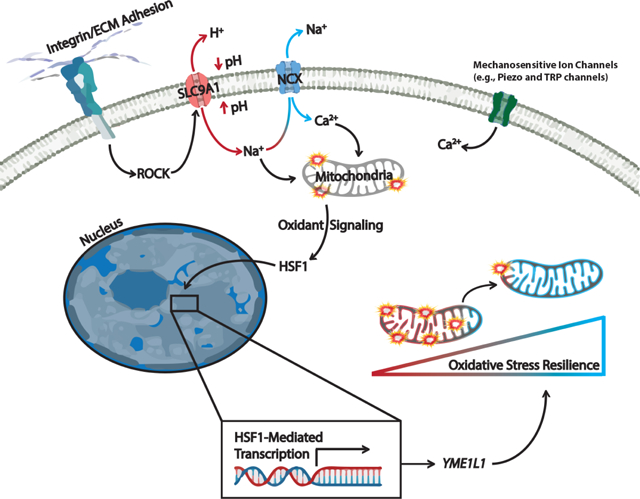
Summary
Mitochondria control eukaryotic cell fate by producing the energy needed to support life and the signals required to execute programmed cell death. The biochemical milieu is known to affect mitochondrial function and contribute to the dysfunctional mitochondrial phenotypes implicated in cancer and the morbidities of aging. However, the physical characteristics of the extracellular matrix are also altered in cancer and in aging tissues. We demonstrate that cells sense the physical properties of the extracellular matrix and activate a mitochondrial stress response that adaptively tunes mitochondrial function via solute carrier family 9 member A1 dependent ion exchange and heat shock factor-1 dependent transcription. Overall, our data indicate that adhesion-mediated mechanosignaling may play an unappreciated role in the altered mitochondrial functions observed in aging and cancer.
Keywords: Mitochondrial Unfolded Protein Response, UPRmt, Proteostasis, Mitochondria, ETC, Metabolism, Cancer, Aging, YME1L1, HSF1, Oxidative Stress, Redox, Integrin, ECM, Extracellular Matrix, Adhesion, Mechanical Stress, Stiffness, Tension, Mechanosignaling, Mechanotabolism, Mechano-Metabolic
Graphical Abstract
eTOC Blurb
Tharp et al. demonstrate that adhesion-mediated mechanosignaling dictates cellular metabolic programming. Mechanosensitive ion exchange and an HSF1-mediated mitochondrial stress response alters metabolic programming of cells actively adapting to mechanically distinct microenvironments. These findings offer one explanation for metabolic phenotypes observed in some epithelial cells residing within fibrotic/stiffened tumor tissue microenvironments.
Introduction
Alterations in mitochondrial function permit cancer cells to rapidly proliferate and metastasize and aging cells to regulate senescence phenotypes induced by DNA damage. Mitochondria provide a privileged metabolic compartment where oxidative phosphorylation (OxPhos) consumes oxygen and reducing equivalents to produce ATP. While OxPhos provides an efficient means to produce ATP, it can create collateral cellular damage through the release of reactive oxygen species (ROS), which can oxidize proteins, lipids, and nucleic acids. In response to elevated mitochondrial ROS exposure, cancer and aging cells activate adaptive stress responses that allow them to harness ROS-mediated proliferation and migration effects without activating ROS-mediated cell death (Balaban et al., 2005; Reczek and Chandel, 2017; Scialò et al., 2016; Wallace, 2012). These types of oxidative stress resilience (OxSR) programs alter cellular metabolism to enhance ROS buffering in the cytosol to limit damage caused by mitochondrial ROS.
The overproduction of ROS via mitochondrial dysfunction is thought to occur because of the biochemical composition of the aged or tumor microenvironment (TME) (Fane and Weeraratna, 2020; Ladiges et al., 2010; Sun et al., 2016; Vyas et al., 2016). However, the physical characteristics of the cellular microenvironment are also altered by cancer and aging - and can affect cell fate, function, and metabolism by regulating the activity of stress- and ROS-associated transcription factors (Fane and Weeraratna, 2020; Miroshnikova et al., 2016; Northcott et al., 2018; Oudin and Weaver, 2016). Sensing of the mechanical properties of the extracellular matrix (ECM) can also affect cellular metabolism by regulating the levels and/or activity of cytoplasmic enzymes responsive to mechanosignaling-induced cytoskeletal dynamics (Papalazarou et al., 2020; Park et al., 2020). Cellular mechanosignaling relies on adhesion receptors, such as integrins, transducing signals that mechanically entrain the cytoskeleton to the ECM, and these cytoskeletal remodeling events can affect the topological distribution of metabolic organelles, cargoes, and enzymes (Anesti and Scorrano, 2006; Northcott et al., 2018; Schedin and Keely, 2011). Cytoskeletal dynamics also play a critical role in the regulation of mitochondrial structure (Helle et al., 2017; Manor et al., 2015; Moore et al., 2016) and mechanosensitive transcription factors can alter mitochondrial gene expression (Tharp et al., 2018). Because mitochondrial structure influences mitochondrial function, we sought to determine if and how adhesion-mediated mechanosignaling affects mitochondrial function (Figure 1A).
Figure 1. Adhesion-mediated mechanosignaling alters mitochondrial structure and function of human mammary epithelial cells (MECs).
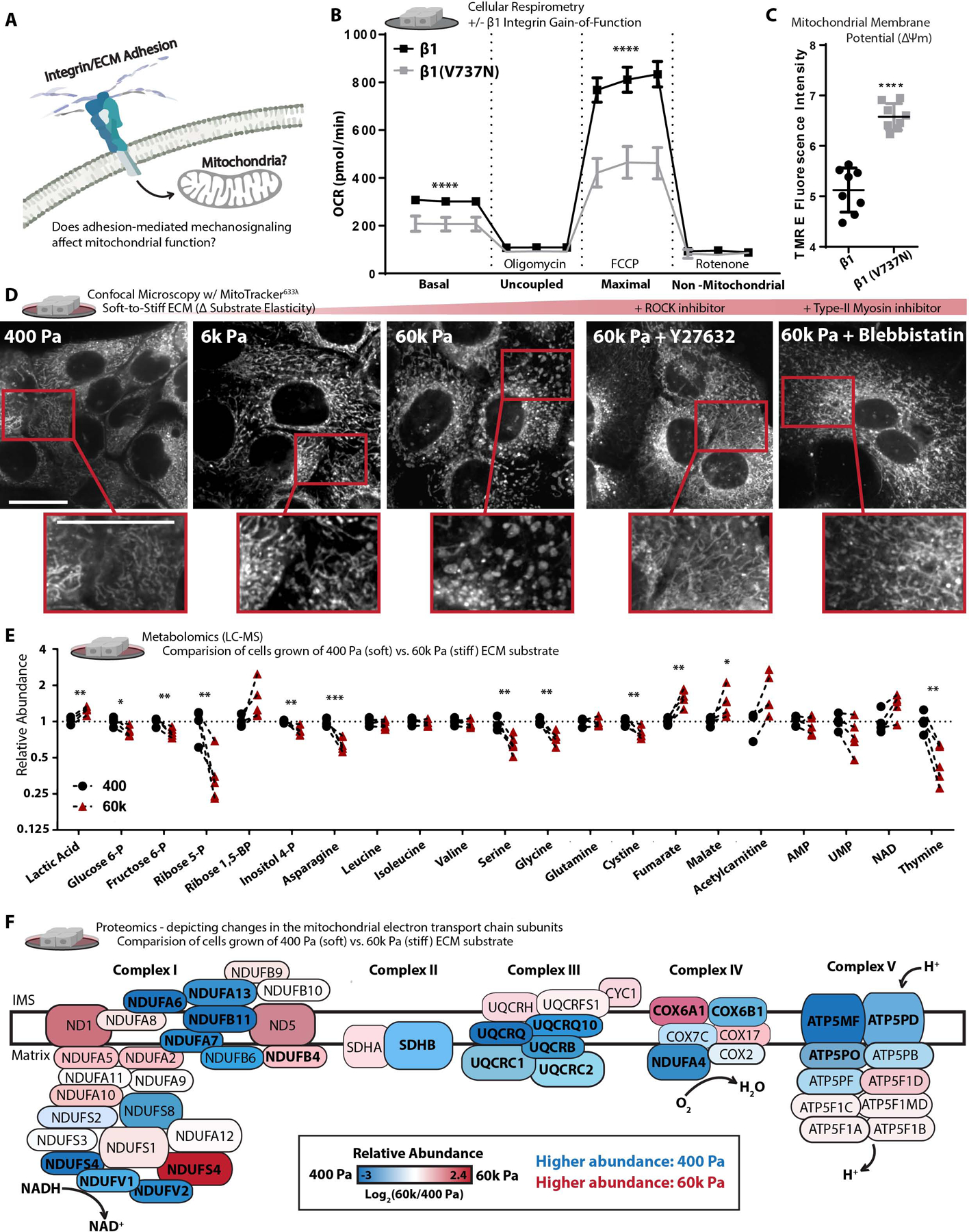
A. Graphical representation of the experimental question.
B. Mitochondrial oxygen consumption rate (OCR) of β1-integrin or β1(V7373N) expressing cells (100k cells per well, n=5 wells, 3 replicate measures, repeated 3 times), mitochondrial stress test conditions: uncoupled = oligomycin [1 μM], maximal = Trifluoromethoxy carbonylcyanide phenylhydrazone (FCCP) [1 μM], non-mitochondrial = antimycin A [1 μM] and rotenone [1 μM].
C. Mitochondrial membrane potential, measured after 1 h treatment of Tetramethylrhodamine ethyl-ester (TMRE) [10 nM] (n = 2 wells, repeated 4 times).
D. Confocal Microscopy depicting mitochondrial network structure in PFA-fixed cells cultured on varied soft-to-stiff fibronectin coated [6 μM/cm2] polyacrylamide hydrogels (soft-to-stiff ECM), for 24 h +/− y27632 [10 μM] or Blebbistatin [10 μM], stained with mitotracker (deep red FM) [100 nM]. (Scale Bar: 10 μm). MitoMAPR Quantification: 400 (18), 6k (20) 60k (7), 60k + Y27632 (15), and 60k + Blebbistatin (12) junctions per network.
E. Selection of metabolites measured with (LC-MS) from MECs cultured on soft of stiff ECM for 24 h, fold change relative to 400 Pa. (n=4–5 biological replicates LC-MS run together, repeated 2 times)
F. Relative abundance (fold change) of mitochondrial ETC subunits measured via timsTOF LC-MS of MECs cultured on soft or stiff ECM for 24 h (n=3 biological replicates). Bolded text indicates *P of < 0.05 or less, locations and sizes of ETC subunits graphically depicted are approximate and not to molecular scale.
Results
Mechanosignaling alters mitochondrial structure and function
We investigated the relationship between adhesion-dependent mechanosignaling and mitochondrial function by exogenously expressing a β1-integrin “gain of function” mechanosignaling model in nonmalignant human mammary epithelial cells (MECs; MCF10A). Expression of the β1-integrin (V737N, point mutation) promotes focal adhesion assembly, phosphorylation of focal adhesion kinase (FAK) (Figure S1A), cytoskeletal remodeling, actomyosin tension (Paszek et al., 2005), and the suppression of mitochondrial oxygen consumption (Figure 1B). Cellular respiration is primarily a product of OxPhos, in which the mitochondrial electron transport chain (ETC) consumes oxygen and reducing equivalents to produce ATP. Contrary to the paradigm that suppressed mitochondrial function (e.g. reduced respiration) occurs due to the loss of the mitochondrial membrane potential (ΔΨm), the β1(V737N)-integrin expressing MECs had higher, not lower, mitochondrial membrane potential (Figure 1C). To ensure that these phenotypes were not due to an indirect effect of β1(V737N)-integrin expression, we varied the surface density coating of fibronectin, an ECM component and integrin adhesion ligand that increases mechanosignaling via β1-integrin (Oria et al., 2017). MECs plated on a higher density of a fibronectin surface coating [60 μM/cm2], also showed repression of mitochondrial oxygen consumption, similar to β1(V737N)-integrin expression (Figure S1B). Furthermore, activating integrin mechanosignaling with acute exposure to manganese (Mn2+) (Lin et al., 2013) suppressed mitochondrial oxygen consumption (Figure S1C). The data indicate that that increased integrin mechanosignaling impacts mitochondrial function.
Integrin mechanosignaling is highly sensitive to the stiffness of the adhesion substrate, which affects mitochondrial respiration (oxygen consumption). Accordingly, we examined the mitochondrial morphology of MECs cultured for 24 hours on fibronectin coated [6 μM/cm2] polyacrylamide hydrogel gels ranging in elasticity (stiffness) between normal breast (400 Pa) and tumor (6k – 60k Pa) ECM (Caliari and Burdick, 2016; Tharp and Weaver, 2018) (note: tissue culture polystyrene elasticity is supraphysiological, ~3G Pa). MECs cultured on this range of biologically relevant ECM elasticities displayed a variety of mitochondrial morphologies, ranging from thin interconnected filaments (400 Pa), to thickened filaments (6k Pa), and then ~300 nM diameter fragments with toroidal shapes (60k Pa) (Figure 1D and S1D–E). Cells respond to ECM stiffness by ligating ECM adhesion receptors that induce Rho-GTPase and Rho-associated protein kinase (ROCK) cytoskeletal remodeling and increase actomyosin tension via type-II myosins (Butcher et al., 2009). We therefore bypassed the adhesion receptor ligation step and induced downstream mechanosignaling via an inducible ROCK, ROCK::ER (Croft and Olson, 2006), which was sufficient to suppress mitochondrial oxygen consumption (Figure S1F & S1A). In contrast, pharmacological inhibition of ROCK with Y27632 or type-II myosins with blebbistatin reduced the prevalence of the thick or toroidal mitochondrial fragments in MECs plated on the stiff ECM (Figure 1D), and restored mitochondrial oxygen consumption in the ROCK::ER cells (Figure S1G). Finally, β1(V737N)-integrin expressing cells displayed a fragmented/toroidal mitochondrial morphology, even on the soft ECM (Figure S1H) in direct contrast to MECs expressing a wild-type β1 integrin. The data indicate that mitochondrial structure and function is sensitive to the stiffness of the ECM through integrin- and ROCK-mediated mechanosignaling.
Mitochondrial function affects many aspects of cellular metabolism, therefore we broadly examined the steady state levels of polar metabolites present in MECs cultured on ECM surfaces that mimic the soft normal ECM (400 Pa) or stiff tumor ECM (60k Pa) (Figure 1E and Video S1–3). MECs cultured on the stiff ECM possessed higher levels of lactate and lower levels of upstream glycolytic or pentose phosphate pathway (PPP) intermediates, which may indicate increased flux through those pathways. We also noted lower levels of serine (Yik-Sham Chung et al., 2018) and increased levels of tricarboxylic acid cycle (TCA cycle) intermediates such as malate and fumarate, an oncometabolite (Sciacovelli et al., 2016), which could indicate that TCA cycle flux has reduced. Indeed, previous studies have indicated that TCA cycle impairment can affect mitochondrial structure (Barasa et al., 1973). Since TCA cycle flux is largely dependent on the activity of the ETC, we mapped the compositional changes in ETC subunit abundance with mass spectrometry based proteomics (Figure 1F). We found that a number of critical ETC subunits changed in abundance due to ECM stiffness and their relative levels can alter properties of mitochondrial function (Jimenez-Blasco et al., 2020). These mechanosignaling induced compositional changes in the ETC could explain the reduction of mitochondrial oxygen consumption, and the increased ΔΨm observed when integrin mechanosignaling is high due to decreased entry of electrons via NADH (Complex I) and proton pumping from the inner membrane space (IMS) into the mitochondrial matrix (MM) through ATP Synthase (Complex V).
Hyperglycemia and stiff ECM facilitate similar mitochondrial responses
Hyperglycemia [>5 mM glucose] is a biochemical stress that induces mitochondrial fragmentation, raises intracellular pH (pHi) (Lindström and Sehlin, 1984), lowers extracellular pH (pHe), and increases mitochondrial membrane potential in cultured cells (Wang et al., 2017). To explore whether the fragmented/toroidal mitochondrial morphologies induced by high cytoskeletal tension, were similar to those induced by hyperglycemia, we increased the media glucose concentration from 5 mM (“low glucose”) to 25 mM (“high glucose”) for MECs plated on the soft ECM (400 Pa) and examined changes in mitochondrial organization. Lattice light sheet microscopy (LLSM), which permits live cell imaging with limited phototoxicity (Chen et al., 2014), revealed that exposing MECs to hyperglycemia induced a rapid transition of mitochondrial morphology from a filamentous network into fragmented/toroidal structures (Figure S1I and Video 4), comparable to MECs cultured on stiff ECM (Figure 1D & S1D). Moreover, cells exposed to hyperglycemia or plated on stiff ECM express similar gene profiles that have been implicated in the mitochondrial unfolded protein response (UPRmt) (Aldridge et al., 2007; Lin and Haynes, 2016) (Figure S1J). Since both hyperglycemia and stiff ECM induce these mitochondrial stress response genes, we hypothesized that the reorganization of the mitochondria may reflect a pro-survival stress response (Sprenger and Langer, 2019; Youle and van der Bliek, 2012).
Because ECM stiffness induces mitochondrial reorganization by inducing cytoskeletal tension (Figure 1D) and the similarities between hyperglycemia and ECM stiffness induced mechanosignaling, we asked if hyperglycemia was sufficient to increase cellular elasticity. Atomic force microscopy (AFM) indentation revealed that hyperglycemia significantly enhanced cortical tension in MECs (Figure S1K). These findings were confirmed in a second cell line, the MDA-MB-231 MECs, which is a model of triple negative human breast cancer. Finally, similar to the reduced respiration rate induced by manipulating cytoskeletal tension, hyperglycemia also reduced mitochondrial oxygen consumption (Figure S1L). These findings demonstrate that biochemical and physical cues appear to stimulate similar changes in mitochondrial structure and function, and these changes may occur through the same stress response.
Mitochondrial fragmentation is thought to coordinate with mitophagy (autophagosome-mediated degradation of mitochondria) to repair dysfunctional mitochondria that have reduced mitochondrial membrane potential or increased reactive oxygen species (ROS) production/leak (Liu and Hajnóczky, 2011; Miyazono et al., 2018; Twig and Shirihai, 2011). However, our data indicated that the mitochondrial fragmentation induced by stiff ECM had elevated mitochondrial membrane potential (Figure 2A–B). Thus, we reasoned that large mitochondrial fragments with toroidal morphologies likely arise through a different mechanism than has been previously described. Mitochondrial membrane potential reflects a pH differential between the mitochondrial inner membrane space and the mitochondrial matrix, but it’s measurement with lipophilic cations (TMRE) can be sensitive to changes in intracellular pH (pHi) (Perry et al., 2011). Since we found higher levels of lactate in the MECs cultured on stiff ECM, we were concerned that these cells may have a transiently lower pHi which could influence the mitochondrial localization of TMRE. To verify that pHi was not confounding our measurement of ΔΨm, we measured pHi of MECs cultured on the stiff ECM or exposed to hyperglycemia and found that both stresses increased pHi (Figure 2C). One possible explanation for the mechanosensitive elevation of pHi despite elevated glycolytic metabolism could be that ROCK, a key mechanosignaling kinase, regulates the activity of SLC9A1 (Na+/H+ Exchanger 1 (NHE1)). SLC9A1 is responsible for the efflux of H+ from the cytoplasm and is responsible for regulating the pH of the adhesion-proximal cytosol to facilitate pH-dependent conformational changes in FAK critical for its phosphorylation and mechanosignaling downstream of integrin adhesions (Choi et al., 2013; Tominaga and Barber, 1998).
Figure 2. SLC9A1 facilitates stiff ECM induced mitochondrial programming.
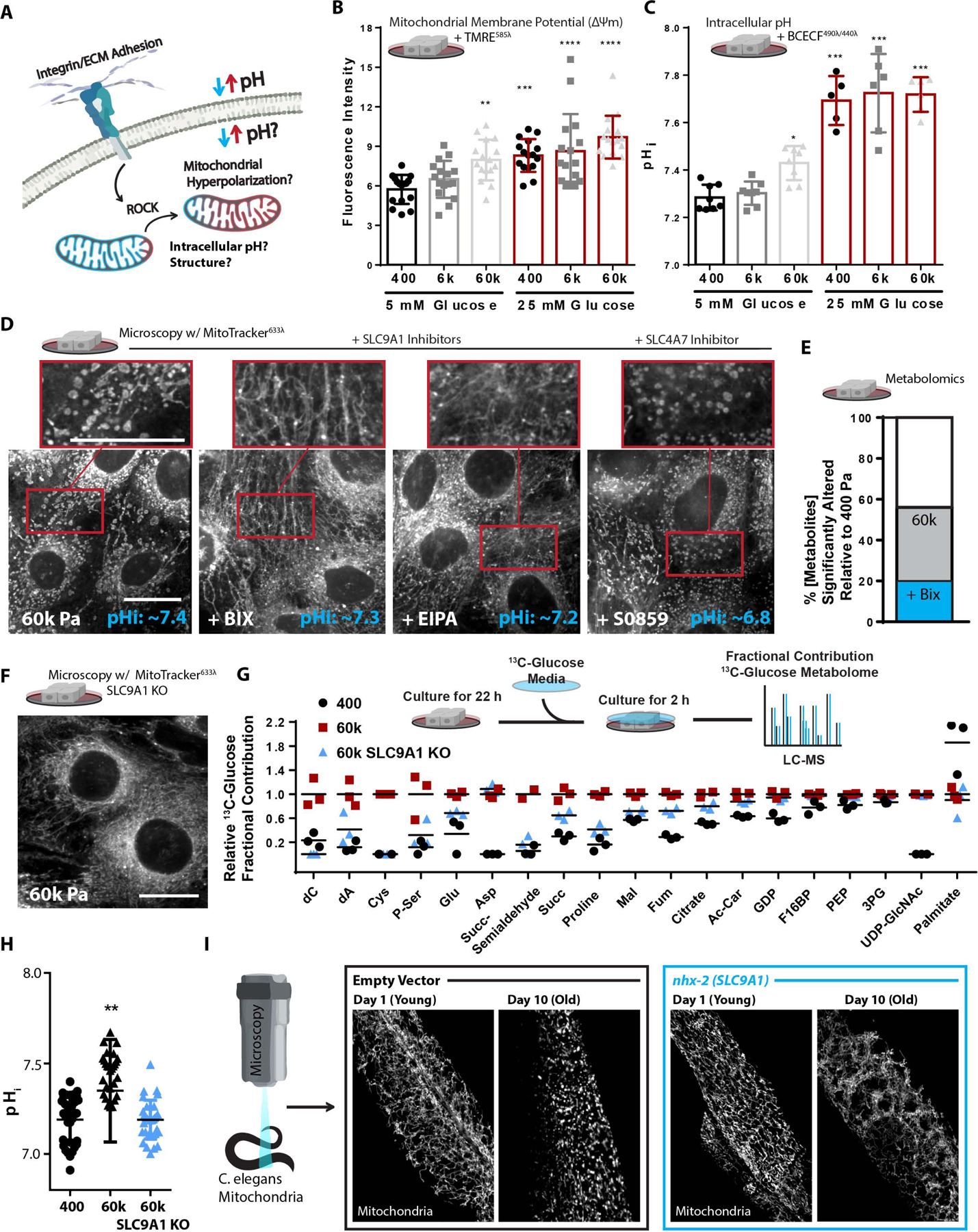
A. Graphical representation of the experimental question.
B. Mitochondrial membrane potential, measured after 1 h treatment of Tetramethylrhodamine ethyl-ester (TMRE) [10 nM] (n = 4 replicate PA gels repeated 4 times, shown together).
C. Intracellular pH (pHi) of cells grown of varied stiffness PA gels +/− glucose [25 mM], measured via 2’,7’-Bis-(2-Carboxyethyl)-5-(and-6)-Carboxyfluorescein, Acetoxymethyl Ester (BCECF) [1 μM] (n=2 replicate PA gels repeated 3–4 times, shown together).
D. Confocal microscopy depicting mitochondrial network structure and caption depicting of pHi measurements (mean of n=5) in MECs on 60k Pa surfaces treated with BIX [500 nM], EIPA [10 μM], S0859 [50 μM], or vehicle for 24 h, mitotracker (deep red FM) [100 nM] and BCECF [1 μM] (n=2 replicate PA gels repeated 3 times). (Scale Bar: 10 μm). MitoMAPR Quantification: 60k (9), BIX (20), EIPA (20), and S0859 (7) junctions per network.
E. Metabolomics (LC-MS) of cells cultured on 400 or 60k ECM for 24 h, % metabolites significantly altered relative to 400 Pa +/− BIX [500 nM]. (n=4–5 biological replicates LC-MS run together, repeated 2 separate times).
F. Confocal microscopy depicting mitochondrial network structure of SLC9A1 KO cells on 60k Pa surfaces for 24 h, mitotracker (deep red FM) [100 nM]. (Scale Bar: 10 μm). MitoMAPR Quantification: WT (8) and SLC9A1 KO (21) junctions per network.
G. Fractional contribution of 13C6-Glucose to a selection of pertinent metabolites. 2 h labeling, (n=3 biological replicates, LC-MS run together)
H. Intracellular pH (pHi) of WT or SLC9A1 KO cells grown of varied stiffness PA gels measured via 2’,7’-Bis-(2-Carboxyethyl)-5-(and-6)-Carboxyfluorescein, Acetoxymethyl Ester (BCECF) [1 μM] (n=6 replicate PA gels, repeated 4 times, shown together).
I. Representative microscopy depicting mitochondrial network structure of live C.elegans expressing MLS::mRuby (mitochondrial matrix) grown on empty vector or nhx-2 (SLC9A1 orthologue) RNAi from hatch of 5 d or 15 d old animals.
SLC9A1-mediated ion exchange affects mitochondrial structure and function
To test if the elevated pHi was responsible for the altered mitochondrial morphology observed in MECs cultured on stiff ECM, we lowered pHi by inhibiting SLC9A1 with BIX and EIPA, and SLC4A7 (Na+/HCO3− cotransporter) with S0859. While all of these interventions lowered pHi in MECs on stiff ECM to levels equivalent or lower than soft ECM, only SLC9A1 inhibition restored the filamentous mitochondrial morphology (Figure 2D & S2A). These data suggest that a pHi-independent effect of SLC9A1 may be responsible for the mitochondrial morphology induced by stiff ECM. SLC9A1 inhibition also restored the concentrations of approximately 60% of the significantly altered polar metabolites we measured in MECs plated on the stiff ECM back to the concentrations observed in MECs on soft ECM (Figure 2E). SLC9A1 inhibition also rescued the impaired mitochondrial oxygen consumption caused by β1(V737N)-integrin or ROCK::ER expression (Figure S2B–D). Since both hyperglycemia and ROCK activity have been show to increase pHi (Lindström and Sehlin, 1984; Tominaga et al., 1998), we assayed if the proportional decrease in pHe (extracellular acidification, H+ pumped out of the cell), and found that the pHi:pHe dynamics induced by hyperglycemia were sensitive to inhibition ROCK or SLC9A1 (Figure S2E).
CRISPR-mediated knockout of SLC9A1 in MECs (SLC9A1 KO) resulted in MECs that maintained a filamentous mitochondrial morphology on stiff ECM (Figure 2G and S2F). SLC9A1 KO was sufficient to normalize mitochondrial respiration in MECs exposed to hyperglycemia, as well as those on high-density fibronectin coating (Figure S2G–H). SCL9A1 KO cells cultured on stiff ECM metabolized glucose similarly to WT cells cultured on soft ECM (Figure 2G) and did not increase pHi when cultured on stiff ECM (Figure 2H). To explore the physiological relevance of these findings, we examined the ability of SLC9A1 to affect mitochondrial morphology in C. elegans, a model organism that is amenable to live microscopy of mitochondria and genetic manipulations (Nehrke and Melvin, 2002) that has been used extensively to study OxSR (Ristow and Schmeisser, 2011). RNAi-mediated knockdown of nhx-2 (SLC9A1 orthologue) prevented the mitochondrial fragmentation/toroidal phenotype typically found in the aged gut epithelium of this organism (Figure 2I) and instead promoted an abundant and hyperfused mitochondrial network (Figure S2I–L).
As a byproduct of H+ efflux (raising pHi, lowering pHe) SLC9A1 facilitates Na+ import that subsequently reverses the directionality of the Na+/Ca2+ exchangers (NCX, SLC8A1–3), a process which ultimately causes mitochondrial ROS production via mitochondrial calcium (Ca2+) overload (Brookes et al., 2004; Giorgi et al., 2018). Additionally, cellular Na+ can affect the solubility of mitochondrial Ca2+ phosphate precipitates, fluidity of the mitochondrial inner membrane, and ROS production from Complex III of the ETC (Hernansanz-Agustín et al., 2020). To determine if adhesion-dependent cytoskeletal tension regulates mitochondrial Ca2+ content through SLC9A1 activity, we measured mitochondrial and total cellular Ca2+ levels in MECs plated on a range of soft-to-stiff ECM. Imaging of Rhod2-AM, Calcium Green-1 AM, and Fura 2 AM revealed that mitochondrial Ca2+ concentration was highest on the stiff ECM, and could be reduced either by inhibiting or knocking out SLC9A1 (Figure 3B and S3A). SLC9A1 inhibition was also able to suppress the mitochondrial ROS production induced by mitochondrial Ca2+ loading in the CGP37157 (7-Chloro-5-(2-chlorophenyl)-1,5-dihydro-4,1-benzothiazepin-2(3H)-one)-treated cells (Figure 3C & S3B–C).
Figure 3. SLC9A1 facilitates mitochondrial oxidative stress.
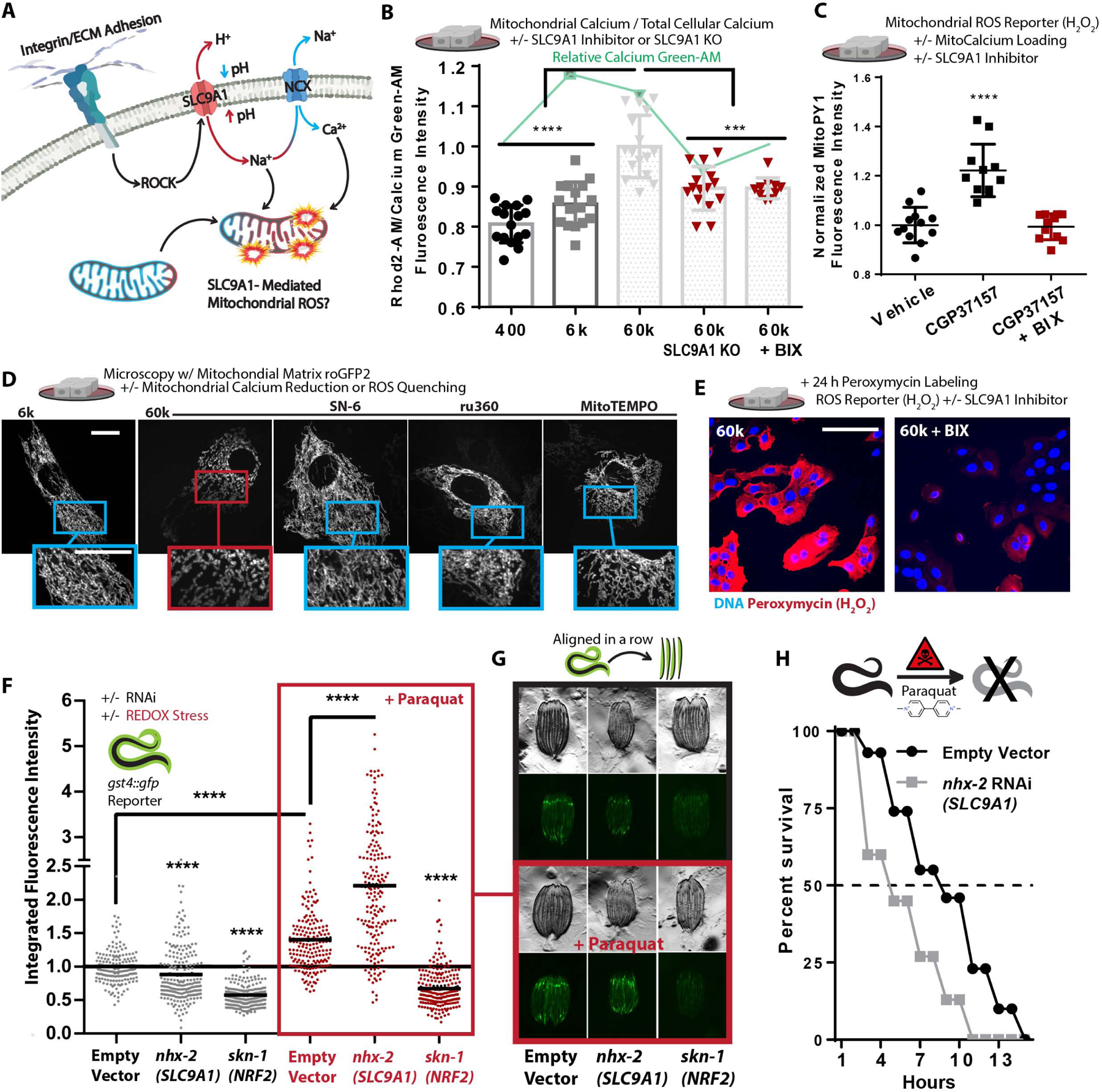
A. Graphical schematic indicating how SLC9A1 affects mitochondrial oxidative stress.
B. Calcium content of MCF10A cells cultured on soft-to-stiff ECM for 24 h, treated with Rhod2-AM [2 μM] (mitochondrial) and Calcium Green-1-AM [2 μM] (intracellular). (n=4 replicates, repeated 4 times).
C. Mitochondrial H2O2 production of cells cultured on 6k Pa surfaces and treated with BIX [500 nM] or vehicle for 24 h and then MitoPy1 [5 μM] and vehicle or CGP37157 [1 μM] for 1 h (n=6, repeated 2 times).
D. Confocal microscopy depicting mitochondrial network structure of PFA fixed MECs on 6k or 60k Pa ECM treated with 1 μM ru360 (MCU inhibitor), 10 μM SN-6 (NCX reverse mode inhibitor, opposite direction of CCP37157), and 2 μM MitoTEMPO for 24 h. (Scale Bar: 10 μm). MitoMAPR Quantification: 6k (21), 60k (6), 60k + SN-6 (27), 60k + Ru360 (13), and 60k + MitoTEMPO (12) junctions per network.
E. Confocal microscopy of peroxymycin (H2O2) (Yik-Sham Chung et al., 2018) staining over 24 h on 60k Pa ECM +/− BIX [500 nM], quantitated in Figure S3I.
F-G. gst-4p::gfp reporter fluorescent intensity of C.elegans measured with a large particle cytometer, +/− paraquat [50 mM] (n=177, 206, 190, 187, 191, and 215 animals in order left to right) with representative images (G) of C. elegans quantified, repeated 3 times.
H. C. elegans survival in 50 mM paraquat at 1 d; animals grown from hatch on nhx-2 RNAi vs empty-vector control (80 worms per condition, repeated 3 times).
Increased fibronectin surface density (Figure S3D–F), hyperglycemia (Figure S3G), and Mn2+ treatment altered mitochondrial structure, increased mitochondrial Ca2+, enhanced mitochondrial ROS production, in part through the activity of SLC9A1 (Figure S3H–I). Consistently, treating cells with CGP37157 or kaempferol (mitochondrial calcium uniporter (MCU) activator), to increase mitochondrial Ca2+ content, was sufficient to induce the fragmented/toroidal mitochondrial morphology in MECs plated on the soft ECM (Figure S3J). Suppression of mitochondrial ROS, with 2-(2,2,6,6-tetramethylpiperidin-1-oxyl-4-ylamino)-2-oxoethyl)-triphenylphosphonium chloride (MitoTEMPO), or suppression of the mitochondrial Ca2+ loading via selective inhibition of the reverse-mode of NCX exchangers, with SN-6 (Ethyl 2-[[4-[(4-nitrophenyl)methoxy]phenyl]methyl]-1,3-thiazolidine-4-carboxylate), and the MCU, with ru360 (oxo-bridged dinuclear ruthenium ammine), were also sufficient to prevent the fragment/toroid formation on stiff ECM (Figure 3D, Video S5–6, and S3K), providing additional evidence that Ca2+ overload and ROS were causative of mitochondrial remodeling.
To directly test whether ECM stiffness could impact ROS production, we next monitored the ROS production in MECs on a range of soft-to-stiff ECM over the course of 24 hours (Yik-Sham Chung et al., 2018). As expected, we found that cells seeded on stiff ECM produced more ROS than those on soft ECM, in an SLC9A1 dependent fashion (Figure 3E & S3L). Additionally, ROS production in ROCK:ER cells was also suppressed by SLC9A1 inhibition (Figure S3M). Finally, to determine whether the functional role of SLC9A1 on ROS-mediated oxidative stress was conserved in a whole organism, we tested its impact on oxidative stress in C. elegans. Using an reporter for oxidative stress (gst-4p::gfp), we found that nhx-2 (SLC9A1) knockdown reduced the expression of the reporter, indicating a lower basal level of ROS production/response in these animals (Figure 3F–G). In agreement with the free radical theory of aging, which postulates that lifespan is shortened due to accumulated oxidation-mediated damage, we found that the lifespan of the C. elegans nhx-2 knockdown was dramatically extended (Figure S3N).
Since oxidative stress primarily regulates the expression of gst-4p through the transcriptional activity of skn-1 (NRF-2 orthologue), which facilitates the canonical Oxidative Stress Response (OSR) by prompting transcription of genes with antioxidant response elements (ARE) in their promoters (e.g. gst-4p), we used a knockdown of skn-1/NRF-2 as a negative control. As a positive control, we treated the gst-4p::gfp reporter animals with paraquat, an herbicide that promotes mitochondrial ROS leak/production (Castello et al., 2007), which increased oxidative stress reporter activity. Surprisingly, paraquat treatment induced a more robust gst-4p::gfp reporter response to paraquat in nhx-2 knockdown animals (Figure 3F–G). This result suggests that nhx-2 knockdown exacerbated paraquat-induced mitochondrial oxidative stress, likely because OxPhos could not be throttled. To test the difference in oxidative stress sensitivity of nhx-2 knockdown animals, we assayed the survival of these animals in response to paraquat-induced oxidative stress. Corroborating the gst-4p::gfp reporter measurements (Figure 3F–G), the nhx-2 knockdown animals were more sensitive to paraquat exposure than control animals (Figure 3H). Accordingly, these results indicate that SLC9A1-activity may induce OxSR via adhesion-mediated production of sub-lethal mitochondrial ROS, which promote mitochondrial reorganization (toroid/fragment formation) and may prepare cells and animals to overcome subsequent oxidative stresses.
The greater induction of gst-4p::gfp and the rapidity of death observed in the nhx-2 knockdown animals suggested that they were less adapted to manage the ROS-mediated oxidative stress induced by paraquat. Since greater SLC9A1 activity appeared to promote mitochondrial ROS production (Figure 3C, 3E, S3G, & S3I–J) we hypothesized that because the nhx-2 knockdown animals experienced lower basal levels of ROS exposure, they were not pre-adapted to survive the paraquat exposure. It has been reported that mitochondrial stresses, particularly ROS production and respiratory dysfunction, promote adaptive reprogramming of mitochondrial function and oxidative stress resilience (OxSR) through a process described as “mitohormesis” (Ma, 2013; Ristow, 2014; Yun and Finkel, 2014). In biological systems, hormesis describes a biphasic dose response in which a stress/signal is moderated by a compensatory response (Mattson, 2008). For example, cancer cells produce more ROS than healthy cells, but the oncogene induced overproduction of ROS elicits compensatory ROS quenching response mediated by the transcriptional activity of NRF2 (nuclear factor erythroid 2–related factor 2). This NRF2-mediated ROS quenching OSR facilitates metabolic remodeling that provides cancer cells with OxSR requisite to harness ROS-mediated proliferation and migration effects without succumbing to ROS-mediated cell death (Reczek and Chandel, 2017). OxSR in cells and animals can be the product of NRF2-mediated OSR or other adaptive programs which are less well defined.
Stiff ECM forces HSF1-mediated mitochondrial reprogramming
Since MECs cultured on stiff ECM experienced a greater amount of ROS stress during as they adapted to the environment via integrin adhesion, we sought to determine if they had induced the NRF2-mediated OSR to survive and adapt (Figure 4A). To test this, we utilized RNA-Seq to characterize the transcriptional state of MECs cultured on soft-to-stiff ECM or soft ECM with hyperglycemia for 24 hours. We compared the transcriptional programs induced by hyperglycemia and adhesion substrate elasticity because they both affect cytoskeletal tension, mitochondrial reorganization (fragment/toroid), and mitochondrial oxygen consumption. Unsupervised hierarchical clustering demonstrated that the greatest transcriptomic signature overlap occurred in MECs plated on the soft ECM (400 Pa) that were exposed to hyperglycemia [25 mM] and MECs plated on the stiff ECM (60k Pa) with physiological glucose [5 mM] (Figure 4B). Gene ontology analysis indicated that the categories that were downregulated in response to stiff ECM included oxidation-reduction process, oxidoreductase activity, ion transport, mitochondrion, and mitochondrial inner membrane (Figure S4A). We also found that of all mitocarta 2.0 (Calvo et al., 2016) annotated mitochondrial genes encoded by the nuclear genome which change (up or down) in response to hyperglycemia or stiff ECM, the vast majority of these changes were conserved between both stresses (Figure 4C). Specifically, a number of ETC subunits were downregulated (NDUFA7, ATP5B, ATP5D, COX6b1 etc.) while mitochondrial-localized chaperones and proteases, which facilitate mitochondrial import, protein folding, and structural remodeling of the mitochondria during UPRmt were upregulated (YME1L1, HSPE1, DNAJC10 (hsp40)), HSPD1, HSPA9, HSPB11, etc.) (Labbadia et al., 2017). With regards to NRF2 and the OSR it facilitates, unexpectedly, cells cultured on stiff ECM had downregulated many canonical ARE-containing target genes (HMOX1, TXN, GPX2, GPX4, and NQ01, etc).
Figure 4: Mechanosignaling facilitates mitochondrial stress response via HSF1.
A. Graphical representation of the paradigm and remaining questions.
B. Heatmap depicting unsupervised hierarchical clustering of RNAseq of cells cultured on soft-to-stiff ECM for 24 h +/− glucose [5 or 25 mM], (n=2 duplicate libraries of 3 biological replicates, ~ 10 million reads per library).
C. Comparison of significantly altered MitoCarta 2.0 catalogued genes from the 400 pa, 60k, and 400 pa + 25 mM glucose conditions shown in Figure 4B.
D.hsp-6::gfp reporter fluorescent intensity representative images of C. elegans quantified, in Figure 4E RNAis were mixed at a 5:1 ratio of ev, ina-1, or pat-3 RNAi to ev or cco-1 RNAi as depicted (hsp-6 is the HSPA9/mtHSP70 orthologue)
E. Quantification of hsp-6::gfp reporter fluorescent intensity of C.elegans measured with a large particle cytometer, +/− cco-1 RNAi (n=387, 309, 377, 326, 312, and 294 animals in order left to right, repeated 3 times).
F. Western blot depicting relative protein abundance of HSF1, electron transport chain components, or β-actin within 5 μg of total protein derived from lysates of cells cultured on soft-to-stiff ECM for 24 h +/− glucose [5 or 25 mM].
G. Stable Isotope mitochondrial proteomics of crude mitochondrial fraction of MCF10A cells grown 400 or 60k Pa ECM for 24 h +/− KRIBB11 [2 μM] [(n=4 biological replicates LC-MS run together, repeated 2 times)
H. Confocal microscopy of 100 nM mitotracker (deep red FM) stained and fixed (PFA) cells cultured on 60k Pa ECM surfaces for 24 h +/− vehicle or KRIBB11 [2 μM]. MitoMAPR Quantification: 60k (7) and 60k + Kribb11 (12) junctions per network.
I. Metabolomics (LC-MS) of cells cultured on 400 or 60k Pa ECM for 24 h, % significantly altered relative to 400 Pa +/− KRIBB11 [2 μM]. (n=4–5 biological replicates LC-MS/MS run together, repeated 2 times)
J. Oxidized/reduced glutathione (NEM protected) measurements of MCF10A cells grown on 400 or 60k Pa ECM for 24 h +/− KRIBB11 [2 μM] [(n=4 biological replicates, repeated 2 times) (n=4–5 biological replicates LC-MS run together, repeated 2 times).
K. Oxidative stress indicator intensity of cells after 1 hour, MCF10A cells cultured on varied 400 or 60k Pa ECM for 24 h +/− vehicle or KRIBB11 [2 μM] prior, measured with 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFDA) [2 μM].
It was paradoxical that genes with AREs in their promoters were not upregulated in MECs cultured on stiff ECM, since they had experienced a greater amount of redox stress. However, this result could indicate that the transcriptomes measured reflected a post OSR state, in which oxidants were not actively produced and therefore ARE-mediated gene expression was downregulated. To address this possibility, we compared the transcriptional signature of MECs cultured on stiff ECM to other known stress responses (Grandjean et al., 2019) that can remediate damage resulting from oxidative stress, such as protein misfolding (Reichmann et al., 2018). We found that the majority of genes which characterize the Integrated Stress Response (ISR) were downregulated, Heat Shock Response (HSR) genes were upregulated, and genes ascribed to the OSR and UPRmt were inconsistently up- and downregulated. However, when comparing the upregulated genes of the UPRmt, OSR, and HSR, we noted that the upregulated genes associated with all of these stress responses were primarily heat shock proteins (HSPs) regulated by heat shock factor 1 (HSF1) (Table S1) (Grandjean et al., 2019).
Activation of UPRmt is thought to occur in response to mitochondrial respiratory dysfunction, so we tested if integrin signaling affected the activation of the UPRmt to the same degree as stiff ECM. UPRmt has been most well defined in C. elegans, so we used an established model of cytochrome c oxidase-1 subunit Vb (cco-1/COX4) knockdown, which robustly induces a fluorescent UPRmt reporter, hsp-6::gfp (HSPA9/mtHSP70 orthologue), in C. elegans. We performed double RNAi of cco-1 in conjunction with knockdown of ina-1 (ITGA orthologue, most similar to ITGA3,6, and 7) or pat-3 (ITGB1 orthologue). We found that pat-3/ITGB1 knockdown robustly attenuated cco1-mediated UPRmt, which suggests that integrin signaling is an important input to the activation of UPRmt (Fig. 4D–E). We then determined that HSF1 is required for the maximal activation of cco1-mediated UPRmt in C.elegans, as RNAi-knockdown of hsf-1 partially suppressed UPRmt induction (Figure S4B). Since C. elegans activate UPRmt primarily through atfs-1 (Nargund et al., 2012), which does not have a conspicuous mammalian orthologue (Fiorese et al., 2016), the data suggests that HSF1 may play a larger role in the mammalian UPRmt (Katiyar et al., 2020) or may resolve a UPRmt-overlapping aspect of mitochondrial dysfunction (Boos et al., 2019) that promotes OxSR.
HSF1 is primarily known to regulate a transcription program that facilitates the survival of cells experiencing heat stress (~43°C), but it may have an unappreciated role in OxSR since its transcriptional activity is regulated by ROS (H2O2) (Ahn and Thiele, 2003). Upregulation of HSF1 expression is an outcome of the NRF2-mediated OSR, because the HSF1 promoter (−1.5k to −1.7k bp) is heavily enriched with AREs (Paul et al., 2018). Indeed, we found that HSF1 abundance increased in response to stiff ECM or hyperglycemia, as was mitochondrial ATP5A, but not the mitochondrial encoded subunit of oxygen consuming ETC complex IV subunit MTC01 (Figure 4F). Inhibiting SLC9A1 in MECs cultured on stiff ECM repressed the expression of HSF1 and its downstream targets (Figure S4C). Treatment with MitoTEMPO, a mitochondria-targeted antioxidant, suppressed stiff ECM or paraquat-induced HSF1 and HSF1-target gene expression (Figure S4D and E). Overall, these data indicate that ROS induced by the stiff ECM via SLC9A1 activity promotes HSF1 expression and activity (Figure 3E and S3I).
We postulated that HSF1 could modify mitochondrial structure/function by influencing the expression of the mitochondrial import machinery, such as mtHSP70 (HSPA9) (Wiedemann and Pfanner, 2017), or by regulating mitochondrial biogenesis in collaboration with peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) (Charos et al., 2012). To test these possibilities, we measured the incorporation of newly synthesized proteins into the mitochondria with stable isotope incorporation mass spectrometry, which revealed that the stiff ECM enhanced the incorporation of newly synthesized proteins in an HSF1-dependent fashion (Figure 4G). Consistent with the hypothesis that an HSF1-mediated response facilitated the stiff ECM induced mitochondrial adaptation, HSF1 inhibition was sufficient to prevent the altered mitochondrial morphology induced by stiff ECM (Figure 4H). Inhibition of HSF1 also restored ~80% of the metabolite concentrations measured in MECs plated on stiff ECM to that of MECs cultured on soft ECM (Figure 4I). Overall, these findings suggest that ECM mechanosignaling alters mitochondrial reorganization and metabolic programming through a heat-stress independent HSF1-mediated program (Mendillo et al., 2012).
To explore if adhesion-mediated mechanosignaling facilitates a HSF1-dependent OxSR program, we quantified the levels of reduced and oxidized glutathione, the primary cellular oxidant detoxification and redox (reduction:oxidation) management system which becomes oxidized in the presence of ROS. MECs cultured on stiff ECM had lower levels of oxidized glutathione than those on soft ECM. HSF1 inhibition was sufficient to significantly increase the levels of oxidized glutathione in MECs on stiff ECM (Figure 4J). Metabolomics allowed us to observe that many metabolite changes that reflect OxSR (LeBoeuf et al., 2020), such as pentose phosphate pathway activity, which generates reduced nicotinamide adenine dinucleotide phosphate (NADPH) required to regenerate reduced glutathione and mitigate oxidative stress, were also elevated in response to stiff ECM, and could be normalized by inhibiting HSF1 (Figure S4F). Indeed, HSF1 inhibition abolished the enhanced reducing capacity of MECs that had been cultured on stiff ECM for 24 hours prior to the measurement of redox stress (Figure 4K). These data indicate that while MECs experience more redox stress in response to stiff-ECM, they adapt and acquire an OxSR through HSF1-dependent changes in cellular metabolism facilitated by mitochondrial reprogramming via compositional changes (Figure 1D, 4E–F).
To examine how HSF1 influences mitochondrial metabolic flux, we traced the metabolic fate of isotopic glucose metabolism in cells grown on soft or stiff ECM with or without HSF1 inhibition (Figure 5A). We allowed the cells to metabolize the labeled glucose for 2 h to ensure robust labeling of mitochondrial TCA cycle intermediates (Jang et al., 2018). Consistently, we found that stiff ECM dramatically alters the relative abundance of the whole metabolome, but also the flux of glucose metabolism (Figure 5B–C). Of note, we observed increases in the fractional labeling (enrichment of isotopic carbon derived from glucose) in many metabolites of the TCA cycle, urea cycle, and purine and pyrimidine metabolism pathway (Figure S5) which could indicate a concerted remodeling of metabolism to support mechanosignaling or OxSR (Figure 6A). Interestingly, the oncometabolite fumarate (Sciacovelli et al., 2016) is a metabolic intermediate between the urea cycle and the TCA cycle that may also be a driver of the altered mitochondrial morphologies observed (Crooks et al., 2021) in cells adapting to stiff ECM.
Figure 5: HSF1 facilitates mechanosignaling-mediated metabolic reprogramming.
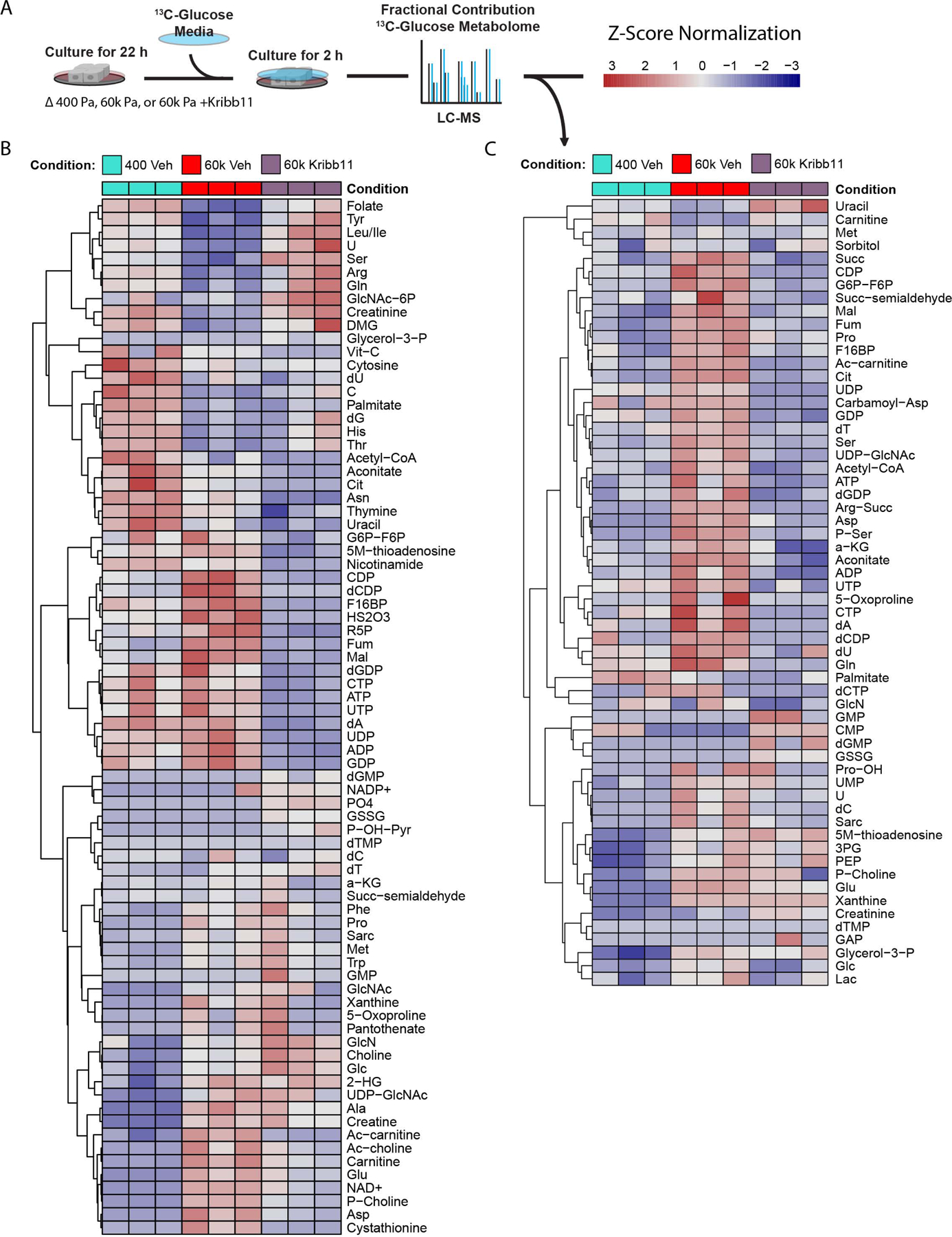
A. Graphical depiction of experimental design.
B. Heat map of relative metabolite levels of MECs cultured on 400 Pa or 60k Pa vehicle (DMSO) treated or 60k Pa ECM with Kribb11 [2 μM] for 22 h followed by media exchanged for 13C6-glucose containing media for 2 h and then harvested for LC-MS analysis. (n=3 biological replicates)
C. Heat map of fractional contributions of 13C6-glucose to the metabolome of MECs cultured on 400 Pa or 60k Pa vehicle (DMSO) treated or 60k Pa ECM with Kribb11 [2 μM] over the course of 2 h. MECs were previously cultured for 22 h in the same conditions with unlabeled glucose media. (n=3 biological replicates, LC-MS analysis)
Figure 6: HSF1 induces mitochondrial reprogramming.
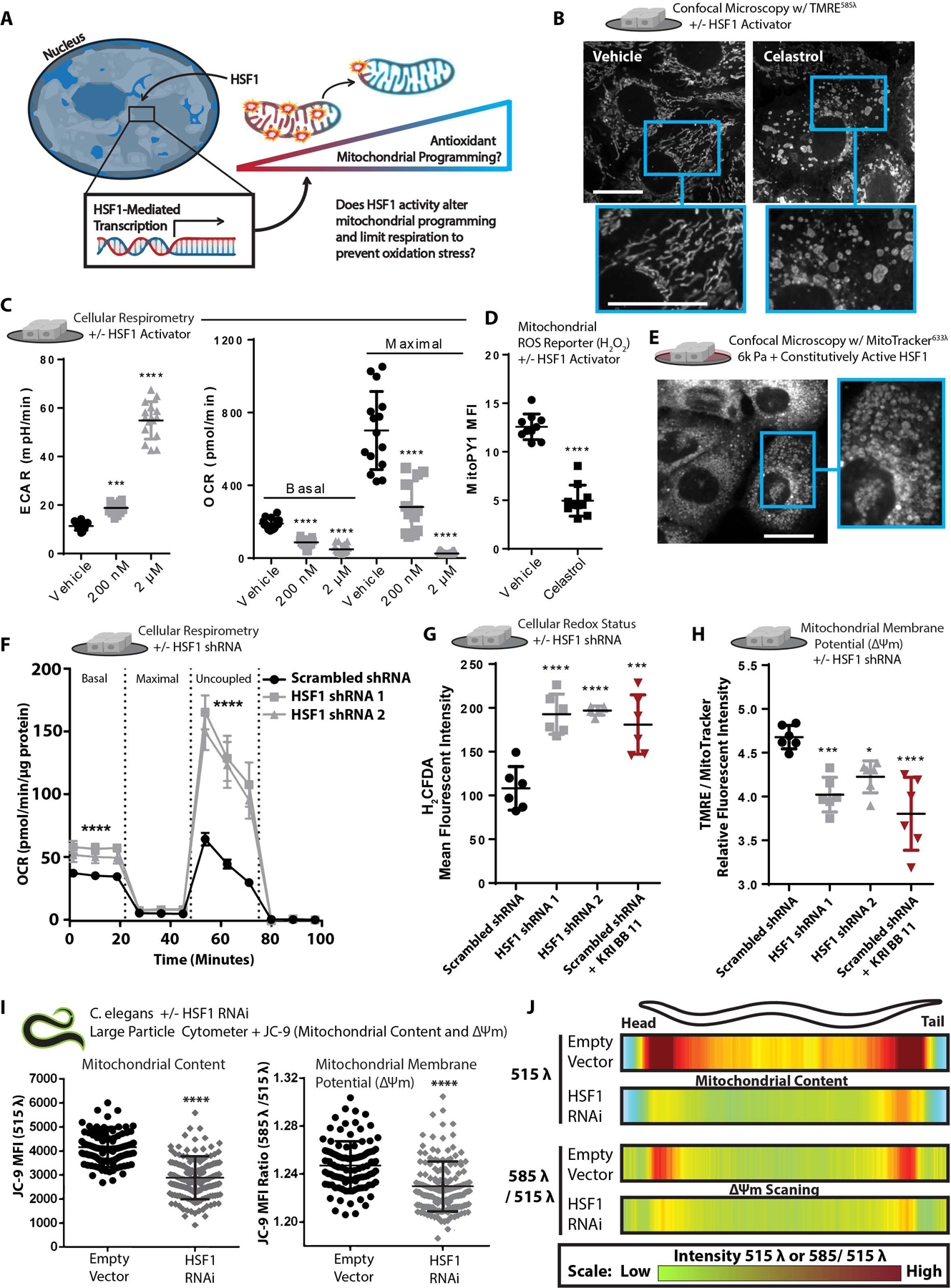
A. Graphical depiction of experimental question.
B. Confocal microscopy depicting morphology and mitochondrial membrane potential staining of live cells via TMRE [10 nM] staining +/− vehicle or Celastrol [2 μM] treatment for 40 minutes prior to imaging. MitoMAPR Quantification: vehicle (10) and celastrol (6) junctions per network.
C. Extracellular acidification rate (ECAR) and OCR of MECs treated +/− vehicle or Celastrol [200 nM] for 24 h or Celastrol [2 μM] for 40 minutes (n=5 wells, 3 replicate measures).
D. Mitochondrial H2O2 production of cells treated with Celastrol [2 μM] treatment for 40 minutes, measured with MitoPY [1 μM] (n=5, repeated 2 times).
E. Confocal microscopy depicting mitochondrial morphology of PFA-fixed cells expressing constitutively-active HSF1 and cultured on 6k Pa ECM for 24 h, stained with 100 nM mitotracker (deep red FM).
F. Oxygen consumption rate (OCR) of MCF10A cells expressing a scrambled shRNA or two different shRNAs targeting HSF1 (n=5 wells, 3 replicate measures, repeated 3 times)
G. Oxidative stress indicator intensity after 1 h in MCF10A cells cultured on TCPS expressing a scrambled shRNA +/− KRIBB11 [2 μM] or two different shRNAs targeting HSF1, measured with 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFDA) [2 μM].(n=6 wells, repeated 3 times).
H. Mitochondrial membrane potential of MCF10A cells cultured on TCPS expressing a scrambled shRNA +/− KRIBB11 [2 μM] or two different shRNAs targeting HSF1, measured with TMRE [1 nM] and mitotracker [100 nM] after 1 h staining (n=6, repeated 3 times).
I. Mean fluorescent intensity of 150 per condition JC-9 stained C. elegans grown on empty vector or hsf-1 RNAi from hatch, depicting mitochondrial mass (515 λ alone) or mitochondrial membrane potential (585 λ / 515 λ), spatially quantified in Figure 5J.
J. Heatmap depicting mitochondrial content (515 λ alone) or mitochondrial membrane potential (585 λ / 515 λ) across the body length (head (left) to tail (right)) of 150 C. elegans animals grown on empty vector or hsf-1 RNAi from hatch; JC-9 staining via administration of JC-9 loaded C.elegans food (E.coli) (repeated 3 times).
HSF1 can be pharmacologically activated using Celastrol, a reactive electrophile derived from the “Thunder of God” vine (Tripterygium wilfordii) (Ma et al., 2015), which was sufficient to induce mitochondrial fragmentation/toroids in MECs on all substrates (Figure 6B and S6A). HSF1 activation also increased extracellular acidification (ECAR), a proxy measure of glycolytic flux, and reduced mitochondrial oxygen consumption (Figure 6C). A mitochondria-localized ROS (H2O2) reporter (MitoPY1) revealed that MECs treated with Celastrol had significantly suppressed mitochondrial ROS production (Figure 6D). Expression of a constitutively active HSF1 induced fragmented/toroidal mitochondria in MECs plated on the soft ECM (Figure 6E and S6B), and increased mitochondrial membrane potential (Figure S6C). Conversely, HSF1 knockdown increased mitochondrial respiration (Figure 6F), induced oxidative stress (Figure 6G & S5D), and decreased mitochondrial membrane potential (Figure 6H & S6E) in both the MCF10A (nonmalignant) and MDA-MB-231 (aggressive and malignant) MECs (Figure S6F–I). The physiological relevance of these findings was confirmed by reducing hsf-1 expression in C. elegans, which decreased mitochondrial content and membrane potential throughout the whole organism (Figure 6I–J). This indicates that increased HSF1 activity reduces mitochondrial oxygen consumption and increases the mitochondrial membrane potential because proton flux from IMS to MM is reduced, which may limit ROS produced as a byproduct of OxPhos, mediating an enhanced OxSR by suppressing the mitochondrial contribution to net oxidative stress.
ECM-mechanosignaling engenders mitochondrial OxSR via HSF1 and YME1L1
Thus far, the data suggested that adhesion-mediated mechanosignaling stimulates a heat-stress independent HSF1 transcriptional program, previously implicated in cancer, that alters mitochondrial structure/function, restricts mitochondrial respiration, and oxidant production. Accordingly, we next stress-tested if the OxSR adaption was sufficient to oppose mitochondrial ROS-mediated apoptosis, a trait associated with many tumors (Reczek et al., 2017). We treated MECs cultured on soft-to-stiff ECM, with paraquat and assayed for apoptosis (Sprenger and Langer, 2019). Consistent with the hypothesis that mechnosignaling promotes OxSR via HSF1, MECs cultured on stiff ECM were less sensitive to paraquat treatment. This OxSR phenotype could be further enhanced via expression of constitutively active HSF1 and ablated by HSF1 knockdown or inhibition (Figure 7A–B & S7A–B). Functional links between mechanosignaling and mitochondrial OxSR adaptation were verified by determining that mitochondrial depletion negated the impact of ECM stiffness on redox sensitivity to paraquat (Figure S6C).
Figure 7: ECM-mediated mechanosignaling controls OxSR via HSF1 and YME1L1.
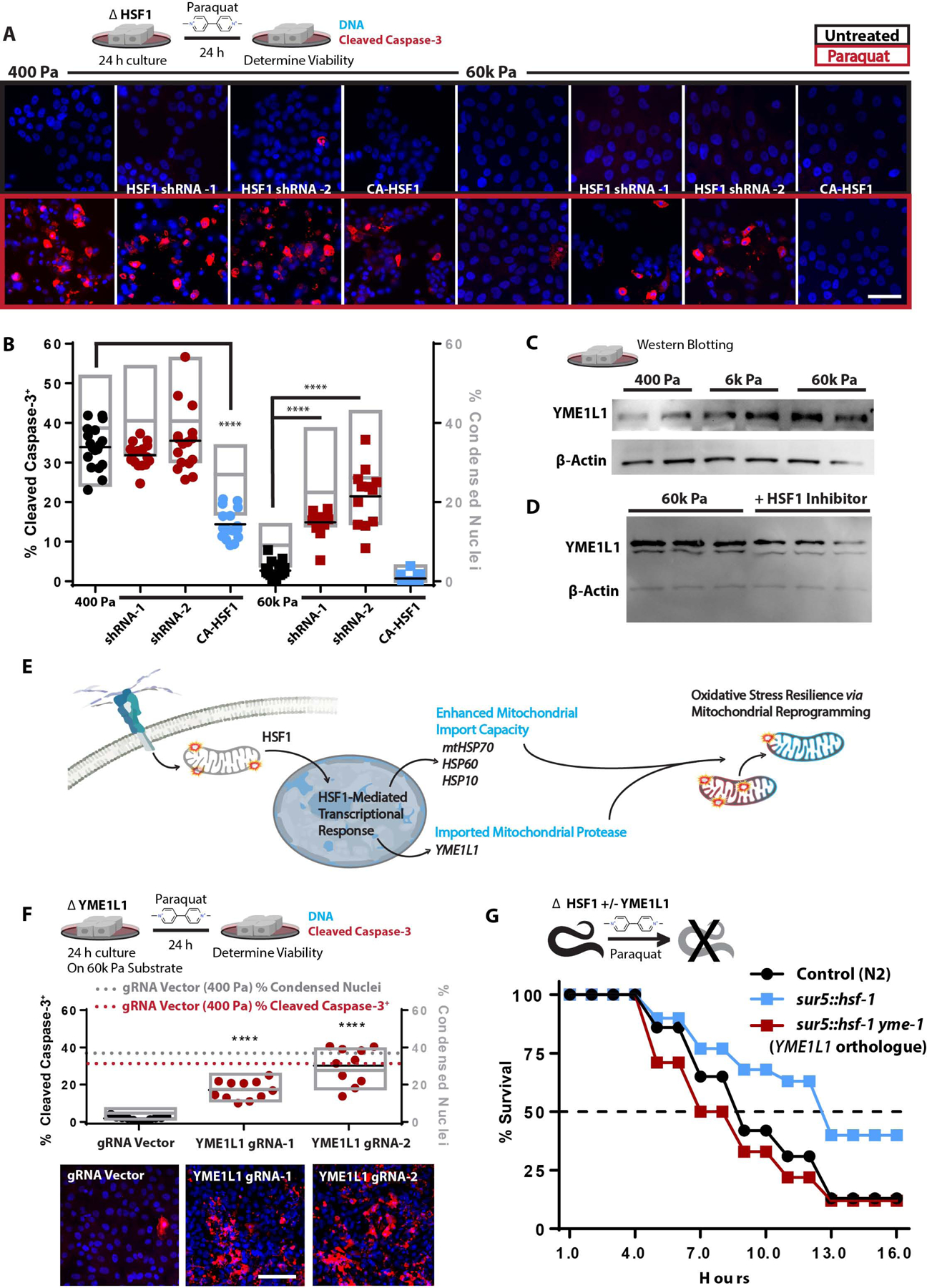
A. Confocal microscopy of indicators of apoptosis with cleaved caspase 3 staining (red) and nuclear condensation (dapi) of MECs cultured on 400 or 60k Pa ECM for 24 h with subsequent 24 h +/− paraquat treatment [10 mM]. 100k cells/well of 24 well plate. (n=4 replicates, repeated 3 separate times)
B. Quantitation of cells from 16 field views depicted in G for condensed nuclei and cleaved caspase-3 positive cells (1653–575 cells counted per condition, repeated 3 times).
C. Western blot of YME1L1 and β-actin from 5 μg of protein derived from cells cultured on soft-to-stiff ECM for 24 h. (2 biological replicates shown, repeated 3 times)
D. Western blot of YME1L1 and β-actin from 5 μg of protein derived from cells cultured on 60k Pa ECM for 24 h +/− KRIBB11 [2 μM] (3 biological replicates shown, repeated 2 times)
E. Graphical representation of the conceptual paradigm pertaining to this figure.
F. Quantitation of MECs with YME1L1 knockdown via CRISPR-I compared to CRISPR-I and empty guide vector expressing cells on 400 Pa (dashed lines) or 60k Pa ECM, 11 field views quantified for condensed nuclei and cleaved caspase-3 positive cells (923–2880 cells counted per condition, repeated 3 times).
G. C. elegans survival in 50 mM paraquat, with C. elegans overexpressing hsf-1 (sur-5p::hsf-1) compared or control line (N2) grown on either empty vector or ymel-1 RNAi from hatch (n=80 animals per condition, repeated 3 times).
Due to the fact that stiff ECM and HSF1 altered mitochondrial protein turnover rates, and mitochondrial import is required for mitochondrial protein turnover, we next assessed if cytoskeletal tension mediated OxSR was dependent upon mitochondrial protein import. Nuclear-encoded mitochondrial proteins are imported into the mitochondria (Schmidt et al., 2010), a process which requires a number HSF1 target genes to efficiently occur (e.g. HSPD1 (HSP60), HSPE1 (HSP10), and HSPA9 (mtHSP70) (Deocaris et al., 2006; Schneider et al., 1994). To test if cytoskeletal tension enhanced mitochondrial import was critical for OxSR, we used JG-98, an inhibitor of HSP70 that enriches in mitochondria (Li et al., 2013; Srinivasan et al., 2018). JG-98 treated cells grown on stiff ECM were not apoptotic, but were hyper-sensitized to paraquat-induced death (Figure S6D). To identify the specific mediators of the HSF1-dependent OxSR we cross-referenced conserved nuclear-encoded mitochondrial genes containing heat-shock elements (HSE) in their promoters with genes whose expression was upregulated by cytoskeletal tension (Figure 3B). We found one likely candidate, mitochondrial escape 1 like 1 (YME1L1), a zinc-dependent metalloprotease of the AAA+ protein family (ATPases with diverse cellular activity), which is a hallmark of UPRmt and regulates mitochondrial morphology (MacVicar and Langer, 2016; MacVicar et al., 2019). We verified that YME1L1 protein levels were regulated by HSF1 and were upregulated in response to mechanosignaling via stiff ECM (Figure 7C–D).
Since HSF1 resolves mitochondrial import stress (Boos et al., 2019) by regulating the expression mitochondrial import machinery, we hypothesized that dysfunctional or ROS-producing mitochondria elicit a adaptive response to potentiate mitochondrial import capacity and facilitate the import of certain nuclear-encoded mitochondrial proteins that mediate mitochondrial repair/reprogramming (Figure 7E). To determine if YME1L1 played a role in stiff ECM induced OxSR, we utilized CRISPR-I to downregulate YME1L1 expression. YME1L1 knockdown sensitized MECs cultured on stiff ECM to paraquat-induced death to a similar degree as cells plated on soft ECM (Figure 7F). Attenuated YME1LI expression in MECs increased cellular redox stress and lowered mitochondrial membrane potential (Figure 7F) and was dependent HSF1-mediated transcription (Figure S7F). To verify that HSF1 activity conferred OxSR through YME1L1, we examined the paraquat sensitivity of hsf-1-overexpressing C. elegans with or without reduced expression of yme-1 (YME1L1 orthologue). Overexpression of hsf-1 rendered C. elegans more resistant to paraquat-induced death, to a greater extent than that observed in other long-lived strains (e.g. daf-2 knockdown) and potentiated the skn-1-mediated adaptation to redox stress (Figure S7G–H). Impressively, yme-1 knockdown completely abolished the OxSR conferred to C. elegans through the overexpression of hsf-1 (Figure 7G). Overall, these data indicate that YME1L1 plays an essential role in the HSF1-mediated OxSR that is induced by stiff ECM induced adhesion-mediated mechanosignaling.
Mechanosignaling has been established as a key driver of aggressive characteristics in breast cancers (Kai et al., 2019) and HSF1 has also been identified as a key driver of aggressive-characteristics of metastatic breast cancer (Mendillo et al., 2012; Santagata et al., 2011; Scherz-Shouval et al., 2014). Therefore, we sought to determine if the fibrotic, stiffened tissue microenvironment that develops in experimental breast tumors regulates the transcriptional activity of HSF1. We examined nuclear localization of HSF1 in the murine PyMT mammary tumors excised from mice treated with and without the lysyl oxidase inhibitor (β-Aminopropionitrile, BAPN) which reduces tissue fibrosis, collagen crosslinking, and stromal stiffening of mammary tumors (Mouw et al., 2014). BAPN-treatment reduced tissue fibrosis (Figure S7I) and nuclear localization of HSF1 in the mammary tumor cells (Figure S7J–K). HSF1 and YME1L1 have been shown to be involved in many tumor types, however - breast cancer cells appear to be most dependent on HSF1 or YME1L1 (Figure S7K).
Discussion
We identified a mechanism whereby the physical properties of the microenvironment alter mitochondrial composition, structure, and function to tune cellular metabolism through a mechanical stress adaptation. We demonstrate that SLC9A1 and HSF1 alter mitochondrial function to support OxSR by regulating the levels of YME1L1 (MacVicar et al., 2019). The mechano-responsiveness of HSF1 and its ability to limit mitochondrial respiration, may explain why oncogene driven Warburg metabolism has been so difficult to observe in vitro. The rigid tissue culture polystyrene substrates (3G Pa) elevates mechanosignaling and chronically activates HSF1, regardless of oncogene-transformation, and this effect obscures any comparative measurements of mitochondrial function in normal and oncogene-transformed cells. Instead, prudent use of model systems with biomimetic properties (physically and chemically similar to the relevant biological system) are needed to uncover oncogene driven alterations in mitochondrial metabolism (Cantor et al., 2017; DelNero et al., 2018).
Our findings here demonstrate that HSF1-driven redox management not only suppresses the production of ROS, by limiting mitochondrial respiration, but it also opposes oxidant damage by promoting mitochondrial biogenesis/protein turnover and enhancing reducing equivalents (reduced glutathione/NADPH). Previous studies have indicated that cell-detachment/attachment-associated signaling elicits redox stress (Radisky et al., 2005; Schafer et al., 2009; Werner and Werb, 2002). With that in mind, coupling redox stress management to a molecular rheology sensor would be a rational design principle to promote cell survival. HSF1 is a logical candidate to serve as such a molecular rheology sensor because it facilitates cellular stress responses to the accumulation of misfolded proteins in the cytosol. Misfolded proteins can accumulate due to changes in pH, ion concentrations, osmolality, osmotic pressure, molecular crowding, adhesion-associated forces (mechanotransduction), enthalpy (heat), entropy (order), and redox balance (Ahn and Thiele, 2003; Dill, 1990; Dill and MacCallum, 2012; Guo et al., 2017; Higuchi-Sanabria et al., 2018) - all of which are cellular conditions associated with HSF1 activation. By surveying the physical state of the proteome, HSF1 is poised to temper diverse environmental perturbations that elicit mitochondrial dysfunction and oxidant leak. Indeed, HSF1 could mitigate the redox stress induced by conditions that deform mitochondrial structure (Helle et al., 2017), such as the physical stresses cells encounter in tumors with high interstitial pressure, mechanically stressful metastatic sites (Hassell et al., 2017), rigid ECMs, or oncogene induced ROCK activity (Irianto et al., 2016; Isermann and Lammerding, 2017; Samuel et al., 2011).
HSF1 levels are elevated in the majority of tumors and is implicated in cancer aggression and metastasis (Mendillo et al., 2012; Santagata et al., 2011; Scherz-Shouval et al., 2014). Because tumors are stiffer than healthy adjacent tissues, our findings offer a tractable explanation for why HSF1 and its target genes are so frequently upregulated in tumors (Acerbi et al., 2015; Maller et al., 2020). The heat shock independent activation of HSF1 and its target genes would provide the tumor cells with a metabolic adaptation to this chronic mechanical stress. Since metastatic cancer cells require redox stress management adaptations to disseminate to metastatic sites (Faubert et al., 2020) and ECM stiffness promotes metastasis (Kai et al., 2019), our findings may describe an important molecular mechanism by which ECM tension promotes metastatic disease. In this regard, therapeutic approaches to disrupt HSF1 and its target genes have focused on cytosolic and nuclear targets, but can incur difficult to tolerate systemic effects in humans (Dai and Sampson, 2016). We postulate that targeting the specific mediators (e.g., YME1L1, HSP60, mtHSP70, etc.) of the metabolic adaptations conferred by HSF1 could be more tractable anti-tumor therapeutic (MacVicar et al., 2019) than inhibition of HSF1 directly. Overall, our data demonstrate that the physical properties of the microenvironment play a critical role in facilitating adaptive stress responses that may contribute to metastatic characteristics of solid tumors {Deberadinins science paper} or altered metabolism and pathology observed structurally altered tissues (e.g., aged or fibrotic).
Limitations of Study
Our studies reveal a critical role for HSF1 in OxSR, however other HSFs (e.g., HSF2) may be involved in regulating these phenotypes (Östling et al., 2007). We performed our in vitro experiments with DMEM:F12 or DMEM based media classically used to culture our chosen cell line models. However, we strongly believe that our experiments would be more informative if we were to use the Human Plasma Like Medium (HPLM) (Cantor, 2019; Cantor et al., 2017; Rossiter et al., 2020). It is clear that the biochemical milieu influences metabolic programming, especially mitochondrial function. We hope to incorporate HPLM in our future work to more faithfully model cellular metabolic responses to physical cues in the cellular microenvironment. In this manuscript, we have demonstrated that hyperglycemia affects cellular mechanosignaling and metabolic adaptations. We postulate that physiologically irrelevant glucose concentrations found in DMEM are likely one of many metabolic substrates that obfuscate our ability to translate in vitro findings to in vivo models or clinical success.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Valerie M. Weaver (Valerie.Weaver@ucsf.edu).
Materials Availability
Cell lines, animal models, and expression vectors used in this manuscript are available from the lead contact upon request.
Data and Code Availability
The accession number for the RNA sequencing data reported in this paper is GEO:GSE171076. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171076
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Cell Culture
MCF10A and MB-MDA-231 cells were sourced from ATCC, routinely tested and found to be free of mycoplasma contamination, and maintained below passage 22. All cells were maintained and in 5% CO2 at 37 °C. MCF10A were cultured in [5 mM] glucose DMEM:F12 (1:1 mixture of F:12 [10mM] glucose and [0 mM] glucose DMEM) (Life Technologies, 11765054 and 11966025) supplemented with 5% Horse Serum (Gibco, 16050–122), 20 ng/mL epidermal growth factor (Peprotech), 10 μg/mL insulin (Sigma), 0.5 μg/mL hydrocortisone (Sigma), 100 ng/mL cholera toxin (Sigma, C8052–2MG), and 1x penicillin/streptomycin (Gibco). MB-MDA-231 tumor cells (ATCC) were grown in 5 mM glucose DMEM supplemented with 10% fetal bovine serum (FBS) (Hyclone) and 1x penicillin/streptomycin. HEK293T cells (ATCC) were maintained in DMEM supplemented with 10% FBS and 1x penicillin/streptomycin and were used to produce lentiviral particles with psPAX2 (Addgene 12260), pMD2.G (Addgene 12259) and various transfer vectors described hereafter (https://www.addgene.org/guides/lentivirus/).
Murine Mammary Tumor Model
FVB/N-Tg-MMTV-PyMT mice (The Jackson Laboratory) were treated with BAPN (3 mg per kg body weight; Spectrum) in the drinking water (n = 6 per group) injected intraperitoneally twice per week. Treatment started at 4 weeks and mice were tissues were harvested at 11 weeks of age. Mammary tumors were excised and fixed in 4% paraformaldehyde, cryosectioned, and immunostained for microscopy.
C. elegans strains and maintenance
All C. elegans strains are derivatives of the Bristol N2 strain from Caenorhabditis Genetics Center (CGC) and are listed below. All worms are maintained at 15 °C on standard nematode growth media (NGM) plates and fed OP50 E. coli B bacteria and are maintained for a maximum of 20–25 generations (weeks). For all experiments, worms are synchronized using a standard bleaching protocol by degrading carcasses with bleach solution (1.8% sodium hypochlorite, 0.375M KOH), then washing eggs four times with M9 solution (22 mM KH2PO4 monobasic, 42.3 mM Na2HPO4, 85.6 mM NaCl, 1 mM MgSO4), followed by L1 arresting synchronization, achieved by floating eggs in M9 overnight in a 20 °C incubator on a rotator for a maximum of 16 hours. L1s are plated on RNAi bacteria (NGM + 1 μM IPTG and 100 μg/mL carbenicillin; HT115 E. coli K strain containing pL4440 vector control or pL4440 with RNAi of interest) until the desired stages of adulthood. All RNAi constructs were isolated from the Vidal library and verified sequences are available below.
hsf-1
CGTCAGCGCGCCCGTACCGGCACATCAAATCCATTTTCCGGGTACTGTTGCTCATTATCAAACAACAAATCCTCGGCTCCATCATAATTCGACGTCTCCGGAGCATTTTCAAGAGCAAGCTGTCTGAGTGGATCCTCGGATCCTTCTTCATCATCATCCAACGGTACATTATTCCCAAAATCATCCCAATTATGATTACTGACTAAATCTCTGAAACTCTCCAATGAAGTATCAGTTCCAGTGAAATATTCTTGAAGTTCTTGAGATAATTGACGATCAAATGATGGAGAGAGTCCGAGAGTTGGAGAATATAGATTTTGATGAGGATCTGCGTTGGTGGATGAGGTGGAAGTCGTTGGATGATGCTGATCTTCTATTGCCATTAGCTTCTGATGCGGTTGAAGGTATTGATGAGATGGTTGATATGGAATCATTGAAGGATCTGAAGGCATGAAGCCACTGTAATTGTTCACAAATCCTCCCGAATAGTCTTGTTGCGGCTGAAAATTTCGAATTTTTAGAC
nhx-2
GTCAACGAAGTTCTTTTTATTGCCGTGTTCGGAGAATCTCTATTGAACGACGGAGTTGCTGTTGTACTTTATCGAATGTTCTTGACCTTCTCTGAAATTGGAACTGAGAATCTGATAACATCTGATTATATCAATGGAGGTGTTTCTTTCCTGGTTGTTGCATTTGGTGGAATTGGAATTGGTCTTCTTTTTGCATTTTTGACAAGTCTTGTTACAAGATTTGCTAGAGATGAAGAGGTCAAAGTGCTCAATTCTGTATTTATTCTCATTCTTCCGTACACTTGTTATCTTTGTGGAGAACTTTTCGGTCTTTCAAGTATTATGGC
ina-1
AACATCAAATGGATACGTTTCAAATGTCGGCGAAAAGGATTATCTGGACTTGACATTCACTGTGGAAAACAAGAAAGAAAAGGCTTATCAAGCGAATTTTTATCTAGAATATAATGAAGAAGAGCTTGAACTTCCACAAGTTCAAGGTTCCAAGAGAATGATTGCTGAAACAATTGGAAAGAATATTGTGCATTTGCCACTTGGAAATCCAATGAATGGAGCATCAAAACATCAATTTACGATCCAATTCAAATTGACTCGTGGAAGAACTGAAGGAATTGGAAAGGCACTCAAATTCATGGCACATGTCAATTCCACGTCACAAGAAACCGAGGAAGAGTTGAAAGATAATAAATGGGAAGCTGAAGTTCAGATTATCAAGAAGGCAGAGCTGGAGATCTATGGAATCAGTGACCCTGATAGAGTATTCTTTGGAGGAAAAGCAAGAGCAGAGTCTGAATTGGAATTGGAAGAAGATATTGGAACAATGGTTAGACATAACTATACAATTATTAATCATGGTCCATGGACTGTTCGAAATGTGGAAGCACACATTTCTTGGCCTTATCAACTCCGTTCTAGGTTTGGAAGAGGAAAGAATGCTCTCTATCTATTGGATGTACCGACTATTACAACAGAATTCACAGATGGAACAAGTGAAGTTAGAAAGTGTTTCATCAAACAACAGTACGAATATGTGAATCCTGCAGAAATTAAATTGAACACTAAATATAGTACTCAAGAGACTGCTCCACATCGGGTAGAGCATAGAATGAAAAGAGAAATTGATGAGGATGAGGAAGAACAATCAGATGATCTAGGAGCCGTGGAAGAGAATATTCCTTGGTTCTCAACAGCTAATTTCTGGAATCTTTTTGCAA TTAAAGGAGGTGATGGACG
pat-3
GATATGGTTTGTGGAGTTTGTCGATGCAAAGGAGGAAATGTTGGAAAATATTGTGAATGTAATAGACCTGGAATGAGTACTGCTGCGCTCAATGAAAAATGCAAAAGAACTAACGAATCAGCAATCTGTGAGGGTCGTGGTGTGTGTAACTGTGGACGTTGTGAATGTAATCCACGTGCCAATCCAGAAGAACAAATCTCTGGAGAATTCTGTGAATGCGACAACTTCAATTGCCCACGACACGATCGTAAAATCTGTGCAGAACACGGTGAATGCAACTGTGGAAAGTGTATTTGTGCACCTGGATGGACTGGAAGAGCCTGTGAATGCCCAATTTCAACTGATTCATGCCTCTCTGCAAATGGAAAAATCTGTAATGGAAAGGGTGAATGTATTTGTGGAAGATGTCGATGCTTCGATTCGCCCGACGGAAATCGATATTCGGGAGCGAAATGCGAAATTTGTCCGACGTGTCCGACGAAATGTGTGGAATACAAGAATTGTGTAATGTGCCAGCAATGGCAGACAGGGCCACTTAATGAGACCGCCTGTGATCAGTGTGAATTCAAAGTTATTCCTGTTGAGGAATTACCCAATCTCAACGAAACTACACCCTGCCAATTTGTGGATCCAGCTGATGATTGTACATTCTATTATCTCTACTATTACGATGAGGCCACAGATAATGCAACAGTCTGGGTCAGAAAACATAAAGATTGTCCTCCACCTGTCCCTGTGCTCGCAATTGTGCTCGGAGTCATTGCGGGTATCGTAATCCTCGGAATTCTTCTCTTGTTGCT
skn-1
TGTACACGGACAGCAATAATAGGAGCTTTGATGAAGTCAACCATCAGCATCAACAAGAACAAGATTTCAATGGCCAATCCAAATATGATTATCCACAATTCAACCGTCCAATGGGTCTCCGTTGGCGTGATGATCAACGGATGATGGAGTATTTCATGTCGAATGGTCCAGTAGAAACTGTTCCAGTTATGCCAATACTCACCGAGCATCCACCAGCATCTCCATTCGGTAGAGGACCATCTACAGAACGTCCAACCACATCATCTCGATACGAGTACAGTTCGCCTTCTCTCGAGGATATCGACTTGATTGATGTGCTATGGAGAAGTGATATTGCTGGAGAGAAGGGCACACGACAAGTGGCTCCTGCTGATCAGTACGAATGTGATTTGCAGACGTTGACA GAGAAATCGACAGTAGCG
ymel-1
TCGATTCAGTTGGCTCAAAACGTGTTTCGAATTCCATCCATCCATATGCAAATCAAACGATTAATCAACTTCTCAGGTAACAATTTGCTCAATTTTGTGCATTAAAAACTCATCTCCTGATGTTTTCAGTGAAATGGATGGCTTCACCCGTAACGAGGGAATCATTGTAATTGCCGCAACAAATCGTGTCGACGACCTC
METHOD DETAILS
ECM coated polyacrylamide hydrogel cell culture surfaces (PA-gels)
Cleaned (10% ClO, 1M HCL, then 100% EtOH) round #1 German glass coverslips (Electron Microscopy Services) were coated with 0.5% v/v (3-Aminopropyl)triethoxysilane (APTES, Sigma, 440140), 99.2% v/v ethanol, and 0.3% v/v glacial acetic acid for 2 h and then cleaned in 100% EtOH on an orbital shaker at 22 °C. APTES activated coverslips were coated with PBS buffered acrylamide / bis-acrylamide (Bio-Rad, 1610140 and 1610142) solutions (3% / 0.05% for 400 Pa, 7.5% / 0.07% for 6k Pa, and 10% / 0.5% for 60k Pa) polymerized with TEMED (0.1% v/v) (Bio-Rad, 1610801) and Potassium Persulfate (0.1% w/v) (Fisher, BP180) to yield a final thickness of ~ 85 μm. PA-gels were washed with 70% EtOH and sterile PBS prior 3,4-dihydroxy-L-phenylalanine (DOPA, CAS 59–92-7, Alfa Aeser, A1131106) coating for 5 min at 22 °C protected from light with sterile filtered DOPA in pH 10 [10 mM] Tris buffer (Wouters et al., 2016). DOPA coated PA-gels were washed 2x with sterile PBS and ECM functionalized with 5 μg/mL human fibronectin (Millipore, FC010) in sterile PBS 1 h at 37 °C to generate an expected fibronectin coating density of 6 μM/cm2.
Immunofluorescence microscopy
Cells or tissues were fixed in 4% paraformaldehyde (Electron Microscopy Services, 15710) in 1X PBS for 30 min at room temperature, washed and blocked with a blocking buffer (HBSS fortified with: 10% FBS (Hyclone), 0.1% BSA (Fischer, BP1600), 0.05% saponin (EMD Millipore, L3771), and 0.1% Tween 20 (Fischer, BP337500). Primary antibodies [1:100–1:200] for 2 h at RT or 24 h at 4 °C, Secondary antibodies [1:1000] for 2 h at 22 °C. Samples were imaged with a Nikon Eclipse Ti spinning disc microscope, Yokogawa CSU-X, Andor Zyla sCMOS, Andor Multi-Port Laser unit, and Molecular Devices MetaMorph imaging suite.
Antibodies used: Cleaved Caspase-3 (Asp175) (Cell Signaling, 9661, AB_2341188), HSF1 (Cell Signaling, 4356, AB_2120258), SLC9A1 (sc-136239, AB_2191254), and HSP60 (LK1, sc-59567, AB_783870).
MitoTacker staining
Mitotracker deep red FM or Mitotracker Green FM (Invitrogen, M22426 and M7514) was solubilized in DMSO to yield 100 μM frozen aliquots which were diluted into media to yield a 10 μM stock which was added directly to cell culture media already in the culture yielding a final concertation of 100 nM (to prevent media change derived fluid flow shear stress) 30 min before 4% PFA fixation or live cell imaging (MitoTracker red FM was used for PFA fixed samples). Three to five images per condition were analyzed using MitoMAPR (Zhang et al., 2019) to describe junctions per network (presented in the figure legend).
TMRE, H2DCFDA, Rhod-2 AM, Fura-2 AM, Calcium Green-1 AM, and MitoPy1
Stained cells were washed twice with PBS and imaged on a SpectraMax i5 Multi-Mode plate reader. Experiments were carried out with 50k or 100k cells per well in 500 μL of media in 24 well format with or without fibronectin coated PA-gels (indicated). 2 μM Rhod-2 AM (Thermo, R1244) and 2 μM Calcium green-1 AM (Life Technologies, C3011MP) was added to culture media and allowed to stain at 22 °C for 20 min prior to imaging (frozen Rhod-2 AM aliquots were used only when the DMSO suspension remained clear), or 2 μM H2DCFDA (Thermo, D399) was added to media 1 or 4 h prior to imaging, 5 μM MitoPy1 (Tocris, 4428), or 1 μM Fura-2 AM (ab120873), or 2 nM TMRE (Fischer, T669) was applied to cells in media without disturbing the existing culture media (similar dilution scheme to MitoTracker) for 1 h prior to microscopy or plate reader assay.
Intracellular pH (pHi)
10 μM BECEF (Invitrogen, B1150) was added to cell culture media for 30 min at 37 °C in 5 % CO2 incubator. Cultures were washed twice and then fluorescent intensities of BCECF was determined with a SpectraMax i5 plate reader in a buffer comprised of 25 mM HEPES, 140 mM NaCl, 5 mM KCl, 1 mM KHPO4, 1 mM MgSO4, 2 mM CaCl2, and 5 or 25 mM glucose. The cultures were then treated with a pH 7.7 buffer containing 10 μM Nigericin (Invitrogen, N1495), 25 mM HEPES, 105 mM KCl, and 1 mM MgCl for 5 min at 22 °C followed by determination of BCECF fluorescent intensities at high pH. The cultures were then treated with a pH 6.6 buffer containing 10 μM Nigericin (Invitrogen, N1495), 25 mM HEPES, 105 mM KCl, and 1 mM MgCl for 5 min at 22 °C followed by determination of BCECF fluorescent intensities at low pH. A linear relationship between pH 6.6 and 7.7 was observed and sample pH was estimated relative to pH standards for each individual culture well.
qPCR
Total RNA was isolated from biological samples with TRIzol (Invitrogen, 15596–018) according to the manufacturer’s instructions. cDNA was synthesized with 1 μg total RNA in 10 μL reaction volume with RNA using M-MLV reverse transcriptase (BioChain, Z5040002–100K) and 5X reaction buffer (BioChain, Z5040002–100K), random hexamers (Roche, 11034731001), dNTPs , and 1U of Ribolock (ThermoFischer, EO0384). RT-thermocycler program: random hexamers and RNA incubated at 70°C for 10 min, then held at 4 °C until the addition of the M-MLV reverse transcriptase, dNTPs, Ribolock, and M-MLV-reverse transcriptase, then 50 °C for 1 h, 95 °C for 5 min, then stored at −20 °C until qPCR was performed. The reverse transcription reaction was then diluted to 50 μL total volume with ddH2O rendering a concentration of 20 ng RNA per 1 μL used in subsequent qPCR reactions. qPCR was performed in triplicate using PerfeCTa SYBR Green FastMix (Quantabio, 95072–05K) with an Eppendorf Mastercycler RealPlex2. qPCR thermocycler program: 95 °C for 10 min, then 40 cycles of a 95 °C for 15 s, 60 °C for 20 s, followed by a melt curve 60–95 °C over 10 min. Melt curves and gel electrophoresis were used to validate the quality of amplified products. The ΔCt values from independent experiments were used to calculate fold change of expression using the 2−ΔΔCt method. For each gene measured, the SEM of the ΔCt values was calculated and used to generate positive and negative error values in the 2−ΔΔCt fold change space. Plots of qPCR data display bars representing the mean fold change ±SEM and individual points representing the fold change value for each experiment relative to the mean.
qPCR primers used:
| 5’-Forward | 5’-Reverse | |
| UCP2 | GGTGGTCGGAGATACCAAAG | CTCGGGCAATGGTCTTGTAG |
| NRF2 | CGGTATGCAACAGGACATTG | ACTGGTTGGGGTCTTCTGTG |
| 18S | GGACACGGACAGGATTGACA | ACCCACGGAATCGAGAAAGA |
| YME1L1 | CCCATGTCTCTGCACAATCC | ACCCCTTCACGAATGATGG |
| PKM1 | CTATCCTCTGGAGGCTGTGC | CCATGAGGTCTGTGGAGTGA |
| PKM2 | CCACTTGCAATTATTTGAGGAA | GTGAGCAGACCTGCCAGACT |
| TFAM | AAGATTCCAAGAAGCTAAGGGTGA | CAGAGTCAGACAGATTTTTCCAGTTT |
| HNRNPA1 | CCTTTGACGACCATGACTCC | ACGACCGAAGTTGTCATTCC |
| ATF5 | CTGGCTCCCTATGAGGTCCTTG | GAGCTGTGAAATCAACTCGCTCAG |
| HSPD1 | GATGCTGTGGCCGTTACAATG | GTCAATTGACTTTGCAACAGTCACAC |
| HSPA9 | CAAGCGACAGGCTGTCACCAAC | CAACCCAGGCATCACCATTGG |
| HSPE1 | TGGCAGGACAAGCGTTTAG | GGTTACAGTTTCAGCAGCAC |
| LONP1 | CATTGCCTTGAACCCTCTC | ATGTCGCTCAGGTAGATGG |
| HSF1 | GCCTTCCTGACCAAGCTGT | AAGTACTTGGGCAGCACCTC |
| XBP1sp | TGCTGAGTCCGCAGCAGGTG | GCTGGCAGGCTCTGGGGAAG |
| CHOP | GCACCTCCCAGAGCCCTCACTCTCC | GTCTACTCCAAGCCTTCCCCCTGCG |
| GAPDH | CGACCACTTTGTCAAGCTCA | AGGGGAGATTCA GTGTGGTG |
| LOX | GAACCAGGTAGCTGGGGTTT | TATGGCTACCACAGGCGATT |
Western blotting
Cells were freeze-thaw lysed (−80 °C) with RIPA buffer (150 mM NaCl, 1% v/v NP-40, 0.5% w/v sodium deoxycholate, 0.1% w/v SDS, and 25 mM tris) containing protease and phosphatase inhibitor coctail (GenDepot, P3100 and P3200). Protein content was determined via BCA (Pierce, 23225) and 5–10 μg of protein was mixed with 5x Laemmli buffer to generate final 1x concentration (50 mM Tris-HCl (Fischer, AAJ2267636) pH 6.8, 4% w/v SDS (Sigma, L3771), 10% v/v glycerol (Fischer, BP229–1), 0.1% w/v bromophenol blue (Bio-Rad, 1610404), 2% v/v β-mercaptoethanol (Bio-Rad, 1610710) and heated to 95 °C for 5 min (no heating of the samples used for the total oxphos (abcam, ab110413) blots). 10%-gels (Bio-Rad, Bulletin_6201) were cast in a PROTEAN Plus multi casting chamer (Bio-Rad). Samples were loaded (~20 μL) and run to completion in Tris Glycine SDS running buffer (25 mM Tris, 192 mM glycine (Fischer, BP381), and 0.1% SDS, pH ~8.6), wet transferred @ 100V for 60 min to methanol (Fischer, A412) activated PVDF (BioRad, 1620177) in Towbin transfer buffer containing (25 mM Tris, 192 mM Glycine, 20% v/v methanol, pH ~8.3). Protein loaded PVDF membranes were washed 2x with TBST (20 mM tris, 150 mM NaCl (S271), 0.1% w/v Tween20) and blocked in 5% milk TBST buffer for 1 h at 22 °C on an orbital shaker.
Antibodies used: HSF1 (Cell Signaling, HSF1, 4356, AB_2120258), YME1L1 (Invitrogen, PA564299, AB_2649732) HSP60 (LK1, sc-59567, AB_783870), HSP70 (3A3, sc32239, AB_627759), mtHSP70 (D-9, sc-133137, AB_2120468), Total OXPHOS WB Antibody Cocktail (Abcam, ab110413, AB_2629281), β-Actin (Sigma, A5441, AB_476744), FAK pY397 (Invitrogen, 44–625G, AB_1500096), FAK (BD, 610088, AB_397495), and pMLCK (Cell Signaling, 3671, AB_330248).
ROCK:ER
pBABEpuro3 ROCK:ER (Croft and Olson, 2006) was generously provided by Dr. Michael F. Olson, packaged into retroviral particles with phoenix cells, and used to generate stable MCF10A cells lines which were activated with 1 μg/mL 4-Hydroxytamoxifen (Sigma).
MTS-roGFP2
COX4L MTS tagged roGFP was expressed under the CMV promoter in a puromycin lentiviral transfer vector generated by the Dillin Lab (Dr. Brant Webster).
ATGCTTGCCACTAGAGTCTTTTCATTGGTAGGTAAAAGGGCCATAAGTACATCAGTCTGCGTGAGAGCCCACACCGGACCGGTCAGCAAGGGCGAGGAGCTGTTCACCGGGGTGGTGCCCATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCGTGTCCGGCGAGGGCGAGGGCGATGCCACCTACGGCAAGCTGACCCTGAAGTTCATCAGCACCACCGGCAAGCTGCCCGTGCCCTGGCCCACCCTCGTGACCACCCTGACCTACGGCGTGCAGTGCTTCAGCCGCTACCCCGACCACATGAAGCGGCACGACTTCTTCAAGTCCGCCATGCCCGAAGGCTACGTCCAGGAGCGCACCATCTTCTTCAAGGACGACGGCAACTACAAGACCCGCGCCGAGGTGAAGTTCGAGGGCGACACCCTGGTGAACCGCATCGAGCTGAAGGGCATCGACTTCAAGGAGGACGGCAACATCCTGGGGCACAAGCTGGAGTACAACTACAACTGCCACAACGTCTATATCATGGCCGACAAGCAGAAGAACGGCATCAAGGTGAACTTCAAGATCCGCCACAACATCGAGGACGGCAGCGTGCAGCTCGCCGACCACTACCAGCAGAACACCCCCATCGGCGACGGCCCCGTGCTGCTGCCCGACAACCACTACCTGAGCACCTGCTCCGCCCTGAGCAAAGACCCCAACGAGAAGCGCGATCACATGGTCCTGCTGGAGTTCGTGACCGCCGCCGGGATCACTCTCGGCATGGACGAGCTGT ACAAGTAA
V737N β1 integrin
Weaver lab generated (Dr. Jonathan Lakins) puromycin lentiviral transfer vector expressing 3x myc tagged V737N β1 integrin using the tetracycline rtTA2(S)-M2 (Urlinger et al., 2000) inducible promoter for 24 h with doxycycline [200 ng/mL] (Sigma, D9891). No respiratory repression of MCF10A or MB-MDA-231 cells was observed with 200 ng/mL, 1 μg/mL, or 2 μg/mL doxycycline. Previous studies have demonstrated that 24 h of 30 μg/mL doxycycline treatment can suppress mitochondrial respiration (Quirós et al., 2017).
ATGAATTTACAACCAATTTTCTGGATTGGACTGATCAGTTCAGTTTGCTGTGTGTTTGCTCAAACAGATGGCGAGCAGAAGCTGATCAGCGAGGAGGACCTGGGCGAGCAGAAGCTGATCAGCGAGGAGGACCTGGGCGAGCAGAAGCTGATCAGCGAGGAGGACCTGGGCGGCGCCCAAACAGATGAAAATCGATGTTTAAAAGCAAATGCCAAATCATGTGGAGAATGTATACAAGCAGGGCCAAATTGTGGGTGGTGCACAAATTCAACATTTTTACAGGAAGGAATGCCTACTTCTGCACGATGTGATGATTTAGAAGCCTTAAAAAAGAAGGGTTGCCCTCCAGATGACATAGAAAATCCCAGAGGCTCCAAAGATATAAAGAAAAATAAAAATGTAACCAACCGTAGCAAAGGAACAGCAGAGAAGCTCAAGCCAGAGGATATTACTCAGATCCAACCACAGCAGTTGGTTTTGCGATTAAGATCAGGGGAGCCACAGACATTTACATTAAAATTCAAGAGAGCTGAAGACTATCCCATTGACCTCTACTACCTTATGGACCTGTCTTACTCAATGAAAGACGATTTGGAGAATGTAAAAAGTCTTGGAACAGATCTGATGAATGAAATGAGGAGGATTACTTCGGACTTCAGAATTGGATTTGGCTCATTTGTGGAAAAGACTGTGATGCCTTACATTAGCACAACACCAGCTAAGCTCAGGAACCCTTGCACAAGTGAACAGAACTGCACCAGCCCATTTAGCTACAAAAATGTGCTCAGTCTTACTAATAAAGGAGAAGTATTTAATGAACTTGTTGGAAAACAGCGCATATCTGGAAATTTGGATTCTCCAGAAGGTGGTTTCGATGCCATCATGCAAGTTGCAGTTTGTGGATCACTGATTGGCTGGAGGAATGTTACACGGCTGCTGGTGTTTTCCACAGATGCCGGGTTTCACTTTGCTGGAGATGGGAAACTTGGTGGCATTGTTTTACCAAATGATGGACAATGTCACCTGGAAAATAATATGTACACAATGAGCCATTATTATGATTATCCTTCTATTGCTCACCTTGTCCAGAAACTGAGTGAAAATAATATTCAGACAATTTTTGCAGTTACTGAAGAATTTCAGCCTGTTTACAAGGAGCTGAAAAACTTGATCCCTAAGTCAGCAGTAGGAACATTATCTGCAAATTCTAGCAATGTAATTCAGTTGATCATTGATGCATACAATTCCCTTTCCTCAGAAGTCATTTTGGAAAACGGCAAATTGTCAGAAGGAGTAACAATAAGTTACAAATCTTACTGCAAGAACGGGGTGAATGGAACAGGGGAAAATGGAAGAAAATGTTCCAATATTTCCATTGGAGATGAGGTTCAATTTGAAATTAGCATAACTTCAAATAAGTGTCCAAAAAAGGATTCTGACAGCTTTAAAATTAGGCCTCTGGGCTTTACGGAGGAAGTAGAGGTTATTCTTCAGTACATCTGTGAATGTGAATGCCAAAGCGAAGGCATCCCTGAAAGTCCCAAGTGTCATGAAGGAAATGGGACATTTGAGTGTGGCGCGTGCAGGTGCAATGAAGGGCGTGTTGGTAGACATTGTGAATGCAGCACAGATGAAGTTAACAGTGAAGACATGGATGCTTACTGCAGGAAAGAAAACAGTTCAGAAATCTGCAGTAACAATGGAGAGTGCGTCTGCGGACAGTGTGTTTGTAGGAAGAGGGATAATACAAATGAAATTTATTCTGGCAAATTCTGCGAGTGTGATAATTTCAACTGTGATAGATCCAATGGCTTAATTTGTGGAGGAAATGGTGTTTGCAAGTGTCGTGTGTGTGAGTGCAACCCCAACTACACTGGCAGTGCATGTGACTGTTCTTTGGATACTAGTACTTGTGAAGCCAGCAACGGACAGATCTGCAATGGCCGGGGCATCTGCGAGTGTGGTGTCTGTAAGTGTACAGATCCGAAGTTTCAAGGGCAAACGTGTGAGATGTGTCAGACCTGCCTTGGTGTCTGTGCTGAGCATAAAGAATGTGTTCAGTGCAGAGCCTTCAATAAAGGAGAAAAGAAAGACACATGCACACAGGAATGTTCCTATTTTAACATTACCAAGGTAGAAAGTCGGGACAAATTACCCCAGCCGGTCCAACCTGATCCTGTGTCCCATTGTAAGGAGAAGGATGTTGACGACTGTTGGTTCTATTTTACGTATTCAGTGAATGGGAACAACGAGGTCATGGTTCATGTTGTGGAGAATCCAGAGTGTCCCACTGGTCCAGACATCATTCCAATTGTAGCTGGTGTTAACGCTGGAATTGTTCTTATTGGCCTTGCATTACTGCTGATATGGAAGCTTTTAATGATAATTCATGACAGAAGGGAGTTTGCTAAATTTGAAAAGGAGAAAATGAATGCCAAATGGGACACGGGTGAAAATCCTATTTATAAGAGTGCCGTAACAACTGTGGTCAATC CGAAGTATGAGGGAAAATGA
WT β1 integrin
Weaver lab generated (Dr. Jonathan Lakins) puromycin lentiviral transfer vector expressing 3x myc tagged β1 integrin using the tetracycline rtTA2(S)-M2 (Urlinger et al., 2000) inducible promoter for 24 h with doxycycline [200 ng/mL] (Sigma, D9891).
ATGAATTTACAACCAATTTTCTGGATTGGACTGATCAGTTCAGTTTGCTGTGTGTTTGCTCAAACAGATGGCGAGCAGAAGCTGATCAGCGAGGAGGACCTGGGCGAGCAGAAGCTGATCAGCGAGGAGGACCTGGGCGAGCAGAAGCTGATCAGCGAGGAGGACCTGGGCGGCGCCCAAACAGATGAAAATCGATGTTTAAAAGCAAATGCCAAATCATGTGGAGAATGTATACAAGCAGGGCCAAATTGTGGGTGGTGCACAAATTCAACATTTTTACAGGAAGGAATGCCTACTTCTGCACGATGTGATGATTTAGAAGCCTTAAAAAAGAAGGGTTGCCCTCCAGATGACATAGAAAATCCCAGAGGCTCCAAAGATATAAAGAAAAATAAAAATGTAACCAACCGTAGCAAAGGAACAGCAGAGAAGCTCAAGCCAGAGGATATTACTCAGATCCAACCACAGCAGTTGGTTTTGCGATTAAGATCAGGGGAGCCACAGACATTTACATTAAAATTCAAGAGAGCTGAAGACTATCCCATTGACCTCTACTACCTTATGGACCTGTCTTACTCAATGAAAGACGATTTGGAGAATGTAAAAAGTCTTGGAACAGATCTGATGAATGAAATGAGGAGGATTACTTCGGACTTCAGAATTGGATTTGGCTCATTTGTGGAAAAGACTGTGATGCCTTACATTAGCACAACACCAGCTAAGCTCAGGAACCCTTGCACAAGTGAACAGAACTGCACCAGCCCATTTAGCTACAAAAATGTGCTCAGTCTTACTAATAAAGGAGAAGTATTTAATGAACTTGTTGGAAAACAGCGCATATCTGGAAATTTGGATTCTCCAGAAGGTGGTTTCGATGCCATCATGCAAGTTGCAGTTTGTGGATCACTGATTGGCTGGAGGAATGTTACACGGCTGCTGGTGTTTTCCACAGATGCCGGGTTTCACTTTGCTGGAGATGGGAAACTTGGTGGCATTGTTTTACCAAATGATGGACAATGTCACCTGGAAAATAATATGTACACAATGAGCCATTATTATGATTATCCTTCTATTGCTCACCTTGTCCAGAAACTGAGTGAAAATAATATTCAGACAATTTTTGCAGTTACTGAAGAATTTCAGCCTGTTTACAAGGAGCTGAAAAACTTGATCCCTAAGTCAGCAGTAGGAACATTATCTGCAAATTCTAGCAATGTAATTCAGTTGATCATTGATGCATACAATTCCCTTTCCTCAGAAGTCATTTTGGAAAACGGCAAATTGTCAGAAGGAGTAACAATAAGTTACAAATCTTACTGCAAGAACGGGGTGAATGGAACAGGGGAAAATGGAAGAAAATGTTCCAATATTTCCATTGGAGATGAGGTTCAATTTGAAATTAGCATAACTTCAAATAAGTGTCCAAAAAAGGATTCTGACAGCTTTAAAATTAGGCCTCTGGGCTTTACGGAGGAAGTAGAGGTTATTCTTCAGTACATCTGTGAATGTGAATGCCAAAGCGAAGGCATCCCTGAAAGTCCCAAGTGTCATGAAGGAAATGGGACATTTGAGTGTGGCGCGTGCAGGTGCAATGAAGGGCGTGTTGGTAGACATTGTGAATGCAGCACAGATGAAGTTAACAGTGAAGACATGGATGCTTACTGCAGGAAAGAAAACAGTTCAGAAATCTGCAGTAACAATGGAGAGTGCGTCTGCGGACAGTGTGTTTGTAGGAAGAGGGATAATACAAATGAAATTTATTCTGGCAAATTCTGCGAGTGTGATAATTTCAACTGTGATAGATCCAATGGCTTAATTTGTGGAGGAAATGGTGTTTGCAAGTGTCGTGTGTGTGAGTGCAACCCCAACTACACTGGCAGTGCATGTGACTGTTCTTTGGATACTAGTACTTGTGAAGCCAGCAACGGACAGATCTGCAATGGCCGGGGCATCTGCGAGTGTGGTGTCTGTAAGTGTACAGATCCGAAGTTTCAAGGGCAAACGTGTGAGATGTGTCAGACCTGCCTTGGTGTCTGTGCTGAGCATAAAGAATGTGTTCAGTGCAGAGCCTTCAATAAAGGAGAAAAGAAAGACACATGCACACAGGAATGTTCCTATTTTAACATTACCAAGGTAGAAAGTCGGGACAAATTACCCCAGCCGGTCCAACCTGATCCTGTGTCCCATTGTAAGGAGAAGGATGTTGACGACTGTTGGTTCTATTTTACGTATTCAGTGAATGGGAACAACGAGGTCATGGTTCATGTTGTGGAGAATCCAGAGTGTCCCACTGGTCCAGACATCATTCCAATTGTAGCTGGTGTGGTTGCTGGAATTGTTCTTATTGGCCTTGCATTACTGCTGATATGGAAGCTTTTAATGATAATTCATGACAGAAGGGAGTTTGCTAAATTTGAAAAGGAGAAAATGAATGCCAAATGGGACACGGGTGAAAATCCTATTTATAAGAGTGCCGTAACAACTGTGGTCAATCCGAAGTATGAGGGAAAATGA
YME1L1 CRISPR-I
Using EF1a-dCas9-KRAB-Blast dCas9 vector and sgRNA lentiviral vectors generously provided by Dr. Michael T McManus and Broad institute GPP sgRNA Design identified sgRNAs targeting YME1L1 (1: 5’-TTCCGTTTCTGGGAGGAGTG, 2: 5’-GCAGTAGCTGTAGGAAGGGG, and 3:CTCCTCCCAGAAACGGAAAA-5’) stable cell lines were generated via sequential selection of blasticidin (CRISPR-I) and puromycin (sgRNA), kill curve and qPCR validated.
In vitro respirometry
Mitochondrial stress tests were performed with a Seahorse XF24e cellular respirometer on non-permeablized cells at ~ 96% confluence (100k cells/well) in V7 microplates, with XF assay medium supplemented with 1 mM pyruvate (Gibco), 2 mM glutamine (Gibco),and 5 or 25 mM glucose (Sigma) at pH 7.4 and sequential additions via injection ports of Oligomycin [1 μM final], FCCP [1 μM final], and Antimycin A/Rotenone [1 μM final] during respirometry (concentrated stock solutions solubilized in 100% ethanol [2.5 mM] for mitochondrial stress test compounds). OCR values presented with non-mitochondrial oxygen consumption deducted.
Atomic force microscopy
AFM and analyses were performed using an MFP3D-BIO inverted optical atomic force microscope mounted on a Nikon TE2000-U inverted fluorescence microscope (Asylum Research). 100k cells were seeded onto fibronectin coated 15 mm2 coverslips and cultured for 24 h. Coverslips were anchored with permanent adhesive dots (Scotch, 00051141908113) to a glass slide that was then magnet-anchored to the stage of the microscope. All samples were measured in media with contact mode using Novascan cantilevers (5 μm radius, Probe 58, k = 0.06 N per m), which were calibrated using the thermal tune method. 36 force measurements were collected over a 250 × 250 μm grid per sample. The resulting force data were converted to elastic modulus values using the Hertz Model program (tissue samples were assumed to be noncompressible, and a Poisson’s ratio of 0.5 was used in the calculation of the Young’s elastic modulus values) in IgorPro v.6.22, supplied by Asylum Research
Knockdown of HSF1
pLKO.1 puro (Addgene #8453) was modified to carry:
Scr insert:
5’-CAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT, shRNA HSF1–1 TRCN0000007481 (HSF1):
5’-CCGGGCAGGTTGTTCATAGTCAGAACTCGAGTTCTGACTATGAACAACCTGCTTTTT, shRNA HSF1–2 TRCN0000318652 (HSF1):
5’-CCGGGCACATTCCATGCCCAAGTATCTCGAGATACTTGGGCATGGAATGTGCTTTTT
Constitutively active HSF1
CD510B-1_pCDH-CMV-MCS-EF1-Puro (SystemBio) vector was modified to carry the hHSF1ΔRD (Δ221–315) transgene (Nakai et al., 2000) under the CMV promoter.
ATGGATCTGCCCGTGGGCCCCGGCGCGGCGGGGCCCAGCAACGTCCCGGCCTTCCTGACCAAGCTGTGGACCCTCGTGAGCGACCCGGACACCGACGCGCTCATCTGCTGGAGCCCGAGCGGGAACAGCTTCCACGTGTTCGACCAGGGCCAGTTTGCCAAGGAGGTGCTGCCCAAGTACTTCAAGCACAACAACATGGCCAGCTTCGTGCGGCAGCTCAACATGTATGGCTTCCGGAAAGTGGTCCACATCGAGCAGGGCGGCCTGGTCAAGCCAGAGAGAGACGACACGGAGTTCCAGCACCCATGCTTCCTGCGTGGCCAGGAGCAGCTCCTTGAGAACATCAAGAGGAAAGTGACCAGTGTGTCCACCCTGAAGAGTGAAGACATAAAGATCCGCCAGGACAGCGTCACCAAGCTGCTGACGGACGTGCAGCTGATGAAGGGGAAGCAGGAGTGCATGGACTCCAAGCTCCTGGCCATGAAGCATGAGAATGAGGCTCTGTGGCGGGAGGTGGCCAGCCTTCGGCAGAAGCATGCCCAGCAACAGAAAGTCGTCAACAAGCTCATTCAGTTCCTGATCTCACTGGTGCAGTCAAACCGGATCCTGGGGGTGAAGAGAAAGATCCCCCTGATGCTGAACGACAGTGGCTCAGCACATGGGCGCCCATCTTCCGTGGACACCCTCTTGTCCCCGACCGCCCTCATTGACTCCATCCTGCGGGAGAGTGAACCTGCCCCCGCCTCCGTCACAGCCCTCACGGACGCCAGGGGCCACACGGACACCGAGGGCCGGCCTCCCTCCCCCCCGCCCACCTCCACCCCTGAAAAGTGCCTCAGCGTAGCCTGCCTGGACAAGAATGAGCTCAGTGACCACTTGGATGCTATGGACTCCAACGAGGATAACCTGCAGACCATGCTGAGCAGCCACGGCTTCAGCGTGGACACCAGTGCCCTGCTGGACCTGTTCAGCCCCTCGGTGACCGTGCCCGACATGAGCCTGCCTGACCTTGACAGCAGCCTGGCCAGTATCCAAGAGCTCCTGTCTCCCCAGGAGCCCCCCAGGCCTCCCGAGGCAGAGAACAGCAGCCCGGATTCAGGGAAGCAGCTGGTGCACTACACAGCGCAGCCGCTGTTCCTGCTGGACCCCGGCTCCGTGGACACCGGGAGCAACGACCTGCCGGTGCTGTTTGAGCTGGGAGAGGGCTCCTACTTCTCCGAAGGGGACGGCTTCGCCGAGGACCCCACCATCTCCCTGCTGACAGGCTCGGAGCCTCCCAAAGCCAAGGACCCCACTGTCTCCTAG
SLC9A1 KO
MCF10A cells were transfected via PEI (https://www.addgene.org/protocols/transfection/) with pSpCas9(BB)-2A-GFP (PX458) - (Addgene #48138) carrying sgRNA for hSLC9A1 5’-GTTTGCCAACTACGAACACG (SLC9A1:HGLibA_45399) and H+-suicide selected (Pouysségur et al., 1984) four separate times to isolate SCL9A1 KOs. ~15% of the cells survived the first H+-suicide selection, ~90% survived the subsequent 4 sections.
C. elegans compound microscopy of mitochondria
Transgenic animals carrying vha-6p::MLS::mRuby (MLS was derived from atp-1: ATGTTGTCCAAACGCATTGTTACCGCTCTTAACACCGCCGTCAAGGTCCAAAATGCCGGAATCGCCA CCACCGCCCGCGGA) were grown from L1 to desired stage of adulthood on standard RNAi plates as described above. Animals were aged by hand-picking adults away from progeny using a pick daily until desired stage of adulthood. For imaging, adult worms are mounted on a glass slide in M9 solution, covered with a cover slip, and imaged immediately for a maximum of 10 minutes per slide. Animals were imaged on a Zeiss AxioObserver 7 LSM900 Airyscan 2 equipped with a 63x/1.4 Plan Aprochromat objective, MA-PMT detector, diode lasers (488 nm, 10mW, laser class 3B; 561 nm, 10 mW, laser class 4B), driven by ZenBlack software. Images were processed using ZEN Module Airyscan for 3D using default software settings. Images were analyzed using MitoMAPR (Zhang et al., 2019) across max projections keeping all parameters constant.
C. elegans Paraquat survival assay
Animals were grown to day 1 adulthood on standard RNAi plates as described above. 10 animals were picked into 75 μL of 100 mM paraquat solution prepared in M9 in a flat-bottom 96-well plate. >8 wells are used per condition for a minimum of 80 animals per replicate. Animals were scored every 2 hours for death. Plates are tapped gently, and any trashing or bending movement is scored as alive. Paraquat survival assays are performed with the experimenter blinded to the strain conditions during scoring and are repeated a minimum of 3 replicates per experiment.
C. elegans stereomicroscopy for fluorescent transcriptional reporters
Transgenic animals carrying gst-4p::GFP were grown on standard RNAi plates as described above until the L4 stage. L4 animals were washed off of plates using M9, centrifuged to pellet, and M9 was replaced with 50 mM paraquat prepared in M9. Animals were incubated rotating in a 20 °C incubator for two hours, and subsequently washed 2x with M9 solution. Animals were then plated on OP50 plates and recovered for 2 hours at 20 °C. For imaging, worms were picked onto a standard NGM plate containing 5 μL of 100 mM sodium azide to paralyze worms. Paralyzed worms were lined up, and imaged immediately on a Leica M250FA automated fluorescent stereomicroscope equipped with a Hamamatsu ORCA-ER camera, standard GFP filter, and driven by LAS-X software.
C. elegans biosorter analysis
For large-scale quantification of fluorescent animals, a Union Biometrica complex object parameter analysis sorter (COPAS) was used (for full details, refer to (Daniele et al., 2017). Briefly, to quantify signal of gst-4p::GFP, animals treated as described above were washed off plates using M9, and run through the COPAS biosort using a 488 nm light source. Integrated fluorescence intensity normalized to the time of flight is collected automatically on the COPAS software, and then normalized again to the extinction to correct for both worm length and worm thickness.
For JC-9 staining, day 1 adult animals were transferred to a plate containing JC-9-treated bacteria (OP50 bacteria were grown during mid-log phase for 4 hours in LB containing 50 μM JC-9 at 37 °C to incorporate JC-9 into bacteria; then bacteria were washed 2x with fresh LB to remove excess JC-9). Animals were grown on JC-9 bacteria for 2 hours at 20 °C to label. After labeling, worms were moved onto standard OP50 plates and grown for an additional 1 hour at 20 °C to remove excess JC-9 from the gut. Animals were then washed off plates and immediately run on a biosorter using a 488 and 561 nm light source. Worm profile data was collected, and run through an orientation and quantification algorithm, LAMPro (Daniele et al., 2017). Briefly, integrated fluorescence intensity is measured throughout the entire profile of the worm and normalized to extinction throughout the length of the worm. Total integrated fluorescence of the entire worm was also calculated by normalizing to the time of flight and integrated extinction of the entire worm. JC-9 fluorescence at 515 nm was used to determine mitochondrial quantity, and the ratio of the fluorescence of 585 nm / 515 nm was used to determine mitochondrial membrane potential.
Lifespan assay
Lifespan measurements were performed on solid NGM plates with RNAi bacteria. Worms were synchronized via bleaching/L1 arrested as described above. Adult animals were moved away from progeny by moving worms onto fresh RNAi plates every day until D7–10 when progeny were no longer visible. Animals were then scored every 1–2 days for death until all animals were scored. Animals with bagging vulval explosion, or other age-unrelated deaths were censored and removed from quantification.
Lattice Light Sheet Microscopy
We used a Custom build lattice light sheet microscope (Chen et al., 2014) to image MCF10A culture on polyacrylamide gels. Polyacrylamide gels (PA-gels) were formed on 5 mm round cover glass (Warner Instruments), coated with fibronectin and seeded with ~ 1000 cells per gel. The samples were cultured for 24 h in MCF10A media prior to imaging in DMEM (5 mM glucose) without phenol red supplemented with 5% Fetal Bovine serum. Samples were illuminated by 561 nm diode laser (0.5W Coherent) or 639 nm diode laser (1W Coherent) using an excitation objective (Special Optics, 0.65 NA with a working distance of 3.74-mm) at 2% AOTF transmittance and output laser power of 100 mW. The measured powers at the back focal plane of the illumination objective were in the range of 0.15–0.2 mW. Order transfer functions were calculated by acquiring Point-spread functions using 200-nm TetraSpeck beads adhered freshly to 5-mm glass coverslips (Invitrogen T7280) for each excitation wavelength and each acquisition filter set. The LLSM was realigned before each experiment.
For illumination we displayed on the spatial light modulator (SLM) a Square lattice generated by an interference pattern of 59 bessels beams separated by 1.67 um and cropped to 0.22 with a 0.325 inner NA and 0.40 outer NA, or by a an interference pattern of 83 bessels beams separated by 1.23 um and cropped to 0.22 with a 0.44 inner NA and 0.55 outer NA The lattice light sheet was dithered 15-25 um to obtain an homogenous illumination with 5% of flyback time. Fluorescent signal was collected by a Nikon detection objective (CFI Apo LWD 25XW, 1.1 NA, 2-mm working distance (WD)), coupled with a 500 mm focal length tube lens (Thorlabs), a set of Semrock filters (BL02-561R-25, BLP01-647R-25, and NF03-405-488-561-635E-25), and a sCMOS camera (Hamamatsu Orca Flash 4.0 v2) with a 103 nm/pixel magnification.
Z-Stacks (Volumes) were acquired by moving the Z-piezo in scanning mode while leaving the lattice light sheet static. The slices of the stacks were taken with an interval of 100-235 nm (S-axis) through ranges of 30-35 um at 20-100 ms exposuretime with 0 - 60 seconds intervals between volumes.
Raw data was flash corrected (Liu et al., 2017) and deconvolved using an iterative Richardson-Lucy algorithm (Chen et al., 2014) on two graphics processing units (GPU) (NVIDIA, GeForce GTX TITAN 4 Gb RAM). Flash calibration, flash correction, channel registration, Order transfer function calculation and Image deconvolution were done using the LLSpy open software (Lambert, 2019). Visualization of the images and volume inspection were done using Spimagine (Github/maweigert/spimagine) and Clear volume (Royer et al., 2015).
For the glucose shock experiment, MCF10A cells were first localized under the LLSM. During the first 30 seconds of the acquisition, the glucose concentration was raised to a final concentration of 25 mM. The glucose infusion caused misalignment of the lattice light sheet microscope which was corrected manually during the first acquisition volume.
Fractal Dimension and Lacunarity
Due to the complex morphology of mitochondria networks, we chose to calculate the fractal dimension and lacunarity to describe its morphology. This analysis has been proven useful to characterize mitochondrial morphology in malignant mesothelioma (Lennon et al., 2016). For the analysis regions of 276×276 pixels containing mitochondrial network were sampled randomly from 4 representative microscopy images for each experimental condition. Regions with more than 25% of its area contained nucleus were excluded from the analysis. Each data point in Figure S1E, represent a sampled region. Each region was first low pass filtered with a radius of 8 pixels and a weight of 0.8 to enhance the mitochondria network signal. After each region was thresholded using the Huang Algorithm. For each binary-masked region we used FracLac (http://rsb.info.nih.gov/ij/plugins/fraclac/FLHelp/Introduction.htm ) to calculate the fractal dimension (Db) and the lacunarity (lambda). For each region we increased linearly the sampled box until a maximum size of 50% of the region area the Db is calculated as the the average of Db from the Box scans. The fractal dimension was calculated using a regression to the logarithmic values of pixels with value 1 versus logarithm of the box size.
RNAseq
Total RNA was isolated using Trizol (Invitrogen), and RNAseq libraries (2 biological replicates per condition comprised of a pool of 4 PA-gel cultures each) prepared using KAPA mRNA HyperPrep Kit (Roche) and IDT dual indexed sequencing adaptors. Multiplexed libraries were sequenced on an Illumina HiSeq4000, and reads were aligned to the human genome (hg19) using RNA STAR (Dobin et al., 2013). Aligned reads were counted using HOMER (Lin et al., 2010) , and hierarchical clustering was performed using Cluster (Eisen et al., 1998) and visualized with Java TreeView. Gene Ontology analysis was performed using Metascape (Zhou et al., 2019).
Mitochondrial ETC proteomics timsTOF
1 million cells were seeded on 50 mM2 varied stiffness ECM coated PA-gels cultured for 24 h. Cells were washed with sterile PBS once, then cells were detached with cold PBS and a cell scraper (rubber policeman). Cell pellets were then suspended in 100 μL urea lysis buffer (ULB: 8M urea, 100 mM Tris, and 75 mM NaCl at pH 8). Probe sonicated 5 times for 2 s on ice. Protein concentrations were determined via BCA, and 100 μg of each sample was alkylated and reduced for 1 h at ~22 °C (protected from light) via the addition of a 10X concentrated stock (ARB: 400 mM 2-Chloroacetamide and 100 mM Tris(2-carboxyethyl)phosphine (TCEP) dissolved in ULB) yielding a final concentration of 40 mM 2-Chloroacetamide and 10 mM TCEP. Samples were then diluted, with a Tris/NaCl buffer (100 mM Tris and 75 mM NaCl at pH 8) containing 1 μg LysC (Promega, Va11A) per sample, to yield a final concertation of 2M urea during a 4 h digestion at ~22 °C (protected from light). Following the LysC digestion, 2 μg of Trypsin (Pierce, 1862746) was added and allowed to react with the sample overnight at 37 °C. The samples were then acidified with Trifluoroacetic acid (TFA) [~1% final] to yield a sample pH of 2. Samples were then desalted on C18 tips (Nest Group), the eluant was lyophilized, and then resuspended in 4% formic acid, 3% acetonitrile at 200 fmol/μL concentration. For each MS analysis, 1 μL of sample was separated over a 25 cm column packed with 1.9 μm Reprosil C18 particles (Dr. Maisch HPLC GmbH) by a nanoElute HPLC (Bruker). Separation was performed at 50 °C at a flow rate of 400 μL/min by the following gradient in 0.1% formic acid: 2% to 17% acetonitrile from 0 to 60 min, followed by 17% to 28% acetonitrile from 60 to 105 min. The eluant was directed electrospray ionized into a Bruker timsTOF Pro mass spectrometer and data was collected using data-dependent PASEF acquisition (Meier et al., 2018). Database searching and extraction of MS1 peptide abundances was performed using the MaxQuant algorithm (Cox and Mann, 2008), and all peptide and protein identifications were filtered to a 1% false-discovery rate. Searches were performed against a protein database of the human proteome (downloaded from Uniprot on 3/21/2018). Lastly quality control analysis was performed via artMS (http://artms.org) and statistical testing was performed with MSstats (Choi et al., 2014).
LC-MS/MS Deuterium Incorporation Proteomics
1 million cells were seeded on 50 mM2 varied stiffness PA-gels cultured for 24 h in 6% D2O culture media in a 5% CO2 incubator humidified with 5% D2O. D2O labeled cells were detached with cold PBS and a cell scraper (rubber policeman), pelleted with centrifugation, and mitochondrial fractions were isolated with the Mitochondria Isolation Kit for Cultured Cells (Thermo, 89874). Protein was isolated by flash freezing and sonication in PBS with 1 mM PMSF, 5 mM EDTA, and 1x Halt protease inhibitor (Thermo, 78440). Protein content was quantified via BCA (Pierce, 23225) and 100 μg of protein from each sample was trypsin (Pierce, 90057) digested overnight after reduction and alkylation with DTT, TFE, and iodoactamide (Russell et al., 2001). Trypsin-digested peptides were analyzed on a 6550 quadropole time of flight (Q-ToF) mass spectrometer equipped with Chip Cube nano ESI source (Agilent Technologies). High performance liquid chromatography (HPLC) separated the peptides using capillary and nano binary flow. Mobile phases were 95% acetonitrile/0.1% formic acid in LC-MS grade water. Peptides were eluted at 350 nL/minute flow rate with an 18 minute LC gradient. Each sample was analyzed once for protein/peptide identification in data-dependent MS/MS mode and once for peptide isotope analysis in MS mode. Acquired MS/MS spectra were extracted and searched using Spectrum Mill Proteomics Workbench software (Agilent Technologies) and a human protein database (www.uniprot.org). Search results were validated with a global false discovery rate of 1%. A filtered list of peptides was collapsed into a nonredundant peptide formula database containing peptide elemental composition, mass, and retention time. This was used to extract mass isotope abundances (M0–M3) of each peptide from MS-only acquisition files with Mass Hunter Qualitative Analysis software (Agilent Technologies). Mass isotopomer distribution analysis (MIDA) was used to calculate peptide elemental composition and curve-fit parameters for predicting peptide isotope enrichment based on precursor body water enrichment (p) and the number (n) of amino acid C-H positions per peptide actively incorporating hydrogen (H) and deuterium (D) from body water. Subsequent data handling was performed using python-based scripts, with input of precursor body water enrichment for each subject, to yield fractional synthesis rate (FSR) data at the protein level. FSR data were filtered to exclude protein measurements with fewer than 2 peptide isotope measurements per protein.
LC-MS Metabolomics
1 million cells were seeded on 50 mm2 varied stiffness ECM coated PA-gels cultured for 24 h. Cells were dissolved in 100% methanol doped with N-Ethylmaleimide (NEM) [8 mM,1 mg/mL] (Sigma-Aldrich, E1271) (Giustarini et al., 2013). Protein concentrations of the methanol extract was determined via BCA (Pierce, 23225) (5 uL transferred into 45 uL RIPA buffer, 5 uL of the RIPA dissolved solution assayed). Data was normalized to 100 μg per sample and polar metabolites were extracted in a total volume of 275 μl of 40:40:20 (acetonitrile:methanol:water) with inclusion of internal standard d3N15-serine (Cambridge Isotope Laboratories, Inc. #DNLM-6863). Extracted samples were centrifuged at 10,000 × g for 10 min and an aliquot of the supernatant was injected onto LC/MS where metabolites were separated by liquid chromatography. Analysis was performed with an electrospray ionization (ESI) source on an Agilent 6430 QQQ LC-MS/MS (Agilent Technologies). The capillary voltage was set to 3.0 kV, and the fragmentor voltage was set to 100 V, the drying gas temperature was 350 °C, the drying gas flow rate was 10 L/min, and the nebulizer pressure was 35 PSI. Polar metabolites were identified by SRM of the transition from precursor to product ions at associated optimized collision energies and retention times (Louie et al., 2016). Quantification of metabolites was performed by integrating the area under the curve and then normalizing to internal standard values. All metabolite levels are expressed as relative abundances compared to the control group.
13C6-Glucose LC-MS Metabolomics
1 million cells were seeded on 50 mm2 varied stiffness ECM coated PA-gels cultured for 22 h in 5 mM glucose DMEM based MCF10A media. The media was exchanged for media with 5 mM 13C6-Glucose (Cambridge Isotope Laboratories Inc., CLM-1396) DMEM based MCF10A media Cells for 2 h. Cells were washed twice with PBS and extracted with mass spectrometry grade 80% methanol (ThermoFisher, A456–1) and 20% water (ThermoFisher, W6500) supplemented with 5 nmol DL-Norvaline (Sigma, N7502). Protein concentrations of the methanol extract was determined via BCA (Pierce, 23225) with no significant variability assessed (5 uL transferred into 45 uL RIPA buffer, 5 uL of the RIPA dissolved solution assayed). Insoluble material was pelleted in a 4°C centrifuge at 16k x g, supernatant was transferred and dried in a Speedvac. Dried metabolites were resuspended in 50% ACN:water and 1/10th of the volume was loaded onto a Luna 3 um NH2 100A (150 × 2.0 mm) column (Phenomenex). The chromatographic separation was performed on a Vanquish Flex (Thermo Scientific) with mobile phases A (5 mM NH4AcO pH 9.9) and B (ACN) and a flow rate of 200 μL/min. A linear gradient from 15% A to 95% A over 18 min was followed by 9 min isocratic flow at 95% A and reequilibration to 15% A. Metabolites were detected with a Thermo Scientific Q Exactive mass spectrometer run with polarity switching (+3.5 kV / −3.5 kV) in full scan mode with an m/z range of 65–975. TraceFinder 4.1 (Thermo Scientific) was used to quantify the targeted metabolites by area under the curve using expected retention time and accurate mass measurements (< 5 ppm). Values were normalized to cell number and sample protein concentration. Relative amounts of metabolites were calculated by summing up the values for all isotopologues of a given metabolite. Fractional contributional (FC) of 13C carbons to total carbon for each metabolite was calculated. Data analysis was accomplished using in-house developed R scripts.
Paraquat survival
10 mM paraquat (Acros Organics, 227320010) was dissolved into media and added to cell culture vessels and allowed to affect the cells for 24 h, experiments were always performed with a fresh suspension of paraquat. After 24 h of paraquat treatment cells were fixed with 4% PFA and stained for cleaved caspase 3 (Cell Signaling, 9661).
QUANTIFICATION AND STATISTICAL ANALYSIS
Data displayed represent at least three independent experiments, unless otherwise specified. Figure legends contain biological and technical replicate information. Plots include each data point, mean, and SEM. Student’s t test with 95% confidence interval was used to determine differences between two comparable groups. ANOVA with 95% confidence interval was used to compare three or more comparable groups. Grubb’s test was used to detect distributions for outliers. Statistical comparisons were completed Graphpad Prism 6 software:
Figure 1. Data shown represents ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, < 0.0001 via two-tailed unpaired Student t test.
Figure 2. Data shown represents ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, < 0.0001 via one-way ANOVA with Tukey test for multiple comparisons to 400 Pa 5 mM Glucose (B and C) or 25 mM glucose (D).
Figure 3. Data shown represents ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, < 0.0001 via two-tailed unpaired Student t test in (D) and one-way ANOVA with Tukey test for multiple comparisons in (B and C).
Figure 4. Data shown represents ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, < 0.0001 via one-way ANOVA with Tukey test for multiple comparisons in (E, G, J and K).
Figure 6. Data shown represents ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, < 0.0001 via two-tailed unpaired Student t test in (D, F, and I) and one-way ANOVA with Tukey test for multiple comparisons in (C, G, and H).
Figure 7. Data shown represents ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, < 0.0001 via one-way ANOVA with Tukey test for multiple comparisons in (B and F).
Figure S1. Data shown represent ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, < 0.0001 via two-tailed unpaired Student t test.
Figure S2. Data shown represent ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, < 0.0001 via two-tailed unpaired Student t test (G–H), one-way ANOVA with Tukey test for multiple comparisons (B, and D–E), or Mann-Whitney (I–L).
Figure S3. Data shown represent ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, < 0.0001 via two-tailed unpaired Student t test (A–E, G, H, K, and M) or one-way ANOVA with Tukey test for multiple comparisons (I and L).
Figure S4. Data shown represent ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, < 0.0001 via two-tailed unpaired Student t test (E) or one-way ANOVA with Tukey test for multiple comparisons (B).
Figure S6. Data shown represent ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, < 0.0001 via two-tailed unpaired Student t test (C, E, and I) or one-way ANOVA with Tukey test for multiple comparisons (B and F–H).
Figure S7. Data shown represent ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, < 0.0001 via one-way ANOVA with Tukey test for multiple comparisons (A–B, E, and H).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Cleaved Caspase-3 (Asp175) | Cell Signaling | AB_2341188 |
| HSF1 | Cell Signaling | AB_2120258 |
| SLC9A1 | SCBT | AB_2191254 |
| HSP60 | SCBT | AB_783870 |
| Total OxPhos WB Antibody Cocktail | Abcam | AB_2629281 |
| YME1L1 | Invitrogen | AB_2649732 |
| HSP70 | SCBT | AB_627759 |
| mtHSP70 | SCBT | AB_2120468 |
| β-Actin | Sigma | AB_476744 |
| FAK pY397 | Invitrogen | AB_1500096 |
| FAK | BD | AB_397495 |
| pMLCK | Cell Signaling | AB_330248 |
| Critical Commercial Assays | ||
| Pierce™ BCA Protein Assay Kit | ThermoFisher | 23225 |
| Bacterial and virus strains | ||
| Bespoke lentiviral particles with psPAX2 and pMD2.G | Addgene | 12260 and 12259 |
| Chemicals, peptides, and recombinant proteins | ||
| 3,4-Dihydroxy-L-phenylalanine (DOPA) | Alfa Aeser | A1131106 |
| Human fibronectin | EMD Millipore | FC010 |
| Paraformaldehyde | Electron Microscopy Services | 15710 |
| Rhod-2 AM | ThermoFisher | R1244 |
| Calcium green-1 AM | Life Technologies | C3011MP |
| H2DCFDA | ThermoFisher | D399 |
| MitoPy1 | Tocris | 4428 |
| Fura-2 AM | Abcam | ab120873 |
| TMRE | ThermoFisher | T669 |
| BECEF | Invitrogen | B1150 |
| Nigericin | Invitrogen | N1495 |
| TRIzol | Invitrogen | 15596–018 |
| M-MLV reverse transcriptase | BioChain | Z5040002 |
| Ribolock | ThermoFisher | EO0384 |
| PerfeCTa SYBR Green FastMix | Quantabio | 95072–05K |
| MitoPy1 | Tocris | 4428 |
| Y-27632 (ROCK inhibitor) | Tocris | 1254 |
| Celastrol | Tocris | 3203 |
| KRIBB11 | Selleckchem | S8402 |
| MitoTempo | Cayman Chemical | 16621 |
| Ru360 | MilliporeSigma | 557440 |
| SN-6 | Tocris | 2184 |
| CGP 37157 | Cayman Chemical | 15611 |
| BIX | Tocris | 5512 |
| EIPA | SigmaAldrich | A3085 |
| Paraquat | MilliporeSigma | 856177 |
| Oligomycin A | MilliporeSigma | 75351 |
| FCCP | MilliporeSigma | C2920 |
| Rotenone | MilliporeSigma | R8875 |
| Antimycin A | MilliporeSigma | A8674 |
| JC-9 | ThermoFisher | D22421 |
| MitoTracker Deep Red FM | ThermoFisher | M22426 |
| MitoTracker Green FM | ThermoFisher | M7514 |
| Peroxymycin | (Yik-Sham Chung et al., 2018) | |
| Deposited data | ||
| RNAseq (Related to Figure 4B) | GEO | GSE171076 |
| Experimental models: cell lines | ||
| HEK293T | ATCC | NA |
| MCF10A (36 year old, Caucasian, female) | ATCC | NA |
| MB-MDA-231 (51 year old, Caucasian, female) | ATCC | NA |
| Experimental models: organisms/strains | ||
| FVB/N-Tg-MMTV-PyMT mice | The Jackson Laboratory | 002374 |
| C. elegans: Bristol (N2) strain as wild type (WT) | CGC | - |
| C. elegans: AGD710: N2, uthIs235[sur-5p::hsf-1, myo-2p::tdTomato] | Dillin Lab | (Baird et al., 2014) |
| C. elegans: AGD2319: N2; unc-119(ed3) III; uthSi62[vha-6p::MLS::mRuby::unc-54 3’UTR, cb-unc-119(+)] IV; | Dillin Lab | This study |
| C. elegans: AGD2490: N2; uthIs235[sur-5p::hsf-1, myo-2p::tdTomato]; dvIs19[pAF15(gst-4p::GFP::NLS)] | Dillin Lab | This study |
| C. elegans: CL2166: N2; gst-4p::GFP | CGC | (Baird et al., 2014) |
| C. elegans: SJ4100: N2; zcIs13(hsp-6p::GFP) | CGC | (Yoneda et al., 2004) |
| Oligonucleotides | ||
| Random hexamers | Roche | 11034731001 |
| qPCR primers, see list in qPCR section, STAR Methods | This paper | N/A |
| shRNA HSF1–1 TRCN0000007481 (HSF1): 5’CCGGGCAGGTTGTTCATAGTCAGAACTCGAGTTCTGACTATGAACAACCTGCTTTTT | Dillin lab and Sigma | TRCN0000007481 |
| shRNA HSF1–2 TRCN0000318652 (HSF1): 5’CCGGGCACATTCCATGCCCAAGTATCTCGAGATACTTGGGCATGGAATGTGCTTTTT | Dillin lab and Sigma | TRCN0000318652 |
| shRNA HSF1–1 TRCN0000007481 (HSF1): 5’CCGGGCAGGTTGTTCATAGTCAGAACTCGAGTTCTGACTATGAACAACCTGCTTTTT | Dillin lab | |
| shRNA HSF1–2 TRCN0000318652 (HSF1): 5’CCGGGCACATTCCATGCCCAAGTATCTCGAGATACTTGGGCATGGAATGTGCTTTTT | Dillin lab | |
| sgRNA YME1L1 1: 5’-TTCCGTTTCTGGGAGGAGTG | This paper | N/A |
| sgRNA YME1L1 2: 5’-GCAGTAGCTGTAGGAAGGGG, and | This paper | N/A |
| sgRNA YME1L1 3: CT CCTCCCAGAAACGGAAAA-5’ | This paper | N/A |
| Recombinant DNA | ||
| V737N β1 integrin, see associated section of STAR Methods for sequence | (Paszek et al., 2005) | |
| β1 integrin, see associated section of STAR Methods for sequence | (Paszek et al., 2005) | |
| MTS-roGFP2, see associated section of STAR Methods for sequence | This study | |
| ROCK:ER | (Croft and Olson, 2006) | |
| vha-6p::MLS::mRuby (C. elegans) ATGTTGTCCAAACGCATTGTTACCGCTCTTAACACCGCCGTCAAGGTCCAAAATGCCGGAATCGCCACCACCGCCCGCGGA | Dillin lab | |
| Constitutively active HSF1, see associated section of STAR Methods for sequence | Dillin lab | |
| Software and algorithms | ||
| RNA STAR | (Dobin et al., 2013) | https://github.com/alexdobin/STAR |
| HOMER | (Lin et al., 2010) | http://homer.ucsd.edu/homer/ngs/rnaseq/index.html |
| Cluster | (Eisen et al., 1998) | http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm |
| Java TreeView | http://jtreeview.sourceforge.net/ | |
| Metascape | (Zhou et al., 2019) | https://metascape.org |
| Seahorse Wave | Agilent | |
| MaxQuant | (Cox and Mann, 2008) | https://www.maxquant.org/ |
| artMS | http://artms.org | http://bioconductor.org/packages/release/bioc/html/artMS.html |
| MSstats | (Choi et al., 2014) | https://www.bioconductor.org/packages/release/bioc/html/MSstats.html |
| FIJI-ImageJ 1.53+ | NIH | https://imagej.net/Fiji |
| MitoMAPR | (Zhang et al., 2019) | |
| FracLac | NIH | https://imagej.ih.gov/ij/plugins/fraclac/FLHelp/Introduction.htm |
| Other | ||
| KAPA RNA HyperPrep Kit | Roche | KK8540 |
Cell-ECM mechanosignaling influences mitochondrial structure and function
Cellular mechanosignaling activates an HSF1-mediated stress response
Stiff ECM alters redox homeostasis and metabolism via mitochondrial reprogramming
Stiff microenvironments confer redox stress resilience to cells via HSF1 and YME1L1
Acknowledgments:
We thank the following people for their support and advice: Diane Barber and Yi Liu, provided advice and reagents pertaining to SLC9A1 and pH regulation. Milos Simic, generated the constitutively active HSF1. Gilberto Garcia, developed the MLS mRuby C.elegans strain. Todd McDevitt, Serah Kang, and Ariel Kauss provided editorial suggestions that significantly improved the quality of the manuscript. Chris Chang and Yik Sham (Clive) Chung for providing us Peroxymycin to use in experiments. Michael McManus and Olga Gulyaeva provided CRISPR-I vectors. Chris Phillart, maintained the cellular respirometer used in these studies. Brant Webster, generated the MTS-roGFP2 vector. Fui Boon Kai, acquired the ROCK:ER construct and generated the ROCK:ER stable cell line. Bram Piersma, provided dry Dutch humor, beneficial laboratory ambiance, and conversation critical to the development of this manuscript.
Funding:
This work was supported by 1F32CA236156-01A1, 5T32CA108462-15, and Sandler Program for Breakthrough Biomedical Research (postdoctoral Independence award) to K.M.T.; R35 CA242447-01A1, R01CA192914, and R01CA222508-01 to V.M.W.; U54 CA210184 to C.F.; 1K99AG065200-01A1 and the Glenn Foundation for Medical Research Postdoctoral Fellowship to R.H.S.; American Diabetes Association 1-19-PDF-058 to G.A.T; 1R01AG055891-01 and Howard Hughes Medical Institute support to A.D ; R01AG059751 to N.J.K. and D.L.S; and National Institutes of Health (NIH) shared Instrumentation grant S10 OD016387.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
Supplemental Videos
Video S1 - Lattice Light Sheet Mitotracker Scan - Soft (400 Pa) - Related to Figure 1
Video S2 - Lattice Light Sheet Mitotracker Scan - Stiff (60k Pa) - Related to Figure 1
Video S3 - Combined Stacks of Mitotracker - 6k Pa PA-gel - Initiates Toroid Formation – Related to Figure 1
Video S4 – Hyperglycemia Induced Mitochondrial Reorganization – Related to Figure S1I
Video S5 - Lattice Light Sheet Mitotracker Scan - Stiff (60k Pa) - Related to Figure 3F
Video S6 - Lattice Light Sheet Mitotracker Scan - Stiff (60k Pa) with Mitotempo - Related to Figure 3F
References
- Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, Chen YY, Liphardt J, Hwang ES, and Weaver VM (2015). Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. (Camb) 7, 1120–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S-G, and Thiele DJ (2003). Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 17, 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge JE, Horibe T, and Hoogenraad NJ (2007). Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One 2, e874–e874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anesti V, and Scorrano L (2006). The relationship between mitochondrial shape and function and the cytoskeleton. Biochim. Biophys. Acta 1757, 692–699. [DOI] [PubMed] [Google Scholar]
- Baird NA, Douglas PM, Simic MS, Grant AR, Moresco JJ, Wolff SC, Yates JR 3rd, Manning G, and Dillin A (2014). HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science 346, 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, and Finkel T (2005). Mitochondria, oxidants, and aging. Cell 120, 483–495. [DOI] [PubMed] [Google Scholar]
- Barasa A, Godina G, Buffa P, and Pasquali-Ronchetti I (1973). Biochemical lesions of respiratory enzymes and configurational changes of mitochondria in vivo. I. The effect of fluoroacetate: a study by phase-contrast microscopy and time-lapse cinemicrography. Z. Zellforsch. Mikrosk. Anat 138, 187–210. [DOI] [PubMed] [Google Scholar]
- Boos F, Krämer L, Groh C, Jung F, Haberkant P, Stein F, Wollweber F, Gackstatter A, Zöller E, van der Laan M, et al. (2019). Mitochondrial protein-induced stress triggers a global adaptive transcriptional programme. Nat. Cell Biol 21, 442–451. [DOI] [PubMed] [Google Scholar]
- Brookes PS, Yoon Y, Robotham JL, Anders MW, and Sheu S-S (2004). Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am. J. Physiol. Cell Physiol 287, C817–C833. [DOI] [PubMed] [Google Scholar]
- Butcher DT, Alliston T, and Weaver VM (2009). A tense situation: forcing tumour progression. Nat. Rev. Cancer 9, 108–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliari SR, and Burdick JA (2016). A practical guide to hydrogels for cell culture. Nat. Methods 13, 405–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, and Mootha VK (2016). MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor JR (2019). The Rise of Physiologic Media. Trends Cell Biol. 29, 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor JR, Abu-Remaileh M, Kanarek N, Freinkman E, Gao X, Louissaint A Jr, Lewis CA, and Sabatini DM (2017). Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell 169, 258–272.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello PR, Drechsel DA, and Patel M (2007). Mitochondria are a major source of paraquat-induced reactive oxygen species production in the brain. J. Biol. Chem 282, 14186–14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charos AE, Reed BD, Raha D, Szekely AM, Weissman SM, and Snyder M (2012). A highly integrated and complex PPARGC1A transcription factor binding network in HepG2 cells. Genome Res. 22, 1668–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B-C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA 3rd, Liu Z, et al. (2014). Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C-H, Webb BA, Chimenti MS, Jacobson MP, and Barber DL (2013). pH sensing by FAK-His58 regulates focal adhesion remodeling. J. Cell Biol 202, 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Chang C-Y, Clough T, Broudy D, Killeen T, MacLean B, and Vitek O (2014). MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526. [DOI] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Croft DR, and Olson MF (2006). Conditional regulation of a ROCK-estrogen receptor fusion protein. Methods Enzymol. 406, 541–553. [DOI] [PubMed] [Google Scholar]
- Crooks DR, Maio N, Lang M, Ricketts CJ, Vocke CD, Gurram S, Turan S, Kim Y-Y, Cawthon GM, Sohelian F, et al. (2021). Mitochondrial DNA alterations underlie an irreversible shift to aerobic glycolysis in fumarate hydratase-deficient renal cancer. Sci. Signal 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, and Sampson SB (2016). HSF1: Guardian of Proteostasis in Cancer. Trends Cell Biol. 26, 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniele JR, Esping DJ, Garcia G, Parsons LS, Arriaga EA, and Dillin A (2017). “High-Throughput Characterization of Region-Specific Mitochondrial Function and Morphology.” Sci. Rep 7, 6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelNero P, Hopkins BD, Cantley LC, and Fischbach C (2018). Cancer metabolism gets physical. Sci. Transl. Med 10, eaaq1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deocaris CC, Kaul SC, and Wadhwa R (2006). On the brotherhood of the mitochondrial chaperones mortalin and heat shock protein 60. Cell Stress Chaperones 11, 116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill KA (1990). Dominant forces in protein folding. Biochemistry 29, 7133–7155. [DOI] [PubMed] [Google Scholar]
- Dill KA, and MacCallum JL (2012). The protein-folding problem, 50 years on. Science 338, 1042–1046. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, and Botstein D (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fane M, and Weeraratna AT (2020). How the ageing microenvironment influences tumour progression. Nat. Rev. Cancer 20, 89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Solmonson A, and DeBerardinis RJ (2020). Metabolic reprogramming and cancer progression. Science 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorese CJ, Schulz AM, Lin Y-F, Rosin N, Pellegrino MW, and Haynes CM (2016). The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr. Biol 26, 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C, Marchi S, and Pinton P (2018). The machineries, regulation and cellular functions of mitochondrial calcium. Nat. Rev. Mol. Cell Biol 19, 713–730. [DOI] [PubMed] [Google Scholar]
- Giustarini D, Dalle-Donne I, Milzani A, Fanti P, and Rossi R (2013). Analysis of GSH and GSSG after derivatization with N-ethylmaleimide. Nat. Protoc 8, 1660–1669. [DOI] [PubMed] [Google Scholar]
- Grandjean JMD, Plate L, Morimoto RI, Bollong MJ, Powers ET, and Wiseman RL (2019). Deconvoluting Stress-Responsive Proteostasis Signaling Pathways for Pharmacologic Activation Using Targeted RNA Sequencing. ACS Chem. Biol 14, 784–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Pegoraro AF, Mao A, Zhou EH, Arany PR, Han Y, Burnette DT, Jensen MH, Kasza KE, Moore JR, et al. (2017). Cell volume change through water efflux impacts cell stiffness and stem cell fate. Proc. Natl. Acad. Sci. U. S. A 114, E8618–E8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassell BA, Goyal G, Lee E, Sontheimer-Phelps A, Levy O, Chen CS, and Ingber DE (2017). Human Organ Chip Models Recapitulate Orthotopic Lung Cancer Growth, Therapeutic Responses, and Tumor Dormancy In Vitro. Cell Rep. 21, 508–516. [DOI] [PubMed] [Google Scholar]
- Helle SCJ, Feng Q, Aebersold MJ, Hirt L, Grüter RR, Vahid A, Sirianni A, Mostowy S, Snedeker JG, Šarić A, et al. (2017). Mechanical force induces mitochondrial fission. Elife 6, e30292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernansanz-Agustín P, Choya-Foces C, Carregal-Romero S, Ramos E, Oliva T, Villa-Piña T, Moreno L, Izquierdo-Álvarez A, Cabrera-García JD, Cortés A, et al. (2020). Na(+) controls hypoxic signalling by the mitochondrial respiratory chain. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi-Sanabria R, Frankino PA, Paul JW 3rd, Tronnes SU, and Dillin A (2018). A Futile Battle? Protein Quality Control and the Stress of Aging. Dev. Cell 44, 139–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Pfeifer CR, Bennett RR, Xia Y, Ivanovska IL, Liu AJ, Greenberg RA, and Discher DE (2016). Nuclear constriction segregates mobile nuclear proteins away from chromatin. Mol. Biol. Cell 27, 4011–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isermann P, and Lammerding J (2017). Consequences of a tight squeeze: Nuclear envelope rupture and repair. Nucleus 8, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C, Chen L, and Rabinowitz JD (2018). Metabolomics and Isotope Tracing. Cell 173, 822–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Blasco D, Busquets-Garcia A, Hebert-Chatelain E, Serrat R, Vicente-Gutierrez C, Ioannidou C, Gómez-Sotres P, Lopez-Fabuel I, Resch-Beusher M, Resel E, et al. (2020). Glucose metabolism links astroglial mitochondria to cannabinoid effects. Nature 583, 603–608. [DOI] [PubMed] [Google Scholar]
- Kai F, Drain AP, and Weaver VM (2019). The Extracellular Matrix Modulates the Metastatic Journey. Dev. Cell 49, 332–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar A, Fujimoto M, Tan K, Kurashima A, Srivastava P, Okada M, Takii R, and Nakai A (2020). HSF1 is required for induction of mitochondrial chaperones during the mitochondrial unfolded protein response. FEBS Open Bio. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Brielmann RM, Neto MF, Lin Y-F, Haynes CM, and Morimoto RI (2017). Mitochondrial Stress Restores the Heat Shock Response and Prevents Proteostasis Collapse during Aging. Cell Rep. 21, 1481–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladiges W, Wanagat J, Preston B, Loeb L, and Rabinovitch P (2010). A mitochondrial view of aging, reactive oxygen species and metastatic cancer. Aging Cell 9, 462–465. [DOI] [PubMed] [Google Scholar]
- Lambert T (2019). (2019). tlambert03/LLSpy: Lattice light-sheet post-processing utility. Zenodo. [Google Scholar]
- LeBoeuf SE, Wu WL, Karakousi TR, Karadal B, Jackson SR, Davidson SM, Wong K-K, Koralov SB, Sayin VI, and Papagiannakopoulos T (2020). Activation of Oxidative Stress Response in Cancer Generates a Druggable Dependency on Exogenous Non-essential Amino Acids. Cell Metab. 31, 339–350.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon FE, Cianci GC, Kanteti R, Riehm JJ, Arif Q, Poroyko VA, Lupovitch E, Vigneswaran W, Husain A, Chen P, et al. (2016). Unique fractal evaluation and therapeutic implications of mitochondrial morphology in malignant mesothelioma. Sci. Rep 6, 24578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, et al. (2009). Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Srinivasan SR, Connarn J, Ahmad A, Young ZT, Kabza AM, Zuiderweg ERP, Sun D, and Gestwicki JE (2013). Analogs of the Allosteric Heat Shock Protein 70 (Hsp70) Inhibitor, MKT-077, as Anti-Cancer Agents. ACS Med. Chem. Lett 4, 1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-F, and Haynes CM (2016). Metabolism and the UPR(mt). Mol. Cell 61, 677–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GL, Cohen DM, Desai RA, Breckenridge MT, Gao L, Humphries MJ, and Chen CS (2013). Activation of beta 1 but not beta 3 integrin increases cell traction forces. FEBS Lett. 587, 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. (2010). A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol 11, 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström P, and Sehlin J (1984). Effect of glucose on the intracellular pH of pancreatic islet cells. Biochem. J 218, 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, and Hajnóczky G (2011). Altered fusion dynamics underlie unique morphological changes in mitochondria during hypoxia-reoxygenation stress. Cell Death Differ. 18, 1561–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Mlodzianoski MJ, Hu Z, Ren Y, McElmurry K, Suter DM, and Huang F (2017). sCMOS noise-correction algorithm for microscopy images. Nat. Methods 14, 760–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie SM, Grossman EA, Crawford LA, Ding L, Camarda R, Huffman TR, Miyamoto DK, Goga A, Weerapana E, and Nomura DK (2016). GSTP1 Is a Driver of Triple-Negative Breast Cancer Cell Metabolism and Pathogenicity. Cell Chem. Biol 23, 567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q (2013). Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol 53, 401–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Xu L, Alberobello AT, Gavrilova O, Bagattin A, Skarulis M, Liu J, Finkel T, and Mueller E (2015). Celastrol Protects against Obesity and Metabolic Dysfunction through Activation of a HSF1-PGC1α Transcriptional Axis. Cell Metab 22, 695–708. [DOI] [PubMed] [Google Scholar]
- MacVicar T, and Langer T (2016). OPA1 processing in cell death and disease - the long and short of it. J. Cell Sci 129, 2297–2306. [DOI] [PubMed] [Google Scholar]
- MacVicar T, Ohba Y, Nolte H, Mayer FC, Tatsuta T, Sprenger H-G, Lindner B, Zhao Y, Li J, Bruns C, et al. (2019). Lipid signalling drives proteolytic rewiring of mitochondria by YME1L. Nature 575, 361–365. [DOI] [PubMed] [Google Scholar]
- Maller O, Drain AP, Barrett AS, Borgquist S, Ruffell B, Zakharevich I, Pham TT, Gruosso T, Kuasne H, Lakins JN, et al. (2020). Tumour-associated macrophages drive stromal cell-dependent collagen crosslinking and stiffening to promote breast cancer aggression. Nat. Mater [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP (2008). Hormesis defined. Ageing Res. Rev 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier F, Brunner A-D, Koch S, Koch H, Lubeck M, Krause M, Goedecke N, Decker J, Kosinski T, Park MA, et al. (2018). Online Parallel Accumulation-Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer. Mol. Cell. Proteomics 17, 2534–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, and Lindquist S (2012). HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150, 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroshnikova YA, Mouw JK, Barnes JM, Pickup MW, Lakins JN, Kim Y, Lobo K, Persson AI, Reis GF, McKnight TR, et al. (2016). Tissue mechanics promote IDH1-dependent HIF1α-tenascin C feedback to regulate glioblastoma aggression. Nat. Cell Biol 18, 1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono Y, Hirashima S, Ishihara N, Kusukawa J, Nakamura K-I, and Ohta K (2018). Uncoupled mitochondria quickly shorten along their long axis to form indented spheroids, instead of rings, in a fission-independent manner. Sci. Rep 8, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouw JK, Yui Y, Damiano L, Bainer RO, Lakins JN, Acerbi I, Ou G, Wijekoon AC, Levental KR, Gilbert PM, et al. (2014). Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat. Med 20, 360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Suzuki M, and Tanabe M (2000). Arrest of spermatogenesis in mice expressing an active heat shock transcription factor 1. EMBO J. 19, 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, and Haynes CM (2012). Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrke K, and Melvin JE (2002). The NHX family of Na+−H+ exchangers in Caenorhabditis elegans. J. Biol. Chem 277, 29036–29044. [DOI] [PubMed] [Google Scholar]
- Northcott JM, Dean IS, Mouw JK, and Weaver VM (2018). Feeling Stress: The Mechanics of Cancer Progression and Aggression. Front. Cell Dev. Biol 6, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oria R, Wiegand T, Escribano J, Elosegui-Artola A, Uriarte JJ, Moreno-Pulido C, Platzman I, Delcanale P, Albertazzi L, Navajas D, et al. (2017). Force loading explains spatial sensing of ligands by cells. Nature 552, 219–224. [DOI] [PubMed] [Google Scholar]
- Östling P, Björk JK, Roos-Mattjus P, Mezger V, and Sistonen L (2007). Heat Shock Factor 2 (HSF2) Contributes to Inducible Expression of hsp Genes through Interplay with HSF1*. J. Biol. Chem 282, 7077–7086. [DOI] [PubMed] [Google Scholar]
- Oudin MJ, and Weaver VM (2016). Physical and Chemical Gradients in the Tumor Microenvironment Regulate Tumor Cell Invasion, Migration, and Metastasis. Cold Spring Harb. Symp. Quant. Biol 81, 189–205. [DOI] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, et al. (2005). Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241–254. [DOI] [PubMed] [Google Scholar]
- Paul S, Ghosh S, Mandal S, Sau S, and Pal M (2018). NRF2 transcriptionally activates the heat shock factor 1 promoter under oxidative stress and affects survival and migration potential of MCF7 cells. J. Biol. Chem 293, 19303–19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SW, Norman JP, Barbieri J, Brown EB, and Gelbard HA (2011). Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques 50, 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouysségur J, Sardet C, Franchi A, L’Allemain G, and Paris S (1984). A specific mutation abolishing Na+/H+ antiport activity in hamster fibroblasts precludes growth at neutral and acidic pH. Proc. Natl. Acad. Sci. U. S. A 81, 4833–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D, Gygi SP, and Auwerx J (2017). Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol 216, 2027–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA, et al. (2005). Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 436, 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek CR, and Chandel NS (2017). The Two Faces of Reactive Oxygen Species in Cancer. Annu. Rev. Cancer Biol 1, 79–98. [Google Scholar]
- Reczek CR, Birsoy K, Kong H, Martínez-Reyes I, Wang T, Gao P, Sabatini DM, and Chandel NS (2017). A CRISPR screen identifies a pathway required for paraquat-induced cell death. Nat. Chem. Biol 13, 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann D, Voth W, and Jakob U (2018). Maintaining a Healthy Proteome during Oxidative Stress. Mol. Cell 69, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M (2014). Unraveling the truth about antioxidants: mitohormesis explains ROS-induced health benefits. Nat. Med 20, 709–711. [DOI] [PubMed] [Google Scholar]
- Ristow M, and Schmeisser S (2011). Extending life span by increasing oxidative stress. Free Radic. Biol. Med 51, 327–336. [DOI] [PubMed] [Google Scholar]
- Rossiter NJ, Huggler KS, Adelmann CH, Keys HR, Soens RW, Sabatini DM, and Cantor JR (2020). CRISPR screens in physiologic medium reveal conditionally essential genes in human cells. BioRxiv 2020.08.31.275107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer LA, Weigert M, Günther U, Maghelli N, Jug F, Sbalzarini IF, and Myers EW (2015). ClearVolume: open-source live 3D visualization for light-sheet microscopy. Nat. Methods 12, 480–481. [DOI] [PubMed] [Google Scholar]
- Russell WK, Park ZY, and Russell DH (2001). Proteolysis in mixed organic-aqueous solvent systems: applications for peptide mass mapping using mass spectrometry. Anal. Chem 73, 2682–2685. [DOI] [PubMed] [Google Scholar]
- Samuel MS, Lopez JI, McGhee EJ, Croft DR, Strachan D, Timpson P, Munro J, Schröder E, Zhou J, Brunton VG, et al. (2011). Actomyosin-mediated cellular tension drives increased tissue stiffness and β-catenin activation to induce epidermal hyperplasia and tumor growth. Cancer Cell 19, 776–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, Schnitt SJ, Whitesell L, Tamimi RM, Lindquist S, et al. (2011). High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc. Natl. Acad. Sci. U. S. A 108, 18378–18383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, and Brugge JS (2009). Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedin P, and Keely PJ (2011). Mammary gland ECM remodeling, stiffness, and mechanosignaling in normal development and tumor progression. Cold Spring Harb. Perspect. Biol 3, a003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R, Santagata S, Mendillo ML, Sholl LM, Ben-Aharon I, Beck AH, Dias-Santagata D, Koeva M, Stemmer SM, Whitesell L, et al. (2014). The reprogramming of tumor stroma by HSF1 is a potent enabler of malignancy. Cell 158, 564–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, and Meisinger C (2010). Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol 11, 655–667. [DOI] [PubMed] [Google Scholar]
- Schneider HC, Berthold J, Bauer MF, Dietmeier K, Guiard B, Brunner M, and Neupert W (1994). Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371, 768–774. [DOI] [PubMed] [Google Scholar]
- Sciacovelli M, Gonçalves E, Johnson TI, Zecchini VR, da Costa ASH, Gaude E, Drubbel AV, Theobald SJ, Abbo SR, Tran MGB, et al. (2016). Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 537, 544–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialò F, Sriram A, Fernández-Ayala D, Gubina N, Lõhmus M, Nelson G, Logan A, Cooper HM, Navas P, Enríquez JA, et al. (2016). Mitochondrial ROS Produced via Reverse Electron Transport Extend Animal Lifespan. Cell Metab. 23, 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger H-G, and Langer T (2019). The Good and the Bad of Mitochondrial Breakups. Trends Cell Biol. 29, 888–900. [DOI] [PubMed] [Google Scholar]
- Srinivasan SR, Cesa LC, Li X, Julien O, Zhuang M, Shao H, Chung J, Maillard I, Wells JA, Duckett CS, et al. (2018). Heat Shock Protein 70 (Hsp70) Suppresses RIP1-Dependent Apoptotic and Necroptotic Cascades. Mol. Cancer Res 16, 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Youle RJ, and Finkel T (2016). The Mitochondrial Basis of Aging. Mol. Cell 61, 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharp KM, and Weaver VM (2018). Modeling Tissue Polarity in Context. J. Mol. Biol 430, 3613–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharp KM, Kang MS, Timblin GA, Dempersmier J, Dempsey GE, Zushin P-JH, Benavides J, Choi C, Li CX, Jha AK, et al. (2018). Actomyosin-Mediated Tension Orchestrates Uncoupled Respiration in Adipose Tissues. Cell Metab. 27, 602–615.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T, and Barber DL (1998). Na-H exchange acts downstream of RhoA to regulate integrin-induced cell adhesion and spreading. Mol. Biol. Cell 9, 2287–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T, Ishizaki T, Narumiya S, and Barber DL (1998). p160ROCK mediates RhoA activation of Na-H exchange. EMBO J. 17, 4712–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, and Shirihai OS (2011). The interplay between mitochondrial dynamics and mitophagy. Antioxid. Redox Signal 14, 1939–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, and Hillen W (2000). Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. U. S. A 97, 7963–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, Zaganjor E, and Haigis MC (2016). Mitochondria and Cancer. Cell 166, 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC (2012). Mitochondria and cancer. Nat. Rev. Cancer 12, 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang M, Torres G, Wu S, Ouyang C, Xie Z, and Zou M-H (2017). Metformin Suppresses Diabetes-Accelerated Atherosclerosis via the Inhibition of Drp1-Mediated Mitochondrial Fission. Diabetes 66, 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner E, and Werb Z (2002). Integrins engage mitochondrial function for signal transduction by a mechanism dependent on Rho GTPases. J. Cell Biol 158, 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, and Pfanner N (2017). Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem 86, 685–714. [DOI] [PubMed] [Google Scholar]
- Wouters OY, Ploeger DTA, van Putten SM, and Bank RA (2016). 3,4-Dihydroxy-L-Phenylalanine as a Novel Covalent Linker of Extracellular Matrix Proteins to Polyacrylamide Hydrogels with a Tunable Stiffness. Tissue Eng. Part C. Methods 22, 91–101. [DOI] [PubMed] [Google Scholar]
- Yik-Sham Chung C, Timblin GA, Saijo K, and Chang CJ (2018). Versatile Histochemical Approach to Detection of Hydrogen Peroxide in Cells and Tissues Based on Puromycin Staining. J. Am. Chem. Soc 140, 6109–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, and Ron D (2004). Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci 117, 4055–4066. [DOI] [PubMed] [Google Scholar]
- Youle RJ, and van der Bliek AM (2012). Mitochondrial fission, fusion, and stress. Science 337, 1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, and Finkel T (2014). Mitohormesis. Cell Metab. 19, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lanjuin A, Chowdhury SR, Mistry M, Silva-García CG, Weir HJ, Lee C-L, Escoubas CC, Tabakovic E, and Mair WB (2019). Neuronal TORC1 modulates longevity via AMPK and cell nonautonomous regulation of mitochondrial dynamics in C. elegans. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA sequencing data reported in this paper is GEO:GSE171076. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171076


