Fig. 6G-6I). In addition, we adoptively transferred mLN CD4+ T cells from these monocolonized R23FR GF mice (Fig. 6G) into E.faecalis monocolonized GF Rag1−/− mice (Fig. 6J). We found that ANSA-Na treatment, but not water, triggered development of colitis in E.faecalis-colonized Rag1−/− mice (Fig. 6K and 6L). These results suggest that ANSA-Na produced by azo-reduction from Red 40 and Yellow 6, induce flares of colitis in primed mice.
Red 40 promotes colitis development in control mice with augmented expression of IL-23
To further validate the findings reported above, we sought next to define if Red 40 could function as a colitogenic agent in non-transgenic mice. To induce stable long-term expression of IL-23, we used hydrodynamic delivery of an IL-23 minicircle DNA (Fig. 7A). Serum IL-23 levels were increased by day 4 post-injection and were sustained over duration of the study (Fig. 7B). Consistent with previous findings(Sherlock et al., 2012), all mice that received the IL-23 minicircle DNA developed ear inflammation, regardless if treated with Red 40 or not (Fig. 7C). In animals expressing systemic levels of IL-23, colitis was only observed in mice that received Red 40 treatment (Fig. 7C-7E).
Fig 7. Red 40 treatment of wild-type mice with dysregulated expression of IL-23 results in development or exacerbation of colitis.
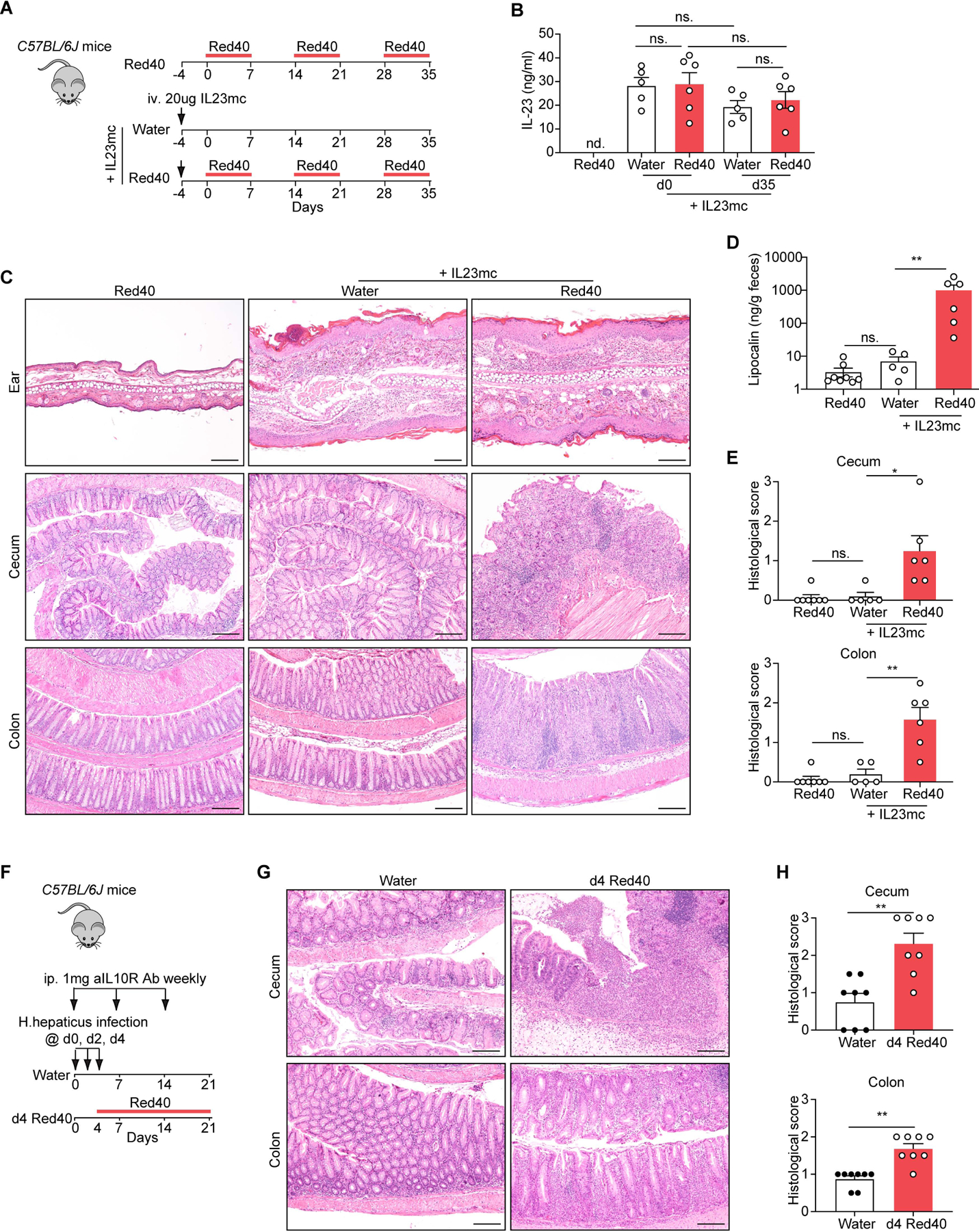
(A) Experimental scheme. 20 μg Mouse IL-23 Minicircle DNA (IL23mc) was injected through the tail vein per mouse into C57BL/6J mice. The mice were treated with or without 0.25 g/L Red 40 in drinking water 4 days after initial injection. (B) Serum IL-23 levels at different time points. (C) Representative H&E-stained sections of the ear, cecum and colon of experimental mice at day 35 described in Fig. 7A. (D,E) Fecal lipocalin-2 levels (D) and histologic scores (E) of the cecum and colon of experimental mice at day 35 described in Fig. 7A. (F) Experimental scheme. C57BL/6J mice were infected with H. hepaticus by oral gravage and treated with anti-IL-10R mAb (1mg) once a week for 3 wk. The mice were treated with or without 0.25 g/L Red 40 in drinking water 4 days after initial infection. (G,H) Representative H&E-stained sections (G) and histologic scores (H) of the cecum and colon of experimental mice at day 21 described in Fig. 7F.
Scale bars in (C) and (G), 50 μm. In (B), (D), (E) and (H), each dot indicates an individual mouse. Error bars indicate SEM. nd, not detected. ns, p >0.05, * p<0.05, ** p<0.01 by nonparametric Mann-Whitney test.
Finally, we evaluated the role of Red 40 in the disease progression in a well-established colitis model involving Helicobacter hepaticus (H. hepaticus) infection and IL-10R blockade. Colitis development in this model is dependent on IL-23 produced by intestinal myeloid cells (Arnold et al., 2016; Kullberg et al., 2006). We treated mice with 0.25g/L Red 40 in drinking water starting 4 days after infection, to coincide with the period in which IL-23 expression is known to be increased (Arnold et al., 2016) (Fig. 7F). We found that the severity of colitis in the Red 40 treated group of mice was significantly higher than that in the water treated group as measured by histology in both colon and cecum (Fig. 7G-7H).
The data altogether indicate that Red 40 treatment of wild-type and transgenic mice with dysregulated expression of IL-23 results in development or exacerbation of colitis.
DISCUSSION
Our studies reveal that food colorants contribute to development of colitis in conditions characterized by increased IL-23 signaling. Disease development in this setting requires commensal bacteria, such as E. faecalis and B. ovatus, to metabolize Red 40 or Yellow 6. Our findings suggest that specific food colorants are risk factors for experimental IBD in conditions of immune-dysregulation.
Although IBD seems to be caused by genetic predisposition and environmental factors, identification of these environmental factors has proven difficult. In mouse models, some food additives such as emulsifiers(Chassaing et al., 2015; Chassaing et al., 2017; Viennois et al., 2017), aluminum(Pineton de Chambrun et al., 2014) and titanium dioxide(Ruiz et al., 2017), have been suggested to represent environmental risk factors (Marion-Letellier et al., 2019; Valadez et al., 2018). Here we identify Red 40 as a novel risk factor for development of colitis. Red 40 is widely present in the human food chain, as colorant in many beverages, food and medicines. Red 40 is not a component of many commercially available rodent diets, and therefore mice are not exposed to it. Similar to what is observed with humans, administration of Red 40 (up to 40mg/kg/day) to control mice, or to R23FR mice before expression of IL-23, does not induce colitis. However, when given to mice with increased IL-23 signaling, it promotes the generation of a T cell mediated response that leads to colitis, even when used at doses that are lower than those consider safe in humans (the acceptable daily intake is 7mg/kg body weight) (Bastaki et al., 2017a), as demonstrated in our studies using common beverages and medicines containing Red 40, such as Pedialyte AdvancedCare cherry punch flavor.
The fact that Red 40 consumption does not lead to development of colitis in the population at large suggests development of immunological tolerance to this compound (or to proteins it binds to). Indeed, our results indicate that exposure to Red 40 prior to elevated IL-23 expression induces a state of tolerance. These observations have important implications because they suggest that exposure to Red 40 (present in food/beverage and medications) in individuals that experience increased IL-23 signaling (due to infections or genetic conditions) may predispose them to develop colitis later on in life. This may be particularly relevant in the case of children when first exposed to food or drink containing these azo-dyes. We hypothesize that increased IL-23 signaling may affect several immunological pathways, including antigen-presentation and/or generation of T effector or T regulatory cell populations, as amply documented in the literature(Harbour et al., 2015; Kullberg et al., 2006). The fact that disease can be induced via transfer of unfractionated CD4 T cells (containing a normal complement of Tregs) from previously primed R23FR mice to lymphopenic mice upon Red 40 re-challenge, suggests that IL-23 may cause disease in part by inhibiting the generation of Treg specifically recognizing Red 40 (or its metabolites)-bound antigens.
The ability of Red 40 to cause colitis is predicated on the expression of IL-23. IL-23 plays an important role in both innate and adaptive immune-driven colitis. IL-23 appears to drive innate intestinal pathology via group 3 innate lymphoid cells (Buonocore et al., 2010; Chen et al., 2015). A role for IL-23 in adaptive immune colitis appears to depend on IL-17+ and/or IL-17+IFN-γ+ CD4+ T cells (Harbour et al., 2015). Here we show that IL-17 and IL-22 cells are present among effector CD4+ T cells, but that the immunopathology driven by Red 40 and IL-23 does not appear to depend on classical Th17 responses, as blocking IL-17A and IL-17F fails to prevent colitis development in R23FR mice. Rather, we found that IFN-γ producing CD4+ T cells were the major population driving disease development. A role for IFN-γ producing CD4+ T cells in experimental colitis was first demonstrated by Powrie and colleagues in their classic study published in 1994(Powrie et al., 1994). We suggest that the development of IFN-γ producing cells is most likely a critical immunopathogenic mechanism elicited by IL-23, as the default immune response to Red 40 administration is the generation of tolerance. Whether IL-23 actively converts antigen-specific Tregs into IFN-γ producing CD4 T cells in this setting is an open issue at present.
Development of colitis in mice expressing IL-23 is episodic and dependent on the administration of Red 40, a small molecular weight compound (molecular weight = 496.43). The fact that disease is triggered by a small molecule, depends on CD4+ T cells, and is independent of B cells(Chen et al., 2018), suggests that IL-23 and Red 40 may induce colitis through a well-established immunopathogenic mechanism, namely, a Type IV mucosal hypersensitivity response or delayed type hypersensitivity (DTH) response. This hypothesis is consistent with the observation that IL-23 is important in skin DTH responses(Ghilardi et al., 2004).
Haptens such as Trinitrobenzene sulfonic acid (TNBS) and oxazolone are known to induce colitis in mouse. Administration of these haptens to susceptible strains of mice results in CD4+ T cell (Wirtz et al., 2017) or NKT cell(Iyer et al., 2018) mediated immunity in TBNS or oxazolone colitis model respectively. We suggest that different from these haptens, Red 40 may work as a pro-hapten, whose biological activity depends on its processing by commensal bacteria. We identified B.ovatus and E.faecalis as a few representative microbes have capacity to metabolize Red 40. These two bacteria do not cover the intestinal miciobiome as a whole, and we acknowledge that other bacterial taxa may have this capability. As shown here 1-amino-2-naphthol-6-sulphonate sodium salt (ANSA-Na) produced by azo-reduction of Red 40, is biologically capable to induce flares of colitis and may represent a biologically relevant hapten. The fact that this moiety is also produced upon processing of Yellow 6, explains why animals primed with Red 40 can develop colitis upon exposure to Yellow 6 and vice versa. These findings suggest that different food colorants may cause hapten-induced immune intestinal responses in predisposed hosts expressing higher levels of IL-23. Identification of proteins or peptides bound to ANSA-Na that form adducts with relevant immunological properties remain warranted.
In sum, our studies show that dysregulated expression of the cytokine IL-23 (in transgenic and non-transgenic settings) combined with recent exposure to Red 40 promote development of colitis in mice. These results may have implications for human health as IL-23 is clearly implicated in development of IBD, and consumption of food colorants such as Red 40 and Yellow 6, is widespread.
Limitations of Study
Our studies were conducted in mice and it remains to be assessed if similar effects will be observed in humans. While our data show that IFN-γ producing CD4+ T cells are the major population driving colitis development, it is unknown how IL-23 induces IFN-γ+ T cells and how IL-23 shifts the overall immune response to Red 40 from tolerance to disease. While we have shown that bacterial processing of Red 40 and Yellow 6 leads to production of ANSA-Na, it is still unknown if further chemical modification of this compound is needed, and whether this metabolite binds bacterial or host proteins to induce an immune response. Lastly, this study is limited by its focus on evaluating the role of food colorants on colitis development in the host with dysregulated expression of IL-23, so future investigation into other IL-23 independent preclinical colitis model will be important to understand the broader impact of food colorants.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sergio A. Lira (Sergio.lira@mssm.edu)
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The 16S rDNA sequencing data generated during this study have been deposited in the NCBI Sequence Read Archive (Accession numbers: PRJNA702431 and PRJNA702461).
Experimental model and subject details
Mouse strains
C57BL/6 (stock # 000664), Rag1−/−(stock # 002216), Ifng−/− (stock # 002287) and CD45.1 (stock # 002014) mice were purchased from The Jackson laboratory (Bar Harbor, ME). R23FR mice(Chen et al., 2018), FR mice(Chen et al., 2018) and Il22−/− mice(Chen et al., 2019) were described previously. All mice were on a C57BL/6 background and had been backcrossed at least ten generations. Mice were maintained under specific pathogen-free (SPF) conditions at the Icahn School of Medicine at Mount Sinai. Germ-free (GF) C57BL/6, Rag1−/− and R23FR mice (Chen et al., 2018) were bred in-house at the Microbiome Translational Center at the Icahn School of Medicine at Mount Sinai. All animal experiments in this study were approved by the Institutional Animal Care and Use Committee of Icahn School of Medicine at Mount Sinai, and were performed in accordance with the approved guidelines for animal experimentation at the Icahn School of Medicine at Mount Sinai.
Growth of bacteria
B. ovatus was grown under anaerobic conditions with no shaking in Vinyl Anaerobic Chambers using 70% N2, 20% CO2, and 10% H2 gas mixture at 37 °C in Brain Heart Infusion (BHI) Broth (Sigma-Aldrich) supplemented with L-cysteine (1 mg/ml), hemin (0.5 mg/L) and NaHCO3 (2%)(Bacic and Smith, 2008). Both E. coli and E. faecalis were cultured under aerobic conditions at 37 °C in Luria Broth (LB) Base (Invitrogen). Aerobic cultures were done in flask bottles using 250rpm shaker.
Methods details
TAM treatment.
All mice were raised on the basal diet 5053, which was purchased from LabDiet (St. Louis, MO). Tamoxifen (TAM) (500mg/kg) (Sigma, St Louis, MO) was added to the Envigo diet 2019 (TD.160647) to produce 2019 TAM diet (TD.130968) (Madison, WI). Tamoxifen (TAM) (500mg/kg) (Sigma, St Louis, MO) was added to the basal diet 5053 to produce 5053 TAM diet (TD.190129), which was purchased from Envigo (Madison, WI).
Food colorant treatment.
Red 40 (Allura Red AC), Red 3 (Erythrosine), Yellow 6 (Sunset Yellow FCF), and Blue 1 (Brilliant Blue FCF) were purchased from Sigma-Aldrich. Mice were exposed to food colorant in drinking water (0.025% w/v, 0.25g/L). The same water was used for the water-treated (control) group. These solutions were changed every week. Red 40 solutions were filtered by 0.2 μm filter for experiments performed in germ-free condition. In some studies, mice were exposed to food colorant in diet (containing 0.25g/kg Red 40, diet 2019, TD.160647).
Separation of ingredients of diet 2019 and test of their colitogenic properties in vivo
The diet 2019 (1 kg) was finely chopped and extracted it with 95% ethanol at room temperature for 2 hrs. The solution was centrifuged at 2,000g for 5 min and the supernatant containing the ethanol-soluble fraction was collected. The pellet was dissolved in water at room temperature for 2 hrs and centrifuged at 4000 g to obtain the water-soluble fraction (Fig S1A). The ethanol-extracted fraction was dried under airflow and reduced pressure. The resulting powder (100g) was mixed with chopped 5053 diet and used to feed R23FR mice for 7 days (Fig S1A). The water-soluble fraction (Fig S1A) was lyophilized and resuspended in 10% of the original diet weight in water (100ml), and subsequently gavaged into R23FR mice at remission (0.5ml/ per time, twice daily for 7 days). After treatment the cecum was taken for histological analysis and feces were collected for Lcn2 ELISA.
Determination of Red 40 concentration.
The Red 40 concentration in the Kool-Aid beverage or Pedialyte cherry punch flavor was determined by comparison of its absorbance against a standard curve generated by serial dilution of Red 40 (Allura Red AC, Sigma) at an absorbance 504 nm(Stevens et al., 2014).
T cell adoptive transfer.
For CD4+ T-cell isolation, mLNs were digested in collagenase as described previously(Chen et al., 2018). CD4+ T cells were enriched by positive immunoselection using CD4-(L3T4) microbeads (Miltenyi Biotec, Bergish Gladbach, Germany). The magnetic-activated cell sorting (MACS) purified CD4+ T cells were used as donor cells in adoptive transfer experiments. One million CD4+ T cells from mLN in remission stage of R23FR mice (d48 after TAM and Red 40 treatment) enriched by using MACS-beads were transferred into SPF Rag1−/− mice or GF Rag1−/− mice by intravenous (i.v.) injection.
Histology.
Tissues were dissected, fixed in 10% phosphate-buffered formalin, and then processed for paraffin sections. Five-micrometer sections were stained with hematoxylin and eosin (H&E) for histological analyses. All the sections were evaluated for a wide variety of histological features that included epithelial integrity, number of goblet cells (mucin production), stromal inflammation, crypt abscesses, erosion, and submucosal edema. Severity of disease was then classified as described before(Chen et al., 2018).
Quantification of fecal Lcn-2 by Enzyme-Linked Immunosorbent Assay
Freshly collected or frozen fecal samples were reconstituted in phosphate-buffered saline containing 0.1% Tween 20 (100 mg/mL) and vortexed for 20 minutes to get a homogenous fecal suspension(Chassaing et al., 2012). These samples were then centrifuged for 10 minutes at 12,000 revolutions per minute and 4°C. Clear supernatants were collected and stored at −20°C until analysis. Lcn-2 levels were estimated in the supernatants using Duoset murine Lcn-2 enzyme-linked immunosorbent assay kit (R&D Systems, Minneapolis, MN).
In vivo antibody treatment.
Blockade of IL-17A, IL-17F, TNF-a and IFN-γ were performed in an adoptive cell transfer model. Briefly, CD4+ T cells from R23FR mice in remission (d48) were adoptively transferred to Rag1−/− mice that were subsequently fed with diet 2019 for two cycles. 1mg of anti-IL-17A (17F3, BioXcell), anti-IL-17F (MM17F8F5.1A9, BioXcell), anti-TNFa (TN3–19.12, BioXcell), anti-IFN-γ (XMG1.2, BioXcell), or their isotype antibodies were administered by intraperitoneal injection at indicated days shown in the figures. Adoptively transferred Rag1−/− mice were sacrificed at day 21, and the large intestine was taken for histological analysis.
Regulatory T cells suppression assay in vivo.
CD4+ T cells were enriched by positive immunoselection using CD4-(L3T4) microbeads (Miltenyi Biotec) from mLN in remission stage of R23FR mice (d48 after TAM and Red 40 treatment) and used as responder cells. Regulatory T cells (Treg), CD45.1+CD4+CD25+ from Red 40-treated or water-treated CD45.1 mice were isolated by negative selection of CD4+ cells followed by positive selection of CD25+ cells with Treg isolation kit (Miltenyi Biotec). Colitis was induced in the recipient Rag1−/− mice by intravenous injection of 106 responder T cells + 4× 105 Treg cells and treatment with Red 40 in the drinking water (0.025% w/v, 0.25g/L).
Hydrodymic gene delivery of IL-23 minicircle DNA.
Mouse IL-23 Pre-made Minicircle DNA (RSV->FLAG-mP40-mP19-pA) (MN651MC-1) was purchased from System Biosciences. Minicircle DNA was introduced into mice using a hydrodynamic tail vein injection. Briefly, 20 μg Mouse IL-23 Pre-made Minicircle DNA was diluted into a solution of sterile saline (0.9%) at a total volume of 10% of the body weight of the mouse and was injected through the tail vein using a 3 ml syringe with a 26-gauge needle. Serum IL-23 in mice after injection was measured using a ELISA MAX™ Deluxe Set Mouse IL-23 kit (433704) from Biolegend.
H. hepaticus infection plus anti-IL10R injection induced colitis.
H. hepaticus were grown on sheep blood agar plates (Remel) under microaerobic gas mixture consisting of 80% N2, 10% H2, and 10% CO2 in a vented jar. The microaerobic jars containing bacterial plates were left at 37 °C for 2–3 days. After culture, the bacteria were collected and suspended in Brucella broth with 20% glycerol and adjusted bacterial density to OD600 nm readings at 1.5 OD/ml, and frozen at −80 °C. For oral infection, 0.2 ml frozen stock aliquots of H. hepaticus was administered to each mouse by oral gavage every other day for three doses. An anti-IL-10R (clone 1B1.2) antibody was injected intraperitoneally (1mg) weekly per mouse for three weeks.
Microbiota transplantation.
Cecal extracts pooled from 3–5 donor SPF mice were suspended in PBS (2.5 ml per cecum) and gavaged (0.1 ml per mouse) immediately to Rag1−/− germ-free mice(Chen et al., 2018). For additional transfer experiments we used a commensal bacteria consortium described before(Britton et al., 2019). This pooled cocktail of cultured strains was gavaged (200–300 μL) into Rag1−/− germ-free mice. Transplanted mice were maintained in sealed positive pressure cages (Allentown) for two weeks after which they were used for the further experiments.
Antibiotic treatment.
SPF mice were treated with vancomycin (500mg/L, Sigma-Aldrich) or polymycin B (100mg/L, Sigma-Aldrich) in the drinking water during the indicated times. These antibiotic solutions were renewed every week.
16S rRNA gene amplicon sequencing.
DNA was extracted by bead-beating followed by QiaQuick columns (Qiagen) and quantified by Qubit assay (Life Technologies). Briefly, mouse fecal pellets (~50 mg), were re-suspended in a solution containing 700μL of extraction buffer [0.5% SDS, 0.5 mM EDTA, 20 mM Tris-Cl] and 200μL 0.1-mm diameter zirconia/silica beads. Cells were then mechanically disrupted using a bead beater (BioSpec Products, Bartlesville, OK; maximum setting for 5 min at room temperature), followed by extraction with QIAquick 96 PCR Purification Kit (Qiagen). Bacterial 16S rRNA genes were amplified using the primers as previously described(Faith et al., 2013). Sample preparation and analysis of 16S rDNA sequence were done as previously described(Chen et al., 2018). The 16S rDNA data were analyzed with MacQIIME 1.9.1. Operational taxonomic units (OTUs) were picked using closed reference OTU picking with 97% sequence similarity using the Greengenes reference database set with a minimum sequence length of 150 bp(Caporaso et al., 2010; McDonald et al., 2012).
Dead B. ovatus solution preparation.
Cultured B. ovatus was centrifuged at 4 °C to remove the culture supernatant and washed with sterile PBS for twice. The bacterial pellet was resuspensed into PBS at the final concentration of 10g/L and sonicated (probe diameter = 12mm) on ice at 60% output using a Sonic Dismembrator (Fisherbrand Model 705, Fisher scientific) for 20 × 20 seconds pluses for 5min interval. No viable bacteria remained in this solution as determined by inoculation into agar plates and culture at 37°C under anaerobic conditions for 3 days. The re-suspended bacterial solution was used as drinking water to feed germ-free mice.
LC-MS/MS sample preparation and method.
Culture supernatants of bacteria with Red 40 were shipped on dry ice to the Proteomic and Metabolomics Facility at Cornell University. 90 μL of the medium was incubated with 400 μL methanol: acetonitrile (1:1) for 10 min on ice and then centrifuged at 16,000 g to precipitate the proteins. 30 μL of 10μM trypan blue (internal standard, IS) was added in the supernatant. 10 μL of this sample was used for analysis. All the standards (Red 40, cresidine-4-sulfonic acid and 1-amino-2-naphthol-6-sulphonate acid) were prepared by dilution with optima water to give 1mM concentration. Samples used by injecting 1mM standards diluted by 10 times with the flow rate of 200 μL/min for optimization of Sciex X500B instrument and LC-MS. Sciex OS1.7 software was used for all identification and quantitation analysis. The analyte/IS area ratio was used for quantification of Red 40, cresidine-4-sulfonic acid and 1-amino-2-naphthol-6-sulphonate acid levels.
Chemical reduction of Red 40.
Sodium 6-hydroxy-5-(2-(2-methoxy-5-methyl-4-sulfonatophenyl)hydrazinyl)naphthalene-2-sulfonate (Dihydro Red 40) was prepared using a modified procedure(Zhang and Wang, 2003). N2H4•H2O (4.5 mL, 92 mmol) was added to a solution of Red 40 (450 mg, 0.91 mmol) in water (23 mL). The reaction was stirred at 60°C for 60 h. The progress of reaction was monitored by color, which changed from red to yellow. The reaction mixture was allowed to cool to room temperature and the solvent was removed under reduced pressure. The product was purified through trituration with a mixture of dichloromethane:methanol (10:1) and dried to give a product as brown solid (390 mg, 86% yield). 1H NMR (DMSO-d6, 600 MHz) 7.87–7.86 (2 H, m), 7.47 (1 H, d, J = 8.5 Hz), 7.16 (1 H, s), 7.09–7.06 (2 H, m), 6.35 (1 H, s), 4.90 (1 H, br), 4.67 (2 H, br), 3.68 (3 H, s), 2.30 (3 H, s); HRMS (ESI-TOF) m/z: [M + 2Na]2+ calculated for C18H18N2O8S2Na2 250.0144; found 250.0842.
Test colitogenic properties of compounds in vivo
Red 40’s metabolites: cresidine-4-sulfonic acid (Aablocks) and 1-amino-2-naphthol-6-sulphonate acid (Carbosynth) were dissolved into H2O (0.5g/L) and the pH adjusted to 7. R23FR mice at remission stage (d49, after TAM and Red 40 treatment) were exposed to metabolites in drinking water for 7 days. After treatment the large intestine was taken for histological analysis.
Quantification and statistical analysis
Except for deep-sequencing data, statistical analyses were performed with GraphPad Prism 7 software (GraphPad, La Jolla, CA). Differences between groups were analyzed with a nonparametric Mann-Whitney test. Statistical tests are indicated throughout the Figure legends. Differences were considered significant when p < 0.05, and levels of significance are specified throughout the Figure legends. Data are shown as mean values ± SEM throughout. No statistical method was used to predetermine sample size.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse IL-17A | BioXcell | Cat# BE0173; RRID: AB_10950102 |
| anti-mouse IL-17F | BioXcell | Cat# BE0303; RRID: AB_2715461 |
| anti-mouse/rat/rabbit TNFα | BioXcell | Cat# BE0244; RRID: AB_2687725 |
| anti-mouse IFNg | BioXcell | Cat# BE0055; RRID: AB_1107694 |
| anti-mouse IL-10R | BioXcell | Cat# BE0050; RRID: AB_1107611 |
| Bacterial Strains | ||
| H.hepaticus | ATCC | Cat#51449 |
| B.ovatus | ATCC | Cat#8483 |
| E.coli | ATCC | Cat#K-12MG1655 |
| E.faecalis | ATCC | Cat#29212 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma Aldrich | Cat#: T5648; CAS: 10540–29-1 |
| Red 40 (Allura Red 40) | Sigma Aldrich | Cat#: 458848; CAS: 25956–17-6 |
| Yellow 6 (Sunset Yellow FCF) | Sigma Aldrich | Cat#: 465224; CAS: 2783–94-0 |
| Red 3 (Erythrosin extra bluish) | Sigma Aldrich | Cat#: E8886; CAS: 16423–68-0 |
| Blue 1 (Brilliant Blue FCF) | Sigma Aldrich | Cat#: 80717; CAS: 3844–45-9 |
| vancomycin | Sigma Aldrich | Cat#: C2002; CAS: 1404–93-9 |
| polymyxin B | Sigma Aldrich | Cat#: P0972; CAS: 1405–20-5 |
| cresidine-4-sulfonic acid | Aablocks | Cat#: AA00IBBN; CAS: 6471–78-9 |
| 1-amino-2-naphthol-6-sulphonate | Carbosynth | Cat#:FA44984; CAS: 5639–34-9 |
| Critical Commercial Assays | ||
| CD4 (L3T4) MicroBeads, mouse | Miltenyi Biotec | Cat#: 130–117-043 |
| CD4+CD25+ Regulatory T cell Isolation Kit, mouse | Miltenyi Biotec | Cat#: 130–091-041 |
| Mouse Lipocalin-2/NGAL DuoSet ELISA | R&D Systems | Cat#: DY1857 |
| ELISA MAX™ Deluxe Set Mouse IL-23 | Biolegend | Cat#: 433704 |
| Deposited Data | ||
| 16S rDNA sequence data | This paper | PRJNA702461, PRJNA702431 |
| Experimental Models: Organisms/Strains | ||
| Mouse: R23FR | Chen et al., 2018 | N/A |
| Mouse: FR | Chen et al., 2018 | N/A |
| Mouse: Il22−/− | Chen et al., 2020 | N/A |
| Mouse: CD45.1 | Jackson Laboratory | Jax: 002014 |
| Mouse: C57BL/6 | Jackson Laboratory | Jax: 000664 |
| Mouse: Rag1−/− | Jackson Laboratory | Jax: 002216 |
| Mouse: Ifng−/− | Jackson Laboratory | Jax: 002287 |
| Mouse: germ-free Rag1−/− | This paper | N/A |
| Mouse: germ-free R23FR | Chen et al., 2018 | N/A |
| Mouse: Ifng−/− Rag1−/− | This paper | N/A |
| Recombinant DNA | ||
| Mouse IL-23 Pre-made Minicircle DNA (RSV->FLAG-mP40-mP19-pA) | System Biosciences | Cat#: MN651MC-1 |
| Software and Algorithms | ||
| Graphpad Prism 7.1 | Graphpad.com | N/A |
| R | r-project.org | N/A |
| Other | ||
| Diet 2019 with TAM (500mg/kg) (Red) | Envigo | TD.130968 |
| Diet 2019 (Red) | Envigo | TD.160647 |
| Diet 2019 (No colorant, grey) | Envigo | TD.130833 |
| Diet 5053 with TAM (500mg/kg) | Envigo | TD.190129 |
Highlights:
Food colorants Red 40 and Yellow 6 induce colitis in mice overexpressing IL-23
Elevated IL-23 induces development of pathogenic CD4+ T cells that produce IFN-γ.
Commensal bacteria such as B.ovatus and E.faecalis metabolize Red 40.
ANSA-Na, a metabolite of Red 40 and Yellow 6, induces relapse of colitis.
Acknowledgments
We thank Alan J. Soto (Biorepository and Pathology CoRE, Sinai) for technical support with histology and Dr. Huaibin M. Ko (Department of Pathology, Sinai) for help with the pathology evaluation. We thank Joseph Eggers (Bongers Lab), Ilaria Mogno and Zhihua Li (Faith Lab) for technology support with anaerobic culture of bacteria. We also thank Drs. Howard C. Hang (Scripps Research) and Gabriel D. Victora (The Rockefeller University) for critical comments. This work was supported by grants from the National Institutes of Health R01 DK 110352 and DK 121009 (to S.A.L.), P30 ES 002109 and P01CA028842 (to J.G.F.) and a Career Development Award (634253) from the Crohn’s & Colitis Foundation of America (CCFA) (to L.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
All authors disclose no conflicts.
References
- Arnold IC, Mathisen S, Schulthess J, Danne C, Hegazy AN, and Powrie F (2016). CD11c(+) monocyte/macrophages promote chronic Helicobacter hepaticus-induced intestinal inflammation through the production of IL-23. Mucosal Immunol 9, 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacic MK, and Smith CJ (2008). Laboratory maintenance and cultivation of bacteroides species. Curr Protoc Microbiol Chapter 13, Unit 13C 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastaki M, Farrell T, Bhusari S, Bi X, and Scrafford C (2017a). Estimated daily intake and safety of FD&C food-colour additives in the US population. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 34, 891–904. [DOI] [PubMed] [Google Scholar]
- Bastaki M, Farrell T, Bhusari S, Pant K, and Kulkarni R (2017b). Lack of genotoxicity in vivo for food color additive Allura Red AC. Food Chem Toxicol 105, 308–314. [DOI] [PubMed] [Google Scholar]
- Bernshtein B, Curato C, Ioannou M, Thaiss CA, Gross-Vered M, Kolesnikov M, Wang Q, David E, Chappell-Maor L, Harmelin A, et al. (2019). IL-23-producing IL-10Ralpha-deficient gut macrophages elicit an IL-22-driven proinflammatory epithelial cell response. Sci Immunol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton GJ, Contijoch EJ, Mogno I, Vennaro OH, Llewellyn SR, Ng R, Li Z, Mortha A, Merad M, Das A, et al. (2019). Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORgammat(+) Regulatory T Cells and Exacerbate Colitis in Mice. Immunity 50, 212–224 e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, and Powrie F (2010). Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 464, 1371–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, and Knight R (2010). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, and Gewirtz AT (2015). Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, and Vijay-Kumar M (2012). Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One 7, e44328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Van de Wiele T, De Bodt J, Marzorati M, and Gewirtz AT (2017). Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut 66, 1414–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, He Z, Iuga AC, Martins Filho SN, Faith JJ, Clemente JC, Deshpande M, Jayaprakash A, Colombel JF, Lafaille JJ, et al. (2018). Diet Modifies Colonic Microbiota and CD4(+) T-Cell Repertoire to Induce Flares of Colitis in Mice With Myeloid-Cell Expression of Interleukin 23. Gastroenterology 155, 1177–1191 e1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, He Z, Slinger E, Bongers G, Lapenda TLS, Pacer ME, Jiao J, Beltrao MF, Soto AJ, Harpaz N, et al. (2015). IL-23 activates innate lymphoid cells to promote neonatal intestinal pathology. Mucosal Immunol 8, 390–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Strohmeier V, He Z, Deshpande M, Catalan-Dibene J, Durum SK, Moran TM, Kraus T, Xiong H, Faith JJ, et al. (2019). Interleukin 22 disrupts pancreatic function in newborn mice expressing IL-23. Nat Commun 10, 4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KT, Stevens SE Jr., and Cerniglia CE (1992). The reduction of azo dyes by the intestinal microflora. Crit Rev Microbiol 18, 175–190. [DOI] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. (2006). A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. (2013). The long-term stability of the human gut microbiota. Science 341, 1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagan BG, Sandborn WJ, D’Haens G, Panes J, Kaser A, Ferrante M, Louis E, Franchimont D, Dewit O, Seidler U, et al. (2017). Induction therapy with the selective interleukin-23 inhibitor risankizumab in patients with moderate-to-severe Crohn’s disease: a randomised, double-blind, placebo-controlled phase 2 study. Lancet 389, 1699–1709. [DOI] [PubMed] [Google Scholar]
- Feng J, Cerniglia CE, and Chen H (2012). Toxicological significance of azo dye metabolism by human intestinal microbiota. Front Biosci (Elite Ed) 4, 568–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremia A, and Jewell DP (2012). The IL-23/IL-17 pathway in inflammatory bowel disease. Expert Rev Gastroenterol Hepatol 6, 223–237. [DOI] [PubMed] [Google Scholar]
- Ghilardi N, Kljavin N, Chen Q, Lucas S, Gurney AL, and De Sauvage FJ (2004). Compromised humoral and delayed-type hypersensitivity responses in IL-23-deficient mice. J Immunol 172, 2827–2833. [DOI] [PubMed] [Google Scholar]
- Harbour SN, Maynard CL, Zindl CL, Schoeb TR, and Weaver CT (2015). Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. P Natl Acad Sci USA 112, 7061–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma M (2015). Evaluation of the in vivo genotoxicity of Allura Red AC (Food Red No. 40). Food and Chemical Toxicology 84, 270–275. [DOI] [PubMed] [Google Scholar]
- Iyer SS, Gensollen T, Gandhi A, Oh SF, Neves JF, Collin F, Lavin R, Serra C, Glickman J, de Silva PSA, et al. (2018). Dietary and Microbial Oxazoles Induce Intestinal Inflammation by Modulating Aryl Hydrocarbon Receptor Responses. Cell 173, 1123–1134 e1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Okamoto S, Hisamatsu T, Kamada N, Chinen H, Saito R, Kitazume MT, Nakazawa A, Sugita A, Koganei K, et al. (2008). IL23 differentially regulates the Th1/Th17 balance in ulcerative colitis and Crohn’s disease. Gut 57, 1682–1689. [DOI] [PubMed] [Google Scholar]
- Kullberg MC, Jankovic D, Feng CG, Hue S, Gorelick PL, McKenzie BS, Cua DJ, Powrie F, Cheever AW, Maloy KJ, and Sher A (2006). IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J Exp Med 203, 2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvedaraite E, Lourda M, Idestrom M, Chen P, Olsson-Akefeldt S, Forkel M, Gavhed D, Lindforss U, Mjosberg J, Henter JI, and Svensson M (2016). Tissue-infiltrating neutrophils represent the main source of IL-23 in the colon of patients with IBD. Gut 65, 1632–1641. [DOI] [PubMed] [Google Scholar]
- Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al. (2015). Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 47, 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion-Letellier R, Amamou A, Savoye G, and Ghosh S (2019). Inflammatory Bowel Diseases and Food Additives: To Add Fuel on the Flames! Nutrients 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, and Hugenholtz P (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6, 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, et al. (2018). Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 390, 2769–2778. [DOI] [PubMed] [Google Scholar]
- Pineton de Chambrun G, Body-Malapel M, Frey-Wagner I, Djouina M, Deknuydt F, Atrott K, Esquerre N, Altare F, Neut C, Arrieta MC, et al. (2014). Aluminum enhances inflammation and decreases mucosal healing in experimental colitis in mice. Mucosal Immunol 7, 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrie F, Leach MW, Mauze S, Menon S, Caddle LB, and Coffman RL (1994). Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity 1, 553–562. [DOI] [PubMed] [Google Scholar]
- Ruiz PA, Moron B, Becker HM, Lang S, Atrott K, Spalinger MR, Scharl M, Wojtal KA, Fischbeck-Terhalle A, Frey-Wagner I, et al. (2017). Titanium dioxide nanoparticles exacerbate DSS-induced colitis: role of the NLRP3 inflammasome. Gut 66, 1216–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandborn WJ, Gasink C, Gao LL, Blank MA, Johanns J, Guzzo C, Sands BE, Hanauer SB, Targan S, Rutgeerts P, et al. (2012). Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N Engl J Med 367, 1519–1528. [DOI] [PubMed] [Google Scholar]
- Sands BE, Chen J, Feagan BG, Penney M, Rees WA, Danese S, Higgins PDR, Newbold P, Faggioni R, Patra K, et al. (2017). Efficacy and Safety of MEDI2070, an Antibody Against Interleukin 23, in Patients With Moderate to Severe Crohn’s Disease: A Phase 2a Study. Gastroenterology 153, 77–86 e76. [DOI] [PubMed] [Google Scholar]
- Sands BE, Sandborn WJ, Panaccione R, O’Brien CD, Zhang H, Johanns J, Adedokun OJ, Li K, Peyrin-Biroulet L, Van Assche G, et al. (2019). Ustekinumab as Induction and Maintenance Therapy for Ulcerative Colitis. N Engl J Med 381, 1201–1214. [DOI] [PubMed] [Google Scholar]
- Sharma V, McKone HT, and Markow PG (2011). A Global Perspective on the History, Use, and Identification of Synthetic Food Dyes. J Chem Educ 88, 24–28. [Google Scholar]
- Sherlock JP, Joyce-Shaikh B, Turner SP, Chao CC, Sathe M, Grein J, Gorman DM, Bowman EP, McClanahan TK, Yearley JH, et al. (2012). IL-23 induces spondyloarthropathy by acting on ROR-gammat+ CD3+CD4-CD8- entheseal resident T cells. Nat Med 18, 1069–1076. [DOI] [PubMed] [Google Scholar]
- Stevens LJ, Burgess JR, Stochelski MA, and Kuczek T (2014). Amounts of artificial food colors in commonly consumed beverages and potential behavioral implications for consumption in children. Clin Pediatr (Phila) 53, 133–140. [DOI] [PubMed] [Google Scholar]
- Valadez D, Malakouti M, and Lunsford T (2018). Food Additives, the Gut Microbiota, and Inflammatory Bowel Disease: Interpreting the Interplay. Pract Gastroenterol 42, 60–68. [Google Scholar]
- Viennois E, Merlin D, Gewirtz AT, and Chassaing B (2017). Dietary Emulsifier-Induced Low-Grade Inflammation Promotes Colon Carcinogenesis. Cancer Res 77, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R, Gingell R, and Murrells DF (1971). Mechanisms of azo reduction by Streptococcus faecalis. I. Optimization of assay conditions. Xenobiotica 1, 221–229. [DOI] [PubMed] [Google Scholar]
- Wirtz S, Popp V, Kindermann M, Gerlach K, Weigmann B, Fichtner-Feigl S, and Neurath MF (2017). Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc 12, 1295–1309. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2017). Evaluation of certain food additives. World Health Organ Tech Rep Ser, 1–162. [PubMed] [Google Scholar]
- Zhang CR, and Wang YL (2003). A simple and efficient method for the reduction of azo compounds. Synthetic Commun 33, 4205–4208. [Google Scholar]
- Zou L, Spanogiannopoulos P, Pieper LM, Chien HC, Cai W, Khuri N, Pottel J, Vora B, Ni Z, Tsakalozou E, et al. (2020). Bacterial metabolism rescues the inhibition of intestinal drug absorption by food and drug additives. Proc Natl Acad Sci U S A 117, 16009–16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rDNA sequencing data generated during this study have been deposited in the NCBI Sequence Read Archive (Accession numbers: PRJNA702431 and PRJNA702461).


